Abstract
Seasonal influenza vaccines offer little protection against pandemic influenza virus strains. It is difficult to create effective pre-pandemic vaccines because it is uncertain which influenza virus subtype will cause the next pandemic. Here, we developed a nucleoside-modified mRNA-LNP vaccine encoding hemagglutinin antigens from all 20 known influenza A and B virus subtypes. This multivalent vaccine elicited high levels of cross-reactive and subtype-specific antibodies in mice and ferrets that reacted to all 20 encoded antigens. Vaccination protected mice and ferrets challenged with matched and mismatched viral strains and this protection was at least partially dependent on antibodies. Our studies indicate that mRNA vaccines can provide protection against antigenically variable viruses by simultaneously inducing antibodies against multiple antigens.
One Sentence Summary:
mRNA vaccines expressing antigens from all known influenza subtypes elicit protective responses in mice and ferrets.
There are at least 18 different influenza A virus (IAV) subtypes that circulate in animal reservoirs and these viruses occasionally enter the human population and cause a pandemic (1). Currently, H1N1 and H3N2 IAVs, as well as one or two antigenically distinct lineages of influenza B viruses (IBVs) circulate seasonally in the human population. Although surveillance programs and modeling studies have increased our knowledge of pandemic risk (2, 3), we cannot accurately predict which influenza subtype will cause the next pandemic. Several universal influenza vaccines are in development to provide protection against diverse influenza virus subtypes (4). Most universal influenza vaccines include a limited number of antigens that possess epitopes that are conserved across different influenza virus subtypes (5-7). An alternative approach for inducing universal immunity is to design multivalent vaccines that encode antigens from every known influenza virus subtype. This approach may be impractical using conventional influenza vaccine technologies, but is now feasible with nucleic-acid based vaccine platforms (8). We previously developed nucleoside-modified mRNA lipid nanoparticle (LNP) vaccines expressing hemagglutinin (HA) antigens from single influenza virus subtypes and found that these vaccines elicit antibodies against both the HA head and stalk in mice and ferrets (9, 10). Here, we generated a nucleoside-modified mRNA-LNP vaccine expressing HA antigens from all known influenza virus subtypes and found that this multivalent vaccine elicits diverse antibodies that protect mice and ferrets against matched and mismatched viral strains.
We prepared 20 different HA-encoding nucleoside-modified mRNAs encapsulated in LNPs as previously described (9). We included a representative HA from each IAV subtype and IBV lineage (fig. S1, A to C). We vaccinated groups of mice intramuscularly with a low dose (3 μg) of each individual HA mRNA vaccine to verify that each mRNA vaccine component was immunogenic. Each individual HA mRNA vaccine elicited antibodies that reacted more efficiently to the encoded HA compared to other HAs that we tested (fig. S1D). There was a low level of cross-reactivity among antibodies elicited by single HA mRNA vaccinations, which is consistent with our previous work (9) demonstrating that higher doses of vaccines are required to elicit antibodies that target conserved epitopes such as the HA stalk.
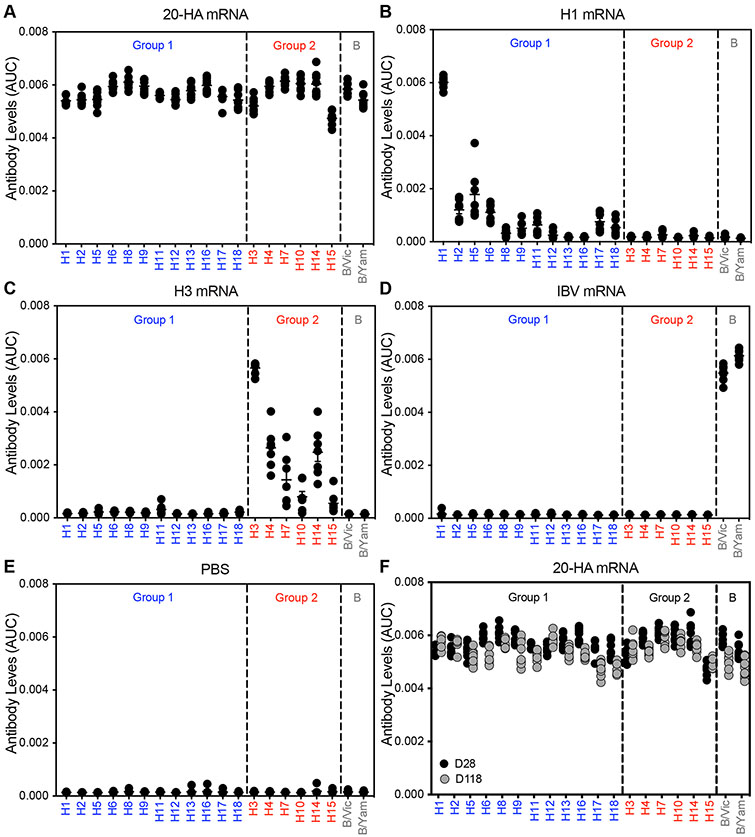
We then vaccinated mice with all 20 HA mRNA-LNPs simultaneously with a combined dose of 50 μg of HA mRNA (2.5 μg of each individual HA mRNA-LNP). As controls, we vaccinated mice with a 50 μg dose of mRNA-LNPs encoding single HAs from H1N1, H3N2, IBV or mRNA-LNPs expressing luciferase. Mice vaccinated with the 20-HA mRNA-LNPs produced antibodies that reacted to all 20 encoded HAs (Fig. 1A; fig. S2) (P<0.05, comparing vaccinated versus PBS control) whereas mice vaccinated with single HA mRNA-LNPs (Fig. 1, B to D), PBS (Fig. 1E), or mRNA-LNPs expressing luciferase (fig. S3) did not. Antibody levels in mice immunized with the 20-HA mRNA-LNP vaccine remained largely unchanged 4 months postvaccination (Fig. 1F). We created a vaccine composed of 50 μg of 20 different recombinant HA proteins (2.5 μg per protein) so that we could compare the 20-HA mRNA-LNP vaccine to a more conventional protein vaccine platform. Mice immunized with the multivalent protein vaccine produced low levels of anti-HA antibodies (fig. S4). We also tested the 20-HA mRNA-LNP vaccine in mice that were previously infected with either antigenically matched or mismatched H1N1 viruses (fig S5). Animals infected with H1N1 viruses possessed antibodies that reacted to H1 and other group 1 HAs prior to vaccination (fig. S5, A to C). H1 antibodies in pre-exposed animals were boosted by the 20-HA mRNA-LNP vaccine, but this did not come at the expense of generating new de novo antibody responses against the other HA components (fig. S5, D to F). Thus, the 20-HA mRNA-LNP vaccine elicits high levels of antibodies against all 20 encoded HAs in mice with and without prior influenza virus exposures.
Fig. 1. 20-HA mRNA-LNP vaccine elicits long-lived antibody responses that react to all 20 HAs.
(A) Mice were simultaneously vaccinated i.m. with 20 different HA mRNA-LNPs (a combined total dose of 50 μg mRNA-LNP including 2.5 μg of each individual HA mRNA-LNP). Other groups of mice were vaccinated i.m. with 50 μg of H1 mRNA-LNP (B), 50 μg of H3 mRNA-LNP (C), 50 μg of IBV HA mRNA-LNP (D), or PBS (E). Sera were collected 28 days (A to E) or 118 days (F) later and antibody reactivity to different HAs were quantified using ELISAs coated with recombinant proteins. Seven or eight mice were included for each experimental group and in some instances data points overlap. Group 1 HAs are shown in blue and group 2 HAs are shown in red. AUC; area under the curve. Data are representative of two independent experiments and are shown as mean ± SEM. Raw ELISA data curves are shown in fig S2.
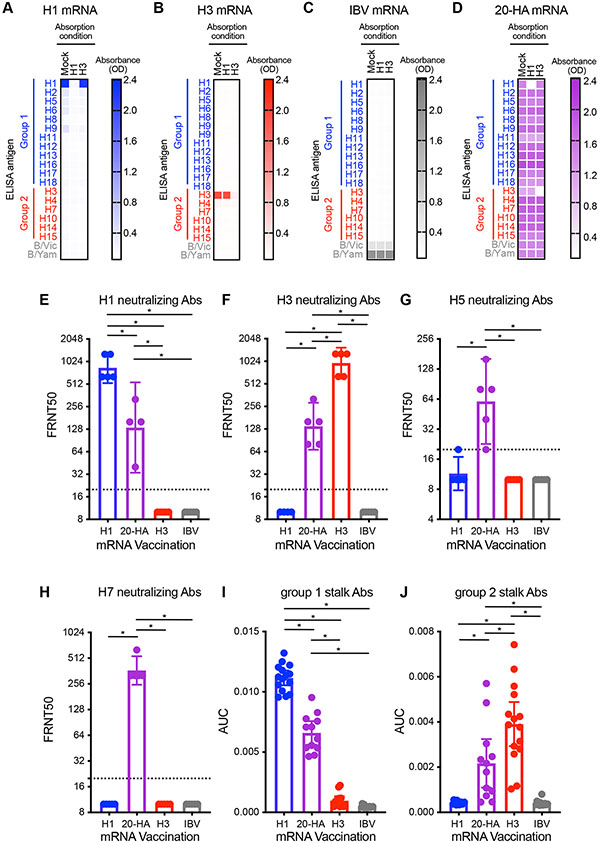
We completed absorption assays to determine the level of cross-subtype reactivity of antibodies elicited by vaccination. mRNA-LNPs encoding single HAs elicited antibodies that were efficiently depleted by beads coupled with the corresponding encoded HA (Fig. 2, A to C). H1-coupled beads efficiently depleted H1-reactive antibodies and H3-coupled beads efficiently depleted H3-reactive antibodies in the serum of mice vaccinated with the 20-HA mRNA-LNP vaccine, but these absorptions did not substantially decrease binding of antibodies reactive to other HAs in our testing panel (Fig. 2D). This indicates that the 20-HA mRNA-LNP vaccine elicits antibodies reactive to distinct HAs, rather than purely cross-reactive antibodies capable of recognizing all HA subtypes. The 20-HA mRNA-LNP vaccine elicited group 1 (H1N1 and H5N1) and 2 (H3N2 and H7N9) neutralizing antibodies (Fig. 2, E to H) and group 1 and 2 HA stalk-reactive antibodies (Fig. 2, I and J). As expected, H1 and H3 neutralizing antibodies were elicited at lower levels in mice receiving the 20-HA mRNA-LNP vaccine (which contained only 2.5 μg of H1 mRNA and 2.5 μg of H3 mRNA) compared to mice receiving 50 μg of H1 or H3 mRNA-LNPs (Fig. 2, E and F). Thus, mRNA vaccines can successfully deliver at least 20 distinct HA antigens that elicit antibodies targeting both variable and conserved epitopes.
Fig. 2. 20-HA mRNA-LNP vaccine elicits diverse antibodies targeting both conserved and variable epitopes.
Serum samples were collected from mice 28 days after (A) H1, (B) H3, (C) IBV, or (D) 20-HA mRNA-LNP vaccination. Samples were absorbed with magnetic beads coupled to recombinant H1, H3, or no HA (mock), and antibody levels remaining in the unabsorbed fraction were quantified by ELISA (A to D). Focus reduction neutralization tests (FRNT) were completed using (E) A/Michigan/45/2015 H1, (F) A/Singapore/INFIMH-16-0019/2016 H3, (G) A/Vietnam/1203/2004 H5, or (H) A/Shanghai/02/1013 H7. Titers are reported as the inverse of the highest dilutions of serum amount required to inhibit 50% of virus infection are reported. HA stalk-reactive antibodies were quantified by ELISA using “headless” (I) group 1 (H1) and (J) group 2 (H3) recombinant proteins. AUC; area under the curve. (A to H) Six mice were included for each experimental group. (I and J) Twelve mice were included for each experimental group. Data are representative of two or three independent experiments and are shown as mean ± SEM. Data in panels A to D are shown as mean. Data in panels E to H are shown as geometric mean ± 95% CI and were compared using a one-way ANOVA with Tukey’s post-test on log-transformed values. *P<0.05.
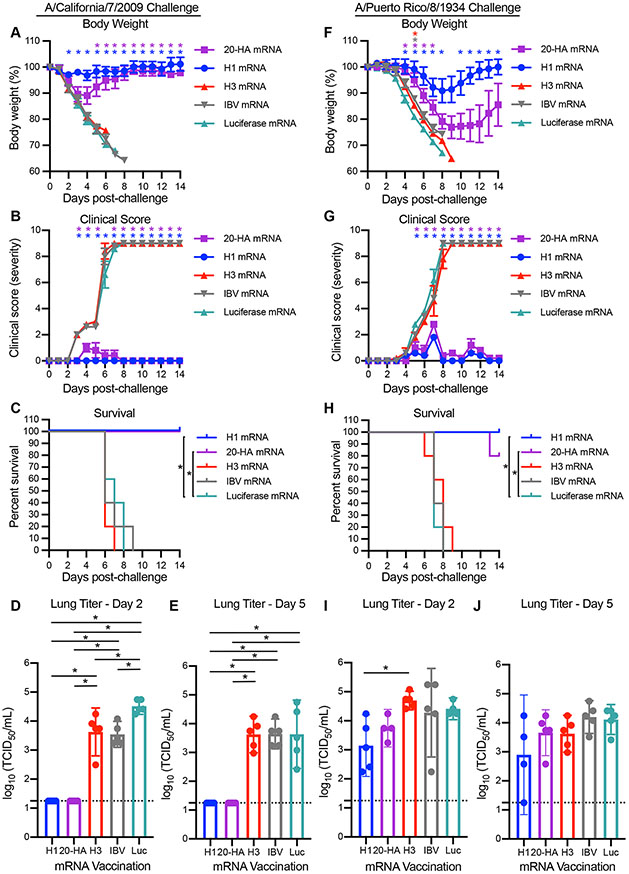
We challenged mice 28 days after vaccination with an H1N1 virus (A/California/07/2009) that was similar (97.2% HA amino acid homology) to the H1 component of the vaccine (A/Michigan/45/2015) or an H1N1 virus (A/Puerto Rico/08/1934) that was antigenically distinct (81.8% HA amino acid homology) compared to the H1 component of the vaccine. Antibodies elicited by the 20-HA mRNA-LNP vaccine and H1 mRNA-LNP vaccine bound to HAs from A/California/07/2009 and A/Puerto Rico/08/1934 (fig. S6, A and B), but only neutralized the A/California/07/2009 virus (fig. S6, C and D). Mice vaccinated with H3 mRNA-LNP, IBV mRNA-LNP, or an mRNA-LNP expressing luciferase rapidly lost weight, displayed severe clinical signs, and died between 7-9 days after infection with either A/California/07/2009 (Fig. 3, A to C) or A/Puerto Rico/8/1934 (Fig. 3, F and G). Mice vaccinated with H1 mRNA-LNP or the 20-HA mRNA-LNP did not lose as much weight following infection with A/California/07/2009 (Fig. 3A), displayed few clinical signs (Fig. 3B), and survived the viral challenge (Fig. 3C). A/California/07/2009 viral titers in the lungs of mice vaccinated with H1 mRNA-LNP or the 20-HA mRNA-LNP were undetectable 2 and 5 days following infection (Fig. 3, D and E). Mice vaccinated with H1 mRNA-LNP or the 20-HA mRNA-LNP initially lost weight following infection with the mismatched A/Puerto Rico/08/1934 virus (Fig. 3F), displayed clinical signs (Fig. 3G), but then began recovering 7-8 days after infection (Fig. 3G) and most of these mice survived (Fig. 3H). A/Puerto Rico/08/1934 viral titers in the lungs were similar between the experimental groups 2 and 5 days following infection (Fig. 3, I and J). Thus, the 20-HA mRNA-LNP vaccine provided mice different degrees of protection against matched and mismatched viral strains.
Fig. 3. The 20-HA mRNA-LNP vaccine protects mice from challenge with antigenically matched and mismatched distinct H1N1 strain.
Mice were vaccinated with mRNA-LNPs encoding H1 (blue), H3 (red), IBV (gray), luciferase (green) or 20 HAs (purple) and then 28 days later they were infected i.n. with A/California/7/2009 (5 LD50) or A/Puerto Rico/8/1934 H1N1 (2 LD50) influenza virus respectively. (A) Weight loss, (B) clinical scores, and (C) survival were monitored for 14 days following A/California/7/2009 infection. Virus levels in lung homogenate samples isolated 2 (D) and 5 (E) days following infection were quantified using TCID50 assays. (F) Weight loss, (G) clinical scores, and (H) survival were monitored for 14 days following A/Puerto Rico/8/1934 H1N1 infection. Virus levels in lung homogenate samples isolated 2 (I) and 5 (J) days following infection were quantified using TCID50 assays. Horizontal dotted lines in D, E, I, and J denote limit of detection of the assay and samples with no detectable titers were assigned a titer at this limit of detection. Five mice were included per group. Data in A, B, F, and G are shown as mean ± SEM and were analyzed by mixed model ANOVA with Geisser-Greehouse correction and Sidak’s multiple comparisons. For animals that died, weight on day prior to death was carried forward for statistical analyses. Differences compared to Luciferase mRNA vaccination are indicated in A, B, F, and G; *P<0.05. Data in C and H were analyzed using a log-rank test *P<0.05. Data in D, E, I, and J are shown as mean ± 95% CI and titers were compared using a one-way ANOVA with Tukey’s post-test; * P <0.05.
To determine if the 20-HA mRNA-LNP vaccine requires all 20 HA components, we vaccinated mice with a combination of every HA mRNA-LNP except H1 mRNA-LNP (19 HA mRNA-LNP vaccine) and then challenged these animals with A/California/07/2009 or A/Puerto Rico/08/1934 H1N1 viruses (fig. S7). Mice vaccinated with the multivalent 19-HA mRNA-LNP lacking the H1 mRNA-LNP rapidly lost weight (Fig. S7A), displayed clinical signs (fig. S7B), and frequently died (fig. S7C) following infection with A/California/07/2009 H1N1. This suggested that the H1 component of the 20-HA mRNA-LNP vaccine was critically important for eliciting protective responses against the A/California/07/2009 H1N1 strain. Mice vaccinated with the 19-HA mRNA-LNP lost weight (fig. S7A), displayed some clinical signs (fig. S7B), but survived following A/Puerto Rico/08/1934 infection (fig. S7C). Although the immunological basis of this finding is unclear, the HA of A/Puerto Rico/08/1934, but not A/California/07/2009, likely shares a conserved epitope with a non-H1 immunogen in the 20-HA mRNA-LNP vaccine.
To further probe the mechanism(s) by which the 20-HA mRNA-LNP vaccine provides protection against different H1N1 virus strains, we depleted CD4+ and CD8+ T cells from mice vaccinated with the 20-HA mRNA-LNP vaccine and then challenged these animals with A/California/07/2009 or A/Puerto Rico/08/1934 (fig. S8, A to D). Vaccinated mice lacking CD4+ and CD8+ T cells had similar survival rates compared to vaccinated mice with intact T cells. Mice that received a passive transfer of serum from 20-HA mRNA-LNP-vaccinated mice survived A/California/07/2009 H1N1 infection, but were not fully protected against A/Puerto Rico/08/1934 H1N1 infection (fig. S8, E to F). We hypothesized that antibodies elicited by the 20-HA mRNA-LNP vaccine contribute to protection against mismatched viral strains through non-neutralizing mechanisms such as antibody-dependent cellular cytotoxicity (ADCC) (11). Antibodies elicited by the 20-HA mRNA-LNP vaccine efficiently mediated ADCC with cells expressing either matched or mismatched HAs (fig. S9). Antibodies elicited by the high dose of the H1 mRNA-LNP vaccine were able to mediate ADCC with cells expressing a mismatched H1 but not with a matched H1 (fig. S9). This was likely because the high dose of the H1 mRNA-LNP vaccine elicited high levels of neutralizing antibodies (fig S6), which have been previously found to inhibit ADCC activity mediated by non-neutralizing antibodies (12, 13).
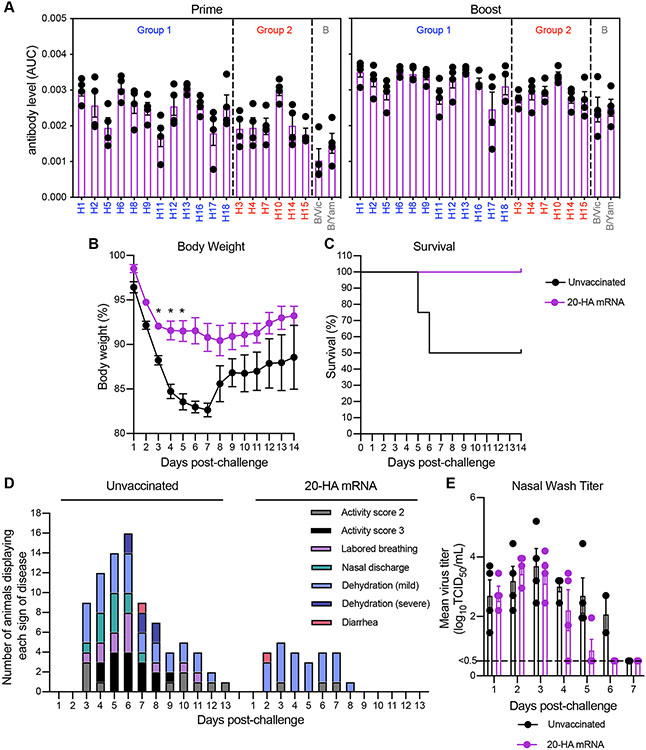
Finally, we completed a prime/boost vaccination experiment in ferrets to mimic the dosing schedule initially employed for SARS-CoV-2 mRNA vaccines (14, 15). Each ferret produced antibodies reactive to all 20 HAs after a single vaccination, and antibody levels increased after the booster vaccination that was delivered 28 days later (Fig. 4A). We challenged vaccinated and unvaccinated ferrets with an avian H1N1 virus (A/Ruddy turnstone/Delaware/300/2009) that was distinct from the H1 HA that was included in the vaccine (81.8% HA amino acid homology with vaccine H1 component) to mimic a pandemic with an unknown viral strain. Unvaccinated animals rapidly lost weight and two out of four animals died after infection, while vaccinated ferrets lost less weight and all animals survived following infection (Fig. 4, B and C). Unvaccinated animals displayed more clinical signs relative to vaccinated animals after infection (Fig. 4D). Viral titers in nasal washes were similar in unvaccinated and vaccinated animals at days 1-4 after infection, but virus was cleared more efficiently in vaccinated animals at days 5 and 6 after infection (Fig. 4E). Thus, the 20-HA mRNA-LNP vaccine protected ferrets against an antigenically mismatched avian H1N1 virus.
Fig. 4. 20-HA mRNA-LNP vaccination protects ferrets from challenge with an antigenically distinct H1N1 strain.
Ferrets were primed with 60 μg of the 20-HA mRNA-LNP vaccine (3 μg of each HA mRNA-LNP) and then boosted with the same vaccine dose 28 days later. (A) Sera were collected 28 days after the first and second vaccinations and antibody reactivity to different HAs were quantified using ELISAs coated with recombinant proteins. Twenty-eight days after the second vaccination, ferrets were infected i.n. with 106 TCID50 of A/Ruddy turnstone/Delaware/300/2009 H1N1 influenza virus. As a control, unvaccinated animals were also infected with the virus. (B) Weight loss (C) survival and (D) signs of disease were monitored for 14 days following infection. The same animal can make multiple contributions to the graph in (D). (E) Virus levels in nasal wash samples isolated 1-7 days following infection were quantified using TCID50 assays. Horizontal dashed line indicates limit of detection. 4 ferrets were included for each experimental group. Data shown are means ± SEM (A, B, and E). Data in B and E are shown as mean ± SEM and were analyzed by mixed model ANOVA with Geisser-Greehouse correction and Sidak’s multiple comparisons; * P <0.05. For animals that died, weight on day prior to death was carried forward for statistical analyses. Data in C were analyzed using a Mantel-Cox log-rank test.
In this report, we present an alternative strategy for inducing universal immunity against distinct influenza virus strains. We previously demonstrated that nucleoside-modified mRNA-LNP vaccines expressing HA and conserved influenza virus antigens are immunogenic in mice (9, 14). Instead of focusing on immunogens to elicit antibodies against epitopes that are conserved among many different influenza virus strains, we designed a vaccine that encodes separate immunogens from all known IAV subtypes and IBV lineages. Previous studies have shown that cocktails of virus-like particles encoding antigens from four different influenza virus subtypes are immunogenic in mice when delivered intranasally (16). We found that antigens from at least 20 distinct influenza viruses can be simultaneously delivered via mRNA-LNPs. The production and standardization of different antigens expressed by mRNA-LNP vaccines is simpler compared to other vaccine approaches (17)(18) and there may be unique properties of mRNA vaccines that allow the induction of immune responses to multiple antigens without noticeable immunodominance biases, even in the context of pre-existing immune responses. For example, we previously reported that mRNA-LNP vaccines induce long-lived germinal center reactions in mice (19), a finding that has been recently found to occur in SARS-CoV2 mRNA-vaccinated humans as well (20, 21). Long-lived germinal centers may facilitate the simultaneous induction of immune responses against multiple epitopes, including epitopes that are usually subdominant.
Further studies will be required to fully elucidate the mechanisms by which the 20-HA mRNA vaccine provides protection. Our studies suggest that protection against antigenically matched strains is mediated by neutralizing antibodies, whereas protection against mismatched viral strains may occur through non-neutralizing mechanisms such as ADCC. Over the course of our studies, we used antigenically matched as well as antigenically mismatched challenge strains to mimic the emergence of a novel pandemic influenza virus strain. It is likely that mRNA influenza vaccines that are imperfectly matched to novel pandemic influenza virus strains will not provide sterilizing immunity, but instead, limit disease severity and protect against death through non-neutralizing mechanisms. This may be occurring with SARS-CoV-2 variant infections in humans immunized with SARS-CoV-2 mRNA vaccines that were developed using spike sequences obtained from viral strains isolated early in the pandemic. In most cases, symptoms and severity are greatly reduced and virus is cleared faster in vaccinated individuals infected with antigenically drifted SARS-CoV-2 variants (22, 23).
Our overall approach will likely be useful for infectious diseases other than influenza viruses. Multivalent mRNA-LNP vaccines may be applied against other variable pathogens, such as coronaviruses and rhinoviruses. For example, vaccine manufacturers around the globe are currently contemplating multivalent SARS-CoV-2 mRNA vaccines to combat antigenically distinct strains (24). Additional studies will be required to determine the maximum number of antigens that can be simultaneously delivered via mRNA-LNP vaccines and the underlying immunological mechanisms that allow the induction of responses against multiple antigens.
Supplementary Material
Acknowledgments:
We thank all members of the Hensley laboratory for helpful discussions related to this project. We thank A. McDermott and B. Graham (Vaccine Research Center at the National Institutes of Health) for providing plasmids to express “headless” H1 and H3 proteins and J. Bloom (Fred Hutchinson Cancer Center) for providing plasmids to generate viruses expressing GFP.
Funding:
This project has been funded in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. 75N93021C00015 (S.E.H.), Contract No. 75N93019C00050 (S.E.H.), 1R01AI108686 (S.E.H.) and R56AI150677 (S.E.H.). C.P.A. was supported by the Training in Emerging Infectious Diseases T32AI055400. S.E.H. holds an Investigators in the Pathogenesis of Infectious Disease Awards from the Burroughs Wellcome Fund.
Footnotes
Competing interests: S.E.H. reports receiving consulting fees from Sanofi Pasteur, Lumen, Novavax, and Merck for work unrelated to this manuscript. S.H.Y.F. and Y.K.T. are employees of Acuitas Therapeutics, a company focused on the development of lipid nanoparticulate nucleic acid delivery systems for therapeutic applications. Y.K.T. is named on patents describing the use of modified mRNA lipid nanoparticles. D.W. is a coinventor on patents that describe the use of nucleoside-modified mRNA as a platform to deliver therapeutic proteins: D.W. and N.P. are coinventors on patents describing the use of modified mRNA in lipid nanoparticles as a vaccine platform. S.E.H. and D.W. filed a provisional patent on this work. T.G. is now an employee at GSK but was working at a postdoctoral fellow at the University of Pennsylvania while completing experiments for this manuscript. C.P.A. and M.J.B. are now employees of Pfizer but were working as graduate students at the University of Pennsylvania while completing experiments for this manuscript.
Data and materials availability: The data that support the findings of this study are included in the manuscript. All materials used in this manuscript are available from the authors upon reasonable request.
Other Supplementary Materials for this manuscript includes the following:
MDAR Reproducibility Checklist
An acknowledgment list for all GISAID sequences used in this study
References and Notes
- 1.Krammer F. et al. , Influenza. Nat Rev Dis Primers 4, 3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson MI et al. , Fogarty International Center collaborative networks in infectious disease modeling: Lessons learnt in research and capacity building. Epidemics 26, 116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrington WN, Kackos CM, Webby RJ, The evolution and future of influenza pandemic preparedness. Exp Mol Med 53, 737 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erbelding EJ et al. , A Universal Influenza Vaccine: The Strategic Plan for the National Institute of Allergy and Infectious Diseases. J Infect Dis 218, 347 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yassine HM et al. , Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med 21, 1065 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Corbett KS et al. , Design of Nanoparticulate Group 2 Influenza Virus Hemagglutinin Stem Antigens That Activate Unmutated Ancestor B Cell Receptors of Broadly Neutralizing Antibody Lineages. mBio 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nachbagauer R. et al. , A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat Med 27, 106 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Graham BS, Mascola JR, Fauci AS, Novel Vaccine Technologies: Essential Components of an Adequate Response to Emerging Viral Diseases. JAMA 319, 1431 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Pardi N. et al. , Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat Commun 9, 3361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willis E. et al. , Nucleoside-modified mRNA vaccination partially overcomes maternal antibody inhibition of de novo immune responses in mice. Sci Transl Med 12, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiLillo DJ, Tan GS, Palese P, Ravetch JV, Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat Med 20, 143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He W. et al. , Epitope specificity plays a critical role in regulating antibody-dependent cell-mediated cytotoxicity against influenza A virus. Proc Natl Acad Sci U S A 113, 11931 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leon PE et al. , Optimal activation of Fc-mediated effector functions by influenza virus hemagglutinin antibodies requires two points of contact. Proc Natl Acad Sci U S A 113, E5944 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freyn AW et al. , A Multi-Targeting, Nucleoside-Modified mRNA Influenza Virus Vaccine Provides Broad Protection in Mice. Mol Ther 28, 1569 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Topol EJ, Messenger RNA vaccines against SARS-CoV-2. Cell 184, 1401 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartzman LM et al. , An Intranasal Virus-Like Particle Vaccine Broadly Protects Mice from Multiple Subtypes of Influenza A Virus. mBio 6, e01044 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pardi N, Hogan MJ, Weissman D, Recent advances in mRNA vaccine technology. Curr Opin Immunol 65, 14 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Pardi N, Hogan MJ, Porter FW, Weissman D, mRNA vaccines - a new era in vaccinology. Nat Rev Drug Discov 17, 261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pardi N. et al. , Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J Exp Med 215, 1571 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turner JS et al. , SARS-CoV-2 infection induces long-lived bone marrow plasma cells in humans. Nature 595, 421 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Lederer K. et al. , Germinal center responses to SARS-CoV-2 mRNA vaccines in healthy and immunocompromised individuals. Cell 185, 1008 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheikh A. et al. , SARS-CoV-2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet 397, 2461 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez Bernal J. et al. , Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N Engl J Med, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLean G. et al. , The Impact of Evolving SARS-CoV-2 Mutations and Variants on COVID-19 Vaccines. mBio 13, e0297921 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.