Abstract
Aging in mammals is accompanied by an imbalance of intestinal homeostasis and accumulation of mitochondrial DNA (mtDNA) mutations. However, little is known about how accumulated mtDNA mutations modulate intestinal homeostasis. We observe the accumulation of mtDNA mutations in the small intestine of aged male mice, suggesting an association with physiological intestinal aging. Using polymerase gamma (POLG) mutator mice and wild-type mice, we generate male mice with progressive mtDNA mutation burdens. Investigation utilizing organoid technology and in vivo intestinal stem cell labeling reveals decreased colony formation efficiency of intestinal crypts and LGR5-expressing intestinal stem cells in response to a threshold mtDNA mutation burden. Mechanistically, increased mtDNA mutation burden exacerbates the aging phenotype of the small intestine through ATF5 dependent mitochondrial unfolded protein response (UPRmt) activation. This aging phenotype is reversed by supplementation with the NAD+ precursor, NMN. Thus, we uncover a NAD+ dependent UPRmt triggered by mtDNA mutations that regulates the intestinal aging.
Subject terms: Mitochondria, Stem cells, Mutation
How age-accumulated mtDNA mutations in the small intestine modulate intestinal homeostasis is unclear. Here, the authors show that increased mtDNA mutation burden triggers an ATF5 dependent UPRmt by NAD+ depletion, and thus regulates intestinal aging through impaired Wnt/β-catenin signaling.
Introduction
The adult intestinal epithelium is composed of differentiated cells forming villi and proliferating cells in crypts. Leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5)-expressing intestinal stem cells (ISCs), residing at the base of the crypt, can differentiate to different epithelial cells (enterocytes, Paneth cells, goblet cells, microfold cells, tuft cells, etc.) and regenerate the epithelium1,2. These differentiated epithelial cells form orderly intestinal structures to maintain intestinal homeostasis and fundamental functions such as absorption, secretion, barrier and antimicrobial function.
ISCs are supported by a niche of accessory cell types to generate mature epithelial cell types, which fuel intestinal renewal, regeneration and development. These niche cells of ISCs include Gli1+, Foxl1+ stromal cells and Paneth cells, and secrete Wnt molecules as self-renewal factors to support intestinal crypts3–5. Signaling pathways underlying the development process of intestinal epithelial cells are well studied. Wnt and Notch signals are essential for maintaining stem cell activity and balanced differentiation between secretory and absorptive cell lineages6,7. The phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt) pathway is also involved in the intestinal differentiation and proliferation8, while the transforming growth factor beta (TGF-β) pathway negatively regulates growth of normal epithelial cells9–12. Notum and Wnt signals play vital roles in the aging process of intestine as marked by reduction of ISC number and alteration of ISC function13,14. However, detailed understanding of the signal pathways regulating intestinal aging is lacking.
Mitochondrial DNA (mtDNA) mutations are widely regarded as an important cause of aging and age-associated diseases15,16 and reportedly induce the aging of multiple organs in mouse such as ovary, heart and liver17–19. In particular, the aging phenotypes of tissues with strong energy requirements such as heart and liver tissues are accelerated by excessive mtDNA mutations, which facilitate mitochondrial dysfunction by compromising oxidative phosphorylation20. The accumulation of mtDNA mutations in aged human clinical intestinal samples21–23 indicates that mtDNA stability may also be important for intestinal aging. However, the sub-cellular mechanisms involved are still unknown while the perturbed epithelial cell crosstalk caused by increased mtDNA mutation content remains to be investigated.
mtDNA-mutator (PolgAMut/Mut) mice exhibit multiple premature aging phenotypes by nine months of age17,24, and decreased lifespan of 13–15 months25,26. As a result, PolgAMut/Mut mice are widely utilized to investigate the roles of mtDNA mutations in the aging process27–31. Using this mouse model, we demonstrate an association between mtDNA mutation content and the intestinal aging phenotypes. We found that increased mtDNA mutation burden triggers an ATF5-dependent mitochondrial unfolded protein response (UPRmt) by NAD+ depletion. These findings reveal a regulatory mechanism whereby the UPRmt mediates the intestinal aging phenotype caused by increased mtDNA mutation burden.
Results
High mtDNA mutation burden induces an aging-like phenotype in the small intestine
Similar to aged human clinical intestinal samples21–23, the small intestine of aged mouse intestine, characterized by decreased intestinal crypt number and increased villus length, higher expression of CDKN1A/p21 (a well-known senescence marker) and shorter telomere length (Supplementary Fig. 1a–c), accumulates more mtDNA mutations, primarily low-frequency (less than 0.05) point mutations (Supplementary Fig. 1d, e). This observation demonstrates a link between mtDNA mutation content and physiological intestinal aging.
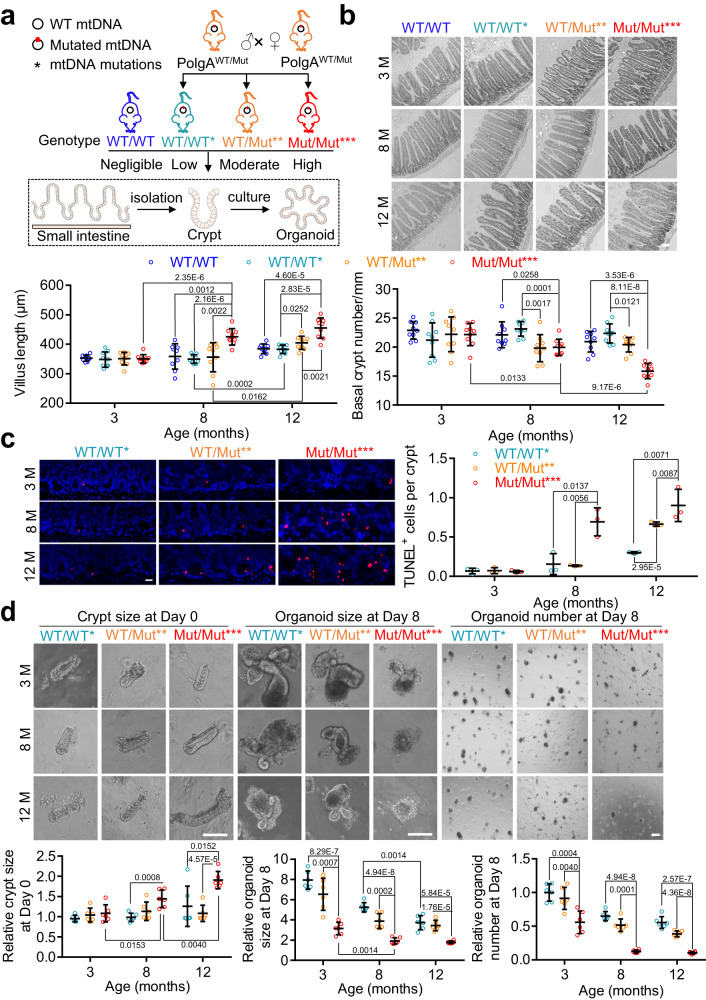
To investigate the effect of mtDNA mutation content on small intestinal aging, we generated mice with four ascribed categories of mtDNA mutational burden: negligible (WT/WT), low (WT/WT*), moderate (WT/Mut**) and high (Mut/Mut***) as described18. We then detected intestinal changes at 3, 8 and 12 months in intestinal sections and using an organoid system (Fig. 1a). Because intestinal crypt number decreases and villus length increases during intestinal aging14,32–34, we first performed hematoxylin-eosin (H&E) staining to examine intestinal architecture. As compared to WT/WT, WT/WT* mice with theoretically maternally transmitted mtDNA mutations showed no significant difference in intestinal architecture during aging, indicating little effect on the intestinal aging. As compared to WT/WT and WT/WT*, Mut/Mut*** mice exhibited clear aging-like changes and accumulation of apoptotic cells in the small intestine at 8 months, which was more pronounced at 12 months (Fig. 1b, c). The higher expression of CDKN1A/ p21 and shorter telomere length in Mut/Mut*** at 8 months also supported exacerbation of small intestine aging with increased mtDNA mutation burden (Supplementary Fig. 1f, g). We also used an in vitro intestinal organoid system to reflect ISC function1,35 and found that organoids derived from Mut/Mut*** mice were fewer and smaller in size versus WT/WT* and WT/Mut** mice (Fig. 1d). The decreased colony formation efficiency in Mut/Mut*** mice indicates mtDNA mutations impair ISC function.
Fig. 1. Increased mtDNA mutation burden induces an aging-like phenotype in the small intestine.
a Diagram depicting the experimental methods for detecting the changes of small intestine and organoids derived from isolated crypts in the mice with four categories of mtDNA mutation burden at 3, 8, and 12 month. The figures of small intestine, intestinal crypt, and organoids were generated with the help of Servier Medical Art, provided by Servier, licensed under a Creative Commons Attribution 4.0 unported license (https://creativecommons.org/licenses/by/4.0/deed.en). PolgAWT/Mut (WT/Mut**) mice were crossed to generate three types of offspring with a theoretically maternally transmitted mtDNA mutations, i.e. PolgAWT/WT (WT/WT*), PolgAWT/Mut (WT/Mut**) and PolgAMut/Mut (Mut/Mut***). PolgAWT/WT mice with negligible level of mtDNA mutations (WT/WT) were used as an additional control. b Representative images of small intestine stained with H&E in WT/WT, WT/WT*, WT/Mut**, and Mut/Mut*** mice at 3, 8, and 12 months of age (Scale bar, 100 μm). The villus length and number of crypts per millimeter of small intestine are quantified below (Data are presented as the mean ± S.D of 9 intestinal segments from 3 mice per group; two-way ANOVA test). c Representative images of intestinal crypts stained with TUNEL in WT/WT*, WT/Mut**, and Mut/Mut*** mice at 3, 8, and 12 months of age (Scale bar, 20 μm). Since no difference was observed during intestinal aging between WT/WT and WT/WT*, WT/WT* mice was used a control. The number of TUNEL-positive cells per crypt is quantified on right (Data are presented as the mean ± S.D and n = 3 mice per group; two-way ANOVA test). d Representative crypt images at Day 0 and organoids at Day 8 in the intestine of WT/WT*, WT/Mut**, and Mut/Mut*** mice at 3, 8, and 12 months of age (Scale bar, 100 μm). Crypt size, organoid size and relative organoid number per well were quantified. Average organoid number per well was normalized to WT/WT* at 3 months. Data are presented as the mean ± S.D of 6 wells from 3 mice per group; two-way ANOVA test. Source data are provided with this paper.
To study the link between the observed aging phenotypes and mtDNA mutation content in detail, we profiled mtDNA mutation types and frequencies in the small intestines of our mouse models. As compared to WT/WT mice, at 8 months Mut/Mut*** mice accumulate significantly more point mutations, primarily as low-frequency (less than 0.05) point mutations; these were also observed in the small intestine of aged wild-type mice (Supplementary Fig. 2a–d). Notably, the intestine of WT/WT* and WT/Mut** mice with little apparent aging phenotype accumulate low quantities of mtDNA mutations (less than 900) at 8 months whereas the intestine of Mut/Mut*** mice accumulates high quantities of mtDNA mutations (average mutation number above 4,000) at 8 months. Interestingly, the brain and liver of WT/Mut** mice did accumulate much more expected mtDNA point mutations but small intestine didn’t, implying a possible mechanism for clearing mutated mtDNAs in the small intestine (Supplementary Fig. 2b–d and Supplementary Fig. 3a, b). Overall, the above results indicate that increased mtDNA mutation burden induces an aging-like phenotype in the small intestine.
mtDNA mutations result in NAD+ depletion during intestinal aging
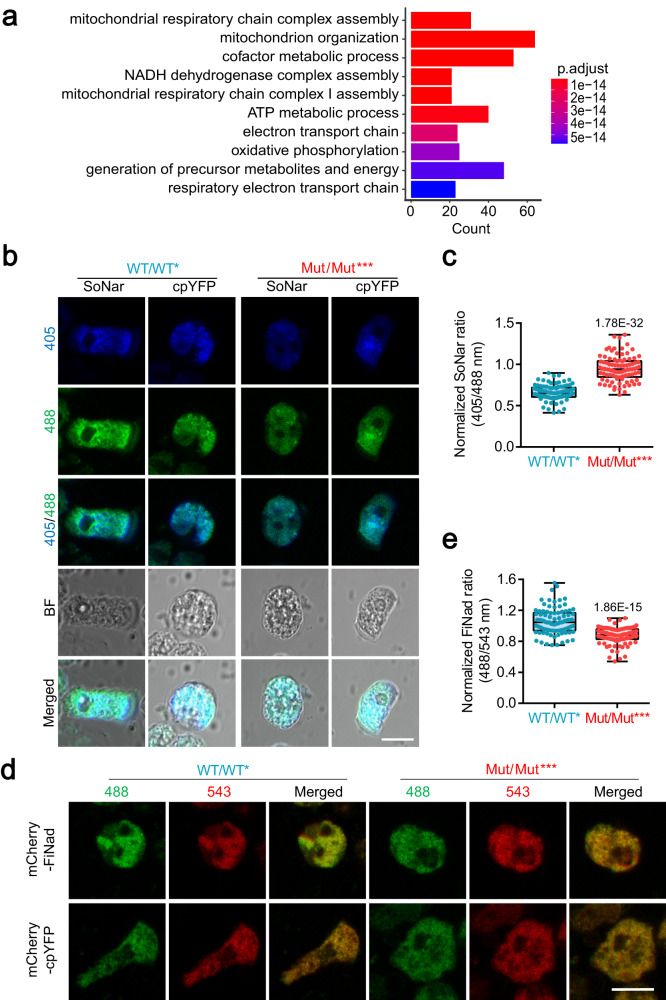
To further explore the mechanism by which increased mtDNA mutation burden regulates intestinal aging, we performed RNA sequencing (RNA-seq) and analyzed 780 upregulated differential genes in the intestines of Mut/Mut*** mice as compared to WT/WT* at 8 months using Gene Ontology (GO) analysis (Supplementary Fig. 4a, b). Most enriched GO terms are involved in mitochondrial respiratory function and metabolic processes, demonstrating an alteration of mitochondrial function caused by mtDNA mutations. As expected, the intestinal crypt cells in Mut/Mut*** mice exhibit a lower mitochondrial inner membrane potential and higher ROS concentrations as compared to WT/WT* mice, indicating that high mtDNA mutation content results in mitochondrial dysfunction during intestinal aging (Supplementary Fig. 4c, d). Notably, the enriched NADH dehydrogenase complex assembly pathway implies an impairment of NADH/NAD+ redox in Mut/Mut*** intestines (Fig. 2a). This perturbed redox potential was confirmed by transfection of intestinal crypt cells with SoNar, a NADH/NAD+ sensor36,37, which showed a higher NADH/NAD+ ratio in Mut/Mut*** mice (Fig. 2b, c). We also transfected intestinal crypt cells with FiNad (a NAD+ sensor38), and found less NAD+ content in the Mut/Mut*** cells (Fig. 2d, e), leading us to investigate the role of NAD+ depletion in the intestinal aging.
Fig. 2. Increased mtDNA mutation burden results in NAD+ depletion during the intestinal aging.
a Gene Ontology (GO) enrichment of differentially upregulated genes in Mut/Mut*** mice at 8 months of age versus WT/WT* mice was performed using clusterProfiler package, and significance was determined by Fisher exact test of adjusted p-value by Benjamini–Hochberg (BH) method (n = 3 mice per group). b Detection of NADH/NAD+ ratio in the intestinal crypts of Mut/Mut*** mice and WT/WT* mice at 8 months of age using SoNar and cpYFP imaging (Scale bar, 10 μm). c Quantification of NADH/NAD+ ratios as in (b) (90 intestinal crypts from 3 mice per group; unpaired comparison using nonparametric Mann–Whitney test). Central line: median, box: interquartile ranges (IQR), whisker: ranges except extreme outliers (>1.5*IQR), circles: individual data points; d Detection of NAD+ content in the intestinal crypts of Mut/Mut*** mice and WT/WT* mice using mCherry-FiNad and mCherry-cpYFP imaging (Scale bar, 10 μm). e Quantification of mCherry-FiNad fluorescence corrected by mCherry-cpYFP as in (d) (90 intestinal crypts from 3 mice per group; unpaired comparison using nonparametric Mann–Whitney test). Central line: median, box: IQR, whisker: ranges except extreme outliers (>1.5*IQR), circles: individual data points. Source data are provided with this paper.
NAD+ repletion alleviates small intestinal aging caused by increased mtDNA mutation content, which is independent of mitophagy
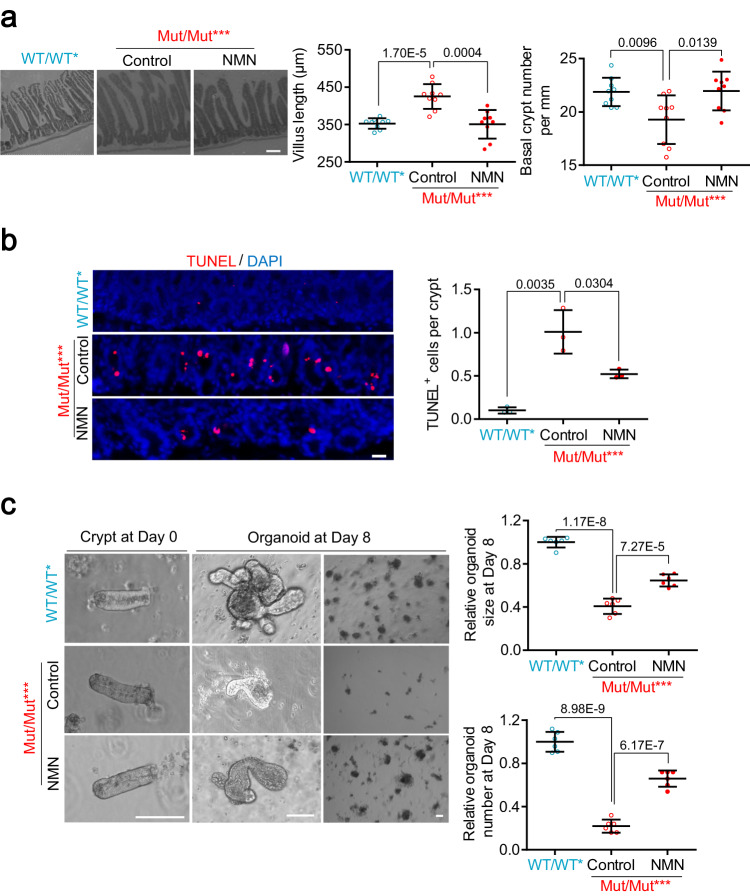
In response to dietary supplementation with the NAD+ precursor, nicotinamide mononucleotide (NMN)39,40, we found increased crypt number, decreased villus length and presence of apoptotic cells in the intestine of Mut/Mut*** mice (Fig. 3a, b). We further validated the upregulation of NAD+ concentration in the intestinal crypts of Mut/Mut*** mice treated with NMN (Supplementary Fig. 5), thus indicating that NAD+ repletion in vivo alleviates the small intestine aging phenotypes caused by mtDNA mutation burden. In parallel, we found NAD+ repletion in vitro could rescue decreased colony formation efficiency in Mut/Mut*** intestinal organoids (Fig. 3c). Together these results indicate that NAD+ depletion is a key mediator by which increased mtDNA mutation burden induces the aging phenotype of small intestine.
Fig. 3. NAD+ repletion alleviates small intestine aging phenotypes caused by mtDNA mutation burden.
a Representative images of small intestine stained with H&E in Mut/Mut*** mice at 8 months of age with NMN or control (water) (Scale bar, 100 μm). Villus length and number of crypts per millimeter are quantified on right; WT/WT* is used as an additional control (Data are presented as the mean ± S.D of 9 intestinal segments from 3 mice per group; unpaired two-tailed Student’s t-test). b Representative images of intestinal crypts stained with TUNEL in Mut/Mut*** mice at 8 months of age with NMN or control (Scale bar, 20 μm). The number of TUNEL-positive cells per crypt is quantified on right (Data are presented as the mean ± S.D and n = 3 mice per group; unpaired two-tailed Student’s t-test). c Representative images of crypts at Day 0 and organoids at Day 8 from the intestine of Mut/Mut*** mice at 8 months of age with 200 μM NMN or water control (Scale bar, 100 μm). Organoid size and relative organoid number per well are quantified at bottom. Average organoid number per well is normalized to the control. Data are presented as the mean ± S.D of 6 wells from 3 mice per group; paired two-tailed Student’s t-test. Source data are provided with this paper.
As NAD+ depletion reportedly compromises mitophagy41,42, it raises the possibility that NAD+ repletion could activate mitophagy to remove mutated mtDNAs, thereby alleviating the small intestine aging phenotypes. By detecting mitophagy using anti-LC3B and anti-TOM20 IF in the intestine crypts of WT/WT*, WT/Mut** and Mut/Mut*** mice at 3, 8 and 12 months, we indeed observed compromised mitophagy in Mut/Mut*** mice at 8 months (Supplementary Fig. 6a). Nevertheless, NAD+ repletion in vivo did not rescue mitophagy deficiency or mtDNA mutation burden in the intestinal crypts of Mut/Mut*** mice at 8 months (Supplementary Fig. 6b, c), indicating that intestinal aging induced by NAD+ depletion is independent of mitophagy.
Increased mtDNA mutation burden impairs Wnt/β-catenin signaling and induces a decline in LGR5-positive intestinal cells through NAD+ depletion
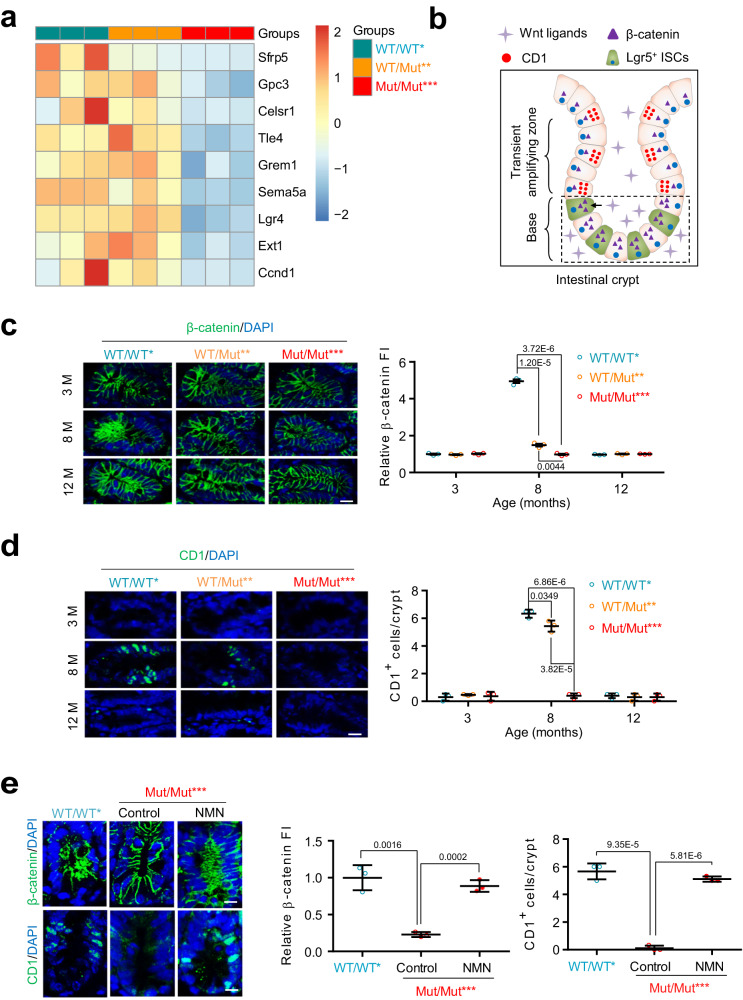
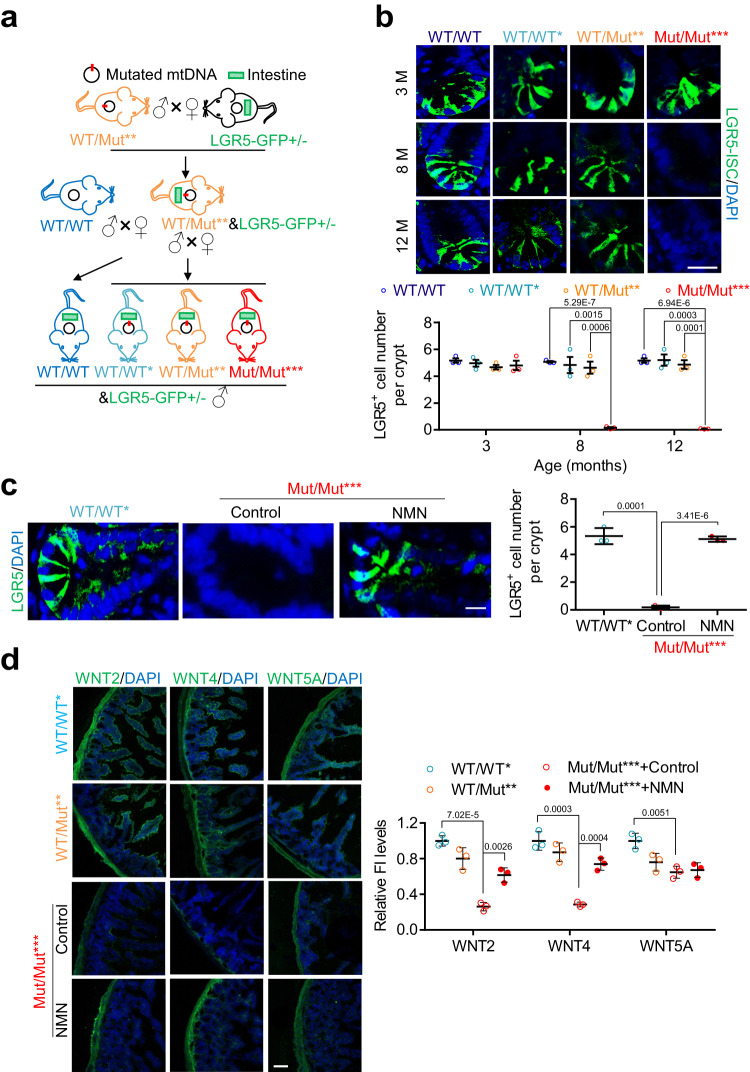
Due to the important roles of signaling pathways including Wnt, Notch, Notum, PI3K/Akt, and TGF-β pathways in regulating the development process of intestinal epithelial cells, we assayed these signaling pathways and their correlation with NAD+ depletion during intestinal aging. Using RNA-seq analysis, we found that only differential genes associated with Wnt/β-catenin signaling were enriched in Mut/Mut*** versus WT/WT* intestine at 8 months, which are downregulated by mtDNA mutations (Fig. 4a). As Wnt/β-catenin signaling is required for LGR5-expressing ISCs to maintain intestinal regeneration, as marked by the presence of Cyclin D1 (CD1)-expressing cells (Fig. 4b), we further performed anti-β-catenin and anti-CD1 immunofluorescence (IF) in the intestinal crypts of WT/WT*, WT/Mut** and Mut/Mut*** mice at 3, 8 and 12 months. We found that β-catenin fluorescence intensity (FI) is decreased at 8 months with fewer CD1-expressing cells in Mut/Mut*** mice versus either WT/WT* or WT/Mut** mice (Fig. 4c, d). These results suggest that increased mtDNA mutation burden impairs the Wnt/β-catenin pathway. Importantly, NAD+ repletion rescues this pathway in the intestine of Mut/Mut*** mice (Fig. 4e), indicating NAD+ depletion underlies the lack of ISC regeneration through the Wnt/β-catenin pathway.
Fig. 4. NAD+ depletion impairs Wnt/β-catenin signaling during the intestinal aging caused by mtDNA mutation burden.
a Heatmap of differential genes associated with Wnt/β-catenin signaling in WT/WT*, WT/Mut** and Mut/Mut*** mice at 8 months of age (n = 3 mice per group). b Schematic illustration of LGR5-expressing ISCs, proliferated epithelial cells and Wnt/β-catenin signaling in the intestinal crypts. The figure of intestinal crypt was generated with the help of Servier Medical Art, provided by Servier, licensed under a Creative Commons Attribution 4.0 unported license (https://creativecommons.org/licenses/by/4.0/deed.en). c, d Anti-β-catenin (c) and anti-CD1 (d) IF in the intestinal crypts of WT/WT*, WT/Mut** and Mut/Mut*** mice at 3, 8 and 12 months of age. Relative FIs of β-catenin and CD1+ cell number per crypt are quantified on right (Scale bar, 10 μm; data are presented as the mean ± S.D and n = 3 mice per group; two-way ANOVA test). e Anti-β-catenin and anti-CD1 IF in the intestinal crypts of Mut/Mut*** mice at 8 months of age treated with NMN or water. Relative FI of β-catenin and CD1+ cell number per crypt are quantified on right (Scale bar, 10 μm; data are presented as the mean ± S.D and n = 3 mice per group; unpaired two-tailed Student’s t-test). Source data are provided with this paper.
We next investigated the effect of mtDNA mutation burden on the number of LGR5-expressing ISCs. Crossing LGR5-GFP+/- (LGR5-eGFP-IRES-CreERT2 reporter)1 and WT/Mut** (PolgAWT/Mut) mice, we generated four genotype LGR5-GFP positive mice with increasing levels of mtDNA mutation burden as depicted in Fig. 5a. We noted that LGR5-positive cells decline sharply at 8 months in Mut/Mut*** mice (Fig. 5b), indicating an adverse effect of mtDNA mutation burden. NAD+ repletion increases the number of LGR5-positive intestinal cells in Mut/Mut*** mice (Fig. 5c).
Fig. 5. Increased mtDNA mutation burden induces ISC decrease through NAD+ depletion.
a Generation of LGR5-GFP reporter mice with four categories of mtDNA mutation burden. LGR5WT/GFP female mice were first crossed to WT/Mut** male mice, and the offspring of LGR5WT/GFP& WT/Mut** mice were further used to generate WT/WT*&LGR5WT/GFP, WT/Mut**&LGR5WT/GFP and Mut/Mut***&LGR5WT/GFP. WT/WT&LGR5WT/GFP mice generated by crossing WT/WT male mice to female LGR5WT/GFP female mice were used as an additional control. The figure was partly generated with the help of Servier Medical Art. b Representative images of LGR5-expressing ISCs in WT/WT&LGR5WT/GFP, WT/WT*&LGR5WT/GFP, WT/Mut**&LGR5WT/GFP and Mut/Mut***&LGR5WT/GFP male mice at 3, 8 and 12 months of age (Scale bar, 20 μm). ISC number is quantified at bottom (Data are presented as the mean ± S.D and n = 3 mice per group; two-way ANOVA test). c Representative images of LGR5-expressing ISCs in Mut/Mut***&LGR5WT/GFP mice at 8 months of age treated with NMN or water control (Scale bar, 10 μm). ISC number is quantified on right (Data are presented as the mean ± S.D and n = 3 mice per group; unpaired two-tailed Student’s t-test). d Anti-WNT2, anti-WNT4 and anti-WNT5A IF in the intestine of WT/WT*, WT/Mut** and Mut/Mut*** mice at 8 months of age treated with NMN or water control. Relative FI is quantified on right (Scale bar, 100 μm; data are presented as the mean ± S.D and n = 3 mice per group; one-way ANOVA test). Source data are provided with this paper.
For the requirement of Wnt molecules, such as WNT243, WNT44 and WNT5A44, secreted from niche cells including Gli1+, Foxl1+ stromal, Paneth and myofibroblast cells as self-renewal factors to support ISCs3–5,43,44, we assayed the expression of these Wnt molecules in the intestinal crypts of WT/WT*, WT/Mut** and Mut/Mut*** mice at 8 months. We found all the tested Wnt proteins are decreased in Mut/Mut*** versus WT/WT* mice (Fig. 5d), showing that increased mtDNA mutation burden inhibits secretion of Wnt molecules. NAD+ repletion partially rescues the reduction of WNT2 and WNT4 protein expression in Mut/Mut*** mice (Fig. 5d and Supplementary Fig. 7a, b), implying that mtDNA mutation burden can inhibit the secretion of these two Wnt molecules through NAD+ depletion. Furthermore, we observed downregulation of Foxl1 but not Gli1 in Mut/Mut*** intestine, implying a possible reduction of Foxl1+ cells caused by mtDNA mutation burden (Supplementary Fig. 7c).
As activation of Notch signaling reportedly induces a subset of Paneth cells to gain stem cell features with subsequently proliferation and differentiation into villus epithelial cells45, we performed anti-Notch1 immunofluorescence. Notch signal activation was observed in the intestinal crypt base of Mut/Mut*** mice at 8 months (Supplementary Fig. 7d), providing a possible explanation for the increase of villus length in the aged intestine lacking the Wnt/β-catenin pathway. NAD+ repletion rescues the Foxl1 downregulation and Notch1 upregulation in Mut/Mut*** mice (Supplementary Fig. 7e), suggesting that mtDNA mutation burden can regulate the function or number of niche cells through NAD+ depletion. Taken together, NAD+ depletion caused by increased mtDNA mutation burden induces the decline of LGR5-positive intestinal cells via impairment of the Wnt/β-catenin pathway.
NAD+ depletion activates ISR to regulate mtDNA mutation-induced aging phenotypes through impairment of the Wnt/β-catenin pathway
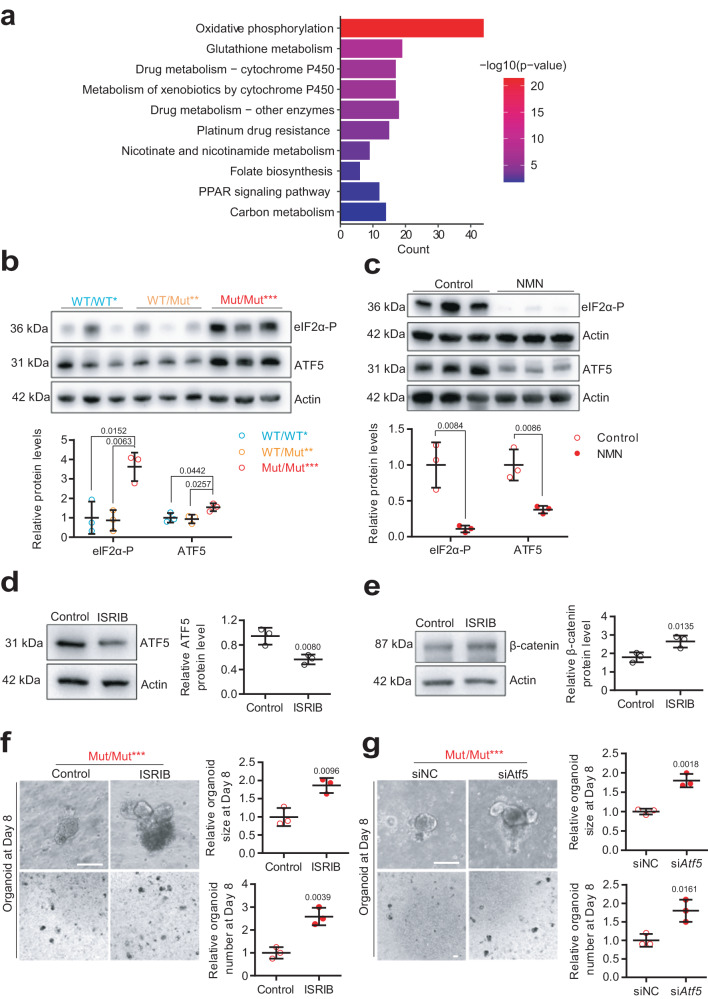
How does NAD+ depletion regulate the Wnt/β-catenin pathway during the intestinal aging caused by mtDNA mutation burden? We performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the upregulated differential genes in the intestines of Mut/Mut*** versus WT/WT* mice at 8 months. Here, increased mtDNA mutation burden triggers a retrograde mitochondria-to-nucleus communication by upregulating ISR including oxidative phosphorylation, glutathione metabolism, nicotinate and nicotinamide metabolism, folate biosynthesis, the PPAR signaling pathway and carbon metabolism (Fig. 6a and Supplementary Fig. 8a).
Fig. 6. NAD+ depletion activates ISR to regulate mtDNA mutation-induced aging phenotypes through impaired Wnt/β-catenin signaling.
a Kyoto Encyclopedia of Genes and Genomes pathway enrichment of differentially upregulated genes in Mut/Mut*** mice at 8 months of age versus WT/WT* mice was performed using clusterProfiler package, and significance was determined by Fisher exact test of adjusted p-value by BH method (n = 3 mice per group). b Protein expression of eIF2α-P and ATF5 by western blot analysis in the intestinal crypts of WT/WT*, WT/Mut** and Mut/Mut*** mice at 8 months of age. Relative band densities quantified using ImageJ are shown at bottom (Data are presented as the mean ± S.D and n = 3 mice per group; one-way ANOVA test). c Protein expression of eIF2α-P and ATF5 by western blot analysis in the intestinal crypts of Mut/Mut*** mice treated with NMN or water. Relative band densities quantified using ImageJ are shown at bottom (Data are presented as the mean ± S.D and n = 3 mice per group; one-way ANOVA test). d Protein expression of ATF5 by western blot analysis in Mut/Mut*** intestinal crypt cells treated with Control or ISRIB. Relative band densities quantified using ImageJ are shown on right (Data are presented as the mean ± S.D and n = 3 mice per group; paired two-tailed Student’s t-test). e Protein expression of β-catenin by western blot analysis in Mut/Mut*** intestinal crypt cells treated with ISRIB or control. Relative band densities quantified using ImageJ are shown on right (Data are presented as the mean ± S.D and n = 3 mice per group; paired two-tailed Student’s t-test). f Representative organoid images at Day 8 from the intestinal crypts of Mut/Mut*** mice at 8 months of age with 500 nM ISRIB or water control (Scale bar, 100 μm). Organoid size and relative organoid number per well are quantified on right, and the average organoid number per well is normalized to the control. (Data are presented as the mean ± S.D and n = 3 mice per group; unpaired two-tailed Student’s t-test) g Representative organoid images at Day 8 from the intestinal crypts of Mut/Mut*** mice at 8 months of age with siAtf5 or control siNC (Scale bar, 100 μm); organoid size and relative organoid number per well are quantified at bottom. Average organoid number per well is normalized to the control. Data are presented as the mean ± S.D and n = 3 mice per group; unpaired two-tailed Student’s t-test. Source data are provided with this paper.
Upon mitochondrial ISR, eukaryotic initiation factor 2 α subunit (eIF2α) is phosphorylated46, which subsequently results in global attenuation of cytosolic translation coincident with preferential translation of mitochondrial stress-responsive transcription factors such as activating transcription factor 4 (ATF4)47–50, ATF551,52 and the C/EBP Homologous Protein (CHOP)53,54 to restore mitochondrial function. Using western blotting, we observed upregulation of eIF2α phosphorylation (eIF2α-P) and ATF5 but not ATF4 and CHOP in the intestine of Mut/Mut*** mice, which was decreased by in vivo NAD+ repletion (Fig. 6b, c and Supplementary Fig. 8b, c). Consistent with direct control of ATF5 translation by phosphorylation of eIF2α in response to stress55, eIF2α phosphorylation inhibitor, ISRIB, suppresses ATF5 upregulation in Mut/Mut*** intestine (Fig. 6d). These results showed that NAD+ depletion triggers ATF5-dependent ISR activation.
ISR inhibition increases β-catenin protein expression in the intestinal crypts of Mut/Mut*** mice (Fig. 6e), implying it regulates intestinal aging by restoring Wnt/β-catenin signaling. To test the roles of ISR activation in intestinal aging, we used ISRIB and siAtf5 to inhibit ISR in Mut/Mut*** intestinal crypts (Supplementary Fig. 8d). We found that both ISRIB and siAtf5 could obviously rescue decreased organoid formation efficiency caused by increased mtDNA mutation burden (Fig. 6f, g).
We next asked how NAD+ depletion regulates ISR? Among NAD+ consuming enzymes, SIRT7 downregulation caused by NAD+ depletion during aging reportedly regulates the UPRmt by suppressing mitochondrial ribosomal protein expression56,57. Despite no reduction of SIRT7 protein in Mut/Mut*** versus WT/WT* mice (Supplementary Fig. 8e), SIRT7 overexpression inhibits eIF2α phosphorylation of Mut/Mut*** intestinal crypt cells (Supplementary Fig. 8f). Consistent with previous reports58,59, these observations support that NAD+ depletion caused by increased mtDNA mutation burden decreases SIRT7 activity to regulate ISR. Taken together, these results indicate that mtDNA mutation burden causes intestinal aging through NAD+ depletion, which activates ATF5-dependent ISR to inhibit Wnt/β-catenin signaling.
The ISR caused by mtDNA mutation burden regulates UPRmt activation
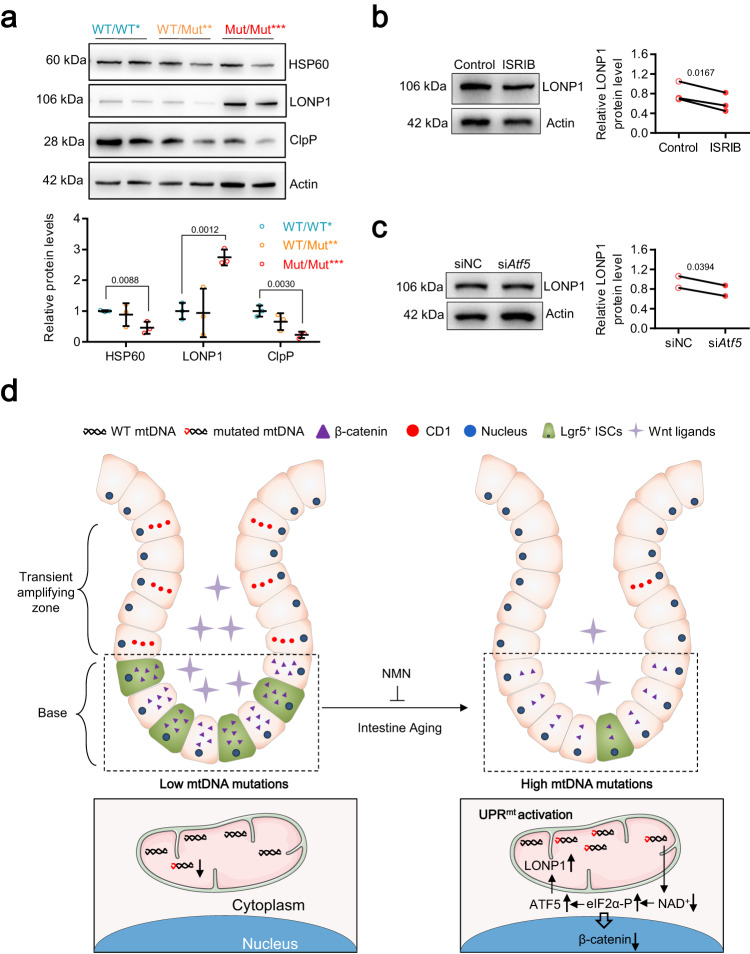
As the ISR regulation factor, ATF5, activate UPRmt by regulating mitochondrial import efficiency60, we assayed the marker protein expression of UPRmt including LONP1, caseinolytic peptidase P (ClpP) and heat shock protein 60 (HSP60) in Mut/Mut*** intestine, which exhibited ATF5-dependent ISR activation. Interestingly, only LONP1, a key mitochondrial protease for clearing unfolded proteins upon mitochondrial stress response61, is obviously increased while its mitochondrial content is not altered in Mut/Mut*** intestinal crypts (Fig. 7a and Supplementary Fig. 9a, b). Both ISRIB and siAtf5 inhibited the upregulation of LONP1 in Mut/Mut*** intestinal crypt cells (Fig. 7b, c), demonstrating that mtDNA mutation burden regulates LONP1 expression by ATF5-dependent ISR. Overall, intestinal aging induced by increased mtDNA mutation burden is mediated through the ATF5-dependent UPRmt activation, which is characterized by LONP1 upregulation.
Fig. 7. The ISR caused by mtDNA mutation burden regulates UPRmt activation.
a Protein expression of HSP60, LONP1, and ClpP by western blot analysis in the intestinal crypts in WT/WT*, WT/Mut**, and Mut/Mut*** mice at 8 months of age. Relative band densities quantified using ImageJ are shown at bottom (Data are presented as the mean ± S.D and n = 3 mice per group; one-way ANOVA test). b Protein expression of LONP1 by western blot analysis in the intestinal crypts of Mut/Mut*** mice treated with ISRIB or control. Relative band densities quantified using ImageJ are shown on right (Data are presented as the mean ± S.D and n = 3 mice per group; paired two-tailed Student’s t-test). c Protein expression of LONP1 by western blot analysis in the intestinal crypts of Mut/Mut*** mice treated with siAtf5 or siNC. Relative band densities quantified using ImageJ are shown on right (Data are presented as the mean ± S.D and n = 2 mice per group; paired two-tailed Student’s t-test). d Summary of mechanism regulating intestinal aging caused by mtDNA mutation burden. The figures of small intestine and mitochondria were generated with the help of Servier Medical Art, provided by Servier, licensed under a Creative Commons Attribution 4.0 unported license (https://creativecommons.org/licenses/by/4.0/deed.en).
Discussion
During the aging process, mtDNA mutations are observed to be accumulated in aged rodent and human tissues21–23,62–65. We showed previously that the significant accumulation of much more low-frequency (less than 0.5%) mtDNA point mutations in human oocytes during aging is linked with impaired blastocyst formation18. We also observed the accumulation of low-frequency point mutations in aged mouse intestine, implying an important role in physiological aging. Notably, the aged intestine of POLG mutator mice show a similar but more rapid accumulation of low-frequency mtDNA point mutations versus wild-type mice aged intestine, providing a suitable model for investigating the roles of mtDNA mutation burden on intestinal aging.
Using this mouse model, we found that increased mtDNA mutation burden resulted in NAD+ depletion during intestinal aging with subsequent activation of the ATF5-dependent UPRmt. These mechanisms regulate mtDNA mutation-induced aging phenotypes and LGR5-positive ISC exhaustion through impaired Wnt/β-catenin signaling (Fig. 7d).
Using in vivo ISC labeling and organoid technology, we defined the landscape of cross-cell signals caused by mtDNA mutation burden in ISCs and niche cells. Wnt/β-catenin signaling is required for ISCs to maintain the balance between proliferation and differentiation7,66,67. High levels of Wnt signaling at the base of intestinal crypts activates β-catenin in ISCs, which upregulates its target genes (c-MYC, Cyclin D1, etc.) to promote ISC differentiation into the transient amplifying (TA) zone68. Our study showed that low mtDNA mutation burden activates the Wnt/β-catenin pathway in the base of intestinal crypts and increases the number of CD1-expressing cells in the TA zone during intestinal aging (Fig. 4). Impairment of this cross-cell communication was observed in the aged intestine caused by increased mtDNA mutation burden (Fig. 4), implying an important role in small intestine aging. These findings provide insights into the intestinal cell differentiation processes. Upon aging, intestinal crypt size, villus length, Paneth cell number, and goblet cell number all increase, and the number and regenerative potential of ISCs are reduced upon aging in mice14,34,69. It is interesting that intestinal villus length increases while ISC number is decreased in the small intestine, requiring further studies.
NAD+ depletion is regarded as a common signature of aging70,71, which is observed in many tissues during aging72–75. It also reportedly compromises mitophagy41,42, which exacerbates accelerated aging41,76–79. Similarly, we observed NAD+ depletion and compromised mitophagy during intestinal aging in response to increased mtDNA mutation burden. We also found that NAD+ depletion activates the ATF5-dependent UPRmt to regulate intestinal aging and that the compromised mitophagy could not be restored by NAD+ repletion. Overall, our study provides a mechanism linking NAD+ depletion and aging, and supports its utility as a biomarker to monitor the aging of different tissues.
Mitochondrial ISR, triggered by mitochondrial stress, modulates protein synthesis and selectively overexpresses a set of stress-responsive genes to activate pathways including one carbon metabolism, serine acid biosynthesis and antioxidant mechanism/redox homeostasis48,49,53,54,80–82. The stress-responsive transcription factors, ATF447–50, ATF551,52 and/or CHOP53,54, reportedly mediate the transcription of mitochondrial ISR genes. We found that increased mtDNA mutation burden triggers ISR activation through NAD+ depletion. Interestingly, ATF5 but not ATF4 and CHOP is upregulated in the aged Mut/Mut*** intestine, demonstrating a preferentially translation of ATF5.
The UPRmt is reportedly activated by various mitochondrial stresses including mitonuclear protein imbalance and mitochondrial import efficiency60,83, which is marked by increased protein expression of LONP1, HSP60 and ClpP. We found that increased mtDNA mutation burden triggers the ATF5-dependent UPRmt activation and resulting LONP1 upregulation, and regulates intestinal aging phenotypes. The specific upregulation of LONP1 protein provides a candidate marker for intestinal aging caused by increased mtDNA mutation burden.
Finally, we showed that NAD+ repletion in vitro and in vivo increases Wnt/β-catenin signaling as well as the number of LGR5-expressing ISCs in the small intestine. Moreover, both siAtf5 and ISRIB enhance the regenerative potential of aged intestinal crypts. These findings identify multiple potential candidates for both treating and preventing intestinal aging.
Methods
Mice
PolgAD257A heterozygous mice (PolgAWT/Mut, Stock no: 017341, USA) and LGR5-EGFP-IRES-creERT2 heterozygous mice (LGR5WT/GFP Stock no: 008875, USA) were purchased from the Jackson Laboratory. The genotypes of siblings were determined by sequencing the genome of mouse tail at 3 weeks old. Wild-type C57BL/6 J mice with different ages (3 months, 8, 12, and 20 months) were purchased from the GemPharmatech co. Ltd (Nanjing, China). PolgAWT/Mut male mice were continuously crossed to WT female mice for maintaining the population of PolgAWT/Mut mice. Then, the male and female PolgAWT/Mut siblings were crossed to generate three genotype mice including WT/WT*, WT/Mut** and Mut/Mut***. Mice were housed in an environment of suitable temperature (25 °C) and humidity (typically 50%) under a 12 h: 12 h light/dark cycle (7 am/7 pm) with accessing to food and water ad libitum. The same batch of male mice was used to perform experiments in accordance with relevant guidelines and regulations, which had been reviewed and approved by Guangzhou Institutes of Biomedicine and Health Ethical Committee (Approve no. 2018040). Mice were euthanized using CO2 inhalation.
Histology
Sections of jejunum, the middle region of small intestine, were dissected from mice and fixed in 4% formalin for 24 h and embedded in paraffin. The jejunum sections were mounted on slides and stained with hematoxylin-eosin (H&E), and observed under a light microscope (Zeiss, Germany).
TUNEL staining
A TUNEL assay kit (In Situ Cell Death Detection Kit, Roche, 45-12156792910) was used to characterize apoptosis in the jejunum sections according to the manufacture’s instruction. After being dewaxed and hydrated, the jejunum sections were placed in 200 mL 0.1 M citric acid buffer (pH 6.0) and irradiated with 750 watts microwave for 1 min. The sections were immediately cooled with double distilled water (~25°C) and washed twice with PBS. After antigen repair, the sections were fixed with 4% paraformaldehyde for 1 h at room temperature, and were subsequently incubated with protease K for 30 min at 37 °C. Then, the slides were immersed in 0.1 M Tris-HCl (pH 7.5) containing 3% bovine serum albumin and 20% normal bovine serum for 30 min at room temperature. The sections were incubated with 50 μL TUNEL reaction mixture in a humid environment under dark for 1 h at 37 °C. After 3 times of PBS washing, the sections were stained with DAPI solution (D9542, Sigma–Aldrich, USA), sealed and visualized under a Zeiss LSM 710 with a 10× objective. The number of TUNEL-positive cells was quantified by ImageJ software.
Detection of telomere length
Mouse telomere repeats were measured by real-time quantitative PCR as described84. Briefly, total genome of jejunum was prepared using a genome extraction kit (TianGen, China, DP304). The PCR reaction was run on a CFX-96 real-time PCR detection system (BioRad, USA) with an SYBR Green QPCR kit (TaKaRa, RR820A). The primers (forward primer: 5′-CGGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT-3′, reverse primer: 5′-GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT-3′) were used to detect telomere. Amplification of 36B4 in the same samples was used as an internal control for detecting telomere length, and the primers (forward primer: 5′-ACTGGTCTAGGACCCGAGAAG-3′, reverse primer: 5′-TCAATGGTGCCTCTGGAGATT-3′) were used to detect 36B4. Relative telomere length was calculated and normalized to values obtained from the amplification of 36B4.
Analysis of mtDNA mutations
Total genome of jejunum was prepared using a genome extraction kit (TianGen, China, DP304). Four pairs of primers were used to amplify region 1611-5657, amplified with a forward primer (5′-AAAAGCAGCCACCAATAAAG-3′) and a reverse primer (5′-GAGAAATGATGGTGGTAGGA-3′), region 5636-9914, amplified with a forward primer (5′-ACTCCTACCACCATCATTTC-3′) and a reverse primer (5′-GAGAAGGCTATGGTGAGGTT-3′), region 9876-14278, amplified with a forward primer (5′-TATGCCATCTACCTTCTTCA-3′) and a reverse primer (5′-ATTTGGACTATTAGGCAGAC-3′) and region 14258-1611, amplified with a forward primer (5′-AGTCTGCCTAATAGTCCAAA-3′) and a reverse primer (5′-CTTTATTGGTGGCTGCTTTT-3′), which covers the whole mtDNA sequence.
PCR of mtDNA using the genome of jejunum as template was performed, and the four amplified fragments were mixed for sequencing. Briefly, 3 μg mixture of mtDNA was used to construct a library as described85 for a better homogeneity of sequencing depth. Then, the library was sent to Berry Genomics. Co., Ltd (Beijing, China) and sequenced on an Illumina Hiseq4000 platform using 150 bp paired end reads for 3 G flux. mtDNA mutations were analyzed as described15.
Small intestinal crypt isolation and organoid culture
Small intestinal crypts were isolated from jejunum sections as described86, and cultured using “ENR” culture medium as described87,88. Mice were sacrificed by cervical dislocation. The jejunum was dissected longitudinally, cleaned with PBS at 4 °C, and cut into tissue blocks with 1 centimeter length. After being cleaned 10–20 times with PBS containing 100 U/mL penicillin/streptomycin, the jejunum segments were incubated with 2 mM EDTA solution (pH 8.0) for 20 min at 4 °C, shaken by a vortex shaker for 2 min and centrifuged at 110 g for 5 min. After the supernatant was removed, the intestinal crypts were gently flicked with DMEM/F-12 medium, centrifuged at 110 g for 5 min, and resuspended with ENR medium. 10 μL intestinal crypt suspension and 60 μL matrigel were mixed, added into a 24-well cell culture plate, and polymerized for 15 min at 37 °C. The intestinal crypts were cultured with ENR medium, and the frequency of organoid formation was analyzed as described35. siAtf5 (Sense strand sequence: 5′-GCTCGTAGACTATGGGAAAdTdT-3′) was used for silencing the Atf5 genes in the intestinal crypt cells following the procedure described in the Lipofectamine™ RNAiMAX transfection kit. After validating silence efficiency, the siRNA and control siNC (5′-TTCTCCGAACGTGTCACGT-3′) were added into the “ENR” medium. Then, isolated intestinal crypts were cultured with this medium for the first 2 days, and organoid formation was visualized with an optical microscope (ECLIPSE Ts2-FL, Nikon, Japan) at Day 8.
Detection of mitochondrial inner membrane potential and ROS
For detecting mitochondrial inner membrane potential, the isolated mouse intestinal crypt cells were incubated with JC-1 (Beyotime, C2003S, China) for 30 min at room temperature in the dark. Then, fluorescence intensity was detected using a BD Fortessa flow cytometer (BD Biosciences, USA) within 1 h and analyzed using FlowJo V10 software. The gating strategy is shown in Supplementary Fig. 10. For detecting ROS, isolated mouse intestinal crypt cells were first seeded on a 20 mm glass-bottom cell culture dish. Then, the cells were incubated with DCF (Invitrogen, D399, USA) for 30 min at room temperature, and imaged using a Zeiss LSM 880 with a 63× objective.
Immunofluorescence
Immunofluorescence was performed on 6 μm thick paraffin or frozen sections. The paraffin sections of jejunum were deparaffinized, rehydrated, and permeabilized in 10 mM sodium citrate buffer. After being restored to room temperature, the frozen sections of jejunum were first soaked in PBS for 10 min and then blocked for 30 min. Then, all sections were incubated with primary antibodies overnight at 4 °C. After 5 times washing with PBS, the sections were incubated with Alexa Fluor 488 and 568 conjugated secondary antibodies (Life Technologies). Finally, the sections were imaged using a Zeiss LSM 900 or a Zeiss LSM 710 with a 10× objective after incubation with DAPI solution (D9542, Sigma–Aldrich, USA) for 10 min. Antibodies used were as follows: anti-HSP60 (Abcam, ab46798, 1:200), anti-Cyclin D1 (Abcam, ab16663, 1:200), anti-Gli1 (Santa Cruz Biotechnology, sc-515751, 1:100), anti-Foxl1 (Santa Cruz Biotechnology, SC-130373, 1:100), anti-LC3B (Cell Signal Technology, 2775, 1:200), anti-TOM20 (Abcam, ab56783, 1:200), anti-β-catenin (Cell Signal Technology, 9562 S, 1:200), anti-Notch1 (Abclonal, A7636, 1:100), anti-WNT2 (Abclonal, A23997, 1:100), anti-WNT5A (Abclonal, A12744, 1:100), anti-WNT4 (Bio-Techne, MAB4751, 1:100), anti-CDKN1A/p21 (Abclonal, A2691, 1:100), anti-Mouse IgG (H + L), Alexa Fluor 488 (Thermo Fisher Scientific, A-11001, 1:500), anti-Rabbit IgG (H + L), Alexa Fluor 488 (Thermo Fisher Scientific, A-11008,1:500) and anti-Rabbit IgG (H + L), Alexa Fluor 568, (Thermo Fisher Scientific, A-11011, 1:500).
Imaging for detecting NADH/NAD+ ratio and NAD+ level
SoNar/cpYFP plasmids for measuring NADH/NAD+ ratio, and mCherry-FiNad/mCherry-cpYFP plasmids for measuring the NAD+ level were gifts from Professor Yi Yang and Yuzheng Zhao (East China University of Science and Technology, China). The intestinal crypt cells were transfected with these plasmids for 24–48 h, and then seeded on a 20 mm glass-bottom cell culture dish (FluoroDish, World Precision Instruments, FD35-100) for imaging using a Zeiss LSM 710 with a 40× objective. The F405 nm/F488 nm ratios of SoNar and the F488 nm/F543 nm ratios of mCherry-FiNad were calculated as described previously32–34.
RNA sequencing
Total RNA of the jejunum sections was extracted using a TRIzol-based protocol. Libraries were prepared following the instructions for the IlluminaTruSeq RNA Sample Prep kit. After sequencing was performed on a Illumina NovaSeq instrument, data were analyzed with RSEM software. Then, DESeq2 was used to identify the differential expressed genes, with p-adjust <0.05 & abs (log2FoldChange)>0.5. Gene Ontology was performed using ClusterProfiler.
NMN and ISRIB supplementation
For NAD+ repletion in vivo, β-NMN (1094-61-7, Sigma–Aldrich, USA) was administered in drinking water to treat mice for 2 weeks as described53. For NAD+ repletion in vitro, β-NMN solution was first prepared into autoclaved water at the certain dose and sterilely filtered, and then added into “ENR” medium for culturing intestinal crypt cells. For inhibiting ISR in vitro, 500 nM ISRIB (S0706, Selleck, USA) was first prepared into autoclaved water at the certain dose and filtering sterilely as described89, and then added into “ENR” medium for culturing intestinal crypt cells.
SIRT7 overexpression
The pLVX-SIRT7-Puro and control pLVX-Puro plasmids were separately transfected into intestinal crypt cells following the instructions for the Lipofectaminew 3000 Reagent kit (L3000015, Invitrogen, USA). After 48 h, intestinal crypt cells were lysed, and then protein expression of SIRT7 and eIF2α-P was assayed by western blotting.
Measurement of NAD+ content
Small intestinal crypts were isolated and sent to Tsinghua University (Beijing, China) for determining NAD+ content using a liquid chromatography-tandem mass spectrometry (LC–MS/MS). The ACQUITY UPLC H-Class system was coupled a 6500plus QTrap mass spectrometer (AB SCIEX, USA), equipped with a heated electrospray ionization (HESI) probe. Extracts were separated by a synergi Hydro-RP column (2.0 × 100 mm, 2.5 μm, phenomenex). The mobile phase consisted of a binary solvent system: mobile phase A (2 mM triisobutylamine adjusted with 5 mM acetic acid in water) and mobile phase B (methanol). A 15-min gradient with flow rate of 250 μL/min was used as follows: 0–1.5 min, 5%B; 1.5–9 min, 5–35% B; 9.5–12 min, 98% B;12.1–15 min, 5%B. Column chamber and sample tray were held at 35 °C and 10 °C, respectively, and data were acquired in multiple reaction monitor (MRM) mode. The nebulizer gas (Gas1), heater gas (Gas2), and curtain gas were set at 50, 50, and 35 psi, respectively. The ion spray voltage was −4500 V in negative ion mode. The optimal probe temperature was determined to be 450 °C. The SCIEX OS 1.6 software was applied for metabolite identification and peak integration.
Western blotting
Equal amounts of total protein extracted from the intestinal crypt cells, were electrophoresed on 12% polyacrylamide/sodium dodecyl sulfate gel and transferred onto polyvinylidene fluoride membranes. After being blocked for 45 min, the membranes were incubated with primary antibodies for 1 h. Subsequently, membranes were incubated with horseradish peroxidase-coupled secondary antibodies for 1 h after 3 times washing with PBS. Finally, the immunoreactivity was detected using Immobilon Western Chemiluminescent HRP Substrate (Millipore, USA, WBKLS0500). The antibodies used were as follows: anti-HSP60 (Abcam, ab46798, 1:1000), anti-LONP1 (Cell Signal Technology, 28020, 1:1000), anti-CDKN1A/p21 (Abclonal, A2691, 1:100), anti-Phospho-eIF2α-S51 (Abclonal, AP0692, 1:1000), anti-ClpP (Abcam, ab124822, 1:1000), anti-ATF4 (Abcam, ab216839, 1:1000), anti-ATF5(Abcam, ab184923, 1:1000), anti-CHOP(Cell Signal Technology, 2895 S, 1:1000), anti-TOM20 (Proteintech, 11802-1-AP, 1:1000), anti-WNT4 (Bio-Techne, MAB4751, 1:1000), anti-SIRT7 (Abclonal, A22735, 1:1000), anti-Actin (HUABIO, ET1702-67, 1:3000), HRP Conjugated Goat anti-Mouse IgG (HUABIO, HA1006, 1:3000) and HRP Conjugated Goat anti-Rabbit IgG (HUABIO, HA1001,1:3000).
Statistical analysis
Data are shown as mean ± Standard Deviation (SD). All statistical analysis was performed using Student’s t-test or ANOVA test from the GraphPad Prism software (GraphPad) as indicated in the figure legends. P values of less than 0.05 were considered as significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This work was financially supported by the National Key Research and Development Program of China (2022YFA1103800), the National Natural Science Foundation projects of China (32025010, 92157202, 32241002, 92254301, 32261160376, 31970709, 32070729, 32100619, 32170747, 32322022, 32370782, 32371007, 32300608, 32300620, 92357302), NSFC/RGC Joint Grant Scheme 2022/2023 (N_CUHK 428/22), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB0480000), the National Key Research and Development Program of China (2022YFE0210100, 2019YFA0904500, 2023YFE0210100), the Key Research Program, CAS (ZDBS-ZRKJZ-TLC003), International Cooperation Program, CAS (154144KYSB20200006), CAS Project for Young Scientists in Basic Research (YSBR-075), Guangdong Province Science and Technology Program (2023B1111050005, 2023A1515030231, 2022A1515110493, 2023B1212060050, 2021A1515012513, 2021B1515020096, 2022A1515012616, 2022A1515110951), Guangzhou Science and Technology Program (202102021037, 202102020827, 202102080066, 202206060002, 2023A04J0414), Health@InnoHK funding support from the Innovation Technology Commission of the Hong Kong SAR and CAS Youth Innovation Promotion Association (to Y. W and K. C). We thank the Experimental Animal Center and Public Instrument Center in Guangzhou Institutes of Biomedicine and Health, and the Metabolomics Facility Center of Metabolomics and Lipidomics in National Protein Science Technology Center of Tsinghua University for LC–MS/MS experiments.
Author contributions
X. Liu initiated and supervised the project. L.Y., Z.R., and X. Lin designed, performed the experiments, and analyzed the data. H.W., Y.X., H.T., Z.H., J.W., Y.Z., and Y.W. participated in the experiments. D.Q., G.L., K.M.L., and W-Y.C. gave suggestions. X. Liu, L.Y., and Z.R. wrote the manuscript.
Peer review
Peer review information
Nature Communications thanks Aleksandra Trifunovic and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The raw RNA-seq and mtDNA-seq data have been deposited in the Genome Sequence Archive (GSA) at the Beijing Institute of Genomics (BIG) Data Center, BIG, Chinese Academy of Sciences (https://bigd.big.ac.cn/gsa), and the accession numbers are CRA012233 for RNA-seq (https://ngdc.cncb.ac.cn/gsa/browse/CRA012233) and CRA012237 for mtDNA-seq (https://ngdc.cncb.ac.cn/gsa/browse/CRA012237) that are publicly accessible. The LC–MS/MS data for measuring NAD+ content has been deposited in the MassIVE data repository (https://massive.ucsd.edu/) and are available with the accession code MSV000093751 (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?accession=MSV000093751). All other relevant data supporting the key findings of this study are available within the article and its supplementary information files. Any additional information is available upon request to the corresponding author (Xingguo Liu, liu_xingguo@gibh.ac.cn). Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Liang Yang, Zifeng Ruan, Xiaobing Lin.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-44808-z.
References
- 1.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 2.Metcalfe C, Kljavin NM, Ybarra R, de Sauvage FJ. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell. 2014;14:149–159. doi: 10.1016/j.stem.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Shoshkes-Carmel M, et al. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature. 2018;557:242–246. doi: 10.1038/s41586-018-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Degirmenci B, Valenta T, Dimitrieva S, Hausmann G, Basler K. GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature. 2018;558:449–453. doi: 10.1038/s41586-018-0190-3. [DOI] [PubMed] [Google Scholar]
- 5.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fevr T, Robine S, Louvard D, Huelsken J. Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol. Cell. Biol. 2007;27:7551–7559. doi: 10.1128/MCB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian H, et al. Opposing activities of Notch and Wnt signaling regulate intestinal stem cells and gut homeostasis. Cell Rep. 2015;11:33–42. doi: 10.1016/j.celrep.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao B, et al. The non-muscle-myosin-II heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells. Nat. Commun. 2015;6:7166. doi: 10.1038/ncomms8166. [DOI] [PubMed] [Google Scholar]
- 9.Fischer JM, et al. Single cell lineage tracing reveals a role for TgfbetaR2 in intestinal stem cell dynamics and differentiation. Proc. Natl Acad. Sci. USA. 2016;113:12192–12197. doi: 10.1073/pnas.1611980113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiener Z, et al. Oncogenic mutations in intestinal adenomas regulate Bim-mediated apoptosis induced by TGF-beta. Proc. Natl Acad. Sci. USA. 2014;111:E2229–E2236. doi: 10.1073/pnas.1406444111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynolds A, et al. Canonical Wnt signals combined with suppressed TGFbeta/BMP pathways promote renewal of the native human colonic epithelium. Gut. 2014;63:610–621. doi: 10.1136/gutjnl-2012-304067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Massague J. TGFbeta in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pentinmikko N, et al. Notum produced by Paneth cells attenuates regeneration of aged intestinal epithelium. Nature. 2019;571:398–402. doi: 10.1038/s41586-019-1383-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nalapareddy K, et al. Canonical Wnt signaling ameliorates aging of intestinal stem cells. Cell Rep. 2017;18:2608–2621. doi: 10.1016/j.celrep.2017.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsson NG. Somatic mitochondrial DNA mutations in mammalian aging. Annu. Rev. Biochem. 2010;79:683–706. doi: 10.1146/annurev-biochem-060408-093701. [DOI] [PubMed] [Google Scholar]
- 16.Payne BA, Chinnery PF. Mitochondrial dysfunction in aging: much progress but many unresolved questions. Biochim. Biophys. Acta. 2015;1847:1347–1353. doi: 10.1016/j.bbabio.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trifunovic A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 18.Yang L, et al. Mitochondrial DNA mutation exacerbates female reproductive aging via impairment of the NADH/NAD(+) redox. Aging Cell. 2020;19:e13206. doi: 10.1111/acel.13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niemann J, et al. An mtDNA mutation accelerates liver aging by interfering with the ROS response and mitochondrial life cycle. Free Radic. Biol. Med. 2017;102:174–187. doi: 10.1016/j.freeradbiomed.2016.11.035. [DOI] [PubMed] [Google Scholar]
- 20.Herbst A, et al. Accumulation of mitochondrial DNA deletion mutations in aged muscle fibers: evidence for a causal role in muscle fiber loss. J. Gerontol. A Biol. Sci. Med. Sci. 2007;62:235–245. doi: 10.1093/gerona/62.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baines HL, et al. Similar patterns of clonally expanded somatic mtDNA mutations in the colon of heterozygous mtDNA mutator mice and ageing humans. Mech. Ageing Dev. 2014;139:22–30. doi: 10.1016/j.mad.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greaves LC, et al. Comparison of mitochondrial mutation spectra in ageing human colonic epithelium and disease: absence of evidence for purifying selection in somatic mitochondrial DNA point mutations. PLoS Genet. 2012;8:e1003082. doi: 10.1371/journal.pgen.1003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greaves LC, et al. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc. Natl Acad. Sci. USA. 2006;103:714–719. doi: 10.1073/pnas.0505903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kujoth GC, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 25.Chen ML, et al. Erythroid dysplasia, megaloblastic anemia, and impaired lymphopoiesis arising from mitochondrial dysfunction. Blood. 2009;114:4045–4053. doi: 10.1182/blood-2008-08-169474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norddahl GL, et al. Accumulating mitochondrial DNA mutations drive premature hematopoietic aging phenotypes distinct from physiological stem cell aging. Cell Stem Cell. 2011;8:499–510. doi: 10.1016/j.stem.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Vermulst M, et al. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat. Genet. 2008;40:392–394. doi: 10.1038/ng.95. [DOI] [PubMed] [Google Scholar]
- 28.Vermulst M, et al. Mitochondrial point mutations do not limit the natural lifespan of mice. Nat. Genet. 2007;39:540–543. doi: 10.1038/ng1988. [DOI] [PubMed] [Google Scholar]
- 29.Hauser, D. N., Primiani, C. T., Langston, R. G., Kumaran, R. & Cookson, M. R. The polg mutator phenotype does not cause dopaminergic neurodegeneration in DJ-1-deficient mice. eNeuro10.1523/ENEURO.0075-14.2015 (2015). [DOI] [PMC free article] [PubMed]
- 30.Nissanka N, Bacman SR, Plastini MJ, Moraes CT. The mitochondrial DNA polymerase gamma degrades linear DNA fragments precluding the formation of deletions. Nat. Commun. 2018;9:2491. doi: 10.1038/s41467-018-04895-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross JM, et al. Germline mitochondrial DNA mutations aggravate ageing and can impair brain development. Nature. 2013;501:412–415. doi: 10.1038/nature12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corazza GR, et al. Proliferating cell nuclear antigen expression is increased in small bowel epithelium in the elderly. Mech. Ageing Dev. 1998;104:1–9. doi: 10.1016/S0047-6374(98)00044-X. [DOI] [PubMed] [Google Scholar]
- 33.Webster SG, Leeming JT. The appearance of the small bowel mucosa in old age. Age Ageing. 1975;4:168–174. doi: 10.1093/ageing/4.3.168. [DOI] [PubMed] [Google Scholar]
- 34.Martin K, Kirkwood TB, Potten CS. Age changes in stem cells of murine small intestinal crypts. Exp. Cell Res. 1998;241:316–323. doi: 10.1006/excr.1998.4001. [DOI] [PubMed] [Google Scholar]
- 35.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y, et al. In vivo monitoring of cellular energy metabolism using SoNar, a highly responsive sensor for NAD(+)/NADH redox state. Nat. Protoc. 2016;11:1345–1359. doi: 10.1038/nprot.2016.074. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y, et al. SoNar, a highly responsive NAD+/NADH sensor, allows high-throughput metabolic screening of anti-tumor agents. Cell Metab. 2015;21:777–789. doi: 10.1016/j.cmet.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zou Y, et al. Illuminating NAD(+) metabolism in live cells and in vivo using a genetically encoded fluorescent sensor. Dev. Cell. 2020;53:240–252 e247. doi: 10.1016/j.devcel.2020.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imai S. A possibility of nutriceuticals as an anti-aging intervention: activation of sirtuins by promoting mammalian NAD biosynthesis. Pharmacol. Res. 2010;62:42–47. doi: 10.1016/j.phrs.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caton PW, Kieswich J, Yaqoob MM, Holness MJ, Sugden MC. Nicotinamide mononucleotide protects against pro-inflammatory cytokine-mediated impairment of mouse islet function. Diabetologia. 2011;54:3083–3092. doi: 10.1007/s00125-011-2288-0. [DOI] [PubMed] [Google Scholar]
- 41.Fang EF, et al. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell. 2014;157:882–896. doi: 10.1016/j.cell.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang EF, et al. NAD(+) augmentation restores mitophagy and limits accelerated aging in Werner syndrome. Nat. Commun. 2019;10:5284. doi: 10.1038/s41467-019-13172-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niec RE, et al. Lymphatics act as a signaling hub to regulate intestinal stem cell activity. Cell Stem Cell. 2022;29:1067–1082 e1018. doi: 10.1016/j.stem.2022.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kondo A, Kaestner KH. FoxL1(+) mesenchymal cells are a critical source of Wnt5a for midgut elongation during mouse embryonic intestinal development. Cells Dev. 2021;165:203662. doi: 10.1016/j.cdev.2021.203662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu S, et al. Paneth cell multipotency induced by notch activation following injury. Cell Stem Cell. 2018;23:46–59.e45. doi: 10.1016/j.stem.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dalton LE, Healey E, Irving J, Marciniak SJ. Phosphoproteins in stress-induced disease. Prog. Mol. Biol. Transl. Sci. 2012;106:189–221. doi: 10.1016/B978-0-12-396456-4.00003-1. [DOI] [PubMed] [Google Scholar]
- 47.Quiros PM, et al. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 2017;216:2027–2045. doi: 10.1083/jcb.201702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo X, et al. Mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway. Nature. 2020;579:427–432. doi: 10.1038/s41586-020-2078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bao, X. R. et al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. Elife10.7554/eLife.10575 (2016). [DOI] [PMC free article] [PubMed]
- 50.Sasaki, K. et al. Mitochondrial translation inhibition triggers ATF4 activation, leading to integrated stress response but not to mitochondrial unfolded protein response. Biosci. Rep.10.1042/BSR20201289 (2020). [DOI] [PMC free article] [PubMed]
- 51.Liou, Y. H. et al. Hepatic Fis1 regulates mitochondrial integrated stress response and improves metabolic homeostasis. JCI Insight10.1172/jci.insight.150041 (2022). [DOI] [PMC free article] [PubMed]
- 52.Gorrell L, Makareeva E, Omari S, Otsuru S, Leikin S. ER, mitochondria, and ISR regulation by mt-HSP70 and ATF5 upon procollagen misfolding in osteoblasts. Adv. Sci. 2022;9:e2201273. doi: 10.1002/advs.202201273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaspar, S. et al. Adaptation to mitochondrial stress requires CHOP-directed tuning of ISR. Sci. Adv.10.1126/sciadv.abf0971 (2021). [DOI] [PMC free article] [PubMed]
- 54.Fessler E, et al. A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature. 2020;579:433–437. doi: 10.1038/s41586-020-2076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou D, et al. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J. Biol. Chem. 2008;283:7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]
- 56.Mohrin M, et al. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science. 2015;347:1374–1377. doi: 10.1126/science.aaa2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He M, et al. An acetylation switch of the NLRP3 inflammasome regulates aging-associated chronic inflammation and insulin resistance. Cell Metab. 2020;31:580–591.e585. doi: 10.1016/j.cmet.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang CL, et al. The mitochondrial unfolded protein response regulates hippocampal neural stem cell aging. Cell Metab. 2023;35:996–1008.e1007. doi: 10.1016/j.cmet.2023.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohkubo R, et al. The hepatic integrated stress response suppresses the somatotroph axis to control liver damage in nonalcoholic fatty liver disease. Cell Rep. 2022;41:111803. doi: 10.1016/j.celrep.2022.111803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fiorese CJ, et al. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr. Biol. 2016;26:2037–2043. doi: 10.1016/j.cub.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao Q, et al. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cortopassi GA, Arnheim N. Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res. 1990;18:6927–6933. doi: 10.1093/nar/18.23.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Piko L, Hougham AJ, Bulpitt KJ. Studies of sequence heterogeneity of mitochondrial DNA from rat and mouse tissues: evidence for an increased frequency of deletions/additions with aging. Mech. Ageing Dev. 1988;43:279–293. doi: 10.1016/0047-6374(88)90037-1. [DOI] [PubMed] [Google Scholar]
- 64.Taylor RW, et al. Mitochondrial DNA mutations in human colonic crypt stem cells. J. Clin. Invest. 2003;112:1351–1360. doi: 10.1172/JCI19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lob M, Hugonnaud C. [Danger of pneumoconiosis due to hard metals and berilosis in the dental technicians who work with metallic prosthesis] Dent Press. 1978;14:21–26. [PubMed] [Google Scholar]
- 66.Acar A, et al. Inhibition of Wnt signalling by Notch via two distinct mechanisms. Sci. Rep. 2021;11:9096. doi: 10.1038/s41598-021-88618-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pinto D, Clevers H. Wnt control of stem cells and differentiation in the intestinal epithelium. Exp. Cell Res. 2005;306:357–363. doi: 10.1016/j.yexcr.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 68.Bastide P, et al. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J. Cell Biol. 2007;178:635–648. doi: 10.1083/jcb.200704152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Igarashi M, et al. NAD(+) supplementation rejuvenates aged gut adult stem cells. Aging Cell. 2019;18:e12935. doi: 10.1111/acel.12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fang EF, et al. NAD(+) in aging: molecular mechanisms and translational implications. Trends Mol. Med. 2017;23:899–916. doi: 10.1016/j.molmed.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yaku K, Okabe K, Nakagawa T. NAD metabolism: implications in aging and longevity. Ageing Res. Rev. 2018;47:1–17. doi: 10.1016/j.arr.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 72.Mills KF, et al. Long-term administration of nicotinamide mononucleotide mitigates age-associated physiological decline in mice. Cell Metab. 2016;24:795–806. doi: 10.1016/j.cmet.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang H, et al. NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science. 2016;352:1436–1443. doi: 10.1126/science.aaf2693. [DOI] [PubMed] [Google Scholar]
- 74.Zhu XH, Lu M, Lee BY, Ugurbil K, Chen W. In vivo NAD assay reveals the intracellular NAD contents and redox state in healthy human brain and their age dependences. Proc. Natl Acad. Sci. USA. 2015;112:2876–2881. doi: 10.1073/pnas.1417921112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fang EF, et al. NAD(+) replenishment improves lifespan and healthspan in ataxia telangiectasia models via mitophagy and DNA repair. Cell Metab. 2016;24:566–581. doi: 10.1016/j.cmet.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fivenson EM, et al. Mitophagy in neurodegeneration and aging. Neurochem. Int. 2017;109:202–209. doi: 10.1016/j.neuint.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fang EF, et al. Mitophagy inhibits amyloid-beta and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat. Neurosci. 2019;22:401–412. doi: 10.1038/s41593-018-0332-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Babbar M, Basu S, Yang B, Croteau DL, Bohr VA. Mitophagy and DNA damage signaling in human aging. Mech. Ageing Dev. 2020;186:111207. doi: 10.1016/j.mad.2020.111207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Onishi M, Yamano K, Sato M, Matsuda N, Okamoto K. Molecular mechanisms and physiological functions of mitophagy. EMBO J. 2021;40:e104705. doi: 10.15252/embj.2020104705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Khan NA, et al. mTORC1 regulates mitochondrial integrated stress response and mitochondrial myopathy progression. Cell Metab. 2017;26:419–428 e415. doi: 10.1016/j.cmet.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 81.Kuhl, I. et al. Transcriptomic and proteomic landscape of mitochondrial dysfunction reveals secondary coenzyme Q deficiency in mammals. Elife10.7554/eLife.30952 (2017). [DOI] [PMC free article] [PubMed]
- 82.Mick, E. et al. Distinct mitochondrial defects trigger the integrated stress response depending on the metabolic state of the cell. Elife10.7554/eLife.49178 (2020). [DOI] [PMC free article] [PubMed]
- 83.Houtkooper RH, et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Callicott RJ, Womack JE, Real-time PCR. assay for measurement of mouse telomeres. Comp. Med. 2006;56:17–22. [PubMed] [Google Scholar]
- 85.Murienne J, et al. PCR-free shotgun sequencing of the stone loach mitochondrial genome (Barbatula barbatula) Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016;27:4211–4212. doi: 10.3109/19401736.2015.1022744. [DOI] [PubMed] [Google Scholar]
- 86.Gjorevski N, et al. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–564. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- 87.Tao S, et al. Wnt activity and basal niche position sensitize intestinal stem and progenitor cells to DNA damage. EMBO J. 2017;36:2920–2921. doi: 10.15252/embj.201797813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Basak O, et al. Induced quiescence of Lgr5+ stem cells in intestinal organoids enables differentiation of hormone-producing enteroendocrine cells. Cell Stem Cell. 2017;20:177–190.e174. doi: 10.1016/j.stem.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 89.Xie SZ, et al. Sphingolipid modulation activates proteostasis programs to govern human hematopoietic stem cell self-renewal. Cell Stem Cell. 2019;25:639–653.e637. doi: 10.1016/j.stem.2019.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw RNA-seq and mtDNA-seq data have been deposited in the Genome Sequence Archive (GSA) at the Beijing Institute of Genomics (BIG) Data Center, BIG, Chinese Academy of Sciences (https://bigd.big.ac.cn/gsa), and the accession numbers are CRA012233 for RNA-seq (https://ngdc.cncb.ac.cn/gsa/browse/CRA012233) and CRA012237 for mtDNA-seq (https://ngdc.cncb.ac.cn/gsa/browse/CRA012237) that are publicly accessible. The LC–MS/MS data for measuring NAD+ content has been deposited in the MassIVE data repository (https://massive.ucsd.edu/) and are available with the accession code MSV000093751 (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?accession=MSV000093751). All other relevant data supporting the key findings of this study are available within the article and its supplementary information files. Any additional information is available upon request to the corresponding author (Xingguo Liu, liu_xingguo@gibh.ac.cn). Source data are provided with this paper.