Abstract
Background
Observational studies and randomized controlled trials have found evidence that higher maternal circulating cortisol levels in pregnancy are associated with lower offspring birth weight. However, it is possible that the observational associations are due to residual confounding.
Methods
We performed two-sample Mendelian Randomisation (MR) using a single genetic variant (rs9989237) associated with morning plasma cortisol (GWAS; sample 1; N = 25,314). The association between this maternal genetic variant and offspring birth weight, adjusted for fetal genotype, was obtained from the published EGG Consortium and UK Biobank meta-analysis (GWAS; sample 2; N = up to 406,063) and a Wald ratio was used to estimate the causal effect. We also performed an alternative analysis using all GWAS reported cortisol variants that takes account of linkage disequilibrium. We also tested the genetic variant’s effect on pregnancy cortisol and performed PheWas to search for potential pleiotropic effects.
Results
The estimated effect of maternal circulating cortisol on birth weight was a 50 gram (95% CI, -109 to 10) lower birth weight per 1 SD higher log-transformed maternal circulating cortisol levels, using a single variant. The alternative analysis gave similar results (-33 grams (95% CI, -77 to 11)). The effect of the cortisol variant on pregnancy cortisol was 2-fold weaker than in the original GWAS, and evidence was found of pleiotropy.
Conclusions
Our findings provide some evidence that higher maternal morning plasma cortisol causes lower birth weight. Identification of more independent genetic instruments for morning plasma cortisol are necessary to explore the potential bias identified.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12884-024-06250-3.
Keywords: UK Biobank, EFSOCH, Cortisol, Birth weight, Mendelian Randomization
Background
Variation in human birth weight is associated with adverse perinatal health outcomes as well as long term health outcomes [1]. In particular, lower than average birth weight is associated with higher neonatal mortality and a higher risk of cardiovascular disease [2], type 2 diabetes [3] and hypertension [4] in adulthood. Understanding mechanisms that influence variation in birth weight could help identify targets for intervention to ensure healthy birth weight.
Experimental studies in animal models and observational studies in humans have demonstrated links between higher fetal glucocorticoid exposure and lower birth weight [5]. Higher maternal cortisol levels are one potential source of increased fetal glucocorticoid exposure, with evidence of higher levels of both maternal plasma [6] and salivary [7] cortisol being associated with lower birth weight infants. Infants exposed to antenatal corticosteroids in a secondary analyses of a randomized controlled trial (RCT) of women at risk of preterm birth also have lower birth weight compared to those randomised to placebo, although this was in part related to also having a shorter gestation [8].
There are challenges to assessing the effect of maternal cortisol levels on offspring birth weight. There are several maternal characteristics that can confound the relationship between maternal cortisol and offspring birth weight, such as maternal smoking and body mass index (BMI) [9], which can be difficult or even impossible to fully account for in conventional observational studies. Also, whilst the RCT evidence was from a large and well conducted study and therefore unlikely to be biased by confounding, it was limited to women at risk of preterm birth only. Furthermore, it was not a direct test of the effect of maternal cortisol on birth weight and the lower birth weight in those randomized to corticosteroids was driven in large part by reduced gestational duration [8]. Mendelian Randomization (MR), uses genetic variants to probe the effect of modifiable exposures (e.g. maternal cortisol levels) on health outcomes (e.g. offspring birth weight) [10]. Given that genetic variation is randomised at conception, MR is less susceptible to being biased by variables that are observationally correlated with the exposure variable but independently impact the outcome via a mechanism independent of the mechanism being tested.
We hypothesized that higher maternal plasma cortisol causes lower offspring birth weight and used MR to test this hypothesis. We used the most recent Genome Wide Association Study (GWAS) of fasting plasma cortisol levels [11] as the source of genetic variant associations with the exposure, and we used the GWAS of offspring birth weight in the Early Growth Genetics (EGG) Consortium and UK Biobank [12] to obtain estimates of maternal genetic effects on birth weight conditional on the fetal genotype. To investigate the plausibility of instrumental variable assumptions, we also tested the genetic variant’s association with cortisol in pregnancy in a European-ancestry birth cohort [13] and searched for potential sources of horizontal pleiotropy using an online database [14, 15].
Methods
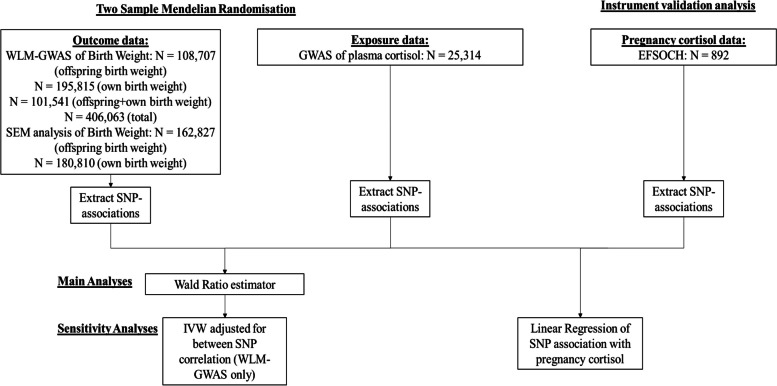
We used two-sample MR to estimate the causal effect of maternal plasma cortisol on offspring birth weight [16]. This method involves using estimates of the single nucleotide polymorphism (SNP)-exposure associations (using SNPs that are robustly associated with the exposure, in this case plasma cortisol) as well as using SNP-outcome associations extracted from a pre-existing data set (in this case offspring birth weight). For each SNP, the SNP-outcome association is divided by the SNP-exposure association. Normally, these ratios would be pooled to give an estimate of the causative effect of the exposure on an outcome. For this study we were limited by the fact that only one genome-wide significant locus for plasma cortisol has been identified. The study design and different sources used are summarised in Fig. 1.
Fig. 1.
Diagram summarising the key data sources and analysis steps for this study
Data sources
A summary of all the cohorts contributing to the GWAS summary statistics used in this study can be found in Table 1.
Table 1.
Summary of studies contributing to a) the circulating cortisol GWAS (CORNET), b) the maternal effects on pregnancy GWAS (EGG) and c) the observational pregnancy circulating cortisol (EFSOCH)
| a) | Study | Country | N | Age, years (SD) | Cortisol, nmol/l (SD) | Sampling time |
| ORCADES | UK | 1974 | 53.5 (15.7) | 765 (315) | 0830-1030 | |
| CROATIA-Korcula | Croatia | 898 | 56.2 (13.9) | 698 (207) | 0800-0900 | |
| CROATIA-Split | Croatia | 496 | 45.0 (14.7) | 979 (404) | 0730-0900 | |
| CROATIA-Vis | Croatia | 892 | 56.4 (15.5) | 622 (230) | 0730-0900 | |
| Rotterdam Study | Netherlands | 6497 | 63.3 (9.6) | 359 (115) | 0800-1100 | |
| HBCS1934-44 | Finland | 451 | 60.61 (2.80) | 393 (120) | 0750-1055 | |
| NFBC1966 | Finland | 1324 | 31.1 (0.3) | 380 (160) | 0800-1100 | |
| ALSPAC | UK | 1567 | 15.43 (0.26) | 486 (174) | 0800-1057 | |
| PIVUS | Sweden | 919 | 70.2 (0.17) | 386 (125) | 0800-1000 | |
| PREVEND | Netherlands | 1151 | 49.4 (13.0) | 442 (201) | 0800-1100 | |
| ET2DS | UK | 1048 | 67.9 (4.2) | 731 (190) | 0800-0830 | |
| Raine Study | Australia | 860 | 17.1 (0.29) | 614 (235) | Awakening (before 1000) | |
| MrOS Sweden | Sweden | 969 | 75.3 (3.2) | 487 (133) | 0700-1000 | |
| VIKING | UK | 2073 | 49.9 (15.2) | 292 (170) | 0800-1030 | |
| SHIP | Germany | 910 | 49.8 (13.8) | * | Before 1300 | |
| TwinsUK | UK | 5654 | 53.3 (13.8) | * | 0800-1200 | |
| KORA | Germany | 1651 | 60.92 (8.7) | * | NA | |
| b) | Study | Country | N | Age, years (SD) | Birthweight, g (SD) | Gestational age, weeks (IQR) |
| UK Biobank | UK | 190,406 | 25.3 (4.5) | 3227 (477) | NA | |
| B58C-WTCCC | UK | 858 | 26.2 (5.2) | 3325 (483) | 40 (40–41) | |
| B58C-T1DGC | UK | 836 | 26.1 (5.4) | 3379 (469) | 40 (40–41) | |
| DNBC-GOYA | Denmark | 1805 | 29.2 (4.2) | 3643 (495) | 40 (39–41) | |
| DNBC-PTB-CONTROL | Denmark | 1656 | 29.9 (4.2) | 3595 (497) | 40 (39–40) | |
| MoBa-2008 | Norway | 650 | 28.5 (3.3) | 3679 (430) | 40 (0.9) | |
| NFBC1966 | Finland | 2035 | 26.5 (3.7) | 3525 (461) | 40 (2) | |
| NTR | Netherlands | 707 | 27.1 (3.7) | 3469 (529) | 40 (38–42) | |
| QIMR | Australia | 892 | 24.5 (4.0) | 3344 (532) | NA | |
| TWINSUK | UK | 1603 | NA | NA | NA | |
| ALSPAC | UK | 6,686 | 28.0 (5.0) | 3468 (475) | 40 (40–41) | |
| HAPO | USA | 1280 | 31.5 (5.3) | 3557 (517) | 40 (1.7) | |
| EFSOCH | UK | 855 | 30.5 (5.9) | 3506 (472) | 40 (37–43) | |
| c) | Study | Country | N | Age, years (SD) | Cortisol, nmol/l (SD) | Sampling time |
| EFSOCH | UK | 892 | 30.4 (5.3) | 1010 (234) | 0900 (within 60 minutes) |
aApart from EFSOCH [13] circulating cortisol (in pregnancy), the data comes from Crawford et al 2021 [11] and Warrington et al 2019 [12]
bThis table only shows the studies that contributed maternal genotype and offspring birthweight data (n = 210,267) to the final WLM-adjusted GWAS of offspring birthweight (n = 406,063). More information regarding offspring genotype and own birthweight data can be found in Warrington et al. 2019
cORCADES Orkney Complex Disease Study, HBCS1934-44 Helsinki Birth Cohort Study 1934-1944, NFBC1966 the Northern Finland 1966 Birth Cohort, ALSPAC Avon Longitudinal Study of Parents and Children; PIVUS Prospective Investigation of the Vasculature in Uppsala Seniors, PREVEND Prevention of Renal and Vascular End-stage Disease, ET2D2 Edinburgh Type 2 Diabetes Study, B58C-T1DGC British 1958 Birth Cohort – Type 1 Diabetes Genetics Consortium, MrOS Sweden Osteoporotic Fractures in Men-Sweden, KORA Cooperative Health Research in the Augsburg Region, SHIP Study of Health in Pomerania, VIKING Viking Health Study-Shetland, B58C-WTCCC British 1958 Birth Cohort – Wellcome Trust Case Control Consortium, DNBC-GOYA Danish National Birth Cohort – Genetics of Overweight Young Adults, DNBC-PTB-CONTROL Danish National Birth Cohort – Preterm Birth-Control Mothers, MoBa-2008 the Norwegian Mother and Baby Cohort, 2008, NTR Netherlands Twin Registry, QIMR Queensland Institute of Medical Research, HAPO Hyperglycaemia and Adverse Pregnancy Outcome Study, NA Not applicable;
Genetic associations with plasma cortisol
SNPs associated with circulating cortisol were identified from the most recent GWAS (N=25,314) [11]. In total, 17 cohorts contributed to the GWAS, and usually measured circulating cortisol levels before 12pm (range 7am to 1pm) [11]. in which four SNPs within one locus (i.e. the SERPINA6/SERPINA1 locus) were associated with fasting plasma cortisol at genome wide significance (p-value ≤ 5e-8) [11]. These four SNPs are in partial linkage disequilibrium (LD) with one another and we selected the SNP most strongly associated with circulating cortisol, rs9989237, as the genetic instrument for our main MR analysis [11]. Details of the identified SNPs are found in Additional file 1 (Additional Table 1).
Genetic associations with birth weight
For our second sample we used the latest maternal GWAS of offspring birth weight from the Early Growth Genetics (EGG) meta-analysis. A total of 406,063 participants contributed to the weighted linear model analyses (WLM, see below) to estimate maternal effects conditional on offspring genotype, and offspring effects conditional on maternal genotype (see Additional file 1 (Methods)). Of these participants, 101,541 were UK Biobank participants who reported their own birth weight and birth weight of their first child, 195,815 were UK Biobank and EGG participants with own birth weight data, and 108,707 were UK Biobank and EGG participants with offspring birth weight data [12].In the UK Biobank and EGG meta-analysis, birth weight was standardized within each of the cohorts so that birth weight in our analyses is measured in SD units and our results were initially the difference in mean birth weight in SD units. We converted these to a difference in mean birth weight in grams by using the SD of birth weight from an earlier EGG paper (1 SD of birthweight = 484g) [17].
Genetic associations with maternal pregnancy cortisol
Cortisol levels in 892 mothers in the EFSOCH cohort [13] were assayed at 28 weeks gestation (Additional file 1 (Methods)). EFSOCH mothers were genotyped in three batches (one in Exeter, two in Bristol) using the Illumina Infinium HumanCoreExome-24 array, and when multiple genotyping batches are used for the same sample, bias can occur due to random differences between those participants assigned to one batch versus another (i.e., a batch effect) [18]. The association between the GWAS identified SNP and pregnancy cortisol in EFSOCH was adjusted for the genotyping chip to guard against batch effects.
Data analyses
Our main analysis was to estimate the effect of maternal plasma cortisol on offspring birth weight in the UK Biobank and EGG meta-analysis. In addition to this, we undertook analyses to assess instrumental variable assumptions, specifically to determine the strength of the cortisol instruments and to explore the possibility of horizontal pleiotropy in the cortisol instrument.
Adjusting for the fetal genotype
To avoid violating the third assumption of MR (i.e. that a genetic instrument affects the outcome only via the associated exposure) due to fetal genetic effects [10], we adjusted for the fetal genotype. For the main analysis, to ensure our analyses considered only the effect of the maternal genotype, and not the correlated fetal genotype, we used SNP-birth weight associations that had been adjusted for fetal genotype using a weighted linear model (WLM) [12]. The WLM is a method that was developed to combine data from disparate study designs to estimate conditional maternal and fetal genetic effects, similar to conditional genetic association analysis in genotyped mother-child pairs (see Additional file 1 (Methods) and references [12, 19]). To verify the WLM-adjusted summary statistics, we also applied the SEM method to obtain the SNP maternal effect on offspring birth weight, adjusted for the fetal genotype using UK Biobank participants (own birth weight N = 186,810; offspring birth weight N = 162,827) and repeated the main MR analysis to check we obtained similar results.
Main MR analyses
We performed two-sample MR using the Wald ratio estimator [20], which was calculated by dividing the SNP’s effect on birth weight by the same SNP’s effect on circulating cortisol. Standard errors were calculated by dividing the standard error of the SNP’s effect on birth weight by the SNP’s effect on cortisol. This was done using SNP-outcome estimates from both the main WLM analysis and from our own SEM analysis. The resulting effect estimates from our MR analyses are reported per 1 SD of log-transformed plasma cortisol levels [11].
IVW analysis adjusting for between SNP correlations
To maximise power, we performed an additional MR analysis incorporating the four SNPs in partial LD at the SERPINA6/SERPINA1 locus, as reported by Crawford et al [11]. Given those SNPs were partially correlated, we used a modified inverse variance weighted (IVW) analyses which accounts for the correlation across genetic instruments using the TwoSampleMR [21] and MendelianRandomisation [22] R packages and a correlation matrix of variants obtained from the 1000 genomes EUR reference panel via TwoSampleMR [21]. The correlation matrix of the R values used for this analysis is presented in Additional file 1 (Additional Table 2).
Testing cortisol instrument strength
An MR assumption is that the genetic instruments are robustly associated with the exposure. In two-sample MR, as undertaken here, weak instrument bias is expected to bias estimates towards the null in the absence of sample overlap. To test the strength of the genetic instruments for cortisol, we calculated the R2 and F-Statistic for all four SNP-cortisol associations reported in the GWAS (see Additional file 1 (Methods) for further details).
Testing cortisol instruments relevance to pregnancy
The cortisol GWAS was performed in a non-pregnant, mixed sex population, therefore it is possible that the instruments detected do not predict variations in circulating cortisol during pregnancy, or if they do, this is with a different magnitude to what we assume when using the GWAS result. We therefore compared the association between SNP rs9989237 and fasting plasma cortisol levels measured in pregnancy in the EFSOCH cohort with the same results from the original GWAS (see Additional file 1 (Methods) for further details).
Exploring the possibility of horizontal pleiotropy in the cortisol instrument
Another core MR assumption is that any effect of the genetic instrument on the outcome is fully mediated by the exposure. If this assumption is violated, the genetic instrument is considered invalid and MR estimates could be biased. Numerous MR methods have been developed that are robust to the presence of invalid instruments e.g. MR-Egger [23], weighted-median [24], Radial MR [25]. However, these methods typically require that multiple genetic instruments from different loci are available for a particular exposure. Given that only one independent SNP was available for our analyses, we explored the plausibility of the assumption of no invalid instruments by assessing the specificity of our genetic instrument in a phenome-wide association (PheWAS) scan using data from the MR-Base platform [14, 15], which has data from a wide range of GWAS that can be easily downloaded via R. To perform the scan, we downloaded every tested association between rs9989237 and an available GWAS variable using the “ieu-gwas-r” package [14], by specifying the p-value threshold at 1. This gave us 19,269 different variables in total. Though all of the variables associated with rs9989237 could result in pleiotropy, we decided to focus our attention on those variables whose p-value passed a Bonferroni threshold of 2.6e-06.
Results
Main results and sensitivity analyses
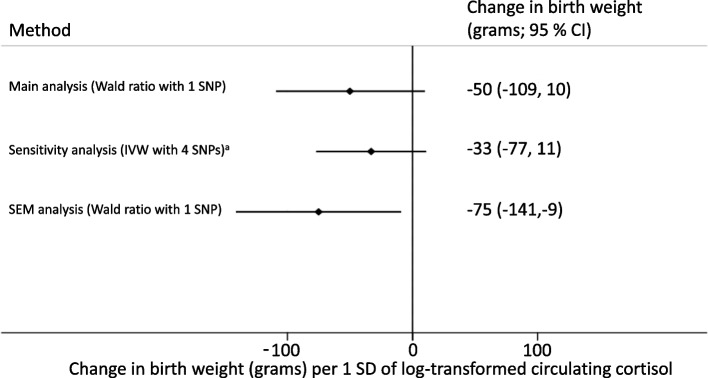
The estimated effect of maternal circulating cortisol was a 50 (95% CI, -109 to 10) grams lower offspring birth weight per 1 SD higher log-transformed maternal circulating cortisol levels. When using all four SNPs in IVW analysis adjusted for correlation between SNPs, the result was similar (-33 (95% CI, -77 to 11). Using the SEM to adjust for the fetal genotype gave similar results (-75 (95% CI, -141 to -9)). All effect estimates are shown in Fig. 2.
Fig. 2.
Mendelian Randomisation causative effect estimates for maternal plasma cortisol on mean birth weight. a) The SNPs used in the sensitivity analyses are correlated with the SNP used in the main analyses and each other. We used a form of IVW analyses that adjusts for between SNP correlations. b) IVW, Inverse Variance Weighted; SEM, Structural Equation Model
SNP validation
Instrument strength and relevance in pregnancy
Using the data from the largest available GWAS, we estimated that the SNP used in the main analyses (rs9989237) explained ~0.2% of the variation in cortisol and had an F-statistic of 62. The R2 values and F-statistics for the other SNPs are shown in Table 2.
Table 2.
R2 and F-statistic results for the genetic variants that were genome wide significant in the original genome wide association study
| Genetic variant (SNP) ID | Number of-participants | Minor allele frequency | Per allele difference (SDs of log-transformed units) in plasma cortisol (95% CI) | R2 | F-Statistic |
|---|---|---|---|---|---|
| rs11620763 | 25314 | 0.19 | 0.09 (0.06 to 0.11) | 0.0023 | 57.49 |
| rs2736898 | 25314 | 0.49 | 0.06 (0.04 to 0.07) | 0.0017 | 43.37 |
| rs7146221 | 25314 | 0.45 | 0.05 (0.03 to 0.07) | 0.0013 | 31.87 |
| rs9989237a | 25314 | 0.21 | 0.09 (0.07 to 0.10) | 0.0024 | 61.83 |
aSNP used in main analysis
bThe R2 and F-statistics for rs11620763, rs2736898 and rs7146221 may be under or overestimated due to linkage disequilibrium with rs9989237
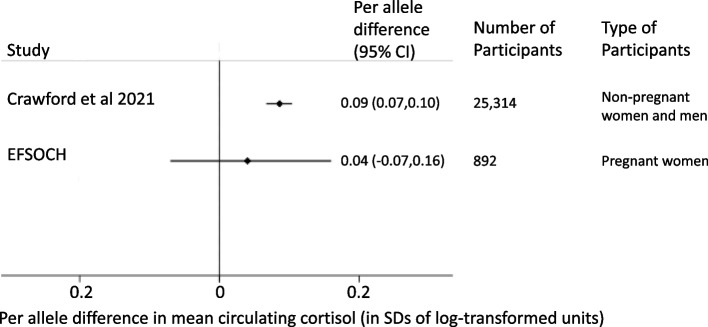
In the EFSOCH study [13], the mean value of women’s fasting plasma cortisol was 1,010 nmol/l (SD; 233 nmol/l) or 3 log-transformed nmol/l (SD; 0.1 log-transformed nmol/l). The SNP used in our main analyses had a considerably (2-fold) weaker association with women’s fasting plasma cortisol levels in pregnancy than seen in the main GWAS of non-pregnant women and men (0.04 (95% CI, -0.07 to 0.16) vs 0.09 (95% CI, 0.07 to 0.10)), though given the small sample size the estimate was imprecise with very wide confidence intervals that included the GWAS point estimate and the null (see Fig. 3).
Fig. 3.
Main SNPs effect on cortisol levels in primary GWAS and EFSOCH pregnancy sample
Possibility of the instrument influencing birth weight through horizontal pleiotropy
In total, 11 variables were associated at Bonferroni significance with rs9989237, and a further 1,516 variables were nominally associated with rs9989237. These associations with the cortisol increasing variant included higher levels of SERPINA1 (beta = 0.123, p = 4.09e-18), 39S ribosomal protein L33 (beta = 0.252, p = 2.82e-17), PH and SEC7 domain-containing protein 1 (beta = 0.200, p = 2.24e-11) and Histidine (beta = 0.026, p = 3e-07), as well as lower levels of Albumin (beta = -0.034, p = 1.18e-28), Synaptosomal-associated protein 25 (beta = -0.18, p = 1.78e-09) and sex-hormone binding globulin (SHBG) both with (beta = -0.005, p = 3.4e-07) and without (beta = -0.005, p = 1.7e-06) adjustment for body mass index (BMI), and in male only GWAS of SHBG (with BMI adjustment, beta = -0.007, p = 2.3e-06; without BMI adjustment, beta = -0.008, p = 6.6e-07). See Table 3 for details of the Bonferroni significant associations and Additional file 1 (Addition Table 3) for details for all nominally significant results.
Table 3.
Bonferroni threshold significant results for IEU-GWAS-R PheWAS of rs9989237
| Trait | N | Units | Per trait raising allele effect size | Per trait raising allele standard error | P |
|---|---|---|---|---|---|
| Albumin levels (inverse rank normalized transformed) | 315268 | Quantiles | -0.03354 | 0.00302 | 1.18E-28 |
| Albumin levels | 315268 | g/L | -0.08683 | 0.00789 | 3.63E-28 |
| SERPINA1 RNA expression in whole blood | 29950 | Z-score matrices | 0.123405 | 0.014223 | 4.09E-18 |
| Expression of 39S ribosomal protein L33, mitochondrial | 3301 | Relative concentration | 0.2515 | 0.0298 | 2.82E-17 |
| Expression of PH and SEC7 domain-containing protein 1 | 3301 | Relative concentration | 0.1998 | 0.0299 | 2.24E-11 |
| Expression of Synaptosomal-associated protein 25 | 3301 | Relative concentration | -0.18 | 0.0299 | 1.78E-09 |
| Histidine levels | 114895 | Z-scores | 0.026138 | 0.005102 | 3.00E-07 |
| Sex hormone-binding globulin levels adjusted for BMI | 368929 | Log-transformed nmol/l | -0.00535 | 0.00103 | 3.40E-07 |
| Sex hormone-binding globulin levels (male only GWAS) | 180726 | Log-transformed nmol/l | -0.00772 | 0.001513 | 6.60E-07 |
| Sex hormone-binding globulin levels | 370125 | Log-transformed nmol/l | -0.00515 | 0.001147 | 1.70E-06 |
| Sex hormone-binding globulin levels adjusted for BMI (male only GWAS) | 180094 | Log-transformed nmol/l | -0.00712 | 0.001419 | 2.30E-06 |
Discussion
We used two-sample MR with a single genetic variant to investigate the effect of maternal plasma cortisol on offspring birth weight. The results of the main analysis, the IVW analysis adjusted for between variant correlation and the SEM analysis were all directionally consistent with the observational association of higher maternal cortisol associating with lower offspring birth weight. However, all three methods of analysis used, provided imprecise estimates, which included values that are potentially of importance, as well as small or zero mean differences. For example, the 50 to 75 gram reductions in birth weight in both the main and SEM secondary analysis, respectively, together with their higher 95% confidence interval levels (both higher than 100g) are likely to be of clinical importance, whereas the lower confidence intervals (of an increase in 10 grams in the main analysis and a decrease of 9 grams in the SEM) are unlikely to be so. Therefore, the evidence of an effect of maternal cortisol on birth weight is uncertain and larger studies are required to identify whether maternal cortisol levels are a modifiable target for supporting healthy fetal growth and hence birth weight. That said, the point estimate for the association between the main genetic variant and cortisol measured in pregnancy may be considerably smaller than that seen in the original GWAS, which could mean our results are biased towards the null. In addition, with just one independent genetic variant we were unable to explore horizontal pleiotropy, using conventional two-sample MR methods and our MR PheWAS suggested that the cortisol increasing variant also related to lower mean levels of SHBG which could result in biased estimates.
A systematic review of the associations of maternal pregnancy cortisol with a range of offspring outcomes identified three studies that explored the association with offspring birth weight [26]. Two of the studies examined associations of maternal saliva cortisol and with birth weight in small numbers (70 and 55 participants). One study, which included 2810 participants, explored the association of maternal serum cortisol with birth weight [6]. Several estimates from the study suggested an inverse association with mean birth weight (ranging from a mean difference of -0.94 (95% CI, -1.75 to -0.12) to -0.07 (95% CI, -0.23 to 0.08) grams per nmol/l), which is directionally consistent with our findings. That study was not our own data, and it used different units of analyses, therefore we cannot directly compare the findings with our MR estimates. Further evidence of an inverse effect of maternal plasma cortisol on offspring birth weight came from a large (N = 1,858), well conducted RCT of antenatal corticosteroids in mothers at risk of preterm birth, found that randomization to antenatal corticosteroids was associated with lower offspring birth weight (mean difference -113.1 (95% CI, -187 to -41.17) grams) compared to placebo [27]. A secondary analysis of that RCT found that at least two thirds of the association could be explained by shorter gestational duration, though an effect was still detected (mean difference -33.5 (95% CI,-66.3 to -0.7) grams) [8]. Neither study reported the change in circulating corticosteroids in the mothers randomised to antenatal corticosteroid treatment compared to placebo, hence these findings cannot be compared with our MR results in the way we have previously compared MR and RCT results [28]. Lower birth weight has been associated with higher circulating cortisol in later life [29]. It is therefore possible that pregnant women with higher cortisol levels may have been smaller at birth and that an association between maternal cortisol and offspring birth weight could arise via the correlation between maternal and offspring size at birth. The birth weight effects of maternal genetic variants considered in our analyses were adjusted for the correlation with fetal genetics [12], so while this possibility remains to be investigated, it would not have influenced our results. A recent MR study on the effect of cortisol on birth weight, which has been published as part of a PhD thesis only (thus not peer-reviewed), found evidence of higher maternal cortisol leading to lower birth weight (-19 (95% CI, -34 to -7) grams per 1 log-transformed SD of cortisol). This was directionally consistent, but with a considerably weaker and more precisely estimated effect than we found. This study used an older, smaller GWAS for selecting genetic instruments than we used in this study [30, 31], which identified different genetic instruments, and used different methods to prepare the variables to adjust for between SNP correlations [32].
Strengths and limitations
This study used a large genome-wide data set of offspring birth weight, the UK Biobank and EGG meta-analyses [12]. However, the UK Biobank and EGG meta-analyses did not adjust for gestational duration, and as maternal cortisol has been associated with gestational duration in observational studies [33], this could be an alternative mechanism by which cortisol effects birth outcomes. We used a number of novel MR techniques to measure the effect of an exposure on an outcome when only a single locus is available. Additionally, we were able to partially validate the effect of the genetic instrument on maternal pregnancy cortisol using data from the EFSOCH cohort [13].
There are two important limitations to our study which relate to the genetic instruments for cortisol. First, despite using results from the largest GWAS to date of cortisol in our main analyses we only had one genetic instrument. Nonetheless, we chose the SNP with the strongest association with cortisol (R2 = 0.2%, F-statistic = 62) for the main analysis. Furthermore, we had near identical results when combining all four genome wide associated SNPs and controlling for their correlation. However, we cannot rule out weak instrument bias resulting in an underestimate of the causative effect [16]. We were not able to undertake conventional sensitivity analyses that are more robust to potential bias due to unbalanced horizontal pleiotropy [10]. The association of the genetic instrument with SHBG, albumin and histidine in MR-Base (at a p-value ≤2.6e-6) might indicate pleiotropic effects of our genetic instrument that may have biased our results. SHBG is produced in the liver and binds to steroid hormones, as does corticosteroid-binding globulin [34], which the SERPINA1/A6 locus encodes [11]. SHBG has been observed to be negatively associated with insulin resistance, type 2 diabetes and gestational diabetes (a cause of higher mean birth weight [35]) even after adjusting for BMI [36]. As the cortisol raising allele was associated with lower circulating levels of SHBG, this could result in masking pleiotropy, meaning our results are an underestimate of a true, stronger inverse effect. Circulating albumin levels are widely seen as a marker of protein sufficiency (lower levels, less sufficient), and low maternal albumin levels have been associated with lower offspring birth weight [37]. Histidine is a precursor to the inflammatory compound histamine [38], and higher maternal circulating levels of histidine have been shown to be associated with lower offspring birth weight in previous MR studies [39]. As the cortisol raising allele was associated with lower albumin levels and higher histidine levels, it could be that the suggestive evidence of a negative effect of the cortisol raising allele on birth weight is due, at least in part to pleiotropy, meaning our results could be biased. Additionally, our genetic instrument was associated with the expression of three proteins, none of which (to the best of our knowledge) has been found to be directly associated with birth weight in humans. In our PheWAS, we used a Bonferroni corrected p-value threshold, which is common in PheWAS exploring potential multiple causal effects of an exposure (e.g. 19,269). However, one could argue that when exploring bias this is less appropriate and we should not make this correction, or at least have a less stringent approach, as here the aim is to be as rigorous as possible in exploring potential biases [40]. Larger GWAS of circulating cortisol levels are needed to identify additional independent genetic instruments.
Our results assume that the effect of the genetic instrument on cortisol observed in the GWAS is the same as that during pregnancy. If the true effect in pregnancy is closer to what we observe in the EFSOCH pregnancy sample, then our MR analyses may be biased towards the null. Further evidence that the genetic instrument may not be valid in pregnancy comes from our PheWAS analysis, which shows the effect of rs9989237 on SHBG is stronger in men than women. However, the EFSOCH population sample is limited (N = 892; all in relative health) and the confidence intervals of the estimate captured the GWAS reported cortisol association. Despite this potential mitigation, the 2-fold difference between the GWAS reported cortisol association and the EFSOCH pregnancy cortisol association means there is legitimate concern that the SERPINA1/A6 locus is a weak instrument for pregnancy cortisol, leading to bias.
Conclusions
In conclusion, we found some evidence that higher maternal plasma cortisol may cause lower birth weight. Despite using the largest GWAS of cortisol to date, we only had one independent genetic locus and considering the potential sources of bias discussed above, more investigations are needed to make robust conclusions about the effect of maternal pregnancy cortisol on offspring birth weight.
Supplementary Information
Acknowledgements
This research has been conducted using the UK Biobank Resource under application number 7036. We would like to thank the participants and researchers from the UK Biobank who contributed or collected data and the families that took part in EFSOCH. We are grateful to the Genetics of Complex traits team at the University of Exeter, for their assistance in learning the methods and navigating the study data. The authors would like to acknowledge the use of the University of Exeter high-performance computing (HPC) facility in carrying out this work. This research was funded in part by the Wellcome Trust [Grant number WT220390]. For the purpose of open access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission.
Abbreviations
- RCT
Randomized Control Trial
- MR
Mendelian Randomization
- GWAS
Genome Wide Association Study
- EGG
Early Growth Genetics
- SNP
Single nucleotide polymorphism
Authors’ contributions
M-CB, DAL, RMF and RMR designed this study, with WDT further developing the design. NMW and DME supervised the development and running of the Structural Equation Model of birth weight to estimate conditional maternal and fetal genetic effects. ATH contributed to the collection and management of EFSOCH data, and TJM oversaw the collection, extraction, preparation and measurement of the EFSOCH pregnancy circulating cortisol data. WDT, RMF, DAL wrote the analysis plan, and WDT undertook most of the analyses with support from JT, M-CB, RB, ARW, NMW, DME, RMF and DAL. WDT wrote the first draft of the paper with support from RMR, M-CB, RMF and DAL; all authors read and made critical revisions to the paper. WDT, RMF, M-CB and DAL act as guarantors for the papers integrity
Funding
This study was supported by the US National Institute of Health (R01 DK10324), the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013) / ERC grant agreement no 669545, the British Heart Foundation (CS/16/4/32482 and AA/18/7/34219) and the NIHR Biomedical Centre at the University Hospitals Bristol NHS Foundation Trust and the University of Bristol. The Exeter Family Study of Childhood Health (EFSOCH) was supported by South West NHS Research and Development, Exeter NHS Research and Development, the Darlington Trust and the Peninsula National Institute of Health Research (NIHR) Clinical Research Facility at the University of Exeter. Genotyping of the EFSOCH study samples was funded by Wellcome Trust and Royal Society grant 104150/Z/14/Z. WDT is supported by the GW4 BIOMED DTP awarded to the Universities of Bath, Bristol, Cardiff and Exeter from the UK Medical Research Council (MRC). M-CB was supported by a UK MRC Skills Development Fellowship (MR/P014054/1). RMF and RNB were funded by a Wellcome Trust and Royal Society Sir Henry Dale Fellowship (WT104150). RMF is supported by a Wellcome Senior Research Fellowship (WT220390). M-CB, RMF and DAL work in / are affiliated with a unit that is supported by the University of Bristol and UK Medical Research Council (MC_UU_00011/6). DAL is a NIHR Senior Investigator (NF-0616-10102). ATH is supported by a NIHR Senior Investigator award and also a Wellcome Trust Senior Investigator award (098395/Z/12/Z). TJM is supported by a National Institute of Health Research Senior Clinical Lectureship (ICA-SCL-2016-02-003). RMR acknowledges the support of the British Heart Foundation (RE/18/5/34216). NMW is funded by an Australian National Health and Medical Research Council Investigator Grant (APP2008723). DME is funded by Australian National Health and Medical Research Council Senior Research Fellowships (APP1137714).The funders had no role in the design of the study, the collection, analysis, or interpretation of the data; the writing of the manuscript, or the decision to submit the manuscript for publication. The views expressed in this paper are those of the authors and not necessarily those of any funder.
Availability of data and materials
Our study uses two-sample Mendelian randomization (MR). We used both published summary results (i.e. taking results from published research papers and websites) and individual participant cohort data as follows:
For the two sample MR, we used genetic variants associated with circulating plasma cortisol. We extracted the exposure associations for these genetic variants from a dataset available to download at the University of Edinburgh DataShare site.
We extracted the outcome associations for these genetic instruments from genome-wide datasets of offspring birth weight adjusted for maternal genotype, available for download from the EGG Consortium.
http://egg-consortium.org/birth-weight-2019.html
The references to the journals that reported data sources are cited in the main paper.
We used individual participant data for the second MR sample and for undertaking sensitivity analyses from the UK Biobank and EFSOCH cohorts.
The data in UK Biobank is fully available, via managed systems, to any researchers. The managed system for both studies is a requirement of the study funders but access is not restricted on the basis of overlap with other applications to use the data or on the basis of peer review of the proposed science.
UK Biobank. Full information on how to access these data can be found here - https://www.ukbiobank.ac.uk/using-the-resource/
EFSOCH. Requests for access to the original EFSOCH dataset should be made in writing in the first instance to the EFSOCH data team via the Exeter Clinical Research Facility crf@exeter.ac.uk.
Declarations
Ethics approval and consent to participate
For UK Biobank, all participants provided written informed consent, including for their collected data to be used by international scientists. UK Biobank has approval from the North West Multi-centre Research Ethics Committee (MREC), which covers the UK. UK Biobank’s research ethics committee and Human Tissue Authority research tissue bank approvals mean that researchers wishing to use the resource do not need separate ethics approval. For EFSOCH, all mothers and fathers gave informed consent and ethical approval was obtained from the North and East Devon Local Research Ethics Committee.
Consent for publication
This study does not use data that could be used as a means of identification.
Competing interests
DAL has received support from Medtronic LTD and Roche Diagnostics for biomarker research that is not related to the study presented in this paper. The other authors report no conflicts.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Freathy RM, Lawlor DA and Borges MC joint senior authors who contributed equally.
References
- 1.Iliodromiti S, Mackay DF, Smith GC, Pell JP, Sattar N, Lawlor DA, Nelson SM. Customised and Noncustomised Birth Weight Centiles and Prediction of Stillbirth and Infant Mortality and Morbidity: a Cohort Study of 979,912 Term Singleton Pregnancies in Scotland. PLoS Med. 2017;14(1):e1002228. doi: 10.1371/journal.pmed.1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang SF, Shu L, Sheng J, Mu M, Wang S, Tao XY, Xu SJ, Tao FB. Birth weight and risk of coronary heart disease in adults: a meta-analysis of prospective cohort studies. J Dev Orig Health Dis. 2014;5(6):408–419. doi: 10.1017/S2040174414000440. [DOI] [PubMed] [Google Scholar]
- 3.Whincup PH, Kaye SJ, Owen CG, Huxley R, Cook DG, Anazawa S, Barrett-Connor E, Bhargava SK, Birgisdottir BE, Carlsson S, et al. Birth weight and risk of type 2 diabetes: a systematic review. JAMA. 2008;300(24):2886–2897. doi: 10.1001/jama.2008.886. [DOI] [PubMed] [Google Scholar]
- 4.de Jong F, Monuteaux MC, van Elburg RM, Gillman MW, Belfort MB. Systematic review and meta-analysis of preterm birth and later systolic blood pressure. Hypertension (Dallas, Tex : 1979) 2012;59(2):226–234. doi: 10.1161/HYPERTENSIONAHA.111.181784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reynolds RM. Glucocorticoid excess and the developmental origins of disease: two decades of testing the hypothesis–2012 Curt Richter Award Winner. Psychoneuroendocrinology. 2013;38(1):1–11. doi: 10.1016/j.psyneuen.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Goedhart G, Vrijkotte TGM, Roseboom TJ, van der Wal MF, Cuijpers P, Bonsel GJ. Maternal cortisol and offspring birthweight: Results from a large prospective cohort study. Psychoneuroendocrinology. 2010;35(5):644–652. doi: 10.1016/j.psyneuen.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Cherak SJ, Giesbrecht GF, Metcalfe A, Ronksley PE, Malebranche ME. The effect of gestational period on the association between maternal prenatal salivary cortisol and birth weight: a systematic review and meta-analysis. Psychoneuroendocrinology. 2018;94:49–62. doi: 10.1016/j.psyneuen.2018.04.023. [DOI] [PubMed] [Google Scholar]
- 8.Murphy KE, Willan AR, Hannah ME, Ohlsson A, Kelly EN, Matthews SG, Saigal S, Asztalos E, Ross S, Delisle MF, et al. Effect of antenatal corticosteroids on fetal growth and gestational age at birth. Obstet Gynecol. 2012;119(5):917–923. doi: 10.1097/AOG.0b013e31825189dc. [DOI] [PubMed] [Google Scholar]
- 9.Bleker LS, Roseboom TJ, Vrijkotte TG, Reynolds RM, de Rooij SR. Determinants of cortisol during pregnancy - The ABCD cohort. Psychoneuroendocrinology. 2017;83:172–181. doi: 10.1016/j.psyneuen.2017.05.026. [DOI] [PubMed] [Google Scholar]
- 10.Lawlor D, Richmond R, Warrington N, McMahon G, Davey Smith G, Bowden J, Evans DM. Using Mendelian randomization to determine causal effects of maternal pregnancy (intrauterine) exposures on offspring outcomes: sources of bias and methods for assessing them. Wellcome Open Res. 2017;2:11–11. doi: 10.12688/wellcomeopenres.10567.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford AA, Bankier S, Altmaier E, Barnes CLK, Clark DW, Ermel R, Friedrich N, van der Harst P, Joshi PK, Karhunen V, et al. Variation in the SERPINA6/SERPINA1 locus alters morning plasma cortisol, hepatic corticosteroid binding globulin expression, gene expression in peripheral tissues, and risk of cardiovascular disease. J Hum Genet. 2021;66(6):625–636. doi: 10.1038/s10038-020-00895-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warrington NM, Beaumont RN, Horikoshi M, Day FR, Helgeland Ø, Laurin C, Bacelis J, Peng S, Hao K, Feenstra B, et al. Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat Genet. 2019;51(5):804–814. doi: 10.1038/s41588-019-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knight B, Shields BM, Hattersley AT. The Exeter Family Study of Childhood Health (EFSOCH): study protocol and methodology. Paediatr Perinat Epidemiol. 2006;20(2):172–179. doi: 10.1111/j.1365-3016.2006.00701.x. [DOI] [PubMed] [Google Scholar]
- 14.Elsworth B, Lyon M, Alexander T, Liu Y, Matthews P, Hallett J, Bates P, Palmer T, Haberland V, Smith GD et al: The MRC IEU OpenGWAS data infrastructure. In.; 2020: 2020.2008.2010.244293.
- 15.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawlor DA. Commentary: Two-sample Mendelian randomization: opportunities and challenges. Int J Epidemiol. 2016;45(3):908–915. doi: 10.1093/ije/dyw127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horikoshi M, Beaumont RN, Day FR, Warrington NM, Kooijman MN, Fernandez-Tajes J, Feenstra B, van Zuydam NR, Gaulton KJ, Grarup N, et al. Genome-wide associations for birth weight and correlations with adult disease. Nature. 2016;538(7624):248–252. doi: 10.1038/nature19806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C, Grennan K, Badner J, Zhang D, Gershon E, Jin L, Liu C. Removing batch effects in analysis of expression microarray data: an evaluation of six batch adjustment methods. PLoS One. 2011;6(2):e17238. doi: 10.1371/journal.pone.0017238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warrington NM, Freathy RM, Neale MC, Evans DM: Using structural equation modelling to jointly estimate maternal and fetal effects on birthweight in the UK Biobank. Int J Epidemiol. 2018;47:1229–41. [DOI] [PMC free article] [PubMed]
- 20.Stephen B, Dylan SS, Simon GT. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2015;26(5):2333–2355. doi: 10.1177/0962280215597579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker VM, Davies NM, Hemani G, Zheng J, Haycock PC, Gaunt TR, Davey Smith G, Martin RM. Using the MR-Base platform to investigate risk factors and drug targets for thousands of phenotypes. Wellcome Open Res. 2019;4:113–113. doi: 10.12688/wellcomeopenres.15334.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46(6):1734–1739. doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40(4):304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davey Smith G, Spiller W, Bowden J, Del Greco MF, Thompson J, Sheehan N, Minelli C. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. Int J Epidemiol. 2018;47(4):1264–1278. doi: 10.1093/ije/dyy101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zijlmans MAC, Riksen-Walraven JM, de Weerth C. Associations between maternal prenatal cortisol concentrations and child outcomes: a systematic review. Neurosci Biobehav Rev. 2015;53:1–24. doi: 10.1016/j.neubiorev.2015.02.015. [DOI] [PubMed] [Google Scholar]
- 27.Murphy KE, Hannah ME, Willan AR, Hewson SA, Ohlsson A, Kelly EN, Matthews SG, Saigal S, Asztalos E, Ross S, et al. Multiple courses of antenatal corticosteroids for preterm birth (MACS): a randomised controlled trial. Lancet. 2008;372(9656):2143–2151. doi: 10.1016/S0140-6736(08)61929-7. [DOI] [PubMed] [Google Scholar]
- 28.Thompson WD, Tyrrell J, Borges M-C, Beaumont RN, Knight BA, Wood AR, Ring SM, Hattersley AT, Freathy RM, Lawlor DA. Association of maternal circulating 25(OH)D and calcium with birth weight: a mendelian randomisation analysis. PLOS Medicine. 2019;16(6):e1002828. doi: 10.1371/journal.pmed.1002828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Montfoort N, Finken MJJ, le Cessie S, Dekker FW, Wit JM. Could cortisol explain the association between birth weight and cardiovascular disease in later life? A meta-analysis. Eur J Endocrinol. 2005;153(6):811–817. doi: 10.1530/eje.1.02050. [DOI] [PubMed] [Google Scholar]
- 30.Bolton JL, Hayward C, Direk N, Lewis JG, Hammond GL, Hill LA, Anderson A, Huffman J, Wilson JF, Campbell H, et al. Genome Wide Association Identifies Common Variants at the SERPINA6/SERPINA1 Locus Influencing Plasma Cortisol and Corticosteroid Binding Globulin. PLoS Genet. 2014;10(7):e1004474. doi: 10.1371/journal.pgen.1004474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crawford AA, Soderberg S, Kirschbaum C, Murphy L, Eliasson M, Ebrahim S, Davey Smith G, Olsson T, Sattar N, Lawlor DA, et al. Morning plasma cortisol as a cardiovascular risk factor: findings from prospective cohort and Mendelian randomization studies. Eur J Endocrinol. 2019;181(4):429–438. doi: 10.1530/EJE-19-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Travers M: Can Racial Disparities in Poor Birth Outcomes Be Partially Attributed to Stress: A Mendellian Randomization Study. CUNY Academic Works 2021. https://academicworks.cuny.edu/sph_etds/72/.
- 33.Giurgescu C. Are maternal cortisol levels related to preterm birth? J Obstet Gynecol Neonatal Nurs. 2009;38(4):377–390. doi: 10.1111/j.1552-6909.2009.01034.x. [DOI] [PubMed] [Google Scholar]
- 34.Hammond GL. Plasma steroid-binding proteins: primary gatekeepers of steroid hormone action. J Endocrinol. 2016;230(1):R13–R25. doi: 10.1530/JOE-16-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kc K, Shakya S, Zhang H. Gestational Diabetes Mellitus and Macrosomia: A Literature Review. Ann Nutr Metab. 2015;66(Suppl. 2):14–20. doi: 10.1159/000371628. [DOI] [PubMed] [Google Scholar]
- 36.Thadhani R, Wolf M, Hsu-Blatman K, Sandler L, Nathan D, Ecker JL. First-trimester sex hormone binding globulin and subsequent gestational diabetes mellitus. Am J Obstet Gynecol. 2003;189(1):171–176. doi: 10.1067/mob.2003.343. [DOI] [PubMed] [Google Scholar]
- 37.Chaudhry ER, Chaudhry ZR, Chaudhry SR. Correlation of Maternal Albumin Levels with Neonatal Birth Weight. J Islam Int Med Coll (JIIMC) 2017;12(2):97–100. [Google Scholar]
- 38.Andersen HH, Elberling J, Arendt-Nielsen L. Human surrogate models of histaminergic and non-histaminergic itch. Acta Derm Venereol. 2015;95(7):771–777. doi: 10.2340/00015555-2146. [DOI] [PubMed] [Google Scholar]
- 39.Zhao J, Freathy R, Evans D, Warrington N, Langenberg C, Stewart I, Lotta L, Pietzner M, Borges MC, Lawlor D. 586Effects of maternal circulating amino acids on offspring birthweight: a Mendelian randomisation analysis. Int J Epidemiol. 2021;50(Supplement_1):dyab168.757.
- 40.Yang Q, Sanderson E, Tilling K, Borges MC, Lawlor DA. Exploring and mitigating potential bias when genetic instrumental variables are associated with multiple non-exposure traits in Mendelian randomization. Eur J Epidemiol. 2022;37:683–700. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Our study uses two-sample Mendelian randomization (MR). We used both published summary results (i.e. taking results from published research papers and websites) and individual participant cohort data as follows:
For the two sample MR, we used genetic variants associated with circulating plasma cortisol. We extracted the exposure associations for these genetic variants from a dataset available to download at the University of Edinburgh DataShare site.
We extracted the outcome associations for these genetic instruments from genome-wide datasets of offspring birth weight adjusted for maternal genotype, available for download from the EGG Consortium.
http://egg-consortium.org/birth-weight-2019.html
The references to the journals that reported data sources are cited in the main paper.
We used individual participant data for the second MR sample and for undertaking sensitivity analyses from the UK Biobank and EFSOCH cohorts.
The data in UK Biobank is fully available, via managed systems, to any researchers. The managed system for both studies is a requirement of the study funders but access is not restricted on the basis of overlap with other applications to use the data or on the basis of peer review of the proposed science.
UK Biobank. Full information on how to access these data can be found here - https://www.ukbiobank.ac.uk/using-the-resource/
EFSOCH. Requests for access to the original EFSOCH dataset should be made in writing in the first instance to the EFSOCH data team via the Exeter Clinical Research Facility crf@exeter.ac.uk.