Abstract
The oral organism Tannerella forsythia is auxotrophic for peptidoglycan amino sugar N-acetylmuramic acid (MurNAc). It survives in the oral cavity by scavenging MurNAc and MurNAc-linked peptidoglycan fragments (muropeptides) secreted by co-habiting bacteria such as Fusobacterium nucleatum with which it forms synergistic biofilms. Muropeptides, MurNAc-L-Ala-D-isoGln (MDP, muramyl dipeptide) and D-γ-glutamyl-meso-DAP (iE-DAP dipeptide), are strong immunostimulatory molecules that activate nucleotide oligomerization domain (NOD)-like innate immune receptors and induce the expression of inflammatory cytokines and antimicrobial peptides. In this study, we utilized an in vitro T. forsythia – F. nucleatum co-culture model to determine if T. forsythia can selectively scavenge NOD ligands from the environment and impact NOD-mediated inflammation. The results showed that NOD-stimulatory molecules were secreted by F. nucleatum in the spent culture broth which subsequently induced cytokine and antimicrobial peptide expression in oral epithelial cells. In the spent broth from T. forsythia-F. nucleatum co-cultures, the NOD-stimulatory activity was significantly reduced. These data indicated that F. nucleatum releases NOD2-stimulatory muropeptides in the environment, and T. forsythia can effectively scavenge the muropeptides released by co-habiting bacteria to dampen NOD-mediated host responses. This proof-of-principle study demonstrated that peptidoglycan scavenging by T. forsythia can impact the innate immunity of oral epithelium by dampening NOD activation.
Keywords: Periodontitis, Tannerella forsythia, Fusobacterium nucleatum, Peptidoglycan, muramyl peptides, NOD
1. INTRODUCTION
Periodontitis (PD) is a chronic inflammation of the tooth supporting tissues that affects over 700 million people worldwide with a financial burden of over $442 billion per year (Hajishengallis et al., 2023; Kassebaum et al., 2014). It results from the dysregulated inflammatory responses of the host against the polymicrobial and dysbiotic subgingival microbial community (Haffajee et al., 1998; Hajishengallis et al., 2023). Tannerella forsythia, a Gram-negative strict anaerobe frequently found in dysbiotic subgingival communities is one of the major pathobionts implicated in the pathogenesis of periodontitis (Tanner & Izard, 2006). T. forsythia as a biomarker in patient saliva can predict the severity of the periodontitis disease (Ji et al., 2023). T. forsythia is auxotrophic for the peptidoglycan amino sugar N-acetylmuramic acid (MurNAc) due the lack of UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) and UDP-N-acetyl enolpyruvoylglucosamine reductase (MurB) - key enzymes involved in the de novo synthesis of MurNAc from simple sugars (Ruscitto & Sharma, 2018). The organism thus survives in the oral cavity by scavenging peptidoglycan fragments released by the co-habiting oral bacteria during their cell wall turnover or death. We have shown that T. forsythia possesses a dedicated peptidoglycan recycling system which helps the bacterium to degrade and utilize the peptidoglycan fragments released by the co-habiting oral bacteria including Fusobacterium nucleatum (Ruscitto et al., 2017). In this context, T. forsythia forms synergistic biofilms with F. nucleatum and induces increasing inflammatory alveolar bone loss in mice when co-infected with F. nucleatum (Settem et al., 2012). Moreover, the two organisms are frequently found in close physical proximity in the human dental plaque (Zijnge et al., 2010). These studies suggest a mutualistic partnership between the two species.
The peptidoglycan provides structural integrity to bacteria and its turnover during cell division releases peptidoglycan fragments (muropeptides) that can function as ligands for the NOD (nucleotide-binding and oligomerization domain)-like intracellular receptors in innate immune cells such as monocytes, macrophages, neutrophils and epithelial cells (Fritz et al., 2006; Girardin, Travassos, et al., 2003). Specifically, oral epithelial cells that come in direct contact with bacteria can recognize peptidoglycan fragments via NOD receptors to mount an innate immune response including the production of antimicrobial peptides, cytokines, and chemokines (Groeger & Meyle, 2019). The D-γ-glutamyl-meso-DAP (iE-DAP) and MurNAc-L-Ala-D-isoGln (MDP) dipeptides released from the breakdown of peptidoglycan act as ligands for NOD1 and NOD2 receptors, respectively (Caruso et al., 2014; Fritz et al., 2006; Jacobs et al., 1994). iE-DAP- and MDP-containing muropeptides are released from bacterial cells during bacterial cell wall recycling due to the action of bacterial amidases and lytic hydrolases (Heidrich et al., 2001). It has been demonstrated that the amounts of NOD1 and NOD2 stimulatory ligands released in the environment by different bacterial species differ considerably and some species are poor producers of NOD stimulatory molecules (Girardin, Boneca, et al., 2003; Hasegawa et al., 2006). In this regard, the NOD mediated response of epithelium constitutes immune homeostasis and surveillance mechanism, whereas its dysregulation can result in dysbiotic inflammation. The objectives of this study were to determine if F. nucleatum releases NOD stimulatory fragments in the environment and if T. forsythia has the potential to scavenge these fragments from the environment and subsequently dampen the NOD -mediated inflammatory responses in oral epithelial cells.
2. MATERIALS AND METHODS
2.1. Culturing T. forsythia and test organisms
T. forsythia ATCC43037, Porphyromonas gingivalis 381, F. nucleatum ATCC25586 and Streptococcus gordonii Challis were streaked from glycerol stocks (stored frozen in liquid nitrogen at the University at Buffalo Microbial Collection) on brain heart infusion (BHI)-agar (37 g/L; Difco Laboratories ,MI, USA) plates containing 10 g/L yeast extract, 1 g/L L-cysteine, 5 μg/mL hemin, 2 μg/ml menadione (Sigma-Aldrich, MO, USA) , 5%(v/v) horse serum (Thermo Fisher Scientific, MA, USA), and 10 μg/mL MurNAc (Sigma-Aldrich, MO, USA) in the case of T. forsythia (Sharma et al., 1998) (Tomek et al., 2014)at 37°C under anaerobic conditions (90% nitrogen, 5% carbon dioxide, 5% hydrogen) in an anaerobe chamber (Forma Scientific). For broth culturing, single colonies were inoculated from agar plates into liquid broth cultures and grown anaerobically at 37°C. P. gingivalis, F. nucleatum and S. gordonii were grown in brain heart infusion medium containing 5 μg/ml hemin, 0.5 μg/ml menadione, 0.1% L-cysteine and 5% fetal bovine serum (BHI broth). T. forsythia was grown in brain heart infusion BHI broth containing 0.001% N -acetylmuramic acid (BHI-MurNAc broth).
2.2. Growth of T. forsythia in the presence of test organisms.
Test organisms (P. gingivalis, F. nucleatum and S. gordonii) were streaked from frozen stocks on BHI-agar plates and single colonies were inoculated into BHI broth (10 ml) and grown at 37°C under anaerobic atmosphere to an O.D. of 1.0 at 600nm. Bacterial cultures were centrifuged at 8000 x g for 10 min at 4°C and the supernatant was filtered through a 0.2 μm cellulose filter and the filtrate (spent broth) was collected. Each spent broth was then inoculated with the T. forsythia early log-phase cells to a final O.D. of 0.05 and cultures grown anaerobically. The bacterial growth was monitored by measuring O.D. at 600 nm for 7 days.
2.3. Validation of NOD2 expression in oral epithelial cells by Western blot
The human gingival epithelial cell line, OBA-9 (gift from Denis Kinane, University of Pennsylvania). was maintained in keratinocyte serum-free medium supplemented with 10 μg/ml of insulin, 5 μg/ml of transferrin, 10 μM 2-mercaptoethanol, 10 μM 2-aminoethanol, 10 nM sodium selenite, 50 μg/ml of bovine pituitary extract, 100 U/ml of penicillin-streptomycin, and 50 ng/ml amphotericin B (Lonza Inc., MA, USA). Cells were lysed in cell lysis buffer (10 mM Tris, pH 7.4, 100 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1) containing a mixture of protease inhibitors (Roche Diagnostics, IN, USA). Protein lysates were separated on 7.5–12% SDS-PAGE gels and transferred to PVDF membranes (EMD Millipore, MA, USA). The membranes were blocked with 5% fat-free milk powder at room temperature for 1 h and immunoblotted overnight at 4°C with primary antibody NOD2 antibody (2D9; Cat # NBP2-8-883, Novus Biologicals, USA). Next, blots were incubated with HRP-conjugated secondary antibody for 1 h at room temperature. After each step, the membranes were washed 5 times with PBS with Tween for 5 min each time. Finally, the blots were developed using the enhanced chemiluminescence (ECL) system (GE Healthcare Life Sciences). A 98-kDa band of expected size for human NOD2 protein was detected with the monoclonal antibody in cell extracts and HEK-hNOD2 (positive control) cells (data not shown). These data confirmed that the OBA-9 cells selected for stimulation with peptidoglycan expressed the NOD2 receptor.
2.4. Transwell co-culture model and assessment of NOD2 ligand levels in the spent broth
We utilized a transwell co-culture model described previously (Honma et al., 2018) to assess the levels of NOD2 stimulatory molecules in the spent broth of T. forsythia and F. nucleatum. Briefly, F. nucleatum and T. forsythia cells were collected from early log cultures and each species was washed twice with BHI broth by repeated centrifugation and resuspension. F. nucleatum and T. forsythia cells were diluted to OD600 of 0.01 and 0.1, respectively. 0.5 mL of cell suspension of each of F. nucleatum and T. forsythia (ATCC 43037 or its isogenic mutant TfΔampG deficient in muropeptide permease AmpG) was loaded into the top and bottom chambers, respectively, of a transwell culture dish with a 0.45 μm filter (Corning, Thermo Fisher) and the culture dish was incubated anaerobically at 37 °C. After 72 hours different aliquots of spent broth were removed from each condition and tested for NOD2 stimulatory molecule using HEK-hNOD2 cells (Invivogen, CA, USA) stably expressing human NOD2 and an NF-κB and AP-1 inducible secreted embryonic alkaline phosphatase (SEAP) reporter. Briefly, HEK-hNOD2 reporter cells were cultured in 48-well plates at a density of 5 × 104 cells in DMEM supplemented with 10% heat-inactivated FCS, 30 μg/ml blasticidin, 100 μg/ml zeocin, 50 U/ml penicillin, 50 μg/ml streptomycin, and 100 μg/ml normocin as suggested by Invivogen at 37°C under 5% CO2 in a humidified incubator. Cells were stimulated overnight with the aliquots of spent broth (not exceeding 20% of total cell culture volume). Muramyl dipeptide (MDP, 0.5ng) was used as a positive control agonist. After stimulation, culture supernatant was collected (typically 50 μl) and mixed with an equal volume of detection reagent. After incubation for 10 min at room temperature, the SEAP activity was estimated by measuring the optical density of the purple/blue color at 620 nm.
2.5. Cytokines and HBD2 levels by ELISA
OBA-9 cells seeded in 48-well plates at a density of 1 × 105 were exposed to F. nucleatum spent broth in the presence of T. forsythia (ATCC 43037 or TfΔampG), with or without prior blocking with a RIPK2 inhibitor (GSK583; Fisher Scientific Company LLC, USA) at 5μM for 16 h. After stimulation, the cell supernatants were collected and stored at −20 °C until further analysis. ELISAs for HBD2 and cytokines (IL-1β and IL-6) were performed using the commercially available kits (R&D Systems, MN, USA) according to manufacturer’s instructions. Levels of cytokine were calculated and presented as pg/mL.
2.6. Statistical analysis
All studies were performed as two or three independent experiments each with three replicates per data point and data were expressed as mean ± standard deviation (SD). Comparison between two groups was made using a Student’s t test and comparison between multiple groups was made using ANOVA, with statistical significance defined as P < 0.05. Data were analyzed with Minitab 17 Statistical Software (Minitab, Inc., PA, USA).
3. RESULTS
3.1. F. nucleatum secretes NOD2-inducing muropeptides that support T. forsythia growth.
iE-DAP - and MDP - containing muropeptides are released during cell wall turnover by the action of amidases and lytic hydrolases (Heidrich et al., 2001; Jacobs et al., 1994). Previous studies have indicated that the levels of NOD1 (iE-DAP like) and NOD2 (MDP like) stimulatory ligands are differentially released in the environment among bacterial species (Girardin, Boneca, et al., 2003; Hasegawa et al., 2006). As mentioned before, T. forsythia is auxotrophic for MurNAc and relies on exogenous sources to scavenge this amino sugar, and in particular, the spent broth from F. nucleatum culture has been shown to support the growth of T. forsythia (Dzink et al., 1987). We wanted to test if muramyl peptide fraction secreted by F. nucleatum contains MDP-like NOD2 stimulatory molecules. In addition, we were interested in determining if the spent broths of P. gingivalis and S. gordonii cultures could support the growth of T. forsythia and possess NOD2-stimulation activity. For this purpose, each test species was grown in BHI broth anaerobically to an optical density of 1.0 at 600 nm. The spent broth was then filtered through a 0.2 μm filter and inoculated with T. forsythia cells to a final O.D. of 0.05. The culture was grown under anaerobic condition for 7 days with continuous monitoring of growth at 600 nm. The data showed that T. forsythia grew successfully in the F. nucleatum spent broth, as expected, but failed to grow in the spent broth from P. gingivalis or S. gordonii (Fig. 1). It remains to be determined if P. gingivalis and S. gordonii release insufficient or non-recyclable muramyl fragments, or produce toxic metabolites because of which T. forsythia is unable to survive. Next, we determined if F. nucleatum, P. gingivalis or S. gordonii produced muramyl peptides with NOD2 stimulatory activity. For this purpose, aliquots of spent broth from each bacterial culture were incubated with the HEK-hNOD2 reporter cells and SEAP induction was monitored as a measure of NOD2 activation. HEK-hNOD2 cells provided a convenient detection and quantitation of NOD2 ligands in the spent medium. The data showed that as compared to the control sterile broth or the spent broth from the P. gingivalis or S. gordonii cultures, the F. nucleatum spent broth stimulated significantly higher expression of NF-kB inducible SEAP reporter enzyme. The data showed that F. nucleatum secreted significantly higher amount of NOD2 stimulatory molecules as compared to that by P. gingivalis or S. gordonii (Fig. 2). To further establish the identity of F. nucleatum NOD2 stimulatory molecules we stimulated the oral epithelial cell line OBA-9 with F. nucleatum spent broth in the presence or absence of an inhibitor targeting the RIPK-2 adaptor (receptor-interacting serine/threonine-protein kinase-2). NOD receptor signaling involves recruitment of RIPK2 adaptor kinase to form large polymers that subsequently activate the downstream kinases and the AP-1 and NF-κB transcription factors (Park et al., 2007). After stimulation, cytokine levels (IL-1β and IL-6) were assayed by ELISA. The data showed that the F. nucleatum spent broth induced IL-1β and IL-6 secretion and the levels of these cytokines were significantly reduced when cells were stimulated with the spent broth in the presence of a RIPK2 inhibitor (Fig. 3). These data confirmed that F. nucleatum secreted muropeptides signal via the NOD receptor to induce proinflammatory cytokine expression.
Fig.1. F. nucleatum spent broth supports T. forsythia growth.

Planktonic growth of T. forsythia was monitored in spent broth from the cultures of F. nucleatum (Fn.SB), P. gingivalis (Pg.SB) or S. gordonii (Sg.SB). For this purpose, each test species was grown in BHI broth anaerobically to an optical density of 1.0 at 600 nm. The spent broth was then filtered through a 0.2 μm filter and inoculated with T. forsythia cells to a final O.D. of 0.05. The culture was grown under anaerobic conditions for 7 days with continuous monitoring of growth at 600 nm. Data present means S.D. of triplicate samples. A representative result from two independent experiments is shown. *, p < 0.05 (Fn.SB compared with BHI.MurNAc, Pg.SB and Sg.SB at each time point).
Fig. 2. F. nucleatum spent medium activates NOD2.

HEK-hNOD2 reporter cells were stimulated for 16 h with bacterial spent broth at a concentration of 5% or 20% of the total cell culture volume. The level of NOD2 activation was determined by assaying for SEAP enzyme activity (OD520 nm). The results shown are mean ± SD of triplicate cultures and representative of three independent experiments. Abbreviations: MDP, muramyl di-peptide (positive control); BHI, brain heart infusion broth (sterile control); Fn.SB, F. nucleatum spent broth; Pg,SB, P. gingivalis spent broth; Sg.SB, S. gordonii spent broth. The small letters above columns indicate significance. The groups labeled with the same letter are not statistically different from each other while the groups labeled with different letters are significantly different from each other (p < 0.05).
Fig. 3. F. nucleatum secreted peptidoglycan fragments induce NOD-dependent cytokine secretion from oral epithelial cells.
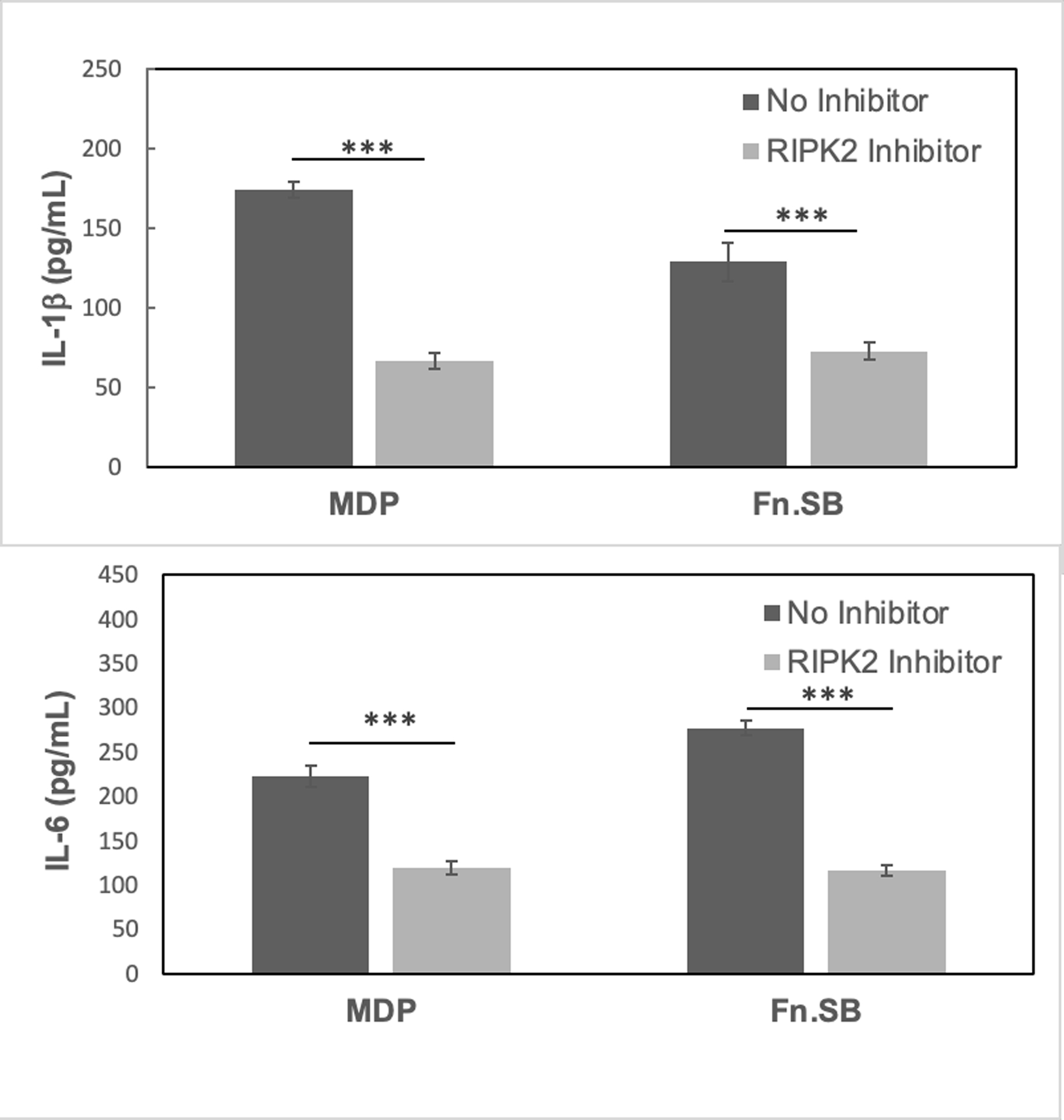
OBA-9 cells were stimulated with F. nucleatum spent broth in the presence or absence of a RIPK2 inhibitor which blocks NOD signaling. The levels of IL-1β and IL-6 in the supernatant were assayed by ELISA. Data are representative of two independent experiments. ***, p < 0.001.
3.2. T. forsythia scavenges F. nucleatum secreted NOD2 stimulatory molecules.
We then interrogated if T. forsythia consumes F. nucleatum NOD2 stimulatory muramyl peptides and dampens the NOD2 mediated response. For this we utilized a transwell co-culture model where the two bacteria were separated by a 0.45 μm filter and the spent broth samples were tested for NOD2 stimulation in HEK-hNOD2 reporter cells. The data showed that when F. nucleatum was co-cultured with T. forsythia, the spent broth from the T. forsythia chambers induced significantly less SEAP enzyme compared to the spent broth from the cultures of F. nucleatum alone (Fig. 4). These data showed that T. forsythia not only grew in the presence of F. nucleatum but specifically scavenged the NOD2 stimulatory molecules rich in MurNAc as a metabolite. The above conclusions were supported by results of the co-culture experiments between F. nucleatum with the TfΔampG mutant deficient in the uptake of muropeptides due to the lack of AmpG permease. In this situation, the mutant grew poorly when co-cultured with F. nucleatum and was unable to reduce the levels of NOD2 molecules from the spent broth.
Fig. 4. T. forsythia scavenges F. nucleatum secreted NOD2 stimulatory molecules.

F. nucleatum and T. forsythia cells were cultured anaerobically in a transwell tissue culture dish with a 0.45 μm filter and after 48 h the spent broth from the T. forsythia well was collected and incubated with HEK-hNOD2 reporter cells (20% spent broth in total cell culture medium) for assaying NOD2 stimulatory activity. As shown the spent broth from the T. forsythia - F. nucleatum co-culture condition (Fn/TfWT) induced significantly less NOD2 activity as compared to the spent broth from the culture of F. nucleatum alone (Fn/-) or the co-culture of F. nucleatum - T. forsythia muropeptide deficient mutant (Fn/TfΔampG). The results shown are mean ± SD of triplicate cultures and representative of three independent experiments. *, p < 0.05; ***, p < 0.001.
3.3. T. forsythia scavenges NOD ligands to dampen antimicrobial peptide and cytokine secretion in epithelial cells.
Here, we determined if muramyl peptide scavenging activity of T. forsythia would dampen the antimicrobial and cytokine response of oral epithelial cells. For this purpose, we treated the OBA-9 cells with the spent broth from the transwell cultures of F. nucleatum alone or co-cultures of F. nucleatum and T. forsythia parental strain or its isogenic mutant TfΔampG. Our data showed that the spent broth from T. forsythia-F. nucleatum co-cultures was significantly less effective in inducing HBD2 (human β-defensin 2) and cytokine expression in OBA-9 cells than the spent broth from F. nucleatum cultures alone (Fig. 5).
Fig. 5. T. forsythia scavenges NOD ligands to dampen cytokine and antimicrobial human β-defensin secretion from oral epithelial cells.

OBA-9 cells were stimulated with spent broth collected as in the Fig. 4 for 16 h. MDP and sterile broth were used as positive and negative controls, respectively. The levels of cytokines and HBD from the culture supernatants were assayed by ELISA. Data are mean ± SD of two independent experiments each performed with three replicates per condition. *, p < 0.05; ***, p < 0.001.
4. DISCUSSION
The data from this study demonstrated that T. forsythia, an oral organism that lacks the capacity to de novo synthesize the amino sugar MurNAc, utilizes the peptidoglycan fragments secreted by F. nucleatum for its survival. Our data showed that F. nucleatum releases muramyl peptides in the environment that have NOD2-stimulatoty activity, triggering a β-defensin and inflammatory cytokine response from oral epithelial cells. Moreover, T. forsythia is able to specifically scavenge NOD2 stimulatory molecules from the environment to dampen the NOD-mediated antimicrobial and cytokine responses of oral epithelial cells.
T. forsythia possesses several lytic hydrolases and muropeptide transporters for the utilization of peptidoglycan fragments released in the environment by the co-habiting oral bacteria including F. nucleatum (Ruscitto et al., 2017) . In this regard, F. nucleatum appears to be a strong partner which provides peptidoglycan fragments (muropeptides) to T. forsythia. This assumption is based on the observations that T. forsythia and F. nucleatum exhibit synergistic partnership in relation to biofilm formation (Sharma et al., 2005) and ability to induce inflammatory alveolar bone loss in mice (Settem et al., 2012). Moreover, the two organisms are found frequently in close proximity in the dental plaque (Zijnge et al., 2010). We also tested if P. gingivalis and S. gordonii could potentially support T. forsythia growth. We selected P. gingivalis since this periodontal pathogen is often associated in disease with T. forsythia. S. gordonii was selected as one of the major oral Gram-positive organisms with abundant cell peptidoglycan found in healthy conditions. To our surprise, the spent culture broth of these organisms could not support T. forsythia growth. We are unclear of the reasons for these observations, but it is likely that these bacteria either produce insufficient or non-utilizable peptidoglycan in the environment not amenable to T. forsythia peptidoglycan recycling system and/or secrete metabolites toxic to the growth of T. forsythia.
Our study explored the impact of NOD2 stimulatory molecule scavenging by T. forsythia on oral epithelial cell responses. We demonstrated the clear ability of T. forsythia to scavenge muramyl di-peptide fragments from the environment and to dampen NOD2 mediated epithelial cell responses. We do not rule out the possibility that T. forsythia might also scavenge iDAP peptides from the environment, and therefore dampens NOD1 response as well. However, given that muramyl fragments are essential for T. forsythia’s survival, we hypothesize that NOD2 signaling would be dominantly impacted in oral epithelial cells with the growth of the bacterium. Moreover, NOD2 has been shown to be important in the inflammatory bone resorption in periodontitis due its role in osteoclast differentiation and macrophage activation in response to Gram-negative bacteria (Souza et al., 2016). However, the role of NOD2 in oral epithelium is underappreciated. Oral epithelium comes in direct contact with colonizing oral bacteria and is perceivably the first barrier against bacteria in the periodontal tissue. NOD receptors are strongly expressed in oral epithelial cells (Sugawara et al., 2006). The role of NOD signaling in gingival epithelium in the context of periodontitis has not been explored in detail. To this end, NOD2 mediated antibacterial peptide and cytokine responses might have a critical role in maintaining immune homeostasis during health. In support of this notion, periodontal inflammation has been reported in NOD2 deficient mice without pathogen challenge but with normal flora (Yuan et al., 2013). These findings can be explained given that NOD2 deficiency can cause breakdown of host immune surveillance, leading to overgrowth of resident bacteria and dysbiotic inflammation. Moreover, it has been shown that NOD2 activation is essential in controlling commensal bacterial growth In the intestinal niche and NOD2 loss-of-function mutations are strong genetic risk factors in the pathogenesis of ileal Crohn’s disease (Petnicki-Ocwieja et al., 2009; Sidiq et al., 2016).
We hypothesize that the mutualistic relationship that exists between T. forsythia and F. nucleatum is dependent in part on the ability of T. forsythia to dampen the NOD2 mediated inflammatory response of oral epithelium. The dampened NOD mediated defenses might allow these organisms to thrive in the dental plaque. It remains to be determined as to what extent T. forsythia scavenging of peptidoglycan fragments released at oral epithelial sites by cohabiting bacteria such as F. nucleatum impacts NOD2 mediated innate immunity. The dampened NOD-mediated inflammatory response of oral epithelium due to the consumption of muropeptides by T. forsythia may help to keep the inflammatory reaction under control and thus contribute to immune homeostasis in the periodontium. Alternatively, the dampening of NOD2-mediated immunity at oral epithelial sites may promote microbial colonization and dysbiosis.
In conclusion, our findings provide proof-of-concept evidence that peptidoglycan scavenging by T. forsythia dampens NOD2-mediated inflammatory responses of oral epithelial cells, which might impact innate immunity, leading to increased susceptibility to periodontitis.
ACKNOWLEDGMENTS
This study was supported by US Public Health Service grant R01DE029497 to A.S. from National Institute of Dental and Craniofacial Research/National Institute of Health.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
REFERENCES
- Caruso R, Warner N, Inohara N, & Nunez G (2014). NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity, 41(6), 898–908. doi: 10.1016/j.immuni.2014.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzink JL, Smith CM, & Socransky SS (1987). Development of a broth medium for Bacteroides forsythus. Journal of Clinical Microbiology, 25(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz JH, Ferrero RL, Philpott DJ, & Girardin SE (2006). Nod-like proteins in immunity, inflammation and disease. Nat Immunol, 7(12), 1250–1257. doi: 10.1038/ni1412 [DOI] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jehanno M, Viala J, … Philpott DJ (2003). Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science, 300(5625), 1584–1587. doi: 10.1126/science.1084677 [DOI] [PubMed] [Google Scholar]
- Girardin SE, Travassos LH, Herve M, Blanot D, Boneca IG, Philpott DJ, … Mengin-Lecreulx D (2003). Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. Journal of Biological Chemistry, 278(43), 41702–41708. doi: 10.1074/jbc.M307198200 [DOI] [PubMed] [Google Scholar]
- Groeger S, & Meyle J (2019). Oral Mucosal Epithelial Cells. Front Immunol, 10, 208. doi: 10.3389/fimmu.2019.00208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffajee AD, Cugini MA, Tanner A, Pollack RP, Smith C, Kent RL Jr., & Socransky SS (1998). Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J Clin Periodontol, 25(5), 346–353. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/9650869 [DOI] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ, & Koo H (2023). Oral polymicrobial communities: Assembly, function, and impact on diseases. Cell Host Microbe, 31(4), 528–538. doi: 10.1016/j.chom.2023.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Yang K, Hashimoto M, Park JH, Kim YG, Fujimoto Y, … Inohara N (2006). Differential release and distribution of Nod1 and Nod2 immunostimulatory molecules among bacterial species and environments. Journal of Biological Chemistry, 281(39), 29054–29063. doi: 10.1074/jbc.M602638200 [DOI] [PubMed] [Google Scholar]
- Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, … Holtje JV (2001). Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Molecular Microbiology, 41(1), 167–178. doi: 10.1046/j.1365-2958.2001.02499.x [DOI] [PubMed] [Google Scholar]
- Honma K, Ruscitto A, & Sharma A (2018). beta-Glucanase Activity of the Oral Bacterium Tannerella forsythia Contributes to the Growth of a Partner Species, Fusobacterium nucleatum, in Cobiofilms. Applied and Environmental Microbiology, 84(1). doi: 10.1128/AEM.01759-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C, Huang LJ, Bartowsky E, Normark S, & Park JT (1994). Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta-lactamase induction. The EMBO journal, 13(19), 4684–4694. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/7925310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S, Kook JK, Park SN, Lim YK, Choi GH, & Jung JS (2023). Characteristics of the Salivary Microbiota in Periodontal Diseases and Potential Roles of Individual Bacterial Species To Predict the Severity of Periodontal Disease. Microbiol Spectr, e0432722. doi: 10.1128/spectrum.04327-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassebaum NJ, Bernabe E, Dahiya M, Bhandari B, Murray CJ, & Marcenes W (2014). Global burden of severe periodontitis in 1990–2010: a systematic review and meta-regression. Journal of Dental Research, 93(11), 1045–1053. doi: 10.1177/0022034514552491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Kim YG, McDonald C, Kanneganti TD, Hasegawa M, Body-Malapel M, … Nunez G (2007). RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. Journal of Immunology, 178(4), 2380–2386. doi: 10.4049/jimmunol.178.4.2380 [DOI] [PubMed] [Google Scholar]
- Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H, & Kobayashi KS (2009). Nod2 is required for the regulation of commensal microbiota in the intestine. Proceedings of the National Academy of Sciences of the United States of America, 106(37), 15813–15818. doi: 10.1073/pnas.0907722106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruscitto A, Honma K, Veeramachineni VM, Nishikawa K, Stafford GP, & Sharma A (2017). Regulation and Molecular Basis of Environmental Muropeptide Uptake and Utilization in Fastidious Oral Anaerobe Tannerella forsythia. Front Microbiol, 8, 648. doi: 10.3389/fmicb.2017.00648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruscitto A, & Sharma A (2018). Peptidoglycan synthesis in Tannerella forsythia: Scavenging is the modus operandi. Molecular oral microbiology, 33(2), 125–132. doi: 10.1111/omi.12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settem RP, El-Hassan AT, Honma K, Stafford GP, & Sharma A (2012). Fusobacterium nucleatum and Tannerella forsythia Induce Synergistic Alveolar Bone Loss in a Mouse Periodontitis Model. Infection and Immunity, 80(7), 2436–2443. doi: 10.1128/iai.06276-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Inagaki S, Sigurdson W, & Kuramitsu HK (2005). Synergy between Tannerella forsythia and Fusobacterium nucleatum in biofilm formation. Oral Microbiology and Immunology, 20(1), 39–42. Retrieved from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15612944 [DOI] [PubMed] [Google Scholar]
- Sharma A, Sojar HT, Glurich I, Honma K, Kuramitsu HK, & Genco RJ (1998). Cloning, expression, and sequencing of a cell surface antigen containing a leucine-rich repeat motif from Bacteroides forsythus ATCC 43037. Infection and Immunity, 66(12), 5703–5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidiq T, Yoshihama S, Downs I, & Kobayashi KS (2016). Nod2: A Critical Regulator of Ileal Microbiota and Crohn’s Disease. Front Immunol, 7, 367. doi: 10.3389/fimmu.2016.00367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza JA, Medeiros MC, Rocha FR, de Aquino SG, Avila-Campos MJ, Spolidorio LC, … Rossa CJ (2016). Role of NOD2 and RIP2 in host-microbe interactions with Gram-negative bacteria: insights from the periodontal disease model. Innate Immun, 22(8), 598–611. doi: 10.1177/1753425916666652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara Y, Uehara A, Fujimoto Y, Kusumoto S, Fukase K, Shibata K, … Takada H (2006). Toll-like receptors, NOD1, and NOD2 in oral epithelial cells. Journal of Dental Research, 85(6), 524–529. doi: 10.1177/154405910608500609 [DOI] [PubMed] [Google Scholar]
- Tanner AC, & Izard J (2006). Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontology 2000, 42, 88–113. Retrieved from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16930308 [DOI] [PubMed] [Google Scholar]
- Yuan H, Zelkha S, Burkatovskaya M, Gupte R, Leeman SE, & Amar S (2013). Pivotal role of NOD2 in inflammatory processes affecting atherosclerosis and periodontal bone loss. Proceedings of the National Academy of Sciences of the United States of America, 110(52), E5059–5068. doi: 10.1073/pnas.1320862110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijnge V, van Leeuwen MB, Degener JE, Abbas F, Thurnheer T, Gmur R, & Harmsen HJ (2010). Oral biofilm architecture on natural teeth. PLoS One, 5(2), e9321. doi: 10.1371/journal.pone.0009321 [DOI] [PMC free article] [PubMed] [Google Scholar]


