ABSTRACT
Toxin production by Clostridioides difficile damages the colonic epithelium and leads to a robust inflammatory response. This disruption of the epithelial barrier markedly alters the nutritional landscape in the C. difficile-infected gut. The impact of toxin-mediated nutritional remodeling during C. difficile infection (CDI) on resident microbiota remains largely unexplored. One group of opportunistic pathogens, the enterococci, thrive during CDI, but it is unclear what strategies they employ to survive in this altered environment. Here, we demonstrate that Enterococcus faecalis, a heme auxotroph, takes advantage of C. difficile toxin-mediated damage to acquire host heme for enhanced fitness. Specifically, heme acquired from the C. difficile-infected gut is used by E. faecalis to populate a heme-dependent cytochrome and aerobically respire. This fitness advantage is specific to C. difficile toxin-mediated damage, as infection with a toxin-null strain of C. difficile does not provide E. faecalis with a fitness advantage. Finally, targeted disruption of the E. faecalis cytochrome (cydABDC) operon leads to a fitness defect in the C. difficile-infected gut. Together, this work demonstrates that C. difficile toxin remodels the gut ecosystem and improves the fitness of E. faecalis in a cydABDC-dependent manner. These data further highlight growing evidence of a cooperative partnership between C. difficile and enterococci that has implications on susceptibility to and severity of CDI.
IMPORTANCE
Clostridioides difficile and Enterococcus faecalis are two pathogens of great public health importance. Both bacteria colonize the human gastrointestinal tract where they are known to interact in ways that worsen disease outcomes. We show that the damage associated with C. difficile infection (CDI) releases nutrients that benefit E. faecalis. One particular nutrient, heme, allows E. faecalis to use oxygen to generate energy and grow better in the gut. Understanding the mechanisms of these interspecies interactions could inform therapeutic strategies for CDI.
KEYWORDS: Clostridioides difficile, Enterococcus, heme, infectious disease, gut microbiome, microbial ecology
INTRODUCTION
Clostridioides difficile is a Gram-positive toxigenic bacterium that causes a wide range of gastrointestinal pathologies and represents an urgent public health threat (1). The primary toxins of C. difficile, TcdA and TcdB, disrupt and damage the host epithelium and reshape the metabolic and nutritional landscape of the gut (2, 3). The mechanisms by which C. difficile toxins cause damage to the colonic epithelium are well established (4); however, it is unclear how toxin-mediated damage and subsequent reshaping of the nutritional landscape impact gut-resident microbiota. Recent studies have demonstrated that C. difficile infection (CDI) leads to increased levels of heme in the intestinal lumen in both mice and humans (5, 6). Heme is an essential cofactor for a variety of cellular processes (7, 8). Paradoxically, heme is also toxic to cells, and bacteria must sense and respond to toxic levels of heme (9). C. difficile encodes a heme efflux pump for efficient detoxification and co-opts heme to help combat oxidative stress during infection (5, 10). The impact of C. difficile-mediated heme influxes on the resident gut microbiota is not known.
Enterococci are members of the microbiota that have been shown to thrive in the C. difficile-infected gut (11–16). They represent both common human gut commensals as well as opportunistic pathogens that pose a serious risk to public health (17). Intrinsic and acquired antibiotic resistance coupled with a diverse metabolic repertoire allow enterococcal outgrowth in the gut during microbial community perturbation (12, 18, 19). Enterococcal domination often precedes translocation across the intestinal epithelium, leading to bacteremia and infection of distal body sites (18, 20). Our work has shown that outgrowth of enterococci enhances the virulence of C. difficile during infection (13). This work suggested that enterococcal outgrowth plays a central role in clinical outcomes of CDI; however, we still lack an understanding of the factors that promote enterococcal fitness in the C. difficile-infected gut. Opportunistic pathogens have been shown to leverage metabolic shifts in the gastrointestinal tract to gain a fitness advantage over commensal organisms (21–28). Specifically, inflammation-associated shifts in nutrient availability allow facultative anaerobes to aerobically respire and enhance growth (26–28). Nearly all facultative anaerobes encode a complete heme biosynthetic pathway in order to generate their own heme for respiration (29), enabling heme-containing cytochromes to establish a proton gradient and reduce molecular oxygen. Interestingly, enterococci are heme auxotrophs (7, 30) and must acquire heme exogenously to perform heme-dependent processes. E. faecalis encodes several heme-dependent enzymes, including a cytochrome bd terminal oxidase (encoded by the cydABDC operon), a catalase (KatA), a heme efflux pump (HatBA), and a heme-dependent transcriptional regulator (FhtR) (31–34). If supplied with exogenous heme, E. faecalis can perform aerobic respiration in the presence of oxygen by incorporating heme cofactors into the cytochrome subunits CydA and CydB. CydD and CydC are necessary for the assembly of the CydAB complex and were recently implicated as the elusive heme importers in E. faecalis (33, 35, 36). In the related pathogen Streptococcus agalactiae, also a heme auxotroph, respiration through a cyd-encoded cytochrome oxidase provides a fitness advantage in human blood (37). However, a functional cydABDC operon seems to impair the fitness of E. faecalis in the bloodstream by sensitizing it to the oxidative burst of immune cells (34, 38). These findings suggest that the conserved cydABDC operon may be beneficial to E. faecalis in its native gastrointestinal tract environment but may be detrimental when the bacteria infect extraintestinal body sites.
CDI results in dysanaerobiosis at the epithelial barrier (39), but it remains unclear whether altered oxygen availability and toxin-dependent liberation of heme support a permissive environment for enterococci to perform aerobic respiration and thrive in the gut. In this study, we demonstrate that heme influx during CDI provides a fitness advantage to E. faecalis. This effect is dependent on C. difficile toxin and specific to CDI-associated inflammation. We propose that C. difficile-mediated damage supports polymicrobial cooperation with the enterococci that exacerbates disease caused by co-infection with these two pathogens.
RESULTS
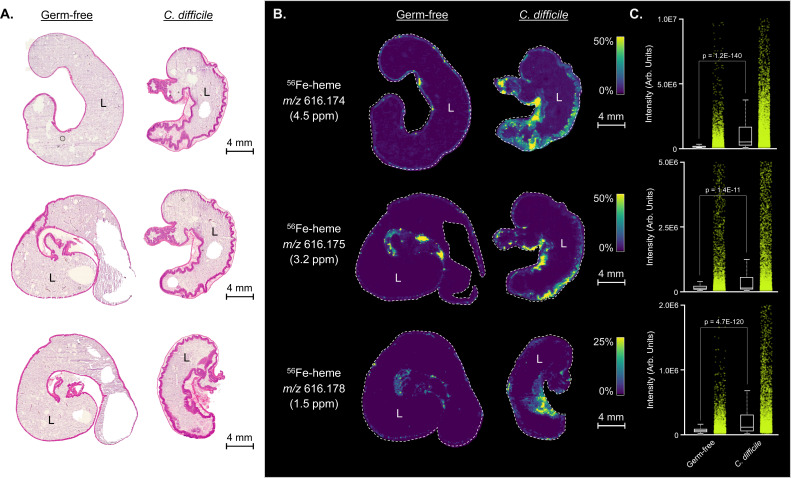
Damage and inflammation associated with CDI leads to a remodeling of the nutritional landscape in the gut. Specifically, heme is highly abundant in the stools of patients with CDI, and hemoglobin is enriched in the cecum of mice infected with C. difficile (5, 6). We sought to confirm that the influx of heme into the tissues and lumen of the gut was directly associated with C. difficile-mediated damage. Using germ-free mice infected with C. difficile, we spatially mapped heme with matrix-assisted laser desorption/ionization (MALDI) imaging mass spectrometry. As opposed to uninfected germ-free mice, mice mono-infected with C. difficile showed an influx of heme into the lumen and surrounding tissue during CDI (Fig. 1). These data demonstrate that C. difficile-mediated damage, independent of endogenous microbiota, increases available heme in the lumen of the gut.
Fig 1.
(A) Hematoxylin and eosin-stained cecal tissue sections from germ-free and C. difficile (CD196) mono-infected mice (n = 3 tissue replicates). L marks the lumen of the cecum. (B) Matched MALDI imaging mass spectrometry ion images of 56Fe-heme displaying heme localization to the mucosa of the infected intestinal tract. (C) Relative quantitation of MALDI imaging mass spectrometry 56Fe-heme intensities between germ-free and C. difficile (CD196) mono-infected mice.
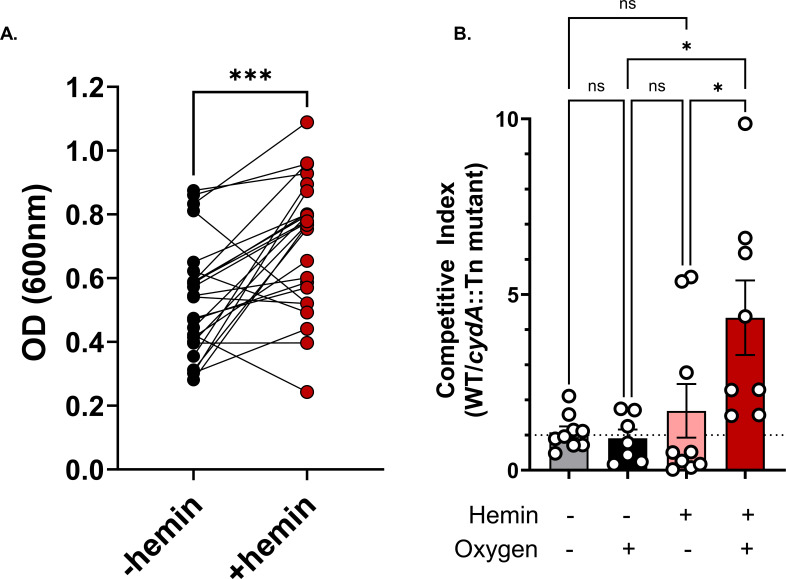
The role that heme plays in shaping the ecosystem and ecology of the gut microbiota during CDI has not been explored. Enterococci are of particular interest, as they play a critical role in shaping C. difficile behavior and virulence (13). We first sought to establish whether the capacity for heme utilization was prevalent in clinical strains of enterococci isolated from adult patients with CDI. We isolated 25 E. faecalis strains from stool samples collected from patients with CDI and grew them in aerobic culture with or without supplemental hemin. About 80% of these CDI-associated E. faecalis strains showed enhanced growth in the presence of hemin and oxygen (Fig. 2A). Next, we sought to determine if increased E. faecalis fitness in the presence of heme was dependent on the cydAB cytochrome. Using a transposon mutant with a disrupted cydA (OG1RF_11666) gene (cydA::Tn), which encodes the subunit containing two heme prosthetic groups, we performed in vitro competition assays with the wild-type OG1RF E. faecalis strain. These experiments demonstrated that cydA confers a fitness advantage for E. faecalis, but only in the presence of both hemin and oxygen (Fig. 2B). This confirms past reports that E. faecalis has the capacity to perform aerobic respiration when supplied with heme and oxygen—conditions experienced in the CDI gut (39).
Fig 2.
(A) E. faecalis isolate growth after overnight aerobic culture at 37°C in the absence (–hemin) or presence (+hemin) of 10 uM hemin. OD600 values are averaged from three replicate measurements and lines connect measurements from the same isolate. N = 25; significance was measured by a paired t-test, *** (P < 0.001). (B) Competitive index of wild-type E. faecalis OG1RF relative to E. faecalis cydA::Tn in vitro after 24 hours of growth in the indicated conditions as measured by selective CFU plating. Data are mean +/− SEM; n = 7. No hemin/aerobic, n = 8 hemin/aerobic, n = 9 for all other groups; Kruskal-Wallis test for significance. ns = not significant, * (P < 0.05).
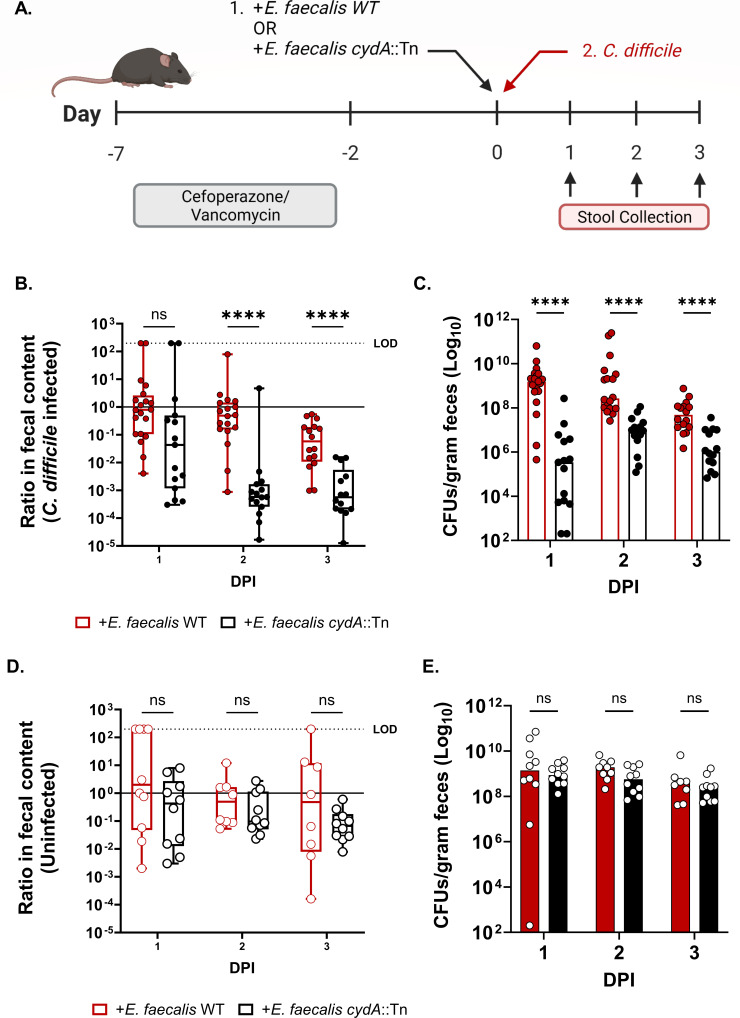
Next, to explore whether the cydABDC operon confers a fitness advantage to E. faecalis during CDI, we co-infected conventional mice with C. difficile and either wild-type E. faecalis or E. faecalis cydA::Tn. Mice were first transiently depleted of their endogenous enterococcal population using vancomycin prior to colonization with E. faecalis. Mice were then infected with C. difficile and the endogenous and exogenous populations of enterococci were tracked over the course of infection (Fig. 3A). Ratios of exogenous E. faecalis to endogenous enterococci were calculated to quantify the fitness of each exogenous strain. Wild-type E. faecalis significantly outperformed E. faecalis cydA::Tn during the course of CDI, with the cydA mutant being rapidly cleared from the CDI gut (Fig. 3B). The mutant strain was also present at a significantly lower absolute abundance during the time course of infection compared to the wild-type strain (Fig. 3C). Notably, CydA was not required for E. faecalis fitness in the absence of CDI, suggesting a specific role for this cytochrome during CDI (Fig. 3D and E). In either case, there was no significant difference in the endogenous enterococcal population between mice colonized with wild-type E. faecalis or E. faecalis cydA::Tn (Fig. S1C and D). In addition to the cytochrome, E. faecalis encodes a heme-dependent catalase (katA, OG1RF_11314). Surprisingly, a transposon mutant lacking catalase function (katA::Tn) does not exhibit a defect during CDI or in healthy mice (Fig. S1B and S2). Together, these data suggest that E. faecalis uses host heme to perform aerobic respiration in CDI gut and gains a competitive advantage against other members of the microbiota.
Fig 3.
(A) Schematic of C. difficile mouse infection model. (B) Ratio of E. faecalis OG1RF or E. faecalis cydA::Tn CFU burdens to endogenous enterococci CFU burdens in the stool of mice treated with cefoperazone and vancomycin and gavaged with each exogenous E. faecalis strain immediately prior to C. difficile infection (CDI), and (C) CFUs of each exogenously introduced E. faecalis strain in C. difficile-infected mice. Data are shown with median; n = 20 mice per day for CDI/E. faecalis WT; n = 15 mice per day for CDI/E. faecalis cydA::Tn. P-values are from multiple Mann-Whitney tests for significance with Bonferroni-Dunn correction for multiple comparisons. (D) Ratio of E. faecalis OG1RF or E. faecalis cydA::Tn CFU burdens to endogenous enterococci CFU burdens in mice similarly treated as above but without CDI, and (E) CFUs of each exogenously introduced E. faecalis strain in the uninfected mice. Data are shown with median; n = 10 per group per day for no CDI; P-values are from multiple Mann-Whitney test for significance with Bonferroni-Dunn correction for multiple comparisons. ns = not significant, **** (P < 0.0001). LOD = limit of detection. Box plots show minimum, maximum, median, and interquartile range. DPI = days post-infection.
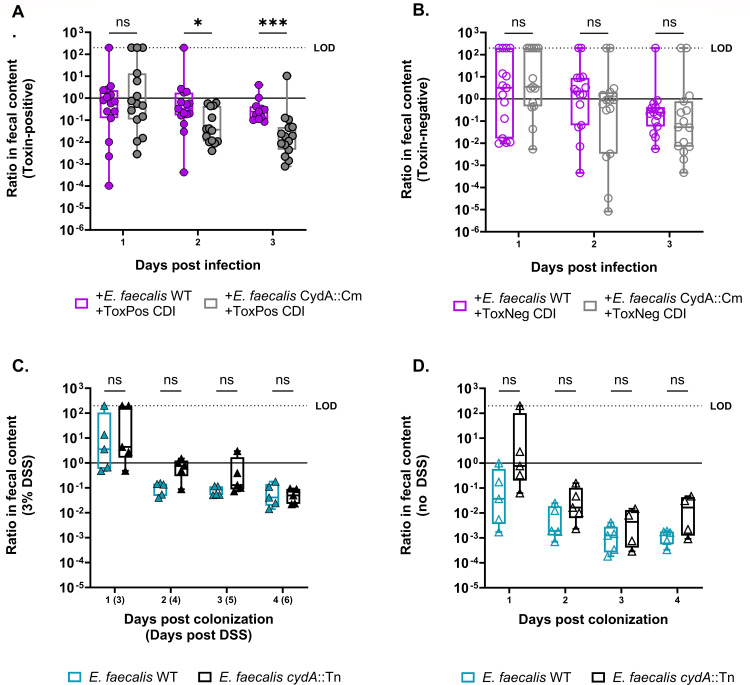
Next, to determine if this phenomenon was dependent on the action of the C. difficile toxins, we performed mouse infections with C. difficile strains incapable of producing toxin. Mice were infected with either a toxin-mutant strain of C. difficile M7404 (tcdA−tcdB−) or a toxin-positive strain (tcdA+tcdB+). M7404 and CD196 are ribotype 027 strains of C. difficile and induce a similar level of disease pathology during infection (Fig. S3). We found that the E. faecalis cydA::Tn mutant only suffered a fitness defect when the C. difficile toxin was present (Fig. 4A and B), suggesting that toxins are critical to liberating heme and enabling cydA-specific advantages to E. faecalis in the CDI gut. We hypothesized that other inflammatory conditions would release heme and cause a similar defect in the cyd mutant. We measured heme by ultra-high-performance liquid chromatography coupled with high-resolution mass spectrometry (UHPLC-HRMS) in the stool of mice infected with toxin-positive C. difficile or toxin-negative C. difficile, or mice treated with 3% dextran sodium sulfate (DSS). We found elevated levels of heme in both the toxin-positive CDI and DSS conditions (Fig. S4A). Notably, the advantage conferred by the E. faecalis cytochrome appears to be relatively specific to CDI-associated damage and inflammation as chemically induced colitis through DSS treatment did not lead to a fitness disadvantage for the cydA::Tn mutant (Fig. 4C and D). While aerobic respiration appears to play an important role for E. faecalis during CDI, these data suggest other factors may shape E. faecalis fitness in the context of chemically induced colitis. Interestingly, metabolomic data from human patients shows that fecal heme levels are significantly higher in children with IBD + CDI compared to IBD alone (Fig. S4B), suggesting that the role of heme during CDI is clinically relevant. Taken together, these results demonstrate that C. difficile toxin-mediated damage provides a fitness advantage for E. faecalis through heme liberation and that this phenomenon is not generalizable to all inflammatory conditions.
Fig 4.
(A) Ratio of E. faecalis OG1RF or E. faecalis cydA::Tn CFU burdens to endogenous enterococci CFU burdens in the stool of mice treated with cefoperazone and vancomycin and gavaged with each exogenous E. faecalis strain immediately prior to infection with C. difficile M7404 tcdA+tcdB + (toxin-positive) or (B) C. difficile M7404 tcdA−tcdB− (toxin-negative). Data are shown with median; n = 15 mice per group per day; multiple Mann-Whitney test for significance with Bonferroni-Dunn method for correction for multiple comparisons, * (P < 0.05), *** (P < 0.001). (C) Ratio of E. faecalis OG1RF or E. faecalis cydA::Tn CFU burdens to endogenous enterococci CFU burdens in the stool of mice treated with vancomycin and gavaged with each exogenous E. faecalis strain with 3% DSS treatment (D) or without. Data are shown with median; n = 5 mice per group per day; multiple Mann-Whitney test for significance with Bonferroni-Dunn method for correction for multiple comparisons. ns = not significant. LOD = limit of detection. Box plots show minimum, maximum, median, and interquartile range. DPI = days post-infection.
Finally, to specifically determine the conservation of heme utilization in gut isolates of enterococci from CDI patients, we analyzed the genomes of strains of E. faecalis described above. Whole-genome sequencing revealed a diverse repertoire of virulence factors, antibiotic resistance markers, and plasmids that varied widely between strains. This indicates that there is a diversity of genetic and phenotypic properties in the enterococcal populations across CDI patients. However, all strains that were sequenced encoded a fully intact cydABDC operon (Fig. S5), suggesting that the capacity to use heme as a resource in the CDI gut is widely conserved. These data demonstrate that the ability to use heme to gain a fitness advantage in the CDI gut is widely conserved across E. faecalis strains found in this perturbed ecosystem.
DISCUSSION
Of the many changes in the gut ecosystem observed during acute CDI, one of the most significant is the increase in opportunistic pathogens in the intestinal microbiome (40). One of the most clinically relevant and important members of the CDI-associated microbiota are the enterococci (13). The mechanisms by which enterococci, particularly E. faecalis, thrive in the CDI gut environment remain unexplored. In this study, we demonstrate that toxin production by C. difficile reshapes the gastrointestinal ecosystem and provides a relative fitness advantage to E. faecalis. Specifically, toxin-mediated damage liberates heme, which E. faecalis incorporates into its cytochrome to perform aerobic respiration. These observations provide an important mechanistic paradigm for the cooperative interactions between these two important and commonly co-occurring pathogens.
Bacterial pathogens leverage changes in the nutritional landscape of the large intestine during inflammation to gain a fitness advantage. For example, the Enterobacteriaceae, a family of commensals and opportunistic pathogens, outcompete commensal bacteria when inflammation and damage liberate alternative electron acceptors into the gut environment (22–24, 27). Specifically, Escherichia coli takes advantage of the altered nutrient pool to respire oxygen, which is known to be present in the gut during DSS treatment (26, 41). Moreover, both Citrobacter and Salmonella produce toxins that disrupt the intestinal epithelium and allow elevated levels of oxygen into the lumen that they can then use to respire (22–24, 27). CDI and toxin production are associated with the release of nutrients such as heme and amino acids into the lumen, as well as disruption of the hypoxic barrier that normally maintains strict anaerobicity in the large intestine (2, 5, 6, 13, 39).
The large intestine is anaerobic in healthy individuals, preventing aerobic respiration and thereby supporting anaerobes that promote homeostasis and host health. Under inflammatory conditions, epithelial damage leads to a PPARγ-dependent shift in colonocyte metabolism that allows molecular oxygen to diffuse into the lumen of the gut (41). Combined with oxygen delivered directly from an influx of red blood cells, the large intestine enters a state of dysanaerobiosis alongside the dysbiosis of the microbiota (24, 42). It has also been shown directly that damage and dysbiosis caused by the C. difficile toxins eliminate the hypoxic barrier in the epithelium and allow oxygen into the gut lumen where it can be utilized by respiring bacteria (39). Together this likely creates an environment conducive to expansion by E. faecalis, but only in the presence of heme (13).
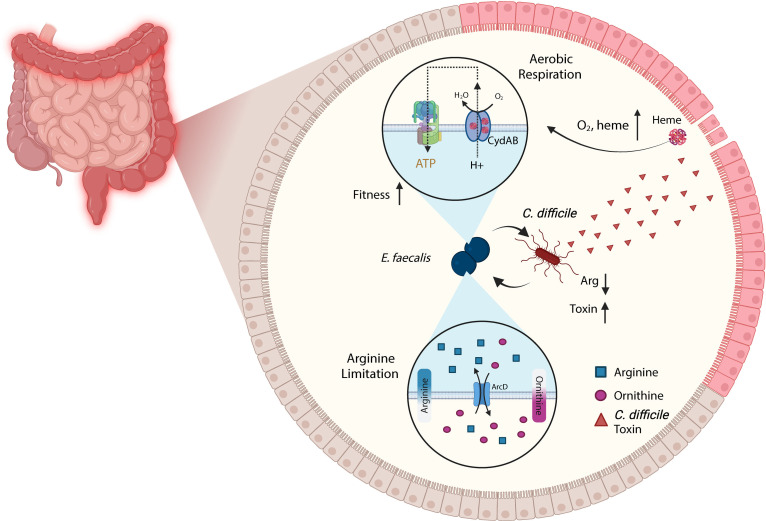
E. faecalis is relatively unique in that its capacity for aerobic respiration is limited by heme auxotrophy. As suggested in other studies and explicitly here, the C. difficile toxins are sufficient to release host heme into the gut lumen (5, 6). Enabling a fitness advantage to endogenous E. faecalis could lead to a host of complications during CDI. For example, we previously showed that enterococci exacerbate CDI by dramatically remodeling the metabolic environment in the gut and consuming luminal arginine (Fig. 5) (13). This acts as a reversible cue to C. difficile to increase toxin production and ultimately leads to increased pathogenesis. Additionally, E. faecalis and C. difficile form robust interspecies biofilms that are protected from antibiotic treatment and could act as a reservoir for persistence and recurrence (13). Moreover, heme utilization in these tight associations enables metronidazole resistance in C. difficile (43, 44). In each case, the ability of C. difficile to support the fitness of E. faecalis could further enhance these pathogenic phenotypes. Staphylococcus aureus and E. coli have been shown to directly cross-feed heme to E. faecalis and bolster its growth in vitro (31, 36). In fact, prior reports showing that respiration actually handicaps E. faecalis within the hemoglobin-rich environment of the bloodstream, along with interspecies biofilms allowing for the robust exchange of heme, support the idea that this functionality evolved for use in the polymicrobial environment of the gut (36, 38).
Fig 5.
Model schematizing known interactions between Enterococcus faecalis and Clostridioides difficile. As described in this study, C. difficile toxins cause damage to the host that releases heme and oxygen into the lumen. E. faecalis uses heme and oxygen to respire and thrive through the cydABDC operon (top). Previous work describes a relationship wherein E. faecalis consumes and limits the luminal arginine pool. This arginine restriction acts as a reversible cue that causes C. difficile to increase toxin production, forming a positive ecological feedback loop that exacerbates disease severity (bottom).
The precise origin of heme utilized by E. faecalis in the context of CDI remains to be discovered. E. faecalis can directly import heme or first degrade hemoglobin and other hemoproteins using its encoded gelatinase (31, 36). However, the ability of E. faecalis to derive heme cofactors from erythrocytes varies across isolates. Hemolysis relies on the cytolysin encoded by the cyl operon (45, 46). Since the strain used in this study, OG1RF, does not encode the cytolysin, it cannot lyse erythrocytes and access heme without assistance from C. difficile. However, clinical isolates of E. faecalis variably encode the cytolysin (Fig. S5). Therefore, the genotype of E. faecalis might impact its ability to use heme and could determine the ecological scenarios under which E. faecalis gains a fitness advantage in vivo.
Though the nutritional requirements to perform aerobic respiration are similarly met in the DSS-treated mice, E. faecalis does not suffer a disadvantage from losing cydA. This would imply that E. faecalis may prioritize other modes of metabolism in this environment. The nutritional state of the gut changes dramatically after DSS treatment (47); however, these data demonstrate that different drivers of inflammation can have distinct impacts on the ecology of the microbiome. The complex relationship between the gut environment and E. faecalis fitness warrants further study.
In our study we did not observe a defect in a KatA-deficient strain of E. faecalis during CDI. E. faecalis encodes three heme-independent peroxidases that could compensate for the activity of KatA (39, 48, 49). It is possible that the two major heme-dependent complexes of E. faecalis, CydABDC and KatA, also synergize during CDI to provide a fitness advantage. During infection, reactive oxygen species (ROS) are produced at the epithelial interface by the host NADPH oxidase NOX1 as a defense mechanism against invading pathogens. E. coli was shown to convert these ROS into molecular oxygen via its catalases and then undergo respiration through a cytochrome bd oxidase (28). With heme supplied by the host during CDI, we suspect that E. faecalis could similarly convert ROS from NOX1/2 via KatA into oxygen for respiration via CydABDC. Moreover, the action of the catalase may not directly benefit E. faecalis, but could provide cross-protection to other microbes that benefit E. faecalis ecologically (50). Further studies are required to fully understand the tripartite interactions between these two pathogens and the host as well as the role of heme in the oxidative stress response of E. faecalis.
The work described in this study details an indirect route by which C. difficile is able to cross-feed heme to E. faecalis. This is also the first reported instance of the cydABDC operon boosting E. faecalis fitness within its host. Even though aerobic respiration seems to impair E. faecalis in the bloodstream (34), the ability to grow to high levels in the gut could potentially allow translocation of heterogenous strains, thereby increasing the risk of enterococcal complications such as bacteremia during CDI. Further studies are required to understand the consequences of this fitness advantage during CDI as well as infections or conditions beyond CDI that enable E. faecalis in this manner. Additionally, this study establishes that the action of the C. difficile toxins remodels the gut environment and provides altered nutrient niches for members of the gut microbiota, opening a broad avenue of ecological interactions during infection. Understanding the ecology between these two pathogens is critical to the development of effective treatment strategies for patients suffering from CDI.
MATERIALS AND METHODS
Bacterial strains and growth conditions
C. difficile and E. faecalis strains were grown at 37°C in an anaerobic chamber (90% nitrogen, 5% hydrogen, 5% carbon dioxide; Coy Lab Products) or aerobically in brain-heart-infusion broth (BD Life Sciences) supplemented with 0.5% yeast extract (BD Life Sciences) and 0.1% L-cysteine (Sigma-Aldrich) (BHIS) unless otherwise stated. Media was supplemented with 50 µM hemin (Sigma-Aldrich) when noted. Strains are listed in Table S1. All strains were confirmed by whole genome sequencing. The location of the transposon was validated in each transposon mutant used in the study, and we confirm no significant off-target mutations (Fig. S1A).
In vitro competition assay
Wild-type E. faecalis and E. faecalis cydA::Tn overnight cultures were mixed at a 1:1 ratio and subcultured at a 1:100 ratio into fresh BHIS media. Mixed cultures were grown under the indicated conditions for 24 hours. Abundance of each strain was quantified as colony-forming units (CFUs) on BHIS agar and BHIS + chloramphenicol (10 µg/mL) agar. The ratio of output was normalized to the ratio of inoculum to quantify competitive index.
Animal models of infection
Animal experiments were approved by the Animal Care and Use Committees of the Children’s Hospital of Philadelphia (IAC 18–001316). For CDIs in conventional facilities, 4- to 8-week-old C57BL/six male mice were purchased from Jackson Laboratories and given one week to equilibrate their microbiota prior to experimentation. All experimental manipulations were performed in a biosafety level two laminar flow hood. Mice were housed in individual cages under the same conditions during the experiment, and all mice were culture-negative for C. difficile prior to infection. For all CDIs, mice were given antibiotics (0.5 mg/mL cefoperazone + 1 mg/mL vancomycin) in drinking water ad libitum for 5 days followed by a 2-day recovery period and subsequent infection. Mice were confirmed culture-negative for endogenous enterococci after vancomycin treatment via selective plating as described below. Mice were infected with 5 × 108 E. faecalis (wild-type strain OG1RF, cydA::Tn, or katA::Tn) cells. E. faecalis cells were grown to stationary phase, washed in cold PBS prior to infection, and orally gavaged. Mice were subsequently co-infected via oral gavage with 1 × 105 spores of C. difficile resuspended in sterile PBS. C. difficile strains CD196, M7404, and M7404 TcdA− TcdB− were used for conventional infections, as described in the text. Mice were monitored for survival and were euthanized after reaching a terminal endpoint of appearing moribund or experiencing weight loss >20% from baseline. C. difficile and enterococcal CFUs were quantified daily from fecal samples. All samples were collected, and all stool-related data are reported, unless animals were too sick to acquire a fresh stool sample. Samples were diluted and homogenized in PBS and serially plated onto taurocholate cycloserine cefoxitin fructose agar (TCCFA) for C. difficile and Bile Esculin agar for total enterococci. E. faecalis OG1RF strains were grown on Bile Esculin agar with rifampicin (200 µg/mL).
For infections involving dextran sodium sulfate (DSS), mice were given vancomycin (1 mg/mL) in drinking water ad libitum for 5 days followed immediately by 3% DSS in drinking water ad libitum for 2 days. Mice were infected by oral gavage with 5 × 108 E. faecalis (wild-type strain OG1RF or cydA::Tn) cells as described above. Mice were continued on the course of DSS throughout the infection and were similarly monitored for survival. Enterococcal CFUs were quantified daily from fecal samples as described above.
Imaging mass spectrometry
Tissue samples to be analyzed by MALDI imaging mass spectrometry were embedded in a 20% mixture (vol/vol) of optimal cutting temperature (OCT) compound and water, shipped to the University of Florida on dry ice, and stored at −80°C until analysis. Tissue sections were prepared at 12 µm thickness using a Leica CM 3050S Research Cryostat (Leica Biosystems) (−30°C object temperature, −28°C chamber temperature) and thaw-mounted onto indium tin oxide-coated microscope slides. Samples to be compared via imaging mass spectrometry were mounted on the same microscope slide to ensure identical sample preparation and facilitate accurate analyte comparisons between tissue types. Slides with mounted tissue sections were then warmed to room temperature in a desiccator for ~30 minutes before application of a 2,5-dihydroxybenzoic acid (DHB; Sigma Aldrich) MALDI matrix solution (50% methanol, 0.1% TFA) using a TM-sprayer (HTX Technologies) (51). The robotic spraying conditions were velocity 1,200 mm/minutes, flow rate 0.1 mL/minutes, spray temperature 85°C, heated tray temperature, 38°C, number of passes 6, track spacing 3 mm, and sheath gas pressure 10 psi.
All imaging mass spectrometry experiments were performed in positive ion mode on a 7T Fourier transform ion cyclotron resonance (FT-ICR) solariX mass spectrometer equipped with a dynamically harmonized ParaCell (Bruker Daltonics). The instrument contained an Apollo II dual MALDI/ESI source that uses a Smartbeam II Nd:YAG MALDI laser (2 kHz, 355 nm). Images were acquired at a pixel spacing of 125 µm in both the x and y dimensions using a ∼25-µm laser beam and a 110-µm Smart Walk (300 laser shots). Tissue data were collected from m/z 200 to 1,200 using a 0.9787 s time-domain transient length, resulting in a resolving power of ∼87,000 (FWHM at m/z 598). Internal calibration using a quadratic fit was performed using common endogenous lipid ions. Ion image distributions and intensity box plots were visualized using SciLS Lab software (Bruker Daltonics). Ion images are displayed without normalization and using pixel interpolation. Ion intensity box plots were generated by extracting .imzML files and converted to .csv format using python version 3.10.9. The statistical analysis was performed using the SciPy module in Python version 3.10.9 (52). P-value was calculated by comparing the t-statistic of the ion intensity data against a theoretical t-distribution. Following image acquisition, serial tissue sections were stained using hematoxylin and eosin (H&E), bright field scanned using a Axio Imager M2 Microscope (Carl Zeiss Microscopy), and visualized using Zen microscopy software (Carl Zeiss Microscopy).
Ultra-high-performance liquid chromatography coupled with high-resolution mass spectrometry
Mouse stool was homogenized in cold PBS, and the supernatant was filter sterilized using a 0.2-µm filter. The sterile supernatant was extracted utilizing a neutral acetone extraction (53). All solvents used for sample preparation were Fisher HPLC grade. Briefly, 800 µL of ice-cold acetone was added to 100 µL of sterile supernatant. The sample and acetone mixtures were vortexed and allowed to rest on ice for 15 minutes, vortexed again, and allowed to rest for another 15 minutes for a total of 30 minutes of rest. The mixture was then centrifuged for 10 minutes at 4°C and 3,260 × g to pellet the proteins. Eight hundred microliter of the supernatant was taken and dried in a microcentrifuge tube under a nitrogen gas stream at room temperature. Dried supernatants were then reconstituted using 100 µL of 25% methanol/75% 0.1% formic acid water and allowed to solubilize for 30 minutes, with vortexing every 15 minutes.
Samples and blanks were analyzed on a UHPLC-HRMS system consisting of a Dionex UltiMate 3000 UHPLC (Thermo Fisher Scientific) coupled to a Q Exactive Orbitrap Mass Spectrometer (Thermo Fisher Scientific). An Avantor ACE C18-PFP column (Catalogue number: EXL-1010–1002U) placed in a column oven set to 50°C was used for separation with a gradient elution method using water with 0.1% formic acid (FA) and methanol as mobile phases A and B, respectively. All solvents used for LC separations were Fisher Optima grade. All analyses were acquired in positive electrospray mode (Table S2).
Inclusion of integrated peak values into the post-processing data analysis was contingent on the following criteria being met: matching peak retention time (within 0.05 minutes of chemical standard), matching the exact mass of the most abundant isotope (5 ppm error 616.1767 ± 0.0031 with respect to theoretical), at least eight scans across the peak at full width at half max (FWHM), and visually matching isotopic distribution with respect to theoretical spectrum. Data analysis was performed using Qual Browser from Thermo Xcalibur (4.3.73.11). Integration was performed using boxcar smoothing with five points, a mass tolerance of 10 ppm, and valley detections with an expected width of 25 seconds.
Human samples
Children’s Hospital of Philadelphia
Subjects were recruited at the Children’s Hospital of Philadelphia (CHOP) from September 2015 to April 2018, and informed consent was acquired (IRB approval number 15-011817), as previously described (6). Groups included healthy children (HC), children with IBD (IBD), and children with IBD and concurrent CDI (IBD + CDI). Healthy children were age matched to those with IBD + CDI. Inclusion and exclusion criteria were described previously (6, 13). Untargeted metabolomics on the stool samples from these patients was performed as previously described (6). No images of human subjects are included in the figures, extended data, or supplementary materials.
University of Pittsburgh Medical Center
Adult patients with toxin-producing CDI were identified through the Enhanced Detection System for Healthcare-Associated Transmission (EDS-HAT) project (54). The Institutional Review Board of the University of Pittsburgh gave ethical approval for this work under Protocol STUDY21040126. Stool samples were stored at −80°C. Approximately 10 uL of each stool sample was thawed and streaked onto bile esculin azide (BEA) agar, and plates were incubated at 30°C for 48 hours. Individual colonies were isolated from plates that showed growth of presumed enterococci (i.e., white, gray, or black colonies that turned the agar underneath them black). Colonies were restreaked onto BEA agar before being stored at −80°C in tryptic soy broth with 16% glycerol.
Whole genome sequencing and genomic analyses
Transposon mutant confirmation
Genomic DNA was extracted from the E. faecalis isolates using the Qiagen DNeasy Blood and Tissue kit. Sequencing library preparation and sequencing on the Illumina platform were performed by the CHOP Microbiome Center Sequencing Core. The whole-genome Average Nucleotide Identity was calculated, and the conserved regions between wild-type genome and cydA transposon genome were visualized using FastANI v1.34 (55). The genome alignment between wild-type and cydA transposon genomes was analyzed using mummer4 (56). The genomes were annotated through Bakta v1.8.2 (57) and SNPs between genomes were called using Snippy v4.6.0 (58). Mauve v2.4.0 was used to arrange the contigs of the genomes using complete E. faecalis OG1RF genome assembly (accession number: NC_017316) as the reference (59). The genome alignment was visualized using BLAST Ring Image Generator v0.95 (60). R package gggenes was used to better visualize the insertion of transposon in the genes (61).
Virulence factor analysis
Genome DNA was extracted from E. faecalis isolates using a Qiagen Dneasy Blood and Tissue Kit. Sequencing library preparation and sequencing on the Illumina platform were performed by SeqCenter, LLC, in Pittsburgh, PA. Genomes were de novo assembled using CLC Genomics Workbench v11.0.1. Fasta files of assembled genomes were analyzed using ResFinder and VirulenceFinder via the Center for Genomic Epidemiology (62–66).
Isolate growth analysis
E. faecalis isolates were grown overnight in brain heart infusion (BHI) media and were then diluted 1:1,000 into fresh BHI or BHI supplemented with 10 μM hemin. Following overnight aerobic growth at 37°C with shaking at 170 rpm, the optical density at 600 nm (OD600) was measured for each culture. Three replicates of each isolate were tested, and OD600 values were background subtracted and averaged across replicates.
ACKNOWLEDGMENTS
We thank the members of the Zackular laboratory for support and critical feedback on this manuscript. We thank the Gary Dunny laboratory for support and providing access to numerous resources, including the Enterococcus faecalis OG1RF transposon library. Some figures were created using BioRender.com using a purchased license.
Funding was from National Institutes of Health grant K22AI7220 (J.P.Z.), National Institutes of Health grant R35GM138369 (J.P.Z.), National Institutes of Health grant U19AI174998 (J.P.Z. and B.M.P.), National Institutes of Health grant R21AI164018 (D.V.T.), National Institutes of Health grant R03AI168491 (D.V.T.), and Young Investigator Award from Eli Lilly and Company (B.M.P.).
A.B.S and J.P.Z. conceived the study. A.B.S. designed and performed the experimentation and animal work with support from K.K.H., A.M.L., Y.L., and K.M.E.. B.M.P., J.T.S., T.R.S., M.W.C., T.J.G., and Y.G. performed the imaging mass spectrometry, UHPLC-HRMS, and analyses. D.V.T. performed phylogenetic analyses and provided experimental support. Q.S. and A.M.M. performed whole-genome sequencing and alignment. Funding was acquired by J.P.Z., B.M.P., and D.V.T. Writing was performed by A.B.S. and J.P.Z., with input from the other authors. J.P.Z. supervised the research.
Contributor Information
Joseph P. Zackular, Email: joseph.zackular@pennmedicine.upenn.edu.
Kimberly A. Kline, Universite de Geneve, Geneva, Switzerland
DATA AVAILABILITY
WGS data for E. faecalis isolates have been deposited in NCBI under BioProject PRJNA996476 with accession numbers listed in Table S1. WGS data for E. faecalis transposon mutants have been deposited in NCBI under BioProject PRJNA1039837.
ETHICS APPROVAL
The Institutional Review Board of the University of Pittsburgh gave ethical approval for this work under Protocol STUDY21040126.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.01656-23.
Circle maps.
Ratio of E. faecalis OG1RF or E. faecalis katA::Tn CFU burdens to endogenous enterococcus CFU burdens.
Weight loss and clinical sickness scores of infected mice.
Relative abundance of heme in stools.
Core genome phylogeny of 25 E. faecalis isolates collected from CDI patient stools.
Supplemental figure legends.
Strains.
Liquid chromatography and mass spectrometry parameters used in the analysis of heme.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Lessa FC, Winston LG, McDonald LC, Emerging Infections Program C. difficile Surveillance Team . 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:2369–2370. doi: 10.1056/NEJMc1505190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fletcher JR, Pike CM, Parsons RJ, Rivera AJ, Foley MH, McLaren MR, Montgomery SA, Theriot CM. 2021. Clostridioides difficile exploits toxin-mediated inflammation to alter the host nutritional landscape and exclude competitors from the gut microbiota. Nat Commun 12:462. doi: 10.1038/s41467-020-20746-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chumbler NM, Farrow MA, Lapierre LA, Franklin JL, Lacy DB. 2016. Clostridium difficile toxins TcdA and TcdB cause colonic tissue damage by distinct mechanisms. Infect Immun 84:2871–2877. doi: 10.1128/IAI.00583-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kordus SL, Thomas AK, Lacy DB. 2022. Clostridioides difficile toxins: mechanisms of action and antitoxin therapeutics. Nat Rev Microbiol 20:285–298. doi: 10.1038/s41579-021-00660-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Knippel RJ, Zackular JP, Moore JL, Celis AI, Weiss A, Washington MK, DuBois JL, Caprioli RM, Skaar EP. 2018. Heme sensing and detoxification by HatRT contributes to pathogenesis during Clostridium difficile infection. PLoS Pathog 14:e1007486. doi: 10.1371/journal.ppat.1007486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bushman FD, Conrad M, Ren Y, Zhao C, Gu C, Petucci C, Kim M-S, Abbas A, Downes KJ, Devas N, et al. 2020. Multi-omic analysis of the interaction between Clostridioides difficile infection and pediatric inflammatory bowel disease. Cell Host Microbe 28:422–433. doi: 10.1016/j.chom.2020.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baureder M, Hederstedt L. 2013. Heme proteins in lactic acid bacteria. Adv Microb Physiol 62:1–43. doi: 10.1016/B978-0-12-410515-7.00001-9 [DOI] [PubMed] [Google Scholar]
- 8. Shimizu T, Lengalova A, Martínek V, Martínková M. 2019. Heme: emergent roles of heme in signal transduction, functional regulation and as catalytic centres. Chem Soc Rev 48:5624–5657. doi: 10.1039/c9cs00268e [DOI] [PubMed] [Google Scholar]
- 9. Anzaldi LL, Skaar EP. 2010. Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infect Immun 78:4977–4989. doi: 10.1128/IAI.00613-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Knippel RJ, Wexler AG, Miller JM, Beavers WN, Weiss A, de Crécy-Lagard V, Edmonds KA, Giedroc DP, Skaar EP. 2020. Clostridioides difficile senses and hijacks host heme for incorporation into an oxidative stress defense system. Cell Host Microbe 28:411–421. doi: 10.1016/j.chom.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roghmann MC, McCarter RJ, Brewrink J, Cross AS, Morris Jr JG. 1997. Clostridium difficile infection is a risk factor for bacteremia due to vancomycin-resistant enterococci (VRE) in VRE-colonized patients with acute leukemia. Clin Infect Dis 25:1056–1059. doi: 10.1086/516112 [DOI] [PubMed] [Google Scholar]
- 12. Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MRM, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smith AB, Jenior ML, Keenan O, Hart JL, Specker J, Abbas A, Rangel PC, Di C, Green J, Bustin KA, et al. 2022. Enterococci enhance Clostridioides difficile pathogenesis. Nature 611:780–786. doi: 10.1038/s41586-022-05438-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zackular JP, Moore JL, Jordan AT, Juttukonda LJ, Noto MJ, Nicholson MR, Crews JD, Semler MW, Zhang Y, Ware LB, Washington MK, Chazin WJ, Caprioli RM, Skaar EP. 2016. Dietary zinc alters the microbiota and decreases resistance to Clostridium difficile infection. Nat Med 22:1330–1334. doi: 10.1038/nm1216-1502d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fujitani S, George WL, Morgan MA, Nichols S, Murthy AR. 2011. Implications for vancomycin-resistant Enterococcus colonization associated with Clostridium difficile infections. Am J Infect Control 39:188–193. doi: 10.1016/j.ajic.2010.10.024 [DOI] [PubMed] [Google Scholar]
- 16. Lesniak NA, Schubert AM, Flynn KJ, Leslie JL, Sinani H, Bergin IL, Young VB, Schloss PD, Blaser MJ. 2022. The gut bacterial community potentiates Clostridioides difficile infection severity. mBio 13:e0118322. doi: 10.1128/mbio.01183-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Van Tyne D, Gilmore MS. 2014. Friend turned foe: evolution of enterococcal virulence and antibiotic resistance. Annu Rev Microbiol 68:337–356. doi: 10.1146/annurev-micro-091213-113003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MRM, Kamboj M, Pamer EG. 2010. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 120:4332–4341. doi: 10.1172/JCI43918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Keith JW, Dong Q, Sorbara MT, Becattini S, Sia JK, Gjonbalaj M, Seok R, Leiner IM, Littmann ER, Pamer EG. 2020. Impact of antibiotic-resistant bacteria on immune activation and Clostridioides difficile infection in the mouse intestine. Infect Immun 88:e00362-19. doi: 10.1128/IAI.00362-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chakraborty R, Lam V, Kommineni S, Stromich J, Hayward M, Kristich CJ, Salzman NH. 2018. Ceftriaxone administration disrupts intestinal homeostasis, mediating noninflammatory proliferation and dissemination of commensal enterococci. Infect Immun 86:e00674-18. doi: 10.1128/IAI.00674-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rivera-Chávez F, Mekalanos JJ. 2019. Cholera toxin promotes pathogen acquisition of host-derived nutrients. Nature 572:244–248. doi: 10.1038/s41586-019-1453-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rivera-Chávez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, Xu G, Velazquez EM, Lebrilla CB, Winter SE, Bäumler AJ. 2016. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe 19:443–454. doi: 10.1016/j.chom.2016.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Bäumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. doi: 10.1038/nature09415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2:204. doi: 10.1016/j.chom.2007.08.002 [DOI] [PubMed] [Google Scholar]
- 25. Cevallos SA, Lee J-Y, Tiffany CR, Byndloss AJ, Johnston L, Byndloss MX, Bäumler AJ. 2019. Increased epithelial oxygenation links colitis to an expansion of tumorigenic bacteria. mBio 10:e02244-19. doi: 10.1128/mBio.02244-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hughes ER, Winter MG, Duerkop BA, Spiga L, Furtado de Carvalho T, Zhu W, Gillis CC, Büttner L, Smoot MP, Behrendt CL, Cherry S, Santos RL, Hooper LV, Winter SE. 2017. Microbial respiration and formate oxidation as metabolic signatures of inflammation-associated dysbiosis. Cell Host Microbe 21:208–219. doi: 10.1016/j.chom.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lopez CA, Miller BM, Rivera-Chávez F, Velazquez EM, Byndloss MX, Chávez-Arroyo A, Lokken KL, Tsolis RM, Winter SE, Bäumler AJ. 2016. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science 353:1249–1253. doi: 10.1126/science.aag3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chanin RB, Winter MG, Spiga L, Hughes ER, Zhu W, Taylor SJ, Arenales A, Gillis CC, Büttner L, Jimenez AG, Smoot MP, Santos RL, Winter SE. 2020. Epithelial-derived reactive oxygen species enable AppBCX-mediated aerobic respiration of Escherichia coli during intestinal inflammation. Cell Host Microbe 28:780–788. doi: 10.1016/j.chom.2020.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dailey HA, Dailey TA, Gerdes S, Jahn D, Jahn M, O’Brian MR, Warren MJ. 2017. Prokaryotic heme biosynthesis: multiple pathways to a common essential product. Microbiol Mol Biol Rev 81:e00048-16. doi: 10.1128/MMBR.00048-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fiore E, Van Tyne D, Gilmore MS. 2019. Pathogenicity of enterococci. Microbiol Spectr 7. doi: 10.1128/microbiolspec.GPP3-0053-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saillant V, Lipuma D, Ostyn E, Joubert L, Boussac A, Guerin H, Brandelet G, Arnoux P, Lechardeur D. 2021. A novel Enterococcus faecalis heme transport regulator (FhtR) senses host heme to control its intracellular homeostasis. mBio 12:e03392-20. doi: 10.1128/mBio.03392-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Frankenberg L, Brugna M, Hederstedt L. 2002. Enterococcus faecalis heme-dependent catalase. J Bacteriol 184:6351–6356. doi: 10.1128/JB.184.22.6351-6356.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Winstedt L, Frankenberg L, Hederstedt L, von Wachenfeldt C. 2000. Enterococcus faecalis V583 contains a cytochrome bd-type respiratory oxidase. J Bacteriol 182:3863–3866. doi: 10.1128/JB.182.13.3863-3866.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Van Tyne D, Manson AL, Huycke MM, Karanicolas J, Earl AM, Gilmore MS. 2019. Impact of antibiotic treatment and host innate immune pressure on enterococcal adaptation in the human bloodstream. Sci Transl Med 11:eaat8418. doi: 10.1126/scitranslmed.aat8418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramsey M, Hartke A, Huycke M. 2014. Edited by Gilmore M. S., Clewell D. B., Ike Y., and Shankar N. Enterococci: from commensals to leading causes of drug resistant infection [PubMed] [Google Scholar]
- 36. Ch’ng J-H, Muthu M, Chong KKL, Wong JJ, Tan CAZ, Koh ZJS, Lopez D, Matysik A, Nair ZJ, Barkham T, Wang Y, Kline KA. 2022. Heme cross-feeding can augment Staphylococcus aureus and Enterococcus faecalis dual species biofilms. ISME J 16:2015–2026. doi: 10.1038/s41396-022-01248-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yamamoto Y, Poyart C, Trieu-Cuot P, Lamberet G, Gruss A, Gaudu P. 2005. Respiration metabolism of Group B Streptococcus is activated by environmental haem and quinone and contributes to virulence. Mol Microbiol 56:525–534. doi: 10.1111/j.1365-2958.2005.04555.x [DOI] [PubMed] [Google Scholar]
- 38. Painter KL, Hall A, Ha KP, Edwards AM. 2017. The electron transport chain sensitizes Staphylococcus aureus and Enterococcus faecalis to the oxidative burst. Infect Immun 85:e00659-17. doi: 10.1128/IAI.00659-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weiss A, Lopez CA, Beavers WN, Rodriguez J, Skaar EP. 2021. Clostridioides difficile strain-dependent and strain-independent adaptations to a microaerobic environment. Microb Genom 7:000738. doi: 10.1099/mgen.0.000738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Antharam VC, Li EC, Ishmael A, Sharma A, Mai V, Rand KH, Wang GP. 2013. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J Clin Microbiol 51:2884–2892. doi: 10.1128/JCM.00845-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Byndloss MX, Olsan EE, Rivera-Chávez F, Tiffany CR, Cevallos SA, Lokken KL, Torres TP, Byndloss AJ, Faber F, Gao Y, Litvak Y, Lopez CA, Xu G, Napoli E, Giulivi C, Tsolis RM, Revzin A, Lebrilla CB, Bäumler AJ. 2017. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357:570–575. doi: 10.1126/science.aam9949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rigottier-Gois L. 2013. Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. ISME J 7:1256–1261. doi: 10.1038/ismej.2013.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boekhoud IM, Sidorov I, Nooij S, Harmanus C, Bos-Sanders I, Viprey V, Spittal W, Clark E, Davies K, Freeman J, Kuijper EJ, Smits WK, COMBACTE-CDI Consortium . 2021. Haem is crucial for medium-dependent metronidazole resistance in clinical isolates of Clostridioides difficile. J Antimicrob Chemother 76:1731–1740. doi: 10.1093/jac/dkab097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu X, Shen W-J, Deshpande A, Olaitan AO, Palmer KL, Garey KW, Hurdle JG. 2021. The integrity of heme is essential for reproducible detection of metronidazole-resistant Clostridioides difficile by agar dilution susceptibility tests. J Clin Microbiol 59:e0058521. doi: 10.1128/JCM.00585-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Van Tyne D, Martin MJ, Gilmore MS. 2013. Structure, function, and biology of the Enterococcus faecalis cytolysin. Toxins (Basel) 5:895–911. doi: 10.3390/toxins5050895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakagawa S, Matsuo Y. 1981. Bacteriocin and hemolysin from Streptococcus faecium. Antimicrob Agents Chemother 20:542–544. doi: 10.1128/AAC.20.4.542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Osaka T, Moriyama E, Arai S, Date Y, Yagi J, Kikuchi J, Tsuneda S. 2017. Meta-analysis of fecal microbiota and metabolites in experimental colitic mice during the inflammatory and healing phases. Nutrients 9:1329. doi: 10.3390/nu9121329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. La Carbona S, Sauvageot N, Giard J-C, Benachour A, Posteraro B, Auffray Y, Sanguinetti M, Hartke A. 2007. Comparative study of the physiological roles of three peroxidases (NADH peroxidase, Alkyl hydroperoxide reductase and Thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis. Mol Microbiol 66:1148–1163. doi: 10.1111/j.1365-2958.2007.05987.x [DOI] [PubMed] [Google Scholar]
- 49. Baureder M, Reimann R, Hederstedt L. 2012. Contribution of catalase to hydrogen peroxide resistance in Enterococcus faecalis. FEMS Microbiol Lett 331:160–164. doi: 10.1111/j.1574-6968.2012.02567.x [DOI] [PubMed] [Google Scholar]
- 50. Rodríguez E, Peirotén Á, Landete JM, Medina M, Arqués JL. 2015. Gut catalase-positive bacteria cross-protect adjacent bifidobacteria from oxidative stress. Microbes Environ 30:270–272. doi: 10.1264/jsme2.ME15025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kihara M, Matsuo-Tezuka Y, Noguchi-Sasaki M, Yorozu K, Kurasawa M, Shimonaka Y, Hirata M. 2017. Visualization of 57Fe-labeled heme isotopic fine structure and localization of regions of erythroblast maturation in mouse spleen by MALDI FTICR-MS imaging. J Am Soc Mass Spectrom 28:2469–2475. doi: 10.1007/s13361-017-1768-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, et al. 2020. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 17:352. doi: 10.1038/s41592-020-0772-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Espinas NA, Kobayashi K, Takahashi S, Mochizuki N, Masuda T. 2012. Evaluation of unbound free heme in plant cells by differential acetone extraction. Plant Cell Physiol 53:1344–1354. doi: 10.1093/pcp/pcs067 [DOI] [PubMed] [Google Scholar]
- 54. Sundermann AJ, Chen J, Kumar P, Ayres AM, Cho ST, Ezeonwuka C, Griffith MP, Miller JK, Mustapha MM, Pasculle AW, Saul MI, Shutt KA, Srinivasa V, Waggle K, Snyder DJ, Cooper VS, Van Tyne D, Snyder GM, Marsh JW, Dubrawski A, Roberts MS, Harrison LH. 2022. Whole-genome sequencing surveillance and machine learning of the electronic health record for enhanced healthcare outbreak detection. Clin Infect Dis 75:476–482. doi: 10.1093/cid/ciab946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. FastANI. 2023. https://github.com/ParBLiSS/FastANI.
- 56. Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. 2018. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol 14:e1005944. doi: 10.1371/journal.pcbi.1005944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schwengers O, Jelonek L, Dieckmann MA, Beyvers S, Blom J, Goesmann A. 2021. Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb Genom 7:000685. doi: 10.1099/mgen.0.000685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. snippy. 2020. https://github.com/tseemann/snippy.
- 59. Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. gggenes. 2023. https://github.com/wilkox/gggenes.
- 62. Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, Philippon A, Allesoe RL, Rebelo AR, Florensa AF, et al. 2020. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 75:3491–3500. doi: 10.1093/jac/dkaa345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zankari E, Allesøe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM. 2017. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother 72:2764–2768. doi: 10.1093/jac/dkx217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52:1501–1510. doi: 10.1128/JCM.03617-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Malberg Tetzschner AM, Johnson JR, Johnston BD, Lund O, Scheutz F. 2020. In silico genotyping of Escherichia coli isolates for extraintestinal virulence genes by use of whole-genome sequencing data. J Clin Microbiol 58:e01269–01220. doi: 10.1128/JCM.01269-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Circle maps.
Ratio of E. faecalis OG1RF or E. faecalis katA::Tn CFU burdens to endogenous enterococcus CFU burdens.
Weight loss and clinical sickness scores of infected mice.
Relative abundance of heme in stools.
Core genome phylogeny of 25 E. faecalis isolates collected from CDI patient stools.
Supplemental figure legends.
Strains.
Liquid chromatography and mass spectrometry parameters used in the analysis of heme.
Data Availability Statement
WGS data for E. faecalis isolates have been deposited in NCBI under BioProject PRJNA996476 with accession numbers listed in Table S1. WGS data for E. faecalis transposon mutants have been deposited in NCBI under BioProject PRJNA1039837.