ABSTRACT
Campylobacter jejuni colonizes the thick mucus layer lining the lower intestinal epithelium of hosts to promote commensalism in birds and animals or diarrheal disease in humans. C. jejuni displays higher swimming velocity as external viscosity increases, which likely benefits colonization and persistence in the intestinal mucus layer of hosts. C. jejuni produces one of the most structurally complex bacterial flagellar motors, which has been proposed to generate high torque for high swimming velocity. However, it is unknown how this flagellar motor alters output to impact swimming velocities in a viscosity-dependent manner. In this work, we identified viscosity-dependent determinant A (VidA) (Cjj81176_0996) and VidB (Cjj81176_1107) in modulating swimming velocity primarily in low-viscosity environments. C. jejuni ΔvidA cells were non-motile or swam slowly in low-viscosity media, but swam with high velocity similar to wild-type cells in both Newtonian and non-Newtonian media with high viscosity. These data indicate that VidA is required for swimming in low-viscosity environments. Suppressor mutants suggested that the lower swimming velocity of ΔvidA in low viscosity was due to unregulated activity of VidB, which we propose has a brake- or clutch-like activity to reduce swimming velocity in low viscosity. Unlike other bacterial flagellar brakes or clutches, we found no evidence for cyclic diguanylate monophosphate influencing VidB activity. Our analyses suggest that the mechanics of the C. jejuni flagellar motor has naturally evolved for high-velocity swimming in milieus with high viscosity and requires the unique combination of VidA and VidB to modulate its activity to slow swimming velocity in low-viscosity environments.
IMPORTANCE
Bacteria can adapt flagellar motor output in response to the load that the extracellular milieu imparts on the flagellar filament to enable propulsion. Bacteria can adapt flagellar motor output in response to the load that the extracellular milieu imparts on the flagellar filament to enable propulsion through diverse environments. These changes may involve increasing power and torque in high-viscosity environments or reducing power and flagellar rotation upon contact with a surface. C. jejuni swimming velocity in low-viscosity environments is comparable to other bacterial flagellates and increases significantly as external viscosity increases. In this work, we provide evidence that the mechanics of the C. jejuni flagellar motor has evolved to naturally promote high swimming velocity in high-viscosity environments. We found that C. jejuni produces VidA and VidB as auxiliary proteins to specifically affect flagellar motor activity in low viscosity to reduce swimming velocity. Our findings provide some of the first insights into different mechanisms that exist in bacteria to alter the mechanics of a flagellar motor, depending on the viscosity of extracellular environments.
KEYWORDS: flagellar motility, viscosity, swimming velocity, Campylobacter jejuni
INTRODUCTION
The bacterial flagellum is a reversible rotary motor for swimming motility and navigation of individual cells through environments (1). The flagellum is composed of many proteins that form three main substructures: a basal body associated with the cytoplasmic membrane and connected to the periplasmic rod, a surface hook attached to the tip of the rod that serves as a universal joint, and an extracellular filament tethered to the hook that rotates to propel bacteria (2–4). The basal body contains core components of a flagellar type III secretion system (fT3SS) that secretes most proteins forming the flagellar structure (5–10). The fT3SS is surrounded by the membrane/supramembrane (MS) ring (a multimer of FliF) in the cytoplasmic membrane (11, 12). The C ring is associated with the cytoplasmic face of the MS ring and contains the rotor (a multimer of FliG), with the switch complex (formed by FliM, FliN, and/or FliY) that alters direction of the rotor and, hence, the flagellum (13–20). FliH attaches to the lower face of the C ring and serves as spokes to integrate the Fli6FliJ complex below the fT3SS to energize its secretory activity (21–25).
Rotation of the flagellar motor is powered by stator units in the cytoplasmic membrane that form a stator ring decorating the circumference of the rotor (26–33). A stator unit is composed of five MotA proteins and two MotB proteins (34, 35). MotA interacts with FliG subunits in the rotor, whereas MotB interacts with cell wall components (26, 36–38). Transport of protons or ions by the stators causes a conformation change in MotA to exert a pushing force on the rotor that generates torque to rotate the flagellum (26, 27, 39–43).
A small number of studies over the last few decades have measured swimming velocities for peritrichous and polar flagellates in both Newtonian and non-Newtonian fluids and estimated or calculate torque generated by the respective flagellar motors (44–48). Whereas water is a Newtonian fluid in which viscosity remains constant when stress is applied, the viscosity of non-Newtonian fluids such as ketchup or quicksand increases or decreases when a force or stress is applied. Although the bacterial species analyzed and viscosity agents employed have not been uniform in these studies, a relatively consistent observation is that the swimming velocity of bacterial flagellates increases to varying levels in non-Newtonian fluids as viscosity increases, but velocities decrease as viscosity increases in Newtonian fluids (44–47).
In comparison to most peritrichous organisms, polar flagellates generally possess higher swimming velocities (45, 48–51). For instance, Campylobacter jejuni, an amphitrichous polar flagellate that produces a single flagellum at each pole, has a similar swimming velocity at low viscosity as Salmonella and Escherichia coli, but swimming velocity increases to a greater extent than Salmonella and E. coli (50–100 μm/s for C. jejuni vs ~35 to 45 µm/s for Salmonella or E. coli) as viscosity in a non-Newtonian fluid increases (49, 50). The flagellar motors of C. jejuni and other polar flagellates, along with Borrelia burgdorferi, which produces polar periplasmic flagella, have been proposed to generate more torque to enable propulsion through viscous milieus, along with higher swimming velocities relative to peritrichous bacteria (49, 51–56).
The amount of torque generated by a flagellar motor to turn the rotor is influenced by the number of stator units incorporated into the motor and the distance the stators are positioned relative to the flagellar rod, the central axle of the motor (57–61). In some species, stator units are dynamic components, with a direct correlation between the number of stators per motor and the extracellular mechanical load upon the filament. In E. coli, one stator unit is sufficient to turn the flagellar motor, but the motor accommodates up to 11 stators as load increases (58, 59, 61, 62). As the external load on the filament increases, such as what occurs upon contact with a surface or in environments with higher viscosity, an increase in stator number increases the amount of power for the motor and torque generated to maintain propulsion of bacterial cells. Additionally, interactions of stator units with cell wall components have been observed to be strengthened by a catch-bond mechanism to retain stators within the motor for a longer period of time to generate more torque (63).
The flagellar motors of B. burgdorferi and many polar flagellates like C. jejuni, Helicobacter pylori, and Vibrio species have evolved wider rotors that enable an increase in stator unit occupancy (52, 56, 64–67). For example, B. burgdorferi incorporates 16 stator units around a 56-nm rotor, and C. jejuni incorporates 17 stator units around a 53-nm rotor, in contrast to E. coli and Salmonella species with up to 11 stator units surrounding a 44-nm rotor (52, 56, 64). This rotor enlargement coincides with the evolution of structural disks or scaffolds that position stators around the rotor at an increased distance from the flagellar rod (52, 54, 55, 64, 65, 68–75). These scaffolding structures of B. burgdorferi and C. jejuni flagellar motors may continuously allow maximal stator occupancy. Thus, stator number may be independent of load and not fluctuate in these motors. Both the increase in number and wider placement of the stators presumably increase the amount of torque generated by these motors that likely translates into increased motor output and the higher swimming velocities observed especially in viscous environments (48–50).
Flagella and flagellar motility are required by C. jejuni to infect the human lower intestinal tract to promote diarrheal disease and colonize the lower intestinal tract of avian species, such as chickens, and mammals for commensalism (76–79). C. jejuni efficiently colonizes the mucus layer atop the epithelia of these hosts. Intestinal mucus and mucus lining the epithelium in other tissues of the host are considered to behave as a non-Newtonian medium (80). In intestinal mucus, C. jejuni encounters low viscosity at the luminal surface of the mucus layer and higher viscosity as it moves deeper in the mucus toward the epithelial surface (79, 81–84). Thus, C. jejuni has evolved to swim efficiently and with high velocity in such a viscous environment to benefit the bacterium in colonizing human, avian, and other hosts. While stator number and a wider rotor may provide an explanation for how the C. jejuni flagellar motor generates more torque for high swimming velocities in viscous milieus relative to many other motile bacteria, other factors that may influence the motor to modulate swimming velocities in a viscosity-dependent manner are unknown. Recently, external viscosity was shown to facilitate the correct wrapping of leading and lagging polar filaments of C. jejuni to promote effective swimming at higher viscosities (85).
In this report, we identify two proteins of C. jejuni—viscosity-dependent determinant A (VidA) and VidB—that are responsible for modulating swimming velocity. However, these proteins primarily impact swimming velocity at low viscosity, with less influence on swimming velocity in high-viscosity Newtonian and non-Newtonian fluids. Further analysis suggested that VidB acts as a potential brake or clutch, while VidA represses this activity. As these auxiliary proteins appear to function at low viscosity, our findings suggest that the C. jejuni flagellar motor has evolved to power flagellar rotation and generate torque for high-velocity swimming in viscous environments by default. Our analysis of these proteins provide new insights into a different mechanism for how a high-torque flagellar motor can be altered to mediate different velocities of motility in a range of viscosities.
RESULTS
Characterization of swimming velocities of C. jejuni populations across viscosities in non-Newtonian fluids
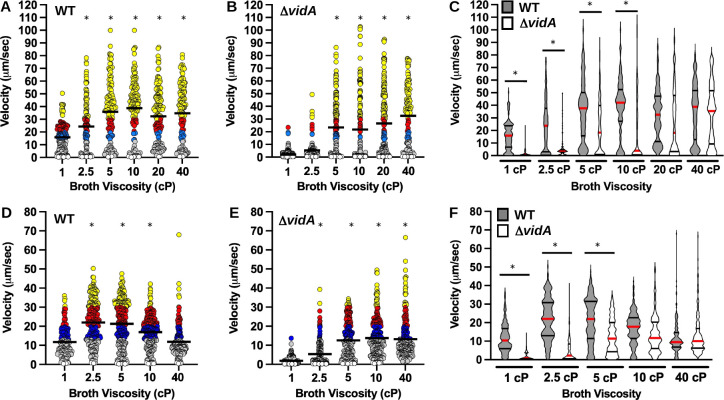
We initially characterized the swimming velocity of wild-type (WT) C. jejuni strain 81–176 in Mueller-Hinton (MH) broth with physiologically relevant viscosities that the bacterium likely encounters in the intestinal lumen and mucus layer atop the intestinal epithelium of hosts. This C. jejuni strain is a clinical isolate from a patient with diarrheal disease that can also colonize the intestinal tract of avian hosts without causing disease (76, 79). Considering that in nature C. jejuni is found in the mucus layer atop the intestines of hosts and mucus has non-Newtonian properties (80), we initially chose to supplement MH broth with methylcellulose, a branched-chain polymer, to increase viscosity and create a non-Newtonian medium to analyze swimming velocities. C. jejuni was grown overnight in standing cultures of MH alone [representing low viscosity at 1 centipoise (cP)] or with increasing concentrations of methylcellulose to increase viscosity from 2.5 to 40 cP (the latter the approximate viscosity of corn oil at room temperature) in microaerobic conditions at 37°C. After growth, the motility tracks of over 100 individual C. jejuni cells were recorded by dark-field microscopy and then converted to swimming velocities. In MH broth alone (1 cP), the swimming velocity of individual WT cells ranged from 0.4 to 50.5 µm/s with a mean velocity of 15.9 µm/s (Fig. 1A and C). As viscosity increased, we observed both an increase in the mean swimming velocity and a wider range of velocities (Fig. 1A and C; Movies S1 and S2). The mean swimming velocity increased to 24.4 µm/s at 2.5 cP and reached an average between 32.3 and 38.8 μm/s at viscosities of 5–40 cP. In these higher viscosities, the majority of cells were swimming higher than 30 µm/s. These data suggest that the C. jejuni flagellar motor promotes a higher swimming velocity as the viscosity of a non-Newtoian fluid increases. This observation is similar to E. coli, which can swim with higher velocity to an extent as the viscosity of a non-Newtonian fluid increases with increasing methylcellulose concentrations (46).
Fig 1.
Swimming velocity of WT C. jejuni and C. jejuni ΔvidA in media with different viscosities. Swimming velocity of WT C. jejuni or C. jejuni ΔvidA populations after growth for 24 h in MH broth alone (1 cP) or MH broth with increasing concentrations of (A–C) methylcellulose or (D–F) Ficoll to increase extracellular viscosity up to 40 cP. Swimming velocities of individual cells (n > 100) were measured by video tracking under dark-field microscopy. Assays were performed in triplicate and combined. For panels A B, D, and (E), each bar represents the mean swimming velocity of a population. Circles represent individual cells with velocities of < µm/s (white), 1–13 μm/s (gray), 13–20 μm/s (blue), 20–30 μm/s (red), and >30 µm/s (yellow). Statistical significance of difference in swimming velocities between WT or ΔvidA at 1 cP to cells grown in media at other viscosities was calculated by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test. *P < 0.05. (C and F) Violin plots of (C) the data presented in panels A and B and (F) the data presented in panels D and E. The red bar represents the median, and black bars represent the 25th and 75th quartiles. Statistical significance of difference in swimming velocities between WT (gray) and ΔvidA (white) cells in cultures grown at each viscosity was calculated by one-way ANOVA followed by Tukey’s multiple comparisons test. *P < 0.05.
Previous studies identified Cjj81176_0996 (Cj0977 in C. jejuni NCTC11168, hereafter referred to as VidA (based on findings described below) as a protein required for WT levels of invasion of C. jejuni into intestinal epithelial cells and colonization of the chick intestinal tract (86–88). The respective gene is a member of the σ28-dependent regulon and co-transcribed during the last stage of flagellar assembly with genes encoding proteins that compose the flagellar filament, such as the major flagellin FlaA (86, 88, 89). A crystal structure for VidA revealed a homodimer of subunits with a hotdog fold and a potential binding site for an acyl-CoA compound (87). However, this structure did not reveal further insights into how VidA may contribute to invasion or colonization. Although mutants lacking vidA showed comparable levels of flagellar motility in semi-solid MH motility media [containing 0.4% agar (86, 88), Fig. 2A)], one report made a cursory observation that a vidA mutant was impaired for motility in liquid media (90). Therefore, we analyzed the swimming velocity of C. jejuni ΔvidA in media with differing viscosities. We found that ΔvidA was severely impaired for motility in MH broth alone (1 cP), with a mean swimming velocity of 2.1 µm/s (Fig. 1B; Movie S3). Only 1.6% of ΔvidA cells in the population demonstrated a velocity equal to or greater than the average for WT cells at 1 cP (15.9 µm/s, Fig. 1B and C). However, the mutant swam faster as viscosity increased with average swimming velocities of 5.3 µm/s at 2.5 cP, 21.7–26.6 μm/s at 5–20 cP, and 32.6 µm/s at 40 cP (Fig. 1B and C; Movie S4). Although no significant difference was observed in mean swimming velocities between WT and ΔvidA in media at 20 and 40 cP, the distributions of velocities of the populations were most similar only at 40 cP (Fig. 1A through C). Examination of flagellar structure by transmission electron microscopy did not detect any differences in flagellar number, length, or curvature between WT C. jejuni and ΔvidA grown in MH broth at 1 or 40 cP (Fig. 2B). In trans complementation of ΔvidA with a constitutive promoter to produce VidA or VidA with an N-terminal FLAG tag (FLAG-VidA) restored swimming velocity at 1 cP to WT levels and production of VidA (Fig. 2C and D). However, overexpression of VidA in WT C. jejuni from a plasmid did not increase swimming velocity in 1- or 40-cP media (data not shown). Our findings indicate that VidA is required for WT levels of swimming velocity in a low-viscosity, non-Newtonian fluid but is not required for WT swimming velocities in a high-viscosity, non-Newtonian fluid.
Fig 2.
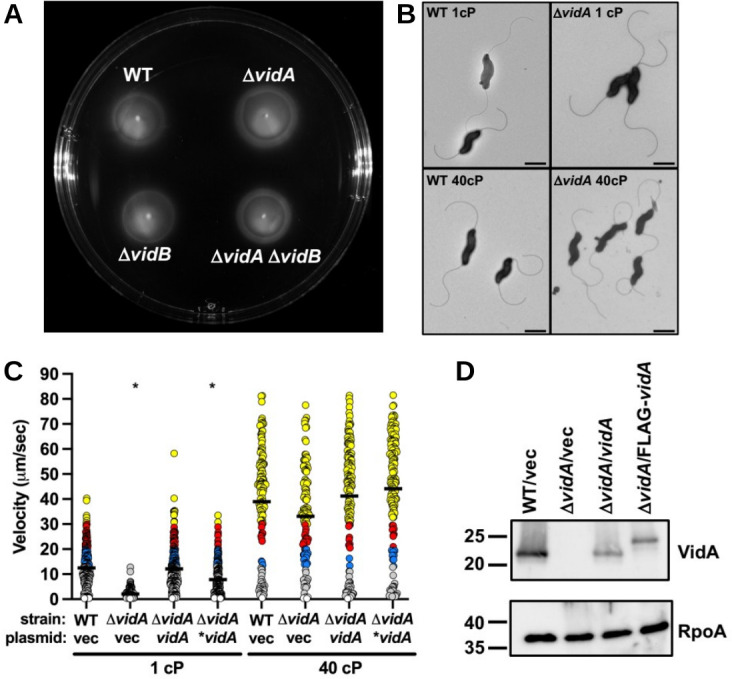
Flagellation and motility phenotypes of C. jejuni ΔvidA. (A) Motility assay in semi-solid motility agar. Motility assays of WT C. jejuni and isogenic ΔvidA and ΔvidB mutants were performed by stabbing cultures of similar optical densities into MH motility medium containing 0.4% agar. Plates were incubated in microaerobic conditions at 37°C for 30 h. (B) Electron micrographs of WT C. jejuni and C. jejuni ΔvidA grown in media of different viscosities. After overnight growth in MH broth alone (1 cP) or MH broth with methylcellulose (40 cP), strains were visualized by transmission electron microscopy. Bar = 1 µm. (C) Swimming velocities of C. jejuni ΔvidA upon in trans complementation with WT vidA. Swimming velocity of WT with vector (vec) alone, ΔvidA with vector alone, ΔvidA with vector encoding WT vidA, and ΔvidA with vector encoding an N-terminal FLAG-tagged VidA (*vidA) after growth for 24 h in MH broth alone (1 cP) or MH broth with methylcellulose to achieve a viscosity of 40 cP. Swimming velocities of individual cells (n > 100) were measured by video tracking under dark-field microscopy. Assays were performed in triplicate and combined. Circles represent individual cells with velocities of <1 µm/s (white), 1–13 μm/s (gray), 13–20 μm/s (blue), 20–30 μm/s (red), and >30 µm/s (yellow). The statistical significance of the difference in swimming velocities between the WT strain with the vector alone and other strains at both 1 and 40 cP was calculated using one-way ANOVA followed by Tukey’s multiple comparisons test. *P < 0.05. (D) Immunoblot analysis of VidA in whole-cell lysates of WT C. jejuni or C. jejuni ΔvidA containing vector (vec) alone or vectors encoding WT vidA or N-terminal FLAG-tagged VidA (*vidA). Specific antiserum to VidA was used. Detection of RpoA serves as a control to ensure equal loading of proteins across strains.
Examination of swimming velocities of WT C. jejuni and C. jejuni ΔvidA with increasing viscosities in Newtonian fluids
Although other bacterial flagellates increase swimming velocity as viscosity in non-Newtonian fluids increases, swimming velocities of these bacteria decrease as viscosity in Newtonian fluids increases (44–47). Considering our observations above, we assessed whether WT C. jejuni increases its swimming velocity in a Newtonian fluid and whether C. jejuni ΔvidA would show similar differences in swimming velocities relative to WT in a Newtonian fluid at different viscosities. We attempted to grow C. jejuni in MH broth supplemented with viscosity agents to give media Newtonian properties, but we were unsuccessful, presumably due to an intoxicant present in the chemical preparation of the reagents. However, we could monitor by dark-field microscopy swimming velocities of C. jejuni strains first grown overnight in MH broth alone and then resuspended in fresh MH broth alone or MH broth with increasing concentrations of Ficoll, a viscosity agent that maintains the Newtonian behavior of MH broth across a range of viscosities.
In MH broth alone (1 cP), the mean swimming velocity of WT cells was 11.8 µm/s (Fig. 1D and F). The swimming velocity of WT C. jejuni almost doubled to 21.2–22.0 µm/s as viscosity increased to 2.5 and 5.0 cP and was still higher at 10 cP (17.0 µm/s, Fig. 1D and F). Unlike MH with methylcellulose, we observed that swimming velocity did decrease in MH with Ficoll at the highest viscosity tested (40 cP) and returned to that observed with MH broth alone. These findings reveal a significant difference from E. coli: WT C. jejuni increases its swimming velocity as viscosity increases, regardless if the fluid has Newtonian or non-Newtonian characteristics. In contrast, E. coli swimming velocity was observed to decrease even at 2.5 cP and continued to decrease as viscosity increases in a Newtonian fluid (44–47).
Similar to our observations of C. jejuni swimming velocity in the non-Newtonian MH broth containing methylcellulose, we found that the mean swimming velocity of C. jejuni ΔvidA increased as viscosity increased (Fig. 1E and F). Compared with a swimming velocity of 1.9 µm/s in MH broth alone, we observed swimming velocities of 5.4 µm/s in MH with Ficoll at 2.5 cP, 12.6 µm/s in 5-cP broth, and 18 µm/s in 10-cP broth (Fig. 1E and F). While the average swimming velocity of ΔvidA was lower than WT in media with Ficoll at 2.5 and 5.0 cP (Fig. 1D through F), swimming velocities of WT and ΔvidA were similar at 10 cP. Like WT C. jejuni, the average swimming velocity of ΔvidA decreased in MH with Ficoll at 40 cP (13.3 µm/s). These data indicate that like in non-Newtonian fluids, VidA is required for WT swimming velocities in low-viscosity media but is not required for motility in high-viscosity environments. Since we observed VidA to mediate these effects on swimming velocities at low viscosities in both Newtonian and non-Newtonian fluids, we conclude that VidA activity is sensitive to the viscosity of the environment rather than whether the fluid has Newtonian or non-Newtonian behavior. Due to the ease in growing C. jejuni in MH broth with methylcellulose and that this media with methylcellulose has non-Newtonian properties analogous to intestinal mucus of hosts where C. jejuni resides in nature, all remaining assays in this work were performed in MH broth with methylcellulose to increase viscosity of the media.
Dynamics of alteration of swimming velocity upon a change in viscosity
With either WT C. jejuni or C. jejuni ΔvidA, we observed differences in swimming velocities after overnight growth in MH media at viscosities of 1 or 40 cP. We investigated whether viscosity-dependent differences in swimming velocities required growth and adaptation over time to the viscosity of the environment, or if changes in swimming velocity upon a change in viscosity occurred instantly. For this investigation, WT and ΔvidA were grown in MH broth alone at 1 cP or MH broth with methylcellulose to achieve 40 cP at 37°C as starting cultures. After overnight growth, cultures were washed, split equally, and then resuspended in fresh MH broth with a viscosity of 1 or 40 cP (Fig. 3A). Thus, bacteria were either introduced into fresh media with the same viscosity in which they were grown or introduced into fresh media at the alternative viscosity. Swimming velocities of standing cultures were then measured over time. As expected, WT and ΔvidA cells that were introduced into fresh media with the same viscosity in which they were grown retained the same swimming velocity over time (Fig. 3B and C). However, when WT or ΔvidA cells were introduced into media of the alternative viscosity, swimming velocities immediately changed. WT cells initially grown at 1 cP immediately swam with higher velocity in media at 40 cP (Fig. 3B). Similarly, WT cells initially grown in media at 40 cP immediately swam slower when given 1-cP media (Fig. 3B). We made similar observations with C. jejuni ΔvidA (Fig. 3C). Overall, these results suggest that modulation of swimming velocity in a viscosity-dependent manner involves mechanics that function on a relatively short time scale. Thus, these viscosity-dependent changes to swimming velocities likely do not require transcriptional changes or the production of new proteins to alter flagellar motor function.
Fig 3.
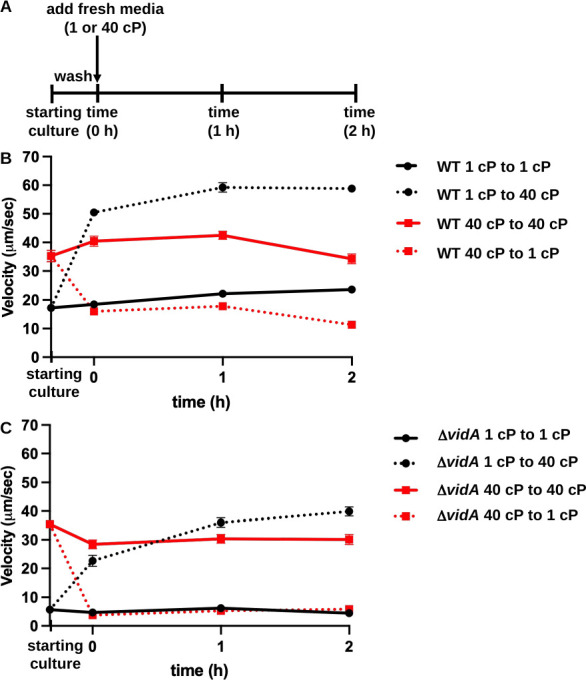
Changes in swimming velocity upon changes in extracellular viscosity over time. (A) Outline of experimental procedures. WT C. jejuni and C. jejuni ΔvidA were grown overnight in MH broth alone (1 cP) or with methylcellulose (40 cP) for starting cultures. Cultures were washed, split, and then resuspended in fresh MH media alone or with methylcellulose (40 cP) and incubated as standing cultures in microaerobic conditions at 37°C. Swimming velocities of individual cells (n > 100) in starting cultures or at times 0, 1, and 2 h after introduction into fresh MH media at 1 or 40 cP were measured by video tracking under dark-field microscopy. (B and C) Swimming velocities of WT C. jejuni (B) and C. jejuni ΔvidA (C) over time. For panels B and C, average swimming velocities (±standard errors) of cultures at each time are shown. Starting cultures represent cells after overnight growth in MH at 1 or 40 cP. Measurements at time 0 h are taken immediately after resuspension of washed cells in fresh MH media alone (1 cP) or MH with methylcellulose for 40 cP. Black solid lines with circles indicate cultures grown overnight in 1-cP media and introduced into fresh media of the same viscosity, whereas solid red lines with squares indicate cultures grown overnight into 40-cP media and introduced into fresh media of the same viscosity. Dotted black lines with squares indicate cultures grown overnight in 1-cP media and then switched to 40-cP media, whereas dotted red lines indicate cultures grown overnight in 40-cP media and then switched to 1-cP media.
Suppressor analysis of C. jejuni ΔvidA for high-velocity swimming at low viscosity
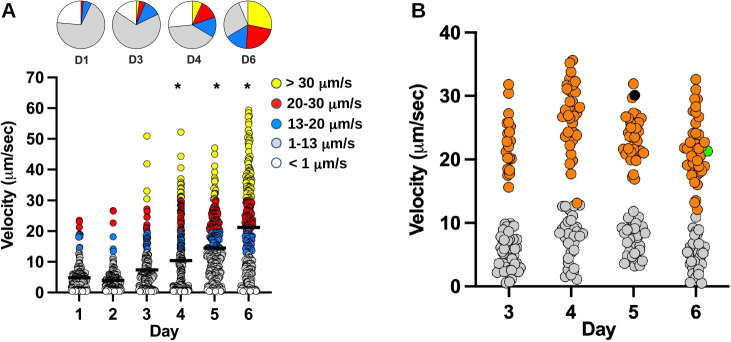
To identify other factors that may modulate swimming velocity in low-viscosity environments, we pursued isolation of suppressor mutants of C. jejuni ΔvidA with higher swimming velocity in low-viscosity MH broth. For this analysis, we grew ΔvidA as standing cultures in MH broth alone (1 cP) for 24 h and passaged the cultures into fresh MH broth (1 cP) each day for up to 6 days. Swimming velocities of individual cells (n > 192) in the population were measured each day, and up to 72 random isolates were saved. During the first 2 days of passaging, the average swimming velocity of the population was between 4.0 and 4.8 μm/s (Fig. 4A). Between days 3 and 6, we observed a gradual increase in the mean swimming velocity of cells (10.4–21.2 µm, Fig. 4A). Suppressors with high swimming velocities above 30 µm/s also emerged during this period.
Fig 4.
Identification of suppressor mutants of C. jejuni ΔvidA with increased swimming velocity upon passaging in low-viscosity media. (A) Swimming velocities of individual C. jejuni ΔvidA cells upon passaging in standing cultures of MH broth (1 cP) for up to 6 days. Swimming velocities of individual cells (n > 192) were measured by video tracking under dark-field microscopy each day. Bar represents the mean swimming velocity of the population. Circles represent individual cells with velocities of <1 µm/s (white), 1–13 μm/s (gray), 13–20 μm/s (blue), 20–30 μm/s (red), and >30 µm/s (yellow). Pie charts above represent portions of the population with different swimming velocities. Statistical significance of difference in swimming velocities of ΔvidA cells after day 1 vs cells from later cultures after passaging were calculated by one-way ANOVA followed by Tukey’s multiple comparisons test. *P < 0.05. (B) Swimming velocity of 72 C. jejuni ΔvidA isolates recovered at days 3–6 during passaging in standing cultures of MH broth (1 cP). Isolates with swimming velocities of >13 µm/s were analyzed by genomic sequencing or PCR to identify the mutation in the chromosome possibly responsible for the increased viscosity. Orange circles indicate individual cells with mutations in vidB. Gray circles represent isolates without vidB mutations, isolates that did not have increased viscosity in MH broth at 1 cP upon further analysis, or isolates that were not genetically analyzed. The black circle at day 5 represents one suppressor mutant with recombination in the flaA and flaB genes, but with WT vidB. The green circle at day 6 represents the suppressor mutant with the VidBS363G point mutation.
We measured the swimming velocities of 284 isolates recovered from days 3–6 of the procedure. We set a velocity of 13 µm/s in MH broth alone (1 cP) as a cut-off for isolates with potentially higher swimming velocities than the C. jejuni ΔvidA parental strain. At day 3, 27.8% of isolates showed velocities greater than 13 µm/s, and this percentage increased to 56.5% by day 6 (Fig. 4B). Overall, 44.7% of the isolates (127 total) demonstrated swimming velocities greater than 13 µm/s (Fig. 4B). Genomic sequencing of 15 of the high swimming velocity suppressor mutants revealed that 14 had alterations in a single gene, Cjj81176_1107, which shares fairly weak homology in a region to genes encoding diguanylate cyclases (DGCs) that generate cyclic diguanylate monophosphate (c-di-GMP) as a secondary messenger for signal transduction in many bacteria (91). The remaining ΔvidA suppressor mutant recombined parts of flaA and flaB, encoding the major and minor flagellins, respectively [Fig. 4B (92–94)]. This suppressor mutation was not analyzed further. Sequencing PCR products for Cjj81176_1107 from the remaining 112 high swimming velocity suppressors revealed that all had mutations in Cjj81176_1107 (Table 1; Fig. 4B). In total, 99.2% of ΔvidA suppressor mutants with a higher swimming velocity in low-viscosity media than the parental ΔvidA strain had alterations in Cjj81176_1107. Due to our observations, we renamed Cjj81176_1107 as vidB to be consistent in designating viscosity-dependent determinants of motility, analogous to our identification and reannotation of vidA.
TABLE 1.
Identification of C. jejuni ΔvidA suppressor mutations
| ΔvidA suppressor mutation in Cjj81176_1107 | Position of mutation | Effect | No. of occurrences |
|---|---|---|---|
| Duplication of 17 nucleotides | 20-bp 5′ to start | Promoter | 1 |
| T47C (L16S)a | T47 | Nonsenseb | 1 |
| G1158A (W386Stop)a | G158 | Missenseb | 1 |
| Deletion of 31 nucleotides | T196-A226 | Frameshift | 1 |
| T6 tract to T7 tract | T231-T236 | Frameshift | 2 |
| A5 tract to A4 tract | A331-A335 | Frameshift | 3 |
| Insertion of 13 nucleotides | T610 | Frameshift | 1 |
| G661T (E221Stop)a | G661 | Missenseb | 1 |
| A7 tract to A8 tract | A728-A734 | Frameshift | 1 |
| A7 tract to A6 tract | A759-A766 | Frameshift | 23 |
| A7 tract to A8 tract | A759-A766 | Frameshift | 4 |
| A4 tract to A3 tract | A882-A885 | Frameshift | 1 |
| AG4 tract to AG3 tract | A951-G958 | Frameshift | 64 |
| AG4 tract to AG5 tract | A951-G958 | Frameshift | 16 |
| T1087C (S363G)a | T1087 | Nonsense | 1 |
| ATTT1 tract to ATTT2 tract | A1110-T1124 | Frameshift | 5 |
| Duplication of 21 nucleotides | T1113 | Frameshift | 1 |
Effect of nonsense or missense mutation on protein sequence is indicated in parentheses.
Nonsense or missense mutation created an unstable protein in lysates as determined by immunoblot analysis.
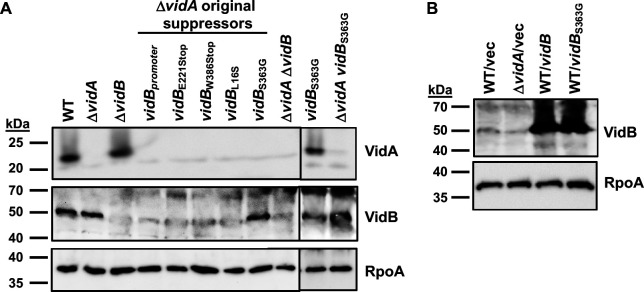
Of the 127 suppressor mutations in vidB, 122 were frameshift mutations that altered the vidB coding sequence to result in a premature stop codon (Table 1). Another mutant had a mutation in the promoter of vidB, eliminating expression of the protein (Fig. 5A). Four other mutants were due to nonsense or missense mutations to result in VidBE221Stop, VidBW386Stop, VidBL16S, and VidBS363G. Analysis of whole-cell lysates (WCLs) from these mutants revealed that only the vidBS363G mutant produced a VidB protein at similar levels as WT VidB (Fig. 4B and 5A). All other mutants failed to produce VidB mutant proteins either at the same level as WT or at the same stability as the WT VidB protein. We also observed that VidA and VidB were not required for the stability of each other as ΔvidA produced WT levels of VidB and ΔvidB produced WT levels of VidA (Fig. 5A).
Fig 5.
VidA and VidB levels in WT C. jejuni and mutants. (A) Immunoblot analysis of VidA and VidB in whole-cell lysates of WT C. jejuni, ΔvidA, ΔvidA suppressor mutants with higher swimming velocities, and vidB mutants constructed in WT C. jejuni or ΔvidA. (B) Immunoblot analysis of VidB in whole-cell lysates of WT C. jejuni or ΔvidA containing vector alone or vector to overexpress vidB or vidBS363G. For panels Aand B, specific antiserum to VidA and VidB was used to detect each protein. Detection of RpoA serves as a control to ensure equal loading of proteins across strains. Note that a background band of ~50 kDa is observed around the same size of VidB in ΔvidB that hinders fully visualizing the absence of VidB in certain mutants.
Validation of VidB for impacting swimming velocity at low viscosity
We pursued a series of analyses to verify that VidB relieves the low swimming velocity of C. jejuni ΔvidA. Whereas the mean swimming velocity of the ΔvidA population was 2.6 µm/s in media at 1 cP, the ΔvidAΔvidB mutant population swam with an average velocity of 27.8 µm/s (Fig. 6; Movie S5), verifying that mutations in vidB suppress the low-velocity defect of ΔvidA at low viscosity. Of note, ΔvidA ΔvidB had an average swimming velocity ~2.5-fold greater than WT C. jejuni at 1 cP (27.8 µm/s vs 11.6 µm/s, Fig. 6). The average swimming velocity of ΔvidAΔvidB was similar and not statistically different in low and high viscosities (Fig. 6; Fig. S1; Movies S5 and S6). In addition, the mean swimming velocities of WT C. jejuni and ΔvidAΔvidB were similar at 40 cP (Fig. 6), and we did not detect any differences in motility of the strains in semi-solid MH motility agar (Fig. 2A). Replacement of WT vidB with vidBS363G in ΔvidA also relieved the swimming velocity defect of ΔvidA at 1 cP (Fig. 6). Multiple assays consistently showed that the ΔvidA vidBS363G population tended to have an average swimming velocity modestly lower than that of ΔvidAΔvidB (~18.2 vs 27.8 µm/s, Fig. 6; data not shown). The average swimming velocities of WT, ΔvidAΔvidB, and ΔvidA vidBS363G were equivalent to each other in media up to 40 cP (Fig. 6; Fig. S1). Although statistical significance varied with repeated assays, deletion or mutation of vidB in ΔvidA tended to cause higher swimming velocity in MH broth at 40 cP compared to the parental ΔvidA mutant.
Fig 6.
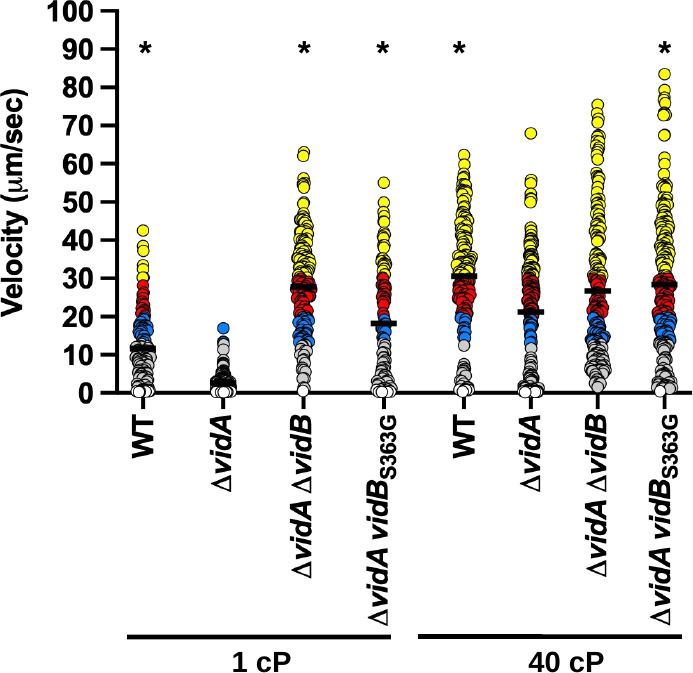
Swimming velocity of C. jejuni ΔvidA with or without vidB mutations at low and high viscosities. Swimming velocity of WT, ΔvidA, ΔvidA ΔvidB, and ΔvidA vidBS363G after growth for 24 h in MH broth alone (1 cP) or MH broth methylcellulose (40 cP). Swimming velocities of individual cells (n > 100) were measured by video tracking under dark-field microscopy. Assays were performed in triplicate and combined. Bar represents the mean swimming velocity of the population. Circles represent individual cells with velocities of <1 µm/s (white), 1–13 μm/s (gray), 13–20 μm/s (blue), 20–30 μm/s (red), and >30 µm/s (yellow). Statistical significance of difference in swimming velocities of ΔvidA at 1 or 40 cP compared to other strains grown in media of the same viscosity was calculated by one-way ANOVA followed by Tukey’s multiple comparisons test. *P < 0.05.
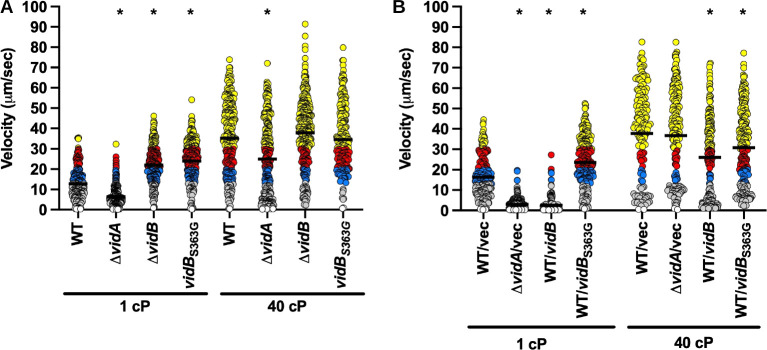
Because mutation of VidB suppressed the swimming velocity defect of ΔvidA, we investigated whether VidB was sufficient to modulate flagellar motor output and swimming velocity in different viscosities for WT C. jejuni. The single ΔvidB or vidBS363G mutant populations showed nearly a 100% increase in the average swimming velocity in media at 1 cP compared to the WT strain (Fig. 7A). However, the swimming velocities of WT and mutant strains in media at 40 cP were equivalent (34.6–37.9 µm/s; Fig. 7A). No apparent defect in motility was observed for ΔvidB relative to the WT strain in semi-solid motility agar (Fig. 2A).
Fig 7.
Swimming velocity of C. jejuni lacking vidB or overexpressing vidB at low and high viscosities. (A and B) Swimming velocity of cells after growth of cultures for 24 h in MH broth alone (1 cP) or MH broth with methylcellulose (40 cP). For panel A, strains included WT, ΔvidA, ΔvidB, and vidBS363G. For panel B, strains included WT with empty vector or vector containing vidB or vidBS363G to overexpress the respective proteins and ΔvidA with the empty vector. Swimming velocities of individual cells (n > 100) were measured by video tracking under dark-field microscopy. Assays were performed in triplicate and combined. Bar represents the mean swimming velocity of the population. Circles represent individual cells with velocities of <1 µm/s (white), 1–13 μm/s (gray), 13–20 μm/s (blue), 20–30 μm/s (red), and >30 µm/s (yellow). Statistical significance of difference in swimming velocities of WT for panel A or WT with empty vector for panel B at 1 or 40 cP compared to other strains grown in media of the same viscosity was calculated by one-way ANOVA followed by Tukey’s multiple comparisons test. *P < 0.05.
We then determined whether overexpression of VidB could cause a reduction in swimming velocity for WT C. jejuni. For this approach, WT vidB or vidBS363G was expressed from a strong promoter for the C. jejuni major flagellin (flaA) on a plasmid, in addition to expression of WT vidB from the native locus on the chromosome. Both the WT and mutant VidB protein were confirmed to be expressed at greater levels in WT C. jejuni than in WT C. jejuni with plasmid alone (Fig. 5B). Overexpression of WT vidB caused a severe reduction in swimming velocity when grown at 1 cP compared to the WT strain with plasmid alone (2.6 µm/s vs 16.2 µm/s, Fig. 7B; Movie S7). This low velocity due to vidB overexpression was comparable to that of ΔvidA with vector alone (Fig. 7B). In support of the S363G mutation in VidB isolated in the suppressor mutant analysis disabling activity of the protein, overexpression of VidBS363G in WT C. jejuni failed to reduce swimming velocity at 1 cP, unlike WT VidB overexpressed in WT (Fig. 7B). Overexpression of WT VidB in WT C. jejuni was able to reduce the mean swimming velocity in media at 40 cP (37.8 µm/s vs 26.0 µm/s) but could not reduce the velocity as low as was observed with WT overexpressing VidB in media at 1 cP (26.0 µm/s vs 2.6 µm/s, Fig. 7B; Movie S8). Our data show that VidB alone, either by its removal in WT C. jejuni or ΔvidA, or by overexpression in WT C. jejuni, is sufficient to modulate swimming velocity, especially in low-viscosity conditions.
Investigation of a linkage between VidB and c-di-GMP signaling for altering swimming velocity
VidB is predicted to be a cytoplasmic protein of 465 amino acids. Structural predictions and homology searches by PHYRE2 and other analyses predict a C-terminal domain (residues 289–444) most similar to the enzymatic domain of DGCs. In some bacterial flagellar systems, specific c-di-GMP binding proteins function as auxiliary brakes or clutches to alter flagellar motor function and swimming velocity under specific conditions (95–100). Clutch proteins of Bacillus subtilis prevent stators from contacting the rotor to decrease power and rotation (98, 100), whereas brake proteins in E. coli contact the rotor (or stators) to slow flagellar rotation (95, 96). However, most residues essential for DGC activity such as GTP and Mg2+ binding and catalysis are not conserved or very poorly conserved in VidB (101). Some c-di-GMP effector proteins have been postulated to be ancestral DGCs that have lost enzymatic activity but retained c-di-GMP binding at an inhibitory site [I site (102, 103)]. These proteins have neofunctionalized as c-di-GMP-dependent effectors to mediate specific biological functions. VidB also lacks a well conserved I site. Bioinformatic analysis did not detect any other regions of VidB with homology to domains with known functions.
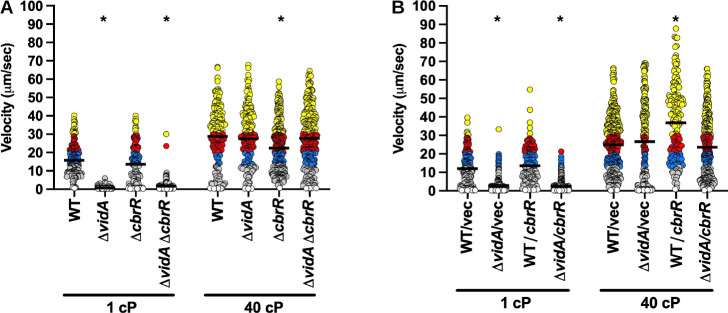
Due to known flagellar brake or clutch proteins being influenced by c-di-GMP and VidB having activities that are consistent with a brake or clutch activity to modulate swimming velocity in a viscosity-dependent manner, we searched for any evidence of involvement in c-di-GMP in modulating VidB activity or swimming velocity in C. jejuni. C. jejuni is predicted to encode only one DGC, CbrR, with no obvious predicted partner phosphodiesterase to degrade c-di-GMP or effector protein evident in the genome. Previous work suggested that CbrR may have a negative influence on flagellar motility in C. jejuni, but it is unclear whether CbrR, which can bind GTP and c-di-GMP, is a bona fide DGC that synthesizes c-di-GMP in the C. jejuni cell (104).
We first analyzed CbrR, VidA, and VidB for in vitro c-di-GMP synthesis, c-di-GMP binding, or GTP binding activity. For this approach, we purified Pseudomonas aeruginosa WspR as a positive control for DGC activity and binding of the GTP substrate and c-di-GMP product (105). In in vitro differential radial capillary action of ligand assays (DRaCALAs), recombinant WspR and CbrR bound both GTP and c-di-GMP, but VidA and VidB did not (Fig. S2A). By thin-layer chromatography analysis of c-di-GMP generation from GTP, we could not observe DGC activity by CbrR or VidB, but we did observe c-di-GMP synthesis by the positive control WspR protein (Fig. S2B).
We analyzed the effect of deleting cbrR (to decrease any possible c-di-GMP synthesis) or overexpressing CbrR (to increase any possible c-di-GMP synthesis) in both WT C. jejuni and ΔvidA on swimming velocity at low and high viscosities. Contrary to a previous report (104), we found that either deletion of cbrR or overexpression of cbrR slightly enhanced flagellar motility in semi-solid motility agar (Fig. S2C). The reason for this discrepancy with the previous report is unknown as the same strain of C. jejuni was used for both studies. Deletion of cbrR had no effect on swimming velocity of WT C. jejuni or ΔvidA in media at 1 cP or in ΔvidA at 40 cP (Fig. 8A). We did observe a slight decrease in swimming velocity upon deletion of cbrR in the WT strain (27.5 µm/s vs 22.4 µm/s) that was statistically significant; however, this difference was not consistently reproducible across replicate experiments (Fig. 8A). We did not observe any effect of overexpression of CbrR in WT C. jejuni or ΔvidA on swimming speeds at 1 cP (Fig. 8B). In media at 40 cP, we did observe an increase in swimming velocity when CbrR was expressed in WT C. jejuni, but this increase was not consistently observed across repeated experiments (Fig. 8B and data not shown). Together, these data suggest that unlike most previously discovered brake and clutch proteins that modulate the activity of the flagellar motor to alter motility in a c-di-GMP-dependent manner (95, 96, 98), VidB functions in a c-di-GMP-independent mechanism with VidA to control swimming velocity in a viscosity-dependent manner.
Fig 8.
Swimming velocity of C. jejuni lacking cbrR or overexpressing cbrR at low and high viscosities. (A and B) Swimming velocity of cells after growth of cultures for 24 h in MH broth alone (1 cP) or MH broth with methylcellulose (40 cP). For panel A, strains included WT, ΔvidA, ΔcbrR, and ΔvidA ΔcbrR. For panel B, strains included WT or ΔvidA with the empty vector or vector containing cbrR for overexpression. Swimming velocities of individual cells (n > 100) were measured by video tracking under dark-field microscopy. Assays were performed in triplicate and combined. Bar represents the mean swimming velocity of the population. Circles represent individual cells with velocities of <1 µm/s (white), 1–13 μm/s (gray), 13–20 μm/s (blue), 20–30 μm/s (red), and >30 µm/s (yellow). Statistical significance in difference in swimming velocities of WT for panel A or WT with empty vector for panel B at 1 or 40 cP compared to other strains grown in media of the same viscosity was calculated by one-way ANOVA followed by Tukey’s multiple comparisons test. *P < 0.05.
Analysis of VidA and VidB for an impact on colonization
C. jejuni efficiently colonizes the mucus layer atop the intestinal epithelium of hosts, which includes the ceca and large intestines of avian species, which usually results in long-term colonization in the absence of disease, and the colon of humans to promote diarrheal disease (79, 81, 82). The bacterium encounters low-viscosity environments in the intestinal lumen and at the luminal surface of the intestines, but experiences higher viscosity as it swims deeper into the mucus layer toward the intestinal epithelial surface. We previously determined that VidA is required for WT levels of colonization of the chick ceca (88). Considering our analysis of the requirement of VidA for WT swimming velocity in low-viscosity conditions, it is unclear whether the colonization defect of ΔvidA is due to slower swimming speeds in low-viscosity environments that the bacterium likely encounters in vivo. In addition, we questioned whether the fairly unregulated high swimming velocity we observed in vitro in ΔvidB or ΔvidAΔvidB may impact commensal colonization. Therefore, we inoculated day-of-hatch chicks with 100 CFU of WT and mutant strains and determined the level of C. jejuni in the chick ceca at day 7 post-infection. As we previously found (88), WT C. jejuni colonized the chick ceca at 1.30 × 109-CFU/g cecal content, and ΔvidA showed a 5.6-fold colonization defect (~2.31 × 108-CFU/g cecal content, Fig. 9). However, deletion of vidB from WT C. jejuni did not alter the colonization capacity. Likewise, the ΔvidAΔvidB mutant only showed a colonization defect twofold lower than ΔvidA. While this colonization defect of ΔvidAΔvidB was statistically significant compared to WT, it did not meet statistical significance when compared to ΔvidA (Fig. 8). These data suggest that the increased, relatively unregulated swimming velocity afforded by deleting vidB in C. jejuni does not impair the ability of the bacterium to efficiently colonize the chick ceca.
Fig 9.
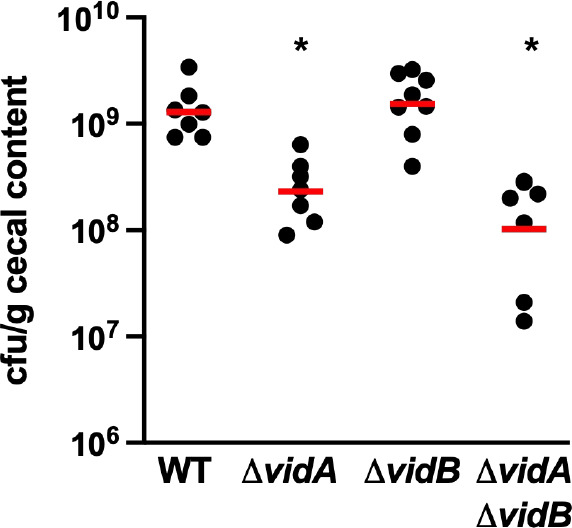
Colonization of WT C. jejuni and isogenic ΔvidA and ΔvidB mutants in the avian intestinal tract. Day of hatch chicks were orally infected with approximately 100 CFU of WT C. jejuni, and ΔvidA, ΔvidB, or ΔvidA ΔvidB mutants. Chicks were sacrificed at day 7 post-infection, and the levels of each C. jejuni strain in the ceca were determined (reported as CFU per gram of content). Each circle represents the level of C. jejuni in a single chick. Horizontal bars represent the geometric mean for each group. Statistical significance of difference in colonization capacity of strains to WT C. jejuni was determined using a two-tailed Mann-Whitney U test. *P < 0.05.
DISCUSSION
As a resident of the lower intestines of many avian and other hosts and a pathogen invading the colonic epithelium of humans to promote diarrheal disease, C. jejuni is expected to have evolved strategies to colonize and maintain a presence in the viscous mucus layer atop the intestinal epithelium of hosts. It was first reported decades ago that the amphitrichous flagella of C. jejuni promote swimming with greater velocities in environments with higher viscosity than in those with lower viscosity (49–51). However, specific factors and mechanisms that modulate C. jejuni flagellar motor output to alter swimming velocity in a viscosity-dependent manner are largely unknown. In a recent investigation, we have reported that increased viscosity of the extracellular environments assists in proper wrapping of the amphitrichous flagella around the C. jejuni cell body to promote efficient motility and higher swimming velocity in higher viscosities compared to lower ones (85).
Other bacterial flagellates—both peritrichous organisms like Salmonella and E. coli and polar flagellates like P. aeruginosa and Vibrio cholerae—are known to swim more rapidly as viscosity increases in non-Newtonian fluids. The exact mechanisms and physical forces of flagella in relation to the viscous non-Newtonian fluid that mediates this increase in swimming velocity are not fully understood, but shear thinning is one factor (46). In contrast, E. coli and Salmonella swimming velocities plateau and steadily decrease as viscosity increases in Newtonian fluids (44, 45, 47). These observations indicate that the non-Newtonian nature of viscous fluids and perhaps not viscosity itself contributes to the ability of bacterial flagellates to swim faster in comparison to low-viscosity fluids.
By analyzing swimming velocities in both Newtonian and non-Newtonian fluids of increasing viscosity, we uncovered aspects of C. jejuni swimming motility that suggest the C. jejuni flagellar motor is mechanistically different from the model flagellar system of E. coli in altering motor output and swimming velocities in a viscosity-dependent manner. First, we found that C. jejuni swimming velocity increased in MH broth of increasing viscosity with either Newtonian or non-Newtonian properties. Thus, this increase in C. jejuni swimming velocity is due to viscosity itself rather than whether the fluid has Newtonian or non-Newtonian behavior. We suspect a combination of the high-torque generated by the C. jejuni flagellar motor due to an increased number of stators positioned at a wide distance from the flagellar motor, the amphitrichous positioning of the flagellar motors, the proper wrapping of the flagella around the C. jejuni cell body, and perhaps the rotation and wobble of the helical C. jejuni cell body all contribute to how the flagella increase swimming velocity in both types of fluids as viscosity increases. Indeed, deeper analyses of the spatial physics and interaction of the C. jejuni cell body and its rotating flagella with the polymers of the viscosity agents will be required to understand how the C. jejuni flagellar motor is mechanistically different from others in viscous environments to promote efficient swimming motility.
Second, we identified two factors—VidA and VidB—that appear to specifically modulate the swimming velocity of C. jejuni in a viscosity-dependent manner independent of whether the medium has Newtonian or non-Newtonian properties. VidA (Cjj81176_0996, Cj0977) was originally identified as a protein encoded within the C. jejuni σ28 regulon along with proteins that compose the flagellar filament and are produced at the last stage in flagellar biogenesis (86, 88, 89). However, VidA was not observed to be required for flagellation or flagellar motility in semi-solid motility agar (86, 88). Instead, VidA was found to be required for invasion of human intestinal epithelial cells and commensal colonization of the chick intestinal tract (86). An additional study observed that C. jejuni ΔvidA had a motility defect in liquid broth, which seemed to counter the original observations that VidA was not required for swimming in semi-solid motility agar (90). In this study, we more thoroughly analyzed the requirement of VidA for motility by using agents to increase the viscosity of liquid media and video-tracking analysis to measure velocity of swimming motility. By this approach, we established that VidA is required by C. jejuni for WT swimming velocity in low-viscosity media (1 cP, MH broth alone). Individual cells in the ΔvidA population rarely swam as fast at WT C. jejuni and the mean swimming velocity was five-fold lower than the WT population. The requirement for VidA for motility at low viscosity appears to be to repress or offset the function of VidB. All but one suppressor mutant in the C. jejuni ΔvidA background that swam with at least WT velocity in low-viscosity media had a mutation in vidB. At the lowest viscosity of media we analyzed (MH broth alone, 1 cP), WT C. jejuni swam faster than ΔvidA. Thus, without VidA counteracting the activity of VidB, VidB has a dominant, negative impact on motility of C. jejuni in low viscosity by reducing swimming velocity to very low levels. However, introducing either WT C. jejuni and ΔvidA to increasing viscosity in both Newtonian or non-Newtonian media resulted in augmented swimming velocities, although the mean swimming velocity of the ΔvidA population was lower than that of WT and ΔvidB in media of the highest viscosity. These findings suggest that viscosity alone does not fully counteract the negative effect of VidB activity on flagellar motor output and swimming velocity. Thus, two factors are required to offset the impact of VidB on the flagellar motor to reduce swimming velocity, the intrinsic VidA protein and the extrinsic viscosity of the environment.
One potential caveat in our analysis of enhanced swimming velocity in the presence of viscosity agents to create Newtonian and non-Newtonian fluids is the possible contamination of the polymer preparations with small carbon sources, which may be metabolized by some bacteria to enhance the proton-motive force and subsequent swimming velocity (46, 47). This was particularly important in previous studies in which dialysis removed impurities from polymers that contributed to enhanced swimming velocity of bacteria in motility buffers lacking nutrients (or very minimal nutrients) (47). We cannot fully discount the presence of impurities impacting the proton-motive force of C. jejuni when polymers were added to media to enhance swimming velocity as we used undialyzed polymers in our analyses. However, one consideration against this point is that the C. jejuni lacking VidB displayed high swimming velocity regardless whether any polymer was added to media, indicating that any such impurity, if present, did not further enhance the proton-motive force to increase swimming velocity. Furthermore, unlike previous analyses (47), our analysis was performed by adding polymers to MH media, which is a complex media that robustly supports C. jejuni growth, and any contaminating impurities in the polymer preparations likely do not significantly boost the proton-motive force to greatly enhance the swimming velocities we observed.
Removal of vidB from WT C. jejuni or ΔvidA resulted in unregulated flagellar motors mediating maximal swimming velocities in media with either low or high viscosities. This finding provided an intriguing revelation for the evolution of the C. jejuni flagellar motor. We propose that C. jejuni has evolved a flagellar motor that, by default, is naturally hardwired for increased power and generation of torque for a high velocity of swimming in viscous environments. Unlike flagellar motors of many other bacterial species that function optimally at low viscosity especially in Newtonian fluids, the normal activity of the C. jejuni flagellar motor allows for rapid propulsion of bacterial cells through viscous environments and actually requires evolution of proteins such as VidA and VidB to modulate flagellar motor output to slow down swimming velocity in low-viscosity environments (48, 49). The evolution of a naturally high-torque flagellar motor that needed to evolve auxiliary proteins for optimal motility in low viscosity likely reflects the natural habitat of C. jejuni. C. jejuni is found mainly in the thick mucus layer lining the lower intestinal tract of avian and other hosts. Its microaerobic growth requirements and an optimal growth temperature of 42°C further support the lower intestinal tracts of avian hosts, which have a body temperature of 42°C, as the primary natural reservoir for C. jejuni. Maintaining persistence in the viscous mucus layer atop the intestinal epithelium of avian and animal hosts for colonization would require a flagellar motor equipped to constantly propel the bacterium in this thick milieu and resist peristalsis for potential excretion by the intestines. Having a flagellar motor naturally hardwired to generate high torque for high swimming speeds in viscous media is consistent with our reported in situ C. jejuni flagellar motor structures revealed by cryo-electron tomography (52). Our previous work showed that C. jejuni produces one of the most complex bacterial flagellar motors with scaffolding structures and a rotor with a wider diameter that allows for an increased number of stator units per motor and their placement at a wider distance from the axle (52). The C. jejuni flagellar motor appears to maintain maximal stator occupancy with 17 stators incorporated per motor, unlike that of E. coli that has a dynamic number of stators from 1 to 11 that directly correlate with the amount of extracellular load on the flagellar filament (58, 59, 61). These alterations in stator number and placement in the C. jejuni flagellar motor presumably contribute to increased power and torque for flagellar rotation. Although C. jejuni can exist outside of hosts in feces, water sources, milk, or surface of foods for transmission from one host to another, these instances are presumably transient and likely at lower viscosities relative to the natural association with the host lower intestines. As such, VidA and VidB may be necessary to slow down the flagellar motor for low-viscosity conditions for relatively brief periods of time in the lifestyle of the organism outside of hosts. Thus, VidA and VidB are not essential for motility in the natural high-viscosity environments encountered by the bacterium. Instead, these proteins are auxiliary factors that modulate swimming velocity in alternative low-viscosity conditions.
Although we propose that VidA and VidB have evolved to slow swimming velocities in low-viscosity conditions, we have yet to observe a fitness disadvantage of a vidB mutant that naturally swims at an unregulated, maximal velocity in a low-viscosity environment. Efficient swimming in low viscosity conceivably requires fewer than the 17 stator units that appear to be consistently associated with the C. jejuni flagellar motor. Thus, fully powering flagellar rotation via proton transport by all 17 stator units in the C. jejuni motor could have a negative impact on the proton motive force and generation of the highest amount of cellular energy possible in low viscosity. For our assays, we grew C. jejuni in rich media with plentiful nutrients for energy, which may compensate for any drain on generating energy due to fueling the powerful flagellar motor of the C. jejuni ΔvidB mutant in low viscosity. It is possible that during transmission from one host to another, ΔvidB swimming at unregulated, maximal velocity may be less fit in a low-viscosity, nutrient-poor reservoir. Thus, we only speculate that VidA and VidB may have evolved to conserve energy in such environments.
While our study has provided original findings into viscosity-dependent modulations of swimming velocity in bacteria, we have been unable to reveal a mechanism by which VidB modulates flagellar motor output to impact swimming velocity and how VidA and external viscosity counteracts the activity of VidB. Since removal of VidB increases swimming velocity and overexpression of VidB in WT reduces swimming speeds, VidB activity is consistent with those of brake and clutch proteins found in flagellar motors of other bacterial species. Thus, VidB might interact with the rotor or stator components to separate connections of the stators and rotor [as a clutch (98–100)] or to exert force on the stators and/or rotor to stop rotation [as a brake (95–97)]. However, we have been unable to detect an interaction between VidB and the FliG rotor protein or the MotA and MotB stator proteins (data not shown). Furthermore, we have not been able to determine interactions of VidB with any other protein due to the inability to add an epitope tag to VidB (on either terminus or internally) for co-immunoprecipitation experiments that maintain VidB function to alter swimming velocity in a viscosity-dependent manner. We also attempted to overexpress VidB in the WT strain and isolate suppressor mutants that acquired the ability to swim at low viscosity, reasoning that we may find a mutation in a protein that is the target of VidB to lower flagellar motor output and swimming velocity. However, suppressor mutants that arose had acquired mutations in vidB itself, even though vidB was present at its normal chromosomal location and also overexpressed from a plasmid in trans. We have been able to co-immunoprecipitate proteins with a functional FLAG-VidA protein, but we did not acquire any evidence that VidA or VidB interact (data not shown). If VidA and VidB compete for docking to a motor component such as the rotor to alter swimming velocity, there may not be a direct interaction between VidA and VidB. Considering that both VidA and the extrinsic viscosity of the environment modulate the VidB activity, we may gain insight into how VidA, VidB, and viscosity impact the structure and components of the flagellar motor, specifically the stator and rotor parts, by analyzing the in situ structures of the C. jejuni flagellum in WT and mutant cells grown in low and high-viscosity media in subsequent exploration.
Since some bacterial flagellar brake and clutch proteins are c-di-GMP-dependent effector proteins (95, 96, 98), we investigated whether there was any role in c-di-GMP in modulating swimming velocity with VidB. This also allowed us to fully explore fairly weak sequence and structural predictions from bioinformatic analysis that VidB may have a C-terminus with an ancestral origin from bacterial DGCs that generate c-di-GMP. Curiously, the ΔvidA suppressor mutant that had a point mutation in VidB that created a stable protein is located near the poorly conserved GGDEF motif that is required to generate c-di-GMP by DGCs (101). However, we found no evidence that recombinant VidB could bind GTP or c-di-GMP as substrates or products of DGC activity. We also detected no in vitro DGC activity for VidB. Only one DGC, CbrR, is predicted to be encoded in the C. jejuni genome, but it is unclear if CbrR actually produces c-di-GMP in C. jejuni and may instead be a c-di-GMP binding effector (104). This previous report also suggested that CbrR might have an impact on flagellar motility, with deletion of cbrR causing increased flagellar motility and overexpression causing reduced flagellar motility in semi-solid motility agar. However, we were unable to replicate these findings as both our cbrR mutant and cbrR overexpressed strain caused modest increases in flagellar motility. There is always the possibility that another enzyme encoded in the C. jejuni genome could generate c-di-GMP or another secondary messenger that modulates VidB activity. However, we observed in this work instantaneous shifts from low to high velocity of swimming motility and vice versa upon a change in viscosity. Thus, there was no time delay for changes that would be expected with adaptation requiring the generation and accumulation of a second messenger to influence VidB activity. The simplest explanation is that modulation of VidB activity is independent of a second messenger and most likely relies on VidA and extrinsic viscosity. This postulation is consistent with the unique combination of VidA and VidB encoded in the genomes of Campylobacter species and not in others.
We analyzed whether the flagellar motors of ΔvidB cells that function at maximal swimming velocity in both low and high viscosity might alter the colonization capacity of C. jejuni for the natural avian hosts. However, compared to the WT strain, the ΔvidB mutant colonized the ceca of chicks at similar levels out to 7 days of infection. We previously observed that ΔvidA did have a commensal colonization defect for the chick ceca (88). Considering that our current study verified that the ΔvidA mutant swims slowly at low viscosity, we analyzed whether deletion of vidB in ΔvidA may rescue the colonization defect since ΔvidAΔvidB swims with maximal swimming velocity regardless of viscosity. Curiously, the ΔvidAΔvidB mutant continued to show a similar colonization defect as ΔvidA. This finding suggests that the low swimming velocity phenotype of ΔvidA is not the reason for the defect in commensal colonization. We think this is a reasonable interpretation as the viscosity in the chick ceca is likely high enough to relieve VidB activity and promote higher swimming velocities in ΔvidA as we observed in our in vitro studies presented here. These findings suggest that VidA has another role for C. jejuni in promoting optimal commensal colonization of the chick intestinal tract. A previous report has indicated that VidA interacts with another protein of C. jejuni, FlgV, whose biological role has not been fully elucidated (106). A crystal structure of VidA suggests a dimer with a binding pocket at the dimer interface for an acyl-CoA-linked metabolite for another function (87), which may be important for VidA activity in commensal colonization of chickens and/or invasion of human colonic epithelial cells.
To our knowledge, we have identified some of the first intrinsic factors that influence a bacterial flagellar motor to modulate swimming velocities specifically in a viscosity-dependent manner. Flagella have been known to participate in mechanosensing by modulating flagellar motor activity and gene expression upon contact with a surface (107–111). Our observations with C. jejuni reported herein may suggest a type of mechanosensing that occurs with viscosity changes in liquid milieus, but perhaps is more akin to rheosensing but without an applied flow rate (112, 113). Currently, there is very little evidence that C. jejuni has a swarming phenotype on solid surfaces like other motile bacteria. What is striking about the structurally complex C. jejuni flagellar motor is that, by default, it generates high torque for a high velocity of swimming in viscous environments that often impede motility by less complex flagellar motors exemplified by model peritrichous organisms such as E. coli or Salmonella species, especially in viscous milieus with Newtonian behavior. Instead of requiring auxiliary proteins to enhance function of the flagellar motor for higher swimming velocities, C. jejuni has apparently evolved proteins to dampen the normal high-torque output of the flagellar motor to swim with lower velocity in less viscous environments. Continued exploration of this complex, high-torque flagellar motor, along with those produced by other bacteria such as Vibrio, Helicobacter, and Borrelia species, will undoubtedly provide new insights into adaptation of the mechanics of bacterial flagellar motors to propel bacteria through environments.
MATERIALS AND METHODS
Bacterial strains and plasmids
C. jejuni 81–176 strains created and used in this study are described in Table 2. All plasmids constructed or involved in creation of strains in these studies are described in Table 3. Prior to each experiment, C. jejuni strains were routinely grown from freezer stocks on MH agar in microaerobic conditions (85% N2, 10% CO2, and 5% O2) at 37°C for 48 h and then restreaked on MH agar and grown for 16 h under identical conditions. As required, antibiotics were added to MH broth or agar at the following concentrations: 10-µg/mL trimethoprim, 15-µg/mL chloramphenicol, 50-µg/mL kanamycin, 30-µg/mL cefoperazone, or 0.5-, 1.0-, 2.0-, or 5.0-mg/mL streptomycin. All C. jejuni strains were stored at −80°C in an 85% MH broth and 15% glycerol solution. Escherichia coli DH5α and DH5α/pRK212.1 were grown on Luria-Bertani (LB) agar or in LB broth containing 100-µg/mL ampicillin, 15-µg/mL chloramphenicol, 50-µg/mL kanamycin, or 12.5-µg/mL tetracycline as appropriate. All E. coli strains were stored at −80°C in an 80% LB broth and 20% glycerol solution.
TABLE 2.
Bacterial strains used in this study
| Strain | Genotype | Source/reference |
|---|---|---|
| E. coli strains | ||
| DH5α | E. coli supE44 ∆lacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Invitrogen |
| DH5α/pRK212.1 | DH5α with conjugation transfer element | (114) |
| Top10 | F− mcrA ∆(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ∆lacX74 recA1 araD139 ∆(ara-leu)7697 galE15 galK16 rpsL (Strr) endA1 nupG | Invitrogen |
| XL1-Blue | F′::Tn10 proA+B+lacIqΔ(lacZ)M15/recA1 endA1 gyrA96 (Nalr) thi hsdR17 (rK− mK+) glnV44 relA1 lac | New England Biolabs |
| Campylobacter jejuni strains | ||
| DRH212 | 81–176 rpsLSm | (115) |
| DRH837 | 81–176 rpsLSm/pECO102 | (116) |
| ABT501 | 81–176 rpsLSmΔvidA | (88) |
| DAR5362 | 81–176 rpsLSm ΔvidA/pECO102 | This study |
| DAR5364 | 81–176 rpsLSmΔvidA/pDAR5476 | This study |
| DAR5615 | 81–176 rpsLSmΔvidA/pDAR5357 | This study |
| DAR6160 | 81–176 rpsLSm cbrR::cat-rpsL | This study |
| DAR6162 | 81–176 rpsLSmΔvidA cbrR::cat-rpsL | This study |
| DAR6236 | 81–176 rpsLSmΔvidA vidB::cat-rpsL | This study |
| DAR6232 | 81–176 rpsLSm vidB::cat-rpsL | This study |
| DAR6249 | 81–176 rpsLSmΔcbrR | This study |
| DAR6307 | 81–176 rpsLSmΔvidAΔcbrR | This study |
| DAR6357 | 81–176 rpsLSmΔvidA ΔvidB | This study |
| DAR6668 | 81–176 rpsLSmΔvidA vidBS363G | This study |
| DAR6408 | 81–176 rpsLSmΔvidB | This study |
| DAR6672 | 81–176 rpsLSm vidBS363G | This study |
| DAR6673 | 81–176 rpsLSm/pDAR1604 | This study |
| DAR6675 | 81–176 rpsLSmΔvidA/pDAR1604 | This study |
| DAR6727 | 81–176 rpsLSm/pDAR6580 | This study |
| DAR6812 | 81–176 rpsLSmΔvidA/pDAR6606 | This study |
| DAR6851 | 81–176 rpsLSm/pDAR6301 | This study |
| DAR6853 | 81–176 rpsLSmΔvidA/DAR6301 | This study |
TABLE 3.
Plasmids used in this study
| Plasmid | Genotype | Source/reference |
|---|---|---|
| pUC19 | AmpR, general cloning vector | New England BioLabs |
| pQE30 | AmpR, expression vector for adding 6XHis-tag to N-terminus of proteins | Qiagen |
| pBAD/myc-HisA | Invitrogen | |
| pRY108 | KanR, E. coli-C. jejuni shuttle vector | (117) |
| pRY109 | Source of cat cassette | (117) |
| pDRH265 | Source of cat-rpsL cassette | (115) |
| pECO102 | CatR, E. coli-C. jejuni shuttle vector containing cat promoter and stop codon for expression of genes for complementation | Wiesner |
| pVL791 | Contains wspR | (114) |
| pDAR964 | CatR, E. coli-C. jejuni shuttle vector containing cat promoter with an in-frame N-terminal FLAG sequence for expression of genes for complementation | (118) |
| pDAR1425 | pRY108 with 206 bases of DNA containing promoter for flaA and start codon cloned into the XbaI and BamHI sites | This study |
| pDAR1604 | pRY108 with 206 bases of DNA containing promoter for flaA and sequence for FLAG tag fused in-frame to the start codon cloned into the XbaI and BamHI sites | (119) |
| pDAR5357 | pDAR964 with codon 2 to stop codon of vidA cloned into the BamHI site of pDAR964 to create a N-terminal FLAG tag fusion protein | This study |
| pDAR5476 | pRY112 with 76 bases of DNA containing promoter for cat and start codon with in-frame BamHI site, FLAG tag epitope sequence, and stop codon cloned into the XbaI and PstI sites for expression of C-terminal FLAG-tagged proteins for complementation | This study |
| pDAR5571 | pDAR5476 with codon 2 to penultimate codon of vidA cloned in-frame in BamHI site | This study |
| pDAR6131 | pUC19 with DNA fragment to create in-frame deletion of codons 47–347 of vidB with 0.8 kb of upstream and downstream sequences cloned in to the EcoRI site | This study |
| pDAR6134 | pQE30 with codon 2 to stop codon of vidB cloned into the BamHI site | This study |
| pDAR6144 | pUC19 with DNA fragment to create in-frame deletion of codon 2 to 375 of cbrR with 0.8 kb of upstream and downstream sequences cloned in to the EcoRI site | This study |
| pDAR6141 | pUC19 with DNA fragment containing cbrR with 0.8 kb of upstream and downstream sequences cloned into the EcoRI site; contains G522A mutation to create an EcoRV site within cbrR | This study |
| pDAR6152 | SmaI cat-rpsL cassette cloned into the EcoRV site of cbrR in pDAR6141 | This study |
| pDAR6158 | pUC19 with DNA fragment containing vidB with 0.8 kb of upstream and downstream sequences cloned into the EcoRI site, contains C890A mutation to create an EcoRV site within vidB | This study |
| pDAR6203 | pBAD/myc-HisA with codon 2 to penultimate codon of cbrR cloned into the NcoI and HindIII sites | This study |
| pDAR6214 | SmaI cat-rpsL cassette cloned into the EcoRV site of vidB in pDAR6158 | This study |
| pDAR6301 | pDAR1425 with cbrR from codon 2 to the stop codon cloned into the BamHI site | This study |
| pDAR6535 | pBAD/myc-HisA with codon 2 to penultimate codon of wspR cloned into the NcoI and HindIII sites | This study |
| pDAR6574 | pUC19 with DNA fragment containing vidBS363G with 0.8 kb of upstream and downstream sequence cloned into the EcoRI site | This study |
| pDAR6580 | pDAR1425 with vidB from codon 2 to the stop codon cloned into the BamHI site | This study |
| pDAR6606 | pDAR1425 with vidBS363G from codon 2 to the stop codon cloned into the BamHI siteco | This study |
Construction of C. jejuni mutants
C. jejuni mutants were constructed by electroporation of plasmid DNA or natural transformation of in vitro methylated plasmid DNA following previously described methods (115, 120). All plasmids were constructed by ligation of DNA fragments into plasmids by T4 DNA ligase or Gibson Assembly Mastermix (New England BioLabs).
For construction of vidB mutants, DNA fragments with approximately 800-bp upstream and downstream of the vidB coding sequence were amplified by PCR from C. jejuni 81–176 genomic DNA. Primers were designed to create a C890A point mutation inside the vidB coding sequence to create an EcoRV restriction site. The DNA fragments were cloned into EcoRI-digested pUC19 to create pDAR6158. A SmaI fragment containing a cat-rpsL cassette from pDRH265 was then inserted into the EcoRV site in vidB in pDAR6158 to result in pDAR6214 (115). pDAR6214 was then introduced into DRH212 or ABT501, and potential transformants that replaced WT vidB with vidB::cat-rpsL were recovered on MH agar with chloramphenicol (88, 115). Mutations were verified by colony PCR to result in DAR6232 (81–176 rpsLSm vidB::cat-rpsL) and DAR6236 (81–176 rpsLSm ΔvidA vidB::cat-rpsL). A plasmid harboring an in-frame deletion of vidB was created by designing primers to amplify approximately 800 bases upstream and downstream of vidB and portions of vidB coding sequence to delete codons 47–347. These DNA fragments were fused and assembled into EcoRI-digested pUC19 to create pDAR6131. For cloning of vidBS363G, primers were used to amplify the vidB sequence from ABT501 suppressors 6–10 with approximately 800 bp of upstream and downstream sequence and cloned into EcoRI-digested pUC19 to create pDAR6574. pDAR6131 and pDAR6574 were introduced into DAR6232 and DAR6236. Transformants were isolated on MH agar with streptomycin and then screened for sensitivity to chloramphenicol. Colony PCR and DNA sequencing verified mutants that had replaced vidB::cat-rpsL with the ΔvidB or vidBS363G on the chromosome to result in DAR6408 (81–176 rpsLSm ΔvidB), DAR6357 (81–176 rpsLSm ΔvidAΔvidB), DAR6672 (81–176 rpsLSm vidBS363G), and DAR6668 (81–176 rpsLSm ΔvidA vidBS363G).
For construction of cbrR mutants, DNA fragments with approximately 800 bp upstream and downstream of the cbrR coding sequence were amplified by PCR from C. jejuni 81–176 genomic DNA. Primers were designed to generate a G522A point mutation inside the cbrR coding sequence to create an EcoRV restriction site. The DNA fragments were cloned into EcoRI-digested pUC19 to create pDAR6141. A SmaI fragment containing a cat-rpsL cassette from pDRH265 was then inserted into the EcoRV site in cbrR in pDAR6141 to result in pDAR6152 (115). pDAR6152 was introduced into DRH212 or ABT501, and potential transformants that replaced WT cbrR with cbrR::cat-rpsL were recovered on MH agar with chloramphenicol (88, 115). Colony PCR verified correct construction of mutants to result in DAR6160 (81–176 rpsLSm cbrR::cat-rpsL) and DAR6162 (81–176 rpsLSm ΔvidA cbrR::cat-rpsL). A plasmid containing an in-frame deletion of cbrR was created by designing primers to amplify approximately 800 bases upstream and downstream of cbrR and portions of cbrR coding sequence to delete codons 2–375 to create pDAR6144. This plasmid was then introduced into DAR6160 and DAR6162. Transformants were isolated on MH agar with streptomycin and then screened for sensitivity to chloramphenicol. Colony PCR verified correct mutants that had replaced cbrR::cat-rpsL with ΔcbrR on the chromosome to result in DAR6249 (81–176 rpsLSm ΔcbrR) and DAR6307 (81–176 rpsLSm ΔvidAΔcbrR).
Construction of plasmids for complementation studies
Multiple plasmids were constructed to allow for expression of genes from promoters of different strengths to result in different levels of proteins with or without epitope tags for complementation of mutants. Primers were designed to amplify by PCR from the genome of C. jejuni 81–176 a 206-bp fragment containing the promoter for flaA (encoding the FlaA major flagellin) and its start codon, which was followed by an in-frame BamHI restriction site. This fragment was cloned into the XbaI and BamHI sties of pRY108 to result in pDAR1425 (117). A similar fragment was amplified by primers so that DNA for an in-frame FLAG tag epitope is encoded after the start codon of flaA, followed by an in-frame BamHI restriction site. This fragment was cloned into the XbaI and BamHI sites of pRY108 to create pDAR1604 (117).
Plasmids containing coding sequences of specific genes were constructed to complement various mutants. Primers were designed to amplify the coding sequence of vidA from codon 2 through the stop codon with both 5′ and 3′ in-frame BamHI sties from the C. jejuni 81–176 genome. The resultant DNA fragment was cloned into the BamHI-digested pECO102 to create pDAR5629 and BamHI-digested pDAR964 to result in pDAR5357, which encodes VidA with an N-terminal FLAG tag (121). Primers were also designed to amplify by PCR the vidB or vidBS363G coding from codon 2 to the stop codon of WT C. jejuni 81–176 or ABT501 suppressors 6–10, respectively, with 5′ and 3′ in-frame BamHI restriction sites. These DNA fragments were then inserted into BamHI-digested pDAR1425 to create pDAR6580 (encoding WT VidB) and pDAR6606 (encoding VidBS363G). Similarly, primers were designed to amplify the cbrR coding sequence from codon 2 to the stop codon with 5′ and 3′ in-frame BamHI restriction sites from C. jejuni 81–176. This DNA fragment was then inserted into BamHI-digested pDAR1425 to create pDAR6301.
All plasmids were transformed into E. coli DH5α/pRK212.1, which served as the donor strain for conjugation into C. jejuni (114). Plasmids were then conjugated into C. jejuni strains as previously described (122).
Measurement of swimming velocity of individual cells
Methylcellulose (4,000 cP, Fisher) was added at various concentrations to MH broth immediately after autoclaving and stirred overnight to achieve a range of viscosities. Final viscosities were determined using a Gilmont falling ball viscometer. For measurement of swimming velocity of individual cells in MH broth of different viscosities, C. jejuni strains were grown under standard conditions overnight for 16 h on MH agar with appropriate antibiotics. Strains were resuspended from plates in MH broth without methylcellulose and diluted to an OD600 0.8. One milliliter of culture was added to 20 mL of MH broth alone with appropriate antibiotics (no methylcellulose, 1 cP) or containing methylcellulose to create viscosities up to 40 cP. Cultures were incubated as standing cultures for 24 h at 37°C in microaerobic conditions. After growth, strains were diluted to an OD600 between 0.1 and 0.4 in the same viscosity of MH that they were grown. Samples of cultures were applied to microscope slides precleaned with 100% ethanol and then covered with a cover slip. Motility tracks of individual cells were visualized using an Olympus BX-40 microscope adapted for dark-field microscopy with a UPlanFL ×100 objective and recorded with an Orca-Spark (Hamamatsu) camera and HCImage Live software. Captured motility tracks of individual cells were subsequently processed for calculation of swimming velocity using ImageJ and an in-house python script. Velocity measurements for at least 100 cells were examined for each strain for each experiment. Each experiment was performed three times.
For analysis of swimming velocities in MH broth with Ficoll to increase viscosity, a 36% Ficoll 400 (Cytiva) stock solution was prepared in MH broth and sterilized. This solution was diluted with MH broth to achieve a range of viscosities as determined by a Gilmont falling ball viscometer. C. jejuni strains were grown as described above in MH broth alone (1 cP) for 24 h at 37°C in microaerobic conditions. After growth, strains were diluted to an OD600 between 0.1 and 0.4. One-milliliter samples were pelleted for 3 min by centrifugation and resuspended in fresh MH broth alone or MH broth containing Ficoll at different concentrations to expose cells to different viscosities. Samples of cultures were then examined by dark-field microscopy as described above.
To assess the temporal scale for adjusting swimming velocities upon a change in viscosity, WT C. jejuni and ΔvidA mutant strains were prepared as above and then grown as standing cultures for 24 h at 37°C in microaerobic conditions in MH broth alone (1 cP) or MH broth with methylcellulose (40 cP) in triplicate. Individual cells in each culture were analyzed by dark-field microscopy for determination of mean swimming velocities of the populations. Each culture was then diluted with 10 volumes of phosphate-buffered saline (PBS), centrifuged for 10 min at 5,500 × g, and washed again with 200 mL of PBS. After resuspension in PBS, cultures were split in half and centrifuged again. One pellet was resuspended in 20 mL of MH broth at the original viscosity, whereas the other pellet was resuspended in 20 mL of MH broth at the alternative viscosity. Cultures were incubated in microaerobic conditions at 37°C as standing cultures and analyzed immediately (time 0) or at 1 and 2 h upon incubation by dark-field microscopy to determine mean swimming velocities of the populations. The experiment was performed three times.
Assessment of flagellar motility in semi-solid motility agar
After overnight growth on MH agar with appropriate antibiotics at 37°C in microaerobic conditions, strains were resuspended in MH broth and diluted to OD600 0.8. Strains were then stabbed into MH motility agar containing 0.4% agar and appropriate antibiotics when required. Agar plates were then incubated at 37°C in microaerobic conditions for 30 h.
Transmission electron microscopy
WT C. jejuni and C. jejuni ΔvidA were grown under standard conditions overnight for 16 h on MH agar with appropriate antibiotics. Strains were resuspended from plates in MH broth without methylcellulose and diluted to an OD600 0.8. One milliliter of culture was added to 20 mL of MH broth alone with appropriate antibiotics (no methylcellulose, 1 cP) or containing methylcellulose to create a viscosity of 40 cP. Cultures were incubated as standing cultures for 24 h at 37°C in microaerobic conditions. One milliliter samples of culture were pelleted for 3 min at 13,200 rpm in a microcentrifuge, resuspended in 2.5% glutaraldehyde in 0.1-M cacodylate and then incubated on ice for 1 h. Copper-coated formvar grids were negatively glow discharged, and bacterial samples were then applied to the grids. The samples were stained with 2% uranyl acetate and visualized with an FEI Technai G2 Spirit Bio TWIN transmission electron microscope.
Isolation of C. jejuni ΔvidA suppressor mutants with increased swimming velocity in low-viscosity media
C. jejuni ΔvidA [ABT501 (88)] was grown under standard conditions overnight for 16 h on MH agar. Bacteria were resuspended from plates in MH broth and diluted to an OD600 of 0.8. One milliliter of culture was added to 20 mL of MH broth alone (without methylcellulose, 1 cP) to achieve an OD600 of ~0.04 and incubated as a standing culture in microaerobic conditions at 37°C. After 24 h, a sample of culture was analyzed by dark-field microscopy to determine the swimming velocity of individual cells (n > 192) and the mean swimming velocity of the population. Dilutions of the culture were plated on MH agar to isolate individual colonies, which were stored at −80°C. Twenty milliliters of fresh MH broth was inoculated with the overnight culture to an OD600 of ~0.04, and the culture was incubated as a standing culture in microaerobic conditions at 37°C for another 24 h. Passaging of cultures into fresh MH broth without methylcellulose and subsequent dark-field microscopy analysis of swimming velocities of the populations were continued for 6 days. At the conclusion of the experiment, up to 72 individual potential suppressor mutants for each day from days 1 to 6 were stored at −80°C. Each potential suppressor mutant was then grown as individual cultures in MH broth alone (no methylcellulose, 1 cP) and analyzed by dark-field microscopy to determine its swimming velocity in MH broth (1 cP) relative to the WT C. jejuni and the parental ΔvidA strain.
Genomic DNA from the parental C. jejuni ΔvidA strain (ABT501) and 15 suppressor mutants that were verified to have a higher swimming velocity in low-viscosity media relative to ΔvidA was prepared. Purified genomic DNA was then treated with RNase If (New England BioLabs) and cleaned using the Zymo Genomic DNA Clean and Concentrator kit. The genomic DNA from above was used to generate bar-coded Bioo NEXTflex DNA libraries at the Indiana University Center for Genomics and Bioinformatics. These libraries were cleaned and verified using an Agilent 2200 TapeStation before pooling and sequencing on the Illumina NextSeq platform. Paired-end reads were de-multiplexed subsequent to analysis. Reads obtained from the parent (ABT501) and suppressor mutant genomes were mapped to the C. jejuni 81–176 reference genome (GCA_000015525.1) using Geneious R10.2.6. Single-nucleotide polymorphisms (SNPs) with a minimum variant frequency of 20% were identified in regions of the parent and suppressor mutant genomes with at least 5× coverage. SNPs identified in suppressor mutant genomes were individually compared to those identified within the parent genome. SNPs that were unique to the suppressor mutant genomes were presumed to be associated with the suppressor mutant phenotypes and investigated further. Identification of other suppressor mutants for which genomic sequencing was not applied involved PCR amplification of one or more suspected genes and then sequencing of PCR products.
Purification of recombinant proteins
The coding sequence from codon 2 to the stop codon of VidB was amplified by PCR from C. jejuni 81–176 and then cloned by Gibson Assembly into BamHI-digested pQE30 to create pDAR6134. This plasmid was then moved to E. coli XL1-Blue for recombinant protein purification. XL1-Blue containing pDAR6134 was grown to mid-log phase in LB broth with shaking at 37°C and then induced with 1-mM isopropyl-beta-D1-thiogalactopyranoside (IPTG) for 3 h at 37°C.
C. jejuni 81–176 cbrR from codon 2 to the stop codon was amplified by PCR and inserted into NcoI and HindIII-digested pBadMyc/HisA by Gibson Assembly to create pDAR6203. P. aeruginosa WspR was amplified from pVL791 and cloned similarly in pBADMyc/HisA to create pDAR6535 (123). The plasmids were moved into E. coli Top10 for induction and purification. After growth to mid-log phase in LB broth, arabinose was added to a final concentration of 0.2% to induce expression of CbrR-Myc-6XHis and WspR-Myc-6XHis for 3 h at 37°C. 6XHis-VidA was induced from pABT522 as previously described (88).
After induction of proteins, cell pellets were collected by centrifugation and washed once with lysis buffer (50-mM NaH2PO4, 300-mM NaCl, 10-mM imidazole, pH 8.0). Cells were passed through an EmulsiFlex-C5 disruptor at 15,000–20,000 lb/in2 to lyse cells. The soluble fraction was recovered by centrifugation at 20,000 × g for 2 h. Recombinant 6XHis-VidB, CbrR-Myc-6XHis, WspR-Myc-6xHis, and 6XHis-VidA were purified via fast protein liquid chromatography (FPLC) using an NGC Chromatography System and Profinity IMAC columns (Bio-Rad) according to manufacturer’s instruction. Glycerol was added to a final concentration of 20% prior to storage at −80°C.
Polyclonal antisera generation
Purified recombinant 6XHis-VidB was used to immunize guinea pigs by standard procedures for antisera generation via a commercial vendor (Cocalico Biologicals). All uses of animals in experimentation were approved by The Institutional Animal Care and Use Committee (IACUC) at the University of Texas Southwestern Medical Center.
Immunoblotting analyses
Preparation of C. jejuni strains for WCLs was performed as previously described (116). All immunoblotting analyses of WCLs or immunoprecipitated proteins were performed with equal amounts of samples from C. jejuni strains after SDS-PAGE by standard procedures. For specific detection of proteins in WCLs, 10 µL of WCLs was analyzed to detect VidA and RpoA, and 20 µL of WCLs was analyzed to detect VidB. Proteins were detected with specific murine or guinea pig antisera generated previously or as described above (88, 124). Primary antisera were used at the following dilutions: VidA M155, 1:500; VidB UTGP296, 1:500; and RpoA M252, 1:2,000. All primary antisera were applied to immunoblots for 2 h. Appropriate horseradish peroxidase-conjugated goat antibodies were used as secondary antibodies to develop immunoblots.
Analysis of c-di-GMP synthesis
Recombinant WspR, CbrR, and VidB were assessed for their ability to synthesize c-di-GMP from GTP following standard protocols (125). Briefly, MgCl2 was added to a final concentration of 10 mM to 40 µg of recombinant proteins. Reactions were started by addition of 10 µCi [α32P]-GTP, incubated at room temperature for 1 h, and then stopped with the addition EDTA to a final concentration of 250 mM. Thin-layer chromatography (TLC, BakerFlex Cellulose) plates were allowed to equilibrize to room temperature, and then 2 µL of the reactions was spotted onto the TLC plate. After spots had dried, the TLC plate was placed inside a sealed beaker containing approximately 250 mL of solvent (2:3 mixture of saturated NH4SO4 and KH2PO4). Once solvent migration had neared the end of the TLC plate, the plate was removed, dried, covered with a clear wrapping, and exposed to a phosphorimager cassette for 2 h or overnight.
GTP and c-di-GMP binding analyses
Recombinant WspR, CbrR, VidA, and VidB were assessed for their ability to bind GTP and c-di-GMP by the DRaCALA (126). Briefly, 10× binding buffer (100-mM Tris-HCl, pH 8, 1-M NaCl, and 50-mM MgCl2), 10 µCi of [α32P]-GTP, and 14 µg of protein were added to a final reaction volume of 20 µL. Two microliters of each reaction was spotted onto nitrocellulose membranes and exposed to a phosphorimager for 30 min. For analysis of c-di-GMP binding, [32P]-labeled c-di-GMP was generated by recombinant WspR as described above and then heated at 95°C for 5 min to inactivate WspR. The reaction was then centrifuged at 20,000 × g for 5 min to separate WspR from [32P]-labeled c-di-GMP. DRaCALA reactions were performed as above except substituting [32P]-labeled c-di-GMP and spotting 6 µL of reaction mixture onto nitrocellulose membranes.
Chick colonization assays
All uses of animals in experimentation were approved by IACUC at the University of Texas Southwestern Medical Center. The ability of wild-type C. jejuni or mutant strains to colonize the ceca of chicks after oral inoculation was determined as previously described (79). Briefly, fertilized chicken eggs (SPAFAS) were incubated for 21 d at 37.8°C with appropriate humidity and rotation in a Sportsman II model 1502 incubator (Georgia Quail Farms Manufacturing Company). Approximately 12–36 h after hatching, chicks were orally infected with 100 µL of PBS containing approximately 100 CFU of WT C. jejuni or an isogenic mutant strain. Strains were prepared for oral gavage by resuspension from MH agar after growth and dilution in PBS to an OD600 of 0.4 followed by serial dilution to obtain the appropriate inoculum for oral gavage of chicks. The number of CFUs of the inoculum was determined by serial dilution on MH agar. At day 7 post-infection, chicks were sacrificed and the cecal contents were recovered, weighed, and suspended in PBS to 0.1-g cecal content/mL PBS. Serial dilutions were spread on MH agar containing trimethoprim (TMP) and cefoperazone. Bacteria were grown for 72 h at 37°C in microaerobic conditions and then counted to determine the CFU per gram of cecal content for each chick. Statistical analyses were performed by the Mann-Whitney U test, with statistically significant differences between wild-type and mutant strains indicated with P values of <0.05.
ACKNOWLEDGMENTS
We thank Thomas G. Ribardo, Jr., for the development of and feedback on the in-house Python script to track swimming bacteria for the calculation of swimming velocities. We also thank Dr. Morgan Beeby for many fruitful discussions.
This work was supported by Public Health National Institutes of Health grants R01AI065539 (D.R.H.), R01HD095830 (D.R.H.), R21AI159140 (D.R.H.), and R01AI166535 (J.J.J.).
Contributor Information
David R. Hendrixson, Email: david.hendrixson@utsouthwestern.edu.
Nina R. Salama, Fred Hutchinson Cancer Center, Seattle, Washington, USA
DATA AVAILABILITY
All data and methodologies are available upon request. Genomic sequences are available at the National Center for Biotechnology Information as part of Bioproject PRJNA987628, with individual genomic sequences assigned the following accession numbers: ABT501 (SRR25146826), ABT501 S1 (SRR25146825), ABT501 S2 (https://www.ncbi.nlm.nih.gov/sra/SRR25146820SRR25146820), ABT501 S3 (SRR25146819), ABT501 S4 (SRR25146818), ABT501 S5 (SRR25146817), ABT501 S6 (SRR25146816), ABT501 S7 (SRR25146815), ABT501 S8 (SRR25146814), ABT501 S9 (SRR25146813), ABT501 S10 (SRR25146824), ABT501 S11 (SRR25146823), ABT501 S12 (SRR25146822), and ABT501 S13 (SRR25146821).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.02544-23.
Figures S1 and S2 and Movie S1-S8 legends.
WT swimming motility at low viscosity (1 cP).
WT swimming motility at high viscosity (40 cP).
vidA mutant swimming motility at low viscosity (1 cP)
vidA mutant swimming motility at high viscosity (40 cP).
vidA vidB mutant swimming motility at low viscosity.
vidA vidB swimming motility at high viscosity (40 cP).
WT C.jejuni overexpressing VidB swimming motility at low viscosity (1 cP).
WT C. jejuni overexpressing VidB swimming motility at high viscosity (40 cP).
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Nakamura S, Minamino T. 2019. Flagella-driven motility of bacteria. Biomolecules 9:279. doi: 10.3390/biom9070279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Erhardt M, Namba K, Hughes KT. 2010. Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb Perspect Biol 2:a000299. doi: 10.1101/cshperspect.a000299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chevance FFV, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465. doi: 10.1038/nrmicro1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johnson S, Furlong EJ, Deme JC, Nord AL, Caesar JJE, Chevance FFV, Berry RM, Hughes KT, Lea SM. 2021. Molecular structure of the intact bacterial flagellar basal body. Nat Microbiol 6:712–721. doi: 10.1038/s41564-021-00895-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Minamino T, Macnab RM. 1999. Components of the salmonella flagellar export apparatus and classification of export substrates. J Bacteriol 181:1388–1394. doi: 10.1128/JB.181.5.1388-1394.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kuhlen L, Abrusci P, Johnson S, Gault J, Deme J, Caesar J, Dietsche T, Mebrhatu MT, Ganief T, Macek B, Wagner S, Robinson CV, Lea SM. 2018. Author correction: structure of the core of the type III secretion system export apparatus. Nat Struct Mol Biol 25:743. doi: 10.1038/s41594-018-0095-8 [DOI] [PubMed] [Google Scholar]
- 7. Abrusci P, Vergara-Irigaray M, Johnson S, Beeby MD, Hendrixson DR, Roversi P, Friede ME, Deane JE, Jensen GJ, Tang CM, Lea SM. 2013. Architecture of the major component of the type III secretion system export apparatus. Nat Struct Mol Biol 20:99–104. doi: 10.1038/nsmb.2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ohnishi K, Fan F, Schoenhals GJ, Kihara M, Macnab RM. 1997. The FliO, fliP, FliQ, and FliR proteins of Salmonella typhimurium: putative components for flagellar assembly. J Bacteriol 179:6092–6099. doi: 10.1128/jb.179.19.6092-6099.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fukumura T, Makino F, Dietsche T, Kinoshita M, Kato T, Wagner S, Namba K, Imada K, Minamino T. 2017. Assembly and stoichiometry of the core structure of the bacterial flagellar type III export gate complex. PLoS Biol 15:e2002281. doi: 10.1371/journal.pbio.2002281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fabiani FD, Renault TT, Peters B, Dietsche T, Gálvez EJC, Guse A, Freier K, Charpentier E, Strowig T, Franz-Wachtel M, Macek B, Wagner S, Hensel M, Erhardt M. 2017. A flagellum-specific chaperone facilitates assembly of the core type III export apparatus of the bacterial flagellum. PLoS Biol 15:e2002267. doi: 10.1371/journal.pbio.2002267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morimoto YV, Ito M, Hiraoka KD, Che Y-S, Bai F, Kami-Ike N, Namba K, Minamino T. 2014. Assembly and stoichiometry of FliF and FlhA in Salmonella flagellar basal body. Mol Microbiol 91:1214–1226. doi: 10.1111/mmi.12529 [DOI] [PubMed] [Google Scholar]
- 12. Johnson S, Fong YH, Deme JC, Furlong EJ, Kuhlen L, Lea SM. 2020. Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation. Nat Microbiol 5:966–975. doi: 10.1038/s41564-020-0703-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Francis NR, Sosinsky GE, Thomas D, DeRosier DJ. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol 235:1261–1270. doi: 10.1006/jmbi.1994.1079 [DOI] [PubMed] [Google Scholar]
- 14. Francis NR, Irikura VM, Yamaguchi S, DeRosier DJ, Macnab RM. 1992. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc Natl Acad Sci U S A 89:6304–6308. doi: 10.1073/pnas.89.14.6304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao R, Amsler CD, Matsumura P, Khan S. 1996. FliG and FliM distribution in the Salmonella typhimurium cell and flagellar basal bodies. J Bacteriol 178:258–265. doi: 10.1128/jb.178.1.258-265.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao R, Pathak N, Jaffe H, Reese TS, Khan S. 1996. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J Mol Biol 261:195–208. doi: 10.1006/jmbi.1996.0452 [DOI] [PubMed] [Google Scholar]
- 17. Notti RQ, Bhattacharya S, Lilic M, Stebbins CE. 2015. A common assembly module in injectisome and flagellar type III secretion sorting platforms. Nat Commun 6:7125. doi: 10.1038/ncomms8125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lam KH, Xue C, Sun K, Zhang H, Lam WWL, Zhu Z, Ng JTY, Sause WE, Lertsethtakarn P, Lau KF, Ottemann KM, Au SWN. 2018. Three SpoA-domain proteins interact in the creation of the flagellar type III secretion system in Helicobacter pylori. J Biol Chem 293:13961–13973. doi: 10.1074/jbc.RA118.002263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Henderson LD, Matthews-Palmer TRS, Gulbronson CJ, Ribardo DA, Beeby M, Hendrixson DR. 2020. Diversification of Campylobacter jejuni flagellar C-ring composition impacts its structure and function in motility, flagellar assembly, and cellular processes. mBio 11:e02286-19. doi: 10.1128/mBio.02286-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lowenthal AC, Hill M, Sycuro LK, Mehmood K, Salama NR, Ottemann KM. 2009. Functional analysis of the Helicobacter pylori flagellar switch proteins. J Bacteriol 191:7147–7156. doi: 10.1128/JB.00749-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, Li Z, Shi J, Tocheva EI, Müller A, Dobro MJ, Jensen GJ. 2011. Structural diversity of bacterial flagellar motors. EMBO J 30:2972–2981. doi: 10.1038/emboj.2011.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Minamino T, Yoshimura SDJ, Morimoto YV, González-Pedrajo B, Kami-Ike N, Namba K. 2009. Roles of the extreme N-terminal region of FliH for efficient localization of the FliH-FliI complex to the bacterial flagellar type III export apparatus. Mol Microbiol 74:1471–1483. doi: 10.1111/j.1365-2958.2009.06946.x [DOI] [PubMed] [Google Scholar]
- 23. Ibuki T, Imada K, Minamino T, Kato T, Miyata T, Namba K. 2011. Common architecture of the flagellar type III protein export apparatus and F- and V-type ATpases. Nat Struct Mol Biol 18:277–282. doi: 10.1038/nsmb.1977 [DOI] [PubMed] [Google Scholar]
- 24. Minamino T, Morimoto YV, Hara N, Namba K. 2011. An energy transduction mechanism used in bacterial flagellar type III protein export. Nat Commun 2:475. doi: 10.1038/ncomms1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ibuki T, Uchida Y, Hironaka Y, Namba K, Imada K, Minamino T. 2013. Interaction between FliJ and FlhA, components of the bacterial flagellar type III export apparatus. J Bacteriol 195:466–473. doi: 10.1128/JB.01711-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kojima S, Blair DF. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041–13050. doi: 10.1021/bi011263o [DOI] [PubMed] [Google Scholar]
- 27. Blair DF, Berg HC. 1990. The mota protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60:439–449. doi: 10.1016/0092-8674(90)90595-6 [DOI] [PubMed] [Google Scholar]
- 28. Kojima S, Blair DF. 2004. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 43:26–34. doi: 10.1021/bi035405l [DOI] [PubMed] [Google Scholar]
- 29. Wilson ML, Macnab RM. 1990. Co-overproduction and localization of the Escherichia coli motility proteins MotA and MotB. J Bacteriol 172:3932–3939. doi: 10.1128/jb.172.7.3932-3939.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chun SY, Parkinson JS. 1988. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science 239:276–278. doi: 10.1126/science.2447650 [DOI] [PubMed] [Google Scholar]
- 31. Dean GE, Macnab RM, Stader J, Matsumura P, Burks C. 1984. Gene sequence and predicted amino acid sequence of the MotA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J Bacteriol 159:991–999. doi: 10.1128/jb.159.3.991-999.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stader J, Matsumura P, Vacante D, Dean GE, Macnab RM. 1986. Nucleotide sequence of the Escherichia coli motB gene and site-limited incorporation of its product into the cytoplasmic membrane. J Bacteriol 166:244–252. doi: 10.1128/jb.166.1.244-252.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou J, Fazzio RT, Blair DF. 1995. Membrane topology of the MotA protein of Escherichia coli. J Mol Biol 251:237–242. doi: 10.1006/jmbi.1995.0431 [DOI] [PubMed] [Google Scholar]
- 34. Deme JC, Johnson S, Vickery O, Aron A, Monkhouse H, Griffiths T, James RH, Berks BC, Coulton JW, Stansfeld PJ, Lea SM. 2020. Structures of the stator complex that drives rotation of the bacterial flagellum. Nat Microbiol 5:1553–1564. doi: 10.1038/s41564-020-0788-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Santiveri M, Roa-Eguiara A, Kühne C, Wadhwa N, Hu H, Berg HC, Erhardt M, Taylor NMI. 2020. Structure and function of stator units of the bacterial flagellar motor. Cell 183:244–257. doi: 10.1016/j.cell.2020.08.016 [DOI] [PubMed] [Google Scholar]
- 36. Kojima S, Takao M, Almira G, Kawahara I, Sakuma M, Homma M, Kojima C, Imada K. 2018. The helix rearrangement in the periplasmic domain of the flagellar stator B subunit activates peptidoglycan binding and ion influx. Structure 26:590–598. doi: 10.1016/j.str.2018.02.016 [DOI] [PubMed] [Google Scholar]
- 37. Zhu S, Takao M, Li N, Sakuma M, Nishino Y, Homma M, Kojima S, Imada K. 2014. Conformational change in the periplamic region of the flagellar stator coupled with the assembly around the rotor. Proc Natl Acad Sci U S A 111:13523–13528. doi: 10.1073/pnas.1324201111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reboul CF, Andrews DA, Nahar MF, Buckle AM, Roujeinikova A. 2011. Crystallographic and molecular dynamics analysis of loop motions unmasking the peptidoglycan-binding site in stator protein MotB of flagellar motor. PLoS One 6:e18981. doi: 10.1371/journal.pone.0018981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Manson MD, Tedesco P, Berg HC, Harold FM, Van der Drift C. 1977. A protonmotive force drives bacterial flagella. Proc Natl Acad Sci U S A 74:3060–3064. doi: 10.1073/pnas.74.7.3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakamura S, Kami-ike N, Yokota JP, Kudo S, Minamino T, Namba K. 2009. Effect of intracellular pH on the torque-speed relationship of bacterial proton-driven flagellar motor. J Mol Biol 386:332–338. doi: 10.1016/j.jmb.2008.12.034 [DOI] [PubMed] [Google Scholar]
- 41. Glagolev AN, Skulachev VP. 1978. The proton pump is a molecular engine of motile bacteria. Nature 272:280–282. doi: 10.1038/272280a0 [DOI] [PubMed] [Google Scholar]
- 42. Morimoto YV, Nakamura S, Kami-ike N, Namba K, Minamino T. 2010. Charged residues in the cytoplasmic loop of MotA are required for stator assembly into the bacterial flagellar motor. Mol Microbiol 78:1117–1129. doi: 10.1111/j.1365-2958.2010.07391.x [DOI] [PubMed] [Google Scholar]
- 43. Morimoto YV, Nakamura S, Hiraoka KD, Namba K, Minamino T. 2013. Distinct roles of highly conserved charged residues at the MotA-FliG interface in bacterial flagellar motor rotation. J Bacteriol 195:474–481. doi: 10.1128/JB.01971-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Berg HC, Turner L. 1979. Movement of microorganisms in viscous environments. Nature 278:349–351. doi: 10.1038/278349a0 [DOI] [PubMed] [Google Scholar]
- 45. Schneider WR, Doetsch RN. 1974. Effect of viscosity on bacterial motility. J Bacteriol 117:696–701. doi: 10.1128/jb.117.2.696-701.1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Qu Z, Breuer KS. 2020. Effects of shear thinning viscosity and viscoleastic stresses on flagellated bacteria motility. Phys Rev Fluids 5:073103. doi: 10.1103/PhysRevFluids.5.073103 [DOI] [Google Scholar]
- 47. Martinez VA, Schwarz-Linek J, Reufer M, Wilson LG, Morozov AN, Poon WCK. 2014. Flagellated bacterial motility in polymer solutions. Proc Natl Acad Sci U S A 111:17771–17776. doi: 10.1073/pnas.1415460111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chaban B, Coleman I, Beeby M. 2018. Evolution of higher torque in Campylobacter-type bacterial flagellar motors. Sci Rep 8:97. doi: 10.1038/s41598-017-18115-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shigematsu M, Umeda A, Fujimoto S, Amako K. 1998. Spirochaete-like swimming mode of Campylobacter jejuni in a viscous environment. J Med Microbiol 47:521–526. doi: 10.1099/00222615-47-6-521 [DOI] [PubMed] [Google Scholar]
- 50. Ferrero RL, Lee A. 1988. Motility of Campylobacter jejuni in a viscous environment: comparison with conventional rod-shaped bacteria. J Gen Microbiol 134:53–59. doi: 10.1099/00221287-134-1-53 [DOI] [PubMed] [Google Scholar]
- 51. Karim QN, Logan RP, Puels J, Karnholz A, Worku ML. 1998. Measurement of motility of Helicobacter pylori, Campylobacter jejuni, and Escherichia coli by real time computer tracking using the Hobson BacTracker. J Clin Pathol 51:623–628. doi: 10.1136/jcp.51.8.623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Beeby M, Ribardo DA, Brennan CA, Ruby EG, Jensen GJ, Hendrixson DR. 2016. Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold. Proc Natl Acad Sci U S A 113:E1917–E1926. doi: 10.1073/pnas.1518952113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Guo S, Xu H, Chang Y, Motaleb MA, Liu J. 2022. FliL ring enhances the function of periplasmic flagella. Proc Natl Acad Sci U S A 119:e2117245119. doi: 10.1073/pnas.2117245119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chang Y, Xu H, Motaleb MA, Liu J, Søgaard-Andersen L. 2021. Characterization of the flagellar collar reveals structural plasticity essential for spirochete motility. mBio 12:e0249421. doi: 10.1128/mBio.02494-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xu H, He J, Liu J, Motaleb MA. 2020. BB0326 is responsible for the formation of periplasmic flagellar collar and assembly of the stator complex in Borrelia burgdorferi. Mol Microbiol 113:418–429. doi: 10.1111/mmi.14428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chang Y, Moon KH, Zhao X, Norris SJ, Motaleb MA, Liu J. 2019. Structural insights into flagellar stator-rotor interactions. Elife 8:e48979. doi: 10.7554/eLife.48979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Blair DF, Berg HC. 1988. Restoration of torque in defective flagellar motors. Science 242:1678–1681. doi: 10.1126/science.2849208 [DOI] [PubMed] [Google Scholar]
- 58. Reid SW, Leake MC, Chandler JH, Lo C-J, Armitage JP, Berry RM. 2006. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc Natl Acad Sci U S A 103:8066–8071. doi: 10.1073/pnas.0509932103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP. 2006. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443:355–358. doi: 10.1038/nature05135 [DOI] [PubMed] [Google Scholar]
- 60. Castillo DJ, Nakamura S, Morimoto YV, Che Y-S, Kami-Ike N, Kudo S, Minamino T, Namba K. 2013. The C-terminal periplasmic domain of MotB is responsible for load-dependent control of the number of stators of the bacterial flagellar motor. Biophysics 9:173–181. doi: 10.2142/biophysics.9.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tipping MJ, Delalez NJ, Lim R, Berry RM, Armitage JP. 2013. Load-dependent assembly of the bacterial flagellar motor. mBio 4:e00551-13. doi: 10.1128/mBio.00551-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lele PP, Hosu BG, Berg HC. 2013. Dynamics of mechanosensing in the bacterial flagellar motor. Proc Natl Acad Sci U S A 110:11839–11844. doi: 10.1073/pnas.1305885110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nord AL, Gachon E, Perez-Carrasco R, Nirody JA, Barducci A, Berry RM, Pedaci F. 2017. Catch bond drives stator mechanosensitivity in the bacterial flagellar motor. Proc Natl Acad Sci U S A 114:12952–12957. doi: 10.1073/pnas.1716002114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chang Y, Zhang K, Carroll BL, Zhao X, Charon NW, Norris SJ, Motaleb MA, Li C, Liu J. 2020. Molecular mechanism for rotational switching of the bacterial flagellar motor. Nat Struct Mol Biol 27:1041–1047. doi: 10.1038/s41594-020-0497-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Qin Z, Lin W-T, Zhu S, Franco AT, Liu J. 2017. Imaging the motility and chemotaxis machineries in Helicobacter pylori by cryo-electron tomography. J Bacteriol 199:e00695–e00616. doi: 10.1128/JB.00695-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhu S, Nishikino T, Takekawa N, Terashima H, Kojima S, Imada K, Homma M, Liu J. 2020. In situ structure of the Vibrio polar flagellum reveals a distinct outer membrane complex and its specific interaction with the stator. J Bacteriol 202:e00592-19. doi: 10.1128/JB.00592-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhu S, Nishikino T, Hu B, Kojima S, Homma M, Liu J. 2017. Molecular architecture of the sheathed polar flagellum in Vibrio alginolyticus. Proc Natl Acad Sci U S A 114:10966–10971. doi: 10.1073/pnas.1712489114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Moon KH, Zhao X, Manne A, Wang J, Yu Z, Liu J, Motaleb MA. 2016. Spirochetes flagellar collar protein FlbB has astounding effects in orientation of periplasmic flagella, bacterial shape, motility, and assembly of motors in Borrelia burgdorferi. Mol Microbiol 102:336–348. doi: 10.1111/mmi.13463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Moon KH, Zhao X, Xu H, Liu J, Motaleb MA. 2018. A tetratricopeptide repeat domain protein has profound effects on assembly of periplasmic flagella, morphology and motility of the lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 110:634–647. doi: 10.1111/mmi.14121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Morris DC, Peng F, Barker JR, Klose KE. 2008. Lipidation of an FlrC-dependent protein is required for enhanced intestinal colonization by Vibrio cholerae. J Bacteriol 190:231–239. doi: 10.1128/JB.00924-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Martinez RM, Dharmasena MN, Kirn TJ, Taylor RK. 2009. Characterization of two outer membrane proteins, FlgO and FlgP, that influence Vibrio cholerae motility. J Bacteriol 191:5669–5679. doi: 10.1128/JB.00632-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Terashima H, Fukuoka H, Yakushi T, Kojima S, Homma M. 2006. The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na-driven flagella and required for stator formation. Mol Microbiol 62:1170–1180. doi: 10.1111/j.1365-2958.2006.05435.x [DOI] [PubMed] [Google Scholar]
- 73. Kojima S, Shinohara A, Terashima H, Yakushi T, Sakuma M, Homma M, Namba K, Imada K. 2008. Insights into the stator assembly of the Vibrio flagellar motor from the crystal structure of MotY. Proc Natl Acad Sci U S A 105:7696–7701. doi: 10.1073/pnas.0800308105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Terashima H, Li N, Sakuma M, Koike M, Kojima S, Homma M, Imada K. 2013. Insight into the assembly mechanism in the supramolecular rings of the sodium-driven Vibrio flagellar motor from the structure of FlgT. Proc Natl Acad Sci U S A 110:6133–6138. doi: 10.1073/pnas.1222655110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Terashima H, Koike M, Kojima S, Homma M. 2010. The flagellar basal body-associated protein FlgT is essential for a novel ring structure in the sodium-driven Vibrio motor. J Bacteriol 192:5609–5615. doi: 10.1128/JB.00720-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Black RE, Levine MM, Clements ML, Hughes TP, Blaser MJ. 1988. Experimental Campylobacter jejuni infection in humans. J Infect Dis 157:472–479. doi: 10.1093/infdis/157.3.472 [DOI] [PubMed] [Google Scholar]
- 77. Lindblom GB, Sjörgren E, Kaijser B. 1986. Natural campylobacter colonization in chickens raised under different environmental conditions. J Hyg (Lond) 96:385–391. doi: 10.1017/s0022172400066146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wassenaar TM, Fry BN, van der Zeijst BA. 1993. Genetic manipulation of Campylobacter: evaluation of natural transformation and electro-transformation. Gene 132:131–135. doi: 10.1016/0378-1119(93)90525-8 [DOI] [PubMed] [Google Scholar]
- 79. Hendrixson DR, DiRita VJ. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol Microbiol 52:471–484. doi: 10.1111/j.1365-2958.2004.03988.x [DOI] [PubMed] [Google Scholar]
- 80. Lai SK, Wang Y-Y, Wirtz D, Hanes J. 2009. Micro- and macrorheology of mucus. Adv Drug Deliv Rev 61:86–100. doi: 10.1016/j.addr.2008.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Beery JT, Hugdahl MB, Doyle MP. 1988. Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni. Appl Environ Microbiol 54:2365–2370. doi: 10.1128/aem.54.10.2365-2370.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Skirrow MB, Blaser MJ. 2000. Clinical aspects of Campylobacter infection, p 69–88. In Nachamkin I, Blaser MJ (ed), Campylobacter, 2nd ed. ASM Press, Washington, DC. [Google Scholar]
- 83. Swidsinski A, Sydora BC, Doerffel Y, Loening-Baucke V, Vaneechoutte M, Lupicki M, Scholze J, Lochs H, Dieleman LA. 2007. Viscosity gradient within the mucus layer determines the mucosal barrier function and the spatial organization of the intestinal microbiota. Inflamm Bowel Dis 13:963–970. doi: 10.1002/ibd.20163 [DOI] [PubMed] [Google Scholar]
- 84. Atuma C, Strugala V, Allen A, Holm L. 2001. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am J Physiol Gastrointest Liver Physiol 280:G922–G929. doi: 10.1152/ajpgi.2001.280.5.G922 [DOI] [PubMed] [Google Scholar]
- 85. Cohen EJ, Nakane D, Kabata Y, Hendrixson DR, Nishizaka T, Beeby M. 2020. Campylobacter jejuni motility integrates specialized cell shape, flagellar filament, and motor, to coordinate action of its opposed flagella. PLoS Pathog 16:e1008620. doi: 10.1371/journal.ppat.1008620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Goon S, Ewing CP, Lorenzo M, Pattarini D, Majam G, Guerry P. 2006. A s28-regulated Nonflagella gene contributes to virulence of Campylobacter jejuni 81-176. Infect Immun 74:769–772. doi: 10.1128/IAI.74.1.769-772.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yokoyama T, Paek S, Ewing CP, Guerry P, Yeo H-J. 2008. Structure of a s28-regulated nonflagellar virulence protein from Campylobacter jejuni. J Mol Biol 384:364–376. doi: 10.1016/j.jmb.2008.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Barrero-Tobon AM, Hendrixson DR. 2012. Identification and analysis of flagellar coexpressed determinants (Feds) of Campylobacter jejuni involved in colonization. Mol Microbiol 84:352–369. doi: 10.1111/j.1365-2958.2012.08027.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Carrillo CD, Taboada E, Nash JHE, Lanthier P, Kelly J, Lau PC, Verhulp R, Mykytczuk O, Sy J, Findlay WA, Amoako K, Gomis S, Willson P, Austin JW, Potter A, Babiuk L, Allan B, Szymanski CM. 2004. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J Biol Chem 279:20327–20338. doi: 10.1074/jbc.M401134200 [DOI] [PubMed] [Google Scholar]
- 90. Novik V, Hofreuter D, Galán JE. 2010. Identification of Campylobacter jejuni genes involved in its interaction with epithelial cells. Infect Immun 78:3540–3553. doi: 10.1128/IAI.00109-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- 92. Logan SM, Trust TJ, Guerry P. 1989. Evidence for posttranslational modification and gene duplication of Campylobacter flagellin. J Bacteriol 171:3031–3038. doi: 10.1128/jb.171.6.3031-3038.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Guerry P, Logan SM, Thornton S, Trust TJ. 1990. Genomic organization and expression of Campylobacter flagellin genes. J Bacteriol 172:1853–1860. doi: 10.1128/jb.172.4.1853-1860.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Guerry P, Alm RA, Power ME, Logan SM, Trust TJ. 1991. Role of two flagellin genes in Campylobacter motility. J Bacteriol 173:4757–4764. doi: 10.1128/jb.173.15.4757-4764.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107–116. doi: 10.1016/j.cell.2010.01.018 [DOI] [PubMed] [Google Scholar]
- 96. Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a "backstop brake" mechanism. Mol Cell 38:128–139. doi: 10.1016/j.molcel.2010.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Pilizota T, Brown MT, Leake MC, Branch RW, Berry RM, Armitage JP. 2009. A molecular brake, not a clutch, stops the Rhodobacter sphaeroides flagellar motor. Proc Natl Acad Sci U S A 106:11582–11587. doi: 10.1073/pnas.0813164106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Subramanian S, Gao X, Dann CE, Kearns DB. 2017. MotI (DgrA) acts as a molecular clutch on the flagellar stator protein MotA in Bacillus subtilis. Proc Natl Acad Sci U S A 114:13537–13542. doi: 10.1073/pnas.1716231114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Guttenplan SB, Blair KM, Kearns DB. 2010. The EpsE flagellar clutch is bifunctional and synergizes with EPS biosynthesis to promote Bacillus subtilis biofilm formation. PLoS Genet 6:e1001243. doi: 10.1371/journal.pgen.1001243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. 2008. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320:1636–1638. doi: 10.1126/science.1157877 [DOI] [PubMed] [Google Scholar]
- 101. Schirmer T. 2016. C-di-GMP synthesis: structural aspects of evolution, catalysis and regulation. J Mol Biol 428:3683–3701. doi: 10.1016/j.jmb.2016.07.023 [DOI] [PubMed] [Google Scholar]
- 102. Krasteva PV, Giglio KM, Sondermann H. 2012. Sensing the messenger: the diverse ways that bacteria signal through c-di-GMP. Protein Sci 21:929–948. doi: 10.1002/pro.2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ryan RP, Tolker-Nielsen T, Dow JM. 2012. When the PilZ don't work: effectors for cyclic di-GMP action in bacteria. Trends Microbiol 20:235–242. doi: 10.1016/j.tim.2012.02.008 [DOI] [PubMed] [Google Scholar]
- 104. Cox CA, Bogacz M, El Abbar FM, Browning DD, Hsueh BY, Waters CM, Lee VT, Thompson SA. 2021. The Campylobacter jejuni response regulator and cyclic-di-GMP binding CbrR is a novel regulator of flagellar motility. Microorganisms 10:86. doi: 10.3390/microorganisms10010086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. D’Argenio DA, Calfee MW, Rainey PB, Pesci EC. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol 184:6481–6489. doi: 10.1128/JB.184.23.6481-6489.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Gao B, Lara-Tejero M, Lefebre M, Goodman AL, Galán JE, Blaser MJ. 2014. Novel components of the flagellar system in epsilonproteobacteria. mBio 5:e01349-14. doi: 10.1128/mBio.01349-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Belas R. 2014. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol 22:517–527. doi: 10.1016/j.tim.2014.05.002 [DOI] [PubMed] [Google Scholar]
- 108. O’Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x [DOI] [PubMed] [Google Scholar]
- 109. McCarter L, Hilmen M, Silverman M. 1988. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell 54:345–351. doi: 10.1016/0092-8674(88)90197-3 [DOI] [PubMed] [Google Scholar]
- 110. Cairns LS, Marlow VL, Bissett E, Ostrowski A, Stanley-Wall NR. 2013. A mechanical signal transmitted by the flagellum controls signalling in Bacillus subtilis. Mol Microbiol 90:6–21. doi: 10.1111/mmi.12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Chawla R, Gupta R, Lele TP, Lele PP. 2020. A skeptic's fuide to bacterial mechanosensing. J Mol Biol 432:523–533. doi: 10.1016/j.jmb.2019.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Padron GC, Shuppara AM, Palalay J-J, Sharma A, Sanfilippo JE. 2023. Bacteria in fluid flow. J Bacteriol 205:e0040022. doi: 10.1128/jb.00400-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Sanfilippo JE, Lorestani A, Koch MD, Bratton BP, Siryaporn A, Stone HA, Gitai Z. 2019. Microfluidic-based transcriptomics reveal force-independent bacterial rheosensing. Nat Microbiol 4:1274–1281. doi: 10.1038/s41564-019-0455-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi: 10.1073/pnas.76.4.1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hendrixson DR, Akerley BJ, DiRita VJ. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol Microbiol 40:214–224. doi: 10.1046/j.1365-2958.2001.02376.x [DOI] [PubMed] [Google Scholar]
- 116. Sommerlad SM, Hendrixson DR. 2007. Analysis of the roles of FlgP and FlgQ in flagellar motility of Campylobacter jejuni. J Bacteriol 189:179–186. doi: 10.1128/JB.01199-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Yao R, Alm RA, Trust TJ, Guerry P. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127–130. doi: 10.1016/0378-1119(93)90355-7 [DOI] [PubMed] [Google Scholar]
- 118. Gulbronson CJ, Ribardo DA, Balaban M, Knauer C, Bange G, Hendrixson DR. 2016. Flhg employs diverse intrinsic domains and influences Flhf Gtpase activity to numerically regulate polar Flagellar Biogenesis in Campylobacter Jejuni. Molecular microbiology 99:291–306. doi: 10.1111/mmi.13231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Luethy PM, Huynh S, Parker CT, Hendrixson DR. 2015. Analysis of the activity and Regulon of the two-component regulatory system composed by Cjj81176_1484 and Cjj81176_1483 of Campylobacter Jejuni. Journal of bacteriology 197:1592–1605. doi: 10.1128/JB.02564-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Beauchamp JM, Leveque RM, Dawid S, DiRita VJ. 2017. Methylation-dependent DNA discrimination in natural transformation of Campylobacter jejuni. Proc Natl Acad Sci U S A 114:E8053–E8061. doi: 10.1073/pnas.1703331114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Wiesner RS, Hendrixson DR, DiRita VJ. 2003. Natural transformation of Campylobacter jejuni requires components of a type II secretion system. J Bacteriol 185:5408–5418. doi: 10.1128/JB.185.18.5408-5418.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Guerry P, Yao R, Alm RA, Burr DH, Trust TJ. 1994. Systems of experimental genetics for Campylobacter species. Methods Enzymol 235:474–481. doi: 10.1016/0076-6879(94)35163-5 [DOI] [PubMed] [Google Scholar]
- 123. Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol 65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Joslin SN, Hendrixson DR. 2009. Activation of the Campylobacter jejuni FlgSR two-component system is linked to the flagellar export apparatus. J Bacteriol 191:2656–2667. doi: 10.1128/JB.01689-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Kazmierczak BI. 2017. Synthesis of [(32)P]-c-di-GMP for diguanylate cyclase and phosphodiesterase activity determinations. Methods Mol Biol 1657:285–289. doi: 10.1007/978-1-4939-7240-1_22 [DOI] [PubMed] [Google Scholar]
- 126. Seminara AB, Turdiev A, Turdiev H, Lee VT. 2019. Differential radial capillary action of ligand assay (DRaCALA). Curr Protoc Mol Biol 126:e84. doi: 10.1002/cpmb.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 and S2 and Movie S1-S8 legends.
WT swimming motility at low viscosity (1 cP).
WT swimming motility at high viscosity (40 cP).
vidA mutant swimming motility at low viscosity (1 cP)
vidA mutant swimming motility at high viscosity (40 cP).
vidA vidB mutant swimming motility at low viscosity.
vidA vidB swimming motility at high viscosity (40 cP).
WT C.jejuni overexpressing VidB swimming motility at low viscosity (1 cP).
WT C. jejuni overexpressing VidB swimming motility at high viscosity (40 cP).
Data Availability Statement
All data and methodologies are available upon request. Genomic sequences are available at the National Center for Biotechnology Information as part of Bioproject PRJNA987628, with individual genomic sequences assigned the following accession numbers: ABT501 (SRR25146826), ABT501 S1 (SRR25146825), ABT501 S2 (https://www.ncbi.nlm.nih.gov/sra/SRR25146820SRR25146820), ABT501 S3 (SRR25146819), ABT501 S4 (SRR25146818), ABT501 S5 (SRR25146817), ABT501 S6 (SRR25146816), ABT501 S7 (SRR25146815), ABT501 S8 (SRR25146814), ABT501 S9 (SRR25146813), ABT501 S10 (SRR25146824), ABT501 S11 (SRR25146823), ABT501 S12 (SRR25146822), and ABT501 S13 (SRR25146821).