Ring sideroblast isolation combined with state-of-the-art multiomics identifies survival mechanisms underlying SF3B1-mutant erythropoiesis and establishes an active role for erythroid differentiation and ring sideroblasts themselves in SF3B1-mutant myelodysplastic syndrome pathogenesis.
Abstract
Myelodysplastic syndromes with ring sideroblasts (MDS-RS) commonly develop from hematopoietic stem cells (HSC) bearing mutations in the splicing factor SF3B1 (SF3B1mt). Direct studies into MDS-RS pathobiology have been limited by a lack of model systems that fully recapitulate erythroid biology and RS development and the inability to isolate viable human RS. Here, we combined successful direct RS isolation from patient samples, high-throughput multiomics analysis of cells encompassing the SF3B1mt stem-erythroid continuum, and functional assays to investigate the impact of SF3B1mt on erythropoiesis and RS accumulation. The isolated RS differentiated, egressed into the blood, escaped traditional nonsense-mediated decay (NMD) mechanisms, and leveraged stress-survival pathways that hinder wild-type hematopoiesis through pathogenic GDF15 overexpression. Importantly, RS constituted a contaminant of magnetically enriched CD34+ cells, skewing bulk transcriptomic data. Mis-splicing in SF3B1mt cells was intensified by erythroid differentiation through accelerated RNA splicing and decreased NMD activity, and SF3B1mt led to truncations in several MDS-implicated genes. Finally, RNA mis-splicing induced an uncoupling of RNA and protein expression, leading to critical abnormalities in proapoptotic p53 pathway genes. Overall, this characterization of erythropoiesis in SF3B1mt RS provides a resource for studying MDS-RS and uncovers insights into the unexpectedly active biology of the “dead-end” RS.
Significance:
Ring sideroblast isolation combined with state-of-the-art multiomics identifies survival mechanisms underlying SF3B1-mutant erythropoiesis and establishes an active role for erythroid differentiation and ring sideroblasts themselves in SF3B1-mutant myelodysplastic syndrome pathogenesis.
Graphical Abstract
Introduction
Myelodysplastic syndromes (MDS) are clonal myeloid malignancies in which one or multiple branches of hematopoiesis are disrupted due to ineffective differentiation of hematopoietic stem and progenitor cells (HSPC), generally resulting in cytopenia (1). MDSs with ring sideroblasts (MDS-RS) are a slowly progressing low-risk MDS subgroup comprising 16% to 24% of all MDS (2). MDS-RS primarily affect the erythroid lineage and are characterized by extensive dyserythropoiesis and perinuclear accumulation of mitochondria loaded with aberrant ferritin–iron complexes, reflected as the RS phenotype (3).
Hematopoietic stem cell-borne mutations in the SF3B1 gene (pre-mRNA-splicing factor 3b subunit 1) are the primary disease driver in more than 80% of MDS-RS cases (4, 5). Moreover, 80% of SF3B1mt patients with MDS-RS display none or few comutations, confirming SF3B1mt as the disease-driving molecular event (6). SF3B1 mutations cause extensive RNA mis-splicing (7, 8), leading to increased nonsense-mediated decay (NMD) of mis-spliced transcripts. Mis-splicing and downstream NMD of ABCB7 and PPOX transcripts in particular is mechanistically associated with RS development by disrupting iron processing (9, 10), which cascades into redox imbalance, obstructed differentiation, and increased apoptosis of erythroid precursors (11, 12).
Despite alternative splicing (AS) and NMD supposedly reducing the survivability of SF3B1mt cells (particularly erythroid cells), the MDS-RS bone marrow (BM) is hyperplastic and usually displays increased erythropoiesis with abundant RS, which often comprise 50% or more of all erythroid cells. The molecular mechanisms that enable RS survival, expansion, and accumulation in the MDS-RS BM, therefore remain unclear.
Exploring how SF3B1mt affects erythropoiesis has been pursued through diverse model systems (5, 13–16), each of which is unable to recapitulate full BM/erythroid biology and/or RS development. Crucially, the isolation of viable human RS has not yet been achieved, complicating direct studies into MDS-RS pathobiology.
By exploiting the accumulation of ferric iron inherent to the RS phenotype, we here isolate RS and investigate the entire course of SF3B1mt erythropoiesis through an integrative multiomic approach. We identify several molecular pathways engaged to minimize the consequences of widespread mis-splicing and extreme oxidative stress, promoting RS survival and accumulation. Finally, we identify increased RNA mis-splicing, decreased NMD, and transcriptome–proteome uncoupling during erythroid differentiation, which altogether demonstrate a role for SF3B1mt erythropoiesis itself in driving disease pathogenesis.
Materials and Methods
Study design, sample collection, and ethical approval
BM and/or peripheral blood (PB) samples were collected from 36 patients with MDS-RS and 3 patients with MDS non-RS evaluated at Karolinska University Hospital, Huddinge, Sweden. Diagnostic procedures were performed according to the European LeukemiaNet recommendation and WHO classification for myeloid neoplasms (17, 18). As the specific purpose was to dissect the pathobiology of SF3B1mt MDS-RS, all patients with MDS-RS belonged to the SF3B1α category (6). RS presence was quantified according to standard clinical practice. Additional samples were collected from a total of 40 healthy normal BM (NBM) donors for control purposes. A deidentified donor and experiment index including clinical and mutational status (5) is provided in Supplementary Data S1. All source material was provided with written informed consent for research use, given in accordance with the Declaration of Helsinki, and the study was approved by the Ethics Research Committee at Karolinska Institutet (2010/427–31/3, 2017/1090–31/4).
BM/PB sample processing and density gradient separation
Samples were separated by Lymphoprep (STEMCELL Technologies) density gradient centrifugation at 400 × g for 30 minutes, room temperature, to derive a mononuclear cell layer (MNC) and an erythrocyte-rich high-density layer (HD). MNC were cryopreserved in 40% RPMI1640 Glutamax (Thermo Fisher Scientific), 50% inactivated FBS (Thermo Fisher Scientific) and 10% dimethyl sulfoxide (DMSO; Sigma-Aldrich). Cryopreserved MNC were thawed in RPMI1640 Glutamax + 20% FBS + 100 U/mL DNase I (Sigma-Aldrich). HD cells were washed with PBSAG [PBS (Sigma-Aldrich) + 1 mg/mL BSA (Sigma-Aldrich) + 2 mg/mL glucose (Sigma-Aldrich)] and stored at 4°C for a maximum of 1 week.
RS isolation and antibody-mediated magnetic-activated cell sorting
For RS isolation, packed HD cells were diluted 1:10 with auto-magnetic-activated cell sorting (MACS) Running Buffer (MACS buffer; Miltenyi Biotec) at 4°C and distributed at 5 mL per LS column (Miltenyi Biotec), or 5 × 106 BM MNC were thawed, resuspended in 5 mL MACS buffer, and distributed in one LS column. LS columns were washed with 20 mL MACS buffer and eluted with 5 mL MACS buffer. For antibody-mediated separation, the manufacturer's protocol (Miltenyi Biotec) was followed for separation using CD34, CD235a/GPA, or CD71 MicroBeads.
Flow cytometry analysis and FACS
Cells were analyzed on a CytoFlex S (Beckman Coulter) or analyzed and sorted on a FACS ARIA II Fusion (Becton Dickinson) at the MedH FACS facility of Karolinska Institutet. All steps were performed in FACS buffer (PBS + 2% FBS + 1 mmol/L EDTA) kept at 4°C. All experiments included fluorescent-minus-one (FMO) and single-stained controls. Antibodies/fluorescent reagents utilized are listed in Supplementary Table S1, including the respective RRIDs. Data were analyzed using FlowJo v. 10.7.2 (Becton Dickinson, RRID: SCR_008520).
Morphologic evaluation and microscopy analysis
Cells were cytospun, fixed in methanol for 15 minutes at room temperature, air-dried, and submitted to Karolinska University Hospital for iron staining. Brightfield micrographs were acquired using a Pannoramic MIDI II slide scanner (3D Histech) at 40× with a Hitachi HV-F22 3CCD SXGA camera (Hitachi Kokusai Electric) using Pannoramic Scanner v. 1.17 (3D Histech). Image analysis was performed using QuPath v. 0.2.0m9 (19) and Fiji v. 2.3.0/1.53f51 (20). Fixed-cell immunofluorescence is detailed in Supplementary Materials and Methods.
Droplet digital PCR
Droplet digital PCR (ddPCR) was performed with probes for the detection of SF3B1 mutations K700E and K666N (Bio-Rad) as described previously (5). QuantaSoft analysis software v.1.7.4 (Bio-Rad) was used to calculate variant allele frequencies (VAF) based on the Poisson distribution. At least one known mutated sample, one wild-type sample, and one H2O sample were included as controls in every run.
Bulk RNA sequencing
CD34+ HSPC samples, mixed GPA+ erythroblast samples, and CD71+ PB reticulocyte samples (RetPB) were isolated through MACS. RS and siderocytes were obtained through MACS+FACS. Cells were lysed in RLT (Qiagen) + 40 mmol/L dithiothreitol (Sigma-Aldrich) and RNA extraction was performed with RNeasy Micro Kit (Qiagen) with RNase-free DNase treatment according to the manufacturer's protocol. RNA integrity numbers (RIN) were estimated using Agilent RNA 6000 Pico Kits (Agilent Technologies). A minimum RIN value of 6.5 was considered adequate. Additional details are provided in Supplementary Materials and Methods. The count matrix and sample metadata are provided in Supplementary Data S2.
Single-cell RNA-seq
Four experiments were conducted: two 10x Genomics experiments (5 SF3B1mt MDS-RS patients and 3 NBM donors), one Smart-seq3 (21) experiment (2 MDS-RS donors) and one Smart-seq3xpress (22) experiment (1 MDS-RS donor). Additional details on sample preparation, sequencing, and downstream analysis are provided in Supplementary Materials and Methods. A complete experimental design for the 10x experiments is provided in Supplementary Fig. S1.
In vitro cell culture and GDF15 measurement
Erythroid culture was initiated by thawing and seeding BM MNCs in polyurethane scaffolds as described previously (14). GDF15 levels were measured in both neat (1:1) and diluted (1:10) culture supernatant using a Human GDF15 ELISA Kit (LSBio), based on a standard curve of known GDF15 concentrations.
CD34+ colony-forming unit assay
CD34+ HSPCs from 3 NBM donors and 5 SF3B1mt patients with MDS-RS were treated with RPMI plus 100 ng/mL recombinant GDF15 (NBP2–76204–20ug; Novus Biologicals) or 0.5% v/v sterile H2O for 1 hour. After preincubation, GDF15 or H2O were added to a projected concentration of 100 ng/mL, cells were resuspended in MethoCult (H4434; STEMCELL Technologies) and plated in culture dishes. NBM and MDS cells were plated at 4,000 cells/dish and 40,000 cells/dish (due to differential CFU-forming ability; ref. 23), cultured as described previously (5) and counted using a Leica DM inverted microscope (Leica Microsystems).
Tandem mass tag proteomics
FACS-separated samples were snap frozen in liquid nitrogen. Cell pellets were lysed with 4% SDS lysis buffer and prepared for MS using a modified version of the SP3 protein clean up and digestion protocol (24). Additional details are provided in Supplementary Materials and Methods and Supplementary Table S2. Expression matrix (centered against a pool of all samples) files, processed files, and downstream analyses are provided in Supplementary Data S3.
Statistical methods
Statistical comparisons were performed through ANOVA, multiple comparison-corrected paired and unpaired t tests (Benjamini–Hochberg), and Fisher exact test. Correlation coefficients were calculated and tested for association through Pearson product–moment correlation. Statistical methods specific to the analysis of each high-throughput data format are detailed in their respective sections. Single-cell RNA sequencing (scRNA-seq) analysis was performed with RStudio Server v.1.3.1056 (RRID:SCR_000432) and R v.3.6.3 (RRID:SCR_001905; ref. 25). All other statistical analyses were performed with RStudio v.1.4.1767 (RRID:SCR_000432), R v.4.0.5 (RRID:SCR_001905), Excel v.2204 (RRID:SCR_016137), and GraphPad Prism v.9.4.0 (RRID:SCR_002798).
Data availability
Preprocessed and deidentified RNA-seq/proteomic data and downstream results generated in this study are available within the article and its supplementary data files. Deidentified raw data for all high-throughput datasets, the corresponding metadata, and intermediate analysis files have been deposited on the Swedish National Data Service's research data repository with restricted access in accordance with the European Union General Data Protection Regulation and are accessible upon reasonable request to the Swedish National Data Service through the respective DOI links (online data descriptions are available in both English and Swedish). Other raw data are available from the corresponding author upon request.
SND-ID 2023–118: Tandem mass tag proteomics of erythroblasts from healthy donors and erythroblasts/ring sideroblasts from patients with MDS-RS. DOI: https://doi.org/10.48723/f2xa-4005
SND-ID 2023–120: Bulk RNA sequencing of BM cells from patients with MDS-RS and healthy donors. DOI: https://doi.org/10.48723/41wa-yv42
SND-ID 2023–121: 10x single-cell RNA sequencing of BM cells from patients with MDS-RS and healthy donors. DOI: https://doi.org/10.48723/nq2a-1e03
SND-ID 2023–122: Smart-seq3 and Smart-seq3xpress single-cell RNA sequencing of BM cells from patients with MDS-RS. DOI: https://doi.org/10.48723/0f0c-p816
Results
SF3B1mt RS remain active during MDS-RS erythropoiesis
To investigate RS development during erythropoiesis, we first pursued erythroblast staging in MNC from MDS-RS and healthy BM samples through co-detection of Band 3 and integrin α4 (Fig. 1A; ref. 26). MDS-RS samples displayed a clear increase in total erythroid frequency, but with relative accumulation at early precursor stages (Pro/Basophilic) and matching depletion at the Orthochromatic stage (Fig. 1B). Morphologic analysis identified a progressive increase in RS frequency with erythroid maturation (Fig. 1C; Supplementary Figs. S2A and S2B, S3, and S4). Assessing mitotic activity showed significantly increased DNA in MDS with no change in Ki-67 expression, indicating uncoupled cell-cycle dynamics corresponding to a dysplastic phenotype (Fig. 1D). Although RS decreased with increasing Ki-67 signal, the median Ki-67 population had equal RS frequencies to diagnostic BM smears (Fig. 1E). Interestingly, these experiments also identified aberrantly decreased CD71 surface expression by MDS precursors as a potential mechanism to avoid iron overload (Fig. 1F). Finally, erythroid enucleation ability was assessed through 3D cell culture of MDS-RS cells (14). RS were found to enucleate at equal rates to normoblasts from the same patients (Fig. 1G; Supplementary Fig. S5), fully recapitulating progression through erythropoiesis.
Figure 1.
Erythroid differentiation and enucleation remain active in SF3B1mt MDS-RS erythroblasts. A, Flow cytometry strategy for staging of erythroblast populations from BM MNC of patients with MDS-RS. Gating steps identify live and terminally differentiating erythroid cells (Lin−7AAD−GPA+), from which erythroblasts (EB) are staged according to Band 3 and integrin α4 expression (26). Integrin α4–negative cells are excluded from quantification to avoid skewing by anucleate cells. B, Mean (±SEM) cell population frequencies within flow cytometry parent populations (singlets > terminally differentiating erythroid > erythroblast subsets), quantified in NBM donors (nNBM = 4) and patients with MDS-RS (nMDS-RS = 6). Erythroid cells are quantified within the singlet population. Erythroblast subsets are quantified within the GPA+ population. C, Mean (±SEM) RS frequencies per sorted erythroblast subset and compared with frequencies in matched diagnostic BM smears (nMDS-RS = 6). D, Gating and quantification [mean (±SEM)] of DNA content (Draq-5) and intracellular Ki-67 abundance in GPA+ magnetically sorted cells (nNBM = 3; nMDS-RS = 6). E, Mean (±SEM) RS frequencies per sorted Ki67-expressing subset and compared with frequencies in matched diagnostic BM smears (nMDS-RS = 6). F, Mean (±SEM) CD71 (transferrin receptor, TFRC) median fluorescence indices per erythroblast subset (nNBM = 4; nMDS-RS = 6). G, Mean (±SEM) enucleation frequencies after 28-day 3D culture of MDS-RS BM MNCs (nMDS-RS = 4) and separated by iron granule visibility upon morphological analysis. Statistical comparison was performed by paired t test analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonstatistically significant.
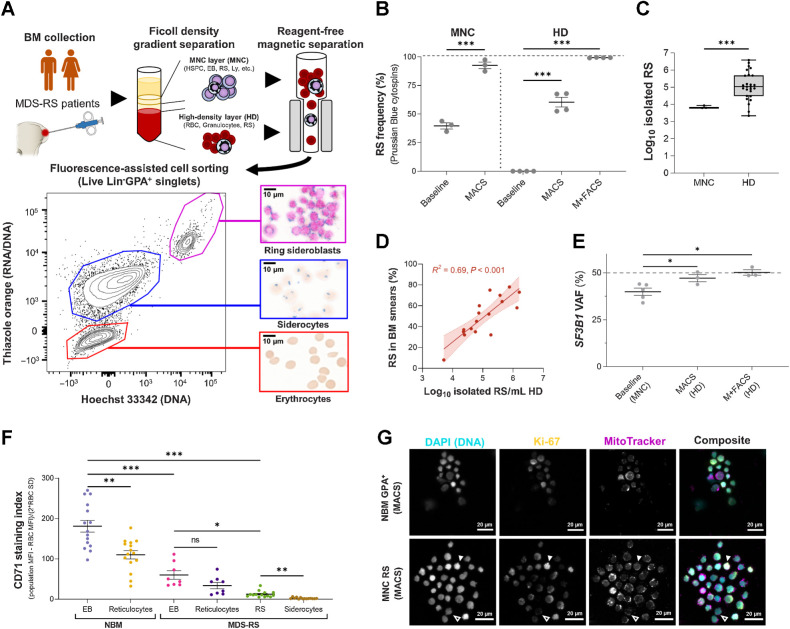
Successful separation of viable SF3B1mt RS/siderocytes enables focused investigations
Further assessment of RS biology was challenging in heterogeneous cell populations. However, as hemozoin-rich red blood cells (RBC) are commonly separated by reagent-free magnetic isolation (MACS; ref. 27), we hypothesized that iron burden could similarly be exploited to isolate SF3B1mt RS/siderocytes (Fig. 2A). MACS was performed on MNC (viably frozen) and high-density (HD, fresh) fractions and achieved significant RS enrichment in both (Fig. 2B). FACS of HD-isolated cells further enriched a high-purity (99.3%) RS population at significantly higher abundance than MNC (Fig. 2C; Supplementary Fig. S6) and was the method utilized throughout this study unless otherwise specified. Isolated RS significantly correlated with morphologic BM RS frequencies (Fig. 2D) and were confirmed as fully clonally involved (Fig. 2E). Validating our previous flow cytometric and morphologic analysis of MDS-RS erythroblasts, purified RS displayed highly significantly decreased surface CD71 (Fig. 2F; Supplementary Figs. S7A and S7B) as well as identifiable nuclear localization of Ki-67 (Fig. 2G; Supplementary Fig. S8).
Figure 2.
Reagent-free MACS enables direct characterization of viable SF3B1mt RS. A, Method for RS and siderocyte purification from BM aspiration material. A representative flow cytometric diagram plots RNA/DNA content (Thiazole Orange) against DNA content (Hoechst 33342) in Lin−GPA+ singlets after MACS of HD cells. Representative micrographs are shown at the right (blue, iron granules; brown, hemoglobin; pink, DNA). Scale bars, 10 μm. B, Mean (±SEM) RS frequencies before and after MACS alone in three MNC and four HD samples, and further purification with FACS (M+FACS) of the same HD samples. HD RS quantification before enrichment steps identifies only 0.1% to 0.001% as potential RS due to high RBC proportions. C, Isolated RS numbers in MACS-enriched cells from 5 × 106 MNCs (n = 3) or M+FACS-enriched HD cells (n = 26, 19 unique biological replicates + 7 repeat visits). D, Correlation of log10-converted isolated RS numbers and RS frequencies in matched BM aspirates (n = 17). E, Mean (± SEM) SF3B1 mutation (SF3B1mt) VAF in unfractionated MNCs (baseline) and MACS-enriched or M+FACS-enriched HD cells, as determined by ddPCR (n = 3 per enrichment method, 5 patients in total). The dashed line indicates complete heterozygosity (VAF = 50%). F, Mean (±SEM) CD71 staining indices (MFI of the cell population – MFI of the CD71-negative RBC population divided by 2 × SD of the RBC population; nNBM = 15, nMDS-RS = 8, nRS = 14). G, Immunofluorescence of Ki-67 detection in NBM erythroblast and an MNC-derived RS isolate, co-labeled for DNA (DAPI; cyan), Ki-67 (yellow), and mitochondria (MitoTracker; magenta). Individual grayscale channels and a composite image of all three markers are shown. A Ki-67neg RS is shown with an outlined arrow; a Ki-67hi RS with a filled arrow. Scale bars, 20 μm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonstatistically significant.
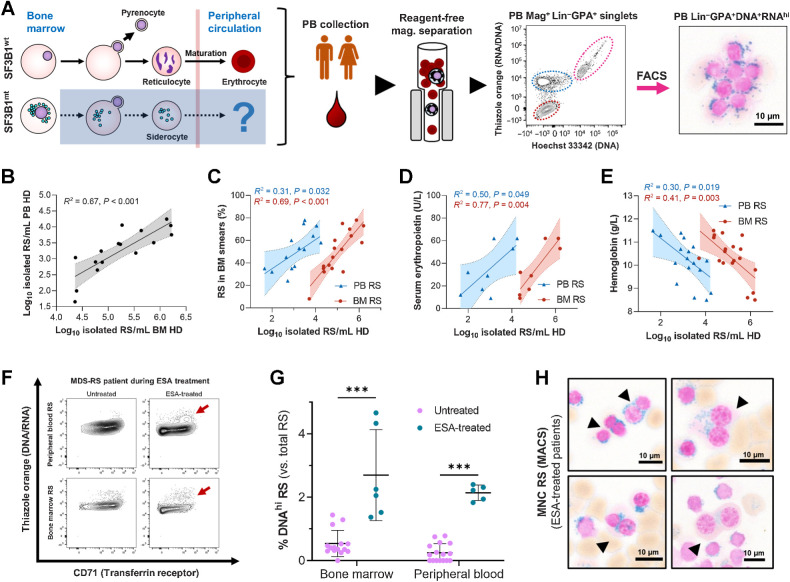
Circulating RS are common and clinically relevant in MDS-RS
Reagent-free magnetic isolation was tested in PB samples to detect circulating siderocytes. Surprisingly, M+FACS assessment of PB HD cells identified a substantial number of RS (Fig. 3A). No previous reports have described appreciable numbers of circulating RS in MDS-RS PB (3). However, peripheral RS were a ubiquitous observation correlating well in abundance with BM RS isolates (Fig. 3B). Where matched clinical data were available, PB and BM RS isolates were positively correlated with diagnostic smear RS (Fig. 3C) and serum erythropoietin (Fig. 3D), and negatively correlated with hemoglobin (Fig. 3E). Interestingly, the identification of PB RS in FC also detected increased DNA content RS in erythropoiesis-stimulating agent (ESA)-treated patients (Fig. 3F). This was morphologically validated in several cases (Fig. 3G and H), identifying a positive effect of ESAs on RS (increasing survival of dysplastic cells or increasing mitosis).
Figure 3.
Peripherally circulating RS are common and clinically relevant in MDS-RS. A, Isolation steps from the PB HD fraction of patients with MDS-RS through reagent-free magnetic separation and representative flow cytometry diagram, where RS are identifiable and validated as present through morphological analysis. B, Correlation of RS abundances isolated from matched BM and PB samples (leftmost subpanel, n = 16). C–E, Correlation of log10-converted isolated RS numbers obtained from anemic (Hb < 12.0 g/dL) patients with MDS-RS with BM RS percentages from clinical BM smears (nPB = 15, nBM = 17; C), serum erythropoietin levels (untreated patients only, nPB = 8, nBM = 8; D), and hemoglobin levels (nPB = 18, nBM = 19; E). F, Flow cytometry example of BM and PB RS with increased DNA content comparing two visits of the same patient to the clinic, before and after ESA treatment. A cell population of increased DNA content is highlighted with dark red arrows. G, Mean (SD) frequency of RS with elevated DNA content, separated by EPO treatment status and cell fraction of origin. Arrows indicate binucleate RS identified during morphological analysis of EPO-treated and RS-enriched samples. Scale bars, 10 μm. H, Morphologic visualization of binucleated RS in ESA-treated, RS-enriched samples. Scale bars, 10 μm. ***, P < 0.001.
SF3B1 mutations have limited impact on the gene expression of true MDS-RS HSPC
We next investigated SF3B1mt erythropoiesis through RNA-seq, comparing MDS-RS cell populations against healthy individuals at bulk (Fig. 4A; Supplementary Table S3) and single-cell level (Fig. 4B; Supplementary Figs. S1, S9, S10A and S10B, S11, and S12). We first focused on CD34+ HSPC, as these constitute the compartment of origin in MDS-RS (5). As reported previously (9, 28), differential gene expression (DE) analysis comparing MACS-enriched CD34+ cells between MDS-RS and healthy individuals identified overexpression of erythroid genes and underexpression of known SF3B1mt targets (Fig. 4C, left; Supplementary Data S4). However, no increased erythroid transcripts were observed in transcriptomically identified HSPCs (Fig. 4C, right). AUCell analysis (29) was then conducted to assess a potential proerythroid signature (top 100 erythroblast development genes; ref. 30) and similarly demonstrated no difference between MDS-RS and NBM HSPC clusters (Fig. 4D).
Figure 4.
SF3B1 mutations have limited impact on the gene expression of true MDS-RS HSPC. A, Principal component analysis (PCA) plots of a full-length bulk RNA-seq experiment encompassing sorted cell populations from NBM donors (nCD34 = 7, nGPA = 4, nRet = 4) and SF3B1mt MDS-RS patients (nCD34 = 6, nGPA = 5, nRS = 4, nSid = 4). Sample distribution along PC1 is visualized against PC2/PC3. B, Global overview of two integrated 10x Genomics scRNA-seq experiments encompassing sorted cell populations from NBM donors and SF3B1mt patients with MDS-RS (nNBM = 3, nMDS-RS = 5, nRS = 1). A UMAP-based bidimensional projection is displayed and separated per sample group, where each cell is visualized as one point. The dotted circle indicates the RS-enriched cell subset, which is absent in NBM. Cell types were annotated according to gene expression signatures per cluster set. Sample and cell type composition in the total dataset are shown below the UMAP plots. C, Volcano plot (left) displaying differentially expressed genes (DEG) in CD34+ MACS-enriched BM cells comparing SF3B1mt MDS-RS versus NBM. Cut-offs for significance were log2 FC > 0.5, adjusted P value <0.01. Genes were overexpressed (OE; red), underexpressed (UE; blue), or not significantly different (NS; gray). Gene set enrichment analysis (GSEA) of overexpressed genes (middle) was performed with the Enrichr Human Gene Atlas. The right UMAP heatmap displays expression of bulk OE genes in scRNA-seq. The dotted rectangle highlights the HSPC transcriptomic cluster. D, AUCell erythroid score [based on erythroid markers from An et al. (30)] mapped in the UMAP overlays and separated by mutational background. The erythroid score is similarly displayed in violin plots (gray, NBM; orange, SF3B1mt) and grouped per cell population (excluding cell subsets unrelated to erythroid development, e.g., macrophages). E, Representative CD34 and GPA FACS plots from CD34+ MACS-separated BM MNCs isolated from an patient with SF3B1mt MDS-RS and from a patient with non-SF3B1mt non-RS MDS. Lin−CD34−GPA+ cells are gated in blue and connected to representative micrographs. Scale bars, 10 μm. F, Mean (±SEM) percentage of Lin−CD34−GPA+ cells in CD34+-enriched cells (n = 3 per group). G, Mean (±SEM) cell frequencies based on morphologic analysis of Lin−CD34−GPA+ in MACS-purified CD34+ MDS-RS samples. H, UMAP projection of CD34 RNA-positive HSPCs in the scRNA-seq dataset. I, Mean (±SEM) frequencies of transcriptomically identifiable HSPC subsets as set out in H and compared between NBM and MDS-RS samples. J, Gene set enrichment analysis results for Gene Ontology biological process (GO BP) enrichment of differentially expressed genes identified in the HSPC cluster between MDS-RS and NBM cells (nNBM = 432, mean 144 cells/donor; nMDS-RS = 510, mean 102 cells/donor). ***, P < 0.001; ns, nonstatistically significant.
In light of the efficiency of RS isolation through reagent-free MACS, we investigated whether antibody-mediated MACS could induce sample contamination. Indeed, CD34+-enriched MDS-RS cells displayed a Lin−CD34−GPA+ population averaging ∼90% RS/siderocytes, which was absent in MDS samples without RS (Fig. 4E–G), highlighting important limitations in interpreting magnetically enriched cell data from MDS-RS. (7–10) To circumvent these, we evaluated the molecular signature of CD34 transcript-positive HSPC in scRNAseq (Fig. 4H; Supplementary Fig. S13; Supplementary Data S5). Transcriptomically defined HSPC subset frequencies were unchanged (Fig. 4I), matching previous data of phenotypical HSPC in MDS-RS (5). Interestingly, few DE genes were detected in CD34+ HSPCs despite these maintaining RNA mis-splicing, and the only enrichment identified was a protein/ER stress-associated response (Fig. 4J). Given these limited HSPC results, we next focused our approach on specifically investigating erythroid biology.
RS actively modulate their gene expression to survive extensive oxidative/splicing stress
RS were used as the primary point of comparison against NBM erythroblasts due to being phenotypically and clonally engaged. Bulk RNA-seq identified extensive underexpression of cell-cycle-related genes, matching our initial characterization (Fig. 5A). Most importantly, overexpressed genes in bulk RNA-seq mapped to an MDS-specific scRNA-seq cluster, which was overrepresented in RS and absent in healthy donors (Fig. 5B). This cluster presented a more aberrant transcriptomic phenotype in purified HD-RS versus MNC-RS (Fig. 5C). However, and beyond this MDS-specific cluster, RS clearly encompassed all stages of erythroid differentiation.
Figure 5.
SF3B1mt RS activate a prosurvival transcriptomic program against oxidative and RNA splicing stress. A, Volcano plot displaying differentially expressed genes in bulk data comparing M+FACS-purified SF3B1mt RS against MACS-purified NBM GPA+ erythroblasts with an absolute log2 FC cut-off of 2 and an adjusted P value cut-off of 10−4. B, UMAP overlays of RS overexpressed transcript percentages per cell (top row) and AUCell scores of RS identity (bottom row), separated by sample type. Transcript percentages are mapped with an initial baseline cut-off of 4% of total transcripts. AUCell scores were based on the RS overexpressed gene set. C, Gene Ontology biological process (GO BP) enrichment analysis of differentially expressed genes identified in the RS-enriched cluster comparing HD fraction-derived RS versus MNC-derived RS (nonspecifically present among MDS-RS MNCs and with presumably decreased iron load). D, Heatmap of all differentially expressed genes between RS from SF3B1mt MDS-RS patients versus NBM, subclustered by cell subset. The top bar above the heatmap identifies each sample type, and cells are further clustered according to cell type, identified by the bottom bar. Dashed lines highlight cell type separation. E, Metascape gene ontology term network generated from all differentially expressed genes identified through comparison of RS and NBM samples at each transcriptomically identified differentiation stage cluster (HSPC, ProEB + Early EB, Differentiating EB, and LateEB + RS-enriched), correcting for differentiation stage skewing. Gene ontology subterms (small circles) are organized and clustered by major functional terms (numbered black circles). Clusters are annotated in the table (right), including adjusted P values from Metascape analysis. F,ABCB7 RNA expression in bulk RNA-seq displayed in log normalized counts from all assayed cell populations (left) and overlaid in the 10x UMAP projection (right). G, Sashimi plots for canonical (normal font) and mis-spliced (bold) read counts of the ABCB7 alternative 3′ splice site associated with targeting by NMD. H,GDF15 expression based on RNA sequencing of purified populations (quantified in log normalized counts; left) and single cells (UMAP overlay; right). I, Mean (±SEM) GDF15 concentration in culture supernatants obtained from 28-day erythroid culture of BM MNCs (nNBM = 3, nMDS-RS = 3), as determined by ELISA. One empty scaffold was kept in the same media and culture conditions to evaluate GDF15 levels in base media. J, Mean (±SD) erythroid and myeloid colony formation from MACS-enriched CD34 cells (nNBM = 3, nMDS-RS = 5), normalized to untreated numbers. Minimum total colonies counted were 254 among NBM donor conditions and 124 among MDS-RS donor conditions. Cells were treated with either recombinant GDF15 peptide at a concentration of 100 ng/mL (gray squares) or with an equal volume of water (vehicle, black circles). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonstatistically significant.
DE analysis was then performed between RS/NBM within each scRNA-seq transcriptomic cluster to define differentiation-independent alterations (Fig. 5D; Supplementary Data S6). This approach identified a major enrichment of stress response pathways (Fig. 5E), particularly related to autophagy, proteasomal processing, and mitophagy. Metabolic changes were also evident, particularly the significant enrichment of several antioxidant pathways (glutamine/ROS response); together, these indicate an active transcriptional state in RS to maintain cell homeostasis against overactive mis-splicing and oxidative stress.
Erythroid-specific genes were also heavily affected in this comparison, with RS displaying loss of expression of hemoglobin chains HBA2 and HBD, a hallmark for iron deficiency anemia (Supplementary Fig. S14; ref. 31), and decreased HBB expression in cells with higher RS-associated transcripts (Supplementary Fig. S15). Conversely, erythroid gene expression was higher in peripherally circulating RS, suggesting that this population is either more differentiated or less phenotypically affected (Supplementary Figs. S16A and S16B and S17).
Focusing on bulk expression of ABCB7, the major SF3B1mt-induced NMD-targeted event in SF3B1mt MDS-RS (9, 28), we found that its abundance was evidently decreased in HSPCs (matching previous reports) but significantly increased in RS/siderocytes both in bulk and scRNA-seq (Fig. 5F; Supplementary Fig. S18) and identified lower levels of the cryptic ABCB7 splice site in RS as compared with CD34+ and GPA+ cells (Fig. 5G).
Both RNA-seq datasets identified overexpression of Growth Differentiation Factor 15 (GDF15) as a major occurrence (Fig. 5H). GDF15 is a stromal/erythroid stress factor previously shown to be increased in MDS-RS patient sera (32) and reported to suppress HSPC growth in culture (33). When in vitro culture was used to induce stress erythropoiesis, cultured SF3B1mt cells still secreted more than 9× GDF15 than normal cells, highlighting GDF15 overexpression is disease-related (Fig. 5I). A colony-forming unit (CFU) experiment to evaluate GDF15 effects on HSPC suppression displayed variable results in MDS-RS samples, but identified a consistent and significant decrease in NBM erythroid/total CFU-forming ability (Fig. 5J), linking GDF15 secretion by RS to an effect on upstream HSPC biology.
SF3B1mt mis-splicing is context-specific and significantly increased in RS
Given the surprising patterns of ABCB7 expression/splicing in RS (Fig. 5F–H), we next focused on a broader analysis of AS in the bulk RNA-seq data comparing healthy individuals and SF3B1mt MDS-RS [HSPC, RS, vs. erythroblasts (nucleated erythroid, N) and siderocytes vs. reticulocytes (anucleate erythroid, A)]. RS presented substantially more differential AS events; 63 genes were AS in both HSPC/RS, and only 7 genes were AS in all three groups (Fig. 6A).
Figure 6.
Distinct RNA dynamics in erythroid differentiation intensify SF3B1mt mis-splicing. A, Proportional Venn diagram of genes undergoing statistically significant AS [FDR < 0.001, absolute difference in percentage spliced in levels (Abs. ΔPSI) > 0.2] in the HSPC (green; MDS-RS CD34+ vs. NBM CD34+), nucleated erythroid [red; MDS-RS RS vs. NBM EB, Erythroid (N)] and anucleate erythroid [blue; MDS-RS Siderocytes vs. NBM RetPB, Erythroid (A)] populations. B, Gene Ontology biological process (GO BP) enrichment analysis results comprising genes mis-spliced in both the HSPC and erythroid (N) populations are shown in the top bar chart. The Human Gene Atlas enrichment results with the lowest adjusted P value are shown in the bottom table. C, Frequency of AS events split by rMATS category in each sample group comparison. SE, skipped exon; RI, retained intron; MXE, mutual exon exclusion; A5SS, alternative 5′ splice site; A3SS, alternative 3′ splice site. Statistical comparisons of A3SS+RI frequencies were performed with Fisher exact test. D, Box plots of percent spliced-in (PSI) values of literature-validated SF3B1mt-induced A3SS events in MDS-RS samples, separated by sample type (CD34, GPA, RS). Known targeting by NMD is indicated with a red circle; in-frame events without a PTC are indicated by a blue circle. E, Box plots of PSI values in newly identified ASEs affecting known driver genes implicated in the pathogenesis of MDS and CSA. Known targeting by NMD is indicated with a red circle; PTC detection with unverified NMD is indicated with an orange circle; in-frame events without a PTC are indicated by a blue circle. F, Distribution of base pair distances from cryptic A3SS sites to canonical splice sites (horizontal axis) in HSPC and Erythroid (N). Further detail is provided in −400 bp to 0 bp for increased contrast. Lines at −30 bp and −10 bp demarcate the interval associated with SF3B1 mis-splicing (38). Additional lines at −140 bp and −330 bp demarcate additional intervals of interest. G, Sequence logos of canonical and A3SS sequences encompassing the 3′ splice site (starting at −35 bp upstream of the AG motif) and statistical comparison through a two-sample logo. H, Frequency of A3SS events per rMATS cell type comparison where the splice site shift remains in-frame (blue) or induces a frameshift event (orange). I, Frequency of exon insertion events per rMATS cell-type comparison where the splice site shift incorporates a new PTC (pink) or remains in-frame with no PTC induction (green). J and K, RNA velocity analysis of transcriptomically identified HSPC and erythroblast subsets in 10x scRNA-seq, visualizing the percentage of spliced transcripts along pseudotime in the total cell populations (violin plots) or separated by sample group (scatter plots). The analysis in J includes all transcripts, whereas K excludes ribosomal and globin transcripts. L, UMAP overlay of FACS-purified HSPC subsets and GPA+ EB from 1 SF3B1mt MDS-RS patient after Smart-seq3xpress (SS3x), visualizing true versus predicted cell-type identity. M, RNA velocity analysis of spliced RNA read percentages in the FACS-sorted SS3× experiment, analyzed independently of the 10x dataset. This graph excludes ribosomal and globin transcripts. N, Mean (±SEM) differences in PSI after 3 hours cycloheximide treatment (70 μg/mL) versus DMSO (1:1,000, vehicle) in MDS-RS CD34 and GPA cells. SF3B1mt-associated NMD-targeted ASEs with sufficient coverage are shown at far left, SF3B1mt-associated in-frame ASEs at middle-left, and endogenous NMD-targeted transcripts at middle-right. The far-right plot visualizes all ASEs. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonstatistically significant.
Shared AS events between HSPC/RS were enriched in RNA splicing-associated genes (Fig. 6B), in line with previous SF3B1mt studies (9, 34). However, genes that were specifically AS in each population significantly enriched for cell type identity, reflecting distinct transcriptomic programs. AS event types also progressively shifted toward predominantly exon skipping events during erythroid differentiation, compared with the initial skew of alternative 3′-splicing and intron retention events in HSPCs (Fig. 6C; Supplementary Figs. S19A and S19B).
Focusing on literature-validated SF3B1mt AS events (9, 35), we found that decreased mis-splicing of ABCB7 was a singular case in RS; in fact, every other AS event displayed a constant or increased median level of the cryptic transcript in RS/erythroblasts as compared with HSPC, including known NMD-targeted AS (Fig. 6D). Similarly, although new AS sites affecting several genes implicated in MDS (36) and congenital sideroblastic anemia (CSA; SLC25A38; ref. 37) were detected in HSPC at very low levels, cryptic mis-spliced transcripts of these genes increased during erythropoiesis to become the predominant transcript form in RS, with many of these inducing frameshifts and/or inclusion of premature termination codons (PTC; Fig. 6E).
Functional analysis of cryptic 3′ site locations in HSPCs confirmed their enrichment between −30 and −10 bp upstream of the canonical 3′ site (Fig. 6F), as reported previously (38). Conversely, RS displayed events in the same interval but had a much larger distance distribution, indicating reduced fidelity for splice site recognition. This is functionally supported by differentially altered RS nucleotide frequencies upstream of and surrounding the cryptic 3′ splice site (Fig. 6G), as well as a much larger proportion of frameshift/PTC events (Fig. 6H and I).
Modified RNA dynamics in erythropoiesis justify the magnified mis-splicing profile of RS
We next hypothesized that the highly aberrant AS profile in RS could be caused by altered splicing/NMD dynamics in erythropoiesis. RNA velocity analysis (39) of spliced transcript percentages in the 10x clusters identified increased splicing rates already in early erythroblasts (Fig. 6J). We were concerned that the high erythroid transcript burden of ribosomal/globin genes could skew these data; however, this trend remained after discarding these transcripts from analysis (Fig. 6K). Importantly, NBM and MDS samples behaved identically, highlighting the role of differentiation and not disease in this process. As velocity analysis of the 10x samples was based solely on clusters identified through computational prediction, we then confirmed the same splicing dynamics in FACS-purified HSPC/erythroblast populations through Smart-seq3xpress (Fig. 6L and M; Supplementary Figs. S20, S21, and S22A–S22C).
Cycloheximide treatment of HSPC and erythroblasts was then pursued to evaluate NMD dynamics as originally reported in HSPC by Shiozawa and colleagues (8). Overall, NMD-targeted cryptic transcripts (either SF3B1mt-associated or of endogenous origin; ref. 40) were increased in frequency by cycloheximide treatment in HSPC alone (Fig. 6N), through which we determine erythroid differentiation displays decreased NMD activity. This is consistent with the significantly higher proportions of NMD-targeted AS events in RS (Fig. 6D). Thus, taken together, we conclude that erythroid differentiation magnifies the downstream effects of SF3B1mt.
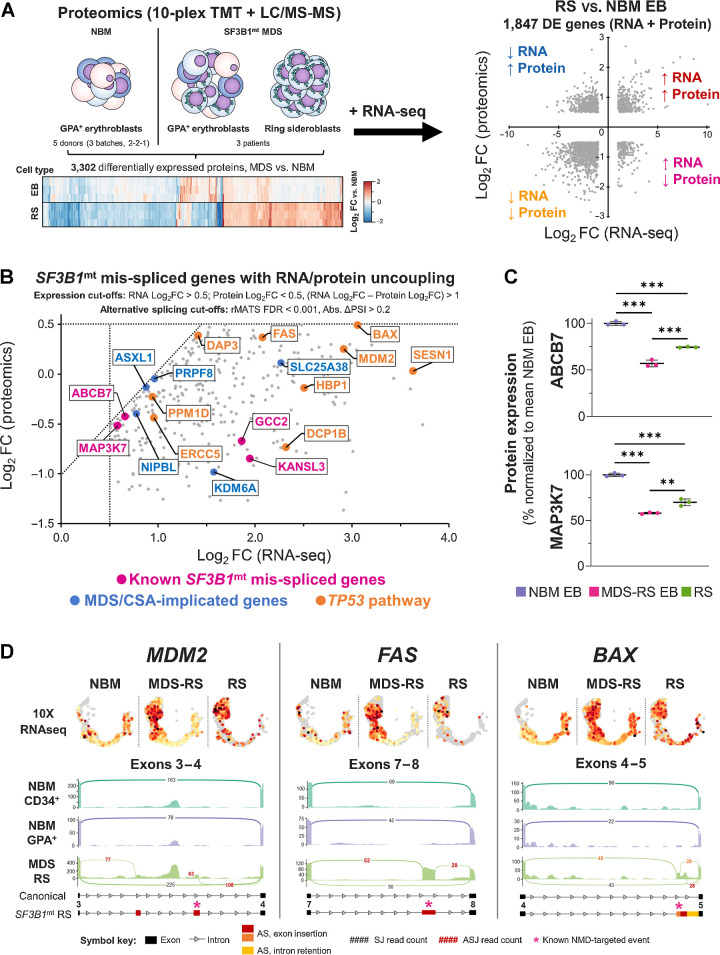
Integrative proteomic analysis of SF3B1mt RS identifies disruption of proapoptotic genes
Semiquantitative proteomics of healthy and MDS-RS erythroblasts/RS were performed and integrated with RNA-seq to functionally assess the outcome of SF3B1mt expression/splicing (Fig. 7A; Supplementary Figs. S23A and 23B). This analysis identified four major signatures of differential RNA/protein expression, which validated our previous scRNA-seq results (Supplementary Table S4). The same pro-survival pathways from scRNA-seq were validated as overexpressed, together with a highly significant decrease in cell cycle–associated proteins; however, the proteomic dataset also identified a specific and significant decrease in NMD target-degrading proteins (Supplementary Fig. S24). Importantly, GDF15 was significantly overexpressed in both erythroblasts and RS, further supporting previous findings (Supplementary Fig. S25).
Figure 7.
Proteomic analysis of SF3B1mt RS defines RNA/protein uncoupling downstream of RNA mis-splicing with severe dysregulation of proapoptotic genes. A, Design of a combined transcriptomic and proteomic analysis of SF3B1mt RS. Erythroblast (EB) samples from five NBM donors (separated into three biologically distinct batches) and paired EB + RS samples from three patients with MDS-RS were subjected to semiquantitative proteomics. RNA-seq DE genes are compared against differentially expressed proteins to obtain four major signatures of differential expression, which are highlighted in each quadrant and correspond to the gene ontology enrichment results provided in Supplementary Table S4. B, Scatter plot of SF3B1mt mis-spliced genes with uncoupled RNA and protein expression in RS as compared with NBM EB. RNA and protein expression cut-offs were applied to limit analysis only to genes with RNA Log2FC > 0.5, Protein Log2FC < 0.5, and a difference between RNA and protein Log2FC values > 1 (RNA Log2FC – Protein Log2FC). AS detection cutoffs were followed as indicated in Fig. 6A (FDR < 0.001; Abs. ΔPSI > 0.2). Pink, known SF3B1mt mis-spliced genes (Fig. 6D). Blue, MDS/CSA-implicated driver genes (Fig. 6E). TP53 pathway/proapoptotic genes were detected through enrichment analysis and are highlighted in orange. C, Mean (±SEM) protein expression levels of ABCB7 and MAP3K7, normalized to mean NBM expression. D, 10x single-cell RNA-seq cross-validation of increased RNA expression for TP53 pathway genes MDM2, BAX, and FAS, with gene expression values overlaid in the HSPC/erythroid UMAP projection and separated by sample type. Below each gene, sashimi plots display major mis-spliced transcript regions. Black, canonical splice junction counts (SJ); red, cryptic SJ counts. A full legend for the sashimi plots is provided below the graph. The asterisks indicate sites corresponding to transcripts canonically targeted by NMD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonstatistically significant.
Cases of RNA/protein uncoupling, that is, genes with increased RNA expression and negative or no correlation between RNA and protein abundance, were particularly interesting as candidate targets of dysfunctional AS events. Indeed, this gene subset encompassed several known SF3B1mt-affected genes (Fig. 7B; refs. 9, 38), as well as several of the newly identified RS-enriched AS events affecting driver genes implicated in MDS/CSA pathogenesis (Fig. 6E). In adition, pathway analysis of this gene set identified specific enrichment of the TP53 pathway, including several proapoptotic effectors; and also the detection of EIF2AK1, identified in a previous report to be highly overexpressed in single-cell RNA-seq data of MDS-RS erythroblasts (41) and yet here underexpressed at protein level in both MDS-RS erythroblasts and RS, potentially due to multiple mis-splicing events (Supplementary Figs. S26A–S26D).
Investigating ABCB7 and MAP3K7 protein levels to further dissect this mechanism demonstrated significant underexpression of both proteins in MDS-RS erythroblasts and RS in comparison with healthy erythroblasts. However, both were significantly increased in RS as compared with MDS-RS erythroblasts (Fig. 7C). Therefore, the increased RNA mis-splicing detection in RS resulting from decreased NMD is partially compensated through RNA overexpression. However, this process fails to completely normalize protein abundance and likely also retains dysfunctional proteins in the cell generated via translation of the nondegraded mis-spliced transcripts.
MDM2, BAX, and FAS were selected as major elements for assessing the downstream consequences of TP53 pathway mis-splicing due to their key roles in apoptosis. All were confirmed to be overexpressed in 10x scRNA-seq (Fig. 7D, top). However, diverse mis-splicing events were identified in both RNA-seq datasets as exclusive to RS (rMATS FDR < 10−5; Fig. 7D, bottom; Supplementary Fig. S27). Several would canonically undergo NMD, and thus are detrimental to production of functional proteins; however, the cryptic transcripts increase in RS to even become the main form in the case of FAS. We thus conclude that RNA mis-splicing affects apoptotic regulation in SF3B1mt RS, and for the first time identify SF3B1mt-associated mis-splicing of TP53 pathway genes.
Discussion
Although in vitro and in vivo studies of SF3B1mt erythropoiesis have proven successful in modeling HSPC pathobiology and recreating RS generation, the molecular mechanisms driving the growth and survival of SF3B1mt RS in the MDS-RS BM and the competitive advantage of SF3B1mt cells have remained elusive (5, 14, 15). Through an integrative multiomics approach, we investigated SF3B1mt MDS-RS erythropoiesis at a cell and molecular level to address these open questions.
We established a novel method for viable RS and siderocyte isolation from patient BM samples, enabling exploration of their disrupted erythroid development and rendering us the unique possibility to explore the entire process of SF3B1mt erythroid development from HSC to RS. This method was also validated in PB samples, identifying circulating RS as a relevant clinical observation correlating in abundance with BM RS burden and hemoglobin/serum EPO levels.
An important outcome associated with reagent-free magnetic isolation of RS was the finding that CD34+ MACS enrichment (a core experimental approach for HSPC isolation; refs. 7–10, 28) indirectly isolates a significant number of RS and siderocytes due to their high iron content, creating an artificial erythroid subpopulation, which was absent in matched MDS samples without RS. This subpopulation fundamentally underlies the bulk transcriptomic erythroid signature conventionally associated with MDS-RS HSPCs, which we validated in our bulk RNA-seq data and yet failed to verify in a detailed scRNA-seq analysis including enriched HSPCs. Indeed, despite showing a clear functional impairment in long-term culture-initiating cell and CFU assays (5), true MDS-RS HSC/MPPs displayed few alterations in gene expression and no detectable differences in commitment, subset frequencies, or erythroid identity. However, a previous study focusing on scRNA-seq and scATAC-seq analysis of MDS-RS HSPCs identified an increase in erythroid versus megakaryocyte commitment of megakaryocyte-erythroid progenitors occurring through differential chromatin usage (41). We thus speculate that combining these insights with additional data regarding RNA splicing and protein production at HSPC subset resolution will be instrumental towards understanding the functional consequences of SF3B1mt and downstream RNA mis-splicing on early hematopoietic development.
In evaluating erythroblast and RS biology, we demonstrated that RS constitute a living, differentiating, and molecularly active population, with decreased cell-cycle progression as the major sign of dysfunction and which may explain the marked abundance of RS in the MDS-RS BM. In comparison with approaches focusing on the general and highly heterogeneous erythroblast population, the single-cell and proteomic analysis of purified RS provides a much increased granularity to the diverse homeostatic mechanisms engaged to survive oxidative and RNA splicing stress, including the surprising protein rescue of known mis-spliced and NMD-targeted genes ABCB7 and MAP3K7 (9).
We explored the molecular mechanisms underlying these changes and define an intensified SF3B1mt mis-splicing panorama in RS. This includes the novel identification of PTC induction and general mis-splicing and dysregulation of several MDS-implicated driver genes (36), each of which may constitute a target for further studies. We thus speculate that modified RNA splicing and NMD dynamics in erythroid cells combine to switch “quality control” from RNA to protein, supported by the finding that RS sensitivity to proteasome inhibition by bortezomib has been demonstrated in a clinical trial (42).
Importantly, we demonstrate that the observed uncoupling of RNA and protein expression enhances detection and functional validation of RNA mis-splicing events and in turn allows us to directly associate SF3B1 mutations with disruption of proapoptotic genes in the TP53 pathway. In addition to providing a potential explanation for why RS accumulate in the BM, this finding may also help to further dissect a major question in the study of MDS: what explains the increased risk conferred by SF3B1 mutations in TP53-mutant settings such as MDS5q−, AML, and other malignancies including chronic lymphocytic leukemia, in comparison with the relatively benign course of MDS-RS with isolated SF3B1 mutations (43, 44)? More specifically, could SF3B1mt mis-splicing of TP53 pathway modulators and proapoptotic genes specifically enhance the cell's adaptability when TP53 function is already compromised? These are complex questions that beckon the need to evaluate how complex genetic profiles and RNA splicing act in tandem to potentiate new interactions beyond the effects of a single driver mutation.
An additional major finding of our multiomics studies was the repeated identification of SF3B1mt RS as a key source of GDF15. Previously reported as a marker of increased erythropoiesis in MDS-RS (45), we here instead confirmed a detrimental impact of this cytokine on wild-type HSPC biology. Chronic myeloproliferative neoplasms develop over decades, and we recently reported preliminary data indicating this is also the case in MDS-RS (46, 47). On the basis of our data, we hypothesize that SF3B1mt cell effects on their surroundings may contribute to clonal expansion and clinical disease evolution. It is thus tantalizing to speculate that RS could function as disease-augmenting “foot soldiers,” similarly to Reed–Sternberg cells in Hodgkin lymphoma (48), and further exploration of how SF3B1mt cells affect their surrounding microenvironment and wild-type cells will be essential.
In conclusion, our characterization of SF3B1mt erythropoiesis constitutes a unique platform for the study of MDS-RS, providing novel insights into the unexpectedly active biology of the “dead-end” RS and enabling further investigation of disease pathogenesis and treatment avenues.
Supplementary Material
Main supplemental file
Descriptive statistics, deidentified clinical and experiment map of patients and healthy donors included in the study
Gene count matrix and experiment metadata for bulk RNAseq
Semi-quantitative TMT raw data, DE analysis, enrichment analysis raw data and predicted NMD analysis, comprising NBM EB, MDS-RS EB and purified RS
DESeq2 differential expression analysis of population-level RNAseq data
Seurat clustering and differential expression analysis between RS/NBM and MDS/NBM cell identity clusters in scRNAseq
STRING and Metascape enrichment analysis results, respectively separated per cluster and global (corrected by erythroid stage)
Acknowledgments
The authors would like to thank the patients and healthy donors for their willingness to participate in this research. The authors would like to acknowledge support from the Karolinska Institutet MedH Flow Cytometry Core Facility for assistance with FACS experiments; the KI Bioinformatics and Expression Analysis Core Facility for assistance with scRNAseq experiments; the facility of Clinical Proteomics Mass Spectrometry at Karolinska University Hospital and Science for Life Laboratory for assistance in mass spectrometry and data analysis; the KI Single-Cell Core Facility (SICOF) for assistance with Smart-seq3 scRNAseq; the KI Research Data Office for assistance with data management and deposition; and support from the National Genomics Infrastructure in Stockholm funded by Science for Life Laboratory, the Knut and Alice Wallenberg Foundation and the Swedish Research Council, and SNIC/Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure. This project received grant support from Cancerfonden / Swedish Cancer Society (grant number 21 0340 to P.L. Moura and 19 0200 to E. Hellström-Lindberg); Vetenskapsrådet/Swedish Research Council (grant number 2020–01902 to V. Lundin and 211133 to E. Hellström-Lindberg); and from the Knut and Alice Wallenberg Foundation (grant number 2017.0359 to E. Hellström-Lindberg).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
P.L. Moura reports grants from Cancerfonden (Swedish Cancer Society) during the conduct of the study. W.W. Kretzschmar reports personal fees from PerkinElmer/Revvity, Inc. outside the submitted work. M. Hagemann-Jensen reports holding shares in Xpress Genomics AB. C. Ziegenhain reports holding shares in Xpress Genomics AB. V. Lundin reports grants from Vetenskapsrådet (Swedish Research Council) during the conduct of the study. P.S. Woll reports grants from Knut and Alice Wallenberg Foundation during the conduct of the study and grants from Cancerfonden and Radiumhemmets Forskningsfonder outside the submitted work. R. Sandberg reports holding shares in Xpress Genomics AB. E. Hellström-Lindberg reports grants from Cancerfonden (Swedish Cancer Society), Knut and Alice Wallenberg Foundation, and Vetenskapsrådet (Swedish Research Council) during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
P.L. Moura: Conceptualization, data curation, software, formal analysis, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. T. Mortera Blanco: Formal analysis, investigation, writing–review and editing. I.J. Hofman: Investigation, writing–review and editing. G. Todisco: Software, formal analysis, investigation, writing–review and editing. W.W. Kretzschmar: Software, writing–review and editing. A. Björklund: Investigation, writing–review and editing. M. Creignou: Investigation, writing–review and editing. M. Hagemann-Jensen: Software, investigation, writing–review and editing. C. Ziegenhain: Software, investigation, writing–review and editing. D. Cabrerizo Granados: Investigation, writing–review and editing. I. Barbosa: Investigation, writing–review and editing. G. Walldin: Investigation, writing–review and editing. M. Jansson: Investigation, writing–review and editing. N. Ashley: Investigation, writing–review and editing. A.J. Mead: Supervision, investigation, writing–review and editing. V. Lundin: Supervision, writing–review and editing. M. Dimitriou: Validation, writing–review and editing. T. Yoshizato: Resources, software, writing–review and editing. P.S. Woll: Resources, supervision, validation, writing–review and editing. S. Ogawa: Supervision, writing–review and editing. R. Sandberg: Supervision, methodology, writing–review and editing. S.E.W. Jacobsen: Resources, supervision, validation, writing–review and editing. E. Hellström-Lindberg: Conceptualization, resources, data curation, supervision, funding acquisition, validation, writing–original draft, project administration, writing–review and editing.
References
- 1. Meyer SE. Splicing together the origins of MDS-RS. Blood 2017;130:841–2. [DOI] [PubMed] [Google Scholar]
- 2. Swerdlow SH. International agency for research on C, World Health O. WHO classification of tumours of haematopoietic and lymphoid tissues. Lyon: International Agency for Research on Cancer 2017. [Google Scholar]
- 3. Cazzola M, Invernizzi R. Ring sideroblasts and sideroblastic anemias. Haematologica 2011;96:789–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011;478:64–9. [DOI] [PubMed] [Google Scholar]
- 5. Mortera-Blanco T, Dimitriou M, Woll PS, Karimi M, Elvarsdottir E, Conte S, et al. SF3B1-initiating mutations in MDS-RSs target lymphomyeloid hematopoietic stem cells. Blood 2017;130:881–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bernard E, Tuechler H, Greenberg PL, Hasserjian RP, Ossa JEA, Nannya Y, et al. Molecular international prognostic scoring system for myelodysplastic syndromes. 2022;1:EVIDoa2200008. [DOI] [PubMed] [Google Scholar]
- 7. Dolatshad H, Pellagatti A, Fernandez-Mercado M, Yip BH, Malcovati L, Attwood M, et al. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia 2015;29:1092–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shiozawa Y, Malcovati L, Galli A, Sato-Otsubo A, Kataoka K, Sato Y, et al. Aberrant splicing and defective mRNA production induced by somatic spliceosome mutations in myelodysplasia. Nat Commun 2018;9:3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dolatshad H, Pellagatti A, Liberante FG, Llorian M, Repapi E, Steeples V, et al. Cryptic splicing events in the iron transporter ABCB7 and other key target genes in SF3B1-mutant myelodysplastic syndromes. Leukemia 2016;30:2322–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nikpour M, Scharenberg C, Liu A, Conte S, Karimi M, Mortera-Blanco T, et al. The transporter ABCB7 is a mediator of the phenotype of acquired refractory anemia with ring sideroblasts. Leukemia 2013;27:889–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Conte S, Katayama S, Vesterlund L, Karimi M, Dimitriou M, Jansson M, et al. Aberrant splicing of genes involved in haemoglobin synthesis and impaired terminal erythroid maturation in SF3B1 mutated refractory anaemia with ring sideroblasts. Br J Haematol 2015;171:478–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jin X, He X, Cao X, Xu P, Xing Y, Sui S, et al. Iron overload impairs normal hematopoietic stem and progenitor cells through reactive oxygen species and shortens survival in myelodysplastic syndrome mice. Haematologica 2018;103:1627–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Obeng EA, Chappell RJ, Seiler M, Chen MC, Campagna DR, Schmidt PJ, et al. Physiologic expression of Sf3b1(K700E) causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell 2016;30:404–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Elvarsdottir EM, Mortera-Blanco T, Dimitriou M, Bouderlique T, Jansson M, Hofman IJF, et al. A three-dimensional in vitro model of erythropoiesis recapitulates erythroid failure in myelodysplastic syndromes. Leukemia 2020;34:271–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clough CA, Pangallo J, Sarchi M, Ilagan JO, North K, Bergantinos R, et al. Coordinated missplicing of TMEM14C and ABCB7 causes ring sideroblast formation in SF3B1-mutant myelodysplastic syndrome. Blood 2022;139:2038–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Asimomitis G, Deslauriers AG, Kotini AG, Bernard E, Esposito D, Olszewska M, et al. Patient-specific MDS-RS iPSCs define the mis-spliced transcript repertoire and chromatin landscape of SF3B1-mutant HSPCs. Blood Adv 2022;6:2992–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Malcovati L, Hellstrom-Lindberg E, Bowen D, Ades L, Cermak J, Del Canizo C, et al. Diagnosis and treatment of primary myelodysplastic syndromes in adults: recommendations from the European LeukemiaNet. Blood 2013;122:2943–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016;127:2391–405. [DOI] [PubMed] [Google Scholar]
- 19. Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: open source software for digital pathology image analysis. Sci Rep 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hagemann-Jensen M, Ziegenhain C, Chen P, Ramskold D, Hendriks GJ, Larsson AJM, et al. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat Biotechnol 2020;38:708–14. [DOI] [PubMed] [Google Scholar]
- 22. Hagemann-Jensen M, Ziegenhain C, Sandberg R. Scalable single-cell RNA sequencing from full transcripts with Smart-seq3xpress. Nat Biotechnol 2022;40:1452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hellstrom-Lindberg E, Schmidt-Mende J, Forsblom AM, Christensson B, Fadeel B, Zhivotovsky B. Apoptosis in refractory anaemia with ringed sideroblasts is initiated at the stem cell level and associated with increased activation of caspases. Br J Haematol 2001;112:714–26. [DOI] [PubMed] [Google Scholar]
- 24. Moggridge S, Sorensen PH, Morin GB, Hughes CS. Extending the compatibility of the SP3 paramagnetic bead processing approach for proteomics. J Proteome Res 2018;17:1730–40. [DOI] [PubMed] [Google Scholar]
- 25. R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2021. [Google Scholar]
- 26. Hu J, Liu J, Xue F, Halverson G, Reid M, Guo A, et al. Isolation and functional characterization of human erythroblasts at distinct stages: implications for understanding of normal and disordered erythropoiesis in vivo. Blood 2013;121:3246–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hackett S, Hamzah J, Davis TM, St Pierre TG. Magnetic susceptibility of iron in malaria-infected red blood cells. Biochim Biophys Acta 2009;1792:93–9. [DOI] [PubMed] [Google Scholar]
- 28. Shiozawa Y, Malcovati L, Galli A, Pellagatti A, Karimi M, Sato-Otsubo A, et al. Gene expression and risk of leukemic transformation in myelodysplasia. Blood 2017;130:2642–53. [DOI] [PubMed] [Google Scholar]
- 29. Aibar S, Gonzalez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 2017;14:1083–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. An X, Schulz VP, Li J, Wu K, Liu J, Xue F, et al. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood 2014;123:3466–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harthoorn-Lasthuizen EJ, Lindemans J, Langenhuijsen MM. Influence of iron deficiency anaemia on haemoglobin A2 levels: possible consequences for beta-thalassaemia screening. Scand J Clin Lab Invest 1999;59:65–70. [DOI] [PubMed] [Google Scholar]
- 32. Ramirez JM, Schaad O, Durual S, Cossali D, Docquier M, Beris P, et al. Growth differentiation factor 15 production is necessary for normal erythroid differentiation and is increased in refractory anaemia with ring-sideroblasts. Br J Haematol 2009;144:251–62. [DOI] [PubMed] [Google Scholar]
- 33. Uchiyama T, Kawabata H, Miura Y, Yoshioka S, Iwasa M, Yao H, et al. The role of growth differentiation factor 15 in the pathogenesis of primary myelofibrosis. Cancer Med 2015;4:1558–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, Aird D, et al. Cancer-associated SF3B1 hotspot mutations induce cryptic 3' splice site selection through use of a different branch point. Cell Rep 2015;13:1033–45. [DOI] [PubMed] [Google Scholar]
- 35. Bondu S, Alary AS, Lefevre C, Houy A, Jung G, Lefebvre T, et al. A variant erythroferrone disrupts iron homeostasis in SF3B1-mutated myelodysplastic syndrome. Sci Transl Med 2019;11:eaav5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ogawa S. Genetics of MDS. Blood 2019;133:1049–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kannengiesser C, Sanchez M, Sweeney M, Hetet G, Kerr B, Moran E, et al. Missense SLC25A38 variations play an important role in autosomal recessive inherited sideroblastic anemia. Haematologica 2011;96:808–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang J, Ali AM, Lieu YK, Liu Z, Gao J, Rabadan R, et al. Disease-causing mutations in SF3B1 alter splicing by disrupting interaction with SUGP1. Mol Cell 2019;76:82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, et al. RNA velocity of single cells. Nature 2018;560:494–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Leclair NK, Brugiolo M, Urbanski L, Lawson SC, Thakar K, Yurieva M, et al. Poison exon splicing regulates a coordinated network of SR protein expression during differentiation and tumorigenesis. Mol Cell 2020;80:648–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Adema V, Ma F, Kanagal-Shamanna R, Thongon N, Montalban-Bravo G, Yang H, et al. Targeting the EIF2AK1 signaling pathway rescues red blood cell production in SF3B1-mutant myelodysplastic syndromes with ringed sideroblasts. Blood Cancer Discov 2022;3:554–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Daher M, Hidalgo Lopez JE, Randhawa JK, Jabbar KJ, Wei Y, Pemmaraju N, et al. An exploratory clinical trial of bortezomib in patients with lower risk myelodysplastic syndromes. Am J Hematol 2017;92:674–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Meggendorfer M, Haferlach C, Kern W, Haferlach T. Molecular analysis of myelodysplastic syndrome with isolated deletion of the long arm of chromosome 5 reveals a specific spectrum of molecular mutations with prognostic impact: a study on 123 patients and 27 genes. Haematologica 2017;102:1502–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jeromin S, Weissmann S, Haferlach C, Dicker F, Bayer K, Grossmann V, et al. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia 2014;28:108–17. [DOI] [PubMed] [Google Scholar]
- 45. Hoeks M, Bagguley T, van Marrewijk C, Smith A, Bowen D, Culligan D, et al. Toxic iron species in lower-risk myelodysplastic syndrome patients: course of disease and effects on outcome. Leukemia 2021;35:1745–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fabre MA, de Almeida JG, Fiorillo E, Mitchell E, Damaskou A, Rak J, et al. The longitudinal dynamics and natural history of clonal haematopoiesis. Nature 2022;606:335–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moura PL, Hofman IJF, Nannya Y, Aliouat A, Mortera-Blanco T, Yoshizato T, et al. Long-term clonal inversion in an MDS-RS case with dual SF3B1 mutations. Blood 2022;140:6898–9. [Google Scholar]
- 48. Rengstl B, Newrzela S, Heinrich T, Weiser C, Thalheimer FB, Schmid F, et al. Incomplete cytokinesis and re-fusion of small mononucleated Hodgkin cells lead to giant multinucleated Reed-Sternberg cells. Proc Natl Acad Sci USA 2013;110:20729–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Main supplemental file
Descriptive statistics, deidentified clinical and experiment map of patients and healthy donors included in the study
Gene count matrix and experiment metadata for bulk RNAseq
Semi-quantitative TMT raw data, DE analysis, enrichment analysis raw data and predicted NMD analysis, comprising NBM EB, MDS-RS EB and purified RS
DESeq2 differential expression analysis of population-level RNAseq data
Seurat clustering and differential expression analysis between RS/NBM and MDS/NBM cell identity clusters in scRNAseq
STRING and Metascape enrichment analysis results, respectively separated per cluster and global (corrected by erythroid stage)
Data Availability Statement
Preprocessed and deidentified RNA-seq/proteomic data and downstream results generated in this study are available within the article and its supplementary data files. Deidentified raw data for all high-throughput datasets, the corresponding metadata, and intermediate analysis files have been deposited on the Swedish National Data Service's research data repository with restricted access in accordance with the European Union General Data Protection Regulation and are accessible upon reasonable request to the Swedish National Data Service through the respective DOI links (online data descriptions are available in both English and Swedish). Other raw data are available from the corresponding author upon request.
SND-ID 2023–118: Tandem mass tag proteomics of erythroblasts from healthy donors and erythroblasts/ring sideroblasts from patients with MDS-RS. DOI: https://doi.org/10.48723/f2xa-4005
SND-ID 2023–120: Bulk RNA sequencing of BM cells from patients with MDS-RS and healthy donors. DOI: https://doi.org/10.48723/41wa-yv42
SND-ID 2023–121: 10x single-cell RNA sequencing of BM cells from patients with MDS-RS and healthy donors. DOI: https://doi.org/10.48723/nq2a-1e03
SND-ID 2023–122: Smart-seq3 and Smart-seq3xpress single-cell RNA sequencing of BM cells from patients with MDS-RS. DOI: https://doi.org/10.48723/0f0c-p816



![Figure 1. Erythroid differentiation and enucleation remain active in SF3B1mt MDS-RS erythroblasts. A, Flow cytometry strategy for staging of erythroblast populations from BM MNC of patients with MDS-RS. Gating steps identify live and terminally differentiating erythroid cells (Lin−7AAD−GPA+), from which erythroblasts (EB) are staged according to Band 3 and integrin α4 expression (26). Integrin α4–negative cells are excluded from quantification to avoid skewing by anucleate cells. B, Mean (±SEM) cell population frequencies within flow cytometry parent populations (singlets > terminally differentiating erythroid > erythroblast subsets), quantified in NBM donors (nNBM = 4) and patients with MDS-RS (nMDS-RS = 6). Erythroid cells are quantified within the singlet population. Erythroblast subsets are quantified within the GPA+ population. C, Mean (±SEM) RS frequencies per sorted erythroblast subset and compared with frequencies in matched diagnostic BM smears (nMDS-RS = 6). D, Gating and quantification [mean (±SEM)] of DNA content (Draq-5) and intracellular Ki-67 abundance in GPA+ magnetically sorted cells (nNBM = 3; nMDS-RS = 6). E, Mean (±SEM) RS frequencies per sorted Ki67-expressing subset and compared with frequencies in matched diagnostic BM smears (nMDS-RS = 6). F, Mean (±SEM) CD71 (transferrin receptor, TFRC) median fluorescence indices per erythroblast subset (nNBM = 4; nMDS-RS = 6). G, Mean (±SEM) enucleation frequencies after 28-day 3D culture of MDS-RS BM MNCs (nMDS-RS = 4) and separated by iron granule visibility upon morphological analysis. Statistical comparison was performed by paired t test analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonstatistically significant.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/b1af/10790130/50d6076a9fe2/211fig1.jpg)
![Figure 4. SF3B1 mutations have limited impact on the gene expression of true MDS-RS HSPC. A, Principal component analysis (PCA) plots of a full-length bulk RNA-seq experiment encompassing sorted cell populations from NBM donors (nCD34 = 7, nGPA = 4, nRet = 4) and SF3B1mt MDS-RS patients (nCD34 = 6, nGPA = 5, nRS = 4, nSid = 4). Sample distribution along PC1 is visualized against PC2/PC3. B, Global overview of two integrated 10x Genomics scRNA-seq experiments encompassing sorted cell populations from NBM donors and SF3B1mt patients with MDS-RS (nNBM = 3, nMDS-RS = 5, nRS = 1). A UMAP-based bidimensional projection is displayed and separated per sample group, where each cell is visualized as one point. The dotted circle indicates the RS-enriched cell subset, which is absent in NBM. Cell types were annotated according to gene expression signatures per cluster set. Sample and cell type composition in the total dataset are shown below the UMAP plots. C, Volcano plot (left) displaying differentially expressed genes (DEG) in CD34+ MACS-enriched BM cells comparing SF3B1mt MDS-RS versus NBM. Cut-offs for significance were log2 FC > 0.5, adjusted P value <0.01. Genes were overexpressed (OE; red), underexpressed (UE; blue), or not significantly different (NS; gray). Gene set enrichment analysis (GSEA) of overexpressed genes (middle) was performed with the Enrichr Human Gene Atlas. The right UMAP heatmap displays expression of bulk OE genes in scRNA-seq. The dotted rectangle highlights the HSPC transcriptomic cluster. D, AUCell erythroid score [based on erythroid markers from An et al. (30)] mapped in the UMAP overlays and separated by mutational background. The erythroid score is similarly displayed in violin plots (gray, NBM; orange, SF3B1mt) and grouped per cell population (excluding cell subsets unrelated to erythroid development, e.g., macrophages). E, Representative CD34 and GPA FACS plots from CD34+ MACS-separated BM MNCs isolated from an patient with SF3B1mt MDS-RS and from a patient with non-SF3B1mt non-RS MDS. Lin−CD34−GPA+ cells are gated in blue and connected to representative micrographs. Scale bars, 10 μm. F, Mean (±SEM) percentage of Lin−CD34−GPA+ cells in CD34+-enriched cells (n = 3 per group). G, Mean (±SEM) cell frequencies based on morphologic analysis of Lin−CD34−GPA+ in MACS-purified CD34+ MDS-RS samples. H, UMAP projection of CD34 RNA-positive HSPCs in the scRNA-seq dataset. I, Mean (±SEM) frequencies of transcriptomically identifiable HSPC subsets as set out in H and compared between NBM and MDS-RS samples. J, Gene set enrichment analysis results for Gene Ontology biological process (GO BP) enrichment of differentially expressed genes identified in the HSPC cluster between MDS-RS and NBM cells (nNBM = 432, mean 144 cells/donor; nMDS-RS = 510, mean 102 cells/donor). ***, P < 0.001; ns, nonstatistically significant.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/b1af/10790130/d2740beaff97/211fig4.jpg)
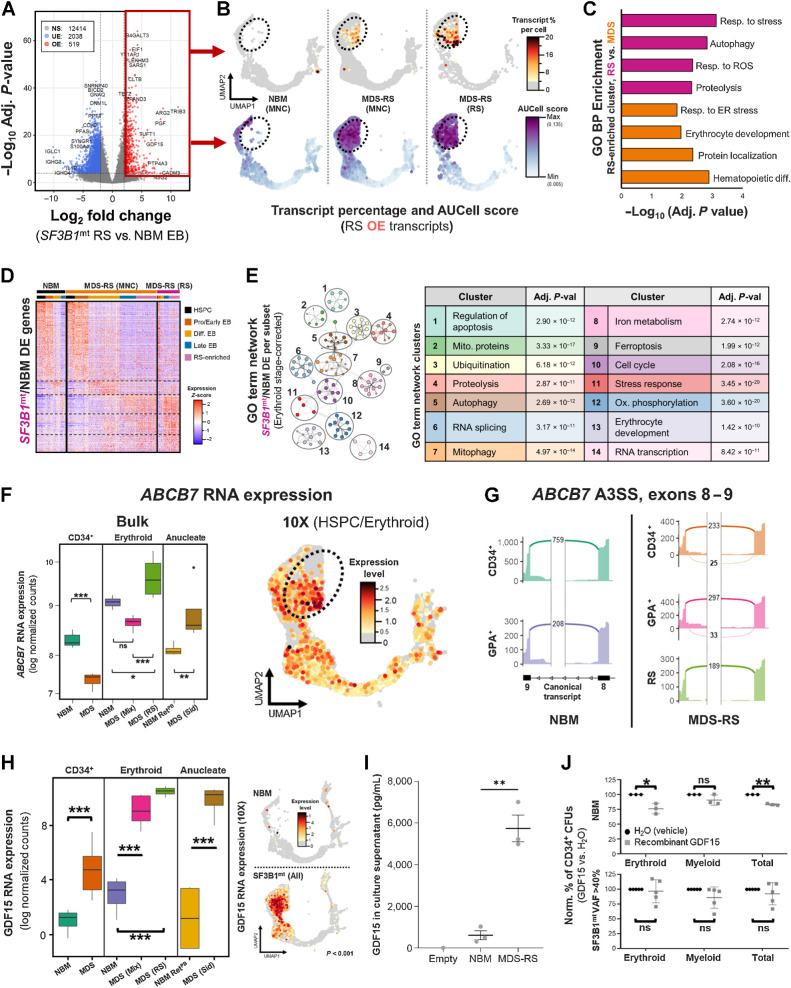
![Figure 6. Distinct RNA dynamics in erythroid differentiation intensify SF3B1mt mis-splicing. A, Proportional Venn diagram of genes undergoing statistically significant AS [FDR < 0.001, absolute difference in percentage spliced in levels (Abs. ΔPSI) > 0.2] in the HSPC (green; MDS-RS CD34+ vs. NBM CD34+), nucleated erythroid [red; MDS-RS RS vs. NBM EB, Erythroid (N)] and anucleate erythroid [blue; MDS-RS Siderocytes vs. NBM RetPB, Erythroid (A)] populations. B, Gene Ontology biological process (GO BP) enrichment analysis results comprising genes mis-spliced in both the HSPC and erythroid (N) populations are shown in the top bar chart. The Human Gene Atlas enrichment results with the lowest adjusted P value are shown in the bottom table. C, Frequency of AS events split by rMATS category in each sample group comparison. SE, skipped exon; RI, retained intron; MXE, mutual exon exclusion; A5SS, alternative 5′ splice site; A3SS, alternative 3′ splice site. Statistical comparisons of A3SS+RI frequencies were performed with Fisher exact test. D, Box plots of percent spliced-in (PSI) values of literature-validated SF3B1mt-induced A3SS events in MDS-RS samples, separated by sample type (CD34, GPA, RS). Known targeting by NMD is indicated with a red circle; in-frame events without a PTC are indicated by a blue circle. E, Box plots of PSI values in newly identified ASEs affecting known driver genes implicated in the pathogenesis of MDS and CSA. Known targeting by NMD is indicated with a red circle; PTC detection with unverified NMD is indicated with an orange circle; in-frame events without a PTC are indicated by a blue circle. F, Distribution of base pair distances from cryptic A3SS sites to canonical splice sites (horizontal axis) in HSPC and Erythroid (N). Further detail is provided in −400 bp to 0 bp for increased contrast. Lines at −30 bp and −10 bp demarcate the interval associated with SF3B1 mis-splicing (38). Additional lines at −140 bp and −330 bp demarcate additional intervals of interest. G, Sequence logos of canonical and A3SS sequences encompassing the 3′ splice site (starting at −35 bp upstream of the AG motif) and statistical comparison through a two-sample logo. H, Frequency of A3SS events per rMATS cell type comparison where the splice site shift remains in-frame (blue) or induces a frameshift event (orange). I, Frequency of exon insertion events per rMATS cell-type comparison where the splice site shift incorporates a new PTC (pink) or remains in-frame with no PTC induction (green). J and K, RNA velocity analysis of transcriptomically identified HSPC and erythroblast subsets in 10x scRNA-seq, visualizing the percentage of spliced transcripts along pseudotime in the total cell populations (violin plots) or separated by sample group (scatter plots). The analysis in J includes all transcripts, whereas K excludes ribosomal and globin transcripts. L, UMAP overlay of FACS-purified HSPC subsets and GPA+ EB from 1 SF3B1mt MDS-RS patient after Smart-seq3xpress (SS3x), visualizing true versus predicted cell-type identity. M, RNA velocity analysis of spliced RNA read percentages in the FACS-sorted SS3× experiment, analyzed independently of the 10x dataset. This graph excludes ribosomal and globin transcripts. N, Mean (±SEM) differences in PSI after 3 hours cycloheximide treatment (70 μg/mL) versus DMSO (1:1,000, vehicle) in MDS-RS CD34 and GPA cells. SF3B1mt-associated NMD-targeted ASEs with sufficient coverage are shown at far left, SF3B1mt-associated in-frame ASEs at middle-left, and endogenous NMD-targeted transcripts at middle-right. The far-right plot visualizes all ASEs. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonstatistically significant.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/b1af/10790130/53a3038b7f64/211fig6.jpg)