Abstract
Abstract Sympoventuriaceae (Venturiales, Dothideomycetes) comprises genera including saprophytes, endophytes, plant pathogens, as well as important animal or human opportunistic pathogens with diverse ecologies and wide geographical distributions. Although the taxonomy of Sympoventuriaceae has been well studied, generic boundaries within the family remain poorly resolved due to the lack of type materials and molecular data. To address this issue and establish a more stable and reliable classification system in Sympoventuriaceae, we performed multi-locus phylogenetic analyses using sequence data of seven genes (SSU, ITS, LSU, act1, tub2, tef1 and rpb2) with increased taxon sampling and morphological analysis. The molecular data combined with detailed morphological studies of 143 taxa resolved 22 genera within the family, including one new genus, eight new species, five new combinations and one new name. Finally, we further investigated the evolutionary history of Sympoventuriaceae by reconstructing patterns of lifestyle diversification, indicating the ancestral state to be saprophytic, with transitions to endophytic, animal or human opportunistic and plant pathogens.
Citation: Wei TP, Zhang H, Zeng XY, et al. 2022. Re-evaluation of Sympoventuriaceae. Persoonia 48: 219–260. https://doi.org/10.3767/persoonia.2022.48.07.. Effectively published online: 17 June 2022 [Received: 2 February 2022; Accepted: 27 April 2022].
Keywords: evolution, lifestyle, multigene analysis, new taxa, systematics, Venturia
INTRODUCTION
Sympoventuriaceae is a large family in Venturiales (Dothideomycetes, Ascomycota) with diverse ecology, wide geographic distribution and rich species diversity (Zhang et al. 2011, Seyedmousavi et al. 2013, Liu et al. 2017, Wijayawardene et al. 2018, Crous et al. 2019a, Shen et al. 2020). Members of this family are usually hyphomycetes with conidia liberated by rhexolytic secession (Seifert et al. 2011, Machouart et al. 2014, Crous et al. 2014, Huanraluek et al. 2019). Sympoventuriaceae is mainly known as a ubiquitous environmental saprobic fungus and plant endophytes or pathogens, while a few species have been documented as opportunistic neurotropic pathogens in vertebrate hosts, including humans (Satow et al. 2008, Seyedmousavi et al. 2014, Kidd et al. 2016, Zhang et al. 2018, Samerpitak et al. 2019, Benavent 2021, Murata et al. 2022). They are also known for their thermophilic properties, such as living in hot springs (Revankar & Sutton 2010, Hao et al. 2013, Samerpitak et al. 2014, 2015b, Wang et al. 2018, Crous et al. 2020).
Sympoventuriaceae was introduced by Zhang et al. (2011) with Sympoventuria (type genus), Veronaeopsis and fusicladium-like species included. The generic organization of sequestrate taxa within the Sympoventuriaceae has long been a subject of debate, due to a high level of morphological plasticity, and the lack of molecular data (Machouart et al. 2014, Samerpitak et al. 2016). Sympoventuria was first described for a venturia-like ascomycete, typified by S. capensis, a species found on decaying leaves of Eucalyptus, which was characterised by its saprobic lifestyle, pseudoparaphyses, and hyaline, symmetrical ascospores and subcylindrical asci (Crous et al. 2007a, b). Veronaeopsis was introduced as a monotypic genus for V. simplex, which was previously separated from Veronaea based on its shorter conidiophores, geniculate rachis and prominent conidiogenous loci (Papendorf 1969, Arzanlou et al. 2007). Morphologically, Sympoventuria is allied to Venturia (Sivanesan 1977, Zhang et al. 2011, Zhang et al. 2016), although their asexual morphs are quite distinct. For instance, Fusicladium (asexual morph of Venturia) was established by Bonorden (1851) to accommodate F. virescens, a well-known pathogen of pears. Fusicladium is characterised by sympodial conidiogenesis, differentiated conidiophores, and melanized conidia with dark basal scars. However, the taxonomy of this genus has continued to be controversial (Baldacci & Ciferri 1937, Schubert et al. 2003, Beck et al. 2005, Koukol 2010). Shen et al. (2020) resolved Fusicladium as asexual morph of Venturia, but also introduced several additional fusicladium-like genera, namely Fuscohilum, Neofusicladium, Parafusicladium and Pinaceicola.
Since its introduction, several genera have either been included or excluded from Sympoventuriaceae, and many mycologists commented that there might be more unrecognized genera within the family (Machouart et al. 2014, Samerpitak et al. 2016). Scolecobasidium (= Ochroconis) and Verruconis are very similar genera that have sympodial conidiogenous cells and T- or Y-shaped to cylindrical or clavate conidia (Abbott 1927, De Hoog & Von Arx 1973). Due to an unusual combination of morphological and ecological characters, their systematic position has historically been controversial. Samerpitak et al. (2014) distinguished these genera based on their ecological and physiological traits and morphological differences. They accommodated mesophilic species with smooth-walled to verruculose conidia in Scolecobasidium (as Ochroconis), and retained the thermophilic taxa with verrucose to coarsely ornamented conidia in Verruconis. Nevertheless, morphological and ecological delimitation of Scolecobasidium and Verruconis is problematic and remains obscure (Samerpitak et al. 2016, Qiao et al. 2019). Acroconidiellina was introduced by Ellis (1971) to accommodate A. arecae, A. chloridis, A. loudetiae (type species) and A. urtiagae. Hernández-Restrepo et al. (2016) further pointed out that Acroconidiellina is allied to the Scolecobasidium/ Ochroconis complex, and belonged to Sympoventuriaceae (Li et al. 2016, Wijayawardene et al. 2020). Furthermore, although Acroconidiellina currently contains four species, the taxonomic placement of only A. arecae has thus far been confirmed based on phylogenetic studies. The monotypic genus Mycosisymbrium was proposed by Carris (1994), re-described by Pratibha & Prabhugaonkar (2016), initially regarded as incertae sedis in the Pezizomycotina, and later placed in Sympoventuriaceae. Following these studies, three interesting genera, Echinocatena, Matsushimaea and Yunnanomyces were analysed phylogenetically suggesting a close relationship to Sympoventuriaceae, each of which formed a monophyletic clade with other genera in this family (Crous et al. 2018a, b, Tibpromma et al. 2018). Pseudosigmoidea (typified by P. cranei) was introduced based on species of Sigmoidea with enteroblastic conidia and phialidic conidiogenesis (Ando & Nakamura 2000), and subsequent studies showed that it also resided in Sympoventuriaceae (Diene et al. 2013, Crous et al. 2019a). Hernández-Restrepo et al. (2020) reassessed the taxonomic placement of Melnikomyces to accommodate an increasing number of emerging species, and placed it in Sympoventuriaceae, together with other genera producing septate conidia from denticulate conidiogenous cells (Crous et al. 2014, Wei et al. 2020).
The classification of Sympoventuriaceae includes a wide range of taxa based on morphological characters, although these are chiefly asexual genera (Zhang et al. 2011, Machouart et al. 2014). However, some genera (e.g., Clavatispora, Neocoleroa) of Sympoventuriaceae were established only based on their sexual morphology, and very few links between sexual and asexual morphs have been confirmed. Sympoventuria capensis was introduced with both a sexual and asexual morph (Crous et al. 2007a, Machouart et al. 2014). Subsequently, Boonmee et al. (2014) introduced Clavatispora based on the sexual morph C. thailandica. Its unique ascospores and bitunicate asci resemble Pleosporales species but differ from most other taxa in Venturiales (Seifert et al. 2011, Hyde et al. 2013, 2020). In addition, the phylogenetic analysis of conserved genes (nuSSU, nuLSU, mtSSU and rpb2) indicated that Verruconis is distinct from Scolecobasidium, while several related sexual morphs were also included in the Sympoventuriaceae (Machouart et al. 2014). Another sexual genus, Neocoleroa (typified by N. sibirica), is characterised by lobed to dichotomously branched, blunt-tipped setae on superficial pseudothecia (Petrak 1934, Johnston & Park 2016). Morphologically, Neocoleroa is most comparable to Wentiomyces (Koorders 1907), and they have had a tangled taxonomic history (Barr 1997, Kirk et al. 2008). It is noteworthy, except for a few species of Clavatispora, Neocoleroa, Scolecobasidium, Sympoventuria and Verruconis, that the sexual morphs of most species of the Sympoventuriaceae are unknown. Moreover, the asexual and sexual morphs of Sympoventuriaceae often develop separately, or only one morph is formed, making it difficult to confirm links between morphs of the same species.
In summary, Sympoventuriaceae has been extensively reviewed in recent years in efforts to clarify the phylogeny and taxonomic relationships of its species and allied fungi, and has resulted in a modern redefinition of the family, which provides a solid foundation to facilitate future DNA phylogenetic studies (Tibpromma et al. 2018, Crous et al. 2019a, Shen et al. 2020). In spite of this, however, many questions remain unresolved about the phylogenetic relationships of some poorly documented taxa, especially genera and species for which molecular data are not yet available. This has justified an urgent need to reconsider the species boundaries for Sympoventuriaceae based on a robust family-wide phylogenetic backbone and framework. Furthermore, Sympoventuriaceae includes approximately 164 species, is a morphologically and ecologically diverse fungal group with different lifestyles and modes of nutrition (MycoBank, April 2022). In order to adapt to changing environmental conditions, their ecological habitat varies from saprobic, animal or human opportunistic and plant pathogens to extremophilic species (thermophilic fungi) (Samerpitak et al. 2019, Benavent 2021, Murata et al. 2022). Apparently, these fungi have evolved different lifestyles to exploit their environment, suggesting that adaptive radiations within Sympoventuriaceae was most likely driven by the ecological diversity (Martin et al. 2016, Haridas et al. 2020). Thus, to know more about the evolutionary importance of this feature, a more detailed study on the co-evolutionary history of this fungal group and its association with the environment is necessary, to elucidate the origin of this family and understand the evolutionary patterns of its lifestyles.
In this study, seven DNA barcodes (SSU, ITS, LSU, act1, tub2, tef1 and rpb2) were sequenced for 33 strains representing Guizhoumyces (two isolates), Matsushimaea (one isolate), Mycosisymbrium (one isolate), Scolecobasidium (27 isolates) and Verruconis (two isolates). In addition, a multi-locus phylogenetic analysis was performed including 143 taxa of Sympoventuriaceae, and ancestral character states of Sympoventuriaceae were reconstructed. Our specific goals were as follows:
i determine the taxonomic position of newly collected strains based on morphological and molecular evidence;
ii provide a revised phylogram for Sympoventuriaceae;
iii clarify the phylogenetic relationship between Scolecobasidium and Verruconis and other similar genera; and
iv to reconstruct the ancestral state and clarify the life strategies during the evolutionary history of Sympoventuriaceae.
MATERIALS AND METHODS
Fungal materials and isolation
The soil, plant and forest litter were collected from China. Each sample or specimen was separately stored in a zip-lock bag or envelope before returning to the laboratory for isolation. Strains were isolated by dilution plate and single spore isolation methods, and subcultured on 2 % potato dextrose agar (PDA) (Crous et al. 2019b). In the present study, 33 strains from 18 species of Scolecobasidium (13 species) and the closely related genera Guizhoumyces (one species), Matsushimaea (one species), Mycosisymbrium (one species) and Verruconis (two species) were collected. The samples include five of the 22 recognised genera in Sympoventuriaceae plus one genus newly described here. The holotype specimens were deposited in the Herbarium of the Department of Plant Pathology, Agricultural College, Guizhou University (HGUP). The ex-type cultures are conserved in the Culture Collection of the Department of Plant Pathology, Agriculture College, Guizhou University, China (GUCC) and the China General Microbiological Culture Collection Center (CGMCC).
DNA extraction, amplification and sequencing
Genomic DNA was extracted after 7 d from fresh mycelial cultures grown on PDA. Approximately 50 mg mycelium was scraped off the surface of the medium and transferred to a 1.5 mL microcentrifuge tube. The DNA was extracted using the BIOMIGA Fungus Genomic DNA Extraction Kit GD2416 (Biomiga, USA) following the manufacturer’s instructions. The partial nucleotide and protein coding genes were subjected to PCR amplification and sequencing of internal transcribed spacer regions and the intervening 5.8S rRNA gene (ITS) of the rDNA operon, 28S rRNA gene (LSU), 18S ribosomal RNA (SSU), actin gene (act1), translation elongation factor 1-alpha (tef1), RNA polymerase II second largest subunit (rpb2) and β-tubulin (tub2). For primers and conditions see Table 1. The PCR products were purified and sequenced by Sangon Biotech, and both directions were sequenced to ensure accuracy. The newly generated sequences in this study were deposited in GenBank, the alignments in TreeBASE (Submission ID S29226), and all the sequences used for phylogenetic analysis are shown in Table 2 and 3.
Table 1.
Primers and PCR conditions.
| Genes | Primers | Sequences / PCR conditions | References |
|---|---|---|---|
| ITS | ITS5 (Fw) | 5′-GGAAGTAAAAGTCGTAACAAGG-3′ | White et al. (1990) |
| ITS4 (Rw) | 5′-TCCTCCGCTTATTGATATGC-3′ | White et al. (1990) | |
| 95 °C 5 min, (95 °C 35 s, 56 °C 30 s, 72 °C 1 min) 35 cycles, 72 °C 4 min | |||
| LSU | LR0R (Fw) | 5′-ACCCGCTGAACTTAAGC-3′ | Vilgalys & Hester (1990) |
| LR5 (Rw) | 5′-TCCTGAGGGAAACTTCG-3′ | Vilgalys & Hester (1990) | |
| 95 °C 5 min, (95 °C 45 s, 56 °C 40 s, 72 °C 2 min) 35 cycles, 72 °C 10 min | |||
| SSU | NS1 (Fw) | 5′-GTAGTCATATGCTTGTCTC-3′ | White et al. (1990) |
| NS24 (Rw) | 5′-AAACCTTGTTACGACTTTTA-3′ | Gargas & Taylor (1992) | |
| 95 °C 5 min, (95 °C 45 s, 56 °C 40 s, 72 °C 2 min) 35 cycles, 72 °C 10 min | |||
| act1 | 512 (Fw) | 5′-ATGTGCAAGGCCGGTTTCGC-3′ | Carbone & Kohn (1999) |
| 783 (Rw) | 5′-TACGAGTCCTTCTGGCCCAT-3′ | Carbone & Kohn (1999) | |
| 95 °C 5 min, (96 °C 45 s, 56 °C 30 s, 72 °C 1 min) 35 cycles, 72 °C 5 min | |||
| tub2 | Bt2a (Fw) | 5-GGTAACCAAATCGGTGCTGCTTTC-3 | Glass & Donaldson (1995) |
| Bt2b (Rw) | 5′-ACCCTCAGTGTAGTGACCCTTGGC-3′ | Glass & Donaldson (1995) | |
| 95 °C 5 min, (95 °C 35 s, 56 °C 50 s, 72 °C 2 min) 35 cycles, 72 °C 7 min | |||
| tef1 | 728 (Fw) | 5′-CATCGAGAAGTTCGAGAAGG-3′ | Carbone & Kohn (1999) |
| 986 (Rw) | 5′-TACTTGAAGGAACCCTTAC-3′ | Carbone & Kohn (1999) | |
| 983 (Fw) | 5′-GCYCCYGGHCAYCGTGAYTTYAT-3′ | Rehner & Buckley (2005) | |
| 2218 (Fw) | 5′-ATGACACCRACRGCRACRGTYTG-3′ | Rehner & Buckley (2005) | |
| 95 °C 5 min, (96 °C 45 s, 56 °C 30 s, 72 °C 45 s) 35 cycles, 72 °C 5 min | |||
| rpb2 | 5 (Fw) | 5′-GAYGAYMGWGATCAYTTYGG-3′ | Liu et al. (1999) |
| 7CR (Rw) | 5′-CCCATRGCTTGYTTRCCCAT-3′ | Liu et al. (1999) | |
| 95 °C 5 min, (96 °C 45 s, 56 °C 30 s, 72 °C 2 min) 35 cycles, 72 °C 5 min |
Table 2.
Strains used in the phylogenetic analysis of Sympoventuriaceae and GenBank accession numbers.
| Species | Strain1 | Host and substrate | Locality | GenBank accession numbers2 | ||||
|---|---|---|---|---|---|---|---|---|
| ITS | LSU | tub2 | tef1 | rpb2 | ||||
| Acroconidiellina arecae | NFCCI 3696 | On little patches on the leaves of Areca Catechu | India | KX306747 | KX306776 | – | – | – |
| Bellamyces quercus | CBS 46217* | Lecanora chlarotera on Quercus trunks | UK | MK810901 | MK810788 | – | MK888726 | MK887796 |
| Clavatispora thailandiaca | MFLUCC 100107 | On dead stems of herbaceous plants | Thailand | MH065721 | KF770458 | – | KF770459 | – |
| Echinocatena arthrinioides | CBS 144202 | Acacia crassicarpa, leaves | Malaysia | MH107890 | MH107937 | – | – | – |
| Fuscohilum rhodensis | CBS 121641* | Ceratonia siliqua, branches | Greece | MK810909 | MK810796 | MK926471 | MK888733 | MK887802 |
| Fu. siciliana | CBS 105.85* | Chamaerops humilis | Italy | MK810910 | MK810797 | MK926472 | MK888734 | MN091924 |
| Guizhoumyces aciculaea | GUCC 18195* | Isolated from soil | China | MZ503724 | MZ503757 | MZ546903 | MZ546870 | MZ546866 |
| GUCC 18152 | From leaf litter | China | MZ503723 | MZ503756 | MZ546902 | MZ546869 | MZ546865 | |
| Helicopsis olivaceum | CBS 728.83 | Dicksonia antarctica, dead petiole | Australia | MH861681 | MH873393 | – | – | – |
| Matsushimaea fasciculata | CBS 167.97* | On dead leaf of Cinnamomum japonicum | Japan | LT962397 | LT962402 | – | – | – |
| GUCC 18239 | Isolated from soil | China | MZ503725 | MZ503758 | MZ546904 | MZ546871 | MZ546867 | |
| Ma. monilioides | CBS 143867* | Garden soil | Spain | LT883468 | LT883469 | – | – | – |
| Melnikomyces longisporum | HUGP 18226* | From forest litter | China | MT731290 | MT731291 | MT739515 | MT739516 | – |
| Me. thailandicus | CBS 145767* | Isolated from soil | Thailand | MN794374 | MN794351 | – | – | – |
| Me. vietnamensis | CBS 136209* | On dry leaves of broadleaved tree | Vietnam | KJ869156 | KJ869213 | – | – | – |
| Mycosisymbrium cirrhosum | MTCC12435 | On dead leaves of Vaccinium macrocarpon | United States | KR259883 | KR259884 | – | – | KR349124 |
| GUCC 1837 | Isolated from decaying Camellia sinensis leaf litter | China | MZ503722 | MZ503755 | MZ546901 | MZ546868 | MZ546864 | |
| Neocoleroa cameroonensis | CBS 129041* | Crematogaster sp. (ant) carton on Barteria nigritana | Cameroon | MK810902 | MK810789 | MN078219 | MK888727 | MK887797 |
| Nc. metrosideri | ICMP 21139* | On living leaves of Metrosideros excelsa | New Zealand | KU131678 | KU131677 | – | – | – |
| Neofusicladium eucalypti | CBS 128216* | Eucalyptus regnans, leaf litter | Australia | MK810903 | MK810790 | MK926468 | MK888728 | MK887798 |
| Nf. eucalypticola | CBS 141301* | Eucalyptus robusta, leaf litter | France | MK810904 | MK810791 | – | MK888729 | MK887799 |
| CBS 143427 | Eucalyptus dunnii, leaves | Australia | MK810905 | MK810792 | – | – | – | |
| Nf. regnans | CBS 143411* | Eucalyptus regnans, leaves | Australia | MG386066 | MG386119 | MG386169 | – | – |
| CBS 144605 | On leaves of Eucalyptus pauciflora (Myrtaceae) | Australia | MK442628 | MK442563 | MK442748 | MK442722 | – | |
| Parafusicladium amoenum | CBS 254.95* | Eucalyptus sp., fallen leaves | Cuba | MK810906 | MK810793 | MK926469 | MK888730 | – |
| Pa. intermedium | CBS 110746* | Eucalyptus sp., leaf litter | Madagascar | MK810907 | MK810794 | MK926470 | MK888731 | MK887800 |
| Pa. paraamoenum | CBS 141322* | Eucalyptus regnans, leaf litter | Australia | MK810908 | MK810795 | – | MK888732 | MK887801 |
| Pinaceicola cordae | Pinus sylvestris, litter needles | Czech Republic | MK810911 | MK810798 | MK926473 | MK888735 | – | |
| CBS 675.82 | Pinus sylvestris, litter needles | Netherlands | MK810912 | MK810799 | MK926474 | MK888736 | – | |
| CBS 143494 | Pinus sylvestris, litter needles | Germany | MK810913 | MK810800 | MK926475 | MK888737 | – | |
| Pi. pini | CBS 463.82* | Pinus sylvestris, litter needles | Netherlands | MK810915 | MK810802 | MK926477 | MK888739 | MK887804 |
| CBS 462.82 | Pinus sp., litter needles | Netherlands | MK810914 | MK810801 | MK926476 | MK888738 | MK887803 | |
| Pseudosigmoidea alnicola | CBS 145034* | Leaf litter of Alnus glutinosa (Betulaceae) | Germany | MK442620 | MK442556 | – | – | – |
| Ps. excentrica | CBS 469.95* | Lauraceae, leaf litter | Cuba | HQ667543 | KF282669 | MK926478 | KF155975 | – |
| Ps. ibarakiensis | NBRC 107891* | Natural forest soil | Japan | LC146758 | LC146759 | – | – | – |
| Scolecobasidium anellii | CBS 284.64* | Stalactite | Italy | FR832477 | KF156138 | KF156184 | KF155995 | KF282684 |
| Sc. anomala | CBS 131816* | Lascaux Cave | France | HE575201 | KF156137 | KF156194 | KF155986 | HE575205 |
| Sc. blechni | CBS 146055* | Leaves of Blechnum capense (Blechnaceae) | South Africa | MN562134 | MN567641 | MN556843 | MN556826 | – |
| Sc. constricta | CBS 211.53* | Soil | Canada: Ontario | HQ667519 | KF156148 | KF156187 | KF156005 | KF282686 |
| Sc. crassihumicola | CBS 120700 | Soil | Papua New Guinea | KJ867429 | KJ867430 | KJ867433 | KJ867428 | – |
| Sc. gamsii | CBS 239.78* | Caryota plumosa, leaf | Sri Lanka | KF156019 | KF156150 | KF156190 | KF155982 | – |
| Sc. icarus | CBS 536.69* | Forest soil | Canada: Ontario | HQ667524 | KF156132 | KF156174 | KF156009 | KF282700 |
| Sc. lascauxensis | CBS 131815* | Black stain on cave sediment | France | FR832474 | KF156136 | KF156183 | KF155994 | FR832481 |
| Sc. longiphorum | CBS 435.76* | In excrement of Insecta, and Quercus | Japan | KF156038 | KF156135 | KF156182 | KF155978 | – |
| Sc. musicola | CBS 144441* | On leaves of Musa sp. (Musaceae) | Malaysia | MH327824 | MH327860 | MH327898 | MH327887 | MH327876 |
| Sc. phaeophora | CBS 206.96* | Leaf in coastal rain forest | Papua New Guinea | KP798631 | KP798634 | KT272062 | KT272098 | KF282692 |
| Sc. tshawytschae | CBS 100438* | On young Oncorhynchus tshawytscha | USA: California | HQ667562 | KF156126 | KF156180 | KF155990 | KF282697 |
| Sc. verrucosa | CBS 383.81* | From soil | India: Kerala | KF156015 | KF156129 | KF156185 | KT272099 | – |
| Sterila eucalypti | CBS 144019* | Eucalyptus sp. | Portugal | MK810918 | MK810805 | – | MK888742 | MK887807 |
| CPC 14942 | Eucalyptus sp. | Portugal | MK810916 | MK810803 | – | MK888740 | MK887805 | |
| CPC 14943 | Eucalyptus sp. | Portugal | MK810917 | MK810804 | – | MK888741 | MK887806 | |
| Sympoventuria capensis | CBS 120136* | On leaf litter of Eucalyptus sp. (Myrtaceae) | South Africa | MK810921 | MK810808 | MK926481 | MK888745 | MK887810 |
| CPC 12839 | On leaf litter of Eucalyptus sp. (Myrtaceae) | South Africa | MK810922 | MK810809 | MK926482 | MK888746 | MK887811 | |
| CPC 12840 | On leaf litter of Eucalyptus sp. (Myrtaceae) | South Africa | MK810923 | MK810810 | MK926483 | MK888747 | MK887812 | |
| Sy. melaleucae | CBS 143407* | Melaleuca sp., leaves | Australia | MG386059 | MG386112 | MG386168 | – | – |
| Troposporella fumosa | CBS 351.94 | On the old bark of Populus tremula (Salicaceae) | Finland | MK810924 | MH874121 | – | – | – |
| Tr. monilipes | MUCL 19867 | On decayed wood of Quercus (Fagaceae) | United States | DQ351723 | AY856871 | – | – | – |
| Veronaeopsis simplex | CBS 588.66* | Acacia karroo, leaf litter | South Africa | EU041820 | EU041877 | – | – | MN091925 |
| Verruconis gallopava | CBS 437.64* | Meleagris gallopavo, brain abscess | United States | HQ667553 | KF156112 | KF156203 | KF155968 | KF282689 |
| Ve. mangrovei | NFCCI-4390* | On decaying wood of Excoecaria agallocha | India | MN782361 | MN241144 | MN848140 | – | – |
| Ve. panacis | CBS 142802* | From the root of Panax notoginseng | China | MF536882 | MF536880 | MF536883 | MF536881 | – |
| Ve. pseudotricladiata | YMF1.04915* | Leaves of a broad-leaf species in a stream | China | MK244396 | MK248270 | MK253013 | MK248273 | – |
| Ve. terricola | CBS 131795* | Isolated from soil | China | MK810925 | MK810811 | – | – | KC337072 |
| Ve. verruculosa | CBS 119775* | Grassland soil | India | KF156014 | KF156106 | KF156193 | KF155974 | – |
| Yunnanomyces pandanicola | MFLUCC 17-2260* | On decaying leaves of Pandanus amaryllifolius | China | MH388369 | MH376743 | – | MH388403 | MH412736 |
| Yu. phoenicis | MFLUCC 19-0254* | On fallen rachides and leaves of Phoenix paludosa | Thailand | – | MK976738 | – | MK986486 | MK986484 |
| MFLUCC 19-0253 | On fallen rachides and leaves of Phoenix paludosa | Thailand | – | MK976737 | – | MK986485 | MK986483 | |
| Tyrannosorus lichenicola | CBS 144018* | Letharia sp. | USA | MK810953 | MK810838 | MK926509 | MK888775 | MK887840 |
| Venturia saliciperda | CBS 480.61* | Salix cordata | Switzerland | MK811007 | MK810891 | MK926558 | MK888825 | MK887886 |
1 CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; CPC: Culture collection of Pedro Crous, housed at Westerdijk Fungal Biodiversity Institute; CGMCC: Chinese General Microbiological Culture Collection Center, Beijing, China; GUCC: Culture Collection of the Department of Plant Pathology, Agriculture College, Guizhou University, China; HGUP: Herbarium of the Department of Plant Pathology, Agricultural College, Guizhou University, China; ICMP: International Collection of Micro-organisms from Plants, Landcare Research, Private Bag 92170, Auckland, New Zealand; IRAN: Fungal Culture Collections of the Iranian Research Institute of Plant Protection; MFLU (CC): Mae Fah Luang University Culture Collection, Chiang Ria, Thailand; MUCL: Universite Catholique de Louvain, Louvain-la-Neuve, Belgium. MTCC: Institute of Microbial Technology, Chandigarh, India; NBRC: Biological Resource Center; NFCCI: National Fungal Culture Collection of India, Pune, India.
2 ITS: internal transcribed spacers and intervening 5.8S nrDNA; LSU: partial large subunit (28S) nrRNA gene; SSU: partial small subunit (18S) nrRNA gene; act1: actin; tub2: partial β-tubulin gene; tef1: partial translation elongation factor 1-alpha gene; rpb2: partial DNA-directed RNA polymerase II second largest subunit gene. Accession numbers of sequences generated in this study are in bold; – indicates unavailable sequences or unknown collection data.
* Ex-holotype or ex-type strains.
Table 3.
Strains used in the phylogenetic analysis of Scolecobasidium and Verruconis and GenBank accession numbers.
| Species | Strain 1 | Host and substrate | Locality | GenBank accession numbers 2 | |||||
|---|---|---|---|---|---|---|---|---|---|
| SSU | ITS | LSU | act1 | tub2 | tef1 | ||||
| S. ailanthi | MFLUCC 17-0923* | On fallen pod of Ailanthus sp. | Thailand | MK347838 | MK347730 | MK347947 | MK412893 | MK412883 | – |
| MFLU 18-2110 | On fallen pod of Ailanthus sp. | Thailand | MK347839 | MK347731 | MK347948 | MK412892 | MK412881 | – | |
| S. anellii | CBS 284.64* | Stalactite | Italy | KF156070 | FR832477 | KF156138 | KF155912 | KF156184 | KF155995 |
| S. anomalum | CBS 131816* | Lascaux Cave | France | KF156065 | HE575201 | KF156137 | KF155935 | KF156194 | KF155986 |
| S. aquaticum | CBS 140316* | Silicone | Germany | KX668260 | KX668258 | KX668259 | – | – | – |
| S. bacilliforme | CBS 100442* | On stainless steel biofilm in drinking water | Germany | KP798638 | KP798632 | KP798635 | KT272051 | KT272059 | KT272070 |
| S. blechni | CBS 146055* | Leaves of Blechnum capense | South Africa | – | MN562134 | MN567641 | – | MN556843 | MN556826 |
| S. camellicola | GUCC 18242* | On decaying Camellia sinensis leaf litter | China | MZ503654 | MZ503728 | MZ503761 | MZ546837 | MZ546907 | MZ546874 |
| GUCC 18243 | On decaying Camellia sinensis leaf litter | China | MZ503655 | MZ503729 | MZ503762 | MZ546838 | MZ546908 | MZ546875 | |
| GUCC 18244 | Isolated from forest litter | China | MZ503656 | MZ503730 | MZ503763 | MZ546839 | MZ546909 | MZ546876 | |
| S. capsici | CBS 142096* | Leaf of Capsicum annuum | Thailand | – | KY173427 | KY173518 | – | – | – |
| S. coiledmyces | GUCC 18245* | Isolated from lawn soil | China | MZ503657 | MZ503731 | MZ503764 | MZ546840 | MZ546910 | MZ546877 |
| S. constrictum | CBS 211.53* | Soil | Canada: Ontario | KF156073 | HQ667519 | KF156148 | KF155941 | KF156187 | KF156005 |
| CBS 131913 | Human, cutaneous mycosis | Thailand | KF156071 | KF156025 | KF156146 | KF155940 | KF156176 | KF156006 | |
| GUCC 18255 | Isolated from dead branches | China | MZ503667 | MZ503741 | MZ503774 | MZ546850 | MZ546920 | MZ546887 | |
| GUCC 18256 | Isolated from forest litter | China | MZ503668 | MZ503742 | MZ503775 | MZ546851 | MZ546921 | MZ546888 | |
| GUCC 18257 | Isolated from soil | China | MZ503669 | MZ503743 | MZ503776 | MZ546852 | MZ546922 | MZ546889 | |
| GUCC 18258 | Isolated from lawn soil | China | MZ503670 | MZ503744 | MZ503777 | MZ546853 | MZ546923 | MZ546890 | |
| S. cordanae | CBS 475.80* | Mauritia minor , leaf litter | Colombia | KF156058 | KF156022 | KF156122 | HQ916976 | KF156197 | KF155981 |
| CBS 412.51 | Not available | United States | KF156056 | HQ667540 | KF156123 | KF155907 | KF156200 | KF155980 | |
| S. crassihumicola | CBS 120700 | Soil | Papua New Guinea | KJ867431 | KJ867429 | KJ867430 | KJ867427 | KJ867433 | KJ867428 |
| S. dracaenae | CBS 141323* | Leaf spots of Dracaena reflexa | USA | – | KX228283 | KX228334 | – | – | KX228377 |
| S. echinulatum | GUCC 18247* | Isolated from soil | China | MZ503659 | MZ503733 | MZ503766 | MZ546842 | MZ546912 | MZ546879 |
| GUCC 18248 | Isolated from soil | China | MZ503660 | MZ503734 | MZ503767 | MZ546843 | MZ546913 | MZ546880 | |
| S. ellipsoideum | CBS 131796* | Soil | China | – | MN077367 | – | – | – | – |
| GUCC 18264 | Isolated from soil | China | MZ503676 | MZ503750 | MZ503783 | MZ546859 | MZ546929 | MZ546896 | |
| GUCC 18265 | Isolated from submerged wood | China | MZ503677 | MZ503751 | MZ503784 | MZ546860 | MZ546930 | MZ546897 | |
| GUCC 18266 | Isolated from forest litter | China | MZ503678 | MZ503752 | MZ503785 | MZ546861 | MZ546931 | MZ546898 | |
| S. ferulica | IRAN3232C* | Root of Ferula ovina | Iran | – | MF186874 | MH400207 | – | – | – |
| S. gamsii | CBS 239.78* | Caryota plumosa , leaf | Sri Lanka | KF156088 | KF156019 | KF156150 | KF155936 | KF156190 | KF155982 |
| S. globale | CBS 119644* | Indoor sample, house | Germany | KF961108 | KF961086 | KF961097 | KF956086 | KF961065 | KF961075 |
| CBS 135924 | Bathroom; black biofilm, sink drain | Germany | KF961107 | KF961092 | KF961104 | KF956092 | KF961070 | KF961079 | |
| GUCC 18249 | From forest humus | China | MZ503661 | MZ503735 | MZ503768 | MZ546844 | MZ546914 | MZ546881 | |
| GUCC 18250 | From soil | China | MZ503662 | MZ503736 | MZ503769 | MZ546845 | MZ546915 | MZ546882 | |
| S. guangxiensis | SS23* | Soil and sugarcane root | China | MK929277 | MK934570 | MK956169 | – | – | – |
| X22 | Soil and sugarcane root | China | MK961269 | MK961215 | MK961247 | – | – | – | |
| S. helicteris | NFCCI 4310* | Living leaves of Helicteris isora | India | – | MK014833 | – | – | MK321318 | – |
| S. humicola | CBS 116655* | Peat soil | Canada: Ontario | KF156068 | HQ667521 | KF156124 | KF155904 | KF156195 | KF155984 |
| S. icarus | CBS 536.69* | Forest soil | Canada: Ontario | KF156084 | HQ667524 | KF156132 | KF155944 | KF156174 | KF156009 |
| CBS 423.64 | Rhizosphere | Netherlands | KF156085 | HQ667523 | KF156131 | KF155943 | KF156173 | KF156008 | |
| S. lascauxense | CBS 131815* | Black stain on cave sediment | France | KF156069 | FR832474 | KF156136 | KF155911 | KF156183 | KF155994 |
| S. leishanicola | HGUP 1808* | Soil | China | MK377071 | MK377301 | MK377073 | – | – | – |
| GUCC 18259 | Isolated from soil | China | MZ503671 | MZ503745 | MZ503778 | MZ546854 | MZ546924 | MZ546891 | |
| S. longiphorum | CBS 435.76* | In excrement of Insecta , and Quercus | Japan | KF156060 | KF156038 | KF156135 | KF155908 | KF156182 | KF155978 |
| S. macrozamiae | CBS 137971* | Macrozamia , leaf litter | Australia | – | KJ869123 | KJ869180 | – | – | – |
| CBS 102491 | On leaf litter of Macrozamia | Australia | KF156092 | KF156021 | KF156152 | KF155938 | KF156191 | KF155983 | |
| S. minimum | CBS 510.71* | Gossypium arboreum , rhizosphere | Nigeria | KF156087 | HQ667522 | KF156134 | KF155945 | KF156172 | KF156007 |
| CBS 119792 | Soil | India | KF156086 | KF156027 | KF156133 | KF155946 | KF156175 | KT272073 | |
| GUCC 18260 | From forest humus | China | MZ503672 | MZ503746 | MZ503779 | MZ546855 | MZ546925 | MZ546892 | |
| S. mirabilis | CBS 413.51* | Breathing regulator for diver | Netherlands | KF156076 | HQ667536 | KF156140 | KF155957 | KF156164 | KF156001 |
| S. musae | CBS 729.95* | Regulator of diver | Netherlands | KF156082 | KF156029 | KF156144 | KF155948 | KF156171 | KF155999 |
| S. musicola | CBS 144441* | On leaves of Musa sp. (Musaceae ) | Malaysia | – | MH327824 | MH327860 | – | MH327898 | MH327887 |
| S. obovoideum | GUCC 18246* | Isolated from forest litter | China | MZ503658 | MZ503732 | MZ503765 | MZ546841 | MZ546911 | MZ546878 |
| S. olivaceum | CBS 137170* | Man, bronchoalveolar lavage fluid | USA: Utah | LM644548 | LM644521 | LM644564 | LM644600 | LM644605 | KT272067 |
| S. pandanicola | CBS 140660* | Pandanus utilis , leaves | France | – | KT950850 | KT950864 | – | – | – |
| S. phaeophorum | CBS 206.96* | Leaf in coastal rain forest | Papua New Guinea | KP798637 | KP798631 | KP798634 | KT272054 | KT272062 | KT272098 |
| S. podocarpi | CBS 143174* | Podocarpus grayae , leaves | Australia | – | MG386032 | MG386085 | – | – | MG386162 |
| S. podocarpicola | CBS 146057* | Leaves of Podocarpus latifolius | South Africa | – | MN562138 | MN567645 | – | – | – |
| S. ramosum | CBS 137173* | Isolated from nail of Homo sapiens | USA: California | LM644551 | LM644524 | LM644567 | LM644603 | LM644608 | KT272069 |
| CBS 137171 | Skin | United States | LM644549 | LM644522 | LM644565 | LM644601 | LM644606 | KT272068 | |
| GUCC 18261 | From forest litter | China | MZ503673 | MZ503747 | MZ503780 | MZ546856 | MZ546926 | MZ546893 | |
| GUCC 18262 | From soil | China | MZ503674 | MZ503748 | MZ503781 | MZ546857 | MZ546927 | MZ546894 | |
| GUCC 18263 | From forest litter | China | MZ503675 | MZ503749 | MZ503782 | MZ546858 | MZ546928 | MZ546895 | |
| S. robustum | CBS 112.97* | Leaf litter of Ouercus ilex | Spain | KP798639 | KP798633 | KP798636 | KT272052 | KT272060 | KT272071 |
| S. sexuale | CBS 135765* | Swabs in a laboratory | South Africa | KF156089 | KF156018 | KF156118 | KF155902 | KF156189 | KF155976 |
| CBS 131965 | Ant | Brazil | KF156090 | KF156017 | KF156119 | KF155903 | KF156188 | KF155977 | |
| S. terreum | CBS 203.27* | From soil | USA: Louisiana | – | HQ667544 | – | – | HQ877665 | – |
| S. tshawytschae | CBS 100438* | On young Oncorhynchus tshawytscha | USA: California | KF156062 | HQ667562 | KF156126 | KF155918 | KF156180 | KF155990 |
| CBS 228.66 | Peat-bog soil | Ireland | KF156064 | KF156016 | KF156128 | KF155915 | KF156179 | KF155992 | |
| GUCC 18251 | From lawn soil | China | MZ503663 | MZ503737 | MZ503770 | MZ546846 | MZ546916 | MZ546883 | |
| GUCC 18252 | From plant litter | China | MZ503664 | MZ503738 | MZ503771 | MZ546847 | MZ546917 | MZ546884 | |
| GUCC 18253 | From soil | China | MZ503665 | MZ503739 | MZ503772 | MZ546848 | MZ546918 | MZ546885 | |
| GUCC 18254 | From soil | China | MZ503666 | MZ503740 | MZ503773 | MZ546849 | MZ546919 | MZ546886 | |
| S. variabile | NBRC 32268 | From soil | Canada: Ontario | EU107353 | DQ307334 | EU107310 | – | – | DQ307356 |
| S. verrucaria | GUCC 18240* | From soil | China | MZ503652 | MZ503726 | MZ503759 | MZ546835 | MZ546905 | MZ546872 |
| S. verrucosum | CBS 383.81* | From soil | India: Kerala | KF156067 | KF156015 | KF156129 | KF155910 | KF156185 | KT272099 |
| S. zunyiense | GUCC 18241* | From forest litter | China | MZ503653 | MZ503727 | MZ503760 | MZ546836 | MZ546906 | MZ546873 |
| V. calidifluminalis | CBS 125818* | Water of a hot stream | Japan | KF156046 | AB385698 | KF156108 | KF155901 | KF156202 | KF155959 |
| CBS 125817 | Water of a hot stream | Japan | KF156045 | AB385699 | KF156107 | KF155900 | KF156201 | KF155958 | |
| V. cylindricalis | GUCC 18299* | From forest humus | China | MZ503680 | MZ503754 | MZ503787 | MZ546863 | MZ546933 | MZ546900 |
| V. gallopava | CBS 437.64* | Meleagris gallopavo , brain abscess | United States | KF156053 | HQ667553 | KF156112 | HQ916989 | KF156203 | KF155968 |
| CBS 118.91 | Man | United States | KF156047 | HQ667551 | KF282655 | KF155932 | HQ877643 | JF440539 | |
| CBS 867.95 | Sputum from patient with cardiac | United States | KF156051 | HQ667561 | KF282657 | KF155928 | KF156213 | KF155972 | |
| CBS 116660 | Human, transplantation | United States | KF156048 | HQ667557 | KF156115 | KF155929 | KF156206 | KF155969 | |
| V. hainanensis | YMF1.04165* | From leaves of a dicotyledonous plant | China | MK248267 | MK244397 | MK248269 | MK248271 | – | MK248272 |
| V. heveae | MFLUCC 17-0092* | On dried latex on bark of Hevea brasiliensis | Thailand | – | MH602349 | MH602348 | – | – | – |
| V. mangrovei | NFCCI-4390* | On decaying wood of Excoecaria agallocha | India | MN241147 | MN782361 | MN241144 | – | MN848140 | – |
| NFCCI-4391 | On decaying wood of Excoecaria agallocha | India | MN241148 | MN782362 | MN241145 | – | MN848141 | – | |
| V. panacis | CBS 142802* | From the root of Panax notoginseng | China | MF536879 | MF536882 | MF536880 | – | MF536883 | MF536881 |
| V. pseudotricladiata | YMF1.04915* | Leaves of a broad-leaf species in a stream | China | MK248268 | MK244396 | MK248270 | – | MK253013 | MK248273 |
| V. terricola | CBS 131795* | Isolated from soil | China | – | MK810925 | MK810811 | – | – | – |
| V. thailandica | CBS 145768* | From soil | Thailand | – | MN794375 | MN794352 | – | – | – |
| GUCC 18267 | Isolated from the humus soil in the stream | China | MZ503679 | MZ503753 | MZ503786 | MZ546862 | MZ546932 | MZ546899 | |
| V. tricladiata | NBRC 30208 | On rotten leaves | Bismarck Archipelago | EU107354 | – | EU107286 | – | – | DQ307352 |
| V. verruculosa | CBS 119775* | Grassland soil | India | KF156055 | KF156014 | KF156106 | KF155919 | KF156193 | KF155974 |
| Pseudosigmoidea excentrica | CBS 469.95* | Lauraceae , leaf litter | Cuba | KF156096 | HQ667543 | KF282669 | KF155934 | MK926478 | KF155975 |
| Sympoventuria capensis | CBS 120136* | Eucalyptus sp., leaf litter | South Africa | KF156094 | MK810921 | MK810808 | – | MK926481 | MK888745 |
1 CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; CPC: Culture collection of Pedro Crous, housed at Westerdijk Fungal Biodiversity Institute; CGMCC: Chinese General Microbiological Culture Collection Center, Beijing, China; GUCC: Culture Collection of the Department of Plant Pathology, Agriculture College, Guizhou University, China; HGUP: Herbarium of the Department of Plant Pathology, Agricultural College, Guizhou University, China; ICMP: International Collection of Micro-organisms from Plants, Landcare Research, Private Bag 92170, Auckland, New Zealand; IRAN: Fungal Culture Collections of the Iranian Research Institute of Plant Protection; MFLU (CC): Mae Fah Luang University Culture Collection, Chiang Ria, Thailand; MUCL: Universite Catholique de Louvain, Louvain-la-Neuve, Belgium. MTCC: Institute of Microbial Technology, Chandigarh, India; NBRC: Biological Resource Center; NFCCI: National Fungal Culture Collection of India, Pune, India.
2 ITS: internal transcribed spacers and intervening 5.8S nrDNA; LSU: partial large subunit (28S) nrRNA gene; SSU: partial small subunit (18S) nrRNA gene; act1: actin; tub2 : partial β-tubulin gene; tef1: partial translation elongation factor 1-alpha gene; rpb2: partial DNA-directed RNA polymerase II second largest subunit gene. Accession numbers of sequences generated in this study are in bold; – indicates unavailable sequences or unknown collection data.
3* Ex-holotype or ex-type strains.
Phylogenetic analyses
The concatenated DNA sequence dataset (SSU, ITS, LSU, act1, tub2, tef1 and rpb2) of 143 taxa was used to infer phylogenetic relationships among the new isolates and other taxa of Sympoventuriaceae. DNA sequence data were initially blast searched to determine the placement of new strains in Sympoventuriaceae. Multiple sequence alignments were carried out with MAFFT v. 7.4.9 (Rozewicki et al. 2019), and then rechecked and adjusted manually as necessary using BioEdit v. 7.1.9 (Hall 1999). The single gene datasets were combined using MEGA X (Kumar et al. 2018). Data were converted from fasta to nexus and phylip format with AliView v. 1.19 for RAxML, MrBayes and PAUP analysis (Larsson 2014). Finally, phylogenetic analyses for the individual data matrix and combined datasets were conducted by employing maximum likelihood (ML), maximum parsimony (MP) and Bayesian inference (BI).
The ML analyses used RAxML-HPC2 on XSEDE v. 8.2.12 (Stamatakis 2014) via the CIPRES Science Gateway platform (Miller et al. 2012). The GTRGAMMA model was chosen and ML bootstrap analyses were estimated with 1 000 replicates. Prior to Bayesian analysis, jModelTest v. 2.1.7 (Darriba et al. 2012) was used to select a best-fit model of nucleotide substitution for each data partition under the output strategy of Akaike information criterion (AIC) (Nylander 2004). The Bayesian posterior probabilities were determined by Markov Chain Monte Carlo sampling (MCMC) in MrBayes v. 3.2.7 (Ronquist et al. 2012). The six simultaneous Markov chains were run for 2 M generations, starting from random trees and sampling trees every 100th generation, and 25 % of ageing samples were discarded, running until the average standard deviation of the split frequencies dropped below 0.01. For MP, the dataset was analysed in PAUP v. 4.0b10 (Swofford 2003) using a heuristic search algorithm with 1000 random addition sequence replicates. One tree was saved at each step during stepwise addition, while tree bisection reconnection (TBR) was used to swap branches, and the maximum number of trees was set to 10000. The ambiguously aligned regions were eliminated and gaps were treated as missing data. The phylogenetic trees were visualised in FigTree v. 1.4.4 and edited using Adobe Illustrator CC 2020.
Estimating transitions in lifestyle evolution
Ancestral character states of the lifestyle (Table 2 and 3) were reconstructed with the Bayesian Binary Method (BBM) of RASP v. 4.2 (Yu et al. 2015, 2020). Because BBM analysis requires a set of phylogenetic trees and a consensus topology, we generated the phylogenetic trees via Bayesian phylogenetic analysis in BEAST v. 2.6.6 (Barido-Sottani et al. 2018) using six DNA loci (SSU, ITS, LSU, act1, tub2 and tef1). The length of the MCMC chain reaction was set as 500 M generations sampled every 100 000 generations; thus, a total of 5 000 trees were kept. Tracer v. 1.7.2 (Rambaut et al. 2018) was used to check that the values of the mean and ESS in the log file were over 200. After removal of a proportion of each run as burn-in, the remaining trees were summarised as maximum clade credibility (MCC) trees in TreeAnnotator v. 2.6.6 (Barido-Sottani et al. 2018). For BBM analysis, the Markov chains were run for 50 000 generations, using 10 chains, with a sample frequency of 100, a temperature of 0.1, state frequencies fixed (JC), and among-site rate variation equal. We subsequently examined the resulting reconstructions of the selected characters to determine if the lifestyle changes were arising convergently or resulted from shared ancestry.
Morphological observations
The microscopic features and colony characteristics of the putative novel and known species were examined. The macro-morphological characters and relevant data (colony colour and diameter, mycelium) of the isolates were examined under a dissecting microscope (Leica S9i, Germany), and images of colonies cultured on three media, malt extract agar (MEA), oatmeal agar (OA) (Crous et al. 2019b) and PDA were captured after 2 wk. The slide culture technique on OA was used for microscopic observation. If sterile on OA, morphological characters produced on other media were described. Measurements and descriptions of reproductive structures were taken from specimens mounted in lactic acid or lactophenol cotton blue. Micrographs were captured with an Olympus BX53 compound microscope. Tarosoft (R) Image Frame Work program was used to measure the lengths and widths of microscopic structures including conidiophores, conidiogenous cells, conidia and chlamydospores per isolate. At least 30 measurements were made for each microscopic structure to calculate the mean value, standard deviation, and minimum–maximum values, with the extreme measurements in parentheses (Giraldo & Crous 2019, Fan et al. 2020, Liu et al. 2022). Descriptions are based on observations after 14 d of incubation at 26 °C. Slow-growing species were allowed to grow longer, for 20–30 d, until sporulation was observed.
RESULTS
Phylogenetic analyses
For the two datasets in the present study (Table 2 and 3), phylogenetic analyses obtained from ML, MP and BI analyses resulted in trees with similar topologies, and the best scoring ML tree was selected to represent and discuss the phylogenetic relationships among taxa (Fig. 1, 2). Sympoventuriaceae phylogeny (Fig. 1): the first tree was based on a concatenated DNA sequence dataset (ITS, LSU, tef1, tub2 and rpb2) used to infer the phylogenetic position of the treated genera and species within the Sympoventuriaceae. The sequence data comprised 69 taxa for Sympoventuriaceae with Tyrannosorus lichenicola and Venturia saliciperda as the outgroup taxa. The dataset consisted of 5 175 characters, of which 2 399 were constant, 2 173 parsimony-informative and 603 parsimony-uninformative. Based on the results of the jModelTest, GTR+I+G was estimated as the optimal nucleotide substitution model under the output strategy of AIC; Scolecobasidium and Verruconis phylogeny (Fig. 2): for the sake of revealing the phylogenetic relationship between Scolecobasidium and Verruconis species, a second analysis was performed on the six gene regions (ITS, LSU, SSU, act1, tub2 and tef1) of 97 taxa within the genus, and Pseudosigmoidea excentrica and Sympoventuria capensis were used as outgroups. The final aligned sequence matrix contained 5 919 characters, of which 3 054 were constant, 2 152 parsimony-informative and 713 parsimony-uninformative. The optimal nucleotide substitution model HKY+I+G was used for the phylogenetic analyses.
Fig. 1.
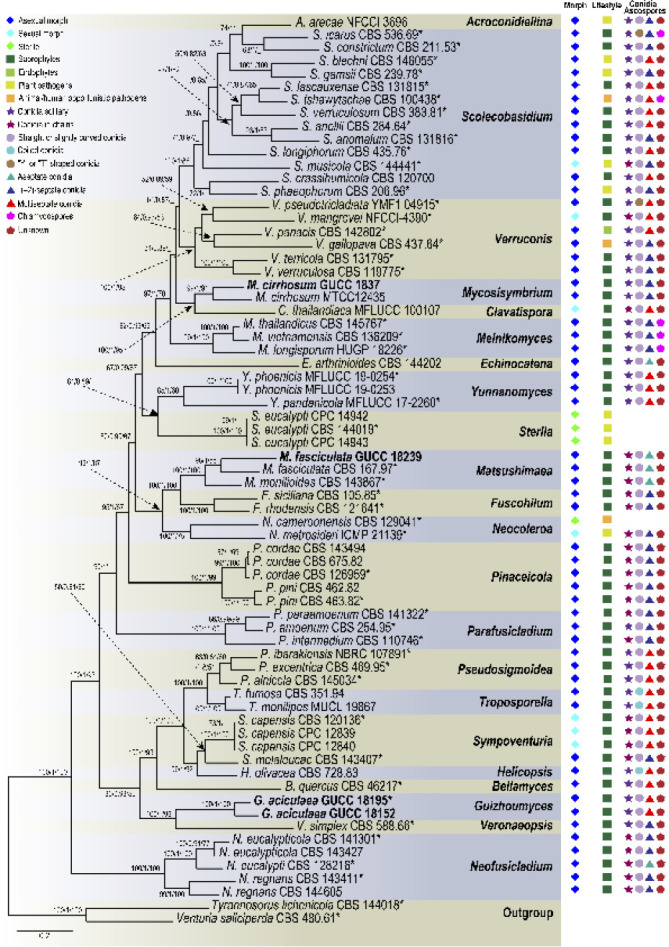
Phylogenetic tree of the family Sympoventuriaceae based on RAxML analyses of combined DNA dataset of ITS, LSU, tef1, tub2 and rpb2 gene sequences. Bootstrap values ≥ 50 % for Maximum parsimony and Maximum likelihood, and Bayesian posterior probabilities ≥ 80 % are presented at the branches (ML/BI/MP). Some branches were shortened to facilitate layout, and the scale bar represents the number of changes. Tyrannosorus lichenicola and Venturia saliciperda are used as outgroup. Those in bold are new taxa or new combinations proposed in the current study and the strains obtained, as well as type strains are marked with an asterisk (*). Lifestyles and typical morphological characteristics of individual strains are shown at the right side of the phylogenetic tree, and the related icons plotted are explained in the legend in the upper left corner.
Fig. 2.
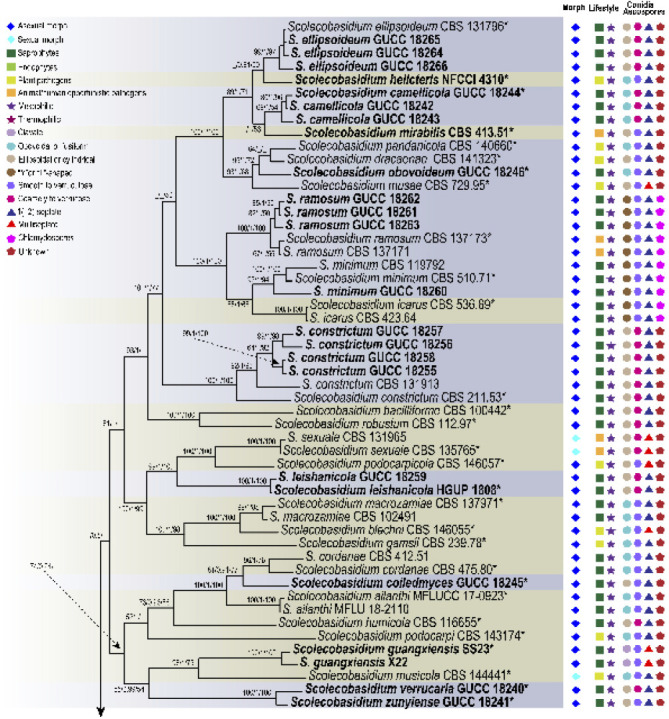
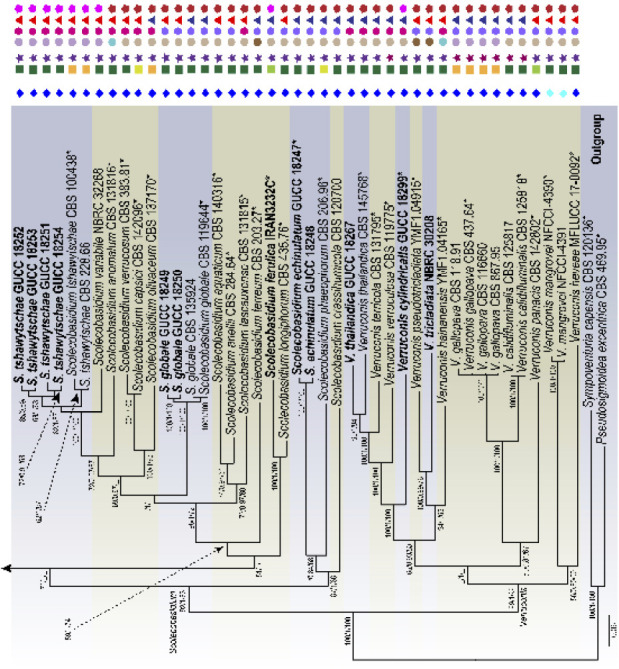
Phylogenetic tree of the genera Scolecobasidium and Verruconis based on RAxML analyses of combined DNA dataset of SSU, ITS, LSU, act1, tub2 and tef1 gene sequences. Bootstrap values ≥ 50 % for Maximum parsimony and Maximum likelihood, and Bayesian posterior probabilities ≥ 80 % are presented at the branches (ML/BI/MP). Some branches were shortened to facilitate layout, and the scale bar represents the number of changes. Pseudosigmoidea excentrica and Sympoventuria capensis are used as outgroup. Those in bold are new taxa or new combinations proposed in the current study and the strains obtained, as well as type strains are marked with an asterisk (*). Lifestyles and typical morphological characteristics of individual strains are shown at the right side of the phylogenetic tree, and the related icons plotted are explained in the legend in the upper left corner.
The phylogenetic tree of Sympoventuriaceae distinguished 22 subclades, each subclade representing a highly supported monophyletic group (Fig. 1). Acroconidiellina was located at the terminal end of the phylogenetic tree, closely related to Scolecobasidium and Verruconis (Fig. 1). Phylogenetic analyses resolved 43 species of Scolecobasidium, which chiefly clustered in five subclades, of which 37 correspond to known species of the genus (Fig. 2). In the Verruconis lineage three major subclades were observed, corresponding closely to the currently recognised 11 species (Fig. 2). The 29 newly collected strains clustered in eight well-supported subclades of Scolecobasidium and Verruconis respectively, including seven new species, as well as six new combinations proposed here (Fig. 2). The monotypic genus Mycosisymbrium consisted of two highly supported clades, which can be distinguished from other genera in this family, and Clavatispora as the sister genus (Fig. 1). Three clades were distinguished in Melnikomyces, corresponding closely to the currently recognized three species (M. longisporus, M. thailandicus and M. vietnamensis), which are saprophytes in soil or plant debris (Fig. 1). Echinocatena, a monotypic genus represented by E. arthrinioides, formed a robust lineage at the base of the Melnikomyces (Fig. 1). Sterila and Yunnanomyces clustered in the same subclade and received moderate to strong support, which was divided into two clades; the first one included the ex-type strain of Y. pandanicola and two Y. phoenicis strains, and the second one included three sterile strains (S. eucalypti) (Fig. 1).
Fuscohilum, Matsushimaea and Neocoleroa grouped together and represent three monophyletic groups; the first group comprising F. rhodense and F. sicilianum, the second group M. fasciculata and M. mtonilioides, and the third group N. cameroonensis and N. metrosideri, which are plant, animal and human pathogens (Fig. 1). Pinaceicola (P. cordae and P. pini) and Parafusicladium (P. amoenum, P. intermedium and P. paraamoenum) formed a well circumscribed clade (Fig. 1). Pseudosigmoidea included P. alnicola, P. excentrica and P. ibarakiensis, which formed a robust clade with four other genera, viz., Bellamyces, Helicopsis, Sympoventuria and Troposporella (Fig. 1). However, the phylogenetic relationship of these genera with the other clades remains poorly unresolved. Guizhoumyces belongs to a monophyletic clade, together with Veronaeopsis, representing a new genus in Sympoventuriaceae (Fig. 1). Neofusicladium encompassed three highly statistically supported subclades at the base of Sympoventuriaceae, which represent N. eucalypti, N. eucalypticola and N. regnans, respectively (Fig. 1). Relationships of the new taxa are discussed in the notes.
Lifestyle evolution analysis
Ancestral states of the lifestyles in Sympoventuriaceae were inferred on the reconstructed phylogeny. We defined the life strategies in four states: saprophytes, endophytes, plant pathogens, and animal/ human opportunistic pathogens. Overall, the results of BBM analysis revealed that the life strategies of Sympoventuriaceae was based on saprophytes as the primitive state, and endophytes, plant pathogens, animal/ human opportunistic pathogens were derived states, correlating with phylogeny (Fig. 3). At the genus level, Bellamyces, Echinocatena, Guizhoumyces, Helicopsis, Neofusicladium, Parafusicladium, Pseudosigmoidea, Sympoventuria, Troposporella and Veronaeopsis were basal in Sympoventuriaceae with a saprotrophic lifestyle, correlating with their ecology. The recently introduced Sterila and Neocoleroa were strongly supported as ingroup taxa of Sympoventuriaceae, and have evolved from saprophytes to become plant pathogens (for S. eucalypti and N. metrosideri) or animal/ human opportunistic pathogens (for N. cameroonensis) (Fig. 3). However, these lifestyles reverted to saprotrophic again in the well-supported Clavatispora, Fuscohilum, Matsushimaea, Melnikomyces and Mycosisymbrium clades. In the Acroconidiellina, Scolecobasidium and Verruconis clades, the number of plant pathogens and animal or human opportunistic pathogens increased from Sterila (one species) to Acroconidiellina (one species), Scolecobasidium (16 species) and Verruconis (one species), whereas the transition to endophytes occurred only twice in Scolecobasidium and Verruconis, respectively (Fig. 3). Thus, we conclude that the outstanding diversification of Scolecobasidium is related to the evolution of derived life strategies, and that the lifestyle of this genus may have been influenced by both plants and animals. Moreover, the common ancestor of the other genera except for Acroconidiellina, Neocoleroa and Sterila was saprophytic (Fig. 3), which is a derived condition from a saprotrophic ancestor of Sympoventuriaceae.
Fig. 3.
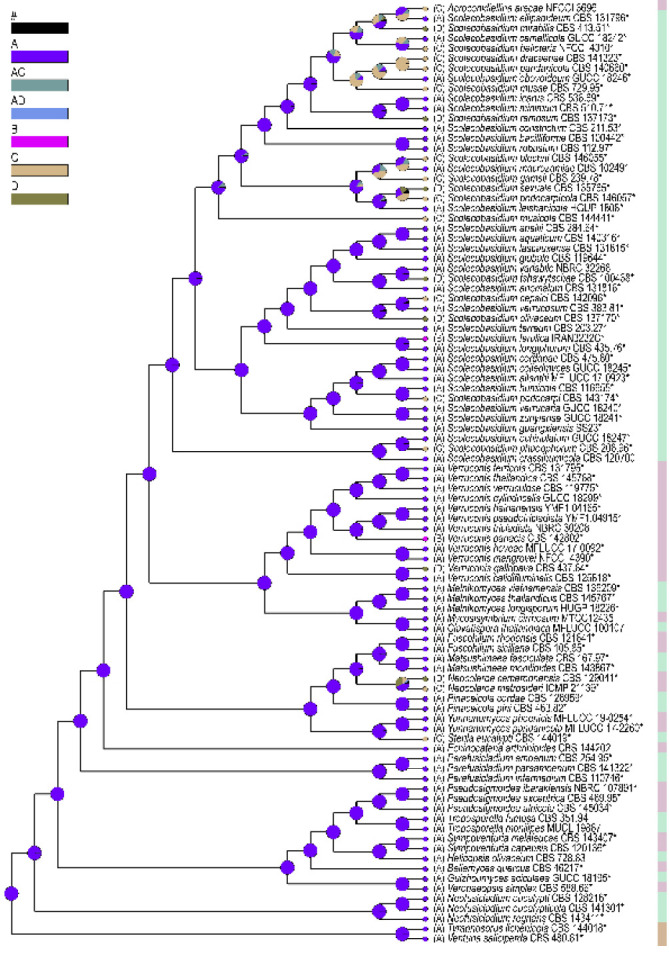
Ancestral character state analysis focusing on lifestyles in Sympoventuriaceae, using Bayesian Binary MCMC as alternative method. The pie chart at each node indicates the relative probabilities of all possible ancestral states from the Bayesian analysis, and black (#) shows the pooled probabilities of estimates that each account for < 5 %. Coloured circles at the tip and letters next to taxa represent their current lifestyle. A. saprophytes; B. endophytes; C. plant pathogens; D. animal/human opportunistic pathogens. The type strains are marked with an asterisk (*). Tyrannosorus lichenicola and Venturia saliciperda are used as outgroup.
TAXONOMY
Sympoventuriaceae Y. Zhang ter et al., Fungal Diversity 51: 255. 2011
Type genus. Sympoventuria Crous & Seifert.
Notes — Sympoventuriaceae was established by Zhang et al. (2011) with Sympoventuria designated as the type genus, which can be distinguished from the Venturiales by its saprophytic lifestyle, presence of pseudoparaphyses, and hyaline, symmetrical ascospores (Arzanlou et al. 2007, Crous et al. 2007a, b, Zhang et al. 2011). Phylogenetically, Sympoventuriaceae forms a well-supported family clade within Venturiales (Zhang et al. 2011, Machouart et al. 2014). Subsequently, Hyde et al. (2013) recognised three lineages (i.e., Sympoventuria, Veronaeopsis and fusicladium-like species) within Sympoventuriaceae and provided more details. Over the past few years, more genera have been accepted in Sympoventuriaceae, such as Ochroconis, Scolecobasidium and Verruconis (Machouart et al. 2014). The taxonomy of Sympoventuriaceae has since been widely studied and dramatically changed (Wijayawardene et al. 2014, Tibpromma et al. 2018). Despite these changes, the phylogenetic placement of many genera in the Sympoventuriaceae remains to be elucidated. Shen et al. (2020) re-described Sympoventuriaceae based on a multigene phylogenetic analysis, morphological and ecological comparisons, and included 15 genera in this family. However, generic boundaries within Sympoventuriaceae are poorly resolved and still controversial, due to lack of type materials and unresolved phylogenies. In this study, we accept 22 genera in Sympoventuriaceae (Fig. 1, Table 4).
Table 4.
Genera accepted in Sympoventuriaceae.
| Zhang et al. (2011) | Wijayawardene et al. (2014) | Tibpromma et al. (2018) | Wijayawardene et al. (2020) | Shen et al. (2020) | This study (2022) |
|---|---|---|---|---|---|
| Fusicladium-like | Clavatispora | Fusicladium | Acroconidiellina | Bellamyces | Acroconidiellina |
| Sympoventuria | Ochroconis | Ochroconis | Clavatispora | Echinocatena | Bellamyces |
| Veronaeopsis | Sympoventuria | Sympoventuria | Fusicladium | Fuscohilum | Clavatispora |
| Veronaeopsis | Scolecobasidium | Matsushimaea | Helicopsis | Echinocatena | |
| Veronaeopsis | Mycosisymbrium | Neocoleroa | Fuscohilum | ||
| Verruconis | Ochroconis | Neofusicladium | Guizhoumyces | ||
| Yunnanomyces | Sympoventuria | Pseudosigmoidea | Helicopsis | ||
| Veronaeopsis | Parafusicladium | Matsushimaea | |||
| Verruconis | Pinaceicola | Melnikomyces | |||
| Yunnanomyces | Sympoventuria | Mycosisymbrium | |||
| Scolecobasidium | Neocoleroa | ||||
| Sterila | Neofusicladium | ||||
| Troposporella | Pseudosigmoidea | ||||
| Veronaeopsis | Parafusicladium | ||||
| Verruconis | Pinaceicola | ||||
| Sympoventuria | |||||
| Scolecobasidium | |||||
| Sterila | |||||
| Troposporella | |||||
| Veronaeopsis | |||||
| Verruconis | |||||
| Yunnanomyces |
Guizhoumyces T.P. Wei & Y.L. Jiang, gen. nov. — MycoBank MB 840922
Etymology. Named after Guizhou, where this species was collected and the Greek name for fungi (myces).
Type species. Guizhoumyces hyalinaea T.P. Wei & Y.L. Jiang.
Mycelium consisting of curved or straight, branched, pale brown, septate, smooth-walled hyphae, frequently forming hyphal coils. Conidiophores subcylindrical, simple, branched, straight or slightly geniculate, pale brown, smooth, septate, sometimes reduced to conidiogenous cells. Conidiogenous cells integrated, simple, polyphialidic, sympodially proliferating, elongate lageniform or ampulliform, terminal or intercalary, pale brown, with an inconspicuous or distinct denticle at the conidiogenous locus after rhexolytic conidial secession. Conidia enteroblastic, solitary, acicular to obclavate or cylindrical, septate, straight or somewhat curved, smooth, thin-walled, subhyaline to pale brown, apex subobtuse to pointed, base truncate to short obconically truncate, with thickened and darkened hilum; anastomosis between mature conidia. Chlamydospores were not observed and sexual morph unknown.
Notes — The genus Guizhoumyces was established to accommodate a new species G. aciculaea. The phylogeny of concatenated ITS, LSU, rpb2, tef1 and tub2 DNA sequences indicated that Guizhoumyces formed a fully supported monophyletic lineage in Sympoventuriaceae, which is sister to Bellamyces, Helicopsis, Sympoventuria and Veronaeopsis. Morphologically, this genus has certain similarities with Pseudosigmoidea and Sigmoidea, but can be distinguished from them by its acicular to obclavate or cylindrical, less than four septate and smaller conidia (Crane 1968, Ando & Nakamura 2000, Crous et al. 2019a). Therefore, based on its unique morphological characteristics and phylogenetic location, a new genus name Guizhoumyces is introduced to accommodate this new fungus.
Guizhoumyces aciculaea T.P. Wei & Y.L. Jiang, sp. nov. — MycoBank MB 840923; Fig. 4
Fig. 4.
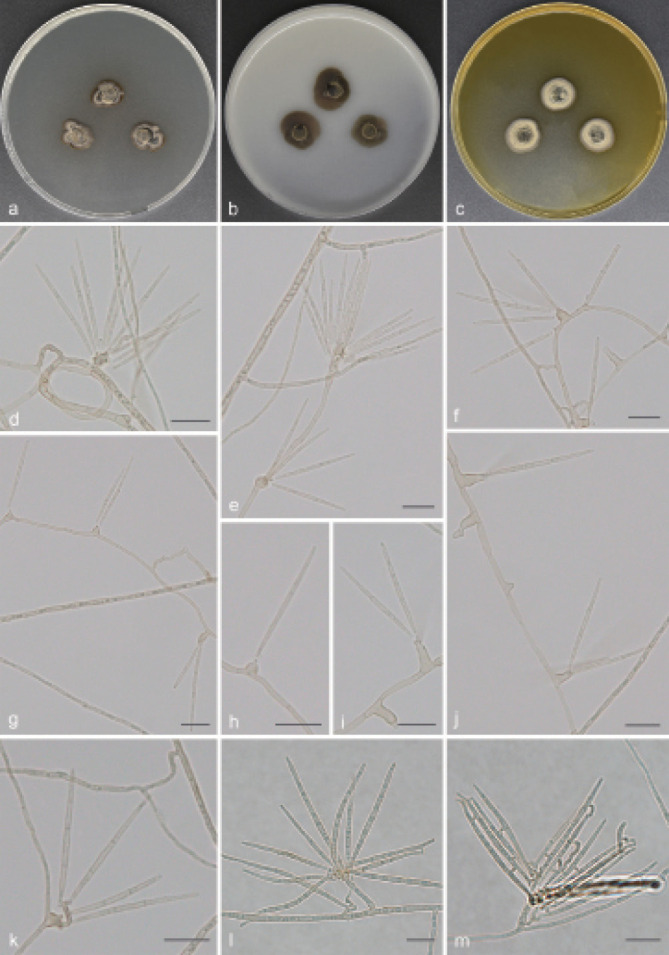
Guizhoumyces aciculaea (culture ex-type GUCC 18195). a–c. Colony on PDA, OA and MEA; d. hyphal coils and conidia; e–j. conidiophores reduced to conidiogenous cells; k–l. conidiophores with conidiogenous cells and conidia; m. anastomosis between mature conidia. — Scale bars: d–m = 10 µm.
Etymology. The epithet refers to the acicular conidia.
Typus. CHINA, Guizhou Province, Shiqian County, Pingshan Township, Fodingshan National Nature Reserve, N27°40'50" E108°07'30", 1100 m a.s.l., isolated from soil, 2 Nov. 2019, T.P. Wei (holotype HGUP 18195, isotype CGMCC 3.20543, culture ex-type GUCC 18195).
Mycelium consisted of branched, pale brown, septate, thick-walled, 2–3 µm diam hyphae, frequently forming hyphal coils. Conidiophores mostly flask-shaped to subcylindrical, simple, straight or slightly geniculate, pale brown, smooth, septate, sometimes reduced to conidiogenous cells, (10–)13–46.5(–48) × 1.5–2.5(–3) µm (av. ± SD = 25.5 ± 12.7 × 2.1 ± 0.4 µm, n = 30). Conidiogenous cells integrated, simple, polyphialidic, sympodially proliferating, elongate lageniform or ampulliform, terminal or intercalary, pale brown, 4–12(–15.5) × 2–3.5 µm (av. ± SD = 7.8 ± 2.7 × 2.7 ± 0.4 µm, n = 30), with one or numerous denticles in the apex, hyaline to pale brown, 1–3 µm long. Conidia separate rhexolytically from conidiogenous cells, enteroblastic, solitary, acicular to obclavate or cylindrical, 0(–3)-septate, straight or somewhat curved, smooth, thin-walled, subhyaline to pale brown, apex subobtuse to pointed, base truncate to short obconically truncate, with thickened and darkened hilum, anastomosis between mature conidia, (19.5–)21.5–37(–39.5) × 1.5–2 µm (av. ± SD = 27.8 ± 4.2 × 1.5 ± 0.1 µm, n = 30).
Culture characteristics — Colonies on PDA reaching up to 14–15 mm diam after 14 d at 26 °C, compact, surface grey brown, slightly raised at centre. On OA reaching 16–19 mm diam, with sparse aerial mycelium, olivaceous grey. On MEA reaching 14–16 mm diam, raised, hairy, with abundant aerial hyphae, grey at the surface, reverse pale brown.
Additional material examined. CHINA, Guizhou Province, Shiqian County, Ganxi Township, Fodingshan National Nature Reserve, N27°40'51" E108°07'21", 1240 m a.s.l., from leaf litter, 10 June 2019, T.P. Wei (HGUP 18152), living culture GUCC 18152 = CGMCC 3.20542.
Notes — Guizhoumyces aciculaea somewhat resembles the type species P. cranei and S. prolifera of Pseudosigmoidea and Sigmoidea in conidial morphology, with enteroblastic conidiogenesis and phialidic conidiogenous cells, which would suggest that our taxon could be accommodated here. Unfortunately, Pseudosigmoidea and Sigmoidea (Halosphaeriaceae, Microascales) are distantly related to G. aciculaea (Fig. 1). Morphologically, G. aciculaea can also be distinguished from P. cranei and S. prolifera. The conidia of P. cranei are scolecoid, 3(–8)-septate and longer (29–116.5 × 1.5–2.5 µm) (Ando & Nakamura 2000); conidia of S. prolifera are scolecoid, 5(–11)-septate, hyaline and larger (44–110 × 2–2.5 µm) (Crane 1968). In contrast, G. aciculaea has acicular to obclavate or cylindrical, 0(–3)-septate, subhyaline to pale brown and smaller conidia (19.5–39.5 × 1.5–2 µm), and anastomosis occurs among mature conidia. Moreover, the ex-type culture of G. hyalinaea and V. simplex clustered in two distinct clades representing two different genera (Fig. 1). Guizhoumyces hyalinaea is clearly distinct from V. simplex, which has oblong to subcylindrical, 0(–1)-septate and very small conidia (6–15 × 2–4 µm) (Arzanlou et al. 2007).
Scolecobasidium E.V. Abbott, Mycologia 19: 30. 1927
Synonym. Ochroconis de Hoog & Arx, Kavaka 1: 57. 1974 ‘1973’.
Type species. Scolecobasidium terreum E.V. Abbott.
Notes — Scolecobasidium was first described by Abbott (1927) to accommodate S. constrictum and S. terreum isolated from cotton and sugarcane soils in Louisiana, USA, with S. terreum designated as the generic type, which has Y-shaped and yellowish conidia. The salient characters of Scolecobasidium are rust-brown to olivaceous colonies producing small, brownish conidiophores bearing small numbers of dark, septate, rough-walled, rhexolytic conidia (Abbott 1927, Ellis 1976). Abbott (1927) pointed out that Scolecobasidium is distinguished from other groups by the shape of its conidia and the way conidia are arranged on its conidiophores. In the following decades, this genus received unanimous support (Barron & Busch 1962, Roy et al. 1962, Graniti 1963). Later, more species with unbranched conidia were described within Scolecobasidium, which led De Hoog & Von Arx (1973) to introduce a separate genus, Ochroconis, typified by O. constricta for hyphomycetous species with unbranched, subspherical to cylindrical or clavate, melanised conidia (Matsushima 1975, 1980, Punithalingam & Spooner 2011). Scolecobasidium was restricted to species with T- or Y-shaped or bilobed, two- to multi-celled conidia (Martin-Sanchez et al. 2012). It is noteworthy that the ex-type strains of both S. terreum (CBS 203.27) and O. constricta (CBS 202.27) are now sterile (Horre et al. 1999, Gams 2015).
Samerpitak et al. (2014) revised Ochroconis and Scolecobasidium using SSU, ITS, LSU, act1, tub2 and tef1 DNA sequences. They found that Ochroconis and Scolecobasidium clustered together, while Scolecobasidium was considered as doubtful because the ex-type culture was sterile (Samerpitak et al. 2017). This opinion, however, was not shared by Gams (2015) who regarded Ochroconis as a synonym of Scolecobasidium, which was supported by Seifert et al. (2011). More recently, Shen et al. (2020) resolved Ochroconis as a synonym of Scolecobasidium based on the multi-locus (ITS, LSU, tef1, tub2 and rpb2) analysis combined with morphology and ecology, with strong support for its monophyly. We agree with Shen et al. (2020) that Solecobasidium equals Ochroconis. As noted by Gams (2015) and Shen et al. (2020), although the ex-type strain of S. terreum is sterile, there are many reliably named cultures of S. terreum globally, which clearly define the identity of this characteristic fungus. Scolecobasidium will always be applied to the clade that includes the type species S. terreum and O. constricta of Scolecobasidium and Ochroconis. This study shows that the clade defined as Scolecobasidium combines monophyly, sexual and asexual morphs, and ecological characters in a coherent way that can logically be recognised at the generic rank. Additionally, many species of Ochroconis for which DNA data are available have since been transferred to Scolecobasidium by Shen et al. (2020), except O. ferulica, O. guang- xiensis, O. helicteris O. mirabilis and O. terricola, which are discussed below.
Scolecobasidium camellicola T.P. Wei & Y.L. Jiang, sp. nov. — MycoBank MB 840926; Fig. 5
Fig. 5.

Scolecobasidium camellicola (culture ex-type GUCC 18242). a–c. Colony on PDA, OA and MEA; d–h. conidiogenous cells giving rise to conidia; i. aging conidia forming conidiogenous loci; j–k. hypha with conidiogenous cells and conidia. — Scale bars: d–k = 10 µm.
Etymology. The epithet refers to Camellia, the host genus from which this fungus was collected.
Typus. CHINA, Guizhou Province, Meitan County, N27°75'09" E107°47'99", 910 m a.s.l., isolated from decaying Camellia sinensis leaf litter, 10 Aug. 2019, T.P. Wei (holotype HGUP 18242, isotype CGMCC 3.20547, culture ex-type GUCC 18242).
Mycelium partly superficial, partly immersed, hyphae branched, pale brown, septate, smooth, thick-walled, 1–2 µm wide. Conidiophores arising directly from superficial hyphae, mostly unbranched, subcylindrical, straight or flexuous, brown, continuous or septate, (10–)11.5–55(–61.5) × 2.5–4 µm (av. ± SD = 19.4 ± 13.7 × 2.6 ± 0.4 µm, n = 30). Conidiogenous cells integrated, polyblastic, intercalary or terminal, sympodial extensions, pale brown to brown, bearing 1–6 conidia at the apex, (6–)7.5–13(–14) × (2–)2.5–3.5 µm (av. ± SD = 9.2 ± 2.1 × 2.6 ± 0.4 µm, n = 30). Conidia secession rhexolytic from conidiogenous cells, subcylindrical or fusoid, 1-septate, minutely echinulate, pale brown, slightly constricted at the septum, with hilum bearing a marginal frill, (7–)7.5–10.5(–11.5) × (2.5–)3–4.5 µm (av. ± SD = 8.7 ± 0.9 × 3.4 ± 0.5 µm, n = 30).
Culture characteristics — Colonies on PDA attaining 21–22 mm diam after 14 d at 26 °C, growing slow, isabelline, raised in the centre. On OA reaching up to 23–24 mm diam, flat, spreading, immersed, dark brown. On MEA reaching 22–24 mm diam, raised, with sparse to moderate aerial hyphae, pale to medium brown.
Additional materials examined. CHINA, Guizhou Province, Meitan County, N27°75'09" E107°47'99", 910 m a.s.l., on decaying Camellia sinensis leaf litter, 10 Aug. 2019, T.P. Wei (HGUP 18243), living culture GUCC 18243 = CGMCC 3.20548; Leishan County, N26°24'02" E107°77'22", 1178 m a.s.l., from forest litter, 12 Mar. 2018, T.P. Wei (HGUP 18244), living culture GUCC 18244 = CGMCC 3.20549.
Notes — Scolecobasidium camellicola is introduced as a new species based on morphological and phylogenetic differences to other Scolecobasidium species. Phylogenetically, S. camellicola shares a sister relationship with S. mirabilis and S. helicteris with high statistical support (Fig. 2). Nevertheless, S. camellicola showed high heterogeneity, forming a well-separated clade, which was genetically distant from all species. Morphologically, S. mirabilis differs from S. camellicola by its smooth-walled to verruculose and larger conidia (9.0–13.5 × 4.8–6.7 µm vs 7–11.5 × 2.5–4.5 µm) (Samerpitak et al. 2014); S. helicteris differs by its smooth to verruculose, ellipsoid or pyriform and smaller conidia (4–8 × 2–3.4 µm vs 7–11.5 × 2.5–4.5 µm) (Singh et al. 2019).
Scolecobasidium coiledmyces T.P. Wei & Y.L. Jiang, sp. nov. — MycoBank MB 840927; Fig. 6
Fig. 6.
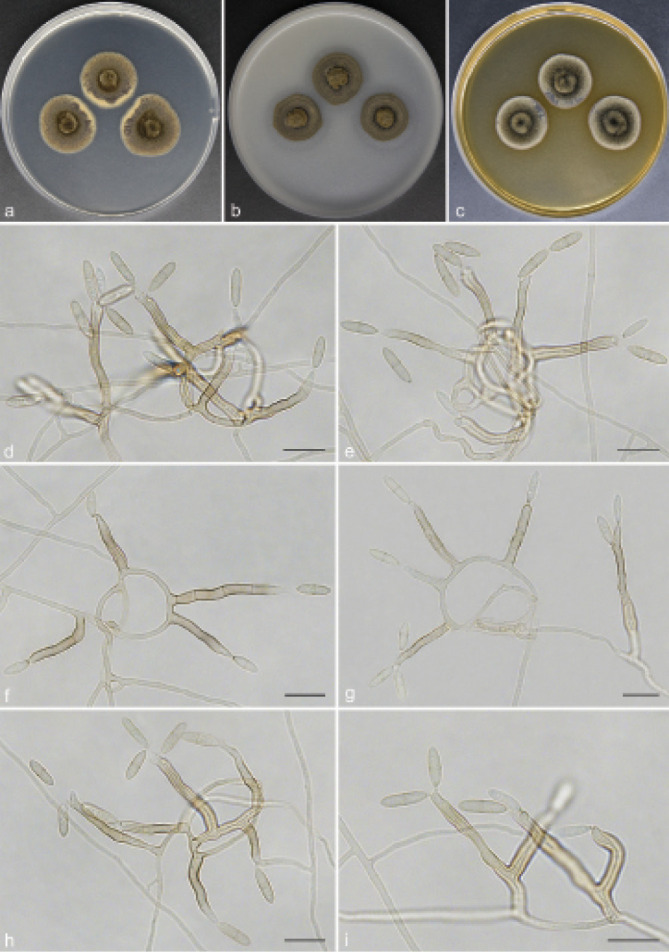
Scolecobasidium coiledmyces (culture ex-type GUCC 18245). a–c. Colony on PDA, OA and MEA; d–g. conidiophores arising from hyphal coils; h. conidiophores with conidiogenous cells and conidia; i. branched conidiophores. — Scale bars: d–i = 10 µm.
Etymology. The epithet refers to the frequently forming hyphal coils.
Typus. CHINA, Guizhou Province, Guiyang City, Huaxi Wetland Park, N26°43'92" E106°67'76", 1140 m a.s.l., isolated from lawn soil, 16 Nov. 2018, T.P. Wei (holotype HGUP 18245, isotype CGMCC 3.20550, culture ex-type GUCC 18245).
Mycelium consisting of smooth, septate, branched, subhyaline or medium brown, 1.5–3 µm diam hyphae, forming hyphal coils. Conidiophores erect, 0(–4)-septate, occasionally branched, brown to dark brown, smooth and thick walled, subcylindrical, (12–)13–43(–56) × 2.5–4(–4.5) µm (av. ± SD = 23.9 ± 9.5 × 3.1 ± 0.5 µm, n = 30). Conidiogenous cells terminal, subhyaline or pale brown, sympodially proliferating, producing several cylindrical denticles in the apical region, (5–)7–17.5(–19) × 2.5–3 µm (av. ± SD = 12.4 ± 3.9 × 2.6 ± 0.3 µm, n = 30). Conidia solitary, medianly 1-septate, subcylindrical, apex obtuse, frills remaining on denticle and on conidial hilum, 0.5 µm long, medium brown, verruculose, released by rhexolytic secession, 8–11.5(–12) × 2.5–3.5 µm (av. ± SD = 9.6 ± 1.1 × 2.7 ± 0.3 µm, n = 30).
Culture characteristics — Colonies on PDA attaining 24– 26 mm diam after 14 d at 26 °C, brown, slightly raised in the centre, with moderate aerial mycelium and smooth. On OA reaching up to 20–22 mm diam, flat, spreading, dark brown. On MEA reaching 21–22 mm diam, raised, with moderate aerial mycelium, olivaceous.
Notes — Scolecobasidium coiledmyces is phylogenetically related to S. cordanae and S. ailanthi, but S. coiledmyces forms a single branch as the sister clade to the other two species with high support from three independent algorithms (Fig. 2). Furthermore, S. coiledmyces is distinct based on its morphology. The conidia of S. cordanae are smaller (5–10 × 2.5–3.5 µm vs 8–12 × 2.5–3.5 µm), obovoidal to broadly fusoid and constricted at the median septum (Samerpitak et al. 2014); S. ailanthic differed from our strain in having fusoid, longitudinally striate and smaller conidia (9–10 × 2.4–2.6 µm vs 8–12 × 2.5–3.5 µm) with a thick septum, as well as unbranched conidiophores (Jayasiri et al. 2019).
Scolecobasidium echinulatum T.P. Wei & Y.L. Jiang, sp. nov. — MycoBank MB 842082; Fig. 7
Fig. 7.
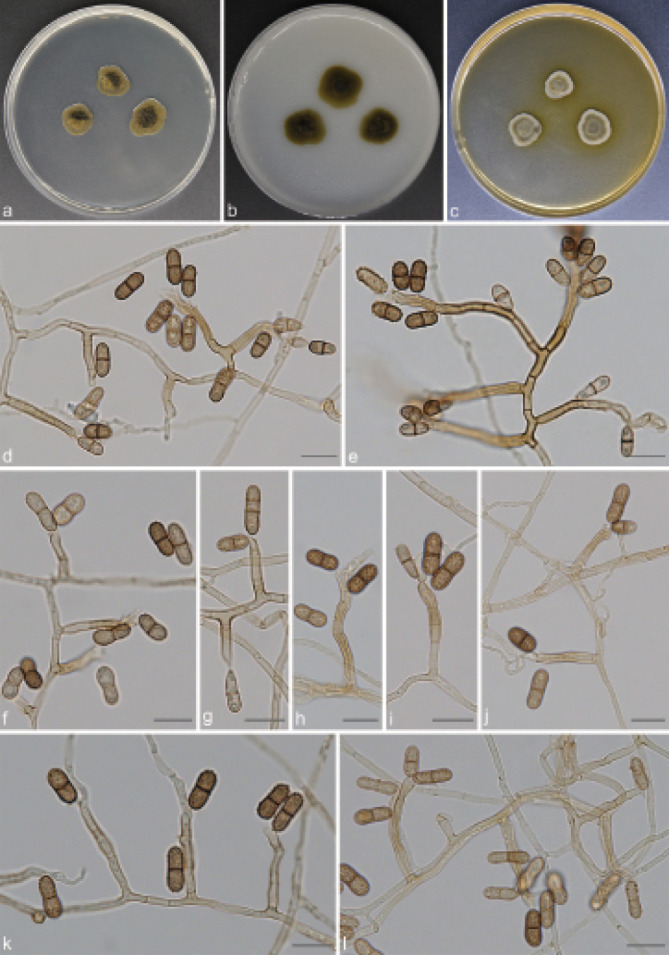
Scolecobasidium echinulatum (culture ex-type GUCC 18247). a–c. Colony on PDA, OA and MEA; d–f. conidial apparatus with rhexolytic conidia, produced from sympodial conidiogenous cells; g. immature and 1(–2) septate conidia; h–l. conidiophores with conidiogenous cells and conidia. — Scale bars: d–l = 10 µm.
Etymology. The epithet refers to the conidia with minutely echinulate cell walls.
Typus. CHINA, Guizhou Province, Guiyang City, Huaxi Wetland Park, N26°43'92" E106°67'76", 1140 m a.s.l., isolated from soil, 16 Nov. 2018, T.P. Wei (holotype HGUP 18247, isotype CGMCC 3.20552, culture ex-type GUCC 18247).
Mycelium superficial or immersed, hyphae brown, smooth, thin- walled, septate, 1.5–3 µm wide. Conidiophores clearly differentiated, arising at right angles from creeping hyphae, branched, erect, straight or slightly flexuous, brown, smooth, septate, (12–) 13.5–70(–87) × (2.5–)3–4 µm (av. ± SD = 29.7 ± 18.1 × 3.1 ± 0.3 µm, n = 30). Conidiogenous cells integrated, terminal or intercalary, elongate to cylindrical, with some scattered denticles in the apical region, pale brown, smooth, (4–)5–15(–20.5) × 2.5–4 µm (av. ± SD = 9.8 ± 3.9 × 3.1 ± 0.4 µm, n = 30). Conidia ellipsoidal to cylindrical, verruculose or minutely echinulate, dark brown to black, 1(–2)-septate, slightly narrower around the middle, frills remaining on denticle and on conidial hilum, released by rhexolytic secession, 8.5–10(–11.5) × 4–5 µm (av. ± SD = 9.5 ± 0.9 × 4.3 ± 0.3 µm, n = 30).
Culture characteristics — Colonies on PDA attaining 13– 18 mm diam after 14 d at 26 °C, spreading, dark olivaceous brown. On OA reaching up to 18–20 mm diam, colonies moderately expanding, immersed, flat, dark olivaceous brown. On MEA reaching 13–15 mm diam, olivaceous, raised, with moderate aerial mycelium.
Additional material examined. CHINA, Guizhou Province, Qingzhen City, Red maple lake scenic area, N26°54'35" E106°38'74", 1272 m a.s.l., from soil, 07 Aug. 2020, T.P. Wei (HGUP 18248), living culture GUCC 18248 = CGMCC 3.20553.
Notes — The proposed new species, S. echinulatum, is phylogenetically related to S. phaeophorum and S. crassihumicola, but they can be distinguished by their morphological characteristics and DNA sequence data. Scolecobasidium echinulatum differs from S. phaeophorum as it has ellipsoidal to cylindrical, verruculose or minutely echinulate, dark brown to black and 1(–2)-septate conidia (the cylindrical to fusoid conidia of S. phaeophorum are smooth-walled, pale brown and 1-septate) (Samerpitak et al. 2015b); S. crassihumicola differs from S. echinulatum by having 1(–3)-septate, non-constricted, ovoid to cylindrical and larger conidia (7.5–13 × 4.2–5.5 µm vs 8.5–11.5 × 4–5 µm) (Matsushima 1971).
Scolecobasidium obovoideum T.P. Wei & Y.L. Jiang, sp. nov. — MycoBank MB 842083; Fig. 8
Fig. 8.
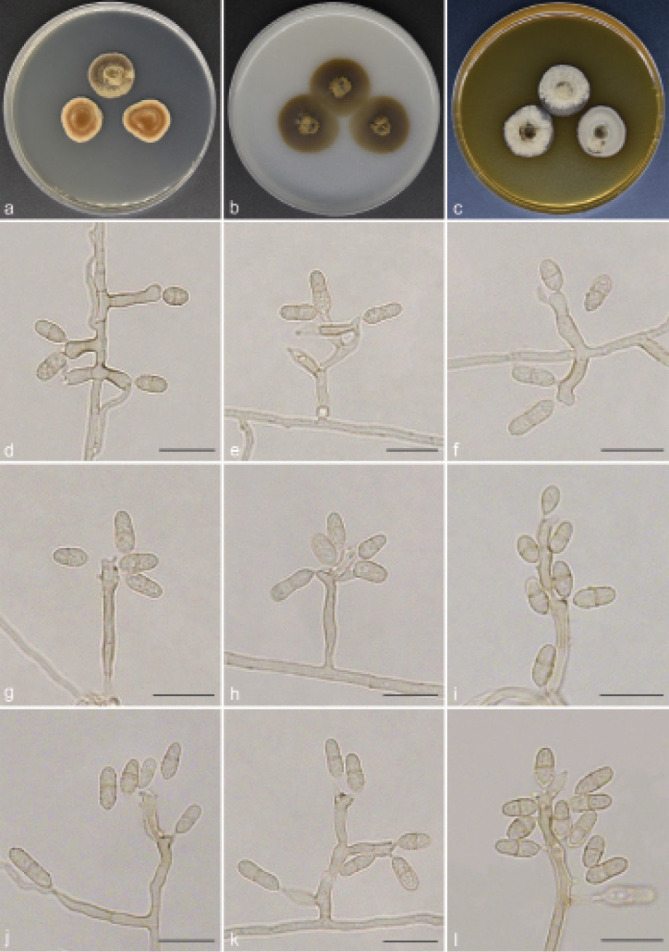
Scolecobasidium obovoideum (culture ex-type GUCC 18246). a–c. Colony on PDA, OA and MEA; d–i. conidial apparatus with rhexolytic conidia, produced from sympodial conidiogenous cells; j–k. branched conidiophores; l. conidiophores with conidiogenous cells and conidia. — Scale bars: d–l = 10 µm.
Etymology. The epithet refers to the obovoidal conidia.
Typus. CHINA, Guizhou Province, Guiyang City, Tianhetan Tourist Holiday Resort, N26°43'95" E106°57'64", 1164 m a.s.l., isolated from forest litter, 16 Dec. 2019, T.P. Wei (holotype HGUP 18246, isotype CGMCC 3.20551, culture ex-type GUCC 18246).
Mycelium composed of hyaline to pale brown, septate, branched, smooth, thick-walled, 1.5–3 µm wide hyphae. Conidiophores arising directly from vegetative hyphae, subcylindrical, branched, multi-septate, brown, erect, straight or flexuous, (7–)9.5–39.5(–43) × (2–)2.5–3.5 µm (av. ± SD = 20.0 ± 9.9 × 2.6 ± 0.4 µm, n = 30). Conidiogenous cells polyblastic, terminal or intercalary, subcylindrical to subclavate, pale brown, producing conidia sympodially on long open denticles, 6–14(–15.5) × 2–3.5 µm (av. ± SD = 9.4 ± 2.1 × 2.6 ± 0.4 µm, n = 30). Conidia solitary, obovoidal to fusoid, sometimes slightly apiculate at the base, finely verruculose, 1-septate, constricted at the septum, brown, released by rhexolytic secession, 5.5–8.5 × 2.5–4 µm (av. ± SD = 7.2 ± 0.6 × 3.1 ± 0.3 µm, n = 30).
Culture characteristics — Colonies on PDA reaching up to 19–20 mm diam after 14 d at 26 °C, with moderate aerial mycelium, dark brown. On OA reaching 24–26 mm diam, flat, immersed, olivaceous. On MEA reaching 19–21 mm diam, raised, aerial mycelium moderate to abundant, olive grey.
Notes — Scolecobasidium obovoideum is phylogenetically closely related to S. pandanicola and S. dracaenae and can be differentiated from that species by DNA sequences of ITS, LSU, SSU, act1, tub2 and tef1 gene regions. Morphologically, S. pandanicola can be distinguished from S. obovoideum as it has subhyaline to hazel brown, thin-walled, fusoid or ellipsoid and larger conidia (6–10 × 3–4.5 µm vs 5.5–8.5 × 2.5–4 µm) (Crous et al. 2015); S. dracaenae differs from S. obovoideum in having subcylindrical and larger conidia (6.5–10 × 3–4 µm vs 5.5–8.5 × 2.5–4 µm), and shorter conidiophores (10–30 × 2–3 µm vs 7–43 × 2–3.5 µm) (Crous et al. 2016). Phylogenetically, our new isolate GUCC 18246 forms a single clade separated from other Scolecobasidium species (Fig. 2).
Scolecobasidium verrucaria T.P. Wei & Y.L. Jiang, sp. nov. — MycoBank MB 840924; Fig. 9
Fig. 9.
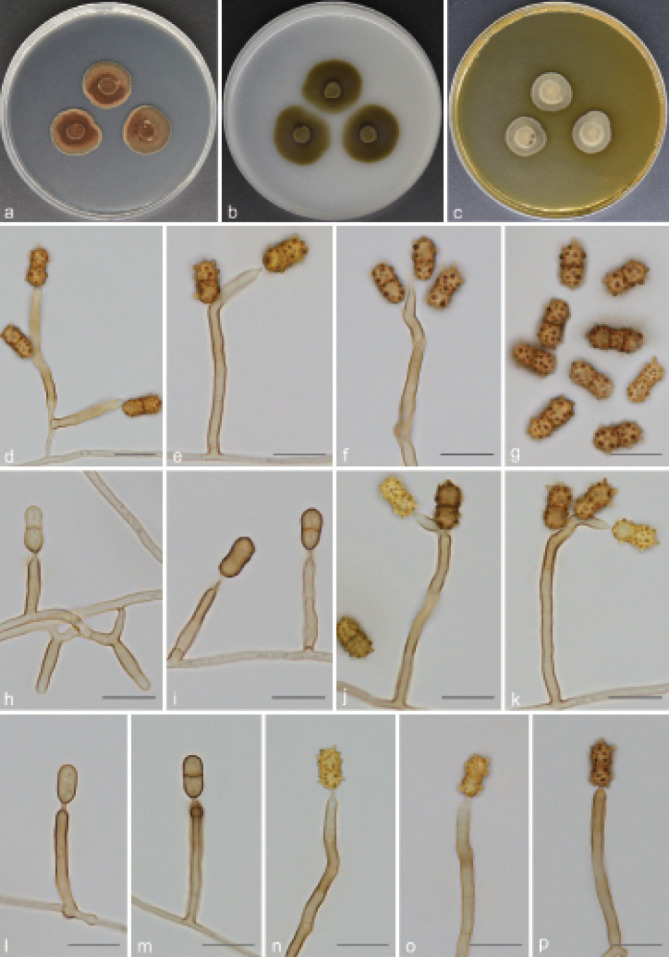
Scolecobasidium verrucaria (culture ex-type GUCC 18240). a–c. Colony on PDA, OA and MEA; d–f. conidiophores with conidiogenous cells and conidia; g. 1(–2) septate and coarsely verrucose conidia; h–p. maturation process of conidia. — Scale bars: d–p = 10 µm.
Etymology. The epithet refers to its verrucose conidia.
Typus. CHINA, Guizhou Province, Qingzhen City, Red maple lake scenic area, N26°54'35" E106°38'74", 1272 m a.s.l., from soil, 16 Apr. 2018, T.P. Wei (holotype HGUP 18240, isotype CGMCC 3.20545, culture ex-type GUCC 18240).
Mycelium mostly superficial or semi-immersed, hyphae pale brown, smooth, branched, 1–2 µm wide. Conidiophores arising directly from vegetative hyphae, occasionally branched, continuous or septate, dark brown, straight or slightly geniculate, cylindrical, (12.5–)14.5–51.5(–54) × 3–3.5 µm (av. ± SD = 30.1 ± 11.8 × 2.9 ± 0.2 µm, n = 30). Conidiogenous cells integrated, terminal, polyblastic, sympodial, cylindrical, subhyaline, (6.5–)7–16.5(–17.5) × 2.5–3.5 µm (av. ± SD = 10.2 ± 2.8 × 2.7 ± 0.3 µm, n = 30), with one or more denticles in the apical region, denticles 1–3 µm long. Conidia acropleurogenous, broadly ellipsoidal, 1(–2)-septate, strongly constricted at the septum, 8.5–11(–11.5) × 4.5–5.5(–6) µm (av. ± SD = 9.3 ± 0.8 × 5.0 ± 0.3 µm, n = 30), the colour of immature conidia changed from brown to yellow to dark brown, and gradually from smooth to coarsely verrucose, verrucous protrusions up to 2.7 µm long.
Culture characteristics — Colonies on PDA reaching up to 20–22 mm diam after 14 d at 26 °C, slightly raised at centre, isabelline, lobate margin. On OA reaching 24–26 mm diam, flat, spreading, immersed, olivaceous. On MEA reaching 16–18 mm diam, raised, hairy, grey brown.
Notes — Scolecobasidium verrucaria was collected from natural forest soil, and has the same lifestyle as some species of Scolecobasidium. Morphologically, the polyblastic sympodial conidiogenous cells, conidial apparatus with rhexolytic conidiogenesis, and olivaceous colonies point to Scolecobasidium. It is noteworthy that the ellipsoidal, 1(–2)-septate, yellow to dark brown, and coarsely verrucose conidia of S. verrucaria differ from other reported members of Scolecobasidium. Phylogenetically, S. verrucaria nests in the Scolecobasidium clade, being closely related to S. zunyiense. However, S. zunyiense can be distinguished from S. verrucaria by its brown grey, verruculose, 0(–1)-septate and larger conidia (8.5–14 × 4–5.5 µm vs 8.5–11.5 × 4.5–6 µm).
Scolecobasidium zunyiense T.P. Wei & Y.L. Jiang, sp. nov. — MycoBank MB 840925; Fig. 10
Fig. 10.

Scolecobasidium zunyiense (culture ex-type GUCC 18241). a–c. Colony on PDA, OA and MEA; d–e. conidiophores and conidiogenous cells bearing conidia; f–g. branched conidiophores; h–o. acropleurogenous, ellipsoidal, 1-septate and verruculose conidia. — Scale bars: d–o = 10 µm.
Etymology. Named after Zunyi, where this species was collected.
Typus. CHINA, Guizhou Province, Zunyi City, Phoenix Mountain National Forest Park, N27°36'27" E106°45'07", 1024 m a.s.l., from forest litter, 10 June 2019, T.P. Wei (holotype HGUP 18241, isotype CGMCC 3.20546, culture ex-type GUCC 18241).
Mycelium consisting of pale brown, smooth, branched, septate, 1.5–2.5 µm diam hyphae, often giving rise to hyphal coils. Conidiophores branched, straight to irregularly curved, solitary or at times two arising from the same basal cell, smooth, subcylindrical, septate, pale brown to brown, (9–)11–60(–78) × 3–4 µm (av. ± SD = 32.1 ± 15.9 × 3.2 ± 0.3 µm, n = 30). Conidiogenous cells polyblastic, sympodial, terminal or intercalary, subhyaline to brown, smooth, subcylindrical, (5–)5.5–15.5(–17.5) × 3–4 µm (av. ± SD = 9.8 ± 2.9 × 3.3 ± 0.3 µm, n = 30), with 1–9 terminal denticles, 1–2 µm. Conidia released by rhexolytic secession, solitary, acropleurogenous, ellipsoidal with rounded ends, 0(–1)-septate, brown grey, verruculose, prominently constricted at the septum, 8.5–13(–14) × 4–5.5 µm (av. ± SD = 10.2 ± 1.2 × 4.4 ± 0.4 µm, n = 30).
Culture characteristics — Colonies on PDA attaining 18– 20 mm diam after 14 d at 26 °C, slightly raised at centre, immersed, isabelline. On OA reaching up to 20–23 mm diam, flat, spreading, dark brown, reverse brown. On MEA reaching 20–22 mm diam, raised, hairy, pale brown at the surface, reverse olivaceous.
Notes — Scolecobasidium zunyiense is phylogenetically related to S. verrucaria, being fully supported in three independent algorithms (Fig. 2). Nevertheless, they can be distinguished based on their morphological characteristics as S. zunyiense is characterised by brown grey, 0(–1)-septate and verruculose conidia, while the conidia of S. verrucaria are yellow to dark brown, 1(–2)-septate, and verrucous protrusions can be up to 2.7 µm long, a feature not observed for S. zunyiense.
Scolecobasidium ferulica (Z. Tazik & K. Rahnama) T.P. Wei & Y.L. Jiang, comb. nov. — MycoBank MB 840930
Basionym. Ochroconis ferulica Z. Tazik & K. Rahnama, Nova Hedwigia 110: 374. 2020.
Description — Tazik et al. (2020).
Notes — Scolecobasidium ferulica was initially reported as an endophyte on roots of Ferula ovina in northeast Iran (Tazik et al. 2020). Multi-locus phylogenetic analyses indicate that this species is sister to S. longiphorum and is fully supported as phylogenetically distinct (Fig. 2). Morphologically, the ellipsoidal and smaller conidia of S. ferulica (8–10.5 × 6–7.5 µm) distinguishes this species from S. longiphorum (12–19 × 3.5–4.5 µm), which is characterised by long cylindrical and larger conidia (Samerpitak et al. 2014).
Scolecobasidium guangxiensis (Xie et al.) T.P. Wei & Y.L. Jiang, comb. nov. — MycoBank MB 840931
Basionym. Ochroconis guangxiensis Xie et al., Mycoscience 61: 308. 2020
Description — Chen et al. (2020).
Notes — Scolecobasidium guangxiensis was collected from rhizosphere soil of sugarcane in Guangxi, China, and was introduced by Chen et al. (2020). Based on the phylogeny presented here, despite the S. guangxiensis shares a sister relationship with S. musicola (Fig. 2), the considerably high number of variable positions in the ITS (88 bp, 17 %) and LSU (55 bp, 6 %) alignments supports the split into two distinct taxa. No SSU, act1, tub2 and tef1 data are currently available for the ex-type of S. guangxiensis and S. musicola. Morphologically, it is characterised by forming flask-shaped or cylindrical conidiophores and smooth-walled to verruculose, yellow brown, clavate or cylindrical, 1(–3)-septate conidia; S. musicola differs by its sexual morph forming fusoid to ellipsoid, hyaline to pale brown and straight to slightly curved ascospores (Crous et al. 2018b), whereas S. guangxiensis lacks a sexual morph.
Scolecobasidium helicteris (Singh et al.) T.P. Wei & Y.L. Jiang, comb. nov. — MycoBank MB 840932
Basionym. Ochroconis helicteris Singh et al., Phytotaxa 427: 192. 2019.
Description — Singh et al. (2019).
Notes — Scolecobasidium helicteris was previously recorded as pathogen associated with leaf spots on Helicteris isora in India (Singh et al. 2019). According to our analysis, S. helicteris resolved as the closest phylogenetic relative to S. ellipsoideum (Fig. 2). Scolecobasidium helicteris is, however, clearly distinguished morphologically by its obovoid to fusoid or pear shaped and smooth to verruculose conidia, and shorter conidiophores (ellipsoidal to oblong and spinulose conidia in S. ellipsoideum) (Fig. 13; Ren et al. 2013).
Fig. 13.

Scolecobasidium ellipsoideum (GUCC 18264). a–c. Colony on PDA, OA and MEA; d–f. conidiophores and conidiogenous cells bearing conidia; g. branched conidiophores; h–p. medium brown, 1-septate and ellipsoidal to cylindrical conidia. — Scale bars: d–p = 10 µm.
Scolecobasidium leishanicola (X. Zhang & Y.L. Jiang) T.P. Wei & Y.L. Jiang, comb. & nom. nov. — MycoBank MB 840929; Fig. 11
Fig. 11.

Scolecobasidium leishanicola (GUCC 18259). a–c. Colony on PDA, OA and MEA; d–e. conidial apparatus with rhexolytic conidia, produced from sympodial conidiogenous cells; f–l. germinating conidia; j–m. maturation process of conidia. — Scale bars: d–m = 10 µm.
Basionym. Ochroconis terricola X. Zhang & Y.L. Jiang, Mycotaxon 135: 146. 2020.
Mycelium consisting of branched, septate, subhyaline to pale brown, smooth-walled, 1.5–2.5 µm diam hyphae. Conidiophores arising directly from vegetative hyphae, erect, subcylindrical, unbranched, medium brown, multi-septate, smooth-walled, straight to flexuous, (9.5–)11–41(–108) × 2.5–4 µm (av. ± SD = 26.1 ± 18.6 × 3.2 ± 0.4 µm, n = 30). Conidiogenous cells integrated, terminal, subcylindrical, subhyaline to pale brown, with one to several sympodial denticle-like loci, (7.5–)9–23.5(–25) × (2.5–)3–4 µm (av. ± SD = 13.4 ± 4.6 × 3.1 ± 0.3 µm, n = 30). Conidia solitary, brown, ellipsoidal to cylindrical or fusoid, 1-septate, minutely echinulate, sometimes slightly constricted at the septum, apex obtuse, base narrowly truncated with hilum bearing a marginal frill, 9.5–12(–13.5) × 3–4.5 µm (av. ± SD = 11.6 ± 1.5 × 3.5 ± 0.4 µm, n = 30).
Culture characteristics — Colonies on PDA attaining 22–24 mm diam after 14 d at 26 °C, effuse, grey brown, reverse dark brown, growing slowly. On OA reaching up to 24–26 mm diam, olivaceous brown, immersed, with sparse aerial mycelium. On MEA reaching 18–23 mm diam, spreading, hairy, isabelline, raised in the centre.
Material examined. CHINA, Guizhou Province, Leishan County, N26°24'02" E107°77'22", 1178 m a.s.l., isolated from soil, 12 Mar. 2018, T.P. Wei (HGUP 18259), living culture GUCC 18259 = CGMCC 3.20564.
Notes — Scolecobasidium leishanicola (as Ochroconis terricola) was previously recorded from soil in China (Zhang et al. 2020). The phylogenetic tree constructed based on six gene loci showed that one isolate (GUCC 18259) from the present study clusters in a clade closely related to S. leishanicola, and was sister to S. sexualis and S. podocarpicola (Fig. 2). Morphologically, the conidial dimensions in the present study fit exactly with those in Zhang et al. (2020). Therefore, this species was placed in Scolecobasidium, for which a new name had to be introduced (S. leishanicola), as S. terricola was already occupied.
Scolecobasidium mirabilis (Samerp. & de Hoog) T.P. Wei & Y.L. Jiang, comb. nov. — MycoBank MB 840933
Basionym. Ochroconis mirabilis Samerp. & de Hoog, Fungal Diversity 65: 114. 2014.
Description — Samerpitak et al. (2014).
Notes — Scolecobasidium musae was originally isolated from the fruit surface of Musa basjoo in Hainan, China (Hao et al. 2013). Almost simultaneously, Samerpitak et al. (2014) described S. mirabilis from the regulator of a scuba diver in the Netherlands. Later, Samerpitak et al. (2015a) compared DNA sequence data between the two species and thought that the LSU sequences of S. mirabilis and S. musae were almost identical. To solve this taxonomic dilemma, they proposed S. mirabilis as a synonym for S. musae. However, our phylogenetic analysis indicates that S. mirabilis and S. musae cluster apart, and are phylogenetically distant (Fig. 2). They are genetically distinct in 2 bp (1 %), 21 bp (3 %), 3 bp (1 %), 10 bp (4 %), 46 bp (10 %) and 16 bp (4 %) in SSU, ITS, LSU, act1, tub2 and tef1 loci. Based on these results as well as their morphology, we thus resurrect S. mirabilis and recognise S. mirabilis and S. musae as two different species of Scolecobasidium.
Scolecobasidium constrictum E.V. Abbott, Mycologia 19:
30. 1927 — Fig. 12
Fig. 12.

Scolecobasidium constrictum (GUCC 18256). a–c. Colony on PDA, OA and MEA; d–m. conidial apparatus with rhexolytic conidia, produced from sympodial conidiogenous cells; n–o. hyphal coil; p. cylindrical to fusoid conidia. — Scale bars: d–p = 10 µm.
Synonym. Ochroconis constricta (E.V. Abbott) de Hoog & Arx, Kavaka 1: 57. 1973.
Mycelium composed of hyaline or pale brown, septate, branched, smooth-walled, 1.5–2 µm diam hyphae. Conidiophores differentiated arising directly from vegetative hyphae, branched, sparingly septate, brown, smooth and thick-walled, cylindrical, mostly geniculate-sinuous, (11–)12–46.5(–56) × (2–)2.5– 3.5 µm (av. ± SD = 23.8 ± 10.3 × 2.5 ± 0.3 µm, n = 30). Conidiogenous cells integrated, terminal, flask-shaped or ampulliform to cylindrical, hyaline or pale brown, with single or several sympodial apical loci with rhexolytic conidiogenesis, (3.5–)5.5–14(–15) × (2–)2.5–4(–4.5) µm (av. ± SD = 8.8 ± 3.1 × 3.0 ± 0.5 µm, n = 30). Conidia solitary, subhyaline to pale brown, 1-septate, broadly ellipsoidal to cylindrical, finely echinulate to verruculose, usually constricted at the septum, frills remaining on denticle and on conidial base, ends obtusely rounded, 9.5–12 × 3.5–4.5 µm (av. ± SD = 10.3 ± 0.6 × 3.9 ± 0.3 µm, n = 30).
Culture characteristics — Colonies on PDA attaining 14–15 mm diam after 14 d at 26 °C, effuse, hairy, olivaceous brown, raised, usually slow-growing. On OA reaching up to 18–20 mm diam, cottony to floccose at centre, glabrous at periphery, grey olivaceous. On MEA reaching 14–15 mm diam, aerial mycelium moderate, olivaceous black, raised in the centre.
Materials examined. CHINA, Guizhou Province, Shibing County, Fodingshan National Nature Reserve, N27°06'47" E108°10'82", 1180 m a.s.l., from dead branches, 12 Sept. 2020, T.P. Wei (HGUP 18255), living culture GUCC 18255 = CGMCC 3.20560; Guiyang City, Huaxi District, N26°42'55" E106°68'07", 1140 m a.s.l., from forest litter, 6 May 2018, T.P. Wei (HGUP 18256), living culture GUCC 18256 = CGMCC 3.20561; Qingzhen City, N26°54'35" E106°38'74", 1272 m a.s.l., from soil, 16 Apr. 2018, T.P. Wei (HGUP 18257), living culture GUCC 18257 = CGMCC 3.20562; Guiyang City, Guanshan Lake Park, N26°64'28" E106°63'06", 1260 m a.s.l., from lawn soil, 5 Apr. 2021, T.P. Wei (HGUP 18258), living culture GUCC 18258 = CGMCC 3.20563.
Notes — Scolecobasidium constrictum was reported by Abbott (1927) as the type species of Scolecobasidium, which is characterised by having ampulliform or cylindrical conidiogenous cells and broadly ellipsoidal to cylindrical conidia. The phylogenetic result shows that our four isolates cluster together with S. constrictum and are well-supported (Fig. 2). Therefore, we identified our isolates as S. constrictum and provide an illustration of the species.
Scolecobasidium ellipsoideum Ren et al., Mycoscience 54: 422. 2013 — Fig. 13
Mycelium partly superficial or semi-immersed, hyphae simple branched, septate, pale brown, smooth-walled, 1.5–2 µm wide. Conidiophores erect, branched, slightly to distinctly geniculate- sinuous, cylindrical, multi-septate, brown, smooth, (10.5–)11–64.5(–77.5) × (2–)2.5–3.5 µm (av. ± SD = 30.6 ± 19.7 × 2.6 ± 0.3 µm, n = 30). Conidiogenous cells terminal or intercalary, subcylindrical, pale to medium brown, producing conidia sympodially on long open denticles, (4–)6–10(–11) × 2–2.5(–3) µm (av. ± SD = 8.2 ± 2.2 × 2.3 ± 0.2 µm, n = 30). Conidia solitary, ellipsoidal to oblong, 1-septate, pale to dark brown, spinulose, apex subobtuse, base truncate with basal marginal frill, sometimes slightly constricted at the septum, released by rhexolytic secession, 7.5–9.5 × 3–4 µm (av. ± SD = 8.2 ± 0.7 × 3.3 ± 0.3 µm, n = 30).
Culture characteristics — Colonies on PDA attaining 17–25 mm diam after 14 d at 26 °C, spreading, yellow brown, with moderate aerial mycelium. On OA reaching up to 26–32 mm diam, immersed, olivaceous, with sparse aerial mycelium. On MEA reaching 20–24 mm diam, raised in the centre, grey olivaceous.
Materials examined. CHINA, Guizhou Province, Leishan County, N26°24'02" E107°77'22", 1178 m a.s.l., from soil, 12 Mar. 2018, X. Zhang (HGUP 18264), living culture GUCC 18264 = CGMCC 3.20569; Meitan County, N27°75'09" E107°47'99", 910 m a.s.l., from submerged wood, 10 Aug. 2019, T.P. Wei (HGUP 18265), living culture GUCC 18265 = CGMCC 3.20570; Zunyi City, N27°36'27" E106°45'07", 1024 m a.s.l., from forest litter, 10 June 2019, T.P. Wei (HGUP 18266), living culture GUCC 18266 = CGMCC 3.20571.
Notes — Scolecobasidium ellipsoideum was originally described as a saprophyte from soils in Guizhou Province, China (Ren et al. 2013). In this study, phylogenetic inference revealed that our three newly collected isolates clustered together with the ex-type strains of S. ellipsoideum (Fig. 2). The morphological characters of our studied specimens fit well with S. ellipsoideum (Fig. 13). Moreover, S. ellipsoideum differs from the phylogenetically related species S. helicteris by nucleotide differences in ITS (7 bp, 1 %) and tub2 (GUCC 18264: 3 bp, 2 %). No LSU, SSU, act1 and tef1 are available for the ex-type of S. ellipsoideum and S. helicteris.
Scolecobasidium globale (Samerpitak et al.) Crous et al., Stud. Mycol. 96: 211. 2020 — Fig. 14
Fig. 14.

Scolecobasidium globale (GUCC 18249). a–c. Colony on PDA, OA and MEA; d–k. hyphae, conidiophores with sympodially proliferating conidiogenous cells and cylindrical to pyriform and rhexolytic succession conidia; l. germinating conidia. — Scale bars: d–l = 10 µm.
Basionym. Ochroconis globalis Samerpitak et al., Mycol. Progr. 14: 3. 2015.
Mycelium superficial or immersed, hyphae septate, hyaline to pale brown, smooth and thin-walled, 1.5–2 µm wide. Conidiophores differentiated arising directly from vegetative hyphae, straight to flexuous, cylindrical, multi-septate, branched, pale to dark brown, (20.5–)27.5–190(–211.5) × (2.5–)3–4.5(–5.5) µm (av. ± SD = 99.7 ± 59.8 × 3.7 ± 0.8 µm, n = 30). Conidiogenous cells terminal or intercalary, proliferating sympodially, with a single or more denticle-like conidiogenous loci, subhyaline to brown, (6.5–)7.5–22(–30.5) × 3.5–4.5(–5) µm (av. ± SD = 12.7 ± 6.8 × 3.7 ± 0.4 µm, n = 30). Conidia solitary, ellipsoidal to cylindrical, brown, 1(–3)-septate, smooth or somewhat verrucose, constricted at the septum, frills remaining visible on denticle and on conidial base, (7–)8.5–10(–11.5) × 4–5.5(–6) µm (av. ± SD = 9.7 ± 1.6 × 5.1 ± 0.7 µm, n = 30).
Culture characteristics — Colonies on PDA attaining 10– 11 mm diam after 14 d at 26 °C, surface yellow brown, growing slowly, with sparse aerial mycelium. On OA reaching up to 13–16 mm diam, moderately expanding, immersed, olivaceous. On MEA reaching 12–13 mm diam, raised, hairy, brownish olive green.
Materials examined. CHINA, Guizhou Province, Shiqian County, Ganxi Township, Fodingshan National Nature Reserve, N27°40'51" E108°07'21", 1240 m a.s.l., from forest humus, 2 Nov. 2019, T.P. Wei (HGUP 18249), living culture GUCC 18249 = CGMCC 3.20554; Guiyang City, Tianhetan Tourist Holiday Resort, N26°43'95" E106°57'64", 1164 m a.s.l., from soil, 26 Oct. 2020, T.P. Wei (HGUP 18250), living culture GUCC 18250 = CGMCC 3.20555.
Notes — Scolecobasidium globale was introduced by Samerpitak et al. (2015a). In this study, two newly collected isolates clustered together with S. globale and were fully supported phylogenetically (Fig. 2). The comparison of morphological characteristics of the three strains found them to be similar. However, our newly obtained isolates differ from the ex-type culture of S. globale in having more septate conidia (mostly 1-septate in CBS 119644 vs up to 1(–3)-septate in GUCC 18249) (Samerpitak et al. 2015a; Fig. 14), which may depend on the state (mature or immature) of the specimen being observed. Importantly, the ex-type culture (CBS 119644) of S. globale had the following nucleotide similarities with the sequences of our newly collected strain (GUCC 18249). On ITS, LSU, SSU, act1, tub2 and tef1, respectively: 677/687 (99 %, including six gaps), 791/794 (99 %, including two gaps), 1463/1464 (99 %, including one gap), 334/348 (96 %, including 12 gaps), 459/488 (94 %, including seven gaps) and 508/513 (99 %, including one gap). Therefore, we identified our isolates as S. globale, and provide an illustration from a different host, representing a new record from China.
Scolecobasidium minimum (Fassat.) Crous et al., Stud.
Mycol. 96: 212. 2020 — Fig. 15
Fig. 15.
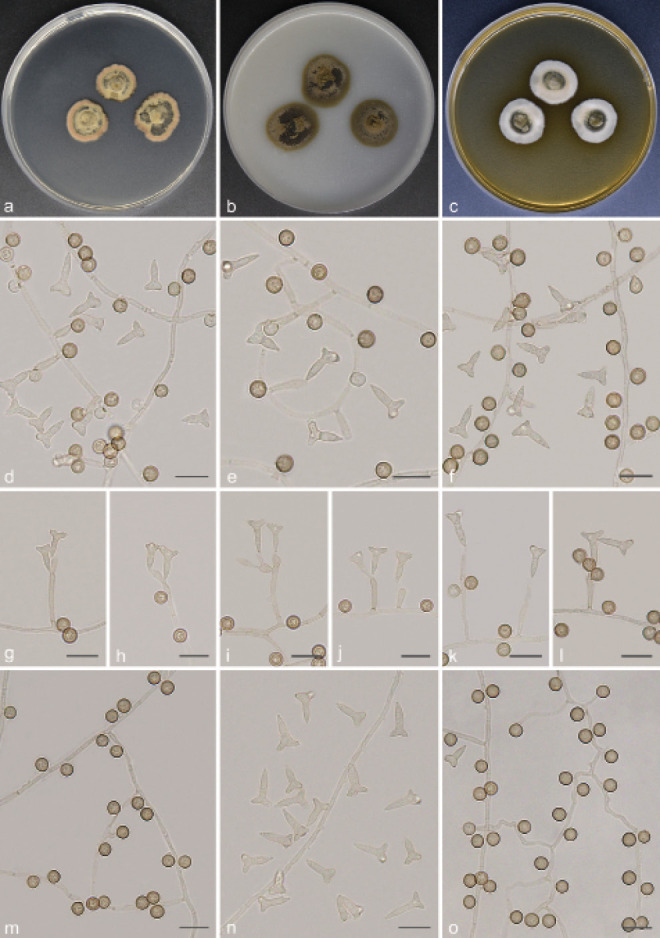
Scolecobasidium minimum (GUCC 18260). a–c. Colony on PDA, OA and MEA; d–l. conidial apparatus with rhexolytic conidia, produced from sympodial conidiogenous cells; n. T- or Y- shaped conidia; m, o. chlamydospores. — Scale bars: d–o = 10 µm.
Basionym. Humicola minima Fassat., Ceská Mykol. 21: 87. 1967.
Synonym. Ochroconis minima (Fassat.) Samerpitak & de Hoog, Fungal Diversity 65: 110. 2013.
Mycelium consisting of hyaline to yellow brown, smooth, septate, branched, 1.5–2.5 µm diam hyphae. Conidiophores hyaline to brown, cylindrical, septate, straight to flexuous, branched, mostly reduced to conidiogenous cells, 7–29(–36.5) × 2.5–4 µm (av. ± SD = 14.6 ± 8.4 × 3.1 ± 0.5 µm, n = 30). Conidiogenous cells flask-shaped to clavate, pale brown, mostly standing at right angles from undifferentiated hyphae, with some scattered denticles in the apical region, 5.5–11.5(–15) × 3–4 µm (av. ± SD = 8.6 ± 2.1 × 3.3 ± 0.4 µm, n = 30). Conidia acrogenous, yellow brown, 1-septate, smooth, somewhat T- or Y- shaped, composed of the main axis and two branches, branch form an angle of 45° with the apex of main axis; main axis 9–14.5 × 3–4 µm (av. ± SD = 10.9 ± 1.3 × 3.3 ± 0.2 µm, n = 30), secondary branches 1.5–5 × 2.5–4.5 µm (av. ± SD = 3.4 ± 0.7 × ٣. 6 ± 0.3 µm, n = ٣٠). Chlamydospores spherical, dark brown, aseptate, smooth, arising directly from vegetative hyphae, 5–6 µm (av. ± SD = 5.4 ± 0.2, n = 30).
Culture characteristics — Colonies on PDA attaining 17– 21 mm diam after 14 d at 26 °C, dark brown at centre, pale yellowish at periphery, fluffy aerial mycelium. On OA reaching up to 20–23 mm diam, pale olivaceous brown, with sparse aerial mycelium. On MEA reaching 19–21 mm diam, raised, centre olivaceous to grey olivaceous and white toward the periphery.
Material examined. CHINA, Guizhou Province, Shiqian County, Fodingshan National Nature Reserve, N27°40'49" E108°07'35", 1100 m a.s.l., from forest humus, 2 Nov. 2019, T.P. Wei (HGUP 18260), living culture GUCC 18260 = CGMCC 3.20565.
Notes — In the current study, the phylogenetic result shows that our new collection GUCC 18260 clusters together with S. minimum, sharing a sister relationship to S. ramosum and S. icarus with high statistical support from three independent algorithms (Fig. 2). All three species have T- or Y-shaped conidia. Chen et al. (2020) obtained S. minimum from sugarcane and banana rhizospheres in Guangxi. Therefore, this is the second report of this species from China.
Scolecobasidium ramosum (Giraldo et al.) Crous et al., Stud. Mycol. 96: 212. 2020 — Fig. 16
Fig. 16.
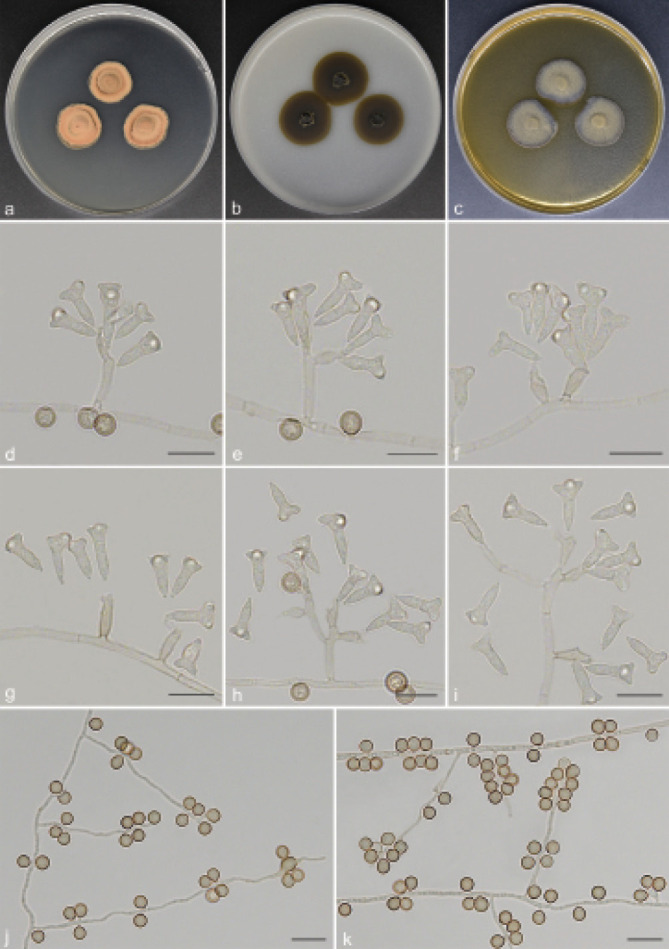
Scolecobasidium ramosum (GUCC 18261). a–c. Colony on PDA, OA and MEA; d–i. hyphae, conidiophores with sympodially proliferating conidiogenous cells and T- or Y- shaped and rhexolytic succession conidia; j–k. chlamydospores. — Scale bars: d–k = 10 µm.
Basionym. Ochroconis ramosa Giraldo et al., J. Clinical Microbiol. 52: 4197. 2014.
Mycelium composed of branched, septate, hyaline to pale brown, smooth-walled, 1.5–2 µm diam hyphae. Conidiophores pale brown, clavate or cylindrical with beaked apex, simple or sympodially branched, often reduced to conidiogenous cells, 7.5–64 (–69.5) × 3–4 µm (av. ± SD = 21.2 ± 14.6 × 3.1 ± 0.3 µm, n = 30). Conidiogenous cells subhyaline to pale brown, bearing one or more denticles in the apical region, ampulliform to cylindrical, 5.5–14.5(–17.5) × 2.5–4 µm (av. ± SD = 9.1 ± 2.9 × 3.1 ± 0.4 µm, n = 30). Conidia solitary, pale brown, 1-septate, trilobate, T- or Y-shaped, smooth or verrucose, base slightly tapered to truncate hilum, main axis 10–13.5 × 3–4 µm (av. ± SD = 11.3 ± 0.9 × 3.5 ± 0.2 µm, n = 30), secondary branches 1.5–6.5 × 3–5.5 µm (av. ± SD = 3.5 ± 1.2 × 4 ± ٠.٤ µm, n = ٣٠), released by rhexolytic secession. Chlamydospores solitary, growing directly on vegetative hyphae, globose or subglobose, aseptate, dark brown, sessile, smooth, 4.5–5.5 µm (av. ± SD = 4.5 ± 0.2 µm, n = 30).
Culture characteristics — Colonies on PDA attaining 17–20 mm diam after 14 d at 26 °C, slightly raised, yellow-brown, spreading. On OA reaching up to 21–23 mm diam, flat, chocolate brown, felty at center, membranous toward the periphery. On MEA reaching 19–20 mm diam, olivaceous grey, velvety to cottony, slightly raised to umbonate.
Materials examined. CHINA, Guizhou Province, Zunyi City, Phoenix Mountain National Forest Park, N27°36'27" E106°45'07", 1024 m a.s.l., from forest litter, 10 June 2019, T.P. Wei (HGUP 18261), living culture GUCC 18261 = CGMCC 3.20566; Qingzhen City, N26°54'35" E106°38'74", 1272 m a.s.l., from soil, 16 Apr. 2018, T.P. Wei (HGUP 18262), living culture GUCC 18262 = CGMCC 3.20567; Leishan County, N26°24'02" E107°77'22", 1178 m a.s.l., from forest litter, 12 Mar. 2018, T.P. Wei (HGUP 18263), living culture GUCC 18263 = CGMCC 3.20568.
Notes — Scolecobasidium ramosum is phylogenetically closely related to S. minimum and S. icarus (Fig. 2). The sequences of ITS, LSU, SSU, act1, tub2 and tef1 gene regions can differentiate S. ramosum from these two species. This species was originally collected from human nails in the USA (Giraldo et al. 2014). Recently, Chen et al. (2020) isolated this species from the soil rhizosphere of sugarcane, and the strain we obtained was derived from forest litter and soil, which indicates that S. ramosum has both a parasitic and saprophytic lifestyle.
Scolecobasidium tshawytschae (Doty & D.W. Slater) McGinnis & Ajello, Trans. Brit. Mycol. Soc. 63: 202. 1974 — Fig. 17
Fig. 17.
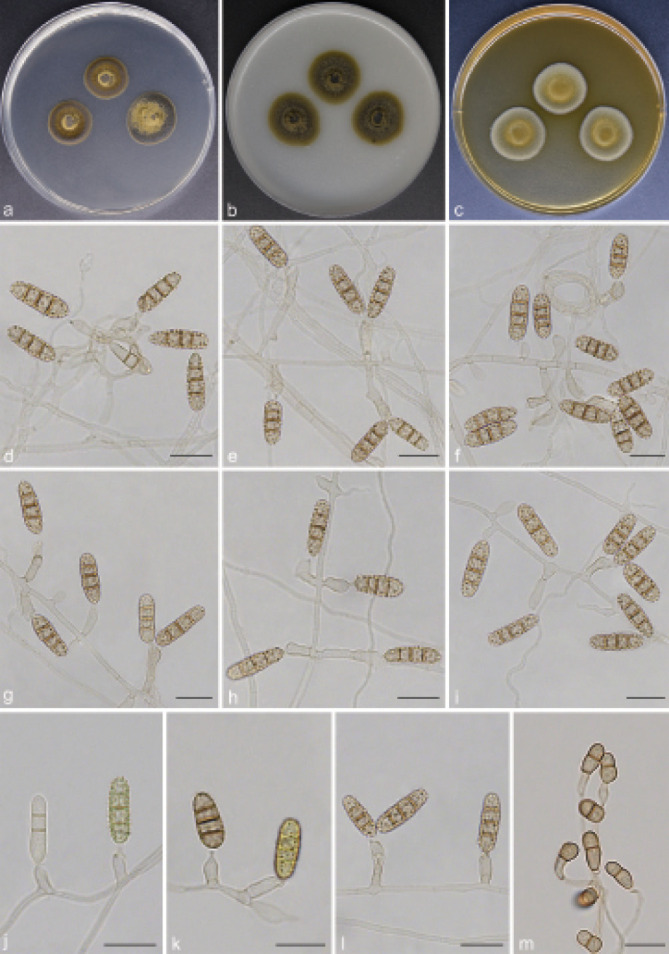
Scolecobasidium tshawytschae (GUCC 18254). a–c. Colony on PDA, OA and MEA; d–h. conidial apparatus with rhexolytic conidia; i. conidiophores reduced to conidiogenous cells; j–l. maturation process of conidia; m. chlamydospores. — Scale bars: d–m = 10 µm.
Basionym. Heterosporium tshawytschae Doty & D.W. Slater, Amer. Midl. Naturalist 36: 663. 1946.
Synonym. Ochroconis tshawytschae (Doty & D.W. Slater) Kiril. & Al-Achmed, Mykrobiol. Zhurn. 39: 305. 1977.
Mycelium superficial or immersed, hyphae branched, septate, hyaline to pale brown, smooth and thin-walled, 1.5–2.5 µm wide. Conidiophores erect, cylindrical, straight to gently curved, septate, unbranched, mostly reduced to conidiogenous cells, (5.5–)6.5–18.5(–24) × 3–3.5(–4) µm (av. ± SD = 11.6 ± 5.1 × 3.2 ± 0.3 µm, n = 30). Conidiogenous cells integrated, terminal, often somewhat inflated, lageniform or ampulliform, with one to several conidiogenous loci, leaving minute collarettes on denticulate loci, 5–7.5(–9) × 4–5 µm (av. ± SD = 6.5 ± 1.1 × 4.1 ± 0.4 µm, n = 30). Conidia solitary, verrucose, pale to dark brown, 1(–3)-septate, cylindrical or slightly clavate, the colour of immature conidia changed from greenish olivaceous to yellow to dark brown, 12.5–22(–24.5) × (4.5–)5–6(–6.5) µm (av. ± SD = 16.4 ± 1.6 × 5.2 ± 0.4 µm, n = 30). Chlamydospores acrogenous, brown, smooth, 0(–1)-septate, ellipsoidal to fusoid, arising directly from vegetative hyphae, 7.5–13 × (4–)4.5–6(–6.5) µm (av. ± SD = 9.7 ± 1.9 × 4.8 ± 0.6 µm, n = 30).
Culture characteristics — Colonies on PDA attaining 15– 18 mm diam after 14 d at 26 °C, olivaceous at centre, dark brown at periphery, slightly raised to umbonate. On OA reaching up to 20–22 mm diam, felty, olivaceous brown to olive, submerged mycelium. On MEA reaching 20–22 mm diam, slightly domed, woolly, with dense dark hairs at the centre and grey loose mycelium near the edge.
Materials examined. CHINA, Guizhou Province, Guiyang City, Guanshan Lake Park, N26°64'28" E106°63'06", 1260 m a.s.l., from lawn soil, 5 Apr. 2021, T.P. Wei (HGUP 18251), living culture GUCC 18251 = CGMCC 3.20556; Zunyi City, N27°36'27" E106°45'07", 1024 m a.s.l., from plant litter, 10 June 2019, T.P. Wei (HGUP 18252), living culture GUCC 18252 = CGMCC 3.20557; Guiyang City, Huaxi District, N26°42'55" E106°68'07", 1140 m a.s.l., isolated from soil, 6 May 2018, T.P. Wei (HGUP 18253), living culture GUCC 18253 = CGMCC 3.20558; Shiqian County, Pingshan Township, N27°40'50" E108°07'30", 1100 m a.s.l., isolated from soil, 2 Nov. 2019, T.P. Wei (HGUP 18254), living culture GUCC 18254 = CGMCC 3.20559.
Notes — Scolecobasidium tshawytschae was originally isolated as the etiologic agent of kidney mycosis in chinook salmon (Oncorhynchus tshawytscha) smolts (Doty & Slater 1946). The four strains (GUCC 18251 to GUCC 18254) obtained in the present study and many strains that have been reported so far originate in soil and plant litter (Barron & Busch 1962, Hamayun et al. 2009, Samerpitak et al. 2014), indicating that this species can also be saprophytic, corroborating the supposition that its infective ability in fish is purely opportunistic.
Verruconis Samerpitak et al., Fungal Diversity 65: 117. 2013 ‘2014’
Type species. Verruconis gallopava (W.B. Cooke) Samerpitak & de Hoog.
Notes — Verruconis was established by Samerpitak et al. (2014) to accommodate thermophilic species separated from Ochroconis (O. calidifluminalis and O. gallopava) and Scolecobasidium (S. verruculosum), with the type species, V. gallopava being an opportunistic neurotropic pathogen (Salkin et al. 1990, Seyedmousavi et al. 2013, Wang et al. 2018, Samerpitak et al. 2019). Therefore, thermophilicity and unbranched conidia were the main characteristics distinguishing this genus from Ochroconis. Subsequently, Zhang et al. (2018) and Qiao et al. (2019) successively placed the mesophilic species V. panacis, V. hainanensis and V. pseudotricladiata with Y- or T-shaped and cylindrical conidia under Verruconis. They found that Verruconis is not limited to thermophilic species with clavate to cylindrical conidia (Hernández-Restrepo et al. 2020). However, the addition of these species blurred the major distinguishing feature between Verruconis and Ochroconis. In contrast, the molecular systematics has played an important role in the taxonomy of these two genera (Machouart et al. 2014, Huanraluek et al. 2019, Shen et al. 2020). Samerpitak et al. (2016) revealed that the concatenated dataset of SSU, ITS, LSU, act1, tub2 and tef1 served as a reference for genus and species delimitations of Ochroconis and Verruconis (Lackner et al. 2014, Al-Hatmi et al. 2016). It is noteworthy that V. mangrovei is the first reported sexual species in Verruconis (Hyde et al. 2020).
Verruconis cylindricalis T.P. Wei & Y.L. Jiang, sp. nov. — MycoBank MB 840928; Fig. 18
Fig. 18.
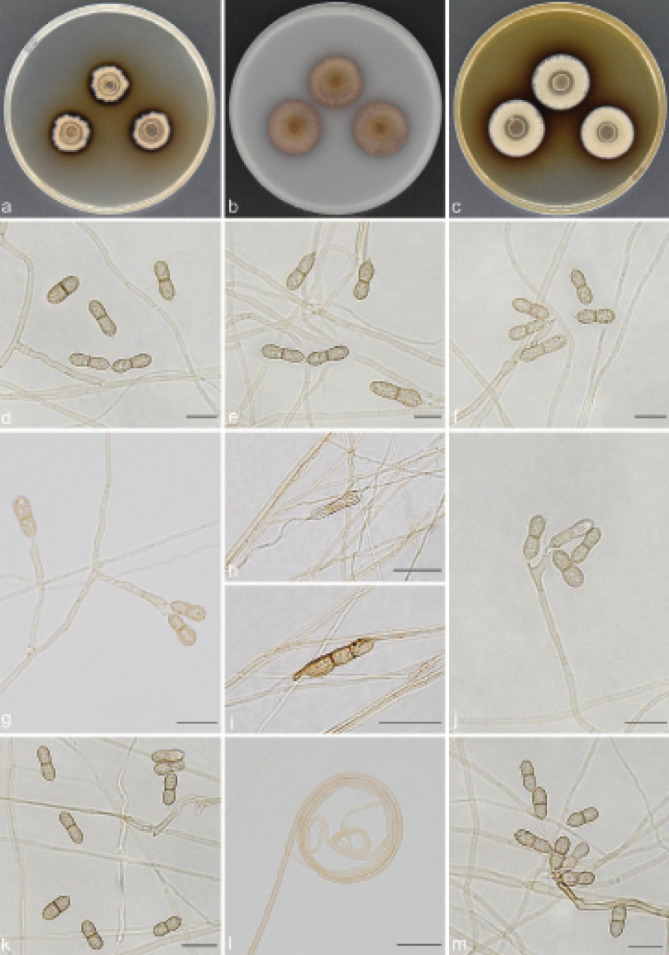
Verruconis cylindricalis (culture ex-type GUCC 18299). a–c. Colony on PDA, OA and MEA; d–g, j–k, m. conidial apparatus with rhexolytic conidia, produced from sympodial conidiogenous cells; h–i. chlamydospores; l. hyphal coil. — Scale bars: h–i, l = 20 µm, all others = 10 µm.
Etymology. The epithet refers to its cylindrical chlamydospores.
Typus. CHINA, Guizhou Province, Shiqian County, Fodingshan National Nature Reserve, N27°40'49" E108°07'35", 1100 m a.s.l., from forest humus, 2 Nov. 2019, T.P. Wei (holotype HGUP 18299, isotype CGMCC 3.20573, culture ex-type GUCC 18299).
Mycelium consisting of branched, subhyaline or pale brown, smooth, 1–3 µm thick hyphae, frequently forming hyphal coils. Conidiophores solitary, straight or flexuous, subcylindrical, pale brown, sparsely septate, (21–)27–57.5(–87.5) × 2.5–3(–3.5) µm (av. ± SD = 44.3 ± 22.2 × 2.6 ± 0.3 µm, n = 30). Conidiogenous cells integrated, terminal, sympodial, denticulate, bearing 1–2 conidium at the apex, (8–)9.5–16(–17.5) × 2.5–3(–3.5) µm (av. ± SD = 12.0 ± 3.6 × 2.5 ± 0.4 µm, n = 30). Conidia sparse on PDA and MEA, broadly ellipsoidal with prominent hila, 1-septate, minutely echinulate, brown to olivaceous brown, slightly constricted at the septum, (7–)7.5–12 × 4–5 µm (av. ± SD = 9.4 ± 1.2 × 4.3 ± 0.2 µm, n = 30). Chlamydospores intercalary, verruculose, cylindrical or clavate, slightly tapered at both ends, 2-septate, often asymmetric with smaller middle cells, olivaceous brown, usually constricted at the septum, 25–29.5 × 4.5–6 µm (av. ± SD = 26.9 ± 2.2 × 5.1 ± 0.6 µm, n = 30).
Culture characteristics — Colonies on PDA attaining 17– 20 mm diam after 14 d at 26 °C, brown, margin irregular, submerged mycelium. On OA reaching up to 23–25 mm diam, velvety, dark brown, woolly at centre. On MEA reaching 20–23 mm diam, hairy, olivaceous brown. On PDA and MEA reverse blood colour with diffuse blood pigment spreading into agar.
Notes — Verruconis cylindricalis shares a few morphological similarities with V. thailandica and V. calidifluminalis in having two-celled conidia with protuberant hila. However, V. thailandica produces smaller, verrucose conidia (5–7 × 2.2–3.1 µm vs 7–12 × 4–5 µm), with a wing-like gelatinous brown sheath (Hernández-Restrepo et al. 2020). The conidia of the V. calidifluminalis are cylindrical to clavate, pale to dark brown and larger (9.5–20.5 × 2.5–5.0 µm vs 7–12 × 4–5 µm) (Yarita et al. 2010). Additionally, V. cylindricalis has longer conidiophores, cylindrical or clavate and 2-septate chlamydospores. These characters form the most notable differences with respect to V. thailandica and V. calidifluminalis. Phylogenetically, V. cylindricalis clustered in a distinct clade with full support from three independent algorithms (Fig. 2).
Verruconis tricladiata (Matsush.) T.P. Wei & Y.L. Jiang, comb. nov. — MycoBank MB 842456
Basionym. Scolecobasidium tricladiatum Matsush., Microfungi Solomon Isl. Papua-New Guinea: 52. 1971.
Description — Matsushima (1971).
Notes — Scolecobasidium tricladiatum was introduced based on its Y- or T-shaped or ellipsoidal to fusoid conidia, a species previously isolated from rotten leaves in Papua New Guinea (Matsushima 1971). Although we did not locate the type specimen, we have examined the DNA sequence data of another culture lodged under this species name in GenBank. Multi-locus phylogenetic analysis showed it to cluster in Verruconis, as sister to V. pseudotricladiata (Fig. 2). Nevertheless, S. tricladiatum and V. pseudotricladiata are genetically distinct in 6 bp (1 %), 87 bp (10 %) and 85 bp (9 %) in SSU, LSU and tef1 loci. No ITS, act1 and tub2 data are currently available for S. tricladiatum. Morphologically, S. tricladiatum differs from V. pseudotricladiata by its moniliform, irregularly branched conidiophores and mostly unbranched, pale olivaceous to brown, verruculose conidia (Matsushima 1971, Qiao et al. 2019). Based on morphological characters and phylogenetic analyses, we therefore transferred S. tricladiatum to Verruconis.
Verruconis thailandica Giraldo López & Crous, Fung. Syst. Evol. 6: 21. 2020 — Fig. 19
Fig. 19.
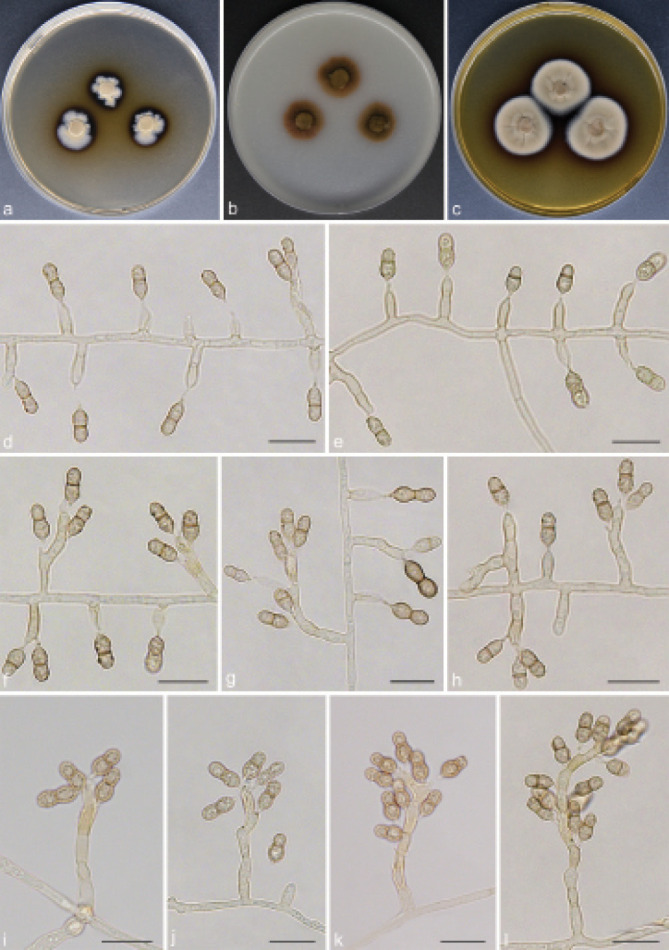
Verruconis thailandica (GUCC 18267). a–c. Colony on PDA, OA and MEA; d–e. conidiophores reduced to conidiogenous cells; f–l. hyphae, conidiophores with sympodially proliferating conidiogenous cells and ellipsoidal and rhexolytic succession conidia. — Scale bars: d–l = 10 µm.
Mycelium composed of branched, septate, pale brown, smooth, thin-walled, 2–2.5 µm diam hyphae. Conidiophores erect, arising directly from vegetative hyphae, sometimes reduced to conidiogenous cells, simple or branched, pale brown, septate, subcylindrical, straight or slightly curved, (7.5–)9.5–45(–52.5) × 3–4 µm (av. ± SD = 19.3 ± 10.9 × 3.2 ± 0.3 µm, n = 30). Conidiogenous cells terminal or intercalary, flask-shaped to clavate, 5–13.5(–14) × 3–4 µm (av. ± SD = 7.5 ± 2.2 × 3.3 ± 0.3 µm, n = 30), producing conidia sympodially on long open denticles; denticles cylindrical, pale brown, up to 1 µm long. Conidia soli- tary, broadly ellipsoidal with a protuberant hilum, strongly constricted at the septum, 1-septate, brown, finely echinulate to verrucose, sometimes with a wing-like gelatinous brown sheath, released by rhexolytic secession, 7–11(–11.5) × 3.5–5 µm (av. ± SD = 8.4 ± 1.0 × 3.9 ± 0.3 µm, n = 30).
Culture characteristics — Colonies on PDA attaining 16–18 mm diam after 14 d at 26 °C, felty, growing slowly, margin irregular, grey olivaceous. On OA reaching up to 17–18 mm diam, cottony to floccose, immersed, dark brown. On MEA reaching 24–26 mm diam, grey olivaceous, aerial mycelium moderate. On PDA and MEA with ochreous diffusible pigment.
Material examined. CHINA, Guizhou Province, Guiyang City, Huaxi Wetland Park, N26°43'92" E106°67'76", 1140 m a.s.l., isolated from the humus soil in the stream, 16 Nov. 2020, T.P. Wei (HGUP 18267), living culture GUCC 18267 = CGMCC 3.20572.
Notes — Multi-locus phylogenetic analyses indicate that our newly obtained isolates GUCC 18267 clustered together with V. thailandica, and were sister to V. terricola, V. verruculosa and V. cylindricalis. Morphologically, V. thailandica can be readily distinguished from other members of Verruconis by its two-celled ellipsoidal conidia with a wing-like gelatinous sheath (Hernández-Restrepo et al. 2020).
Matsushimaea Subramanian, Kavaka 5: 96. 1977 ‘1978’
Type species. Matsushimaea fasciculata (Matsush.) Subramanian.
Notes — Matsushimaea was introduced by Subramanian (1977) to accommodate species segregated from Torula (T. fasciculata), which are characterised by the production of sessile, branched and aseptate conidia from polyblastic sympodial coni- diogenous cells. This genus was formerly placed in the Pezizomycotina as incertae sedis (Castañeda-Ruiz et al. 1996, Matsushima 1996). Later, Crous et al. (2018b) using the rDNA (ITS and LSU) sequence data elucidated the phylogenetic position of Matsushimaea and placed it in Sympoventuriaceae. Presently, Matsushimaea includes four species, M. fasciculata, M. fertilis, M. magna and M. monilioides (Castañeda-Ruiz et al. 1996, Matsushima 1996, Crous et al. 2018b). Among them, M. fertilis and M. magna lack authentic cultures and DNA sequence data, thus their phylogenetic position remains unknown.
Matsushimaea fasciculata (Matsush.) Subram., Kavaka 5: 96. 1977 — Fig. 20
Fig. 20.

Matsushimaea fasciculata (GUCC 18239). a–c. Colony on PDA, OA and MEA; d–l. conidia in simple or branched chains arising from conidiogenous cells; m–o. olivaceous to plate brown, subglobose or pyriform and smooth-walled conidia. — Scale bars: d, n = 20 µm, all others = 10 µm.
Basionym. Torula fasciculata Matsush., Icon. Microfung. Matsush. Lect. (Kobe): 153. 1975.
Mycelium superficial or immersed, hyphae branched, septate, pale brown, smooth, occasionally with irregular swellings, 1.5–2 µm wide. Conidiophores reduced to conidiogenous cells arising directly from vegetative hyphae. Conidiogenous cells integrated, terminal or intercalary, mono- or polyblastic, pale brown, cylindrical to inflated, smooth-walled, 4–5.5 × 4–5 µm (av. ± SD = 4.4 ± 0.4 × 4.1 ± 0.3 µm, n = 30). Conidia catenate, usually formed in branched chains, straight to sometimes curved, up to (9.5–)14–50.5(–53) µm (av. ± SD = 27.5 ± 9.5 µm, n = 30) long; cells subglobose or ellipsoidal to somewhat pyriform, sessile or on short protrusions, aseptate, smooth, pale to dark brown, 3.5–5 × 3.5–5 µm (av. ± SD = 3.9 ± 0.4 × 4.1 ± 0.4 µm, n = 30).
Culture characteristics — Colonies on PDA attaining 14–15 mm diam after 14 d at 26 °C, velvety, olivaceous at centre, dark brown at periphery, margin entire. On OA reaching up to 15–19 mm diam, dark brown, dusty, flat, with sparse aerial mycelium. On MEA reaching 15–16 mm diam, olivaceous grey, floccose to loosely cottony.
Material examined. CHINA, Guizhou Province, Guiyang City, Tianhetan Tourist Holiday Resort, N26°43'95" E106°57'64", 1164 m a.s.l., from soil, 16 Apr. 2018, T.P. Wei (HGUP 18239), living culture GUCC 18239 = CGMCC 3.20544.
Notes — In this study, the phylogenetic analysis is partly consistent with the morphological comparison, and our isolate GUCC 18239 and M. fasciculata have basically the same morphological characteristics (Fig. 1). All members of Matsushimaea have been isolated from forest litter and soil, which indicates that the lifestyle of this genus is probably saprophytic (Fig. 3). Moreover, this second report of this poorly known taxon extends its distribution to southwest China from its original location in Japan (Matsushima 1975).
Mycosisymbrium Carris, Mycologia 86: 132. 1994
Type species. Mycosisymbrium cirrhosum Carris.
Notes — The monotypic genus Mycosisymbrium was first described by Carris (1994), based on M. cirrhosum collected from dead leaves of Vaccinium macrocarpon in Massachusetts. It is characterised by discrete aggregates of conidiophores terminating in sterile, filiform appendages and brown, 1-septate conidia. Initially, this genus was treated as incertae sedis in Pezizomycotina. Pratibha & Prabhugaonkar (2016) confirmed the phylogenetic placement of Mycosisymbrium, which is a well-supported sister genus to Ochroconis and Verruconis in Sympoventuriaceae. It is noteworthy that since Mycosisymbrium was described, only Pratibha & Prabhugaonkar (2016) have reported on this species.
Mycosisymbrium cirrhosum Carris, Mycologia 86: 132. 1994 — Fig. 21
Fig. 21.
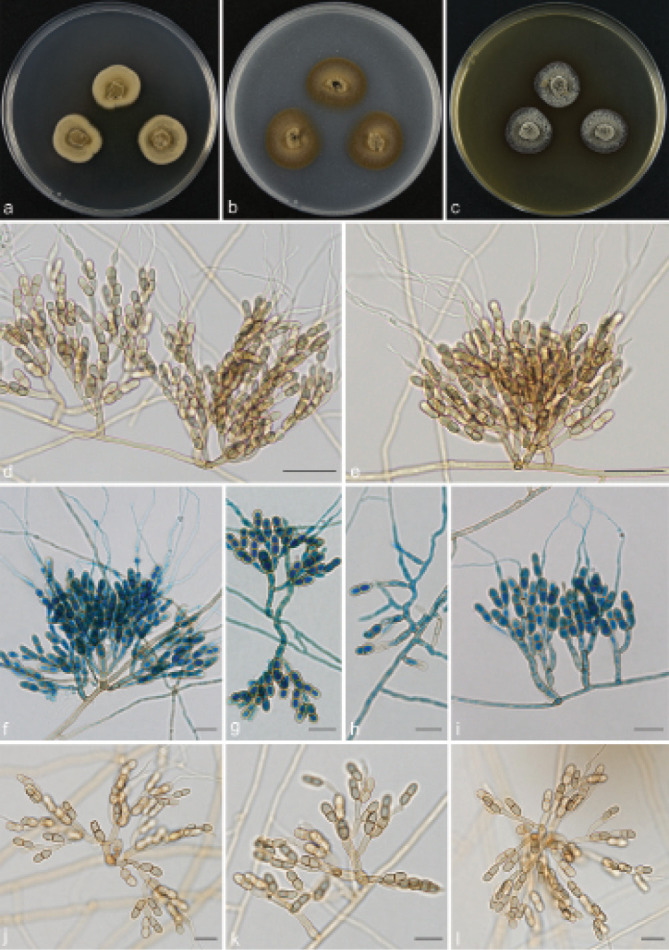
Mycosisymbrium cirrhosum (GUCC 1837). a–c. Colony on PDA, OA and MEA; d–e. filiform appendages terminating from conidiophores; f–i. conidia stained with lactic acid phenol cotton blue; j–l. conidiophore aggregates with conidia. — Scale bars: d–e = 20 µm, all others = ١٠ µm.
Mycelium consisting of brown, septate, branched, smooth, thick-walled, 1.5–2 µm diam hyphae. Conidiophores in determinate clusters, discrete, infundibuliform, pale to dark brown, smooth, branched, each branch terminating in a filiform appendage, (21.5–)24–50(–57.5) × 3–4 µm (av. ± SD = 37.9 ± 9.3 × 3.2 ± 0.3 µm, n = 30); appendages hyaline, flexuous, up to (29–)47–85.5(–90.5) µm (av. ± SD = 63.4 ± 16.9 µm, n = 30) long. Conidiogenous cells terminal or intercalary, mono- to polyblastic, cylindrical, pale brown, with one to two denticle-like conidiogenous loci inconspicuous to slightly prominent, (4.5–)5.5–10.5(–11) × (2–)2.5–3.5(–4) µm (av. ± SD = 7.8 ± 1.2 × 2.6 ± 0.3 µm, n = 30). Conidia solitary, oblong, 1-septate, smooth-walled, with bluntly rounded ends, constricted at median septum, 9–12 × (3.5–)4–5 µm (av. ± SD = 9.8 ± 0.8 × 4.0 ± 0.3 µm, n = 30).
Culture characteristics — Colonies on PDA attaining 24–27 mm diam after 14 d at 26 °C, margin effuse, brown, with moderate aerial mycelium. On OA reaching up to 16–25 mm diam, mycelium immersed, dark brown, felty or granulose. On MEA reaching 23–24 mm diam, woolly to loosely cottony, olivaceous grey.
Material examined. CHINA, Guizhou Province, Meitan County, N27°41'08" E107°25'41", 910 m a.s.l., isolated from decaying Camellia sinensis leaf litter, 10 Aug. 2019, T.P. Wei (HGUP 1837), living culture GUCC 1837 = CGMCC 3.20541.
Notes — Multi-locus phylogenetic analyses indicate that our isolate GUCC 1837 clusters with M. cirrhosum with high statistical support. Furthermore, the two strains are morphologically similar (Fig. 21). This is the third report of M. cirrhosum, which was previously isolated in Massachusetts, USA (Carris 1994) and Goa, India (Pratibha & Prabhugaonkar 2016). In our study we extended its distribution to southwest China and added another new host record.
DISCUSSION
The controversy of Ochroconis, Scolecobasidium and Verruconis
Ochroconis, Scolecobasidium and Verruconis are morphologically and phylogenetically similar. Historically, there is some disagreement and incongruence about the phylogenetic placement, circumscription and classification of these three genera (De Hoog & Von Arx 1973, Ren et al. 2013, Giraldo et al. 2014, Samerpitak et al. 2014). Although several taxonomic revisions of these genera have been made based on morphology and phylogeny, their species boundaries have not been completely resolved due to historical confusion and limited molecular data (Samerpitak et al. 2016, Qiao et al. 2019, Shen et al. 2020). In this study, our multi-locus phylogenetic analyses indicated that Scolecobasidium and Ochroconis are synonymous and sister to Verruconis, and reside in Sympoventuriaceae (Fig. 1,2). Morphologically, Verruconis and Scolecobasidium are distinguished mainly based on their conidial shape; in Verruconis the conidia are pale to dark brown, verrucose to coarsely ornamented, with protuberant hila, while in Scolecobasidium the conidial shape is more variable from ellipsoidal, cylindrical, bilobate, to T- or Y-shaped, and conidia are smooth-walled to verruculose. Additionally, the dark brown diffuse red colony pigmentation on PDA of Verruconis also distinguishes this genus from Scolecobasidium, which tends to have a pale luteus pigment. The chlamydospores seem to be another taxonomically important feature, in Scolecobasidium they are spherical or subcylindrical, smooth-walled, but in Verruconis they are cylindrical or clavate, verruculose and larger. Furthermore, Verruconis includes thermophilic and mesophilic species, while Scolecobasidium only has mesophilic species.
The species boundaries drawn in this study are mainly based on the multi-locus phylogeny as well as morphology. Compared to previous studies, more samples and additional gene markers were used to provide a better understanding of the phylogenetic relationships among species of Scolecobasidium and Verruconis. By comparing morphological characteristics and related DNA sequence data, seven new species were proposed in Scolecobasidium and Verruconis, namely S. camellicola, S. coiledmyces, S. echinulatum, S. obovoideum, S. verrucaria, S. zunyiense and V. cylindricalis. We also proposed six new combinations in Scolecobasidium and Verruconis as S. ferulica, S. guangxiensis, S. helicteris, S. leishanicola, S. mirabilis and V. tricladiata. The number of species in Scolecobasidium and Verruconis has increased significantly over the years (Crous et al. 2019c, Shen et al. 2020). Scolecobasidium and Verruconis are rather common genera of saprotrophic soil hyphomycetes, some of which are opportunistic neurotropic pathogens of humans, fish or other animals, and some are also known for their thermophilic properties (Samerpitak et al. 2014, 2019). Scolecobasidium is the largest genus of Sympoventuriaceae with 88 accepted species names recorded in MycoBank (http://www.mycobank.org, April 2022), 39 of which are devoid of DNA sequence data in GenBank. Therefore, to resolve the phylogenetic position of these species in Scolecobasidium, they need to be re-collected, sequenced and epitypified.
Genera of Sympoventuriaceae
Sympoventuriaceae was originally established for three groups, namely Sympoventuria, Veronaeopsis and fusicladium-like species (Zhang et al. 2011). However, as discussed by Zhang et al. (2011), the fusicladium-like morphs are polyphyletic and include species residing in another different family, Venturiaceae. Machouart et al. (2014) transferred Scolecobasidium (= Ochroconis) and Verruconis to the Sympoventuriaceae, thereby expanding the concept of the family. Subsequent phylogenetic studies further expanded the concept of Sympoventuriaceae, making it the largest family of Venturiales (Johnston & Park 2016, Tibpromma et al. 2018, Crous et al. 2019a, Shen et al. 2020). Although these advances have allowed the resolution of several long-standing questions concerning the generic boundaries of Sympoventuriaceae, many questions remain unresolved about the phylogenetic relationships of some taxa, especially genera and species for which molecular data are not yet available. To better define the generic boundaries and reveal the evolutionary relationship of Sympoventuriaceae, we carried out a more comprehensive analysis of this group based on a hitherto most complete sequence dataset consisting of seven loci (SSU, ITS, LSU, act1, tub2, tef1 and rpb2). The present results resolved 22 well-supported monophyletic lineages, representing 22 genera (Fig. 1), viz., Acroconidiellina, Bellamyces, Clavatispora, Echinocatena, Fuscohilum, Guizhoumyces, Helicopsis, Matsushimaea, Melnikomyces, Mycosisymbrium, Neocoleroa, Neofusicladium, Parafusicladium, Pinaceicola, Pseudosigmoidea, Scolecobasidium, Sterila, Sympoventuria, Troposporella, Veronaeopsis, Verruconis and Yunnanomyces. These included fungi with a broad spectrum of morphology, lifestyles and modes of nutrition, accommodating saprophytes, endophytes, plant pathogens, and animal or human opportunistic pathogens (Samerpitak et al. 2014, 2019, Wang et al. 2018, Crous et al. 2020).
The number of known taxa in Sympoventuriaceae is increasing at a steady pace as more geographic areas and habitats are investigated, and its taxonomy has also changed dramatically (Wijayawardene et al. 2014, Tibpromma et al. 2018, Shen et al. 2020). In this study, in addition to the 15 genera previously placed in Sympoventuriaceae (Table 4), we also reassessed species of Acroconidiellina, Clavatispora, Guizhoumyces, Matsushimaea, Melnikomyces, Mycosisymbrium and Yunnanomyces. The morphological characters and molecular data of these genera provide significant evidence for their taxonomic placement in Sympoventuriaceae. In the multi-locus phylogenetic tree of the current study, A. arecae is allied with Scolecobasidium at the top of the clade (Fig. 1). Nevertheless, A. arecae can be distinguished morphologically from the species in Scolecobasidium by its multi-septate, obclavate, large conidia with a truncate base and macronematous conidiophores (Li et al. 2016). The placement of Acroconidiellina in Sympoventuriaceae contradicts earlier placements in the order Pleosporales due to similarities in the type of conidiophores and conidium morphology (Ellis 1971, Zhang et al. 2009, Bhat 2010). Therefore, our results are more in line with the proposal of Hernández-Restrepo et al. (2016) about a closer affinity with members of the Sympoventuriaceae. Furthermore, our findings support the placement of Mycosisymbrium in Sympoventuriaceae, as suggested by previous molecular studies (Pratibha & Prabhugaonkar 2016). The extended taxon sampling and the use of five markers allow us to strongly corroborate these findings. Species of two genera were included in the analyses, Mycosisymbrium and Clavatispora, which formed two different sister subclades (Fig. 1). Mycosisymbrium can be distinguished by having discrete aggregates of conidiophores terminating in sterile, filiform appendages and 1-septate conidia (Fig. 21), while Clavatispora has 1(–3)-septate, guttulate conidia that are deeply constricted at their septa (Boonmee et al. 2014). Clavatispora also produced a sexual morph in culture characterised by ostiolate ascomata, bitunicate asci and muriformly septate ascospores.
According to the multigene phylogeny and morphology in the present study, a new genus, Guizhoumyces, is now recognised within Sympoventuriaceae (Fig. 1). Morphologically, members of Guizhoumyces are quite distinct from those of Sympoventuriaceae, having straight or curved, smooth and acicular to obclavate conidia, which contrasts with the mostly verrucose to denticulate and subcylindrical to fusoid-ellipsoidal conidia typical for species of other genera; as well as by the presence of anastomosis between mature conidia in Guizhoumyces. Moreover, Matsushimaea forms a robust clade with another two genera, Fuscohilum and Neocoleroa (Fig. 1). As we mentioned before, our results confirm its placement within Sympoventuriaceae, although the DNA sequences of M. fertilis and M. magna were not available. The polyblastic and sympodial conidiogenous cells of several other genera in Sympoventuriaceae share similar morphologies with those of Matsushimaea. However, Matsushimaea differs in having sessile, branched and aseptate conidia arising directly from vegetative hyphae (Crous et al. 2018b; Fig. 20), which strongly supports the genus as monophyletic. Based on the general characteristics of Melnikomyces, previous studies hypothesized its close relationship with Scolecobasidiella and Scolecobasidium (Crous et al. 2014, Hernández-Restrepo et al. 2020). In our phylogenetic tree, Melnikomyces formed a separate clade in Sympoventuriaceae distinct from other genera (Fig. 1). Morphologically, this genus has certain similarities with Scolecobasidiella and Scolecobasidium, but can be distinguished from them by its fusoid-ellipsoidal conidia and the chlamydospores that are subglobose, occurring in branched chains (Wei et al. 2020). Thus, our analysis shows that the phylogenetic isolation of Melnikomyces, Scolecobasidiella and Scolecobasidium may be supported by the unique morphological features and genetic differences. Yunnanomyces is characterised by its globose to broadly oval, yellow to brown and muriformly septate conidia (Tibpromma et al. 2018). This genus is phylogenetically related to the monotypic genus Sterila, and formed a well-supported clade within Sympoventuriaceae (Fig. 1). Morphologically it is not possible to compare these two genera, as the latter is sterile in culture. Nonetheless, Yunnanomyces differs from its closest phylogenetic neighbour Sterila by unique fixed alleles in four loci of their type species, by 6 bp in ITS (3 %), 52 bp in LSU (7 %), 278 bp in tub2 (57 %) and 191 bp in rpb2 (24 %). More specifically, these two genera represent significantly different lineages in Sympoventuriaceae.
Sympoventuria currently includes three species, namely S. africana, S. capensis and S. melaleucae, with the sexual species S. capensis designated as the generic type (Crous et al. 2007a, 2017). Sympoventuria is phylogenetically closely related to the asexual genus Helicopsis with high support from three independent algorithms (Fig. 1). Sympoventuria is, however, clearly distinguished from other asexual morphs by its fusoid-ellipsoid or cylindrical, simple or branched conidial chains (helically coiled conidia with thick conidial filaments in Helicopsis) (Karsten 1888, Crous et al. 2007b, Tsui & Berbee 2010). Furthermore, Neocoleroa metrosideri, a little-known sexual morph is here shown to be closely related to the sterile N. cameroonensis. However, N. metrosideri differs from its closest phylogenetic neighbour N. cameroonensis by unique fixed alleles in two loci, by 56 bp in ITS (14 %) and 26 bp in LSU (3 %). Notably, species with sexual morphs were scattered throughout the phylogenetic tree, which indicates that sexual reproduction may have evolved more than once within the family (Fig. 1). Overall, the results of this study have provided a robust overview of the species boundaries in Sympoventuriaceae. This is based largely on seven gene regions inferring an updated phylogram of all concerned genera in the family. On the other hand, multi-gene phylogenetic analyses combined with morphological features provide a robust means to delimit fungal species boundaries (Woudenberg et al. 2017, Lücking et al. 2020, Crous et al. 2021). Results of this study revealed that SSU, ITS, LSU, act1, tub2, tef1 and rpb2 gene regions can provide stable and reliable resolution for species delimitation in Sympoventuriaceae. Although each of these loci proved to be suitable barcoding markers for species identification, a combined analysis is highly recommended.
Evolution of lifestyles in Sympoventuriaceae
The transition from saprophytes to endophytes, plant pathogens and animal or human opportunistic pathogens is a notable feature within Sympoventuriaceae (Fig. 3). Studies have shown that the saprotrophic fungal ancestors experienced a large-scale loss of plant cell wall degrading enzymes, and obtained effector-like secreted proteins to fit a plant-fungal associated lifestyle (Bödeker et al. 2014, Kohler et al. 2015, Martin et al. 2016, Haridas et al. 2020, Shen et al. 2020, Benavent 2021). In our reconstruction analyses, although the Sympoventuriaceae clade most conspicuously includes saprophytes, members of the Neocoleroa and Sterila clades convergently evolved towards plant pathogens and animal/human opportunistic pathogens, with a shift towards endophytes in Verruconis (Fig. 3). In addition, we found the Scolecobasidium clade to have a high species richness, including saprophytes, endophytes, animal/human opportunistic and plant pathogens, and to be associated with significant increasing shifts in diversification rate (Fig. 3). It is worth noting that transition from saprophytes to plant pathogens and animal or human opportunistic pathogens has independently occurred multiple times during the evolution of Sympoventuriaceae, which is possibly driven by habitat selection (Fig. 3). The present study has provided a wealth of data about the lifestyle and phylogeny of Sympoventuriaceae, showing some trends in the evolution of species in the family. For example, saprophytes were mainly at the early diverged clades in phylogenetic trees, e.g., Neofusicladium, Parafusicladium, Pseudosigmoidea and Veronaeopsis. In con- trast, the endophytes, plant pathogens and animal/human opportunistic pathogens were only found in the Acroconidiellina, Neocoleroa, Scolecobasidium, Sterila and Verruconis species in more recently diverged clades, supporting a strong correlation between the evolution of Sympoventuriaceae life strategies and its phylogeny.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (32060009).
The authors declare that there is no conflict of interest.
REFERENCES
- Abbott EV. 1927. Scolecobasidium, a new genus of soil fungi. Mycologia 19: 29–31. [Google Scholar]
- Al-Hatmi AMS, Gerrits van den Ende AHG, Stielow JB, et al. 2016. Evaluation of two novel barcodes for species recognition of opportunistic pathogens in Fusarium. Fungal Biology 120: 231–245. [DOI] [PubMed] [Google Scholar]
- Ando K, Nakamura N. 2000. Pseudosigmoidea: a new genus for a hyphomycete (ATCC 16660) formerly identified as Sigmoidea prolifera. The Journal of General and Applied Microbiology 46: 51–57. [DOI] [PubMed] [Google Scholar]
- Arzanlou M, Groenewald JZ, Gams W, et al. 2007. Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Studies in Mycology 58: 57–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldacci E, Ciferri R. 1937. Un nuovo genere di micete parassita del pioppo Pollaccia radiosa (Lib.) Baldacci e Ciferri, Revisione dei G. Stigmella e Stigmina. I. Pollaccia radiosa (Lib.) Baldacci e Ciferri. Atti dell’Istituto Botanico della Universita e Laboratorio Crittogamico di Pavia 10: 55–72. [Google Scholar]
- Barido-Sottani J, Bošková V, Plessis LD, et al. 2018. Taming the BEAST – A community teaching material resource for BEAST 2. Systematic Biology 67: 170–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr ME. 1997. Notes on some ‘Dimeriaceous’ fungi. Mycotaxon 64: 149–171. [Google Scholar]
- Barron GL, Busch LV. 1962. Studies on the soil hyphomycete Scolecobasidium. Canadian Journal of Botany 40: 77–84. [Google Scholar]
- Beck A, Ritschel A, Schubert K, et al. 2005. Phylogenetic relationships of the anamorphic genus Fusicladium s. lat. as inferred by ITS nrDNA data. Mycological Progress 4: 111–116. [Google Scholar]
- Benavent IG. 2021. Macro-and microevolutionary perspectives on Seynesiella juniperi, a fungus in the Venturiales (Dothideomycetes, Ascomycota). Botanica complutensis 45: 115–126. [Google Scholar]
- Bhat DJ. 2010. Fascinating microfungi (hyphomycetes) of Western Ghats, India. Department of Botany, Goa University. [Google Scholar]
- Bödeker IT, Clemmensen KE, De Boer W, et al. 2014. Ectomycorrhizal Cortinarius species participate in enzymatic oxidation of humus in northern forest ecosystems. New Phytologist 203: 245–256. [DOI] [PubMed] [Google Scholar]
- Bonorden HF. 1851. Handbuch der allgemeinen Mykologie. Schweizerbart’sche Verlagshandlung, Stuttgart. [Google Scholar]
- Boonmee S, Bhat DJ, Maharachchikumbura SS, et al. 2014. Clavatispora thailandica gen. et sp. nov., a novel taxon of Venturiales (Dothideomycetes) from Thailand. Phytotaxa 176: 92–101. [Google Scholar]
- Carbone I, Kohn L. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- Carris LM. 1994. Vaccinium fungi: Mycosisymbrium cirrhosum gen. et sp. nov. Mycologia 86: 31–133. [Google Scholar]
- Castañeda-Ruiz RF, Guarro J, Cano J. 1996. Notes on conidia fungi. Two new dematiaceous hyphomycetes from Cuba. Mycotaxon 57: 463–469. [Google Scholar]
- Chen Y, Xie L, Long Y, et al. 2020. A new species and two new Chinese records of Ochroconis from sugarcane and banana rhizosphere in Guangxi, China. Mycoscience 61: 307–314. [Google Scholar]
- Crane JL. 1968. Freshwater hyphomycetes of the northern Appalachian Highland including New England, and three coastal plain states. American Journal of Botany 55: 996–1002. [Google Scholar]
- Crous PW, Carlier J, Roussel V, et al. 2020. Pseudocercospora and allied genera associated with leaf spots of banana (Musa spp.). Fungal Systematics and Evolution 7: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Lombard L, Sandoval-Denis M, et al. 2021. Fusarium: more than a node or a foot-shaped basal cell. Studies in Mycology 98: 100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Mohammed C, Glen M, et al. 2007a. Eucalyptus microfungi known from culture. 3. Eucasphaeria and Sympoventuria genera nova, and new species of Furcaspora, Harknessia, Heteroconium and Phacidiella. Fungal Diversity 25: 19–36. [Google Scholar]
- Crous PW, Schubert K, Braun U, et al. 2007b. Opportunistic, human-patho-genic species in the Herpotrichiellaceae are phenotypically similar to saprobic or phytopathogenic species in the Venturiaceae. Studies in Mycology 58: 185–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schumacher RK, Akulov A, et al. 2019a. New and interesting fungi. 2. Fungal Systematics and Evolution 3: 57–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schumacher RK, Wingfield MJ, et al. 2018a. New and interesting fungi. 1. Fungal Systematics and Evolution 1: 169–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Shivas RG, Quaedvlieg W, et al. 2014. Fungal Planet description sheets: 214–280. Persoonia 32: 184–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkley GJM, Groenewald JZ, et al. 2019b. Fungal Biodiversity. [Westerdijk Laboratory Manual Series no.1.] Utrecht, Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands. [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, et al. 2017. Fungal Planet description sheets: 625–715. Persoonia 39: 270–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, et al. 2018b. Fungal Planet description sheets: 716–784. Persoonia 40: 239–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Le Roux JJ, et al. 2015. Fungal Planet description sheets: 371–399. Persoonia 35: 264–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Lombard L, et al. 2019c. Fungal Planet description sheets: 951–1041. Persoonia 43: 223–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Richardson DM, et al. 2016. Fungal Planet description sheets: 400–468. Persoonia 36: 316–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, et al. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hoog GS, Von Arx JA. 1973. Revision of Scolecobasidium and Pleurophragmium. Kavaka 1: 55–60. [Google Scholar]
- Diene O, Wang W, Narisawa K. 2013. Pseudosigmoidea ibarakiensis sp. nov., a dark septate endophytic fungus from a cedar forest in Ibaraki, Japan. Microbes and Environments 28: 381–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doty MS, Slater DW. 1946. A new species of Heterobasidium tshawytschae pathogenic on young chinook salmon. American Midland Naturalist 36: 663–665. [Google Scholar]
- Ellis MB. 1971. Dematiaceous Hyphomycetes. Commonwealth Mycological Institute, Kew, Surrey. [Google Scholar]
- Ellis MB. 1976. More Dematiaceous Hyphomycetes. Commonwealth Mycological Institute, Kew, England. [Google Scholar]
- Fan XL, Bezerra JDP, Tian CM, et al. 2020. Cytospora (Diaporthales) in China. Persoonia 45: 1–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gams W. 2015. An ex-type culture cannot always tell the ultimate truth. IMA Fungus 6: A69. [Google Scholar]
- Gargas A, Taylor JW. 1992. Polymerase chain reaction (PCR) primers for amplifying and sequencing 18S rDNA from lichenized fungi. Mycologia 84: 589–592. [Google Scholar]
- Giraldo A, Crous PW. 2019. Inside plectosphaerellaceae. Studies in Mycology 92: 227–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo A, Sutton DA, Samerpitak K, et al. 2014. Occurrence of Ochroconis and Verruconis species in clinical specimens from the United States. Journal of Clinical Microbiology 52: 4189–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied & Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graniti A. 1963. Scolecobasidium anellii nsp. agente di annerimenti superficiali di stalattiti (Scolecobasidium anellii n. sp. causing a superficial darkening of stalacitites). Nuovo Giornale Botanico Italiano 69: 360–365. [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hamayun M, Khan SA, Kim HY, et al. 2009. Gibberellin production and plant growth enhancement by newly isolated strain of Scolecobasidium tshawytschae. Journal of Microbiology and Biotechnology 19: 560–565. [PubMed] [Google Scholar]
- Hao L, Chen C, Zhang R, et al. 2013. A new species of Scolecobasidium associated with the sooty blotch and flyspeck complex on banana from China. Mycological Progress 12: 489–495. [Google Scholar]
- Haridas S, Albert R, Binder M, et al. 2020. 101 Dothideomycetes genomes: a test case for predicting lifestyles and emergence of pathogens. Studies in Mycology 96: 141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Restrepo M, Giraldo A, Van Doorn R, et al. 2020. The genera of fungi - G6: Arthrographis, Kramasamuha, Melnikomyces, Thysanorea, and Verruconis. Fungal Systematics and Evolution 6: 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Restrepo M, Schumacher RK, Wingfield MJ, et al. 2016. Fungal Systematics and Evolution: FUSE 2. Sydowia 68: 193–230. [Google Scholar]
- Horré R, De Hoog GS, Kluczny C. et al. 1999. rDNA diversity and physiology of Ochroconis and Scolecobasidium species reported from humans and other vertebrates. Studies in Mycology 43: 194–205. [Google Scholar]
- Huanraluek N, Phukhamsakda C, Senwanna C, et al. 2019. Verruconis heveae, a novel species from Hevea brasiliensis in Thailand. Phytotaxa 403: 47–54. [Google Scholar]
- Hyde KD, Dong Y, Phookamsak R, et al. 2020. Fungal diversity notes 1151-1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100: 5–277. [Google Scholar]
- Hyde KD, Jones EBG, Liu JK, et al. 2013. Families of Dothideomycetes. Fungal Diversity 63: 1–313. [Google Scholar]
- Jayasiri SC, Hyde KD, Jones EBG, et al. 2019. Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 10: 1–186. [Google Scholar]
- Johnston PR, Park D. 2016. Neocoleroa metrosideri sp. nov. (Sympoventuriaceae, Venturiales). Phytotaxa 253: 214–218. [Google Scholar]
- Karsten PA. 1888. Diagnoses fungorum nonnullorum novorum, in Fennia detectorum. Revue Mycologique Toulouse 10: 73–75. [Google Scholar]
- Kidd S, Halliday C, Alexiou H, et al. 2016. Descriptions of medical fungi (3rd ed.). Newstyle Printing, Adelaide, Australia. [Google Scholar]
- Kirk PM, Cannon PF, Minter DW, et al. 2008. Ainsworth & Bisby’s dictionary of the fungi. CAB International, Wallingford. [Google Scholar]
- Kohler A, Kuo A, Nagy LG, et al. 2015. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nature Genetics 47: 410–415. [DOI] [PubMed] [Google Scholar]
- Koorders SH. 1907. Botanische Untersuchungen. Verhandelingen Koninklijke Nederlandse Akademie van Wetenschappen Afdeling Natuurkunde 13: 1–264. [Google Scholar]
- Koukol O. 2010. Revision of “Septonema ochraceum” revealed three new species of Venturiaceae and Herpotrichiellaceae. Mycological Progress 9: 369–378. [Google Scholar]
- Kumar S, Stecher G, Li M, et al. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner M, De Hoog GS, Yang L, et al. 2014. Proposed nomenclature for Pseudallescheria, Scedosporium and related genera. Fungal Diversity 67: 1–10. [Google Scholar]
- Larsson A., 2014. AliView: a fast and lightweight alignment viewer and editor for large data sets. Bioinformatics 30: 3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Bhat DJ, Phookamsak R, et al. 2016. Sporidesmioides thailandica gen. et sp. nov. (Dothideomycetes) from northern Thailand. Mycological Progress 15: 1169–1178. [Google Scholar]
- Liu F, Ma ZY, Hou LW, et al. 2022. Updating species diversity of Colletotrichum, with a phylogenomic overview. Studies in Mycology 101: 1–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JK, Hyde KD, Jeewon R, et al. 2017. Ranking higher taxa using divergence times: a case study in Dothideomycetes. Fungal Diversity 84: 75–99. [Google Scholar]
- Liu YJ, Whelen S, Hall BD. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- Lücking R, Aime MC, Robbertse B, et al. 2020. Unambiguous identification of fungi: where do we stand and how accurate and precise is fungal DNA barcoding. IMA Fungus 11: 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machouart M, Samerpitak K, De Hoog GS, et al. 2014. A multigene phylogeny reveals that Ochroconis belongs to the family Sympoventuriaceae (Venturiales, Dothideomycetes). Fungal Diversity 65: 77–88. [Google Scholar]
- Martin F, Kohler A, Murat C, et al. 2016. Unearthing the roots of ectomycorrhizal symbioses. Nature Reviews Microbiology 14: 760–773. [DOI] [PubMed] [Google Scholar]
- Martin-Sanchez PM, Nováková A, Bastian F, et al. 2012. Two new species of the genus Ochroconis, O. lascauxensis and O. anomala isolated from black stains in Lascaux Cave, France. Fungal Biology 116: 574–589. [DOI] [PubMed] [Google Scholar]
- Matsushima T. 1971. Microfungi of the Solomon Islands and Papua-New Guinea. Kobe, Japan. [Google Scholar]
- Matsushima T. 1975. Icones microfungorum: a Matsushima lectorum. Kobe, Japan. [Google Scholar]
- Matsushima T. 1980. Saprophytic microfungi from Taiwan part 1. Hyphomycetes. Matsushima Mycological Memoirs 1: 1–82. [Google Scholar]
- Matsushima T. 1996. Matsushima Mycological Memoirs 9: 1–30. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2012. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans, LA. CA, San Diego Supercomputer Center: 1–8. [Google Scholar]
- Murata K, Ogawa Y, Kusama K, et al. 2022. Disseminated Verruconis gallopava infection in a patient with systemic lupus erythematosus in Japan: A case report, literature review, and autopsy case. Medical Mycology Case Reports 35: 35–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA. 2004. MrModeltest v2. 2. Program distributed by the author: 2. Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- Papendorf MC. 1969. New South African soil fungi. Transactions of the British Mycological Society 52: 483–489. [Google Scholar]
- Petrak F. 1934. Mykologische Beiträge zur Flora von Sibirien. Hedwigia 74: 30–78. [Google Scholar]
- Pratibha J, Prabhugaonkar A. 2016. Distribution and phylogeny of Mycosisymbrium cirrhosum. Mycosphere 7: 44–50. [Google Scholar]
- Punithalingam E, Spooner BM. 2011. A new fungicolous Scolecobasidium (hyphomycetes) and Caducirostrum gen. nov. (coelomyces) from leaf litter in the UK and Italy. Kew Bulletin 66: 309–324. [Google Scholar]
- Qiao M, Tian W, Castañeda-Ruiz RF, et al. 2019. Two new species of Verruconis from Hainan, China. MycoKeys 48: 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, et al. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1. 7. Systematic Biology 67: 901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehner SA, Buckley E. 2005. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–98. [DOI] [PubMed] [Google Scholar]
- Ren J, Jie CY, Zhou QX, et al. 2013. Molecular and morphological data reveal two new species of Scolecobasidium. Mycoscience 54: 420–425. [Google Scholar]
- Revankar SG, Sutton D. 2010. Melanized fungi in human disease. Clinical Microbiology Reviews 23: 884–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van der Mark P, et al. 2012. MrBayes 3. 2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy RY, Dwivedi RS, Mishra RR. 1962. Two new species of Scolecobasidium from soil. Lloydia 25: 164–166. [Google Scholar]
- Rozewicki J, Li S, Amada KM, et al. 2019. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Research 47: 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salkin IF, Dixon DM, Kemna ME, et al. 1990. Fatal encephalitis caused by Dactylaria constricta var. gallopava in a snowy owl chick (Nectea scandiaca). Journal of Clinical Microbiology 28: 2845–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samerpitak K, Alfjorden A, Seyedmousavi S, et al. 2019. Ochroconis globalis infecting Atlantic salmon (Salmo salar), with a review of Ochroconis species in cold-blooded animals. Journal of Fish Diseases 42: 947–957. [DOI] [PubMed] [Google Scholar]
- Samerpitak K, Duarte APM, Attili-Angelis D. et al. 2015a. A new species of the oligotrophic genus Ochroconis (Sympoventuriaceae). Mycological Progress 14: 1–10. [Google Scholar]
- Samerpitak K, Gerrits van den Ende BH, Stielow JB, et al. 2016. Barcoding and species recognition of opportunistic pathogens in Ochroconis and Verruconis. Fungal Biology 120: 219–230. [DOI] [PubMed] [Google Scholar]
- Samerpitak K, Gerrits van den Ende AH, Menken SBJ. et al. 2015b. Three new species of the genus Ochroconis. Mycopathologia 180: 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samerpitak K, Gloyna K, De Hoog GS. 2017. A novel species of the oligotrophic genus Ochroconis colonizing indoor wet cells. Mycoscience 58: 290–296. [Google Scholar]
- Samerpitak K, Van der Linde E, Choi HJ, et al. 2014. Taxonomy of Ochroconis genus including opportunistic pathogens on humans and animals. Fungal Diversity 65: 89–126. [Google Scholar]
- Satow MM, Attili-Angelis D, De Hoog GS, et al. 2008. Selective factors involved in oil flotation isolation of black yeast from the environment. Studies in Mycology 61: 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert K, Ritschel A, Braun U. 2003. A monograph of Fusicladium s. lat. (Hyphomycetes). Schlechtendalia 9: 1–132. [Google Scholar]
- Seifert KA, Morgan-Jones G, Gams W, et al. 2011. The genera of hyphomycetes. CBS-KNAW Fungal Biodiversity Centre, Utrecht, the Netherlands. [Google Scholar]
- Seyedmousavi S, Guillot J, De Hoog GS. 2013. Phaeohyphomycoses, emerging opportunistic diseases in animals. Clinical Microbiology Reviews 26: 19–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedmousavi S, Samerpitak K, Rijs AJMM, et al. 2014. Antifungal susceptibility patterns of opportunistic fungi in the genera Verruconis and Ochroconis. Antimicrobial Agents and Chemotherapy 58: 3285–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M, Zhang JQ, Zhao LL, et al. 2020. Venturiales. Studies in Mycology 96: 185–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Singh NK, Singh PN, et al. 2019. Additions to Ochroconis from India. Phytotaxa 427: 186–199. [Google Scholar]
- Sivanesan A. 1977. The taxonomy and pathology of Venturia species. Lubrecht & Cramer Ltd, Vaduz. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian CV. 1977. Revisions of Hyphomycetes I. Kavaka 5: 93–98. [Google Scholar]
- Swofford DL. 2003. PAUP* 4.0b10. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, MA, USA. [Google Scholar]
- Tazik Z, Rahnama K, Iranshahi M, et al. 2020. Ochroconis ferulica sp. nov. (Venturiales), a fungal endophyte from Ferula ovina. Nova Hedwigia 110: 369–381. [Google Scholar]
- Tibpromma S, Hyde KD, McKenzie EH, et al. 2018. Fungal diversity notes 840–928: micro-fungi associated with Pandanaceae. Fungal Diversity 92: 1–160. [Google Scholar]
- Tsui CKM, Berbee ML. 2010. Transfer of two Helicoma species to Troposporella based on molecular and morphological data. Mycoscience 51: 144–148. [Google Scholar]
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Cai W, Gerrits van den Ende AH, et al. 2018. Indoor wet cells as a habitat for melanized fungi, opportunistic pathogens on humans and other vertebrates. Scientific Reports 8: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei TP, Zhang X, Ren PP, et al. 2020. A novel Melnikomyces species from forest litter in China. Phytotaxa 471: 61–68. [Google Scholar]
- White TJ, Bruns T, Lee S, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ. et al. (eds), PCR Protocols: a guide to methods and applications: 315–322. Academic Press, San Diego, California, USA. [Google Scholar]
- Wijayawardene NN, Crous PW, Kirk PM, et al. 2014. Naming and outline of Dothideomycetes – 2014 including proposals for the protection or suppression of generic names. Fungal Diversity 69: 1–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayawardene NN, Hyde KD, Al-Ani LKT, et al. 2020. Outline of fungi and fungus-like taxa. Mycosphere 11: 1060–1456. [Google Scholar]
- Wijayawardene NN, Hyde KD, Lumbsch HT, et al. 2018. Outline of ascomycota: 2017. Fungal Diversity 88: 167–263. [Google Scholar]
- Woudenberg JHC, Hanse B, Van Leeuwen GCM, et al. 2017. Stemphylium revisited. Studies in Mycology 87: 77–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarita K, Sano A, Samerpitak K, et al. 2010. Ochroconis calidifluminalis, a sibling of the neurotropic pathogen O. gallopava, isolated from hot spring. Mycopathologia 170: 21–30. [DOI] [PubMed] [Google Scholar]
- Yu Y, Blair C, He XJ. 2020. RASP 4: Ancestral State Reconstruction Tool for Multiple Genes and Characters. Molecular Biology and Evolution 37: 604–606. [DOI] [PubMed] [Google Scholar]
- Yu Y, Harris AJ, Blair C, et al. 2015. RASP (Reconstruct Ancestral State in Phylogenies): a tool for historical biogeography. Molecular Phylogenetics and Evolution 87: 46–49. [DOI] [PubMed] [Google Scholar]
- Zhang JQ, Dou ZP, Zhou YP, et al. 2016. Venturia chinensis sp. nov. a new venturialean ascomycete from Khingan Mountains. Saudi Journal of Biological Sciences 23: 592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TY, Yu Y, Zhang MY, et al. 2018. Verruconis panacis sp. nov., an endophyte isolated from Panax notoginseng. International Journal of Systematic and Evolutionary Microbiology 68: 2499–2503. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang KY, Ren PP, et al. 2020. Ochroconis terricola sp. nov. from China. Mycotaxon 135: 143–150. [Google Scholar]
- Zhang Y, Crous PW, Schoch CL, et al. 2011. A molecular, morphological and ecological re-appraisal of Venturiales – a new order of Dothideomycetes. Fungal Diversity 51: 249–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Schoch CL, Fournier J, et al. 2009. Multi-locus phylogeny of Pleosporales: a taxonomic, ecological and evolutionary re-evaluation. Studies in Mycology 64: 85–102. [DOI] [PMC free article] [PubMed] [Google Scholar]


