Abstract
Primary ovarian insufficiency (POI) is a common condition leading to the pathological decline of ovarian function in women of reproductive age, resulting in amenorrhea, hypogonadism, and infertility. Biochemical premature ovarian insufficiency (bPOI) is an intermediate stage in the pathogenesis of POI in which the fertility of patients has been reduced. Previous studies suggest that granulosa cells (GCs) play an essential role in the pathogenesis of POI, but their pathogenetic mechanisms remain unclear. To further explore the potential pathophysiological mechanisms of GCs in POI, we constructed a molecular long non-coding RNA (lncRNA)-microRNA (miRNA)-messenger RNA (mRNA) network using GC expression data collected from biochemical premature ovarian failure (bPOI) patients in the GEO database. We discovered that the GCs of bPOI patients had differential expression of 131 mRNAs, 191 lncRNAs, and 28 miRNAs. By systematic network analysis, we identified six key genes, including SRSF1, PDIA5, NEURL1B, UNK, CELF2, and CFL2, and five hub miRNAs, namely hsa-miR-27a-3p, hsa-miR-24-3p, hsa-miR-22-3p, hsa-miR-129-5p, and hsa-miR-17-5p, and the results suggest that the expression of these key genes may be regulated by two hub miRNAs, hsa-miR-27a-3p and hsa-miR-17-5p. Additionally, a POI model in vitro was created to confirm the expression of a few important genes. In this study, we discovered a unique lncRNA-miRNA-mRNA network based on the ceRNA mechanism in bPOI for the first time, and we screened important associated molecules, providing a partial theoretical foundation to better understand the pathogenesis of POI.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10815-023-02937-2.
Keywords: Primary ovarian insufficiency (POI), Biochemical premature ovarian failure (bPOI), Granulosa cells (GCs), Competitive endogenous RNA network (ceRNA network), Key genes, Bioinformatics analysis
Introduction
Premature ovarian failure (POI) is defined as the loss of ovarian function before age 40, affecting some women of childbearing age [1]. In clinics, POI is divided into three stages, namely the insidious, biochemical, and evident (previously known as premature ovarian failure). Regular menstrual cycles are observed in patients with biochemical premature ovarian failure (bPOI), but they also have high FSH levels and reduced fertility [2]. The available studies suggest that POIs are multi-layered and sophisticated pathogenic processes involving genetic, autoimmune, metabolic disorders, and infections [3]. To date, the pathogenesis of POI is not clear.
Some researchers found that apoptosis of granulosa cells is present in all cases of premature ovarian failure [4]. Granulosa cells (GCs), essential follicle somatic components, are crucial to the process of folliculogenesis. Recent studies suggest that dysfunction of GCs in patients with POI may contribute to the development of ovarian insufficiency [5–7]. Wang et al. investigated the lncRNA expression profile in GCs of bPOI patients and discovered that the down-regulated lncRNA HCP5 impairs the granulosa cells’ DNA damage repair (DDR) process [8]. Zhang et al. found that MiR-127-5p overexpression in GCs of bPOI patients had negative impacts on GC proliferation and DNA repair activity [9]. However, the crosstalk regulatory gene networks and possible therapeutic targets of GCs in bPOI are still poorly studied.
With a more profound understanding of genomics, long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), these non-coding RNAs (ncRNAs) of the human transcriptome have received increasing attention from researchers [10–12]. Numerous studies have confirmed that microRNAs serve as pragmatic markers for diagnosis and play an essential role in the pathogenesis of various diseases [13, 14]. Besides, ncRNAs are involved in multiple complex networks of many molecular mechanisms that regulate genomic function and competitive RNA-RNA interactions [15]. Competitive endogenous RNAs (ceRNAs) are a recently discovered mechanism involving complex gene regulation between ncRNAs and mRNAs. Competing for shared miRNAs allows transcripts in the ceRNA network to control one another at the post-transcriptional level, indicating that the interaction between lncRNA, miRNA, and mRNA has a significant impact on many diseases [16].
Recent studies have constructed ceRNA networks in various diseases that help reveal the pathogenesis of diseases and provide potential therapeutic sites for disease treatment [17, 18]. However, no report has investigated the lncRNA-miRNA-mRNA regulatory networks in POI development. By analyzing the expression data of human granulosa cells from bPOI patients in the Gene expression Omnibus (GEO) database, we constructed an lncRNA-miRNA-mRNA ceRNA network in the current study to further reveal the ceRNAs interaction underlying human POI. We also identified key molecules in the granulosa cells of patients with biochemical premature ovarian failure (bPOI) in an effort to offer new insights for future studies on the POI pathological underpinnings.
Materials and methods
Microarray data
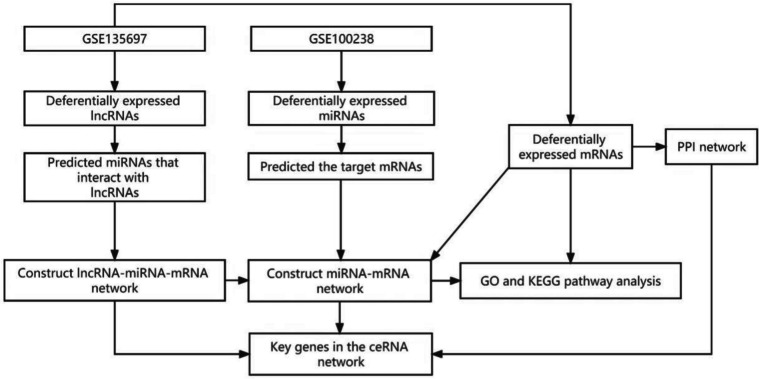
The Gene Expression Omnibus (GEO) library of the National Center for Biotechnology Information (NCBI) was searched for gene expression profile datasets of MGCs from women with bPOI and healthy women without bPOI (https://www.ncbi.nlm.nih.gov/geo/). Data from chip, second-generation sequencing, and other high-throughput sequencing techniques are kept in the GEO repository, a public database. The research was carried out under the Helsinki Declaration (as revised in 2013). Premature ovarian insufficiency, bPOI, granulosa cells, and homo sapiens were the search terms. GSE100238 and GSE135697 were chosen for additional examination (Fig. 1). Ten samples from the dataset GSE100238 (platform: GPL19128 Exiqon miRCURY LNA microRNA array) are bPOI samples, whereas ten other samples are not. Ten bPOI samples and ten non-bPOI samples make up the GSE135697 dataset (platform: GPL16956 Agilent-045997 Arraystar human lncRNA microarray V3).
Fig. 1.
Flowchart of data processing and analysis. LncRNA: long non-coding RNA; miRNA: microRNA; mRNA: messenger RNA; ceRNA network: competitive endogenous RNA network; PPI: protein-protein interaction; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes
Identification of differentially expressed mRNAs and non-coding RNAs
Using GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r), we examined the mRNAs and non-coding RNAs that were differently expressed between the POI and non-POI samples in these two selected datasets (GSE100238 and GSE135697), respectively. RNAs that have differential expression were those with |log FC| > 1 and p < 0.05.
Functional and pathway enrichment analysis
Metascape (https://metascape.org/) was used to analyze DEGs by performing GO functional annotation and KEGG analysis. For both GO keywords and KEGG pathways, statistical significance was set at p < 0.05.
Prediction and construction of the miRNA-mRNA network
Three miRNA target prediction databases—MiRTarBase (https://maayanlab.cloud/Harmonizome/resource/MiRTarBase), TargetScan (http://www.targetscan.org/vert 72/), and miRDB (http://mirdb.org/)—were used to assess the target genes of differentially expressed miRNAs. The use of Perl (version 5.32.0) was made for data analysis. The differential expression of target genes and miRNAs exhibits divergent patterns of change. The miRNA-mRNA regulatory network was seen using Cytoscape (http://www.cytoscape.org/).
Constructing the lncRNA-miRNA-mRNA ceRNA network
Based on the lncRNA target prediction databases presented below, miRcode (http://www.mircode.org/) and Perl were used to evaluate lncRNA-miRNA interactions (version 5.32.0). The miRNAs and lncRNAs have opposing patterns of expression change and are differently expressed. After that, the competing triplets that were co-expressed were put together to create lncRNA-miRNA-mRNA networks that were differentially expressed. To visualize the network, Cytoscape was utilized. The hub miRNAs and hub genes in the ceRNA network were also found using the CytoHubba software.
Protein-protein interaction (PPI) network generation and module analysis
Using STRING (https://string-db.org/), the PPI network of the DEGs was examined. A cut-off threshold of more than 0.15 was established. Cytoscape was used to display the PPI network. By using Cytoscape cytoHubba, hub genes were ranked [19]. Modules in the PPI network were screened by Cytoscape MCODE (degree cut-off = 2, max. depth = 100, k-core = 2, and node score cut-off = 0.2) to identify the key genes among the DEGs [19]. In order to analyze the PPI network modules and find the key genes among the DEGs, the MCODE program in Cytoscape was used (standard: degree cut-off = 2, max. depth = 100, k-core = 2, and node score cut-off = 0.2). Modules that have more than three MCODE points and five nodes were chosen.
Key gene identification and validation
By considering the intersection of hub genes in the ceRNA network and PPI network, the key genes were identified. The key genes were then retrieved for further investigation. In this study, we used GraphPad Prism 9 (version 9.0.0) to show key DEGs across several datasets.
Next, we used cisplatin (Selleck, USA) treatment of the human granulosa-like tumor cell line KGN (MEISEN CELL, China) to construct a POI model in vitro to verify the expression of these key genes. The cells grew in DMEM /F12 medium (Gibco, USA) supplemented with 10% FBS (Gibco, USA) and penicillin-streptomycin (Invitrogen, USA) at 37 °C in an atmosphere of 5% CO2 [20]. KGN cells (1.5 × 105 cells/well) in logarithmic growth were seeded onto a 6-well plate. The POI group was incubated with a concentration of 50 μM cisplatin (Selleck, USA) for 24 h, and the control group was the absence of cisplatin. After 24 h, the cells are collected to extract RNA [21]. RNAiso Plus (TaKaRa, Japan) was used to extract total RNA in accordance with the manufacturer’s instructions. The purity and concentration of all RNA samples were then determined using the NanoDrop2000 (TermoScientifc, USA). PrimeScript RT Master Mix was used to synthesize cDNA (Complementary DNA) from 1 ug of total RNA (TaKaRa, Japan). SYBR Premix ExTaq kit was used to run Q-PCR on a QuantStudio Dx (ABI, America) (Takara, Japan). The relative expression was calculated by the 2−△△Ct method with GAPDH reference control. Meanwhile, triplicates of each Q-PCR experiment were performed. The following list of primers was produced using the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/nucleotid).
SRSF1: 5`-TATCCGCGACATCGACCTCAAG-3` (forward) and 5`-AAACTCCACCCGCAGACGGTAC-3` (reverse);
PDIA5: 5`-GGAGCCAAAGATGTTGTCCACC-3` (forward) and 5`-GAAATGCGGCATCATCCTCTTGC-3` (reverse);
NEURL1B: 5`-ATGTCTACGGCATCACCGACGA-3` (forward) and 5`-ACCACCTGGTTGTTCTCGAGCT-3` (reverse);
UNK: 5`-CTGGCAGCTATAAGAAGGCTCC-3` (forward) and 5`-AGCAGAGGCTGCTCTTGACTCC-3` (reverse);
CELF2: 5`-CCAGCACCAATGCAAACCCTCT-3` (forward) and 5`-GAGAGGTCAAGGAGTTCATGGC-3` (reverse);
CFL2: 5`-TTGTGAAGTTGCTACCTCTGAATG-3` (forward) and 5`-TTAAAGGTGCACTTTCAGGAGCC-3` (reverse).
GAPDH: 5`-GTCTCCTCTGACTTCAACAGCG-3` (forward) and 5`-ACCACCCTGTTGCTGTAGCCAA-3` (reverse).
Statistical analysis
The Wilcoxon test was used to conduct statistical comparisons between two sets of data in accordance with the test condition, and p < 0.05 was utilized to denote statistical significance.
Results
Differentially expressed lncRNAs, miRNAs, and mRNAs
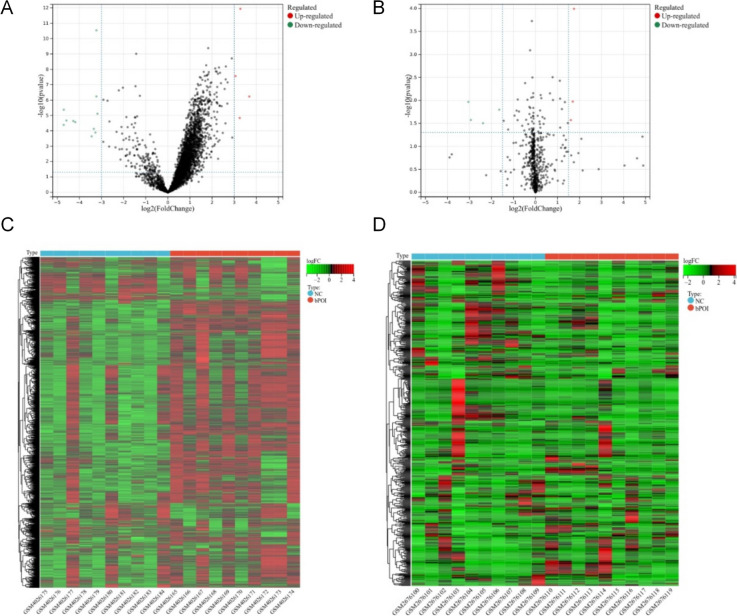
Between the GCs of patients with bPOI and those of healthy controls, 131 mRNAs, 191 lncRNAs, and 28 miRNAs displayed differential expression (Supplementary Table S1). In the GSE135697 dataset, which also contained lncRNA and mRNA data, 191 DELs (146 of which showed up-regulation and 44 of which showed down-regulation) and 131 DEmRNAs (61 of which showed up-regulation and 70 of which showed down-regulation) were found by comparing the gene expression data between bPOI and control samples. From GC samples, including those from bPOI and healthy individuals, 28 DEmiRs of the GSE100238 dataset were found—15 of which were upregulated and 13 of which were down-regulated. The volcano map and heatmap for differentially expressed lncRNAs, miRNAs, and mRNAs were created using GSE135697’s cut-off criteria (log |fold change|>3, adjust p 0.05) and GSE100238’s cut-off criteria (log |fold change|>1.5, adjust p 0.05) (Fig. 2A–D).
Fig. 2.
Differentially expressed analysis of LncRNAs, miRNAs, and mRNAs. (A) Volcano map of GSE135697, lncRNAs, and mRNAs. (B) Volcano map of GSE100238, miRNAs. (C) heatmap analysis of GSE135697, lncRNAs, and mRNAs. (D) Heatmap analysis of GSE100238, miRNAs. The data from healthy women were compared with those from patients with bPOI (n = 10). Differentially expressed LncRNA and mRNA molecules were screened under the cut-off criteria log |fold change| > 3 and adjust p < 0.05. Differentially expressed miRNA molecules were screened under the cut-off criteria log |fold change| > 1.5 and adjust p < 0.05. LncRNA: long non-coding RNA; miRNA: microRNA; mRNA: messenger RNA; DEGs: differentially expressed genes; NC: negative control; bPOI: biochemical premature ovarian failure
Table 1 show the GO terms and KEGG pathways of DEGs in GSE135697. The most significantly enriched BPs were membrane protein ectodomain proteolysis, regulation of RNA splicing response to amphetamine, proteolysis involved in cellular protein catabolic process, protein autophosphorylation, muscle system process, chemical synaptic transmission, positive regulation of the catabolic process, regulation of response to DNA damage stimulus, and sulfur compound biosynthetic process. The KEGG pathways with the highest levels of enrichment included the VEGFA-VEGFR2 signaling pathway, the post-chaperonin tubulin folding pathway, protein-protein interactions at synapses, Rho GTPase signaling, and recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double-strand breaks.
Table 1.
GO/KEGG analysis of DEGs from the GSE135697 dataset
| Term of GO/KEGG analysis | Count | P-value | Genes |
|---|---|---|---|
| WP3888: VEGFA-VEGFR2 signaling pathway | 10 | 1.7378E-05 | ABL1,ACACA,HBD,LDHA,TBCA,TKT,ADAM9,ATF6,EPN1,ACKR3 |
| GO:0006509⁓membrane protein ectodomain proteolysis | 3 | 9.12011E-05 | ADAM9,RBMX,APH1A,HDAC2,NEURL1B |
| R-HSA-389977: post-chaperonin tubulin folding pathway | 3 | 0.000104713 | TBCA,TUBA4A,TUBA1B,ABL1,HBD,HDAC2,PSMA5,RTN3,ATF6,APH1A,RAB18,SRGAP3,UPF3A,SNAP23,VRK2,ALG3,SCFD1,ST8SIA5,CLDN14 |
| GO:0043484⁓regulation of RNA splicing | 6 | 0.000109648 | CLK1,FUS,SRSF1,CELF2,RBMX,NSRP1,ECD,LSM1,NT5C3B,UPF3A |
| GO:0001975⁓response to amphetamine | 3 | 0.000269153 | DBH,EDNRA,HDAC2,ABL1,ACKR2,P2RY1,ACKR3,GPR37L1 |
| GO:0051603⁓proteolysis involved in cellular protein catabolic process | 10 | 0.000331131 | PSMA5,TRIM25,CUL4A,HERC2,ATF6,RCHY1,UCHL5,HECW2,RNF217,TRIM72,SAE1,NEURL1B,KLHL36,ABL1,KDM4A,TUBA4A,SNAP23,TUBA1B,COLEC12 |
| R-HSA-6794362: protein-protein interactions at synapses | 4 | 0.000489779 | PPFIA2,DLGAP1,NRXN2,RTN3,TUBA4A,TUBA1B |
| R-HSA-194315: signaling by Rho GTPases | 10 | 0.000831764 | ABL1,TUBA4A,VRK2,SNAP23,SRGAP3,TUBA1B,KDM4C,SCFD1,RBMX,ARHGAP15 |
| GO:0046777⁓protein autophosphorylation | 5 | 0.001202264 | ABL1,CLK1,GRK5,VRK2,TRPM7,PPFIA2,ADAM9 |
| GO:0003012⁓muscle system process | 6 | 0.001258925 | EDNRA,HDAC2,P2RY1,P2RX6,KDM4A,TRIM72,ABL1,HBD,SRGN,TUBA4A,TUBA1B,PDE10A,AKAP6 |
| GO:0007268⁓chemical synaptic transmission | 7 | 0.001513561 | DBH,SNAP23,P2RX6,DLGAP1,NRXN2,SV2C,RIMBP2 |
| GO:0009896⁓positive regulation of catabolic process | 8 | 0.001513561 | NRDC,ADAM9,ATF6,RCHY1,LSM1,HECW2,NT5C3B,RNF217,HDAC2,APH1A,UCHL5 |
| GO:2001020⁓regulation of response to DNA damage stimulus | 6 | 0.001949845 | ABL1,FUS,CUL4A,UCHL5,ACKR3,MEAF6 |
| GO:0044272⁓sulfur compound biosynthetic process | 4 | 0.002951209 | ACACA,MTRR,CSGALNACT1,DSEL,EDNRA |
| GO:0051259⁓protein complex oligomerization | 5 | 0.003801894 | ACACA,FUS,RBMX,TRPM7,TRIM72 |
| GO:2000736⁓regulation of stem cell differentiation | 3 | 0.003801894 | ABL1,HDAC2,KDM4C,GPR37L1,KDM4A,VRK2,MCM3AP,MEAF6,UCHL5 |
| R-HSA-5693565: recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double-strand breaks | 3 | 0.004265795 | ABL1,HERC2,KDM4A |
| GO:1901361⁓organic cyclic compound catabolic process | 6 | 0.005011872 | DBH,SNX17,PDE10A,LSM1,UPF3A,NT5C3B,PURA,RTN3,UNK,FUS,PSMA5,SRSF1,RPP14,RBMX |
| GO:0006897⁓endocytosis | 7 | 0.005248075 | ABL1,ACKR2,SNX17,EPN1,NEURL1B,ACKR3,COLEC12 |
| GO:0001701⁓in utero embryonic development | 6 | 0.005623413 | EDNRA,CUL4A,KDM4C,EPN1,NSRP1,UNK |
GO Gene Ontology, KEGG Kyoto Encyclopedia of Genes and Genomes, DEGs differentially expressed genes
PPI network of the DEGs and module analysis
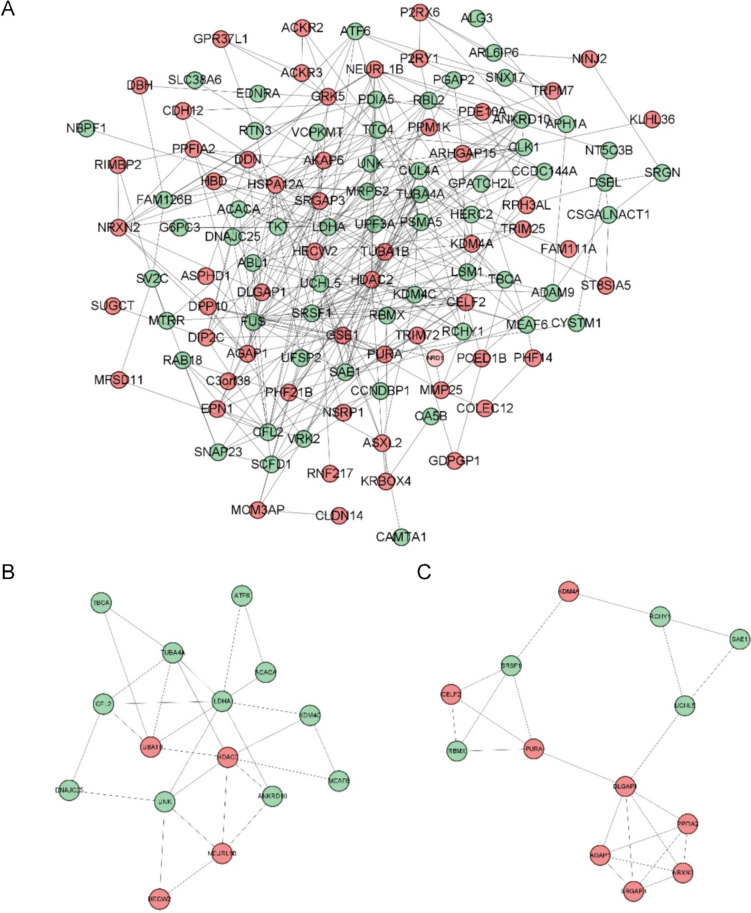
Using STRING, the PPI network of the 131 DEGs from the GSE135697 dataset was created. There were 357 edges and 115 nodes in it (Fig. 3A). HDAC2, CUL4A, FUS, TUBA1B, SRSF1, SAE1, LDHA, UCHL5, PSMA5, and KDM4A were shown to be hub genes by overlapping the top 10 genes in cytoHubba according to the degree and maximum clique centrality (MCC) approaches. From the PPI network, three clusters were found (Fig. 3B,C). Module 1 had 15 nodes and 28 edges, whereas module 2 had 13 nodes and 23 edges. Peptidyl-lysine modification, histone modification, and covalent chromatin modification were shown to be the most substantially enriched BPs in module 1 according to enrichment studies. The KEGG pathways in module 1 were mainly enriched in propanoate metabolism, pyruvate metabolism, gap junction, glucagon signaling pathway, and apoptosis. For module 2, the most significantly enriched BPs included RNA splicing via transesterification reactions with bulged adenosine as the nucleophile, mRNA splicing via spliceosome, and RNA splicing via transesterification reactions. Spliceosome and ubiquitin-mediated proteolysis were primarily abundant in the KEGG pathways in module 2.
Fig. 3.
PPI analysis. (A) The PPI network of DEGs from the GSE135697 dataset. Disconnected nodes in the network are hidden; (B) module 1 (score: 4); (C) module 2 (score: 3.833); modules with an MCODE score >3 and nodes >5 were selected. PPI: protein-protein interaction; DEGs: differentially expressed genes. Green represents down-regulation, and red shows up-regulation
Analysis of the miRNA-mRNA network
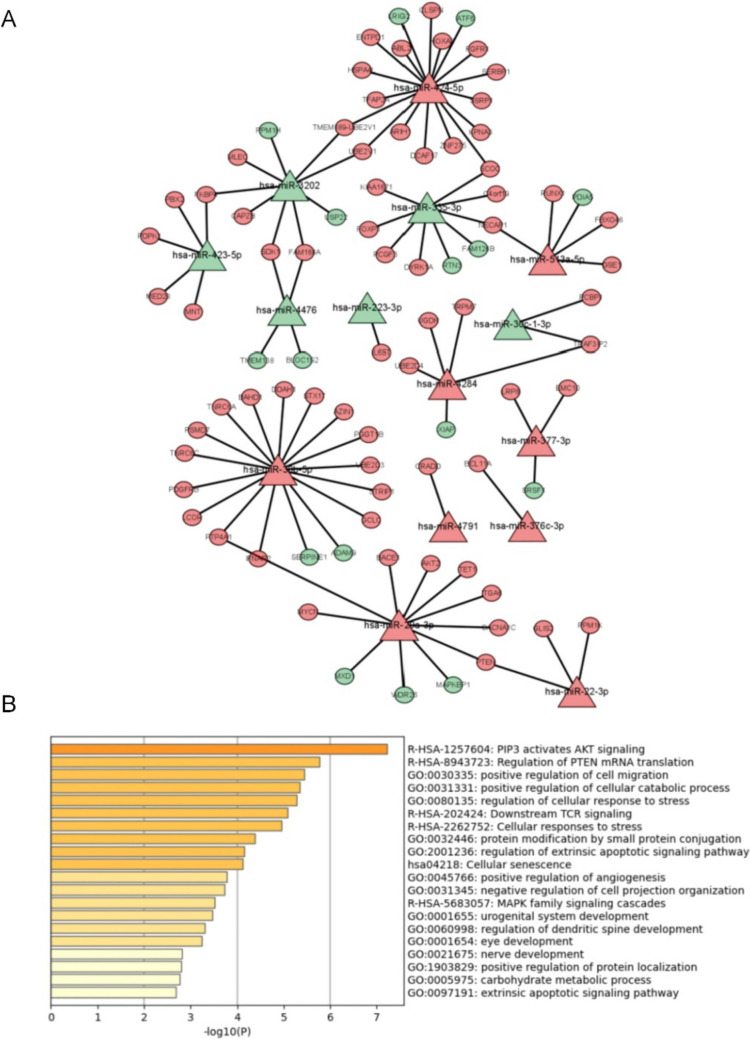
From the GSE100238 dataset, we identified 28 miRNAs that were differently expressed. Among these, 13 miRNAs were down-regulated, while 15 miRNAs were upregulated. The projected targets of these differentially expressed miRNAs and the DEGs from the GSE135697 dataset have 84 genes in common. These genes were associated with 15 different miRNAs. There is now a network of 94 miRNA-mRNA couples that regulate each other (Fig. 4A). The most GO enrichment included positive regulation of cell migration, positive regulation of the cellular catabolic process, regulation of cellular response to stress, protein modification by small protein conjugation, and regulation of extrinsic apoptotic signaling pathway (Fig. 4B, Table 2). The KEGG pathway showed significant enrichment in PIP3 activates AKT signaling, regulation of PTEN mRNA translation, downstream TCR signaling, cellular responses to stress, and cellular senescence (Fig. 4B, Table 2).
Fig. 4.
miRNA-mRNA network and GO/KEGG analysis of DEGs. (A) miRNA-mRNA network; (B) GO enrichment and KEGG enrichment of DEGs in miRNA-mRNA network; rectangular nodes represent miRNAs, and oval-shaped nodes represent mRNA. Green represents down-regulation, and red shows up-regulation. miRNA: microRNA; mRNA: messenger RNA; DEGs: differentially expressed genes; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes
Table 2.
GO/KEGG analysis of 84 overlapping genes between the predicted target genes of DEmiRs and the DEGs from the GSE135697 dataset
| Term of GO/KEGG analysis | Count | P-value | Genes |
|---|---|---|---|
| R-HSA-1257604:PIP3 activates AKT signaling | 9 | 6.0256E-08 | AKT2,XIAP,FGFR1,PDGFRB,PDPK1,PSMD7,PTEN,TNRC6A,TNRC6C,CACNA1C,RTN3,ATF6,BACE1,CLSPN,ENTPD1,IFNAR2,GCLC,KPNA3,RUNX1,DYRK1A |
| R-HSA-8943723: regulation of PTEN mRNA translation | 3 | 1.7378E-06 | PTEN,TNRC6A,TNRC6C,RUNX1,AKT2,FKBP4,PDPK1,SSRP1,CRADD,DYRK1A,TRAF3IP2,SERBP1,PSMD7 |
| GO:0030335⁓positive regulation of cell migration | 10 | 3.54813E-06 | AKT2,FGFR1,ITGA6,SERPINE1,PDGFRB,PDPK1,PTP4A1,ADAM9,FOXP1,EMC10,PTEN,ABL2,XIAP,RUNX1,IFNAR2,IL6ST,DYRK1A,TRPM7,PSMD7,BCL11A,STX17,CLSPN,LRP8,CRADD,GCLC,CACNA1C |
| GO:0031331⁓positive regulation of cellular catabolic process | 9 | 4.57088E-06 | AKT2,GCLC,PTEN,ADAM9,ATF6,ARIH1,TNRC6A,TNRC6C,SCOC,XIAP,BACE1,RUNX1,SERPINE1,CRADD,LRIG2,AZIN1,UBE2V1 |
| GO:0080135⁓regulation of cellular response to stress | 11 | 5.24807E-06 | XIAP,DYRK1A,PTEN,UBE2V1,LRIG2,ATF6,MAPKBP1,FAM168A,USP22,DDAH1,EMC10,FGFR1,PDGFRB,ADAM9 |
| R-HSA-202424: downstream TCR signaling | 5 | 8.12831E-06 | PDPK1,PSMD7,PTEN,UBE2V1,PEDS1-UBE2V1,UBE2D3,ABL2,AKT2,IFNAR2,IL6ST,KPNA3,ARIH1,CAPZB,UBE2D4,FKBP4 |
| R-HSA-2262752: cellular responses to stress | 11 | 1.12202E-05 | AKT2,CAPZB,FKBP4,GCLC,PSMD7,UBE2D3,PDIA5,HSPA4L,ATF6,TNRC6A,TNRC6C |
| GO:0032446⁓protein modification by small protein conjugation | 10 | 4.16869E-05 | XIAP,UBE2D3,UBE2V1,PCGF3,TRAF3IP2,USP22,ARIH1,UBE2D4,BCL11A,DCAF17,HSPA4L,ATF6,PSMD7,WDR26,AKT2 |
| GO:2001236⁓regulation of extrinsic apoptotic signaling pathway | 5 | 6.91831E-05 | FGFR1,GCLC,ITGA6,SERPINE1,PTEN,MNT,CRADD |
| hsa04218: cellular senescence | 5 | 7.58578E-05 | AKT2,SERPINE1,PTEN,TRAF3IP2,TRPM7,LRIG2,HOXA3,PDGFRB,EMC10,MAPKBP1,BACE1,FAM126B |
| GO:0045766⁓positive regulation of angiogenesis | 5 | 0.000165959 | RUNX1,SERPINE1,PDPK1,DDAH1,EMC10,XIAP |
| GO:0031345⁓negative regulation of cell projection organization | 5 | 0.000190546 | CAPZB,FKBP4,PTEN,LRIG2,BCL11A,ABL2,ITGA6,LRP8,DYRK1A,GCLC,TET1 |
| R-HSA-5683057: MAPK family signaling cascades | 6 | 0.000295121 | FGFR1,IL6ST,PDGFRB,PSMD7,TNRC6A,TNRC6C,AKT2,PCGF3,IFNAR2 |
| GO:0001655⁓urogenital system development | 6 | 0.000346737 | FKBP4,PDGFRB,PTEN,TFAP2A,TRAF3IP2,GLIS2,XIAP,CRADD,ITGA6,ATF6,BACE1 |
| GO:0060998⁓regulation of dendritic spine development | 3 | 0.000489779 | PTEN,LRP8,SDK1 |
| GO:0001654⁓eye development | 6 | 0.00057544 | CACNA1C,PBX2,PDGFRB,TFAP2A,ATF6,SDK1,ITGA6 |
| GO:0021675⁓nerve development | 3 | 0.001513561 | HOXA3,TFAP2A,LRIG2 |
| GO:1903829⁓positive regulation of protein localization | 6 | 0.001584893 | AKT2,PDPK1,UBE2D3,ADAM9,LRIG2,GLIS2,XIAP,FGFR1,TET1 |
| GO:0005975⁓carbohydrate metabolic process | 6 | 0.001698244 | AKT2,GCLC,IL6ST,PTEN,UGDH,MLEC,TFAP2A,BACE1 |
| GO:0097191⁓extrinsic apoptotic signaling pathway | 3 | 0.002041738 | PDPK1,CRADD,BLOC1S2 |
GO Gene Ontology, KEGG Kyoto Encyclopedia of Genes and Genomes, DEGs differentially expressed genes, DEmiRs differentially expressed miRNAs
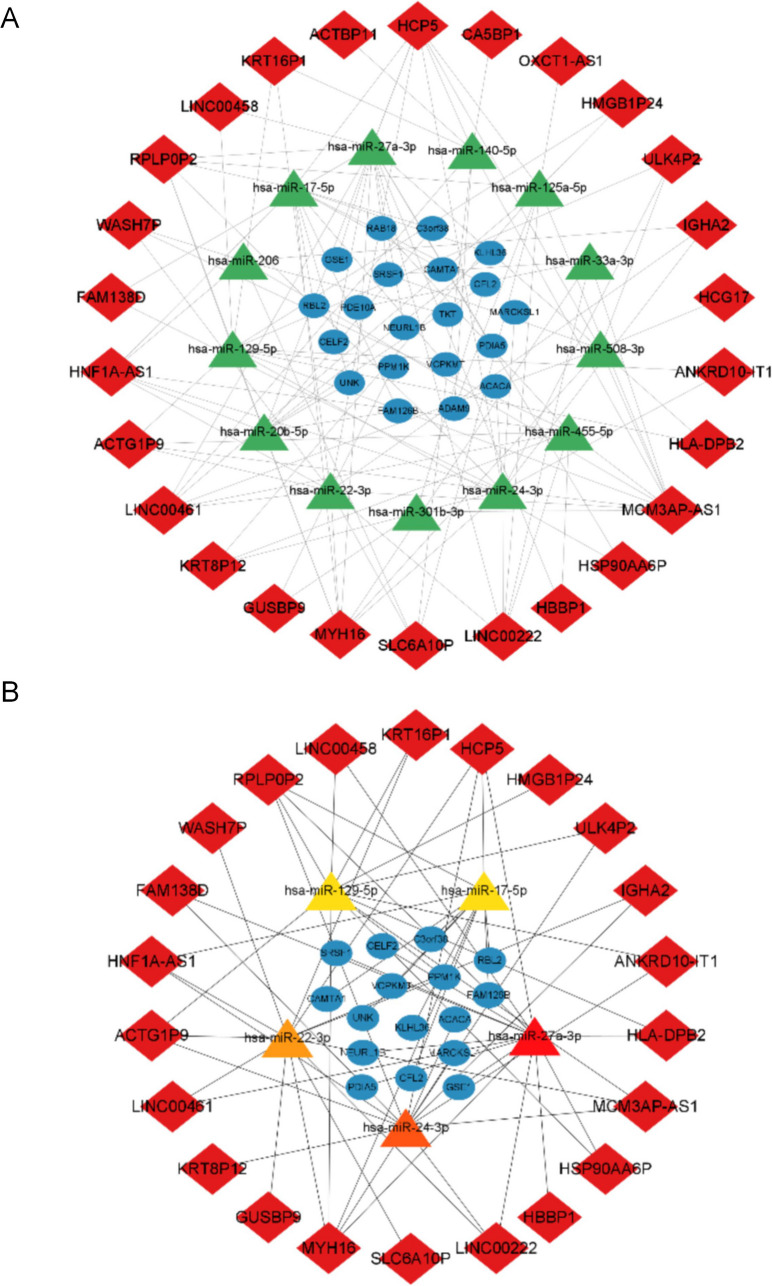
Analysis of the lncRNA-miRNA-mRNA ceRNA network
The interactions between the differentially expressed lncRNAs, miRNAs, and mRNAs served as the foundation for our lncRNA-miRNA-mRNA ceRNA network. The ceRNA network has 123 edges, 27 lncRNA nodes, 13 miRNA nodes, and 20 mRNA nodes, and it may be shown in Fig. 5A. A functional study using Metascape was performed on DEGs in the ceRNA network. The metabolism of ribose phosphate and the synthesis of organophosphate were the BPs that were most dramatically enriched. VEGFA-VEGFR2 signaling pathway was the most prominent pathway enriched in KEGG (Table 3). The top five hub miRNAs were found to be hsa-miR-27a-3p, hsa-miR-24-3p, hsa-miR-22-3p, hsa-miR-129-5p, and hsa-miR-17-5p by using the degree and MCC techniques in cytoHubba. Figure 5B displays the subnetworks of these five hub miRNAs and their first neighbors. In the sub-network, we chose five hub miRNAs for further analysis: hsa-miR-27a-3p, hsa-miR-24-3p, hsa-miR-22-3p, hsa-miR-129-5p, and hsa-miR-17-5p; 22 lncRNAs and 16 mRNAs interacted with these five miRNAs. Similar to this, the most enriched BPs comprised the organophosphate biosynthetic and ribose phosphate metabolic processes (Table 3). The VEGFA-VEGFR2 signaling pathway was significantly enriched in the KEGG pathway (Table 3).
Fig. 5.
The lncRNA-miRNA-mRNA ceRNA network and GO/KEGG analysis of DEGs. (A) The whole lncRNA-miRNA-mRNA ceRNA network; (B) the sub networks of five hub miRNAs with their first neighbors. Diamond-shaped nodes represent lncRNAs; rectangular nodes represent miRNAs; oval-shaped nodes represent mRNAs. LncRNA: long non-coding RNA; miRNA: microRNA; mRNA: messenger RNA; ceRNA network: competitive endogenous RNA network
Table 3.
GO/KEGG analysis of DEGs in lncRNA-miRNA-mRNA ceRNA network
| Term of GO/KEGG analysis | Count | P-value | Genes |
|---|---|---|---|
| GO:0019693⁓ribose phosphate metabolic process | 3 | 0.001949845 | ACACA,TKT,PDE10A |
| WP3888: VEGFA-VEGFR2 signaling pathway | 3 | 0.002884032 | ACACA,TKT,ADAM9 |
| GO:0090407⁓organophosphate biosynthetic process | 3 | 0.005128614 | ACACA,TKT,FAM126B |
GO Gene Ontology, KEGG Kyoto Encyclopedia of Genes and Genomes, DEGs differentially expressed gene, LncRNA long non-coding RNA, miRNA microRNA, mRNA messenger RNA, ceRNA network competitive endogenous RNA network
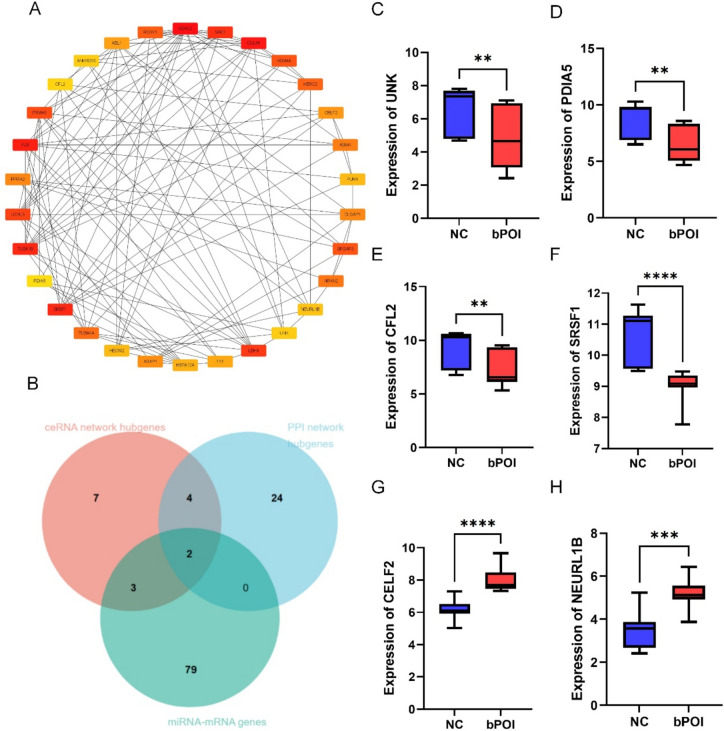
Identification and validation of key genes
The 16 hub genes of the ceRNA network that were discovered utilizing the cytoHubba degree, closeness, and betweenness centrality algorithms (Fig. 5B). When the ceRNA network’s DEGs were uploaded to the STRING website, 116 DEGs used to build a PPI network included 357 interaction connections. In Fig. 6A, the top 30 DEGs of the PPI network, as determined by cytoHubba, were displayed. The Venn diagram of the ceRNA network and PPI network revealed that the six important genes were SRSF1, PDIA5, NEURL1B, UNK, CELF2, and CFL2. Additionally, the Venn diagram of the ceRNA network, PPI network, and miRNA-mRNA network identified two important genes (Fig. 6B). In the established network of hub miRNAs, hsa-miR-17-5p, which regulates SRSF1, PDIA5, NEURL1B, UNK, CELF2, and CFL2, these miRNAs can also influence other miRNAs (Fig. 5B). The GSE135697 datasets revealed the underlying processes for important genes by showing up-regulation of NEURL1B and CELF2 and down-regulation of SRSF1, PDIA5, UNK, and CFL2 (Fig. 6C–H).
Fig. 6.
Difference analysis of key genes. (A) Relationship network diagram of hub genes from PPI network; (B) Venn diagram of key genes (PPI hub genes, ceRNA hub genes, miRNA-mRNA genes); (C) difference analysis box plot of key genes including UNK (C), PDIA5 (D), CFL2 (E), SRSF1 (F), CELF2 (G), and NEURL1B (H). miRNA: microRNA; mRNA: messenger RNA; ceRNA network: competitive endogenous RNA network; PPI: protein-protein interaction; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; NC: negative control; bPOI: biochemical premature ovarian failure. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
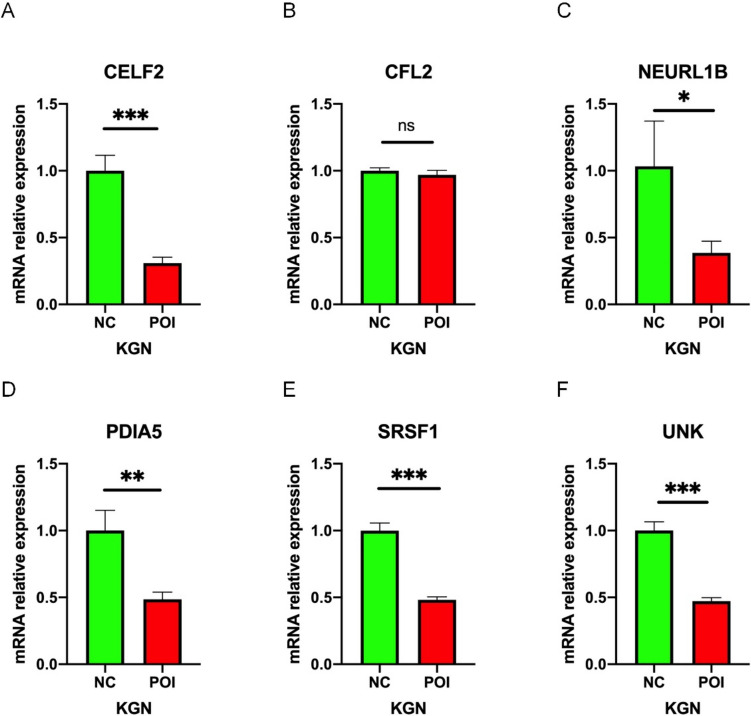
To confirm these results, we measured the expression of six key genes in a vitro model of POI treated with cisplatin. It was found that NEURL1B, CELF2, SRSF1, PDIA5, and UNK were down-regulated, while CFL2 had no significant change (Fig. 7C–H). Of these, the expression levels of SRSF1, PDIA5, and UNK between the POI and control groups were consistent with the results of bioinformatics analysis.
Fig. 7.
Validation of key genes in the ceRNA network. The effect of cisplatin on the mRNA level of CELF2 (A), CFL2 (B), NEURL1B (C), PDIA5 (D), SRSF1 (E), and UNK (F) and KGN cells was cultured in the presence or absence of cisplatin (50 μM) for 24 h. The mRNA levels of CELF2, CFL2, NEURL1B, PDIA5, SRSF1, and UNK were determined by qPCR and normalized to GAPDH levels. ceRNA network: competitive endogenous RNA network; NC: negative control; POI: premature ovarian failure; qPCR: quantitative polymerase chain reaction. The data is expressed as the means±SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
Discussion
We found that the GCs of women with bPOI and healthy women differed in the expression of 131 mRNAs, 191 lncRNAs, and 28 miRNAs in this study. Hub genes among the DEGs were found to include SRSF1, PDIA5, NEURL1B, UNK, CELF2, and CFL2. The top five hub miRNAs were filtered from the PPI network: hsa-miR-27a-3p, hsa-miR-24-3p, hsa-miR-22-3p, hsa-miR-129-5p, and hsa-miR-17-5p. Then, we found in the ceRNA sub-network that hsa-miR-27a-3p may regulate PDIA5 and SRSF1, NEURL1B, and CELF2, while hsa-miR-17-5p may regulate UNK and CFL2 in human bPOI. Furthermore, we examined the expression of these key genes in a vitro model of POI with cisplatin treatment and found that the expression levels of SRSF1, PDIA5, and UNK were consistent with the microarray results.
One of the most prevalent reproductive endocrine diseases, primary ovarian insufficiency (POI), affects 1% of women before the age of 40 [22]. Common causes of POI include genetic factors, autoimmune abnormalities, medically induced injuries, and infections [3]. There is a clear link between apoptosis of granulosa cells and premature ovarian failure, as demonstrated by several studies indicating apoptosis of granulosa cells occurs in conjunction with the etiology of early ovarian failure [23, 24].
Previous studies mainly focused on the influence of exon-encoded protein genes on ovarian function and the related mechanism of POI. The role of non-coding RNA (ncRNA) has progressively come to light with the advancement of microarray technology and high-throughput sequencing [25]. Recent studies have shown that some microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) play an essential role in the occurrence and development of POI [26, 27]. Li et al. found that down-regulation of lncRNA ZNF674-AS1 in GCs of patients with bPOI inhibited the proliferation of GCs [28]. Apoptosis of GCs in POI-induced follicular atresia, ultimately leading to impaired ovarian function.
LncRNAs coordinate gene regulation at several levels, according to recent findings. Dysregulation of lncRNAs may also have an impact on the development and incidence of pathogenic diseases [16]. According to research, lncRNAs play an essential regulatory role in the process of major human diseases such as cancer and neurological diseases [29]. Another non-coding RNA-small molecule, ribonucleic acids (microRNAs) are widely present in various tissues and body fluids of the human body and regulate gene expression at the post-transcriptional level [30]. Numerous studies have demonstrated the presence of various interactions between RNA molecules, including those between lncRNAs and mRNAs, miRNAs and mRNAs, and lncRNAs and lncRNAs. These molecules work together to create a dynamic regulatory network of lncRNAs that serves to balance out competing endogenous RNAs (ceRNAs) [31]. In fact, lncRNAs play a key role in the ceRNA network, controlling mRNA expression by sponging miRNAs, which makes them less efficient in binding to mRNA targets [32]. Moreover, ceRNA crosstalk is widespread in many environments and can be observed under both normal and pathological conditions [33]. Therefore, the lncRNA-miRNA-mRNA ceRNAs network we have created will aid in our understanding of the etiology of POI.
Our results showed that SRSF1, PDIA5, UNK, NEURL1B, CELF2, and CFL2 were identified as hub genes and were validated in KGN cells treated with cisplatin.
SRSF1 (SF2/ASF) is a typical serine/arginine (SR)-rich protein involved in exon skipping, control of alternative, and identification of splice sites at intron-exon junctions, all of which are crucial for cell survival [34]. Wang et al. found that SRSF1 can regulate porcine granulosa cell apoptosis and follicular atresia. Moreover, SRSF1 was down-regulated in granulosa cells of porcine atretic follicles, consistent with our findings [35]. The rest of the five hub genes have not been reported in POI or granulosa cells. The PDI family protein PDIA5, which plays a role in the development of gliomas, is significantly expressed in these tumors. PDIA5 overexpression is linked to immune infiltration in gliomas and might represent a viable therapeutic target for glioma immunotherapy [36]. UNK is also poorly understood and is reported to be associated with the development of gastric cancer [37]. The RBP CELF2 is a member of the CELF protein family and is connected to follicle development as well as certain types of carcinogenesis [38–40]. Human neuralized 2 (NEURL1B), one of the homologs of E3 ubiquitin ligase, is a crucial gene mainly localized in the cytoplasm and intensively expressed during embryogenesis. It is highly expressed in peripheral tissues such as the heart, liver, and testis [41, 42]. Last but not least, CFL2, a member of the actin depolymerizing factor/cofilin protein family that is exclusive to skeletal muscle, is responsible for controlling actin filament dynamics [43]. CFL2 has been shown to have an impact on human myopathies, and through controlling myoblast proliferation, it is essential for myoblast differentiation [44, 45].
To summarize the above, these essential genes have been reported in some other diseases but rarely in POI, suggesting that these genes may possess an unclarified vital role in the pathogenesis of POI. In addition, our experimental results showed that changes in the expression levels of SRSF1, PDIA5, and UNK between the POI and control groups were the same as the analytical results. To some extent, we identified these key genes from the ceRNA network that were altered in the granulosa cells of bPOI patients.
Moreover, we found in the ceRNA sub-network that hsa-miR-27a-3p may regulate PDIA5 and SRSF1, NEURL1B, and CELF2, while hsa-miR-17-5p may regulate UNK and CFL2. Therefore, hsa-miR-27a-3p and hsa-miR-17-5p screened from the PPI network may play an important role in the pathogenesis of POI. However, research on hsa-miR-27a-3p and hsa-miR-129-5p is scant. Dang et al. found that miR-22-3p and miR-24-3p were substantially down-regulated in the plasma of POI patients relative to healthy controls for other hub miRNAs [46]. Additionally, Ding et al. discovered that the miRNA-17-5p in human umbilical cord mesenchymal stem cell exosomes might enhance ovarian function in individuals with premature ovarian failure [47]. By controlling the level of important genes’ expression, these hub miRNAs may also contribute to the development of POI; 22 lncRNAs with differential expression that interact with hub miRNAs in the ceRNA network were also discovered by this investigation. There are not many studies on other lncRNAs, except from HCP5’s role in POI.
It is critical to be aware of some of this study’s shortcomings. Even while our work offers crucial details for understanding how bPOI develops, these particular processes still need to be thoroughly shown. The expression of all important genes was also not totally compatible with the findings of the bioinformatics study since the cell model we used for validation might not be exactly equivalent to the status of granulosa cells in bPOI patients. Therefore, in order to further confirm our findings, we need to identify additional appropriate POI models.
Conclusion
In conclusion, we completed the validation of essential genes and discovered a new lncRNA-miRNA-mRNA network based on the ceRNA mechanism in bPOI. Novel genes and non-coding RNAs that may contribute to the emergence and progression of POI were discovered. Future investigations on the underlying pathogenic processes of POI with the ceRNA network may have a theoretical foundation thanks to our results, which offered new information.
Supplementary information
(DOCX 54 kb)
(XLSX 127 kb)
(XLSX 814 kb)
Acknowledgements
This work was supported by the Department of Obstetrics and Gynecology, Shanghai Tenth People’s Hospital, and Tongji University. The results published here are in whole or partly based on data obtained from the Gene expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/).
Author contribution
BL and LL contributed equally to this work. BL and LL contributed to the idea of the manuscript, completed the experiment, analyzed the data, and wrote this manuscript. BL and LL completed the revision of the manuscript. ZS, CW, JZ, and LW helped interpret the data. ZC and SL conceived the study. All authors read and approved the manuscript. BL and LL contributed equally to this work.
Funding
This project was supported by the National Nature Science Foundation of China (NO. 31900522).
Data availability
The datasets generated and analyzed during the current study are available in the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database (Accession Number: GSE135697, GSE100238).
Declarations
Ethics approval
Not applicable.
Consent to participate
Written informed consent was obtained from all participants included in the study.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Biting Liu and Li Liu contributed equally to this work.
Contributor Information
Shupeng Liu, Email: lshup@tongji.edu.cn.
Zhongping Cheng, Email: mdcheng18@tongji.edu.cn.
References
- 1.Jiao X, Ke H, Qin Y, Chen ZJ. Molecular genetics of premature ovarian insufficiency. Trends Endocrinol Metab. 2018;29:795–807. doi: 10.1016/j.tem.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Lew R. Natural history of ovarian function including assessment of ovarian reserve and premature ovarian failure. Best Pract Res Clin Obstet Gynaecol. 2019;55:2–13. doi: 10.1016/j.bpobgyn.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Huhtaniemi I, Hovatta O, La Marca A, Livera G, Monniaux D, Persani L, et al. Advances in the molecular pathophysiology, genetics, and treatment of primary ovarian insufficiency. Trends Endocrinol Metab. 2018;29:400–419. doi: 10.1016/j.tem.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 4.Tu J, Chen Y, Li Z, Yang H, Chen H, Yu Z. Long non-coding RNAs in ovarian granulosa cells. J Ovarian Res. 2020;13:63. doi: 10.1186/s13048-020-00663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pankiewicz K, Laudański P, Issat T. The Role of Noncoding RNA in the Pathophysiology and Treatment of Premature Ovarian Insufficiency. Int J Mol Sci. 2021;22(17):9336. 10.3390/ijms22179336 [DOI] [PMC free article] [PubMed]
- 6.Veitia RA. Primary ovarian insufficiency, meiosis and DNA repair. Biomed J. 2020;43:115–123. doi: 10.1016/j.bj.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu P, Zhang X, Hu J, Cui L, Zhao S, Jiao X, et al. Dysregulated cytokine profile associated with biochemical premature ovarian insufficiency. Am J Reprod Immunol. 2020;84:e13292. doi: 10.1111/aji.13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Zhang X, Dang Y, Li D, Lu G, Chan WY, et al. Long noncoding RNA HCP5 participates in premature ovarian insufficiency by transcriptionally regulating MSH5 and DNA damage repair via YB1. Nucleic Acids Res. 2020;48:4480–4491. doi: 10.1093/nar/gkaa127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Dang Y, Liu R, Zhao S, Ma J, Qin Y. MicroRNA-127-5p impairs function of granulosa cells via HMGB2 gene in premature ovarian insufficiency. J Cell Physiol. 2020;235:8826–8838. doi: 10.1002/jcp.29725. [DOI] [PubMed] [Google Scholar]
- 10.Dahariya S, Paddibhatla I, Kumar S, Raghuwanshi S, Pallepati A, Gutti RK. Long non-coding RNA: classification, biogenesis and functions in blood cells. Mol Immunol. 2019;112:82–92. doi: 10.1016/j.molimm.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018;109:2093–2100. doi: 10.1111/cas.13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu H, Jia Q, Xi J, Zhou B, Li Z. Integrated analysis of lncRNA, miRNA and mRNA reveals novel insights into the fertility regulation of large white sows. BMC Genomics. 2020;21:636. doi: 10.1186/s12864-020-07055-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren GL, Zhu J, Li J, Meng XM. Noncoding RNAs in acute kidney injury. J Cell Physiol. 2019;234:2266–2276. doi: 10.1002/jcp.27203. [DOI] [PubMed] [Google Scholar]
- 15.Niu ZS, Wang WH, Dong XN, Tian LM. Role of long noncoding RNA-mediated competing endogenous RNA regulatory network in hepatocellular carcinoma. World J Gastroenterol. 2020;26:4240–4260. doi: 10.3748/wjg.v26.i29.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panni S, Lovering RC, Porras P, Orchard S. Non-coding RNA regulatory networks. Biochim Biophys Acta Gene Regul Mech. 2020;1863:194417. doi: 10.1016/j.bbagrm.2019.194417. [DOI] [PubMed] [Google Scholar]
- 17.Caponnetto A, Battaglia R, Ferrara C, Vento ME, Borzì P, Paradiso M, et al. Down-regulation of long non-coding RNAs in reproductive aging and analysis of the lncRNA-miRNA-mRNA networks in human cumulus cells. J Assist Reprod Genet. 2022;39:919–931. doi: 10.1007/s10815-022-02446-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H, Cheng S, Xiong W, Tan X. The lncRNA-miRNA-mRNA ceRNA network in mural granulosa cells of patients with polycystic ovary syndrome: an analysis of Gene Expression Omnibus data. Ann Transl Med. 2021;9:1156. doi: 10.21037/atm-21-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishi Y, Yanase T, Mu Y, Oba K, Ichino I, Saito M, et al. Establishment and characterization of a steroidogenic human granulosa-like tumor cell line, KGN, that expresses functional follicle-stimulating hormone receptor. Endocrinology. 2001;142:437–445. doi: 10.1210/endo.142.1.7862. [DOI] [PubMed] [Google Scholar]
- 21.Wu Y, Ma C, Zhao H, Zhou Y, Chen Z, Wang L. Alleviation of endoplasmic reticulum stress protects against cisplatin-induced ovarian damage. Reprod Biol Endocrinol. 2018;16:85. doi: 10.1186/s12958-018-0404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Webber L, Davies M, Anderson R, Bartlett J, Braat D, Cartwright B, et al. ESHRE guideline: management of women with premature ovarian insufficiency. Hum Reprod. 2016;31:926–937. doi: 10.1093/humrep/dew027. [DOI] [PubMed] [Google Scholar]
- 23.Laven JS. Primary ovarian insufficiency. Semin Reprod Med. 2016;34:230–234. doi: 10.1055/s-0036-1585402. [DOI] [PubMed] [Google Scholar]
- 24.Dang Y, Wang X, Hao Y, Zhang X, Zhao S, Ma J, et al. MicroRNA-379-5p is associate with biochemical premature ovarian insufficiency through PARP1 and XRCC6. Cell Death Dis. 2018;9:106. doi: 10.1038/s41419-017-0163-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vishnoi A, Rani S. MiRNA biogenesis and regulation of diseases: an overview. Methods Mol Biol. 2017;1509:1–10. doi: 10.1007/978-1-4939-6524-3_1. [DOI] [PubMed] [Google Scholar]
- 26.Bouckenheimer J, Fauque P, Lecellier CH, Bruno C, Commes T, Lemaître JM, et al. Differential long non-coding RNA expression profiles in human oocytes and cumulus cells. Sci Rep. 2018;8:2202. doi: 10.1038/s41598-018-20727-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo Y, Sun J, Lai D. Role of microRNAs in premature ovarian insufficiency. Reprod Biol Endocrinol. 2017;15:38. doi: 10.1186/s12958-017-0256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li D, Wang X, Li G, Dang Y, Zhao S, Qin Y. LncRNA ZNF674-AS1 regulates granulosa cell glycolysis and proliferation by interacting with ALDOA. Cell Death Discov. 2021;7:107. doi: 10.1038/s41420-021-00493-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malmuthuge N, Guan LL. Noncoding RNAs: regulatory molecules of host-microbiome crosstalk. Trends Microbiol. 2021;29:713–724. doi: 10.1016/j.tim.2020.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Rinn JL, Chang HY. Long noncoding RNAs: molecular modalities to organismal functions. Annu Rev Biochem. 2020;89:283–308. doi: 10.1146/annurev-biochem-062917-012708. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Lu B. The Roles of ceRNAs-mediated autophagy in cancer chemoresistance and metastasis. Cancers (Basel). 2020;12(10):2926. 10.3390/cancers12102926 [DOI] [PMC free article] [PubMed]
- 32.Liao J, Wang J, Liu Y, Li J, Duan L. Transcriptome sequencing of lncRNA, miRNA, mRNA and interaction network constructing in coronary heart disease. BMC Med Genomics. 2019;12:124. doi: 10.1186/s12920-019-0570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi X, Lin Y, Chen J, Shen B. Decoding competing endogenous RNA networks for cancer biomarker discovery. Brief Bioinform. 2020;21:441–457. doi: 10.1093/bib/bbz006. [DOI] [PubMed] [Google Scholar]
- 34.Liu H, Gong Z, Li K, Zhang Q, Xu Z, Xu Y. SRPK1/2 and PP1α exert opposite functions by modulating SRSF1-guided MKNK2 alternative splicing in colon adenocarcinoma. J Exp Clin Cancer Res. 2021;40:75. doi: 10.1186/s13046-021-01877-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, Zhang Y, Zhang J, Du X, Li Q, Pan Z. circSLC41A1 resists porcine granulosa cell apoptosis and follicular atresia by promoting SRSF1 through miR-9820-5p sponging. Int J Mol Sci. 2022;23(3):1509. 10.3390/ijms23031509 [DOI] [PMC free article] [PubMed]
- 36.Zhang H, He J, Dai Z, Wang Z, Liang X, He F, et al. PDIA5 is correlated with immune infiltration and predicts poor prognosis in gliomas. Front Immunol. 2021;12:628966. doi: 10.3389/fimmu.2021.628966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casali PG, Blay JY, Abecassis N, Bajpai J, Bauer S, Biagini R, et al. Gastrointestinal stromal tumours: ESMO-EURACAN-GENTURIS Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2022;33:20–33. doi: 10.1016/j.annonc.2021.09.005. [DOI] [PubMed] [Google Scholar]
- 38.Yeung YT, Fan S, Lu B, Yin S, Yang S, Nie W, et al. CELF2 suppresses non-small cell lung carcinoma growth by inhibiting the PREX2-PTEN interaction. Carcinogenesis. 2020;41:377–389. doi: 10.1093/carcin/bgz113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie SC, Zhang JQ, Jiang XL, Hua YY, Xie SW, Qin YA, et al. LncRNA CRNDE facilitates epigenetic suppression of CELF2 and LATS2 to promote proliferation, migration and chemoresistance in hepatocellular carcinoma. Cell Death Dis. 2020;11:676. doi: 10.1038/s41419-020-02853-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piqué L, Martinez de Paz A, Piñeyro D, Martínez-Cardús A, Castro de Moura M, Llinàs-Arias P, et al. Epigenetic inactivation of the splicing RNA-binding protein CELF2 in human breast cancer. Oncogene. 2019;38:7106–7112. doi: 10.1038/s41388-019-0936-x. [DOI] [PubMed] [Google Scholar]
- 41.Liu J, Liu Z, Zhang X, Yan Y, Shao S, Yao D, et al. Aberrant methylation and microRNA-target regulation are associated with downregulated NEURL1B: a diagnostic and prognostic target in colon cancer. Cancer Cell Int. 2020;20:342. doi: 10.1186/s12935-020-01379-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vollrath B, Pudney J, Asa S, Leder P, Fitzgerald K. Isolation of a murine homologue of the Drosophila neuralized gene, a gene required for axonemal integrity in spermatozoa and terminal maturation of the mammary gland. Mol Cell Biol. 2001;21:7481–7494. doi: 10.1128/MCB.21.21.7481-7494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen MT, Min KH, Lee W. Palmitic acid-induced miR-429-3p impairs myoblast differentiation by downregulating CFL2. Int J Mol Sci. 2021;22(20):10972. 10.3390/ijms222010972 [DOI] [PMC free article] [PubMed]
- 44.Nguyen MT, Lee W. Role of MiR-325-3p in the regulation of CFL2 and myogenic differentiation of C2C12 myoblasts. Cells. 2021;10(10):2725. 10.3390/cells10102725 [DOI] [PMC free article] [PubMed]
- 45.Fattori F, Fiorillo C, Rodolico C, Tasca G, Verardo M, Bellacchio E, et al. Expanding the histopathological spectrum of CFL2-related myopathies. Clin Genet. 2018;93:1234–1239. doi: 10.1111/cge.13240. [DOI] [PubMed] [Google Scholar]
- 46.Dang Y, Zhao S, Qin Y, Han T, Li W, Chen ZJ. MicroRNA-22-3p is down-regulated in the plasma of Han Chinese patients with premature ovarian failure. Fertil Steril. 2015;103:802–807.e801. doi: 10.1016/j.fertnstert.2014.12.106. [DOI] [PubMed] [Google Scholar]
- 47.Ding C, Zhu L, Shen H, Lu J, Zou Q, Huang C, et al. Exosomal miRNA-17-5p derived from human umbilical cord mesenchymal stem cells improves ovarian function in premature ovarian insufficiency by regulating SIRT7. Stem Cells. 2020;38:1137–1148. doi: 10.1002/stem.3204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 54 kb)
(XLSX 127 kb)
(XLSX 814 kb)
Data Availability Statement
The datasets generated and analyzed during the current study are available in the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database (Accession Number: GSE135697, GSE100238).