Abstract
Proteus mirabilis, a cause of complicated urinary tract infection, expresses urease when exposed to urea. While it is recognized that the positive transcriptional activator UreR induces gene expression, the levels of expression of the enzyme during experimental infection are not known. To investigate in vivo expression of P. mirabilis urease, the gene encoding green fluorescent protein (GFP) was used to construct reporter fusions. Translational fusions of urease accessory gene ureD, which is preceded by a urea-inducible promoter, were made with gfp (modified to express S65T/V68L/S72A [B. P. Cormack et al. Gene 173:33–38, 1996]). Constructs were confirmed by sequencing of the fusion junctions. UreD-GFP fusion protein was induced by urea in both Escherichia coli DH5α and P. mirabilis HI4320. By using Western blotting with antiserum raised against GFP, expression level was shown to correlate with urea concentration (tested from 0 to 500 mM), with highest induction at 200 to 500 mM urea. Fluorescent E. coli and P. mirabilis bacteria were observed by fluorescence microscopy following urea induction, and the fluorescence intensity of GFP in cell lysates was measured by spectrophotofluorimetry. P. mirabilis HI4320 carrying the UreD-GFP fusion plasmid was transurethrally inoculated into the bladders of CBA mice. One week postchallenge, fluorescent bacteria were detected in thin sections of both bladder and kidney samples; the fluorescence intensity of bacteria in bladder tissue was higher than that in the kidney. Kidneys were primarily infected with single-cell-form fluorescent bacteria, while aggregated bacterial clusters were observed in the bladder. Elongated swarmer cells were only rarely observed. These observations demonstrate that urease is expressed in vivo and that using GFP as a reporter protein is a viable approach to investigate in vivo expression of P. mirabilis virulence genes in experimental urinary tract infection.
Urinary tract infection (UTI) with Proteus mirabilis may lead to serious complications that include renal stones, acute pyelonephritis, catheter obstruction, and bacteremia (20, 35, 39). Urease is recognized as a major virulence factor for P. mirabilis by virtue of its ability to rapidly generate ammonia from the hydrolysis of urea present at 400 to 500 mM in urine (13). Elevated pH results in ion precipitation in the form of struvite or carbonate-apatite kidney or bladder stones. Ammonia may also have a direct cytotoxic effect upon kidney cells in cultures (13, 23, 30, 32–34). Production of urease appears to be one reason that Proteus infections cause more severe histological damage than Escherichia coli infections (20, 35, 40).
P. mirabilis urease, a nickel-metalloenzyme, resides in the cytosol of the bacterium (21). The urease gene cluster is comprised of eight contiguous genes. The structural genes, ureABC, which encode subunits of the enzyme, are flanked immediately upstream by the ureD and downstream by the ureEFG genes. These seven genes are transcribed on the same mRNA transcript (18, 21, 22, 37). The four accessory genes (ureDEFG) are necessary for the insertion of nickel ions into the apoenzyme and required for assembly of a catalytically active urease. The ureR gene lies 400 bp upstream of ureD, is oriented opposite the other seven genes, and acts as a positive regulator in the presence of urea to activate transcription of urease structural and accessory genes via sequences upstream of ureD (36).
To evaluate the contribution of urease to virulence, a urease-negative mutant was previously constructed and the virulence was analyzed by our lab, using the CBA mouse model of ascending UTI (20, 23). After 48 h of infection, the number of mutant bacteria recovered from urine, bladder, and both kidneys was significantly (100-fold) lower than that of the parent strain. After 1 week of infection, the mutant concentration was 1 million times less than that of the parent, which produced significantly more severe pathology than the mutant. The urease-negative mutant had a 50% infective dose of >2.7 × 109 CFU, a value more than 1,000-fold greater than that of the parent strain (2.2 × 106 CFU).
To assess urease expression in situ, green fluorescent protein (GFP) from Aequorea victoria was used as a reporter protein in this study. In comparison with products of other reporter genes (e.g., lacZ, lux, or cat), GFP does not require addition of a cofactor or substrate to permit observation of its expression, merely excitation with UV light. GFP is stable in bacterial cells, is not photobleached by prolonged exposure to UV light, and does not require lysis of bacterial or host cell for accurate detection (7, 11, 24, 38). The GFP variant used in these studies [GFP(S65T/V68L/S72A)] has its peak excitation band shifted from the wild-type position of 395 nm (470-nm shoulder) to 481 nm, as well as improved folding efficiency (9). Consequently, it exhibits enhanced brightness when expressed in bacteria compared to the wild-type protein (9). The emission maximum of the mutant GFP is 507 nm.
For studies of pathogenic bacteria, GFP has thus far been used to assess whether promoters are active in mycobacteria (11, 25) or Salmonella typhimurium (41, 43) within macrophages. S. typhimurium and Yersinia pseudotuberculosis expressing GFP have been sorted by fluorescence-activated cell sorting (42, 43). As well, bacterium-plant interactions (12), aquatic survival (26), and other applications (28) have been studied by using GFP expression. In our study, an in-frame UreD-GFP translational fusion was constructed and fluorescent bacteria from both in vitro cultures and tissue from experimentally challenged mice were detected by fluorescence microscopy. We examined the feasibility of studying gene expression by using GFP in an experimental infection model of ascending UTI.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
P. mirabilis HI4320 (urease positive; produces MR/P, Pmf, and ATF fimbriae; hemolysin positive) was isolated from an elderly woman with urinary catheter-associated bacteriuria (34). E. coli DH5α [supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] was used as a recipient for transformations. Luria broth (10 g of tryptone, 5 g of yeast extract, and 10 g of NaCl per liter) and L agar (Luria broth containing 1.5% agar) were used as culture media. Nonswarming agar (10 g of tryptone, 5 g of yeast extract, 5 ml of glycerol, 0.4 g of NaCl, and 29 g of agar per liter) was used to prevent swarming of P. mirabilis (6). The mutant gfp gene was kindly provided by B. Cormack (Stanford University).
Recombinant DNA techniques.
Chromosomal DNA was isolated by the method of Maniatis et al. (27). Plasmid DNA was isolated by using Qiagen columns as specified by the manufacturer (Qiagen, Inc.). Electroporation, transformation, and other genetic techniques were performed by standard methods (4, 27) or according to manufacturers’ instructions.
Nucleotide sequencing.
Sequencing was performed by the dideoxy-chain termination method (4, 27) with double-stranded DNA as the template. Reagents from the Prism Ready Reaction Dye Deoxy Termination kit (Applied Biosystems) were used in conjunction with Taq polymerase (Boehringer Mannheim Corporation). Reaction were run on a model 373 DNA sequencer (Applied Biosystems).
PCR.
PCR was used to amplify gfp sequence from plasmid pKEN-GFPMut2 (9). Primers were synthesized by the phosphorimidite method on Applied Biosystems automated DNA synthesizer (model 380B). Reactions were carried out in a thermocycler (The Minicycler, model PTC-150-16; MJ Research, Inc.), using Vent DNA polymerase (Biolabs) or Taq DNA polymerase (Boehringer Mannheim). The thermocycler was programmed for 30 cycles of 94°C for 45 s, 52°C for 45 s, and 72°C for 45 s. An upstream primer (5′ AGGATCCCTGCAGGTAAAGGAGAAGAACTT 3′) contains a BamHI site and covers sequence encoding the second, third, and fourth amino acid residues of the N terminus of GFP. The downstream primer (5′ TTGGAATTCTTATTTGTATAGTTCATCC 3′) contains an EcoRI site and includes the sequence encoding the last three residues of the C terminus of GFP. Amplification resulted a 736-bp PCR product which was ligated into the pCRScript SrfI site.
Western blotting.
Soluble protein from whole-cell French press lysates of E. coli DH5α or P. mirabilis HI4320 containing plasmids was electrophoresed on a sodium dodecyl sulfate (SDS)–12% polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore). Western blots were incubated with polyclonal antiserum to GFP (1:10,000; Clontech) raised in rabbits against recombinant GFP isolated from transformed E. coli; this was followed by incubation with a goat anti-rabbit immunoglobulin G (1:2,000; Sigma) coupled to alkaline phosphatase; the blot was developed with 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium (Sigma) as a chromogenic substrate for alkaline phosphatase.
Fluorescence microscopy.
An overnight Luria broth culture of strains carrying the gfp fusion plasmid was diluted 1:100 in Luria broth containing ampicillin (50 μg/μl) and grown to an optical density at 600 nm of 0.1. Bacteria were induced with urea (0 to 500 mM) and harvested after 3 h of additional growth.
For fluorescence microscopy, induced bacteria were washed twice in phosphate-buffered saline (8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, and 0.24 g of KH2PO4 per liter [pH 7.4]). A slide was prepared by air drying a drop of culture on the surface. A Zeiss Axiophot epifluorescence microscope with filter sets for fluorescein isothiocyanate fluorescence was used. Images were recorded on Ektachrome color slide film (ASA 400; Kodak).
Spectrophotofluorimetry.
Bacteria from induced cultures (100 ml) were washed twice in 4 ml of 10 mM Tris-HCl (pH 7.4)–100 mM NaCl–1 mM MgCl2–10 mM dithiothreitol and suspended in 4 ml of the buffer. Cells were lysed in a French pressure cell (20,000 lb/in2), and the lysate was centrifuged (5,000 × g, 5 min, 4°C). The supernatant was collected and centrifuged (27,000 × g, 15 min, 4°C) (7). The emission (470-nm excitation wavelength, emission at 490 to 590 nm, 2-nm slits) and corrected excitation (330 to 530-nm excitation wavelength, emission at 550 nm) spectra of the supernatant from the high-speed centrifugation were obtained on a Spectronics AB-2 spectrophotofluorimeter (Spectronics, Inc., Rochester, N.Y.).
CBA mouse model of ascending UTI.
A modification (19) of the mouse model of ascending UTI originally developed by Hagberg et al. (14) was used. Female mice (20 to 22 g, 6 to 8 weeks old; Jackson Laboratory, Bar Harbor, Maine) tested for the absence of bacteriuria were anesthetized with methoxyflurane and inoculated with P. mirabilis HI4320 (107 CFU suspended in 0.05 ml of phosphate-buffered saline) through a sterile polyethylene catheter inserted into the bladder through the urethra. Mice were provided with drinking water containing ampicillin (250 μg/ml). After 1 week, the mice were sacrificed by administration of an overdose of methoxyflurane. Urine was collected, and the bladder and both kidneys were removed. Half of the bladder or kidney samples were embedded in OCT (Tissue-Tek; Miles Inc.), frozen on dry ice, and cryosectioned into 5- to 10-μm sections for fluorescence microscopic analysis. The remaining half of each sample was quantitatively cultured, and viable counts were determined as CFU/milliliter of urine or CFU/gram of tissue.
RESULTS
Construction of ureD-gfp translational fusion.
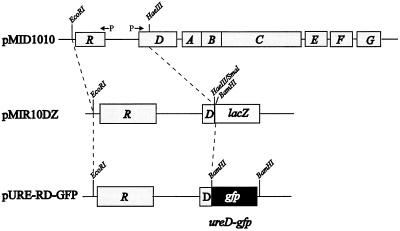
Urease expression is regulated at the transcriptional level. ureD is upstream of structural subunits ureABC and is transcribed on the same mRNA as ureABC, under control of the same promoter (18, 22, 37) (Fig. 1). Thus, the level of UreD-GFP expression reflects urease apoenzyme expression. Full-length ureR and the first 108 bp of ureD were cloned on an EcoRI/BamHI fragment from pMIR10DZ (18), which is a subclone of pMID1010, into pBluescript to form pURE-RD. A fragment carrying the gfp open reading frame amplified by PCR from pGFPmut2 (9) was cloned into pCRScript; the BamHI fragment from this plasmid was isolated and ligated into BamHI-digested pURE-RD. The resultant plasmid, designated pURE-RD-GFP (Fig. 1), was isolated, and insertion of the proper fragment was confirmed by restriction enzyme digestion. The in-frame translational fusion was confirmed by nucleotide sequencing of the junction (data not shown).
FIG. 1.
Construction of a ureD-gfp fusion. Intact ureR and part of ureD were cloned as an EcoRI/BamHI fragment from pMIR10DZ (18), which is a subclone of pMID1010, into pBluescript to form pURE-RD. A fragment carrying the gfp open reading frame (see text) amplified by PCR from pGFPmut2 (9) was cloned into pCRScript; the BamHI fragment from this plasmid was isolated and ligated into BamHI-digested pURE-RD. The final construct was designated pURE-RD-GFP.
Urea induction and Western blotting.
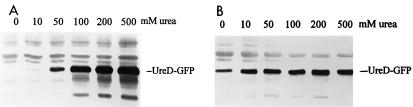
To demonstrate that synthesis of UreD-GFP could be induced by urea, E. coli DH5α(pURE-RD-GFP) was grown in Luria broth at 37°C. When exponentially growing cultures reached an optical density at 600 nm of 0.1, urea (0 to 500 mM) was added. After 2 h of induction, bacteria were collected and lysed by passage through a French pressure cell. Western blots of soluble protein were used to assess expression of the UreD-GFP fusion. Soluble protein from either E. coli DH5α or P. mirabilis HI4320 transformed with pURE-RD-GFP, induced or uninduced by urea, was electrophoresed on an SDS–12% polyacrylamide gel. Proteins were transferred to nitrocellulose and reacted with rabbit anti-GFP. The UreD-GFP fusion protein was predicted to contain the first 37 amino acids of UreD and all but the first two of a total of 238 amino acids of GFP. The fusion protein, therefore, was predicted to contain 275 amino acids and have a molecular size of 30.0 kDa. Western blot analysis showed that urea-induced E. coli(pURE-RD-GFP) cells produced a polypeptide of 30 kDa, consistent with the predicted size (Fig. 2A). The expression level correlated with the urea concentration, with maximal induction at 200 and 500 mM. The band was missing from the uninduced sample, indicating that the induction of fusion GFP synthesis occurs only in the presence of urea in a E. coli background. Although P. mirabilis(pURE-RD-GFP) exhibited similar urea concentration-dependent expression, it also produced a low level of UreD-GFP fusion protein in the absence of urea (Fig. 2B).
FIG. 2.
Western blot analysis of UreD-GFP fusion protein induction by urea. E. coli (A) and P. mirabilis (B) carrying pURE-RD-GFP were uninduced or induced by urea (10, 50, 100, 200, and 500 mM). Soluble protein from these strains was electrophoresed on an SDS–12% polyacrylamide gel. A polyclonal antiserum raised in rabbits against recombinant GFP was used for Western blotting.
Fluorescence determination of GFP expression.
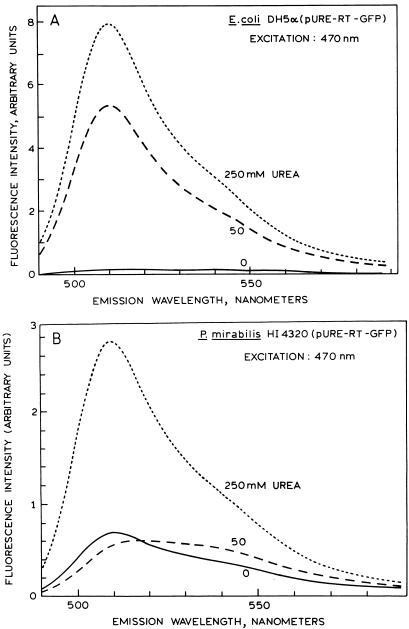
The level of UreD-GFP fusion protein expression was also quantitated by spectrophotofluorimetry. Soluble cell extracts were prepared from uninduced or urea-induced (50 and 250 mM) bacterial cultures. Both fluorescence excitation (data not shown) and emission spectra of the induced soluble cell extracts from E. coli (Fig. 3A) and P. mirabilis (Fig. 3B) were very similar to those observed previously for this GFP variant (9), suggesting that GFP had indeed been expressed. Upon induction, both species exhibited dramatic, urea-dependent increases in fluorescence compared with the corresponding uninduced strains. In particular, E. coli displayed a 33-fold increase in intensity at 509 nm when induced with 50 mM urea and a 50-fold increase when induced with 250 mM urea (Fig. 3A). Similarly, P. mirabilis exhibited a fourfold increase in emission at 509 nm when induced with 250 mM urea and a negligible change in emission when induced with 50 mM urea (Fig. 3B). The differences in the degree of fluorescence enhancement are attributable to both lower fluorescence background in E. coli and more substantial expression of GFP.
FIG. 3.
Fluorescence emission spectra of soluble extracts from E. coli DH5α(pURE-RD-GFP) (A) and P. mirabilis HI4320(pURE-RD-GFP) (B). Bacteria were induced with 0 (——), 50 (———), and 250 (–––) mM urea.
Visible fluorescence of bacteria after urea induction.
Following urea induction in vitro, E. coli(pURED-GFP) and P. mirabilis(pURED-GFP) were also quantitatively assessed by fluorescence microscopy. For E. coli containing the construct, without induction there were no visible fluorescent bacteria. At 10 mM urea, all bacteria were very weakly fluorescent; at ≥100 mM, all bacteria were brightly fluorescent. For P. mirabilis containing the construct, without induction, the vast majority of bacteria showed no fluorescence; a very small percentage of bacteria, however, were brightly fluorescent. At 10 mM urea, all of the bacteria were at least weakly fluorescent and some were brightly fluorescent; at ≥100 mM, all bacteria were brightly fluorescent but did not display the same intensity as did the transformed E. coli.
In vivo expression of GFP in experimental ascending UTI.
To determine whether GFP could be used as a reporter for urease expression in vivo and whether the expression level was sufficient during infection to visualize bacteria, CBA mice were inoculated transurethrally with 107 CFU of P. mirabilis HI4320(pURED-GFP). After 1 week, the geometric means of log10 concentrations of bacteria in urine, bladder, and kidney were determined and found to be typical of previous experimental infections (5): urine, 7.79 CFU/ml; bladder, 6.22 CFU/g; and kidneys, 5.15 CFU/g. Thus, GFP expression did not compromise survival of the challenge strains.
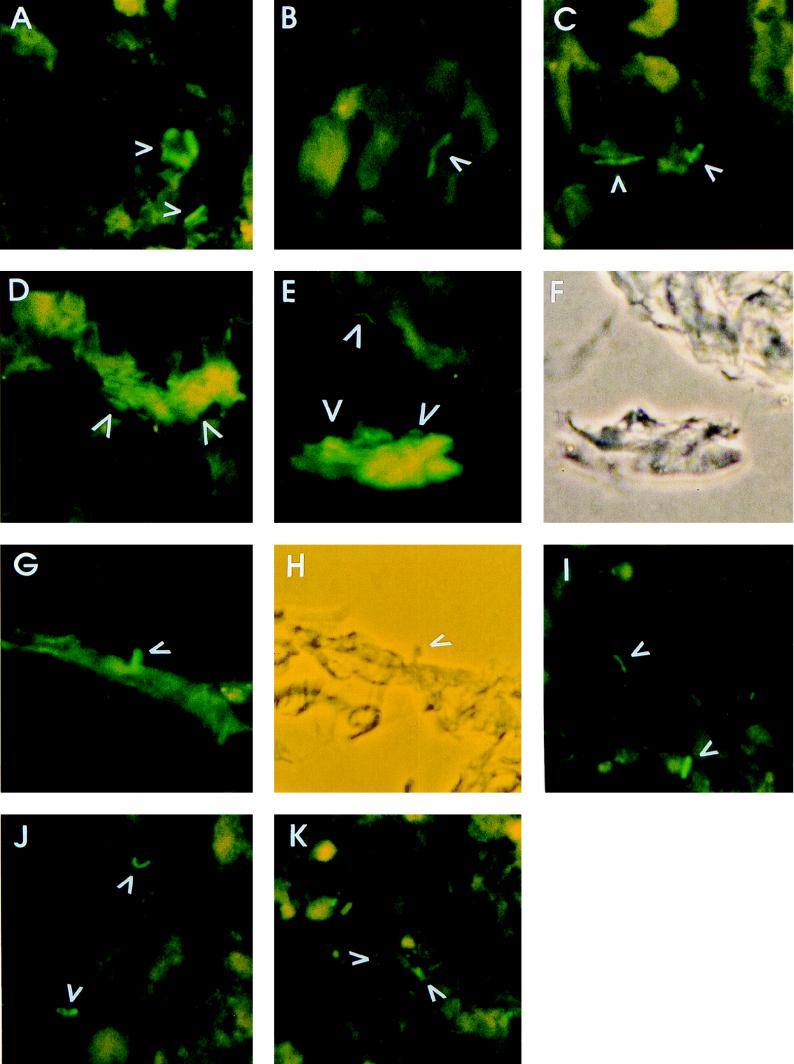
To search for fluorescent bacteria, frozen thin-sectioned bladder (Fig. 4A to H) or kidney (Fig. 4I to K) samples from infected animals were observed by fluorescence microscopy. Numerous green fluorescent bacteria were detected in both bladder and kidney samples. The fluorescence intensity of bacteria from bladder appeared qualitatively higher than that from kidney, suggesting a higher level urease expression in bladder than in kidney. While some single bacteria attached to the surface of bladder epithelium by the pole of the organism (Fig. 4G), other aggregated bacteria, apparently covered in biofilm, were loosely attached to the bladder tissue (Fig. 4D and E). Such large aggregates were commonly observed in the bladder. While an occasional elongated swarmer cell was observed in the bladder sections (Fig. 4B and C), only single vegetative forms of bacteria were found scattered around the kidney tissue samples. The results clearly indicate that the urea concentration was high enough to induce the GFP-UreD fusion protein levels sufficient to make individual bacteria easily visible by fluorescence microscopy.
FIG. 4.
In vivo expression of UreD-GFP by P. mirabilis infecting the bladders and kidneys of CBA mice. Thin sections of bladders (A to H) and kidneys (I to K) obtained from CBA mice infected with P. mirabilis HI4320(pURE-RD-GFP) were observed by fluorescence microscopy. Fluorescent bacteria are identified by arrows. (F and H) Phase-contrast images of panels E and G, respectively.
DISCUSSION
An in-frame translational fusion of ureD with gfp (S65T/V68L/S72A) was successfully constructed and confirmed by nucleotide sequencing of the fusion junction. Using Western blotting and spectrophotofluorimetry, we found expression of UreD-GFP only in the samples with urea induction in E. coli; the expression level was correlated with urea concentration, with highest induction at 200 to 500 mM, an observation consistent with previous reports (37). We have learned that urease genes are indeed expressed in vivo by P. mirabilis during UTI, confirming an assertion for which significant circumstantial evidence exists. More importantly, however, we have demonstrated that GFP can be used successfully to study expression of virulence genes in an experimental model of ascending UTI.
While we are confident that expression of the fusion protein represents an accurate proxy for measurement of urease activity, it should be stressed that we are measuring expression of ureD (encodes an accessory protein that is not part of the enzyme), a gene directly upstream of ureA (encodes the smallest subunit of the apoenzyme) (Fig. 1). It has been determined previously that ureD and ureA are transcribed on the same urea-inducible mRNA (18, 21, 22, 37). Nevertheless, expression of the UreD-GFP fusion is an indirect measurement of urease enzyme expression.
In these studies, we noted that uninduced cultures of P. mirabilis(pURED-GFP) produced a low level of the fusion protein whereas E. coli carrying the same multicopy plasmid maintains tight regulation of enzyme synthesis in the absence of urea. This was observed both on Western blots (Fig. 2) and by spectrophotofluorimetry (Fig. 3) (compare levels of uninduced production of fusion proteins and fluorescence intensity for P. mirabilis and E. coli). This finding is consistent with the fact that uninduced P. mirabilis produces a low level of urease (21, 31). These observations suggest that P. mirabilis may have an additional tier of regulation beyond UreR-mediated transcriptional activation. Allison et al. (3) provided evidence for this reporting that expression of urease-specific mRNA is increased during swarming, suggesting that expression of urease goes beyond simple urea induction. Indeed, it is logical to always produce some enzyme; a low level of urease may be necessary for adequate nitrogen metabolism in P. mirabilis in the bowel or outside the host. In some bacterial species, like Morganella morganii, urease is synthesized constitutively to ensure that some enzyme is always produced (17).
Before the S65T/V68L/S72A variant of gfp was available, both wild-type gfp and gfp (S65T) (8, 15) were fused by us to ureD (data not shown). In both cases, however, no strong fluorescence emitted from either E. coli or P. mirabilis carrying the pURED-GFP plasmid could be observed by fluorescence microscopy or spectrophotofluorimetry. Therefore, we looked for expression of the GFP fusion protein in whole-cell extract by Western blotting using polyclonal anti-GFP. Western blotting demonstrated that the fusion protein was expressed and that levels of induction correlated with urea concentration. However, by separating inclusion bodies from whole-cell extract, we noted that most of the fusion protein partitioned with the inclusion bodies. This finding is consistent with what has been reported by several groups, specifically that overexpressed GFP in inclusion bodies of bacteria does not generate the internal chromophore and is therefore nonfluorescent (16). The newest variant of gfp (S65T/V68L/S72A) appears to overcome the folding problem in bacteria and also has increased fluorescence intensity. Therefore, this version of GFP, unlike previous versions, is suitable for in vivo studies in the urinary tract.
In vivo expression of UreD-GFP was assessed in experimental ascending UTI. Urea output in mouse urine (24.3 mg/24 h; range of volume, 0.9 to 2.9 ml; therefore, the urea concentration range is 140 to 450 mM [10]) is similar to that of humans and is high enough to fully induce GFP in P. mirabilis transformed with pURED-GFP encoding the translational fusion. Fluorescent bacteria were detected as single cells in both bladder and kidney, indicating that the P. mirabilis urease gene was induced in both tissues. In the bladder, some interesting phenomena were observed. First, adherence of single bacteria to the bladder epithelium, which may be the first step of colonization of the host, was mediated by one end of the cell, suggesting the polarized distribution of the adhesin structures or an intimate attachment by the bacterium. Second, aggregates and multiple layers of bacteria appeared to be embedded in biofilm (polysaccharide matrix) that was loosely attached to surface of the bladder epithelium. Interestingly, bacteria clustered inside a protective biofilm have been implicated in chronic bacterial UTI and bladder stone formation and may have the advantage of being more resistant to host defenses and antibiotic therapy (29, 40). Third, vegetative forms of P. mirabilis (single bacterial cells as opposed to elongated swarming cells) were most often observed in the bladder tissue. Fourth, elongated swarming cells (5 to 10 cell lengths) were occasionally found in bladder; the role of these cells, however, is unclear. In the kidneys, extracellular bacteria were fluorescent and tended to remain as single cells in a vegetative form. Since the fluorescence was qualitatively weaker in the kidney than in the bladder, this finding suggested that either the urea concentration was lower in the kidney or that bacteria in the kidney were less accessible to urine because they had invaded more deeply into tissue. The fact that the elongated swarming cells were not observed in kidneys does not necessarily mean that swarming cells do not play a role in infection. These cells may have invaded kidney cells where the urea concentration was too low to induce the fusion protein (1, 2) and thus make themselves visible. Nevertheless, we have demonstrated for the first time that the enhanced GFP can be used to study expression of virulence genes by P. mirabilis in a mouse model of ascending UTI.
ACKNOWLEDGMENTS
This work was supported in part by Public Health Service grants AI23328 and DK47920 from the National Institutes of Health.
REFERENCES
- 1.Allison C, Coleman N, Johns P L, Hughes C. Ability of Proteus mirabilis to invade human urothelial cells is coupled to motility and swarming differentiation. Infect Immun. 1992;60:4740–4746. doi: 10.1128/iai.60.11.4740-4746.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison C, Emody L, Coleman N, Hughes C. The role of swarm cell differentiation and multicellular migration in the uropathogenicity of Proteus mirabilis. J Infect Dis. 1994;169:1155–1158. doi: 10.1093/infdis/169.5.1155. [DOI] [PubMed] [Google Scholar]
- 3.Allison C, Lai H-C, Hughes C. Co-ordinate expression of virulence genes during swarm cell differentiation and population migration of Proteus mirabilis. Mol Microbiol. 1992;6:1583–1591. doi: 10.1111/j.1365-2958.1992.tb00883.x. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 5.Bahrani F K, Massad G, Lockatell C V, Johnson D E, Russell R, Warren J W, Mobley H L T. Construction of an MR/P fimbrial mutant of Proteus mirabilis: role in virulence in a mouse model of ascending urinary tract infection. Infect Immun. 1994;62:3363–3371. doi: 10.1128/iai.62.8.3363-3371.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belas R, Erskine D, Flaherty D. Transposon mutagenesis in Proteus mirabilis. J Bacteriol. 1991;173:6289–6293. doi: 10.1128/jb.173.19.6289-6293.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chalfie M, Yuan T, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 8.Clontech. Living colors™ enhanced GFP vectors. CLONTECHniques. 1996;11(2):2–3. [Google Scholar]
- 9.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 10.Crispens C G., Jr . Handbook on the laboratory mouse. Springfield, Ill: Charles C Thomas; 1975. p. 130. [Google Scholar]
- 11.Dhandayuthapani S, Via L E, Thomas C A, Horowitz P M, Deretic D, Deretic V. Green fluorescent protein as a marker for gene expression and cell biology of mycobacterial interaction with macrophages. Mol Microbiol. 1995;17:901–912. doi: 10.1111/j.1365-2958.1995.mmi_17050901.x. [DOI] [PubMed] [Google Scholar]
- 12.Gage D J, Bobo T, Long S R. Use of green fluorescence protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa. J Bacteriol. 1996;178:7159–7166. doi: 10.1128/jb.178.24.7159-7166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffith D P, Musher D M, Itin C. Urease: the primary cause of infection-induced urinary stones. Invest Urol. 1976;13:346–350. [PubMed] [Google Scholar]
- 14.Hagberg L, Engberg I, Freter R, Lam J, Olling S, Svanborg-Eden C. Ascending unobstructed urinary tract infection in mice cause by pyelonephritogenic Escherichia coli of human origin. Infect Immun. 1983;40:273–283. doi: 10.1128/iai.40.1.273-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heim R, Cubitt A B, Tsien R Y. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 16.Heim R, Prasher D C, Tsien R Y. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu L-T, Nicholson E B, Jones B D, Lynch M J, Mobley H L T. Morganella morganii urease: purification, characterization, and isolation of gene sequences. J Bacteriol. 1990;172:3073–3080. doi: 10.1128/jb.172.6.3073-3080.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Island M, Mobley H L T. Proteus mirabilis urease: operon fusion and linker insertion analysis of ure gene organization, regulation, and function. J Bacteriol. 1995;177:5653–5600. doi: 10.1128/jb.177.19.5653-5660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson D E, Lockatell C V, Hall-Craigs M, Mobley H L T, Warren J W. Uropathogenicity in rats and mice of Providencia stuartii from long-term catheterized patients. J Urol. 1987;138:632–635. doi: 10.1016/s0022-5347(17)43287-3. [DOI] [PubMed] [Google Scholar]
- 20.Johnson D E, Russell R G, Lockatell C V, Zulty J C, Warren J W, Mobley H L T. Contribution of Proteus mirabilis urease to persistence, urolithiasis, and acute pyelonephritis in a mouse model of ascending urinary tract infection. Infect Immun. 1993;61:2748–2754. doi: 10.1128/iai.61.7.2748-2754.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones B D, Mobley H L T. Proteus mirabilis urease: genetic organization, regulation, and expression of structural genes. J Bacteriol. 1988;170:3342–3349. doi: 10.1128/jb.170.8.3342-3349.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones B D, Mobley H L T. Proteus mirabilis urease: nucleotide sequence determination and comparison with jack bean urease. J Bacteriol. 1989;171:6414–6422. doi: 10.1128/jb.171.12.6414-6422.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones B D, Lockatell C V, Johnson D E, Warren J W, Mobley H L T. Construction of a urease-negative mutant of Proteus mirabilis: analysis of virulence in a mouse model of ascending urinary tract infection. Infect Immun. 1990;58:1120–1123. doi: 10.1128/iai.58.4.1120-1123.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kain S R, Adams M, Kondepudi A, Yang T T, Ward W W, Kitts P. Green fluorescent protein as a reporter of gene expression and protein localization. BioTechniques. 1995;19:650–655. [PubMed] [Google Scholar]
- 25.Kremer L, Baulard A, Estaquier J, Poulain-Godefroy O, Locht C. Green fluorescence protein as a new expression marker in mycobacteria. Mol Microbiol. 1995;17:913–922. doi: 10.1111/j.1365-2958.1995.mmi_17050913.x. [DOI] [PubMed] [Google Scholar]
- 26.Leff L G, Leff A A. Use of green fluorescent protein to monitor survival of genetically engineered bacteria in aquatic environments. Appl Environ Microbiol. 1996;62:3486–3488. doi: 10.1128/aem.62.9.3486-3488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 28.Matthysse A G, Stretton S, Dandie C, McClure N C, Goodman A E. Construction of GFP vectors for use in gram-negative bacteria other than E. coli. FEMS Microbiol Lett. 1996;145:87–94. doi: 10.1111/j.1574-6968.1996.tb08561.x. [DOI] [PubMed] [Google Scholar]
- 29.McLean R J C, Downey J A, Lablans A L, Clark J M, Dumanski A J, Nickel J C. Modeling biofilm-associated urinary tract infections in animals. Int J Biodeterior Biodegrad. 1992;30:201–216. [Google Scholar]
- 30.Mobley H L T, Chippendale G R. Hemagglutinin, urease, and hemolysin production by Proteus mirabilis from clinical sources. J Infect Dis. 1990;161:525–530. doi: 10.1093/infdis/161.3.525. [DOI] [PubMed] [Google Scholar]
- 31.Mobley H L T, Chippendale G R, Swihart K G, Welch R. Cytotoxicity of the HpmA hemolysin and urease of Proteus mirabilis and Proteus vulgaris against cultured human renal proximal tubular epithelial cells. Infect Immun. 1991;59:2036–2042. doi: 10.1128/iai.59.6.2036-2042.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mobley H L T, Island M D, Hausinger R P. Molecular biology of microbial urease. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mobley H L T, Hausinger R P. Microbial ureases: significance, regulation, and molecular characterization. Microbiol Rev. 1989;53:85–108. doi: 10.1128/mr.53.1.85-108.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mobley H L T, Warren J W. Urease positive bacteriuria and obstruction of long term urinary catheters. J Clin Microbiol. 1987;25:2216–2219. doi: 10.1128/jcm.25.11.2216-2217.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mobley H L T. Virulence of Proteus mirabilis. In: Mobley H L T, Warren J W, editors. Urinary tract infection: molecular pathogenesis and clinical management. Washington, D.C: ASM Press; 1996. pp. 245–271. [Google Scholar]
- 36.Nicholson E B, Concaugh E A, Foxall P A, Island M D, Mobley H L T. Proteus mirabilis urease: transcriptional regulation by ureR. J Bacteriol. 1993;175:465–473. doi: 10.1128/jb.175.2.465-473.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicholson E B, Concaugh E A, Mobley H L T. Proteus mirabilis urease: use of a ureA-lacZ fusion demonstrates that induction is highly specific for urea. Infect Immun. 1991;64:5332–5340. doi: 10.1128/iai.59.10.3360-3365.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prasher D C, Eckenrode V K, Ward W W, Prendergast F G. Primary structure of the Aequorea victoria green fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 39.Rubin R, Tolkoff-Rubin N, Cotran R. Urinary tract infection, pyelonephrititis and reflux nephropathy. In: Brenner G, Rector F Jr, editors. The kidney. 3rd ed. Philadelphia, Pa: The W. B. Saunders Co.; 1986. pp. 1085–1141. [Google Scholar]
- 40.Salyers A A, Whitt D D. Host defenses against bacterial-pathogens: defenses of body surfaces. In: Salyers A A, Whitt D D, editors. Bacterial pathogenesis: a molecular approach. Washington, D.C: ASM Press; 1994. pp. 3–15. [Google Scholar]
- 41.Valdivia R H, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 42.Valdivia R H, Hromockyj A E, Monack D, Ramakrishnan L, Falkow S. Applications for green fluorescent protein (GFP) in the study of host-pathogen interaction. Gene. 1996;173:47–52. doi: 10.1016/0378-1119(95)00706-7. [DOI] [PubMed] [Google Scholar]
- 43.Valdivia R H, Falkow S. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol Microbiol. 1996;22:367–378. doi: 10.1046/j.1365-2958.1996.00120.x. [DOI] [PubMed] [Google Scholar]