Abstract
Some probiotics including lactobacilli, colonize host animal cells by targeting glycosaminoglycans (GAGs), such as heparin, located in the extracellular matrix. Recent studies have shown that several lactic acid bacteria degrade GAGs. Here we show the structure/function relationship of Lacticaseibacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase (KduI) crucial for metabolism of unsaturated glucuronic acid produced through degradation of GAGs. Crystal structures of ligand-free and bound KduIs were determined by X-ray crystallography and the enzyme was found to consist of six identical subunits and adopt a β-helix as a basic scaffold. Ligands structurally similar to the substrate were bound to the cleft of each enzyme subunit. Several residues located in the cleft interacted with ligands through hydrogen bonds and/or C-C contacts. In addition to substrate analogs, a metal ion coordinated to four residues, His198, His200, Glu205, and His248, in the cleft, and the enzyme activity was significantly inhibited by a chelator, ethylenediaminetetraacetic acid. Site-directed mutants in Arg163, Ile165, Thr184, Thr194, His200, Arg203, Tyr207, Met262, and Tyr269 in the cleft exhibited little enzyme activity, indicating that these residues and the metal ion constituted an active site in the cleft. This is the first report on the active site structure of KduI based on the ligand-bound complex.
Keywords: glycosaminoglycan, KduI, Lacticaseibacillus, unsaturated glucuronic acid, X-ray crystallography
Abbreviations
GAG, glycosaminoglycan; ∆GlcUA, unsaturated glucuronic acid; GlcNAc, N-acetylglucosamine; UGL, unsaturated glucuronyl hydrolase; DHU, 4-deoxy-L-threo-5-hexosulose-uronic acid; DK-II, 3-deoxy-D-glycero-2,5-hexodiulosonate; KDG, 2-keto-3-deoxy-D-gluconate; KduI, 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase; KduD, 2-keto-3-deoxygluconate 5-dehydrogenase; ∆GalUA, unsaturated galacturonic acid; LrhKduI, Lacticaseibacillus rhamnosus KduI; LrhKduD, L. rhamnosus KduD; EcoKduI, Escherichia coli KduI; LB, Luria-Bertani; SDS, sodium dodecyl sulfate; PAGE, polyacrylamide gel electrophoresis; CBB, coomassie brilliant blue; DEH, 4-deoxy-L-erythro-5-hexoseulose uronic acid; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; MES, 2-morpholin-4-ylethanesulfonic acid; MOPS, 3-morpholinopropane-1-sulfonic acid; EfaKduI, Enterococcus faecalis KduI; PCR, polymerase chain reaction; WT, wild type.
INTRODUCTION
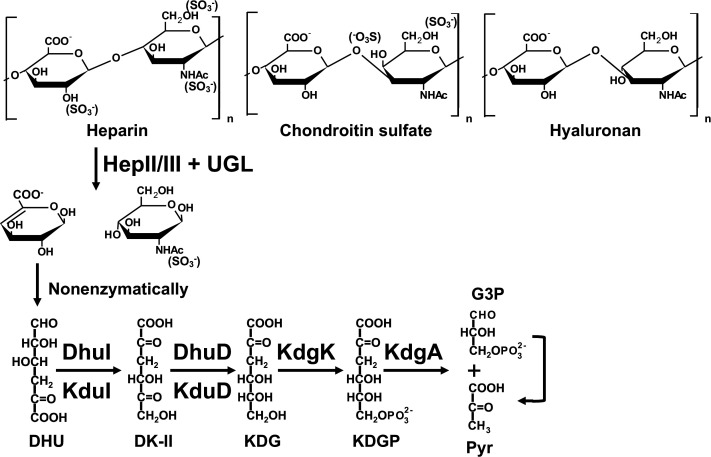
The animal extracellular matrix provides physicochemical support, connects neighboring cells, and plays important roles in cell-cell communication and cell proliferation and differentiation.1) Glycosaminoglycans (GAGs), the major extracellular matrix components, are highly negatively charged polysaccharides. They are composed of typical repeating disaccharide units consisting of an uronic acid residue (glucuronic or iduronic acid) and a frequently N-acetylated amino sugar residue (glucosamine or galactosamine).2) According to their sugar composition, mode of glycoside bond, and sulfate pattern, GAGs are classified as hyaluronan, chondroitin sulfate, dermatan sulfate, keratan sulfate, heparin, and heparan sulfate3),4) (Fig. 1). Except for hyaluronan, most GAGs covalently bind core proteins, forming proteoglycans.5)
Fig. 1. Structure of GAGs and bacterial degradation/metabolism pathway of heparin.
Heparin is depolymerized into an unsaturated disaccharide by heparin lyases II and/or III (HepII/III). The resultant unsaturated disaccharide is degraded into its two constituent monosaccharides [unsaturated glucuronic acid (∆GlcUA) and N-acetylglucosamine (GlcNAc)] by unsaturated glucuronyl hydrolase (UGL). The metabolism of ∆GlcUA is detailed in the text.
Some indigenous and/or pathogenic bacteria target GAGs to adhere to and/or infect host cells.6),7) The Streptococcus species, typical indigenous bacteria (some of them are opportunistic pathogens), produce GAG (heparan sulfate)-binding proteins on their cell surface8) and degrade hyaluronan.9),10) Streptococci depolymerize hyaluronan into an unsaturated disaccharide on the cell surface through a β-elimination reaction catalyzed by hyaluronate lyase (HysA).11) Our previous papers documented how streptococci transport, degrade, and metabolize the resultant unsaturated disaccharide.12),13),14) The phosphotransferase system (PTS) transfers the disaccharide to the cytoplasm where the cytoplasmic unsaturated glucuronyl hydrolase (UGL) further degrades it into its two constituent monosaccharides [unsaturated glucuronic acid (∆GlcUA) and N-acetylglucosamine (GlcNAc)]. The resultant ∆GlcUA is nonenzymatically converted to 4-deoxy-L-threo-5-hexosulose-uronic acid (DHU), which is metabolized to glyceraldehyde-3-phosphate (G3P) and pyruvate (pyr) via 3-deoxy-D-glycero-2,5-hexodiulosonate (DK-II), 2-keto-3-deoxy-D-gluconate (KDG), and 2-keto-3-deoxy-6-phosphogluconate (KDGP) by successive reactions catalyzed by an isomerase (DhuI), an NADH-dependent reductase (DhuD), a kinase (KdgK), and an aldolase (KdgA) (Fig. 1). These proteins responsible for depolymerization, import, degradation, and metabolism of hyaluronan are encoded in a GAG genetic cluster (Fig. S1A; see the J. Appl. Glycosci. Web site).
Several probiotics (including Lacticaseibacillus recently reclassified but known as lactobacilli,15) Lactobacillus, and Enterococcus species) with a similar GAG genetic cluster (Fig. S1B; see the J. Appl. Glycosci. Web site) degrade heparin, and a few of them may bind to human intestinal cells via heparin.16) However, the molecular mechanism of heparin metabolism remains unclear in probiotics. 4-Deoxy-L-threo-5-hexosulose-uronate ketol-isomerase (KduI) and 2-keto-3-deoxygluconate 5-dehydrogenase (KduD) were first identified as a hexuronate-metabolizing isomerase and reductase for pectin metabolism, respectively.17) Unsaturated galacturonic acid (∆GalUA), produced from pectin reacting successively with polysaccharide lyase18) and unsaturated galacturonyl hydrolase,19) is nonenzymatically converted to DHU, because the lack of a hydroxy group at the C4 position of ∆GlcUA and ∆GalUA makes them identical. Thus, KduI converts DHU into DK-II (Fig. 1), and KduD converts DK-II into KDG. KduI and KduD, therefore, correspond to DhuI and DhuD, respectively. In fact, Lacticaseibacillus rhamnosus KduI (LrhKduI) and KduD (LrhKduD) metabolize ∆GlcUA derived from chondroitin sulfate.16) Although the enzymatic properties of bacterial KduI are well documented20),21) and a crystal structure of Escherichia coli KduI (EcoKduI) is available,22) little knowledge on the structure/function relationship of KduI has emerged. This article deals with the active site structure of LrhKduI through structure determination of ligand-free/bound LrhKduIs and site-directed mutagenesis.
MATERIALS AND METHODS
Materials.
Rhamnogalacturonan-I (RG-I) derived from potato pectin was purchased from Megazyme International Ireland (Bray, Ileland). Oligonucleotides were synthesized by Hokkaido System Science Co., Ltd. (Sapporo, Japan). DNA-modifying enzymes were purchased from TOYOBO Co., Ltd. (Osaka, Japan). All other analytical grade chemicals used in this study were commercially available.
Bacteria and cell culture.
As the host for plasmid amplification, E. coli strain DH5α (TOYOBO) cells were routinely cultured at 37 ˚C in Luria-Bertani (LB) medium [1 % (w/v) tryptone, 0.5 % (w/v) yeast extract, and 1 % (w/v) NaCl] containing sodium ampicillin (0.1 mg/mL). E. coli strain BL21(DE3) (Novagen/Merck KGaA, Darmstadt, Germany) was used as the host for the expression of LrhKduI. For expression in E. coli, cells were aerobically precultured at 30˚C in LB medium supplemented with sodium ampicillin (0.1 mg/mL). When the turbidity reached about 0.5 at 600 nm, isopropyl-β-D-thiogalactopyranoside was added to the culture at a final concentration of 0.1 mM, and the cells were further cultured at 16 ˚C for 48 h.
Expression and purification of LrhKduI.
E. coli cells harboring the plasmid containing the LrhKduI gene were harvested by centrifugation at 11,200 × g and 4 ˚C for 10 min, resuspended in 20 mM Tris-HCl (pH 7.5), and ultrasonically disrupted at 0 ˚C and 9 kHz for 20 min (Insonator Model 201M, Kubota Corporation, Tokyo, Japan). After centrifugation at 17,400 × g and 4 ˚C for 20 min, the supernatant was used as the cell extract.
LrhKduI was further purified from the cell extract by affinity chromatography (TALON Metal Affinity Resin: Clontech Laboratories, Inc., Mountain View, CA, USA) and gel filtration chromatography (Superdex 200 prep grade: GE Healthcare Japan Corporation, Tokyo, Japan) as described previously.16) Protein purity was assessed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE),23) followed by staining with Coomassie Brilliant Blue (CBB). The purified LrhKduI was dialyzed at 4 ˚C for 3 h against 20 mM Tris-HCl (pH 7.5) and the dialysate was subjected to the enzyme assay or crystallization.
Enzyme assay.
The isomerase assay of LrhKduI was carried out as described previously.16) Briefly, 0.5 mM unsaturated chondroitin disaccharide with a sulfate group at C6 site of N-acetylgalactosamine residue was incubated in 50 mM Tris-HCl (pH 7.5) with 0.03 mg/mL Streptococcus agalactiae UGL,12) 0.2 mg/mL LrhKduI, and 0.2 mg/mL LrhKduD in the presence of 0.2 mM NADH. In the case of active isomerase, NADH was oxidized by KduD according to reduction of the reaction product derived from the KduI reaction.
Preparation of DHU.
To obtain enzyme/substrate complex, a substrate of KduI, DHU, was prepared from readily available RG-I using RG-I lyase YesX24) and unsaturated galacturonyl hydrolase YesR19) because DHU is generated from GAG and RG-I. The resultant DHU was a mixture with rhamnose, and the DHU concentration was determined by thiobarbituric acid method.20)
Crystallization and X-ray diffraction.
LrhKduI was crystallized at 20 °C by the sitting-drop vapor-diffusion method using crystallization kits purchased from Hampton Research Corp. (Aliso Viejo, CA, USA), Jena Bioscience GmbH (Jena, Germany), Emerald Biostructures Inc. (Bainbridge Island, WA, USA), Qiagen GmbH (Hilden, Germany), and Molecular Dimensions, Inc. (Holland, OH, USA). LrhKduI (1 μL) was mixed with the reservoir solution (1 μL) to form a drop and incubated at 20 ˚C. The enzyme was crystallized in a drop containing 10 % polyethylene glycol 8,000 and 0.2 M magnesium acetate [crystallization condition (i)]. LrhKduI in the presence of 1 mM DHU was also crystallized under conditions of (ii) 10 % polyethylene glycol 6,000 and 0.1 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (pH 6.5), (iii) 12 % polyethylene glycol 4,000 and 0.1 M 2-morpholin-4-ylethanesulfonic acid (MES) (pH 6.5), and (iv) 12 % polyethylene glycol 4,000 and 0.1 M 3-morpholinopropane-1-sulfonic acid (MOPS) (pH 6.5). For cryoprotection, a protein crystal was soaked in the drop solution containing 20 % glycerol. A crystal was picked up from the soaking solution with a mounted nylon loop (Hampton Research) and placed directly into a cold nitrogen-gas stream at −173 °C. X-ray diffraction images were collected with a MAR225HE detector (Rayonix, L.L.C., Evanston, IL, USA) with synchrotron radiation at wavelength 1.00 Å on the beamline BL26B1, BL38B1, or BL44XU of SPring-8 (Hyogo, Japan). The data were indexed, integrated, and scaled using the HKL-2000 program.25) The structure was determined through molecular replacement method with the Molrep program26) in the CCP4 program package using structure of Enterococcus faecalis KduI (EfaKduI) (PDB ID, 1YWK) as a search model. Structure refinement was conducted using the phenix refine program in the PHENIX program package.27) After each refinement cycle, the model was adjusted manually with the winCoot program.28) The atomic coordinates and structure factors [PDB ID, 7VGK for LrhKduI under the crystallization condition (i), 7E4S for LrhKduI under the crystallization condition (HEPES) (ii), 7YE3 for LrhKduI under the crystallization condition (MES) (iii), and 7YRS for LrhKduI under the crystallization condition (MOPS) (iv)] were deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Alignment of amino acid sequences of KduI.
The amino acid sequences of KduI orthologs of other species were obtained from Uniprot KB (https://www.uniprot.org/help/uniprotkb). The obtained sequences were subjected to multiple alignment using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/).
Site-directed mutagenesis.
To substitute Arg163, Ile165, Thr184, Thr194, His200, Arg203, Tyr207, Met262, and Tyr269 of LrhKduI with Ala or Phe, oligonucleotides shown in Table S1 (see J. Appl. Glycosci. Web site) were used. Site-directed mutagenesis was performed using plasmid pET21b-KduI16) as a template and synthetic oligonucleotides as sense and antisense primers using the methods described in a QuickChange site-directed mutagenesis kit manual (Stratagene Co., San Diego, CA, USA), except that KOD-Plus-Neo was used as DNA polymerase for polymerase chain reaction (PCR). The above-described DNA manipulations such as subcloning, transformation, and gel electrophoresis were performed as previously described.29) The resultant plasmids with mutations were used for mutant expression. Mutations were confirmed by DNA sequencing.30) E. coli host strain BL21(DE3) cells were transformed using each plasmid. Similarly, the mutant proteins were purified as described above.
RESULTS AND DISCUSSION
Structure determination of Lacticaseibacillus KduI.
Although crystal structure of EcoKduI has first been determined as a hexuronate-metabolizing enzyme,22) no structure of enzyme/substrate complex has been reported and structure/function relationship of KduI remains to be clarified. We have recently identified LrhKduI as 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase responsible for metabolism of GAGs.16) Thus, LrhKduI was crystallized to determine its structure by X-ray crystallography.
The enzyme was crystallized in the absence of the substrate DHU under the condition (i) as described above. The crystal of LrhKduI was diffracted to 3.1 Å resolution (Table 1). Crystal structure of ligand-free LrhKduI was determined by molecular replacement using coordinates of EfaKduI (PDB ID, 1YWK) as a search model, and statistics of diffraction and refinement data are shown in Table 1. No report on crystal structure of EfaKduI has been published. The final model of LrhKduI was refined to Rwork of 19.0 % and Rfree of 23.9 % up to a resolution of 3.10 Å (Fig. 2A, Table 1). Ramachandran plot analysis revealed that 95.0 % of residues were in the favored regions and 5.0 % were in the additional allowed regions. Due to thin electron density map, residues (Thr194-Arg203) are missing in the crystal structure. Six enzyme molecules are included in the asymmetric unit of the crystal (Fig. 2B). In combination with the crystal packing, the enzyme (32 kDa as a monomer) was eluted as around 150 kDa protein by gel filtration column chromatography, suggesting that LrhKduI consisted of homohexamer. Because, similar to EcoKduI and EfaKduI, LrhKduI has a β-helix as a basic scaffold, the enzyme is considered to be a member of all β proteins class/double-stranded β-helix fold/RmlC-cupin superfamily/KduI-like family in the SCOP (Structural Classification of Proteins) database (Figs. 2A, B).
Table 1. Data collection and refinement statistics.
| LrhKduI | LrhKduI | LrhKduI | LrhKduI | |
| _HEPES | _MES | _MOPS | ||
| PDB | 7VGK | 7E4S | 7YE3 | 7YRS |
| Data collection | ||||
| Wavelength (Å) | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Space group | P212121 | C2 | C2 | C2 |
| Unit-cell parameters a, b, c (Å) β (°) | a = 94.2 | a = 94.3 | a = 93.0 | a = 90.4 |
| b = 131 | b = 189.3 | b = 189.1 | b = 187.4 | |
| c = 157 | c = 117.8 | c = 114.9 | c = 108.9 | |
| β = 106.2 | β = 107.1 | β = 112.5 | ||
| Resolution range (Å) | 50-3.10 (3.15-3.10)a |
50-2.80 (3.15-3.10)a |
50-2.55 (2.97-2.80)a |
50-2.80 (2.97-2.80)a |
| Total observed reflections | 202706 | 148144 | 403935 | 158079 |
| Unique reflections | 35643 | 89592 | 119595 | 78223 |
| Redundancy | 5.7 (5.7)a | 1.65 (1.66)a | 3.38 (3.51)a | 2.02 (1.99)a |
| Completeness (%) | 99.4 (99.4)a | 92.3 (93.6)a | 98.3 (97.9)a | 96.2 (96.1)a |
| I/σ(I) | 20.7 (3.10)a | 6.59 (0.80)a | 17.34 (2.22)a | 6.36 (1.63)a |
| Rmerge (%) | 8.6 (52.9)a | 13.8 (133.1)a | 5.00 (63.0)a | 10.2 (39.1)a |
| Refinement | ||||
| Resolution range (Å) | 45.12-3.10 (3.20-3.10)a |
47.31-2.80 (3.02-2.80)a |
47.47-2.55 (2.59-2.55)a |
49.55-2.80 (3.02-2.80)a |
| R work (%) | 18.96 (27.66)a | 16.46 (38.60)a | 20.55 (36.66)a | 18.51 (31.78)a |
| R free (%) | 23.85 (36.57)a | 23.82 (44.63)a | 27.04 (47.02)a | 26.44 (43.18)a |
| Residues/HOH/GOL/Ligand/Zn | 1539/0/0/0/0 | 1646/31/0/6/5 | 1644/9/0/4/6 | 1646/0/0/4/6 |
| Root-mean-square deviation | ||||
| Bond lengths (Å) | 0.009 | 0.008 | 0.013 | 0.011 |
| Bond angles (°) | 1.2 | 1.1 | 1.6 | 1.7 |
a The highest resolution shell is shown in parentheses.
Fig. 2. Crystal structure of LrhKduI.
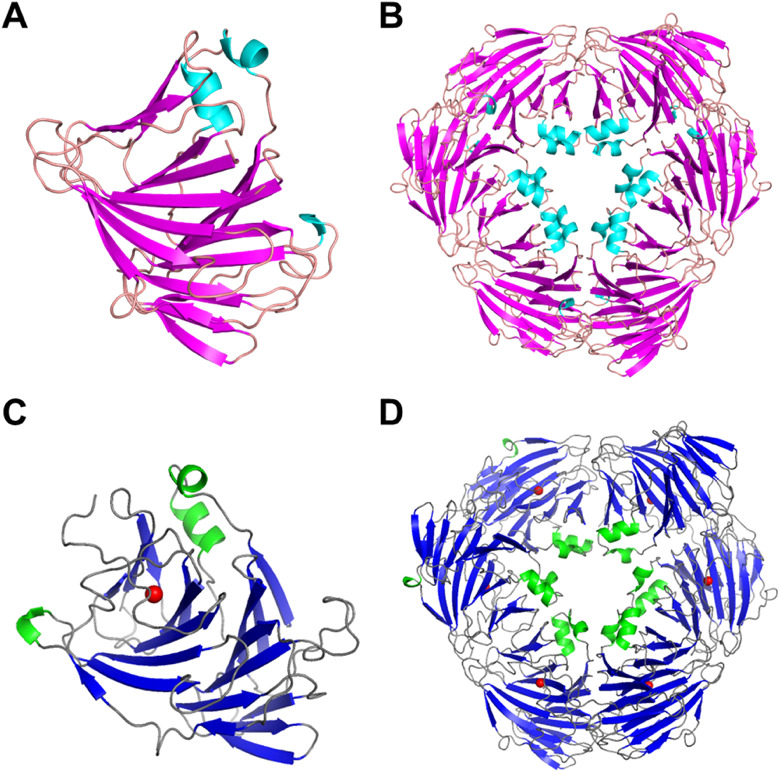
(A) Crystal structure of the ligand-free LrhKduI monomer. (B) Crystal structure of the ligand-free active LrhKduI hexamer. (C) Crystal structure of metal ion (red ball)-bound LrhKduI monomer. (D) Crystal structure of metal ion (red ball)-bound LrhKduI hexamer.
Structures of LrhKduI in complex with substrate analogs.
To clarify the structure/function relationship of KduI, we tried to determine the three-dimensional structure of LrhKduI in complex with DHU. LrhKduI was crystallized in the presence of DHU under various conditions. As a result, high-resolution diffraction data were collected from three crystals obtained under (ii), (iii), and (iv) conditions as described above. Crystal structures of LrhKduI under (ii), (iii), and (iv) conditions were refined at 2.80, 2.55, and 2.80 Å, respectively, after molecular replacement by using the ligand-free enzyme coordinates as a search model. Statistics of diffraction and refinement data are also shown in Table 1. Thr194-Arg203 residues, missing in the ligand-free LrhKduI, showed the electron density maps in these structures.
On the basis of molecular surface model, a large cleft was observed in the center of each enzyme monomeric molecule (Figs. 3A-D). The volume of the cleft in the ligand-free enzyme was estimated to be 4,140 Å3 by a program of ProFunc.31) Structural refinement of three crystals obtained in the presence of DHU indicated that another electron densities of the small molecule distinct from the enzyme molecule were observed inside the cleft in the three structures. The electron density map (Fo-Fc) at 3.0 σ in each structure is shown in Figs. 3A-C. The small molecule was found to be a component of crystallization buffer such as HEPES, MES, and MOPS based on the size and the shape of each map. The temperature factors of these buffer components showed high values (HEPES, 120.5 Å2; MES, 120.8 Å2; and MOPS, 96.8 Å2), indicating the low affinity of the buffer components with LrhKduI. The reason why no DHU was bound to the enzyme was considered to be a reactive feature of DHU. The molecule DEH (4-deoxy-L-erythro-5-hexoseulose uronic acid) derived from unsaturated guluronic acid obtained through alginate degradation is highly reactive and toxic to bacterial and yeast cells due to the presence of aldehyde group.32) Because both of DHU and DEH are classified to be α-keto acids containing aldehyde group, DHU is suggested to be highly reactive and unstable.
Fig. 3. Crystal structures of LrhKduI in complex with substrate analogs.
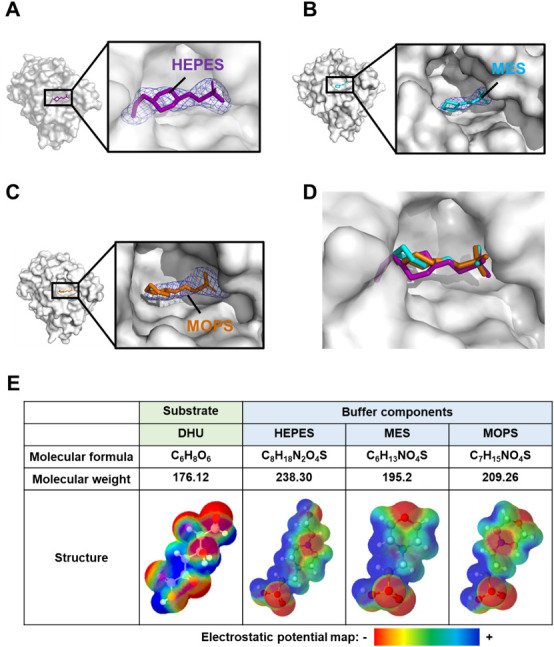
(A) Molecular surface model of HEPES-bound LrhKduI. (B) Molecular surface model of MES-bound LrhKduI. (C) Molecular surface model of MOPS-bound LrhKduI. Electron density map at 3.0 σ (Fo-Fc). (D) Superimposition of HEPES (purple), MES (cyan), and MOPS (orange) bound to LrhKduI. (E) Comparison of DHU, HEPES, MES, and MOPS.
Superimposition of the three buffer components revealed that all molecules were located at similar positions inside the cleft of LrhKduI (Fig. 3D). We investigated the similarities between buffer components and the substrate DHU. Figure 3E shows electrostatic potential maps created by a program of Jmol. Although the molecular weight of DHU is slightly smaller than that of buffer components, the linear structure due to opening of the pyranose ring and the negative charge at both ends and the center of the molecule are similar to those of buffer components. Therefore, these buffer components seem to behave as a substrate analog of LrhKduI, suggesting that the substrate analog-binding cleft acts as a catalytic site.
Metal ion in the cleft.
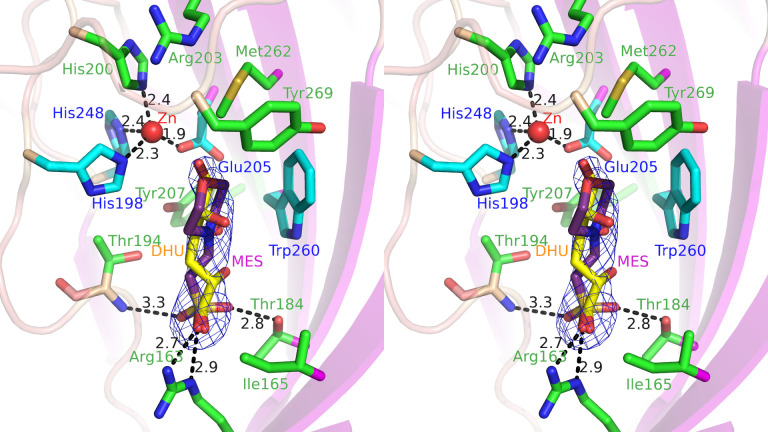
As described above, KduI belongs to the cupin superfamily. Most proteins/enzymes classified to the superfamily include metal ions such as iron, copper, zinc, cobalt, nickel, or manganese ion in the active site and these metal ions function as a cofactor.33) In fact, EcoKduI contains zinc ion and four residues (His196, His198, Glu203, and His245) are coordinated to the zinc ion.34) In the case of LrhKduI, in addition to substrate analogs, an electron density map corresponding to a metal ion was also observed at the vicinity of the substrate analogs in the cleft of the enzyme (Figs. 2C, D and 4A). Four residues, His198, His200, Glu205, and His248 are coordinated to the metal ion (Fig. 4B). These four residues of LrhKduI are well superimposed onto those of zinc ion-bound EcoKduI (Fig. 4C), suggesting that the metal ion bound to LrhKduI is a zinc ion. The enzyme activity of LrhKduI was significantly inhibited by ethylenediaminetetraacetic acid (EDTA) at 5 mM, indicating that the metal ion was involved in the catalytic reaction or construction of the active site structure.
Fig. 4. Active site in LrhKduI.
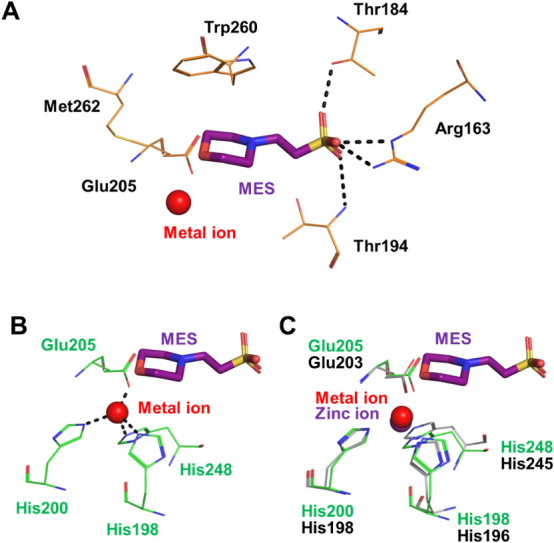
(A) LrhKduI residues interacting with MES. (B) Four residues coordinated to metal ion. (C) Superimposition of metal ion-bound LrhKduI (green) with zinc ion-bound EcoKduI (gray).
Catalytic residues in the cleft.
To identify catalytic residues in the cleft, distances between amino acid residues of LrhKduI (a certain subunit of hexamer) and buffer components were calculated (Table 2). From this result, some amino acid residues, such as Arg163, Thr184, Thr194, His198, Arg203, Glu205, Phe258, Trp260, Met262, and Tyr269, were estimated to interact with the substrate. In other subunits, Ile165 and Tyr207 were also found to interact with the substrate analog. These amino acid residues are highly conserved in some bacterial KduIs (Fig. S2: see J. Appl. Glycosci. Web site). The metal ion-binding residue His200 is also well conserved. In order to investigate the role of such residues in the KduI reaction, seven mutants, R163A, I165A, T184A, T194A, H200A, R203A, and M262A with Arg163, Ile165, Thr184, Thr194, His200, Arg203, and Met262 residues substituted with Ala and two mutants, Y207F and Y269F, with Tyr207 and Tyr269 residues substituted with Phe were constructed and purified to homogeneity (Fig. 5A). The mutants were subsequently subjected to the enzyme assay. Because the NADH-dependent enzyme KduD reduced the reaction product by KduI, the absorbance at 340 nm in the reaction mixture composed of DHU prepared from chondroitin sulfate disaccharide by S. agalactiae UGL, LrhKduI [wild-type enzyme (WT), R163A, I165A, T184A, T194A, H200A, R203A, Y207F, M262A, and Y269F], LrhKduD, and NADH was measured (Fig. 5B). In the reaction mixture using LrhKduI WT, its absorbance at 340 nm was drastically decreased, indicating that NADH was oxidized to NAD+ and subsequent reactions by KduI and KduD occurred. On the other hand, the reaction mixture containing each of LrhKduIs R163A, I165A, T184A, T194A, H200A, R203A, Y207F, M262A, and Y269F showed little decrease of the absorbance at 340 nm, demonstrating that all mutants of LrhKduIs exhibited little enzyme activity (Fig. 5B). This result supported that the cleft constituted by residues such as Arg163, Ile165, Thr184, Thr194, His200, Arg203, Tyr207, Met262, and Tyr269 contained an active site (Fig. 6). Arg163 and Thr184 were considered to be important for binding to substrates based on formation of hydrogen bonds and electrostatic interactions with buffer components. Ile165, Thr194, Arg203, Tyr207, Met262, and Tyr269 were suggested to play a role in fixing the position of substrates through van der Waals interactions. This site-directed mutagenesis indicated the active site located in the cleft of LrhKduI postulated through structural analysis of the enzyme/substrate analog complexes.
Table 2. Interactions between LrhKduI and buffer components.
| HEPES | MES | MOPS | ||||||
| Atoms | Amino Acid | Distance (Å) | Atoms | Amino Acid | Distance (Å) | Atoms | Amino Acid | Distance (Å) |
| Hydorogen bond (<3.4 Å) | Hydorogen bond (<3.4 Å) | Hydorogen bond (<3.4 Å) | ||||||
| O1S | 194T N | 3.2 | O1S | 163R NE | 2.9 | O2 | 184T OG1 | 3.1 |
| O2S | 184T OG1 | 3.1 | O1S | 163R NH2 | 2.7 | O3 | 163R NE | 3.3 |
| O8 | 203R NH2 | 3.3 | O2S | 184T OG1 | 2.8 | |||
| O3S | 194T N | 3.3 | ||||||
| C-C contact (<4.4 Å) | C-C contact (<4.4 Å) | C-C contact (<4.4 Å) | ||||||
| C2 | 260W CE2 | 4.3 | C2 | 205E CD | 4.2 | C1 | 258F CE2 | 4.2 |
| C3 | 269Y CD2 | 4.1 | C2 | 260W CD2 | 3.9 | C1 | 258F CZ | 3.7 |
| C3 | 269Y CE2 | 3.5 | C2 | 260W CE2 | 4.1 | C3 | 260W CD1 | 4.0 |
| C5 | 205E CD | 3.5 | C2 | 260W CE3 | 3.9 | C3 | 260W CE2 | 4.0 |
| C5 | 260W CD2 | 3.6 | C2 | 260W CH2 | 4.2 | C5 | 198H CE1 | 4.3 |
| C5 | 260W CE2 | 4.0 | C2 | 260W CZ2 | 4.2 | C6 | 260W CD2 | 3.9 |
| C5 | 260W CE3 | 3.8 | C2 | 260W CZ3 | 4.0 | C6 | 260W CE2 | 4.2 |
| C5 | 260W CG | 4.0 | C2 | 262M CE | 3.5 | C6 | 260W CE3 | 3.6 |
| C5 | 260W CZ3 | 4.2 | C3 | 205E CD | 4.0 | C6 | 260W CH2 | 3.8 |
| C5 | 262M CE | 3.7 | C3 | 260W CD1 | 4.2 | C6 | 260W CZ2 | 4.2 |
| C6 | 205E CD | 4.1 | C3 | 260W CD2 | 3.9 | C6 | 260W CZ3 | 3.5 |
| C6 | 260W CB | 4.3 | C3 | 260W CE2 | 4.1 | C6 | 262M CE | 3.8 |
| C6 | 260W CD1 | 3.9 | C3 | 260W CE3 | 4.3 | C6 | 269T CE2 | 4.1 |
| C6 | 260W CD2 | 3.9 | C3 | 260W CG | 4.0 | C7 | 260W CD1 | 4.2 |
| C6 | 260W CE2 | 4.1 | C7 | 260W CD2 | 3.4 | |||
| C6 | 260W CG | 3.8 | C7 | 260W CE2 | 3.6 | |||
| C7 | 205E CD | 4.0 | C7 | 260W CE3 | 3.5 | |||
| C7 | 262M CE | 3.8 | C7 | 260W CG | 3.8 | |||
| C8 | 262M CE | 3.5 | C7 | 260W CH2 | 4.1 | |||
| C8 | 269Y CD2 | 3.7 | C7 | 260W CZ2 | 4.0 | |||
| C8 | 269Y CE2 | 3.9 | C7 | 260W CZ3 | 3.9 | |||
Fig. 5. Enzyme activity of site-directed mutants of LrhKduI.
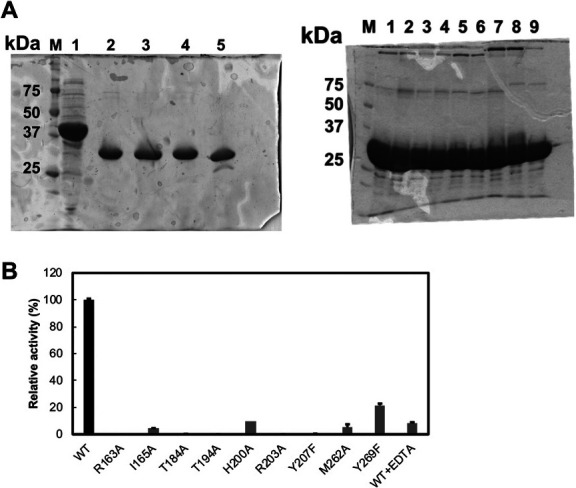
(A) SDS-PAGE profile of the enzymes used in the assay. (left) M, marker; 1, S. agalactiae UGL; 2, LrhKduI WT; 3, LrhKduI R163A; 4, LrhKduI H200A; and 5, LrhKduD. (right) M, marker; 1, LrhKduI WT; 2, LrhKduI R163A; 3, LrhKduI I165A; 4, LrhKduI T184A; 5, LrhKduI T194A; 6, LrhKduI R203A; 7, LrhKduI Y207F; 8, LrhKduI M262A; and 9, LrhKduI Y269F. (B) Enzyme activity of LrhKduI mutants. WT activity was taken as 100 %. WT + EDTA indicates the LrhKduI WT enzyme in the presence of EDTA. Each data represents the average of triplicate individual experiments, except for H200A.
Fig. 6. The estimated DHU-binding site and the mutated residues (stereo-view).
The carbons in the mutated residues are drawn in green. Those of the ligated residues of probable Zn (red) and Trp260 are drawn in cyan. Those of MES and DHU are drawn in purple and yellow, respectively. The DHU is fitted to the density of MES by Coot assuming the carboxyl group occupies the phosphate group of MES. The MES omit map (Fo-Fc map) is drawn with 2.5 σ. The root-mean-square deviation between MES and DHU was calculated to be 0.71 Å for common 9 atoms.
In conclusion, the active site and catalytic residues were first identified in GAG-metabolizing enzyme KduI through structural and functional studies.
CONFLICTS OF INTEREST
The authors declare no conflict of interests.
ACKNOWLEDGMENTS
We thank Drs. S. Baba, N. Mizuno, G. Ueno, and H. Okumura of the Japan Synchrotron Radiation Research Institute (JASRI) for their kind help in data collection. Diffraction data were collected at beamlines BL26B1, BL38B1, and BL44XU of SPring-8 (Hyogo, Japan) with the approval of JASRI (projects 2017B2592, 2019B2557, and 2020A2577). This work was supported in part by JSPS KAKENHI Grant Numbers 15H04629, 18H02166, and 21H02156 (W.H.) and Research Grants (W.H.) from Yakult Bio-science Foundation and Japan Foundation for Applied Enzymology. The authors would like to thank Enago (www.enago.com) for the English language review.
REFERENCES
- 1).Frantz C., Stewart K.M., and Weaver V.M.: The extracellular matrix at a glance. J. Cell Sci., 123, 4195-4200 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Gandhi N.S. and Mancera R.L.: The structure of glycosaminoglycans and their interactions with proteins. Chem. Biol. Drug Des., 72, 455-482 (2008). [DOI] [PubMed] [Google Scholar]
- 3).Sugahara K., Mikami T., Uyama T., Mizuguchi S., Nomura K., and Kitagawa H.: Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate. Curr. Opin. Struct. Biol., 13, 612-620 (2003). [DOI] [PubMed] [Google Scholar]
- 4).Smith P.D., Coulson-Thomas V.J., Foscarin S., Kwok J.C., and Fawcett J.W.: “GAG-ing with the neuron”: The role of glycosaminoglycan patterning in the central nervous system. Exp. Neurol., 274, 100-114 (2015). [DOI] [PubMed] [Google Scholar]
- 5).Souza-Fernandes A.B., Pelosi P., and Rocco P.R.: Bench-to-bedside review: the role of glycosaminoglycans in respiratory disease. Crit. Care, 10, 237 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).García B., Merayo-Lloves J., Martin C., Alcalde I., Quirós L.M., and Vazquez F.: Surface proteoglycans as mediators in bacterial pathogens infections. Front. Microbiol., 7, 220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Martín R., Martín C., Escobedo S., Suárez J.E., and Quirós L.M.: Surface glycosaminoglycans mediate adherence between HeLa cells and Lactobacillus salivarius Lv72. BMC Microbiol., 13, 210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Baron M.J., Bolduc G.R., Goldberg M.B., Aupérin T.C., and Madoff L.C.: Alpha C protein of group B Streptococcus binds host cell surface glycosaminoglycan and enters cells by an actin-dependent mechanism. J. Biol. Chem., 279, 24714-24723 (2004). [DOI] [PubMed] [Google Scholar]
- 9).Stern R. and Jedrzejas M.J.: Hyaluronidases: their genomics, structures, and mechanisms of action. Chem. Rev., 106, 818-839 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Marion C., Stewart J.M., Tazi M.F., Burnaugh A.M., Linke C.M., Woodiga S.A., and King S.J.: Streptococcus pneumoniae can utilize multiple sources of hyaluronic acid for growth. Infect. Immun., 80, 1390-1398 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Rigden D.J. and Jedrzejas M.J.: Structures of Streptococcus pneumoniae hyaluronate lyase in complex with chondroitin and chondroitin sulfate disaccharides. Insights into specificity and mechanism of action. J. Biol. Chem., 278, 50596-50606 (2003). [DOI] [PubMed] [Google Scholar]
- 12).Maruyama Y., Nakamichi Y., Itoh T., Mikami B., Hashimoto W., and Murata K.: Substrate specificity of streptococcal unsaturated glucuronyl hydrolases for sulfated glycosaminoglycan. J. Biol. Chem., 284, 18059-18069 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Maruyama Y., Oiki S., Takase R., Mikami B., Murata K., and Hashimoto W.: Metabolic fate of unsaturated glucuronic/iduronic acids from glycosaminoglycans: Molecular identification and structure determination of streptococcal isomerase and dehydrogenase. J. Biol. Chem., 290, 6281-6292 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Oiki S., Nakamichi Y., Maruyama Y., Mikami B., Murata K., and Hashimoto W.: Streptococcal phosphotransferase system imports unsaturated hyaluronan disaccharide derived from host extracellular matrices. PLoS One, 14, e0224753 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Zheng J., Wittouck S., Salvetti E., Franz C.M.A.P., Harris H.M.B., Mattarelli P., O'Toole P.W., Pot B., Vandamme P., Walter J., Watanabe K., Wuyts S., Felis G.E., Gänzle M.G., and Lebeer S.: A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol., 70, 2782-2858 (2020). [DOI] [PubMed] [Google Scholar]
- 16).Kawai K., Kamochi R., Oiki S., Murata K., and Hashimoto W.: Probiotics in human gut microbiota can degrade host glycosaminoglycans. Sci. Rep., 8, 10674 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Hugouvieux-Cotte-Pattat N., Condemine G., Nasser W., and Reverchon S.: Regulation of pectinolysis in Erwinia chrysanthemi. Annu. Rev. Microbiol., 50, 213-257 (1996). [DOI] [PubMed] [Google Scholar]
- 18).Shevchik V.E., Condemine G., Robert-Baudouy J., and Hugouvieux-Cotte-Pattat N.: The exopolygalacturonate lyase PelW and the oligogalacturonate lyase Ogl, two cytoplasmic enzymes of pectin catabolism in Erwinia chrysanthemi 3937. J. Bacteriol., 181, 3912-3929 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Itoh T., Ochiai A., Mikami B., Hashimoto W., and Murata K.: A novel glycoside hydrolase family 105: The structure of family 105 unsaturated rhamnogalacturonyl hydrolase complexed with a disaccharide in comparison with family 88 enzyme complexed with the disaccharide. J. Mol. Biol., 360, 573-585 (2006). [DOI] [PubMed] [Google Scholar]
- 20).Preiss J. and Ashwell G.: Polygalacturonic acid metabolism in bacteria. II. Formation and metabolism of 3-deoxy-D-glycero-2,5-hexodihulosonique acid. J. Biol. Chem., 238, 1577-1583 (1963). [PubMed] [Google Scholar]
- 21).Rothe M., Alpert C., Loh G., and Blaut M.: Novel insights into E. coli's hexuronate metabolism: KduI facilitates the conversion of galacturonate and glucuronate under osmotic stress conditions. PLoS One, 8, e56906 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Crowther R.L. and Georgiadis M.M.: The crystal structure of 5-keto-4-deoxyuronate isomerase from Escherichia coli. Proteins, 61, 680-684 (2005). [DOI] [PubMed] [Google Scholar]
- 23).Laemmli U.K.: Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680-685 (1970). [DOI] [PubMed] [Google Scholar]
- 24).Ochiai A., Itoh T., Kawamata A., Hashimoto W., and Murata K.: Plant cell wall degradation by saprophytic Bacillus subtilis strains: gene clusters responsible for rhamnogalacturonan depolymerization. Appl. Environ. Microbiol., 73, 3803-3813 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Otwinowski Z. and Minor W.: Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307-326 (1997). [DOI] [PubMed] [Google Scholar]
- 26).Vagin A.A. and Isupov M.N.: Spherically averaged phased translation function and its application to the search for molecules and fragments in electron-density maps. Acta Crystallogr. D. Biol. Crystallogr., 57, 1451-1456 (2001). [DOI] [PubMed] [Google Scholar]
- 27).Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C., Richardson J.S., Terwilliger T.C., and Zwart P.H. PH: PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr., 66, 213-221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Emsley P., Lohkamp B., Scott W., and Cowtan K.: Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr., 66, 486-501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Sambrook J., Fritsch E.F., and Maniatis T.: Molecular cloning. A laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. (1989). [Google Scholar]
- 30).Sanger F., Nicklen S., and Coulson A.R., A.R.: DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA., 74, 5463-5467 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Laskowski R.A., Watson J.D., and Thornton J.M.: ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res., 33, W89-93 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Hashimoto W., Kawai S., and Murata K.: Bacterial supersystem for alginate import/metabolism and its environmental and bioenergy applications. Bioeng. Bugs, 1, 97-109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Dunwell J.M., Purvis A., and Khuri S.: Cupins: the most functionally diverse protein superfamily? Phytochemistry, 65, 7-17 (2004). [DOI] [PubMed] [Google Scholar]
- 34).Vorobjeva N.N., Kurilova S.A., Petukhova A.F., Nazarova T.I., Kolomijtseva G.Y., Baykov A.A., and Rodina E.V.: A novel, cupin-type phosphoglucose isomerase in Escherichia coli. Biochim. Biophys. Acta, 1864, 129601 (2020). [DOI] [PubMed] [Google Scholar]