Abstract
The alternative complement pathway (ACP) functions as a surveillance mechanism by which microorganisms are opsonized with C3b in the absence of specific antibodies. The effectiveness of the ACP relies on its ability to distinguish self from non-self. This recognition function is mediated by C3 regulatory proteins including serum factor H, membrane cofactor protein (MCP), and membrane decay-accelerating factor (DAF). H activity against bound C3b can be increased by host components such as sialic acid and decreased by microbial polysaccharides. DAF and MCP may also recognize cell surface changes such as the presence of viral glycoproteins, since some virus-infected and tumor cells activate the ACP. In the present study, liposomes containing wild-type and mutant Salmonella minnesota lipopolysaccharide (LPS) were tested for ACP activation in serum. LPS-containing liposomes with bound C3b were then tested for their susceptibility to C3 convertase regulation by H and membrane DAF and for the sensitivity of their bound C3b to the cofactor activity of H. The results indicate that while the shortest mutant, Re595 LPS, did not induce ACP activation, R7 LPS containing an additional disaccharide did. This activation was poorly regulated by DAF but was inhibited by H. The regulatory activity of H for liposome-bound C3b, however, decreased when LPS of greater polysaccharide size was present in the membrane. In contrast the ACP activation induced by the phospholipid phosphatidylethanolamine was effectively inhibited by DAF but only poorly inhibited by H.
The alternative complement pathway (ACP) is a primary host defense system that can be triggered by a wide variety of bacteria, parasites, virus-infected cells, and tumor cells. In contrast to the classical pathway, which relies on immunoglobulins or other recognition molecules, the ACP performs a constant surveillance function that does not depend on acquired immunity. Continuous low-grade interaction of C3 and factors B, D, and P cleaves C3 to C3a and C3b (34, 35). The nascent C3b that is generated contains a reactive thioester which can bind covalently to cell surfaces (16). Activation of the ACP is achieved following formation of the critical C3 convertase enzyme, C3bBb, which generates additional C3b in a positive feedback loop. The result is opsonization for clearance by phagocytic cells bearing C3 receptors and/or formation of the lytic membrane attack complex.
Normal human cells and tissues have specific regulatory proteins that block complement activation by inhibiting the formation of C3bBb and by promoting the breakdown of the convertase and C3b (2). These proteins share structural and functional characteristics and are encoded by a gene cluster termed the regulators of complement activation locus (13). The genes in this cluster encode serum factor H and C4b binding protein, as well as the cell surface-associated decay-accelerating factor (DAF), membrane cofactor protein (MCP), and complement receptors CR1 and CR2. DAF and MCP are responsible for protecting host cells from autologous complement-mediated attack. They have wide tissue distributions and function intrinsically by destabilizing C3 convertases (DAF) or by serving as cofactors for the cleavage of deposited C3b by factor I (MCP) (17, 26, 27, 37). CR1 and CR2 have more limited tissue distributions and function extrinsically as receptors for C3b/C4b (CR1) or C3dg (CR2) deposited on complement activators (1). Since in general foreign cells do not possess C3 regulatory molecules, these proteins provide a mechanism for distinguishing self from non-self (2).
Factor H is the principal serum regulator of the ACP. It inhibits convertase formation both in the fluid phase and on cells by binding C3b in place of factor B and acting as a cofactor for C3b cleavage. With respect to C3b on cells, H is capable of discriminating human cells and pathogens by virtue of exhibiting higher affinity for bound C3b when it jointly recognizes sialic acid or polyanions on human cell surfaces (6, 15, 20). The ability to decrease H affinity for C3b and thereby activate the ACP has been demonstrated with microbial cell wall polysaccharides, such as zymosan and bacterial lipopolysaccharide (LPS), consistent with the role of restricted H function in ACP-mediated defense (7, 33).
Complement regulatory function on human cells can be restricted and human cells can be converted to activators of the ACP by virus infection or malignant transformation. For example, some human lymphoid cell lines have been shown to activate the ACP in human serum despite the presence of membrane DAF and MCP (5). Activation of the ACP by viral glycoproteins expressed on infected cells has also been demonstrated (24). Three previous reports have provided evidence that DAF, like H, has reduced regulatory activity on some surfaces which activate the ACP (22, 25, 31). However, the mechanism by which DAF activity is overcome on ACP activators is unknown.
Studies by Pangburn (29, 30) of C3b in the fluid phase have established that occupancy of the C3 covalent binding site by acceptor groups in certain polysaccharides can decrease H binding. More recently, analysis of deletion mutants of H has identified at least three sites on H that can promote its attachment to C3b on sheep erythrocytes (38). These authors reported that H binding to C3b on zymosan was less sensitive to deletion of two of these sites, suggesting that the lack of participation of these additional binding sites may account for the decreased affinity of H for C3b bound to ACP activators. Our previous studies indicate that DAF remains functional in the presence of phospholipid ACP activators but is a poor regulator of ACP activation by LPS (25). Binding of C3b to structures such as viral glycoproteins at a distance from the cell membrane might decrease the regulatory activity of membrane-bound DAF.
This study was undertaken to compare the regulatory activities of DAF and H on membranes containing ACP activators. A liposome model was used to compare regulation of C3bBb on membranes containing LPS molecules differing in length as prototypic polysaccharide activators and on membranes containing phospholipids which activate the ACP. The results indicate differences in restriction of H and DAF function by ACP activators, consistent with the concept that these regulators have complementary roles in the regulation of complement activation on cell membranes.
MATERIALS AND METHODS
Buffers.
The following buffers were used: VBS (5 mM Veronal, 0.15 M NaCl [pH 7.4]), GVB (VBS containing 0.1% gelatin), GVB-Ni (GVB containing 1 mM NiCl2), GGVB (GVB diluted with an equal volume of 5% glucose), and PBS (phosphate-buffered saline; 10 mM phosphate buffer, 0.15 NaCl [pH 7.4]).
Preparation of liposomes.
Materials used for the preparation of liposomes were synthetic dipalmitoylphosphatidylcholine (DPPC; Sigma, St. Louis, Mo.), dipalmitoylphosphatidylethanolamine (DPPE) and cholesterol (CHOL) (Calbiochem, La Jolla, Calif.), and phenol-extracted LPS from wild-type and mutant (Re595, R7, and R5) serotypes of Salmonella minnesota and wild-type Salmonella typhosa. Lipids were dissolved in chloroform at 10 mM and stored under nitrogen at −20°C.
All liposomes used in these studies were prepared with a 2:1 molar ratio of DPPC and CHOL except for phosphatidylethanolamine (PE)-containing liposomes (PE liposomes), which were prepared with 6:4:5 molar ratios of DPPC, DPPE, and CHOL. LPS-containing liposomes (LPS liposomes) were formed by adding 12.5 μg of LPS (at 1 mg/ml in 80% methanol–20% chloroform) per μmol of DPPC. Lipids and LPS were mixed, coated onto the side of a pear-shaped flask by rotary evaporation, and dried under vacuum for 1 h. VBS was added to make the phospholipid concentration 10 mM, and the mixture was incubated for 1 h at 45°C. Multilamellar liposomes were then formed by vigorously vortexing the flask for 1 min followed by sonication for 1 h in an 80-W Ultramet III sonicator (Buehler, Evanston, Ill.). The resulting liposome preparations were pelleted by centrifugation at 12,000 × g for 5 min and then resuspended in VBS to a concentration of 10 mM phospholipid.
Quantitation of available LPS on liposomes.
LPS at 1 mg/ml in 100 μl of VBS was mixed with an equal amount of Immunopure photoactivatable biotin (Pierce Chemical, Rockford, Ill.) in the dark. Mixtures were irradiated with three pulses of a Vivitar photoflash to induce cross-linking, incubated for 15 min on ice, and then dialyzed against VBS. The biotin-labeled LPS was serially diluted from 0.05 mg/ml in VBS, and 100-μl standards were incubated on microtiter wells overnight at 4°C. The wells were blocked with 0.5% bovine serum albumin (BSA) in PBS and developed with horseradish peroxidase (HRP)-conjugated streptavidin (BRL Life Technologies, Gaithersburg, Md.) diluted 1/1,000 in 0.1% BSA in PBS and ABTS [2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid)] substrate (1 mg of ABTS per ml and 0.01% H2O2 in citrate-phosphate buffer [pH 4.6]). Liposome samples to be tested were prepared as described above except that 25% of the total LPS used was biotin labeled. After preparation, duplicate 50-μl samples of each liposome preparation were pelleted by centrifugation and incubated for 1 h at room temperature in 100 μl of HRP-streptavidin. Liposomes were transferred to new tubes, washed three times with 1 ml of VBS, and incubated with 100 μl of ABTS substrate. All reactions were stopped after 1 h by adding 50 μl of 0.25 M oxalic acid. Liposomes were pelleted, supernatants were transferred to enzyme-linked immunosorbent assay (ELISA) plates containing the corresponding LPS standards, and the absorbance at 405 nm was determined. The concentration of LPS exposed on liposomes was calculated from standards after subtraction of values for nonlabeled liposome controls.
Preparation of complement reagents.
Normal human serum (NHS) was obtained by centrifugation of blood collected from healthy adult volunteers and stored in aliquots at −70°C. To inhibit activation of the classical pathway, NHS was treated for 5 min on ice with 10 mM EGTA and 2.5 mM MgCl2 (MgEGTA). To inhibit both pathways of activation, NHS was treated with 10 mM EDTA.
C3 was purified by the method of Hammer et al. (11). Human factors B (14), D (4), H (12), I (11), and P (39) were purified by published methods. Factor I was further purified on an affinity matrix prepared with goat anti-factor I (Quidel, San Diego, Calif.) and CarboLink coupling gel (Pierce). Affinity-purified erythrocyte DAF was prepared as previously described (41). Purified C3 was radiolabeled with Na125I (ICN, Costa Mesa, Calif.) to a specific activity of ∼250,000 cpm/μg, using Iodo-beads (Pierce).
Complement activation by liposomes.
Liposomes (20 μl at 10 mM phospholipid in VBS) were incubated in a 60-μl reaction mixture containing 20 μl of MgEGTA-treated NHS and 20 μl of GVB for 30 min at 37°C. Reactions were carried out in duplicate, and control samples were incubated in EDTA-treated NHS to assess nonspecific C3 binding, which was subtracted. The liposomes were washed three times in 1 ml of cold GVB and lysed with 150 μl of 0.5% Triton X-100 in PBS. Liposome samples were tested for C3 deposition by using a competitive ELISA as previously described (23).
C3 convertase inhibition by DAF.
In these experiments the Ni2+-stabilized convertase was used to deposit C3b (8). All liposome types (4 μmol of phospholipid washed in GVB-Ni) were precoated with equal levels of bound C3b by incubation in 150 μl of preformed convertase solution (1 mg of C3 per ml, 0.6 mg of B per ml, 16 μg of D per ml, and 8 mM NiCl2 in VBS, incubated for 5 min at room temperature) and C3 (0.1 mg/μmol of phospholipid) for 30 min at 37°C. Control liposomes were incubated in 150 μl of GVB in place of convertase to assess nonspecific C3 binding. Aliquots (400 nmol of phospholipid) were analyzed by competitive ELISA for bound C3, and the amount of C3b deposition on sample liposomes was determined by subtracting C3 binding on controls.
DAF was incorporated into C3b-bearing liposomes by incubation at 37°C for 30 min in DAF diluted in VBS to a Triton X-100 concentration of 0.003% or less. Liposomes were washed three times in 1 ml of GVB-Ni and resuspended at 200 nmol of phospholipid in GVB-Ni. Liposomes were incubated in a 38-μl reaction mixture of 2 mM NiCl2, 127 μg of B per ml, 2 μg of D per ml, 20 μg of C3 containing 200,000 cpm of 125I-C3, and GGVB for 45 min at 37°C. Following the incubation, total counts per minute per tube was determined, liposomes were transferred to new tubes and washed three times with 1 ml of GVB, and the counts per minute bound to liposome pellets was determined. All reactions were carried out in triplicate and in parallel with control liposomes incubated with C3 alone to determine nonspecific binding, which was subtracted. The level of DAF inhibition of convertase activity was calculated as the percent decrease in the amount of C3b specifically bound in the presence of DAF compared to controls without DAF.
C3 convertase inhibition by H.
Liposomes were prepared with equal amounts of bound C3b as described above. The ability of increasing H concentrations to regulate C3 deposition on liposomes by the ACP C3 convertase was determined as described for DAF except that H was added at the same time as convertase components and 125I-C3.
H cofactor activity.
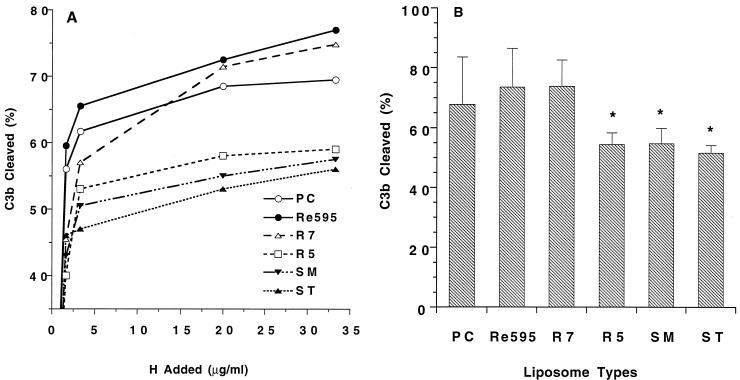
Phosphatidylcholine (PC)-containing liposomes (PC liposomes) and LPS liposomes with bound 125I-C3b were prepared by a three-step procedure. In the first step, liposomes containing 1.6 μmol of DPPC were washed with 1 ml of GVB-Ni and incubated for 30 min at 37°C in a 60-μl reaction volume containing 0.15 mg of C3/μmol of DPPC and 20 μl of preformed convertase (0.65 mg of C3 per ml, 0.42 mg of B per ml, 10.6 μg of D per ml, 40.6 μg of P per ml, and 5 mM NiCl2 in VBS, incubated for 5 min at room temperature). In the second and third steps, liposomes were washed with 1 ml of GVB-Ni and reincubated for 30 min at 37°C in 300 μl of GGVB with 0.15 mg of 125I-C3/μmol of DPPC (6 × 106 cpm) and 0.1 mg of B per ml, 5 μg of D per ml, 30 μg of P per ml, and 5 mM NiCl2. Following the third incubation, the 125I-C3b liposomes were washed three times in 1 ml of GVB and adjusted to 10 mM DPPC in VBS. The 125I-C3b liposomes were divided into 20-μl samples and incubated for 45 min at 37°C with increasing concentrations of H and 0.5 μg of factor I per ml in a volume of 30 μl. Reaction mixtures were boiled for 10 min in 25 μl of reducing sample buffer (0.125 M Tris [pH 6.8], 4% sodium dodecyl sulfate [SDS], 20% glycerol, 10% 2-mercaptoethanol, 0.001% bromophenol blue) and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on 5 to 15% gels. Gels were dried and analyzed by using a PhosphorImager and ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.). The percent C3b cleaved was calculated from the ratio of the 68-kDa iC3b band divided by the sum of the 101-kDa C3b band and the 68-kDa iC3b band (36). Samples incubated with H or I only showed no cleavage of C3b.
Assay for DAF in liposomes.
The level of DAF incorporation into liposomes was determined by using a direct ELISA. Microtiter wells were coated overnight at 4°C with DAF standards diluted in VBS. Wells were blocked with 0.5% BSA in PBS and incubated for 1 h with anti-DAF monoclonal antibody IA10 at 10 μg/ml in 0.1% BSA in PBS. ELISA wells were developed with HRP-conjugated rabbit anti-mouse immunoglobulin G (Cappel, Durham, N.C.) and ABTS substrate. Liposomes containing 300 nmol of phospholipid in VBS were pelleted and incubated sequentially with monoclonal antibody IA10, secondary antibody, and ABTS substrate as described above except in microcentrifuge tubes. After the final incubation, liposomes were pelleted and the absorbance at 405 nm of supernatants was determined after transfer to ELISA plates. Reactions were carried out in duplicate and in parallel with control liposomes.
RESULTS
ACP activation by liposomes.
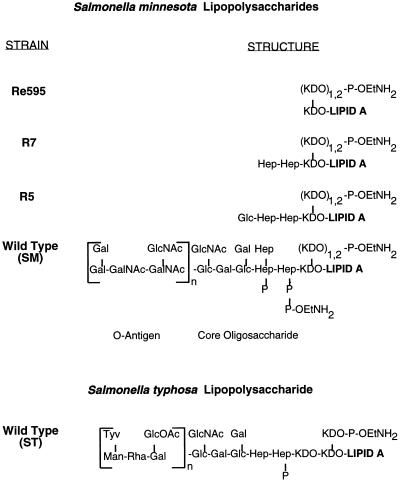
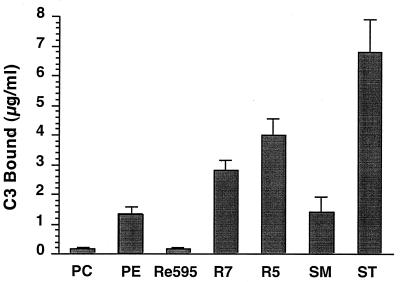
SM LPS and ST LPS (LPS from wild-type S. minnesota and S. typhosa, respectively) from smooth strains consist of a hydrophobic lipid A domain covalently linked to a conserved core oligosaccharide and a unique O antigen that may be repeated from 1 to 40 times (Fig. 1). These forms of LPS together with LPS from rough mutant strains of S. minnesota (Re595, R7, and R5) provided a series of LPS types differing in polysaccharide length which could be incorporated into membranes through their hydrophobic lipid A domains (10). Lipid vesicles of DPPC and CHOL were prepared with or without 12.5 μg of LPS per μmol of phospholipid and tested for ACP activation in MgEGTA-chelated serum (Fig. 2). The amount of LPS on liposome surfaces was approximately equal to the amount of LPS added during liposome preparation. As shown in Fig. 2, liposomes containing either SM, ST, R5, or R7 LPS activated the ACP, whereas liposomes containing Re595 LPS and control PC liposomes did not. ACP activation by the various LPS liposomes is consistent with previous results obtained by using micellular aggregates of LPS (40). As previously reported, liposomes containing 40% PE also activated the ACP (Fig. 2).
FIG. 1.
Structures of S. minnesota LPS (40) and general structure of S. typhosa LPS. Abbreviations: Gal, galactose; Glc, glucose; GlcNac, N-acetylglucosamine; GlcOAc, O-acetylglucose; Hep, l-glycero-d-mannoheptose; KDO, 3-deoxy-d-manno-octulosonic acid; Tyv, tyvelose; Rha, rhamnose; Man, mannose; P-OEtNH2, phosphoethanolamine; P, phosphate (n = 1 to 40).
FIG. 2.
ACP activation by liposomes containing LPS or PE. PC control liposomes were prepared with 2:1 molar ratios of DPPC and CHOL. LPS liposomes were prepared by addition of 12.5 μg of LPS of the indicated serotype/μmol of phospholipid. PE liposomes were prepared with 6:4:5 molar ratios of DPPC, DPPE, and CHOL. Liposomes (200 nmol of phospholipid) were incubated with MgEGTA-treated NHS, and C3 binding was measured by ELISA. Data presented are the mean ± SEM from three or more experiments.
C3 deposition by purified C3, B, and D in the absence of regulatory proteins was nearly equal for all liposome types (data not shown). This result suggested that increased C3 deposition on LPS and PE liposomes incubated in serum resulted from decreased complement regulation rather than increased C3 binding sites.
Effect of DAF and H on ACP activation by LPS.
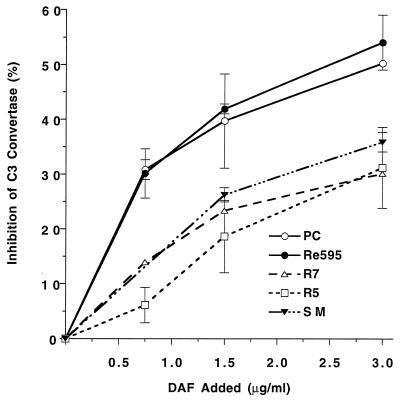
C3, B, and D were used to deposit equal amounts of C3b (51 ± 6 ng of C3b/μmol of phospholipid) on liposomes containing mutant and wild-type LPS. DAF was then incorporated, and its ability to inhibit C3bBb activity was determined. The amount of DAF incorporated did not differ significantly among the different liposome types. The results illustrated in Fig. 3 demonstrate that DAF regulatory activity was reduced when R5, R7, or SM LPS was present in the membrane. In contrast, DAF activity was not affected by the addition of the nonactivating Re595 LPS, which has a lipid A moiety identical to that of the other LPS molecules. The inhibition of C3 convertase activity by DAF at 1.5 μg/ml was significantly decreased on R5, R7, or SM LPS liposomes compared to PC or Re595 LPS liposomes (P < 0.05).
FIG. 3.
Effect of LPS on DAF regulation of ACP C3 convertase activity. Liposomes with equal amounts of bound C3b were incubated with DAF to allow incorporation, washed, and incubated with 125I-C3, B, D, and NiCl2 to form the ACP C3 convertase. Inhibition of convertase activity by DAF was determined as the percent decrease in bound 125I-C3b observed in the presence of DAF. Results are the means ± SEM from two experiments using triplicate samples. t tests on the 1.5-μg/ml DAF values indicate that DAF inhibition of the C3 convertase did not differ on the PC and Re595 liposomes but that DAF inhibition was significantly less when R5, R7, or SM LPS liposomes were used (P < 0.05).
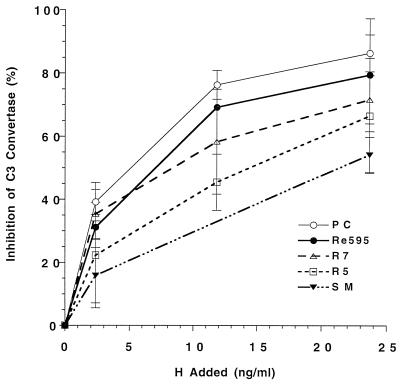
We then used the same assay to examine the effect of the incorporated LPS molecules on H function. Liposomes bearing equal amounts of C3b (57 ± 6 ng of C3b/μmol of phospholipid) were incubated with C3, B, and D in the presence of H. Although both DAF and H were inhibited by LPS forms which activate the ACP in serum, H inhibition was correlated with LPS length whereas DAF inhibition was equivalent for all LPS molecules except the nonactivating Re595 LPS (Fig. 4). The results in Fig. 4 indicate a negative correlation between the functional activity of H and the length of the LPS polysaccharide in the liposomes.
FIG. 4.
Effect of LPS on H regulation of ACP C3 convertase activity. Liposomes with equal amounts of bound C3b were incubated with 125I-C3, B, D, and NiCl2 to form the ACP C3 convertase in the presence and absence of H. Inhibition of convertase activity by H was determined as the percent decrease in bound 125I-C3b observed in the presence of H. Results are the means ± SEM from two or more experiments with triplicate samples. t tests on the 2.4-μg/ml H values indicate that H inhibition of the C3 convertase was significantly greater for PC or Re595 LPS liposomes than for SM LPS liposomes (P < 0.05). H inhibition of liposomes prepared with R5 and R7 LPS was intermediate and not significantly different from H inhibition of either PC or SM LPS liposomes.
Effect of LPS on the cofactor function of H.
In the previous assay, H inhibition could have occurred on fluid-phase as well as liposome-bound C3b. To eliminate this possibility and to further examine the effect of LPS on H, the cofactor activity of H for factor I cleavage of C3b was determined on liposomes containing LPS. Liposomes containing ST, SM, R5, R7, or Re595 LPS or no LPS were prepared, and equal amounts of 125I-C3b (32 ng/μmol of phospholipid) were deposited by incubation with C3, B, D, and P. The 125I-C3b liposomes were then incubated with increasing concentrations of H and 0.5 μg of I per ml followed by SDS-PAGE on 5 to 15% gels under reducing conditions. H cofactor activity was determined by quantitating the amount of C3b cleaved to iC3b with a PhosphorImager. Figure 5A shows the results from a representative set of experiments with different concentrations of H. The results indicate that despite the presence of equal amounts of C3b, a greater amount of C3b was cleaved on PC, Re595, and R7 liposomes than on liposomes containing R5, SM, or ST LPS. Figure 5B shows the mean ± standard error of the mean (SEM) from three separate experiments for the percentage of cleaved C3b at the highest H concentration. Consistent with the results of the convertase inhibition assay, a greater fraction of C3b sites were resistant to H and I on R5 and wild-type LPS liposomes than on control PC liposomes or liposomes containing the shortest mutant LPS (Re595 and R7 LPS).
FIG. 5.
Cofactor activity of H for factor I cleavage of C3b on liposomes. (A) Liposomes with equal amounts of bound 125I-C3b were incubated with increasing concentrations of H in the presence of 0.5 μg of factor I per ml. Reaction mixtures were separated by SDS-PAGE on 5 to 15% gels under reducing conditions. Radioactive bands were analyzed with a PhosphorImager to determine the amount of C3b cleaved to iC3b. The percent C3 cleaved was determined from the quantity of 68-kDa iC3b α chain divided by the sum of the 68-kDa α chain and the 101-kDa C3b α chain. Less than 2% C3b cleavage was observed in the absence of H. Results of single experiments for each liposome type are shown. (B) Percentage of C3b cleaved on liposomes by I (0.5 μg/ml) in the presence of the maximum H concentration (33.3 μmol/mol of phospholipid). The level of C3b cleaved on liposomes was determined as described for panel A. Results presented are the mean ± SEM for three experiments. ∗, mean value significantly different from that for PC control liposomes (P < 0.05).
Effect of phospholipids on DAF and H.
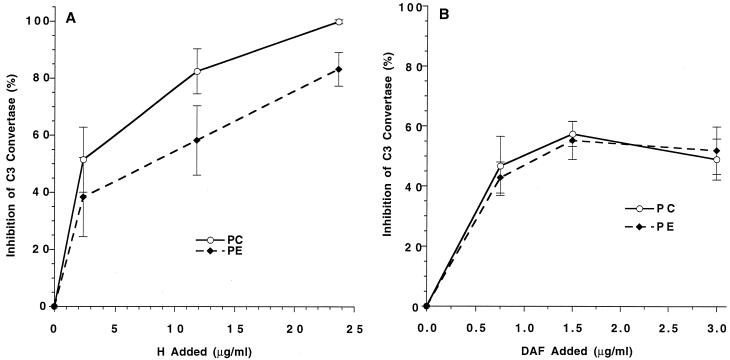
The addition of PE converts nonactivating PC liposomes to vesicles capable of activating the ACP (Fig. 2). Previous studies have shown that this activation is associated with decreased binding of H to C3b on the liposomes (23) but that DAF can regulate this activation in the presence of H in serum (25). Based on these results, it was of interest to compare the effect of PE directly on DAF and H regulation, using the purified ACP convertase. In convertase inhibition assays, whereas DAF behaved with equivalent efficiency in PC and PE liposomes (Fig. 6B), H was less effective on PE liposomes (Fig. 6A).
FIG. 6.
Effect of phospholipid composition on H and DAF regulation of ACP C3 convertase activity. PC and PE liposomes were prepared as for Fig. 1. (A) Liposomes with equal amounts of bound C3b were tested for inhibition of C3 convertase activity by DAF as described for Fig. 3. (B) Liposomes with equal amounts of bound C3b were tested for inhibition of C3 convertase activity by H as described for Fig. 4. Data presented are the means ± SEM of two or more experiments using triplicate samples.
DISCUSSION
Activation of the ACP depends on a positive feedback mechanism in which bound C3b interacts with B and D to form the C3 convertase, C3bBb. The specificity of this system resides in the selective inability of complement regulatory proteins to prevent formation of the C3 convertase on foreign or altered host cell surfaces. H is the primary regulator of the ACP which functions in serum, and the relative affinity of H for bound C3b is a major determining factor in ACP activation on surfaces (6, 7, 15, 33). The affinity of H for C3b is increased when C3b is bound to surfaces with sialic acid or other polyanions due to one or more polyanion binding sites on H (3, 20, 32, 38). The affinity of H for C3b is lower when C3b is bound to ACP activators such as zymosan or LPS (30, 33). DAF and MCP are found on most host cell membranes and are involved in preventing complement activation by host cells physiologically (17, 27). The role of these membrane regulatory proteins in ACP activation by altered host cells is less clear. In this report, the functional activities of DAF and factor H were compared in assays using liposomes modified to activate the ACP by the addition of various structurally defined LPS molecules or phospholipid. The results indicate that there are distinct differences in the effectiveness of the two regulatory proteins in the presence of different types of ACP-activating molecules.
DAF is a 70-kDa cell surface protein which functions to protect host tissue from autologous complement activation by destabilizing both classical and alternative pathway C3 convertases, C4b2a and C3bBb (9, 26). DAF functions only intrinsically and is attached to the cell surface via a C-terminal glycophosphatidylinositol (GPI) anchor (19, 41). A key property of GPI-anchored proteins is their ability when purified to reintegrate into lipid bilayers (18) and, once reintegrated, to function comparably to endogenously expressed proteins (17). While DAF normally acts in the context of other integral membrane proteins, studies using pure lipid vesicles have indicated that DAF functions independently of other membrane proteins to accelerate the decay of the ACP C3 convertase (25).
It has also been shown that ACP activation in serum by liposomes containing LPS is poorly regulated by DAF (25). Others have reported reduced DAF regulation of C3bBb on the human ACP activators, zymosan and rabbit and guinea pig erythrocytes (21, 31). On the other hand, DAF apparently lacks the polyanion-binding ability of H (31), and DAF activity was not increased in membranes containing sialated glycoproteins (25). Inhibitory effects of membrane constituents on DAF activity are suggested by the ability of transformed cells and cells expressing viral glycoproteins to activate the ACP despite the presence of DAF and MCP (5, 24). It is possible that difference in the interaction of C3bBb on ACP activators with DAF as well as H functions as a mechanism to promote ACP activation on altered host cells and facilitate their destruction.
We have used liposomes containing LPS chemotypes of known structure and activating phospholipids to further analyze the functions of DAF and H on C3b bound to ACP activators. The shortest LPS studied from serotype Re595 failed to activate the ACP or to regulate DAF or H activity. The failure of Re595 LPS to affect DAF function indicates that the insertion of lipid A into the membrane does not interfere with the function of GPI-anchored DAF. The presence of the additional disaccharide made up of two l-glycero-d-mannoheptose residues in the R7 mutant was sufficient to result in ACP activation in serum. DAF was a poor regulator of C3bBb on liposomes containing R7 or larger LPS molecules, and there was no difference in DAF regulatory activity for the different-length LPS (Fig. 3).
H activity was also decreased on liposomes containing R7, R5, and wild type LPS but not by 3-deoxy-d-manno-octulosonic acid–lipid A. Results obtained with H differed from those obtained with DAF in that decreased H function correlated with increasing LPS length (Fig. 4). The R7 mutant consistently restricted DAF function but was ineffective in limiting H (Fig. 5). The R5 LPS, in which glucose is attached to two l-glycero-d-mannoheptose residues, and especially the wild-type LPS molecules were more effective than R7 in inhibiting H but equivalent to R7 in their effects on DAF. Thus, H is able to discriminate among LPS structures, consistent with its role in regulating ACP activation by microorganisms. Pangburn described a similar effect of polysaccharide size on inhibition of H binding to C3b in solution (30). Covalent binding of C3b to a polysaccharide of at least tetrameric size was required, and an interaction between the polysaccharide and an H binding site on C3b was postulated. Although we observed high-molecular-weight bands consistent with covalent binding of 125I-C3b to LPS on liposomes after serum incubation, we did not observe these bands after incubation of LPS liposomes with purified components (data not shown). This finding suggests that when both C3b and LPS are bound to the same surface, LPS can affect H activity in the absence of covalent C3b binding to LPS.
Although DAF was a poor regulator of the ACP on liposomes containing LPS, it was able to effectively regulate ACP activation by liposomes containing PE. These liposomes had previously been shown to activate the ACP in serum and to show reduced H binding (23). In the present study, regulation of the assembled ACP convertase, C3bBb, by H was decreased on PE liposomes (Fig. 6). DAF was equally effective in nonactivating PC and activating PE membranes, consistent with its role in preventing complement activation on membrane surfaces. We have previously observed normal DAF function in sickled erythrocytes which activate the ACP due to exposure of PE and phosphatidylserine (42).
The results of our experiments indicate different restriction of H and DAF regulatory function by ACP activators. The mechanism of this restriction is unknown. It is possible that engagement of sites on C3b by molecules on the surface of activators could play a role. The lack of a requirement for C3b covalent binding to LPS and the different effects of PE and R7 on DAF and H make this possibility less likely. In addition, DAF and H show different binding specificities for C3b and C3bBb (9, 28). Alternatively, it is possible that DAF like H have an ability to recognize structures on ACP activators and nonactivators. Further experiments are needed to clarify these issues.
The results presented in this report provide further evidence for the selective inhibition of DAF in membranes containing ACP-activating polysaccharides. However, they point out differences in the recognition of ACP activators by DAF and H consistent with their roles as fluid-phase and membrane regulatory proteins. Selective inhibition of DAF function could be adaptive in that it could help convert virus-infected or transformed human cells expressing new or altered surface molecules to ACP activators, thereby aiding in their elimination.
ACKNOWLEDGMENTS
This work was supported by grants AI23598, DK38181 (to M.E.M.), and AR42538 (to C.M.) from the National Institutes of Health.
REFERENCES
- 1.Ahearn J M, Fearon D T. Structure and function of the complement receptors, CR1 (CD35) and CR2 (CD21) Adv Immunol. 1989;46:183–219. doi: 10.1016/s0065-2776(08)60654-9. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson J P, Farries T. Separation of self from non-self in the complement system. Immunol Today. 1987;8:212–215. doi: 10.1016/0167-5699(87)90167-8. [DOI] [PubMed] [Google Scholar]
- 3.Blackmore T K, Sadlon T A, Ward H M, Lublin D M, Gordon D L. Identification of a heparin binding domain in the seventh short consensus repeat of complement factor H. J Immunol. 1996;157:5422–5427. [PubMed] [Google Scholar]
- 4.Catana E, Schifferli J A. Purification of human complement factor D from the peritoneal fluid of patients on chronic ambulatory peritoneal dialysis. J Immunol Methods. 1991;138:265–271. doi: 10.1016/0022-1759(91)90175-f. [DOI] [PubMed] [Google Scholar]
- 5.Caudwell V, Porter F, Calender A, Pangburn M K, Halbwachs-Mecarelli L. Complement alternative pathway activation and control on membranes of human lymphoid B cell lines. Eur J Immunol. 1990;20:2643–2650. doi: 10.1002/eji.1830201218. [DOI] [PubMed] [Google Scholar]
- 6.Fearon D T. Regulation by membrane sialic acid of β1H-dependent decay-dissociation of amplification C3 convertase of the alternative complement pathway. Proc Natl Acad Sci USA. 1978;75:1971–1975. doi: 10.1073/pnas.75.4.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fearon D T, Austen K F. Activation of the alternative complement pathway due to resistance of zymosan-bound amplification convertase to endogenous regulatory mechanisms. Proc Natl Acad Sci USA. 1977;74:1683–1687. doi: 10.1073/pnas.74.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fishelson Z, Pangburn M K, Müller-Eberhard H J. C3 convertase of the alternative complement pathway. Demonstration of an active, stable C3b,B,(Ni) complex. J Biol Chem. 1983;258:7411–7415. [PubMed] [Google Scholar]
- 9.Fujita T, Inoue T, Ogawa K, Iida K, Tamura N. The mechanism of action of decay-accelerating factor (DAF). DAF inhibits the assembly of C3 convertases by dissociating C2a and Bb. J Exp Med. 1987;166:1221–1228. doi: 10.1084/jem.166.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grossman N, Leive L. Complement activation via the alternative pathway by purified Salmonella lipopolysaccharide is affected by its structure but not its O-antigen length. J Immunol. 1984;132:376–385. [PubMed] [Google Scholar]
- 11.Hammer C H, Wirtz G H, Renfer L, Gresham H D, Tack B F. Large scale isolation of functionally active components of the human complement system. J Biol Chem. 1981;256:3995–4006. [PubMed] [Google Scholar]
- 12.Hong K, Kinoshita T, Dohi Y, Inoue K. Effect of trypsinization on the activity of human factor H. J Immunol. 1982;129:647–652. [PubMed] [Google Scholar]
- 13.Hourcade D, Holers V M, Atkinson J P. The regulators of complement activation (RCA) gene cluster. Adv Immunol. 1989;45:381–416. doi: 10.1016/s0065-2776(08)60697-5. [DOI] [PubMed] [Google Scholar]
- 14.Hunsicker L G, Ruddy S, Austen K F. Alternative complement pathway: factors involved in cobra venom factor (CoVF) activation of the third component of complement (C3) J Immunol. 1973;110:128–138. [PubMed] [Google Scholar]
- 15.Kazatchkine M D, Fearon D T, Austen K F. Human alternative complement pathway: membrane-associated sialic acid regulates the competition between B and β1H for cell-bound C3b. J Immunol. 1979;122:75–81. [PubMed] [Google Scholar]
- 16.Law S K, Lichtenberg N A, Levine R P. Evidence for an ester linkage between the labile binding site of C3b and receptive surfaces. J Immunol. 1979;123:1388–1394. [PubMed] [Google Scholar]
- 17.Medof M E, Kinoshita T, Nussenzweig V. Inhibition of complement activation on the surface of cells after incorporation of decay-accelerating factor (DAF) into their membranes. J Exp Med. 1984;160:1558–1578. doi: 10.1084/jem.160.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medof M E, Nagarajan S, Tykocinski M L. Cell-surface engineering with GPI-anchored proteins. FASEB J. 1996;10:574–586. doi: 10.1096/fasebj.10.5.8621057. [DOI] [PubMed] [Google Scholar]
- 19.Medof M E, Walter E I, Roberts W L, Haas R, Rosenberry T L. Decay-accelerating factor of complement is anchored to cells by a C-terminal glycolipid. Biochemistry. 1986;25:6740–6747. doi: 10.1021/bi00370a003. [DOI] [PubMed] [Google Scholar]
- 20.Meri S, Pangburn M K. Discrimination between activators and nonactivators of the alternative pathway of complement: regulation via a sialic acid/polyanions binding site on factor H. Proc Natl Acad Sci USA. 1990;87:3982–3986. doi: 10.1073/pnas.87.10.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meri S, Pangburn M K. Regulation of alternative pathway complement activation by glycosaminoglycans: specificity of the polyanion binding site on factor H. Biochem Biophys Res Commun. 1994;198:52–59. doi: 10.1006/bbrc.1994.1008. [DOI] [PubMed] [Google Scholar]
- 22.Miyagawa S, Shirakura R, Matsumiya G, Nakata S, Matsuda H, Hatanaka M, Matsumoto M, Kitamura H, Seya T. Test for ability of decay-accelerating factor (DAF,CD55) and CD59 to alleviate complement-mediated damage of xeno-erythrocytes. Scand J Immunol. 1993;38:37–44. doi: 10.1111/j.1365-3083.1993.tb01691.x. [DOI] [PubMed] [Google Scholar]
- 23.Mold C. Effect of membrane phospholipids on activation of the alternative complement pathway. J Immunol. 1989;143:1663–1668. [PubMed] [Google Scholar]
- 24.Mold C, Bradt B M, Nemerow G R, Cooper N R. Activation of the alternative complement pathway by EBV and the viral envelope glycoprotein, gp350. J Immunol. 1988;140:3867–3874. [PubMed] [Google Scholar]
- 25.Mold C, Walter E L, Medof M E. The influence of membrane components on regulation of alternative pathway activation by decay-accelerating factor. J Immunol. 1990;145:3836–3841. [PubMed] [Google Scholar]
- 26.Nicholson-Weller A, Burge J, Fearon D T, Weller P F, Austen K F. Isolation of a human erythrocyte membrane glycoprotein with decay-accelerating activity for C3 convertases of the complement system. J Immunol. 1982;129:184–189. [PubMed] [Google Scholar]
- 27.Oglesby T J, Allen C J, Liszewski M K, White D J G, Atkinson J P. Membrane cofactor protein (CD46) protects cells from complement-mediated attack by an intrinsic mechanism. J Exp Med. 1992;175:1547–1551. doi: 10.1084/jem.175.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pangburn M K. Differences between the binding sites of the complement regulatory proteins DAF, CR1, and factor H on C3 convertases. J Immunol. 1986;136:2216–2221. [PubMed] [Google Scholar]
- 29.Pangburn M K. Analysis of the mechanism of recognition in the complement alternative pathway using C3b-bound low molecular weight polysaccharides. J Immunol. 1989;142:2759–2765. [PubMed] [Google Scholar]
- 30.Pangburn M K. Analysis of recognition in the alternative pathway of complement. Effect of polysaccharide size. J Immunol. 1989;142:2766–2770. [PubMed] [Google Scholar]
- 31.Pangburn M K. Reduced activity of DAF on complement enzymes bound to alternative pathway activators. Similarity with factor H. Immunology. 1990;71:598–600. [PMC free article] [PubMed] [Google Scholar]
- 32.Pangburn M K, Atkinson M A L, Meri S. Localization of the heparin-binding site on complement factor H. J Biol Chem. 1991;266:16847–16853. [PubMed] [Google Scholar]
- 33.Pangburn M K, Morrison D C, Schreiber R D, Müller-Eberhard H J. Activation of the alternative complement pathway: recognition of surface structures on activators by bound C3b. J Immunol. 1980;124:977–982. [PubMed] [Google Scholar]
- 34.Pangburn M K, Müller-Eberhard H J. Relation of a putative thioester bond in C3 to activation of the alternative pathway and the binding of C3b to biological targets of complement. J Exp Med. 1980;152:1102–1114. doi: 10.1084/jem.152.4.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pangburn M K, Schreiber R D, Müller-Eberhard H J. Formation of the initial C3 convertase of the alternative complement pathway. Acquisition of C3b-like activities by spontaneous hydrolysis of the putative thioester in native C3. J Exp Med. 1981;154:856–867. doi: 10.1084/jem.154.3.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ross G D, Lambris J D, Cain J A, Newman S L. Generation of three different fragments of bound C3 with purified factor I or serum. I. Requirements for factor H vs CR1 cofactor activity. J Immunol. 1982;129:2051–2060. [PubMed] [Google Scholar]
- 37.Seya T, Turner J R, Atkinson J P. Purification and characterization of a membrane protein (gp45-70) that is a cofactor for cleavage of C3b and C4b. J Exp Med. 1986;163:837–855. doi: 10.1084/jem.163.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma A K, Pangburn M K. Identification of three physically and functionally distinct binding sites for C3b in human complement factor H by deletion mutagenesis. Proc Natl Acad Sci USA. 1996;93:10996–11001. doi: 10.1073/pnas.93.20.10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith C A, Pangburn M K, Vogel C-W, Müller-Eberhard H J. Molecular architecture of human properdin, a positive regulator of the alternative pathway of complement. J Biol Chem. 1984;259:4582–4588. [PubMed] [Google Scholar]
- 40.Vukajlovich S W, Hoffman J, Morrison D C. Activation of human serum complement by bacterial lipopolysaccharides: structural requirements for antibody independent activation of the classical and alternative pathways. Mol Immunol. 1987;24:319–331. doi: 10.1016/0161-5890(87)90173-8. [DOI] [PubMed] [Google Scholar]
- 41.Walter E I, Roberts W L, Rosenberry T L, Ratnoff W D, Medof M E. Structural basis for variations in the sensitivity of human decay accelerating factor to phosphatidylinositol-specific phospholipase C cleavage. J Immunol. 1990;144:1030–1036. [PubMed] [Google Scholar]
- 42.Wang R H, Phillips G, Jr, Medof M E, Mold C. Activation of the alternative complement pathway by exposure of phosphatidylethanolamine and phosphatidylserine on erythrocytes from sickle cell disease patients. J Clin Invest. 1993;92:1326–1335. doi: 10.1172/JCI116706. [DOI] [PMC free article] [PubMed] [Google Scholar]