Abstract
Introduction:
As machine learning (ML) and artificial intelligence (AI) expand to many segments of our society, they are increasingly being used for drug discovery. Recent deep learning models offer an efficient way to explore high-dimensional data and design compounds with desired properties, including those with antibacterial activity.
Areas covered:
This review covers key frameworks in antibiotic discovery, highlighting physicochemical features and addressing dataset limitations. The deep learning approaches here described include discriminative models such as convolutional neural networks, recurrent neural networks, graph neural networks, and generative models like neural language models, variational autoencoders, generative adversarial networks, normalizing flow, and diffusion models. As the integration of these approaches in drug discovery continues to evolve, this review aims to provide insights into promising prospects and challenges that lie ahead in harnessing such technologies for the development of antibiotics.
Expert opinion:
Accurate antimicrobial prediction using deep learning faces challenges such as imbalanced data, limited datasets, experimental validation, target strains, and structure. The integration of deep generative models with bioinformatics, molecular dynamics, and data augmentation holds the potential to overcome these challenges, enhance model performance, and utlimately accelerate antimicrobial discovery.
1. Introduction
The introduction of antibiotics significantly improved the quality of health in modern society by extending human lifespan by as much as one to two decades [1]. Although antibiotics have provided countless benefits in healthcare, agriculture, zootechnics, and other areas, their unlimited and improper use has led to the development of pan-drug resistant microbes, which were responsible for more than 4 million deaths globally in 2019 alone [2,3]. The number of deaths caused by SARS-CoV-2 prior to vaccination campaigns in 2020, approximately 3 million, is comparable to the current number of deaths caused by drug-resistant infections [4]. The World Health Organization (WHO) has elaborated a global action plan that aims to improve the surveillance and understanding of antimicrobial resistance, optimize the use of antimicrobial drugs, and, particularly, ensure sustained funding in countering antibiotic resistance by supporting the Member States to develop national action strategies [5]. Besides the rapid evolution of resistance, traditional methods for the discovery and development of new antibiotics are no longer cost- or time-effective. In fact, it takes an average of 15 years to bring a new drug to the market, a timeline that includes drug discovery and development, preclinical and clinical trials, drug review, and post-market safety monitoring by the U.S. Food and Drug Administration (FDA) [6]. Shortening this timeline represents an orthogonal challenge that laboratories, companies, and political institutions need to address. Investments in both research and development, along with innovation and a viable marketplace, are key to tackling the antibiotic crisis.
Historically, new antibiotics were identified by screening microbes from the soil and looking for secondary metabolites. Starting from these molecules, many derivatives have been synthesized; these derivatives have improved bioactivities and are more likely than the original molecules to meet clinical standards [7]. More recent discovery approaches involve screening large libraries by using high-throughput methods. However, the unexplored chemical space for finding drug-like molecules is immense, and screening innumerable probable combinations is costly and time-consuming [8]. Ultimately, the discovery of a new antibiotic has always involved a large degree of chance and serendipity.
In this context, artificial intelligence (AI) and machine learning (ML) represent an innovative, emerging approach for speeding up the identification of new antimicrobial drugs [9–13]. AI is defined as the capability of computers to perform tasks that usually require human intelligence, while ML is a subcategory of AI that uses models to learn from datasets. Thus, the combination of computer science and large data sets is used to train ML algorithms, generating a system applicable to several functions, such as the design of new drugs, the prediction of their activity, the optimization of previously known molecules, and the identification of antimicrobial resistance (AMR) markers. Machine-learning algorithms could eventually be used for the diagnosis of diseases and the prescription of antibiotics [9,14].
Computer-aided drug discovery techniques are becoming essential in the preliminary stages of drug development. Recently, over 2,603 encrypted peptide antibiotics were discovered through a computational method able to search the human proteome for antimicrobial sequences [15–19]. Fifty-six of these sequences showed potent anti-infective activity in two different mouse models. Several of these encrypted peptides also synergized with each other at nanomolar concentrations to target pathogens. Other studies based on genetic algorithms demonstrated the ability of machines to design effective antibiotics [20]. Moreover, quantitative structure-activity relationship (QSAR) methods have been extensively used to predict the activity of new antimicrobial molecules by correlating biological properties and molecular descriptors [21–23]. Such methods present multiple advantages over non-quantitative methods, such as the wide or random exploration of sequence space, because quantitative explorations yields results that can be tracked and learnt heuristically and computationally, serving as the basis for guided design [24–26]. The predictions rely on the computational representation of antibiotic candidates, which can be used as input for the ML drug discovery pipeline. Topological properties and descriptors derived from quantum mechanics are usually employed. Hundreds of measurements have been extracted and combined in online platforms to describe amino acid residues and other small molecules (Table 1) [23].
Table 1.
Online tools for the extraction of molecular descriptors.
| Antibiotic compounds | Molecular descriptor tools | References |
|---|---|---|
|
| ||
| Peptides/proteins | Z-scales | [83,84] |
| VHSE | [83,85] | |
| T-scales | [83,86] | |
| ST-scales | [83,87] | |
| MS-WHIM | [83,88] | |
| FASGAI | [83,89] | |
| BLOSUM | [83,90] | |
| ProtFP | [83,91] | |
| Aaaindex | [92,93] | |
| Small molecules | alvaDesc | [94] |
| ChemAxon | [95] | |
| ISIDA/QSPR | [96] | |
| MOLD2 | [97] | |
| QuBiLS-MIDAS | [98] | |
| Rdkit | [99,100] | |
Based on the adopted methodologies, the computational tools developed so far for antibiotic discovery can be grouped into two main categories. The first category includes traditional ML-based predictors, while the second category comprises deep-learning based methods, which draw upon larger sets of data and longer training compared to the former and can automatically learn high-level features from raw data. Here, we provide an overview of the main features applied as descriptors for peptides and small molecules, describe the primary deep-learning (DL) approaches used to develop new antibiotics and conclude with a section highlighting the main limitations encountered so far in this emerging field. This review offers valuable insights into how AI’s transformative role in antibiotic discovery by expediting new drug development, optimizing molecule design, and combating drug-resistant infections.
2. Deep learning models for antimicrobial drug discovery
2.1. Input representation
Before selecting which DL model to employ, it is important, even more than the algorithm itself, to choose the right representation for the input data [27]. Indeed, the input representation drives how the model learns about the molecules, thus playing an important role in the algorithm coding. Translating a molecule into vectors of numbers is called “molecular featurization” [27]. There are three main categories that describe the representation of the input data: one-dimensional [(1D), e.g., string-based], two-dimensional [(2D), e.g., graphical-based], and three-dimensional [(3D), e.g., coordinate-based]. The most common string-based representation is SMILES (Simplified Molecular Input LineEntry System), a sequence of characters based on symbols of atoms or, more specifically, amino acids, which are easily represented by computers [28–31]. Other 1D-like annotations such as Hierarchical Editing Language for macromolecules (HELM) have been also introduced to describe a broad range of macromolecules (e.g., peptides, antibodies, chemical modifiers)[32]. Although string-based sequences such as SMILES annotation are the easiest way to represent molecules, they cannot directly describe the molecular structures of compounds. Alternatively, it is possible to represent molecules in 2D by using neural network-based graphs where each node of the network describes an atom and the neural edges correspond to the bonds. Lastly, it is possible to represent molecules in 3D using point clouds, which capture not only atomic bonds, but also details about conformational preferences. More complex biophysical information is contained in 2D and 3D representations than in 1D representations. Although in the past few years, 2D and 3D representations have been applied to generative models, there are disadvantages associated with these types of input representations. For example, they cannot easily be applied to large molecules because they are computationally expensive. Also, graphs and point clouds used in autoregressive generative models, which impose a canonical ordering among atoms, can introduce bias into the algorithm, potentially limiting the model’s performance. This bias arises from the discrepancy between constitutional isomers. As a result, accurately representing diverse molecular structures becomes challenging, impacting the overall effectiveness of the model [28]. In several prediction tasks, peptides are extensively represented by their amino acid composition and physicochemical properties [33] (Table 1). Another common way to represent peptide molecules is through their primary structure, which refers to their amino acid sequence. A peptide with length L can be naturally encoded by using a string of characters or integers that is L units long. In this scenario, a peptide can be represented by a matrix with dimensions of L x n, where each amino acid corresponds to a unique n-dimensional vector able to capture peptide properties. Alternatively, studies based on model architectures such as variational autoencoders, generative adversarial networks, or attention networks, used the latent representation learned from deep neural networks as peptide representation [33].
2.2. Deep-learning approaches
Computers can use different algorithmic approaches to learn subjects from data and to provide outputs. We can distinguish two main types of models: discriminative and generative models. Whereas the first literally separates data points into two different classes that it then uses for classification or regression, the latter generates a probability based on the distribution of a dataset [34]. In the following sections, we describe the structure underlying DL-based discriminative and generative frameworks, and list examples of their application for antibiotic design.
2.2.1. Discriminative DL-models
2.2.1.1. Architecture and applications of NNs in predicting antibiotic activity
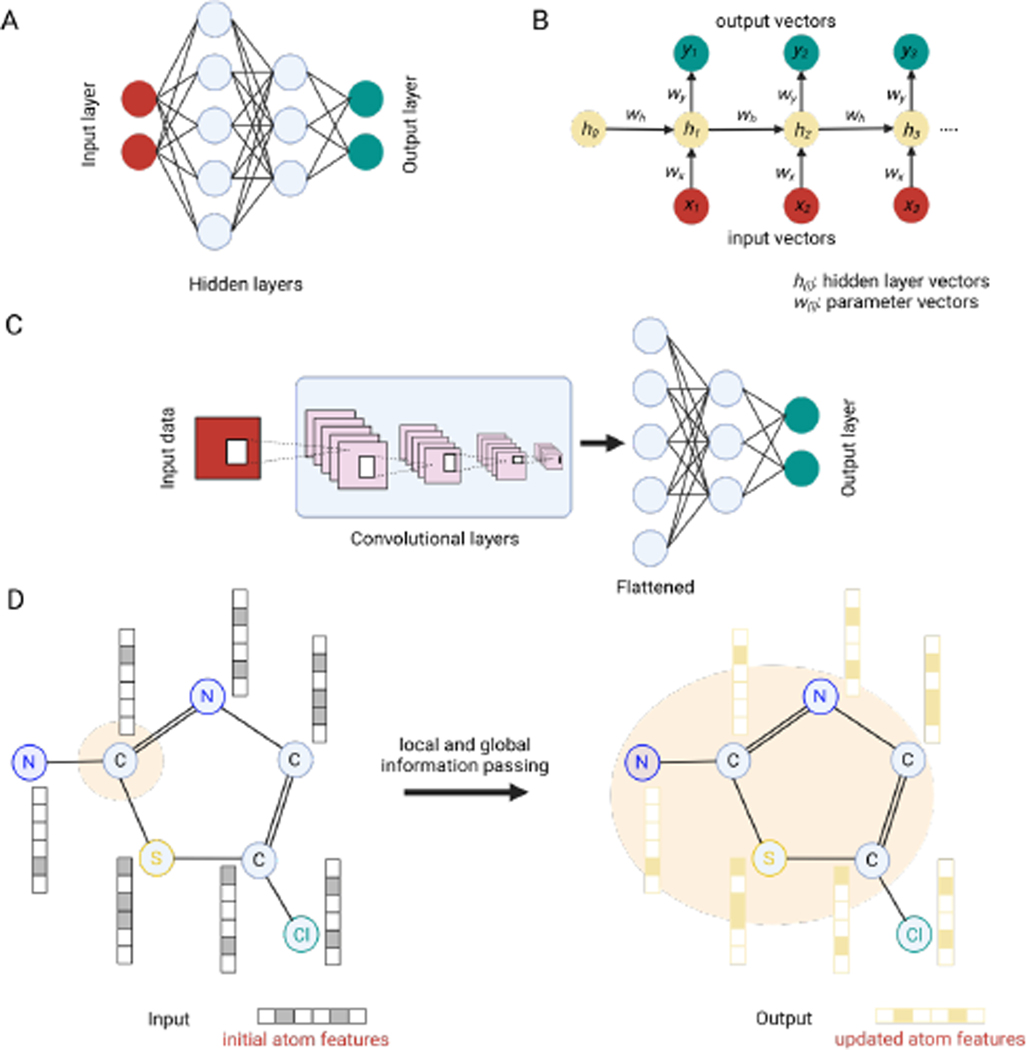
Historically, the architecture of NNs is composed of a fully-connected input layer, several hidden layers, and an output layer (Figure 2A). For antibiotic discovery, these layers are used to capture complex relationships between the structure and activity of antibiotics using nonlinear transformations [33,35,36]. The hidden layers are constructed from a set of nodes that serve to process input data (e.g., usually vectors) and combine and transmit the signals to the output layer. The model predictions are evaluated by the comparison of predicted and expected values and the iterative backpropagation of errors from the output to hidden layers to minimize the loss, i.e., the difference between predicted and expected values, through the optimization of network parameters, such as weight and bias values. Despite computational costs, the capacity (modeling power) of NNs usually grows with their sizes (i.e., number of hidden layers and hidden nodes) when the amount of data is large. The so-called deep-neural network (DNN) is trained on big datasets and used for predictions over the vast chemical spaces of small molecules and macromolecules, such as peptides [35].
Figure 2. Overview of neural network models.
(A) Fully-connected, (B) recurrent, (C) convolutional, and (D) graph neural networks. Models are frequently used for the prediction and design of antimicrobial peptides (AMPs) and small molecules. This figure was created in BioRender.com.
Currently implemented deep learning (DL) models to predict antimicrobial peptides (AMPs) and small molecules mainly rely on the convolution, recurrent, and graphical NN approaches [37,38]. Depending on their algorithms, DL models learn appropriate feature encoding methods to most accurately predict which peptides and small molecules have antimicrobial activity [33,36].
2.2.1.2. Convolutional neural networks (CNNs)
For convolutional neural networks (CNNs) [39], the raw input dataset is encoded in the form of multidimensional arrays and passed through convolutional layers composed of multiple 2D-, 3D- and N-dimensional kernel matrices of tunable width and height (Figure 2C). Convolution operations between kernels and inputs in each layer are applied to extract features sequentially; thus, the hidden features learned from the last few CNN layers can expressively represent the given input with respect to the modeling task of interest [17,18]. Additionally, the CNNs may contain optional pooling layers used to merge and reduce the size or resolution of feature maps. Finally, the feature representations of the last convolutional layer may be flattened and taken as input to fully connected layers to yield predicted values [33,36]. By adopting small kernels instead of fully-connected networks, CNNs exploit the local structures of inputs and are suitable for structural data whose local elements are highly correlated with each other, including images, sequences, and languages.
In the case of AMPs, each amino acid residue is represented by a single character or one-hot encoding feature in the context of 20-bit binary vectors to encode amino acid compositional information. To train the CNN models, the width and height of the matrices can represent amino acids and their physico-chemical properties as numerical values that are tunable hyper-parameters. In this respect, Deep-AmPEP30, a two-layered CNN model with additional pooling layers, was developed based on PseKRAAC reduced amino acid composition: experimentally validated by screening the genome of Candida glabrata, short-length peptides were predicted to have antimicrobial activity with an accuracy of 77% [40]. Another proposed CNN model, constructed from an embedding layer for encoding peptide sequences and 64 multi-scale convolutional layers followed by maximum pooling layers, resulted in an accuracy of 92% on the APD3 benchmark dataset [41]. In this approach, the embedding layer vector dimension was 128 and the peptide sequences were encoded as numerical vectors of real numbers (0–20) with a length of 200 amino acid residues. Also, the multiple kernel layers had various filter lengths to ensure that all latent features were learned. Moreover, introducing attention layers in the architecture of NN models has resulted in the identification of antimicrobial peptides from the human gut microbiome with significant efficacy in a mouse model of bacterial lung infection [42]. Moreover, a combined model of CNNs, regression, and decision tree was employed to optimize small molecules for inhibiting RNA translation in Mycobacterium tuberculosis, revealing important features for molecule-RNA binding and demonstrating the reliability of CNN as a predictor. Two representations, simplified molecular-input line-entry system (SMILES) and low-resolution images, were used showing that CNNs can reliably predict small molecule inhibitors for RNA translation in M. tuberculosis using either input type, with accurate results obtained from low-resolution images [43].
2.2.1.2. Recurrent neural networks (RNNs)
Recurrent neural networks (RNNs) have been developed to process inputs having sequential structures (e.g., time-series or biological sequences), [44]. In RNNs, sequential information is captured by enabling the nodes in a hidden layer to receive the information from both the current inputs and their previous nodes (Figure 2B) [44]. RNN models have resulted in excellent performance in sequence data analysis of AMPs of various sizes. In the RNN algorithms, each amino acid is considered as a word in a sentence and demonstrated by one-dimensional vectors. In addition, one-hot encoding may be used to construct numerical information. For instance, in 2D-convolution modules of AMP sequences with binary 2-D matrices, all variables are filled up with zeros, except the position of the corresponding amino acid [33,36]. Also, an RNN model with a stack-augmented GRU was used to discover new metronidazole drugs by generating precise SMILES strings [45]. The model was trained on ~ 1.6 million small molecules, utilized transfer learning to generate structurally similar molecules and finally generated generated 20 metronidazole-like compounds, with 19 potentially novel ones, that demonstrated activity in vitro against E. coli, P. aeruginosa, B. subtilis, and S. aureus strains [45].
2.2.1.3. Graph neural networks (GNNs)
Compared with these architectures, graph neural networks (GNNs) have been frequently used to extract complex feature vectors and predict the biochemical functions of antimicrobial peptides and small molecules from their 2D or structures [8,46]. In the GNNs, biomolecule can be interpreted as consisting of nodes (e.g., atoms and amino acids) and edges, (corresponding to their connectivities, e.g., bonds and geometric distances) (Figure 2D). Nodes and edges are associated with feature descriptors summarizing the types of atoms, amino acids, or bond types, as well as physicochemical and geometric information. For each node and edge, NNs are used to gather information from their neighbor edges and nodes and update the feature descriptors (Figure 2D). This procedure is known as message passing and enables the nodes and edges to capture their 2D/3D structural information in the graph. By combining all feature descriptors updated via several rounds of message passing, the structural fingerprint of the biomolecules is extracted and mapped to their properties using an end-to-end classification/regression neural network. Stokes et al. trained a GNN with 2,335 antimicrobial molecules selected from natural compounds and a library of FDA-approved drugs [8]. The algorithm was applied to predict antibiotic compounds from the Drug Repurposing Hub. Among the highly ranked molecules, halicin was experimentally identified and validated to demonstrate potent bactericidal activity.
2.2.1.4. Attention function
When predicting a certain property given an input x (e.g., an antibiotic), only a fraction of that input (e.g., important amino acid sites in an AMP or substructures in a small molecule antibiotic) contributes to the property formation, while the rest is irrelevant and can even confuse prediction models. The attention mechanism is employed to automatically identify important parts of the data, addressing the issue of irrelevant information in predictions [47].
To compare deep learning models with and without an attention mechanism we use an example, which involves an antibiotic, denoted as x, consisting of N elements such as amino acids or atoms. The deep learning model without attention is characterized by M layers of nonlinear transformations, which process the antibiotic x to generate a corresponding prediction y. The model is represented as follows:
Here, G1(), G2(), …, and GM() correspond to the M nonlinear transformations (i.e., each layer itself can be fully-connected, CNN and RNN), and Out() is a linear function to map the extracted hidden features to final output prediction y.
To introduce attention mechanism into above deep learning model, we can rewrite the above process as:
Out() is defined the same as mentioned above. Att1(), Att2(),..., and AttM() are neural networks that accept the antibiotic or extracted features from the antibiotic as inputs. The output comprises an N-dimensional vector, which signifies the importance of each element in the antibiotic, considering that the antibiotic contains N elements in our specific example. Functions G1(), G2(), …, and GM() (implemented as neural networks) will take not only the antibiotic or its extracted features, but also the N-dimensional vectors highlighting the significant region of the antibiotic as input to make predictions. Altogether, utilizing the “attention” function in NNs, a large dataset of sequences is readily memorized by ignoring less important parts of the input data; thus, more focus is devoted to small but important part of the data [47].
The deep NNs for the prediction of antimicrobial compounds differ from other ML predictors in that, for the deep NNs, the high-level description vectors can automatically be generated and learned while less prior understanding of biology is required [33,48–50]. In fact, the model performance of the ML algorithms depends on how exactly the feature vectors are selected and extracted. In comparison, extracting the feature vectors is a less hand-coded and complicated part in DL techniques compared to other ML algorithms, which is less time-consuming in developing feature extractors. Nonetheless, the model performance and benchmarking in the DL tools to predict antibacterial molecules may be affected by the choice of datasets used for training [51]. Therefore, providing researchers with explicit information about the performance of the predictor, including code sharing, is suggested for fair model benchmarking [51].
2.2.2. Generative DL-models and their applications in antibiotic design
2.2.2.1. Neural language models (NLMs)
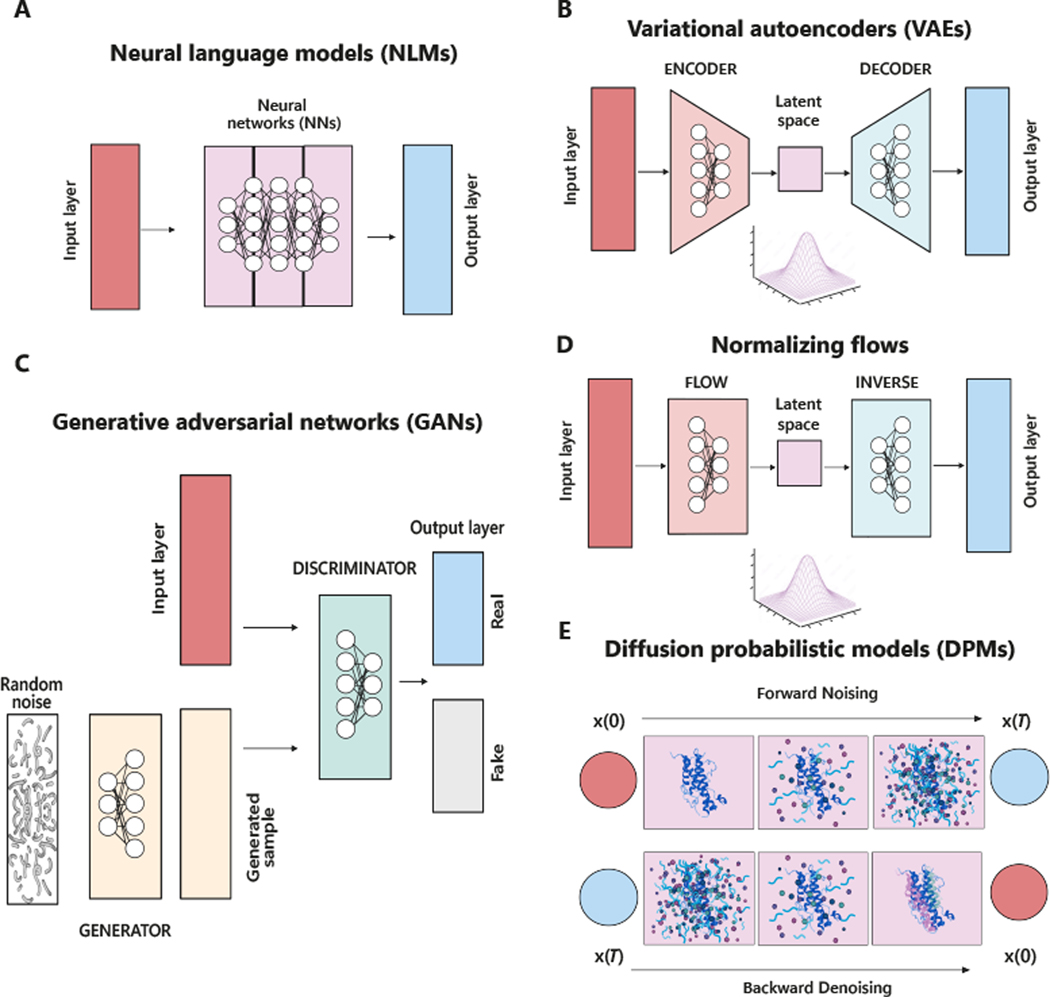
Given sequences of words, language models estimate their probability distribution. In the context of computational biology, primary sequences of DNAs, RNAs, proteins and peptides, as well as small molecules in SMILES representations, can all be represented in sequence forms. In these cases, the corresponding nucleic acids, amino acids, and SMILES characters are considered as words. Neural language models, equipped with deep neural networks, are widely used to take partial sequence inputs and estimate the likelihood of the missing input parts (Figure 3A). Data generation is achieved by completing the missing parts with the most likely words predicted by the DNN (Figure 3A). RNNs, graph neural networks, and attention models are commonly applied to build NLMs for biomolecules. NLMs have been extensively used to generate potentially antimicrobial compounds. Indeed, several studies have employed RNNs and public datasets of antimicrobial compounds to predict drug candidates. Moreover, additional filters (e.g., classifiers or prediction servers) have been applied to adapt the model to antibiotic discovery and remove undesired molecules.
Figure 3. Schematic of the main deep-learning architectures applied in antibiotic discovery.
(A) Neural language models (NLMs) use neural networks (NNs) to model the probability distribution of molecules predicted to have antimicrobial activity based on previously generated outputs. (B) Variational autoencoders (VAEs) encode and decode information to create a latent space, whose variables are used to generate new drug candidates. (C) In generative adversarial networks (GANs), a generator creates synthetic samples while a discriminator distinguishes real generated samples from fake. (D) Normalizing flows (NF) transform simple probability distribution into complex tractable distributions (i.e., input data) through a series of invertible mappings and vice versa. (E) Diffusion probabilistic models (DPMs) gradually add noise to the input data (x0) through a series of T steps, and then a trained neural network recovers the data by reversing the process. The model can generate new data through the denoising course. All elements present within the figure have been created in Biorender.com and assembled using Adobe Illustrator version 27.7.
Capecchi et al. identified eight short non-hemolytic AMPs by training a RNN with data from the DBAASP (Database of Antimicrobial Activity and Structure of Peptides) [52]. Besides an AMP activity classifier, these authors also applied a hemolysis classifier to select peptide sequences predicted to be non-hemolytic. As a result, they obtained peptide antibiotics with low cytotoxicity profiles that were active against drug-resistant bacterial strains. Additionally, Nagarajan et al. designed new AMPs by using an RNN language model [53]. The RNN was based on the correlation of frequencies and distribution of amino acid residues with antimicrobial sequences, as an underlying grammar of antimicrobial function. In their study, ten AMPs, identified and tested for activity against bacterial pathogens, targeted the bacterial membranes of methicillin-resistant Staphylococcus aureus and β-lactamase-resistant and carbapenem-resistant strains.
2.2.2.2. Variational autoencoders
Variational autoencoders (VAEs) present a model architecture similar to that of the classic autoencoders typically trained to compress and decompress inputs through an encoder and a decoder neural network. Unlike classic autoencoders, however, a VAE is a probabilistic modeling framework that interprets the latent representations (i.e., the compressed inputs) as random variables following some distribution (e.g., Gaussian distribution) (Figure 2B). Data generation is achieved by sampling latent representations followed by decoding and/or decompression.
Dean et al. demonstrated the potential utility of a VAE in designing de novo AMP sequences [54]. The latent space (i.e., the low-dimensional space that the latent representations lie in) extrapolated by the VAE learned from thousands of known and scrambled AMP sequences from APD3 highlighted the amphipathicity (mainly represented by hydrophobic moment) as the main difference between the active sequences generated and the scrambled random sequences. The obtained computational tool allowed the authors to generate new peptide sequences, whose antimicrobial activity has been experimentally confirmed in vitro. Indeed, in 2021 Dean et al. published a new study in which the improved model, named PepVAE, was demonstrated to generate new peptide sequences with low antimicrobial activity against Escherichia coli, S. aureus, and Pseudomonas aeruginosa strains [54,55].
Another interesting application of VAE has been shown by Das et al. [50,56] . The authors demonstrated that a VAE trained on a large number of peptide sequences, combined with ML-based predictions for properties such as antimicrobial activity or toxicity and molecular dynamics simulations, could be used to quickly design novel antimicrobial therapeutic molecules: 20 peptide candidates were identified and experimentally validated in 48 days. Among the tested compounds, two AMPs highly were active against both Gram-positive and Gram-negative bacterial species and had little likelihood of selecting for drug-resistant phenotypes [50].
Recently, Szymczak et al. proposed HydrAMP, a generative model based on a multitasking conditional VAE framework. HydrAMP generated not only effective analogues of existing peptide antibiotics but also de novo antimicrobial structures. To train the model, the authors used a curated data set from UniProt of non-redundant peptide sequences known to be antimicrobial. Besides standard encoder and decoder network-based models, HydrAMP also uses a Classifier, which is an extra pre-trained neural network used to determine whether a given sequence is or is not an AMP [57].
2.2.2.3. Generative adversarial networks (GANs)
GANs, which are built of two generative and discriminative neural networks, are trained in an adversarial way to predict antibacterial peptides [33,36,58,59]. Specifically, the generative module captures the input data but is trained to generate fake instances to fool the discriminative network, whereas the discriminator module obtains both real training data and the fake instances from the generator, which then determines whether the input data is real or not (Figure 2C). In training GANs for the AMPs prediction, the generator and discriminator accomplish adversarial functions against each other, resulting in the network modules [33,36,58,59]. PepGAN uses active and non-active datasets from various sources and controls the probability distribution of generated sequences to distinguish real antibacterial sequences from non-active ones. In this regard, a peptide activity-aware generative model, PepGAN, was designed to produce a sequence with antibacterial activity higher than that of ampicillin in vitro [60].. Also, AMPGAN models were utilized as bidirectional conditional networks for the design of AMPs with improved activity profiles [61]. These encoder networks can be used to structurally diversify and manipulate the generated peptide sequences for selective bacterial cell targeting [36,58]. The utilization of GANs accelerated the discovery of A-222, an a-helical peptide with low homology to well-established AMPs, that was strongly effective against the emerging global opportunistic strains Stenotrophomonas maltophilia WH 006 and P. aeruginosa PAO1 [59].
2.2.2.4. Normalizing flow and diffusion models
In addition to the three types of generative models described above, normalizing flow (NF) and diffusion models are also gaining increasing attention [28]. While resembling VAE in terms of framework (Figure 3D), NF utilizes invertible transformation functions (implemented as some neural networks) to convert complex data to random variables drawn from a simple distribution (i.e., encoding process) and vice versa (i.e., decoding process). While VAEs are trained by optimizing the lower bound of data likelihood, the use of invertible transformations enables NF to be optimized by directly maximizing the likelihood of data. For instance, in 2019 Madhawa et al. proposed GraphNVP, the first invertible NF-based model able to efficiently generate molecular graphs [62]. Interestingly, these authors empirically demonstrated that the learned latent space can be used to search for molecules with maximized chemical properties with respect to the parent compounds. In addition, Shi et al. proposed and experimentally validated GraphAF, a flow-based model for generating molecular graphs, which was found to generate chemical-valid and optimized molecules both with and without chemical knowledge rules [63].
Diffusion-based models have also been explored and applied to address molecular design tasks. Diffusion models describe a sequence of possible events, whose probability only depends on the previous events, by adding random noise to data (Figure 3E) until the inputs become a random noise (from some random distribution). Those models built the desired samples by learning how to reverse the diffusion process from the noise. Unlike the VAE and NF, the latent variable (the random noise) will have the same dimensionality as the original input. Corso et al. recently developed DiffDock, a diffusion generative model able to address molecular docking tasks [64]. After training the model with a variety of ligand and protein structures, DiffDock identifies new binding sites within the protein pocket, highlighting new 3D coordinates for the ligands. Molecular docking, a critical task in drug design, focuses on predicting the binding structure between a small molecule ligand and a protein. Although recent deep learning approaches have successfully reduced runtime when compared to traditional search-based methods, they have not yielded notable improvements in accuracy when treating docking as a regression problem. In contrast, DiffDock stands out from previous methods by showcasing significantly higher precision, offering fast inference times, and providing confidence estimates with a high level of selective accuracy [64]. This tool demonstrated to accelerate drug discovery compared to traditional methods. Also, Watson et al. introduced RoseTTAFold (RF) Diffusion, a new diffusion model able to generate proteins with minimal knowledge using simple molecular specifications [65]. They simulated the noise (for instance data - images or text - corrupted with Gaussian noise) on structure of protein from Protein Data Bank (PDB) to generate training inputs. To reverse the noising process, RF Diffusion was then trained by approximating the loss between the predicted shape and the true protein structure. According to Watson et al., the proteins generated by RF Diffusion can potentially treat infection, fight cancer, reprogram autoimmune disorders, or serve as building blocks for a new generation of materials [65].
5. Expert opinion
5.1. Physicochemical features associated with deep learning (DL) models
The use of physicochemical properties as quantitative descriptors for the guided design of antimicrobial agents has been significantly improved by the application of computational methods. The usual types of features used for drug design are classified by their dimensionality. 1D descriptors are scalar properties, such as molecular weight, net charge, hydrophobicity, number of hydrogen bond donors and acceptors, and aromaticity, which are by far the most frequently used physicochemical descriptors because they can be easily assessed computationally or empirically. 2D descriptors are related to properties that describe molecular topology, for example, topological polar surface area, rotatable bonds, molecular connectivity indices, and autocorrelation functions. Similarly to 1D descriptors, the 2D descriptors are easy to obtain computationally and can represent various molecules ranging from small molecules to polymeric or complex supramolecular structures. 3D descriptors, on the other hand, are much more difficult to obtain and are not usually related to small molecules or planar medium-sized molecules. The calculation of thse physicochemical descriptors requires a considerable computational cost. These descriptors represent most structural and electronic features of the molecules, such as molecular surface area, dipole moment, electrostatic potential, and solvent-accessible surface area.
DL and other computational methods have allowed the calculation of more complex and accurate features like 3D descriptors and other properties that are combinations of them, which are not related to structure or empirical characterization of molecules or their parts. The main disadvantage is that it is also costly and time demanding to include such complex features [66]. Some of the existing DL-based methods do not consider physicochemical features directly for the learning process and generation of bioactive molecules. Alternatively, deep learning models can be integrated with encoding techniques that utilize sequence-based characteristics to generate feature vectors based on the molecule’s composition. Among the most widely used feature-encoding methods, we highlight one-hot encoding [67], general and pseudo amino acid composition [68], and reduced amino acid composition [69].
AI4AMP, for instance, is a feature-encoding method based on general amino acid composition. The model was trained on sequences encoded with PC6 [70], a deep learning model for predicting AMP sequence based on a combined physicochemical property matrix. The features were selected from a hierarchical clustering and included, volume of side chains, polarity, hydrophobicity, pH at the isoelectric point, pKa, and the net charge index of the side chains, i.e., properties that play a role in the interaction of the compound with the target microbe. The PC6 encoding method combined with a DL architecture consists of three layers (convolutional, LSTM, and dense layers) and performed similarly to the word embedding-based model [71].
Other examples are Deep-ABPpred [72] and sAMP-PFPDeep [73]. Deep-ABPpred uses bidirectional long short-term memory [72] and the word embedding-based model coupled with amino acid level features and support vector machine and random forest models with peptide-level features, including physicochemical properties, to identify AMPs (accuracy of 97%). sAMP-PFPDeep consists of a DL method with three feature-encoding aspects: (1) position, (2) frequency, and (3) one or more of the following 12 physicochemical features: hydropathy index, volume, polarity, pKa side chain, percentage of exposed residues, hydrophilicity, refractivity, local flexibility, accessible surface area folded, mass, solvent exposed area, and accessible surface area. sAMP-PFPDeep was able to identify AMPs up to 30 residues in length with an accuracy of 84%.
In the future, we envision that the accurate embedding of multiple physicochemical features will lead to models that can generate highly active compounds. Some success in identifying anticancer peptides has been achieved with DNN models such as ENNAACT [74] which has eight dense layers with physicochemical features [75] as input. ENNAACT presented a 98.3% accuracy at 10-fold cross-validation and performed better than random forest and support vector machines with linear and radial basis function (RBF) kernels in the same dataset. Another DNN model that considers physicochemical properties for predicting the activity of antiviral peptides is ENNAVIA [76] (accuracy of 95.7%), which uses three dense layers with augmentative and alternative communication, data predictive control, AAindex [77], and physicochemical properties as input features. Deep-AntiFP [78] is a DL model with three dense layers: composite physicochemical properties [79], quasi-sequence order [80], and reduced amino acid alphabet [81], combined with physicochemical features. Recently, Deep-AntiFP predicted antifungal peptides with an accuracy of 94.23%. These models show that it is possible to combine physicochemical features with DL architecture for the accurate prediction and generation of active anticancer or antimicrobial agents.
5.2. Limitations related to datasets and experimental validation of deep learning models
There have been several major achievements in DL-based recognition and the prediction of antibacterial peptides and other small molecules; nevertheless, this field still requires development [37]. Indeed, the performance of DL models is significantly influenced by the quality rather than the size of the training dataset, which should consist of both active and inactive compounds [82,83]. In fact, the sources of the antibacterial datasets often suffer from inconsistency and heterogeneity in the activity evaluation of collected data, and lack of sufficient information on their mechanismsof action.
Besides data curation, public access to the negative datasets (i.e., peptides lacking activity against pathogens) is limited, as this information is rarely released [84]. Nevertheless, the negative data for the training of DL models to predict antibacterial peptides are often extracted as random sequences from available collections of proteins (e.g., Uniprot), which can be larger in data size than a few thousand true positive data. Most DL models have difficulty handling highly imbalanced antibacterial peptidesor small molecule datasets to prevent biased predictions [33]. Furthermore, the activities of AMPs are very sensitive to changes in peptide composition and length, and their activity varies greatly depending on the target bacterial strain [85,86]. These limitations pose problems especially for the prediction and recognition of antibacterial sequences. Additionally, depending on the selected DL algorithms, appropriate feature-encoding methods need to be selected over the training course of classifiers [36,37]. It is still a challenging task to consider expensive feature vectors relevant to the 3D-structure of peptides and small molecules to improve the prediction of new sequences or chemical scaffolds. Owing to the distinct selectivity of various antibiotic and antibacterial sequences for Gram-negative and Gram-positive bacteria, as well as the diversity of the activity determination protocols used under various laboratory conditions, sorting, and prioritization of predicted compounds for wet-lab activity determination and validation are still unsatisfactory, even though they are often used by binary classifiers [87]. Moreover, it is still unclear how to design classifiers that predict peptides that are resistant to protease degradation or that can model enzymatic cleavage sites in the peptide, to design more stable peptides [88]. Alternatively, there are models that leverage D-amino acid substitution, non-canonical amino acid residues, and cyclization to increase peptide stability [89]. Thus, the bioavailability of antimicrobial sequences remains an open area of research. Finally, the undesired concentration-dependent side-effects of antibacterial peptides need to be considered when designing DL predictors [37,87].
5.3. Data availability for DL models
The biggest hurdle to developing DL models that are accurate in generating peptides with biological activity is to have enough data for training the models. Whereas small organic compounds have databases such as ZINC [90], SciFinder, Reaxys, and PubChem, which count with millions to hundreds of millions of molecules but mostly with no information on bioactive activities, bioactive peptides databases, such as APD3 [91], DBAASP [92], CAMP [93], Hemolytik [94], CPPsite [95], DRAMP [96], and dbAMP [97], count no more than a couple of tens of thousands of data points. It is worth mentioning that databases such as UniProt or GenBank house extensive collections of peptides and proteins, yet it should be noted that these repositories do not exhibit antimicrobial activities and are still untapped sources for antimicrobially active molecules.
Overcoming the data availability issue relies on combining deep generative models with bioinformatic tools [98] or molecular dynamics simulations [50], or doing data augmentation [99] to increase the performance of the models. Bioinformatic tools and molecular dynamics simulations provide an enhanced sampling of sought-after properties for the biological activity desired. Among the most important features retrieved to enrich the datasets are the physicochemical features that can be extracted from high throughput simulations with a high level of detail. On the other hand, data augmentation will not generate new information on the dataset, but it will increase the number of examples in the training set while also introducing more variety in what the model sees and learns from. Both bioinformatic tools and molecular dynamics simulations are valid strategies for different biological purposes. We envision that coupling these strategies to high performing computing will improve quality sampling with less experimental input needed and may accelerate the discovery of potent antimicrobial agents.
5.4. Summary
In spite of advances in drug design and pharmacology [100] (Table 2), the development of new antibiotics is still time-consuming and costly, while the need for them is becoming ever more urgent. DL approaches have revealed promising results in accelerating antimicrobial discovery, thus attracting the attention of many researchers and companies in the field. In fact, the combination of generative models and DNNs demonstrates the ability to generate and predict molecules with desired bioactivities. Furthermore, these models can potentially reduce the time and expense of antibiotic discovery and increase the chance of success. Although there is no unique and standardized DL framework that yields the best results in terms of antibiotic prediction, all the machine architectures described here set the stage for the development of efficient platforms for discovering small molecules and peptides with antimicrobial properties. Particularly, well-trained DL models have been demonstrated to generate outputs with properties similar to those of training data, assign a probability of new data points within data distribution, and define important features. We envision that DL tools combined with larger datasets and more refined molecular descriptors will change the way that synthetic antibiotics will be designed in the near future.
Table 2.
Models and their application in antimicrobial peptides discovery.
| Model | Application | References |
|---|---|---|
|
| ||
| ABP-Finder | Screen peptidomes and identify antibacterial peptides | [111] |
| rAMPage | To mine amphibian and insect transciptomes for antimicrobial peptides | [111,112] |
| AI4AMP | To predict antimicrobial potential of a given seqeunce and perform proteome screening | [62] |
| Deep-ABPpred | Identify antibacterial peptides within protein seqeunces | [65] |
| sAMP-PFPDeep | To predict short (<30 residues long) antimicrobial peptide sequences | [113] |
| Deep-AntiFP | To identify antifungal peptide seqeunces | [70] |
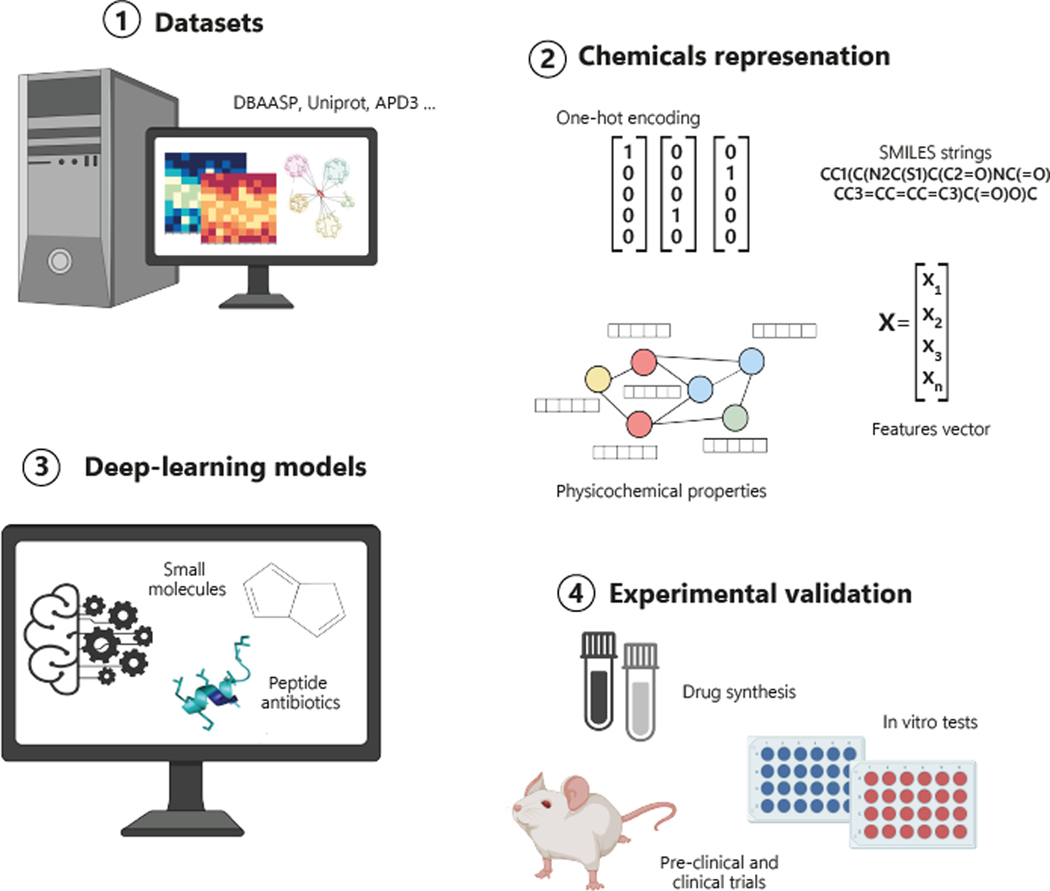
Figure 1. Application of AI methods in antibiotic discovery.
(1) Datasets containing experimental information are selected for model training. (2) The data are translated into machine-readable representations. (3) Subsequently, the algorithms are trained and then used to predict new active molecules. (4) Wet-lab experiments are performed to validate the antimicrobial activity of computationally predicted candidate drugs. All elements present within the figure have been created in BioRender.com and assembled using Adobe Illustrator version 27.7. The peptide molecule shown in the figure was rendered using the PyMOL Molecular Graphics System, Version 2.5.2 Schrödinger, LLC.
Article highlights box.
AI and ML provide innovative ways to expedite antibiotic discovery by optimizing molecular design.
The successful use of discriminative and generative deep learning models is directly impacted by the algorithm and influences the modeĺs ability to represent diverse molecular structures.
Discriminative deep learning models are computational frameworks for predicting antibiotic activity, leveraging their specific architectures and approaches.
Generative deep learning models are applied in antibiotic design utilizing the composition of molecules to generate potent drug candidates.
Deep learning models yet face challenges due to data quality and availability limitations.
Acknowledgements:
The authors thank Dr. K Pepper for editing the manuscript and de la Fuente Lab members for insightful discussions
Funding:
C de la Fuente-Nunez holds a Presidential Professorship at the University of Pennsylvania, is a recipient of the Langer Prize by the AIChE Foundation, and acknowledges funding from the IADR Innovation in Oral Care Award, the Procter & Gamble Company and United Therapeutics. Funding is also acknowledged via a BBRF Young Investigator Grant, the Nemirovsky Prize, Penn Health-Tech Accelerator Award, the Dean’s Innovation Fund from the Perelman School of Medicine at the University of Pennsylvania, the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM138201, and the Defense Threat Reduction Agency (DTRA; HDTRA11810041, HDTRA1-21-1-0014, and HDTRA1-23-1-0001).
Footnotes
Reviewer Disclosures:
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Declaration of Interest:
C de la Fuente-Nunez provides consulting services to Invaio Sciences and is a member of the Scientific Advisory Boards of Nowture S.L. and Phare Bio. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
- [1].Cesaro A, Lin S, Pardi N, de la Fuente-Nunez C, Advanced delivery systems for peptide antibiotics, Adv Drug Deliv Rev. 196 (2023). 10.1016/j.addr.2023.114733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Magana M, Pushpanathan M, Santos AL, Leanse L, Fernandez M, Ioannidis A, Giulianotti MA, Apidianakis Y, Bradfute S, Ferguson AL, Cherkasov A, Seleem MN, Pinilla C, de la Fuente-Nunez C, Lazaridis T, Dai T, Houghten RA, Hancock REW, Tegos GP, The value of antimicrobial peptides in the age of resistance, Lancet Infect Dis. 20 (2020) e216–e230. 10.1016/S1473-3099(20)30327-3. [DOI] [PubMed] [Google Scholar]
- [3].Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Aguilar G. Robles, Gray A, Han C, Bisignano C, Rao P, Wool E, Johnson SC, Browne AJ, Chipeta MG, Fell F, Hackett S, Haines-Woodhouse G, Hamadani B.H. Kashef, Kumaran EAP, McManigal B, Agarwal R, Akech S, Albertson S, Amuasi J, Andrews J, Aravkin A, Ashley E, Bailey F, Baker S, Basnyat B, Bekker A, Bender R, Bethou A, Bielicki J, Boonkasidecha S, Bukosia J, Carvalheiro C, Castañeda-Orjuela C, Chansamouth V, Chaurasia S, Chiurchiù S, Chowdhury F, Cook AJ, Cooper B, Cressey TR, Criollo-Mora E, Cunningham M, Darboe S, Day NPJ, De Luca M, Dokova K, Dramowski A, Dunachie SJ, Eckmanns T, Eibach D, Emami A, Feasey N, Fisher-Pearson N, Forrest K, Garrett D, Gastmeier P, Giref AZ, Greer RC, Gupta V, Haller S, Haselbeck A, Hay SI, Holm M, Hopkins S, Iregbu KC, Jacobs J, Jarovsky D, Javanmardi F, Khorana M, Kissoon N, Kobeissi E, Kostyanev T, Krapp F, Krumkamp R, Kumar A, Kyu HH, Lim C, Limmathurotsakul D, Loftus MJ, Lunn M, Ma J, Mturi N, Munera-Huertas T, Musicha P, Mussi-Pinhata MM, Nakamura T, Nanavati R, Nangia S, Newton P, Ngoun C, Novotney A, Nwakanma D, Obiero CW, Olivas-Martinez A, Olliaro P, Ooko E, Ortiz-Brizuela E, Peleg AY, Perrone C, Plakkal N, Ponce-de-Leon A, Raad M, Ramdin T, Riddell A, Roberts T, Robotham JV, Roca A, Rudd KE, Russell N, Schnall J, Scott JAG, Shivamallappa M, Sifuentes-Osornio J, Steenkeste N, Stewardson AJ, Stoeva T, Tasak N, Thaiprakong A, Thwaites G, Turner C, Turner P, van Doorn HR, Velaphi S, Vongpradith A, Vu H, Walsh T, Waner S, Wangrangsimakul T, Wozniak T, Zheng P, Sartorius B, Lopez AD, Stergachis A, Moore C, Dolecek C, Naghavi M, Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis, The Lancet. 399 (2022) 629–655. 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].The true death toll of COVID-19: estimating global excess mortality, (2023). https://www.who.int/data/stories/the-true-death-toll-of-covid-19-estimating-global-excess-mortality (accessed July 30, 2023).
- [5].Comprehensive review of the WHO Global Action Plan on Antimicrobial Resistance : Evaluation brief – September 2021, Https://Www.Who.Int/Publications/m/Item/Comprehensive-Review-of-the-Who-Global-Action-Plan-on-Antimicrobial-Resistance-Evaluation-Brief-September-2021. (2021).
- [6].Hutchings MI, Truman AW, Wilkinson B, Antibiotics: past, present and future, Curr Opin Microbiol. 51 (2019) 72–80. 10.1016/j.mib.2019.10.008. [DOI] [PubMed] [Google Scholar]
- [7].Wang G, Vaisman II, van Hoek ML, Machine Learning Prediction of Antimicrobial Peptides, in: 2022: pp. 1–37. 10.1007/978-1-0716-1855-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. **Stokes JM, Yang K, Swanson K, Jin W, Cubillos-Ruiz A, Donghia NM, MacNair CR, French S, Carfrae LA, Bloom-Ackermann Z, Tran VM, Chiappino-Pepe A, Badran AH, Andrews IW, Chory EJ, Church GM, Brown ED, Jaakkola TS, Barzilay R, Collins JJ, A Deep Learning Approach to Antibiotic Discovery, Cell. 180 (2020) 688–702.e13. 10.1016/j.cell.2020.01.021. This paper used a graph neural network to identify halicin, a previously unrecognized antibacterial small molecule with broad-spectrum activity against various bacterial pathogens.
- [9].Youn J, Rai N, Tagkopoulos I, Knowledge integration and decision support for accelerated discovery of antibiotic resistance genes, Nat Commun. 13 (2022) 2360. 10.1038/s41467-022-29993-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sandiford SK, What is an ideal antibiotic and what does this mean for future drug discovery and design?, Expert Opin Drug Discov. 18 (2023) 485–490. 10.1080/17460441.2023.2198701. [DOI] [PubMed] [Google Scholar]
- [11].Maasch JRMA, Torres MDT, Melo MCR, de la Fuente-Nunez C, Molecular de-extinction of ancient antimicrobial peptides enabled by machine learning, Cell Host Microbe. 31 (2023) 1260–1274. 10.1016/j.chom.2023.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wong F, de la Fuente-Nunez C, Collins JJ, Leveraging artificial intelligence in the fight against infectious diseases, Science. 381 (2023) 164–170. 10.1126/science.adh1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fernandes FC, Cardoso MH, Gil-Ley A, Luchi LV, da Silva MGL, Macedo MLR, de la Fuente-Nunez C, Franco OL, Geometric deep learning as a potential tool for antimicrobial peptide prediction, Frontiers in Bioinformatics. 3 (2023). 10.3389/fbinf.2023.1216362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Anahtar MN, Yang JH, Kanjilal S, Applications of Machine Learning to the Problem of Antimicrobial Resistance: an Emerging Model for Translational Research, J Clin Microbiol. 59 (2021). 10.1128/JCM.01260-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15]. **Cesaro A, Torres MDT, Gaglione R, Dell’Olmo E, Di Girolamo R, Bosso A, Pizzo E, Haagsman HP, Veldhuizen EJA, de la Fuente-Nunez C, Arciello A, Synthetic Antibiotic Derived from Sequences Encrypted in a Protein from Human Plasma, ACS Nano. (2022) acsnano.1c04496. 10.1021/acsnano.1c04496. The paper demonstrates the antimicrobial activity of encrypted peptides derived from human plasma.
- [16]. **Torres MDT, Melo MCR, Flowers L, Crescenzi O, Notomista E, de la Fuente-Nunez C, Mining for encrypted peptide antibiotics in the human proteome, Nat Biomed Eng. 6 (2022) 1451–1451. 10.1038/s41551-022-00967-2. This paper presents the first exploration of the human proteome for antibiotics, leading to the identification of thousands of new encrypted peptide antibiotics.
- [17].Cesaro A, Gaglione R, Chino M, De Luca M, Di Girolamo R, Lombardi A, Filosa R, Arciello A, Novel Retro-Inverso Peptide Antibiotic Efficiently Released by a Responsive Hydrogel-Based System, Biomedicines. 10 (2022) 1301. 10.3390/biomedicines10061301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gaglione R, Cesaro A, Dell’Olmo E, Di Girolamo R, Tartaglione L, Pizzo E, Arciello A, Cryptides Identified in Human Apolipoprotein B as New Weapons to Fight Antibiotic Resistance in Cystic Fibrosis Disease, Int J Mol Sci. 21 (2020) 2049. 10.3390/ijms21062049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gaglione R, Cesaro A, Dell’Olmo E, Della Ventura B, Casillo A, Di Girolamo R, Velotta R, Notomista E, Veldhuizen EJA, Corsaro MM, De Rosa C, Arciello A, Effects of human antimicrobial cryptides identified in apolipoprotein B depend on specific features of bacterial strains, Sci Rep. 9 (2019) 6728. 10.1038/s41598-019-43063-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. **Porto WF, Irazazabal L, Alves ESF, Ribeiro SM, Matos CO, Pires ÁS, Fensterseifer ICM, Miranda VJ, Haney EF, Humblot V, Torres MDT, Hancock REW, Liao LM, Ladram A, Lu TK, de la Fuente-Nunez C, Franco OL, In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design, Nat Commun. 9 (2018). 10.1038/s41467-018-03746-3. This paper describes the use of a genetic algorithm to computationally design a peptide antibiotic, called guavanin 2, with anti-infective activity in a preclinical mouse infection model.
- [21].Murcia-Soler M, Pérez-Giménez F, García-March FJ, Salabert-Salvador MT, Díaz-Villanueva W, Castro-Bleda MJ, Villanueva-Pareja A, Artificial neural networks and linear discriminant analysis: A valuable combination in the selection of new antibacterial compounds, J Chem Inf Comput Sci. 44 (2004). 10.1021/ci030340e. [DOI] [PubMed] [Google Scholar]
- [22].Cherkasov A, Hilpert K, Jenssen H, Fjell CD, Waldbrook M, Mullaly SC, Volkmer R, Hancock REW, Use of artificial intelligence in the design of small peptide antibiotics effective against a broad spectrum of highly antibiotic-resistant superbugs, ACS Chem Biol. 4 (2009). 10.1021/cb800240j. [DOI] [PubMed] [Google Scholar]
- [23].Cesaro A, Torres MDT, de la Fuente-Nunez C, Methods for the design and characterization of peptide antibiotics, in: Methods Enzymol, Academic Press, 2022: pp. 303–326. 10.1016/bs.mie.2021.11.003. [DOI] [PubMed] [Google Scholar]
- [24].Torres MDT, Cao J, Franco OL, Lu TK, de la Fuente-Nunez C, Synthetic Biology and Computer-Based Frameworks for Antimicrobial Peptide Discovery, ACS Nano. 15 (2021) 2143–2164. 10.1021/acsnano.0c09509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Torres MDT, Pedron CN, Higashikuni Y, Kramer RM, Cardoso MH, Oshiro KGN, Franco OL, Silva Junior PI, Silva FD, Oliveira Junior VX, Lu TK, de la Fuente-Nunez C, Structure-function-guided exploration of the antimicrobial peptide polybia-CP identifies activity determinants and generates synthetic therapeutic candidates, Commun Biol. 1 (2018) 221. 10.1038/s42003-018-0224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Torres MDT, de la Fuente-Nunez C, Reprogramming biological peptides to combat infectious diseases, Chemical Communications. 55 (2019) 15020–15032. 10.1039/c9cc07898c. [DOI] [PubMed] [Google Scholar]
- [27].Lavecchia A, Deep learning in drug discovery: opportunities, challenges and future prospects, Drug Discov Today. 24 (2019). 10.1016/j.drudis.2019.07.006. [DOI] [PubMed] [Google Scholar]
- [28].Bilodeau C, Jin W, Jaakkola T, Barzilay R, Jensen KF, Generative models for molecular discovery: Recent advances and challenges, WIREs Computational Molecular Science. 12 (2022). 10.1002/wcms.1608. [DOI] [Google Scholar]
- [29].Qin D, Bo W, Zheng X, Hao Y, Li B, Zheng J, Liang G, DFBP: a comprehensive database of food-derived bioactive peptides for peptidomics research, Bioinformatics. 38 (2022) 3275–3280. 10.1093/bioinformatics/btac323. [DOI] [PubMed] [Google Scholar]
- [30].Rajpoot S, Solanki K, Kumar A, Zhang KYJ, Pullamsetti SS, Savai R, Faisal SM, Pan Q, Baig MS, In-Silico Design of a Novel Tridecapeptide Targeting Spike Protein of SARS-CoV-2 Variants of Concern, Int J Pept Res Ther. 28 (2022) 28. 10.1007/s10989-021-10339-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang F, Sangfuang N, McCoubrey LE, Yadav V, Elbadawi M, Orlu M, Gaisford S, Basit AW, Advancing oral delivery of biologics: Machine learning predicts peptide stability in the gastrointestinal tract, Int J Pharm. 634 (2023) 122643. 10.1016/j.ijpharm.2023.122643. [DOI] [PubMed] [Google Scholar]
- [32].David L, Thakkar A, Mercado R, Engkvist O, Molecular representations in AI-driven drug discovery: a review and practical guide, J Cheminform. 12 (2020). 10.1186/s13321-020-00460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wan F, Kontogiorgos-Heintz D, de la Fuente-Nunez C, Deep generative models for peptide design, Digital Discovery. 1 (2022) 195–208. 10.1039/D1DD00024A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ulusoy I, Bishop CM, Generative versus discriminative methods for object recognition, in: Proceedings - 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, CVPR 2005, 2005. 10.1109/CVPR.2005.167. [DOI] [Google Scholar]
- [35].Lecun Y, Bengio Y, Hinton G, Deep learning, Nature 2015 521:7553. 521 (2015) 436–444. 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- [36].Chen X, Li C, Bernards MT, Shi Y, Shao Q, He Y, Sequence-based peptide identification, generation, and property prediction with deep learning: A review, Mol Syst Des Eng. 6 (2021). 10.1039/d0me00161a. [DOI] [Google Scholar]
- [37].Yan J, Cai J, Zhang B, Wang Y, Wong DF, Siu SWI, Recent Progress in the Discovery and Design of Antimicrobial Peptides Using Traditional Machine Learning and Deep Learning, Antibiotics (Basel). 11 (2022). 10.3390/ANTIBIOTICS11101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38]. **García-Jacas CR, Pinacho-Castellanos SA, García-González LA, Brizuela CA, Do deep learning models make a difference in the identification of antimicrobial peptides?, Brief Bioinform. 23 (2022). 10.1093/BIB/BBAC094. This reference explores machine learning models for antimicrobial peptide activity prediction, questioning the widely accepted superiority of deep learning over shallow learning for AMP identification.
- [39].Cong S, Zhou Y, A review of convolutional neural network architectures and their optimizations, Artif Intell Rev. 56 (2023) 1905–1969. 10.1007/S10462-022-10213-5/FIGURES/13. [DOI] [Google Scholar]
- [40].Yan J, Bhadra P, Li A, Sethiya P, Qin L, Tai HK, Wong KH, Siu SWI, Deep-AmPEP30: Improve Short Antimicrobial Peptides Prediction with Deep Learning, Mol Ther Nucleic Acids. 20 (2020) 882–894. 10.1016/J.OMTN.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Su X, Xu J, Yin Y, Quan X, Zhang H, Antimicrobial peptide identification using multi-scale convolutional network, BMC Bioinformatics. 20 (2019). 10.1186/S12859-019-3327-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ma Y, Guo Z, Xia B, Zhang Y, Liu X, Yu Y, Tang N, Tong X, Wang M, Ye X, Feng J, Chen Y, Wang J, Identification of antimicrobial peptides from the human gut microbiome using deep learning, Nature Biotechnology 2022 40:6. 40 (2022) 921–931. 10.1038/s41587-022-01226-0. [DOI] [PubMed] [Google Scholar]
- [43].Grimberg H, Tiwari VS, Tam B, Gur-Arie L, Gingold D, Polachek L, Akabayov B, Machine learning approaches to optimize small-molecule inhibitors for RNA targeting, J Cheminform. 14 (2022) 4. 10.1186/s13321-022-00583-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Chen N, Yang L, Ding N, Li G, Cai J, An X, Wang Z, Qin J, Niu Y, Recurrent neural network (RNN) model accelerates the development of antibacterial metronidazole derivatives, RSC Adv. 12 (2022) 22893–22901. 10.1039/D2RA01807A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chen N, Yang L, Ding N, Li G, Cai J, An X, Wang Z, Qin J, Niu Y, Recurrent neural network (RNN) model accelerates the development of antibacterial metronidazole derivatives, RSC Adv. 12 (2022) 22893–22901. 10.1039/D2RA01807A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Yan K, Lv H, Guo Y, Peng W, Liu B, sAMPpred-GAT: prediction of antimicrobial peptide by graph attention network and predicted peptide structure, Bioinformatics. 39 (2023). 10.1093/BIOINFORMATICS/BTAC715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hernández A, Amigó JM, Attention Mechanisms and Their Applications to Complex Systems, Entropy. 23 (2021) 283. 10.3390/e23030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Müller AT, Hiss JA, Schneider G, Recurrent Neural Network Model for Constructive Peptide Design, J Chem Inf Model. 58 (2018). 10.1021/acs.jcim.7b00414. [DOI] [PubMed] [Google Scholar]
- [49].Melo MCR, Maasch JRMA, de la Fuente-Nunez C, Accelerating antibiotic discovery through artificial intelligence, Commun Biol. 4 (2021). 10.1038/s42003-021-02586-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50]. **Das P, Sercu T, Wadhawan K, Padhi I, Gehrmann S, Cipcigan F, Chenthamarakshan V, Strobelt H, dos Santos C, Chen P-Y, Yang YY, Tan JPK, Hedrick J, Crain J, Mojsilovic A, Accelerated antimicrobial discovery via deep generative models and molecular dynamics simulations, Nat Biomed Eng. 5 (2021) 613–623. 10.1038/s41551-021-00689-x. This paper provides a comprehensive study, also experimentally validated, on the use of a variational autoencoder model for antimicrobial drug discovery.
- [51].Sidorczuk K, Gagat P, Pietluch F, Kała J, Rafacz D, Bąkała L, Słowik J, Kolenda R, Rödiger S, Fingerhut LCHW, Cooke IR, MacKiewicz P, Burdukiewicz M, Benchmarks in antimicrobial peptide prediction are biased due to the selection of negative data, Brief Bioinform. 23 (2022) 1–12. 10.1093/BIB/BBAC343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Capecchi A, Cai X, Personne H, Köhler T, van Delden C, Reymond JL, Machine learning designs non-hemolytic antimicrobial peptides, Chem Sci. 12 (2021). 10.1039/d1sc01713f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Nagarajan D, Nagarajan T, Roy N, Kulkarni O, Ravichandran S, Mishra M, Chakravortty D, Chandra N, Computational antimicrobial peptide design and evaluation against multidrug-resistant clinical isolates of bacteria, Journal of Biological Chemistry. 293 (2018). 10.1074/jbc.M117.805499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Dean SN, Walper SA, Variational autoencoder for generation of antimicrobial peptides, ACS Omega. 5 (2020). 10.1021/acsomega.0c00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Dean SN, Alvarez JAE, Zabetakis D, Walper SA, Malanoski AP, PepVAE: Variational Autoencoder Framework for Antimicrobial Peptide Generation and Activity Prediction, Front Microbiol. 12 (2021). 10.3389/fmicb.2021.725727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Das Payel, Wadhawan Kahini, Chang Oscar, Sercu Tom, dos Santos Cicero Nogueira, Riemer Matthew, Padhi Inkit, Chenthamarakshan Vijil, Mojsilovic Aleksandra, PepCVAE: Semi-Supervised Targeted Design of Antimicrobial Peptide Sequences, ArXiv. abs/1810.07743 (2018). [Google Scholar]
- [57]. **Szymczak P, Możejko M, Grzegorzek T, Jurczak R, Bauer M, Neubauer D, Sikora K, Michalski M, Sroka J, Setny P, Kamysz W, Szczurek E, Discovering highly potent antimicrobial peptides with deep generative model HydrAMP, Nat Commun. 14 (2023) 1453. 10.1038/s41467-023-36994-z. This paper provides a comprehensive study on the use of a deep learning model for the discovery of new antimicrobial peptides.
- [58].Lin E, Lin CH, Lane HY, De Novo Peptide and Protein Design Using Generative Adversarial Networks: An Update, J Chem Inf Model. 62 (2022) 761–774. 10.1021/ACS.JCIM.1C01361. [DOI] [PubMed] [Google Scholar]
- [59].Cao Q, Ge C, Wang X, Harvey PJ, Zhang Z, Ma Y, Wang X, Jia X, Mobli M, Craik DJ, Jiang T, Yang J, Wei Z, Wang Y, Chang S, Yu R, Designing antimicrobial peptides using deep learning and molecular dynamic simulations, Brief Bioinform. 24 (2023). 10.1093/BIB/BBAD058. [DOI] [PubMed] [Google Scholar]
- [60].Tucs A, Tran DP, Yumoto A, Ito Y, Uzawa T, Tsuda K, Generating Ampicillin-Level Antimicrobial Peptides with Activity-Aware Generative Adversarial Networks, ACS Omega. 5 (2020) 22847–22851. 10.1021/ACSOMEGA.0C02088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].van Oort CM, Ferrell JB, Remington JM, Wshah S, Li J, AMPGAN v2: Machine Learning-Guided Design of Antimicrobial Peptides, J Chem Inf Model. 61 (2021) 2198–2207. 10.1021/ACS.JCIM.0C01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Madhawa K, Ishiguro K, Nakago K, Abe M, GraphNVP: An Invertible Flow Model for Generating Molecular Graphs, (n.d.). [Google Scholar]
- [63].Shi C, Xu M, Zhu Z, Zhang W, Zhang M, Tang J, GraphAF: a Flow-based Autoregressive Model for Molecular Graph Generation, (n.d.). https://github.com/DeepGraphLearning/GraphAF (accessed April 11, 2023).
- [64].Corso G, Stärk H, Jing B, Barzilay R, Jaakkola T, DiffDock: Diffusion Steps, Twists, and Turns for Molecular Docking, (2022). [Google Scholar]
- [65].Watson JL, Juergens D, Bennett NR, Trippe BL, Yim J, Eisenach HE, Ahern W, Borst AJ, Ragotte RJ, Milles LF, Wicky BIM, Hanikel N, Pellock SJ, Courbet A, Sheffler W, Wang J, Venkatesh P, Sappington I, Torres SV, Lauko A, De Bortoli V, Mathieu E, Barzilay R, Jaakkola TS, DiMaio F, Baek M, Baker D, Broadly applicable and accurate protein design by integrating structure prediction networks and diffusion generative models, BioRxiv. (2022). [Google Scholar]
- [66].Jenssen H, Descriptors for antimicrobial peptides, Expert Opin Drug Discov. 6 (2011) 171–184. 10.1517/17460441.2011.545817. [DOI] [PubMed] [Google Scholar]
- [67].Li J, Pu Y, Tang J, Zou Q, Guo F, DeepAVP: A Dual-Channel Deep Neural Network for Identifying Variable-Length Antiviral Peptides, IEEE J Biomed Health Inform. 24 (2020) 3012–3019. 10.1109/JBHI.2020.2977091. [DOI] [PubMed] [Google Scholar]
- [68].Chou K-C, Prediction of protein cellular attributes using pseudo-amino acid composition, Proteins: Structure, Function, and Genetics. 43 (2001) 246–255. 10.1002/prot.1035. [DOI] [PubMed] [Google Scholar]
- [69].Weathers EA, Paulaitis ME, Woolf TB, Hoh JH, Reduced amino acid alphabet is sufficient to accurately recognize intrinsically disordered protein, FEBS Lett. 576 (2004) 348–352. 10.1016/j.febslet.2004.09.036. [DOI] [PubMed] [Google Scholar]
- [70].Lin T-T, Yang L-Y, Lu I-H, Cheng W-C, Hsu Z-R, Chen S-H, Lin C-Y, AI4AMP: an Antimicrobial Peptide Predictor Using Physicochemical Property-Based Encoding Method and Deep Learning, MSystems. 6 (2021). 10.1128/mSystems.00299-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ganguly D, Roy D, Mitra M, Jones GJF, Word Embedding based Generalized Language Model for Information Retrieval, in: Proceedings of the 38th International ACM SIGIR Conference on Research and Development in Information Retrieval, ACM, New York, NY, USA, 2015: pp. 795–798. 10.1145/2766462.2767780. [DOI] [Google Scholar]
- [72].Schuster M, Paliwal KK, Bidirectional recurrent neural networks, IEEE Transactions on Signal Processing. 45 (1997) 2673–2681. 10.1109/78.650093. [DOI] [Google Scholar]
- [73].Sharma R, Shrivastava S, Singh S. Kumar, Kumar A, Saxena S, Singh R. Kumar, Deep-ABPpred: identifying antibacterial peptides in protein sequences using bidirectional LSTM with word2vec, Brief Bioinform. 22 (2021). 10.1093/bib/bbab065. [DOI] [PubMed] [Google Scholar]
- [74].Timmons PB, Hewage CM, ENNAACT is a novel tool which employs neural networks for anticancer activity classification for therapeutic peptides, Biomedicine & Pharmacotherapy. 133 (2021) 111051. 10.1016/j.biopha.2020.111051. [DOI] [PubMed] [Google Scholar]
- [75].Müller AT, Gabernet G, Hiss JA, Schneider G, modlAMP: Python for antimicrobial peptides, Bioinformatics. 33 (2017) 2753–2755. 10.1093/bioinformatics/btx285. [DOI] [PubMed] [Google Scholar]
- [76].Timmons PB, Hewage CM, ENNAVIA is a novel method which employs neural networks for antiviral and anti-coronavirus activity prediction for therapeutic peptides, Brief Bioinform. 22 (2021). 10.1093/bib/bbab258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Kawashima S, AAindex: Amino Acid index database, Nucleic Acids Res. 28 (2000) 374–374. 10.1093/nar/28.1.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Ahmad A, Akbar S, Khan S, Hayat M, Ali F, Ahmed A, Tahir M, Deep-AntiFP: Prediction of antifungal peptides using distanct multi-informative features incorporating with deep neural networks, Chemometrics and Intelligent Laboratory Systems. 208 (2021) 104214. 10.1016/j.chemolab.2020.104214. [DOI] [Google Scholar]
- [79].Nath A, Karthikeyan S, Enhanced Prediction and Characterization of CDK Inhibitors Using Optimal Class Distribution, Interdiscip Sci. 9 (2017) 292–303. 10.1007/s12539-016-0151-1. [DOI] [PubMed] [Google Scholar]
- [80].Akbar S, Rahman AU, Hayat M, Sohail M, cACP: Classifying anticancer peptides using discriminative intelligent model via Chou’s 5-step rules and general pseudo components, Chemometrics and Intelligent Laboratory Systems. 196 (2020) 103912. 10.1016/j.chemolab.2019.103912. [DOI] [Google Scholar]
- [81].Etchebest C, Benros C, Bornot A, Camproux A-C, de Brevern AG, A reduced amino acid alphabet for understanding and designing protein adaptation to mutation, European Biophysics Journal. 36 (2007) 1059–1069. 10.1007/s00249-007-0188-5. [DOI] [PubMed] [Google Scholar]
- [82].Rives A, Meier J, Sercu T, Goyal S, Lin Z, Liu J, Guo D, Ott M, Zitnick CL, Ma J, Fergus R, Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences, Proc Natl Acad Sci U S A. 118 (2021) e2016239118. 10.1073/PNAS.2016239118/SUPPL_FILE/PNAS.2016239118.SAPP.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Xu J, Li F, Leier A, Xiang D, Shen HH, Lago T.T. Marquez, Li J, Yu DJ, Song J, Comprehensive assessment of machine learning-based methods for predicting antimicrobial peptides, Brief Bioinform. 22 (2021). 10.1093/BIB/BBAB083. [DOI] [PubMed] [Google Scholar]
- [84].Sidorczuk K, Gagat P, Pietluch F, Kała J, Rafacz D, Bąkała L, Słowik J, Kolenda R, Rödiger S, Fingerhut LCHW, Cooke IR, Mackiewicz P, Burdukiewicz M, Benchmarks in antimicrobial peptide prediction are biased due to the selection of negative data, Brief Bioinform. 23 (2022). 10.1093/BIB/BBAC343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Liu Z, Brady A, Young A, Rasimick B, Chen K, Zhou C, Kallenbach NR, Length Effects in Antimicrobial Peptides of the (RW)n Series, Antimicrob Agents Chemother. 51 (2007) 597. 10.1128/AAC.00828-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Tan P, Lai Z, Zhu Y, Shao C, Akhtar MU, Li W, Zheng X, Shan A, Multiple Strategy Optimization of Specifically Targeted Antimicrobial Peptide Based on Structure-Activity Relationships to Enhance Bactericidal Efficiency, ACS Biomater Sci Eng. 6 (2020) 398–414. 10.1021/ACSBIOMATERIALS.9B00937/SUPPL_FILE/AB9B00937_SI_001.PDF. [DOI] [PubMed] [Google Scholar]
- [87].Wang C, Garlick S, Zloh M, Deep Learning for Novel Antimicrobial Peptide Design, Biomolecules. 11 (2021) 1–17. 10.3390/BIOM11030471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Corrochano AR, Cal R, Kennedy K, Wall A, Murphy N, Trajkovic S, O’Callaghan S, Adelfio A, Khaldi N, Characterising the efficacy and bioavailability of bioactive peptides identified for attenuating muscle atrophy within a Vicia faba-derived functional ingredient, Curr Res Food Sci. 4 (2021) 224–232. 10.1016/J.CRFS.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Cavaco M, Valle J, Flores I, Andreu D, Castanho MARB, Estimating peptide half‐life in serum from tunable, sequence‐related physicochemical properties, Clin Transl Sci. 14 (2021) 1349–1358. 10.1111/cts.12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Irwin JJ, Tang KG, Young J, Dandarchuluun C, Wong BR, Khurelbaatar M, Moroz YS, Mayfield J, Sayle RA, ZINC20—A Free Ultralarge-Scale Chemical Database for Ligand Discovery, J Chem Inf Model. 60 (2020) 6065–6073. 10.1021/acs.jcim.0c00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Wang G, Li X, Wang Z, APD3: the antimicrobial peptide database as a tool for research and education, Nucleic Acids Res. 44 (2016) D1087–D1093. 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Pirtskhalava M, Amstrong AA, Grigolava M, Chubinidze M, Alimbarashvili E, Vishnepolsky B, Gabrielian A, Rosenthal A, Hurt DE, Tartakovsky M, DBAASP v3: database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics, Nucleic Acids Res. 49 (2021) D288–D297. 10.1093/nar/gkaa991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Gawde U, Chakraborty S, Waghu FH, Barai RS, Khanderkar A, Indraguru R, Shirsat T, Idicula-Thomas S, CAMPR4: a database of natural and synthetic antimicrobial peptides, Nucleic Acids Res. 51 (2023) D377–D383. 10.1093/nar/gkac933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Gautam A, Chaudhary K, Singh S, Joshi A, Anand P, Tuknait A, Mathur D, Varshney GC, Raghava GPS, Hemolytik: a database of experimentally determined hemolytic and non-hemolytic peptides, Nucleic Acids Res. 42 (2014) D444–D449. 10.1093/nar/gkt1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Agrawal P, Bhalla S, Usmani SS, Singh S, Chaudhary K, Raghava GPS, Gautam A, CPPsite 2.0: a repository of experimentally validated cell-penetrating peptides, Nucleic Acids Res. 44 (2016) D1098–D1103. 10.1093/nar/gkv1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Shi G, Kang X, Dong F, Liu Y, Zhu N, Hu Y, Xu H, Lao X, Zheng H, DRAMP 3.0: an enhanced comprehensive data repository of antimicrobial peptides, Nucleic Acids Res. 50 (2022) D488–D496. 10.1093/nar/gkab651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Jhong J-H, Yao L, Pang Y, Li Z, Chung C-R, Wang R, Li S, Li W, Luo M, Ma R, Huang Y, Zhu X, Zhang J, Feng H, Cheng Q, Wang C, Xi K, Wu L-C, Chang T-H, Horng J-T, Zhu L, Chiang Y-C, Wang Z, Lee T-Y, dbAMP 2.0: updated resource for antimicrobial peptides with an enhanced scanning method for genomic and proteomic data, Nucleic Acids Res. 50 (2022) D460–D470. 10.1093/nar/gkab1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Kaushik AC, Wei D-Q, An Accurate Bioinformatics Tool For Anti-Cancer Peptide Generation Through Deep Learning Omics, BioRxiv. (2019). [Google Scholar]
- [99].Lee B, Shin MK, Hwang I-W, Jung J, Shim YJ, Kim GW, Kim ST, Jang W, Sung J-S, A Deep Learning Approach with Data Augmentation to Predict Novel Spider Neurotoxic Peptides, Int J Mol Sci. 22 (2021) 12291. 10.3390/ijms222212291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Agüero-Chapin G, Antunes A, Marrero-Ponce Y, A 2022 Update on Computational Approaches to the Discovery and Design of Antimicrobial Peptides, Antibiotics. 12 (2023) 1011. 10.3390/antibiotics12061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Erjavac I, Kalafatovic D, Mauša G, Coupled encoding methods for antimicrobial peptide prediction: How sensitive is a highly accurate model?, Artificial Intelligence in the Life Sciences. 2 (2022) 100034. 10.1016/J.AILSCI.2022.100034. [DOI] [Google Scholar]
- [102].Pripp AH, Isaksson T, Stepaniak L, Sørhaug T, Ardö Y, Quantitative structure activity relationship modelling of peptides and proteins as a tool in food science, Trends Food Sci Technol. 16 (2005) 484–494. 10.1016/J.TIFS.2005.07.003. [DOI] [Google Scholar]
- [103].Mei H, Liao ZH, Zhou Y, Li SZ, A new set of amino acid descriptors and its application in peptide QSARs, Biopolymers - Peptide Science Section. 80 (2005). 10.1002/bip.20296. [DOI] [PubMed] [Google Scholar]
- [104].Tian F, Zhou P, Li Z, T-scale as a novel vector of topological descriptors for amino acids and its application in QSARs of peptides, J Mol Struct. 830 (2007) 106–115. 10.1016/J.MOLSTRUC.2006.07.004. [DOI] [Google Scholar]
- [105].Yang L, Shu M, Ma K, Mei H, Jiang Y, Li Z, ST-scale as a novel amino acid descriptor and its application in QSAM of peptides and analogues, Amino Acids. 38 (2010). 10.1007/s00726-009-0287-y. [DOI] [PubMed] [Google Scholar]
- [106].Gancia E, Bravi G, Mascagni P, Zaliani A, Global 3D-QSAR methods: MS-WHIM and autocorrelation, J Comput Aided Mol Des. 14 (2000). 10.1023/A:1008142124682. [DOI] [PubMed] [Google Scholar]
- [107].Liang G, Yang L, Chen Z, Mei H, Shu M, Li Z, A set of new amino acid descriptors applied in prediction of MHC class I binding peptides, Eur J Med Chem. 44 (2009) 1144–1154. 10.1016/J.EJMECH.2008.06.011. [DOI] [PubMed] [Google Scholar]
- [108].van Westen GJ, Wegner J, IJzerman A, van Vlijmen H, Bender A, Molecular bioactivity extrapolation to novel targets by support vector machines, J Cheminform. 2 (2010). 10.1186/1758-2946-2-s1-o3. [DOI] [Google Scholar]
- [109].van Westen GJP, Swier RF, Cortes-Ciriano I, Wegner JK, Overington JP, Jzerman API, van Vlijmen HWT, Bender A, Benchmarking of protein descriptor sets in proteochemometric modeling (part 2): Modeling performance of 13 amino acid descriptor sets, J Cheminform. 5 (2013). 10.1186/1758-2946-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Kawashima S, AAindex: Amino Acid index database, Nucleic Acids Res. 28 (2000) 374–374. 10.1093/nar/28.1.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Xu Y, Verma D, Sheridan RP, Liaw A, Ma J, Marshall NM, McIntosh J, Sherer EC, Svetnik V, Johnston JM, Deep Dive into Machine Learning Models for Protein Engineering, J Chem Inf Model. 60 (2020) 2773–2790. 10.1021/acs.jcim.0c00073. [DOI] [PubMed] [Google Scholar]
- [112].Mauri A, alvaDesc: A tool to calculate and analyze molecular descriptors and fingerprints, in: Methods in Pharmacology and Toxicology, 2020. 10.1007/978-1-0716-0150-1_32. [DOI] [Google Scholar]
- [113].Warszycki D, Struski Ł, Śmieja M, Kafel R, Kurczab R, Pharmacoprint: A Combination of a Pharmacophore Fingerprint and Artificial Intelligence as a Tool for Computer-Aided Drug Design, J Chem Inf Model. 61 (2021). 10.1021/acs.jcim.1c00589. [DOI] [PubMed] [Google Scholar]
- [114].Ruggiu F, Solov’Ev V, Marcou G, Horvath D, Graton J, le Questel JY, Varnek A, Individual hydrogen-bond strength QSPR modelling with ISIDA local descriptors: A step towards polyfunctional molecules, Mol Inform. 33 (2014). 10.1002/minf.201400032. [DOI] [PubMed] [Google Scholar]
- [115].Hong H, Xie Q, Ge W, Qian F, Fang H, Shi L, Su Z, Perkins R, Tong W, Mold2, molecular descriptors from 2D structures for chemoinformatics and toxicoinformatics, J Chem Inf Model. 48 (2008). 10.1021/ci800038f. [DOI] [PubMed] [Google Scholar]
- [116].García-Jacas CR, Marrero-Ponce Y, Acevedo-Martínez L, Barigye SJ, Valdés-Martiní JR, Contreras-Torres E, QuBiLS-MIDAS: A parallel free-software for molecular descriptors computation based on multilinear algebraic maps, J Comput Chem. 35 (2014). 10.1002/jcc.23640. [DOI] [PubMed] [Google Scholar]
- [117].Ståhl N, Falkman G, Karlsson A, Mathiason G, Boström J, Deep Convolutional Neural Networks for the Prediction of Molecular Properties: Challenges and Opportunities Connected to the Data, J Integr Bioinform. 16 (2018). 10.1515/jib-2018-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Bento AP, Hersey A, Félix E, Landrum G, Gaulton A, Atkinson F, Bellis LJ, De Veij M, Leach AR, An open source chemical structure curation pipeline using RDKit, J Cheminform. 12 (2020). 10.1186/s13321-020-00456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Ruiz-Blanco YB, Agüero-Chapin G, Romero-Molina S, Antunes A, Olari LR, Spellerberg B, Münch J, Sanchez-Garcia E, ABP-Finder: A Tool to Identify Antibacterial Peptides and the Gram-Staining Type of Targeted Bacteria, Antibiotics. 11 (2022). 10.3390/antibiotics11121708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Lin D, Sutherland D, Aninta SI, Louie N, Nip KM, Li C, Yanai A, Coombe L, Warren RL, Helbing CC, Hoang LMN, Birol I, Mining Amphibian and Insect Transcriptomes for Antimicrobial Peptide Sequences with rAMPage, Antibiotics. 11 (2022) 952. 10.3390/antibiotics11070952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Hussain W, sAMP-PFPDeep: Improving accuracy of short antimicrobial peptides prediction using three different sequence encodings and deep neural networks, Brief Bioinform. 23 (2022). 10.1093/bib/bbab487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The biggest hurdle to developing DL models that are accurate in generating peptides with biological activity is to have enough data for training the models. Whereas small organic compounds have databases such as ZINC [90], SciFinder, Reaxys, and PubChem, which count with millions to hundreds of millions of molecules but mostly with no information on bioactive activities, bioactive peptides databases, such as APD3 [91], DBAASP [92], CAMP [93], Hemolytik [94], CPPsite [95], DRAMP [96], and dbAMP [97], count no more than a couple of tens of thousands of data points. It is worth mentioning that databases such as UniProt or GenBank house extensive collections of peptides and proteins, yet it should be noted that these repositories do not exhibit antimicrobial activities and are still untapped sources for antimicrobially active molecules.
Overcoming the data availability issue relies on combining deep generative models with bioinformatic tools [98] or molecular dynamics simulations [50], or doing data augmentation [99] to increase the performance of the models. Bioinformatic tools and molecular dynamics simulations provide an enhanced sampling of sought-after properties for the biological activity desired. Among the most important features retrieved to enrich the datasets are the physicochemical features that can be extracted from high throughput simulations with a high level of detail. On the other hand, data augmentation will not generate new information on the dataset, but it will increase the number of examples in the training set while also introducing more variety in what the model sees and learns from. Both bioinformatic tools and molecular dynamics simulations are valid strategies for different biological purposes. We envision that coupling these strategies to high performing computing will improve quality sampling with less experimental input needed and may accelerate the discovery of potent antimicrobial agents.