Abstract
Neuropsychiatric disorders (NPD) are prevalent and devastating, posing an enormous socioeconomic burden to modern society. Recent genetic studies of NPD have identified a plethora of common genetic risk variants with small effect sizes and rare risk variants of high penetrance. While exciting, there is a pressing need to translate these genetic discoveries into better understanding of disease biology and more tailored clinical interventions. Human induced pluripotent stem cell (hiPSC)-derived 2D and 3D neural cultures are becoming a promising cellular model for bridging the gap between genetic findings and disease biology for NPD. Leveraging the accessibility of patient biospecimen to convert into stem cells and the power of genome editing technology to engineer disease risk variants, hiPSC model holds the promise to disentangle the disease polygenicity, model genetic interaction with environmental factors, and uncover convergent gene pathways that may be targeted for more tailored clinical intervention.
Keywords: brain organoids, genomics, induced neurons, induced pluripotent stem cell, neuropsychiatric disorder
Neuropsychiatric disorders (NPD) are mental illnesses that have neurological basis and impact brain function, emotion, and mood. NPD mainly include schizophrenia (SZ), bipolar disorder (BD), autism spectrum disorder (ASD), major depressive disorder (MDD) and attention deficit hyperactivity disorder (ADHD). These illnesses are prevalent (about 0.64 % of adults for SZ, 2.8 % of adults for BD, and 2.8 % of children for ASD, 4.4 % of adults for ADHD, and 8.4 % of adults for MDD in US alone) and highly debilitating (Data from US National Institute of Mental Health: www.nimh.nih.gov/health/statistics). Based on 2021 National Survey on Drug Use and Health (NSDUH), more than one in five (20 %) U.S. adults (57.8 million) are estimated to have a mental illness. Given the well-known environmental risk factors such as stress, the high prevalence of NPD may be further elevated by the time-lag effects of Coronavirus Disease 2019 (COVID-19) pandemic [1]. Despite years of research efforts, the available effective treatments for these disorders remain scarce and largely rely on old drugs. For instance, most antipsychotic drugs for treating positive symptoms of SZ still target only dopamine D2 receptors (DRD2), a discovery that was made half a century ago [2]. The field is in dire need of a better mechanistical understanding of how one develops NPD and more effective druggable targets.
Understanding individual susceptibility to NPD will benefit from our knowledge on risk genes involved. It has been known for a long time that NPD are heritable, i.e., run in families. However, it is not until recently that the psychiatric field has witnessed the success in identifying genetic risk variants that predispose an individual to NPD. Through unbiased genome-wide interrogation of millions of genetic markers, i.e., single nucleotide polymorphisms (SNPs), in large population case/control samples, genome-wide association studies (GWAS) of SZ and other NPD have identified hundreds of risk loci with common genetic risk variants. For instance, the most recent SZ GWAS conducted by Psychiatric Genomics Consortium (PGC3) reported 287 independent risk loci each encompassing one to multiple genes with small effect sizes (odds ratios [OR] < 1.2) [3]. On the other side of the SZ genetic risk spectrum, there are multiple rare Copy Number Variants (CNVs, i.e., genomic segments that are duplicated or deleted, such as 22q11.2 deletion) as well as a handful genes with ultra-rare loss-of-function (LoF) mutations of high penetrance [4]. These genetic findings provide unprecedented opportunities for the neuropsychiatric field to better understand disease biology of NPD. However, the polygenicity, i.e., the involvement of many risk genes of small population effect sizes, has hindered the mechanistic understanding of disease pathogenesis of NPD. The pressing questions is how can we disentangle disease polygenicity of NPD at the molecular and cellular level and identify the convergence that may guide the clinical intervention in precision?
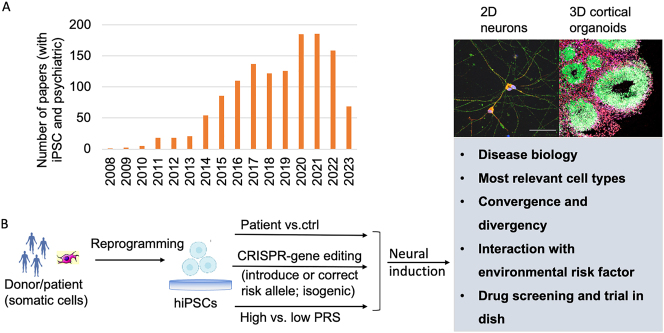
The key to address this question is to have an experimental model system that can recapitulate disease-relevant neural or brain activity and mimic the genetic perturbation. Human induced pluripotent stem cells (hiPSCs)-based neural cultures in a Petri dish have becoming a promising cellular model. There has been an exponential increase of use of iPSCs for studying psychiatric disorders during the last decade (Figure 1A). In essence, any somatic cells like blood cells from patients or healthy controls can be reprogramed into pluripotent stem cells (i.e., hiPSCs) simply by exogenously expressing some transcription factors (i.e., Yamanaka factors: Oct3/4, Sox2, Klf4, c-Myc) [5], and iPSCs can then be re-differentiated into different brain cell types that are relevant to NPD (Figure 1B). HiPSC-derived neural cells can be cultured not only in relatively pure monolayer (i.e., two dimension, 2D), but also in the form of brain spheroids or organoids (i.e., 3D) that have anatomical structures reminiscent of the developing human brain [6] (Figure 1B). For 2D models, iPSC can be efficiently differentiated into all major brain cell types, such as neural progenitor cells (NPCs), glutamatergic, GABAergic, dopaminergic, and cholinergic neurons, oligodendrocyte, astrocyte, and microglia, that are relevant to NPDs (see details in review [7]). However, despite the relatively high purity of these iPSC-differentiated cell types, their brain region or subtype identities are not clearly defined and their maturation is often comprised. In comparison, the 3D organoids are especially promising, because they can partially recapitulate not only morphological pattern but also epigenomic and transcriptomic signatures of human neurodevelopmental trajectory from early neurodevelopment to mid-or later-fetal stages [8]. Currently, organoids of different brain regions can be successfully differentiated from iPSC and assembled into models that can better recapitulate the in vivo neurodevelopment and neural circuits [6, 7, 9]. iPSC model thus allows to study disease-relevant molecular and cellular phenotypic changes in a temporal and cell type-specific manner. More importantly, empowered by Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated (Cas) nuclease-mediated genome editing system, iPSC models are amenable to genetic modification or epigenomic perturbation, e.g., by “knock-in” or “correct” a disease risk allele (Figure 1B).
Figure 1:
The use of hiPSC in studying neuropsychiatric disorders. (A) The rapid increase of studies of psychiatric disorders using iPSC in the past 15 years. The numbers of papers were searched in PubMed by using terms of “iPSC” and “Psychiatric”. (B) Both 2D neural cultures and 3D brain organoids can be generated from patient-specific iPSC lines and used for modeling different aspects of neuropsychiatric disorders towards a better understanding of disease biology and developing more tailored treatments. hiPSC, human induced pluripotent stem cell; CRISPR, Clustered Regularly Interspaced Short Palindromic Repeats; PRS, polygenic risk score.
HiPSC model has been used for studying the functional impacts of both rare (allele frequency < 1 % in population) and common (allele frequency > 1 %) NPD risk variants [7, 9]. Modeling rare risk variants is more straightforward, because such rare risk variants often alter protein coding sequences and have high penetrance, and the resultant cellular phenotypes are more interpretable. If modeling rare risk variants can be considered as lowing-hanging fruit, studying common GWAS risk variants of NPD in iPSC models is much more challenging. This is largely due to the polygenicity of NPD and the small effect sizes of common risk variants. As a result of purifying selection against deleterious mutations in the population, the small effect sizes of common risk variants are expected to yield small magnitude of biological effects that may be challenging to detect. However, functional interpretation of common GWAS risk variants of NPD is important because most known NPD genetic risk variants are common. We and others have successfully used iPSC-derived neurons as a neurodevelopmental cellular model to unravel cell-type-specific molecular and cellular impacts of common GWAS risk variants [10], [11], [12]. In addition to allowing to examine cell-type-specific effects, iPSC models can also enable the study of other context-dependent effects of genetic risk variants [13], e.g., under stress or neural activation that may mimic the interplay between specific genetic risk variants or polygenic risk backgrounds and environmental risk factors for NPD.
Despite the initial success of studying NPD using hiPSC models, our understanding of novel disease biology underlying the recent exciting genetic discoveries remain limited. A big conundrum for the field is that while the polygenic risk factors of small effects and the clinical heterogeneity of NPD require a sizable sample size in iPSC modeling study, scaling up the iPSC work is currently technically and economically a daunting challenge. The current state of iPSC modeling is somewhat reminiscent of the early stage of the candidate gene association studies of NPD when a small sample size and a small number of interrogated genetic markers often yield false-positive findings and leave the evidence hanging in the balance. Although selecting donor lines with common clinical manifestations, with rare genetic variants of high penetrance, or with extreme polygenic risk scores (PRS) may help improve study power, only scaled assays with sufficiently large number of donor lines and genetic risk variants/genes can help disentangle the biology underpins of disease polygenicity and identify convergent gene pathways that may guide precision drug screening. In this regard, it is noteworthy that National Institute of Mental Health (NIMH; USA) is teaming up different assay centers to form a Scalable and Systematic Neurobiology of Psychiatric and Neurodevelopmental Disorder Risk Genes (SSPsyGene) Consortium (https://www.nimh.nih.gov/funding/opportunities-announcements/sspsygene-consortium-frequently-asked-questions-faqs) that will leverage scalable technologies to functionally characterize about 100–250 null alleles of NPD risk genes.
Bearing in mind that each experimental model has its own pros and cons, iPSC-derived 2D and 3D models are advantageous to the animal models or postmortem brain tissues in that they enable the access of brain cell types that are relatively close to those in living human brains, which is extremely important for studying NPD. Moreover, because NPD are not caused by single variants/genes rather than many genes likely acting as gene networks that also interact with non-genetic risk factors, iPSC-derived neural cultures provide a tractable model for studying the multi-omics effect of multiplex genetic perturbation and for ascertaining the gene x environmental risk factors (e.g., stress) relevant to NPD. It is also noteworthy that given different genetic risk spectrum across NPDs, iPSC model may be more suitable for studying certain NPD than others; for instance, compared to SZ, ASD has rarer and highly penetrant disease risk variants that may manifest as early neurodevelopmental deficits which would be more suitable for iPSC modeling. Finally, although the current iPSC modeling of NPD is still primarily at the stage of understanding disease biology, potential bench-to-bedside translational applications, such as identifying more tailored drug targets based on convergent risk gene pathways and testing for genotype-based clinical drug response, are quite foreseeable. In sum, albeit a variety of limitations, iPSC-derived 2D and 3D neural models are becoming a powerful and tractable experimental model system for understanding disease biology underlying neuropsychiatric genetic findings, modeling gene x environmental risk factors, and promoting precision medicine for NPD.
Footnotes
Research funding: This work is supported by National Institute of Health (NIH) grants R01MH106575, R01MH116281, RM1MH133065 and R01AG081374, and by Charles. R. Walgreen family.
Author contributions: The author has accepted responsibility for the entire content of this manuscript and approved its submission.
Competing interests: Authors state no conflict of interest.
Informed consent: Not applicable.
Ethical approval: The local Institutional Review Board deemed the study exempt from review.
References
- 1.Penninx B, Benros ME, Klein RS, Vinkers CH. How COVID-19 shaped mental health: from infection to pandemic effects. Nat Med. 2022;28:2027–37. doi: 10.1038/s41591-022-02028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snyder SH. The dopamine hypothesis of schizophrenia: focus on the dopamine receptor. Am J Psychiatr. 1976;133:197–202. doi: 10.1176/ajp.133.2.197. [DOI] [PubMed] [Google Scholar]
- 3.Trubetskoy V, Pardiñas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604:502–8. doi: 10.1038/s41586-022-04434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh T, Poterba T, Curtis D, Akil H, Al Eissa M, Barchas JD, et al. Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature. 2022;604:509–16. doi: 10.1038/s41586-022-04556-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 6.Paşca SP. Assembling human brain organoids. Science. 2019;363:126–7. doi: 10.1126/science.aau5729. [DOI] [PubMed] [Google Scholar]
- 7.Muhtaseb AW, Duan J. Modeling common and rare genetic risk factors of neuropsychiatric disorders in human induced pluripotent stem cells. Schizophr Res. 2022;S0920–9964:00156–6. doi: 10.1016/j.schres.2022.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon A, Yoon SJ, Tran SS, Makinson CD, Park JY, Andersen J, et al. Long-term maturation of human cortical organoids matches key early postnatal transitions. Nat Neurosci. 2021;24:331–42. doi: 10.1038/s41593-021-00802-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang M, Zhang L, Gage FH. Modeling neuropsychiatric disorders using human induced pluripotent stem cells. Protein Cell. 2020;11:45–59. doi: 10.1007/s13238-019-0638-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang S, Zhang H, Zhou Y, Qiao M, Zhao S, Kozlova A, et al. Allele-specific open chromatin in human iPSC neurons elucidates functional disease variants. Science. 2020;369:561–5. doi: 10.1126/science.aay3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schrode N, Ho SM, Yamamuro K, Dobbyn A, Huckins L, Matos MR, et al. Synergistic effects of common schizophrenia risk variants. Nat Genet. 2019;51:1475–85. doi: 10.1038/s41588-019-0497-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forrest MP, Zhang H, Moy W, McGowan H, Leites C, Dionisio LE, et al. Open chromatin profiling in hiPSC-derived neurons prioritizes functional noncoding psychiatric risk variants and highlights neurodevelopmental loci. Cell Stem Cell. 2017;21:305–18. doi: 10.1016/j.stem.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang S, Sanders AR, Duan J. Modeling PTSD neuronal stress responses in a dish. Nat Neurosci. 2022;25:1402–4. doi: 10.1038/s41593-022-01172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]