Abstract
Bacteriophages in the Agtrevirus genus are known for expressing multiple tail spike proteins (TSPs), but little is known about their genetic diversity and host recognition apart from their ability to infect diverse Enterobacteriaceae species. Here, we aim to determine the genetic differences that may account for the diverse host ranges of Agrevirus phages. We performed comparative genomics of 14 Agtrevirus and identified only a few genetic differences including genes involved in nucleotide metabolism. Most notably was the diversity of the tsp gene cluster, specifically in the receptor-binding domains that were unique among most of the phages. We further characterized agtrevirus AV101 infecting nine diverse Extended Spectrum β-lactamase (ESBL) Escherichia coli and demonstrated that this phage encoded four unique TSPs among Agtrevirus. Purified TSPs formed translucent zones and inhibited AV101 infection of specific hosts, demonstrating that TSP1, TSP2, TSP3, and TSP4 recognize O8, O82, O153, and O159 O-antigens of E. coli, respectively. BLASTp analysis showed that the receptor-binding domain of TSP1, TSP2, TSP3, and TSP4 are similar to TSPs encoded by E. coli prophages and distant related virulent phages. Thus, Agtrevirus may have gained their receptor-binding domains by recombining with prophages or virulent phages. Overall, combining bioinformatic and biological data expands the understanding of TSP host recognition of Agtrevirus and give new insight into the origin and acquisition of receptor-binding domains of Ackermannviridae phages.
Keywords: Bacteriophages, Ackermannviridae, Agtrevirus, host range analysis, tail spike proteins, O-antigen receptors
Agtrevirus phage AV101 expresses four unique tail spike proteins that recognize different O-antigens of E. coli including strains producing Extended Spectrum β-Lactamases (ESBL).
Abbreviations
- ESBL
Extended Spectrum β-Lactamase
- ETEC
Enterotoxigenic E. coli
- HMdU
Hydroxymethyluracil
- LB
Luria–Bertani
- LPS
Lipopolysaccharide
- NAMPT
Nicotinamide phosphoribosyl transferase
- NCBI
National Center for Biotechnology Information
- PFU
Plaque formation unit
- RPPK
Ribose-phosphate pyrophosphokinase
- TEM
Transmission electron microscopy
- TSP
Tail spike protein
- VriC
Virulence-associated protein
Introduction
The recent establishment of genome-based taxonomy of phages has led to the classification of an increasing number of phage families and genera (Lefkowitz et al. 2018). Thus, based on genetic similarity, the Ackermannviridae family was established in 2017 and currently includes two subfamilies (Cvivirinae and Aglimvirinae) and 10 genera (Kuttervirus, Agtrevirus, Limestonevirus Taipeivirus, Tedavirus, Nezavisimistyvirus, Miltonvirus, Campanilevirus, Vapseptimavirus, and Kujavirus) (Kropinski et al. 2017, Adriaenssens et al. 2018). In the Ackermannviridae family, phages exhibit a conserved genome architecture including gene synteny in genomes of substantial size of ~150 kb. In addition, these phages encode hydroxymethyluracil (HMdU) synthase leading to substitution of thymine with hydroxylmethyl uracil in their genomes (Adriaenssens et al. 2012b, Kutter et al. 2011, 2012a, Hsu et al. 2013). This nucleotide substitution has been suggested to prevent cleavage of the phage genomes by host-encoded restriction enzymes, which has been proposed to allow Ackermannviridae phages to infect a broad range of Enterobacteriaceae species (Adriaenssens et al. 2012b, Kutter et al. 2011, 2012a, Hsu et al. 2013).
The most well-studied feature of Ackermannviridae phages is their receptor binding properties arising from expressing up to four diverse tail spike proteins (TSPs). These four TSPs form a complex protruding from the baseplate in a star-like morphology that can be visually observed in transmission electron micrographs (TEMs) (Adriaenssens et al. 2012a, Plattner et al. 2019). The TSPs are hinged together in a complex, and to the baseplate, by protein interactions between the conserved N-terminal modules of each TSPs (Plattner et al. 2019, Chao et al. 2022). In contrast, the receptor-binding domain of these TSPs are highly diverse and binds to polysaccharide receptors like the O-antigen or K-antigen through conserved folds including a β-helix commonly observed for TSPs in the Caudoviricetes order (Barbirz et al. 2008, Andres et al. 2010, Lee et al. 2017a, Olszak et al. 2017, Prokhorov et al. 2017, Kunstmann et al. 2018). As the phages express multiple TSPs, each capable of binding to a specific O-antigen or K-antigen, their host ranges are broader compared to phages only expressing a single TSP targeting such polysaccharide receptor (Steinbacher et al. 1996, Plattner et al. 2019, Sørensen et al. 2021, Witte et al. 2021). For example, Kuttervirus CBA120 infects Salmonella O21, Escherichia coli O157, E. coli O77, and E. coli O78 strains through the specific binding of TSP1, TSP2, TSP3, and TSP4, respectively (Plattner et al. 2019). Both E. coli and Salmonella are common hosts for Kuttervirus and Agtrevirus phages, but these bacteria express more than 185 and 46 diverse O-antigens, respectively (Liu et al. 2014, 2020). Thus, Ackermannviridae phages match the diversity of O-antigens expressed the bacterial hosts by encoding diverse TSP. For example, we previously analyzed 374 TSPs encoded by 99 Ackermannviridae phages and found 96 diverse TSP subtypes each carrying unique receptor-binding domains (Sørensen et al. 2021). Furthermore, the TSP subtypes were strongly associated with phage genera. In addition, further analysis allowed us to predict the host recognition of TSP subtypes encoded by phages of the Kuttervirus, Limestonevirus, and Taipei genera, but due to lack of biological data, no information of Agtrevirus phages could be revealed (Sørensen et al. 2021).
The Agtrevirus genus is not very well studied in terms of host range and receptor recognition. The phages are known to infect Gram-negative bacteria like Salmonella, E. coli, Enterobacter, and Shigella. But, unlike other members of the Ackermannviridae family, such as phages in Kuttervirus genus that infect both E. coli and Salmonella, Agtrevirus phages are only known to infect one bacterial species each (Anany et al. 2011, Heyse et al. 2015, Soffer et al. 2016, Akter et al. 2019, Thanh et al. 2020, Kwon et al. 2021, Imklin et al. 2022). We recently isolated agtrevirus AV101 infecting Extended Spectrum β-Lactamase (ESBL) E. coli (Vitt et al. 2023). A large host range analysis of 198 ESBL E. coli strains showed that Agtrevirus phage AV101 had a narrow host range only infecting 9 of 198 strains tested (Vitt et al. 2023). In addition, we previously observed that five Agtrevirus phages express unique TSPs without similarity to other phages in the genus or Ackermannviridae family (Sørensen et al. 2021). Thus, the diverse hosts infected by Agtrevirus phages may be due to genetic differences between the phages including unique receptor-binding domains of the TSPs.
Here, we investigated the genetic diversity of the growing number of phages belonging to the Agtrevirus genus and further characterized the host binding capabilities of agtrevirus phage AV101. Whole-genome comparison of 14 phages in the Agtrevirus genus showed that the tsp gene cluster represented the most diverse region. In silico analysis of the TSPs showed that the receptor-binding domains of phage AV101 were not observed in any Agtrevirus phages, but the receptor-binding domains share amino acid sequence similarity towards other phages and prophages. We further determined the host recognition of the four TSPs which correlates with the O-antigens of the bacterial hosts. Our work expands the understanding of TSP host recognition in the Agtrevirus genus and give new insight into exchange of receptor-binding domain between phages in different families and genera.
Material and methods
Bacterial strains and phages
All phage genomes used for genomic analysis are presented in Table S1 (Supporting Information). A list of the bacterial strains used in the study is presented in Table S4 (Supporting Information).
Bioinformatic analysis
All genomes of phages from the Agtrevirus genus and unclassified Aglimvirinae subfamily were extracted from NCBI. Phylogenetic analysis of all Agtevirus phages was performed using whole genomes sequence in CLC genomics version 22 (Qiagen) with default settings (date 04/01–23). To align and visualize all the genomes as well as the tsp gene cluster Easyfig version 2.2.5 or CLC genomics were used (Sullivan et al. 2011). 0.4 minimum identity was chosen as the BLAST setting. To identify homolog TSPs of the four TSPs of AV101 (TSP1: WJJ54142.1, TSP2: WJJ54143.1, TSP3: WJJ54145.1, and TSP4: WJJ54146.1), BLASTp was used with standard settings. Afterwards, the genomes identified was used to analysis of prophage in the E. coli genomes and was carried out with PHAge Search Tool Enhanced Release (PHASTER) with standard settings (Arndt et al. 2016).
Phage propagation
Agtrevirus AV101 was propagated on E. coli strain ESBL058 as described earlier (Sørensen et al. 2021). A single colony of E. coli ESBL058 was inoculated into LB media (Lysogeny Broth, Merck, Darmstadt, Germany) and incubated until exponential phase at 37°C at 180 rpm. A previous phage stock of AV101 (1.6 × 1010 PFU ml−1) was 10-fold diluted followed mixing 100 µl of the dilutions with 100 µl of the ESBL058. On a LA plate (LB with 1.2% agar), 4 ml of molten top agar (LBov; LB broth with 0,6% Agar bacteriological no.1, Oxoid) was applied. 5 ml of SM buffer (0.1 M NaCl, 8 mM MgSO47H2O, 50 mM Tris-HCl, pH 7.5) was added to the plates after an overnight incubation at 37°C, and the plates were then incubated at 4°C at 50 rpm. The overnight samples were collected, centrifuged for 15 min at 11 000 rpm, then passed through a 0.2-M filter. By using a phage plaque assay (described below), the new phage stock was used to estimate the phage titre.
Phage DNA isolation
(Gencay et al. 2019) 1 ml of phage lysate was filtered three times with 0.22 µM syringe filters. RNase (10 µg ml−1) and DNase (20 µg ml−1) were added to the filtered lysate and incubated for 1 h at 37°C. The degradation was stopped by adding sterile EDTA (pH 8) at a final concentration of 20 mM. Afterwards Proteinase K was added (50 µg ml−1) and incubate for 2 h at 56°C followed by cooling the sample to room temperature. To isolate the phage DNA, we used Genomic DNA clean and concentrator™ (Zymo research) following the manufactures instructions. The DNA concentration was measured using Qubit 2.0 Fluorometer (Invitrogen).
Transmission electron microscopy
To visualize phage AV101 with transmission electron microscopy we used a previously described method (Ackermann 2009). Briefly, bacteriophages from high-titer stock were sedimented at 12 000 g for 60 min at 4°C and washed three times with Ammonium Acetate (0.1 M, pH 7). Final sediment was used for imaging. 200 mesh copper coated carbon grids (Ted Pella, Inc.) were made hydrophilic by glow discharging the grids using a Leica Coater ACE 200 for 30 s at 10 mA. A volume of 6 µl of phages at a PFU ml−1 of 1012 were pipetted on the grids and incubated for 30 s. All liquid was removed with a Whatman filter paper. The phages were stained by incubating the grid with 6 µl of 2% uranyl acetate for 30 s. In a washing step, 6 µl of ddH2O was pipetted on the grid, incubated for 30 s, and removed with a Whatman paper. The phages where imaged using a CM100 microscope with a Bio TWIN objective lens and a LaB6 emitter. Pictures were taken with an Olympus Veleta camera and analyzed using ImageJ to determine phage particle measurements.
Phage host range analysis
The newly prepared phage stock (3.4 × 1012 PFU ml−1) was used to determine the host range of AV101 (Gencay et al. 2019). Bacterial strains were inoculated into 5 ml LB medium (Lysogeny Broth, Merck) and grown for 5 h. Afterwards, 100 µl of the strain were suspended into 4 ml of top-agar and poured onto LB plates left to solidify (~30 min). 10-fold serial dilutions of the phage stock were prepared, and three times of 10 µl of each dilution were spotted on the bacterial lawn. Subsequently the spots were dried the plates were incubated overnight at 37°C. The next day, dilutions with single plaques were counted to calculate the PFU ml−1.
TSP cloning
Purified AV101 genomic DNA was used as a template for tsp cloning using in vivo assembly cloning (García-Nafría et al. 2016, Sørensen et al. 2021). The tsp genes [tsp1- (2202 bp.), tsp2- (2289 bp.), tsp3- (1947 bp.), and tsp4- (3279 bp.)] were individually amplified with Phusion™ High-Fidelity DNA polymerase (Thermo Scientific™) following the manufacture instructions with primers carrying homologous overhangs to the expression vector pET-28a (+). Furthermore, the expression vector with primers to linearize the vector were also added to each PCR reaction. The primers amplifying the tsp genes would allow for homologous recombination between the HinCII and Eco52KI restriction sites in the multiple cloning site of the vector. Primers (Table 1) were ordered from TAG Copenhagen A/S. Furthermore, the cloning site in the pET-28a (+) also express a his-tag upstream so the expressed TSPs could be purified with affinity chromatography. In order to degrade the parental pET28a-(+) plasmid, 1 µl of FastDigest DpnI enzyme (Thermo ScientificTM) was added to the PCR reactions after the amplification of the tsp genes and the expression vector. Each of the PCR samples were transformed into competent E. coli StellarTM cells (Takara Bio). The next day, pET_AV101_TSP1-4 extracted with GeneJET Plasmid Miniprep Kit (Thermo Scientific™). Vectors carrying the correct tsp insert (pET_AV101_TSP1-4) were confirmed by Sanger sequencing (Eurofins Genomics).
Table 1.
Overview of primers for cloning of the four tsp genes.
| Primers | Sequences (5’–3’) |
|---|---|
| AV101_TSP1_F | GCAAGCTTGTCGACGGAGTTACTGGATAACAATACTACACTCCATATCTGAAACAAC |
| AV101_TSP1_R | CGCGGATCCGAATTCGAGATGAACGAAATGTTCTCACAAGGTGGTAAAG |
| AV101_TSP2_F | GCAAGCTTGTCGACGGAGTTAATTACTAATCTTACCACGTATGGATAGCTCAAGAGC |
| AV101_TSP2_R | CGCGGATCCGAATTCGAGATGACCAGAAACGTCGAGAGCATC |
| AV101_TSP3_F | GCAAGCTTGTCGACGGAGTTATAGCTGGCAAGTAAAGCTCAAGTCTATTGATGC |
| AV101_TSP3_R | CGCGGATCCGAATTCGAGATGATTTCTCAATTCAATCAACCACGCGG |
| AV101_TSP4_F | GCAAGCTTGTCGACGGAGTTATATTACTGACGTCACAGTTCCAGATATAGACGC |
| AV101_TSP4_R | CGCGGATCCGAATTCGAGATGGCTAATAAACCAACACAGCCTGTTTTC |
| pET-28a(+)_F | CTCGAATTCGGATCCGCG |
| pET-28a(+)_R | CTCCGTCGACAAGCTTGC |
TSP purification
pET_AV101_TSP1-4 were transformed into electrocompetent E. coli BL21 cells for the expression of the TSPs (Sørensen et al. 2021). A single colony of each of transformants carry one of the four pET_AV101_TSP1-4 was inoculated into LB medium with 50 µg ml−1 kanamycin and incubated overnight at 37°C and 170 rpm. Next day, 10 ml of the starter culture was added to 1 l of LB medium with 50 µg ml−1 kanamycin at 37°C and incubated until an OD600 value of 0.6. Then, a final concentration of 0.5 mM of isopropyl-b-d-thiogalactopyranoside was added to induce protein expression. The culture was incubated for an entire night at a reduced temperature of 16°C. The culture was centrifuged the following morning for 10 min at 13 000 g, and the pellet was then resuspended in 9 ml of lysis buffer (0.5 M NaCl, 20 mM Na2HPO4, 50 mM Imidazole, pH 7.4). Sonication was used to disrupt the cells, with a program of nine cycles lasting 30 s each at 80% power. Centrifugation at 9500 g for 30 min at 4°C separated cell debris. Using 0.22 mm filters, the supernatant containing the expressed proteins was filtered. HisGraviTrapTM (GE Healthcare) was then used to purify the proteins using an elution buffer (0.5 M NaCl, 20 mM Na2HPO4, 0.5 M Imidazole, pH 7.4). To transfer the TSPs into a new buffer [20 mM HEPES (pH 7.4)], Amicon Ultra-15 Centrifugal Filter Units with a 50 kDa cutoff (Merck Milipore) were utilized. Using a Qubit 2.0 Fluorometer and the QubitTM Protein Assay Kit, protein concentration was determined (Invitrogen).
TSP spot assay and inhibition assay
The TSP spot and inhibition assay was performed as previously described (Sørensen et al. 2021). Shortly, bacterial strains were grown to an OD600 value of 0.6. A volume of 100 µl of the bacterial culture was then added to 4 ml top-agar and poured onto an LB agar plate. After the bacterial lawns were solidified, 1.5 µg of the four TSPs were spotted onto the bacterial lawn and left to dry for 30 min. Phage AV101 and the protein buffer [20 mM HEPES (pH 7.4)] were used as a positive and negative control, respectively. The plates were incubated overnight at 37°C and the next day the presence of a translucent zone was evaluated. To further validate the spot assay, the inhibitory effect of the TSPs on the infectivity of the bacterial strains were evaluated. The four strains ESBL-038, -040, -058, and -144 were used as they represented strains that TSP1-4 recognize, respectively. A single colony of the bacterial strains was incubated in LB medium at 37°C at 170 rpm until OD600 reached 0.3. The cells were then cooled on ice before 100 µl of the cells were added to 5 mg of the TSPs in individually tubes. The cell-TSP suspension was then preincubated at 37°C for 20 min. Afterwards, the suspension was added to 4 ml top agar and poured onto an LB agar plate and left to solidify. Three times 10 µl phage AV101 dilutions (10−1–10−8) was spotted on top of the plate and incubated overnight at 37°C. Next day, the inhibitory effect of the TSPs were evaluated by comparing the PFU ml−1 of the phage sensitive strains with the PFU ml−1 with strains incubated with the respective TSPs. The inhibition assay was carried out in triplicates and the results were visualized in Graphpad Prism9 with the mean standard deviations shown in the figure.
Alphafold2 prediction
Models for the TPSs were predicted in a Nvidia Quadro RTX 8000 using Alphafold2-multimer (version 3.2.1) (Jumper et al. 2021, Evans et al. 2022). We utilized global search for the multiple sequence alignment, five recycling rounds, and the amber relaxation was skipped. The best model was ranked based on the iptm+ptm score and was the one selected for the analysis.
Results
Comparative genomics of Agtrevirus phages
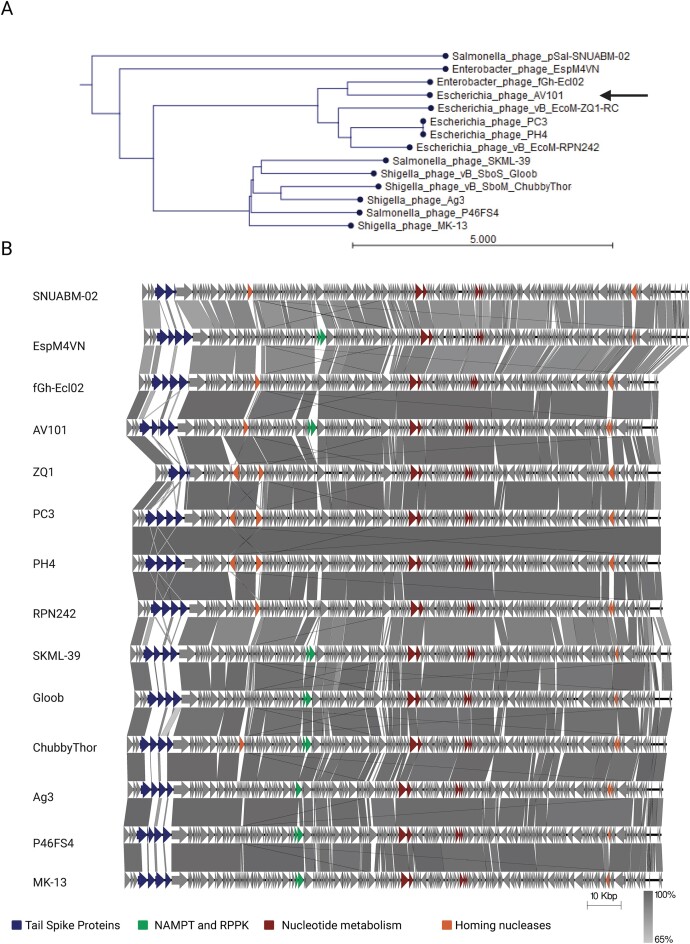
Agtrevirus phages are known to infect species within the Enterobacteriaceae family like Shigella, Salmonella, Enterobacter, and E. coli including ESBL, however, each phage are only known to infect one of the species (Akter et al. 2019, Thanh et al. 2020, Kwon et al. 2021, Imklin et al. 2022, Vitt et al. 2023). To investigate the genetic differences that may account for such diverse host ranges, we extracted all Agtrevirus genomes as well as unclassified Aglimvirinae genomes, not to miss any potential Agtrevirus phages (Table S1, Supporting Information). We did not include the unclassified Aglimvirinae Dickeya phages: phiDP10.3 and phiDP23.1 as they have previously been suggested to belong to the Limestonevirus genus in the Aglimvirinae subfamily (Czajkowski et al. 2015). We confirmed that all phages in the genus as well as the unclassified phages belonged to Agtrevirus by aligning all genomes and demonstrate an overall average nucleotide identity (ANI) between 86% and 97% (Figure S1, Supporting Information). As phages PH4 and PC3 are 99.99% identical, the same phage may have been isolated twice. Phylogenetic analysis using whole genome sequences demonstrated grouping of phages infecting Salmonella and Shigella, whereas E. coli phages formed a separate group with Enterobacter phage fGh-Ecl02. In contrast, Enterobacter phage EspM4VN and Salmonella phage SNUABM-02 did not group with any other phages (Fig. 1A).
Figure 1.
Agtrevirus phages are genetic similar with a few expectations. (A) ANI phylogenetic analysis of phage AV101 and the other 13 extracted Agtrevirus phages using CLC workbench 22. The analysis showed that AV101 closes relative is Enterobacter phage fGh-Ecl02. (B) Based on the phylogenetic analysis the Agtrevirus genomes were aligned using EasyFig. Genetic differences are highlighted with colours; Genes encoding nicotinamide phosphoribosyl transferase (NAMPT) and ribose-phosphate pyrophosphokinase (RPPK) (green), other nucleotide metabolism genes (red), homing nucleases (orange), and the tsp gene cluster (blue). Phage genomes and the variable genes are listed in Table S2 (Supporting Information).
To further investigate the genetic differences of the Agtrevirus phages, we aligned all genomes using EasyFig (Sullivan et al. 2011). Overall, the phage genomes showed similar genetic organization, and all encode HMdU transferase genes used for synthesis of hydroxylmethyl uracil replacing thymine as previously described (Fig. 1B; Table S2, Supporting Information) (Adriaenssens et al. 2012a,b, Hsu et al. 2013). Interestingly, other nucleotide metabolism genes like nicotinamide phosphoribosyl transferase (NAMPT) and ribose-phosphate pyrophosphokinase (RPPK) were detected in eight of the 14 phage genomes analyzed (Fig. 1B; Table S2, Supporting Information). These genes were associated with the group of phages classified as Salmonella and Shigella phages, but also in E. coli phage AV101 and Enterobacter phage EspM4VN. Beside genes involved nucleotide metabolism, we observed diversity within genes encoding homing nucleases, yet with no corelation to the phylogenetic groups (Fig. 1B; Table S2, Supporting Information). Several other genes varied between phages, but all were annotated as hypothetical proteins. Finally, the gene cluster encoding TSPs expected to be responsible for host recognition exhibited the highest diversity (Fig. 1B). Our overall genomic analysis showed that the Agtrevirus phages grouped into two phylogeny groups with two outliners and that the phages are highly conserved, except for the tsp gene cluster.
Agtrevirus phages encode highly diverse TSPs
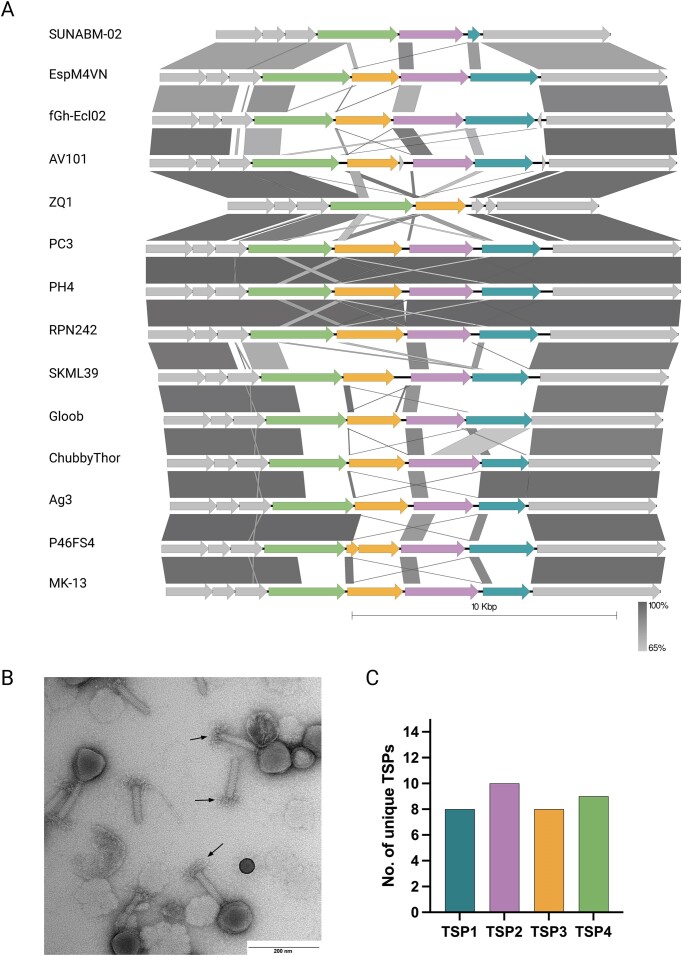
To further analyse the diversity of TSPs of Agtrevirus phages, we compared the tsp gene cluster of the 14 phages (Fig. 2A). While most Agtrevirus phages, like AV101, encode four tsp genes, phage ZQ1 and pSal-SNUABM-2 only encode two tsp genes. In phage AV101, we further confirmed the presence of multiple TSPs by performing transmission electron microscopy showing the expected star-like complex protruding the baseplate (Fig. 2B).
Figure 2.
The majority of tsp genes in Agtrevirus phages encode unique receptor-binding domains. (A) Alignment of the tsp gene cluster located between the Virulence associated gene (VriC) and baseplate wedge genes (grey) of all available Agtrevirus phages showed high diversity of the tsp genes among the phages. Colours; Turkish: TSP1, purple: TSP2, yellow: TSP3, green: TSP4, and grey: hypothetical proteins, baseplate wedge, and VriC. B) TEM photo of AV101 showing virion morphology similar to other Agtrevirus phages. The arrows points to the distinctive star-like TSP complex. (C) All tsp1, tsp2, tsp3, and tsp4 genes were individually aligned and the number of dissimilar genes of Agtrevirus tsp genes were visualized plotted.
Analyzing the tsp sequences in more detail demonstrated that most tsp genes were similar in the N-termini, which are known to be conserved within phage genera due to their importance for the TSP complex formation and hinging to the baseplate (Plattner et al. 2019, Chao et al. 2022) (Fig. 2A). Furthermore, we observed a small region of nucleotide identity in the N-termini immediately upstream the β-helix responsible for receptor binding in tsp1, tsp3, and tsp4 genes of some phages (Fig. 2A). This small region of nucleotide identity coincides with the tandem repeat domain previously suggested as a location for recombination between receptor-binding domains of tsp genes of Kuttervirus phages (Sørensen et al. 2021).
Even though comparative genomics revealed two groups of Agtrevirus (Fig. 1A), no apparent correlation between tsp genes and phylogenetic groups were observed. For instance, the group of E. coli phages encode diverse tsp genes expect from phage PH4 and PC3 (99.9% identical) and phage RPN242 that encode identical tsp clusters (Figs 1A and 2A; Figure S1, Supporting Information). In the group of Shigella and Salmonella phages, Shigella phage Ag3 encode tsp4 with similarity to Salmonella phage P46FS4 and tsp1 with similarity to Shigella phage ChubbyThor (Fig. 2B). Interestingly, the region coding for the receptor-binding domain of ChubbyThor tsp2 was similar to the receptor-binding domain of Shigella phage Gloob tsp1 gene (Fig. 2A). Thus, within this group, some of the tsp genes are similar but overall, most of the receptor-binding domains of the tsps are diverse. Furthermore, aligning all tsp1, tsp2, tsp3, and tsp4 genes individually confirmed that Agtrevirus phages mainly express receptor-binding domains unique to each phage, thus suggesting a total of 35 different receptor recognitions (Fig. 2C). In summary, our analysis showed that most Agtrevirus phages encode tsp genes that only show N-terminus sequence similarity.
The TSPs of AV101 recognize specific O-antigens of E. coli hosts
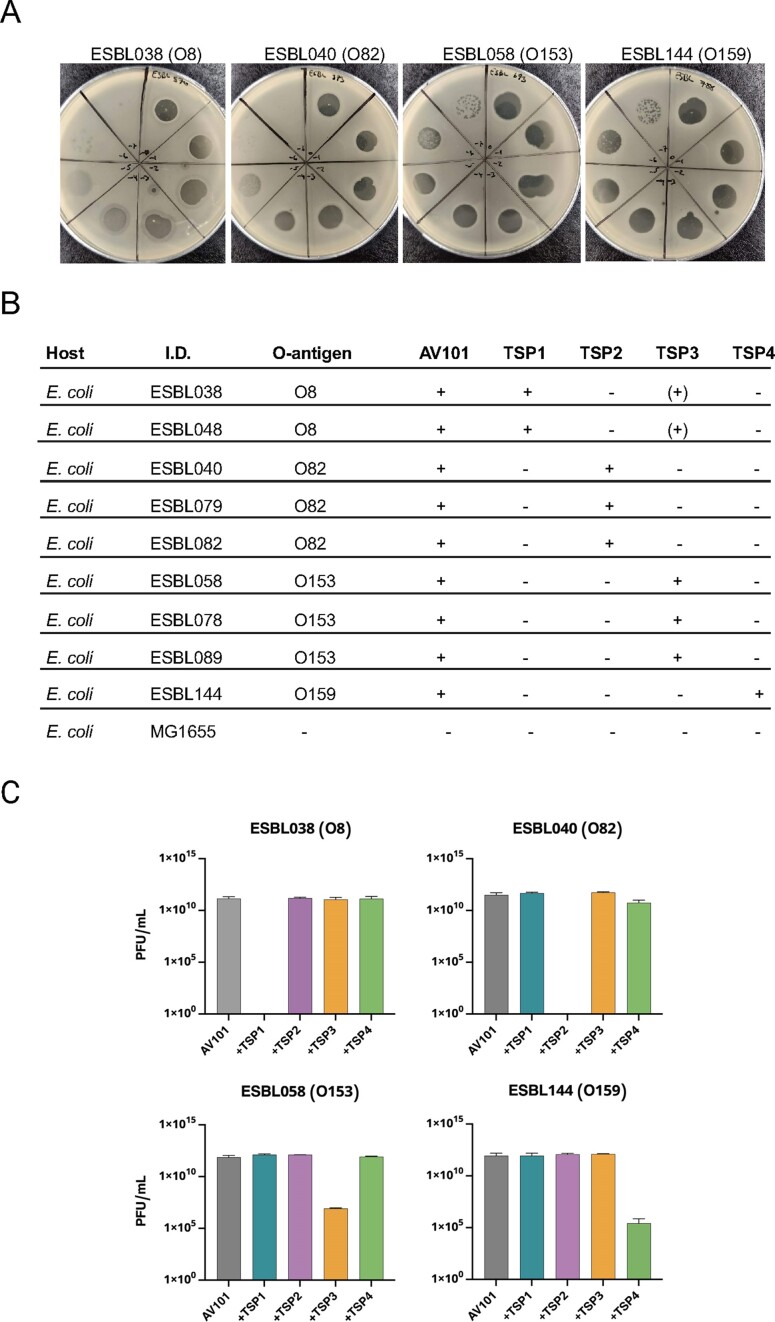
The unique receptor-binding domains of Agtrevirus TSPs suggest that they recognize different bacterial receptors. To further investigate host recognition of Agtrevirus, we used AV101 as an example. AV101 was previously shown to infect 9 out of 198 ESBL E. coli strains tested (Vitt et al. 2023). To further investigate the host range, we spotted a serial dilution of phage AV101 on the E. coli ECOR strain collection (n = 72) as well as representative strains of Salmonella enterica subspecies Derby, Typhimurium, Enteritidis, Seftenberg, Anatum, Odersepoort, and Minnesota. None of the tested strains could be infected by phage AV101, as no single plaques could be observed by performing a standard plaque assay (data now shown). The host range of AV101 was thus limited to the previous identified nine E. coli hosts (Fig. 3A and B).
Figure 3.
The four TSPs of AV101 recognize different O-antigens of ESBL E. coli. (A) Phage AV101 infection on different bacterial hosts showing diverse plaque morphologies. (B) Successful phage infection or detection of a clear translucent zone on bacterial lawns are indicated with plus sign. TSP3 makes small translucent zones on two E. coli strains expressing the O8 O-antigen (+). Big clear translucent zone is indicated with a + sign. Small translucent zone is indicated with (+) sign. (C) Inhibition of AV101 infectivity on ESBL E. coli host after individual TSPs was preincubated with the ESBL hosts. TSP1 and TPS2 were able to completely block the infection of AV101 on their respective hosts, whereas only a partial inhibition with TSP3 and TSP4 could be observed.
To identify the receptors of the individual TSPs encoded by Agtrevirus phage AV101, we cloned, expressed, and purified the four TSPs and spotted each of them on the nine known E. coli hosts. After incubation we noted if the TSPs were able to degrade the O-antigen by forming translucent zones on the bacterial lawns (Fig. 3B; Figure S2, Supporting Information). By this analysis, we observed a correlation between the TSP type and the O-antigen encoded by the individual bacterial hosts as TSP1, TSP2, TSP3, and TSP4 formed translucent zones only on O8, O82, O153, and O159 E. coli strains, respectively (Fig. 3B). Yet, TSP1 were not able to make translucent zones on the E. coli strains ECOR7 and ECOR72 expressing O8 O-antigen (data not shown). Moreover, phage AV101 is not able to infect these strains either, which may be due to modification of the O-antigen (Knirel et al. 2015, Liu et al. 2020). Surprisingly, TSP3 also formed a translucent zone on O8 hosts although smaller compared to TSP1 (Figure S2, Supporting Information), suggesting that TSP3 may be able to degrade both O153 and O8 antigens, even though the two O-antigens does not share any similarity in their sugar composition (Liu et al. 2020).
To show that binding of the individual TSPs to specific E. coli O-antigens are important for AV101 infection, we carried out an inhibition assay. For this experiment, we chose ESBL038 (O8), ESBL040 (O82), ESBL058 (O153), and ESBL144 (O159) to represent hosts recognized by TSP1, TSP2, TSP3, and TSP4, respectively. The strains were grown to exponential phase and mixed with the individually TSPs, allowing the TSPs to bind to and degrade their receptor before plating to form a lawn. Afterwards, AV101 were spotted on the lawn to evaluate the ability of the phage to access the receptor and subsequently form plaques. TSP1 and TSP2 completely abolished the infection of AV101 on ESBL038 and ESBL040, respectively (Fig. 3C). In contrast, TSP3 and TPS4 had an inhibitory effect on infection of ESBL058 and ESBL144, respectively, leading to ~5-log reduction of the phage titre (Fig. 3C). The differences in the ability of the TSPs to inhibit infection may suggest different kinetics of the enzymatic activity of the receptor-binding domains. Finally, while we observed small translucent zones on ESBL038 (O8 O-antigen) when spotting TSP3, the protein did not inhibit infection of phage AV101 of ESBL038. This suggest that TSP3 does not degrade the O8 O-antigen but may degrade another surface polysaccharides of this strain, thus forming a translucent zone unrelated to O-antigen degradation. Overall, our results demonstrate phage AV101 express four TSPs that are unique for the Agtrevirus genus and each of the TSPs recognize distinct O-antigens required for infection of E. coli.
The receptor binding module of phage AV101 TSPs show similarity to prophages and virulent phages
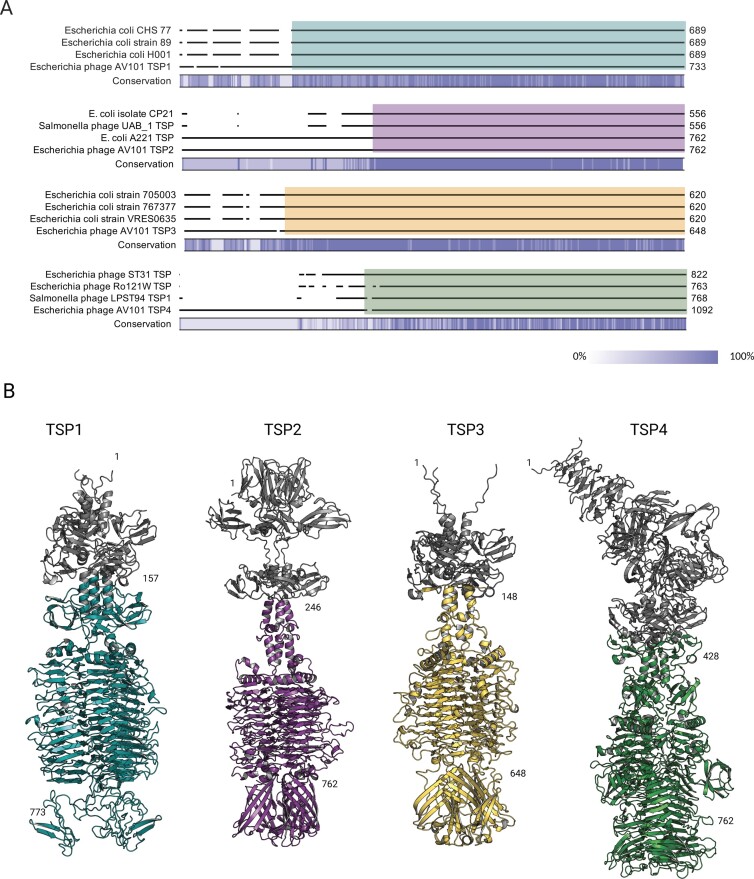
While we only observed little tsp sequence similarity between AV101 and other Agtrevirus phages, it is known that the receptor-binding domain of TSPs may be subjected to horizontal gene transfer between phages from distant related families (Pires et al. 2016, Latka et al. 2019, Sørensen et al. 2021). Thus, to further investigate if the receptor-binding domain could be found in other phages than Agtrevirus, we extracted the amino acid sequence and conducted a BLASTp analysis of the four TSPs of phage AV101. TSP1, TSP2, and TSP3 did not show overall similarity to any virulent phage genomes except for the conserved N-termini of TPSs of Ackermannviridae phages (data not shown). Instead, similarities in the C-terminal were found to E. coli genome sequences, suggesting that these TSPs share sequences of the receptor-binding domain to diverse prophages (Fig. 4A; Table S3, Supporting Information). To identify and further classify the corresponding prophages, we extracted and analyzed these E. coli genomes using PHASTER (Arndt et al. 2016). Indeed, the analysis identified prophages in all the E. coli genomes that were either intact or questionable (Table S4, Supporting Information), suggesting that these prophages express TSP with similar receptor-binding domains. Furthermore, we investigated if the TSPs shared receptor-binding domains in prophages encoded by our own ESBL E. coli strain collection (n = 198). However, none of the four TSPs shared similarity to prophages in our collection (data not shown). In contrast, TSP4 showed similarity to virulent phages infecting E. coli as well as Salmonella including kuttervirus LPST94, kayfunavirus ST31, and phapecoctavirus Ro121c4YLVW (Fig. 4B). The phages ST31 and Ro121c4YLVW infects E. coli and are only distantly related to Agtrevirus, whereas LPST94 belongs to Ackermannviridae and infects Salmonella enterica subspecies (Liu et al. 2018, Yan et al. 2020, Khalifeh et al. 2021). Still, the receptors recognized by these TSPs have not been identified (Yan et al. 2020). To get a better understanding of the similarity in relation to the structural domains, we used Alphafold2-multimer to predict the structure of the four TSP of AV101 (Fig. 4B). All the TSPs shared a modular fold, with an anchor domain in the N-terminus, and a receptor-binding domain close to the C-terminal. Beside the β-helix, the N-terminal head-binding domains of the TSPs had a low prediction score (Figures S3 and S4, Supporting Information). When we compared the alignment of TSPs and the structures, we observed that the amino acid similarity coincides with the β-helix carrying the receptor-binding domain (Fig. 4A and B). Thus, we expect that the prophages and virulent phages bind to the same O-antigen receptors as the TSPs of phage AV101.
Figure 4.
The receptor-binding domain of AV101 TSPs show similarity to prophages and distant-related lytic phages. (A) BLASTp analysis of the four TSPs showed that TSP1, TSP2, and TSP3 share similarity towards proteins found in E. coli strains. Furthermore, TSP2 and TSP4 had similarity towards distant-related lytic phages. A detailed overview of the sequence similarity and the accession numbers of the BLASTp hits can be found in Table S3 (Supporting Information). The coloured boxes represent the region of similarity visualized in the structures. (B) AlphaFold 2 was used to prediction and visualize the four TSPs of AV101. The four TSPs folds like other known TSPs with the conserved β-helix serving as the receptor-binding domain. Moreover, the analysis showed that the amino acid sequence similarity to other TSPs were found in the receptor-binding domains, thus are highlighted in the same colours as in (A). The grey colour represents the N-termini structural domains; TSP1 amino acids 1–157; TSP2 amino acids 1–246; TSP3 amino acids 1–148, and TSP4: amino acids 1–428.
Discussion
With the rise of genome-based phage taxonomy, the investigation of biological functions of a single phage can provide a general understanding of other phages within the same family or genus (Turner et al. 2021). Agtrevirus phages infect different Enterobacteriaceae species, but not much is known about the genetic differences between the phages allowing them to infect such diverse hosts (Akter et al. 2019, Thanh et al. 2020, Kwon et al. 2021, Imklin et al. 2022). Receptor-binding proteins are responsible for the initial binding to the bacterial hosts and the characterization of such proteins can provide important biological information, crucial for understanding phage host ranges. Here we investigated phages belonging to the Agtrevirus genus and observed that while all phages show high nucleotide identity and a similar genome structure, the genes encoding TSPs were highly diverse. Furthermore, as an example, we investigated the specific host recognition of the four TSPs encoded by Agtrevirus phage AV101 previously isolated on ESBL E. coli (Vitt et al. 2023).
To investigate similarities and difference among Agtrevirus phages, we carried out a comparative in silico analysis of all available Agtrevirus genomes. Besides observing diversity in the tsp gene region, we also observed that eight of the phages encoded NAMPT and RPPK. These genes have also been observed in distant related phages including Babavirus Baba19a, Schizotequatrovirus KVP40, and polybotosvirus Atu_ph7 phages (Lee et al. 2017b, Attai et al. 2018, Nilsson et al. 2019). RPPK convert ribose 5-phosphate and ATP to phosphoribosyl pyrophosphate (PRPP) and AMP. PRPP is a precursor of purine and pyrimindine that is used by ribonucleotide reductases, also encoded by all Agtrevirus phages, for nucleotide production during the phage replication (Nilsson et al. 2019). Given that not all Agtrevirus phages encode these genes, they may not be required for phage genome replication but provide other advantages, yet to be discovered, during certain conditions or when infecting specific hosts.
Previously, we have characterized TSPs encoded by phages of the Ackermannviridae family and observed that receptor-binding domains were conserved between phages especially within the Kuttervirus genus (Sørensen et al. 2021). In contrast, we showed that the receptor-binding domain are not as well conserved within the Agtrevirus genus, as most phages encode unique domains, indicating specific host recognition. Instead, our protein alignment and AlphaFold2 predictions showed that the receptor-binding domain of phage AV101 TSPs share sequence similarity and are structural similar to distant related virulent phages as well as prophages. Likewise, the receptor-binding domain of TSP1 of kuttervirus CBA120 show nucleotide similarity to TSP encoded by a prophage found in a Salmonella Minnesota strain (Plattner et al. 2019) and nucleotide similar receptor-binding domain sequences were identified in kuttervirus Det7 and temperate lederbergvirus P22 (Walter et al. 2008). Thus, more receptor-binding domains of TSPs found in the Ackermannviridae family may share receptor binding abilities as prophages suggesting exchange of structural domains between phages and prophages. Furthermore, the receptor-binding domain of TSP4 of AV101 had amino acid sequence similarity to TSP1 encoded by phage Kuttervirus LPST94, demonstrating exchange of receptor-binding domains between phages of different genera within the Ackermannviridae family. So far, host range analysis demonstrated that phage LPST94 infects several Salmonella enterica subspecies expressing different O-antigens (Yan et al. 2020), yet phage AV101 could not infect any of these Salmonella enterica subspecies. In addition, phage LPST94 could infect six E. coli strains, but information about their O-antigens was not included in the study (Yan et al. 2020). Still, our study suggests that TSP1 of phage LPST94 may recognize O8 O-antigen, but it remains to be experimental verified. Overall, combining bioinformatic, structural predictions and biological data of phage AV101 TSPs gives an insight into the origin and acquisition of the receptor binding abilities of Ackermannviridae phages.
ESBL E. coli are of high concern as they encode ESBLs conferring resistance to antibiotics such as penicillin and cephalosporins commonly used in treatment human infections (Paterson and Bonomo 2005, Benz et al. 2021). To propose alternative solutions targeting ESBL E. coli, we recently established a collection of phages, including phage AV101, infecting ESBL E. coli (Vitt et al. 2023). Using this collection, we composed phage cocktails that prevented growth of ESBL E. coli, thus suggesting that phages may indeed be promising alternative antimicrobials (Vitt et al. 2023). Phage AV101 was not included in these cocktails, but may be used to target ESBL E. coli expressing O8, O82, O153, and O159 O-antigens due to the specificities of the four TSP. It should be noted though, that AV101 did not infect two other E. coli strains tested (ECOR7 and ECOR72) carrying the O8 O-antigen, suggesting that internal defence mechanism or modification of the O-antigen, like glycosylation or acetylation may influence phage infection (Knirel et al. 2015, Egido et al. 2022). While we have tested AV101 on ESBL E. coli strains, the four O-antigens are indeed expressed in diverse pathogenic and commensal E. coli strains, hence AV101 most likely also infect commensal E. coli strains expressing the same four O-antigens. (Tamaki et al. 2005a, Marin et al. 2022). For example, O159 O-antigen are often expressed by enterotoxigenic E. coli (ETEC) causing diarrhoea in humans (Tamaki et al. 2005b, Linnerborg et al. 1999), suggesting that AV101 may be used in phage therapy or biocontrol of such strains. Thus, identification of receptor recognition of AV101 may allow design of phage applications dedicated to pathogenic E. coli like O159 ETEC or O8, O82, and O153 E. coli including ESBL-producing strains.
Purified TSPs have also demonstrated promising potential as therapeutics or diagnostic tools (Wang et al. 2023). For instance, oral administration of purified TSP of Lederbergvirus phage P22 dramatically decreased Salmonella colonization in the chicken gut and reduced penetration into internal organs (Waseh et al. 2010). TSPs from bacteriophages 9NA and P22 have been used as tools to detect S. typhimurium (Schmidt et al. 2016) and TSP3 of kuttervirus Det7 was used as a biosensor using surface plasmon resonance to detect S. typhimurium (Hyeon et al. 2020). Thus, instead of using phage AV101 as an antibacterial agent, AV101 TSPs targeting these specific O-antigens on E. coli or O159 of ETEC strains could be of interest. Furthermore, other phage-based solutions for combating pathogenic bacteria based on exploring the knowledge of binding abilities TSPs proteins may be developed. For example, pyocins are phage tail-like particles encoded by Pseudomonas aeruginosa showing bactericidal activity against other Pseudomonas strains (Ge et al. 2020). Such pyocins have been engineered to successful kill other bacteria by exchanging the receptor binding protein of the native pyocin with phage receptor binding proteins (Williams et al. 2008, Scholl et al. 2009). Similarly, TSPs targeting specific O-antigens, like those encoded by AV101, may be used to retarget pyocins to other hosts. TSPs may also be utilized for developing Innolysins, which are novel antibacterials fusing a receptor binding protein or domain to an endolysin, allowing the engineered enzyme to target gram negative bacteria (Zampara et al. 2020). So far Innolysins have been created and shown to kill specifically Campylobacter jejuni or commensal E. coli (Zampara et al. 2020, 2021), and the use of TSP4 could allow development of novel Innolysins targeting E. coli O159 strains including ETEC. Thus, investigation of the receptor recognition of TSPs is not only crucial for understanding the host range of phages but can be further exploited for the development of therapeutics against pathogenic bacteria.
Supplementary Material
Acknowledgements
This work was supported by the Danish Council for Independent Research (9041–00159B). We acknowledge the Core Facility for Integrated Microscopy, Faculty of Health and Medical Sciences, University of Copenhagen.
Contributor Information
Anders Nørgaard Sørensen, Department of Veterinary and Animal Sciences, University of Copenhagen, Stigbøjlen 4, 1870 Frederiksberg C, Denmark.
Dorottya Kalmár, Department of Veterinary and Animal Sciences, University of Copenhagen, Stigbøjlen 4, 1870 Frederiksberg C, Denmark.
Veronika Theresa Lutz, Department of Veterinary and Animal Sciences, University of Copenhagen, Stigbøjlen 4, 1870 Frederiksberg C, Denmark.
Victor Klein-Sousa, Structural Biology of Molecular Machines Group, Protein Structure & Function Program, Novo Nordisk Foundation Center for Protein Research, Faculty of Health and Medical Sciences, University of Copenhagen, Blegdamsvej 3B, 2200 Copenhagen, Denmark.
Nicholas M I Taylor, Structural Biology of Molecular Machines Group, Protein Structure & Function Program, Novo Nordisk Foundation Center for Protein Research, Faculty of Health and Medical Sciences, University of Copenhagen, Blegdamsvej 3B, 2200 Copenhagen, Denmark.
Martine C Sørensen, Department of Veterinary and Animal Sciences, University of Copenhagen, Stigbøjlen 4, 1870 Frederiksberg C, Denmark.
Lone Brøndsted, Department of Veterinary and Animal Sciences, University of Copenhagen, Stigbøjlen 4, 1870 Frederiksberg C, Denmark.
Author contribution
Anders Nørgaard Sørensen (Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing—original draft, Writing—review & editing), Dorottya Kalmár (Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing—review and editing), Veronika Theresa Lutz (Investigation, Formal analysis, Writing—review and editing), Victor Klein-Sousa (TSPs modelling, Writing—review & editing), Nicholas M. I. Taylor (Writing—review & editing), Martine C. Sørensen (Conceptualization, Visualization, Writing—review & editing, Funding acquisition), Lone Brøndsted (Conceptualization, Project administration, Supervision, Visualization, Writing—review & editing, Project administration, Funding acquisition).
Conflict of interest
The authors declare no competing interests.
Data Availability
The genome sequence data of phage AV101 has been submitted to the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/nuccore/) under accession number OQ973471.
References
- Ackermann H-W. Basic phage electron microscopy. In: Clokie Martha RJ, Kropinski AM (eds.), Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions. Totowa: Humana Press, 2009, 113–26. [DOI] [PubMed] [Google Scholar]
- Adriaenssens EM, Ackermann HW, Anany H et al. A suggested new bacteriophage genus: “viunalikevirus”. Arch Virol. 2012a;157:2035–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adriaenssens EM, van Vaerenbergh J, Vandenheuvel D et al. T4-related bacteriophage LIMEstone isolates for the control of soft rot on potato caused by “Dickeya solani”. PLoS ONE. 2012b;7:e33227. 10.1371/journal.pone.0033227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adriaenssens EM, Wittmann J, Kuhn JH et al. Taxonomy of prokaryotic viruses: 2017 update from the ICTV Bacterial and Archaeal Viruses Subcommittee. Arch Virol. 2018;163:1125–9. [DOI] [PubMed] [Google Scholar]
- Akter M, Brown N, Clokie M et al. Prevalence of Shigella boydii in Bangladesh: isolation and characterization of a rare phage MK-13 that can robustly identify shigellosis caused by Shigella boydii type 1. Front Microbiol. 2019;10. 10.3389/fmicb.2019.02461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anany H, Lingohr EJ, Villegas A et al. A Shigella boydii bacteriophage which resembles Salmonella phage ViI. Virol J. 2011;8. 10.1186/1743-422X-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres D, Hanke C, Baxa U et al. Tailspike interactions with lipopolysaccharide effect DNA ejection from phage P22 particles in vitro. J Biol Chem. 2010;285:36768–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt D, Grant JR, Marcu A et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attai H, Boon M, Phillips K et al. Larger than life: isolation and genomic characterization of a jumbo phage that infects the bacterial plant pathogen, Agrobacterium tumefaciens. Front Microbiol. 2018;9. 10.3389/fmicb.2018.01861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbirz S, Müller JJ, Uetrecht C et al. Crystal structure of Escherichia coli phage HK620 tailspike: podoviral tailspike endoglycosidase modules are evolutionarily related. Mol Microbiol. 2008;69:303–16. [DOI] [PubMed] [Google Scholar]
- Benz F, Huisman JS, Bakkeren E et al. Plasmid- and strain-specific factors drive variation in ESBL-plasmid spread in vitro and in vivo. ISME J. 2021;15:862–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao KL, Shang X, Greenfield J et al. Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N). Sci Rep. 2022;12. 10.1038/s41598-022-06073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajkowski R, Ozymko Z, De Jager V et al. Genomic, proteomic and morphological characterization of two novel broad host lytic bacteriophages PdblPD10.3 and PdblPD23.1 infecting pectinolytic Pectobacterium spp. and Dickeya spp. PLoS ONE. 2015;10:e0119812. 10.1371/journal.pone.0119812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egido JE, Costa AR, Aparicio-Maldonado C et al. Mechanisms and clinical importance of bacteriophage resistance. FEMS Microbiol Rev. 2022;46. 10.1093/femsre/fuab048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R, O'Neill M, Pritzel A et al. Protein complex prediction with AlphaFold-multimer. Biorxiv. 2022. 10.1101/2021.10.04.463034. [DOI] [Google Scholar]
- García-Nafría J, Watson JF, Greger IH. IVA cloning: a single-tube universal cloning system exploiting bacterial in vivo assembly. Sci Rep. 2016;6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge P, Scholl D, Prokhorov NS et al. Action of a minimal contractile bactericidal nanomachine. Nature. 2020;580:658–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gencay YE, Gambino M, Prüssing TF et al. The genera of bacteriophages and their receptors are the major determinants of host range. Environ Microbiol. 2019;21:2095–111. [DOI] [PubMed] [Google Scholar]
- Heyse S, Hanna LF, Woolston J et al. Bacteriophage cocktail for biocontrol of Salmonella in dried pet food. J Food Prot. 2015;78:97–103. [DOI] [PubMed] [Google Scholar]
- Hsu CR, Lin TL, Pan YJ et al. Isolation of a bacteriophage specific for a new capsular type of Klebsiella pneumoniae and characterization of its polysaccharide depolymerase. PLoS ONE. 2013;8:e70092. 10.1371/journal.pone.0070092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyeon SH, Lim WK, Shin HJ. Novel surface plasmon resonance biosensor that uses full-length Det7 phage tail protein for rapid and selective detection of Salmonella enterica serovar typhimurium. Biotechnol Appl Biochem. 2020;68:1–8. [DOI] [PubMed] [Google Scholar]
- Imklin N, Sriprasong P, Thanantong N et al. Characterization and complete genome analysis of a novel Escherichia phage, vB_EcoM-RPN242. Arch Virol. 2022;167:1675–9. [DOI] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalifeh A, Kraberger S, Dziewulska D et al. Complete genome sequence of a phapecoctavirus isolated from a pigeon cloacal swab sample. Microbiol Resour Announc. 2021;10:e01471–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knirel YA, Prokhorov NS, Shashkov AS et al. Variations in O-antigen biosynthesis and O-acetylation associated with altered phage sensitivity in Escherichia coli 4 s. J Bacteriol. 2015;197:905–12. 10.1128/JB.02398-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropinski AM, Anany H, Kuhn JH et al. 2017.001B.A.v1.Ackermannviridae. ICTV. 2017.
- Kunstmann S, Scheidt T, Buchwald S et al. Bacteriophage Sf6 tailspike protein for detection of Shigella flexneri pathogens. Viruses. 2018;10:431. 10.3390/v10080431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter EM, Skutt-Kakaria K, Blasdel B et al. Characterization of a ViI-like phage specific to Escherichia Coli O157:H7. Virol J. 2011;8:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J, Kim SG, Kim HJ et al. Bacteriophage as an alternative to prevent reptile-associated Salmonella transmission. Zoonoses Public Health. 2021;68:131–43. [DOI] [PubMed] [Google Scholar]
- Latka A, Leiman PG, Drulis-Kawa Z et al. Modeling the architecture of depolymerase-containing receptor binding proteins in Klebsiella phages. Front Microbiol. 2019;10. 10.3389/fmicb.2019.02649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IM, Tu IF, Yang FL et al. Structural basis for fragmenting the exopolysaccharide of Acinetobacter baumannii by bacteriophage LaB6 tailspike protein. Sci Rep. 2017a;7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Li Z, Miller ES. Vibrio phage KVP40 encodes a functional NAD+ salvage pathway. J Bacteriol. 2017b;199. 10.1128/JB.00855-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz EJ, Dempsey DM, Hendrickson RC et al. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018;46:D708–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnerborg M, Weintraub A, Widmalm G. Structural studies utilizing 13C-enrichment of the O-antigen polysaccharide from the enterotoxigenic Escherichia coli 0159 cross-reacting with Shigella dysenteriae type 4. Eur J B ochem. 1999;266:246–51. [DOI] [PubMed] [Google Scholar]
- Liu B, Furevi A, Perepelov AV et al. Structure and genetics of Escherichia coli O antigens. FEMS Microbiol Rev. 2020;44:655–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Knirel YA, Feng L et al. Structural diversity in Salmonella O antigens and its genetic basis. FEMS Microbiol Rev. 2014;38:56–89. [DOI] [PubMed] [Google Scholar]
- Liu H, Xiong Y, Liu X et al. Complete genome sequence of a novel virulent phage ST31 infecting Escherichia coli H21. Arch Virol. 2018;163:1993–6. [DOI] [PubMed] [Google Scholar]
- Marin J, Clermont O, Royer G et al. The population genomics of increased virulence and antibiotic resistance in Human commensal Escherichia coli over 30 years in France. Appl Environ Microb. 2022;88. 10.1128/aem.00664-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson E, Li K, Fridlund J et al. Genomic and seasonal variations among aquatic phages infecting the Baltic Sea Gammaproteobacterium rheinheimera sp. strain BAL341. Appl Environ Microbiol. 2019;85;e01003–19. 10.1128/AEM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak T, Shneider MM, Latka A et al. The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence. Sci Rep. 2017;7:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18:657–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires DP, Oliveira H, Melo LDR et al. Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl Microbiol Biotechnol. 2016;100:2141–51. [DOI] [PubMed] [Google Scholar]
- Plattner M, Shneider MM, Arbatsky NP et al. Structure and function of the branched receptor-binding complex of bacteriophage CBA120. J Mol Biol. 2019;431:3718–39. [DOI] [PubMed] [Google Scholar]
- Prokhorov NS, Riccio C, Zdorovenko EL et al. Function of bacteriophage G7C esterase tailspike in host cell adsorption. Mol Microbiol. 2017;105:385–98. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Rabsch W, Broeker NK et al. Bacteriophage tailspike protein based assay to monitor phase variable glucosylations in Salmonella O-antigens. BMC Microbiol. 2016;16:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl D, Cooley M, Williams SR et al. An engineered R-type pyocin is a highly specific and sensitive bactericidal agent for the food-borne pathogen Escherichia coli O157:H7. Antimicrob Agents Chemother. 2009;53:3074–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soffer N, Abuladze T, Woolston J et al. Bacteriophages safely reduce Salmonella contamination in pet food and raw pet food ingredients. Bacteriophage. 2016;6:e1220347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen AN, Woudstra C, Sørensen MCH et al. Subtypes of tail spike proteins predicts the host range of Ackermannviridae phages. Comput Struct Biotechnol J. 2021;19:4854–67. 10.1016/j.csbj.2021.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbacher S, Baxa U, Miller S et al. Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proc Natl Acad Sci USA. 1996;93:10584–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki Y, Narimatsu H, Miyazato T et al. The relationship between O-antigens and pathogenic genes of diarrhea-associated Escherichia coli. Jpn J Infect Dis. 2005a;58:65–9. [PubMed] [Google Scholar]
- Tamaki Y, Narimatsu H, Miyazato T et al. The relationship between O-antigens and pathogenic genes of diarrhea-associated Escherichia coli. Jpn J Infect Dis. 2005b;58:65–69. [PubMed] [Google Scholar]
- Thanh NC, Nagayoshi Y, Fujino Y et al. Characterization and genome structure of virulent phage EspM4VN to control Enterobacter sp. M4 isolated from plant soft Rot. Front Microbiol. 2020;11. 10.3389/fmicb.2020.00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D, Kropinski AM, Adriaenssens EM. A roadmap for genome-based phage taxonomy. Viruses. 2021;13:506. 10.3390/v13030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitt AR, Sørensen AN, Bojer MS et al. A collection of diverse bacteriophages for biocontrol of ESBL- and AmpC-β-lactamase-producing E. coli. bioRxiv. 2023. 10.1101/2023.09.14.557699. [DOI] [PMC free article] [PubMed]
- Walter M, Fiedler C, Grassl R et al. Structure of the receptor-binding protein of bacteriophage Det7: a podoviral tail spike in a myovirus. J Virol. 2008;82:2265–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu Y, Bai C et al. Translating bacteriophage-derived depolymerases into antibacterial therapeutics: challenges and prospects. Acta Pharm Sin B. 2023. 10.1016/j.apsb.2023.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waseh S, Hanifi-Moghaddam P, Coleman R et al. Orally administered P22 phage tailspike protein reduces Salmonella colonization in chickens: prospects of a novel therapy against bacterial infections. PLoS ONE. 2010;5:e13904. 10.1371/journal.pone.0013904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SR, Gebhart D, Martin DW et al. Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl Environ Microb. 2008;74:3868–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte S, Zinsli LV, Gonzalez-Serrano R et al. Structural and functional characterization of the receptor binding proteins of Escherichia coli O157 phages EP75 and EP335. Comput Struct Biotechnol J. 2021;19:3416–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T, Liang L, Yin P et al. Application of a novel phage LPST94 for biological control of Salmonella in foods. Microorganisms. 2020;8:400. 10.3390/microorganisms8030400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampara A, Sørensen MCH, Gencay YE et al. Developing Innolysins against Campylobacter jejuni using a novel prophage receptor-binding protein. Front Microbiol. 2021;12. 10.3389/fmicb.2021.619028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampara A, Sørensen MCH, Grimon D et al. Exploiting phage receptor binding proteins to enable endolysins to kill Gram-negative bacteria. Sci Rep. 2020;10. 10.1038/s41598-020-68983-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequence data of phage AV101 has been submitted to the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/nuccore/) under accession number OQ973471.