ABSTRACT
The initial phase of the COVID-19 pandemic changed the nature of course delivery from largely in-person to exclusively remote, thus disrupting the well-established pedagogy of the Genomics Education Partnership (GEP; https://www.thegep.org). However, our web-based research adapted well to the remote learning environment. As usual, students who engaged in the GEP’s Course-based Undergraduate Research Experience (CURE) received digital projects based on genetic information within assembled Drosophila genomes. Adaptations for remote implementation included moving new member faculty training and peer Teaching Assistant office hours from in-person to online. Surprisingly, our faculty membership significantly increased and, hence, the number of supported students. Furthermore, despite the mostly virtual instruction of the 2020–2021 academic year, there was no significant decline in student learning nor attitudes. Based on successfully expanding the GEP CURE within a virtual learning environment, we provide four strategic lessons we infer toward democratizing science education. First, it appears that increasing access to scientific research and professional development opportunities by supporting virtual, cost-free attendance at national conferences attracts more faculty members to educational initiatives. Second, we observed that transitioning new member training to an online platform removed geographical barriers, reducing time and travel demands, and increased access for diverse faculty to join. Third, developing a Virtual Teaching Assistant program increased the availability of peer support, thereby improving the opportunities for student success. Finally, increasing access to web-based technology is critical for providing equitable opportunities for marginalized students to fully participate in research courses. Online CUREs have great potential for democratizing science education.
KEYWORDS: democratizing science education, Course-based Undergraduate Research Experience (CURE), genomics, online, web-based, virtual teaching assistants
PERSPECTIVE
How does one support the democratization of science education during a pandemic?
Democratizing science education increases the number of scientifically literate citizens who can knowledgeably participate in political decisions regarding scientific information (1). The need for a scientifically literate society was underscored by the COVID-19 pandemic, in which misconceptions, vivid anecdotes, and magical thinking vied with medical science and data-informed public health policies. How, then, do we train more citizens to think like scientists? In previous reports (e.g., 2), the Genomics Education Partnership (GEP; https://www.thegep.org) has argued that Course-based Undergraduate Research Experiences (CUREs) provide opportunities for students to grapple with scientific thinking and judgment. Here we recount how the GEP, a consortium devoted to incorporating research participation into the undergraduate science curriculum, succeeded in sustaining and enlarging its efforts to democratize science education, during the COVID-19 pandemic, by engaging undergraduates in genomics and bioinformatics research via CUREs. We acknowledge the limits imposed by inequitable access to computers and internet connectivity. However, we suggest that genomics- and bioinformatics-based CUREs promote the inclusion of faculty and students across a diversity of institutions in the scientific community.
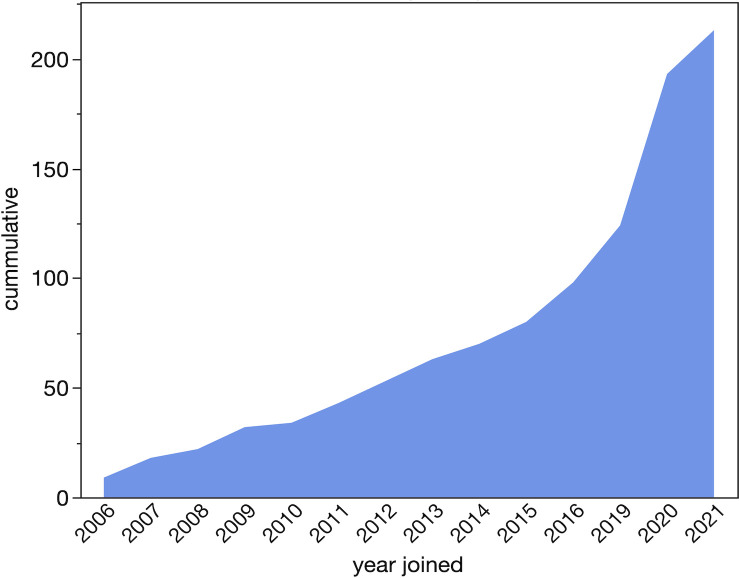
Founded in 2006, the GEP provides opportunities for students to perform original research by applying bioinformatic analyses to generate high-quality gene annotation models. The models are then analyzed collectively to derive and publish novel insights into the evolution of genes and genomes. Students now also have the opportunity to publish their individual gene models in microPublication Biology (e.g., 3). To date, more than 1,100 students have co-authored (4, 5). As of September 2021, the GEP membership consisted of 213 faculty members from 181 institutions providing instruction to over 3,945 students annually. The growth of the GEP consortium by cumulative membership is shown in Fig. 1.
Fig 1.
Cumulative GEP growth from inception to December 2021.
Efforts to democratize science during a pandemic
During the 2020–2021 academic year, the pandemic forced many institutions to halt in-person learning. Fully remote instruction caused concern throughout academia as educators worried about the loss of student learning and engagement. Silverberg expressed the concern most dramatically by entitling a report “The Pandemic Defeated My CURE (6).” Silverberg described the loss of in-person laboratory work and the substitution of a literature review project that could be completed remotely. Wang and colleagues (7) took advantage of an ongoing longitudinal observation of students who pivoted from in-person CUREs to online learning and reported a decline in situational interest (evoked by activities).
Since the curriculum of the GEP CURE is exclusively web-based, requiring no special software, it was ideal for virtual learning and allowed the program to easily transition to remote instruction. Details of the project workflow are recounted elsewhere (e.g., 8), but the key concept is that undergraduate students are provided with digitized projects, based on the genomes of Drosophila species, that require description and annotation of gene sequences. Bioinformatics and other computer-based research projects have several inherent advantages. There is a large repository of publicly accessible raw data and web-based tools; there are no lab safety issues, and the lab is open access 24/7. Consequently, the material lends itself to peer instruction, a huge plus that provides a program multiplier. Equally important, student mistakes are inexpensive in both time and money, giving students more freedom to try things possible in an actual wet lab or field study. We suggest that many more research projects could be designed for undergraduates, whether the setting for learning is in-person or online, making use of the online resources that are so abundant in the current age.
Democratizing science: lessons learned
Since its founding in 2006, the majority of new GEP faculty members were trained in-person at Washington University in St. Louis. After training at the central location, faculty led their courses at their home institutions with undergraduate Teaching Assistants (TAs) as a regular feature. These TAs were also trained in person. Students, in turn, generally experienced the GEP curriculum in person. In fact, despite the web-based nature of the GEP CURE, it established itself as a strong research experience in the in-person modality. The program has been shown to improve comprehension of genomics concepts and increased engagement in science over a diverse group of undergraduate institutions (9, 10).
During the Spring 2020 semester and the 2020–2021 academic year, continuing members of the GEP converted to online instruction. If the students had access to basic computers and internet connections, then the core web-based genomics CURE continued successfully. Instruction changed from in-person to remote, and the GEP introduced an innovative “Virtual TA” (VTA) option. VTAs were available via Zoom video conferencing for all GEP students, ameliorating the loss of in-person teaching assistants.
Anticipating a need for faculty eager to provide their students with genuine research experiences despite social isolation, the GEP trained additional faculty between May 2020 and August 2021 online. The GEP also established other innovations in response to the pandemic, including transitioning the annual week-long professional development workshop for GEP faculty to a virtual platform, and broadening the associate educational resources to include 30+ additional videos on the GEP website leading to the development of the GEP YouTube channel which was launched in January 2021 (https://www.youtube.com/c/genomicseducationpartnership).
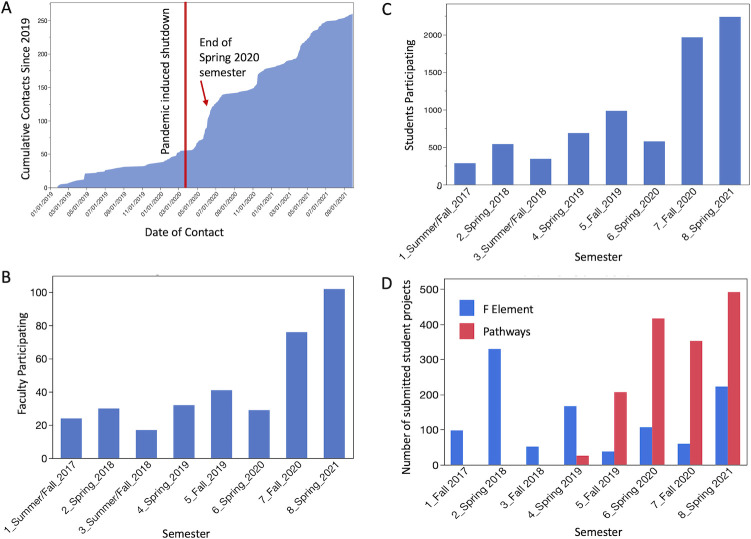
In the context of the pandemic, the accessibility of the GEP program may be measured by observing its growth. Figure 2 illustrates a growing interest based on the number of faculty contacting the GEP for training during 2020. A noticeable increase in interest developed in May 2020 (Fig. 2A) following the online presentation at the virtual TAGC (The Allied Genetics Conference) in April of that year. The GEP made a concerted effort to enhance visibility at the conference, including multiple presentations given by the GEP community, a featured talk by the Program Director, and a virtual exhibitor booth. We hypothesized that since the conference was virtual and free, awareness of the program at Minority-Serving Institutions (MSIs) and Community Colleges (CCs) would be enhanced relative to what a paid, in-person conference may have achieved. During a time of online instruction, the membership increase suggests that genomics- or bioinformatics-based CUREs are a feasible alternative to in-person, hands-on research experiences for a broad group of institutions. The trend lines suggest that the numbers of both faculty and students actively engaged in the curriculum increased (Fig. 2B and C). Notably, between January 2020 and June 2021 the composition of GEP changed as participation increased from “current” to “new” faculty, respectively, among many affiliations—including MSI (23–29%), Hispanic-Serving Institutions (HSIs) (15–17%), and CCs (9–13%)—suggesting that multiple demographics are attracted to the GEP CURE in the online format.
Fig 2.
Growth of the GEP. (A) The number of faculty contacting GEP to inquire about training increased at the time of COVID-19 lockdown. (B) The number of faculty members actively implementing GEP projects and (C) the number of students participating per semester have increased starting in Fall 2020. (D) The number of student research projects submitted per semester remained high during the pandemic. It shows the number of projects submitted for two investigations within the CURE, the F-element Project that analyzes the role of chromatin structure on gene expression and evolution in Drosophila (4, 5), and the Pathways Project that studies regulatory evolution of metabolic and signaling pathway genes (e.g., insulin signaling) across the Drosophila genus (11).
In addition to faculty interest and student participation, normal operation of the GEP can be measured by counting the number of research projects completed and submitted by students to the central GEP data collection server during remote learning. The introduction of a new “Pathways” Project to the ongoing “F-element” Project contributed to a growth of submitted student projects in Spring 2020 (Fig. 2D), and the number of projects continued to grow throughout the 2020–2021 academic year.
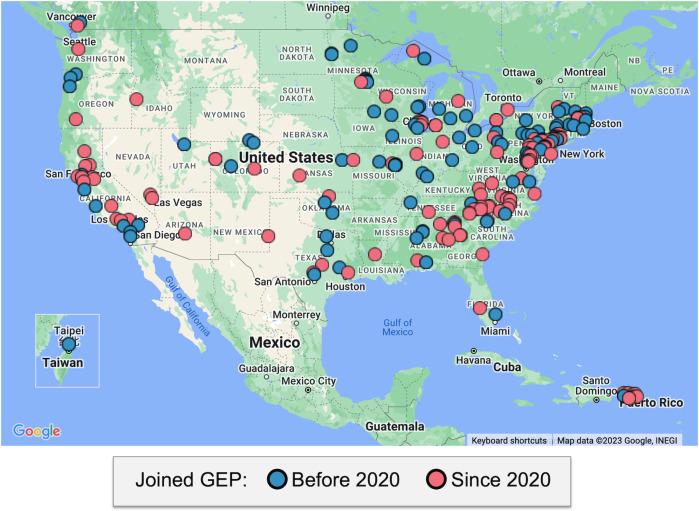
Taken together, the increase in faculty interest, the persistence of faculty/student participation, and the consistency in the number of research projects submitted per time-period all support the assertion that genomics-based CUREs are as robust online as they are in person. The implication is that a remote research experience can successfully increase accessibility to science learning for students at distributed geographical locations (Fig. 3).
Fig 3.
Map showing the geographic distribution of GEP institutions. Color code shows the year that faculty member joined the GEP.
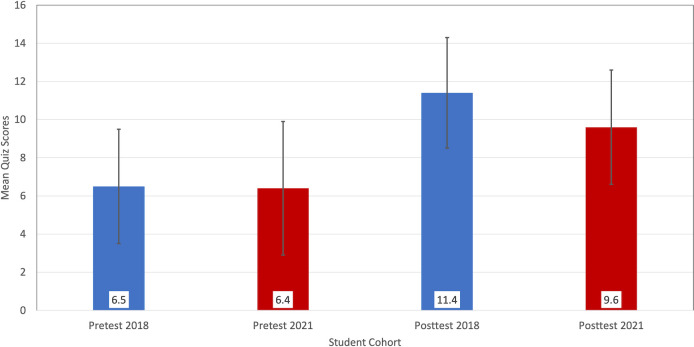
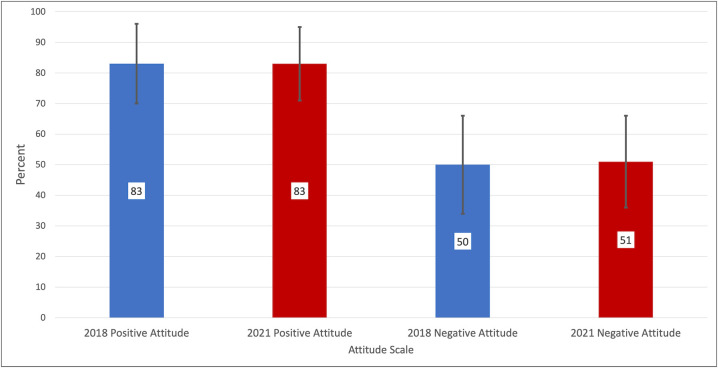
The GEP has a history of measuring student learning with a multi-operational approach that includes a direct measure of gene-structure annotation knowledge and self-reported measures of perceived benefits and attitudes. GEP students who participated from 2020 to 2021 provided data on these measures that was comparable to data provided by a pre-pandemic student cohort. The results of a 20-item quiz based on the annotation experience are shown in Fig. 4. In addition to the quiz, students were asked to evaluate five items concerning personal agency for doing science (positive attitude) and six items concerning the epistemology of science (negative attitude) on a scale of 1 (strongly disagree) to 5 (strongly agree). Examples of positive and negative attitude statements that students evaluated follow, respectively, “I can do well in science courses” versus “Creativity does not play a role in science.” Previous work had established that each set of items could be summed to give the student one positive attitude score and one negative attitude score (12). To examine if the online delivery affected these student attitudes, the post-experience attitude scales were compared for students from the 2017–2018 academic year (in-person instruction) and the 2020–2021 academic year (online instruction). The comparison is shown in Fig. 5. Despite the difference in delivery methods, the data reflect no remarkable differences in either positive or negative attitudes.
Fig 4.
Mean scores for pretest and posttest annotation quizzes for a pre-pandemic, in-person instruction year (2017–2018, blue; N = 401) and a pandemic, remote learning year (2020–2021, red; N = 621). The error bars represent one standard deviation above and below the means. The 2018 posttest mean (11.4) is slightly higher than the 2021 posttest mean (9.6); however, the difference is not meaningful (i.e., less than two quiz points). Analysis using the TOST (two one-sided test) procedure found that the 2021 means were within the equivalence margin of the 2018 means.
Fig 5.
Positive and negative attitudes toward science are compared for a pre-pandemic, in-person instruction year (2017–2018, blue; N = 809 and a pandemic, remote learning year (2020–2021,red, N = 1035). The mean scores are presented as percentages of the maximum possible score because the positive attitude included five items while the negative attitude included six items. The error bars represent one standard deviation above and below the mean percentages. There is no significant difference in student attitudes between pre-pandemic in-person instruction and during-pandemic online instruction. Analysis using the TOST procedure found that the 2021 percentages were with the equivalence margin of the 2018 percentages.
To compensate for the loss of in-person TAs during the pandemic, the GEP organized a novel Virtual Teaching Assistant (VTA) program in which peer, undergraduate mentors were employed and available approximately 60 h per week via Zoom video conferencing to all GEP students across the country. To assess the effectiveness of the VTAs, GEP students were asked on the post-experience survey if they had employed the VTAs, and, if so, to evaluate their experience. While 259 students were aware of the resource but did not use it, 137 students reported using the virtual TA resource. The former group reported that they did not use the VTA resource because they did not feel they needed the assistance (N = 168), the meeting times were not convenient (N = 38), or they did not feel comfortable talking to a student TA they had not met in person (N = 21). Usage of the resource was most frequently 2–4 times during the term (N = 75). The students who did use the VTA resource were asked to evaluate their experience on nine items rated on a scale of 1 (strongly disagree) to 5 (strongly agree). These items and the mean and median responses are shown in Table 1. Overall, students responded favorably to the VTA initiative. Based on these reports, we conclude that the VTA program increases student comfort and supports student success in our genomics CURE.
TABLE 1.
Student responses to propositions about the Virtual Teaching Assistantsa
| Item | Mean | Median |
|---|---|---|
| I felt comfortable asking the virtual TA questions | 4.3 | 5 |
| I felt more comfortable asking the virtual TA questions than my instructor | 3.65 | 4 |
| The virtual TA was knowledgeable about the GEP content material | 4.3 | 4 |
| The virtual GEP TA provided clear and understandable explanations | 4.25 | 4 |
| A virtual TA was available and responsive when I needed help | 4.3 | 4 |
| The virtual TA enabled me to successfully complete my GEP project | 4.23 | 4 |
| I would recommend other students completing GEP projects to utilize the virtual TAs | 4.38 | 5 |
| The virtual TAs are an important part of the GEP program and should be continued | 4.43 | 5 |
| I think all courses (and students) would benefit from having a nationwide set of virtual TAs associated with them | 4.4 | 5 |
Items were evaluated on a scale of 1 (strongly disagree) to 5 (strongly agree). N = 137.
The means reported in Table 1 support the proposition that VTAs are a valuable resource. Our data suggests that online delivery of the novel VTA program allowed the GEP CURE to remain effective through the transition from onsite to online: the GEP CURE is resilient. This resilience aids in the democratization of science education by making research experiences available to all (13); however, achieving the full potential for Virtual TA programs relies on the assumption that the appropriate web access is available to every student.
Strategies for the democratization of science
The SARS-CoV-2 virus continues to infect students and faculty. Should another deadly viral variant evolve, our data indicate that a genomics- or bioinformatics-based CURE can provide research training where other options are not available: especially for remote learners and other non-traditional students. For institutions transitioning from remote back to in-person instruction, we observe that several pedagogic tactics cultivated from remote learning continue to be employed by instructors in combination with in-person learning, including (i) enhancing communication on genome annotation via screen-sharing, (ii) recorded lectures and help sessions on common challenges allowing more instructional time for discussion, (iii) an increase in “flipped classroom” teaching, and (iv) hybrid delivery of help sessions featuring both in-person and virtual teaching assistants, thus, allowing coverage of late-night hours.
In addition to these pedagogical tactics, our experience suggests four broader strategies that show promise for the democratization of science education.
First, the increased accessibility of online professional conferences appears to increase faculty participation and dissemination of teaching opportunities. As shown in Fig. 2, shortly after the pandemic triggered remote learning and a free, virtual TAGC Conference, an organization that took 13 years to grow to 100 members, began a steady increase in numbers. Within 1 year, we had grown from 125 members to more than 200. We suspect faculty who would not have otherwise been able to attend due to financial limitations (e.g., registration fee, travel costs) were able to access the event and learn about the advanced, web-based, genomics CURE, in part due to this free event. Indeed, as the pandemic has stretched over multiple years, many faculty and students have attended conferences that switched from onsite to online or hybrid venues with fee reductions and no travel costs. Maintaining this trend to include at least some cost-free, virtual participation for STEM education conferences going forward may facilitate STEM democratization efforts.
Second, we should continue to exploit virtual training sessions for faculty and students. Prior to the pandemic, GEP members were clustered in geographic subgroups—Regional Nodes—that reduced travel barriers to training opportunities. Development of Regional Nodes reflected our conception that onsite/in-person training was essential to learning the genome browser skills necessary for faculty to run the GEP CURE in classrooms. Similarly, we were convinced students needed to be in person for their training and research experiences. COVID-19 upended our conceptions forcing us to train fully remotely, irrespective of Regional Node placement. Removing many logistical barriers, including travel, allowed for accelerated growth in GEP membership, which broadened student access to genomics instruction and research experiences despite the pandemic. GEP students continued submitting authentic research projects (Fig. 2D) while also making learning and attitudinal gains (Fig. 4 and 5), thus demonstrating that research continued. With the plethora of bioinformatic databases available in the twenty-first century, building more web-based CUREs, that only require access to a web browser (no special software) could be a valid method to increase faculty and student accessibility to CUREs in remote and/or under-resourced locations. The wealth of data available to investigate is staggering.
Third, the pandemic inspired the innovative implementation of the Virtual Teaching Assistant (VTA) program. To compensate for the loss of in-person TAs during the 2020–2021 academic year of remote learning, the GEP initiated its national VTA program to provide increased assistance to students: supplementing the work of course instructors. While only a subset of students took advantage of the VTAs during their inaugural season, student feedback suggests that this is a welcome addition to the GEP program. As many students work jobs and have classes that clash with in-person faculty office hours or TA work sessions, it may be that VTAs could be successfully employed for other Undergraduate Research Experiences or national education programs. Having bi- or multi-lingual virtual peers with availability on evenings and weekends could also dramatically increase student’s access to STEM education through advanced CURE initiatives.
Fourth, we recognize that broader participation in undergraduate research experiences may affect democratization of science education when measured by demographic characteristics of faculty and students. The benefit of remote research experiences may be constrained by unequal access to the appropriate technology. While we observed a 2–6% increase in participation amongst Minority-Serving Institutions (MSIs), Hispanic-Serving Institutions (HSIs), and Community Colleges (CCs) (legend describing Fig. 2), we queried the students about access. In response to a survey question, most GEP students (84%) reported adequate connectivity with the internet to participate fully in course instruction; however, the 16% who did not have adequate connectivity is troubling. Erickson and colleagues (14) reported that many students at remote sites were concerned about “technological issues” during the early stage of the COVID-19 pandemic when attempting to participate in the online Research Experience for Undergraduate (REU) programs (14). As broadband infrastructure reaches remote areas, the extension of high-quality, advanced research education provided by web-based CUREs could reach an increasing number of students, increasing the percentage of scientifically literate citizens.
At least 3,945 undergraduates participated in the GEP program during the 2020–2021 academic year. While we do not expect that all GEP students will continue in paths toward STEM careers, we are optimistic that the experience of conducting a research project, including in many cases defending their work in a report and possibly co-authoring in a publication, imprints students with the understanding that scientific habits of mind and judgment are broadly applicable. It also develops a necessary perspective for understanding our world of science. In general, genomics-/bioinformatics-based CUREs have the potential to democratize access to science education.
ACKNOWLEDGMENTS
This manuscript is based upon work supported by the National Science Foundation (#1915544) and National Institute of General Medical Sciences of the National Institutes of Health (#R25GM130517) to L.K.R. The GEP currently operates with an IRB Authorization Agreement (IAA) between all participating institutions and The University of Alabama at Tuscaloosa; the work has exempt status.
Members of The Genomics Education Partnership co-authors: Abby E. Hare-Harris (Department of Biology, Commonwealth University, Bloomsburg, PA); Adam Haberman (Department of Biology, University of San Diego, San Diego, CA); Adam J. Kleinschmit (Department of Natural and Applied Sciences, University of Dubuque, Dubuque, IA); Alder Yu (Department of Biology, University of Wisconsin-La Crosse, La Crosse, WI); Alexa Sawa (Department of Sciences and Engineering, College of the Desert, Palm Desert, CA); Alexis Nagengast (Departments of Chemistry and Biochemistry, Widener University, Chester, PA); Alisha Howard (Department of Biological and Environmental Sciences, East Central University, Ada, OK); Alma E. Rodriguez Estrada (School of Natural Sciences, Technology and Mathematics, Aurora University, Aurora, IL); Amy T. Hark (Biology Department, Muhlenberg College, Allentown, PA); Ana Almeida (Department of Biological Sciences, California State University East Bay, Hayward, CA); Andrew M. Arsham (Department of Biology, Bemidji State University, Bemidji, MN); Ann K. Corsi (Department of Biology, The Catholic University of America, Washington, DC); Anna K. Allen (Department of Biology, Howard University, Washington, DC); Anthony D. Aragon (Department of Math and Sciences, Dodge City Community College, Dodge City, KS); Aparna Sreenivasan (Department of Biology and Chemistry, California State University, Monterey Bay, Seaside, CA); Brian Yowler (Department of Biology, Grove City College, Grove City, PA); Carina E. Howell (Department of Biological Sciences, Lock Haven University of Pennsylvania, Lock Haven, PA); Catherine Reinke (Department of Biology, Linfield University McMinnville, OR); Chelsey C. McKenna (Department of Biological Science, College of Southern Nevada, North Las Vegas, NV); Christine M. Fleet (Department of Biology, Emory & Henry College, Emory, VA); Christopher J. Jones (Department of Biological Sciences, Moravian University, Bethlehem, PA); Cindy Arrigo (Biology Department, New Jersey City University, Jersey City, NJ); Cindy Wolfe (Biology Program, Kentucky Wesleyan College, Owensboro, KY); Claudia Uhde-Stone (Department of Biological Sciences, California State University, East Bay, Hayward, CA); Daron Barnard (Biology Department, Worcester State University, Worcester, MA); Don Paetkau (Department of Biology, Saint Mary’s College, Notre Dame, IN); Enrique Rodriguez-Borrero (Department of Biology, University of Puerto Rico-Cayey, Cayey, PR); Evan Merkhofer (Division of Natural Sciences, Mount Saint Mary College, Newburgh, NY); Eve M. Mellgren (Department of Biology, Elmhurst University, Elmhurst, IL); Farida Safadi-Chamberlain (Department of Biology, Colorado State University, Fort Collins, CO); Geoffrey D. Findlay (Department of Biology, College of the Holy Cross, Worcester, MA); Gerard McNeil (Department of Biology, York College / CUNY, Jamaica, NY); Heidi S. Bretscher (Department of Genetics, Cell Biology and Development, University of Minnesota-Twin Cities, Minneapolis, MN); Hemayet Ullah (Department of Biology, Howard University, Washington, DC); Hemlata Mistry (Departments of Biology and Biochemistry, Widener University, Chester, PA); H. Howard Xu (Department of Biological Sciences, California State University, Los Angeles, Los Angeles, CA); Indrani Bose (Department of Biology, Western Carolina University, Cullowhee, NC); Jack Vincent (Division of Sciences and Mathematics, University of Washington Tacoma, Tacoma, WA); Jacob D. Kagey (Biology Department, University of Detroit Mercy, Detroit, MI); Jacqueline K. Wittke-Thompson (Department of Natural & Health Sciences, University of St. Francis, Joliet, IL); James E. J. Bedard (Department of Biology, University of the Fraser Valley, Abbotsford, BC); James S. Godde (Department of Biology, Monmouth College, Monmouth, IL); James V. Price (Department of Biology, Utah Valley University, Orem, UT); Jamie O. Dyer (Department of Biology, Rockhurst University, Kansas City, MO); Jennifer A. Roecklein-Canfield (Department of Chemistry, Simmons University, Boston, MA); Jennifer Jemc (Department of Biology, Loyola University Chicago, Chicago, IL); Jennifer Kennell (Department of Biology, Vassar College, Poughkeepsie, NY); Jeroen Gillard (Department of Biology, California State University Bakersfield, Bakersfield, CA); John M. Braverman (Department of Biology, Saint Joseph’s University, Philadelphia, PA); John P. Stanga (Department of Biology, Mercer University, Macon, GA); Joyce Stamm (Department of Biology, University of Evansville, Evansville, IN); Juan C. Martínez-Cruzado (Department of Biology, University of Puerto Rico-Mayagüez, Mayagüez, PR); Judith Leatherman (Department of Biological Sciences, University of Northern Colorado, Greeley, CO); Justin R. DiAngelo (Division of Science, Penn State Berks, Reading, PA); Justin Thackeray (Department of Biology, Clark University, Worcester, MA); Karen L. Schmeichel (Department of Biology, Oglethorpe University, Brookhaven, GA); Katherine C. Teeter (Department of Biology, Northern Michigan University, Marquette, MI); Kayla Bieser (Department of Physical and Life Sciences, Nevada State College, Henderson, NV); Kellie S. Agrimson (Department of Biology, St. Catherine University, St. Paul, MN); Kenneth Saville (Department of Biology, Albion College, Albion, MI); Leocadia Paliulis (Department of Biology, Bucknell University, Lewisburg, PA); Lindsey J. Long (Department of Biology, Oklahoma Christian University, Oklahoma City, OK); Lisa Kadlec (Department of Biology, Wilkes University, Wilkes-Barre, PA); M. Logan Johnson (Department of Natural Sciences, University of Pittsburgh at Greensburg, Greensburg, PA); Maire K. Sustacek (School of Science and Mathematics, Minneapolis Community and Technical College, Minneapolis, MN); Maria Santisteban (Department of Biology, University of North Carolina at Pembroke, Pembroke, NC); Marie Montes-Matias (Division of STEM, Union College, Cranford, NJ); Martin G. Burg (Departments of Biomedical Sciences & Cell and Molecular Biology, Grand Valley State University, Allendale, MI); Mary Ann V. Smith (Department of Biology, Penn State Schuylkill, Schuylkill Haven, PA); Matthew Skerritt (Science Department, SUNY Corning Community College, Corning, NY); Matthew Wawersik (Department of Biology, College of William & Mary, Williamsburg, VA); Melinda A. Yang (Department of Biology, University of Richmond, Richmond, VA); Michael R. Rubin (Department of Biology, University of Puerto Rico-Cayey, Cayey, PR); Michele Eller (Department of Life Science, Southern Wesleyan University, Central, SC); Monica L. Hall-Woods (Department of Biology, St. Charles Community College, Cottleville, MO); Natalie Minkovsky (Department of Biology, Community College of Baltimore County, Baltimore, MD); Nicole Salazar Velmeshev (Department of Biology, San Francisco State University, San Francisco, CA); Nighat P. Kokan (Department of Natural Sciences, Cardinal Stritch University, Milwaukee, WI); Nikolaos Tsotakos (Department of Biology, Penn State Harrisburg, Middletown, PA); Norma Velazquez-Ulloa (Department of Biology, Lewis & Clark College, Portland, OR); Paula Croonquist (Community College - Coon Rapids Campus, Coon Rapids, MN); Rivka L. Glaser (Department of Biological Sciences, Stevenson University, Owings Mills, MD); Robert A. Drewell (Biology Department, Clark University, Worcester, MA); Sarah C. R. Elgin (Department of Biology, Washington University in St. Louis, St. Louis, MO); Sarah Justice (Department of Biology, Taylor University, Upland, IN); Scott Tanner (Division of Natural Sciences and Engineering, University of South Carolina - Upstate, Spartanburg, SC); Shallee T. Page (Division of Health and Natural Sciences, Franklin Pierce University, Rindge, NH); Siaumin Fung (Biology Department, Vanguard University, Costa Mesa, CA); Solomon Tin Chi Chak (Department of Biological Sciences, SUNY Old Westbury, Old Westbury, NY); Srebrenka Robic (Department of Biology, Agnes Scott College, Decatur, GA); Stephanie Toering Peters (Department of Biology, Wartburg College, Waverly, IA); Stephen Klusza (Department of Biology, Clayton State University, Morrow, GA); Susan Parrish (Biology Department, McDaniel College, Westminster, MD); Takrima Sadikot (Department of Biology, Washburn University, Topeka, KS); Tara A. Pelletier (Biology Department, Radford University, Radford, VA); Thomas C. Giarla (Biology Department, Siena College, Loudonville, NY); Vida Mingo (Department of Biology, Columbia College, Columbia, SC); Vidya Chandrasekaran (Biology Department, Saint Mary’s College of California, Moraga, CA); Virginia Rebecca Falkenberg (Department of Life & Earth Sciences, Perimeter College at Georgia State University, Clarkston, GA); A. Zeynep Ozsoy (Department of Biological Sciences, Colorado Mesa University, Grand Junction, CO).
AFTER EPUB
[This article was published on 22 September 2023 with an error in the author affiliations. The error was corrected in the current version, posted on 9 November 2023.]
Contributor Information
S. Catherine Silver Key, Email: ckey@nccu.edu.
Laura K. Reed, Email: lreed1@ua.edu.
Pamela Ann Marshall, Arizona State University, Glendale, Arizona, USA.
The Genomics Education Partnership:
Abby E. Hare-Harris, Adam Haberman, Adam J. Kleinschmit, Alder Yu, Alexa Sawa, Alexis Nagengast, Alisha Howard, E Alma, Rodriguez Estrada, Amy T. Hark, Ana Almeida, Andrew M. Arsham, Ann K. Corsi, Anna K. Allen, Anthony D. Aragon, Aparna Sreenivasan, Brian Yowler, Carina E. Howell, Catherine Reinke, Chelsey C. McKenna, Christine M. Fleet, Christopher J. Jones, Cindy Arrigo, Cindy Wolfe, Claudia Uhde-Stone, Daron Barnard, Enrique Rodriguez-Borrero Don Paetkau, Evan Merkhofer, Eve M. Mellgren, Farida Safadi-Chamberlain, Geoffrey D. Findlay, Gerard McNeil, Heidi S. Bretscher, Hemayet Ullah, Hemlata Mistry, H. Howard Xu, Indrani Bose, Jack Vincent, Jacob D. Kagey, Jacqueline K. Wittke-Thompson, James E. J. Bedard, James S. Godde, James V. Price, Jamie O. Dyer, Jennifer A. Roecklein-Canfield, Jennifer Jemc, Jennifer Kennell, Jeroen Gillard, John M. Braverman, John P. Stanga, Joyce Stamm, Juan C. Martínez-Cruzado, Judith Leatherman, Justin R. DiAngelo, Justin Thackeray, Karen L. Schmeichel, Katherine C. Teeter, Kayla Bieser, Kellie S. Agrimson, Kenneth Saville, Leocadia Paliulis, Lindsey J. Long, Lisa Kadlec, M. Logan Johnson, Maire K. Sustacek, Maria Santisteban, Marie Montes-Matias, Martin G. Burg, Mary Ann V. Smith, Matthew Skerritt, Matthew Wawersik, Melinda A. Yang, Michael R. Rubin, Michele Eller, Monica L. Hall-Woods, Natalie Minkovsky, Nicole Salazar Velmeshev, Nighat P. Kokan, Nikolaos Tsotakos, Norma Velazquez-Ulloa, Paula Croonquist, Rivka L. Glaser, Robert A. Drewell, Sarah C. R. Elgin, Sarah Justice, Scott Tanner, Shallee T. Page, and Siaumin Fung
REFERENCES
- 1. Guston D. 2004. Forget Politicizing science. Let’s democratize science. Issues Sci Technol. https://issues.org/p_guston-3/. [Google Scholar]
- 2. Lopatto D, Rosenwald AG, DiAngelo JR, Hark AT, Skerritt M, Wawersik M, Allen AK, Alvarez C, Anderson S, Arrigo C, Arsham A, Barnard D, Bazinet C, Bedard JEJ, Bose I, Braverman JM, Burg MG, Burgess RC, Croonquist P, Du C, Dubowsky S, Eisler H, Escobar MA, Foulk M, Furbee E, Giarla T, Glaser RL, Goodman AL, Gosser Y, Haberman A, Hauser C, Hays S, Howell CE, Jemc J, Johnson ML, Jones CJ, Kadlec L, Kagey JD, Keller KL, Kennell J, Key SCS, Kleinschmit AJ, Kleinschmit M, Kokan NP, Kopp OR, Laakso MM, Leatherman J, Long LJ, Manier M, Martinez-Cruzado JC, Matos LF, McClellan AJ, McNeil G, Merkhofer E, Mingo V, Mistry H, Mitchell E, Mortimer NT, Mukhopadhyay D, Myka JL, Nagengast A, Overvoorde P, Paetkau D, Paliulis L, Parrish S, Preuss ML, Price JV, Pullen NA, Reinke C, Revie D, Robic S, Roecklein-Canfield JA, Rubin MR, Sadikot T, Sanford JS, Santisteban M, Saville K, Schroeder S, Shaffer CD, Sharif KA, Sklensky DE, Small C, Smith M, Smith S, Spokony R, Sreenivasan A, Stamm J, Sterne-Marr R, Teeter KC, Thackeray J, Thompson JS, Peters ST, Van Stry M, Velazquez-Ulloa N, Wolfe C, Youngblom J, Yowler B, Zhou L, Brennan J, Buhler J, Leung W, Reed LK, Elgin SCR. 2020. Facilitating growth through frustration: using genomics research in a course-based undergraduate research experience. J Microbiol Biol Educ 21:21.1.6. doi: 10.1128/jmbe.v21i1.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lose B, Myers A, Fondse S, Alberts I, Stamm J, Youngblom JJ, Rele CP, Reed LK. 2021. Drosophila Yakuba - Tsc1. MicroPubl Biol. doi: 10.17912/micropub.biology.000407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leung W, Shaffer CD, Chen EJ, Quisenberry TJ, Ko K, Braverman JM, Giarla TC, Mortimer NT, Reed LK, Smith ST, Robic S, McCartha SR, Perry DR, Prescod LM, Sheppard ZA, Saville KJ, McClish A, Morlock EA, Sochor VR, Stanton B, Veysey-White IC, Revie D, Jimenez LA, Palomino JJ, Patao MD, Patao SM, Himelblau ET, Campbell JD, Hertz AL, McEvilly MF, Wagner AR, Youngblom J, Bedi B, Bettincourt J, Duso E, Her M, Hilton W, House S, Karimi M, Kumimoto K, Lee R, Lopez D, Odisho G, Prasad R, Robbins HL, Sandhu T, Selfridge T, Tsukashima K, Yosif H, Kokan NP, Britt L, Zoellner A, Spana EP, Chlebina BT, Chong I, Friedman H, Mammo DA, Ng CL, Nikam VS, Schwartz NU, Xu TQ, Burg MG, Batten SM, Corbeill LM, Enoch E, Ensign JJ, Franks ME, Haiker B, Ingles JA, Kirkland LD, Lorenz-Guertin JM, Matthews J, Mittig CM, Monsma N, Olson KJ, Perez-Aragon G, Ramic A, Ramirez JR, Scheiber C, Schneider PA, Schultz DE, Simon M, Spencer E, Wernette AC, Wykle ME, Zavala-Arellano E, McDonald MJ, Ostby K, Wendland P, DiAngelo JR, Ceasrine AM, Cox AH, Docherty JEB, Gingras RM, Grieb SM, Pavia MJ, Personius CL, Polak GL, Beach DL, Cerritos HL, Horansky EA, Sharif KA, Moran R, Parrish S, Bickford K, Bland J, Broussard J, Campbell K, Deibel KE, Forka R, Lemke MC, Nelson MB, O’Keeffe C, Ramey SM, Schmidt L, Villegas P, Jones CJ, Christ SL, Mamari S, Rinaldi AS, Stity G, Hark AT, Scheuerman M, Silver Key SC, McRae BD, Haberman AS, Asinof S, Carrington H, Drumm K, Embry T, McGuire R, Miller-Foreman D, Rosen S, Safa N, Schultz D, Segal M, Shevin Y, Svoronos P, Vuong T, Skuse G, Paetkau DW, Bridgman RK, Brown CM, Carroll AR, Gifford FM, Gillespie JB, Herman SE, Holtcamp KL, Host MA, Hussey G, Kramer DM, Lawrence JQ, Martin MM, Niemiec EN, O’Reilly AP, Pahl OA, Quintana G, Rettie EAS, Richardson TL, Rodriguez AE, Rodriguez MO, Schiraldi L, Smith JJ, Sugrue KF, Suriano LJ, Takach KE, Vasquez AM, Velez X, Villafuerte EJ, Vives LT, Zellmer VR, Hauke J, Hauser CR, Barker K, Cannon L, Parsamian P, Parsons S, Wichman Z, Bazinet CW, Johnson DE, Bangura A, Black JA, Chevee V, Einsteen SA, Hilton SK, Kollmer M, Nadendla R, Stamm J, Fafara-Thompson AE, Gygi AM, Ogawa EE, Van Camp M, Kocsisova Z, Leatherman JL, Modahl CM, Rubin MR, Apiz-Saab SS, Arias-Mejias SM, Carrion-Ortiz CF, Claudio-Vazquez PN, Espada-Green DM, Feliciano-Camacho M, Gonzalez-Bonilla KM, Taboas-Arroyo M, Vargas-Franco D, Montañez-Gonzalez R, Perez-Otero J, Rivera-Burgos M, Rivera-Rosario FJ, Eisler HL, Alexander J, Begley SK, Gabbard D, Allen RJ, Aung WY, Barshop WD, Boozalis A, Chu VP, Davis JS, Duggal RN, Franklin R, Gavinski K, Gebreyesus H, Gong HZ, Greenstein RA, Guo AD, Hanson C, Homa KE, Hsu SC, Huang Y, Huo L, Jacobs S, Jia S, Jung KL, Wai-Chee Kong S, Kroll MR, Lee BM, Lee PF, Levine KM, Li AS, Liu C, Liu MM, Lousararian AP, Lowery PB, Mallya AP, Marcus JE, Ng PC, Nguyen HP, Patel R, Precht H, Rastogi S, Sarezky JM, Schefkind A, Schultz MB, Shen D, Skorupa T, Spies NC, Stancu G, Vivian Tsang HM, Turski AL, Venkat R, Waldman LE, Wang K, Wang T, Wei JW, Wu DY, Xiong DD, Yu J, Zhou K, McNeil GP, Fernandez RW, Menzies PG, Gu T, Buhler J, Mardis ER, Elgin SCR. 2017. Retrotransposons are the major contributors to the expansion of the Drosophila ananassae Muller F element. G3 (Bethesda) 7:2439–2460. doi: 10.1534/g3.117.040907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leung W, Shaffer CD, Reed LK, Smith ST, Barshop W, Dirkes W, Dothager M, Lee P, Wong J, Xiong D, Yuan H, Bedard JEJ, Machone JF, Patterson SD, Price AL, Turner BA, Robic S, Luippold EK, McCartha SR, Walji TA, Walker CA, Saville K, Abrams MK, Armstrong AR, Armstrong W, Bailey RJ, Barberi CR, Beck LR, Blaker AL, Blunden CE, Brand JP, Brock EJ, Brooks DW, Brown M, Butzler SC, Clark EM, Clark NB, Collins AA, Cotteleer RJ, Cullimore PR, Dawson SG, Docking CT, Dorsett SL, Dougherty GA, Downey KA, Drake AP, Earl EK, Floyd TG, Forsyth JD, Foust JD, Franchi SL, Geary JF, Hanson CK, Harding TS, Harris CB, Heckman JM, Holderness HL, Howey NA, Jacobs DA, Jewell ES, Kaisler M, Karaska EA, Kehoe JL, Koaches HC, Koehler J, Koenig D, Kujawski AJ, Kus JE, Lammers JA, Leads RR, Leatherman EC, Lippert RN, Messenger GS, Morrow AT, Newcomb V, Plasman HJ, Potocny SJ, Powers MK, Reem RM, Rennhack JP, Reynolds KR, Reynolds LA, Rhee DK, Rivard AB, Ronk AJ, Rooney MB, Rubin LS, Salbert LR, Saluja RK, Schauder T, Schneiter AR, Schulz RW, Smith KE, Spencer S, Swanson BR, Tache MA, Tewilliager AA, Tilot AK, VanEck E, Villerot MM, Vylonis MB, Watson DT, Wurzler JA, Wysocki LM, Yalamanchili M, Zaborowicz MA, Emerson JA, Ortiz C, Deuschle FJ, DiLorenzo LA, Goeller KL, Macchi CR, Muller SE, Pasierb BD, Sable JE, Tucci JM, Tynon M, Dunbar DA, Beken LH, Conturso AC, Danner BL, DeMichele GA, Gonzales JA, Hammond MS, Kelley CV, Kelly EA, Kulich D, Mageeney CM, McCabe NL, Newman AM, Spaeder LA, Tumminello RA, Revie D, Benson JM, Cristostomo MC, DaSilva PA, Harker KS, Jarrell JN, Jimenez LA, Katz BM, Kennedy WR, Kolibas KS, LeBlanc MT, Nguyen TT, Nicolas DS, Patao MD, Patao SM, Rupley BJ, Sessions BJ, Weaver JA, Goodman AL, Alvendia EL, Baldassari SM, Brown AS, Chase IO, Chen M, Chiang S, Cromwell AB, Custer AF, DiTommaso TM, El-Adaimi J, Goscinski NC, Grove RA, Gutierrez N, Harnoto RS, Hedeen H, Hong EL, Hopkins BL, Huerta VF, Khoshabian C, LaForge KM, Lee CT, Lewis BM, Lydon AM, Maniaci BJ, Mitchell RD, Morlock EV, Morris WM, Naik P, Olson NC, Osterloh JM, Perez MA, Presley JD, Randazzo MJ, Regan MK, Rossi FG, Smith MA, Soliterman EA, Sparks CJ, Tran DL, Wan T, Welker AA, Wong JN, Sreenivasan A, Youngblom J, Adams A, Alldredge J, Bryant A, Carranza D, Cifelli A, Coulson K, Debow C, Delacruz N, Emerson C, Farrar C, Foret D, Garibay E, Gooch J, Heslop M, Kaur S, Khan A, Kim V, Lamb T, Lindbeck P, Lucas G, Macias E, Martiniuc D, Mayorga L, Medina J, Membreno N, Messiah S, Neufeld L, Nguyen SF, Nichols Z, Odisho G, Peterson D, Rodela L, Rodriguez P, Rodriguez V, Ruiz J, Sherrill W, Silva V, Sparks J, Statton G, Townsend A, Valdez I, Waters M, Westphal K, Winkler S, Zumkehr J, DeJong RJ, Hoogewerf AJ, Ackerman CM, Armistead IO, Baatenburg L, Borr MJ, Brouwer LK, Burkhart BJ, Bushhouse KT, Cesko L, Choi TYY, Cohen H, Damsteegt AM, Darusz JM, Dauphin CM, Davis YP, Diekema EJ, Drewry M, Eisen MEM, Faber HM, Faber KJ, Feenstra E, Felzer-Kim IT, Hammond BL, Hendriksma J, Herrold MR, Hilbrands JA, Howell EJ, Jelgerhuis SA, Jelsema TR, Johnson BK, Jones KK, Kim A, Kooienga RD, Menyes EE, Nollet EA, Plescher BE, Rios L, Rose JL, Schepers AJ, Scott G, Smith JR, Sterling AM, Tenney JC, Uitvlugt C, VanDyken RE, VanderVennen M, Vue S, Kokan NP, Agbley K, Boham SK, Broomfield D, Chapman K, Dobbe A, Dobbe I, Harrington W, Ibrahem M, Kennedy A, Koplinsky CA, Kubricky C, Ladzekpo D, Pattison C, Ramirez RE, Wande L, Woehlke S, Wawersik M, Kiernan E, Thompson JS, Banker R, Bartling JR, Bhatiya CI, Boudoures AL, Christiansen L, Fosselman DS, French KM, Gill IS, Havill JT, Johnson JL, Keny LJ, Kerber JM, Klett BM, Kufel CN, May FJ, Mecoli JP, Merry CR, Meyer LR, Miller EG, Mullen GJ, Palozola KC, Pfeil JJ, Thomas JG, Verbofsky EM, Spana EP, Agarwalla A, Chapman J, Chlebina B, Chong I, Falk IN, Fitzgibbons JD, Friedman H, Ighile O, Kim AJ, Knouse KA, Kung F, Mammo D, Ng CL, Nikam VS, Norton D, Pham P, Polk JW, Prasad S, Rankin H, Ratliff CD, Scala V, Schwartz NU, Shuen JA, Xu A, Xu TQ, Zhang Y, Rosenwald AG, Burg MG, Adams SJ, Baker M, Botsford B, Brinkley B, Brown C, Emiah S, Enoch E, Gier C, Greenwell A, Hoogenboom L, Matthews JE, McDonald M, Mercer A, Monsma N, Ostby K, Ramic A, Shallman D, Simon M, Spencer E, Tomkins T, Wendland P, Wylie A, Wolyniak MJ, Robertson GM, Smith SI, DiAngelo JR, Sassu ED, Bhalla SC, Sharif KA, Choeying T, Macias JS, Sanusi F, Torchon K, Bednarski AE, Alvarez CJ, Davis KC, Dunham CA, Grantham AJ, Hare AN, Schottler J, Scott ZW, Kuleck GA, Yu NS, Kaehler MM, Jipp J, Overvoorde PJ, Shoop E, Cyrankowski O, Hoover B, Kusner M, Lin D, Martinov T, Misch J, Salzman G, Schiedermayer H, Snavely M, Zarrasola S, Parrish S, Baker A, Beckett A, Belella C, Bryant J, Conrad T, Fearnow A, Gomez C, Herbstsomer RA, Hirsch S, Johnson C, Jones M, Kabaso R, Lemmon E, Vieira CMDS, McFarland D, McLaughlin C, Morgan A, Musokotwane S, Neutzling W, Nietmann J, Paluskievicz C, Penn J, Peoples E, Pozmanter C, Reed E, Rigby N, Schmidt L, Shelton M, Shuford R, Tirasawasdichai T, Undem B, Urick D, Vondy K, Yarrington B, Eckdahl TT, Poet JL, Allen AB, Anderson JE, Barnett JM, Baumgardner JS, Brown AD, Carney JE, Chavez RA, Christgen SL, Christie JS, Clary AN, Conn MA, Cooper KM, Crowley MJ, Crowley ST, Doty JS, Dow BA, Edwards CR, Elder DD, Fanning JP, Janssen BM, Lambright AK, Lane CE, Limle AB, Mazur T, McCracken MR, McDonough AM, Melton AD, Minnick PJ, Musick AE, Newhart WH, Noynaert JW, Ogden BJ, Sandusky MW, Schmuecker SM, Shipman AL, Smith AL, Thomsen KM, Unzicker MR, Vernon WB, Winn WW, Woyski DS, Zhu X, Du C, Ament C, Aso S, Bisogno LS, Caronna J, Fefelova N, Lopez L, Malkowitz L, Marra J, Menillo D, Obiorah I, Onsarigo EN, Primus S, Soos M, Tare A, Zidan A, Jones CJ, Aronhalt T, Bellush JM, Burke C, DeFazio S, Does BR, Johnson TD, Keysock N, Knudsen NH, Messler J, Myirski K, Rekai JL, Rempe RM, Salgado MS, Stagaard E, Starcher JR, Waggoner AW, Yemelyanova AK, Hark AT, Bertolet A, Kuschner CE, Parry K, Quach M, Shantzer L, Shaw ME, Smith MA, Glenn O, Mason P, Williams C, Key SCS, Henry TCP, Johnson AG, White JX, Haberman A, Asinof S, Drumm K, Freeburg T, Safa N, Schultz D, Shevin Y, Svoronos P, Vuong T, Wellinghoff J, Hoopes LLM, Chau KM, Ward A, Regisford EGC, Augustine L, Davis-Reyes B, Echendu V, Hales J, Ibarra S, Johnson L, Ovu S, Braverman JM, Bahr TJ, Caesar NM, Campana C, Cassidy DW, Cognetti PA, English JD, Fadus MC, Fick CN, Freda PJ, Hennessy BM, Hockenberger K, Jones JK, King JE, Knob CR, Kraftmann KJ, Li L, Lupey LN, Minniti CJ, Minton TF, Moran JV, Mudumbi K, Nordman EC, Puetz WJ, Robinson LM, Rose TJ, Sweeney EP, Timko AS, Paetkau DW, Eisler HL, Aldrup ME, Bodenberg JM, Cole MG, Deranek KM, DeShetler M, Dowd RM, Eckardt AK, Ehret SC, Fese J, Garrett AD, Kammrath A, Kappes ML, Light MR, Meier AC, O’Rouke A, Perella M, Ramsey K, Ramthun JR, Reilly MT, Robinett D, Rossi NL, Schueler MG, Shoemaker E, Starkey KM, Vetor A, Vrable A, Chandrasekaran V, Beck C, Hatfield KR, Herrick DA, Khoury CB, Lea C, Louie CA, Lowell SM, Reynolds TJ, Schibler J, Scoma AH, Smith-Gee MT, Tuberty S, Smith CD, Lopilato JE, Hauke J, Roecklein-Canfield JA, Corrielus M, Gilman H, Intriago S, Maffa A, Rauf SA, Thistle K, Trieu M, Winters J, Yang B, Hauser CR, Abusheikh T, Ashrawi Y, Benitez P, Boudreaux LR, Bourland M, Chavez M, Cruz S, Elliott G, Farek JR, Flohr S, Flores AH, Friedrichs C, Fusco Z, Goodwin Z, Helmreich E, Kiley J, Knepper JM, Langner C, Martinez M, Mendoza C, Naik M, Ochoa A, Ragland N, Raimey E, Rathore S, Reza E, Sadovsky G, Seydoux M-IB, Smith JE, Unruh AK, Velasquez V, Wolski MW, Gosser Y, Govind S, Clarke-Medley N, Guadron L, Lau D, Lu A, Mazzeo C, Meghdari M, Ng S, Pamnani B, Plante O, Shum YKW, Song R, Johnson DE, Abdelnabi M, Archambault A, Chamma N, Gaur S, Hammett D, Kandahari A, Khayrullina G, Kumar S, Lawrence S, Madden N, Mandelbaum M, Milnthorp H, Mohini S, Patel R, Peacock SJ, Perling E, Quintana A, Rahimi M, Ramirez K, Singhal R, Weeks C, Wong T, Gillis AT, Moore ZD, Savell CD, Watson R, Mel SF, Anilkumar AA, Bilinski P, Castillo R, Closser M, Cruz NM, Dai T, Garbagnati GF, Horton LS, Kim D, Lau JH, Liu JZ, Mach SD, Phan TA, Ren Y, Stapleton KE, Strelitz JM, Sunjed R, Stamm J, Anderson MC, Bonifield BG, Coomes D, Dillman A, Durchholz EJ, Fafara-Thompson AE, Gross MJ, Gygi AM, Jackson LE, Johnson A, Kocsisova Z, Manghelli JL, McNeil K, Murillo M, Naylor KL, Neely J, Ogawa EE, Rich A, Rogers A, Spencer JD, Stemler KM, Throm AA, Van Camp M, Weihbrecht K, Wiles TA, Williams MA, Williams M, Zoll K, Bailey C, Zhou L, Balthaser DM, Bashiri A, Bower ME, Florian KA, Ghavam N, Greiner-Sosanko ES, Karim H, Mullen VW, Pelchen CE, Yenerall PM, Zhang J, Rubin MR, Arias-Mejias SM, Bermudez-Capo AG, Bernal-Vega GV, Colon-Vazquez M, Flores-Vazquez A, Gines-Rosario M, Llavona-Cartagena IG, Martinez-Rodriguez JO, Ortiz-Fuentes L, Perez-Colomba EO, Perez-Otero J, Rivera E, Rodriguez-Giron LJ, Santiago-Sanabria AJ, Senquiz-Gonzalez AM, delValle FRS, Vargas-Franco D, Velázquez-Soto KI, Zambrana-Burgos JD, Martinez-Cruzado JC, Asencio-Zayas L, Babilonia-Figueroa K, Beauchamp-Pérez FD, Belén-Rodríguez J, Bracero-Quiñones L, Burgos-Bula AP, Collado-Méndez XA, Colón-Cruz LR, Correa-Muller AI, Crooke-Rosado JL, Cruz-García JM, Defendini-Ávila M, Delgado-Peraza FM, Feliciano-Cancela AJ, Gónzalez-Pérez VM, Guiblet W, Heredia-Negrón A, Hernández-Muñiz J, Irizarry-González LN, Laboy-Corales ÁL, Llaurador-Caraballo GA, Marín-Maldonado F, Marrero-Llerena U, Martell-Martínez HA, Martínez-Traverso IM, Medina-Ortega KN, Méndez-Castellanos SG, Menéndez-Serrano KC, Morales-Caraballo CI, Ortiz-DeChoudens S, Ortiz-Ortiz P, Pagán-Torres H, Pérez-Afanador D, Quintana-Torres EM, Ramírez-Aponte EG, Riascos-Cuero C, Rivera-Llovet MS, Rivera-Pagán IT, Rivera-Vicéns RE, Robles-Juarbe F, Rodríguez-Bonilla L, Rodríguez-Echevarría BO, Rodríguez-García PM, Rodríguez-Laboy AE, Rodríguez-Santiago S, Rojas-Vargas ML, Rubio-Marrero EN, Santiago-Colón A, Santiago-Ortiz JL, Santos-Ramos CE, Serrano-González J, Tamayo-Figueroa AM, Tascón-Peñaranda EP, Torres-Castillo JL, Valentín-Feliciano NA, Valentín-Feliciano YM, Vargas-Barreto NM, Vélez-Vázquez M, Vilanova-Vélez LR, Zambrana-Echevarría C, MacKinnon C, Chung H-M, Kay C, Pinto A, Kopp OR, Burkhardt J, Harward C, Allen R, Bhat P, Chang JH-C, Chen Y, Chesley C, Cohn D, DuPuis D, Fasano M, Fazzio N, Gavinski K, Gebreyesus H, Giarla T, Gostelow M, Greenstein R, Gunasinghe H, Hanson C, Hay A, He TJ, Homa K, Howe R, Howenstein J, Huang H, Khatri A, Kim YL, Knowles O, Kong S, Krock R, Kroll M, Kuhn J, Kwong M, Lee B, Lee R, Levine K, Li Y, Liu B, Liu L, Liu M, Lousararian A, Ma J, Mallya A, Manchee C, Marcus J, McDaniel S, Miller ML, Molleston JM, Diez CM, Ng P, Ngai N, Nguyen H, Nylander A, Pollack J, Rastogi S, Reddy H, Regenold N, Sarezky J, Schultz M, Shim J, Skorupa T, Smith K, Spencer SJ, Srikanth P, Stancu G, Stein AP, Strother M, Sudmeier L, Sun M, Sundaram V, Tazudeen N, Tseng A, Tzeng A, Venkat R, Venkataram S, Waldman L, Wang T, Yang H, Yu JY, Zheng Y, Preuss ML, Garcia A, Juergens M, Morris RW, Nagengast AA, Azarewicz J, Carr TJ, Chichearo N, Colgan M, Donegan M, Gardner B, Kolba N, Krumm JL, Lytle S, MacMillian L, Miller M, Montgomery A, Moretti A, Offenbacker B, Polen M, Toth J, Woytanowski J, Kadlec L, Crawford J, Spratt ML, Adams AL, Barnard BK, Cheramie MN, Eime AM, Golden KL, Hawkins AP, Hill JE, Kampmeier JA, Kern CD, Magnuson EE, Miller AR, Morrow CM, Peairs JC, Pickett GL, Popelka SA, Scott AJ, Teepe EJ, TerMeer KA, Watchinski CA, Watson LA, Weber RE, Woodard KA, Barnard DC, Appiah I, Giddens MM, McNeil GP, Adebayo A, Bagaeva K, Chinwong J, Dol C, George E, Haltaufderhyde K, Haye J, Kaur M, Semon M, Serjanov D, Toorie A, Wilson C, Riddle NC, Buhler J, Mardis ER, Elgin SCR. 2015. Drosophila Muller F elements maintain a distinct set of genomic properties over 40 million years of evolution. G3 (Bethesda) 5:719–740. doi: 10.1534/g3.114.015966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Silverberg LJ. 2020. The pandemic defeated my CURE: Replacement with a student project of a literature review of the syntheses of small molecule drugs. J. Chem. Educ 97:3450–3454. doi: 10.1021/acs.jchemed.0c00555 [DOI] [Google Scholar]
- 7. Wang C, Bauer M, Burmeister AR, Hanauer DI, Graham MJ. 2020. College student meaning making and interest maintenance during COVID-19: from course-based undergraduate research experiences (cures) to science learning being off-campus and online. Front. Educ 5. doi: 10.3389/feduc.2020.590738 [DOI] [Google Scholar]
- 8. Shaffer CD, Alvarez CJ, Bednarski AE, Dunbar D, Goodman AL, Reinke C, Rosenwald AG, Wolyniak MJ, Bailey C, Barnard D, Bazinet C, Beach DL, Bedard JEJ, Bhalla S, Braverman J, Burg M, Chandrasekaran V, Chung H-M, Clase K, DeJong RJ, DiAngelo JR, Du C, Eckdahl TT, Eisler H, Emerson JA, Frary A, Frohlich D, Gosser Y, Govind S, Haberman A, Hark AT, Hauser C, Hoogewerf A, Hoopes LLM, Howell CE, Johnson D, Jones CJ, Kadlec L, Kaehler M, Silver Key SC, Kleinschmit A, Kokan NP, Kopp O, Kuleck G, Leatherman J, Lopilato J, MacKinnon C, Martinez-Cruzado JC, McNeil G, Mel S, Mistry H, Nagengast A, Overvoorde P, Paetkau DW, Parrish S, Peterson CN, Preuss M, Reed LK, Revie D, Robic S, Roecklein-Canfield J, Rubin MR, Saville K, Schroeder S, Sharif K, Shaw M, Skuse G, Smith CD, Smith MA, Smith ST, Spana E, Spratt M, Sreenivasan A, Stamm J, Szauter P, Thompson JS, Wawersik M, Youngblom J, Zhou L, Mardis ER, Buhler J, Leung W, Lopatto D, Elgin SCR, O’Dowd DK. 2014. A course-based research experience: how benefits change with increased investment in instructional time. LSE 13:111–130. doi: 10.1187/cbe-13-08-0152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elgin SCR, Hauser C, Holzen TM, Jones C, Kleinschmit A, Leatherman J, Genomics Education Partnership . 2017. The GEP: crowd-Sourcing big data analysis with undergraduates. Trends Genet 33:81–85. doi: 10.1016/j.tig.2016.11.004 [DOI] [PubMed] [Google Scholar]
- 10. Shaffer CD, Alvarez C, Bailey C, Barnard D, Bhalla S, Chandrasekaran C, Chandrasekaran V, Chung H-M, Dorer DR, Du C, Eckdahl TT, Poet JL, Frohlich D, Goodman AL, Gosser Y, Hauser C, Hoopes LLM, Johnson D, Jones CJ, Kaehler M, Kokan N, Kopp OR, Kuleck GA, McNeil G, Moss R, Myka JL, Nagengast A, Morris R, Overvoorde PJ, Shoop E, Parrish S, Reed K, Regisford EG, Revie D, Rosenwald AG, Saville K, Schroeder S, Shaw M, Skuse G, Smith C, Smith M, Spana EP, Spratt M, Stamm J, Thompson JS, Wawersik M, Wilson BA, Youngblom J, Leung W, Buhler J, Mardis ER, Lopatto D, Elgin SCR. 2010. The Genomics education partnership: successful integration of research into laboratory classes at a diverse group of undergraduate institutions. CBE Life Sci Educ 9:55–69. doi: 10.1187/09-11-0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rele CP, Williams J, Reed LK, Youngblom JJ, Leung W. 2021. Drosophila grimshawi - Rheb. MicroPubl Biol. doi: 10.17912/micropub.biology.000371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lopatto D, Rosenwald AG, Burgess RC, Silver Key C, Van Stry M, Wawersik M, DiAngelo JR, Hark AT, Skerritt M, Allen AK, Alvarez C, Anderson S, Arrigo C, Arsham A, Barnard D, Bedard JEJ, Bose I, Braverman JM, Burg MG, Croonquist P, Du C, Dubowsky S, Eisler H, Escobar MA, Foulk M, Giarla T, Glaser RL, Goodman AL, Gosser Y, Haberman A, Hauser C, Hays S, Howell CE, Jemc J, Jones CJ, Kadlec L, Kagey JD, Keller KL, Kennell J, Kleinschmit AJ, Kleinschmit M, Kokan NP, Kopp OR, Laakso MM, Leatherman J, Long LJ, Manier M, Martinez-Cruzado JC, Matos LF, McClellan AJ, McNeil G, Merkhofer E, Mingo V, Mistry H, Mitchell E, Mortimer NT, Myka JL, Nagengast A, Overvoorde P, Paetkau D, Paliulis L, Parrish S, Toering Peters S, Preuss ML, Price JV, Pullen NA, Reinke C, Revie D, Robic S, Roecklein-Canfield JA, Rubin MR, Sadikot T, Sanford JS, Santisteban M, Saville K, Schroeder S, Shaffer CD, Sharif KA, Sklensky DE, Small C, Smith S, Spokony R, Sreenivasan A, Stamm J, Sterne-Marr R, Teeter KC, Thackeray J, Thompson JS, Velazquez-Ulloa N, Wolfe C, Youngblom J, Yowler B, Zhou L, Brennan J, Buhler J, Leung W, Elgin SCR, Reed LK. 2022. Student attitudes contribute to the effectiveness of a genomics CURE. J Microbiol Biol Educ 23:e00208-21. doi: 10.1128/jmbe.00208-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gerrity E. 2020. The future of STEM education: engaging our undergraduates in doing science. https://biology.wustl.edu/news/future-stem-education-engaging-our-undergraduates-doing-science.
- 14. Erickson OA, Cole RB, Isaacs JM, Alvarez-Clare S, Arnold J, Augustus-Wallace A, Ayoob JC, Berkowitz A, Branchaw J, Burgio KR, Cannon CH, Ceballos RM, Cohen CS, Coller H, Disney J, Doze VA, Eggers MJ, Farina S, Ferguson EL, Gray JJ, Greenberg JT, Hoffmann A, Jensen-Ryan D, Kao RM, Keene AC, Kowalko JE, Lopez SA, Mathis C, Minkara M, Murren CJ, Ondrechen MJ, Ordoñez P, Osano A, Padilla-Crespo E, Palchoudhury S, Qin H, Ramírez-Lugo J, Reithel J, Shaw CA, Smith A, Smith R, Summers AP, Tsien F, Dolan EL. 2022. “"How do we do this at a distance?!" A descriptive study of remote undergraduate research programs during COVID-19”. CBE Life Sci Educ 21:ar1. doi: 10.1187/cbe.21-05-0125 [DOI] [PMC free article] [PubMed] [Google Scholar]