Graphical abstract
Keywords: Pixian douban, Umami peptides, Feature-based molecular networking (FBMN), Molecular docking
Highlights
-
•
Data-driven strategy for rapid identification and molecular docking of umami peptides.
-
•
Identification of 18 novel peptides using molecular networking.
-
•
QIVK with a high umami threshold of 0.3215 ± 0.1607 mmol/L.
-
•
QIVK exhibits stronger binding to T1R3 than T1R1 in docking.
Abstract
This study presents an innovative cloud-based approach, using Pixian Douban, a well-known Chinese fermented seasoning, as a case study, to improve the identification of umami peptides and explore their interactions with the T1R1/T1R3 receptor. A feature-based molecular networking method was utilized to rapidly identify a total of eighteen peptides, including seven previously unrecorded ones. Notably, the umami threshold of QIVK in an aqueous solution was determined to be 0.3215 mmol/L, surpassing the majority of peptides reported in the past three years. Molecular docking analysis further revealed the strong binding of QIVK to T1R3 receptor residues through hydrogen bonds, as well as interactions via salt bridges and electrostatic attractions. As a result, this research significantly contributes to the efficient screening of umami peptides and the elucidation of the molecular basis of umami sensory perception in complex food systems.
1. Introduction
The term umami, derived from the Japanese language, refers to a “delicious savory taste”. It was officially recognized as the fifth primary taste in 2002 (Nakamura, 2011), and has since been identified in various food sources, including fish, meat, and numerous fermented products (Anand Singh et al., 2023). Recently, umami peptides have gained significant attention due to their multifaceted properties, such as enhancing taste, reducing salt consumption, and offering nutritional benefits (L. Zhang et al., 2023a). The perception of umami taste is initiated by specific chemical structures that activate umami receptor proteins located within taste buds. Among these receptors, the T1R1/T1R3 heterodimer is widely acknowledged as the primary receptor responsible for umami taste perception (Wang et al., 2020).
Pixian Douban (PXDB) is a renowned seasoning widely used in Sichuan-style cuisine in China, particularly in dishes such as hotpot. It is produced through the fermentation of broad beans and chili peppers, during which the proteins in the raw ingredients undergo various peptide transformations under the action of microbial enzymes, contributing to its unique flavor profile (Li, Wang, et al., 2023). However, despite its popularity, the structure and taste mechanism of the umami peptides in PXDB remain largely unknown. This lack of understanding is partly attributed to the significant limitations associated with the conventional approach to identifying umami peptides. The method heavily relies on the capacity of mass spectrometry databases and can only detect peptides included in the database, thereby restricting the range of identified peptides. Furthermore, conducting direct sensory evaluations for numerous candidate peptides raises potential food safety concerns. Consequently, the need to establish a high-throughput approach for efficiently identifying and assessing potential umami peptides is urgent.
Cloud-based computational approaches play a crucial role in advancing scientific research in food chemistry (Li, Mei, et al., 2023). A notable instance in this field is the utilization of Feature-Based Molecular Networking (FBMN), an analytical tool seamlessly integrated into the Global Natural Products Social Molecular Networking (GNPS) platform(Aron et al., 2020; Li, Wu, et al., 2023). By harnessing the computational prowess of cloud platforms, FBMN efficiently consolidates essential data, encompassing liquid chromatography retention time, MS1, and MS2 information, to create molecular families, enabling the elucidation of chemical structures for previously unidentified components (Nothias et al., 2020). Moreover, Umami-MRNN tool introduces a novel machine learning model specifically designed for the rapid and accurate screening of umami peptides (Qi et al., 2023). Moreover, predicting the interactions between umami peptides and T1R1/T1R3 receptors aids in comprehending why certain substances are perceived as having an umami taste. CB-Dock2, an advanced network-based protein–ligand blind docking platform, has been intricately crafted to significantly improve the precision of identifying binding sites and predicting binding poses. In previous test datasets, CB-Dock2 achieved an impressive 85.9 % prediction success rate, marking it as the top performer among similar methods (Y. Liu et al., 2022).
This study presents an innovative approach for efficient umami peptide screening, using PXDB as an illustrative example. We initiated the process by employing liquid chromatography-mass spectrometry (UHPLC-Q-TOFMS) to detect peptides in PXDB. Specifically, in conjunction with FBMN, we explored previously undiscovered peptides within PXDB. These identified peptides then underwent a high-throughput umami threshold assessment using the umami-MRNN tool. Additionally, molecular docking studies were conducted using the CB-Dock2 tool to gain insights into ligand–protein interactions. This cloud-based computational strategy offers a more comprehensive and efficient method for screening umami peptides in PXDB, with potential implications for advancing our understanding of umami taste.
2. Material and methods
2.1. Materials and reagents
The PXDB samples were sourced from Sichuan Pixian Douban Co., Ltd., located in Chengdu, China. Two batches of PXDB with fermentation periods of 1 year and 2 years were employed. Methanol and acetonitrile of chromatographic grade (purity ≥ 99.00 %) were obtained from Sigma Aldrich (St. Louis, MO, USA). Gln-Ile-Val-Lys (5 mg × 5), with a purity of ≥ 95.00 %, was synthesized by Dan Gang Biological Technology Co., Ltd. (Wuhan, Hubei, China). Oasis MCX (30 mg, 3 cc) solid-phase extraction (SPE) cartridges were provided by Waters Corporation (Milford, MA, USA). All reagents utilized in this study, unless otherwise specified, were of analytical grade (purity of ≥ 98.00 %) from Shanghai Yien Chemical Technology Co., Ltd. (Shanghai, China).
2.2. Sample preparation
The sample extraction method followed a previously established procedure (Li, Mei, et al., 2023). Initially, 0.5 g of PXDB samples were accurately weighed and mixed with 10 mL of a 40 % methanol solution. Subsequently, the samples were homogenized using an automatic homogenizer (IKA, Staufen, Switzerland) at 10,000 rpm for 3 min. Ultrasonic extraction was conducted at 25 °C for 30 min using equipment from NingBo Scientz Biotechnology (Ningbo, Zhejiang, China). Following ultrasonic extraction, the PXDB extracts underwent centrifugation (Shuke Instrument, Chengdu, China) at 4,000 g for 15 min. The upper extract was then collected and filtered through a 0.22 μm nylon membrane for further purification.
Sample purification was achieved using an MCX solid-phase extraction (SPE) cartridge (3 cc, 60 mg, 60 μm, Waters, Milford, MA, USA), following a previously protocol (Chen et al., 2022). In brief, 2.0 mL of the PXDB sample extracts were loaded onto the cartridges and subjected to sequential washing with 1.5 mL of a 2 % formic acid solution, followed by 1.5 mL of MeOH. The desired peptides were eluted using 2.0 mL of a 2 % ammonium hydroxide solution in MeOH, followed by nitrogen drying at room temperature. The resulting dry components were dissolved in 200 μL of a 50 % methanol solution and then transferred into a vial for subsequent UHPLC-Q-TOF analysis.
2.3. UHPLC-Q-TOF conditions
The UHPLC analysis was conducted utilizing an LC30 system (Shimadzu Co., Tokyo, Japan) equipped with an HSS T3 C18 (100 mm × 2.1 mm × 1.8 μm) column sourced from Waters Co. (Milford, MA, USA). The mobile phases consisted of solvent A (0.1 % formic acid in water) and solvent B (acetonitrile). The elution program was optimized based on a previous protocol (Chen et al., 2021): 0–2 min, 5.0 % B; 2–5 min, 5 %-10 % B; 5–14 min, 10 %-40 % B; 14–24 min, 40 %-100 % B; 24–26.5 min, 100 %-100 % B; 26.5–27 min, 100 %-5% B. A re-equilibration step was carried out for three minutes at 5 % B. The system operated at a flow rate of 0.3 mL/min, with an injection volume of 5 µL and a column temperature of 40 °C.
Mass spectrometry (SCIEX Co., Framingham, MA, USA) was performed in positive ion mode, with a mass range of 50–1000 m/z. The following settings were used for data collection: declustering potential at 50 V, collision energy at 10 V, and a temperature of 450 °C, curtain gas 35 psi, nebulizer gas (nitrogen) pressure at 2 bar, capillary voltage at 4,200 V (L. Zhang et al., 2023b). The MS/MS data collection was carried out in IDA mode, where the top 10 ions of each spectrum were fragmented using collision-induced dissociative energy of 35 ± 10 eV (Li, Zhou, et al., 2023).
2.4. FBMN analysis
The raw data obtained from UHPLC-Q-TOF analysis were processed using MSDIAL 4.9 software to convert them into formats compatible with FBMN. The converted files were subsequently uploaded to the GNPS webserver. For the Molecular Network analysis, the parameterized configuration was set to the default values of the system. In FBMN, feature finding processes MS/MS data by accessing their respective chromatographic retention times, resulting in a yield of each chromatographic peak abundance. Furthermore, a single representative MS/MS spectrum is chosen per feature (Oak & Jha, 2019). The resulting molecular networks can be accessed at (https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task = e72ab67f0687432e93d4cbb4e34ebdb6). The analysis results were visualized using Cytoscape 3.9.1 (Ramabulana et al., 2021).
2.5. Umami activity prediction of the identified peptides
The Umami-MRNN web-based tool, which integrates the Multi-layer Perceptron and Recurrent Neural Network models, was employed to predict the umami activity of the identified peptides (Qi et al., 2023).
2.6. Electronic tongue and sensory evaluation
The electronic tongue (Baosheng Co., Ltd., Shanghai, China) used in this study consists of a sensor array with six interaction-sensitive electrodes: platinum (1), gold (2), palladium (3), titanium (4), tungsten (5), and silver (6). In the sensor array, each individual interaction-sensing electrode functions akin to taste buds on the tongue. Subsequently, a specialized algorithm on the workstation is employed to assess the taste components of food. The testing protocol was conducted following the previously established method (Dang et al., 2019). The peptide samples are formulated at a concentration of 0.2058 mmol/L and subsequently transferred into a designated beaker for electronic tongue analysis, employing a sampling time of 180 s. The detection process is repeated five times for each sample.
The sensory evaluation in this study was conducted following the method described by Shan et al (Shan et al., 2022). The panel comprised 5 males and 5 females, aged between 22 and 25 years, selected from Xihua University. Before the assessment, the panelists underwent training in sensory evaluation, adhering to the International Standard Method for Training and Monitoring of Assessors (ISO-8586–1, 2012). The sensory experiments took place at a controlled temperature of 24 ± 1 °C and a humidity of 60 %. To determine the umami threshold of QIVK, we initially prepared peptide solutions at a concentration of 10.2881 mmol/L. These solutions were subsequently diluted into a range of different concentrations. The sensory evaluation experiment employed the three-point discrimination test, which entails the simultaneous presentation of three coded samples. Among these samples, two are identical, and the third differs. Assessors are required to identify the distinct sample in each trial (S. Zhao et al., 2024).
2.7. Molecular docking of T1R1/T1R3
This receptor's binding region forms a “ Venus Flytrap Domain (VFD)” structure, which serves as the primary site for ligand-receptor interactions (Nuemket et al., 2017). The ligand-binding region of the umami receptor was analyzed using the metabotropic glutamate receptor as a template with PDB ID 1EWK (Dang et al., 2019). The plausibility of the model was assessed using the Laplace diagram. For protein–ligand blind docking, the CB-Dock2 server was utilized. The protein file (.pdb) and the structure file of the homology model (.mol2) were uploaded to the CB-Dock server.
The evaluation of docking energy is conducted based on the work of Chen et al.(Chen et al., 2021). Water molecules were removed, and hydrogen atoms were added. The active center coordinates for T1R1 were x = 32.328758, y = -3.792158, and z = 33.820896, while for T1R3, the coordinates were x = 17.070000, y = 1.132591, and z = 20.321409. The active cavity had a radius of 10 Å.
2.8. Statistical analysis
All analytical experiments were performed with five replicates in duplicate (n = 5), and the results were presented as the mean ± standard deviation (SD) using Origin software (version 2021, Northampton, USA). The statistical analysis in our sensory study initially employs ANOVA. Upon detecting significance, we further analyze using Tukey's test method via SPSS software (version 25.0; SPSS Inc., Chicago, IL, USA).
3. Results and discussion
3.1. Construction of peptide molecular families through FBMN
The composition of Pixian Douban (PXDB), which includes lipids, polyphenols, and various other components (Yang et al., 2021), presents a significant challenge for peptide identification in these samples. To address this challenge, MCX solid-phase columns were utilized to enrich peptides and reduce impurities, as previously described (Cheng et al., 2023). Furthermore, FBMN employs advanced algorithms to swiftly filter and process large datasets obtained from multidimensional high-resolution mass spectrometry. This rapid data filtering is essential for isolating peptide-related signals amidst the complex background (Mannochio-Russo et al., 2022). This integration enhances the precision and depth of our peptide identification efforts.
Consequently, molecular networks were established, comprising a total of 22,571 mass spectral nodes. Fig. 1 visually represents the distribution of these nodes across various PXDB samples, presenting the data in a pie chart format. Each node within this network corresponds to a unique feature molecule, and these nodes were subsequently categorized into 885 molecular families (nodes ≥ 2). The larger circles represented the small peptides identified, and the green and purple segments in the chart specifically denote samples that underwent one-year and two-year fermentation, respectively. Through a search using the GNPS database, we have successfully identified 11 peptides (Table S2), each belonging to one of five distinct peptide molecular families: MN1 (Ile-Ile-Arg, Ile-Arg, Phe-Arg); MN2 (Pro-Phe, His-Pro, Ile-His, Leu-His); MN3 (Ile-Ile-Arg); MN4 (Phe-Phe, Phe-Leu); MN5 (Tyr-Phe).
Fig. 1.
Five Peptide molecular families obtained from the feature-based molecular networking workflow (the large circles represent the small peptides identified).
Peptides with similar MS2 fragmentation patterns tend to form peptide molecular families. Consequently, we unveiled the chemical structural relationships within the MN1-MN5 peptide families by manually analyzing the fragmentation patterns of the 11 aforementioned peptides (Fig. 2). This forms the basis for the rapid inference of peptides that are not present in the GNPS database.
Fig. 2.
The chemical structural relationships within the MN1-MN5 peptide families were determined through manual analysis of the fragmentation patterns.
Within the MN1 family (Fig. 2A), three peptides with m/z values of 401.1286, 288.2018, and 322.1865 were identified as Ile-Ile-Arg, Ile-Arg, and Phe-Arg, respectively. As shown in the figure, all three peptides exhibited a shared m/z loss of 175, which corresponds to the removal of the arginine residue. Additionally, m/z 113.0829 was attributed to an isoleucine residue, while m/z 147.0662 corresponded to a phenylalanine residue.
In the MN2 molecular family (Fig. 2B), three peptides were found to possess m/z 156.0771, m/z 156.0773 and m/z 156.0740, indicating the presence of a histidine (His) residue. Notably, within the peptide network of MN2, a pair of isomeric peptides was identified: m/z 269.1600 (Leu-His) and m/z 269.1597 (Ile-His).
In the MN3 molecular family, a tripeptide Lys-Ile-Ile was identified, with m/z 147.1134 assigned to a lysine residue and m/z 113.0839 corresponding to an isoleucine residue (Fig. 2C). Regarding the MN4 and MN5 molecular families (Fig. 2DE), their networks mainly consist of peptides that contain m/z 166.0863, representing a phenylalanine residue.
The integration of FBMN and UHPLC-Q-TOF represents an advanced and robust methodology for conducting a comprehensive analysis of peptides within PXDB. FBMN sorts and organizes peptides with structural or functional similarities by leveraging the similarities and correlations identified within the mass spectrometry data. This organization enables the creation of networks or clusters, providing an intuitive and informative platform by visually representing complex data sets. For instance, the application of FBMN resulted in the rapid identification and characterization of twenty-five novel peptides derived from cultures of three distinct bifidobacterial strains (Chen et al., 2022), demonstrating the potency of this approach in swiftly identifying structurally and functionally related peptides within complex biological samples.
3.2. Inference of peptides not present in the database
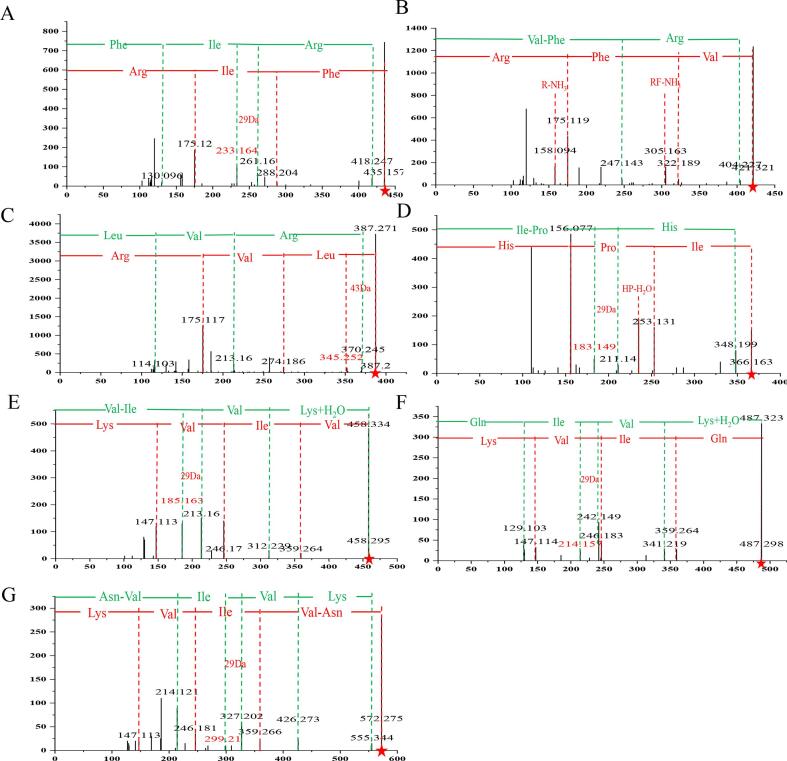
The principle of FBMN revolves around clustering molecules with comparable MS2 fragmentation patterns into molecular families (Nothias et al., 2020). Applying this principle, peptides with known structures can act as “seeds” to infer the structures of unknown peptides within the same molecular family, utilizing the fragmentation patterns illustrated in Fig. 2. As a result, we have successfully derived seven peptides that have not been previously reported (Fig. 3).
Fig. 3.
Inference of Peptides Not Present in the Database.
In MN1, the characteristic feature is the presence of an arginine residue. Collision-induced dissociation (CID) of peptide fragments generates b and y series ions, which exhibit complementary fragmentation patterns, facilitating accurate peptide identification (Jiang et al., 2022). Isomeric peptides containing Leu/Ile residues can be differentiated based on the N-terminal Leu or Ile and the characteristic fragmentation patterns resulting in the loss of C3H7 (-43 Da) or C2H5 (-29 Da) fragments (Samgina et al., 2023, Xiao et al., 2016).
At m/z 261.1600, an ion at m/z 233.1642 was observed due to the neutral loss of C2H5 (-29 Da), indicating the presence of an Ile residue. Consequently, peptide A was identified as Phe-Ile-Arg (Fig. 3A). Similarly, peptide C at m/z 387.2649 was provisionally identified as Leu/Ile-Val-Arg. The presence of m/z 345.2521 was attributed to the loss of C3H7 (-43 Da) from the precursor ion, suggesting the possibility of peptide C being Leu-Val-Arg. Peptide B at m/z 421.2542 was identified as Val-Phe-Arg (Fig. 3B).
In MN2, peptide D (m/z 366.2138) was identified as Ile-Pro-His based on the analysis shown in Fig. 3D. The y-series peptide ions provided insights into the peptide composition, with y1 (m/z 155.0772) corresponding to the His residue, y2 (m/z 97.0542) representing Proline, and y3 (m/z 113.0824) suggesting the presence of Leu/Ile. In the b-series peptide ions, an examination of the N-terminal end of Leu/Ile-Pro revealed a neutral loss of C2H5 (-29 Da), resulting in the formation of the fragment ion at m/z 183.1492, which confirmed the presence of an Ile residue.
In the MN3 peptide molecular family, peptides E (Val-Ile-Val-Lys), F (Gln-Ile-Val-Lys), and G (Asn-Val-Ile-Val-Lys) exhibited a common fragment at m/z 359 (Leu/Ile-Val-Lys). Further analysis of the b-series ions for these peptides revealed additional fragments resulting from the neutral loss of C2H5 (-29 Da) at m/z 185.1634, m/z 214.1573, and m/z 299.2100, indicating the presence of an Ile residue. Peptide ions at m/z 487.3224 and m/z 572.3792 both contained the fragment ion m/z 359 (-Ile-Val-Lys), supporting their identification, as illustrated in Fig. 3F and Fig. 3G, respectively.
In recent years, umami peptides have become a focal point in umami studies due to their enriched nutritional value and flavor-enhancing properties. However, current screening methods for these peptides heavily rely on database searches (Qi et al., 2022), resulting in a significant oversight of peptides that are not included in these databases. This study presents an innovative approach by proposing a new strategy for the rapid screening of umami peptides. Utilizing this pioneering method, researchers successfully identified seven peptides that were previously undocumented in existing databases. Of particular note, the study uncovered six novel peptides associated with PXDB.
3.3. Prediction of umami threshold of peptides
Traditional methods for identifying umami peptides often involve time-consuming and labor-intensive procedures. To address this challenge, we employed Umami-MRNN (Qi et al., 2023), a deep machine learning-based tool, to evaluate candidate umami peptides. The assessment of the 18 identified peptides, as shown in Table 1, led to the successful selection of six novel peptides with umami properties. These peptides, ranked in ascending order of umami threshold, include Gln-Ile-Val-Lys, Leu-His, Ile-Pro-His, Val-Ile-Val-Lys, Ile-His, and Asn-Val-Ile-Val-Lys. Notably, Gln-Ile-Val-Lys (QIVK) exhibited the most pronounced umami taste with a threshold of 21.38 mmol/L.
Table 1.
Identification Data for Pixian Douban Peptides and Umami Threshold Prediction with Umami-MRNN.
| No | Peptide | m/z (Da) | MS2 main fragments 1 | Sample 2 | Similarity 3 | Confidence level 4 | Umami Threshold 5 |
|---|---|---|---|---|---|---|---|
| 1 | His-Pro | 235.1171 | 154.0740/235.1171 | 1 | 0.82 | 2 | non-umami |
| 2 | Pro-Phe | 245.1284 | 115.0550/245.1284 | 2 | 0.82 | 2 | non-umami |
| 3 | Ile-His | 269.1597 | 156.077/269.1597 | 1, 2 | 0.90 | 2 | 35.14 mmol/L |
| 4 | Leu-His | 269.1600 | 156.077/269.1600 | 2 | 0.79 | 2 | 25.48 mmol/L |
| 5 | Phe-Leu | 279.1698 | 132.104/279.1698 | 1 | 0.72 | 2 | non-umami |
| 6 | Ile-Arg | 288.2018 | 175.1190/288.2020 | 2 | 0.75 | 2 | non-umami |
| 7 | Phe-Arg | 322.1865 | 175.1180/322.1870 | 1 | 0.77 | 2 | non-umami |
| 8 | Tyr-Phe | 329.1500 | 166.0860/329.1500 | 1, 2 | 0.79 | 2 | non-umami |
| 9 | Phe-Phe | 331.1639 | 166.0861/331.1639 | 2 | 0.96 | 2 | non-umami |
| 10 | Ile-Pro-His | 366.2138 | b:211.1400/348.1992 y:156.0771/253.1312/366.1634 | 1 | – | 2 | 27.96 mmol/L |
| 11 | Ile-Ile-Lys | 373.2820 | 147.113/260.197/373.2820 | 1, 2 | 0.73 | 2 | non-umami |
| 12 | Leu-Val-Arg | 387.2649 | b:114.1032/213.1600/370.2453 y:175.1171/247.1862/387.2000 | 2 | – | 2 | non-umami |
| 13 | Ile-Ile-Arg | 401.1286 | 175.1210/288.206/401.1286 | 2 | 0.74 | 2 | non-umami |
| 14 | Val-Phe-Arg | 421.2542 | b:247.1432/404.2274 y:175.1190/322.1892/421.3215 | 2 | – | 2 | non-umami |
| 15 | Phe-Ile-Arg | 435.2718 | b:130.0965/261.1600/418.2470 y:175.1190/288.2042/435.1571 | 2 | – | 2 | non-umami |
| 16 | Val-Ile-Val-Lys | 458.3337 | b:213.1600/321.2293/458.2953 y:147.1135/246.1700/359.2644/458.2953 | 2 | – | 2 | 31.41 mmol/L |
| 17 | Gln-Ile-Val-Lys | 487.3224 | b:129.1031/242.1494/341.2193/477.2982 y:147.1142/246.1831/359.2643/487.2982 | 2 | – | 1 | 21.38 mmol/L |
| 18 | Asn-Val-Ile-Val-Lys | 572.3792 | b:214.1214/327.2023/426.2731/555.3442 y:147.1134/246.1811/359.2664/572.2751 | 1, 2 | – | 2 | 36.71 mmol/L |
Note:1. The b and y ions are obtained through the molecular networking approach; 2. “1″ represents samples fermented for 1 year, while ”2″ represents samples fermented for 2 years; 3. The similarity between mass spectrometry data is represented by cosine values; 4. Level 1 involves standard sample comparative identification, while Level 2 is based on compound fragment characteristics. 5. Umami Threshold Prediction with Umami-MRNN.
3.4. Electronic tongue analysis and sensory characterization
Human sensory evaluation can be influenced by various environmental and individual physiological factors. To enhance the consistency and objectivity of taste assessment, the electronic tongue technique was initially employed to determine the umami threshold of QIVK. This technique utilizes an array of six inert metal electrodes designed to simulate human taste perception (Guo et al., 2023). The response profile of QIVK was acquired by measuring its interaction with the six sensors of the electronic tongue (Fig. 4A). Indeed, sensors 1 and 4 demonstrated significant responses, while sensors 2, 3, 5, and 6 exhibited weaker responses (Table S3). The sensory profile of QIVK distinctly showcased pronounced umami and thickness tastes (Fig. 4B). Additionally, the electronic tongue analysis models predicted the umami threshold for QIVK to be 1.1630 ± 0.068 mmol/L.
Fig. 4.
Electronic tongue analysis (A) and sensory characterization (B) of QIVK.
To ensure the accuracy and reliability of umami threshold, human sensory evaluations were conducted in parallel with electronic tongue analysis (Table S4). The umami threshold of QIVK in an aqueous solution was determined to be 0.3215 ± 0.1607 mmol/L, significantly lower than that of monosodium glutamate (1.56 mmol/L), a commonly used flavor enhancer. These results confirm the pronounced umami taste of QIVK, consistent with the assessment conducted using the electronic tongue. Importantly, the umami threshold of QIVK surpasses that of most umami peptides reported in the past 3 years with a range of 0.07–10.04 mmol/L (Chang et al., 2023, Fu et al., 2024). In the future, synthetic biology techniques could be employed for targeted production of QIVK, creating opportunities for its application in the food industry.
The significant discovery of QIVK exclusively in PXDB fermented for two years, at a concentration of 0.9248 mg/g, marks a milestone in our understanding of the umami components in PXDB (Table S6). Interestingly, the near absence of QIVK in PXDB fermented for one year corroborates these findings, highlighting a direct correlation between peptide presence and fermentation duration. This observed correlation aligns with empirical sensory evaluations, which consistently emphasize the notion that the depth and richness of umami flavor in PXDB are heightened with extended fermentation periods. These findings significantly contribute to our comprehension of the molecular constituents responsible for umami sensation, indicating the potential to influence and optimize the production processes of PXDB in order to tailor its umami characteristics to meet diverse preferences or industrial requirements. Further exploration into the dynamic interplay between fermentation duration and peptide composition promises to deepen our understanding of umami enhancement in PXDB, potentially unveiling novel avenues for optimizing its flavor profile in the future. This research has the potential to significantly impact the food industry by informing the development of improved umami-enhanced products.
3.5. Constructing and evaluating the T1R1/T1R3 homology model
For an in-depth investigation into the activation mechanisms of QIVK, we employed the SWISS-MODEL (Waterhouse et al., 2018), to construct the T1R1/T1R3 homology model, as illustrated in Fig. S1A. To assess the model's reliability, we utilized established techniques, including Raman plotting and Verify-3D analysis. Fig. S1B illustrates that the majority of amino acid residues (98.9 %) fall within acceptable ranges, with 94.95 % residing in the Ramachandran favored region. Only 1.37 % and 1.46 % were identified as Ramachandran outliers and rotamer outliers, respectively. Furthermore, Verify-3D analysis results (Fig. S1C) indicated that approximately 82.0 %±0.05 of the residues achieved a 3D/1D average score of ≥ 0.2, with a significant proportion exhibiting an average score of ≥ 0.6. In line with established standards (J. Zhao et al., 2023), a homology model meeting the 90 % critical evaluation criteria and featuring over 80 % of residues with a Verify score ≥ 0.2 demonstrates a reasonable distribution of dihedral angles and three-dimensional collisions, ensuring a high level of model reliability.
3.6. Molecular docking of QIVK into T1R1/T1R3 receptors
In the investigation of QIVK's binding regions with T1R1/T1R3, the CB-Dock2 network server was employed for virtual docking. This advanced tool seamlessly integrates cavity detection, docking, and homologous template fitting, significantly enhancing precision in identifying the binding pocket. Based on the docking scores, the most likely docking site for QIVK with the T1R3 receptor has been confirmed (Fig. 5A). QIVK interacts with a total of 71 active residues within the T1R3 receptor, including SER20, TYR42, ARG46, TRP78, GLY129, PRO130, GLY131, SER132, SER133, ALA136, ALA152, TYR153, SER154, ALA155, THR156, SER157, ASP176, GLN179, TYR204, CYS259, GLU260, SER285, ASP286, GLY287, ALA289, ARG291, ILE308, LYS309, LEU310, ASP375, and LYS377 etal.—approximately twice the number found in the T1R1 receptor (Fig. S2).
Fig. 5.
Prediction of QIVK Binding Pockets and Sites on T1R3 Receptors Using CB-Dock2 (A); Molecular docking of QIVK and T1R3 by DS19 (B); different interaction modes between QIVK and T1R3 receptors: 3D Interaction Plot of Hydrogen Bond Surface Map (C) and 2D Interaction Plot (D).
Fig. 5B illustrates the various interaction modes between QIVK and the surrounding amino acid residues. QIVK's docking energies with T1R1 and T1R3 were computed as −88.3638 kcal/mol and −105.0730 kcal/mol, respectively (Table S1). The lower docking energy values observed for QIVK binding to T1R3 indicate a higher likelihood of peptide binding, resulting in the formation of stable conformations, receptor activation, and umami taste perception (Bu et al., 2021). To further support this, three-dimensional hydrogen bond surface maps reveal the affinity of umami peptides for the binding pocket of T1R3 (Fig. 5C). This affinity is influenced by the closed conformation of T1R1 (Fig. S2B), which results in a less accessible binding cavity (Cheng et al., 2023, Dang et al., 2019, López Cascales et al., 2010).
Analysis of interaction forces and modes reveals that QIVK forms hydrogen bonds with specific amino acid residues within the T1R3 subunit, including SER263, SER166, SER112, ASN235, LYS260, TYR262, and GLY234. Additionally, QIVK establishes salt bridges and electrostatic attractions with Glu292, His111, Glu233, and Glu238 at the binding sites (Fig. 5D). Remarkably, the average distance of these strong hydrogen bonds formed between QIVK and the active sites of T1R3 is 2.2 Å, significantly shorter than the standard hydrogen bond length of 3.5 Å (W. Liu et al., 2021).
The relationship between umami peptides and their structure–function is a current focal point in umami exploration. QIVK contains a glutaminyl moiety in its structure. Previous research has established the significant impact of glutaminyl on the umami taste. For instance, glutathione (GSH) represents the archetypal γ-glutamyl tripeptide, demonstrating diverse biological roles like antioxidative properties, immune enhancement, and remarkable abilities to enhance taste and extend flavors in food (Rae & Williams, 2017). Suzuki et al. developed a method for synthesizing γ-glutamyl compounds and discovered that γ-glutamylation can mitigate the bitterness of phenylalanine, valine, leucine, and histidine, while imparting acidity and enhancing the palatability of food (Suzuki et al., 2003). Furthermore, previous analyses have revealed that among the 98 cataloged umami peptides, those conveying umami taste often encompass glutamic acid, aspartic acid, and other hydrophilic amino acids. For lengthy peptides, their umami properties are not solely determined by amino acid composition; instead, their umami taste relies on their spatial structure, presence of umami, hydrophilic, and hydrophobic amino acids (J. Zhang et al., 2019). The heightened activity of QIVK is intrinsically tied to its unique structural features, which could prompt different conformations or functionally relevant arrangements upon receptor binding, potentially impacting its stereochemical properties (J. Zhang et al., 2019). This implies that QIVK's structural attributes and receptor binding mode significantly influence its high activity. Importantly, the way QIVK binds to receptors may vary from other umami peptides. Molecular docking analyses suggest QIVK might display stronger affinity or specificity towards residues such as Ser, Gln, Asn, Glu, Gly, and Asp within T1R3, which could affect its efficiency or intensity in receptor activation. Thus, a thorough understanding of QIVK's structural characteristics and receptor binding mode sheds light on why QIVK exhibits heightened activity. These receptor interactions could be tailored in the future to modify mouthfeel and flavor, addressing diverse consumer taste preferences. This offers substantial potential applications within the food industry.
4. Conclusion
In summary, this research underscores the innovative utilization of a cloud-based computational approach, employing PXDB as the research model. The successful combination of database matching and FBMN-assisted identification revealed 18 previously undiscovered peptides within PXDB. Following this, subsequent virtual screening and sensory evaluation conclusively verified the strong umami taste associated with the QIVK peptide. Molecular docking analysis further elucidated QIVK's enhanced binding affinity for the VTFD pocket in the T1R3 receptor in contrast to the T1R1 receptor. Additionally, specific residues within T1R3, such as Ser, Gln, Asn, Glu, Gly, and Asp, were identified as potential contributors to umami perception. This coherent amalgamation of molecular networks, molecular docking, and advanced computational tools has unveiled significant potential in deciphering the intricate complexities inherent in food metabolomes. As future investigations continue, the strategic application of this approach to explore umami peptides across various food sources holds promise for advancing our understanding of umami taste perception.
CRediT authorship contribution statement
Sen Mei: Writing – review & editing, Writing – original draft, Methodology. Shanshan Yao: Methodology. Jingjing Mo: Methodology. Yi Wang: Resources, Investigation. Jie Tang: Resources, Conceptualization. Weili Li: Software, Resources. Tao Wu: Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are grateful for the financial support from Sichuan Technology Development Program, China (2020YFN0056, 2021ZHCG0051, 2020YFN0094, 2021YFN0048, 2020YFN0151).
Declaration of Interest Statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2023.101098.
Contributor Information
Weili Li, Email: liweili1207@126.com.
Tao Wu, Email: wutaobox@gmail.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Anand Singh T., Nongthombam G., Goksen G., Singh H.B., Rajauria G., Kumar Sarangi P. Hawaijar – An ethnic vegan fermented soybean food of Manipur, India: A comprehensive review. Food Research International. 2023;170 doi: 10.1016/j.foodres.2023.112983. [DOI] [PubMed] [Google Scholar]
- Aron A.T., Gentry E.C., McPhail K.L., Nothias L.-F., Nothias-Esposito M., Bouslimani A.…Dorrestein P.C. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nature Protocols. 2020;15(6):1954–1991. doi: 10.1038/s41596-020-0317-5. [DOI] [PubMed] [Google Scholar]
- Bu Y., Liu Y., Luan H., Zhu W., Li X., Li J. Characterization and structure–activity relationship of novel umami peptides isolated from Thai fish sauce. Food & Function. 2021;12(11):5027–5037. doi: 10.1039/D0FO03326J. [DOI] [PubMed] [Google Scholar]
- Chang J., Li X., Liang Y., Feng T., Sun M., Song S., Yao L., Wang H., Hou F. Novel umami peptide from Hypsizygus marmoreus hydrolysate and molecular docking to the taste receptor T1R1/T1R3. Food Chemistry. 2023;401 doi: 10.1016/j.foodchem.2022.134163. [DOI] [PubMed] [Google Scholar]
- Chen S., Fu Y., Bian X., Zhao M., Zuo Y., Ge Y., Xiao Y., Xiao J., Li N., Wu J.-L. Investigation and dynamic profiling of oligopeptides, free amino acids and derivatives during Pu-erh tea fermentation by ultra-high performance liquid chromatography tandem mass spectrometry. Food Chemistry. 2022;371 doi: 10.1016/j.foodchem.2021.131176. [DOI] [PubMed] [Google Scholar]
- Chen S., Huang G., Liao W., Gong S., Xiao J., Bai J., Wendy Hsiao W.L., Li N., Wu J.-L. Discovery of the bioactive peptides secreted by Bifidobacterium using integrated MCX coupled with LC–MS and feature-based molecular networking. Food Chemistry. 2021;347 doi: 10.1016/j.foodchem.2021.129008. [DOI] [PubMed] [Google Scholar]
- Cheng K., Wang S., Wang Y., Bao Y., Gao P., Lei L., Liang H., Zhang S., Dong L. Modification of a Novel Umami Octapeptide with Trypsin Hydrolysis Sites via Homology Modeling and Molecular Docking. Journal of Agricultural and Food Chemistry. 2023;71(13):5326–5336. doi: 10.1021/acs.jafc.2c08646. [DOI] [PubMed] [Google Scholar]
- Dang Y., Hao L., Zhou T., Cao J., Sun Y., Pan D. Establishment of new assessment method for the synergistic effect between umami peptides and monosodium glutamate using electronic tongue. Food Research International. 2019;121:20–27. doi: 10.1016/j.foodres.2019.03.001. [DOI] [PubMed] [Google Scholar]
- Fu B., Wu D., Cheng S., Xu X., Zhang L., Wang L., El-Seedi H.R., Liu H., Du M. Three novel umami peptides derived from the alcohol extract of the Pacific oyster (Crassostrea gigas): Identification, characterizations and interactions with T1R1/T1R3 taste receptors. Food Science and Human Wellness. 2024;13(1):146–153. doi: 10.26599/FSHW.2022.9250012. [DOI] [Google Scholar]
- Guo W., Zhang Y., Long Z., Fu X., Ren K. Study on the taste active compounds in Douchi using metabolomics method. Food Chemistry. 2023;412 doi: 10.1016/j.foodchem.2022.135343. [DOI] [PubMed] [Google Scholar]
- Jiang S., Shi J., Li Y., Zhang Z., Chang L., Wang G., Wu W., Yu L., Dai E., Zhang L., Lyu Z., Xu P., Zhang Y. Mirror proteases of Ac-Trypsin and Ac-LysargiNase precisely improve novel event identifications in Mycolicibacterium smegmatis MC2 155 by proteogenomic analysis. Frontiers in Microbiology. 2022;13 doi: 10.3389/fmicb.2022.1015140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Li H., Wen Y., Liu Y., Wang J., Sun B. Molecular Mechanism for the α-Glucosidase Inhibitory Effect of Wheat Germ Peptides. Journal of Agricultural and Food Chemistry. 2021;69(50):15231–15239. doi: 10.1021/acs.jafc.1c06098. [DOI] [PubMed] [Google Scholar]
- Liu Y., Yang X., Gan J., Chen S., Xiao Z.-X., Cao Y. CB-Dock2: Improved protein–ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Research. 2022;50(W1):W159–W164. doi: 10.1093/nar/gkac394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López Cascales J.J., Oliveira Costa S.D., de Groot B.L., Walters D.E. Binding of glutamate to the umami receptor. Biophysical Chemistry. 2010;152(1–3):139–144. doi: 10.1016/j.bpc.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Mannochio-Russo H., de Almeida R.F., Nunes W.D.G., Bueno P.C.P., Caraballo-Rodríguez A.M., Bauermeister A., Dorrestein P.C., Bolzani V.S. Untargeted Metabolomics Sheds Light on the Diversity of Major Classes of Secondary Metabolites in the Malpighiaceae Botanical Family. Frontiers in Plant Science. 2022;13 doi: 10.3389/fpls.2022.854842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura E. One Hundred Years since the Discovery of the “Umami” Taste from Seaweed Broth by Kikunae Ikeda, who Transcended his Time. Chemistry – An Asian Journal. 2011;6(7):1659–1663. doi: 10.1002/asia.201000899. [DOI] [PubMed] [Google Scholar]
- Nothias L.-F., Petras D., Schmid R., Dührkop K., Rainer J., Sarvepalli A.…Dorrestein P.C. Feature-based molecular networking in the GNPS analysis environment. Nature Methods. 2020;17(9):905–908. doi: 10.1038/s41592-020-0933-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuemket N., Yasui N., Kusakabe Y., Nomura Y., Atsumi N., Akiyama S., Nango E., Kato Y., Kaneko M.K., Takagi J., Hosotani M., Yamashita A. Structural basis for perception of diverse chemical substances by T1r taste receptors. Nature Communications. 2017;8(1):15530. doi: 10.1038/ncomms15530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oak S.J., Jha R. The effects of probiotics in lactose intolerance: A systematic review. Critical Reviews in Food Science and Nutrition. 2019;59(11):1675–1683. doi: 10.1080/10408398.2018.1425977. [DOI] [PubMed] [Google Scholar]
- Qi L., Du J., Sun Y., Xiong Y., Zhao X., Pan D., Zhi Y., Dang Y., Gao X. Umami-MRNN: Deep learning-based prediction of umami peptide using RNN and MLP. Food Chemistry. 2023;405 doi: 10.1016/j.foodchem.2022.134935. [DOI] [Google Scholar]
- Qi L., Gao X., Pan D., Sun Y., Cai Z., Xiong Y., Dang Y. Research progress in the screening and evaluation of umami peptides. Comprehensive Reviews in Food Science and Food Safety. 2022;21(2):1462–1490. doi: 10.1111/1541-4337.12916. [DOI] [PubMed] [Google Scholar]
- Ramabulana A.-T., Petras D., Madala N.E., Tugizimana F. Metabolomics and Molecular Networking to Characterize the Chemical Space of Four Momordica Plant Species. Metabolites. 2021;11(11):763. doi: 10.3390/metabo11110763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samgina T.Y., Vasilieva I.D., Kozhevnikov A.Y., Meng Z., Zubarev R.A., Lebedev A.T. Mass spectrometry in de novo sequencing of the skin peptides from Arkhangelsk, Russia Rana temporaria: The variability of secreted AMPs in different populations. International Journal of Mass Spectrometry. 2023;484 doi: 10.1016/j.ijms.2022.116984. [DOI] [Google Scholar]
- Shan Y., Pu D., Zhang J., Zhang L., Huang Y., Li P., Xiong J., Li K., Zhang Y. Decoding of the Saltiness Enhancement Taste Peptides from the Yeast Extract and Molecular Docking to the Taste Receptor T1R1/T1R3. Journal of Agricultural and Food Chemistry. 2022;70(47):14898–14906. doi: 10.1021/acs.jafc.2c06237. [DOI] [PubMed] [Google Scholar]
- Wang W., Zhou X., Liu Y. Characterization and evaluation of umami taste: A review. TrAC Trends in Analytical Chemistry. 2020;127 doi: 10.1016/j.trac.2020.115876. [DOI] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Research. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Vecchi M.M., Wen D. Distinguishing between Leucine and Isoleucine by Integrated LC–MS Analysis Using an Orbitrap Fusion Mass Spectrometer. Analytical Chemistry. 2016;88(21):10757–10766. doi: 10.1021/acs.analchem.6b03409. [DOI] [PubMed] [Google Scholar]
- Yang Y., Niu C., Shan W., Zheng F., Liu C., Wang J., Li Q. Physicochemical, flavor and microbial dynamic changes during low-salt doubanjiang (broad bean paste) fermentation. Food Chemistry. 2021;351 doi: 10.1016/j.foodchem.2020.128454. [DOI] [PubMed] [Google Scholar]
- Zhang J., Sun-Waterhouse D., Su G., Zhao M. New insight into umami receptor, umami/umami-enhancing peptides and their derivatives: A review. Trends in Food Science & Technology. 2019;88:429–438. doi: 10.1016/j.tifs.2019.04.008. [DOI] [Google Scholar]
- Zhao J., Liao S., Han J., Xie Y., Tang J., Zhao J., Shao W., Wang Q., Lin H. Revealing the Secret of Umami Taste of Peptides Derived from Fermented Broad Bean Paste. Journal of Agricultural and Food Chemistry. 2023;71(11):4706–4716. doi: 10.1021/acs.jafc.2c09178. [DOI] [PubMed] [Google Scholar]
- Zhao S., Ma S., Zhang Y., Gao M., Luo Z., Cai S. Combining molecular docking and molecular dynamics simulation to discover four novel umami peptides from tuna skeletal myosin with sensory evaluation validation. Food Chemistry. 2024;433 doi: 10.1016/j.foodchem.2023.137331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.