Abstract
Inborn errors of immunity (IEI) are a collection of diseases resulting from genetic causes that impact the immune system through multiple mechanisms. Natural killer cell deficiency (NKD) is one such IEI where natural killer (NK) cells are the main immune lineage affected. Though rare, the deficiency of several genes has been described as underlying causes of NKD, including MCM4, GINS1, MCM10, and GINS4, all of which are involved in the eukaryotic CMG helicase. The CMG helicase is made up of CDC45 – MCM – GINS and accessory proteins including MCM10. The CMG helicase plays a critical role in DNA replication by unwinding the double helix and enabling access of polymerases to single-stranded DNA, and thus helicase proteins are active in any proliferating cell. Replication stress, DNA damage, and cell cycle arrest are among the cellular phenotypes attributed to loss of function variants in CMG helicase proteins. Despite the ubiquitous function of the CMG helicase, NK cells have an apparent susceptibility to the deficiency of helicase proteins. This review will examine the role of the CMG helicase in inborn errors of immunity through the lens of NKD and further discuss why natural killer cells can be so strongly affected by helicase deficiency.
Keywords: inborn errors of immunity, natural killer cell deficiency, CMG helicase, DNA replication
Introduction
Inborn errors of immunity (IEI) occur as the result of inherited monogenic variants that affect human immune function and lead to immunological disease (1). More than 500 individual genes have been ascribed to various immune conditions that result from IEI, including combined immunodeficiencies, immune dysregulation, and autoinflammatory conditions. Natural killer cell deficiency (NKD) accounts for a small subset of IEI where the primary immune lineage that is affected are natural killer cells (2). NKD presents with a lack of NK cells or a lack of cytotoxic function and is accompanied by severe recurrent viral infection (2). Viruses in the Herpesviridae family, including varicella zoster virus (VZV), human cytomegalovirus (HCMV), and Epstein-Barr virus (EBV) among others, are particularly susceptible to NK cell-mediated immunity (3). It is thought that herpes viruses act to evade cytotoxic T lymphocyte responses primarily by downregulating HLA-I expression in infected host cells, which consequently renders them susceptible to killing by natural killer cells (3, 4). Alternatively, other viruses such as influenza and parainfluenza can be recognized by binding to Natural Cytotoxicity Receptors (NCRs) like haemagglutinin or haemagglutinin-neuraminidase, respectively, to NKp46 on NK cells (5).
While there are few monogenic causes of NKD, an unexpected source of NKD is variants in the CMG helicase that forms the DNA replisome (6, 7, 8, 9, 10, 11). Despite the ubiquitous expression of, and requirement for, this complex in cellular function, the sole or primary immune defect in patients with these variants is frequently within the NK cell subset. The identification of these variants has provided new insight into the requirements for NK cell differentiation and homeostasis and led to a closer examination of the differential requirements for innate lymphocyte development and activation. Here, we will review the latest findings on the role of the CMG helicase in human immune cell function, primarily informed by studying variants associated with NKD.
The CMG Helicase
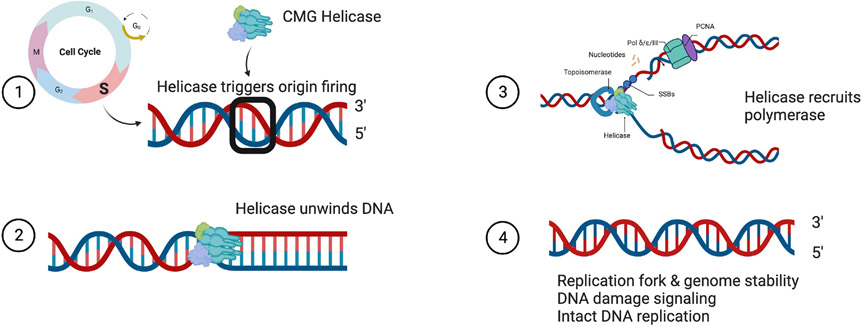
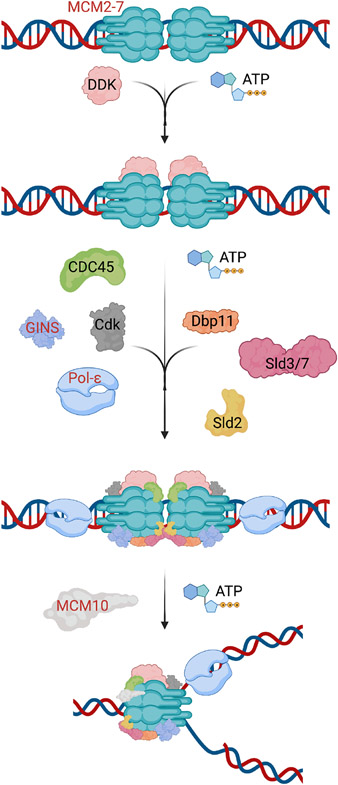
Faithful DNA replication during S phase occurs only once per cell cycle and is critical for maintaining genomic stability. The CMG helicase is composed of 11 subunits of the CDC45-MCM-GINS complexes (12) and is responsible for unwinding double-stranded DNA and recruiting polymerases for DNA replication (Fig 1). CDT1, CDC6, and the double hexamer MCM2–7 complex are recruited to the origin recognition complex (ORC) during G1 phase of the cell cycle; CDC45 and GINS1–4 are subsequently recruited to complete the formation of the CMG helicase. The MCM complex is the catalytic activity center of the CMG helicase as each individual subunit is an ATPase (13). The intact helicase can trigger origin firing and unwinding of the DNA double helix, allowing DNA polymerase to begin priming the DNA (12, 14, 15). The engagement of MCM10 and other factors including PCNA facilitates elongation of the replication fork and aids in overall stability during replication (16, 17, 18). Assembly of the CMG helicase is dynamic and involves the assistance of accessory proteins for proper function and stability. These accessory proteins could also potentially be involved in the development of NKD (Fig 2). CMG helicase function is required in any actively proliferating cell, and the reason why NK cells are seemingly particularly susceptible to damaging variants has not been elucidated.
Figure 1.
The CMG helicase primarily functions in S phase (1) to trigger origin firing and DNA unwinding (2) to facilitate DNA replication (3). An intact helicase maintains replication fork and genomic stability, participates in DNA damage signaling, and is critical for DNA replication to occur (4).
Figure 2. A model of CMG helicase assembly during G1/S phase.
Subunits shown in red font have been identified as helicase complex members and related proteins that have been implicated in NKD.
The CMG helicase, both as individual components and as a functional complex, is highly regulated during DNA replication and, by extension, the cell cycle. Multiple kinases, including Cyclin A-Cdk2 and Dbf4-Cdc7, function to activate the helicase at entry to S phase (19, 20). Another form of regulation lies with CDC45 protein expression, which in mammalian cells has been shown to be the rate-limiting factor in DNA replication (21, 22). Further regulation of the helicase complex is found through the amount of protein actively used. It is thought that an excess of MCM protein is loaded onto chromatin while lacking colocalization with replication forks to ensure ample coverage of origins of replication, a paradigm commonly called the MCM paradox (23). However, recent work has highlighted a possible role for this excess of MCM protein in replication fork speed and in response to replication stress (24). Thus, tight control of the CMG helicase during canonical DNA replication is critical to maintaining genome stability, accurate replication of the genome, and cell cycle progression.
Noncanonical roles for helicase proteins
In addition to the canonical role that helicase proteins play as part of a functional complex in DNA replication, several CMG proteins have auxiliary roles in other cellular functions unrelated to replication. CDC45, for example, binds to single-stranded DNA (ssDNA) (25) and single-strand/double-strand DNA junction sites (26). CDC45 is also responsible for loading replication protein A (RPA) onto nascent ssDNA at the replication fork through binding to the RPA70A subdomain (27). In yeast, Cdc45 is involved in checkpoint signaling during fork stalling by binding with Rad53 (28). As such, CDC45 may play an additional specific role in the sensing of ssDNA and signaling for the DNA damage response.
MCM2–7 proteins likewise can have supplementary roles separate from replication (29). MCM proteins, particularly MCM2, have been implicated in transcriptional control, and blocking MCM2 activity with antibodies inhibits RNA polymerase II (Pol II) transcriptional machinery (30). MCM5 has also been found to bind directly to the transcription factor STAT1 to increase transcription activation during interferon-λ stimulation, an important avenue of MCM function in immunity (31, 32). Mcm7 likewise has been shown to bind with proteins critical to cellular functions, namely retinoblastoma tumor suppressor protein (Rb), which regulates cell cycle and proliferation through E2F (33) and E6 oncoprotein, likely contributing to the development of cancer (34). Finally, MCM proteins may play a role in chromatin regulation and maintenance, including binding of Mcm2 to histone H3 (35) and MCM10 in partnership with MUS81 (11).
The GINS complex, likely through helicase activity in replication and proliferation, is also involved in mammalian development of the nervous system and early embryogenesis (36, 37). Gins2 is required for zebrafish development through interaction with Gins4 protein and promotes overall CMG helicase stability (38). Outside of CMG helicase activity, increased expression of Gins4 has been associated with advanced gastric cancer. Gins4 binds to and actives the Rac1/CDC42 complex, which is a signaling pathway related to cell proliferation and migration (39). Another divergent role for Gins is involvement in chromosomal alignment during mitosis (40). GINS4, also called Sld5, recruits PCM-1, and loss of Sld5 function leaves chromosomes susceptible to fragmentation.
Helicase variants and NKD
Work by our lab and others have defined variants in the eukaryotic DNA helicase complex, including MCM4, GINS1, MCM10, and GINS4, that result in NKD. Specifically, NKD due to helicase variants manifests as impaired NK cell differentiation and maturation leading to significant functional impairment and resulting viral infections in affected individuals (Table 1). At the cellular level, these mutations lead to impaired cell cycle progression, replication stress, and increased DNA damage and DNA damage responses (6, 7, 8, 9, 10) (Table 2).
Table 1.
Clinical Phenotype.
| Gene | Reported variant type |
Patient identifiers |
Clinical phenotype | Viral Infections | Reference |
|---|---|---|---|---|---|
| MCM4 | Homozygous c.70_71insG (frameshift) | 6 + 8 | Viral infection, short stature, adrenal insufficiency | P1: CMV P2: none P1: EBV, HSV, VZV P2: respiratory infections, EBV, HSV P3: EBV, HSV P4: respiratory infections, EBV, HSV, VZV P1: none P2: none P3: none P4: none P5: none P6: pneumonitis P7: none P8: none |
(8, 10) |
| GINS1 | Compound heterozygous P1-2: c.−48C>G (new donor splice site) c.247C>T p.R83C (missense) P3: c.−60A>G (new donor splice site)P4: c.455G>A p.C152Y | 5 | Viral infection, intrauterine growth retardation, neutropenia | P1: CMV, chest infection P2: chest infection, rotavirus P3: CMV, VZV, adenovirus, RSV, rotavirus P4: VZV, HSV, chest infection P5: VZV, influenza |
(9) |
| MCM10 | Compound heterozygous c.1276C>T p.R426C (missense) c.1744C>T p.R582X (stop gain) | 1 | Viral infections | CMV | (7) |
| GINS4 | Compound heterozygous c.511G>C p.V171L (missense); c.C571T p.Q191X (stop gain) | 2 | Viral infections, intrauterine growth restriction and growth delay, neutropenia | P1: BCG infection after vaccination, CMV, VZV, HSV P2: VZV, HSV |
(6) |
| POLA1 | Hypomorphic X-linked c.328G>A | 5 | XLPDR, viral infections, intrauterine and postnatal growth retardation, hypogonadism | Respiratory infections | (79) |
| POLE2 | Homozygous splice-site SNP c.1Ex13G>T | 1 | Respiratory viruses, BCG and Candida albicans infections, diphtheria and tetanus toxoid, combined immunodeficiency, hypothyroidism, facial dysmorphism | BCG infection after vaccination, respiratory infections | (81) |
CMV: Cytomegalovirus
EBV: Epstein-Barr virus
HSV: Herpes simplex virus
VZV: Varicella zoster virus
RSV: respiratory syncytial virus
BCG: Bacillus Calmette-Guerin vaccine
SNP: single nucleotide polymorphism
Table 2.
Cellular Phenotype.
| Gene | NK cell phenotype | Cell cycle | Mouse Model | Reference |
|---|---|---|---|---|
| MCM4 | ↓ NK cell frequency ↓ CD56dim NK cells ↓ CD57+ NK cells ↓ Perforin+ NK cells ↓ Cell proliferation ↑ Apoptosis (CD56bright & CD56dim) |
Fewer cells in G1/S phases, more cells in G2/M phases |
Chaos3 (hypomorphic); Mcm4−/− (lethal). ↓ NK cells No growth retardation Chromosomal instability |
(8, 10) |
| GINS1 | ↓ CD56bright & CD56dim NK cells Unresponsive to cytokine stimulation |
Gins1−/− (embryonic lethal) | (9) | |
| MCM10 | ↓ NK cells ↓ CD56bright NK cells S phase arrest (MCM10-KD cell line) ↑ doubling time (MCM10-KD cell line) ↑ γH2AX foci Normal cytotoxic function (MCM10-KD cell line; K562 target cells) |
(7) | ||
| GINS4 | ↓ NK cells ↑ CD56bright NK cells ↓ CD57+ NK cells ↓ CD94+ NK cells ↓ cytotoxic function (K562 target cells) ↓ IFN-γ production ↓ survival when stimulated with PMA |
Mild mitotic exit defect | (6) | |
| POLA1 | ↓ NK cell frequency ↓ lymphocytes ↓ CD56dim NK cells ↓ CD57+ NK cells Normal perforin & granzyme ↓ cytotoxic function (721.221 cells) Normal proliferation Normal TNF+ or IFNγ+ cells ↓ polarization of lytic granules ↑ Degranulation ↓ cytotoxic function (MCM4 and POLA1 KD NK cell lines) |
Not investigated | None | (79) |
| POLE2 | ↓ NK cell frequency ↑ CD56bright NK cells |
↓ proportion of cells in S phase | None | (81) |
KD: knockdown
PMA: phorbol 12-myristate 13-acetate
Human NK cell development
Insights into the specific effect of helicase variants on NK cells can be gained by careful study of human NK cell development. NK cell and innate lymphoid cell (NK/ILC) precursors are found in lymphoid tissues including fetal liver, bone marrow, thymus, and secondary lymphoid tissues (41, 42, 43). In the adult, NK cells are derived from bone marrow precursors and further mature in lymph nodes (41). Studies of the in vitro generation of NK cells has shown that cytotoxic NK cells are successfully generated from CD34+ precursor cells from cord blood, bone marrow, tonsil, and lymph node in the presence of cytokines (IL-3, IL-7, IL-15, SCF, and Flt3-L) and stromal cells (44, 45, 46).
Circulating human NK cells are commonly identified as CD3− CD56/16+ by flow cytometry. Although recent single cell and scRNA-seq analysis have revealed that NK cells are highly heterogeneous (47, 48, 49, 50), the two major populations of peripheral blood NK cells are frequently defined as CD56brightCD16− (~10% of NK cells), and CD56dim CD16+ subsets (~90%) (51). CD56brightCD16− NK cells have lower basal cytotoxicity but secrete high amounts of IFN-γ in response to exogenous cytokines including IL-12 and IL-18, which are derived from activated dendritic cells and macrophages in the context of inflammation and infection. CD56bright CD16− NK cells also express cytokine receptors including CD25 (high-affinity IL-2Rα), CD117 (SCF receptor), and CD127 (IL-7Rα), which are not highly expressed on CD56dim NK cells. Cytotoxic function in CD56bright CD16− NK cells is decreased relative to CD56dim NK cells, but they can acquire cytotoxic function in response to IL-2 and IL-15 in concert with rapid proliferation (52). In contrast, CD56dim CD16+ NK cells have higher basal cytotoxic capacity, but their proliferation potential is limited except for the CD62L+ CD57− subset, which is considered a developmental intermediate between CD56bright and CD56dim (53).
In vivo and in vitro studies seem to demonstrate that CD56bright NK cells are precursors of CD56dim cells. CD56bright cells have a longer telomere length than CD56dim, and IL-2 or IL-15-activated CD56bright cells acquire CD16 and killer immunoglobulin-like receptors (KIR) during their proliferation (53, 54). The differentiation of CD56bright to CD56dim is also confirmed in humanized mice (55) as well as reconstituted NK cells in hematopoietic stem cell transplantation recipients (56, 57) and scRNA-seq studies (47, 58). While human NK cell precursors are thought to be continuously generated from bone marrow, secondary lymphoid tissues such as lymph nodes also have a role in the generation and differentiation of NK cells (59). Ultimately, the sites and homeostatic dynamics of human NK cell differentiation remain poorly understood.
NK cell kinetics and NKD
In addition to their different functions, the two major subsets of peripheral blood NK cell have different homeostatic turnover rates. In vivo kinetics studies showed that circulating NK cells more rapidly proliferate and die compared to T cells, even without infections (60). Further, immature CD56bright CD16− NK cells generated from bone marrow proliferate faster than CD56dim subsets (60). Even though CD56dim NK cells are less proliferative, a long-lived and self-renewing adaptive NK cell pool is consistently maintained (60). Therefore, an imbalanced CD56bright / CD56dim ratio, such as is often found in NKD, may not be due to increased CD56bright NK cells from a block in terminal maturation but can instead suggest that the long-lived adaptive NK cell pool is not stably differentiated from short-lived CD56bright NK cells.
In addition to inherited variants, de novo mutations can cause an NKD phenotype in adults. One such case is heterozygous GATA2 mutations which can cause the loss of circulating dendritic cells, monocytes, B cells, and NK cells (61, 62, 63, 64, 65). While the cellular phenotype of GATA2 can be quite variable, the selective loss of the CD56bright subset of NK cells is a highly conserved phenotype and, in some individuals with GATA2 deficiency, NKD is the predominant immune phenotype (66, 67, 68). This finding was confirmed in patients with paroxysmal nocturnal hemoglobinuria (PNH) (69). In this study, newly produced glycosyl-phosphatidylinositol (GPI)-negative immune cells from hematopoietic stem and progenitor cells with somatic PIGA mutations make chimerism with GPI-positive cells, and they are clearly discriminated by flow cytometry analysis of GPI expression. While T and B lymphocytes are maintained and dominantly GPI-positive, in the short-lived neutrophil population the GPI-negative ratio increases over time as they are continuously produced from the precursor cells in the bone marrow. Interestingly, the fraction of GPI-negative CD56bright CD16− NK cells correlate with neutrophils, which implies that turnover of this NK cell subset is rapid and that these cells are not differentiating into CD56dim NK cells. In contrast, GPI-positive populations are dominant in CD56dim NK cells, and the fraction is persistent over time compared with T and B cells, suggesting that these cells are long-lived. It may also be speculated that this impaired generation of mature NK cells, including CD56dim and adaptive NK cell subsets, is what leaves individuals with NKD susceptible to recurrent viral infection. The study of human NK cells in the context of naturally occurring variations demonstrates different homeostatic properties and ontogenies of the CD56bright and CD56dim subsets and suggests differential mechanisms for the generation and maintenance of these subsets that can be further informed by considering the effect of helicase deficiencies on NK cells.
Helicase variants and IEI
MCM4
MCM4 was the first helicase complex member associated with NKD when multiple families with a variant in MCM4 (c.71-1insG) were identified (70, 71). These individuals present with adrenal insufficiency, growth retardation and developmental abnormalities, NKD, and recurrent viral infections such as herpes simplex virus (HSV) and varicella zoster virus (VZV). In 2012, another group published a clinical description of eight additional patients from an endogamous Irish traveler cohort with the same variant in MCM4 and a similar clinical phenotype (8). Gineau et al investigated the mechanisms underlying MCM4 deficiency by testing the variant in patient-derived fibroblasts and determined it results in a prematurely truncated protein at the N-terminal domain (10). However, cell extracts from patient fibroblasts showed interaction of truncated MCM4 with other MCM complex proteins as well as binding to chromatin, suggesting it could retain some function in activities critical to DNA replication, including initiation and elongation.
Further investigation into DNA replication found a decreased proportion of MCM4-deficient fibroblasts in G1 and S phase compared to controls with a concomitant increase of cells found in G2 and M phases (10). DNA content by propidium iodide labeling of patient fibroblasts was also higher than that of controls, indicating a disruption in the synchronization of replication and an overall cell cycle defect. This phenotype was amplified by aphidicolin treatment, which is an inhibitor of DNA replication by binding to polymerase-α. Assessment of genomic stability was also analyzed by calculating the average number of chromosome breaks in metaphase and demonstrating that patient fibroblasts had significantly more breaks. The addition of aphidicolin to cells with truncated MCM4 protein resulted in reduced DNA damage repair efficiency relative to those with full-length MCM4, collectively demonstrating that the truncated MCM4 variant leads to decreased cell cycle progression resulting in genomic instability and increased sensitivity to DNA damage.
Despite the ubiquitous function of the CMG complex in DNA replication and cell cycle, there was no impact on myeloid subsets or T cells in affected individuals, but a slight decrease in CD27+CD19+ memory B cells and a significant decrease in natural killer cell numbers was noted (10). Specifically, CD56dim NK cells were decreased, resulting in an increased ratio of CD56bright to CD56dim NK cells and decreased proportions of perforin-positive NK cells and terminally mature CD57+ NK cells (10). CD56bright NK cells did not proliferate in response to IL-2 or IL-15, which are cytokines required for NK cell maturation, proliferation, and survival. Patient CD56bright NK cells had increased apoptosis that could be rescued by the addition of IL-2 and IL-15, while CD56dim cells did not respond to stimulation. Notably, NK cells are also affected in the mouse model that are hypomorphic for the Chaos3 (Mcm4) allele, and this defect is found in addition to global genomic instability, defects in the adrenal gland, and severe growth failure (10). Gineau et al speculated that since MCM4 has higher expression in CD56bright NK cells, a deficiency in this protein results in a block of terminal maturation into the CD56dim subset through an accumulation of chromosomal damage during proliferation (10).
GINS1
The tetrameric GINS complex, including GINS1–4, is required for initiation and elongation of DNA replication by unwinding DNA and preferentially binding to single-stranded DNA (15, 72). Five individuals with a compound heterozygous mutation in GINS1 have been described (9, 70, 71). Consistent with the MCM4 clinical phenotype, these individuals have growth restriction, susceptibility to viral infections, and low NK cell numbers in peripheral blood (9). Unlike the MCM4 patients, the five GINS1-deficient individuals do not have adrenal insufficiency, though chronic neutropenia is present, and two patients also had transient disruptions in T and B cells, particularly CD8+ T cells and memory B cells (70, 71). Cottineau et al further investigated the role that GINS1 plays in NKD (9). Importantly, GINS1 patients had low proportions of both CD56bright and CD56dim subsets, which did not respond to stimulation with IL-2 or IL-15, unlike the MCM4 patients (9). However, while both NK cell subsets were decreased, there was a greater relative decrease of CD56dim NK cells, suggesting that a similar block in NK cell terminal maturation may also be present as in MCM4. In addition, certain ILC subsets were also decreased in these patients, suggesting that a common precursor may be affected.
GINS3 and GINS4 protein expression levels are also decreased in GINS1-patient derived cells, indicating that GINS complex stability and function are affected by loss of GINS1 protein expression (9). Additionally, Cottineau et al. showed that primary fibroblasts from patients exhibited abnormal nuclear morphology, including increased nuclear area and disproportionate misshapen nuclei, often with multi-lobes of varying sizes, indicating a failure to maintain genome integrity and signify mis-segregation of chromosomes. Further investigation into the cell cycle of primary fibroblasts showed increased percentages of cells in G2 and M phases for patient cells, with a concurrent decrease of cells in G1 phase. Patient cells were slow to re-enter cell cycle after synchronization, but eventually the proportion of cells in S phase was maintained. DNA fiber analysis determined that patient cells had disruptions in replication patterns, including decreased numbers of replication clusters and bidirectional forks as well as an increase in stalled replication forks. Replication fork speed was determined to be faster than controls, likely as a mechanism to compensate for the decreased number of active replication forks. Thus, in GINS1-variant cells, DNA replication can be initiated but not all replication origins are activated or maintained. In addition, patient fibroblasts proliferate slower than controls and have a senescent phenotype. DNA damage in response to hydroxyurea was higher in patient E6/E7-fibroblasts as indicated by γH2AX and 53BP1 foci (9). While the DNA damage response through CHK1 and RPA phosphorylation was decreased, CHK2 phosphorylation was normal, indicating that the ATR signaling pathway is affected by GINS1 deficiency, not the ATM signaling pathway. As in the case of MCM4 deficiency, selective NKD is suspected to be the result of negatively impacted NK cell survival. It is likely that the phenotypes of impaired cell cycle progression, replication dynamics, and DNA damage contribute to decreased survival of specific NK cell subsets and neutrophils, although the basis for neutropenia found in GINS1, but not MCM4 deficiency, is not understood.
MCM10
Several additional components are necessary for optimal function of the CMG helicase. MCM10, while distinct from the MCM2–7 helicase, is a highly conserved component required for assembly of the CMG helicase (14) and binding to both single- and double-stranded DNA during origin unwinding (18). Importantly, MCM10 is also essential for the recruitment of polymerase-α and its association with chromatin (18), thus MCM10 is necessary for elongation of DNA synthesis as well as initiation.
Like MCM4 and GINS1, a compound heterozygous variant in MCM10 was determined to be causative of NKD in a single patient with fatal cytomegalovirus (CMV) infection (7) and decreased circulating NK cells in peripheral blood with an increased percentage of the CD56bright cells. While MCM10 protein expression wasn’t affected, patient variants displayed increased chromatin association indicative of a disruption in cell cycle and impaired function of MCM10 protein (7). Like the irregular nuclear morphology of MCM4 and GINS1 variant cells, MCM10 patient fibroblasts displayed increased nuclear area and increased frequencies of γH2AX foci. An increased frequency of patient cells in S phase further indicated that MCM10 variants have a cellular phenotype of replication stress and disrupted cell cycle dynamics. The cell cycle defect was maintained in NK cell lines, with an increased percentage of cells found in S phase as well as an increased doubling time. Consistent with the patient-derived fibroblasts and other published helicase mutations, MCM10 knockdown in the NK92 NK cell line led to more γH2AX foci at baseline and in response to irradiation. It should also be noted that cytotoxic function was unaffected in these cells, indicating that MCM10 does not play a direct role in killing functions, but likely only in maturation and proliferative responses of NK cells. To investigate the role of MCM10 in development of natural killer cells, MCM10 was knocked down in CD34+ precursor cells from a healthy donor using shRNA. When cultured to make in vitro NK cells, the MCM10 deficient precursors led to an increased proportion of immature subsets with a concurrent decrease in CD16+ mature NK cells. NSG mice were injected with CD34+ precursors that were produced from patient-derived fibroblasts reprogrammed into induced pluripotent stem cells (iPSCs). This mouse model reconstituted peripheral blood and splenic NK cells, with increased CD56bright cells and increased γH2AX foci. Using both in vitro models and in vivo reconstitution, the MCM10 protein was determined to be a critical component required for NK cell maturation and extended the phenotype found in core CMG complex members to other regulators of cell cycle and genomic stability.
GINS4
GINS4 has recently been identified as the fourth monogenic cause of NKD related to the CMG helicase (6). GINS4 is component of the GINS tetrameric complex and thus plays a similar role to the GINS1 protein in initiation and elongation during DNA replication. In Drosophila, GINS4, also known as Sld5, interacts with GINS1 and GINS2 as well as MCM10 to preserve genomic integrity (73). In humans, GINS4, which is structurally similar to GINS1, functions as part of the GINS complex to recruit additional components required for DNA replication, including CDC45 and polymerases (74). The absence of Sld5 expression causes cell cycle delays in M and S phase in Drosophila cells, while overexpression of this protein has been associated with human colorectal (75), lung (76), and bladder (77) cancers.
Low numbers of NK cells were detected in two siblings with inherited biallelic GINS4 variants, with a higher frequency of CD56bright cells present in peripheral blood (6). There was also noted to be intrauterine growth restriction and susceptibility to viral infection, a phenotype that resembles other helicase variants that cause NKD. Like the GINS1 patients, GINS4 individuals had neutropenia that responded productively to granulocyte colony-stimulating factor (G-CSF), while other immune subsets such as T and B cells were normal, including γδ T cells, and mucosal-associated invariant T (MAIT) cells. Patients had decreased NK cell cytotoxic function in a standard four-hour chromium release experiment against K562 tumor cells and NK cell cytotoxic function was not rescued by exogenous IL-2. Like the MCM10 study, GINS4 was knocked down in CD34+ precursors to examine the effect of GINS4 protein loss on the development of mature NK cells. Mature NK cells were significantly decreased compared to controls after in vitro differentiation.
The two siblings express ~15% of GINS4 protein relative to unaffected individuals, which affects assembly of the GINS complex as well as expression levels of other GINS proteins, namely GINS1 and GINS3. Interestingly, there were no significant changes to the cell cycle profile of patient-derived B cell lines, yet proliferation studies of patient-derived cells showed they were slower to re-enter cell cycle (6). Gene expression analysis further confirmed no differentially expressed genes related to cell cycle were present in GINS4 deficient NK cells. Further, GINS4 variant B cell lines and GINS4-deficient RPE hTERT cell lines had decreased cells entering S phase compared to healthy controls. This indicates a role for GINS4 protein in mitotic exit. Further diverging from DNA damage and DNA damage response phenotype of other helicase mutations, the GINS4-deficient cell lines displayed only mild increases in DNA damage as evidenced by γH2AX foci, and little or no changes to DNA damage response indicated by ubiquitinated PCNA and pCHK1 compared to healthy controls. GINS4 patient BLCLs also lacked changes in replication fork symmetry or speed as analyzed by DNA fiber analysis, unlike similar experiments in other helicase variants. Although this study lacked an in vivo mouse model, GINS4 contributed to our understanding of the role of the CMG helicase in NK cell development and further highlighted the heterogeneity seen in helicase variants and NKD. It also identified neutropenia to be a common feature resulting from GINS complex variants.
POLA1
Though not a component of the CMG helicase, DNA polymerase-α (Pol-α) is required for DNA replication initiation. In eukaryotes, the catalytic subunit Pol-α is thought to interact with the helicase through binding to MCM10, triggering replication by generating de novo primers for both the leading and lagging strands (16). Partial hypomorphic Pol-α deficiency has been shown to result in X-linked reticulate pigmentary disorder (XLPRD) (78, 79). These patients have overlapping phenotypes with the previously described helicase variant patients, including growth retardation, recurrent infection, hypogonadism, and reduced NK cells with relatively higher frequency of the CD56bright subset (79). siRNA against POLA1, the gene encoding for Pol-α, in both primary NK cells and the NK92 cell line led to decreased cytotoxicity against K562 tumor cells (79). Further investigation found that POLA1 and MCM4 expression were linked and together are causative of decreased cytotoxicity of NK cells. However, no DNA damage was indicated in patient cells according to γH2AX or 53BP1 staining, and there was no impairment of proliferation or increase in apoptosis in patient NK cells.
Though the cellular phenotype of DNA damage and proliferative or cell cycle defects was not studied, Pol-α deficiency was linked to a disruption in lytic granule localization to the microtubule organizing center (MTOC) and the immune synapse (IS), which resulted in decreased cytotoxicity (79). This is the first time a helicase deficiency has been linked directly to cytotoxic function, although this phenotype may also reflect a global impairment in NK cell activation leading to a defect in granule convergence during immune synapse formation. Pol-α deficiency further builds on our understanding of how NK cell development and maturation depends critically on DNA replication machinery.
POLE2
DNA polymerase epsilon (Pol-ε) is involved in synthesis of the leading strand after Pol-α has primed the DNA (80). In addition to the role of leading strand replication, Pol-ε functions in proofreading DNA, regulating cell cycle, and maintaining epigenetics of DNA, and thus is critical for maintaining genome stability (80, 81). Pol-ε closely interacts with several members of the CMG helicase, including the GINS complex and CDC45 during replication (82). The POLE2 gene encodes an important accessory component of pol-ε that is approximately 59 kDa (81, 83). In a brief report by Frugoni et al, an individual was reported to have a homozygous single polymorphic nucleotide (SNP) splice-site mutation at position 1 of intron 13 on POLE2 (81). This individual displayed hypothyroidism and combined immunodeficiency, with T cell lymphopenia, neutropenia, NK cell deficiency with an increased ratio of CD56Bright/CD56Dim, and the absence of circulating B cells, among other immune issues (81). Recurrent respiratory infections were present in this individual as well as systemic BCG infection after vaccination. Though NK cell specific investigation was not conducted, the authors found that T cell stimulation of peripheral blood mononuclear cells (PBMCs) with anti-CD3 with or without anti-CD28 antibodies lead to an increase proportion of cells undergoing apoptosis by Annexin V staining (81). Further work on this individual’s PBMCs and fibroblasts found a decreased percentage of cells found in S phase, indicating that cell cycle progression is impacted (81). The cell cycle phenotype of patient fibroblasts was partially rescued using wild-type POLE2 lentivirus transfection, resulting in an increased proportion of cells in S phase, despite the percentage of cells in G2M phase remaining high (81). Despite the absence of detailed examination in the NK cell compartment in this individual, the role that various polymerases play in human disease is crucial to our understanding of replication and proliferation in lymphocytes development and function.
Other genes
Other genes involved in the CMG helicase have been reported to result in human disease. MCM2 deficiency, through autosomal dominant inheritance, results in deafness through selective apoptosis of cells in the ear (84), while MCM3 and MCM5 deficiencies, through autosomal recessive inheritance, can cause Meier-Gorlin syndrome, a disorder characterized by short stature, small ears, and no or small patellae (85, 86). Primary fibroblasts from one MCM5 deficient individual displayed a dysregulation of progression through S phase using a BrdU pulse-chase experiment. The addition of hydroxyurea to induce replication stress resulted in patient cells remaining arrested in the cell cycle, possibly due to impairment of dormant origins. CDC45 deficiency results in Meier-Gorlin syndrome (87), though further investigation is required to connect CDC45 with a molecular mechanism resulting in disease. More recently, both GINS2 and GINS3 deficiency were also described to result in Meier-Gorlin syndrome with autosomal recessive inheritance patterns (88, 89). Though the GINS2 case study did not investigate the stability or expression of other GINS proteins, GINS3 deficiency resulting in MGS also had decreased GINS1 protein expression. Modeling the GINS2 patient variant in yeast cells did not reveal a growth or cell cycle defect when exposed to hydroxyurea, but replication stress was strongly induced in the presence of histone deacetylase inhibitor NAM (89). It should be noted that none of the genes in this section resulted in an immunodeficiency phenotype, despite displaying similar molecular mechanisms to the genes underlying NKD. However, in patients with severe syndromic disease, relatively milder immune deficiencies may not be noted and an in-depth study of the immune profiles of patients with Meier-Gorlin syndrome has not been reported. Finally, a single case study of a homozygous mutation in RTEL1 identified NKD with fatal VZV infection, decreased NK cells in peripheral blood and decreased function without impact on other lymphocyte populations (90, 91). RTEL1, regulator of telomere elongation helicase 1, functions similarly to the CMG helicase in that DNA is stabilized and unwound during telomere elongation. This is supportive evidence natural killer cells are sensitive to a requirement for and reliant upon different types of helicases for maturation and function.
Abnormal NK cell differentiation in RAG deficient T- B- NK+ SCID and related disorders
Finally, interesting insight can be gained from the study of hypomorphic RAG variants leading to T− B− NK+ severe combined immune deficiency (SCID). Dysfunction of V(D)J rearrangement genes (RAG1, RAG2, DCLRE1C, NHEJ1, and LIG4) causes a broad spectrum of clinical phenotypes dependent on recombinase activities, including T− B- SCID, Omenn syndrome (OS), atypical SCID (AS), and combined immune deficiency with granuloma and/or autoimmunity (CID-G/A) (92, 93, 94, 95). Mutations in these genes can also affect NK cell differentiation and maturation despite the lack of RAG-mediated antigen receptor rearrangement in NK cells, and NK cells from these individuals often have immature phenotypes compared to age-matched healthy subjects (96). Specifically, the frequency of CD56bright NK cells is higher and CD56dim CD16+ are lower in SCID and CID (96). Studies in mice using RAG reporter and fate-mapping systems have demonstrated that RAG expression generates heterogeneity of NK cell origin as some NK cells, but not all, are derived from Rag1+ early progenitors (97). In vitro differentiation of human NK cells from RAG1:GFP iPSC reporter lines also showed that early RAG1+ cells give rise to NK cells, myeloid, and erythroid cells in addition to T/B lymphoid cells (98). While RAG expression could simply be a marker of lymphoid potential, recombinase activity of RAG is not limited to V(D)J recombination during T and B lymphocyte development but also regulates DNA damage recovery and fitness in NK cells (99). RAG-deficient mice have normal NK cell numbers but intrinsic hyporesponsiveness and impaired expansion and survival of NK cells in response to CMV infection (99). Taken together, these data suggest that, while not directly linked to helicase or DNA damage function, activation of DNA damage repair pathways through expression of RAG can confer NK cell fitness and functionality and further link the NK cell phenotype in hypomorphic RAG patients with impaired activation of these pathways.
Why NK cells are particularly affected by helicase deficiencies
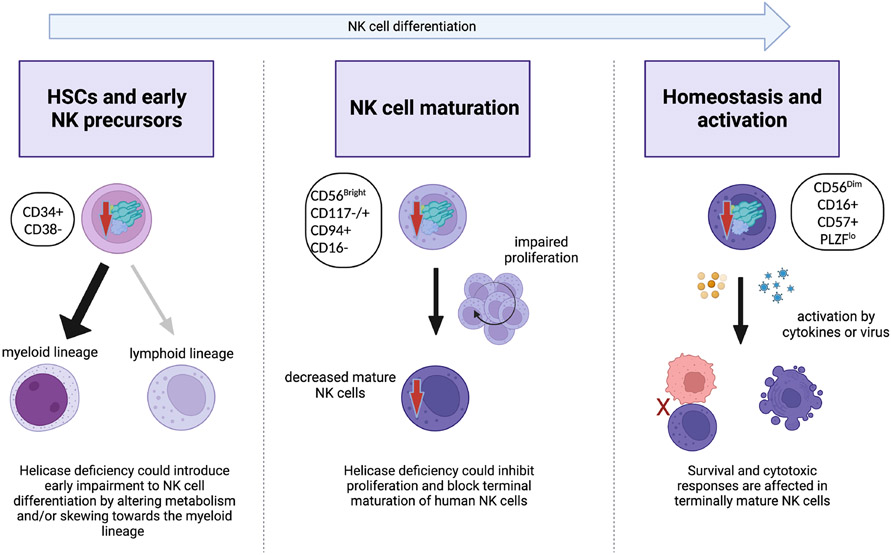
It is still not fully understood why natural killer cells are the primary immune cell subset that is affected by helicase variants in IEI. Proposed mechanisms for this unusual susceptibility include a particular requirement for proliferation in NK cell terminal maturation from the CD56bright to CD56dim stage (6, 7, 8, 9, 10). However, this has not been formally demonstrated, and the limited availability of material from rare individuals has made this hard to test. While NKD is a common feature of individuals with helicase deficiencies, there are cell type-specific effects manifested in the various clinical and cellular phenotypes of the disease. All variants reported to date are partial loss-of-function, and in GINS1 deficiency, the severity of damage by different variants correlated with the clinical severity of the disease, suggesting a dose-dependent effect of the availability of GINS1 protein (9). This, combined with the tight regulation of CMG protein expression, suggests that there may be a dose-dependent window in which NK cells are affected while sparing other immune lineages. Perhaps more severe variants would affect other lymphoid cell subsets, a hypothesis that is supported by the T cell lymphopenia reported in some patients (70, 71). While still speculation at this point, here we summarize this and other potential models for the effect of limited helicase function on NK cell development or homeostasis. We propose that CMG variants could impact NK cells in three potentially overlapping mechanisms; namely, by dysregulating mature NK cell subset homeostasis, by impacting NK cell terminal maturation, or by introducing lineage bias at earlier stages of hematopoiesis (Fig 3). Alternatively, mechanisms of DNA fitness related to induction of DNA damage pathways, as described above in the context of hypomorphic RAG phenotypes, may contribute to NK cell specific effects.
Figure 3.
Proposed model of the effect of CMG helicase functional impairment on NK cell development and maturation.
As the underlying mechanism of NK cell dysfunction in helicase deficiencies is still unclear, it is not known what underlies the viral susceptibility identified in these individuals. However, given that deletion of MCM10 in an NK cell line had no direct effect on NK cell cytotoxic function (7), it is most likely that impaired NK cell development results in both fewer numbers of effective NK cells and intrinsically impaired NK cell function on a single cell basis. While this mechanism is likely not specific to helicase deficiencies and instead applies whenever NK cell function or numbers are compromised, a greater understanding of the true nature of impaired NK cell development in the context of helicase deficiency will help better connect the NK cell phenotype with clinical phenotypes.
In addition to increased susceptibility to viral infections, NKD confers an increased risk of cancer associated with viruses such as EBV, though the direct impact of NKD on cancer incidence is largely unstudied (100). While no individuals with NKD resulting from helicase mutations have described malignancies, this may be due to the small cohort of patients described to date. Several helicase protein deficiencies have been associated with genomic instability, a hallmark of cancer, in patient cells and through experimental modeling. Namely, MCM10 deficiency resulted in increased replication stress which may contribute to genome instability and telomere erosion, and GINS1 deficiency leads to abnormal nuclear structures and investigation of MCM4 deficiency found chromosome breaks (7, 9, 10, 11). This is bolstered by in vivo data from the mouse model Mcm4Chao3, which displayed high levels of chromosomal instability with increased micronuclei and the development of mammary adenocarcinomas (101). Similarly, deficiency of MCM2 in a mouse model displays the development of lymphomas and shortened lifespans (102). Conversely, the overexpression of virtually all helicase protein components have also been found in cancer, details of which are outside the scope of this review (12). The small cohort of patients with helicase deficiency makes it difficult to understand if there is increased susceptibility to cancer and the mechanisms underlying this phenomenon.
The most intuitive mechanism to explain an NK cell specific effect of these variants is that committed NK cell progenitors rely more acutely on DNA replication machinery than other cell types. As mentioned before, NK cells proliferate more rapidly than T cells in the absence of infection (60). Patients with PNH with somatic PIGA mutations clearly demonstrate that CD56bright NK cells and neutrophils are the two most proliferative hematopoietic populations (69). Congruently, a frequent immune phenotype that occurs along with NKD is neutropenia (Table 1). These data suggest that the variants are dysregulating homeostasis of the most proliferative cell types. Further supporting this idea are variants found in DNA synthesis related genes outside of the CMG helicase, POLA1 and POLE2, which also manifest in NKD and other common CMG variant phenotypes (78, 79, 81). In a parallel example, a variant in RTEL1, a gene involved in helicase activity during telomere elongation, can also result in decreased NK cells in PBMC (91). The converging phenotype of NKD in variants of S phase related genes demonstrate that cell cycle dysregulation is an important mechanism. This effect could be due to differential proliferative kinetics or differential regulation of replisome protein expression. Further studies must be carried out to understand what makes NK cell proliferation more vulnerable to these changes.
The PNH example with somatic mutation highlights that in NK cells from adult peripheral blood, CD56bright cells, but not the CD56dim cells, are actively proliferating. In many of the NKD patients, whose variants are germline mutations, both CD56bright and CD56dim populations are affected, suggesting additional mechanisms for subtype specific cellular fitness that may arise throughout development. The decrease of CD56dim NK cells in peripheral blood suggests that CD56dim cells are not being produced as effectively in patients with helicase deficiencies, and in vitro and humanized mouse models of patient cells suggest that NK cell terminal maturation is affected (6, 7). In this model, helicase defects would manifest most prominently during the final steps of NK cell maturation. For example, MCM4 is more highly expressed in CD56bright NK cells, so its insufficiency results in DNA damage and may ultimately reduce, but not completely block, maturation into the CD56dim subset (10). Alternatively, diminishment of mature NK cells may reflect the normal generation but impaired survival of a specific CD56dim subset. The previously described differential survival and turnover of adaptive NK cells could support a model in which adaptive NK cells, or some other subset, were either selectively retained or selectively impaired.
The idea that NK cell differentiation is unaffected up until terminal maturation is challenged by the decrease in absolute number of circulating NK cells in addition to the change in CD56dim to CD56bright ratio found in most affected individuals and the reduced frequency of ILC precursors in GINS1 patients (9). Current evidence on adjacent lineages which can shed light on more developmental phenotype of NKD, such as ILC and unconventional T cells, is limited to only a few patients. Study of the GINS1 variants with decreased ILC and GINS4 variants with unaffected T cells, including γδ T and MAIT cells, suggest that the effects are more specific to NK/ILC common precursors. Further investigation on how other helicase variants affect ILCs and all T cell lineages would be important to understand the lineage bias more comprehensively. However, these cell types are fundamentally tissue-resident, making the accessibility a major challenge. An in vitro or animal model system may be needed to understand these lineage biases with sufficient resolution.
While it is more difficult to analyze the requirements for early human NK cell differentiation due to its localization in tissue, we can consider the effects of helicase deficiency on hematopoiesis and early innate lymphoid cell differentiation. One potential mechanism by which helicase variants could affect NK cells during hematopoiesis would be skewing of lineage biases in hematopoietic precursors or hematopoietic stem cells (HSCs). In mice, delayed cell cycle progression has been observed in HSCs from aged mice, accompanied by lower MCM4 and MCM6 expression and impaired HSC functionality (103). Along with decreased cell cycle, many groups have reported that aged HSCs have a decreased potential for lymphoid lineages while myeloid potential is intact (104, 105, 106, 107). This suggests that decreased CMG expression, including in aging, may position HSCs to be biased against NK cells and other innate lymphoid cells that require continuous regeneration. That said, aging is a complex process that manifests in additional aspects of HSC lineage output, including DNA damage, clonal selection, epigenetic drift, and polarity shift (108). Therefore, further studies must be done to understand how the phenotypes and mechanisms of aged HSCs compare to those of HSCs with helicase variants.
One could speculate that a relatively mild lineage bias in HSCs could be combined with additional defects in proliferation or cell cycle regulation that may be a feature of later stages of NK cell development, leading to the dysregulated NK cell subsets found in the peripheral blood of patients. Further, such a model could also account for what seem to be environmental factors that can contribute to clinical severity, most notably in the GINS4 cohort where the most clinically affected sibling had a significant CMV infection early in life (6). Disruption of stem cell or precursor homeostasis due to severe infection is reminiscent of the effect of recurrent inflammation on HSC function and could link triggering viral events with subsequently limited NK or innate lymphoid cell lineage potential (109). However, the question remains of why NK cells are most affected and why patients don’t exhibit signs of other HSC dysregulation, such as bone marrow failure. Careful manipulation of helicase protein function, combined with molecular studies that link HSC function to downstream innate lymphoid lineage potential, will be necessary to answer these questions.
Given the decreased numbers of NK cells in circulation and the rarity of affected individuals, many studies described above have used patient or control fibroblasts, CD34+ cells, NK cell lines, or B cell lines to understand the mechanisms of helicase variants in cell cycle and DNA damage. However, as shown by disease phenotypes, the effects of the helicase variant appear to be cell type specific and potentially affecting multiple steps throughout hematopoiesis. Human induced pluripotent stem cells (iPSCs) provide a unique opportunity to model the pathogenesis of such diseases in an NK cell specific way. Several studies provide methods for NK cell generation by modeling embryonic or postnatal NK cell development (110, 111, 112). Utilizing patient-derived iPSCs is one tractable model to circumvent technical hurdles associated with the study of specifically human immunity and address questions of lineage specification, albeit one that makes it difficult to study mature NK cell homeostasis and generation of adaptive NK cells.
Finally, we can consider the non-canonical roles of helicase proteins. Multiple helicase components are involved in different aspects of regulating replication, including genomic stability, DNA damage signaling, and replication fork stalling. Additionally, most helicase proteins have alternative binding partners outside of the CMG helicase, which could account for the variability between helicase variants seen in these NKD patients and in vitro studies. While similarities between patient and cellular phenotypes between multiple CMG protein deficiencies suggests that replisome dysfunction is at the heart of NK cell deficiency, better modeling of different aspects of canonical and non-canonical CMG protein functions can be informative of both NK cell biology and the role of replisome function in immune cell generation, homeostasis, and disease.
Summary
The principal activity of the CMG helicase is facilitating DNA replication while maintaining genomic integrity. Partial loss of function of helicase proteins, including MCM4, GINS1, MCM10 and GINS4, results in replication stress and DNA damage, ultimately leading to NKD. Helicase proteins clearly play a critical role in human natural killer cells, thus understanding the role of the CMG helicase through the lens of NKD contributes to our knowledge of NK cell development and function and could potentially unlock novel therapeutic routes for targeting immunodeficiencies.
Funding:
This study was funded in part by R01AI137275 (EMM).
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
References
- 1.Tangye SG, Al-Herz W, Bousfiha A, Cunningham-Rundles C, Franco JL, Holland SM, et al. Human Inborn Errors of Immunity: 2022 Update on the Classification from the International Union of Immunological Societies Expert Committee. J Clin Immunol. 2022;42(7):1473–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mace EM, Orange JS. Emerging insights into human health and NK cell biology from the study of NK cell deficiencies. Immunol Rev. 2019;287(1):202–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Della Chiesa M, De Maria A, Muccio L, Bozzano F, Sivori S, Moretta L. Human NK Cells and Herpesviruses: Mechanisms of Recognition, Response and Adaptation. Front Microbiol. 2019;10:2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orange JS. Natural killer cell deficiency. J Allergy Clin Immunol. 2013;132(3):515–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandelboim O, Lieberman N, Lev M, Paul L, Arnon TI, Bushkin Y, et al. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409(6823):1055–60. [DOI] [PubMed] [Google Scholar]
- 6.Conte MI, Poli MC, Taglialatela A, Leuzzi G, Chinn IK, Salinas SA, et al. Partial loss-of-function mutations in GINS4 lead to NK cell deficiency with neutropenia. JCI Insight. 2022;7(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mace EM, Paust S, Conte MI, Baxley RM, Schmit MM, Patil SL, et al. Human NK cell deficiency as a result of biallelic mutations in MCM10. J Clin Invest. 2020;130(10):5272–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes CR, Guasti L, Meimaridou E, Chuang CH, Schimenti JC, King PJ, et al. MCM4 mutation causes adrenal failure, short stature, and natural killer cell deficiency in humans. J Clin Invest. 2012;122(3):814–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cottineau J, Kottemann MC, Lach FP, Kang YH, Vely F, Deenick EK, et al. Inherited GINS1 deficiency underlies growth retardation along with neutropenia and NK cell deficiency. J Clin Invest. 2017;127(5):1991–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gineau L, Cognet C, Kara N, Lach FP, Dunne J, Veturi U, et al. Partial MCM4 deficiency in patients with growth retardation, adrenal insufficiency, and natural killer cell deficiency. J Clin Invest. 2012;122(3):821–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baxley RM, Leung W, Schmit MM, Matson JP, Yin L, Oram MK, et al. Bi-allelic MCM10 variants associated with immune dysfunction and cardiomyopathy cause telomere shortening. Nat Commun. 2021;12(1):1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seo YS, Kang YH. The Human Replicative Helicase, the CMG Complex, as a Target for Anti-cancer Therapy. Front Mol Biosci. 2018;5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ying CY, Gautier J. The ATPase activity of MCM2–7 is dispensable for pre-RC assembly but is required for DNA unwinding. EMBO J. 2005;24(24):4334–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan Z, Georgescu R, Bai L, Zhang D, Li H, O’Donnell ME. DNA unwinding mechanism of a eukaryotic replicative CMG helicase. Nat Commun. 2020;11(1):688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, O’Donnell ME. The Eukaryotic CMG Helicase at the Replication Fork: Emerging Architecture Reveals an Unexpected Mechanism. Bioessays. 2018;40(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.C Lee BT. Eukaryotic DNA Polymerase-alpha. Encyclopedia of Biological Chemistry: Second Edition. 2013;1:241–4. [Google Scholar]
- 17.Mimura S, Masuda T, Matsui T, Takisawa H. Central role for cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells. 2000;5(6):439–52. [DOI] [PubMed] [Google Scholar]
- 18.Thu YM, Bielinsky AK. Enigmatic roles of Mcm10 in DNA replication. Trends Biochem Sci. 2013;38(4):184–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diffley JF. Regulation of early events in chromosome replication. Curr Biol. 2004;14(18):R778–86. [DOI] [PubMed] [Google Scholar]
- 20.Remus D, Diffley JF. Eukaryotic DNA replication control: lock and load, then fire. Curr Opin Cell Biol. 2009;21(6):771–7. [DOI] [PubMed] [Google Scholar]
- 21.Kohler C, Koalick D, Fabricius A, Parplys AC, Borgmann K, Pospiech H, et al. Cdc45 is limiting for replication initiation in humans. Cell Cycle. 2016;15(7):974–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong PG, Winter SL, Zaika E, Cao TV, Oguz U, Koomen JM, et al. Cdc45 limits replicon usage from a low density of preRCs in mammalian cells. PLoS One. 2011;6(3):e17533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das M, Singh S, Pradhan S, Narayan G. MCM Paradox: Abundance of Eukaryotic Replicative Helicases and Genomic Integrity. Mol Biol Int. 2014;2014:574850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sedlackova H, Rask MB, Gupta R, Choudhary C, Somyajit K, Lukas J. Equilibrium between nascent and parental MCM proteins protects replicating genomes. Nature. 2020;587(7833):297–302. [DOI] [PubMed] [Google Scholar]
- 25.Krastanova I, Sannino V, Amenitsch H, Gileadi O, Pisani FM, Onesti S. Structural and functional insights into the DNA replication factor Cdc45 reveal an evolutionary relationship to the DHH family of phosphoesterases. J Biol Chem. 2012;287(6):4121–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szambowska A, Tessmer I, Kursula P, Usskilat C, Prus P, Pospiech H, et al. DNA binding properties of human Cdc45 suggest a function as molecular wedge for DNA unwinding. Nucleic Acids Res. 2014;42(4):2308–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szambowska A, Tessmer I, Prus P, Schlott B, Pospiech H, Grosse F. Cdc45-induced loading of human RPA onto single-stranded DNA. Nucleic Acids Res. 2017;45(6):3217–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Can G, Kauerhof AC, Macak D, Zegerman P. Helicase Subunit Cdc45 Targets the Checkpoint Kinase Rad53 to Both Replication Initiation and Elongation Complexes after Fork Stalling. Mol Cell. 2019;73(3):562–73 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol Mol Biol Rev. 2004;68(1):109–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yankulov K, Todorov I, Romanowski P, Licatalosi D, Cilli K, McCracken S, et al. MCM proteins are associated with RNA polymerase II holoenzyme. Mol Cell Biol. 1999;19(9):6154–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DaFonseca CJ, Shu F, Zhang JJ. Identification of two residues in MCM5 critical for the assembly of MCM complexes and Stat1-mediated transcription activation in response to IFN-gamma. Proc Natl Acad Sci U S A. 2001;98(6):3034–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang JJ, Zhao Y, Chait BT, Lathem WW, Ritzi M, Knippers R, et al. Ser727-dependent recruitment of MCM5 by Stat1alpha in IFN-gamma-induced transcriptional activation. EMBO J. 1998;17(23):6963–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sterner JM, Dew-Knight S, Musahl C, Kornbluth S, Horowitz JM. Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol Cell Biol. 1998;18(5):2748–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kukimoto I, Aihara S, Yoshiike K, Kanda T. Human papillomavirus oncoprotein E6 binds to the C-terminal region of human minichromosome maintenance 7 protein. Biochem Biophys Res Commun. 1998;249(1):258–62. [DOI] [PubMed] [Google Scholar]
- 35.Ishimi Y, Ichinose S, Omori A, Sato K, Kimura H. Binding of human minichromosome maintenance proteins with histone H3. J Biol Chem. 1996;271(39):24115–22. [DOI] [PubMed] [Google Scholar]
- 36.Jia W, Hsieh HY, Kidoya H, Takakura N. Embryonic expression of GINS members in the development of the mammalian nervous system. Neurochem Int. 2019;129:104465. [DOI] [PubMed] [Google Scholar]
- 37.Mohri T, Ueno M, Nagahama Y, Gong ZY, Asano M, Oshima H, et al. Requirement of SLD5 for early embryogenesis. PLoS One. 2013;8(11):e78961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varga M, Csalyi K, Bertyak I, Menyhard DK, Poole RJ, Cerveny KL, et al. Tissue-Specific Requirement for the GINS Complex During Zebrafish Development. Front Cell Dev Biol. 2020;8:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Z, Yu Z, Rong Z, Luo Z, Zhang J, Qiu Z, et al. The novel GINS4 axis promotes gastric cancer growth and progression by activating Rac1 and CDC42. Theranostics. 2019;9(26):8294–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaur M, Devi R, Ghosh T, Khan MM, Kumar P, Priyanka, et al. Sld5 Ensures Centrosomal Resistance to Congression Forces by Preserving Centriolar Satellites. Mol Cell Biol. 2018;38(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freud AG, Yu J, Caligiuri MA. Human natural killer cell development in secondary lymphoid tissues. Semin Immunol. 2014;26(2):132–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bar-Ephraim YE, Mebius RE. Innate lymphoid cells in secondary lymphoid organs. Immunol Rev. 2016;271(1):185–99. [DOI] [PubMed] [Google Scholar]
- 43.Yang Q, Bhandoola A. The development of adult innate lymphoid cells. Curr Opin Immunol. 2016;39:114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCullar V, Oostendorp R, Panoskaltsis-Mortari A, Yun G, Lutz CT, Wagner JE, et al. Mouse fetal and embryonic liver cells differentiate human umbilical cord blood progenitors into CD56-negative natural killer cell precursors in the absence of interleukin-15. Exp Hematol. 2008;36(5):598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grzywacz B, Kataria N, Sikora M, Oostendorp RA, Dzierzak EA, Blazar BR, et al. Coordinated acquisition of inhibitory and activating receptors and functional properties by developing human natural killer cells. Blood. 2006;108(12):3824–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller JS, Verfaillie C, McGlave P. The generation of human natural killer cells from CD34+/DR- primitive progenitors in long-term bone marrow culture. Blood. 1992;80(9):2182–7. [PubMed] [Google Scholar]
- 47.Crinier A, Dumas PY, Escaliere B, Piperoglou C, Gil L, Villacreces A, et al. Single-cell profiling reveals the trajectories of natural killer cell differentiation in bone marrow and a stress signature induced by acute myeloid leukemia. Cell Mol Immunol. 2021;18(5):1290–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang C, Siebert JR, Burns R, Gerbec ZJ, Bonacci B, Rymaszewski A, et al. Heterogeneity of human bone marrow and blood natural killer cells defined by single-cell transcriptome. Nat Commun. 2019;10(1):3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith SL, Kennedy PR, Stacey KB, Worboys JD, Yarwood A, Seo S, et al. Diversity of peripheral blood human NK cells identified by single-cell RNA sequencing. Blood Adv. 2020;4(7):1388–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med. 2013;5(208):208ra145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caligiuri MA. Human natural killer cells. Blood. 2008;112(3):461–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner JA, Rosario M, Romee R, Berrien-Elliott MM, Schneider SE, Leong JW, et al. CD56bright NK cells exhibit potent antitumor responses following IL-15 priming. J Clin Invest. 2017;127(11):4042–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Juelke K, Killig M, Luetke-Eversloh M, Parente E, Gruen J, Morandi B, et al. CD62L expression identifies a unique subset of polyfunctional CD56dim NK cells. Blood. 2010;116(8):1299–307. [DOI] [PubMed] [Google Scholar]
- 54.Romagnani C, Juelke K, Falco M, Morandi B, D’Agostino A, Costa R, et al. CD56brightCD16- killer Ig-like receptor- NK cells display longer telomeres and acquire features of CD56dim NK cells upon activation. J Immunol. 2007;178(8):4947–55. [DOI] [PubMed] [Google Scholar]
- 55.Huntington ND, Legrand N, Alves NL, Jaron B, Weijer K, Plet A, et al. IL-15 trans-presentation promotes human NK cell development and differentiation in vivo. J Exp Med. 2009;206(1):25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shilling HG, McQueen KL, Cheng NW, Shizuru JA, Negrin RS, Parham P. Reconstitution of NK cell receptor repertoire following HLA-matched hematopoietic cell transplantation. Blood. 2003;101(9):3730–40. [DOI] [PubMed] [Google Scholar]
- 57.Dulphy N, Haas P, Busson M, Belhadj S, Peffault de Latour R, Robin M, et al. An unusual CD56(bright) CD16(low) NK cell subset dominates the early posttransplant period following HLA-matched hematopoietic stem cell transplantation. J Immunol. 2008;181(3):2227–37. [DOI] [PubMed] [Google Scholar]
- 58.Seo S, Mace EM. Diversity of human NK cell developmental pathways defined by single-cell analyses. Curr Opin Immunol. 2022;74:106–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Freud AG, Becknell B, Roychowdhury S, Mao HC, Ferketich AK, Nuovo GJ, et al. A human CD34(+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity. 2005;22(3):295–304. [DOI] [PubMed] [Google Scholar]
- 60.Lutz CT, Karapetyan A, Al-Attar A, Shelton BJ, Holt KJ, Tucker JH, et al. Human NK cells proliferate and die in vivo more rapidly than T cells in healthy young and elderly adults. J Immunol. 2011;186(8):4590–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hsu AP, Sampaio EP, Khan J, Calvo KR, Lemieux JE, Patel SY, et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood. 2011;118(10):2653–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med. 1989;320(26):1731–5. [DOI] [PubMed] [Google Scholar]
- 63.Vinh DC, Patel SY, Uzel G, Anderson VL, Freeman AF, Olivier KN, et al. Autosomal dominant and sporadic monocytopenia with susceptibility to mycobacteria, fungi, papillomaviruses, and myelodysplasia. Blood. 2010;115(8):1519–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bigley V, Haniffa M, Doulatov S, Wang XN, Dickinson R, McGovern N, et al. The human syndrome of dendritic cell, monocyte, B and NK lymphoid deficiency. J Exp Med. 2011;208(2):227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dickinson RE, Griffin H, Bigley V, Reynard LN, Hussain R, Haniffa M, et al. Exome sequencing identifies GATA-2 mutation as the cause of dendritic cell, monocyte, B and NK lymphoid deficiency. Blood. 2011;118(10):2656–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spinner MA, Ker JP, Stoudenmire CJ, Fadare O, Mace EM, Orange JS, et al. GATA2 deficiency underlying severe blastomycosis and fatal herpes simplex virus-associated hemophagocytic lymphohistiocytosis. J Allergy Clin Immunol. 2016;137(2):638–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schlums H, Jung M, Han H, Theorell J, Bigley V, Chiang SC, et al. Adaptive NK cells can persist in patients with GATA2 mutation depleted of stem and progenitor cells. Blood. 2017;129(14):1927–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mace EM, Hsu AP, Monaco-Shawver L, Makedonas G, Rosen JB, Dropulic L, et al. Mutations in GATA2 cause human NK cell deficiency with specific loss of the CD56(bright) subset. Blood. 2013;121(14):2669–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Corat MA, Schlums H, Wu C, Theorell J, Espinoza DA, Sellers SE, et al. Acquired somatic mutations in PNH reveal long-term maintenance of adaptive NK cells independent of HSPCs. Blood. 2017;129(14):1940–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bernard F, Picard C, Cormier-Daire V, Eidenschenk C, Pinto G, Bustamante JC, et al. A novel developmental and immunodeficiency syndrome associated with intrauterine growth retardation and a lack of natural killer cells. Pediatrics. 2004;113(1 Pt 1):136–41. [DOI] [PubMed] [Google Scholar]
- 71.Eidenschenk C, Jouanguy E, Alcais A, Mention JJ, Pasquier B, Fleckenstein IM, et al. Familial NK cell deficiency associated with impaired IL-2- and IL-15-dependent survival of lymphocytes. J Immunol. 2006;177(12):8835–43. [DOI] [PubMed] [Google Scholar]
- 72.Aparicio T, Guillou E, Coloma J, Montoya G, Mendez J. The human GINS complex associates with Cdc45 and MCM and is essential for DNA replication. Nucleic Acids Res. 2009;37(7):2087–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gouge CA, Christensen TW. Drosophila Sld5 is essential for normal cell cycle progression and maintenance of genomic integrity. Biochem Biophys Res Commun. 2010;400(1):145–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choi JM, Lim HS, Kim JJ, Song OK, Cho Y. Crystal structure of the human GINS complex. Genes Dev. 2007;21(11):1316–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rong Z, Luo Z, Zhang J, Li T, Zhu Z, Yu Z, et al. GINS complex subunit 4, a prognostic biomarker and reversely mediated by Kruppel-like factor 4, promotes the growth of colorectal cancer. Cancer Sci. 2020;111(4):1203–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang R, Liu N, Chen L, Jiang Y, Shi Y, Mao C, et al. LSH interacts with and stabilizes GINS4 transcript that promotes tumourigenesis in non-small cell lung cancer. J Exp Clin Cancer Res. 2019;38(1):280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamane K, Naito H, Wakabayashi T, Yoshida H, Muramatsu F, Iba T, et al. Regulation of SLD5 gene expression by miR-370 during acute growth of cancer cells. Sci Rep. 2016;6:30941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Starokadomskyy P, Gemelli T, Rios JJ, Xing C, Wang RC, Li H, et al. DNA polymerase-alpha regulates the activation of type I interferons through cytosolic RNA:DNA synthesis. Nat Immunol. 2016;17(5):495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Starokadomskyy P, Wilton KM, Krzewski K, Lopez A, Sifuentes-Dominguez L, Overlee B, et al. NK cell defects in X-linked pigmentary reticulate disorder. JCI Insight. 2019;4(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Henninger EE, Pursell ZF. DNA polymerase epsilon and its roles in genome stability. IUBMB Life. 2014;66(5):339–51. [DOI] [PubMed] [Google Scholar]
- 81.Frugoni F, Dobbs K, Felgentreff K, Aldhekri H, Al Saud BK, Arnaout R, et al. A novel mutation in the POLE2 gene causing combined immunodeficiency. J Allergy Clin Immunol. 2016;137(2):635–8 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Burgers PMJ, Kunkel TA. Eukaryotic DNA Replication Fork. Annu Rev Biochem. 2017;86:417–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Loeb LA, Monnat RJ Jr. DNA polymerases and human disease. Nat Rev Genet. 2008;9(8):594–604. [DOI] [PubMed] [Google Scholar]
- 84.Gao J, Wang Q, Dong C, Chen S, Qi Y, Liu Y. Whole Exome Sequencing Identified MCM2 as a Novel Causative Gene for Autosomal Dominant Nonsyndromic Deafness in a Chinese Family. PLoS One. 2015;10(7):e0133522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Knapp KM, Jenkins DE, Sullivan R, Harms FL, von Elsner L, Ockeloen CW, et al. MCM complex members MCM3 and MCM7 are associated with a phenotypic spectrum from Meier-Gorlin syndrome to lipodystrophy and adrenal insufficiency. Eur J Hum Genet. 2021;29(7):1110–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vetro A, Savasta S, Russo Raucci A, Cerqua C, Sartori G, Limongelli I, et al. MCM5: a new actor in the link between DNA replication and Meier-Gorlin syndrome. Eur J Hum Genet. 2017;25(5):646–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fenwick AL, Kliszczak M, Cooper F, Murray J, Sanchez-Pulido L, Twigg SR, et al. Mutations in CDC45, Encoding an Essential Component of the Pre-initiation Complex, Cause Meier-Gorlin Syndrome and Craniosynostosis. Am J Hum Genet. 2016;99(1):125–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McQuaid ME, Ahmed K, Tran S, Rousseau J, Shaheen R, Kernohan KD, et al. Hypomorphic GINS3 variants alter DNA replication and cause Meier-Gorlin syndrome. JCI Insight. 2022;7(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nabais Sa MJ, Miller KA, McQuaid M, Koelling N, Wilkie AOM, Wurtele H, et al. Biallelic GINS2 variant p.(Arg114Leu) causes Meier-Gorlin syndrome with craniosynostosis. J Med Genet. 2022;59(8):776–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Etzioni A, Eidenschenk C, Katz R, Beck R, Casanova JL, Pollack S. Fatal varicella associated with selective natural killer cell deficiency. J Pediatr. 2005;146(3):423–5. [DOI] [PubMed] [Google Scholar]
- 91.Hanna S, Beziat V, Jouanguy E, Casanova JL, Etzioni A. A homozygous mutation of RTEL1 in a child presenting with an apparently isolated natural killer cell deficiency. J Allergy Clin Immunol. 2015;136(4):1113–4. [DOI] [PubMed] [Google Scholar]
- 92.Delmonte OM, Schuetz C, Notarangelo LD. RAG Deficiency: Two Genes, Many Diseases. J Clin Immunol. 2018;38(6):646–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Buck D, Moshous D, de Chasseval R, Ma Y, le Deist F, Cavazzana-Calvo M, et al. Severe combined immunodeficiency and microcephaly in siblings with hypomorphic mutations in DNA ligase IV. Eur J Immunol. 2006;36(1):224–35. [DOI] [PubMed] [Google Scholar]
- 94.O’Driscoll M, Cerosaletti KM, Girard PM, Dai Y, Stumm M, Kysela B, et al. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol Cell. 2001;8(6):1175–85. [DOI] [PubMed] [Google Scholar]
- 95.Ege M, Ma Y, Manfras B, Kalwak K, Lu H, Lieber MR, et al. Omenn syndrome due to ARTEMIS mutations. Blood. 2005;105(11):4179–86. [DOI] [PubMed] [Google Scholar]
- 96.Dobbs K, Tabellini G, Calzoni E, Patrizi O, Martinez P, Giliani SC, et al. Natural Killer Cells from Patients with Recombinase-Activating Gene and Non-Homologous End Joining Gene Defects Comprise a Higher Frequency of CD56(bright) NKG2A(+++) Cells, and Yet Display Increased Degranulation and Higher Perforin Content. Front Immunol. 2017;8:798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Igarashi H, Gregory SC, Yokota T, Sakaguchi N, Kincade PW. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity. 2002;17(2):117–30. [DOI] [PubMed] [Google Scholar]
- 98.Motazedian A, Bruveris FF, Kumar SV, Schiesser JV, Chen T, Ng ES, et al. Multipotent RAG1+ progenitors emerge directly from haemogenic endothelium in human pluripotent stem cell-derived haematopoietic organoids. Nat Cell Biol. 2020;22(1):60–73. [DOI] [PubMed] [Google Scholar]
- 99.Karo JM, Schatz DG, Sun JC. The RAG recombinase dictates functional heterogeneity and cellular fitness in natural killer cells. Cell. 2014;159(1):94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moon WY, Powis SJ. Does Natural Killer Cell Deficiency (NKD) Increase the Risk of Cancer? NKD May Increase the Risk of Some Virus Induced Cancer. Front Immunol. 2019;10:1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, Munroe RJ, et al. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet. 2007;39(1):93–8. [DOI] [PubMed] [Google Scholar]
- 102.Pruitt SC, Bailey KJ, Freeland A. Reduced Mcm2 expression results in severe stem/progenitor cell deficiency and cancer. Stem Cells. 2007;25(12):3121–32. [DOI] [PubMed] [Google Scholar]
- 103.Flach J, Bakker ST, Mohrin M, Conroy PC, Pietras EM, Reynaud D, et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014;512(7513):198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pang WW, Price EA, Sahoo D, Beerman I, Maloney WJ, Rossi DJ, et al. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc Natl Acad Sci U S A. 2011;108(50):20012–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rossi DJ, Bryder D, Zahn JM, Ahlenius H, Sonu R, Wagers AJ, et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci U S A. 2005;102(26):9194–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Min H, Montecino-Rodriguez E, Dorshkind K. Effects of aging on the common lymphoid progenitor to pro-B cell transition. J Immunol. 2006;176(2):1007–12. [DOI] [PubMed] [Google Scholar]
- 107.Montecino-Rodriguez E, Kong Y, Casero D, Rouault A, Dorshkind K, Pioli PD. Lymphoid-Biased Hematopoietic Stem Cells Are Maintained with Age and Efficiently Generate Lymphoid Progeny. Stem Cell Reports. 2019;12(3):584–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Akunuru S, Geiger H. Aging, Clonality, and Rejuvenation of Hematopoietic Stem Cells. Trends Mol Med. 2016;22(8):701–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.King KY, Goodell MA. Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat Rev Immunol. 2011;11(10):685–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dege C, Fegan KH, Creamer JP, Berrien-Elliott MM, Luff SA, Kim D, et al. Potently Cytotoxic Natural Killer Cells Initially Emerge from Erythro-Myeloid Progenitors during Mammalian Development. Dev Cell. 2020;53(2):229–39 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ditadi A, Sturgeon CM. Directed differentiation of definitive hemogenic endothelium and hematopoietic progenitors from human pluripotent stem cells. Methods. 2016;101:65–72. [DOI] [PubMed] [Google Scholar]
- 112.Knorr DA, Ni Z, Hermanson D, Hexum MK, Bendzick L, Cooper LJ, et al. Clinical-scale derivation of natural killer cells from human pluripotent stem cells for cancer therapy. Stem Cells Transl Med. 2013;2(4):274–83. [DOI] [PMC free article] [PubMed] [Google Scholar]