ABSTRACT
Following an oral inoculation, Chlamydia muridarum descends to the mouse large intestine for long-lasting colonization. However, a mutant C. muridarum that lacks the plasmid-encoded protein pGP3 due to an engineered premature stop codon (designated as CMpGP3S) failed to do so even following an intrajejunal inoculation. This was because a CD4+ T cell-dependent immunity prevented the spread of CMpGP3S from the small intestine to the large intestine. In the current study, we found that mice deficient in IL-22 (IL-22−/−) allowed CMpGP3S to spread from the small intestine to the large intestine on day 3 after intrajejunal inoculation, indicating a critical role of IL-22 in regulating the chlamydial spread. The responsible IL-22 is produced by CD4+ T cells since IL-22−/− mice were rescued to block the CMpGP3S spread by donor CD4+ T cells from C57BL/6J mice. Consistently, CD4+ T cells lacking IL-22 failed to block the spread of CMpGP3S in Rag2−/− mice, while IL-22-competent CD4+ T cells did block. Furthermore, mice deficient in cathelicidin-related antimicrobial peptide (CRAMP) permitted the CMpGP3S spread, but donor CD4+ T cells from CRAMP−/− mice were still sufficient for preventing the CMpGP3S spread in Rag2−/− mice, indicating a critical role of CRAMP in regulating chlamydial spreading, and the responsible CRAMP is not produced by CD4+ T cells. Thus, the IL-22-producing CD4+ T cell-dependent regulation of chlamydial spreading correlated with CRAMP produced by non-CD4+ T cells. These findings provide a platform for further characterizing the subset(s) of CD4+ T cells responsible for regulating bacterial spreading in the intestine.
KEYWORDS: chlamydial spreading to large intestine, IL-22+ CD4+ T cells
INTRODUCTION
Chlamydia trachomatis, a common cause of sexually transmitted bacterial disease in humans (1), is frequently detected in the gastrointestinal (GI) tract (2–9). However, it remains unclear how and why C. trachomatis colonizes the human gut. Interestingly, C. muridarum, a mouse-adapted species, which has been used to study chlamydial pathogenic mechanisms in the female genital tract (10–15), was also found to colonize the mouse GI tract (4, 16–19). Recent studies have revealed that GI tract C. muridarum can significantly impact chlamydial pathogenicity in the genital tract. On one hand, C. muridarum in the GI tract can promote chlamydial pathogenicity in the upper genital tract after C. muridarum spreads from the genital tract into the GI tract (20–22); on the other, when a naive mouse is first exposed to C. muridarum in the GI tract, the mouse is orally immunized against subsequent chlamydial infections in extra-gut tissues (23–27). Thus, investigating the mechanisms of C. muridarum’s interactions with the GI tract may both promote our understanding of chlamydial pathogenic mechanisms and facilitate the development of oral vaccines against chlamydial infection in the genital tract.
Using the C. muridarum-mouse gut interaction model, various chlamydial factors have been found to promote C. muridarum colonization of the mouse GI tract (20, 28–31). C. muridarum deficient in the plasmid-encoded pGP3 due to an engineered premature stop codon in the pgp3 gene (designated as CMpGP3S) is both attenuated in genital pathogenicity and defective in colonizing the GI tract (29). Orally inoculated CMpGP3S failed to appear in the rectal swabs. However, C. muridarum which lacks the entire plasmid (designated as CMpf) still reached the large intestine and appeared in the rectal swab (28). This phenotype was reproduced following intrajejunal inoculation (30, 32, 33), indicating that CMpGP3S is unable to spread from the small intestine to the large intestine, while CMpf can. Since CMpGP3S is more invasive than CMpf in the mouse genital tract (34), the above observations led to the hypothesis that CMpGP3S might be prevented from spreading into the large intestine by an intestinal barrier that could be activated by CMpGP3S but not CMpf. This hypothesis is supported by the observation that both CMpGP3S and CMpf successfully colonized the colon following intracolonic inoculation (30). Furthermore, when CMpGP3S and CMpf were co-inoculated into the same jejunum, no live chlamydial organism was recovered from the large intestine, although the intrajejunally inoculated CMpf alone still reached the large intestine. Clearly, CMpGP3S might activate an intestinal barrier function for blocking both its own spreading and the spreading of the co-inoculated CMpf (32, 33). Consistently, CMpGP3S still carries the remaining seven plasmid genes, which may render it more stimulatory or invasive than CMpf lacking all plasmid genes (35). Although wild-type C. muridarum must also activate the intestinal barrier, it is still able to spread to the large intestine. This is because wild-type C. muridarum may use its functional pGP3 to evade the intestinal barrier since pGP3 is a known virulence factor that can block immune effector mechanisms, including neutralizing the anti-chlamydial activity of the cathelicidin-related antimicrobial peptide or CRAMP (29, 32, 36–41). Obviously, the above observations raise many interesting questions. For example, how is the intestinal barrier induced by plasmid-encoded non-pGP3 factors? How does the plasmid-encoded pGP3 help the chlamydial organisms evade the intestinal barrier? What is the immunological basis of the intestinal barrier?
By taking advantage of the failure of the intrajejunally inoculated CMpGP3S to spread to the large intestine as a readout, we have recently identified the immunological basis of the intestinal barrier (33). We found that mice deficient in TLR/MyD88- or STING-mediated signaling pathways still prevented CMpGP3S from spreading, while Rag1−/− mice failed to do so, suggesting a critical role of conventional lymphocytes. CD4−/− but not CD8−/− nor μ−/− mice failed to block the spread of CMpGP3S, demonstrating the dependence of the intestinal barrier on CD4+ cells. Consistently, CD4+ T cells but not CD8+ T cells nor B cells restored the intestinal barrier function in CD4−/− or Rag1−/− mice. Thus, CD4+ T cells are both necessary and sufficient for the intestinal barrier to regulate chlamydial spread in the intestine.
The current study was designed to further determine the effector mechanisms for the CD4+ T cell-dependent immunity using the CMpGP3S-jejunal interaction model. Mice deficient in IL-22 allowed the spread of CMpGP3S, while C57BL/6J mice did not, indicating a critical role of IL-22 in regulating the chlamydial spread. The responsible IL-22 is likely produced by CD4+ T cells since the IL-22-deficient mice were rescued to block the CMpGP3S spread by donor CD4+ T cells isolated from C57BL/6J mice. Furthermore, CD4+ T cells that lack IL-22 failed to restore Rag2-deficient mice to inhibit the spread of CMpGP3S, while IL-22-competent CD4+ T cells did successfully. Finally, mice deficient in CRAMP rescued the spreading of CMpGP3S, which correlated with the IL-22-producing CD4+ T cell-dependent regulation of chlamydial spreading. These observations together lay a foundation for further revealing the mechanisms of T cell interaction with enteric bacteria.
RESULTS
IL-22 is essential for inhibiting the spread of pGP3-deficient C. muridarum (CMpGP3S) from the small intestine to the large intestine
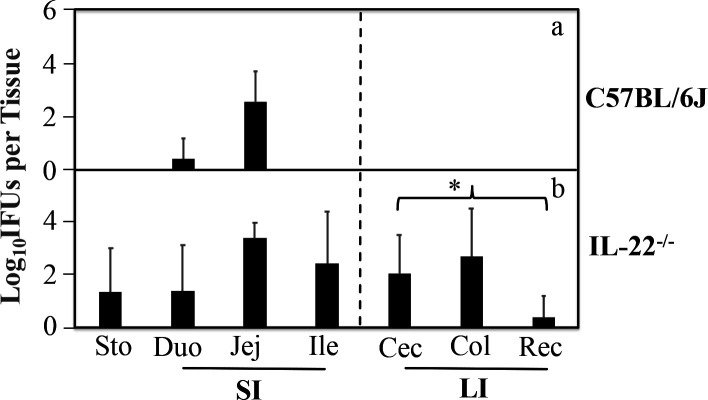
Since IL-22 is required for maintaining intestinal homeostasis (42, 43) and a major cytokine secreted by CD4+ T subset called Th22 (27, 44), we hypothesized that IL-22 might be important for the CD4+ T cell-dependent intestinal barrier to block the spreading of CMpGP3S. To test this hypothesis, CMpGP3S was intrajejunally inoculated to wild-type C57BL/6J and IL-22-deficient (IL-22−/−) mice followed by monitoring live chlamydial organism recoveries from the large intestinal tissues on day 3 (Fig. 1). IL-22−/− but not C57BL/6J mice were detected with significant numbers of live chlamydial organisms in the large intestine (P < 0.05), although live chlamydial organisms were detected in the small intestinal tissues of both groups of mice. All IL-22−/− mice were positive for live chlamydial organisms in the colon with an average chlamydial burden of ~2,500 inclusion-forming units (IFUs) per colon, while none of the C57BL/6J mice was found positive for a live chlamydial organism in the colon. These results have demonstrated that IL-22 is essential for regulating chlamydial spreading from the small intestine to the large intestine.
Fig 1.
The effect of IL-22 deficiency on the spreading of pGP3-deficient C. muridarum from the small intestine into the large intestine. C57BL/6J mice without (panel a, n = 4) or with deficiency in IL-22 (b, IL-22−/−, n = 4) were inoculated with 1 × 105 inclusion forming units of pGP3-deficient C. muridarum (CMpGPG3S) via intrajejunal injection. Three days after the inoculation, mice were sacrificed for collecting stomach (Sto), small intestine (SI) tissues [duodenum (Duo), jejunum (Jej), and ileum (Ile)], and large intestine (LI) tissues [Cecum (Cec), colon (Col) and rectum (Rec)] as listed along the X-axis. Live CMpGP3S organisms were recovered from each tissue sample and expressed as log10 IFUs per tissue. The data came from two independent experiments. Note that significant levels of live chlamydial organisms were detected in the large intestine of IL-22−/− mice. * denotes an observed P-value <0.05, based on a Wilcoxon rank-sum test, between C57 and IL-22−/− in IFU recovery from the overall large intestinal tissues.
CD4+ T cells from C57BL/6J mice are sufficient for rescuing IL-22-deficient mice to block the spread of CMpGP3S from the small intestine to the large intestine
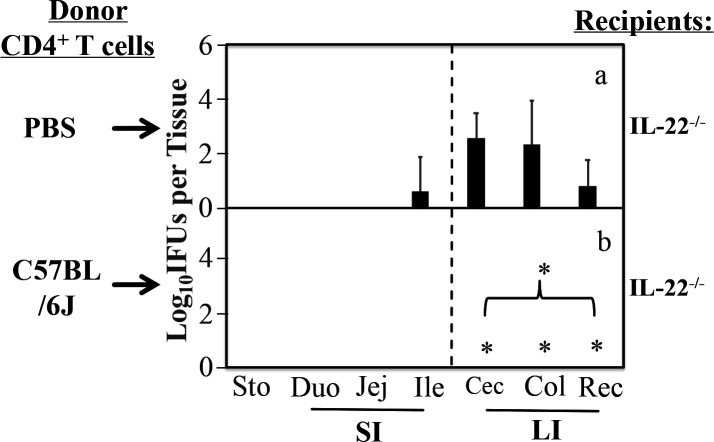
Although the spreading of CMpGP3S from the small intestine to the large intestine was blocked by CD4+ T cells (32, 33) and IL-22 (Fig. 1 of the current study), respectively, there is still a lack of direct evidence on whether the responsible IL-22 is secreted by CD4+ T cells. Thus, wild-type CD4+ T cells were evaluated for their ability to block the spread of CMpGP3S in IL-22−/−-recipient mice following an adoptive transfer (Fig. 2). The adoptive transfer was carried out twice via retro-orbital injection, 3 days prior to and 1 day after intrajejunal infection, respectively. It was found that after receiving donor CD4+ T cells (purified from the spleen of naïve wild-type C57BL/6J mice), IL-22−/− mice re-established the intestinal barrier for blocking the spread of the intrajejunally inoculated CMpGP3S into the large intestine. As a result, no live chlamydial organisms were recovered from the large intestinal tissues of these IL-22−/− mice. However, the group of IL-22−/− mice receiving PBS buffer alone displayed significant numbers of live chlamydial organisms in the large intestine (P < 0.05). These observations have demonstrated that the donor CD4+ T cells produce IL-22 to compensate for the IL-22 function lost in IL-22−/− mice. However, it is also possible that the donor CD4+ T cells may inhibit the chlamydial spread via IL-22-independent mechanisms.
Fig 2.
Effect of CD4+ T cells from C57BL/6J mice as donor cells on the spreading of pGP3-deficient C. muridarum from the small intestine into the large intestine in the IL-22−/−-recipient mice. IL-22−/−-recipient mice were adoptively transferred without (panel a, n = 4) or with (b, n = 5) 1 × 106 donor CD4+ T cells purified from C57BL/6J mice. The transfer was carried out via retro-orbital injection twice 3 days before and 1 day after infection, respectively. Mice were infected via intrajejunal inoculation with 1 × 105 IFUs of pGP3-deficient C. muridarum (CMpGP3S). Three days after inoculation, mice were sacrificed for collecting gastrointestinal tissues as listed along the X-axis. Live CMpGP3S organisms were recovered from each tissue sample and expressed as log10 IFUs per tissue. The data came from two independent experiments. Note that IL-22−/− mice receiving wild-type donor CD4+ T cells completely prevented the spreading of live chlamydial organisms into the large intestine. * denotes an observed P-value <0.05, based on a Wilcoxon rank-sum test, a comparison of the live chlamydial organisms in each of the large intestinal tissues and overall.
CD4+ T cells from IL-22-deficient mice are insufficient for rescuing the Rag2-deficient mice to block the spread of CMpGP3S
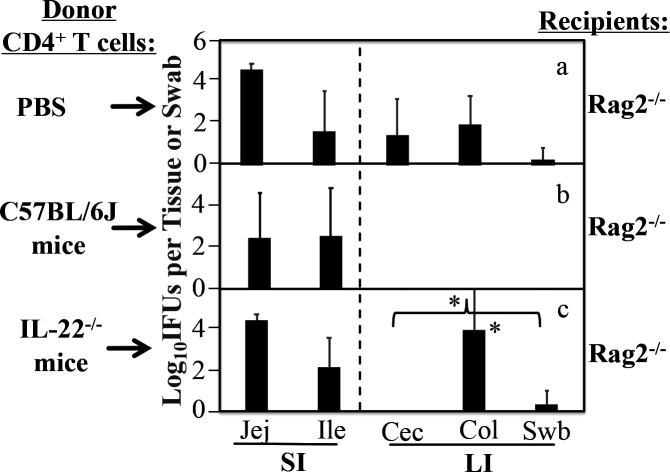
To further determine whether IL-22 secreted by CD4+ T cells is necessary for blocking the chlamydial spreading, we compared IL-22-competent and IL-22-deficient CD4+ T cells as donor cells for inhibiting the spread of CMpGP3S from the small intestine into the large intestine in Rag2-deficient mice (Fig. 3). In this experiment, mice deficient in Rag2 (Rag2−/−) were used as recipients since they are defective in inhibiting the chlamydial spread but can be rescued with an adoptive transfer of wild-type CD4+ T cells (33). It was found that although Rag2−/− mice receiving PBS alone allowed a significant spread of CMpGP3S from the small intestine into the large intestine, adoptive transfer of wild-type CD4+ T cells blocked the chlamydial spread. Importantly, when CD4+ T cells purified from the spleen of IL-22−/− mice were used as donor cells, the Rag2−/− mice still allowed significant spread of CMpGP3S. The chlamydial spreading in these mice paralleled that observed in Rag2−/− mice receiving buffer only. The number of live chlamydial organisms recovered from the large intestine samples of these two groups was significantly higher than that recovered from the group receiving wild-type CD4+ T cells (P < 0.05). These results have demonstrated that IL-22 production is necessary for CD4+ T cells to block chlamydial spread from the small intestine to the large intestine.
Fig 3.
Effect of IL-22-deficient CD4+ T cells as donor cells on pGP3-deficient C. muridarum spreading from small intestine into large intestine in Rag2−/− mice. Mice deficient in Rag2 (Rag2−/−) listed on the right as recipient mice were adoptively transferred without (panel a, n = 5) or with 1 × 106 donor CD4+ T cells purified from C57BL/6J (b, n = 5) or IL-22−/− (c, n = 4) mice. The transfer was performed via retro-orbital injection twice, 3 days before and 1 day after infection, respectively. Mice were infected via intrajejunal injection with 1 × 105 IFUs of CMpGP3S. Three days after inoculation, rectal swabs (Swb) were collected, and all mice were sacrificed for collecting gastrointestinal tissues as listed along the X-axis. Live CMpGP3S organisms were recovered from each sample and expressed as log10 IFUs per tissue or swab. The data came from two or three independent experiments. Note that IL-22-deficient CD4+ T cells were insufficient for rescuing Rag2−/− mice to block the spreading of live chlamydial organisms into the large intestine. * denotes an observed P-value <0.05, based on a Wilcoxon rank-sum test, comparison of the live chlamydial organisms in the colon and large intestinal tissues overall between panels b and c.
The cathelicidin-related antimicrobial peptide is essential for inhibiting the spread of CMpGP3S from the small intestine to the large intestine
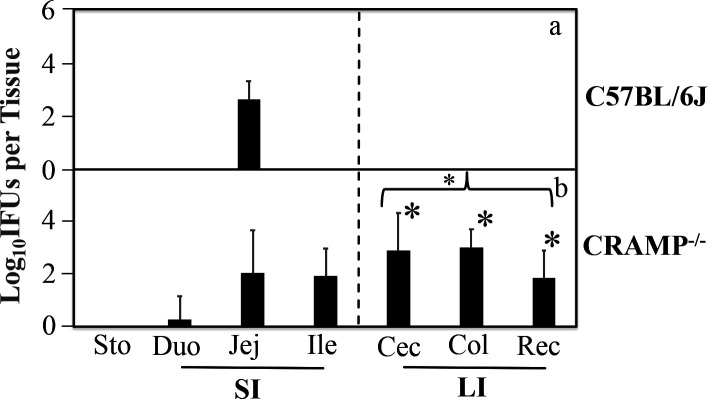
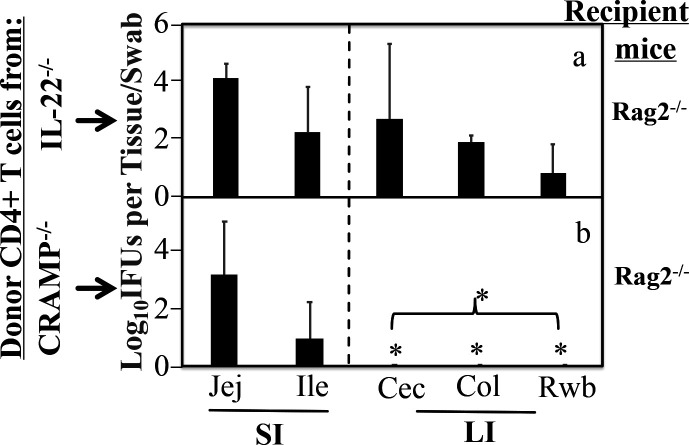
Since CRAMP is an important antimicrobial peptide in the intestinal barrier (45–48) and its anti-chlamydial activity is blocked by the chlamydia-secreted Pgp3 (38, 39), we hypothesized that CRAMP might be important for the IL-22-producing CD4+ T cell-dependent intestinal barrier to block the spreading of CMpGP3S from the small intestine into the large intestine. To test this hypothesis, we intrajejunally infected wild-type C57BL/6J and CRAMP-deficient (CRAMP−/−) mice with CMpGP3S and compared the number of live chlamydial organisms recovered from the large intestinal tissues of both groups on day 3 after the intrajejunal inoculation (Fig. 4). The CRAMP−/− mice had significant numbers of live chlamydial organisms in the large intestine, while C57BL/6J mice had none (P < 0.05), demonstrating an essential role of CRAMP in regulating chlamydial spreading from the small intestine to the large intestine. To further determine the cellular source of CRAMP, we compared CD4+ T cells from CRAMP−/− versus IL-22−/− mice as donor cells for rescuing Rag2−/−-recipient mice to block the CMpGP3S spreading (Fig. 5). It was found that CRAMP−/− but not IL-22−/− donor CD4+ T cells inhibited the spreading of Pgp3-deficient C. muridarum from the small intestine into the large intestine in the Rag2−/−-recipient mice. These results have demonstrated that the CRAMP responsible for inhibiting CMpGP3S spreading is not produced by CD4+ T cells, suggesting that CRAMP may be produced by non-CD4+ T cells for inhibiting CMpGP3S in response to the signaling from IL-22-producing CD4+ T cells. However, further studies are required to identify the precise cell type responsible for producing CRAMP and to determine whether directly delivering CRAMP to the CMpGP3S inoculation site is sufficient for blocking the spread of CMpGP3S without the signals from IL-22-producing CD4+ T cells.
Fig 4.
Effect of CRAMP deficiency on the spreading of pGP3-deficient C. muridarum from the small intestine into the large intestine. C57BL/6J mice without (panel a, n = 4) or with a deficiency in CRAMP (b, CRAMP−/−, n = 7) were inoculated with 1 × 105 IFUs of CMpGP3S via intrajejunal injection. Three days after the inoculation, mice were sacrificed to collect stomach (Sto), small intestine (SI) tissues [duodenum (Duo), jejunum (Jej), and ileum (Ile)], and large intestine (LI) tissues [cecum (Cec), colon (Col), and rectum (Rec)] as listed along the X-axis. Live CMpGP3S organisms were recovered from each tissue sample and expressed as log10 IFUs per tissue. The data came from two independent experiments. Note that significant levels of live chlamydial organisms were detected in the large intestine of CRAMP−/− mice. * denotes an observed P-value <0.05, based on a Wilcoxon rank-sum test, between C57 and CRAMP−/− in IFU recovery from Cec, Col, and Rec, respectively, as well as the overall large intestinal tissues.
Fig 5.
CRAMP-deficient CD4+ T cells can still inhibit the spreading of Pgp3-deficient C. muridarum from the small intestine into the large intestine in Rag2−/−-recipient mice. Rag2−/−-recipient mice were adoptively transferred with 1 × 106 donor CD4+ T cells purified from IL-22−/− (panel a, # of recipient mice or n = 4) or CRAMP−/− mice (panel b, n = 4). The transfer was carried out retro-orbitally twice 3 days before and 1 day after infection, respectively. Mice were infected via intrajejunal inoculation with 1 × 105 IFUs of CMpGP3S. Three days after the inoculation, rectal swabs (Rwb) were collected, and mice were sacrificed to collect small intestine (SI) tissues [jejunum (Jej) and ileum (Ile)] and large intestine (LI) tissues [cecum (Cec) and Colon (Col)] as listed along the X-axis. Live CMpGP3S organisms were recovered from each tissue sample and expressed as log10 IFUs per tissue or swab. The data came from two independent experiments. Note that Rag2−/− mice were rescued to block the spreading of CMpGP3S into the large intestine by CRAMP−/− but not IL-22−/− donor CD4+ T cells. * denotes an observed P-value <0.05 based on a Wilcoxon rank-sum test, comparing the live chlamydial organisms in the Cec, Col, and Rwb as well as overall large intestinal tissues between Rag2−/− mice receiving CRAMP−/− and IL-22−/− donor CD4+ T cells.
DISCUSSION
The frequent detection of the obligate intracellular bacterium C. trachomatis in the human GI tract (5–9) begs an urgent question about the medical significance of the GI tract C. trachomatis. Since the mouse-adapted C. muridarum colonizes the mouse GI tract (4, 16, 49, 50), the murine model has been used for investigating the significance of GI tract Chlamydia. The mouse studies have led to the conclusion that GI tract C. muridarum can significantly impact C. muridarum pathogenicity in the genital tract depending on the order of tissue exposure to C. muridarum. When the first exposure to C. muridarum is in the genital tract, the GI tract C. muridarum may promote genital C. muridarum pathogenicity (20), while an initial oral exposure to C. muridarum becomes oral vaccination, leading to the protection against C. muridarum pathogenicity in the genital tract (23). These interesting findings have motivated further investigations of the mechanisms of C. muridarum-mouse gut interactions using various approaches including C. muridarum mutants, knockout mice, depletion/blockade, and adoptive transfers (19). By taking advantage of the failure of the CMpGP3S mutant to spread from the small intestine to the large intestine, a CD4+ T cell-dependent immunity was identified for regulating chlamydial spread in the intestine (32, 33). The current study has further characterized as the CD4+ T cell-dependent immunological barrier function by determining the roles of IL-22 and CRAMP in inhibiting the CMpGP3S spread. First, IL-22−/− mice reproduced the phenotype of CD4−/− mice or Rag−/− mice (33) by allowing significant spread of CMpGP3S. Second, IL-22-competent CD4+ T cells rescued IL-22−/− mice to block the spread. Third, CD4+ T cells that can no longer produce IL-22 failed to block the CMpGP3S spread in Rag2−/− mice, while IL-22-competent CD4+ T cells successfully restored the blockade. Thus, IL-22+ CD4+ T cells may play a critical role in regulating bacterial spreading into the large intestine. Finally, the IL-22+ CD4+ T cell-dependent regulation of chlamydial spreading further correlated with CRAMP-mediated blockade of the CMpGP3S spread.
The discovery of an immunological barrier for regulating chlamydial spread in the intestine was made from a surprising observation: plasmid-free C. muridarum (CMpf) reached the large intestine while the pGP3-deficient C. muridarum (CMpGP3S) failed to do so following oral or intrajejunal inoculation. Since CMpGP3S is more invasive than CMpf in the genital tract (34) and both CMpGP3S and CMpf can colonize the large intestine well following intracolon inoculation, the above observation has led us to hypothesize that CMpGP3S must activate an immune response for blocking its own spreading into the large intestine. The current study has revealed that IL-22+ CD4+ T cells and CRAMP are critical components of the CMpGP3S-activated barrier. CRAMP is likely downstream of the IL-22+ CD4+ T cells since CD4+ T cells lacking CRAMP can still confer the inhibition of the CMpGP3S spread in the Rag2−/−-recipient mice. However, the precise relationship between IL-22+ CD4+ T cells and the CRAMP-producing cells is to be addressed. In addition, many questions remain. How is the IL-22+ CD4+ T cell-mediated immunity induced by CMpGP3S? What is the nature of the responsible IL-22+ CD4+ T cells? What is the effector mechanism by which the IL-22+ CD4+ T cell-mediated immunity blocks the CMpGP3S spread? How is the IL-22+ CD4+ T cell-mediated immunity evaded by wild-type Chlamydia?
Although the responsible IL-22+ CD4+ T cells are conventional lymphocytes, their function for regulating the chlamydial spread may not depend on their antigen receptor recognition of chlamydial epitopes since CD4+ T cells isolated from native C57BL/6 J mice are sufficient for inhibiting the CMpGP3S spread on day 3 after CMpGP3S inoculation. In other words, it may be the innate function of the conventional lymphocytes that is responsible for blocking CMpGP3S spread. Intracellular bacteria have been shown to re-activate CD4+ T cells via a TCR-independent or non-cognate mechanism (51, 52). The non-cognate mechanism is largely dependent on signaling from cytokine receptors and/or pattern recognition receptors (53). Chlamydial organisms or their components have been shown to directly bind to lymphocytes (54–56). It will be interesting to test whether the intrajejunally injected chlamydial organisms can rapidly activate IL-22+ CD4+ T lymphocytes in the small intestine.
We are aware that the knowledge learned from the mouse model may not directly apply to C. trachomatis infection in humans. Although C. trachomatis is detected in the human GI tracts (5–9), it is unclear whether genital C. trachomatis can spread to the GI tract. Nevertheless, the mechanisms by which C. muridarum interacts with mouse mucosal tissues may still be useful for revealing how C. trachomatis interacts with human gut mucosal tissues. It is worth noting that the focus of the current study is on C. muridarum spreading along the mouse gut lumen. It is possible that Chlamydia may spread in the gut via different pathways, such as the hematogenous route (50). Clearly, more efforts are required to further investigate the mechanisms of enteric bacterial spread from the small intestine to the large intestine.
MATERIALS AND METHODS
Chlamydial organisms
The chlamydial organisms used in the current study are pGP3-deficient Chlamydia muridarum produced previously (34, 57, 58). Briefly, to knock out the plasmid-encoded pGP3, the plasmid-free Chlamydia muridarum (CMpf) clone CMUT3.G5 was derived from C. muridarum strain Nigg3 (GenBank accession no. CP009760.1). CMUT3.G5 was further used as a recipient strain for transformation with a plasmid that carries a premature stop codon in the pgp3 gene to produce the pGP3-deficient clone CMpGP3S. The CMpGP3S organisms were grown in HeLa cells (human cervical carcinoma epithelial cells; ATCC# CCL-2) and purified as elementary bodies (EBs) using discontinuous density centrifugation as previously described (59, 60). The purified EBs were stored in aliquots in sucrose-phosphate-glutamic acid (SPG) buffer (0.2 M sucrose, 20 mM sodium phosphate at pH 7.4, and 5 mM glutamic acid) at −80°C. A frozen aliquot was used for titrating the infectious particles in the form of IFUs.
Mouse infection
All mice used in the current study were 6–8-week-old females or males. They were wild-type C57BL/6J (stock# 000664, Jackson Laboratories, Inc., Bar Harbor, Maine) and mice deficient in Rag2 (Rag2−/− or lacking adaptive immunity, #033562), IL-22 (IL22−/−, IL-22Cre homozygous, #027524), or CRAMP (CRAMP−/−, #017799). Mice were intrajejunally inoculated with CMpGP3S organisms as described below. Three days after the intrajejunal inoculation, mice were sacrificed, and infectious chlamydial organisms in tissue samples were titrated. In some experiments, mice received donor CD4+ T cells via adoptive transfer 3 days before and 1 day after the intrajejunal inoculation.
Intrajejunal inoculation
Mice were anesthetized using a mix of isoflurane and oxygen. Once the mice were unconscious, the abdomen was shaved and sterilized with 70% ethanol. A small incision (approximately 0.25–0.5 inches) was made in the abdomen using a pair of scissors (#503708–12, World Precision Instruments Inc, Sarasota, FL). The jejunum was identified within the body and then partially pulled out from the body cavity using curved tweezers. To the middle of the jejunum, 1 × 105 IFUs of CMpGP3S in 100 µL of SPG were inoculated using a 30-GA needle (#7803–07, 30GA, Removable needles, Hamilton Company, Reno, NV) and a 1-mL syringe (#7654–01, Hamilton Company, Reno, NV). Care was taken not to remove excess intestine from the animal, pierce the intestine completely, or inject air bubbles. After the injection, the jejunum was placed back into the cavity, and the wound was closed using three to four surgical staples (#ACS- KIT, Braintree Scientific, Inc., Braintree, MA). Mice were resuscitated by placing them on a warm heating pad and supplying them with fresh air. Once the mouse regained consciousness and was able to walk, meloxicam (#459550250, ThermoScientific, Waltham, MA) was intraperitoneally administered as an analgesic. Meloxicam was prepared by first dissolving in 100% ethanol (<10% of the total end volume) and then diluted to a concentration of 0.75 mg/mL with sterile water. This solution was further sterilized by filtering through a Steriflip (#SCGP00525, Millipore Sigma, Burlington, MA). The same analgesic was administered intraperitoneally every day thereafter until sacrifice. Since the intrajejunal inoculation surgery is time-consuming, we were only able to evaluate a few mice in a group for an experiment.
Adoptive transfer
Donor mice were sacrificed with overdose isoflurane (#1064728455960, Piramal Critical Care, Bethlehem, PA) followed by cervical dislocation. The spleen was collected for isolating CD4+ T cells using the Mojosort CD4+ T Cell Isolation Kit (#480033, BioLegend, San Diego, CA) according to the manufacturer’s instruction. Briefly, the spleen was placed in a 70-µm mesh cell strainer in a small petri dish (#25010, Corning, Corning, NY) containing 1× Mojosort buffer. The spleen was disrupted using the plunger of a syringe (#7654–01, Hamilton Company, Reno, NV 89502), occasionally raising and lowering the mesh to disperse cells from the strainer. Once the spleen fully disappeared, transfer the spleen cells into a 15-mL conical tube (#8FYE3, Grainger Inc, Lake Forest, IL) using a sterile 1 mL transfer pipette (#13–711-20, Fischer Scientific, Waltham, MA). Additional ~3 mL 1× Mojosort buffer was used to rinse the petri dish into the same 15-mL tube. The rinse was repeated until there was a total of 14 mL of cell suspension in the tube. The tube was then centrifuged at 3,000 rpm for 5 minutes to pellet the cells. The cell pellet was resuspended in 500 µL of 1× Mojosort buffer, and the cell suspension was then transferred to a sterile flow cytometry or fluorescence-activated cell sorting (FACS) tube (#352054, Corning, Corning, NY), followed by adding 50 µL of the MojoSort Mouse CD4 Biotin-Antibody Cocktail, included in the Mojosort Mouse CD4+ T Cell Isolation Kit (#480033, BioLegend, San Diego, CA). After mixing, the tube was incubated on ice for 15 minutes. Then, 50 µL of the magnetic nanobead solution (which had been briefly vortexed) was added to the tube and mixed. The tube was incubated on ice for another 15 minutes. Finally, 2.5 mL of 1× Mojosort buffer was added to the FACS tube, and with the cap removed, the tube was placed into an EasySep cell separator magnet and incubated at room temperature for 5 minutes. The tube was then carefully inverted, and free CD4+ T cells in the suspension were poured into a new sterile 15-mL conical tube. An aliquot of 5 µL was used to count the cell number. The CD4+ T cells were adjusted to a final concentration of 1 × 106 cells per 100 µL using sterile PBS. For each adoptive transfer, 100 µL of PBS alone or 1 × 106 CD4+ T cells was injected retro-orbitally.
Titrating live chlamydial organisms from swabs and tissue homogenates
Rectal swabs were taken using a rayon swab (#P25-800R, Harmony Lab and Safety Supplies, Garden Grove, CA). The swab was wet with sterile SPG and then inserted into the rectum and rotated 20 times. The swab was placed into a 1.5-mL tube containing 500 µL SPG and four glass beads. The swab handle was then cut so that the Eppendorf tube can be capped for vortexing (on high for 2 minutes) to free EBs. Tubes were then placed on ice for titration. Mouse tissues were collected after sacrificing with an overdose of isoflurane and cervical dislocation. The spleen, jejunum, duodenum, ileum, colon, colon lumen, cecum, and cecum lumen were all isolated and placed into SPG with 5 mL SPG for jejunum, 2 mL for all other organs. The tissues were homogenized using the OMNI Tissue Homogenizer (#TH115 OMNI International Kennesaw, GA), followed by sonication using the VC130 sonicator fashioned with a microtip (#VC 130 Sonics and Materials INC, Newton, CT). The sonication was carried out at 40 Amps for 10 seconds, while the sample tube was immersed in ice water. Between each sample, the tip was sterilized by running the sonicator in sterile water and 70% ethanol.
After sonication, all samples were centrifuged at 3,000 rpm for 5 minutes to pellet remaining debris, and the supernatant was used to measure live chlamydial organisms by making the following serial dilutions. Swabs were serially diluted 1, 1:4, 1:16, and 1:64, while tissues were serially diluted 1:10, 1:230, 1:540, and 1:1270. After sample dilution, HeLa cells (prepared the day prior, around 80% confluency at the time of inoculation) in a 96-well plate were prepared for infection by removing the cell media and incubating the cells in 100 µL of diethylaminoethyl (DEAE) dextran [prepared by adding 5 mL of DEAE (#D9885, Sigma Aldrich Inc, St. Louis, MO)] to Dulbecco’s modified Eagle medium (DMEM, #11995065, Thermo Fischer Scientific, Waltham, MA) at 37°C for 10 minutes. After incubation, the DEAE solution was removed, and 100 µL of each dilution was inoculated onto a HeLa cell monolayer. The plate was then centrifuged at 1,000 rpm for 1 hour at room temperature. After the inoculum was removed, 200 µL of DMEM (#11995065 Thermo Fischer Scientific, Waltham, MA) with 10% fetal calf serum (#35-010-CV, Corning, Corning, NY) supplemented with antibiotics gentamicin (#G38000-250, Research Products Int, Mount Prospect, IL) and cycloheximide (#C7698-1G, Millipore Sigma, Burlington, MA) was added to each well. The plates were then incubated at 37°C for 24 hours before fixing for immunofluorescence staining.
Immunofluorescence microscopy
The overnight infected HeLa cells were fixed by adding 100 µL of 4% paraformaldehyde (#ICN15014601, Fischer Scientific, Waltham, MA) to each well and incubating for 1 hour at room temperature. The paraformaldehyde was then removed, and 100 µL of 0.1% Triton (#BP151-500, Fischer Scientific, Waltham, MA) in PBS was added to each well and incubated for 10 minutes at room temperature to permeabilize both HeLa and chlamydial membranes. After removing the Triton solution, 100 µL of 3% BSA (#700–100P, Gemini Bio-Products, Sacramento, CA) was added to each well and incubated at room temperature for 1 hour to prevent non-specific binding. After removing the BSA solution, 50 µL of primary antibody (rabbit anti-Chlamydia muridarum antibody R1064, diluted at 1:1,000 in 1% BSA-PBS) was added to each well and incubated at 37°C for 1 hour. After removing the primary antibody, the plate was washed three times with 1× PBS, and 50 µL of secondary antibody (Goat anti-rabbit IgG conjugated with Cy2, #115-165-003, Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) mixed with Hoechst (#14530-500MG, Sigma-Aldrich Inc., St. Louis, MO), with final concentrations of 1:200 and 1:1,000, respectively, in 1% BSA-PBS was added to each well and incubated at 37°C for 1 hour. The secondary antibody was then removed, and plates were washed five times with 1× PBS after which the wells were all filled with 1× PBS to prevent drying of the cells. The inclusion-forming units were counted using an Olympus IX-81 fluorescence microscope (Olympus, Melville, NY) within a week as described below.
Counting inclusions and calculating IFUs
For each well, IFUs from five random views were counted under an objective lens using the appropriate magnification and averaged. If one or fewer IFUs per view can be found using a 10× objective lens, the entire well is counted. Meanwhile, if 15 or more IFUs can be counted in one view, the magnification should be increased. Based on the number of views per well and the magnification of the objective lens used, the total number of IFUs per well was calculated. To determine the IFUs contained within the sample, the average IFUs/view derived from the five views was multiplied by the number of views possible in the total well per magnification, the dilution, and the factor reflecting the portion of sample used for titration. After completing this for each dilution where IFUs are visible, the average number of IFUs was calculated and expressed as log10 transformed IFUs for statistical analyses. It is worth noting that if tissue samples are toxic to HeLa cells and destroy the cell monolayer, this will affect the accuracy of the measurement. Thus, wells with disrupted HeLa cell monolayers should be excluded from the calculations.
Statistics
For these experiments, the Wilcoxon rank-sum test was used to compare both individual tissue types as well as overall large intestinal chlamydial burden. Furthermore, a Fisher’s exact test was performed to compare the frequency of infection between groups of mice.
ACKNOWLEDGMENTS
This work was supported partly by grants (to G.Z.) from the US National Institutes of Health (R01AI047997).
Contributor Information
Jie Wang, Email: wj0988@163.com.
Guangming Zhong, Email: Zhongg@UTHSCSA.edu.
Sunny Shin, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania, USA.
ETHICS APPROVAL
All animal experiments were carried out following the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Committee on the Ethics of Laboratory Animal Experiments of the University of Texas Health Science Center at San Antonio.
REFERENCES
- 1. CDC . 2020. Sexually transmitted diseases. Atlanta, GA: Services USDoHaH. https://www.cdc.gov/std/statistics/2020. [Google Scholar]
- 2. Yang R, Jacobson C, Gardner G, Carmichael I, Campbell AJD, Ryan U. 2014. Longitudinal prevalence and faecal shedding of Chlamydia pecorum in sheep. Vet J 201:322–326. doi: 10.1016/j.tvjl.2014.05.037 [DOI] [PubMed] [Google Scholar]
- 3. Pospischil A, Borel N, Chowdhury EH, Guscetti F. 2009. Aberrant chlamydial developmental forms in the gastrointestinal tract of pigs spontaneously and experimentally infected with Chlamydia suis. Vet Microbiol 135:147–156. doi: 10.1016/j.vetmic.2008.09.035 [DOI] [PubMed] [Google Scholar]
- 4. Zhang Q, Huang Y, Gong S, Yang Z, Sun X, Schenken R, Zhong G. 2015. In vivo and ex vivo imaging reveals a long-lasting chlamydial infection in the mouse gastrointestinal tract following genital tract inoculation. Infect Immun 83:3568–3577. doi: 10.1128/IAI.00673-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Craig AP, Kong FYS, Yeruva L, Hocking JS, Rank RG, Wilson DP, Donovan B. 2015. Is it time to switch to doxycycline from azithromycin for treating genital chlamydial infections in women? modelling the impact of autoinoculation from the gastrointestinal tract to the genital tract. BMC Infect Dis 15:200. doi: 10.1186/s12879-015-0939-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peters RPH, Dubbink JH, van der Eem L, Verweij SP, Bos MLA, Ouburg S, Lewis DA, Struthers H, McIntyre JA, Morré SA. 2014. Cross-sectional study of genital, rectal, and pharyngeal chlamydia and gonorrhea in women in rural South Africa. Sex Transm Dis 41:564–569. doi: 10.1097/OLQ.0000000000000175 [DOI] [PubMed] [Google Scholar]
- 7. Gratrix J, Singh AE, Bergman J, Egan C, Plitt SS, McGinnis J, Bell CA, Drews SJ, Read R. 2015. Evidence for increased chlamydia case finding after the introduction of rectal screening among women attending 2 Canadian sexually transmitted infection clinics. Clin Infect Dis 60:398–404. doi: 10.1093/cid/ciu831 [DOI] [PubMed] [Google Scholar]
- 8. Musil K, Currie M, Sherley M, Martin S. 2016. Rectal chlamydia infection in women at high risk of chlamydia attending canberra sexual health centre. Int J STD AIDS 27:526–530. doi: 10.1177/0956462415586317 [DOI] [PubMed] [Google Scholar]
- 9. Gratrix J, Singh AE, Bergman J, Egan C, McGinnis J, Drews SJ, Read R. 2014. Prevalence and characteristics of rectal chlamydia and gonorrhea cases among men who have sex with men after the introduction of nucleic acid amplification test screening at 2 Canadian sexually transmitted infection clinics. Sex Transm Dis 41:589–591. doi: 10.1097/OLQ.0000000000000176 [DOI] [PubMed] [Google Scholar]
- 10. Vlcek KR, Li W, Manam S, Zanotti B, Nicholson BJ, Ramsey KH, Murthy AK. 2016. The contribution of Chlamydia-specific CD8+ T cells to upper genital tract pathology. Immunol Cell Biol 94:208–212. doi: 10.1038/icb.2015.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de la Maza LM, Peterson EM. 2002. Vaccines for Chlamydia trachomatis infections. Curr Opin Investig Drugs 3:980–986. [PubMed] [Google Scholar]
- 12. Morrison RP, Caldwell HD. 2002. Immunity to murine chlamydial genital infection. Infect Immun 70:2741–2751. doi: 10.1128/IAI.70.6.2741-2751.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lu C, Peng B, Li Z, Lei L, Li Z, Chen L, He Q, Zhong G, Wu Y. 2013. Induction of protective immunity against Chlamydia muridarum intravaginal infection with the chlamydial immunodominant antigen macrophage infectivity potentiator. Microbes Infect 15:329–338. doi: 10.1016/j.micinf.2013.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson RM, Kerr MS, Slaven JE. 2014. An atypical CD8 T-cell response to Chlamydia muridarum genital tract infections includes T cells that produce interleukin-13. Immunology 142:248–257. doi: 10.1111/imm.12248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rockey DD, Wang J, Lei L, Zhong G. 2009. Chlamydia vaccine candidates and tools for chlamydial antigen discovery. Expert Rev Vaccines 8:1365–1377. doi: 10.1586/erv.09.98 [DOI] [PubMed] [Google Scholar]
- 16. Perry LL, Hughes S. 1999. Chlamydial colonization of multiple mucosae following infection by any mucosal route. Infect Immun 67:3686–3689. doi: 10.1128/IAI.67.7.3686-3689.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yeruva L, Spencer N, Bowlin AK, Wang Y, Rank RG. 2013. Chlamydial infection of the gastrointestinal tract: a reservoir for persistent infection. Pathog Dis 68:88–95. doi: 10.1111/2049-632X.12052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang L, Zhang Q, Zhang T, Zhang Y, Zhu C, Sun X, Zhang N, Xue M, Zhong G. 2016. The Chlamydia muridarum organisms fail to auto-inoculate the mouse genital tract after colonization in the gastrointestinal tract for 70 days. PLoS One 11:e0155880. doi: 10.1371/journal.pone.0155880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhong G. 2021. Chlamydia overcomes multiple gastrointestinal barriers to achieve long-lasting colonization. Trends Microbiol 29:1004–1012. doi: 10.1016/j.tim.2021.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhong G. 2018. Chlamydia spreading from the genital tract to the gastrointestinal tract - A two-hit hypothesis. Trends Microbiol 26:611–623. doi: 10.1016/j.tim.2017.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tian Q, Zhou Z, Wang L, Abu-Khdeir A-M, Huo Z, Sun X, Zhang N, Schenken R, Wang Y, Xue M, Zhong G. 2020. Gastrointestinal coinfection promotes chlamydial pathogenicity in the genital tract. Infect Immun 88:e00905-19. doi: 10.1128/IAI.00905-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou Z, Tian Q, Wang L, Sun X, Zhang N, Xue M, Xu D, Zhong G. 2022. Characterization of pathogenic CD8+ T cells in Chlamydia-infected OT1 mice. Infect Immun 90:e0045321. doi: 10.1128/IAI.00453-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang L, Zhu C, Zhang T, Tian Q, Zhang N, Morrison S, Morrison R, Xue M, Zhong G, Roy CR. 2018. Nonpathogenic colonization with Chlamydia in the gastrointestinal tract as oral vaccination for inducing transmucosal protection. Infect Immun 86:e00630-17. doi: 10.1128/IAI.00630-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhu C, Lin H, Tang L, Chen J, Wu Y, Zhong G. 2018. Oral Chlamydia vaccination induces transmucosal protection in the airway. Vaccine 36:2061–2068. doi: 10.1016/j.vaccine.2018.03.015 [DOI] [PubMed] [Google Scholar]
- 25. Morrison SG, Giebel AM, Toh E, Banerjee A, Nelson DE, Morrison RP. 2020. A genital infection-attenuated Chlamydia muridarum mutant infects the gastrointestinal tract and protects against genital tract challenge. mBio 11:e02770-20. doi: 10.1128/mBio.02770-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou Z, Tian Q, Wang L, Zhong G. 2022. Chlamydia deficient in plasmid-encoded glycoprotein 3 (pGP3) as an attenuated live oral vaccine. Infect Immun 90:e0047221. doi: 10.1128/IAI.00472-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. He R, Torres CA, Wang Y, He C, Zhong G. 2023. Type-I interferon signaling protects against Chlamydia trachomatis infection in the female lower genital tract. Infect Immun 91:e0015323. doi: 10.1128/iai.00153-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shao L, Melero J, Zhang N, Arulanandam B, Baseman J, Liu Q, Zhong G. 2017. The cryptic plasmid is more important for Chlamydia muridarum to colonize the mouse gastrointestinal tract than to infect the genital tract. PLoS One 12:e0177691. doi: 10.1371/journal.pone.0177691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shao L, Zhang T, Melero J, Huang Y, Liu Y, Liu Q, He C, Nelson DE, Zhong G. 2018. The genital tract virulence factor pGP3 is essential for Chlamydia muridarum colonization in the gastrointestinal tract. Infect Immun 86:e00429-17. doi: 10.1128/IAI.00429-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koprivsek JJ, Zhang T, Tian Q, He Y, Xu H, Xu Z, Zhong G. 2019. Distinct roles of chromosome- versus plasmid-encoded genital tract virulence factors in promoting Chlamydia muridarum colonization in the gastrointestinal tract. Infect Immun 87:e00265-19. doi: 10.1128/IAI.00265-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang T, Huo Z, Ma J, He C, Zhong G. 2019. The plasmid-encoded pGP3 promotes Chlamydia evasion of acidic barriers in both stomach and vagina. Infect Immun 87:e00844-18. doi: 10.1128/IAI.00844-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huo Z, He C, Xu Y, Jia T, Wang J, Zhong G. 2020. Chlamydia deficient in plasmid-encoded pGP3 is prevented from spreading to large intestine. Infect Immun 88:e00120-20. doi: 10.1128/IAI.00120-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He C, Xu Y, Huo Z, Wang J, Jia T, Li X-D, Zhong G. 2021. Regulation of Chlamydia spreading from the small intestine to the large intestine via an immunological barrier. Immunol Cell Biol 99:611–621. doi: 10.1111/imcb.12446 [DOI] [PubMed] [Google Scholar]
- 34. Liu Y, Huang Y, Yang Z, Sun Y, Gong S, Hou S, Chen C, Li Z, Liu Q, Wu Y, Baseman J, Zhong G. 2014. Plasmid-encoded Pgp3 is a major virulence factor for Chlamydia muridarum to induce hydrosalpinx in mice. Infect Immun 82:5327–5335. doi: 10.1128/IAI.02576-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhong G. 2017. Chlamydial plasmid-dependent pathogenicity. Trends Microbiol 25:141–152. doi: 10.1016/j.tim.2016.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li Z, Chen D, Zhong Y, Wang S, Zhong G. 2008. The chlamydial plasmid-encoded protein pgp3 is secreted into the cytosol of Chlamydia-infected cells. Infect Immun 76:3415–3428. doi: 10.1128/IAI.01377-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen D, Lei L, Lu C, Galaleldeen A, Hart PJ, Zhong G. 2010. Characterization of Pgp3, a Chlamydia trachomatis plasmid-encoded immunodominant antigen. J Bacteriol 192:6017–6024. doi: 10.1128/JB.00847-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hou S, Dong X, Yang Z, Li Z, Liu Q, Zhong G. 2015. Chlamydial plasmid-encoded virulence factor Pgp3 neutralizes the antichlamydial activity of human cathelicidin LL-37. Infect Immun 83:4701–4709. doi: 10.1128/IAI.00746-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hou S, Sun X, Dong X, Lin H, Tang L, Xue M, Zhong G. 2019. Chlamydial plasmid-encoded virulence factor Pgp3 interacts with human cathelicidin peptide LL-37 to modulate immune response. Microbes Infect 21:50–55. doi: 10.1016/j.micinf.2018.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hou S, Xu R, Zhu C, Shan S, Han L, Wang H. 2018. Chlamydial pasmid-encoded protein pGP3 inhibits development of psoriasis-like lesions in mice. Med Sci Monit 24:5159–5167. doi: 10.12659/MSM.910472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Galaleldeen A, Taylor AB, Chen D, Schuermann JP, Holloway SP, Hou S, Gong S, Zhong G, Hart PJ. 2013. Structure of the Chlamydia trachomatis immunodominant antigen Pgp3. J Biol Chem 288:22068–22079. doi: 10.1074/jbc.M113.475012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ahlfors H, Morrison PJ, Duarte JH, Li Y, Biro J, Tolaini M, Di Meglio P, Potocnik AJ, Stockinger B. 2014. IL-22 fate reporter reveals origin and control of IL-22 production in homeostasis and infection. J Immunol 193:4602–4613. doi: 10.4049/jimmunol.1401244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Castleman MJ, Dillon SM, Purba CM, Cogswell AC, Kibbie JJ, McCarter MD, Santiago ML, Barker E, Wilson CC. 2019. Commensal and pathogenic bacteria indirectly induce IL-22 but not IFNγ production from human colonic ILC3s via multiple mechanisms. Front Immunol 10:649. doi: 10.3389/fimmu.2019.00649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bunjun R, Omondi FMA, Makatsa MS, Keeton R, Wendoh JM, Müller TL, Prentice CSL, Wilkinson RJ, Riou C, Burgers WA. 2021. Th22 cells are a major contributor to the mycobacterial CD4+ T cell response and are depleted during HIV infection. J Immunol 207:1239–1249. doi: 10.4049/jimmunol.1900984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ren Z, Pan LL, Huang Y, Chen H, Liu Y, Liu H, Tu X, Liu Y, Li B, Dong X, Pan X, Li H, Fu YV, Agerberth B, Diana J, Sun J. 2021. Gut microbiota-CRAMP axis shapes intestinal barrier function and immune responses in dietary gluten-induced enteropathy. EMBO Mol Med 13:e14059. doi: 10.15252/emmm.202114059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yoshimura T, McLean MH, Dzutsev AK, Yao X, Chen K, Huang J, Gong W, Zhou J, Xiang Y, H Badger J, O’hUigin C, Thovarai V, Tessarollo L, Durum SK, Trinchieri G, Bian X-W, Wang JM. 2018. The antimicrobial peptide CRAMP is essential for colon homeostasis by maintaining microbiota balance. J Immunol 200:2174–2185. doi: 10.4049/jimmunol.1602073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jia L, Li J, Zhang M, Liu H, Ren Z, Dong XL, Pan X, Qiu J, Pan L-L, Sun J. 2022. Cathelicidin-related antimicrobial peptide protects against enteric pathogen-accelerated type 1 diabetes in mice. Theranostics 12:3438–3455. doi: 10.7150/thno.61433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liang W, Enée E, Andre-Vallee C, Falcone M, Sun J, Diana J. 2022. Intestinal cathelicidin antimicrobial peptide shapes a protective neonatal gut microbiota against pancreatic autoimmunity. Gastroenterology 162:1288–1302. doi: 10.1053/j.gastro.2021.12.272 [DOI] [PubMed] [Google Scholar]
- 49. Rank RG, Yeruva L. 2014. Hidden in plain sight: chlamydial gastrointestinal infection and its relevance to persistence in human genital infection. Infect Immun 82:1362–1371. doi: 10.1128/IAI.01244-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dai J, Zhang T, Wang L, Shao L, Zhu C, Zhang Y, Failor C, Schenken R, Baseman J, He C, Zhong G. 2016. Intravenous inoculation with Chlamydia muridarum leads to a long-lasting infection restricted to the gastrointestinal tract. Infect Immun 84:2382–2388. doi: 10.1128/IAI.00432-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. O’Donnell H, McSorley SJ. 2014. Salmonella as a model for non-cognate Th1 cell stimulation. Front Immunol 5:621. doi: 10.3389/fimmu.2014.00621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Srinivasan A, Salazar-Gonzalez R-M, Jarcho M, Sandau MM, Lefrancois L, McSorley SJ. 2007. Innate immune activation of CD4 T cells in Salmonella-infected mice is dependent on IL-18. J Immunol 178:6342–6349. doi: 10.4049/jimmunol.178.10.6342 [DOI] [PubMed] [Google Scholar]
- 53. Ondondo BO, Brunham RC, Harrison WG, Kinyari T, Sheth PM, Mugo NR, Cohen CR. 2009. Frequency and magnitude of Chlamydia trachomatis elementary body- and heat shock protein 60-stimulated interferon gamma responses in peripheral blood mononuclear cells and endometrial biopsy samples from women with high exposure to infection. J Infect Dis 199:1771–1779. doi: 10.1086/599095 [DOI] [PubMed] [Google Scholar]
- 54. Bard J, Levitt D. 1986. Chlamydia trachomatis (L2 serovar) binds to distinct subpopulations of human peripheral blood leukocytes. Clin Immunol Immunopathol 38:150–160. doi: 10.1016/0090-1229(86)90134-0 [DOI] [PubMed] [Google Scholar]
- 55. Fitzpatrick DR, Wie J, Webb D, Bonfiglioli R, Gardner ID, Mathews JD, Bielefeldt-Ohmann H. 1991. Preferential binding of Chlamydia trachomatis to subsets of human lymphocytes and induction of interleukin-6 and interferon-gamma. Immunol Cell Biol 69:337–348. doi: 10.1038/icb.1991.49 [DOI] [PubMed] [Google Scholar]
- 56. Räsänen L, Lehtinen M, Lehto M, Paavonen J, Leinikki P. 1986. Polyclonal response of human lymphocytes to Chlamydia trachomatis. Infect Immun 54:28–31. doi: 10.1128/iai.54.1.28-31.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lei L, Chen J, Hou S, Ding Y, Yang Z, Zeng H, Baseman J, Zhong G. 2014. Reduced live organism recovery and lack of hydrosalpinx in mice infected with plasmid-free Chlamydia muridarum. Infect Immun 82:983–992. doi: 10.1128/IAI.01543-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu Y, Chen C, Gong S, Hou S, Qi M, Liu Q, Baseman J, Zhong G. 2014. Transformation of Chlamydia muridarum reveals a role for Pgp5 in suppression of plasmid-dependent gene expression. J Bacteriol 196:989–998. doi: 10.1128/JB.01161-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mukhopadhyay S, Clark AP, Sullivan ED, Miller RD, Summersgill JT. 2004. Detailed protocol for purification of Chlamydia pneumoniae elementary bodies. J Clin Microbiol 42:3288–3290. doi: 10.1128/JCM.42.7.3288-3290.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fan T, Lu H, Hu H, Shi L, McClarty GA, Nance DM, Greenberg AH, Zhong G. 1998. Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J Exp Med 187:487–496. doi: 10.1084/jem.187.4.487 [DOI] [PMC free article] [PubMed] [Google Scholar]