ABSTRACT
Corynebacterium is a predominant genus in the skin microbiome, yet its genetic diversity on the skin is incompletely characterized and underrepresented in public databases. We investigated the distribution of Corynebacterium species on the skin and expanded the existing genome reference catalog. We used extant V1-V3 16S rRNA gene sequencing data from 14 body sites of 23 healthy volunteers to characterize Corynebacterium diversity across human skin. Corynebacterium tuberculostearicum, recently proposed to belong to a species complex, is frequently found on human skin. We identified two distinct C. tuberculostearicum ribotypes (A and B) that can be distinguished by variation in the 16S rRNA V1-V3 sequence: ribotype A is distributed across all skin sites, while B is found primarily on the feet. We performed whole genome sequencing of 40 C. tuberculostearicum isolates cultured from the skin of five individuals across seven skin sites. We generated five closed C. tuberculostearicum genomes and determined that they are largely syntenic and carry a diversity of methylation patterns, plasmids, and CRISPR/Cas systems. The pangenome of C. tuberculostearicum is open with a core genome size of 1,806 genes and a pangenome size of 5,451 genes. This expanded pangenome enabled the mapping of 24% more C. tuberculostearicum reads from skin metagenomes. We demonstrated differential growth phenotypes of C. tuberculostearicum ribotypes A and B on rich and skin-like media, suggesting functional differences. Finally, while the genomes from this study fall within the C. tuberculostearicum species complex, we propose that ribotype B isolates constitute a putative new species.
IMPORTANCE
Amplicon sequencing data combined with isolate whole genome sequencing have expanded our understanding of Corynebacterium on the skin. Healthy human skin is colonized by a diverse collection of Corynebacterium species, but Corynebacterium tuberculostearicum predominates on many skin sites. Our work supports the emerging idea that C. tuberculostearicum is a species complex encompassing several distinct species. We produced a collection of genomes that help define this complex, including a potentially new species we term Corynebacterium hallux based on a preference for sites on the feet, whole-genome average nucleotide identity, pangenomic analysis, and growth in skin-like media. This isolate collection and high-quality genome resource set the stage for developing engineered strains for both basic and translational clinical studies.
KEYWORDS: Corynebacterium, tuberculostearicum, genomics, pangenome
INTRODUCTION
The human body sustains a diverse collection of microbiota. Its resident taxa are adapted to unique niches, wherein they fulfill important functional roles. The genus Corynebacterium is one such taxa colonizing the human skin and nares. Foundational studies using 16S rRNA gene sequencing and shotgun metagenomics by our lab (1, 2) and others (3) have established Corynebacterium as common members of the skin microbiome. While Corynebacterium has been positively correlated with the resolution of dysbiosis associated with eczema flares (4), the importance of the Corynebacterium spp. is less defined for skin disease severity in primary immune deficient patients (5, 6). Corynebacterium spp. are predominant members of the human aerodigestive tract microbiome (nares, oral cavity, and respiratory tract) (3) and participate in microbe-microbe interactions with members of the nasal microbiome (7, 8). Corynebacterium has been shown to engage with the host immune system, specifically Corynebacterium accolens-promoted IL23-dependent inflammation in mice on a high-fat diet (9). Corynebacterium bovis and Corynebacterium mastiditis have been shown to predominate the microbiome of an ADAM10-deficient mouse model (10) as well as an ADAM17-deficient mouse model of eczema (11). Finally, Corynebacterium tuberculostearicum (ATCC 35533) has been shown to induce inflammation in human epidermal keratinocyte cell cultures (12). These studies establish Corynebacterium spp. as key members of the skin microbiome capable of both microbe-microbe and microbe-host interactions.
A critical resource for understanding the biology of Corynebacterium on the skin is a robust collection of complete reference genomes, including isolates collected from a variety of individuals and body sites. Previously published genome collections from skin- or nares-resident species include Staphylococcus epidermidis (13), Cutibacterium acnes (14), and the recent comparative analysis of Dolosigranulum pigrum (15). Of note, while emerging bioinformatic methods and pipelines are now being employed to extract nearly complete genomes (metagenome assembled genomes [MAGs]) from metagenomic assemblies of skin samples (16), MAGs are not yet a substitute for genomes from cultured isolates to understand strain level or pangenomic diversity. In addition to functional prediction, comparative genomics are increasingly being used to augment conventional microbiological methods to define or redefine taxonomic boundaries (17, 18), as well as describe the full extent of diversity within these boundaries (19). A pangenome, which encompasses the complete set of genes present within a set of genome sequences, enables the characterization of gene-level heterogeneity within a taxonomic group. The pangenome is commonly subdivided into the “core” genome, referring to genes present in all strains, and the “accessory” or “dispensable” genome, referring to those present in only one or some isolates. (The accessory pangenome can be further subdivided to reflect a wider range of gene uniqueness, e.g., singletons.) Thorough characterization of taxa is limited by the availability of representative and high-quality genome assemblies. Unfortunately, with the exceptions of clinically relevant Corynebacterium spp. (e.g., Corynebacterium diphtheriae, Corynebacterium striatum, and Corynebacterium pseudotuberculosis), the genus is inadequately sequenced, with 75% of species having fewer than six genomes. This includes common skin-associated species like C. tuberculostearicum with just five unique isolate genomes, only two of which are from the skin.
C. tuberulostearicum was first validated as a species in 2004 (20) and named for its production of tuberculostearic acid. It was described as a Gram-positive rod that was non-motile, lipophilic, oxidase-negative, and catalase-positive. Like other lipophilic Corynebacterium spp., C. tuberulostearicum must obtain lipids from external sources like sebum and the stratum corneum. Recently, C. tuberculostearicum was proposed as a member of a species complex that includes Corynebacterium kefirresidentii as well as other potentially new species (21, 22).
In this study, we first characterized the distribution of Corynebacterium spp. across 14 skin sites from 23 healthy volunteers (HVs) using extant 16S rRNA amplicon sequencing data. We identify C. tuberculostearicum as the predominant skin Corynebacterium species. We sequenced 23 distinct C. tuberculostearicum species complex strains (n = 40 genomes before dereplication), resulting in a fourfold increase in the number of publicly available, unique genomes. In addition to short-read assemblies, we generated five complete genomes which, along with the type strain (DSM44922), demonstrate that C. tuberculostearicum genomes are largely syntenic and carry a number of methylation systems as well as a CRISPR/Cas system. Genes from the C. tuberculostearicum species complex genomes in our collection fall into 5,451 gene clusters comprising the species pangenome. This expanded pangenome, as compared to existing public references, improved the mapping of metagenomic reads from unrelated HVs. In addition, we identified a distinct clade of C. tuberculostearicum species complex genomes that is highly enriched on the feet and may represent a new species, tentatively designated Corynebacterium hallux.
RESULTS
Corynebacterium spp. are prominent members of the healthy skin microbiome
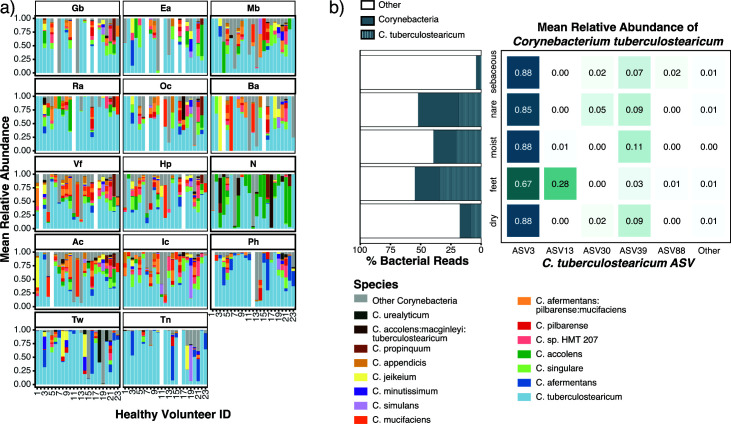
To explore the tropism of Corynebacterium, we surveyed the microbial diversity of healthy human skin using existing 16S rRNA V1-V3 amplicon sequencing data (5, 23). Clinical samples were obtained from 23 HVs across 14 body sites: sebaceous (back, Ba; occiput, Oc; external auditory canal, Ea; retroauricular crease, Ra; manubrium, Mb; glabella, Gb), moist (inguinal crease, Ic; antecubital crease, Ac), dry (hypothenar palm, Hp; volar forearm, Vf), foot (toe nail, Tn; toe web, Tw; plantar heel, Ph), and (N)ares. An average of 10,000 sequences per sample were analyzed which yielded a total of 8,334 amplicon sequence variants (ASVs), or unique 16S rRNA gene signatures. After rarefying the data set to an even depth, 5,967 ASVs remained. As expected, the top three ASV’s taxonomic assignments, present in 94% of skin samples, were Cutibacterium (41% of reads, ASV1 corresponds to C. acnes), Staphylococcus (9% of reads, ASV2 corresponds to S. epidermidis), and Corynebacterium (9% of reads, ASV3 corresponds to C. tuberculostearicum). The genus Corynebacterium was present in 96% of the skin sites sequenced, averaging 17% of reads. With a preference for moist over sebaceous skin sites (Fig. S1), Corynebacterium thrives in the humid, temperate environments of the feet and nares.
C. tuberculostearicum is the most common skin Corynebacterium
A variety of marker gene approaches have been employed to determine the phylogenetic relationships between Corynebacterium species including combinations of 16S rRNA, rpoB, rpoC, and gyrA genes (for review see reference 24). In general, it is difficult to accurately classify Corynebacterium at the species level using amplicon data and standard reference databases. The Human Oral Microbiome Database (3) is a curated database that includes a training set with a supraspecies taxonomic level enabling the assignment of sequences to multiple species where ambiguity exists. In our case, >99.5% of sequences classified as C. tuberculostearicum using the Refseq classification were also classified as C. tuberculostearicum (part of the accolens/macginleyi/tuberculostearicum superspecies) by expanded Human Oral Microbiome Database (eHOMD).
While a variation in species composition was observed between individuals, some sites and habitats displayed species enrichment at specific locations across multiple individuals (Fig. 1a). We observed that C. accolens was enriched in the nares, with a prevalence of 74% across nares samples and constituted an average of 33%–41% of Corynebacterium reads. Corynebacterium afermentans were enriched across feet sites, where they were present in 54% of samples and comprised an average of 17% of Corynebacterium reads. Most notably, however, we found that C. tuberculostearicum was present in 94% of skin sites and was often the most abundant Corynebacterium. C. tuberculostearicum reads represented 67% of corynebacterial reads in the feet, 47% in dry sites, 58% in sebaceous sites, and 46% in the nares.
Fig 1.
Corynebacterium species relative abundance in normal human skin microbiome. (a) Relative abundance of the 15 major Corynebacterium species across 14 skin sites: sebaceous (back, Ba; occiput, Oc; external auditory canal, Ea; retroauricular crease, Ra; manubrium, Mb; glabella, Gb), moist (inguinal crease, Ic; antecubital crease, Ac), dry (hypothenar palm, Hp; volar forearm, Vf), foot (toe nail, Tn; toe web, Tw; plantar heel, Ph), and nares (N). Relative abundances were determined by sequencing of the V1–V3 region of the 16S rRNA gene and subsetting to Corynebacterium reads. (b) Percent of total bacterial reads attributed to Corynebacterium and C. tuberculostearicum in each skin habitat. Of the six ASVs assigned to C. tuberculostearicum, mean relative abundance across skin habitats.
The majority of reads assigned to C. tuberculostearicum belonged to two predominant 16S rRNA sequence variants, ASV3 and ASV13 which differed by a single nucleotide polymorphism (SNP) and a single-base insertion/deletion (indel). ASV3 constituted 83% of C. tuberculostearicum-classified reads (compared to <8% for all other ASVs of this species) and showed a cosmopolitan distribution across body sites (Fig. 1b). Found in 100% of HVs (N = 23) and 87% (254/293) of skin samples, ASV3 was predominant and prevalent across human skin. Relative abundance analysis revealed ASV3 abundance >85% within all habitats except foot sites, where it made up 66% of C. tuberculostearicum-classified reads. As of this writing, the existing C. tuberculostearicum National Center for Biotechnology Information (NCBI) reference genomes containing complete V1–V3 sequences are all ASV3 as are 100% of 16S rRNA gene C. tuberculostearicum references in the SILVA reference database.
In contrast to cosmopolitan ASV3, ASV13 was enriched primarily on feet, constituting 28% of C. tuberculostearicum-classified reads from the Ph, Tw, and Tn sites (8%, 9%, and 70%, respectively). In 9 of 23 HVs, ASV13 constituted over 90% of C. tuberculostearicum-mapped reads within a single foot site (Fig. S2); notably, much of this enrichment was observed in Tn sites. In addition, we observed that some individuals exhibited within-site predominance by other less common ASVs, with some individuals colonized by a single non-dominant ASV across multiple body sites. In HV 12, for example, 52%–100% of C. tuberculostearicum-mapped reads in each body site excluding the toenail are classified as ASV39. We also noted that, while sites on the feet (Ph, Tn, and Tw) were often colonized by multiple ASVs, other body sites tended to be colonized by a single ASV.
We searched the SILVA database (v138.1) for perfect matches to the ASV13 sequence and found 152 matches, all associated with uncultured Corynebacterium. The majority of them were from our own full-length 16S rRNA gene sequencing of skin microbiome samples (1). This observation, combined with the fact that all the existing C. tuberculostearicum reference genomes had a more common ASV3 sequence variant, led us to hypothesize that the ASV13 sequence, which we hereafter refer to as ribotype B, could be associated with an unrecognized species or subspecies. For the purposes of the current work, we will use the term C. tuberculostearicum species complex (21, 22) to refer to all C. tuberculostearicum-like isolates found on the skin. Additionally, we will refer to the predominant ASV3 OTU as ribotype A.
Expanding the C. tuberculostearicum complex reference catalog
Prior to this study, only five C. tuberculostearicum isolates had been sequenced and submitted to NCBI. Only two of those were from human skin and neither was a closed genome. To expand the C. tuberculostearicum complex reference genome catalog, we sequenced isolates from five different HVs (Table S1). To enrich for the previously unsequenced ribotype B Corynebacterium, we screened our collection of skin-associated isolates with full-length 16S rRNA gene sequences. Of the 45 C. tuberculostearicum isolates screened, we identified eight ribotype B isolates for further study. In total, we shotgun sequenced 40 isolates in the C. tuberculostearicum complex—30 from ribotype A, 8 from ribotype B, and 2 from other ASVs. Initial genome clustering using mash indicated that some of the isolates we sequenced were closely related. Therefore, we used dRep (25) to identify groups of highly similar genomes (ANImf > 99.5%) and chose the best representative genome for each genome set based on sequence assembly statistics: maximal N50, minimal number of contigs, and maximal genome size. In cases of comparable assembly quality, genomes were selected to increase body site representation. Applying these dereplication criteria resulted in 23 strain-level genomes that differ by 1,000 s of single nucleotide variants and are distributed across three ribotypes: 18 from ribotype A, 4 from ribotype B, and 1 from ASV30.
Whole-genome features of five complete C. tuberculostearicum complex genomes
In addition to a paucity of C. tuberculostearicum reference genomes at the time of this study, the ones that did exist were not associated with publications describing their general features. To address this, we selected five of the dereplicated genomes, three ribotype A and two ribotype B, for long-read sequencing on the PacBio platform. The subsequent finished or complete C. tuberculostearicum complex genomes revealed four copies of the 16S rRNA gene in each genome. For each genome, we performed a multi-sequence alignment containing each V1–V3 region copy along with the predominant ribotype A sequence first identified in our amplicon sequencing data set. Within a genome, copies of the V1–V3 region are almost entirely identical across alignment, with ribotype A C. tuberculostearicum species complex genomes carrying four copies of ASV3. One exception was a single nucleotide variant identified in one copy of 16S rRNA 5′ region of CTNIH12 (Fig. S3). Notably, no variation was found in the ribotype B genomes, which both carried four identical gene copies marked by the two characteristic sequence variants as identified in the amplicon sequencing data set. The within-genome homogeneity of 16S rRNA genes confirmed its usefulness as a marker.
These complete C. tuberculostearicum complex genomes also enabled us to directly compare the type strain (DSM 44922/FDAARGOS_1117; human bone marrow) to our ribotype A and B isolates without the ambiguity introduced by unfinished genomes. Fig. S4 shows that the five PacBio genomes from this study were largely co-linear, with >80% of the genome in large syntenic blocks, with the type strain DSM 44922. All five of the genomes in this study had a 440 kb region that was reorganized relative to the type strain. This region, comprising around 17.6% of the genome, encoded 392 genes (387 coding). The breakpoints for inversions or translocated blocks in the reference were marked by mobile element families (e.g., IS3, IS256, IS481, and IS6) that suggest a mechanism for this rearrangement.
We extracted the methylation profiles from the PacBio reads of our five genomes (Table S2). The most common methylation pattern, found in all five genomes, was N6-methyladenine modification (m6A) of GATC motifs (~16,000 sites/genome; 99% methylated), typically associated with the Dam methylase. A second motif AAAAC was also found to be methylated (m6A) in all five genomes (~75% methylated). In addition to these two common methylation patterns, ribotype A isolate CTNIH10 had two additional methylated motifs present in hundreds of copies (GGCANNNNNATC and GATDNNNNTGCC). CTNIH20 had an additional three methylated motifs present at 520–1,626 sites/genome. Finally, CTNIH23 had evidence of an additional five methylation motifs across the genome that were all >98% methylated and present at 225–2,159 sites/genome. Methylation systems are important for horizontal gene transfer (HGT), phage resistance, and potential recombinant engineering of these strains.
The presence of CRISPR-Cas as well as other phage defense systems poses additional barriers to HGT. We detected an eight-gene Type I-E Cas gene cluster and two large repeat arrays (24 and 19 spacers) in the CTNIH20 ribotype B genome, but not in any of the other full-length genomes from this study. Additional CRISPR-Cas systems were detected in the short-read assemblies of CTNIH9 (ribotype A; Type I-E Cas gene cluster, eight spacer CRISPR) and CTNIH22 (ribotype B; Type I-E Cas gene cluster, 12 spacer CRISPR). Prior to this, the only public C. tuberculostearicum species complex reference genome with a CRISPR-Cas system was strain SK141 (ACVP01). A variety of other defense systems including restriction modification systems were also identified using the DefenseFinder tool (Table S2) (26, 27).
Plasmids are important for the mobilization of virulence factors, antibiotic resistance genes, and as tools for recombinant engineering. At the time of this study, a single 4.2 kb plasmid was deposited in the public databases associated with the C. tuberculostearicum species complex, p1B146 (NC_014912) (28). Across the five long-read genomes sequenced here, we detected five plasmids, none of which aligned to p1B146. Two plasmids, pCT3-020e and pCT4-9116 from CTNIH23 and CTNIH12, had the same backbone as the C. diphtheriae plasmid pNG2 (ORF9-traA-ORF11-parAB-repA) but lacked the erythromycin-resistance cassette. CTNIH20 carries three plasmids ranging in size from 21.2 to 27.3 kb. All three carried a traA/recD2 ortholog encoding a relaxase/helicase but are otherwise unrelated. While most of the proteins on these three plasmids were annotated as hypothetical, pCT1-afe7 carried an ebrB efflux pump and pCT1-0563 carried an stp (spectinomycin/tetracycline) efflux pump predicted to be involved in resistance to dyes and antibiotics. All five plasmids were characterized by the presence of a TraA/RecD2 encoding gene, suggesting a common mobility mechanism. Furthermore, we found fragments of these plasmids in many of the contig-level genomes. For instance, the C. tuberculostearicum CIP 102622 genome (JAEHFL01) carried both the stp gene and a nearby transcription factor (>99.6% identity) on a 9.8 kb contig, showing the value of these plasmid references for identifying HGT elements.
Taxonomic structure of the skin-associated C. tuberculostearicum species complex
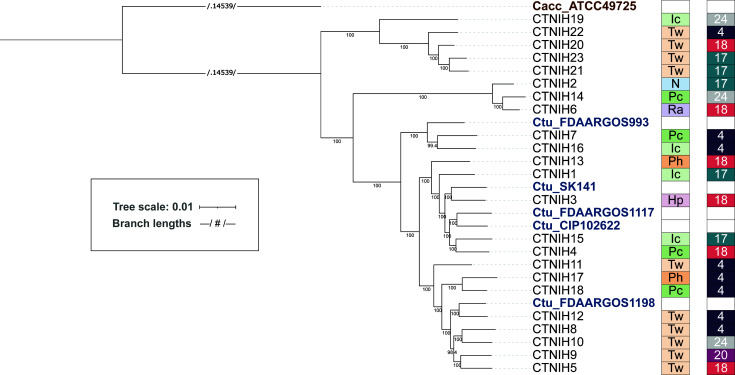
While 16S rRNA amplicon sequencing enabled us to group C. tuberculostearicum species complex isolates into two predominant ASVs, the dereplicated genomes enable further high-resolution taxonomic analysis of this species complex. We used GET_HOMOLOGUES to extract core genes and build a phylogenetic tree based on core genome SNPs (Fig. 2). We noted additional taxonomic structure, particularly amongst ribotype A isolates. The five public reference genomes were in the ribotype A-dominated portion of the tree as expected based on their 16S rRNA gene sequence.
Fig 2.
A maximum-likelihood phylogenetic tree of C. tuberculostearicum species complex genomes from this study and publicly available, calculated from 1,315 core gene cluster alignments. Bootstrap values (located along internal nodes) were calculated from 1,000 replicates. Clustering was generated using GET_HOMOLOGUES OrthoMCL v1.4 option with minimum coverage 90% in BLAST pairwise alignments. The tree was rooted on outgroup C. accolens ATCC 49725. On the right of tree, boxes depict site (body site locations defined in Fig. 1) and individual (HV) from which each isolate was cultured. Sites are colored by niche type, with moist in shades of green; feet in shades of orange; dry in pink; sebaceous in lavender; and nares in blue. Individuals are randomly but consistently colored.
While there was a good correlation between 16S rRNA ASVs and the core SNP tree, we noted a single isolate, CTNIH19, which carried a ribotype A allele but localized with ribotype B isolates on the tree. CTNIH19 was isolated from the inguinal crease and is the most basal member of this clade. Work by Cappelli and colleagues recently defined a number of new Corynebacterium species, and CTNIH19 was >99.9% identical to a species they designated Corynebacterium curieae (29). We calculated the average nucleotide identity (ANI) across isolates using pyANI (Fig. S5) and determined that ribotype B isolates share ANI > 97% with themselves and <94% with other C. tuberculostearicum complex genomes, including C. curieae. When whole-genome ANI is used to define a putative new species, 95% identity is a generally accepted species boundary (18, 30). We also submitted our ribotype B isolate genomes to the DSMZ type strain genome server (TYGS) (31) to obtain a taxonomic/nomenclature assignment. TYGS predicted that ribotype B genomes belong to a new species in both the whole genome and 16S rDNA trees. The closest TYGS references were C. tuberculostearicum DSM 44922 and C. kefirresidentii. C. keffiresidentii was first described in 2017 (32) after isolation from kefir grains but has not been accepted as an official species yet. Three of our isolates (CTNIH2, CTNIH6, and CTNIH14) from three different HVs were 98% identical to the C. kefirresidentii reference, calling into question whether kefir is the only natural host for this bacterium (21, 22). We are proposing that the isolates belonging to clade ribotype B be designated C. hallux, given their association with feet and, particularly, the toenail.
Pangenome of the skin-derived C. tuberculostearicum complex
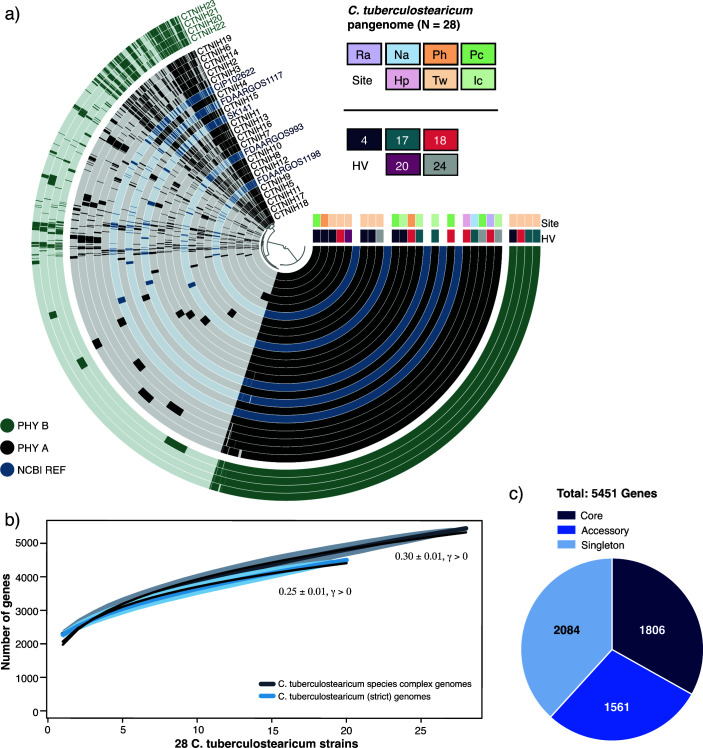
We performed a pangenomic analysis to describe the coding diversity of the C. tuberculostearicum species complex (Fig. 3). We generated an anvi’o (33) pangenomic map to illustrate genomic variation across the combined set (N = 28) of NCBI reference genomes and our dereplicated genomes. (Fig. 3a). Pangenome openness was estimated for the complex using the Heap’s law model (Fig. 3b) as proposed by Tettelin et al. (34). The model indicated an open pangenome (0.30 ± 0.01, γ > 0), predicting that the C. tuberculostearicum complex pangenome would increase with more genomes analyzed. Despite a lower degree of pangenome-wide diversity, the strict set (N = 20) of genomes representing C. tuberculostearicum species also demonstrated an open pangenome (0.25 ± 0.01, γ > 0). With our additional 23 genomes, the species complex pangenome size increases from 3,080 genes to 5,451 genes, resulting in an expansion of the non-core, or accessory genome by over 300% (Fig. 3c). We performed a functional characterization of 23 lab-sequenced and 5 NCBI-derived C. tuberculostearicum species complex genomes using the eggNOG-mapper annotation tool (Fig. S6), which returned annotations for 83.2% of orthologous gene clusters (of which 21% are annotated COG category “S,” Function Unknown). Interestingly, among the non-core genes, inorganic ion metabolism and transport-related genes were among the most abundant. We performed a principal components analysis (PCA) of gene presence/absence data describing our 23 genomes and two skin-derived reference sequences (Fig. 4a). We observed site-specific clustering of genomes isolated from the feet and moist environments. In agreement with the core phylogenetic clustering, we also observed distinct clustering of ribotype B isolates away from other foot-derived C. tuberculostearicum complex genomes. In addition, we identified 11 genes that were unique to and carried by every member of a ribotype (A = 2 and B = 9; Supplementary Table. S3), four of which we were able to assign functional annotation using the UniprotKB sequence similarity search tool, including a bacteriocin and ferric uptake protein.
Fig 3.
The C. tuberculostearicum pangenome. (a) Anvi’o pangenomic map for 28 C. tuberculostearicum genomes (including five NCBI reference genomes). Genomic rings are annotated by skin site and HV metadata and ordered by pyANI ANIb. Genome margins are manually adjusted for clarity. (b) Heap’s Law estimates of pangenome openness for C. tuberculostearicum complex (N = 28) and C. tuberculostearicum species (N = 20) genomes. Rarefaction curves show the total number of genes accumulated with the addition of new genome sequences in random order with 1,000 permutations. Shaded regions represent the 95% CI. A Heap’s law model was fit to the resultant complex and species curves to calculate γ values (0.30 ± 0.01 and 0.25 ± 0.01, respectively). (c) Number of core (belonging to all genomes), accessory (belonging to two or more genomes), and singleton (belonging to only one genome) genes. The expanded pangenome contains 5,451 genes using 90% sequence identity as a cutoff parameter.
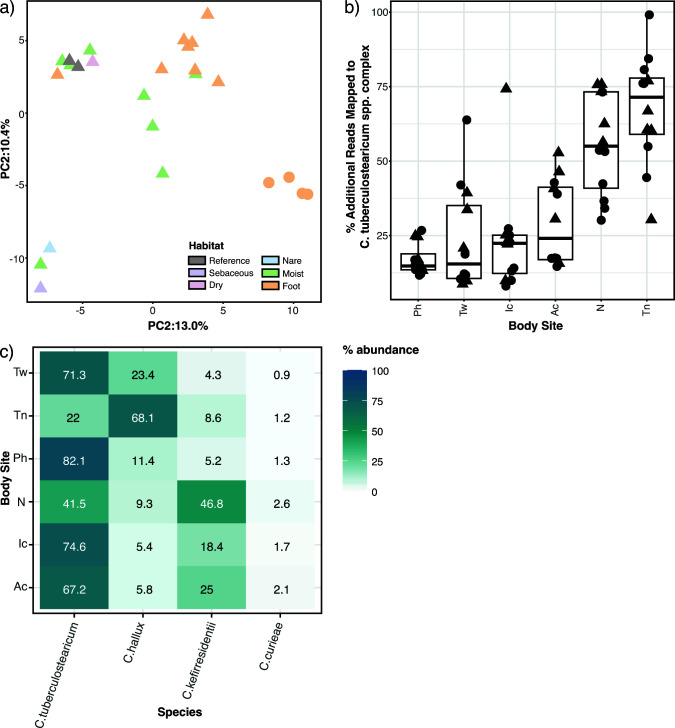
Fig 4.
C. tuberculostearicum complex pangenome clustering and improved metagenomic read mapping. (a) PCA of orthologous gene clustering. The gene presence/absence data for 25 genomes (including two NCBI references, shown in gray) was analyzed using PCA. Ribotype B genomes are shown as circles; other genomes are triangles. (b) Improvement in shotgun metagenomic read mapping with a 28 member C. tuberculostearicum database as compared to the five member NCBI database. Percent increase in mapped C. tuberculostearicum reads by body site. Each point is an HV. Triangles mark HVs that contributed one or more isolates to the expanded mapping database. (c) Relative abundance of C. tuberculostericum species complex members, including the newly proposed species C. hallux, based on bracken-corrected kraken2 analysis.
Improved metagenomic read mapping using an expanded C. tuberculostearicum pangenome
We tested whether the expanded genomic reference set could improve the rate of C. tuberculostearicum read mapping in a set of metagenomic data sets from 12 HVs at six body sites (2). Reads were mapped with bowtie2 (35) against a genome database consisting of the five unique NCBI references or a database of the NCBI references plus the dereplicated genomes from this study. Overall, 27% more reads were assigned to C. tuberculostearicum using the expanded genome set as compared to NCBI references alone. While the five HVs with isolate genomes in the expanded database showed slightly better classification, median improvement of 32%, over those without isolates in the database (median = 24%), the difference was not statistically significant, showing the broad utility of these genomes. Furthermore, when broken down by body site, toenail (Tn) sites showed the largest improvement in C. tuberculostearicum read assignment (72%) while nares, which only contributed a single genome to the expanded database, improved by 55% (Fig. 4b). To control for spurious read mapping to repetitive elements or other assembly artifacts, these analyses were repeated using only the predicted gene catalogs, rather than the whole genomes, and very similar improvements in C. tuberculostearicum read mapping were observed, 25% median improvement and a similar site-dependence. In addition to improved read mapping, read classification also supports the niche specificity of C. tuberculostearicum species complex members. Fig. 4c shows that within the species complex, sites on the feet (e.g., toenail) are enriched in C. hallux as was seen in the analysis of amplicon data (Fig. 1b), while C. kefirresidentii is most abundant in the nares.
Growth of skin-derived C. tuberculostearicum in sweat media
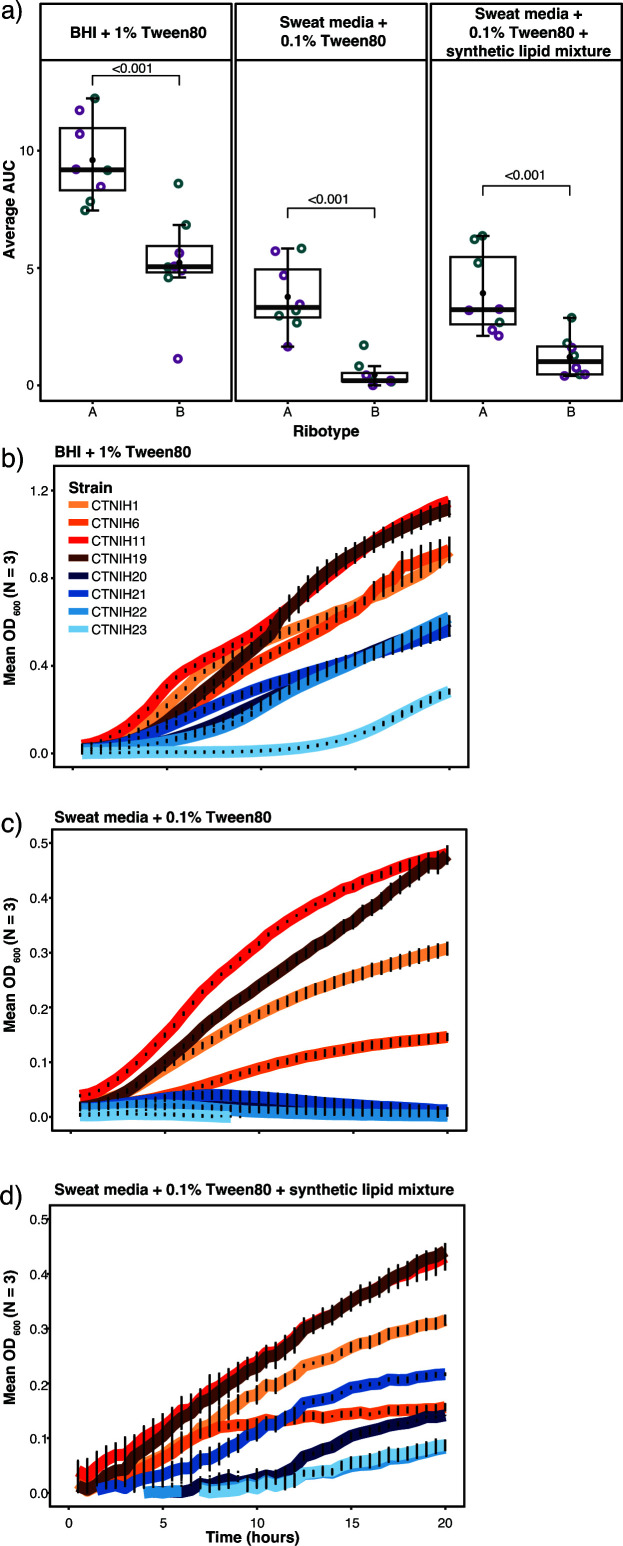
Members of the C. tuberculostearicum species complex are widely distributed across the skin’s microenvironments. Differences in body site physiology and nutrient composition inherent to each niche may provide selective growth advantages (and disadvantages) to a subset of strains. We performed a pilot experiment to investigate differential growth phenotypes of C. tuberculostearicum species complex ribotypes in skin-like media (Fig. 5). Corynebacterium are often cultured on brain-heart infusion (BHI) media plates supplemented with 1% Tween-80 (BHI + 1% Tween80), so this medium was used as a positive control for growth in liquid medium (Fig. 5b). Isolates were cultured on two types of medium consisting of a complex mixture of amino acids, lipids, and other metabolites that mimic human eccrine sweat, with one medium supplemented to include a sebum-like synthetic lipid mixture. Both simulated sweat media were supplemented with 0.1% Tween-80. A collection of eight skin-derived Corynebacterium strains consisting of four ribotype A and four ribotype B strains were grown for 20 hours in triplicate and in two separate experiments for each strain and medium condition (N = 6; Fig. 5b–d). In all three growth conditions, ribotype B isolates demonstrated a lower mean OD600 over time than ribotype A isolates (Fig. 5a). Using analysis of variance (ANOVA) and the Tukey method, we determined that the area-under-the-curve (AUC) difference between the two ribotypes is statistically significant (P < 0.0001) for all media conditions. This pattern was particularly pronounced in the BHI + 1% Tween80 and Sweat media + 0.1% Tween80 conditions. We observed that the addition of synthetic lipid mixture to the eccrine sweat-like medium attenuated, however, still maintained the growth difference between ribotype B and other strains, suggesting lipid-limited growth for members of ribotype B.
Fig 5.
Growth phenotypes of select C. tuberculostearicum complex strains in synthetic sweat media. (a) Empirical area under curve comparison of C. tuberculostearicum species complex strains from ribotype A and ribotype B, with biological replicates grouped by color. Strains were grown in BHI + 1% Tween; Sweat media + 0.1% Tween80; Sweat media + 0.1% Tween80 + synthetic lipid mixture. Medium composition is described in further detail in methods. (b–d) Selected growth curves from a representative experiment plotted with standard error. Ribotype B isolates are shown in shades of blue. Ribotype A isolates are shown in shades of red.
DISCUSSION
In this study, we investigated the genomic diversity of the predominant yet under-sequenced Corynebacterium genus. Our survey of microbial diversity across human skin revealed niche-specific enrichment of Corynebacterium species and identified C. tuberculostearicum as a predominant and widespread species on human skin. Our analysis of existing amplicon sequencing data identified a site-specific novel 16S rRNA gene ribotype which led to an expanded sequencing of the C. tuberculostearicum species complex. In total, we sequenced 23 distinct isolates belonging to the C. tuberculostearicum species complex including C. tuberculostearicum (n = 15), C. kefirresidentii (n = 3), C. curieae (n = 1), and a novel species we are calling C. hallux (n = 4). Discovery of C. kefirresidentii on human skin and nares suggests that humans are a natural host for this species.
C. hallux is likely a new species of skin-associated Corynebacterium and merits further work to formally name it. It was cultured from three different HVs, detected by amplicon sequencing in most HVs, represented in the recently published SMGC (SMGC_122) (16), and detected in public 16S rRNA gene database entries associated with the skin. In our HVs, it was enriched in sites on the feet, particularly the toenail and toe web. Microbial communities on the feet are highly diverse and relatively unstable (2) subject to temperature fluctuations and invasion by environmental microorganisms.
This study helps to resolve the diversity of C. tuberculostearicum species complex strains and provides an important genetic resource for future study. Our whole-genome sequencing effort uncovers substantial genetic and functional diversity across the complex and improves read-mapping overall by >24%, which will in turn bolster future sequencing efforts and further characterization of Corynebacterium across human skin. While this work has greatly expanded the non-core genome, a significant proportion of these genes are putative or lack functional annotation. Overall, we did not detect pronounced differences between ribotype B and other strains’ genes that would explain the observed patterns in skin site distribution and growth on synthetic media.
Eleven genes (ribotype A = 2 and B = 9) perfectly segregated the two ribotypes; however, the roles of these genes remain unclear due to the limitations of annotation tools. Nevertheless, we identified two examples of genes with the potential to affect within-niche competition. One of the genes specific to ribotype B shared sequence similarity with a Lactococcin 972 family bacteriocin. The bactericidal activity of ribotype B against closely related strains could contribute to patterns of within-site dominance as observed between ribotypes (Fig. S2). Bactericidal peptides have recently gained interest as a possible therapeutic intervention for gastrointestinal disease (36). Furthermore, Corynebacterium was shown to be enriched in a recent study (37) of post-operative, healing wounds, suggesting an opportunity for biotherapeutic applications. We also identified a ribotype B-unique copy of a gene encoding ferrous iron transport protein B, a major regulator of bacterial iron uptake. Iron is an essential nutrient for survival, requiring the development of highly efficient sequestering mechanisms by pathogenic and avirulent bacteria alike (38, 39). Under conditions of limited nutrient bioavailability, enhanced ferric uptake may prove to be a determining factor of intraspecies competition.
On both rich and skin-like media, we observed that ribotype B isolates grew less robustly compared to other strains. Thus, the topographical distribution of ribotypes across the skin’s surface may emerge from selective growth advantages including different nutritional requirements or nutrient acquisition mechanisms between strains. The mechanism(s) of this variability may have important clinical implications. For example, characterizing the nutritional limits for sustained growth could inform the development of prebiotic therapeutics to maintain healthy microbial composition within a given microenvironment. Moreover, some strains may perform anti-virulent functions within their respective niches. Work from Ramsey et al. (8) supports the idea that the pathobiont Staphylococcus aureus shifts from virulence to commensalism in the presence of commensal Corynebacterium spp. in the skin/nares. Thus, identifying genomic functions that distinguish C. tuberculostearicum on healthy vs diseased skin could provide the impetus for engineering site-specific, microbe-based drug delivery systems. Understanding the roles and requirements of host-associated microbial communities in maintaining skin health will provide insight into the emergence of skin disorders in addition to novel therapeutic interventions to combat them.
MATERIALS AND METHODS
Subject recruitment and sampling
Adult male and female HVs 18–40 years of age were recruited from the Washington, DC, metropolitan region. This natural history study was approved by the Institutional Review Board of the National Human Genome Research Institute (clinicaltrials.gov/NCT00605878) and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (https://clinicaltrials.gov/ct2/show/NCT02471352), and all subjects provided written informed consent prior to participation. Sampling was performed as described previously (23).
16S rRNA gene sequencing
16S rRNA gene amplicon sequencing of these samples has been described previously (5). Briefly, each DNA sample was amplified with universal primers flanking variable regions V1 (27F, 5′-AGAGTTTGATCCTGGCTCAG) and V3 (534R, 5′-ATTACCGCGGCTGCTGG). For each sample, the universal primers were tagged with unique indexes to allow for multiplexing/demultiplexing (40). The following PCR conditions were used: 2 µL 10× AccuPrime Buffer II, 0.15 µL Accuprime Taq (Invitrogen, Carlsbad, CA), 0.04 µL adapter +V1_27F (100 µM), 2 µL primer V3_354R + barcode (2 µM), and 2 µL of isolated microbial genomic DNA. PCR was performed in duplicate for 30 cycles followed by PCR-clean up and amplicon pooling of ~10 ng DNA. Duplicate amplicons were combined, purified (Agencourt AMPure XP-PCR Purification Kit [Beckman Coulter, Inc., Brea, CA]), and quantified (QuantIT dsDNA High-Sensitivity Assay Kit [Invitrogen, Carlsbad, CA]). An average of ~8 ng DNA of 94 amplicons was pooled together, purified [MinElute PCR Purification Kit (Qiagen, Valencia, CA)], and sequenced on a Roche 454 GS20/FLX platform with Titanium chemistry (Roche, Branford, Connecticut). Flow-grams were processed with the 454 Basecalling pipeline (v2.5.3).
16S rRNA gene amplicon analysis
Sequencing data were processed using DADA2 v1.20.0 (41). Sequences were filtered and trimmed as recommended by the software developers and truncated to 375 nt: filterAndTrim[fnFs, filtFs,maxN = 0, maxEE = c (2), truncQ = 2,truncLen = c(375)]. Sample inference was performed using the learnErrors (randomize = TRUE) and the dada (HOMOPOLYMER_GAP_PENALTY=-1, BAND_SIZE = 32) commands. Chimeras were removed using removeBimeraDenovo (method = “consensus,” allowOneOff = TRUE). Taxonomy was assigned using assignTaxonomy (minBoot = 70) command in DADA2 with the Refseq (https://zenodo.org/record/3266798) or eHOMD v15.1 V1V3 (3) training set databases. The resulting ASVs, taxonomy, and sample metadata were used to build a phyloseq (42) object that was used for further analysis.
Bacterial culturing
Corynebacterium isolates were cultured from HVs as described previously (16). Briefly, skin samples were collected with eSWabs (COPAN e480C) in liquid Amies. Corynebacterium was grown on BHI agar with 10% Tween 80 ± 40 µg/mL fosfomycin at 35°C; isolates from this study were grown aerobically. Potential Corynebacterium isolates were taxonomically classified by amplifying and Sanger sequencing the full-length 16S rRNA gene with primers 8F (5′-AGAGTTTGATCCTGGCTCAG) and 1391R(5′-GACGGGCGGTGWGTRCA).
Bacterial whole genome sequencing
Genomic DNA was purified for each isolate, from which Nextera XT (Illumina) libraries were generated. Each isolate was sequenced using a 2 × 151 paired-end dual index run on an Illumina NovaSeq 6000. The reads were subsampled to achieve 80×–100× coverage using seqtk (version 1.2), assembled with SPAdes (version 3.14.1) (43), and polished using bowtie2 (version 2.2.6) and Pilon (version 1.23) . To achieve full reference genomes for select isolates, genomic DNA was sequenced on the PacBio Sequel II platform (version 8M SMRTCells, Sequel II version 2.0 sequencing reagents, 15 hour movie collection). The subreads were assembled using Canu v2.1 and polished using the pb_resequencing workflow within PacBio SMRTLink v.9.0.0.92188. Genome annotation was performed using NCBI Prokaryotic Genome Annotation Pipeline (https://www.ncbi.nlm.nih.gov/genome/annotation_prok/). Methylation patterns for the assembled genomes were determined using the pb_basemods workflow in SMRTLink v.9.0.0.92188. Whole genome and plasmid alignments were generated in mummer (v3.9.4alpha) and visualized in R.
Full-length 16S rRNA gene copies were extracted from each PacBio complete genome. Briefly, reference Corynebacterium 16S rRNA sequences were downloaded from the RDP database (good quality, >1,200 nt) and used as a BLAST database to identify the coordinates of the four copies in the genome. To detect intragenomic variation in the 16S rRNA gene, all copies within each genome were compared against each other using the EMBL-EBI Multiple Sequence Alignment Tool (MUSCLE). Whole genome alignments were generated in Mauve v 2.4.0.
Phylogenetic analysis
Publicly available genomes were downloaded from NCBI including C. tuberculostearicum (CP068156, CP06979, CP065972, ACVP01, and JAEHFL01), C. kefirresidentii (CP067012, JAHXPF01), C. curieae (JAKMUU01), and C. accolens (ACGD01). GET_HOMOLOGUES (v09212021) was used to cluster protein sequences from 29 genomes (28 C. tuberculostearicum and 1 C. accolens) into orthologous groups and generate a core gene alignment. Prokka GBK files were used as input for clustering. The OrthoMCL (v1.4) option was used to group sequences utilizing the Markov Clustering Algorithm with a minimum coverage value of 90% in blast pairwise protein alignments. A strict core consensus genome was generated by calculating the intersection of single-copy genes present in all 29 genomes. The accompanying GET_PHYLOMARKERS (v. 2.2.9.1) pipeline was used to identify markers for phylogenetic inference. IQTREE (v 2.1.2) was used to generate a maximum-likelihood phylogenetic tree from marker gene cluster alignments with 1,000 bootstrap replicates and a mean branch support value cutoff of 0.7. The top-scoring tree was visualized and annotated using the web-based program interactive Tree of Life (iTOL v6). The ANI matrix for all sequences was plotted and annotated using the package heatmap.2/R.
Gene calling and annotation
The Prokka (v1.14.6) pipeline was used for gene calling and annotation. GFF3- and GBK-format annotations were generated for 28 C. tuberculostearicum sequences derived from 23 lab isolates and 5 NCBI references (CIP 102622, FDAARGOS 993, FDAARGOS 1117, FDAARGOS 1198, and SK141), in addition to the C. accolens representative genome (ATCC 49725) sequence.
Pangenome calculation
Three C. tuberculostearicum pangenomes (for all reference sequences, skin-derived reference sequences and lab sequences, and all sequences) were calculated from Prokka-derived GFF3 files using Panaroo on sensitive mode with a sequence identity threshold of 90% and otherwise default parameters. The resultant gene presence/absence tables were used for downstream analysis.
Pangenome visualization
A pangenomic map was created using anvi’o (v. 7.1) with imported Prokka gene calling information and annotations (GBK format) for 23 lab-sequenced and 5 NCBI reference C. tuberculostearicum genomes. Strains were annotated with sample metadata including skin site, general skin habitat, HV ID, as well as phylogenetic grouping from GET_HOMOLOGUES analysis. ANIb of aligned regions was calculated within anvi’o using pyANI. In addition, eggNOGG (v. 2.1.7) gene annotations were used for gene cluster annotation with the NCBI COG Database (2020) and visualized using ggplot2/R. Core, accessory, and singleton gene counts were derived from gene presence/absence tables for (1) all reference sequences and (2) all sequences. Counts were visualized as pie charts.
A gene rarefaction curve for the C. tuberculostearicum pangenome was found by applying the Vegan/R specaccum() function to the gene presence/absence table, with a random order of additions of genomes permuted 1,000 times. A Heap’s law power law model was fitted to the curve using the nls function in stats/R to calculate constants K and α. The curve was visualized using ggplot2/R.
A PCA was performed on the gene presence/absence table using the prcomp function in stats(v 3.6.2)/R. The resultant object was visualized using ggplot2/R.
Unique genes
Scoary (v. 1.6.16) was used to identify genes unique to ribotype A and B for 23 lab-sequenced C. tuberculostearicum complex isolates. Gene sequences were queried using the UniProtKB online sequence similarity BLAST tool (https://www.uniprot.org/blast).
Metagenomic read mapping
Metagenomic reads from 12 HVs at six body sites, adapter trimmed and host subtracted as described in references 2, 44, were aligned to a bowtie2 (v 2-2.4.5) database built from five NCBI C. tuberculostearicum genomes (CIP_102622, DSM 44922, FDAARGOS_1198, FDAARGOS_993, SK141) with or with supplementation with 23 non-redundant genomes from this study; default bowtie2 parameters were used: --end-to-end --sensitive. Reads were classified using kraken2 v 2.1.2 (--confidence 0.1) against a database of C. tuberculostearicum species complex members (45), and corrected abundances were calculated using bracken v 2.5 (46).
Growth curve starter cultures
Isolates for differential growth analysis were selected on the basis of (i) reliable growth in Brain-Heart-Infusion+Tween80 and (ii) coverage of the phylogenetic tree. C. tuberculostearicum isolates were grown in overnight liquid culture consisting of BHI broth (Sigma-Aldrich), augmented with 1% RPI Tween80, and 40 µg/mL fosfomycin (BHI-T-F) at 37°C with shaking at 220 rpm. To make the “Sweat media + 0.1% Tween80” media, we filter-sterilized Pickering Artificial Eccrine Perspiration Cat. No. 1700-0023 (pH 6.5) and RPI Tween80 (1%). This medium was then vortex-combined with 1% (vol) of synthetic apocrine sweat (Pickering Cat. No. 1700-070X) to produce the “Sweat media + 0.1% Tween80 + synthetic lipid mixture” medium.
Differential growth experiments
C. tuberculostearicum liquid cultures were pelleted, washed, and diluted 10-fold in diH2O to an OD600 of ~0.1. Differential media were inoculated with diluted culture at a concentration of 100:1 and plated in triplicate across a 96-well microplate. Bacterial growth was recorded using the Epoch 2 Microplate. OD600 readings were taken at 30-minute intervals throughout a 24-hour time span. The experiment was performed in duplicate. OD600 measurements were exported, corrected via blank subtraction, and plotted using ggplot2/R. The package growthcurver/R was used to calculate empirical AUC for all isolate:media combinations. Statistical significance testing for ribotype:media interactions was performed using ANOVA and a post-hoc Tukey test.
ACKNOWLEDGMENTS
We thank Tommy Hiller Tran for valuable bioinformatics discussions. We thank Dr. Matthew Kelly for discussions of Corynebacterium biology. The computational resources of the NIH High-Performance Computation Biowulf Cluster (http://hpc.nih.gov) were used for this study.
This work was supported by the Intramural Research Programs of the National Human Genome Research Institute and the National Institute of Arthritis and Musculoskeletal and Skin Diseases. We thank Julie Fekecs for help with final figures.
N.A. performed bioinformatics and growth curve analyses, prepared figures, and wrote the manuscript; P.J. provided technical support for growth curve experiments; C.D. cultured bacteria and prepared DNA-sequencing libraries; NISC sequenced amplicon and whole genome libraries; K.P.L. and H.H.K. discussed results and provided subject-matter expertise; J.A.S. and S.C. conceived the overall study and were responsible for the final version of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Sean Conlan, Email: conlans@mail.nih.gov.
Promi Das, University of California San Diego, La Jolla, California, USA.
NISC Comparative Sequencing Program:
Beatrice B. Barnabas, Sean Black, Gerard G. Bouffard, Shelise Y. Brooks, Juyun Crawford, Holly Marfani, Lyudmila Dekhtyar, Joel Han, Shi-Ling Ho, Richelle Legaspi, Quino L. Maduro, Catherine A. Masiello, Jennifer C. McDowell, Casandra Montemayor, James C. Mullikin, Morgan Park, Nancy L. Riebow, Karen Schandler, Brian Schmidt, Christina Sison, Sirintorn Stantripop, James W. Thomas, Pamela J. Thomas, Meghana Vemulapalli, and Alice C. Young
DATA AVAILABILITY
Genome data are deposited under the NCBI BioProjects PRJNA854648, PRJNA694925, and PRJNA854648 (see Table S1). Some amplicon data were published previously (n = 145; PRJNA46333) (5), and the remainder are new to this study (n = 168; PRJNA46333). Scripts, R code for figures, and the PhyloSeq object are deposited at https://github.com/skinmicrobiome/Ahmed_Corynebacterium_2023.
ETHICS APPROVAL
This natural history study was approved by the Institutional Review Board of the National Human Genome Research Institute (clinicaltrials.gov/NCT00605878) and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (https://clinicaltrials.gov/ct2/show/NCT02471352), and all subjects provided written informed consent prior to participation.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msystems.00632-23.
Relative abundances of 4 major bacterial phyla.
Relative abundance of the major C. tuberculostearicum ASVs.
A schematic representation of 16S rRNA intra-genome variation.
Alignment of the C. tuberculostearicum finished genomes.
Average nucleotide identity (ANI) of C. tuberculostearicum species complex.
Functional classifications of orthologous gene clusters.
Tables S1–S3.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, NISC Comparative Sequencing Program, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324:1190–1192. doi: 10.1126/science.1171700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Oh J, Byrd AL, Park M, NISC Comparative Sequencing Program, Kong HH, Segre JA. 2016. Temporal stability of the human skin microbiome. Cell 165:854–866. doi: 10.1016/j.cell.2016.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Escapa IF, Chen T, Huang Y, Gajare P, Dewhirst FE, Lemon KP. 2018. New insights into human nostril microbiome from the expanded human oral microbiome database (eHOMD): a resource for the microbiome of the human aerodigestive tract. mSystems 3:e00187-18. doi: 10.1128/mSystems.00187-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, Nomicos E, Polley EC, Komarow HD, NISC Comparative Sequence Program, Murray PR, Turner ML, Segre JA. 2012. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res 22:850–859. doi: 10.1101/gr.131029.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oh J, Freeman AF, NISC Comparative Sequencing Program, Park M, Sokolic R, Candotti F, Holland SM, Segre JA, Kong HH. 2013. The altered landscape of the human skin microbiome in patients with primary immunodeficiencies. Genome Res 23:2103–2114. doi: 10.1101/gr.159467.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smeekens SP, Huttenhower C, Riza A, van de Veerdonk FL, Zeeuwen P, Schalkwijk J, van der Meer JWM, Xavier RJ, Netea MG, Gevers D. 2014. Skin microbiome imbalance in patients with STAT1/STAT3 defects impairs innate host defense responses. J Innate Immun 6:253–262. doi: 10.1159/000351912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brugger SD, Eslami SM, Pettigrew MM, Escapa IF, Henke MT, Kong Y, Lemon KP, D’Orazio SEF. 2020. Dolosigranulum pigrum cooperation and competition in human nasal microbiota. mSphere 5:e00852-20. doi: 10.1128/mSphere.00852-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP. 2016. Staphylococcus aureus shifts toward commensalism in response to Corynebacterium species. Front Microbiol 7:1230. doi: 10.3389/fmicb.2016.01230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ridaura VK, Bouladoux N, Claesen J, Chen YE, Byrd AL, Constantinides MG, Merrill ED, Tamoutounour S, Fischbach MA, Belkaid Y. 2018. Contextual control of skin immunity and inflammation by Corynebacterium. J Exp Med 215:785–799. doi: 10.1084/jem.20171079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sakamoto K, Jin S-P, Goel S, Jo J-H, Voisin B, Kim D, Nadella V, Liang H, Kobayashi T, Huang X, Deming C, Horiuchi K, Segre JA, Kong HH, Nagao K. 2021. Disruption of the endopeptidase ADAM10-notch signaling axis leads to skin dysbiosis and innate lymphoid cell-mediated hair follicle destruction. Immunity 54:2321–2337. doi: 10.1016/j.immuni.2021.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kobayashi T, Glatz M, Horiuchi K, Kawasaki H, Akiyama H, Kaplan DH, Kong HH, Amagai M, Nagao K. 2015. Dysbiosis and Staphylococcus aureus colonization drives inflammation in atopic dermatitis. Immunity 42:756–766. doi: 10.1016/j.immuni.2015.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Altonsy MO, Kurwa HA, Lauzon GJ, Amrein M, Gerber AN, Almishri W, Mydlarski PR. 2020. Corynebacterium tuberculostearicum, a human skin colonizer, induces the canonical nuclear factor-ΚB inflammatory signaling pathway in human skin cells. Immun Inflamm Dis 8:62–79. doi: 10.1002/iid3.284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Conlan S, Mijares LA, NISC Comparative Sequencing Program, Becker J, Blakesley RW, Bouffard GG, Brooks S, Coleman H, Gupta J, Gurson N, Park M, Schmidt B, Thomas PJ, Otto M, Kong HH, Murray PR, Segre JA. 2012. Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol 13:R64. doi: 10.1186/gb-2012-13-7-r64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tomida S, Nguyen L, Chiu B-H, Liu J, Sodergren E, Weinstock GM, Li H. 2013. Pan-genome and comparative genome analyses of propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome. mBio 4:e00003–13. doi: 10.1128/mBio.00003-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Flores Ramos S, Brugger SD, Escapa IF, Skeete CA, Cotton SL, Eslami SM, Gao W, Bomar L, Tran TH, Jones DS, Minot S, Roberts RJ, Johnston CD, Lemon KP, Segata N. 2021. Genomic stability and genetic defense systems in Dolosigranulum pigrum, a candidate beneficial bacterium from the human microbiome. mSystems 6:mSystems 6:e0042521. doi: 10.1128/mSystems.00425-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Saheb Kashaf S, Proctor DM, Deming C, Saary P, Hölzer M, NISC comparative sequencing program, Taylor ME, Kong HH, Segre JA, Almeida A, Finn RD. 2022. Integrating cultivation and metagenomics for a multi-kingdom view of skin microbiome diversity and functions. Nat Microbiol 7:169–179. doi: 10.1038/s41564-021-01011-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Caputo A, Fournier P-E, Raoult D. 2019. Genome and pan-genome analysis to classify emerging bacteria. Biol Direct 14:5. doi: 10.1186/s13062-019-0234-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. doi: 10.1038/s41467-018-07641-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, et al. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus Agalactiae: Implications for the microbial “Pan-genome". Proc Natl Acad Sci U S A 102:13950–13955. doi: 10.1073/pnas.0506758102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Feurer C, Clermont D, Bimet F, Candréa A, Jackson M, Glaser P, Bizet C, Dauga C. 2004. Taxonomic characterization of nine strains isolated from clinical and environmental specimens, and proposal of Corynebacterium tuberculostearicum sp. nov. Int J Syst Evol Microbiol 54:1055–1061. doi: 10.1099/ijs.0.02907-0 [DOI] [PubMed] [Google Scholar]
- 21. Salamzade R, Cheong JZA, Sandstrom S, Swaney MH, Stubbendieck RM, Starr NL, Currie CR, Singh AM, Kalan LR. 2023. Evolutionary investigations of the biosynthetic diversity in the skin microbiome using lsaBGC. Microb Genom 9:mgen000988. doi: 10.1099/mgen.0.000988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Salamzade R, Swaney MH, Kalan LR. 2023. Comparative genomic and metagenomic investigations of the Corynebacterium tuberculostearicum species complex reveals potential mechanisms underlying associations to skin health and disease. Microbiol Spectr 11:e0357822. doi: 10.1128/spectrum.03578-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, Kong HH, Segre JA, NIH intramural sequencing center comparative sequencing program . 2013. Topographic diversity of fungal and bacterial communities in human skin. Nature 498:367–370. doi: 10.1038/nature12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oliveira A, Oliveira LC, Aburjaile F, Benevides L, Tiwari S, Jamal SB, Silva A, Figueiredo HCP, Ghosh P, Portela RW, De Carvalho Azevedo VA, Wattam AR. 2017. Insight of genus Corynebacterium: ascertaining the role of pathogenic and non-pathogenic species. Front Microbiol 8:1937. doi: 10.3389/fmicb.2017.01937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Olm MR, Brown CT, Brooks B, Banfield JF. 2017. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J 11:2864–2868. doi: 10.1038/ismej.2017.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tesson F, Hervé A, Touchon M, d’Humières C, Cury J, Bernheim A. 2021. Systematic and quantitative view of the antiviral arsenal of prokaryotes. bioRxiv. doi: 10.1101/2021.09.02.458658 [DOI] [PMC free article] [PubMed]
- 27. Abby SS, Néron B, Ménager H, Touchon M, Rocha EPC. 2014. MacSyfinder: a program to mine genomes for molecular systems with an application to CRISPR-CAS systems. PLoS One 9:e110726. doi: 10.1371/journal.pone.0110726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wieteska Ł, Szewczyk EM, Szemraj J. 2011. Characterization of novel plasmid P1B146 from Corynebacterium tuberculostearicum. J Microbiol Biotechnol 21:796–801. doi: 10.4014/jmb.1101.01011 [DOI] [PubMed] [Google Scholar]
- 29. Cappelli EA, Ksiezarek M, Wolf J, Neumann-Schaal M, Ribeiro TG, Peixe L. 2023. Expanding the bacterial diversity of the female urinary microbiome: description of eight new Corynebacterium species. Microorganisms 11:388. doi: 10.3390/microorganisms11020388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu X-W, De Meyer S, Trujillo ME. 2018. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466. doi: 10.1099/ijsem.0.002516 [DOI] [PubMed] [Google Scholar]
- 31. Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, Göker M. 2022. TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res 50:D801–D807. doi: 10.1093/nar/gkab902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blasche S, Kim Y, Patil KR. 2017. Draft genome sequence of Corynebacterium Kefirresidentii SB, isolated from Kefir. Genome Announc 5:e00877-17. doi: 10.1128/genomeA.00877-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eren AM, Kiefl E, Shaiber A, Veseli I, Miller SE, Schechter MS, Fink I, Pan JN, Yousef M, Fogarty EC, et al. 2021. Community-led, integrated, reproducible multi-Omics with Anvi'O. Nat Microbiol 6:3–6. doi: 10.1038/s41564-020-00834-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tettelin H, Riley D, Cattuto C, Medini D. 2008. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol 11:472–477. doi: 10.1016/j.mib.2008.09.006 [DOI] [PubMed] [Google Scholar]
- 35. Langmead B, Salzberg SL. 2012. Fast Gapped-read alignment with bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lopetuso LR, Giorgio ME, Saviano A, Scaldaferri F, Gasbarrini A, Cammarota G. 2019. Bacteriocins and bacteriophages: therapeutic weapons for gastrointestinal diseases? Int J Mol Sci 20:183. doi: 10.3390/ijms20010183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gupta S, Poret AJ, Hashemi D, Eseonu A, Yu SH, D’Gama J, Neel VA, Lieberman TD. 2023. Cutaneous surgical wounds have distinct microbiomes from intact skin. Microbiol Spectr 11:e0330022. doi: 10.1128/spectrum.03300-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nairz M, Schroll A, Sonnweber T, Weiss G. 2010. The struggle for iron - a metal at the host-pathogen interface. Cell Microbiol 12:1691–1702. doi: 10.1111/j.1462-5822.2010.01529.x [DOI] [PubMed] [Google Scholar]
- 39. Lau CKY, Krewulak KD, Vogel HJ. 2016. Bacterial ferrous iron transport: the feo system. FEMS Microbiol Rev 40:273–298. doi: 10.1093/femsre/fuv049 [DOI] [PubMed] [Google Scholar]
- 40. Lennon NJ, Lintner RE, Anderson S, Alvarez P, Barry A, Brockman W, Daza R, Erlich RL, Giannoukos G, Green L, et al. 2010. A Scalable, fully automated process for construction of sequence-ready Barcoded libraries for 454. Genome Biol 11:R15. doi: 10.1186/gb-2010-11-2-r15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DATA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McMurdie PJ, Holmes S. 2013. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20:714–737. doi: 10.1089/cmb.2013.0084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with kraken 2. Genome Biol 20:257. doi: 10.1186/s13059-019-1891-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lu J, Breitwieser FP, Thielen P, Salzberg SL. 2017. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci 3:e104. doi: 10.7717/peerj-cs.104 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative abundances of 4 major bacterial phyla.
Relative abundance of the major C. tuberculostearicum ASVs.
A schematic representation of 16S rRNA intra-genome variation.
Alignment of the C. tuberculostearicum finished genomes.
Average nucleotide identity (ANI) of C. tuberculostearicum species complex.
Functional classifications of orthologous gene clusters.
Tables S1–S3.
Data Availability Statement
Genome data are deposited under the NCBI BioProjects PRJNA854648, PRJNA694925, and PRJNA854648 (see Table S1). Some amplicon data were published previously (n = 145; PRJNA46333) (5), and the remainder are new to this study (n = 168; PRJNA46333). Scripts, R code for figures, and the PhyloSeq object are deposited at https://github.com/skinmicrobiome/Ahmed_Corynebacterium_2023.