Abstract
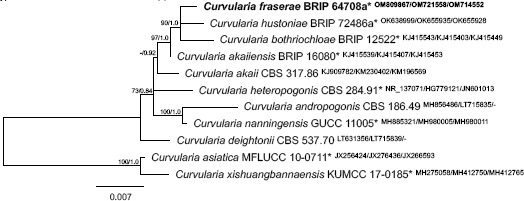
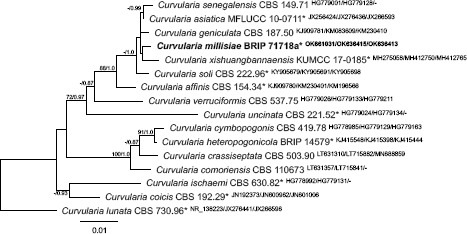
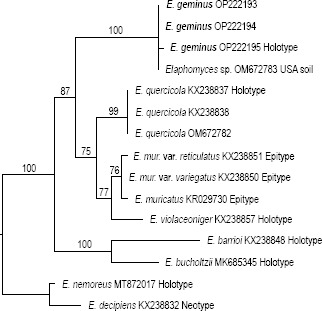
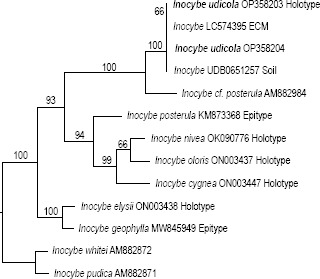
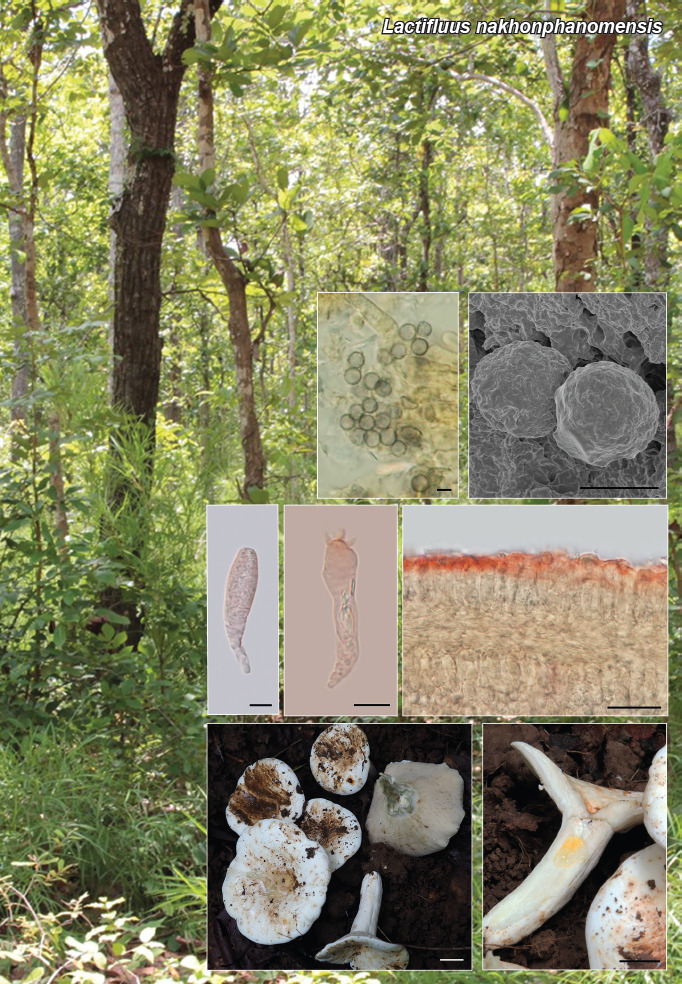
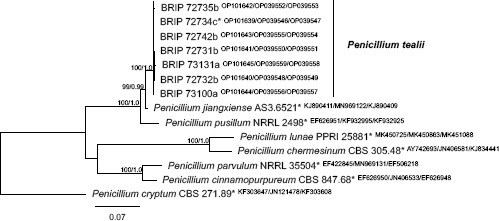
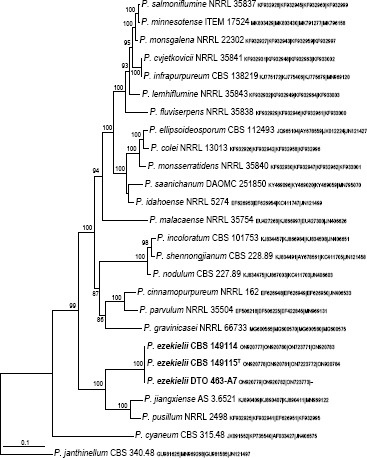
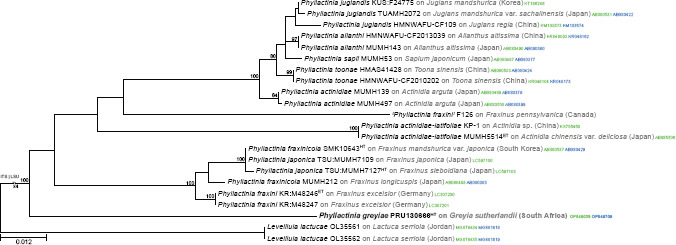
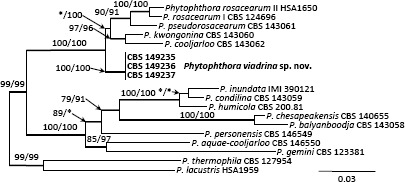
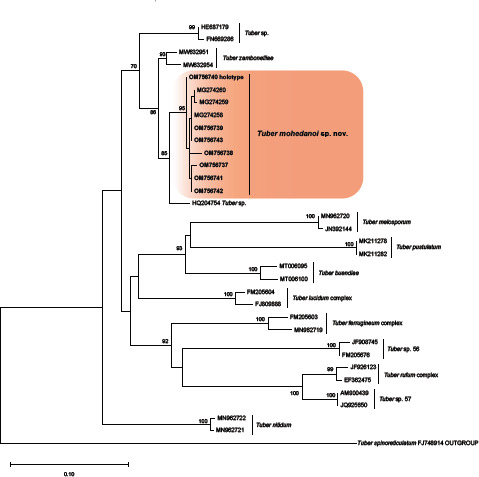
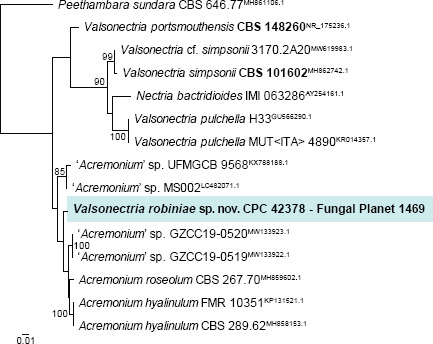
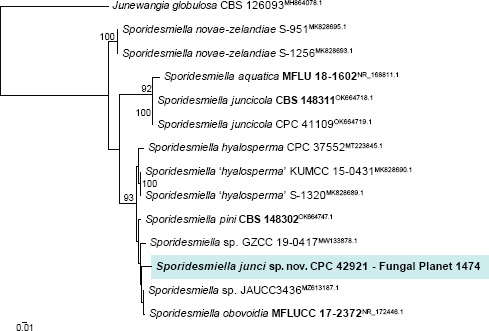
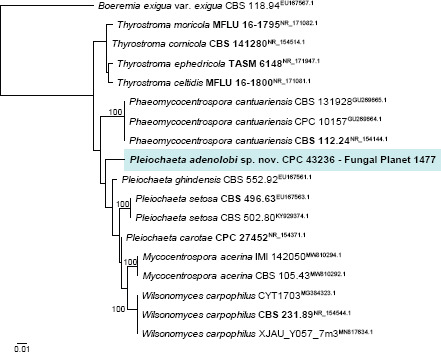
Novel species of fungi described in this study include those from various countries as follows: Argentina, Colletotrichum araujiae on leaves, stems and fruits of Araujia hortorum. Australia, Agaricus pateritonsus on soil, Curvularia fraserae on dying leaf of Bothriochloa insculpta, Curvularia millisiae from yellowing leaf tips of Cyperus aromaticus, Marasmius brunneolorobustus on well-rotted wood, Nigrospora cooperae from necrotic leaf of Heteropogon contortus, Penicillium tealii from the body of a dead spider, Pseudocercospora robertsiorum from leaf spots of Senna tora, Talaromyces atkinsoniae from gills of Marasmius crinis-equi and Zasmidium pearceae from leaf spots of Smilaxglyciphylla. Brazil, Preussia bezerrensis from air. Chile, Paraconiothyrium kelleni from the rhizosphere of Fragaria chiloensis subsp. chiloensis f. chiloensis. Finland, Inocybe udicola on soil in mixed forest with Betula pendula, Populus tremula, Picea abies and Alnus incana. France, Myrmecridium normannianum on dead culm of unidentified Poaceae. Germany, Vexillomyces fraxinicola from symptomless stem wood of Fraxinus excelsior. India, Diaporthe limoniae on infected fruit of Limonia acidissima, Didymella naikii on leaves of Cajanus cajan, and Fulvifomes mangroviensis on basal trunk of Aegiceras corniculatum. Indonesia, Penicillium ezekielii from Zea mays kernels. Namibia, Neocamarosporium calicoremae and Neocladosporium calicoremae on stems of Calicorema capitata, and Pleiochaeta adenolobi on symptomatic leaves of Adenolobus pechuelii. Netherlands, Chalara pteridii on stems of Pteridium aquilinum, Neomackenziella juncicola (incl. Neomackenziella gen. nov.) and Sporidesmiella junci from dead culms of Juncus effusus. Pakistan, Inocybe longistipitata on soil in a Quercus forest. Poland, Phytophthora viadrina from rhizosphere soil of Quercus robur, and Septoria krystynae on leaf spots of Viscum album. Portugal (Azores), Acrogenospora stellata on dead wood or bark. South Africa, Phyllactinia greyiae on leaves of Greyia sutherlandii and Punctelia anae on bark of Vachellia karroo. Spain, Anteaglonium lusitanicum on decaying wood of Prunus lusitanica subsp. lusitanica, Hawksworthiomyces riparius from fluvial sediments, Lophiostoma carabassense endophytic in roots of Limbarda crithmoides, and Tuber mohedanoi from calcareus soils. Spain (Canary Islands), Mycena laurisilvae on stumps and woody debris. Sweden, Elaphomyces geminus from soil under Quercus robur. Thailand, Lactifluus chiangraiensis on soil under Pinus merkusii, Lactifluus nakhonphanomensis and Xerocomus sisongkhramensis on soil under Dipterocarpus trees. Ukraine, Valsonectria robiniae on dead twigs of Robinia hispida. USA, Spiralomyces americanus (incl. Spiralomyces gen. nov.) from office air. Morphological and culture characteristics are supported by DNA barcodes.
Citation: Tan YP, Bishop-Hurley SL, Shivas RG, et al. 2022. Fungal Planet description sheets: 1436–1477. Persoonia 49: 261–350. https://doi.org/10.3767/persoonia.2022.49.08
Keywords: ITS nrDNA barcodes, LSU, new taxa, systematics
Acknowledgements
The work of Pedro Crous and colleagues benefitted from funding by the European Union’s Horizon 2020 research and innovation program (RISE) under the Marie Sklodowska-Curie grant agreement No. 101008129, project acronym ‘Mycobiomics’, and the Dutch NWO Roadmap grant agreement No. 2020/ENW/00901156, project ‘Netherlands Infrastructure for Ecosystem and Biodiversity Analysis – Authoritative and Rapid Identification System for Essential biodiversity information’ (acronym NIEBA-ARISE). Don Cowan gratefully acknowledges funding support from the National Research Foundation (grant number 137954) and from the University of Pretoria. Jose G. Maciá-Vicente acknowledges support from the German Research Foundation under grant MA7171/1-1, and from the Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz (LOEWE) of the state of Hesse within the framework of the Cluster for Integrative Fungal Research (IPF). Cecilia Santelices and colleagues acknowledge financial support from the FONDECYT INICIACIÓN project No. 11191074 from the Chilean National Agency for Research and Development (ANID) and to the FONDEQUIP Program from ANID (grant: EQM200205) for funding a platform of equipment for preservation of microbial genetic resources. Hanna Stepniewska and colleagues were financed by the Ministry of Science and Higher Education of the Republic of Poland (SUB/040013/D019). Saúl De la Peña-Lastra and colleagues would like to thank the Secretaria Regional do Ambientee Alterações Climáticas Açores for the permission granted for sampling (Licença n° 16/2021/DRAAC). The research of P.K. Divakar and colleagues was supported by the Spanish Ministry of Science and Innovation (PID2019-105312GB-I00/AEI/10.13039/501100011033). Furthermore, Luis Alberto Parra is acknowledged for his help on nomenclature, and Donald H. Pfister for his revision of the text. Antonio Mateos and colleagues would like to thank the great naturalist Cristóbal Burgos Morillo for his help in processing the images and Amalio Gutiérrez for his collaboration, company and cooperation on surveys. Gustavo Hernán Ramírez and colleagues acknowledge German Barros and Martín Bonacci for their help with molecular studies. Frances E. Guard and colleagues thank the curation staff at RBGV, PERTH and BRI for their help with loans and processing of specimens. Ángel Bañares and colleagues thank Pablo Alvarado ALVALAB and Fernanda Rodríguez Fariña Unidade de Bioloxía Molecular (SAI) Universidade da Coruña who assisted with the generation of the ITS sequence data. The research of D. Kidanemariam and D.K. Berger was supported by the National Research Foundation of South Africa (grant nr 120398) and the University of Pretoria. Cobus M. Visagie and co-authors are grateful for the advice received from Uwe Braun and thank Konstanze Bensch (MycoBank) for Latin assistance. Steffen Bien and colleagues thank Ulrike Damm from the Senckenberg Natural History Museum Görlitz for technical support. Teresa Lebel and colleagues acknowledge the Royal Botanic Gardens Victoria and the Friends of the Royal Botanic Gardens Victoria for funding and support for the Northern Territory field and lab work; Northern Territory Herbarium staff for space and lab access and help with collection curation; State Herbarium of South Australia for support of the undergraduate intern J. Broadbridge; their research was also supported through funding from Australian Biological Resources Study grant (4-EHOJ161) to the State Herbarium of South Australia. Asunción Morte is grateful to the Fundación Séneca - Agencia de Ciencia y Tecnología de la Región de Murcia (20866/PI/18) for financial support. Malarvizhi Kaliyaperumal and Sugantha Gunaseelan would like to acknowledge DST, SERB-EMR (EMR/2016/003078) for the financial assistance. Malarvizhi Kaliyaperumal and Kanaga valli Raja are grateful to TANSCST, DOTE, Chennai (Student Scheme Project, 2018 (BS-014)) for providing financial aid. Attihalli S. Savitha and co-authors thank Bettadapura R. Nuthan, Department of Studies in Microbiology, for his kind support in culture maintenance and laboratory assistance. Shivannegowda Mahadevakumar thanks the Director of KSCSTE – Kerala Forest Research Institute for support. Mikael Jeppson, Jukka Vauras and Ellen Larsson thank the Swedish Taxonomy Initiative (SLU Artdatabanken, Uppsala) (grant to E. Larsson dha.2019.4.3-13). The study of Daniel Torres-Garcia and colleagues was partially supported by the Spanish Ministerio de Economía, Industria y Competitividad (grant PID2021-128068NB-100). Shivannegowda Mahadevakumar and co-authors thank Bettadapura R. Nuthan, Department of Studies in Microbiology, University of Mysore and Bettadapura R. Meghavarshinigowda, Department of Studies in Botany, University of Mysore, for their assistance in the laboratory. Umpawa Pinruan and colleagues were financially supported by the Platform Technology Management Section, National Center for Genetic Engineering and Biotechnology (BIOTEC), Project Grant No. P19-50231. Antônio Sérgio Ferreira-Sá and colleagues thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for granting PhD scholarships (Finance code – 001) to ASFS and LLS.
REFERENCES
- Akata I, Halici MG, Uzun Y. 2011. Additional macrofungi records from Trabzon province for the mycobiota of Turkey. Turkish Journal of Botany 35: 309–314. [Google Scholar]
- Álvarez VI, Raymundo T, Valenzuela R. 2016. Hongos histerioides (Dothideomycetes, Ascomycota) del bosque tropical caducifolio en el Parque Nacional Lagunas de Chacahua, Oaxaca, México. Acta Botánica Mexicana 116: 49–64. [Google Scholar]
- An YY, Zeng XY, Geng K, et al. 2019. One new species and one new record of Zasmidium in China. Biodiversity Data Journal 9: e59001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen M, Skrede I, Jaklitsch WM, et al. 2021. Multi-locus phylogenetic analysis of lophiostomatoid fungi motivates a broad concept of Lophiostoma and reveals nine new species. Persoonia 46: 240–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin V, Noordeloos M. 2010. A monograph of marasmioid and collybioid fungi in Europe. Eching, IHW-Verlag. [Google Scholar]
- Archana S, Kharwar RN, Raghvendra S, et al. 2014. A new species of Zasmidium (Mycosphaerellaceae) from India. Sydowia 66: 309–312. [Google Scholar]
- Arenal F, Platas G, Peláez F. 2007. A new endophytic species of Preussia (Sporormiaceae) inferred from morphological observations and molecular phylogenetic analysis. Fungal Diversity 25: 1–17. [Google Scholar]
- Ariyawansa HA, Tanaka K, Thambugala KM, et al. 2014. A molecular phylogenetic reappraisal of the Didymosphaeriaceae (= Montagnulaceae). Fungal Diversity 68: 69–104. [Google Scholar]
- Arzanlou M, Groenewald JZ, Gams W, et al. 2007. Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Studies in Mycology 58: 57–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgari B, Zare R. 2010. Two new species of Preussia from Iran. Nova Hedwigia 90: 533–548. [Google Scholar]
- Bandini D, Oertel B, Eberhardt U. 2021a. Noch mehr Risspilze (3): Einundzwanzig neue Arten der Familie Inocybaceae. Mycologica Bavarica 22: 31–138. [Google Scholar]
- Bandini D, Oertel B, Eberhardt U. 2021b. A fresh outlook on the smooth-spored species of Inocybe: type studies and 18 new species. Mycological Progress 20: 1019–1114. [Google Scholar]
- Bandini D, Oertel B, Eberhardt U. 2022. Noch mehr Risspilze (3): Einundzwanzig neue Arten der Familie Inocybaceae. Mycologia Bavarica 22: 31–138. [Google Scholar]
- Bao D-F, McKenzie EHC, Bhat DJ, et al. 2020. Acrogenospora (Acrogenosporaceae, Minutisphaerales) appears to be a very diverse genus. Frontiers in Microbiology 11: 1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral HO, Quijada L. 2020. Nomenclatural novelties. Index Fungorum 10: 1–3. [Google Scholar]
- Bashir H, Chen J, Jabeen S, et al. 2021. An overview of Agaricus section Hondenses and Agaricus section Xanthodermatei with description of eight new species from Pakistan. Scientific Reports 11: 12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A. 2005. An illustrated guide to the coprophilous Ascomycetes of Australia. CBS Biodiversity Series (Utrecht) 3: 1–173. [Google Scholar]
- Bezerra JDP, Sandoval-Denis M, Paiva LM, et al. 2017. New endophytic Toxicocladosporium species from cacti in Brazil, and description of Neocladosporium gen. nov. IMA Fungus 8: 77–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien S, Kraus C, Damm U. 2020. Novel collophorina-like genera and species from Prunus trees and vineyards in Germany. Persoonia 45: 46–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm EWA, Mugambi GK, Miller AN, et al. 2009a. A molecular phylogenetic reappraisal of the Hysteriaceae, Mytilinidiaceae and Gloniaceae (Pleosporomycetidae, Dothideomycetes) with keys to world species. Studies in Mycology 64: 49–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm EWA, Schoch CL, Spatafora JW. 2009b. On the evolution of the Hysteriaceae and Mytilinidiaceae (Pleosporomycetidae, Dothideomycetes, Ascomycota) using four nuclear genes. Mycological Research 113: 461–479. [DOI] [PubMed] [Google Scholar]
- Bon M. 1998. Novitates, 1, Marasmiaceae et Dermolomataceae comb., st. et sp. nov. / Novitates – Genre Inocybe, sous-genre Clypeus Britz., Documents Mycologiques 28: 1–45. [Google Scholar]
- Bonito GM, Gryganskyi AP, Trappe JM, et al. 2010. A global meta-analysis of Tuber ITS rDNA sequences: species diversity, host associations and long-distance dispersal. Molecular Ecology 19: 4994–5008. [DOI] [PubMed] [Google Scholar]
- Bradshaw M, Braun U, Pfister D. 2022. Phylogeny and taxonomy of the genera of the Erysiphaceae, part 1, Golovinomyces. Mycologia 114(6): 964–993. [DOI] [PubMed] [Google Scholar]
- Braun U, Cook RTA. 2012. Taxonomic manual of the Erysiphales (powdery mildews). CBS Biodiversity Series 11. CBS, Utrecht, The Netherlands. [Google Scholar]
- Braun U, Crous PW, Nakashima C. 2014. Cercosporoid fungi (Mycosphaerellaceae) 2. Species on monocots (Acoraceae to Xyridaceae, excluding Poaceae). IMA Fungus 5: 203–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess TI, Simamora AV, White D, et al. 2018. New species from Phytophthora Clade 6a: evidence for recent radiation. Persoonia 41: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro de Almeida DA, Pascholati Gusmão LF, Miller AN. 2014. A new genus and three new species of hysteriaceous ascomycetes from the semiarid region of Brazil. Phytotaxa 176: 298–308. [Google Scholar]
- Ceruti A, Fontana A, Nosenzo C. 2003. Le specie europee del genere Tuber: una revisione storica. Vol. 37. Italy, Turin, Museo Regionale di Scienze Naturali. [Google Scholar]
- Chakraborty D, Parihar A, Mehta N, et al. 2017. A new species of Xerocomus (Boletaceae) from India. Mycosphere 8: 44–50. [Google Scholar]
- Chevallier FF. 1826. Flore générale des environs de Paris, Vol I. Ferra Librairie-Editeur, Paris, France. [Google Scholar]
- Chupp C. 1954. A monograph of the fungus genus Cercospora. Published by the author, Ithaca, New York, USA. [Google Scholar]
- Cock MJW, Evans HC. 1984. Possibilities for biological control of Cassia tora and C. obtusifolia. Tropical Pest Management 30: 339–350. [Google Scholar]
- Crespo A, Ferencova Z, Pérez-Ortega S, et al. 2010. Austroparmelina, a new Australasian lineage in parmelioid lichens (Parmeliaceae, Ascomycota): a multigene and morphological approach. Systematics and Biodiversity 8: 209–221. [Google Scholar]
- Crossland FLS. 1900. New and critical British Fungi found in West Yorkshire. The Naturalist. 1900: 5–10. [Google Scholar]
- Crous PW, Boers J, Holdom D, et al. 2022. Fungal Planet description sheets: 1383–1435. Persoonia 48: 261–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Cowan DA, Maggs-Kölling G, et al. 2020a. Fungal Planet description sheets: 1112–1181. Persoonia 45: 251–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Cowan DA, Maggs-Kölling G, et al. 2021a. Fungal Planet description sheets: 1182–1283. Persoonia 46: 313–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Osieck ER, Jurjević Ž, et al. 2021b. Fungal Planet description sheets: 1284–1382. Persoonia 47: 178–374. [DOI] [PubMed] [Google Scholar]
- Crous PW, Schumacher RK, Wingfield MJ, et al. 2018. New and interesting fungi. 1. Fungal Systematics and Evolution 1: 169–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Shivas RG, Quaedvlieg W, et al. 2014. Fungal Planet description sheets: 214–280. Persoonia 32: 184–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Chooi YH, et al. 2020b. Fungal Planet description sheets: 1042–1111. Persoonia 44: 301–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Schumacher RK, et al. 2020c. New and interesting fungi. 3. Fungal Systematics and Evolution 6: 157–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Cannon PF, Liu F, et al. 2013. The Colletotrichum orbiculare species complex: Important pathogens of field crops and weeds. Fungal Diversity 61: 29–59. [Google Scholar]
- Damm U, Cannon PF, Woudenberg JHC, et al. 2012. The Colletotrichum boninense species complex. Studies in Mycology 73: 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Verkley GJ, Crous PW, et al. 2008. Novel Paraconiothyrium species on stone fruit trees and other woody hosts. Persoonia 20: 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Beer ZW, Marincowitz S, Duong TA, et al. 2016. Hawksworthiomyces gen. nov. (Ophiostomatales), illustrates the urgency for a decision on how to name novel taxa known only from environmental nucleic acid sequences (ENAS). Fungal Biology 120: 1323–1340. [DOI] [PubMed] [Google Scholar]
- De Leo F, Urzì C, De Hoog GS. 1999. Two Coniosporium species from rock surfaces. Studies in Mycology 43: 70–79. [Google Scholar]
- Dennis RWG. 1955. Ascomycetes from Tristan da Cunha. Results of the Norwegian Scientific Expedition to Tristan da Cunha (1937–1938) 36–38: 1–10. [Google Scholar]
- Dennis RWG. 1981. British Ascomycetes. Vaduz, Cramer. [Google Scholar]
- Divakar PK, Crespo A, Kraichak E, et al. 2017. Using a temporal phylogenetic method to harmonize family- and genus-level classification in the largest clade of lichen-forming fungi. Fungal Diversity 84: 101–117. [Google Scholar]
- Divakar PK, Kauff F, Crespo A, et al. 2013. Understanding phenotypical character evolution in parmelioid lichenized fungi (Parmeliaceae, Ascomycota). PLoS ONE 8: e83115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doveri F. 2004. Fungi Fimicoli Italici – a guide to the recognition of Basidiomycetes and Ascomycetes living on faecal material. Associazione Micologica Bresadola, 1st ed. [Google Scholar]
- Ellis MB. 1972. Dematiaceous Hyphomycetes. XI. Mycological Papers 131: 1–25. [Google Scholar]
- Ellis MB, Ellis JP. 1985. Microfungi on land plants. An identification handbook. Macmillan Publishing Company. [Google Scholar]
- Fan YG, Wu RH, Bau T. 2018. Two new species and eight newly recorded species of subg. Inocybe from China. Journal of Fungal Research 16: 70–83. [Google Scholar]
- Farr DF, Rossman AY. 2022. Fungal Databases, U.S. National Fungus Collections, ARS, USDA. https://nt.ars-grin.gov/fungaldatabases/ [Retrieved 25 Sept. 2022]. [Google Scholar]
- Fisher PJ. 1985. The anamorph of Claussenomyces atrovirens. Transactions of the British Mycological Society 85: 759–760. [Google Scholar]
- Gamundi IJ, Giaiotti AL. 1995. A new species of Claussenomyces (Helotiales) from southern South America. New Zealand Journal of Botany 33: 513–517. [Google Scholar]
- Gilliam MS. 1976. The genus Marasmius in the northeastern United States and adjacent Canada. Mycotaxon 4: 1–144. [Google Scholar]
- Goh TK, Hyde KD, Tsui KM. 1998. The hyphomycete genus Acrogenospora, with two new species and two new combinations. Mycological Research 102: 1309–1315. [Google Scholar]
- Gonçalves MFM, Aleixo A, Vicente TFL, et al. 2019. Three new species of Neocamarosporium isolated from saline environments: N. aestuarinum sp. nov., N. endophyticum sp. nov. and N. halimiones sp. nov. Mycosphere 10: 608–621. [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Hale ME, Jr. 1973. Fine structure of the cortex in the lichen family Parmeliaceae viewed with the scanning-electron microscope. Smithsonian Contributions to Botany 10: 1–92. [Google Scholar]
- Hale ME, Jr. 1976. A monograph of the lichen genus Pseudoparmelia Lynge (Parmeliaceae). Smithsonian Contributions to Botany 31: 1–62. [Google Scholar]
- Hale ME, Jr. 1980. Taxonomy and distribution of the Parmelia flaventior group (Lichens: Parmeliaceae). Journal of the Hattori Botanical Laboratory 47: 75–84. [Google Scholar]
- Hansen EM, Wilcox WF, Reeser PW, et al. 2009. Phytophthora rosacearum and P. sansomeana, new species segregated from the Phytophthora megasperma “complex”. Mycologia 101: 129–135. [DOI] [PubMed] [Google Scholar]
- Hattori T, Sakayaroj J, Jones EBG, et al. 2014. Three species of Fulvifomes (Basidiomycota, Hymenochaetales) associated with rots on mangrove tree Xylocarpus granatum in Thailand. Mycoscience 55: 344–354. [Google Scholar]
- He MQ, Chuankid B, Hyde KD, et al. 2018. A new section and species of Agaricus subgenus Pseudochitonia from Thailand. MycoKeys 40: 53–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann P. 1978. Essai d’une clé de determination des genres Agaricus et Micropsalliota. Sydowia 30: 6–37. [Google Scholar]
- Houbraken J, Kocsubé S, Visagie CM, et al. 2020. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Studies in Mycology 95: 5–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Husbands DR, Henkel TW, Bonito G, et al. 2013. New species of Xerocomus (Boletales) from the Guiana Shield, with notes on their mycorrhizal status and fruiting occurrence. Mycologia 105: 422–435. [DOI] [PubMed] [Google Scholar]
- Hyde KD, Jones EBG, Liu J-K, et al. 2013. Families of Dothideomycetes. Fungal Diversity 63: 1–313. [Google Scholar]
- Hyde KD, Tennakoon DS, Jeewon R, et al. 2019. Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 96: 1–242. [Google Scholar]
- Jaklitsch W, Fournier J, Voglmayr H. 2018. Two unusual new species of Pleosporales: Anteaglonium rubescens and Atrocalyx asturiensis. Sydowia 70: 129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowiak R, Stępniewska H, Bilański P. et al. 2014. Occurrence of Phytophthora plurivora and other Phytophthora species in oak forests of southern Poland and their association with site conditions and the health status of trees. Folia Microbiologica 59: 531–542. [DOI] [PubMed] [Google Scholar]
- Jayasiri SC, Hyde KD, Jones EBG, et al. 2018. Taxonomic novelties of hysteriform Dothideomycetes. Mycosphere 9: 803–837. [Google Scholar]
- Jayasiri SC, Hyde KD, Jones EBG, et al. 2019. Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 10: 1–186. [Google Scholar]
- Jayasiri SC, Jones EBG, Kang J-C, et al. 2016. A new species of genus Anteaglonium (Anteagloniaceae, Pleosporales) with its asexual morph. Phytotaxa 263: 233–244. [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, et al. 2017. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics 9: 286–298. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy AH, Schoch CL, Marrero G, et al. 2022. Publicly available and validated DNA reference sequences are critical to fungal identification and global plant protection efforts: A use-case in Colletotrichum. Plant Disease 106: 1573–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MB, Naseer A, Aqdus F, et al. 2022. Inocybe quercicola sp. nov. (Agaricales, Inocybaceae), from Pakistan. Microbial Biosystems 6: 22–29. [Google Scholar]
- Kimura M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16: 111–120. [DOI] [PubMed] [Google Scholar]
- Kong HZ, Liang ZQ. 2003. A new Penicillium species isolated from Jiangxi, China. Mycosystema 22: 4–5. [Google Scholar]
- Kornerup A, Wanscher JH. 1978. Methuen handbook of colour, 3rd ed. Eyre Methuen, London. [Google Scholar]
- Kornerup A, Wanscher JH. 1981. Metheun’s handbook of colours, 3rd edn. Metheun & Co. Ltd., London. [Google Scholar]
- Kruys Å. 2015. New species of Preussia with 8-celled ascospores (Sporormiaceae, Pleosporales, Ascomycota). Phytotaxa 234: 143–150. [Google Scholar]
- Kumar S, Stecher G, Li M, et al. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyper TW. 1986. A revision of the genus Inocybe in Europe. I. Subgenus Inosperma and the smooth-spored species of subgenus Inocybe. Persoonia Supplement 3: 1–247. [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, et al. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34: 772–773. [DOI] [PubMed] [Google Scholar]
- Lebel T, Cooper JA, Castellano MA, et al. 2021. Three independent evolutionary events of sequestrate Lactifluus species in Australasia. Fungal Systematics and Evolution 8: 9–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZL, Hyde KD, Liu JK, et al. 2019. Freshwater Sordariomycetes. Fungal Diversity 99: 451–660. [Google Scholar]
- Lygis V, Vasiliauskas R, Larsson KH, et al. 2005. Wood-inhabiting fungi in stems of Fraxinus excelsior in declining ash stands of northern Lithuania, with particular reference to Armillaria cepistipes. Scandinavian Journal of Forest Research 20: 337–346. [Google Scholar]
- Maciá-Vicente JG, Ferraro V, Burruano S, et al. 2012. Fungal assemblages associated with roots of halophytic and non-halophytic plant species vary differentially along a salinity gradient. Microbial Ecology 64: 668–679. [DOI] [PubMed] [Google Scholar]
- Mackey AP, Miller EN, Palmer WA. 1997. Sicklepod (Senna obtusifolia) in Queensland. Queensland. Department of Natural Resources, Dept. of Natural Resources, Woolloongabba, Qld. [Google Scholar]
- Man in ‘t Veld WA, Rosendahl K, Van Rijswick PCJ, et al. 2019. Multiple Halophytophthora spp. and Phytophthora spp. including P. gemini, P. inundata, and P. chesapeakensis sp. nov. isolated from the seagrass Zostera marina in the Northern Hemisphere. European Journal of Phytopathology 153: 341–357. [Google Scholar]
- Manamgoda DS, Cai L, McKenzie EHC, et al. 2012. Two new Curvularia species from northern Thailand. Sydowia 64: 255–266. [Google Scholar]
- Marin-Felix Y, Groenewald JZ, Cai L, et al. 2017. Genera of phytopathogenic fungi: GOPHY 1. Studies in Mycology 86: 99–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheny BP, Swenie RA. 2018. The Inocybe geophylla group in North America: a revision of the lilac species surrounding I. lilacina. Mycologia 110: 618–634. [DOI] [PubMed] [Google Scholar]
- McNeill J, Barrie FF, Buck WR, et al. (eds). 2012. International Code of Nomenclature for algae, fungi, and plants (Melbourne Code). Regnum Vegetabile no. 154.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010, New Orleans, LA: 1–8. [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, et al. 2020. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37: 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Maximov TC, Bryanin S, et al. 2022. Host phylogeny is the primary determinant of ectomycorrhizal fungal community composition in the permafrost ecosystem of eastern Siberia at a regional scale. Fungal Ecology 55: 101117. [Google Scholar]
- Molia A, Larsson E, Jeppson M, et al. 2020. Elaphomyces section Elaphomyces (Eurotiales, Ascomycota) – taxonomy and phylogeny of North European taxa, with the introduction of three new species. Fungal Systematics and Evolution 5: 283–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moral C. 2004. Filogenia y caracterización molecular de hongos basidiomicetos del género Mycena. PhD thesis, Universidad Autónoma de Madrid, Spain. [Google Scholar]
- Mugambi GK, Huhndorf SM. 2009. Parallel evolution of hysterothecial ascomata in ascolocularous fungi (Ascomycota, Fungi). Systematics and Biodiversity 7: 453–464. [Google Scholar]
- Nag Raj TR, Kendrick WB. 1975. A monograph of Chalara and allied genera. Wilfred Laurier University Press, Waterloo, Ontario, Canada. [Google Scholar]
- Nguyen L-T, Schmidt HA, Von Haeseler A, et al. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Santana B, Lodge DJ, Baroni TJ, et al. 2007. Boletes from Belize and the Dominican Republic. Fungal Diversity 27: 247–416. [Google Scholar]
- Ouellette GB, Korf RP. 1979. Three new species of Claussenomyces from Macaronesia. Mycotaxon 10: 255–264. [Google Scholar]
- Paz A, Bellanger JM, Lavoise C, et al. 2017. The genus Elaphomyces (Ascomycota, Eurotiales): a ribosomal DNA-based phylogeny and revised systematics of European ‘deer truffles’. Persoonia 38: 197–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegler DN, Young TWK. 1972. Studies on African Agaricales II. Kew Bulletin 23: 219–249. [Google Scholar]
- Quaedvlieg W, Verkley GJM, Shin H-D, et al. 2013. Sizing up Septoria. Studies in Mycology 75: 307–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quijada L, Baral HO, Jaen-Molina R, et al. 2014. Phylogenetic and morphological circumscription of the Orbilia aurantiorubra group. Phytotaxa 175: 001–018. [Google Scholar]
- Raper KB, Fennell DI. 1948. New species of Penicillium. Mycologia 40: 507–585. [PubMed] [Google Scholar]
- Robich G. 2006. A revised key to the species of Mycena section Fragilipedes of the Northern Hemisphere. Persoonia 19: 1–43. [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, et al. 2012. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Larga Model Space. Systematics Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman A, Crous PW, Hyde KD, et al. 2015. Recommended names for pleomorphic genera in Dothideomycetes. IMA Fungus 6: 507–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryberg M, Nilsson RH, Kristiansson E, et al. 2008. Mining metadata from unidentified ITS sequences in GenBank: a case study in Inocybe (Basidiomycota). BMC Evolutionary Biology 8: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador-Montoya CA, Popoff OF, Reck M, et al. 2018. Taxonomic delimitation of Fulvifomes robiniae (Hymenochaetales, Basidiomycota) and related species in America: F. squamosus sp. nov. Plant Systematics and Evolution 304: 445–459. [Google Scholar]
- Schoch CL, Shoemaker RA, Seifert KA, et al. 2006. A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 98: 1041–1052. [DOI] [PubMed] [Google Scholar]
- Scholler M, Schmidt A, Meeboon J, et al. 2018. Phyllactinia fraxinicola, another Asian fungal pathogen on Fraxinus excelsior (common ash) introduced to Europe? Mycoscience 59: 85–88. [Google Scholar]
- Silva M, Barreto RW, Pereira OL, et al. 2016. Exploring fungal mega-diversity: Pseudocercospora from Brazil. Persoonia 37: 142–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer R. 1976. Marasmieae (Basidiomycetes - Tricholomataceae). Flora Neotropica 17: 1–347. [Google Scholar]
- Sivanesan A. 1987. Graminicolous species of Bipolaris, Curvularia, Drechslera, Exserohilum and their teleomorphs. Mycological Papers 158: 1–261. [Google Scholar]
- Sivanesan A, Alcorn JL, Shivas RG. 2003. Three new graminicolous species of Curvularia (anamorphic fungi) from Queensland, Australia. Australian Systematic Botany 16: 275–278. [Google Scholar]
- Smith G. 1939. Some new species of mould fungi. Transactions of the British Mycological Society 22: 252–256. [Google Scholar]
- Spegazzini CL. 1880. Fungi argentini Pugillus 2. Anales de la Sociedad Científica Argentina 10: 5–33. [Google Scholar]
- Spegazzini CL. 1910. Mycetes Argentinenses. Series V. Anales del Museo Nacional de Buenos Aires, Series 3, 13: 329–467. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology 57: 758–771. [DOI] [PubMed] [Google Scholar]
- Stangl J. 1989. Die Gattung Inocybe in Bayern. Hoppea 46: 5–388. [Google Scholar]
- Summerbell RC, Gueidan C, Schroers HJ, et al. 2011. Acremonium phylogenetic overview and revision of Gliomastix, Trichothecium and Sarocladium. Studies in Mycology 68: 139–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart L, Crous PW, Denman S, et al. 1998. Fungi occurring on Proteaceae. I. South African Journal of Botany 64: 137–142. [Google Scholar]
- Swofford DL. 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods). Version 4.0b10. Sinauer Associates, Sunderland, Massachusetts. [Google Scholar]
- Swofford DL. 2003. PAUP* 4.0: Phylogenetic Analysis Using Parsimony (*and other methods). Sinauer Associates, Sunderland, Massachusetts. [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. 2013. MEGA6: Molecular evolutionary genetics analysis v. 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YP, Bransgrove KL, Marney TS, et al. 2022. Nomenclatural novelties. Index Fungorum 511: 1–8. [Google Scholar]
- Tao G, Liu ZY, Liu F, et al. 2013. Endophytic Colletotrichum species from Bletilla ochracea (Orchidaceae), with descriptions of seven new species. Fungal Diversity 61: 139–164. [Google Scholar]
- The Royal Horticultural Society . 2015. RHS colour chart, sixth revised edition. Royal Horticultural Society, London. [Google Scholar]
- Thongklang N, Nawaz R, Khalid AN, et al. 2014. Morphological and molecular characterization of three Agaricus species from tropical Asia (Pakistan, Thailand) reveals a new group in section Xanthodermatei. Mycologia 106: 1220–1232. [DOI] [PubMed] [Google Scholar]
- Tibpromma S, Hyde KD, McKenzie EHC, et al. 2018. Fungal diversity notes 840–928: micro-fungi associated with Pandanaceae. Fungal Diversity 93: 1–160. [Google Scholar]
- Trifinopoulos J, Nguyen L-T, Von Haeseler A, et al. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research 44: W232–W235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulasne LR, Tulasne C. 1851. Fungi Hypogaei, Histoire et Monographie des Champignons Hypogés. Klincksieck F. (ed.), Paris, France. [Google Scholar]
- Unterseher M, Peršoh D, Schnittler M. 2013. Leaf-inhabiting endophytic fungi of European Beech (Fagus sylvatica L.) co-occur in leaf litter but are rare on decaying wood of the same host. Fungal Diversity 60: 43–54. [Google Scholar]
- Verkley G, Silva M, Wicklow D, et al. 2004. Paraconiothyrium, a new genus to accommodate the mycoparasite Coniothyrium minitans, anamorphs of Paraphaeosphaeria, and four new species. Studies in Mycology 50: 323–335. [Google Scholar]
- Verkley GJM, Quaedvlieg W, Shin H-D, et al. 2013. A new approach to species delimitation in Septoria. Studies in Mycology 75: 213–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergren T. 1896. Bidrag till kännedomen om Götlands svampflora. Bihang till Kungliga Svenska Vetenskaps-Akademiens Handlingar 22: 1–29. [Google Scholar]
- Videira SIR, Groenewald JZ, Nakashima C, et al. 2017. Mycosphaerellaceae – chaos or clarity? Studies in Mycology 87: 257–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waipara NW, Winks CJ, Gianotti AF, et al. 2006. Surveys for potential biocontrol agents for moth plant in New Zealand and Argentina. New Zealand Plant Protection 59: 18–22. [Google Scholar]
- Wang J, Shao S, Liu C, et al. 2021. The genus Paraconiothyrium: species concepts, biological functions, and secondary metabolites. Critical Reviews in Microbiology 47: 781–810. [DOI] [PubMed] [Google Scholar]
- Wang M, Liu F, Crous PW, et al. 2017. Phylogenetic reassessment of Nigrospora: Ubiquitous endophytes, plant and human pathogens. Persoonia 39: 118–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayawardene NN, Crous PW, Kirk PM, et al. 2014. Naming and outline of Dothideomycetes–2014 including proposals for the protection or suppression of generic names. Fungal Diversity 69: 1–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G. 1883. L. Rabenhorstii fungi Europaei et extraeuropaei exsiccati, Klotzschii herbarii vivi mycologici continuatio. Editio nova. Series secunda. Cent. 30: 2901–3000. [Google Scholar]
- Wu G, Li Y-C, Zhu Z-T, et al. 2016. One hundred noteworthy boletes from China. Fungal Diversity 81: 25–188. [Google Scholar]
- Yanna Hyde KD. 2002. New saprobic fungi on fronds of palms from northern Queensland, Australia. Australian Systematic Botany 15: 755–764. [Google Scholar]
- Yeh Y-H, Kirschner R. 2019. A new species of Alpestrisphaeria (Dothideomycetes) with monodictys-like anamorph and revision of three Monodictys species. Mycological Progress 18: 703–711. [Google Scholar]
- Zhou JL, Su SY, Su HY, et al. 2016. A description of eleven new species of Agaricus sections Xanthodermatei and Hondenses collected from Tibet and the surrounding areas. Phytotaxa 257: 99–121. [Google Scholar]
- Zhou Y, Gong G, Zhang S, et al. 2014. A new species of the genus Trematosphaeria from China. Mycological Progress 13: 33–43. [Google Scholar]
- Zogg H. 1962. Die Hysteriaceae s.str. und Lophiaceae, unter besonderer Berücksichtigung der mitteleuropäischen Formen. Beiträge zur Kryptogamenflora der Schweiz, Band 11: 1–190. [Google Scholar]