Abstract
The major outer membrane protein (OMP) of Actinobacillus actinomycetemcomitans is an OmpA homolog that demonstrates electrophoretic heat modifiability. The gene encoding this protein was isolated from a genomic library of A. actinomycetemcomitans NCTC 9710 by immunoscreening with serum from a patient with localized juvenile periodontitis. Expression of the cloned gene in Escherichia coli and subsequent Western blot analysis revealed a protein with an approximate molecular mass of 34 kDa. The amino acid sequence predicted from the cloned gene demonstrated that the mature protein had a molecular mass of 34,911 Da and significant identity to members of the OmpA family of proteins. We have named the major OMP of A. actinomycetemcomitans Omp34, and its corresponding gene has been named omp34.
Actinobacillus actinomycetemcomitans is a gram-negative coccobacillus which has been strongly implicated as the etiological agent in localized juvenile periodontitis (LJP) (33, 34). A. actinomycetemcomitans harbors a host of virulence factors (30), several of which are surface associated. These include an antiproliferative protein (29), an osteolytic protein (11), and components which can induce cytokine expression (21, 22). Another possible virulence mechanism associated with A. actinomycetemcomitans is the production of proteins that bind the Fc portion of immunoglobulins (26). These receptors are thought to interfere with complement or antibody-dependent host immune mechanisms (14). Recently, the major Fc-binding protein of A. actinomycetemcomitans has been identified as a heat-modifiable outer membrane protein (OMP) which demonstrates significant identity to the outer membrane protein A (OmpA) of Escherichia coli and other related OmpA-like proteins of gram-negative bacteria (17).
It is now well established that individuals with LJP and other forms of periodontitis such as adult periodontitis have elevated levels of serum antibodies to A. actinomycetemcomitans (7, 23, 25) and to the surface-associated material from this organism (15, 16, 29). More specifically, the heat-modifiable OMP of A. actinomycetemcomitans mentioned above has been identified as a major target for immunoglobulin G antibodies in sera from LJP patients (2, 18, 31, 32).
In this study, we constructed a genomic library from A. actinomycetemcomitans NCTC 9710. To identify genes which code for surface-associated antigens, we screened the library with serum from a patient with LJP. Here, we describe the cloning and molecular characterization of the gene coding for the heat-modifiable OMP of A. actinomycetemcomitans. Its relationship to the family of OmpA-like proteins is discussed.
Bacterial strains and plasmids.
A. actinomycetemcomitans NCTC 9710 was cultured at 37°C in a CO2-enriched atmosphere on brain heart infusion agar (Oxoid) supplemented with 5% (vol/vol) horse blood. Bacteria were grown for 48 h, harvested by using saline, and centrifuged at 3,000 × g for 20 min, and the pellet was stored at −70°C. E. coli JM109 was used for all cloning studies and cultured on Luria-Bertani (LB) medium.
Construction of a genomic library of A. actinomycetemcomitans.
Chromosomal DNA was extracted from the bacterial cells by standard methods as described by Sambrook et al. (24), partially digested with Sau3A (Sigma), and size fractionated on a 0 to 40% sucrose gradient. DNA in the size range of 2 to 10 kb was ligated, by using T4 DNA ligase (Promega), into pUC18 which had previously been cleaved with the enzyme BamHI and dephosphorylated with calf intestinal alkaline phosphatase. Recombinant DNA was transformed into E. coli JM109, which was made competent by the method of Hanahan (9). The cells were plated on LB agar containing 50 μg of ampicillin ml−1, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and IPTG (isopropyl-β-d-thiogalactopyranoside) and incubated for 24 h at 37°C.
Serum samples.
One serum sample was used for the immunoscreening; it was selected from sera derived from 16 patients diagnosed as having LJP by standard criteria, including radiographic evidence of bone loss and first permanent molar or incisor pocket depths of 5 mm or more. Selection was made by comparing the sera for their ability to bind A. actinomycetemcomitans proteins as assessed by Western blot analysis (29).
Immunological screening of the genomic library.
Recombinant white colonies were replica plated onto nylon Hybond-N membranes (Amersham). A. actinomycetemcomitans and E. coli JM109/pUC18 colonies were also spotted onto each membrane as positive and negative controls, respectively. Colonies were not lysed, to facilitate the isolation of recombinants expressing surface-associated antigens. Instead, membranes were rinsed in phosphate-buffered saline containing 1% Triton X-100 for 1 h and then transferred to blocking buffer (phosphate-buffered saline containing 0.1% Triton X-100 and 3% low-fat milk powder [Safeway, Aylesford, United Kingdom]) for 1 h. Membranes were incubated in serum from a patient with LJP diluted 1:500 for 2 h. The serum had been extensively adsorbed with 100 μg of E. coli JM109 whole-cell lysate ml−1 for 1 h at 37°C. After washing, the membranes were incubated for 1 h in goat anti-human immunoglobulin G horseradish peroxidase conjugate (Sigma), diluted 1:1,000, and developed in 3,3′-diaminobenzidine tetrahydrochloride solution (10 mg in 15 ml of Tris-buffered saline [pH 7.6]) containing 12 μl of 30% hydrogen peroxide. Approximately 1,000 E. coli transformants were screened for expression of A. actinomycetemcomitans antigens. Colonies which reacted positively to the serum from an LJP patient on the initial screen were subcultured and screened again in the same way. This procedure identified three immunologically reactive colonies, clones containing plasmids pAAL89, pAAL90, and pAAL91.
Western blot analysis of positive clones.
Recombinant bacteria, harvested from LB agar plates containing 50 μg of ampicillin ml−1, X-Gal, and IPTG were centrifuged and resuspended in 50 mM Tris (pH 8.0) containing 2% sodium dodecyl sulfate (SDS) and boiled for 10 min. Cell debris was removed by centrifugation, and 10 μl of the supernatant was mixed with 10 μl of sample buffer (0.06 M Tris, 10% glycerol, 1% SDS, 5% 2-mercaptoethanol, 0.05% bromophenol blue [pH 6.8]) and boiled for a further 2 min. Samples were separated by SDS-polyacrylamide gel electrophoresis on a 12% gel with a Bio-Rad mini-Protean II system. Gels were either stained with colloidal Coomassie blue (Sigma) or immunoblotted, as described previously (29). Western blot analysis of the three immunologically reactive clones, with the same serum sample as that used in the screening procedure, revealed that the clone harboring plasmid pAAL91 (designated JM109/pAAL91) contained a recombinant protein with a molecular mass of approximately 34 kDa (data not shown). This protein band did not appear to be present in the E. coli control lysate (JM109/pUC18) or the remaining two recombinants (JM109/pAAL89 and JM109/pAAL90) but was present in the A. actinomycetemcomitans whole-cell lysate.
Plasmid analysis.
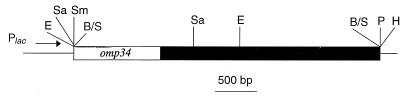
Plasmid-containing cells were grown overnight at 37°C in 5 ml of LB broth supplemented with 50 μg of ampicillin ml−1 (Sigma), and plasmid DNA was recovered with the Magic Miniprep DNA purification system (Promega). All plasmids were cut with HindIII and EcoRI to assess the size of the insert. Fragments were separated by electrophoresis in 0.8% (wt/vol) agarose gels and visualized by staining with ethidium bromide. Plasmids pAAL89, pAAL90, and pAAL91 contained DNA inserts of 500, 2,600, and 3,700 bp, respectively. Plasmid pAAL91 was further digested with a range of restriction endonucleases to produce a restriction map (Fig. 1). The cloned fragment contained restriction sites for the enzymes EcoRI and SacI.
FIG. 1.
Partial restriction map of pAAL91. A 3.7-kb genomic fragment was cloned into the BamHI site of pUC18. The positions of the plasmid sequences are shown by the horizontal narrow lines. A. actinomycetemcomitans sequences are represented by open and closed boxes, with the open box indicating the position of the omp34 gene. The position of the lac promoter (Plac) in pUC18 is also shown. Restriction site abbreviations: E, EcoRI; B/S, BamHI/Sau3A; H, HindIII; P, PstI; Sa, SacI; Sm, SmaI.
Characterization of the gene encoding the 34-kDa OMP.
The DNA sequence of the OMP gene, contained within the 3.7-kb fragment from pAAL91, was determined in both directions with a Sequenase 2.0 kit (United States Biochemical Corp.) by using synthetic primers. Translation of the 1,080-bp DNA sequence by use of the program NIP of Dear and Staden (5) revealed an open reading frame of 1,041 bp (Fig. 2). This open reading frame encodes a protein of 346 amino acid residues with a calculated Mr of 36,905. Analysis of the predicted N-terminal amino acid sequence revealed the presence of a leader peptide rich in hydrophobic domains and with alanine residues present at positions 19 and 21. Both features are characteristic of signal peptides used for protein export. The sequence of the N-terminal region of the major OMP of A. actinomycetemcomitans has been reported on two occasions (17, 31) and is identical to residues 22 to 34 of the deduced amino-terminal sequence of the gene encoding the 34-kDa OMP. Thus, subsequent localization of the signal peptidase cleavage site between amino acids 21 and 22 indicated that the mature protein had an Mr of 34,911. This mass is consistent with the size of the protein expressed by JM109/pAAL91, which was identified by Western blot analysis (data not shown). We have named the major OMP of A. actinomycetemcomitans Omp34, and its corresponding gene has been named omp34.
FIG. 2.
Nucleotide and derived amino acid sequences of Omp34 of A. actinomycetemcomitans. The signal peptide is underlined.
Omp34 is a member of the OmpA family of proteins.
Further analysis of the predicted amino acid sequence of Omp34 of A. actinomycetemcomitans clearly demonstrated that it had significant identity to members of the OmpA family of proteins. The closest relative of Omp34 identified to date is the outer membrane protein P5 (OmpP5) of Haemophilus influenzae (19); these two proteins have 67% identity. Omp34 also has 57.1 and 56.7% identity to the two OmpA homologs, MOMP and OmpA2, of Haemophilus ducreyi, respectively (12). H. ducreyi is not unique in this respect since Aeromonas salmonicida also has two genes which code for OmpA homologs (3). Omp34 also demonstrates 52.9% identity to both E. coli and Salmonella typhimurium OmpAs (1, 8). The OmpA-like protein of Pasteurella multocida has recently been characterized; 10 of the first 12 amino acid residues are identical to the predicted N-terminal sequence of Omp34 (27). Figure 3 shows the sequence alignment of Omp34, OmpP5 of H. influenzae, and OmpA of E. coli. The sequence relatedness between these proteins is greater in the C-terminal region; this is due, in part, to a putative peptidoglycan binding motif (NX2LSX2RAX2VX3L) that is conserved throughout the OmpA family (6, 13).
FIG. 3.
Sequence comparison of A. actinomycetemcomitans (Aa) Omp34 (accession no. AF005079 [this study]), H. influenzae (Hi) OmpP5 (accession no. L20309 [19]), and E. coli (Ec) OmpA (accession no. V00307 [1]). The protein sequences were aligned with the Clustal program (10). The leader peptide sequences are underlined. The α-helical motif, proposed as a fingerprint for OmpA-related proteins (13), is indicated in each sequence by bold type. Identical amino acids are indicated by an asterisk; conservative amino acid substitutions are indicated by a period. Substitutions were designated conservative if all amino acids fell within one of the following exchange groups: T, S, A, G, and P; R, K, and H; F, W, and Y; D, E, Q, and N; I, L, M, and V; and C (4).
A possible virulence role for Omp34 of A. actinomycetemcomitans is indicated by both its strong immunogenic potential and its Fc binding properties (17, 32). Given the high level of identity between OmpP5 of H. influenzae and Omp34, it would be interesting to test OmpP5 for its ability to bind Fc. Moreover, if other members of the OmpA family also possess Fc binding activity, this could potentially explain why they are often identified as major antigens. There is growing evidence that the OmpA of E. coli contributes to pathogenicity (28). Recently, Prasadarao et al. (20) demonstrated that invasion of epithelial cells by E. coli was reduced for a mutant lacking OmpA. The present study may help to elucidate the possible role of Omp34 in the pathogenesis of LJP.
Nucleotide sequence accession number.
The GenBank accession number of the omp34 gene sequence determined in this study is AF005079.
Acknowledgments
We are grateful to the Royal Society, London, United Kingdom, for providing funding to P.A.W.
REFERENCES
- 1.Beck E, Bremer E. Nucleotide sequence of the ompA gene coding the outer membrane protein II* of Escherichia coli K-12. Nucleic Acids Res. 1980;8:3011–3024. doi: 10.1093/nar/8.13.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolstad A I, Kristoffersen T, Olsen I, Preus H R, Vasstrand E N, Bakken V. Outer membrane proteins of Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus studied by SDS-PAGE and immunoblotting. Oral Microbiol Immunol. 1990;5:155–161. doi: 10.1111/j.1399-302x.1990.tb00414.x. [DOI] [PubMed] [Google Scholar]
- 3.Costello G M, Vipond R, MacIntyre S. Aeromonas salmonicida possesses two genes encoding homologs of the major outer membrane protein, OmpA. J Bacteriol. 1996;178:1623–1630. doi: 10.1128/jb.178.6.1623-1630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dayhoff M O, Schwartz R M, Orcott B L. A model of evolutionary change in proteins. In: Dayhoff M O, editor. Atlas of protein sequence and structure. Vol. 5. 1978. pp. 345–352. , suppl. 3. National Biomedical Research Foundation, Washington, D.C. [Google Scholar]
- 5.Dear S, Staden R. A sequence assembly and editing program for efficient management of large projects. Nucleic Acids Res. 1991;19:3907–3911. doi: 10.1093/nar/19.14.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Mot R, Vanderleydon J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol. 1994;12:333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 7.Ebersole J L, Cappelli D. Gingival crevicular fluid antibody to Actinobacillus actinomycetemcomitans in periodontal disease. Oral Microbiol Immunol. 1994;9:335–344. doi: 10.1111/j.1399-302x.1994.tb00283.x. [DOI] [PubMed] [Google Scholar]
- 8.Freudl R, Cole S T. Cloning and molecular characterization of the OmpA gene from Salmonella typhimurium. Eur J Biochem. 1983;134:497–502. doi: 10.1111/j.1432-1033.1983.tb07594.x. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:570–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 10.Higgins D G, Bleasby A J, Fuchs R. Clustal V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 11.Kirby A C, Meghji S, Nair S P, White P A, Reddi K, Nishihara T, Nakashima K, Willis A C, Sim R, Wilson M, Henderson B. The potent bone-resorbing mediator of Actinobacillus actinomycetemcomitans is homologous to the molecular chaperone GroEL. J Clin Invest. 1995;96:1185–1194. doi: 10.1172/JCI118150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klesney-Tait J, Hiltke T J, Maciver I, Spinola S M, Radolf J D, Hansen E J. The major outer membrane protein of Haemophilus ducreyi consists of two OmpA homologs. J Bacteriol. 1997;179:1764–1773. doi: 10.1128/jb.179.5.1764-1773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koebnik R. Proposal for a peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell surface proteins. Mol Microbiol. 1995;16:1269–1270. doi: 10.1111/j.1365-2958.1995.tb02348.x. [DOI] [PubMed] [Google Scholar]
- 14.Langone J J. Protein A from staphylococcus and related immunoglobulin receptors produced by streptococci and pneumococci. Adv Immunol. 1982;32:157–252. [PubMed] [Google Scholar]
- 15.Meghji S, Henderson B, Kirby A C, Newman H N, Wilson M. Serum antibody response to surface-associated material from periodontopathogenic bacteria. FEMS Immunol Med Microbiol. 1995;10:101–108. doi: 10.1111/j.1574-695X.1995.tb00017.x. [DOI] [PubMed] [Google Scholar]
- 16.Meghji S, Henderson B, Wilson M. High-titer antisera from patients with periodontal disease inhibit bacterial capsule-induced bone breakdown. J Periodontal Res. 1993;8:115–121. doi: 10.1111/j.1600-0765.1993.tb01058.x. [DOI] [PubMed] [Google Scholar]
- 17.Mintz K P, Fives-Taylor P M. Identification of an immunoglobulin Fc receptor of Actinobacillus actinomycetemcomitans. Infect Immun. 1994;62:4500–4505. doi: 10.1128/iai.62.10.4500-4505.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller D, Poolman T J, Bernadina W E, van Kol P J, Ruitenberg E J. Characterisation of outer membrane proteins from Actinobacillus actinomycetemcomitans. Microb Pathog. 1990;9:227–233. doi: 10.1016/0882-4010(90)90011-e. [DOI] [PubMed] [Google Scholar]
- 19.Munson R S, Grass S, West R. Molecular cloning and sequence of the gene for the outer membrane protein P5 of Haemophilus influenzae. Infect Immun. 1993;61:4017–4020. doi: 10.1128/iai.61.9.4017-4020.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prasadarao N V, Wass C A, Weiser J N, Stins M F, Huang S-H, Kim K S. Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect Immun. 1996;64:146–153. doi: 10.1128/iai.64.1.146-153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddi K, Wilson M, Poole S, Meghji S, Henderson B. Relative cytokine-stimulating activities of surface components of the oral periodontopathic bacterium Actinobacillus actinomycetemcomitans. Cytokine. 1995;7:534–541. doi: 10.1006/cyto.1995.0072. [DOI] [PubMed] [Google Scholar]
- 22.Reddi K, Nair S P, White P A, Hodges S, Tabona P, Meghji S, Poole S, Wilson M, Henderson B. Surface-associated material from the bacterium Actinobacillus actinomycetemcomitans contains a peptide which, in contrast to lipopolysaccharide, directly stimulates fibroblast interleukin-6 gene transcription. Eur J Biochem. 1996;236:871–876. doi: 10.1111/j.1432-1033.1996.00871.x. [DOI] [PubMed] [Google Scholar]
- 23.Saito S, Hosaka Y, Nakagawa T, Seida K, Yamada S, Takazoe I, Okuda K. Significance of serum antibody against surface antigens of Actinobacillus actinomycetemcomitans in patients with adult periodontitis. Oral Microbiol Immunol. 1993;8:146–153. doi: 10.1111/j.1399-302x.1993.tb00657.x. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 25.Sims T J, Moncla B J, Darveau R P, Page R C. Antigens of Actinobacillus actinomycetemcomitans recognized by patients with juvenile periodontitis and periodontally normal subjects. Infect Immun. 1991;59:913–924. doi: 10.1128/iai.59.3.913-924.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tolo K, Helgeland K. Fc-binding components: a virulence factor in Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 1991;6:373–377. doi: 10.1111/j.1399-302x.1991.tb00509.x. [DOI] [PubMed] [Google Scholar]
- 27.Vasfi-Marandi M, Mittal K R. Characterization of an outer membrane protein of Pasteurella multocida belonging to the OmpA family. Vet Microbiol. 1996;53:303–314. doi: 10.1016/s0378-1135(96)01219-9. [DOI] [PubMed] [Google Scholar]
- 28.Weiser J N, Gotschlich E C. Outer membrane protein A (OmpA) contributes to serum resistance and pathogenicity of Escherichia coli. Infect Immun. 1991;59:2252–2258. doi: 10.1128/iai.59.7.2252-2258.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White P A, Wilson M, Nair S P, Kirby A C, Reddi K, Henderson B. Characterization of an anti-proliferative surface-associated protein from Actinobacillus actinomycetemcomitans which can be neutralized by sera from a proportion of patients with localized juvenile periodontitis. Infect Immun. 1995;63:2612–2618. doi: 10.1128/iai.63.7.2612-2618.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson M, Henderson B. Virulence factors of Actinobacillus actinomycetemcomitans relevant to the pathogenesis of inflammatory periodontal disease. FEMS Microbiol Rev. 1995;17:365–379. doi: 10.1111/j.1574-6976.1995.tb00220.x. [DOI] [PubMed] [Google Scholar]
- 31.Wilson M E. The heat-modifiable outer membrane protein of Actinobacillus actinomycetemcomitans: relationship to OmpA proteins. Infect Immun. 1991;59:2505–2507. doi: 10.1128/iai.59.7.2505-2507.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson M E, Hamilton R G. Immunoglobulin G subclass response of juvenile periodontitis subjects to principle outer membrane proteins of Actinobacillus actinomycetemcomitans. Infect Immun. 1995;63:1062–1069. doi: 10.1128/iai.63.3.1062-1069.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zambon J J. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol. 1985;12:1–20. doi: 10.1111/j.1600-051x.1985.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 34.Zambon J J, Umemoto T, De Nardin E, Nakazawa F, Christersson L A, Genco R J. Actinobacillus actinomycetemcomitans in the pathogenesis of human periodontal disease. Adv Dent Res. 1988;2:269–274. doi: 10.1177/08959374880020021101. [DOI] [PubMed] [Google Scholar]