Spatial analysis identifies IFN-induced MHC-IhiGal9+ CAFs that form a trap for CD8+ T cells, providing insights into the complex networks in the tumor microenvironment that regulate T-cell infiltration and function.
Abstract
Although immunotherapy can prolong survival in some patients with head and neck squamous cell carcinoma (HNSCC), the response rate remains low. Clarification of the critical mechanisms regulating CD8+ T-cell infiltration and dysfunction in the tumor microenvironment could help maximize the benefit of immunotherapy for treating HNSCC. Here, we performed spatial transcriptomic analysis of HNSCC specimens with differing immune infiltration and single-cell RNA sequencing of five pairs of tumor and adjacent tissues, revealing specific cancer-associated fibroblast (CAF) subsets related to CD8+ T-cell infiltration restriction and dysfunction. These CAFs exhibited high expression of CXCLs (CXCL9, CXCL10, and CXCL12) and MHC-I and enrichment of galectin-9 (Gal9). The proportion of MHC-IhiGal9+ CAFs was inversely correlated with abundance of a TCF1+GZMK+ subset of CD8+ T cells. Gal9 on CAFs induced CD8+ T-cell dysfunction and decreased the proportion of tumor-infiltrating TCF1+CD8+ T cells. Collectively, the identification of MHC-IhiGal9+ CAFs advances the understanding of the precise role of CAFs in cancer immune evasion and paves the way for more effective immunotherapy for HNSCC.
Significance:
Spatial analysis identifies IFN-induced MHC-IhiGal9+ CAFs that form a trap for CD8+ T cells, providing insights into the complex networks in the tumor microenvironment that regulate T-cell infiltration and function.
Introduction
Immunotherapy provides patients with advanced or metastatic cancer with an alternative treatment. Although immunotherapy prolongs overall survival and progression-free survival, it still presents a low response rate in head and neck squamous cell carcinoma (HNSCC; refs. 1, 2). Infiltration into tumor regions, including the tumor bed and tumor nests, is the primary step related to cytotoxic CD8+ T-cell function and one of the limiting factors for immunotherapy, especially for chimeric antigen receptor T-cell immunotherapy (CAR-T) efficacy in solid tumors (2, 3). However, the mechanisms for restricting CD8+ T-cell infiltration are poorly understood.
To enhance the infiltration of CD8+ T cells and ultimately the response to immunotherapy, better comprehension of the tumor microenvironment (TME) is necessary (2, 4). Cancer-associated fibroblasts (CAF) are one of the key components of the TME and are reported to be involved in immune regulation and drug resistance in tumors (5, 6). CAFs are regarded as crucial regulators of the malignant environment. These cells recruit and maintain tumor-associated macrophages, polymorphonuclear myeloid-derived suppressor cells and regulatory T cells (Treg) via the secretion of chemokines and cytokines, such as IL6, CXCL1, and CCL2 (7–9). In addition, CAFs block antitumor CD8+ T-cell infiltration into malignant islets by extracellular matrix (ECM) secretion and remodeling (10). However, the heterogeneity of CAFs makes it complicated to modulate these cells as a whole. Simply depletion of αSMA+ CAFs or inhibition of stromal proliferation would further decrease survival time in pancreatic ductal adenocarcinoma (PDAC) mouse models (11, 12). Previous studies on CAFs have identified the major clusters as inflammatory CAFs (iCAF) and myofibroblasts (myCAF). In addition, a subpopulation of CAFs expressing MHC-II and CD74 was identified as antigen-presenting CAFs (apCAF) that contribute to the regulation of immunity in PDAC. Recent work by Hutton and colleagues (13) on the lineage of CAFs found that CD105 could demark two pancreatic fibroblast lineages when CD105-negative fibroblasts restrict tumor growth in a conventional dendritic cell–dependent manner. It was also confirmed that these fibroblasts could differentiate into both iCAFs and myCAFs regardless of the status of CD105. These results indicated that discrimination of iCAFs, myCAFs or apCAFs was important but not sufficient for developing strategies for improving immunotherapy based on CAF modulation.
Tumor-infiltrating immune cells are responsible for the efficacy of immunotherapy, and CD8+ T cells are the critical effectors in antitumoral immunity. The interactions between CD8+ T cells and other types of cells, including CAFs, determine how CD8+ T cells function in this dynamic TME. The pre-effector CD8+ T cells expressing GZMK were indicated to be the intermediate state for cytotoxic and dysfunctional states (14), but the direction-determining factors for the transformation are unclear. TCF1+PD-1+ CD8+ T cells function as stem-like CD8+ T cells and limit tumor growth in response to immune checkpoint blockade (15, 16). However, it is still unknown how these cells are inhibited in primary malignancies. Dysfunctional CD8+ T cells should account for most CD8+ T cells in the TME. Although multiple exhaustion markers for dysfunctional CD8+ T cells, such as T-cell immunoglobulin and mucin domain-containing protein 3 (TIM3) and lymphocyte activation gene 3 protein (LAG3), have been identified by single-cell technologies, the underlying stimuli and mechanisms of T-cell dysfunction remain poorly understood.
In HNSCC, the heterogeneity of CAFs is poorly defined, and the interactions between CAF subclusters and immune cells are rarely reported. Here, we conducted the spatial transcriptomes of different immune types of HNSCC to evaluate previously reported factors that affect the distribution and status of CD8+ T cells. And with single-cell RNA sequencing (scRNA-seq) of tumor and adjacent normal tissues, we found a cluster of CAFs that secreted CXCL9/CXCL10/CXCL12 and expressed high levels of MHC class I molecules (HLA-A/B/C and B2M) and galectin-9 (MHC-IhiGalectin-9+ CAFs) involved in restricting CD8+ T-cell infiltration and further confirmed that these cells promoted tumor growth by restricting the effector differentiation of pre-effector CD8+ T cells (TCF1+GZMK+CD8+ T cells). Our work emphasized the role of MHC class I on CAFs in the distribution of CD8+ T cells and further elucidated galectin-9 as the critical molecules that induced the dysfunctional differentiation of memory-like CD8+ T cells (TCF1+ CD8+ T cells).
Materials and Methods
Patient specimens
The 10 surgery samples (five tumoral tissues and five paired adjacent normal tissues) for scRNA sequencing were obtained from patients with HNSCC treated at the Shanghai Ninth People's Hospital in 2019. The formalin-fixed, paraffin-embedding (FFPE) samples for multiplex immunofluorescent staining and the primary tumors of HNSCC for CAFs isolation were obtained from patients of HNSCC who underwent surgery in 2021. The tissue microarray for galectin-9 expression analysis was obtained from patients of HNSCC who underwent surgery of resection from 2014 to 2020.
Mice
C3H/He mice and nude mice were purchased from Shanghai Bikai Keyi Biotechnology Co., Ltd. All the mice were bred and housed in the animal facility of Shanghai Ninth People's Hospital under specific pathogen-free conditions. All animal experiments were randomized and conducted in accordance with the Guide for the Care and Use of Laboratory Animals. All the animal experiments have been approved by the Laboratory Animal Ethics Committee in Ninth People's Hospital Affiliated to Shanghai Jiao Tong University School of Medicine.
Cell lines and primary cells
CAL27, HN6, and HN30 cells were obtained from the Shanghai Research Institute of Stomatology (Shanghai Ninth People's Hospital) and cultured in DMEM containing 10% v/v FBS and 1% v/v penicillin–streptomycin (PS). CAFs were isolated from primary HNSCC tumoral tissues, cultured in DMEM containing 10% v/v FBS and 1% v/v PS and used for in vitro experiments within five passages. MTFs were isolated from tongues of C3H/He mice and cultured in DMEM culture medium (DMEM containing 10% v/v FBS and 1% v/v PS). All cells were maintained at 37°C in a 5% CO2 setting and routinely tested for Mycoplasma with the GMyc-PCR Mycoplasma Test Kit (Yeasen, cat. #40601ES20).
IHC
Paraffin sections of HNSCC clinical samples were maintained in 100%, 100%, 100% v/v dimethylbenzene, 100%, 90%, 70% v/v ethanol and pure water sequentially for dewaxing to water. Heat-induced epitope retrieval process was conducted by microwave oven in sodium citrate antigen retrieval buffer (Proteintech, PR30001) or Tris-EDTA antigen retrieval buffer (Proteintech, PR30002). 10% v/v goat serum and 3% v/v H2O2 were sequentially applied to sections to block endogenous peroxidase activity and Fc binding. Primary antibodies (anti-CD8α antibody, Cell Signaling Technology, #85336, RRID: AB_2800052; anti-CD4 antibody, Cell Signaling Technology, #25229, RRID: AB_2798898; anti-CD20 antibody, Cell Signaling Technology, #48750) were diluted at 1:200 and applied to tissue sections in 4°C overnight (∼15 hours). Goat anti-Mouse/Rabbit poly-horseradish peroxidase Secondary Antibody (Proteintech, PR30009, RRID: AB_2934294) was applied to tissue sections after three times of washes with PBST (0.2% Tween in PBS). The DAB detection Kit (Absin, abs9210) was used to visualization of CD4/CD8α/CD20. The nuclei were stained with hematoxylin (Absin, abs9214). These slides were scanned with Automated Quantitative Pathology System Vectra (PerkinElmer) after dehydration with a serial of 70%, 90%, 100% v/v ethanol, 100%, 100%, 100% v/v dimethylbenzene and mounting with neutral balsam (Absin, abs9177).
Spatial transcriptome sequencing and analysis
10x Visium spatial RNA-seq data preprocessing The Space Ranger software pipeline (version 1.0.0) provided by 10×Genomics was used to process Visium spatial RNA-seq output and brightfield microscope images to detect tissue, align reads using the STAR2 aligner, generate feature-spot matrices, perform clustering and gene expression analysis, and place spots in spatial context on the slide image. We processed the unique molecular identifier (UMI) count matrix using the R package Seurat (version 3.1.1). We first normalized the data with sctransform4 to account for variance in sequencing depth across data points, detecting high-variance features, and store the data in the SCT assay. The average expression and dispersion were calculated for each gene, genes were subsequently placed into x bins based on expression. Principal component analysis was performed to reduce the dimensionality on the log-transformed gene-barcode matrices of top variable genes. Cells were clustered on the basis of a graph-based clustering approach, and were visualized in 2-dimension using Uniform Manifold Approximation and Projection (UMAP).
scRNA-seq and analysis
The five HNSCC tumoral samples and paired adjacent normal samples were dissected from tissues of patients with HNSCC who underwent complete surgery dissection. The 10 samples were temporally stored in ice-cold storage buffer (RPMI-1640 + 0.04% BSA) before washed twice with storage buffer and cut into pieces approximately 0.5 mm3. Tissue pieces were incubated in a fresh-made enzyme mixture at 37°C for 30–60 minutes and filtered with 40-μm cell strainers. After centrifuge (4°C, 300 × g for 5 minutes), an equal volume of 1X Red Blood Cell Lysis Buffer (MACS, 130–094–183) were added to cell pellets and maintained in 4°C for 10 minutes before centrifuge (4°C, 300 × g for 5 minutes). Cells were washed and resuspended RPMI-1640. Then cDNA libraries were constructed with 10×Genomics Chromium Next GEM Single Cell 3′ Reagent Kits v3.1 (1000268) following the manufacturer's instructions and sequencing was performed on Illumina Nova 6000 PE150 platform.
The Cell Ranger software pipeline (version 3.1.0) provided by 10×Genomics was used to demultiplex cellular barcodes, map reads to the genome and transcriptome using the STAR aligner, and down-sample reads as required to generate normalized aggregate data across samples, producing a matrix of gene counts versus cells. We processed the UMI count matrix using the R package Seurat (version 3.1.1). To remove low-quality cells and likely multiplet captures, which is a major concern in microdroplet-based experiments, a set of criteria were conducted: Cells were filtered by (i) UMI/gene numbers out of the limit of mean value ± 2-fold of standard deviations, assuming that each cell's UMI/gene numbers have a Gaussian distribution, (ii) percentage of mitochondrial RNA UMIs (proportion of UMIs mapped to mitochondrial genes >25%). After applying these QC criteria, 91,624 single cells were included in downstream analyses. To obtain the normalized count, library size normalization was processed using NormalizeData function in Seurat. Specifically, the global-scaling normalization method “LogNormalize” normalized the gene expression measurements for each cell by total expression, multiplied by a scaling factor (10,000 by default), and the results were log-transformed.
Top variable genes across single cells were identified and selected by FindVariableGenes function (mean.function = FastExpMean, dispersion.function = FastLogVMR) in Seurat. Differentially expressed genes (DEG) were identified using the FindMarkers function (test.use = MAST) in Seurat. A P value of <0.05 and log2|foldchange| > 0.58 was set as the threshold for significantly differential expression. Gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of DEGs were, respectively, performed using R based on the hypergeometric distribution. The development pseudotime was determined with the Monocle2 package (v2.9.0) and DiffusionMap algorithm. The SCENIC analysis was run using the motifs database for RcisTarget and GRNboost (SCENIC version 1.1.2.2, which corresponds to RcisTarget 1.2.1 and AUCell 1.4.1) with the default parameters. The cell communication analysis was performed using the CellChat (v 1.1.3) R package. The CellPhoneDB (v2.0) was used to identify biologically relevant ligand–receptor interactions and R packages Igraph and Circlize were used to display the cell–cell communication networks.
HNSCC tumors dissociation for primary culture or flow cytometry
Tumoral tissues dissected from surgery were temporally maintained in ice-cold Tissue Storage Solution (Miltenyi). For primary CAFs isolation, tumoral tissues were washed three times with DMEM containing 10% PS before further disaggregations. Tumoral tissues were then cut into small pieces of approximately 1 mm3 before treated with a human Tumor Dissociation Kit (Miltenyi). Small pieces of tumoral tissues were transferred to gentleMACS C Tube containing the enzyme mix (the mix of enzyme R, enzyme H, enzyme A from human Tumor Dissociation Kit). Then the tissues went through dissociation using the gentleMACS Octo Dissociator (Miltenyi). After dissociation, the tissue suspension went through a 100-μm cell strainer, and washed with DMEM before centrifuged at 300 × g for 3 minutes. Finally, cell pellets were resuspended with DMEM culture medium or ice-cold PBS for primary cells culture or flow cytometry, respectively.
Lentivirus transfection
CAFs, MTFs, and MEFs (mouse embryonic fibroblast) were seeded at concentration of 105 cells per well and cultured at 37°C in a 5% CO2 setting. After cultured for 12 hours, previous culture medium was discarded and treated with 1 mL pre-mixed DMEM culture medium (1 × HiTransG P in DMEM containing 10% FBS and 1% PS) with a total of 5 × 105 TU lentivirus for MOI = 50. After 8 hours, another DMEM containing 10% FBS and 1% PS was added to each well. The culture medium was sequentially replaced by normal DMEM culture medium, 2.5 μg/mL puromycin in DMEM culture medium and normal DMEM culture medium at time point of 16, 48 hours, and 4 days after transfection. Transfected cells were passaged and expanded as usual.
Subcutaneous co-transplant models
Syngeneic female C3H/He mice of 10–12 weeks of age were used for subcutaneous co-transplant model. SCC VII and fibroblasts in 10-cm dishes were washed by pre-cold PBS and digested by 0.25% trypsin, and terminated by 10% FBS/DMEM. Cells were collected by centrifuging at 300 × g for 3 minutes and supernatant was discarded before washed twice by pre-cold PBS. Concentrations of each cell types were determined and required number of SCC VII and fibroblasts were combined in 0.6-mL microcentrifuge tubes. Cells were collected by centrifuging. Then, cell pellets were resuspended in Matrigel (Corning, cat. #354262) at concentration of each cell type of 5,000 cells per μL and mixed gently on ice. Insulin Syringe/Needles (VWR, 9151117) were used to subcutaneously inject 20 μL of cell/Matrigel mix (a total number of 105 fibroblasts and 105 SCC VII cells) into the back of mice. The tumor volume was calculated as V = 1/2 × length × width2.
Mouse-bearing tumor disaggregation and tumor-infiltrated lymphocytes isolation
Before disaggregation, a triple-enzyme mixture and percoll solutions were required preparation. For the stock solution of 10 × triple-enzyme mix, 0.1 g of collagenase IV, 0.1 g of hyaluronidase, and 75000 U DNase I were dissolved in 25-mL RPMI-1640 and filtered via 0.22 μm of pore mesh. 10 × triple-enzyme mix was diluted with RPMI-1640 to prepare 1 × triple-enzyme working solution. To prepare isotonic percoll solution, 45-mL percoll was mixed with 5 mL 10 × PBS. The isotonic percoll solution was then mixed with PBS or RPMI-1640 to prepare 60% and 30% v/v percoll solutions, respectively. Isolated mouse-bearing tumors were first flushed clear with ice-cold PBS and cut into small pieces. These small pieces of tumoral tissues were then treated with the working solution of the triple-enzyme mix that consisting of collagenase IV (0.4 mg/mL), hyaluronidase (0.4 mg/mL), and DNase I (30 U/mL) for 30 minutes. After digestion, tissue suspension was required to go through a 70-μm cell strainer to make cell suspension. Cell suspension was centrifuged at 300 × g for 3 minutes and supernatant was discarded. Each sample of the cell pellet was resuspended with 4 mL 30% v/v percoll and gently placed on 4 mL 60% v/v percoll solution in a 15-mL centrifuge tube. The 15-mL centrifuge tubes went centrifuged at 300 × g for 20 minutes (the rates of acceleration and spinning down were set at zero). After centrifuge, cells at interface of percoll solutions were collected in a new 15-mL centrifuge tube. Cells were washed twice with ice-cold PBS and resuspended in PBS for further analysis.
Coculture of primary CD8+ T cells and CAFs
Cocultured CD8+ T cells and CAFs were isolated from the metastasis lymph nodes and tumors, respectively, from the same patients. CD8+ T cells were cultured and expanded upon CD3/CD28/IL2 stimulations. And CAFs were transfected with B2M-/LGALS9 KD lentivirus. CAFs were seeded in plates 12 hours before coculturing with CD8+ T cells 10 times in quantity. After 48 hours of coculture, cells were fixed for immunofluorescent staining and CD8+ T cells were harvested for flow cytometry.
Multiplexed immunofluorescence
Dewaxing and antigen epitope retrieval of paraffin sections of HNSCC samples and mouse-bearing tumors were processed as mentioned in IHC. The permeabilization process for tissue sections was conducted with 0.5% v/v TritonX-100 in PBST and Fc-blocking was conducted with 10% v/v goat serum in PBST. Interaction of primary antibodies and fluorescence-conjugated secondary antibodies was conducted following general procedure: Sections were incubated with primary antibodies diluted at 1:200, in 4°C, overnight and washed with PBST three times before incubated with fluorescence-conjugated secondary antibodies at room temperature, in the dark, for 2 hours, and unconjugated secondary antibodies were removed by PBST before another primary antibody incubation. For nuclei staining, tissue sections were incubated at room temperature with hoechst 33342 (Absin, abs813337) diluted in PBST at 1 μg/mL for 15 minutes. All immunofluorescence-staining slides were mounted with Fluoromount-G (Yeasen, 36307ES25). Fluorescent microscopy images capture and scanning were conducted with FV3000 (Olympus) and images were processed with Imaris (Oxford Instruments).
Flow cytometry
To determine the galectin-9 and MHC-I molecules in CAFs from clinical tumoral tissues, cell suspension was prepared following the human tumors dissociation as mentioned above. Cells were pelleted and resuspended in 100 μL PBS containing Zombie Green (1:1,000, BioLegend, #423112) at room temperature, in the dark, for 15 minutes. 100 μL of staining buffer containing 0.25-μL Fc block was added to cell pellets after washed twice with PBS containing 0.5% v/v BSA. Then a mix of fluorochrome-conjugated primary antibody, including 5 μL anti–EpCAM-FITC (BioLegend, #324204, RRID: AB_756078), 5 μL anti–CD45-FITC (BD Pharmingen, #555482, RRID: AB_395874), 1 μL anti–CD31-FITC (BD Pharmingen, #555445), 1 μL anti–PDPN-APC (eBioscience, #17–9381–42), 1 μL anti–galectin-9-PE (BioLegend, #348906, RRID: AB_10613466), 1 μL anti–HLA-A/B/C-PerCP-Cy5.5 (BioLegend, #311420, RRID: AB_10709732), were added to cells and gently mixed. After 30 minutes on the ice in the dark, cells were pelleted at 300 × g for 5 minutes and supernatant was discarded. Cells were washed twice in ice-cold PBS before resuspended in 4% v/v paraformaldehyde. These samples were analyzed with Beckman CytoFlex S (Beckman). For flow cytometry of in vitro cells (CAL27, HN6, HN30, and CAFs), cells were harvested with TrypLE Express (Thermo Fisher Scientific, 12604021) and washed with ice-cold PBS. These cells were stained with anti–Galectin-9-PE (1:100, BioLegend) and anti–HLA-A/B/C-APC (1:100, BioLegend). For coculture analysis of CAFs and HN6, cells were stained with anti–PDPN-APC (1:100, BioLegend), anti–EpCAM-FITC (1:100, BioLegend), and anti–HLA-A/B/C-PerCP-Cy5.5 (1:100, BioLegend). Cells were washed with ice-cold PBS twice before being fixed with 4% v/v paraformaldehyde. These cells were analyzed with BD FACSCalibur (BD Biosciences).
RNA extraction and real-time reverse transcription PCR analysis
CAFs and HN6 cells were seeded in 6-well plates at a concentration of a total of 2.5×105 cells per well and cultured at 37°C in a 5% CO2 setting overnight. Different cytokines (IFNα, IFNβ, IFNγ, TGFβ1, TGFβ2, TGFβ3, TNFα, and POSTN) were diluted into 100 ng/mL with DMEM containing 10% FBS and 1% PS and added to CAFs and HN6. After 48 hours of treatment, the culture medium was discarded and cells were washed with ice-cold PBS twice. Then the RNA was extracted with the RNA-Quick Purification Kit (ES Science). Specifically, lysis buffer was added to cells for 500 μL per well and transferred to 1.5-mL microcentrifuge. 500-μL ethanol was added to the lysates, mixed well, and added to the RNA centrifuge columns before centrifuged at 4,000 × g for 1 minute. Then the RNA centrifuge columns were washed by 500 μL wash buffer and RNA was collected in 50-μL elution buffer. Reverse transcription was conducted with Evo M-MLV RT Master (Accurate Biology) at 37°C for 15 minutes and 85°C for 5 seconds. Finally, real-time PCR was conducted with Hieff qPCR SYBR Green Master Mix (Yeasen) and detected by StepOnePlus Real-time PCR System (Applied Biosystems).
Immunoblotting
CAFs and HN6 cells seeded in 6-well plates at a concentration of a total of 2.5×105 cells per well treated with different cytokines (IFNα, IFNβ, IFNγ, TGFβ1, TGFβ2, TGFβ3, TNFα, and POSTN) at a concentration of 100 ng/mL. After 48 hours of treatment, the culture medium was discarded and cells were washed with ice-cold PBS twice. Then lysis buffer was added to each well for 100 μL per well and cells were scraped and harvested into a 1.5-mL microcentrifuge. After heated at 105°C for 10 minutes, the protein concentrations of cell lysates were measured by the Pierce BCA Assay Kit (Thermo Fisher Scientific) following the manufacturer's protocol. The protein concentration was adjusted to 2 μg/μL with lysis buffer and 5X SDS loading buffer before boiled for 15 minutes. Protein was added to SurePAGE protein gel (GenScript) for 20 μg per well before separated at 180 V for 60 minutes. Then, the proteins in the gel were transferred to a polyvinylidene difluoride membrane (0.2 μm, Millipore) with a transfer apparatus at 90 V for 2 hours. After blocking with 5% non-fat milk, the membranes were incubated with primary antibodies for 15 hours at 4°C and washed three times with TBST (0.2% tween in Tis-balanced solution) before incubated with the secondary antibody at room temperature for an hour. Finally, targeted proteins were visualized with Enhanced Chemiluminescent regent (NCM Biotech) by Amersham Imager 600 (GE).
Quantitative analysis of multiplexed immunofluorescence images
Frozen slides, from the same samples for spatial transcriptome RNA-seq, were used for multiplexed immunofluorescent staining with TG TSA Multiplex IHC Assay Kits (TissueGnostics Asia-Pacific Ltd.). Image scanning was performed and visualized with TissueFAXS Spectra Systems (TissueGnostics GmbH), and the images were quantitatively analyzed (quantities of specific cell types and distances between different types of cells) with StrataQuest analysis software (Version 7.1.129, TissueGnostics GmbH). The multiplexed immunofluorescent staining of the 5 paired infiltrated and excluded FFPE slides were stained with the AlphaTSA Multiplex IHC Kit (AXT37100031) by AlphaPainter X30, scanned by ZEISS AXIOSCAN 7 and analyzed by Halo (3.4, Indica Laboratories).
Statistical analysis and reproducibility
Statistical analyses and data plotting were performed in the R Statistical Software and Prism (version 7, GraphPad Software Inc.). The flow cytometry results were analyzed with FlowJo (version 10.4, BD Life Sciences) or CytoExpert (version 10.0.4, Beckman). The unpaired t test assuming two-tailed distribution or one-way ANOVA was used for comparing differences between groups where p/adjusted P value of <0.05 was considered statistically significant for each comparison.
Data availability
The data analyzed in this study were obtained from Tumor Immune Estimation Resource 2.0 (TIMER 2.0) at http://timer.cistrome.org/. The Spatial and single-cell transcriptomics data generated in this study are publicly available in the Genome Sequence Archive, HRA005551 (https://bigd.big.ac.cn/gsa-human/browse/HRA005551) and HRA0005576 (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA005576) and nonprofit research will be approved for access for these data. All other raw data are available upon request from the corresponding author.
Results
CD8+ T cells are maintained in the tumor stroma
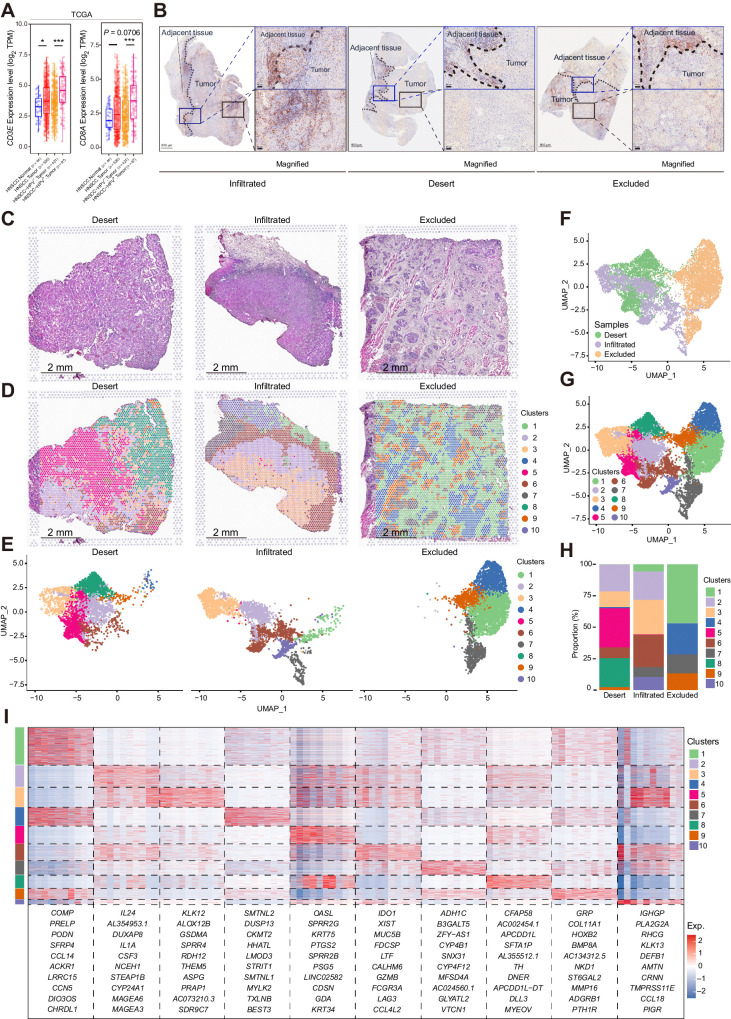
We first investigated CD8+ T-cell infiltration by analyzing CD3E and CD8A expressions between HNSCC tumor and normal samples from The Cancer Genome Atlas (TCGA) database (Fig. 1A). The higher TPM values for CD3E and CD8A in tumoral tissues indicated that CD8+ T-cell infiltration was increased in HNSCC tumoral tissues. In addition, CD3E and CD8A transcript levels were much higher in HPV-positive HNSCC than in HPV-negative HNSCC (Fig. 1A), which suggested higher infiltration of CD8+ T cells. To precisely characterize the spatial distribution of CD8+ T cells in HNSCC, surgical tissues from patients with HNSCC with different immune subtypes were assessed for CD8α staining by IHC (Fig. 1B). The immune infiltration type (infiltrated, desert, and excluded) was also determined by CD4/CD20 analysis (Supplementary Fig. S1A). To compare the tumor and adjacent normal tissues, sections containing both tissue types were selected specifically. In all three immune infiltration types, the infiltration of lymphocytes was limited in the fibrotic areas of the invasive margin (Fig. 1B; Supplementary Fig. S1A). In the tumor bed of immune-infiltrated tumors, some CD8α+ cells infiltrated the nests, although CD8α+ cells preferred fibrotic areas (Fig. 1B, left). For immune-desert tumors, abundant CD8α+ cells were also detected in the fibrotic areas of the tumor invasive margin, and as expected, few CD8α+ cells were found in the tumor bed (Fig. 1B, middle). Interestingly, with abundant CD8α+ cells in the fibrotic areas of the immune-excluded tumors, few CD8α+ cells could infiltrate into the tumor nests (Fig. 1B, right; Supplementary Fig. S1B). These results indicated that the majority of CD8+ T cells were confined to the stroma rather than infiltrating tumor nests.
Figure 1.
CD8α+ cells were maintained in the stroma of all three immune types of HNSCC. A, The CD8A and CD3E expression results of HNSCC bulk RNA sequencing data from TCGA database. The results are displayed in range and mean ± SEM, and statistics are accessed by the Wilcoxon test. *, P < 0.05; ***, P < 0.001. B, IHC staining of CD8α of the infiltrated, desert, and excluded immune type of HNSCC. Representative sites for the margins of tumor invasion and the tumor beds are labeled with dark blue and black rectangles, respectively, and magnified. C and D, H&E staining and spatial distribution of different clusters for spots throughout the tumor (with adjacent) tissues in the 10x Visium capture slides from desert, infiltrated, and excluded immune-subtype specimens. E–G, UMAP distribution of spots captured with the 10x Visium capture slides from desert, infiltrated, and excluded immune-subtype specimens. E, The captured spots are split by sample origins and labeled according to clusters. The spots are also summarized and labeled according to different immune-subtype specimens (F) and clusters (G). H, Proportion of the spots of different clusters in each immune-subtype specimen. I, Heat map of top markers for each cluster of spatial spots. The top 10 genes are listed. The rank of the gene markers was accessed with “findmarker” algorithm.
To further explore the factors that limit CD8+ T cell infiltration, we performed spatial transcriptome sequencing of the three immune types of HNSCC tissues. These tissues were from the same patients but different pieces of samples that were used for the determination of the “immune subtypes” in Fig. 1B. Specimens were processed with hematoxylin and eosin (H&E) staining (Fig. 1C), and spots captured by 10x Visium of all three specimens were grouped into 10 clusters and aligned to the H&E images (Fig. 1D–G). As expected, the three immune types of HNSCC confirmed by spatial expression of lymphocyte-associated genes consisted of multiple clusters of spots and presented heterogeneity among these specimens (Supplementary Fig. S1C). When the infiltrated and desert specimens were compared, the desert specimen exclusively had spots of clusters 5 and 8, and the excluded specimen exclusively had spots of clusters 4 and 9 (Fig. 1H). Cluster 4 was the cluster consisting of muscle cells (Fig. 1D). Cluster 9, which had high levels of COL11A1, GRP, HOXCB2, BMP8A, and NKD1 (Fig.1I; Supplementary Fig. S1D), primarily consisted of fibroblasts surrounding tumor nests according to the alignment of clusters on H&E images (Fig. 1D). Recently, Grout and colleagues (10) identified that, with an ECM reprogram that had a preference for collagen XI and collagen XII, αSMA+ CAFs played a major role in T-cell marginalization in human lung tumors. Herein, we explored the expression and spatial distribution of COL11A1, COL11A2, and COL12A1 in the three immune types. Consistently, the stroma of the excluded-type specimen was enriched with COL11A1 and COL12A1, and the infiltrated-type specimen lacked COL11A1, COL11A2, and COL12A1, whereas the desert-type specimen had high expression of COL12A1 but low expression of COL11A1 and COL11A2 (Supplementary Fig. S1E). However, we also noticed that in the excluded-type specimen, COL11A1 and COL12A1 were also expressed in spots of cluster 1 beyond cluster 9. We further assessed the spatial relationships among αSMA+ CAFs, CD8α+ cells and tumor nests in immune-excluded human HNSCC. Most CD8α+ cells were limited to areas enriched in αSMA+ regions, but we also noticed that even in the actual boundary between αSMA+ fibrotic regions and tumor nests, CD8α+ cells never infiltrated into the tumor nests (Supplementary Fig. S1B). These results indicated that CD8+ T cells are limited in the tumor stroma, and beyond reprogramming of the ECM, there are other molecular mechanisms that confine CD8+ T cells to the stroma.
Single-cell analysis uncovers the heterogeneity of CAFs in HNSCC
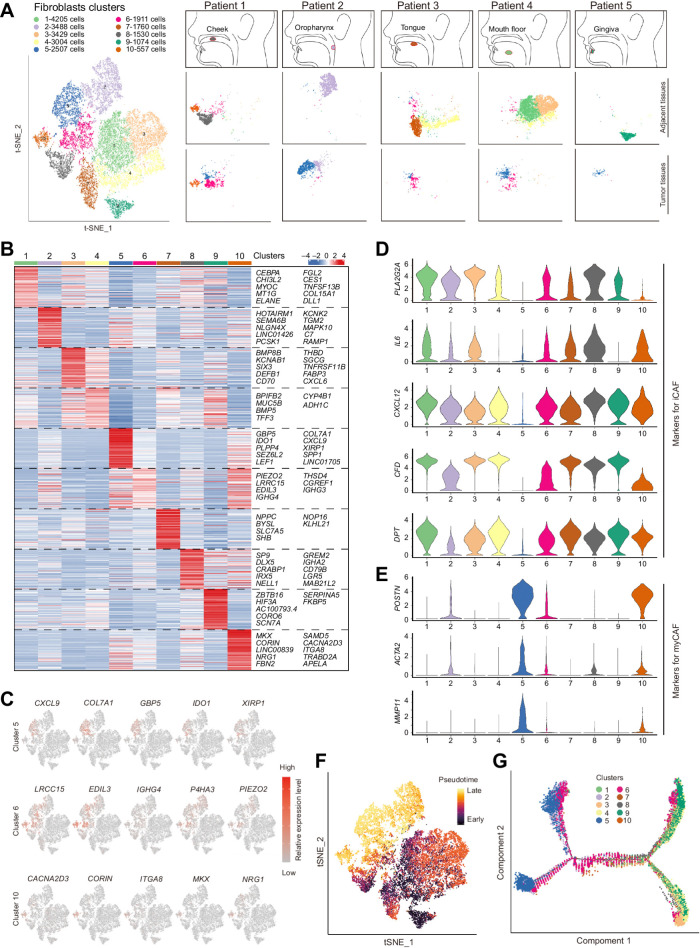
To further investigate the surrounding stroma of HNSCCs, we defined the heterogeneity of fibroblasts by scRNA-seq. Samples of tumor and adjacent normal tissues from five patients with HNSCC from different locations were digested into single-cell suspensions, and viable cells were captured for scRNA-seq (Supplementary Fig. S2A). After quality control and batch correction, 23,485 cells with signatures of fibroblasts were selected for downstream analysis (Supplementary Fig. S2A). We first compared the fibroblast clusters from tumor and adjacent tissues with dimensionality reduction of t-distributed Stochastic Neighbor Embedding, and 10 groups of fibroblasts were identified (Fig. 2A). Consistent with previous assumptions, fibroblasts from tumor, and adjacent normal tissues were diverse. Clusters of fibroblasts from different samples of adjacent normal tissues are robustly different, which could be the result of distinct anatomical sites from which these tissues are harvested (Fig. 2A and B). Except for clusters 1 and 4, the other five normal fibroblast (NF) clusters were barely shared among these five normal-adjacent tissues (Supplementary Fig. S2B–S2D). In contrast, fibroblasts from tumoral tissues that barely existed in adjacent normal tissues, so-called CAFs, presented as only three clusters, cluster 5, 6, and 10 (Fig. 2A; Supplementary Fig. S2B). Clusters 5 and 6 could be identified in all five tumoral tissues, and cluster 5 was the only cluster that barely existed in normal-adjacent tissues and was almost exclusively present in all five tumor samples. To further identify these clusters, we determined markers for these fibroblasts and tried to reveal a possible role of each cluster. However, in contrast with the diversity of fibroblasts, markers for all these fibroblast clusters were not unique, and several clusters shared similar expressions of these markers (Fig. 2C and D). For the three clusters of CAFs, the top markers (such as COL7A1 for cluster 5, LRCC15 for cluster 6 and CACNA2D3 for cluster 10) were also shared by the other clusters (Fig. 2C). Actually, there were few markers that could specifically mark each cluster of CAFs. These results indicated that the phenotypes of CAFs are much dependent on the microenvironment but not their lineage.
Figure 2.
Global analysis of fibroblasts from tumoral and adjacent tissues of HNSCC. A, Clinical information of collected tumoral and adjacent samples for single-cell RNA sequencing and each sample's t-SNE plot results for fibroblasts. B, Heat map of markers for 10 clusters of fibroblasts. The rank of the gene markers is accessed with “findmarker” algorithm. C, The expression of the top 5 markers for CAFs (clusters 5, 6, and 10) were overlaid in the t-SNE plots and also indicated in the violin plots. D and E, Violin plots of gene expression for iCAF- (D) or myCAF (E)-associated genes in fibroblasts. F and G, Pseudotime analysis for fibroblasts. F, Cells were labeled with the pseudodifferentiation time in the t-SNE plots. G, Cells labeled according to different clusters were also assigned to different pseudotime trajectories.
In addition, to compare CAF clusters from HNSCCs with CAFs from other types of tumors (17–19), we assessed the expression of markers identified in previous studies of PDAC, breast cancer, and bladder urothelial carcinoma (Fig. 2D; Supplementary Fig. S2E). Most markers described in other tumors had expression patterns similar to the CAFs in clusters 5, 6, and 10 (Supplementary Fig. S2E). However, we found that cluster 6 had higher expression of several markers for iCAFs, such as IL6 and CXCL12, than cluster 5, whereas cluster 5 expressed markers for myCAFs, such as POSTN and ACTA2 (Fig. 2D and E). Although markers for iCAFs (e.g., PLA2G2A, IL6, CXCL12, CFD, and DPT) could distinguish cluster 6 from cluster 5, NFs express similar or higher levels of these iCAF-related genes than cluster 6. For cluster 10, with the lowest quantity among the three clusters (Supplementary Fig. S2D), these cells expressed both markers for iCAFs and myCAFs (Fig. 2D and E), and their expression patterns were positioned in the middle of clusters 5 and 6 (Supplementary Fig. S2F), which suggested a transitional and transient status of cluster 10. In addition, pseudotime analysis of fibroblasts also confirmed the transitional status of cluster 10 (Fig. 2F and G). We also noticed that both clusters 5 and 6 showed two trajectories in pseudotime analysis (Fig. 2G), which could be the result of different lineages of cells in the clusters. CD105, recently identified as one of the critical markers for different lineages of CAFs (13), did not distinguish the two trajectories of CAFs (Supplementary Fig. S2G), which indicated that other lineage-determining molecules need to be identified.
High-level expression of MHC-I and CXCL molecules linked CAFs with CD8+ T cells
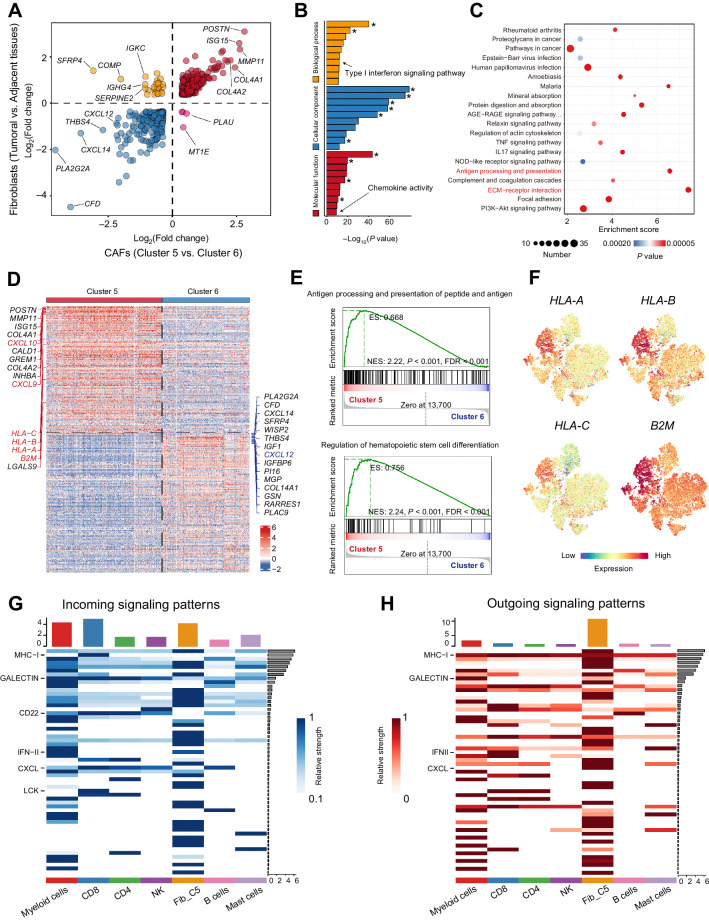
To further investigate and identify the differences between CAFs and NFs, we compared the expression profiles of CAFs and NFs from adjacent normal tissues. Regarding the multiple clusters of fibroblasts mentioned above, we first took CAFs and NFs as wholes and made the comparison. By comparing the expression profiles of clusters 5 and 6, we found that almost all the top DEGs from the comparison of CAFs and NFs were enriched in cluster 5 (Fig. 3A), which indicated that cluster 5 plays a determinant role in the difference between CAFs and NFs. Consistent with prior studies, CAFs had a unique expression pattern and expressed a set of tumor-promoting genes, such as POSTN. GO analysis of the set of DEGs revealed enrichment of terms related to the ECM, emphasizing that CAFs contributed to tumorigenesis via TME remodeling and interactions with cells in the TME (Fig. 3B). In addition, enrichment analysis indicated that CAFs played a role in cytokine signaling (e.g., IL17) and ECM–receptor interaction (Fig. 3C). Interestingly, we noticed that the CAFs also presented enrichment of genes related to T-cell receptor signaling and antigen processing and presentation, with enrichment scores ranked only after ECM–receptor interaction (Fig. 3C; Supplementary Fig. S3A).
Figure 3.
High expression levels of CXCLs and MHC class I molecules linked cluster 5 CAFs and CD8+ T cells. A, Plots of DEGs (fold change >1) between fibroblasts from tumoral and adjacent tissues and between clusters 5 and 6 CAFs. B and C, Gene ontology analysis (B) and GSEA (C) for expression differences between fibroblasts from tumoral and adjacent tissues. Asterisk (*), terms related to extracellular matrix or collagen. D–F, Expression difference between two CAF clusters (clusters 5 and 6). D, Heat map of top DEGs between cluster 5 and cluster 6 CAFs. E, GSEA analysis of DEGs between clusters 5 and 6 in the terms of “Antigen processing and presentation of peptide antigen via MHC class I” and “Regulation of hemataopoietic stem cell differentiation.” F, MHC class I molecules (HLA-A, HLA-B, HLA-C, and B2M) expression in the t-SNE plots of fibroblasts. G and H, Cell interaction analysis of cluster 5 CAFs and immune cells in HNSCC.
Then, we explored the two main clusters of CAFs, clusters 5 and 6, to validate the enrichment of gene sets in the previous comparison. MHC-I molecules, including HLA-A, HLA-B, HLA-C, and B2M, presented higher expression levels in cluster 5 (Fig. 3D). Gene set enrichment analysis (GSEA) further confirmed that the genes associated with antigen processing and presentation had higher expression in cluster 5 (Fig. 3E). Because previous work revealed a population of antigen-presenting CAFs in PDAC (20), we also explored CD74 and HLA-DRA (MHC-II) expressions in our scRNA-seq data but did not find any enrichment in cluster 5 (Supplementary Fig. S3B). Conversely, we confirmed the increased expression of MHC-I molecules (including B2M, HLA-A, HLA-B, and HLA-C) in cluster 5 (Fig. 3F). In addition, in the three immune types of HNSCC specimens, tumor beds had a higher expression level of MHC class I molecules, and spots with high expression of MHC class I molecules were mostly located around tumor nests in the excluded-type specimen (Supplementary Fig. S3C).
Because cluster 5 had more active interactions with immune cells and presented enrichment of the gene set for the regulation of hematopoietic cell differentiation (Fig. 3E; Supplementary Fig. S4A), we further assessed the interaction between cluster 5 CAFs and different types of immune cells in our scRNA-seq data. Myeloid cells and CD8+ T cells were the top-ranked cells in terms of the number of ligand–receptor interactions with cluster 5, although internal interactions among CAFs (clusters 5 and 6) were the most frequent (Supplementary Fig. S4A). To quantitatively analyze the interaction, we considered the expression levels of ligands and receptors and assessed incoming and outgoing signals (Fig. 3G and H). Myeloid cells and CAFs in cluster 5 exhibited a strong network of incoming and outgoing signals. For CD8+ T cells, MHC-I, galectin, CD22, LCK, and CXCL were the strongest incoming signals (Fig. 3G). In terms of outgoing signals, cluster 5 expressed high levels of MHC-I and CXCL for signaling (Fig. 3H). In terms of the CXCL interaction, the CXCL12–CXCR4/CKR3 axes that are reported to be involved in the maintenance of T cells (21) were the major contributors (Supplementary Fig. S4B). In addition, we noticed that CXCL9 and CXCL10, which were identified as critical chemokines for CD8+ T-cell recruitment in a recent study of cutaneous fibroblasts (22), were highly expressed in cluster 5 rather than cluster 6 (Fig. 3D). As expected, correlation analysis by GEPIA (23) revealed a significant positive correlation between CD8A expression and CXCL9/CXCL10/CXCL12 (Supplementary Fig. S4C), confirming the chemotactic effects of CXCL9/CXCL10/CXCL12 on CD8+ T cells. Interestingly, we noticed that CXCL9, CXCL10, and CXCL12 had special expression patterns in different immune types of HNSCC. CXCL9 and CXCL10 were abundant in desert and infiltrated specimens, and their expression was limited and concentrated or negative in the stroma of excluded specimens (Supplementary Fig. S4D and S4E). In contrast, the desert specimen had little expression of CXCL12, and the excluded specimen had a high level of CXCL12 (Supplementary Fig. S4F). The infiltrated specimen showed expression of CXCL12 in the malignant regions but had a higher level of CXCL12 in the adjacent normal regions (Supplementary Fig. S4F).
These results indicated that cluster 5 is responsible for the distinct expression profiles of CAFs and had strong and frequent interactions with CD8+ T cells mediated by MHC-I molecules and CXCL chemokines.
High expression of MHC-I in CAFs limits the antitumor function of CD8+ T cells
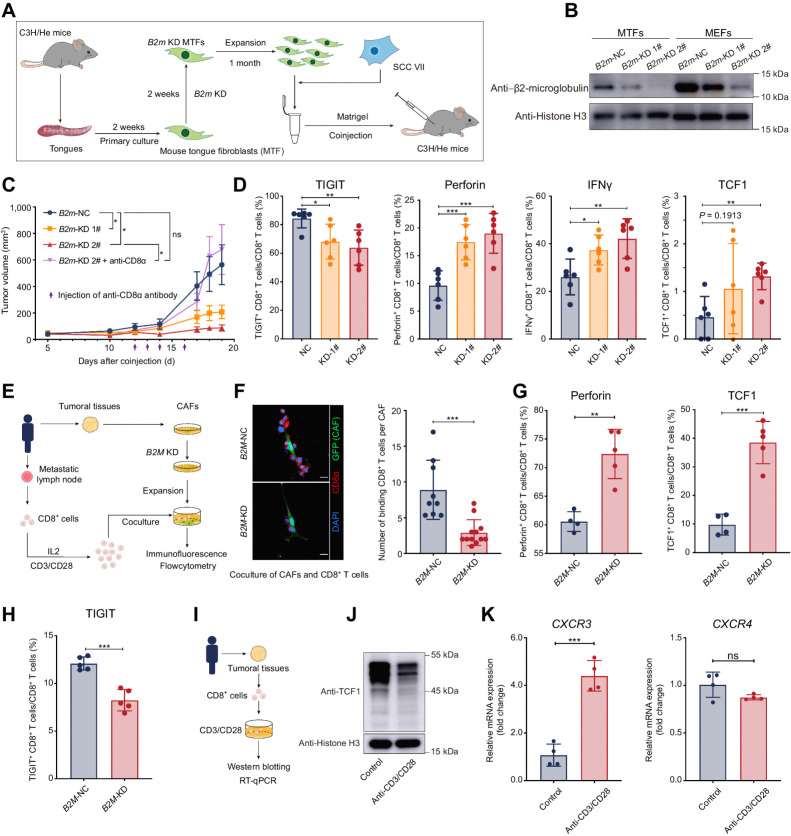
To further clarify the significance of increased expression of MHC-I molecules in CAFs, we first analyzed the contribution of MHC molecules to the survival of patients with HNSCC via the TIMER (24). In contrast with our expectation, in patients with HPV-positive HNSCC whose cumulative survival was positively correlated with immune infiltration, a higher level of MHC class I molecules (HLA-A, HLA-B, HLA-C, and B2M) was related to a worse outcome (Supplementary Fig. S5A). Then, we isolated CAFs from HNSCC tumoral tissues and knocked down their expression for B2M (Supplementary Fig. S5B–S5D), and coinjected them with HN6 in nude mice (deficient in adaptive immunity). There was no significant change in tumor growth or histological morphology (Supplementary Fig. S5E–S5G), suggesting that B2M in CAFs did not directly change the tumor growth.
To further determine whether fibroblasts with different expression levels of MHC-I influence tumor growth and antitumor immunity in vivo, mouse models with an intact immune system were applied for further investigation. With the limitation of spontaneous tumorigenic mouse models of HNSCC, subcutaneous tumor-bearing models formed by SCC VII (a squamous carcinoma cell line constantly applied in HNSCC research) and fibroblasts were used in this study. We used NFs isolated from mouse tongues (mouse tongue fibroblasts, MTFs) that contributed to CAFs as substitutes (Fig. 4A). We first knocked down the expression of B2m in MTFs (Fig. 4B) and confirmed that knocking down B2m did not influence the viability of SCC VII cells (Supplementary Data; Supplementary Fig. S5H). Then, we subcutaneously coinjected SCC VII and fibroblasts into C3H/He mice. Interestingly, compared with the control group, tumors formed by SCC VII and B2m-KD MTFs grew slower and much smaller at the endpoint and depletion of CD8+ T cells rescued the observed phenotype (Fig. 4C; Supplementary Fig. S5I). To investigate how B2m-KD fibroblasts restrict tumor growth, we isolated subcutaneous tumors for further analysis. Exhaustion markers and immune checkpoint molecules, including T-cell immunoglobulin and mucin domain containing-3 (TIM3), lymphocyte activation gene-3 (LAG3), cytotoxic lymphocyte-associated antigen-4 (CTLA4), and T-cell immunoreceptor with Ig and ITIM domains (TIGIT), were examined in CD8+ T cells from subcutaneous tumors and showed a minor decrease of TIGIT in B2m-KD tumors but did not exhibit significant changes in others (Fig. 4D; Supplementary Fig. S5J). In contrast, tumors with B2m-KD MTFs contained a larger number of CD8+ T cells that secreted antitumor molecules (perforin and IFNγ) and had a minor increase of CD8+ T cells expressing granzyme B (Fig. 4D; Supplementary Fig. S5J). We also noticed that memory-like CD8+ T cells (TCF1+CD8+ T cells) also had a slight increase in B2m-KD tumors (Fig. 4D). We also coinjected SCC VII and B2m-KD MEFs into C3H/He mice and obtained similar results (Supplementary Fig. S5K). Then, we isolated and expanded CD8+ T cells from the metastasis lymph nodes and coculture them with CAFs from the same patients (Fig. 4E). We found that the number of CAFs-binding CD8+ T cells decreased in the coculture with B2M-KD CAFs (Fig. 4F) and the expression for perforin and TCF1 also increased (Fig. 4G) when the expression for TIGIT decreased (Fig. 4H) in the B2M-KD group. Then, we activated CD8+ T cells isolated from patients with HNSCC tumors with anti-CD3/CD28 antibodies (Fig. 4I). We confirmed that TCF1 in CD8+ T cells from HNSCC tumors decreased after activation (Fig. 4J) and noticed that CXCR3 (the receptor for CXCL9 and CXCL10) was significantly increased and CXCR4 (the receptor for CXCL12) remained unchanged (Fig. 4K), indicating that CD8+ T cells became more sensitive to CXCL9 and CXCL10 signals that were highly expressing by cluster 5 CAFs (Fig. 3D). These findings indicated that CAFs with high expression of MHC-I could restrict the antitumor ability of CD8+ T cells.
Figure 4.
Higher expression of MHC class I limited the antitumor function of CD8+ T cells. A, Schematic diagram of coinjection of subcutaneous tumor models for mouse tongue fibroblasts and SCC VII. B, Western blotting of β2-microglobulin in MTFs and MEFs. C, Tumor growth of subcutaneous coinjection of SCC VII and B2m knocked-down (B2m-KD) MTFs. Purple arrows, intratumoral injection time points of the anti-CD8α antibody or saline. D, The proportion of CD8+ T-expressing cytotoxic cytokines (perforin and IFNγ), the immune checkpoint (TIGIT), and TCF1 in CD8+ T cells isolated from subcutaneous tumors formed by coinjection of SCC VII and MTFs. E, A diagrammatic sketch for coculture experiments of B2M-KD CAFs and CD8+ T cells from the same patients. F, Left, immunofluorescent results for the CD8+ T cells binding to B2M-KD CAFs. Right, the average numbers of CD8+ T cells binding to one CAF are summarized. G, The flow cytometry results of expression for perforin and TCF1 in CD8+ T cells after coculturing with CAFs. H, The flow cytometry results of expression for TIGIT on CD8+ T cells after coculturing with CAFs. I, A diagrammatic sketch for isolation of CD8+ cells from HNSCC tumoral tissues and activation of TCR signaling in CD8+ cells by anti-CD3/CD8 antibody. J, Western blotting of TCF1 in CD8+ cells isolated from HNSCC tumoral tissues after TCR signaling activation. K, The mRNA expression changes of CXCR3 and CXCR4 in CD8+ T cells isolated from HNSCC tumoral tissues treated with or without anti-CD3/CD28 antibody. Statistics are shown in mean ± SEM (C) or mean± SD (D, F–H, and K) accessed by the unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonsignificant.
The checkpoint molecule ligand galectin-9 was enriched in MHC-ihi CAFs
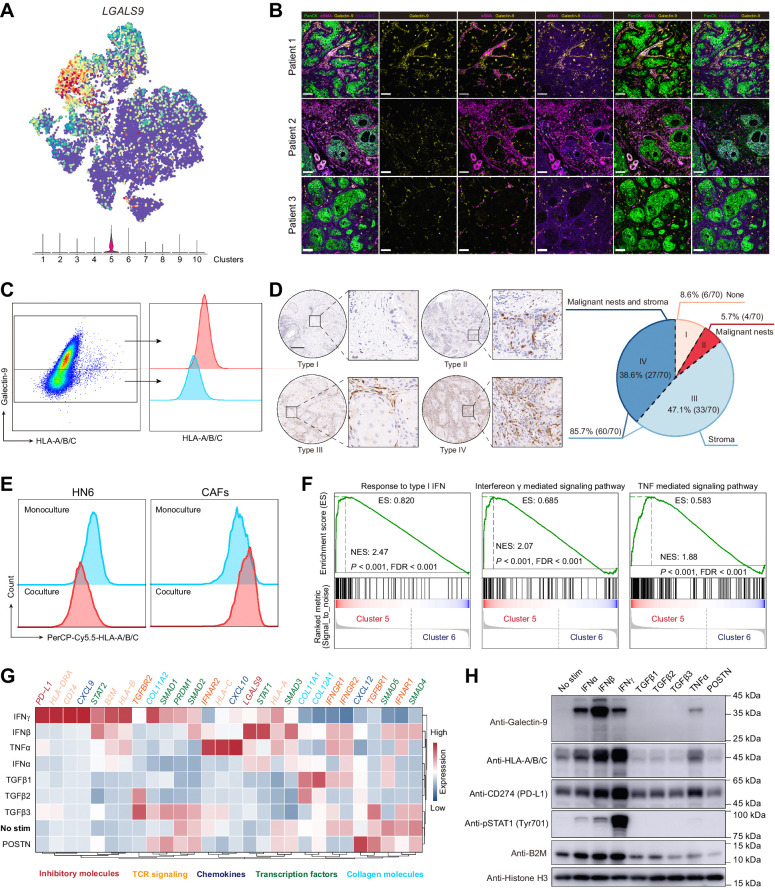
Previous work by Shields and colleagues (25) revealed an antigen-specific deletion of CD8+ T cells of CAFs with FASL and PD-L2 in an MHC-I-dependent manner. Thus, we further examined the expression of the ligand of immune checkpoints in fibroblasts from HNSCC. Interestingly, among all the checkpoint ligands, galectin-9 was the only molecule that was enriched in MHC-Ihi CAFs (cluster 5; Fig. 5A; Supplementary Fig. S6A). To validate the expression of galectin-9 in MHC-Ihi CAFs, several tumor samples of HNSCCs were harvested for flow cytometry and immunofluorescence staining. Indeed, we found that galectin-9 expressed in αSMA+ CAFs (Fig. 5B; Supplementary Fig. S6B) and CAFs expressing galectin-9 had higher levels of MHC-I molecules (Fig. 5C; Supplementary Fig. S6C). LGALS9 (the gene encoding galectin-9) was found in all three immune types of HNSCC specimens (Supplementary Fig. S6D). To further determine the distribution of galectin-9 in tumors, galectin-9 expression was examined in a larger cohort of 70 patients with HNSCC. Among these patients, we divided the expression patterns of galectin-9 into four categories (Fig. 5D): (i) Both the stromal cells and malignant islets did not express galectin-9 (6/70, 8.57%); (ii) galectin-9 only appeared in the tumor nests but not the stroma (4/70, 5.71%); (iii) galectin-9 only existed in the stroma (33/70, 47.14%); and (iv) galectin-9 could be found in both stroma and tumor nests (27/70, 38.57%). In addition, we noticed that CAFs lost the expression of galectin-9 after two weeks of culture in vitro (Supplementary Fig. S6E), and they exhibited a higher level of HLA-A/B/C on the cell surface when cultured with HN6 (an HNSCC cell line), and HLA-A/B/C on HN6 was decreased upon coculture (Fig. 5E), suggesting that the phenotype of CAFs with high expression of HMC-I and positive expression of galectin-9 (MHC-IhiGal9+ CAFs) is dependent on the signaling network in the TME.
Figure 5.
The immune checkpoint ligand molecule, galectin-9, was enriched in cluster 5 of CAFs. A, The t-SNE result of fibroblasts overlaid with LGALS9 (the gene encoding galectin-9). B, Representative immunofluorescence results of human HNSCC tumor samples stained for pan-cytokeratin (PanCK; green), αSMA (wine), galectin-9 (yellow), HLA-A/B/C (purple). Scale bar, 80 μm. C, Galectin-9 and HLA-A/B/C expressions in CAFs from human HNSCC tumor samples. The gating strategy for CAFs is presented in Supplementary Fig. S7C. D, Left, representative images of HNSCC tissue chip for different distribution patterns of galectin-9. Scale bar, 200 μm. Right, the proportions for four types of galectin-9 distribution are also summarized. E, Flow cytometry of HLA-A/B/C expression for monocultured or cocultured HN6 and CAFs. F, GSEA for cytokines-associated pathway enrichment of differentially expressed genes between clusters 5 and 6 CAFs. G and H, Gene expressions in CAFs treated with different cytokines (IFNα, IFNβ, IFNγ, TGFβ1, TGFβ2, TGFβ3, TNFα, and POSTN; the concentration of cytokines was 100 ng/mL). G, CAFs treated with cytokines for 24 hours were harvested for mRNA isolation and qRT-PCR. The data were pooled from three independent experiments and the relative expressions of targeted genes were analyzed relative to GAPDH or ACTB. H, CAFs treated with cytokines for 48 hours were harvested for Western blotting.
To subsequently investigate the contribution of the TME signaling network to MHC-IhiGal9+ CAF formation, scenic analysis was used to identify critical transcription factors in the scRNA-seq data. As expected, clusters of CAFs had different regulons with high activity scores, and MHC-IhiGal9+ CAFs exhibited high activity scores for the regulons of STAT1, PRDM1, and STAT2, which also showed high scores for regulon specificity (Supplementary Fig. S6F–S6H). In addition, in the comparison of clusters of CAFs, the GSEA results indicated that IFN and TNF signaling pathways were active in MHC-IhiGal9+ CAFs (Fig. 5F). Then, we cultured CAFs in vitro to further explore the contribution of the signaling network (Fig. 5G and H). CAFs had higher expression of galectin-9 and MHC class I molecules (HLA/B/C and B2M) with IFN and TNFα treatment (Fig. 5G and H). Specifically, IFNβ induced the highest fold change in galectin-9 expression, whereas IFNγ induced a high fold change in the expression of MHC class I molecules (Fig. 5G and H). As expected, IFN and TNF signals correlated with high expression of CXCL9 and CXCL10 when POSTN signals induced a high level of CXCL12 (Fig. 5G). We also detected the change in PD-L1 expression in CAFs treated with different cytokines. However, the change in PD-L1 expression was not completely consistent with the change in galectin-9 expression in CAFs stimulated with IFNs. Consistent with previous reports, TGFβ signaling was associated with collagen expression (COL11A1, COL11A2, and COL12A1; Fig. 5G).
We also assessed MHC class I molecules and galectin-9 expression in malignant cell lines (CAL27, HN6, and HN30) treated with IFN and TNFα. As expected, HLA-A/B/C and galectin-9 expressions also increased in all three cell lines but at different levels (Supplementary Fig. S6I and S6J). Among these cells, HN6 cells were the most sensitive cells to IFN, and their reactions to IFN and TNFα were similar to those of CAFs (Supplementary Fig. S6K).
These results indicated that galectin-9, the only immune checkpoint ligand enriched in MHC-Ihi CAFs and induced by IFN signals in the TME, was commonly expressed by CAFs across patients with HNSCC.
Gal9+ CAFs induced the dysfunctional differentiation of TCF1+GZMK+CD8+ T cells
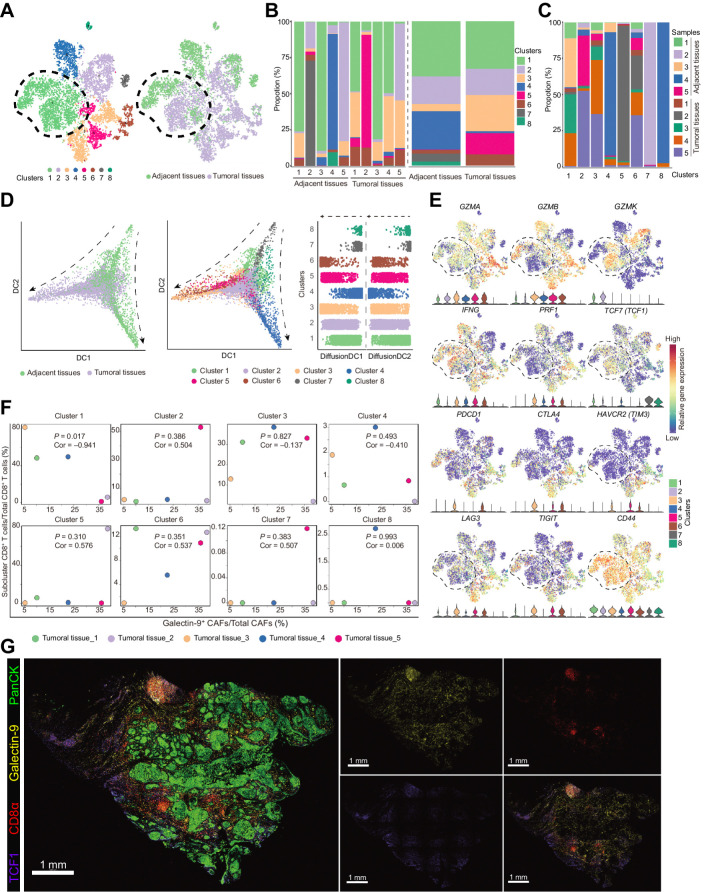
To further identify the role of Gal9+ CAFs and define the subcluster of CD8+ T cells that interacted with Gal9+ CAFs, the scRNA-seq results of CD8+ T cells from the previous five paired tumoral-adjacent normal tissues were further analyzed (Supplementary Fig. S7A and S7B). These CD8+ T cells clustered into eight groups, and clusters 1 and 2 were shared within tumoral tissues and normal tissues (Fig. 6A–C; Supplementary Fig. S7A). Tumoral tissues had more cells in clusters 3, 5, and 6 (Fig. 6B). Clusters 4, 7, and 8 were almost exclusive to normal tissues (Fig. 6C). According to pseudotime analysis, CD8+ T cells from the tumoral and adjacent tissues had different differentiation trajectories, and cluster 1 was positioned at the primary transition stage in CD8+ T cells from tumoral tissues (Fig. 6D). To define the status of each CD8+ T-cell cluster, we examined the expression of genes encoding cytotoxins (GMZA, GMZB, GMZK, IFNG, and PRF1), checkpoint molecules (CTLA4, HAVCR2, PDCD1, LAG3, and TIGIT), and TCF1 in the eight clusters (Fig. 6E). Compared with other clusters, clusters 1 and 2 of CD8+ T cells presented with high expression of GZMK and TCF1 (Fig. 6E), indicating that clusters 1 and 2 were pre-effector CD8+ T cells, as previously reported (26, 27). Clusters 3–6, expressing high levels of GZMA, GZMB, IFNG, and PRF1, were effector CD8+ T cells (Fig. 6E).
Figure 6.
TCF1+GZMK+CD8+ T cells were negatively correlated with galectin-9+ CAFs. A, t-SNE plots of single-cell RNA sequencing result for the CD8+ T cells from the previous 5 paired tumoral and adjacent normal tissues. Cells were clustered into 8 populations (left) and origins (right) are also indicated in the t-SNE plots. B, Bar plots show cluster proportions in tumoral and adjacent tissues (left) and cluster proportions (right) in each sample. C, Bar plots show sample fraction in each cluster. D, Pseudotime analysis of CD8+ T cells by “DiffusionMap” algorithm. Cells were labeled by tissue origins (left) or clusters (middle). Right, the top two major diffusion compounds distribution of cells from different clusters. Arrows, differentiation directions. E, Regression analysis of the fraction of galectin-9+ CAFs in total CAFs and the fraction of eight clusters of CD8+ T cells from the 5 tumoral tissues. F, Expression of genes defining cytotoxic functions (GZMA, GZMB, GZMK, IFNG, and PRF1) and status (TCF1, PDCD1, CTLA4, HAVCR2, LAG3, and TIGIT) of CD8+ T cells and the genes coding the receptors of galectin-9 (CD44, HAVCR2) in the 8 clusters of CD8+ T cells. G, Representative immunofluorescence results of human HNSCC tumor samples stained for PanCK (green), galectin-9 (yellow), CD8α (red), and TCF1 (purple). Scale bar, 1 mm.
To primarily define clusters that interact with Gal9+ CAFs, the five tumoral tissues were selected for analysis of the correlation between Gal9+ CAFs and the eight clusters of CD8+ T cells (Fig. 6F). We found that cluster 1 of CD8+ T cells was the only cluster that had a significant correlation with Gal9+ CAFs (P = 0.017, Cor = −0.941; Fig. 6F). These cells presented with higher levels for CXCR3/CXCR4 and receptors for galectin-9, CD44, and P4HB (Fig. 6E; Supplementary Fig. S7C). In addition, the expression of galectin-9 was also positively correlated with checkpoint molecules (TIM3, CTLA4, TIGIT, LAG3, and PDCD1), which further suggested that the interaction between Gal9+ CAFs and CD8+ T cells was related to the dysfunction of CD8+ T cells (Supplementary Fig. S7D). In addition, the similar spatial distribution of TCF1+CD8+ T cells and galectin-9 also confirmed the interaction potential of Gal9+ CAFs with TCF1+CD8+ T cells (Fig. 6G).
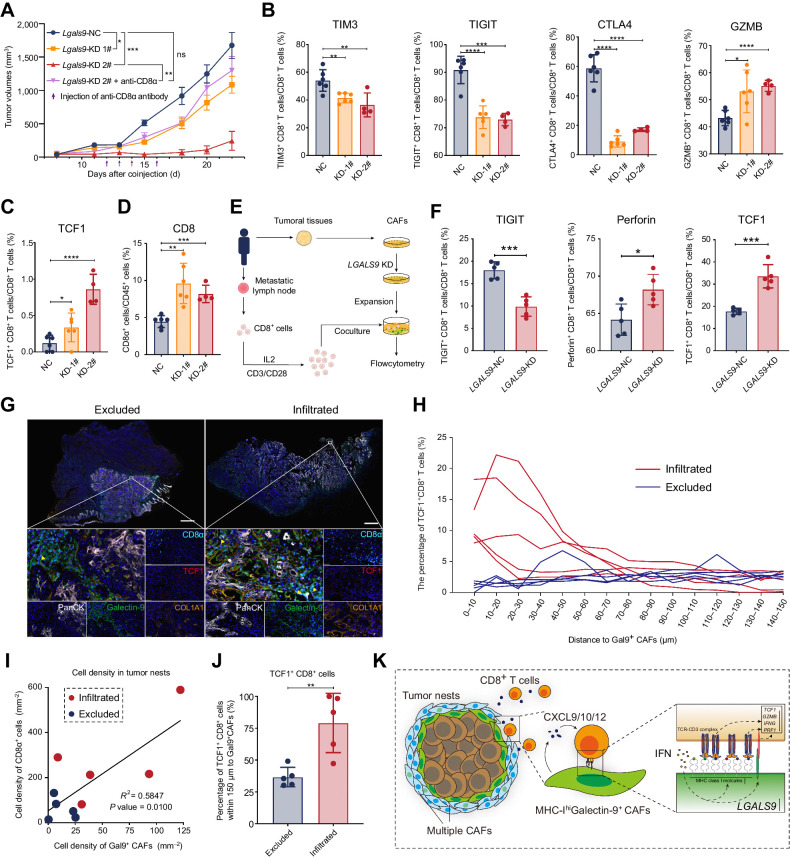
To decipher the function of galectin-9 in fibroblasts to CD8+ T cells, we conducted animal models in which we coinjected Lgals9-knocking-down MTFs with SCC VII and analyzed the change of infiltrated CD8+ T cells (Fig. 7A). Specifically, we analyzed the expression of immune checkpoint molecules (TIM3, TIGIT, and CTLA4) and GZMB (Fig. 7B). As expected, TIM3, TIGIT, and CTLA4 were downregulated in CD8+ cells with cytotoxic cytokines upregulated in the Lgals9-KD group (Fig. 7B). Also, downregulated galectin-9 also induced upregulation of the proportion of CD8+ T cells and TCF1+ CD8+ T cells in the tumors (Fig. 7C and D). Furthermore, we conducted coculture experiments of LGALS9-KD CAFs and CD8+ T cells derived from the same patients, which had similar results to the animal experiments (Fig. 7E and F). The number of binding CD8+ T cells to CAFs did not change in the LGALS9-KD group (Supplementary Fig. S8A). These results suggested that galectin-9 (coded by Lgals9) contributed to the dysfunction of CD8+ T cells.
Figure 7.
Galectin-9+ CAFs were associated with dysfunctional differentiation of TCF1+ CD8+ T cells. A, Tumor growth of subcutaneous coinjection of SCC VII and Lgals9-KD MTFs. Purple arrows, intratumoral injection time points of the anti-CD8α antibody or saline. B and C, The flow cytometry results of the proportion of CD8+ T expressing the immune checkpoint (TIM3, TIGIT, and CTLA4), the cytotoxic cytokine (granzyme B, GZMB) and TCF1, which are isolated from subcutaneous tumors formed by coinjection of SCC VII and MTFs. D, The proportion of CD8+ T cells in CD45+ cells in subcutaneous tumors formed by coinjection of SCC VII and MTFs. E, A diagrammatic sketch for coculture experiments of LGALS9-KD CAFs and CD8+ T cells from the same patients. F, The flow cytometry results of expression for TIGIT, perforin, and TCF1 in CD8+ T cells after coculturing with CAFs. G, Representative multiple immunofluorescent staining results of excluded or infiltrated HNSCC tumor samples stained for PanCK (white), galectin-9 (green), CD8α (cyan), TCF1 (red), and COL1A1 (orange). Yellow arrows, TCF1+CD8+ T cells around Gal9+ CAFs. Scale bars, respectively, indicate 2 mm in the large-scaled images and 50 μm in the magnified images. H, Distribution of the distances between TCF1+ CD8+ cells and galectin-9+ CAFs nuclei for 5 pairs of excluded and infiltrated HNSCC tumors. I, The percentage of TCF1+ CD8+ cells within 150 μm to galectin-9+ CAFs in the excluded and infiltrated HNSCC tumors. J, Correlation between cell density of CD8α+ cells and galectin-9+ CAFs in the 10 HNSCC tumors. K, Illustration of mechanisms for restriction of TCF1+GZMK+CD8+ T cells by MHC-IhiGal9+ CAFs. Statistics are shown in mean ± SEM (A) or mean ± SD (B–D, F, and J) accessed by unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Then, we quantitatively evaluated the spatial relationships between Gal9+ CAFs and TCF1+CD8+ T cells in multiplexed immunofluorescence staining of the three immune types of HNSCC specimens (Fig. 7G–J; Supplementary Fig. S8B) and conducted another multiple IHC analysis for the location correlation between CD8+ T cells and Gal9+ CAFs in FFPE of excluded and infiltrated tumors from a new cohort of patients (n = 2*5; Fig. 7G; Supplementary Fig. S8C). Infiltrated and excluded tumors had higher proportions of Gal9+ CAFs (Supplementary Fig. S8D, left), and these cells were specifically located around tumor nests (PanCK+ regions; Supplementary Fig. S8E). The percentage of TCF1+CD8+ T cells (CD3ε+CD8α+TCF1+ cells) in CD8+ T cells and the ratio of CD8+ T cells to Gal9+ CAFs were higher in the infiltrated and excluded types (Supplementary Fig. S8D, middle and right). And accordingly, in the tumor nests, galectin-9+ CAFs that had a higher level of MHC-I as previously demonstrated in our data positively correlated with a higher density of CD8+ cells (Fig. 7I). The distance between TCF1+CD8+ T cells and Gal9+ CAFs was short, and TCF1+CD8+ T cells preferred Gal9+ CAFs in the infiltrated type (Fig. 7H and J). Along with the higher CXCR3/CXCR4 expression in TCF1+ CD8+ T cells (Supplementary Fig. S7C) and the higher CXCL9/CXCL10 expression in Gal9+ CAFs (Fig. 3D), these results further validated the preference of TCF1+ CD8+ T cells for Gal9+ CAFs in spatial distribution.
All the results taken together suggest that Gal9+ CAFs that have high levels of MHC class I molecules trap and interact with pre-effector CD8+ T cells and correlate with the dysfunctional differentiation of these CD8+ T cells (Fig. 7K).
Discussion
Prior studies have demonstrated that CAFs are involved in the regulation of CD8+ T-cell exhaustion and exclusion from tumor nests via the secretion of cytokines (e.g., TGFβ) and collagen (6, 10). However, the contradictory effects of the depletion of CAFs in different types of malignancies suggest the heterogeneity of CAFs and the binary functions of the dense collagen matrix that limits both T-cell infiltration and tumor growth (10, 12). Thus, enhancing antitumor immunity requires further understanding and precise modulation of the function of CAF subpopulations, and the optimized targets of subpopulations and characteristics of CAFs in HNSCCs remain elusive. Here, we identified a cluster of CAFs expressing high levels of CXCL9/CXCL10/CXCL12 and MHC class I molecules that restricted the infiltration of pre-effector CD8+ T cells into the stroma and presented high galectin-9 expression, contributing to CD8+ T-cell dysfunction. Rather than restricting the infiltration of CD8+ T cells via dense collagen networks, this mechanism emphasizes the chemotactic relationship and direct interaction between CAFs and CD8+ T cells, and it provides a new clue for exploring CD8+ T-cell–based immunotherapy.
Multiple studies have illustrated the correlation between the infiltration of CD8+ T cells and the prognosis of patients with HNSCC, and a few of them further investigated the factors influencing the spatial distribution of CD8+ T cells in tumoral tissues. Although previous studies highlighted that the dense ECM supported by CAFs facilitates the exclusion of CD8+ T cells (10, 28), the mechanisms by which these CD8+ T cells remain in the stroma and become dysfunctional remain unclear.
With IHC staining of CD8α, spatial transcriptome analysis of the three immune types (desert, infiltrated, and excluded) of HNSCC specimens and multiplex immunofluorescence staining, we identified unique stromal regions, especially the stroma that surrounded tumor nests in the excluded specimens, which trapped CD8+ T cells to prevent infiltration of the tumor region. Then, through scRNA-seq of tumoral and adjacent tissues from patients with HNSCCs, we divided fibroblasts into 10 clusters in which CAFs clustered into three subpopulations. Two clusters of these CAFs could be aligned to myCAFs and iCAFs, as illustrated in previous studies (29, 30), and the other small cluster had markers for both myCAFs and iCAFs. We found that the myCAF-like subset of CAFs accounted for the major differences between the fibroblasts of tumoral and adjacent tissues. This myCAF-like cell subset also represented the cells that surrounded the tumor nests, as indicated in the spatial transcriptome results, and these cells limited CD8+ T-cell infiltration into tumor nests. Consistently, a recent study highlighted that myCAFs impede CD8+ T-cell function (31), but the concrete mechanisms remain elusive. Of note, this cluster of CAFs expressed a high level of MHC class I molecules and secreted CXCL9/10/12, which are related to CD8+ T-cell chemotaxis. The secretion of CXCL9/10/12 might contribute to the increased number of CD8+ T cells in the tumor tissues (22, 28), although it seems that these CD8+ T cells are mostly present in the stroma. A recent study found that increased MHC-I and PD-L1 levels in malignant cells promoted lymph node metastasis by evading NK cell recognition and suppressing T cells. In contrast, in primary HNSCC, MHC-I molecules were increased in CAFs rather than malignant cells. We found that the increased level of MHC-I in CAFs protected malignant islets from CD8+ T cells and limited the antitumor function of CD8+ T cells. The current model of MHC–peptide–TCR complex formation indicated that low-affinity peptides, mostly self-peptide molecules, are also involved in TCR interactions and pave the way for peptides with high affinity, suggesting that high expression of MHC class I molecules in CAFs prolongs the maintenance period of CD8+ T cells even when presenting with low-affinity peptides (32). Consistently, it has also been shown that immune-infiltrated tumors have higher levels of MHC-I in tumor nests, whereas immune-excluded tumors have higher expression of MHC-I in the stroma (33). A previous study found that CD8+ T cells could be exhausted by PD-L2/FASL in an antigen-specific way (25). However, in the MHC-Ihi CAFs or other clusters of CAFs in HNSCC, we did not find any enrichment, if expressed, for PD-L2 or FASL. In contrast, galectin-9, the ligand of TIM3 on CD8+ T cells, was significantly enriched in the CAF cluster expressing a high level of MHC class I molecules (MHC-IhiGal9+ CAFs). Furthermore, with paraffin slides of a cohort of 70 patients with HNSCCs, we confirmed the universality of galectin-9 expression in the stroma. CAFs highly expressed galectin-9 in more than 80% of patients with HNSCC, suggesting the potential benefit of agents targeting galectin-9 for immune regulation in patients with HNSCCs.
CAFs have multiple cell-type origins, and the lineages of CAFs have a remarkable influence on their effect on antitumor immunity and tumor growth (13, 34). In this study, we identified that MHC-IhiGal9+ CAFs have multiple cell origins and that maintenance of the phenotype (high expression of MHC class I molecules and galectin-9) is highly dependent on cytokine signals in the TME. Although TGFβ signaling is important for the formation of myCAF-like patterns (29), TGFβ had a minimal effect or inhibitory effect on galectin-9 and MHC-I expression, whereas IFN seemed to induce galectin-9 and MHC-I expression in most cases. These results support that CAFs interact with a subset of CD8+ T cells that have high expression of IFNG.
Here, we found two clusters that were both positive for TCF1 and GZMK and negative for PDCD1 and CTLA4. In tumoral tissues, the proportion of Gal9+ CAFs, which also exhibited a higher level of MHC-I, was negatively correlated with the proportion of TCF1+GZMK+CD8+ T cells that had a higher expression level of IFNG and GZMB. Of note, clonal exhausted-like GZMK+ CD8+ T cells that lack TCF1 also accumulate in tissues with aging and they introduce an inflammatory phenotype to fibroblasts via GZMK (35). The CD8+ T cells that were positive for GZMK but negative for PDCD1 and CTLA4 were also the cells that did not respond to presurgery immune checkpoint blockade treatment (anti–PD-1/CTLA4; ref. 36). Thus, patients, especially aged patients, might further benefit from introducing and maintaining TCF1 in GZMK+CD8+ T cells. Considering the expression of cytotoxic molecules (GZMA, GZMB, GZMK, and IFNG) and the potential transformation of TCF1+GZMK+CD8+ T cells into effector cells, we hypothesized that treatments stimulating the proliferation and effector transformation of these CD8+ T cells would enhance antitumor immunity.
In addition, with the integration of spatial transcriptomic and tissue cytometry of single-cell spatial resolution, we revealed quantitative spatial distributions of Gal9+ CAFs and TCF1+CD8+ T cells in three immune types of HNSCC tumoral tissues, which indicated their potential interactions. The negative correlation between Gal9+ CAFs and pre-effector CD8+ T cells, which are negative for TIM3, further suggested that other receptors on pre-effector CD8+ T cells could interact with galectin-9 and limit the proliferation or activation of CD8+ T cells. In contrast, CD44, another ligand for galectin-9, presented a relatively high level of expression in these TCF1+GZMK+CD8+ T cells. CD44 on CD4+ T cells interacts with galectin-9 to strengthen the TGFβ signal in a Smad3-dependent manner to enforce iTreg cell differentiation and maintenance (37). Although it is known that TGFβ signaling inhibits the effector transformation and proliferation of CD8+ T cells (38, 39), the interaction effect of galectin-9 and CD44 on CD8+ T cells remains unknown and requires further investigation. These receptors could be targets for CD8+ T-cell–based immunotherapy.
Overall, our study identified a cluster of CAFs that traps TCF1+GZMK+CD8+ T cells in a CXCL- and MHC-I–dependent manner and further induces dysfunctional transformation via galectin-9. Our work provided new insights into the factors influencing the distribution and dysfunction of CD8+ T cells and has the potential to be applied to promote the efficacy of immunotherapy.
Supplementary Material
This file includes Figure S1-S8 and their figure legends.
Acknowledgments
This study was supported by grants from the Project of National Natural Science Foundation of China (grant no. 82173148 and 81972573 to J. Zhang; 82272983 to H. Ma; 82273267 and 82073036 to H. Guo; 32000552 to X. Wang), The Innovative Research Team of High-level Local Universities in Shanghai (SHSMU-ZLCX20212300 and SHSMU-ZLCX20212301 to Z. Zhang), Shanghai Clinical Research Center for Oral Diseases (grant/award no. 19MC1910600 to Z. Zhang), Shanghai Municipal Key Clinical Specialty (grant/award no. shslczdzk01601 to Z. Zhang), Shanghai's Top Priority Research Center (grant/award no. 2022ZZ01017 to Z. Zhang), CAMS Innovation Fund for Medical Sciences CIFMS (grant/award no. 2019-I2M-5–037 to Z. Zhang). We appreciate excellent imaging technical assistance from Dr. J. Wang and Jin Wang from Imaging Core at Shanghai Institute of Immunology, Shanghai Jiao Tong University School of Medicine, and equipment supports of Imaging Core at Shanghai Institute of Immunology. We would like to thank OE Biotech Co., Ltd. (Shanghai, China) for providing single-cell RNA sequencing, spatial transcriptome sequencing, and assistance with data analysis, and Dr. Y. Ba for assistance with bioinformatics analysis. We also thank TissueGnostics Asia Pacific limited (Beijing, China) and Alpha X (Beijing) Biotech Co., Ltd. for their technical support for multiplexed immunofluorescence staining, image scanning, and analysis, and the help of Xiaojing Liu, the technical engineer from TissueGnostics Asia Pacific limited (Beijing, China).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
C. Li: Conceptualization, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. H. Guo: Data curation, formal analysis, funding acquisition, investigation. P. Zhai: Data curation, validation, investigation. M. Yan: Conceptualization, investigation. C. Liu: Formal analysis, validation. X. Wang: Funding acquisition, methodology. C. Shi: Methodology. J. Li: Conceptualization, methodology. T. Tong: Investigation. Z. Zhang: Conceptualization, supervision, funding acquisition. H. Ma: Conceptualization, supervision, funding acquisition, writing–review and editing. J. Zhang: Conceptualization, supervision, funding acquisition.
References
- 1. Carlisle JW, Steuer CE, Owonikoko TK, Saba NF. An update on the immune landscape in lung and head and neck cancers. CA Cancer J Clin 2020;70:505–17. [DOI] [PubMed] [Google Scholar]
- 2. Ruffin AT, Li H, Vujanovic L, Zandberg DP, Ferris RL, Bruno TC. Improving head and neck cancer therapies by immunomodulation of the tumour microenvironment. Nat Rev Cancer 2023;23:173–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rafiq S, Hackett CS, Brentjens RJ. Engineering strategies to overcome the current roadblocks in CAR T-cell therapy. Nat Rev Clin Oncol 2020;17:147–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Park J, Hsueh PC, Li Z, Ho PC. Microenvironment-driven metabolic adaptations guiding CD8+ T-cell antitumor immunity. Immunity 2023;56:32–42. [DOI] [PubMed] [Google Scholar]
- 5. Davidson S, Coles M, Thomas T, Kollias G, Ludewig B, Turley S, et al. Fibroblasts as immune regulators in infection, inflammation, and cancer. Nat Rev Immunol 2021;21:704–17. [DOI] [PubMed] [Google Scholar]
- 6. Lavie D, Ben-Shmuel A, Erez N, Scherz-Shouval R. Cancer-associated fibroblasts in the single-cell era. Nat Cancer 2022;3:793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu G, Cheng P, Pan J, Wang S, Ding Q, Jiang Z, et al. An IL6-adenosine positive feedback loop between CD73+ γδTregs and CAFs promotes tumor progression in human breast cancer. Cancer Immunol Res 2020;8:1273–86. [DOI] [PubMed] [Google Scholar]
- 8. Kumar V, Donthireddy L, Marvel D, Condamine T, Wang F, Lavilla-Alonso S, et al. Cancer-associated fibroblasts neutralize the antitumor effect of CSF1 receptor blockade by inducing PMN-MDSC infiltration of tumors. Cancer Cell 2017;32:654–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luo H, Xia X, Huang LB, An H, Cao M, Kim GD, et al. Pan-cancer single-cell analysis reveals the heterogeneity and plasticity of cancer-associated fibroblasts in the tumor microenvironment. Nat Commun 2022;13:6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grout JA, Sirven P, Leader AM, Maskey S, Hector E, Puisieux I, et al. Spatial positioning and matrix programs of cancer-associated fibroblasts promote T-cell exclusion in human lung tumors. Cancer Discov 2022;12:2606–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Özdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell 2014;25:719–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell 2014;25:735–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hutton C, Heider F, Blanco-Gomez A, Banyard A, Kononov A, Zhang X, et al. Single-cell analysis defines a pancreatic fibroblast lineage that supports antitumor immunity. Cancer Cell 2021;39:1227–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schalck A, Sakellariou-Thompson D, Forget MA, Sei E, Hughes TG, Reuben A, et al. Single-cell sequencing reveals trajectory of tumor-infiltrating lymphocyte states in pancreatic cancer. Cancer Discov 2022;12:2330–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eberhardt CS, Kissick HT, Patel MR, Cardenas MA, Prokhnevska N, Obeng RC, et al. Functional HPV-specific PD-1+ stem-like CD8 T cells in head and neck cancer. Nature 2021;597:279–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Siddiqui I, Schaeuble K, Chennupati V, Fuertes Marraco SA, Calderon-Copete S, Pais Ferreira D, et al. Intratumoral Tcf1+PD-1+CD8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 2019;50:195–211. [DOI] [PubMed] [Google Scholar]
- 17. Chen Z, Zhou L, Liu L, Hou Y, Xiong M, Yang Y, et al. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat Commun 2020;11:5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Costa A, Kieffer Y, Scholer-Dahirel A, Pelon F, Bourachot B, Cardon M, et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell 2018;33:463–79. [DOI] [PubMed] [Google Scholar]
- 19. Dominguez CX, Müller S, Keerthivasan S, Koeppen H, Hung J, Gierke S, et al. Single-cell RNA sequencing reveals stromal evolution into LRRC15+ myofibroblasts as a determinant of patient response to cancer immunotherapy. Cancer Discov 2020;10:232–53. [DOI] [PubMed] [Google Scholar]
- 20. Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov 2019;9:1102–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pitt LA, Tikhonova AN, Hu H, Trimarchi T, King B, Gong Y, et al. CXCL12-producing vascular endothelial niches control acute T-cell leukemia maintenance. Cancer Cell 2015;27:755–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu Z, Chen D, Hu Y, Jiang K, Huang H, Du Y, et al. Anatomically distinct fibroblast subsets determine skin autoimmune patterns. Nature 2022;601:118–24. [DOI] [PubMed] [Google Scholar]
- 23. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 2017;77:e108–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lakins MA, Ghorani E, Munir H, Martins CP, Shields JD. Cancer-associated fibroblasts induce antigen-specific deletion of CD8+ T cells to protect tumour cells. Nat Commun 2018;9:948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bassez A, Vos H, Van Dyck L, Floris G, Arijs I, Desmedt C, et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat Med 2021;27:820–32. [DOI] [PubMed] [Google Scholar]
- 27. Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 2018;564:268–72. [DOI] [PubMed] [Google Scholar]
- 28. Hornburg M, Desbois M, Lu S, Guan Y, Lo AA, Kaufman S, et al. Single-cell dissection of cellular components and interactions shaping the tumor immune phenotypes in ovarian cancer. Cancer Cell 2021;39:928–44. [DOI] [PubMed] [Google Scholar]
- 29. Biffi G, Oni TE, Spielman B, Hao Y, Elyada E, Park Y, et al. IL1-induced JAK/STAT signaling is antagonized by TGFβ to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov 2019;9:282–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 2017;171:1611–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Krishnamurty AT, Shyer JA, Thai M, Gandham V, Buechler MB, Yang YA, et al. LRRC15+ myofibroblasts dictate the stromal setpoint to suppress tumour immunity. Nature 2022;611:148–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van der Merwe PA, Dushek O. Mechanisms for T-cell receptor triggering. Nat Rev Immunol 2011;11:47–55. [DOI] [PubMed] [Google Scholar]
- 33. Desbois M, Udyavar AR, Ryner L, Kozlowski C, Guan Y, Dürrbaum M, et al. Integrated digital pathology and transcriptome analysis identifies molecular mediators of T-cell exclusion in ovarian cancer. Nat Commun 2020;11:5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer 2020;20:174–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mogilenko DA, Shpynov O, Andhey PS, Arthur L, Swain A, Esaulova E, et al. Comprehensive profiling of an aging immune system reveals clonal GZMK+ CD8+ T cells as conserved hallmark of inflammaging. Immunity 2021;54:99–115. [DOI] [PubMed] [Google Scholar]
- 36. Luoma AM, Suo S, Wang Y, Gunasti L, Porter CBM, Nabilsi N, et al. Tissue-resident memory and circulating T cells are early responders to pre-surgical cancer immunotherapy. Cell 2022;185:2918–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu C, Thalhamer T, Franca RF, Xiao S, Wang C, Hotta C, et al. Galectin-9–CD44 interaction enhances stability and function of adaptive regulatory T cells. Immunity 2014;41:270–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gorelik L, Flavell RA. Transforming growth factor-beta in T-cell biology. Nat Rev Immunol 2002;2:46–53. [DOI] [PubMed] [Google Scholar]
- 39. Thomas DA, Massagué J. TGF-beta directly targets cytotoxic T-cell functions during tumor evasion of immune surveillance. Cancer Cell 2005;8:369–80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file includes Figure S1-S8 and their figure legends.
Data Availability Statement
The data analyzed in this study were obtained from Tumor Immune Estimation Resource 2.0 (TIMER 2.0) at http://timer.cistrome.org/. The Spatial and single-cell transcriptomics data generated in this study are publicly available in the Genome Sequence Archive, HRA005551 (https://bigd.big.ac.cn/gsa-human/browse/HRA005551) and HRA0005576 (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA005576) and nonprofit research will be approved for access for these data. All other raw data are available upon request from the corresponding author.