ABSTRACT
Carbapenemase-producing Enterobacterales (CPE) pose a major public health threat. Despite active infection prevention efforts, the incidence of KPC-2 carbapenemase-producing Enterobacterales (KPC2-CPE) continues to increase worldwide. In this study, we performed genome sequencing of 135 KPC2-CPE isolates obtained from multiple sources (clinical, hospital environments, and surface water) in Germany between 2013 and 2019 and analyzed them for epidemiological clues regarding transmission. For 92% (124/135) of all isolates, which comprised 14 different species such as Klebsiella, Escherichia, Citrobacter, Enterobacter, Raoultella, and Serratia, KPC-2 was present on an IncN[pMLST15] plasmid. All plasmids carried a novel non-Tn4401-element harboring an aac (3)-IId-bla TEM-1B-bla KPC-2–cassette (designated NTE KPC -Y) that was co-transferred with an adjacent region carrying 12 further antibiotic resistance genes. Identical plasmids were also detected in KPC2-CPE isolates from environmental samples. These plasmids were remarkably stable and were maintained in individual patients colonized with KPC-2 CPEs over a long-term period (>1 year). Thus, a predominant broad host range signature IncN[pMLST15] plasmid mediates transmission of both KPC-2 and associated multiple antimicrobial drug resistance genes in Germany. These data underline the need for in-depth characterization of plasmid carriers of CPE in surveillance and outbreak studies as well as in microbiomes from patients and the environment to identify hidden transmission reservoirs. This information will be essential for the development and implementation of effective infection control and prevention measures to disrupt dissemination of KPC2- CPEs in healthcare and associated environmental settings.
IMPORTANCE
Current infection control protocols assume that the spread of KPC-2 carbapenemase-producing Enterobacterales (KPC2-CPE) by detected carriers to other in-house patients is through clonal transmission and can be restricted by implementing containment measures. We examined the presence of the bla KPC-2 gene in different genera and species of Enterobacterales isolated from humans at different hospitals and surface waters between 2013 and 2019 in Germany. We found that a single IncN[pMLST15] plasmid carrying the bla KPC-2 gene on a novel non-Tn4401-element (NTEKPC-Y), flanked by an adjacent region encoding 12 other antibiotic resistance genes, was uniquely present in multiple species of KPC2-CPE isolates. These findings demonstrate the selective impact of specific IncN plasmids as major drivers of carbapenemase dissemination and suggest “plasmid-based endemicity” for KPC2-CPE. Studies on the dynamics of plasmid-based KPC2-CPE transmission and its presence in persistent reservoirs need to be urgently considered to implement effective surveillance and prevention measures in healthcare institutions.
KEYWORDS: KPC-2, IncN, plasmid-mediated transmission, non-Tn4401-element, carbapenem resistance, Enterobacterales
INTRODUCTION
Infections caused by carbapenemase-producing Enterobacterales (CPE) are increasing, and the lack of safe and efficacious treatment options presents a challenge for healthcare systems and patient safety around the globe (1). Klebsiella pneumoniae carbapenemase (KPC) is one of the leading causes of carbapenem resistance in nosocomial infections and CPE outbreaks worldwide (2 – 4). Current data suggest that these outbreaks are usually locally limited in nature and tend to be restricted to medical facilities, often subsiding following the implementation of infection prevention and control interventions (5). Nevertheless, the incidence of KPC-2-dependent infections worldwide remains high and continues to increase (6, 7).
Dissemination of bla KPC-2 involves horizontal transfer of bla KPC-2-encoding genetic elements and plasmids (8). A retrospective cohort study in Singapore suggested that plasmid-mediated transmission accounted for ~45% of all CPE carriers (9). Plasmids were predominantly of the incompatibility groups (Inc) N and FII, followed by R, A/C2, P, L/M, Q1, FIB, U, FIA, X3, Q2, and N3 (10, 11). Conjugal transfer of carbapenemase-encoding plasmids occurs in different species that have adapted to various ecological niches (12). Inanimate environments and particularly the aqueous environment represent reservoirs for facilitating plasmid-based transmission of carbapenemase genes in hospital settings (13). Dissemination of antibiotic resistance genes (ARGs) via plasmids represents a significant but poorly understood challenge in community health and hospital infection control.
The bla KPC-2 gene is often found embedded within a ~ 10-kb transposon Tn4401 (14 – 17), which was, for example, present in 35.5% (72/192) of all KPC-2 plasmids in the study by Brandt et al. (10). This composite transposon is predominant in epidemiologically successful K. pneumoniae ST258 complex clones (15, 18). However, reports of the presence of bla KPC-2 within non-Tn4401-elements (NTE KPC ) are now frequent and increasing (5, 17, 19). Unlike the transposable Tn4401 element, NTE KPC elements lack the flanking ISKpn7 and the Tn3 family transposase and resolvase. By using additional genes adjacent to bla KPC-2 for classification, three NTE KPC groups, viz., NTE KPC -I, NTE KPC -II and NTE KPC -III, have been described (17). Members of the NTE KPC -II group (with its three subgroups IIa, IIb, and IIc) carry a truncated bla TEM sequence upstream of bla KPC-2 flanked by a partial ISKpn6 element, downstream. These NTE KPC -II subgroups are often associated with different plasmid types (17, 20). Here, we report for the first time on the detection of an NTEKPC variant harboring both bla KPC-2 and a complete bla TEM-1B gene on a unique group of IncN plasmids, which concomitantly also transfers multiple adjacent antibiotic resistance genes to generate pan-resistant CPEs.
RESULTS
Origins and genetic diversity of the KPC-2 carbapenemase-producing Enterobacterales (KPC2-CPE)
In a 3-year genome-based surveillance study between 2017 and 2019 of 61 hospitals in the German Federal State of Hessen, we obtained a total of 589 carbapenem-resistant Gram-negative bacteria isolates. Of these, 346 were carbapenemase-producing Enterobacterales (CPEs). The genotypes detected were OXA-48-like (40.5%), KPC- (31.6%), NDM- (17.8%), and VIM- (15.1%). Notably, carbapenemase genes were detected in 149 of 188 K. pneumoniae isolates and 86 of 105 E. coli isolates, as well as in 49 of 52 Citrobacter, 15 of 41 Enterobacter, four of four Raoultella, and 20 of 21 Serratia isolates (21).
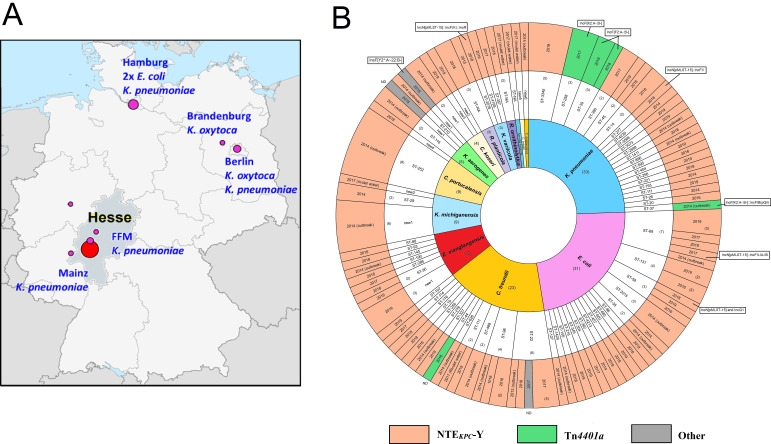
KPC-2 carbapenemase-producing CPE (KPC2-CPE) represented ≈ 25% (n = 85) of all CPEs, and this percentage remained annually constant over the entire study period. For comparison, we included 37 KPC2-CPE isolates from a nosocomial outbreak from 2013/14 in Southern Hesse, Germany (22) and eight additional KPC2-CPE isolates obtained from patients in the German Federal States of Hesse, Hamburg, Berlin, and Brandenburg obtained between 2014 and 2015 (Fig. 1A). Five KPC2-CPEs collected from the surface water (2017) of a rivulet where the suspected index patient of an outbreak in the Rhine-Main region suffered a near-drowning incident were also included in the study (23).
Fig 1.
(A) Geographic location of bla KPC-2-encoding IncN[pMLST15] plasmids identified from the outbreak and various hospitals in Hesse as well as in the rivulet water in Germany. (The base map is reproduced from https://commons.wikimedia.org/wiki/File:Germany_location_map.svg#/media/File:Germany_location_map.svg, by NordNordWest, licensed under the Creative Commons Attribution-Share Alike 3.0 Unported license.) (B) Overview of the 135 KPC-2 carbapenemase-producing isolates with characteristics of the taxonomic and genome sequence types as well as the distribution of the genetic elements surrounding the gene bla KPC-2 (NTEKPC-Y and Tn4401a). All the NTEKPC-Y-harboring bla KPC-2–plasmids identified as Inc-type IncN[pMLST15] in single replicon (n = 120) or as co-integrates of multiple replicon types (n = 4). Most of the Tn4401a –containing bla KPC-2–plasmids belonged to IncF(K) or IncF(II) groups. The year of isolation of the respective isolates is depicted in the outermost ring.
KPC-2-producing isolates included those from rectal swabs (n = 73), urine samples (n = 12), respiratory samples (n = 12), bloodstream-infections (n = 2), and other sites (n = 21). Isolates from the hospital environment associated with the outbreak (n = 10) and rivulet water samples (n = 5) were included in the study. Detailed information of all KPC2-CPE isolates, including their genomic characteristics, plasmid Inc types, and antimicrobial resistance genes, is presented in Table S1.
Genomic taxonomy and multi-locus sequence typing (MLST) analysis revealed high taxonomic and genetic diversity among the 135 KPC2-CPE isolates. We detected 14 species comprising the genera Klebsiella, Escherichia, Citrobacter, Enterobacter, Raoultella, and Serratia that were distributed into 74 individual MLST types (Fig. 1B). The most commonly occurring species were K. pneumoniae (22 STs, n = 33), E. coli (16 STs, n = 31), C. freundii (10 STs, n = 23), and E. xiangfangensis (7 STs, n = 10).
A predominant IncN[pMLST15] plasmid carrying a novel NTEKPC-Y cassette
For 124 isolates of KPC2-CPEs, the bla KPC-2 gene was located on IncN plasmids that were subtyped as pMLST15. Of these, 121 were single-replicon IncN[pMLST15] plasmids and three were cointegrates of this plasmid with IncF-, IncFII-, and IncQ1 plasmids (Table 1; Table S2). In eight of the remaining 11 KPC2-CPE isolates, the bla KPC-2 gene was located on IncF plasmids. Seven of them were associated with a Tn4401a element, whereas in three isolates, no definite assignment of the location on either the chromosome or plasmid was possible (Table S3). Within the surveillance period, IncN[pMLST15] was detected in 75 of 85 KPC2-CPE from 10 different species and associated with more than 46 MLST types. These isolates were obtained from 45 patients in 19 hospitals distributed across the State of Hesse.
TABLE 1.
Contribution of the bla KPC-2 containing NTEKPC-Y-carrying IncN[pMLST15] plasmids based on antimicrobial resistance gene (ARG) profiles in Germany from 2013 to 2019 (n = 124) c , d
| Study category and sampling period (year) | SurvCARE Hesse (2017–2019) | Outbreak in Hesse (2013–2014) | Nation-wide KPC-2 CPE (2014–2015) |
Rivulet water screening (2017) |
|---|---|---|---|---|
| Sequenced KPC-2-carrying isolates | 85 | 37 | 8 | 5 |
| IncN[pMLST15] plasmids carrying NTEKPC-Y | 75 a | 36 | 8 | 5 |
| Percentages | 88% | 97% | 100% | 100% |
| Occurrence in species below: no. of isolates and (ST-Types): | ||||
| C. amalonaticus | - | 1 (ND) | - | - |
| C. braakii | 2 (ND) | - | - | 1 (ND) |
| C. freundii | 14 (9) | 6 (5) | - | 1 (1) |
| C. koseri | 2 (ND) | 2 (ND) | - | - |
| C. portucalensis | 1 (1) | 8 (1) | - | - |
| E. coli | 21 (11) | 7 (5) | 2 (2) | - |
| E. cloacae | 3 (3) | - | - | - |
| E. xiangfangensis | 6 (4) | 1 (1) | - | - |
| K. aerogenes | - | 3 (2) | - | - |
| K. michiganensis | - | 6 (1) | 2 (2) | 1 (1) |
| K. pneumoniae | 22 (14) | 2 (2) | 3 (3) | - |
| K. variicola | 3 (3) | - | - | - |
| R. ornithinolytica | 1 (ND) | - | - | 1 (ND) |
| R. planticola | - | 1 (ND) | - | |
| S. fonticola | 2 (ND) | - | - | 1 (ND) |
| Sampling sources: | ||||
| No. of isolates from humans | 75 | 24 | 8 | 0 |
| No. of patients | 50 | 19 | 8 | 0 |
| No. of hospitals | 19 | 3 | 8 | - |
| No. of isolates from the environment | 0 | 12 | 0 | 5 |
| ARG profile b | ||||
| A | 45 | 31 | 8 | 5 |
| B | 2 | - | - | - |
| C | 7 | - | - | - |
| D | 2 | - | - | - |
| E | 13 | 4 | - | - |
| F | 2 | 1 | - | - |
| Various | 4 | - | - | - |
Includes multi-replicon plasmids of IncN[pMLST15] co-integrated either with IncF[F36+31:A4:B10], IncF(K2:A-:B-]+IncR,, or IncF[K9:A-:B-].
The details of antibiotic resistance genes of ARG profiles are shown in Table 2.
SurvCARE Hesse: a regional genome-based CRE surveillance study (State of Hesse).
ND: not determined.
Overview of bla KPC-2-bearing plasmids characterized by long-read Sequencing
For detailed analysis of bla KPC-2-bearing plasmids, we subjected a subset of the KPC2-CPE (n = 34) to long-read sequencing. The isolates selected for sequencing broadly covered species and multi-locus sequence types together with the year of collection, i.e., between 2013 and 2019, and their KPC-2-encoding plasmids are given in Table 3. This plasmid data set was from 11 species that included eight E. coli of six STs (ST58, ST69, ST127, ST131, ST362, and ST2015), six K. pneumoniae (ST14, ST37, ST111, ST277, ST1411, and ST3345), six C. freundii (ST22, ST98, ST278, and ST327), three K. variicola of ST357, ST20178, and ST4199, as well as two- of E. xiangfangensis, C. koseri, C. portucalensis and one- each of C. amalonaticus, K. aerogenes, R. ornithinolytica, and R. planticola. Isolates were obtained in 2013 (1), 2014 (9), 2016 (1), 2017 (5), 2018 (12), and 2019 (6). Of the 34 long-read-sequenced KPC2-CPEs, 32 carried a unique IncN[pMLST15] plasmid replicon with the allele combination repN_7/korA_3/traJ_6, and the remaining two were IncF plasmids (Table 3). The plasmids with a single IncN[pMLST15] replicon were generally 78 kb in size, but plasmid variants with sizes ranging from 52 kb to 89 kb were also detected.
TABLE 3.
Genomic characteristics of the bla KPC-2-bearing plasmids based on long-read sequencing
| Plasmid name | Size (kb) | Inc type and pMLST | bla KPC-2-harboring genetic elements | ARG-profile | Species | Genome accession no. |
|---|---|---|---|---|---|---|
| pCF13141-KPC2 | 78 | IncN[pMLST15] | NTEKPC-Y | A | C. freundii | VKMY01000050 |
| pCF08698-KPC2 | 78 | IncN[pMLST15] | NTEKPC-Y | A | C. freundii | VKMD01000077 |
| pCP13069-KPC2 | 78 | IncN[pMLST15] | NTEKPC-Y | A | C. portucalensis | VKMZ01000118 |
| pCK13142-KPC2 | 78 | IncN[pMLST15] | NTEKPC-Y | A | C. koseri | VKNB01000047 |
| pKP37361-KPC2 | 78 | IncN[pMLST15] | NTEKPC-Y | A | K. pneumoniae | CP104944 |
| pEClo_Surv151-KPC2 | 78 | IncN[pMLST15] | NTEKPC-Y | A | E. xiangfangensis | CP104949 |
| pKV30046-KPC2 | 78 | IncN[pMLST15] | NTEKPC-Y | A | K. variicola | CP104940 |
| pCPsc18-2783-KPC2 | 78 | IncN[pMLST15] | NTEKPC-Y | A | C. portucalensis | CP104956 |
| pEC16155-KPC2 | 79 | IncN[pMLST15] | NTEKPC-Y | A | E. coli | VKML01000077 |
| pCA13304-KPC2 | 77 | IncN[pMLST15] | NTEKPC-Y | A | C. amalonaticus | VKME01000146 |
| pEXva18-1651-KPC2 | 79 | IncN[pMLST15] | NTEKPC-Y | A | E. xiangfangensis | CP104948 |
| pCK_Surv347-KPC2 | 79 | IncN[pMLST15] | NTEKPC-Y | A | C. koseri | CP104957 |
| pEC32446-KPC2 | 69 | IncN[pMLST15] | NTEKPC-Y | A a | E. coli | CP104950 |
| pEC14408-2-KPC2 | 88 | IncN[pMLST15] | NTEKPC-Y | A | E. coli | LT599827 |
| pCFur18-0060-KPC2 | 89 | IncN[pMLST15] | NTEKPC-Y | A | C. freundii | CP104958 |
| pCF37969-KPC2 | 89 | IncN[pMLST15] | NTEKPC-Y | A | C. freundii | CP104960 |
| pEC32009-KPC2 | 195 | IncN[pMLST15]:: IncF[F36 +31:A4:B10] | NTEKPC-Y | A | E. coli | CP104951 |
| pKV_Surv097-KPC2 | 217 | IncN[pMLST15]:: IncFII(K2:A-:B-]::IncR | NTEKPC-Y | A | K. variicola | CP104941 |
| pCF38954-KPC2 | 80 | IncN[pMLST15] | NTEKPC-Y | D | C. freundii | CP104959 |
| pCF_Surv457-KPC2 | 83 | IncN[pMLST15] | NTEKPC-Y | B a | C. freundii | CP104961 |
| pRP_Surv085-KPC2 | 73 | IncN[pMLST15] | NTEKPC-Y | B | R. planticola | CP104937 |
| pEC_Surv265-KPC2 | 69 | IncN[pMLST15] | NTEKPC-Y | D | E. coli | CP104953 |
| pKP38941-KPC2 | 69 | IncN[pMLST15] | NTEKPC-Y | C | K. pneumoniae | CP104943 |
| pKP41623-KPC2 | 164 | IncN[pMLST15]::IncFII[K9:A-:B-] | NTEKPC-Y | D | K. pneumoniae | CP104942 |
| pRO41724-KPC2 | 71 | IncN[pMLST15] | NTEKPC-Y | E | R. ornithinolytica | CP104938 |
| pKP_Surv398-KPC2 | 59 | IncN[pMLST15] | NTEKPC-Y | E | K. pneumoniae | CP104946 |
| pKP_Surv434-KPC2 | 59 | IncN[pMLST15] | NTEKPC-Y | E | K. pneumoniae | CP104945 |
| pKV33665-KPC2 | 59 | IncN[pMLST15] | NTEKPC-Y | E | K. variicola | CP104939 |
| pEC_Surv291-KPC2 | 59 | IncN[pMLST15] | NTEKPC-Y | E | E. coli | CP104952 |
| pCF12908-KPC2 | 59 | IncN[pMLST15] | NTEKPC-Y | E | C. freundii | VKMA01000036 |
| pEC_Surv190-KPC2-1 | 52 | IncN[pMLST15] | NTEKPC-Y | E | E. coli | CP104954 |
| pEC_Surv190-KPC2-2 | 23 | IncQ1-like | NTEKPC-Y | E b | E. coli | CP104955 |
| pEC11992-KPC2 | 48 | IncN[pMLST15]∆ | NTEKPC-Y | F | E. coli | VKMQ01000091 |
| pKA36387-KPC2 | 166 | IncF[Y2 a :A ~ 22:B-] | Other c | Other d | K. aerogenes | CP104947 |
| pKP11394-KPC2 | 241 | IncF[K2:A-:B-]::IncFIB(pQil) :IncFIB(pQil) | Tn4401a | Other e | K. pneumoniae | VKNL01000120 |
these both plasmids carried only one copy of sul1.
pEC_Surv190-KPC2-2 harbored an E-like profile without strA,, but sul2.
pKA36387-KPC2 harbored neither a NTEKPC-Y nor Tn4401.
bla KPC-2 and bla TEM-1 are not adjacent to each other.
pKP11394-KPC2 harbored two Tn4401a with bla KPC-2.
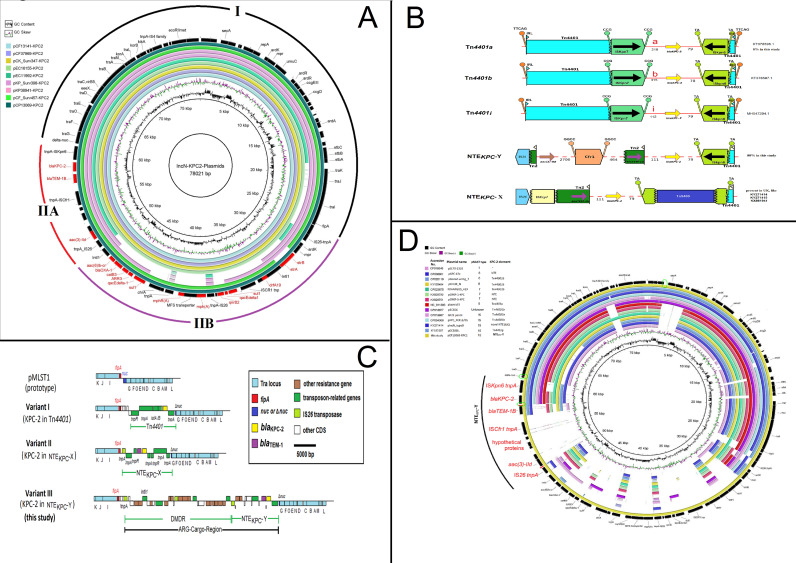
The composite structure of the IncN[pMLST15] plasmids consists of a 43-kb IncN-characteristic backbone (region I), which includes the replication region, a conjugation (tra) system, a stability operon, and an anti-restriction system, similar to that of pNL194, an IncN(pMLST18) plasmid (24). The tra genes are separated into three sections TRA-I, TRA-II, and TRA-III (24). The region between TRA-II and TRA-III (comprising fipA and nuc) is separated by the insertion of an acquired segment of ~35 kb (region II) into the gene nuc, and this region II can in turn be divided into two segments, region IIA and IIB. Region IIA (11 kb) comprises a section carrying three ARGs including bla KPC-2 on a non-Tn4401-element (described in detail as follows), which is commonly present in all IncN[pMLST15] plasmids regardless of size (Fig. 2A).
Fig 2.
(A) Genetic map of the representative complete IncN[pMLST15] plasmids with the common length of 78,021 bp depicting the backbone (region I), the unique genetic environment NTEKPC-Y (region IIA), and the dynamic multi-drug resistance region (DMDR; Region IIB). Antimicrobial resistance genes (ARGs) are marked in red. The genetic environment of the NTEKPC-Y (region IIA), encoding bla KPC-2–bla TEM-1B and acc (3)IId, was present in all plasmids; the DMDR (region IIB) differed in some plasmids, representing different antimicrobial resistance profiles. (B) Comparison of the non-Tn4401-KPC-element NTEKPC-Y (this study), NTEKPC-X (from United Kingdom), and Tn4401 variants from the KPC-2-encoding IncN[pMLST15] plasmids. (C) Insertion sites of the ARG-bearing cargo-region of different IncN[pMLST15] variants. Variant I, bla KPC-2 located in Tn4401 (CP004366, CP004367, CP018963, KX154765, KX276209, KX062091, KX397572, KF182187, and KF181264); variant II, bla KPC-2 located in and NTEKPC-X (KX88194, KY27414, and KY27415); and variant III, the bla KPC-2, was located in NTEKPC-Y (this study). (D) Comparison of different KPC-2 IncN pMLST types. The pCP13069-KPC2 contained a unique region, assigned as NTEKPC-Y. The green circles on the ring maps symbolize the areas for the specific PCR primers.
Region IIB (24 kb) is a mosaic structure that carries a complex array of 12 additional ARGs conferring resistance to nine antibiotic classes, including aminoglycosides (strA; strB), beta-lactams (bla OXA-1), fluoroquinolones (aac(6´)-Ib-cr, qnrB2), macrolides [mph(A)], phenicoles (catB3), rifampicin (ARR-3), sulfonamides (sul1, two copies), trimethoprim (dfrA19), and quaternary ammonium compounds (qacEΔ_1). These ARGs are associated with numerous insertion sequences (ISs) and transposons, for example, two IS26, ISCR1, IS6100, ΔTn5393, ΔTn402, and ΔTnch and a class 1 integron (Fig. 2A). Region IIB is prone to deletions and can also harbor transposon insertions that account for variability in plasmid length, as, for example, with Tn5075 (1327 bp) in pExva18651-KPC2 and Tn5403 (2564 bp) in pEcSurv291-KPC2. In addition, an IS903 flanked by direct repeat sequences (GCGCATGGC) and a truncated TnPa38 (Tn3 family transposon) were present in region I.
Plasmid transfer experiments with the donor strains CF08098 and CP13069 showed bla KPC-2 IncN[pMLST15] transconjugants with plasmids of different sizes (~78 kb and ~60 kb), indicating transfer of resistance to multiple antibiotic classes as well as an inherent instability of the ARGs structure in region II (Fig. S1).
A novel non-Tn4401-KPC2-element (NTEKPC-Y)
The region surrounding bla KPC-2 was unique and differed from both typical Tn4401 structures (tnpR Tn4401 -tnpA Tn4401 -ISkpn7–bla KPC-2-ISkpn6) as well as from other known non-Tn4401 elements (NTE KPC -I, II, and III) from previous studies (19, 25). We provisionally assigned it as NTE KPC -Y, to distinguish it from a newly described NTE KPC -X that is present on an IncN[pMLST15] plasmid in isolates from the United Kingdom (6). Fig. 2B illustrates the differences between the Tn4401-isoforms (a, b, and j) and the non-Tn4401 elements, NTE KPC -Y and NTE KPC -X. In the Tn4401-isoforms, a Tn4401-resolvase, a Tn4401-transposase, and an ISKpn7-transposase are present upstream of the bla KPC-2 gene, while for NTE KPC -Y, an IS26 transposase, aac (3)-IId, three CDSs of unknown function, an ISCfr1, and a Tn2-associated bla TEM-1B gene are present. In NTE KPC -X, an IS26 transposase, ISEcp1 element, and a Tn2-associated bla TEM-1B gene are followed by an intact ISKpn6 downstream of the bla KPC-2 gene, whereby the ISKpn6 is split by an IS5403 element. However, the combination of the aac3-IId, bla TEM-1B, and blaKPC-2 resistance genes on NTE KPC -Y is new and has not been hitherto detected. Fig. 3C shows the common insertion site of the bla KPC-2 with its surrounding elements and ARGs of the three KPC-2-bearing variants I, II, and III. The IncN[pMLST15] variant III plasmids in this study also harbored additional ARGs within its dynamic multi-drug resistance region (DMDR [Fig. 2C]). We designed specific PCR primer sets based on this unique genetic feature of the IncN[pMLST15]-plasmid and combined it with primer pairs for the pMLST15-backbone (Table S5). This composite PCR assay allowed unambiguous detection of this novel IncN[pMLST15] plasmid in diagnostic reference laboratories.
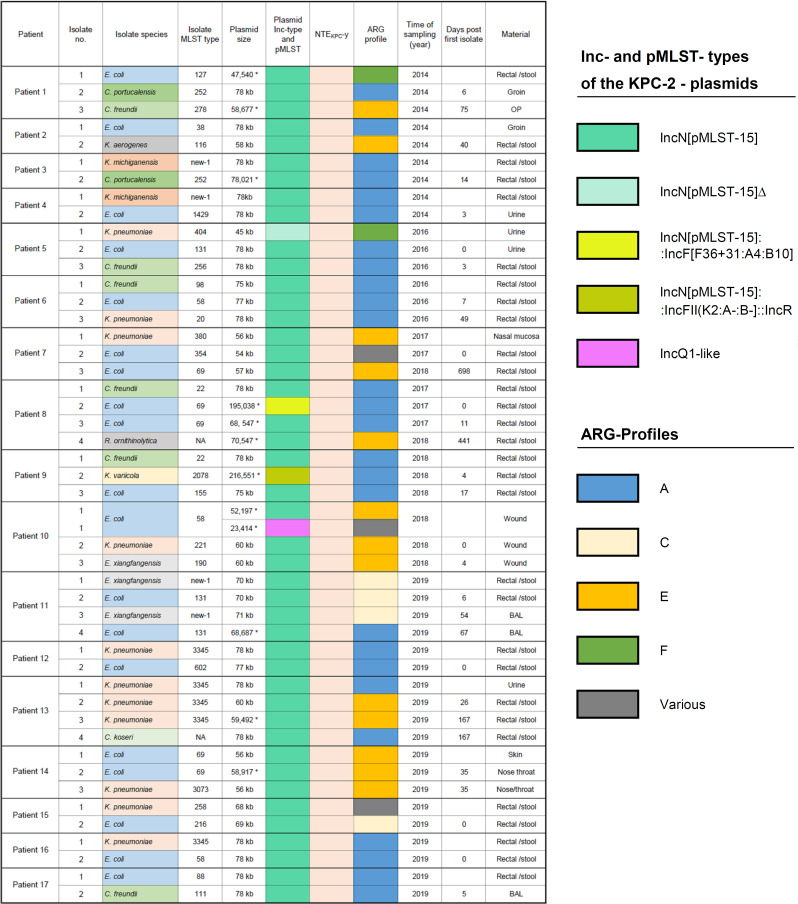
Fig 3.
Patients with multiple KPC-2-producing species carrying the IncN[pMLST15] plasmid with the NTEKPC-Y element.
Several ARG profiles were clustered with respect to the 35-kb inserted segment (Regions IIA + IIB, Tables 2 and 3; Fig. 2). ARG-Profile A with 15 ARGs was the most common type (71%; 89/124) and found in 78% of the 77 KPC2-CPE-patients in 30 hospitals (Table S1). ARG-Profile E containing five ARGs was the next common type (14%; 17/124) and was found in 10 patients in seven hospitals. The remaining minor ARG profiles designated B, C, D, and F and others were detected in 18/124 isolates.
TABLE 2.
ARG profiles of the bla KPC-2-bearing IncN[pMLST15] plasmids
| ARG profile | Antibiotic resistance genes | Median size of single IncN replicon plasmid (Min–Max) in bps | Size of cointegrates (IncN plus IncF/IncR) in bps |
|---|---|---|---|
| A | bla KPC-2, bla TEM-1B, bla OXA-1, strA, strB, aac(6´)-Ib-cr, aac (3)-IId, qnrB2, mph(A), catB3, aar-3, sul1 (1–2x), dfrA19 and qacE∆−1 | 78,021 (68,547–89,053) | 195,038 or 216,551 |
| B | bla KPC-2, bla TEM-1B, bla OXA-1, aac(6´)-Ib-cr, aac (3)-IId, qnrB2, mph(A), catB3, aar-3, sul1 (1–2x), dfrA19, and qacE∆−1; | 73,409–83,074 | |
| C | bla KPC-2, bla TEM-1B, bla OXA-1, strA, strB, aac(6´)-Ib-cr, aac (3)-IId, catB3, aar-3, sul1 (1–2x), dfrA19, and qacE∆−1 | 69,943 (64,969–71,469) | |
| D | bla KPC-2, bla TEM-1B, bla OXA-1, aac(6´)-Ib-cr, aac (3)-IId, catB3, aar-3, sul1 (1–2x), dfrA19, and qacE∆−1 | 67,754 | 164,049 |
| E | bla KPC-2, bla TEM-1B, strA, strB, and aac (3)-IId | 59,478 (58,446–70,547) | |
| F | bla KPC-2, bla TEM-1B, and aac (3)-IId | 47,540 |
Plasmids carrying ARG profile A were significantly associated with outbreaks. They were predominant in the large multi-species outbreak in 2013/2014 and were recovered six times between 2016 and 2019, generally involving two–three patients colonized with different species at different hospitals, as well as in a single-species outbreak in 2019 (Table S4). Of note, all five IncN[pMLST15] plasmids in the rivulet water samples that were collected downstream of a sewage treatment plant had cluster profile A. In contrast, plasmids with ARG profile E were rarely associated with outbreaks but frequently occurred in multiple-species-colonization profiles of individual patients, indicating interspecies spread. The other profiles occurred mostly sporadically, suggesting that they were occasional minority variants resulting from deletions.
Concurrent presence of IncN[pMLST15] plasmids in several species within individual patients or single samples
We observed that the NTE KPC -Y IncN[pMLST15] plasmid was present in multiple bacterial species in 17 individual patients, i.e., in 47 isolates from 10 different species (Fig. 3). Four patients were sampled during the 2013/14 outbreak, and the other 13 were obtained during the surveillance period. The number of the isolates per case was between two and four isolates comprising two to three species, often with a co-occurrence of E. coli and K. pneumoniae. The time span between the first and last isolate ranged between 4 and 167 days, and in two cases, it was even longer than a year (441 and 698 days). In four cases, different species with identical plasmids were detected from single samples such as rectal/stool swab (n = 3) and wound (n = 1). The IncN[pMLST15] plasmids within a patient frequently harbored identical ARG-profiles, mostly profile A.
The IncN[pMLST15] plasmid harboring NTEKPC-Y is unique to Germany
To examine the global distribution of the bla KPC-2-bearing IncN[pMLST15] plasmid with the NTEKPC-Y-element, we compared it to sequences extracted from the NCBI Genbank and other publicly available databases. We subjected data for 476 wholly sequenced IncN plasmids from public databases (NCBI and PLSDB, 34,513 entries) to further analysis. Of these, 258/476 carry repN_1 (true IncN plasmids) and are typable by pMLST. The most common IncN type was the pMLST7 (n = 64), followed by pMLST5, pMLST1, pMLST6, pMLST9, and pMLST15, with 58, 41, 40, 26, and 16 entries, respectively. We found 17 publicly available IncN[pMLST15] plasmids harboring a bla KPC-2. These originated mainly from clinical K. pneumoniae or E. coli isolates from Brazil, Israel, the United Kingdom, and France (Table S5). Comparative genomic analysis demonstrated that all plasmids carried bla KPC-2/-3 at a specific site inserted between a truncated nuc (endonuclease) gene and the fipA gene (Fig. 2D).
Surprisingly, all of the previously described IncN[pMLST15] plasmids are carriers of various bla KPC-2 elements, i.e., Tn4401-based or otherwise, regardless of their geographical origin (Table S6). We note that three similar plasmids with the novel NTE KPC -Y element were recently found in healthcare institutions in the Czech Republic that borders with Germany (accession no. CP070531, CP070538, and CP070545), suggesting regional dispersion.
DISCUSSION
In this study, we detected the presence of blaKPC-2 in different genera and species of Enterobacterales isolated from human and inanimate environments between 2013 and 2019 in Germany. This study analyzed isolates from 61 hospitals in the Federal State of Hesse serving a population of ~6.3 million inhabitants together with additional isolates from other regions in Germany including samples obtained from surface waters. We detected broad species-based dissemination of the blaKPC-2 gene facilitated by intra- and inter-species horizontal gene transfer (HGT) of a single IncN[pMLST-15] plasmid. This transfer also led to the emergence of highly antibiotic-resistant bacteria that carry up to 14 other ARGs encoded on the same plasmid (Table 2). Previously, an association of KPC-2 with plasmids of different incompatibility (Inc) groups, such as FII, N, R, A/C2, P, L/M, Q1, FIB, U, FIA, X3, Q2, and N3, was reported, with FII and N being the most common (10). However, the proportion of the previously mentioned plasmids reported that are in fact cointegrates with an IncN plasmid, as detected in this study, is not known.
Currently, 36% of all reported plasmids worldwide carry the blaKPC-2 gene on the transposon element Tn4401 (10), indicating the importance of transposons in transmission of this resistance. In our study, 94% were associated with NTE KPC -Y, a novel non-Tn4401-KPC2-element carried on IncN[pMLST15] plasmids, while only 6% of all isolates carry KPC-2 on a Tn4401a transposable element. The NTE KPC -Y element is unlikely to be mobile because it is always coupled to the presence of the IncN[MLST15] plasmid, that is, present even in cointegrates. Thus, the NTE KPC -Y element relies on the replication and maintenance properties of the broad host range IncN plasmid for effective dissemination and persistence in a range of different environments (Fig. 4).
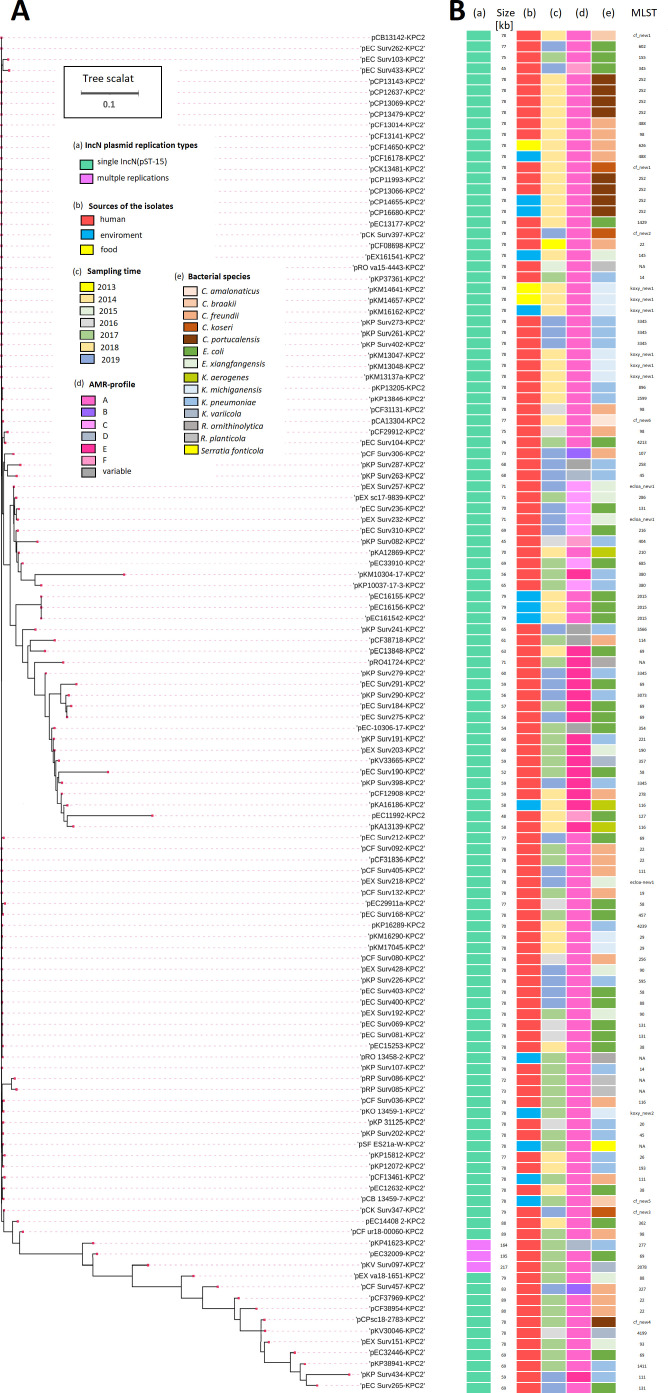
Fig 4.
(A) Phylogenetic tree of the 124 IncN-KPC2-plamids either with a single IncN[pMLST-15] replication protein type (n = 121) or with a multiple replication protein type in co-integrated IncN[pMLST-15] (n = 3), which are IncN[pMLST-15]::IncF[F36-F31:A4:B10], IncN[pMLST-15]::IncF[K9:A -:B-], and IncN[pMLST-15]::IncF[K2:A-:B-]:IncR. Phylogenetic analysis showed that 84 of them were highly homologous, regardless of the time of isolation, source, and bacterial species. (B) Shown are the types, sizes, and AMR profiles of the IncN plasmids, as well as the sources of the isolates, the time of sampling, and the bacterial species.
In the 3-year SurvCARE surveillance study, IncN[pMLST15] comprised 88% of all KPC2-CPEs detected (Table 1). Plasmids carrying the ARG profile A significantly contributed to outbreaks, causing a large multi-species outbreak in southern Hesse in 2013/2014 (Table 1), six smaller multi-species outbreaks in several hospitals between 2016 and 2019, as well as a single-species outbreak in 2019 (Table S4). A largely identical NTE KPC -Y IncN[pMLST15] plasmid was previously reported from isolates obtained during outbreaks from hospitals in Berlin in 2016 (26) and more recently in Cologne (2020–2021) (Xanthopoulou K et al. ECCMID 2022, oral presentation O0234). Cumulatively, these data suggest that hitherto undetected transmission events may have resulted in the spread and persistence of this plasmid over large distances and time periods. The presence of the plasmid in environmental KPC2-CPE from surface water is deeply concerning as it emphasizes the presence of reservoirs in diverse ecological habitats. More recently, the detection of similar IncN[pMLST15] plasmids with NTE KPC -Y in C. freundii and E. hormaechei isolates from Ostrov, Koli, and Prague in the Czech Republic suggests multi-species regional, transboundary spread.
While the NTE KPC -Y element is clearly unique to Germany, similar NTEKPC-2 elements with unique compositions have been described in other countries such as Argentina, Brazil, China, Russia, and the United Kingdom (6, 17, 19). Their frequent association with broad host range IncN plasmids, which provide a versatile platform for the capture of these genes and its genetic plasticity, would facilitate transfer of large sets of ARGs even when antibiotics unrelated to carbapenems are used during therapy. An additional layer of complexity introduced by multi-species outbreaks exploits the ability of these plasmids to find appropriate bacterial hosts, e.g., Citrobacter, Enterobacter, and Raoultella, which are present in a wide variety of near-patient and community environments. This distribution is concerning as there are currently no effective strategies to force the elimination of these plasmids from their various hosts.
Our study has several limitations. The isolates submitted were part of mandatory reporting to regional healthcare authorities as well as voluntary contributions to the National Reference Center, and the study was limited in terms of access to clinical data of patients in the individual hospitals and healthcare institutions based on data protection considerations. The systematic surveillance study was geographically restricted to the state of Hesse and covers a catchment area of 21.115 km². In addition, environmental sampling at the individual institutions was not performed.
We provide evidence that the HGT properties of a single plasmid rather than clonal expansion of successful genetic lineages dominated the dissemination of blaKPC-2 in Germany. Its location on a novel non-transposable Tn4401 element (NTE KPC -Y) suggests that plasmid-encoded factors contribute to the endemicity and long-term persistence of KPC-2 in community-based, hospital, and environmental reservoirs. Data from studies that examine for the presence of plasmids of KPC2-CPE in community and hospital microbiomes, both in patients and the environment, are urgently needed to inform and complement current containment measures to enable the development of effective transmission-control protocols. These results emphasize the need, already now, to be aware of a prominent role of transmissible plasmids during CPE surveillance and their contribution to transmission events.
MATERIALS AND METHODS
Source of the KPC-2-carbapenemase-producing Enterobacterales isolates (KPC-CPE)
In the present study, we collected and sequenced 135 KPC-CPE isolates. Twenty-four isolates derived from 19 patients during a nosocomial outbreak in southern Hesse (2013–2014), twelve isolates from outbreak-associated hospital settings, 75 isolates from 59 patients during a Hesse-wide surveillance project (SurvCARE, 2017–2019), eight isolates from eight patients harboring a KPC-2-IncN-plasmid detected in the NRZ-collection (2014–2015), and five isolates from a screening project of carbapenem-resistant bacteria in surface water in Hesse 2017 (23).
Sample size
To estimate the statistical power of our study, we used data on CPE prevalence from the National Reference Center for Enterobacterales at the Robert-Koch Institute in Germany between 2017 and 2019 (27 – 29). Based on these data, we expected to achieve a statistical power of 95% with a significance level (α) of 0.05 if we had a sample size of 145 isolates. Here, we performed genome sequencing of all 346 CPE samples obtained in this study, which increased the statistical power of our study to more than 99% with the same significance level.
Phenotypic characterization of the bacterial isolates and plasmid conjugation assay
All bacterial isolates were identified using mass spectrometry MALDI-TOF MS (VITEK MS, Biomerieux, Nürtingen Germany), characterized for their phenotypical antimicrobial susceptibility using the automated VITEK2 system (Biomerieux, Nürtingen, Germany), and interpreted following EUCAST guidelines. For plasmid conjugation assays, the E. coli strain J53 was used as a recipient, as previously described (30).
Whole-genome sequencing (WGS)
All isolates underwent WGS (Illumina MiSeq or NextSeq 500). A subset of the isolates was re-sequenced by long-read sequencing using the PacBio Single-Molecular-Real-Time (SMRT) or Nanopore technology to complete the genome. The Illumina library preparation and sequencing were carried out as previously described (31). Briefly, DNA sequencing libraries were prepared using the Nextera XT kit (Illumina MiSeq system (Illumina, Netherlands BV, Eindhoven, the Netherlands) according to the manufacturer’s introductions and sequenced either on a MiSeq instrument with 2 × 300 cycles or on a NextSeq instrument with 2 × 150 cycles. The SMRT-sequencing was carried out on a PacBio RSII machine (Pacific Biosciences, MenloPark, CA, USA), as described earlier (31). Sequencing using Nanopore technology with a MinION sequencer was performed as described previously (32).
Genome assembly and plasmid-sequence completion
The Illumina-sequenced reads were assembled de novo using CLC Genomics Workbench version 8.0.1 (Qiagen, Aarhus A/S, Denmark) and/or SPAdes genome Assembler (33). For PacBio SMRT sequencing, the assembly was performed either using RS HGAP Assembly 3 or SMRT-Link Microbial Assembly v.10.1.0, using default parameters. The validity of each assembly was cross-checked using the RS_Bridgemapper.1 protocol, and each replicon was circularized independently. Finally, the circulated genome sequence was error-corrected by mapping of Illumina reads to the finished genomes using BWA (34), with subsequent variant calling using VarScan (35). A consensus concordance of QV60 was obtained for all of the genomes. For Nanopore sequencing, hybrid assembly was carried out using Unicycler v0.4.6.
For sequence finishing (completion) of the bla KPC-2-bearing plasmids of the isolates sequenced only by Illumina, contig-mapping and read-mapping against closed bla KPC-2-bearing plasmid-genomes from the SMRT-sequencing as references, by using SeqManPro (10.0), MAUVE (2.3.1), and CLC-Workbenches (8.0.1), were performed to complete the bla KPC-2-encoding plasmids of the remaining isolates that were only sequenced by short-read-sequencing.
Genomic analyses
For genome-based species determination and MLST of the isolates, the in-house developed ASA³P pipeline was used (36). It was supplemented by using the websites PubMLST (https://pubmlst.org/databases/) and BIGSdb (https://bigsdb.pasteur.fr/cgi-bin/bigsdb/). Plasmid incompatibility groups (Inc), pMLST, antimicrobial resistance genes (ARG), and insertion sequences (IS) were identified using the Center for Genomic Epidemiology website (https://cge.cbs.dtu.dk/) (37, 38) and ISFinder (39). To annotate mobile antibiotic resistance genes and mobile elements, Galileo AMR of ARC Bio was used (40).
To determine the phylogenetic variation of the IncN plasmids, a maximum likelihood phylogeny was constructed using the Neighbor Joining Method based on the whole sequence alignment (CLC Genomics Workbench v.10.1.1) using default parameter settings. The phylogenetic tree was generated by MEGA6 (41).
Plasmid sequence comparison with the sequence of pCP13069-KPC2 as the reference was generated by using BLAST Ring Image Generator (BRIG) (42).
The NCBI Genbank database and other available complete sequence data were used to search the global distribution of the bla KPC-2-bearing IncN[pMLST15] plasmid with the specific non-Tn4401-KPC-element. IncN plasmid sequences (n = 476) and information from the plasmid database (PLSDB, https://ccb-microbe.cs.uni-saarland.de/plsdb/, as of 15 August 2022) were subjected to further analysis.
ACKNOWLEDGMENTS
The authors thank the administrative authorities at the Hessisches Ministerium für Social Affairs und Integration (HMSI), A. Carstens (Gesundheitsamt Kreis Groß-Gerau), Y. Dragneva, R. Bill (Gesundheits- und Pflegezentrum Rüsselsheim GPR), M. Mielke (Robert -Koch-Institut, Berlin), and C. Wendt (Labor Limbach, Heidelberg) for assistance in coordinating and providing samples. We thank Michael Frowein and Gudrun Bettge-Weller (HLPUG) for the primary isolate identification and Christina Gerstmann, Alexandra Amend, Sylvia Krämer, Martina Hudel, Nicole Heyer, and Simone Severitt for excellent technical support.
This study was supported by grants from the German Center for Infection Research (DZIF) to T.C. and C.I. through the German Federal Ministry of Education and Research (BMBF) (grant nos 8032808811 and 8032808824), the HMSI which financed SurvCARE Hessen, and the Hessian Ministry of Higher Education, Research and Arts within the project HuKKH (Hessisches universitaeres Kompetenzzentrum Krankenhaushygiene).
Y.Y. analyzed and interpreted data, conceived, drafted, and wrote the manuscript. L.F. participated in data analysis and manuscript writing. Y.R. participated in plasmid conjugation experiments and data analysis. J.F. managed genome sequencing data and performed analysis. C.I. contributed to study design, data interpretation, participated in manuscript writing and obtained funding. T.C. supervised the study, interpreted data, conceived, and wrote manuscript drafts with Y.Y. and obtained funding.
The IncN Study Group included the following additional members: Anja M. Hauri and Petra Heinmüller (Hessisches Landesprüfungs- und Untersuchungsamt im Gesundheitswesen - HLPUG, Dillenburg, Germany); Eugen Domann, Hiren Ghosh, Alexander Goesmann, and Stefan Janssen (Justus Liebig University Giessen, Germany); Sören Gatermann, Martin Kaase and Niels Pfennigwerth (German National Reference Center for Multidrug-Resistant Gram-Negative Bacteria, Ruhr-University Bochum, Bochum, Germany); Martin Exner (University of Bonn, Bonn, Germany); Jörg Overmann, Boyke Bunk, and Cathrin Spröer (Leibniz-Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany). They contributed to the recruitment of study participants, provision of bacterial isolates and epidemiological data, long-read genome sequencing, or data analysis.
Contributor Information
Can Imirzalioglu, Email: can.imirzalioglu@mikrobio.med.uni-giessen.de.
Trinad Chakraborty, Email: trinad.chakraborty@mikrobio.med.uni-giessen.de.
Monika Kumaraswamy, University of California, San Diego, La Jolla, California, USA .
the IncN Study Group:
Anja M. Hauri, Petra Heinmüller, Eugen Domann, Hiren Ghosh, Alexander Goesmann, Stefan Janssen, Sören Gatermann, Martin Kaase, Niels Pfennigwerth, Martin Exner, Jörg Overmann, Boyke Bunk, and Cathrin Spröer
DATA AVAILABILITY
All sequencing data have been deposited at NCBI GenBank under BioProject accession numbers PRJNA552260 and PRJNA692829.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.02564-23.
Table S1 (Characteristics of KPC-2 type carbapenemase producers), Table S2 (Molecular characteristics of a novel non-Tn4401-Element [NTEKPC-Y]); Table S3 (Molecular characteristics of the KPC-2-producing Enterobacterales isolates with Tn4401 and other NTE elements); Table S4 (Distribution of the plasmids with ARG-Profile A); Table S5 (PCR-Primers used for stepwise detection of the IncN[pMLST15] plasmid); Table S6 (IncN[pMLST15] and other related plasmids carry the replicase allele repN_7 with an insertion hot spot between their fipA and nuc [truncated] genes); Fig. S1 (Diversity of blaKPC-2-positive plasmids and conjugation studies).
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Logan LK, Weinstein RA. 2017. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis 215:S28–S36. doi: 10.1093/infdis/jiw282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med 18:263–272. doi: 10.1016/j.molmed.2012.03.003 [DOI] [PubMed] [Google Scholar]
- 4. Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev 25:682–707. doi: 10.1128/CMR.05035-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen Y, Marimuthu K, Teo J, Venkatachalam I, Cherng BPZ, De Wang L, Prakki SRS, Xu W, Tan YH, Nguyen LC, Koh TH, Ng OT, Gan Y-H. 2020. Acquisition of plasmid with carbapenem-resistance gene blaKPC2 in hypervirulent Klebsiella pneumoniae, Singapore. Emerg Infect Dis 26:549–559. doi: 10.3201/eid2603.191230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Villa L, Feudi C, Fortini D, Brisse S, Passet V, Bonura C, Endimiani A, Mammina C, Ocampo AM, Jimenez JN, Doumith M, Woodford N, Hopkins K, Carattoli A. 2017. Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb Genom 3:e000110. doi: 10.1099/mgen.0.000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bush K. 2018. Past and present perspectives on β-lactamases. Antimicrob Agents Chemother 62:e01076-18. doi: 10.1128/AAC.01076-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Navon-Venezia S, Kondratyeva K, Carattoli A. 2017. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev 41:252–275. doi: 10.1093/femsre/fux013 [DOI] [PubMed] [Google Scholar]
- 9. Marimuthu K, Venkatachalam I, Koh V, Harbarth S, Perencevich E, Cherng BPZ, Fong RKC, Pada SK, Ooi ST, Smitasin N, et al. 2022. Whole genome sequencing reveals hidden transmission of carbapenemase-producing Enterobacterales. Nat Commun 13:3052. doi: 10.1038/s41467-022-30637-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brandt C, Viehweger A, Singh A, Pletz MW, Wibberg D, Kalinowski J, Lerch S, Müller B, Makarewicz O. 2019. Assessing genetic diversity and similarity of 435 KPC-carrying plasmids. Sci Rep 9:11223. doi: 10.1038/s41598-019-47758-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hendrickx APA, Landman F, de Haan A, Borst D, Witteveen S, van Santen-Verheuvel MG, van der Heide HGJ, Schouls LM, Dutch CPE surveillance Study Group . 2020. Plasmid diversity among genetically related Klebsiella pneumoniae blaKPC-2 and blaKPC-3 isolates collected in the Dutch national surveillance. Sci Rep 10:16778. doi: 10.1038/s41598-020-73440-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Conlan S, Thomas PJ, Deming C, Park M, Lau AF, Dekker JP, Snitkin ES, Clark TA, Luong K, Song Y, et al. 2014. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 6:254ra126. doi: 10.1126/scitranslmed.3009845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, Abudahab K, Goater R, Giani T, Errico G, Aspbury M, Sjunnebo S, Feil EJ, Rossolini GM, Aanensen DM, Grundmann H, EuSCAPE Working Group, ESGEM Study Group . 2019. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol 4:1919–1929. doi: 10.1038/s41564-019-0492-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chmelnitsky I, Shklyar M, Leavitt A, Sadovsky E, Navon-Venezia S, Ben Dalak M, Edgar R, Carmeli Y. 2014. Mix and match of KPC-2 encoding plasmids in Enterobacteriaceae-comparative genomics. Diagn Microbiol Infect Dis 79:255–260. doi: 10.1016/j.diagmicrobio.2014.03.008 [DOI] [PubMed] [Google Scholar]
- 15. Baraniak A, Izdebski R, Fiett J, Herda M, Derde LPG, Bonten MJM, Adler A, Carmeli Y, Goossens H, Hryniewicz W, Brun-Buisson C, Gniadkowski M, MOSAR WP2, WP3, and WP5 Study Groups . 2015. KPC-like carbapenemase-producing Enterobacteriaceae colonizing patients in Europe and Israel. Antimicrob Agents Chemother 60:1912–1917. doi: 10.1128/AAC.02756-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheruvanky A, Stoesser N, Sheppard AE, Crook DW, Hoffman PS, Weddle E, Carroll J, Sifri CD, Chai W, Barry K, Ramakrishnan G, Mathers AJ. 2017. Enhanced Klebsiella pneumoniae carbapenemase expression from a novel Tn4401 deletion. Antimicrob Agents Chemother 61:e00025-17. doi: 10.1128/AAC.00025-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. 2014. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 22:686–696. doi: 10.1016/j.tim.2014.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Andrade LN, Curiao T, Ferreira JC, Longo JM, Clímaco EC, Martinez R, Bellissimo-Rodrigues F, Basile-Filho A, Evaristo MA, Del Peloso PF, Ribeiro VB, Barth AL, Paula MC, Baquero F, Cantón R, Darini AL da C, Coque TM. 2011. Dissemination of blaKPC-2 by the spread of Klebsiella pneumoniae clonal complex 258 clones (ST258, ST11, ST437) and plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae species in Brazil. Antimicrob Agents Chemother 55:3579–3583. doi: 10.1128/AAC.01783-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bispo Beltrão EM, de Oliveira ÉM, Dos Santos Vasconcelos CR, Cabral AB, Rezende AM, Souza Lopes AC. 2020. Multidrug-resistant Klebsiella aerogenes clinical isolates from Brazil carrying IncQ1 plasmids containing the blaKPC-2 gene associated with non-Tn4401 elements (NTEKPC-IId). J Glob Antimicrob Resist 22:43–44. doi: 10.1016/j.jgar.2020.05.001 [DOI] [PubMed] [Google Scholar]
- 20. Cerdeira LT, Lam MMC, Wyres KL, Wick RR, Judd LM, Lopes R, Ribas RM, Morais MM, Holt KE, Lincopan N. 2019. Small IncQ1 and col-like plasmids harboring blaKPC-2 and non-Tn4401 elements (NTEKPC-IId) in high-risk lineages of Klebsiella pneumoniae CG258. Antimicrob Agents Chemother 63:e02140-18. doi: 10.1128/AAC.02140-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yao Y, Falgenhauer L, Falgenhauer J, Hauri AM, Heinmüller P, Domann E, Chakraborty T, Imirzalioglu C. 2021. Carbapenem-resistant Citrobacter spp. as an emerging concern in the hospital-setting: results from a genome-based regional surveillance study. Front Cell Infect Microbiol 11:744431. doi: 10.3389/fcimb.2021.744431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. RKI . 2014. Plasmid-vermittelter multispezies-ausbruch MIT carbapenem-resistenten Enterobacteriacea. Epidemiol Bull 47:455–459. [Google Scholar]
- 23. Heudorf U, Voigt K, Westphal T, Steul K, Schmithausen R, Exner M. 2018. Multiresistente erreger in oberflächengewässern-ein fallbeispiel aus frankfurt am main und seine folgen. Umwelt - Hyg - Arbeitsmed 23:373–379. [Google Scholar]
- 24. Miriagou V, Papagiannitsis CC, Kotsakis SD, Loli A, Tzelepi E, Legakis NJ, Tzouvelekis LS. 2010. Sequence of pNL194, a 79.3-kilobase IncN plasmid carrying the blaVIM-1 metallo-beta-lactamase gene in Klebsiella pneumoniae. Antimicrob Agents Chemother 54:4497–4502. doi: 10.1128/AAC.00665-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen L, Chavda KD, Mediavilla JR, Jacobs MR, Levi MH, Bonomo RA, Kreiswirth BN. 2012. Partial excision of blaKPC from Tn4401 in carbapenem-resistant Klebsiella pneumoniae. Antimicrob Agents Chemother 56:1635–1638. doi: 10.1128/AAC.06182-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schweizer C, Bischoff P, Bender J, Kola A, Gastmeier P, Hummel M, Klefisch F-R, Schoenrath F, Frühauf A, Pfeifer Y. 2019. Plasmid-mediated transmission of KPC-2 carbapenemase in Enterobacteriaceae in critically Ill patients. Front Microbiol 10:276. doi: 10.3389/fmicb.2019.00276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pfenningwerth N. 2018. Bericht des Nationalen Referenzzentrums für gramnegative Krankenhauserreger zeitraum 1. Januar 2017 - 31. Dezember 2017., 2017. Epid Bull 28:263–267. [Google Scholar]
- 28. Pfenningwerth N. 2019. Bericht des Nationalen Referenzzentrums für gramnegative Krankenhauserreger, 2018. Epid Bull 31:289–294. [Google Scholar]
- 29. Pfenningwerth N. 2020. Bericht des Nationalen Referenzzentrums für gramnegative Krankenhauserreger, 2019. Epid Bull 26:3–10. [Google Scholar]
- 30. Falgenhauer L, Ghosh H, Guerra B, Yao Y, Fritzenwanker M, Fischer J, Helmuth R, Imirzalioglu C, Chakraborty T. 2017. Comparative genome analysis of IncHI2 VIM-1 carbapenemase-encoding plasmids of Escherichia coli and Salmonella enterica isolated from a livestock farm in Germany. Vet Microbiol 200:114–117. doi: 10.1016/j.vetmic.2015.09.001 [DOI] [PubMed] [Google Scholar]
- 31. Falgenhauer L, Ghosh H, Doijad S, Yao Y, Bunk B, Spröer C, Kaase M, Hilker R, Overmann J, Imirzalioglu C, Chakraborty T. 2017. Genome analysis of the carbapenem- and colistin-resistant Escherichia coli isolate NRZ14408 reveals horizontal gene transfer pathways towards panresistance and enhanced virulence. Antimicrob Agents Chemother 61:e02359-16. doi: 10.1128/AAC.02359-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chakraborty T, Sadek M, Yao Y, Imirzalioglu C, Stephan R, Poirel L, Nordmann P. 2021. Cross-border emergence of Escherichia coli producing the carbapenemase NDM-5 in switzerland and Germany. J Clin Microbiol 59:e02238-20. doi: 10.1128/JCM.02238-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22:568–576. doi: 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schwengers O, Hoek A, Fritzenwanker M, Falgenhauer L, Hain T, Chakraborty T, Goesmann A. 2019. ASA3P: an automatic and scalable pipeline for the assembly, annotation and higher level analysis of closely related bacterial isolates. bioRxiv. doi: 10.1101/654319 [DOI] [PMC free article] [PubMed]
- 37. Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–6. doi: 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Partridge SR, Tsafnat G. 2018. Automated annotation of mobile antibiotic resistance in Gram-negative bacteria: the multiple antibiotic resistance annotator (MARA) and database. J Antimicrob Chemother 73:883–890. doi: 10.1093/jac/dkx513 [DOI] [PubMed] [Google Scholar]
- 41. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 (Characteristics of KPC-2 type carbapenemase producers), Table S2 (Molecular characteristics of a novel non-Tn4401-Element [NTEKPC-Y]); Table S3 (Molecular characteristics of the KPC-2-producing Enterobacterales isolates with Tn4401 and other NTE elements); Table S4 (Distribution of the plasmids with ARG-Profile A); Table S5 (PCR-Primers used for stepwise detection of the IncN[pMLST15] plasmid); Table S6 (IncN[pMLST15] and other related plasmids carry the replicase allele repN_7 with an insertion hot spot between their fipA and nuc [truncated] genes); Fig. S1 (Diversity of blaKPC-2-positive plasmids and conjugation studies).
Data Availability Statement
All sequencing data have been deposited at NCBI GenBank under BioProject accession numbers PRJNA552260 and PRJNA692829.