Abstract
During an oomycete survey in December 2015, 10 previously unknown Halophytophthora taxa were isolated from marine and brackish water of tidal ponds and channels in saltmarshes, lagoon ecosystems and river estuaries at seven sites along the Algarve coast in the South of Portugal. Phylogenetic analyses of LSU and ITS datasets, comprising all described Halophytophthora species, the 10 new Halophytophthora taxa and all relevant and distinctive sequences available from GenBank, provided an updated phylogeny of the genus Halophytophthora s.str. showing for the first time a structure of 10 clades designated as Clades 1–10. Nine of the 10 new Halophytophthora taxa resided in Clade 6 together with H. polymorphica and H. vesicula. Based on differences in morphology and temperature-growth relations and a multigene (LSU, ITS, Btub, hsp90, rpl10, tigA, cox1, nadh1, rps10) phylo-geny, eight new Halophytophthora taxa from Portugal are described here as H. brevisporangia, H. celeris, H. frigida, H. lateralis, H. lusitanica, H. macrosporangia, H. sinuata and H. thermoambigua. Three species, H. frigida, H. macrosporangia and H. sinuata, have a homothallic breeding system while the remaining five species are sterile. Pathogenicity and litter decomposition tests are underway to clarify their pathological and ecological role in the marine and brackish-water ecosystems. More oomycete surveys in yet undersurveyed regions of the world and population genetic or phylogenomic analyses of global populations are needed to clarify the origin of the new Halophytophthora species.
Citation: Maia C, Horta Jung M, Carella G, et al. 2022. Eight new Halophytophthora species from marine and brackish-water ecosystems in Portugal and an updated phylogeny for the genus. Persoonia 48: 54 – 90. https://doi.org/10.3767/persoonia.2022.48.02..
Keywords: breeding system, ecological role, evolution, lifestyle, oomycetes, Peronosporaceae, Phytophthora
INTRODUCTION
The aquatic genus Halophytophthora is a sister genus to Phytophthora and Nothophytophthora within the Peronosporaceae, class Oomycota, kingdom Straminipila. The first species, originally described as Phytophthora vesicula, was discovered in 1969 in a marine habitat near Vancouver (Anastasiou & Churchland 1969). In 1990, Ho & Jong transferred P. vesiculatogether with eight other marine Phytophthoraspecies, i.e., P. avicennae , P. bahamensis,P. batemanensis,P. epistomium,P. mycoparasitica,P. operculata,P. polymorphicaand P. spinosa (Fell & Master 1975, Pegg & Alcorn 1982, Gerrettson-Cornell & Simpson 1984), to the newly established genus Halophytophthora. Between 1990 and 2003 a further six Halophytophthoraspecies were described, including H. elongata,H. exoprolifera,H. kandeliae,H. masteri,H. porrigovesicaand H. tartarea (Ho et al. 1991, 1992, 2003, Nakagiri et al. 1994, 2001). However, recent phylogenetic studies revealed the polyphyletic nature of Halophytophthora(Lara & Belbahri 2011, Nigrelli & Thines 2013, Jung et al. 2017d) causing a reassignment of numerous Halophytophthoraspecies to other genera. Halophytophthora kandeliaewas transferred to Phytopythium(Thines 2014), H. tartareato Salisapilia (Hulvey et al. 2010), H. spinosato Salispina(Li et al. 2016) and H. operculatato Calycofera, a genus established to accommodate this species (Bennett et al. 2017a). Furthermore, H. exoproliferawas shown to belong to a yet undescribed sister genus of Halophytophthoras.str. (Jung et al. 2017d). Recently, Bennet & Thines (2019) revised Salisapiliaand included another five Halophytophthoraspecies, H. bahamensis,H. elongata,H. epistomia,H. masteriand H. mycoparasitica. Currently, Halophytophthoras.str. comprises seven described species, H. avicennae,H. batemanensis,H. fluviatilis,H. insularis, H. polymorphica,H. souzae and H. vesicula(Yang & Hong 2014, Jung et al. 2017d, Bennet & Thines 2019, Jesus et al. 2019), and the informally designated taxa Halophytophthorasp. Zostera (Govers et al. 2016), Halophytophthorasp. 1 and 2 (Nigrelli & Thines 2013), and Halophytophthorasp-1 and sp-3 (Man in ‘t Veld et al. 2019) from the North Sea, Halophytophthorasp. 1 and 2 from Georgia, USA (Hulvey et al. 2010) and Halophytophthorasp-4 from Southern France (Man in ‘t Veld et al. 2019). The phylogenetic position and taxonomic status of H. porrigovesica, originally described from mangrove stands in Japan and Thailand (Nakagiri et al. 2001), is unclear.
Although Halophytophthoraspecies are mainly found in marine, mangrove and estuarine habitats, from the tropics to the south-eastern bight of the North Sea (Nigrelli & Thines 2013), they are well adapted to a wide range of temperature and salinity with some species occurring in low salinity and even freshwater habitats (Nakagiri 2000, Reeser et al. 2011, Hüberli et al. 2013, Yang & Hong 2014, Caballol et al. 2021). Halophytophthoraspecies share many features of their morphology and lifecycle with Phytophthora(Sullivan et al. 2018). In contrast to Phytophthora, which comprises primary plant pathogens (Erwin & Ribeiro 1996, Lamour 2013, Jung et al. 2018), Halophytophthoraspecies are usually considered as saprophytes, playing an important role in decomposition and secondary production (Nakagiri 2000). However, recent studies (Govers et al. 2016, Man in ‘t Veld et al. 2019) suggested that several marine Phytophthoraand Halophytophthoraspecies, including Halophytophthora sp. Zostera, might be involved in the widespread decline of the seagrass Zostera marina (eelgrass), an important marine foundation species that has been suffering from recurring wasting disease outbreaks since 1930 (Muehlstein et al. 1988). Almost 99 % of tested Z. marina seeds were infected, resulting in reduced germination rates and devastating consequences for restauration efforts (Govers et al. 2016).
Despite being known since more than 50 years, knowledge on the diversity, distribution and ecology of Halophytophthoraspecies is still scarce. Therefore, in December 2015, a survey was carried out in marine and brackish-water ecosystems along the Algarve coast in the South of Portugal which unveiled a high diversity of Halophytophthora species, most of them new to science.
In this study, morphological and physiological characteristics were used in combination with DNA sequence data from six nuclear and three mitochondrial gene regions to characterise and officially describe eight new Halophytophthoraspecies from the Algarve as H. brevisporangia sp. nov., H. celerissp. nov., H. frigida sp. nov., H. lateralissp. nov., H. lusitanicasp. nov., H. macrosporangiasp. nov., H. sinuata sp. nov. and H. thermoambigua sp. nov.
MATERIAL AND METHODS
Sampling and Halophytophthora isolation
Sampling was performed in December 2015 at seven different marine and brackish-water sites, including tidal ponds and channels in saltmarshes which did not dry out during low tides, lagoons and river estuaries (Fig. 1), along the Algarve coast in the South of Portugal using a leaf baiting method adopted from Jung et al. (2017a). Fly mesh and styrofoam were used to create 25 × 30 cm baiting-bag rafts able to float on the water surface. Unwounded leaves of Ceratonia siliqua,Quercus suber,Q. rubraand Citrusspp. were placed inside the rafts and used as baits. Two rafts were placed on each site and collected after three to six days. The leaves were washed with distilled water, blotted dry and 2 × 2 mm pieces of discoloured and/or necrotic tissues were plated onto selective PARPNH agar (V8 juice agar (V8A) amended with 10 µ g/mL pimaricin, 200 µg/mL ampicillin, 10 µg/mL rifampicin, 25 µg/mL pentachloronitrobenzene, 50 µg/mL nystatin and 50 µg/mL hymexazol) and incubated at 20 °C in the dark (Jung et al. 1996). After 16 to 48 h, Petri dishes were observed under the dissecting microscope and axenic cultures obtained by transferring single hyphal tips to V8 juice agar (V8A; 16 g agar, 3 g CaCO3, 100 mL Campbell’s V8 juice, 900 mL distilled water). Stock cultures were maintained on carrot juice seawater agar (sCA; 16 g agar, 3 g CaCO3, 100 mL carrot juice, 450 mL distilled water, 450 mL seawater) at 4–8 °C in the dark.
Fig. 1.

Sampling sites along the Algarve coast in the South of Portugal; a. tidal zone of the Rio Séqua near the estuary in Tavira; b–d. Parque Natural da Ria Formosa; b. saltmarsh near Almancil during high tide with flooded tidal channels and ponds; c. tidal channel in a saltmarsh near Santa Luzia during low tide, with baiting raft; d. saltwater pond in a saltmarsh near Quelfes during low tide, with baiting raft; e. marine lagoon in Ria de Alvor, with baiting raft (arrow) floating among the marine algal vegetation; f. baiting raft tied to an iron stick in the estuary of the Rio Guadiana near Sapal de Castro Marim.
DNA isolation, amplification and sequencing
For Halophytophthora and Phytophthora isolates obtained in this study and several additional Phytophthora and Nothophytophthora isolates used in the phylogenetic analyses the internal transcribed spacer region (ITS1-5.8S-ITS2) of the ribosomal RNA gene (ITS) and the 5’ terminal domain of the large subunit (LSU) of the nuclear ribosomal RNA gene were amplified and sequenced. In addition, for all isolates of the new Halophytophthora species used in the phylogenetic analyses (Table 1) four additional nuclear loci, i.e., heat shock protein 90 (hsp90), β-tubulin (Btub), 60S ribosomal protein L10 (rpl10) and tigA (a locus containing two genes, a triosephosphate isomerase and glyceraldehyde-3-phosphate dehydrogenase, fused into a single transcriptional unit), and the three mitochondrial genes cytochrome-c oxidase 1 (cox1), subunit 1 of NADH dehydrogenase (nadh1) and 40S ribosomal protein S10 (rps10) were amplified and sequenced.
Table 1.
Details of oomycete isolates included in this study. GenBank numbers for sequences obtained in the present study are printed in italics.
| Species | Isolate numbersa | Origin | GenBank accession numbers | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| International collections | Local collections | Source | Location; yearf | Collector; reference | LSU | ITS | coxI | Btub hsp90 | tigA 60S rpl10 | nadh1 40S rps10 | |
| Halophytophthora avicennae b | CBS 188.85; ATCC 64709 | DAR 50187 | Fallen Avicennia marina leave in Clyde River estuary | Bateman’s Bay, AU; 1982 | J. Simpson; Gerrettson-Cornell & Simpson 1984 | AY598668 | HQ643147 | HQ708219 | OK091259 OK091315 | n.a. OK091426 | KY788594 OK091535 |
| H. avicennae | – | BD88 | Baiting; brackish river water, Ribeira de Odelouca | Silves, PT; 2014 | T. Jung; this study | n.a. | OK040996 | OK091587 | n.a. | n.a. | n.a. |
| H. avicennae | – | BD629 | Baiting; Rio Séqua estuary, Ria Formosa | Tavira, PT; 2015 | T. Jung; this study | n.a. | OK040997 | n.a. | n.a. | n.a. | n.a. |
| H. avicennae;b,c,d | – | BD633 | Baiting; Rio Séqua estuary, Ria Formosa | Tavira, PT; 2015 | T. Jung; this study | OK033574 | OK033632 | OK091197 | OK091252 OK091308 | OK091363 OK091419 | n.a. OK091528 |
| H. avicennae | – | BD635 | Baiting; Rio Séqua estuary, Ria Formosa | Tavira, PT; 2015 | T. Jung; this study | OK033575 | OK033633 | OK091198 | OK091253 OK091309 | OK091364 OK091420 | n.a. OK091529 |
| H. avicennae b,c,d | – | BD636 | Baiting; Rio Séqua estuary, Ria Formosa | Tavira, PT; 2015 | T. Jung; this study | n.a. | OK040998 | n.a. | n.a. | n.a. | n.a. |
| H. avicennae b,c,d | – | BD670 | Baiting; coastal lagoon, Ria de Alvor | Alvor, PT; 2015 | T. Jung; this study | OK033576 | OK033634 | OK091199 | OK091254 OK091310 | OK091365 OK091421 | n.a. OK091530 |
| H. avicennae | – | BD671 | Baiting; coastal lagoon, Ria de Alvor | Alvor, PT; 2015 | T. Jung; this study | n.a. | OK040999 | n.a. | n.a. | n.a. | n.a. |
| H. avicennae b,c | – | BD682 | Baiting; coastal lagoon, Ria Formosa | Almancil, PT; 2015 | T. Jung; this study | OK033577 | OK033635 | OK091200 | OK091255 OK091311 | OK091366 OK091422 | n.a. OK091531 |
| H. avicennae b,c | – | BD687 | Baiting; tidal pond in salt marsh, Ria Formosa | Almancil, PT; 2015 | T. Jung; this study | OK033578 | OK033636 | OK091201 | OK091256 OK091312 | OK091367 OK091423 | n.a. OK091532 |
| H. avicennae | – | BD689 | Baiting; estuary of Rio Guadiana | Castro Marim, PT; 2015 | T. Jung; this study | n.a. | OK041001 | n.a. | n.a. | n.a. | n.a. |
| H. avicennaeb,c | – | BD690 | Baiting; estuary of Rio Guadiana | Castro Marim, PT; 2015 | T. Jung; this study | OK033579 | OK033637 | OK091202 | OK091257 OK091313 | OK091368 OK091424 | n.a. OK091533 |
| H. avicennaeb,c | – | BD697 | Baiting; estuary of Rio Guadiana | Castro Marim, PT; 2015 | T. Jung; this study | OK033580 | OK033638 | OK091203 | OK091258 OK091314 | OK091369 OK091425 | n.a. OK091534 |
| H. batemanensisb, ex-type | CBS 679.84; WPC P11617; IMI 327602; NBRC 32616 | DAR 41559 | Fallen Avicennia marina leave in Clyde River estuary | Bateman’s Bay, AU; 1982 | J. Simpson; Gerrettson-Cornell & Simpson 1984 | HQ665286 | HQ643148 | HQ171166 | KY788513 n.a. | n.a. | n.a. |
| H. brevisporangiab,c,d, ex-type | CBS 147238 | BD662 | Baiting; tidal pond in salt marsh, Ria Formosa | Quelfes, PT; 2015 | T. Jung; this study | OK033583 | OK033641 | OK091206 | OK091262 OK091318 | OK091372 OK091429 | OK091481 OK091538 |
| H. brevisporangia b,c | – | BD644 | Baiting; tidal channel in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033581 | OK033639 | OK091204 | OK091260 OK091316 | OK091370 OK091427 | OK091479 OK091536 |
| H. brevisporangia b,c,d | – | BD658 | Baiting; tidal pond in salt marsh, Ria Formosa | Quelfes, PT; 2015 | T. Jung; this study | OK033582 | OK033640 | OK091205 | OK091261 OK091371 | OK091480 OK091537 | |
| H. brevisporangia b,c,d | CBS 147239 | BD695 | Baiting; estuary of Rio Guadiana | Castro Marim, PT; 2015 | G. Carella; this study | OK033584 | OK033642 | OK091207 | OK091263 OK091319 | OK091373 OK091430 | OK091482 OK091539 |
| H. brevisporangia b | – | BD887 | Baiting; tidal channel in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033585 | OK033643 | OK091208 | OK091264 OK091320 | OK091374 OK091431 | OK091483 OK091540 |
| H. celerisb,c,;d, ex-type | CBS 147240 | BD885 | Baiting; tidal channel in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033587 | OK033645 | OK091210 | OK091266 OK091322 | OK091376 OK091433 | OK091485 OK091542 |
| H. celeris b,c,d | CBS 147241 | BD646 | Baiting; tidal channel in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033586 | OK033644 | OK091209 | OK091265 OK091321 | OK091375 OK091432 | OK091484 OK091541 |
| H. celeris b,c,d | – | BD886 | Baiting; tidal channel in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033588 | OK033646 | OK091211 | OK091267 OK091323 | OK091377 OK091434 | OK091486 OK091543 |
| H. celeris b | – | BD985 | Baiting; tidal channel in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033589 | OK033647 | OK091212 | OK091268 OK091324 | OK091378 OK091435 | OK091487 OK091544 |
| H. celerisb | – | BD987 | Baiting; tidal channel in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033590 | OK033648 | OK091213 | OK091269 OK091325 | OK091379 OK091436 | OK091488 OK091545 |
| H. fluviatilisb, ex-type | ATCC MYA-4961 | 57A9 | Baiting; Flint Run Stream | Virginia, US; 2011 | X. Yang; Yang & Hong 2014 | KX252673 | KF734963 | n.a. | KX252669 KX252672 | KX252674 KX252668 | n.a. |
| H. fluviatilis b | – | 59J1 | Baiting; Rappahannock River | Virginia, US; 2012 | X. Yang; Yang & Hong 2014 | n.a. | KF734968 | n.a. | n.a. | n.a. | n.a. |
| H. frigidab,c,d, ex-type | CBS 147235 | BD655 | Baiting; tidal pond in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033594 | OK033652 | OK091217 | OK091273 OK091329 | OK091383 OK091440 | OK091492 OK091549 |
| H. frigida b,c,d | – | BD641 | Baiting; tidal channel in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033591 | OK033649 | OK091214 | OK091270 OK091326 | OK091380 OK091437 | OK091489 OK091546 |
| H. frigida b,c,d | – | BD647 | Baiting; tidal pond in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033592 | OK033650 | OK091215 | OK091271 OK091327 | OK091381 OK091438 | OK091490 OK091547 |
| H. frigidab,c,d | CBS 147236 | BD650 | Baiting; tidal pond in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033593 | OK033651 | OK091216 | OK091272 OK091328 | OK091382 OK091439 | OK091491 OK091548 |
| H. frigidac,d | – | BD654 | Baiting; tidal pond in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | n.a. | OK041002 | OK091589 | n.a. | n.a. | n.a. |
| H. frigida b,c | – | BD675 | Baiting; coastal lagoon, Ria de Alvor | Alvor, PT; 2015 | T. Jung; this study | OK033595 | OK033653 | OK091218 | OK091274 OK091330 | OK091384 OK091441 | OK091493 OK091550 |
| H. frigida b,c | – | BD676 | Baiting; coastal lagoon, Ria de Alvor | Alvor, PT; 2015 | T. Jung; this study | OK033596 | OK033654 | OK091219 | OK091275 OK091331 | OK091385 OK091442 | OK091494 OK091551 |
| H. insularisb, ex-type | CCIBt 4114 | Submerged leaf of Laguncularia racemosa; Perequê river | Ilha do Cardoso,BR; 2012 | A.V. Marano, A.L. Jesus & C.L.A. Pires-Zottarelli; Jesus et al. 2019 | KY327272 | KY320204 | n.a. | n.a. | n.a. | n.a. | |
| H. insularis b | – | AJM 74 | Submerged leaf of Laguncularia racemosa; Perequê river | Ilha do Cardoso, BR; 2012 | A.V. Marano, A.L. Jesus & C.L.A. Pires-Zottarelli; Jesus et al. 2019 | KY327270 | KY320202 | KY327277 | n.a. | n.a. | n.a. |
| H. lateralisb,c,d, ex-type | CBS 147233 | BD657 | Baiting; tidal pond in salt marsh, Ria Formosa | Quelfes, PT; 2015 | T. Jung; this study | OK033597 | OK033655 | OK091220 | OK091276 OK091332 | OK091386 OK091443 | OK091495 OK091552 |
| H. lateralis b,c,d | – | BD660 | Baiting; tidal pond in salt marsh, Ria Formosa | Quelfes, PT; 2015 | T. Jung; this study | OK033598 | OK033656 | OK091221 | OK091277 OK091333 | OK091387 OK091444 | OK091496 OK091553 |
| H. lateralis b,c,d | – | BD665 | Baiting; tidal pond in salt marsh, Ria Formosa | Quelfes, PT; 2015 | T. Jung; this study | OK033599 | OK033657 | OK091222 | OK091278 OK091334 | OK091388 OK091445 | OK091497 OK091554 |
| H. lateralis b,c,d | CBS 147234 | BD680 | Baiting; coastal lagoon, Ria Formosa | Almancil, PT; 2015 | T. Jung; this study | OK033600 | OK033658 | OK091223 | OK091279 OK091335 | OK091389 OK091446 | OK091498 OK091555 |
| H. lusitanicab,c,d, ex-type | CBS 147231 | BD686 | Baiting; tidal pond in salt marsh, Ria Formosa | Almancil, PT; 2015 | T. Jung; this study | OK033605 | OK033663 | OK091228 | OK091284 OK091340 | OK091394 OK091451 | OK091503 OK091560 |
| H. lusitanica b,c,d | CBS 147232 | BD628 | Baiting; Rio Séqua estuary, Ria Formosa | Tavira, PT; 2015 | T. Jung; this study | OK033601 | OK033659 | OK091224 | OK091280 OK091336 | OK091390 OK091447 | OK091499 OK091556 |
| H. lusitanica c,d | – | BD632 | Baiting; Rio Séqua estuary, Ria Formosa | Tavira, PT; 2015 | T. Jung; this study | n.a. | OK041003 | OK091590 | n.a. | n.a. | n.a. |
| H. lusitanica b,c,d | – | BD634 | Baiting; Rio Séqua estuary, Ria Formosa | Tavira, PT; 2015 | T. Jung; this study | OK033602 | OK033660 | OK091225 | OK091281 OK091337 | OK091391 OK091448 | OK091500 OK091557 |
| H. lusitanica | – | BD638 | Baiting; Rio Séqua estuary, Ria Formosa | Tavira, PT; 2015 | T. Jung; this study | n.a. | OK041004 | n.a. | n.a. | n.a. | n.a. |
| H. lusitanica b,c | – | BD679 | Baiting; coastal lagoon, Ria Formosa | Almancil, PT; 2015 | T. Jung; this study | OK033603 | OK033661 | OK091226 | OK091282 OK091338 | OK091392 OK091449 | OK091501 OK091558 |
| H. lusitanica b,c,d | – | BD681 | Baiting; coastal lagoon, Ria Formosa | Almancil, PT; 2015 | T. Jung; this study | OK033604 | OK033662 | OK091227 | OK091283 OK091339 | OK091393 OK091450 | OK091502 OK091559 |
| H. lusitanica | – | BD934 | Baiting; estuary of Rio Guadiana | Castro Marim, PT; 2015 | T. Jung; this study | n.a. | OK041005 | OK091591 | n.a. | n.a. | n.a. |
| H. macrosporangiab,c,d, ex-type | CBS 147290 | BD639 | Baiting; tidal channel in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033606 | OK033664 | OK091229 | OK091285 OK091341 | OK091395 OK091452 | OK091504 OK091561 |
| H. macrosporangia b,d | CBS 147291 | BD642 | Baiting; tidal channel in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033607 | OK033665 | OK091230 | OK091286 OK091342 | OK091396 OK091453 | OK091505 OK091562 |
| H. macrosporangia b,c,d | – | BD643 | Baiting; tidal channel in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033608 | OK033666 | OK091231 | OK091287 OK091343 | OK091397 OK091454 | OK091506 OK091563 |
| H. macrosporangia c,d | – | BD645 | Baiting; tidal channel in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | n.a. | OK041006 | OK091592 | n.a. | n.a. | n.a. |
| H. macrosporangia b,c,d | – | BD649 | Baiting; tidal pond in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033609 | OK033667 | OK091232 | OK091288 OK091344 | OK091398 OK091455 | OK091507 OK091564 |
| H. macrosporangia b,c,d | – | BD659 | Baiting; tidal pond in salt marsh, Ria Formosa | Quelfes, PT; 2015 | T. Jung; this study | OK033610 | OK033668 | OK091233 | OK091289 OK091345 | OK091399 OK091456 | OK091508 OK091565 |
| H. macrosporangia b,c,d | – | BD664 | Baiting; tidal pond in salt marsh, Ria Formosa | Quelfes, PT; 2015 | T. Jung; this study | OK033611 | OK033669 | OK091234 | OK091290 OK091346 | OK091400 OK091457 | OK091509 OK091566 |
| H. polymorphicab, ex-type | CBS 680.84; NBRC 32619; ATCC 56966 | DAR 41562 | Fallen Eucalyptus sp. leave in Clyde River estuary | Bateman’s Bay, AU; 1982 | J. Simpson; Gerrettson-Cornell & Simpson 1984 | AY598669 | HQ643313 | HQ708363 | OK091291 n.a. | OK091401 OK091458 | n.a. OK091567 |
| H. polymorphica b | CCIBt 4112 | AJM 33 | Submerged leaves of Laguncularia racemosa; Perequê river | Ilha do Cardoso, BR; 2012 | A.L. Jesus, A.V. Marano & C.L.A. Pires-Zottarelli; Jesus et al. 2019 | KT455404 | KT455391 | KT897699 | n.a. | n.a. | n.a. |
| H. sinuatab,c,d, ex-type | CBS 147237 | BD656 | Baiting; tidal pond in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033613 | OK033671 | OK091236 | OK091293 OK091348 | OK091403 OK091460 | OK091511 OK091569 |
| H. sinuata b,c,d | – | BD640 | Baiting; tidal channel in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033612 | OK033670 | OK091235 | OK091292 OK091347 | OK091402 OK091459 | OK091510 OK091568 |
| H. sinuata b,c,d | – | BD941 | Baiting; tidal channel in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033614 | OK033672 | OK091237 | OK091294 OK091349 | OK091404 OK091461 | OK091512 OK091570 |
| H. sinuata b,c,d | – | BD942 | Baiting; tidal channel in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033615 | OK033673 | OK091238 | OK091295 OK091350 | OK091405 OK091462 | OK091513 OK091571 |
| H. sinuata b,c,d | CBS 147292 | BD943 | Baiting; tidal channel in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033616 | OK033674 | OK091239 | OK091296 OK091351 | OK091406 OK091463 | OK091514 OK091572 |
| H. sinuata c | – | BD944 | Baiting; tidal pond in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | n.a. | OK041007 | OK091593 | n.a. | n.a. | n.a. |
| H. souzaeb, ex-type | CCIBt 4113 | Submerged leaf of Laguncularia racemosa; Perequê river | Ilha do Cardoso, BR; 2012 | A.L. Jesus, A.V. Marano & C.L.A. Pires-Zottarelli; Jesus et al. 2019 | KY327269 | KY320200 | KY327275 | n.a. | n.a. | n.a. | |
| H. souzaeb | – | AJM 23 | Submerged leaf of Laguncularia racemosa; Perequê river | Ilha do Cardoso, BR; 2012 | A.L. Jesus, A.V. Marano & C.L.A. Pires-Zottarelli; Jesus et al. 2019 | KY327268 | KY320199 | KY327274 | n.a. | n.a. | n.a. |
| H. thermoambiguab,c,d, ex-type | CBS 147229 | BD651 | Baiting; tidal pond in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033622 | OK033680 | OK091244 | OK091300 OK091355 | OK091411 OK091469 | OK091520 OK091577 |
| H. thermoambigua b,c | – | BD91 | Baiting; brackish river water, Ribeira de Odelouca | Silves, PT; 2014 | T. Jung; this study | OK033617 | OK033675 | OK091240 | n.a. | OK091407 OK091464 | OK091515 OK091573 |
| H. thermoambigua b,c | – | BD93 | Baiting; brackish river water, Ribeira de Odelouca | Silves, PT; 2014 | T. Jung; this study | OK033618 | OK033676 | OK091241 | n.a. | OK091408 OK091465 | OK091516 OK091574 |
| H. thermoambiguab | – | BD630 | Baiting; Rio Séqua estuary, Ria Formosa | Tavira, PT; 2015 | T. Jung; this study | OK033619 | OK033677 | OK091242 | OK091297 OK091352 | OK091409 OK091466 | OK091517 OK091575 |
| H. thermoambigua c | – | BD631 | Baiting; Rio Séqua estuary, Ria Formosa | Tavira, PT; 2015 | T. Jung; this study | n.a. | OK041008 | n.a. | n.a. | n.a. | n.a. |
| H. thermoambigua b,c,d | CBS 147230 | BD637 | Baiting; Rio Séqua estuary, Ria Formosa | Tavira, PT; 2015 | T. Jung; this study | OK033620 | OK033678 | OK091243 | OK091298 OK091353 | OK091410 OK091467 | OK091518 OK091576 |
| H. thermoambigua b | – | BD648 | Baiting; tidal pond in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033621 | OK033679 | n.a. | OK091299 OK091354 | n.a. OK091468 | OK091519 n.a. |
| H. thermoambigua b,e | – | BD668 | Baiting; coastal lagoon, Ria de Alvor | Alvor, PT; 2015 | T. Jung; this study | OK033623 | OK033681 | OK091245 | OK091301 OK091356 | OK091412 OK091470 | OK091521 OK091578 |
| H. thermoambigua | – | BD669 | Baiting; coastal lagoon, Ria de Alvor | Alvor, PT; 2015 | T. Jung; this study | n.a. | OK041009 | n.a. | n.a. | n.a. | n.a. |
| H. thermoambigua c | – | BD673 | Baiting; coastal lagoon, Ria de Alvor | Alvor, PT; 2015 | T. Jung; this study | n.a. | OK041010 | n.a. | n.a. | n.a. | n.a. |
| H. thermoambigua b,c | – | BD674 | Baiting; coastal lagoon, Ria Formosa | Alvor, PT; 2015 | T. Jung; this study | OK033624 | OK033682 | OK091246 | OK091302 OK091357 | OK091413 OK091471 | OK091522 OK091579 |
| H. thermoambigua | – | BD683 | Baiting; tidal pond, Ria Formosa | Almancil, PT; 2015 | T. Jung; this study | n.a. | OK041011 | n.a. | n.a. | n.a. | n.a. |
| H. thermoambigua | – | BD684 | Baiting; tidal pond, Ria Formosa | Almancil, PT; 2015 | T. Jung; this study | n.a. | OK041012 | n.a. | n.a. | n.a. | n.a. |
| H. thermoambigua b | – | BD685 | Baiting; tidal pond, Ria Formosa | Almancil, PT; 2015 | T. Jung; this study | OK033625 | OK033683 | OK091247 | OK091303 OK091358 | OK091414 OK091472 | OK091523 OK091580 |
| H. thermoambigua | – | BD692 | Baiting; estuary of Rio Guadiana | Castro Marim, PT; 2015 | T. Jung; this study | n.a. | OK041013 | n.a. | n.a. | n.a. | n.a. |
| H. thermoambigua b | – | BD693 | Baiting; estuary of Rio Guadiana | Castro Marim, PT; 2015 | T. Jung; this study | OK033626 | OK033684 | OK091248 | OK091304 | OK091415 | OK091524 |
| H. thermoambigua | – | BD696 | Baiting; estuary of Rio Guadiana | Castro Marim, PT; 2015 | T. Jung; this study | n.a. | OK041014 | n.a. | n.a. | n.a. | n.a. |
| H. thermoambigua b,c | – | BD698 | Baiting; estuary of Rio Guadiana | Castro Marim, PT; 2015 | T. Jung; this study | OK033627 | OK033685 | OK091249 | OK091305 OK091360 | OK091416 OK091474 | OK091525 OK091582 |
| H. vesiculab, ex-type | CBS 393.81; NBRC 32216 | – | Baiting; Horseshoe Bay | British Columbia; CA; 1968 | C.J. Anastasiou; Anastasiou & Churchland 1969 | JX436352 | JF750389 MG019397 | n.a. | n.a. | n.a. | |
| H. ‘vesicula’b | CBS 152.96 | – | Rhizophora mangle sub- merged, decaying leaves | Florida, Miami, US; 1985 | n.a. | HQ232463 | HQ232472 n.a. | n.a. | n.a. | n.a. | |
| H. ‘vesicula’b | CCIBt 4142 | AJM 124 | Submerged leaf of Laguncularia racemosa; Perequê river | Ilha do Cardoso,BR; 2013 | A.L. Jesus, A.V. Marano & C.L.A. Pires-Zottarelli; Jesus et al. 2019 | KT455407 | KT455395 n.a. | n.a. | n.a. | n.a. | |
| H. ‘vesicula’b | CCIBt 4143 | AJM 126 | Submerged leaf of Laguncularia racemosa; Perequê river | Ilha do Cardoso, BR; 2013 | A.L. Jesus, A.V. Marano & C.L.A. Pires-Zottarelli; Jesus et al. 2019 | KT455408 | KT455396 n.a. | n.a. | n.a. | n.a. | |
| H. ‘vesicula’b | CCIBt 4144 | AJM 133 | Submerged leaf of Laguncularia racemosa; Perequê river | Ilha do Cardoso, BR; 2013 | A.L. Jesus, A.V. Marano & C.L.A. Pires-Zottarelli; Jesus et al. 2019 | KT455409 | KT455397 n.a. | n.a. | n.a. | n.a. | |
| H. ‘vesicula’b | CCIBt 4146 | AJM 137 | Submerged leaf of Rhizophora mangle, Perequê river | Ilha do Cardoso,BR; 2013 | A.L. Jesus, A.V. Marano & C.L.A. Pires-Zottarelli; Jesus et al. 2019 | KT455411 | n.a. n.a. | n.a. | n.a. | n.a. | |
| H. ‘vesicula’b | – | IMB147 | Mangrove fallen yellowish green leaves | Haomeili, TW; 2012 | n.a. | n.a. | KM205201 n.a. | n.a. | n.a. | n.a. | |
| H. sp. Portugal_9b | – | BD694 | Baiting; estuary of Rio Guadiana | Castro Marim, PT; 2015 | T. Jung; this study | n.a. | OK041015 n.a. | n.a. | n.a. | n.a. | |
| H. sp. thermoambigua-likeb | – | BD652 | Baiting; tidal pond in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033628 | OK033686 OK091250 | OK091306 OK091361 | OK091417 OK091475 | OK091526 OK091583 | |
| H. sp. thermoambigua-likeb | – | BD653 | Baiting; tidal pond in salt marsh, Ria Formosa | Santa Luzia, PT; 2015 | T. Jung; this study | OK033629 | OK033687 OK091251 | OK091307 OK091362 | OK091418 OK091476 | OK091527 OK091584 | |
| H. sp.1 KC-2014b | – | 1104-4-5A | Washington state stream | Washington, US; 2001 | n.a. | n.a. | KF889754 n.a. | n.a. | n.a. | n.a. | |
| H. sp.2 KC-2014b | – | 1110-2-5A | Washington state stream | Washington, US; 2001 | n.a. | n.a. | KF889755 n.a. | n.a. | n.a. | n.a. | |
| H. sp. 1 LT6430b | – | LT6430 | Marsh grass (Spartina alterniflora) leaf litter | Saint Simon’s Island, Georgia, USA; 2009 | J. Hulvey; Hulvey et al. 2010 | HQ232456 | HQ232465 KJ654026 | n.a. | n.a. | n.a. | |
| H. sp. 2 LT6465b | – | LT6465 | Marsh grass (Spartina alterniflora) leaf litter | Sapelo Island, Georgia, USA; 2009 | J. Hulvey; Hulvey et al. 2010 | HQ232460 | HQ232469 n.a. | n.a. | n.a. | n.a. | |
| H. sp-1b | CBS 140651 | Zostera marina (seeds) | Sylt, DE; 2014 | K. Rosendahl; Man In’t Veld et al. 2019 | KX364106 n.a. | n.a. | n.a. | n.a. | |||
| H. sp-3b | CBS 140657 | Zostera marina (seeds) | Sylt, DE; 2014 | K. Rosendahl; Man in ‘t Veld et al. 2019 | KX364108 n.a. | n.a. | n.a. | n.a. | |||
| H. sp-4b | – | PD6234625 | Zostera marina (seeds), Thau lagoon | FR; 2015 | K. Rosendahl; Man in ‘t Veld et al. 2019 | n.a. | KX364110 n.a. | n.a. | n.a. | n.a. | |
| H. sp. 1 EMTD7b | CBS 133933 | EMTD7 | Leaf litter and organic debris | Schleswig-Holstein, DE; 2010 | M. Thines; Nigrelli & Thines 2013 | n.a. | JX910903 n.a. | n.a. | n.a. | n.a. | |
| H. sp. 1 EMTD10b | CBS 133859 | EMTD10 | Leaf litter and organic debris | Schleswig-Holstein, DE; 2010 | M. Thines; Nigrelli & Thines 2013 | n.a. | JX910917 n.a. | n.a. | n.a. | n.a. | |
| H. sp. 2 EMTD12b | – | EMTD12 | Leaf litter and organic debris | Schleswig-Holstein, DE; 2010 | M. Thines; Nigrelli & Thines 2013 | n.a. | JX910916 n.a. | n.a. | n.a. | n.a. | |
| H. sp. 2 EMTS19b | CBS 133863 | EMTS19 | Leaf litter and organic debris | Schleswig-Holstein, DE; 2010 | M. Thines; Nigrelli & Thines 2013 | n.a. | JX910916 n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate BBGYB1b | – | USTCMS 4125 n.a. | PH | n.a. | n.a. | MT178024 | n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate BBGYB2Bb | – | USTCMS 4136 n.a. | PH | n.a. | MT178010 | MT178029 | n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate BBGYB5b | – | USTCMS 4129 n.a. | PH | n.a. | MT178016 | n.a. | n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate BCCYBL1b | – | USTCMS 4158 n.a. | PH | n.a. | MT178013 | MT178042 | n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate BCCYGL1b | – | USTCMS 4159 n.a. | PH | n.a. | MT178003 | n.a. | n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate CLE33b | – | CLE33 Zostera marina | California, US | n.a. | MN944508 | n.a. | n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate DGPBL2b | – | USTCMS 4161 n.a. | PH | n.a. | MT177998 | MT178039 | n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate DGPYB2b | – | USTCMS 4162 n.a. | PH | n.a. | MT178018 | MT178041 | n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate PNGLYBEb | – | USTCMS 4123 n.a. | PH | n.a. | MT177993 | MT178043 | n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate PQ1YBb | – | USTCMS 4108 n.a. | PH | n.a. | MT178019 | n.a. | n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate PSQNBL3b | – | USTCMS 4153 n.a. | PH | n.a. | MT177995 | n.a. | n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate PSQNBL4b | – | USTCMS 4152 n.a. | PH | n.a. | MT178000 | n.a. | n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate PSQNYB1b | – | USTCMS 4154 n.a. | PH | n.a. | MT178004 | MT178037 | n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate PSQNYB2b | – | USTCMS 4155 n.a. | PH | n.a. | MT177996 | n.a. | n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate PSQNYL1b | – | USTCMS 4156 n.a. | PH | n.a. | MT178014 | MT178037 | n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate PSQNYL2b | – | USTCMS 4157 n.a. | PH | n.a. | MT178002 | MT178031 | n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate TBJBL1.3b | – | USTCMS 4110 n.a. | PH | n.a. | MT178017 | MT178022 | n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate TTLBL3.2Ab | – | USTCMS 4116 n.a. | PH | n.a. | MT178015 | MT178033 | n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate TTLBL4.2b | – | USTCMS 4118 n.a. | PH | n.a. | MT178001 | MT178034 | n.a. | n.a. | n.a. | n.a. | |
| H. sp. isolate TWBL1b | – | USTCMS 4160 n.a. | PH | n.a. | MT178007 | MT178038 | n.a. | n.a. | n.a. | n.a. | |
| H. sp. NBRC 32444b | NBRC 32444; ATCC 90462 | AN-1063 Fallen leaf, Bruguiera gymnorrhyza | JP | n.a.; Rahman et al. 2014 | n.a. | AB688423 | AB688315 | n.a. | n.a. | n.a. | |
| H. sp. NBRC 32445b | NBRC 32445 | AN-1132 Fallen leaf, Rhizophora stylosa | JP | n.a. | n.a. | AB688424 | AB688316 | n.a. | n.a. | n.a. | |
| H. sp. Zosterab | CBS 140648 | – | Zostera marina (seeds) | Sylt, DE; 2014 | K. Rosendahl; Govers et al. 2016 | KT986007 | n.a. | n.a. | n.a. | n.a. | |
| Phytophthora condilina | – | BD661 Baiting; tidal pond in salt marsh, Ria Formosa | Quelfes, PT; 2015 | T. Jung; Botella & Jung 2021 | n.a. | MW830150 | OK091594 | n.a. | n.a. | MW836949 n.a. | |
| P. condilina | – | BD677 Baiting; coastal lagoon, | Alvor, PT; 2015 | T. Jung; this study | n.a. | OK041016 | OK091595 | n.a. | n.a. | n.a. | |
| P. gonapodyides | – | BD666 Baiting; tidal pond in salt marsh, Ria Formosa | Quelfes, PT; 2015 | T. Jung; this study | n.a. | OK041017 | n.a. | n.a. | n.a. | n.a. | |
| P. inundata | – | BD672 Baiting; coastal lagoon, Ria de Alvor | Alvor, PT; 2015 | T. Jung; this study | n.a. | OK041018 | n.a. | n.a. | n.a. | n.a. | |
| P. plurivora | – | BD691 Baiting; estuary of Rio Guadiana | Castro Marim, PT; 2015 | T. Jung; this study | n.a. | OK041019 | n.a. | n.a. | n.a. | n.a. | |
| P. pseudocryptogea | – | BD688 Baiting; tidal pond, Ria Formosa | Almancil, PT; 2015 | T. Jung; this study | n.a. | OK041020 | n.a. | n.a. | n.a. | n.a. | |
| P. sp. Clade06a New PT | – | BD678 Baiting; coastal lagoon, Ria Formosa | Almancil, PT; 2015 | T. Jung; this study | n.a. | n.a. | OK091596 | OK091597 n.a. | n.a. | OK091598 OK091599 | |
| P. castanetorumb, ex-type | CBS 142299 | BD292 Castanea sativa | Monchique, PT; 2015 | T. Jung; Jung et al. 2017b | OK033631 | MF036182 | MZ736427 | MZ736453 MF036240 | MZ736480 OK091478 | MF036292 OK091586 | |
| P. × cambivorab, ex-type | CBS 141218 | TJ197; IT 5-3 | Quercus pubescens | Sicily, IT; 2013 | T. Jung; Jung et al. 2017c | OK033630 | KU899179 MZ736422 | KU899255 KU899412 | MZ736475 OK091477 | KU899497 OK091585 | |
| Nothophytophthora amphigynosab, ex-type | CBS 142348 | BD268 | Stream baiting; atlantic forest | Sintra, PT; 2015 | T. Jung; Jung et al. 2017d | OK047740 | KY788382 – | – | – | – | |
| N. intricatab, ex-type | CBS 142354 | TJ275; RK113-1s | Aesculus hippocastanum | Wiesbaden, DE; 2011 | T. Jung; Jung et al. 2017d | OK047739 | KY788413 – | – | – | – | |
| ‘Halophytophthora’ exoproliferab, ex-paratype | CBS 252.93; ATCC 76607 | AN-1065 | Bruguiera gymnorrhyza, submerged yellow leaf | Okinawa, JP; 1988 | A. Nakagiri; Ho et al. 1992 | HQ665174 | HQ643132 – | – | – | – | |
| ‘Halophytophthora’ porrigovesicab | WPC P15166; NBRC 33162 | MT-95 | Submerged decaying leaf of Avicennia alba in a mangrove | Ranong, TH; 1999 | A. Nakagiri; Nakagiri et al. 2001 | n.a. | GU258844 n.a. | n.a. | n.a. | n.a. | |
| Calycofera operculatab, ex-type | CBS 241.83; ATCC 44952; IMI 249911 | – | Decaying leaf, Avicennia marina | Moreton Bay, Queens Island, AU | K.G. Pegg & J.L. Alcorn; n.a. | JX115217 | KJ128038 – | – | – | – | |
| Phytopythium mirpurenseb | CBS 124524 | DAOM 238992; LEV 3078 | Water pond | Sindh, Mirpurkhas, PK; 2006 | A.M. Lodhi; n.a. | KJ831614 | KJ831614 – | – | – | – | |
| Ph. ostracodesb | CBS 768.73 | – | Clay soil | Ibiza, ES; 1972 | A.J. van der Plaats-Niterink; n.a. | HQ665295 | AY598663 – | – | – | – | |
| Pythium angustatumb | CBS 522.74 | – | Soil | Oostelijk Flevoland, NL; 1971 | A.J. van der Plaats-Niterink; n.a. | AY598623 | HQ643437 – | – | – | – | |
| Py. myriotylumb | CBS 254.70 | MUCL 16166 | Arachis hypogaea | IL | Z.R. Frank; n.a. | AY598678 | AY598678 – | – | – | – | |
| Elongisporangium anandrumb | CBS 285.31 | – | Rheum rhaponticum | n.a. | C. Drechsler; n.a. | HQ665185 | HQ643435 – | – | – | – | |
| E. dimorphumb, ex-type | CBS 406.72; ATCC 22843 | – | Pinus taeda, dead root | Louisiana, US; 1971 | F.F. Hendrix Jr. & W.A. Campbell; n.a. | HQ665229 | HQ643525 – | – | – | – | |
| Globisporangium paroecandrumb | CBS 157.64 | BPIC 1297 | Loamy nursery soil | South Australia, Adelaide, AU; 1962 | O. Vaartaja; n.a. | AY598644 | AY598644 – | – | – | – | |
| G. spinosum | CBS 275.67 | – | Compost | Baarn, Cantons- park, NL | A.J. van der Plaats-Niterink; n.a. | HQ665181 | HQ643793 – | – | – | – | |
| Salisapilia epistomiumb, ex-type | CBS 590.85; ATCC 28923; IMI 330183 | – | Decaying leaf | Florida, Miami, Bear Cut, US; 1970 | I.M. Master & J.W. Fell; Bennett & Thines 2019 | HQ665279 | HQ643220 – | – | – | – | |
| S. nakagiriib, ex-type | CBS 127947 | LT6456 | Leaf litter of marsh grass (Spartina alternifolia) | Georgia, Sapelo Island, US; 2009 | J. Hulvey; Hulvey et al. 2010 | HQ232458 | HQ232467 – | – | – | – | |
| Aphanomyces euteichesb | CBS 156.73; IMI 170485 | – | Pisum sativum, root | NO | L. Sundheim; n.a. | HQ665132 | HQ643117 – | – | – | – | |
= not available.
a Abbreviations of isolates and culture collections: ATCC = American Type Culture Collection, Manassas, USA; CBS = Centraalbureau voor Schimmelcultures, Utrecht, Netherlands; IMI = CABI Bioscience, UK; PD = Phytophthora Database (http://www.phytophthoradb.org); WPC = World Phytophthora Collection, University of California Riverside, USA; other isolate names and numbers are as given by the collectors and on GenBank, respectively.
b Isolates used in the phylogenetic studies.
c Isolates used in the morphological studies.
d Isolates used in the temperature-growth studies.
e Isolate only used for sporangial measurements.
f Abbreviations of country names: AU = Australia; BR = Brazil; CA = Canada; DE = Germany; ES = Spain; FR = France; IL = Israel; IT = Italy; JP = Japan; NL = Netherlands; NO = Norway; PH = Philippines; PK = Pakistan; PT = Portugal; TH = Thailand; TW = Taiwan; US = United States of America.
DNA was extracted from mycelium scraped from 1–3-wk-old V8A cultures, placed into 2 mL homogenisation tubes (MP Biomedicals, Irvine, USA) and disrupted using a Precellys Evolution instrument (Bertin Technologies, Montigny-le-Bretonneux, France). 300 µL of lysis buffer (Lamour & Finley 2006) supplemented by 3 µL of RNase A (New England Biolabs, Ipswich, USA) was added to each sample and incubated at 65 °C for 20 min (Lamour & Finley 2006). Each sample was centrifuged at 20 000 g for 3 min and the supernatant transferred to a fresh tube. Each sample was then mixed with 150 µL of 5 M potassium acetate, stored at -20 °C for at least 30 min followed by 30 min of centrifugation at 20000 g (Lamour & Finley 2006). The supernatant was transferred to a 2 mL tube and further purified using Monarch ® PCR & DNA Cleanup Kit (New England Biolabs, Ipswich, USA) following the manufacturer´s protocol. DNA was eluted with 80 µL of pre-warmed elution buffer and concentration was estimated by spectrophotometry using QUICKDROP (Molecular Devices, San Jose, USA). DNA was preserved at -80 °C for long-term storage.
PCR amplifications were performed using a LightCycler 480 II instrument (Roche, Basel, Switzerland) and PCR conditions were optimised for each locus. Primers were synthetized by Elizabeth Pharmacon spol. s.r.o. (Brno, Czech Republic) and their annealing temperatures were estimated using Tm calculator (http://tmcalculator.neb.com/#!/main and adjusted empirically, according to observed PCR amplification rates. Table 2 provides a comprehensive overview of the PCR methodology and primers used.
Table 2.
Overview of PCR conditions and details of primers used for amplification and sequencing of oomycete isolates.
| Locus | Primer names | Primer sequences (5’-3’) | Orientation | Annealing temperature (°C) | Extension time (s) | Reference for primer sequences | |
|---|---|---|---|---|---|---|---|
| LSUa,b | CTB6 | GCATATCAATAAGCGGAGG | Forward | 53 | 20 | Garbelotto et al. (1997); | |
| LR3e | CCGTGTTTCAAGACGGG | Reverse | Hopple & Vilgalys (1994) | ||||
| LR3Re | GTCTTGAAACACGGACC | Forward | |||||
| LR7 | TACTACCACCAAGATCT | Reverse | |||||
| ITSa | ITS1 | TCCGTAGGTGAACCTGCGG | Forward | 63 | 12 | White et al. (1990); | |
| ITS4 | TCCTCCGCTTATTGATATGC | Reverse | Cooke et al. (2000 | ||||
| ITS6f | GAAGGTGAAGTCGTAACAAGG | Forward | |||||
| btub a | TUBUF2 | CGGTAACAACTGGGCCAAGG | Forward | 68 | 12 | Kroon et al. (2004) | |
| TUBUR1 | CCTGGTACTGCTGGTACTCAG | Reverse | |||||
| hsp90 a | HSP90_F1int | CAAGGTGATCCCGGACAAGGC | Forward | 66 | 15 | Blair et al. (2008) | |
| HSP90R1 | ACACCCTTGACRAACGACAG | Reverse | |||||
| tigA c | Tig_FY | TCGTGGGCGGYAAYTGGAA | Forward | 60 | 120 | Blair et al. (2008) | |
| Tig_reve | CCGAAKCCGTTGATRGCGA | Reverse | |||||
| G3PDH_fore | TCGCYATCAACGGMTTCGG | Forward | |||||
| G3PDH_rev | GCCCCACTCRTTGTCRTACCAC | Reverse | |||||
| rpl10 a | 60SL10_for | GCTAAGTGTTACCGTTTCCAG | Forward | 64 | 7 | Martin & Tooley (2003) | |
| 60SL10_rev | ACTTCTTGGAGCCCAGCAC | Reverse | |||||
| cox1 | OomCoxI-Levupa | TCAWCWMGATGGCTTTTTTCAAC | Forward | 60 | 10 | Robideau et al. (2011) | |
| OomCoxI-Levloa | CYTCHGGRTGWCCRAAAAACCAAA | Reverse | |||||
| COXF4Nc | GTATTTCTTCTTTATTAGGTGC | Forward | 50 | 65 | Kroon et al. (2004) | ||
| COXR4Nc | c | CGTGAACTAATGTTACATATAC | Reverse | ||||
| nadh1 c | NADHF1 | CTGTGGCTTATTTTACTTTAG | Forward | 50 | 65 | Kroon et al. (2004) | |
| NADHR1 | CAGCAGTATACAAAAACCAAC | Reverse | |||||
| rps10 d | rps10_DB_FOR | GTTGGTTAGAGYARAAGACT | Forward | 48 | 30 | Foster et al. (2021) | |
| rps10_DB_REV | RTAYACTCTAACCAACTGAGT | Reverse |
a PCR protocol 1: 20 µL volume containing 10.4 µL H2O, 4 µL Q5 Reaction Buffer (5X), 1 µL of each primer (10 μM), 0.4 µL deoxynucleotide (dNTP) mixture (Meridian Bioscience, Memphis, USA) (2.5 mM each), 0.2 µL of Q5 High-Fidelity DNA Polymerase (2 U/μL) (New England Biolabs, Ipswich, USA), and 3 µL of gDNA (3–60 ng). Initial denaturation for 30 s at 98 °C; 35 cycles consisting of 5 s at 98 °C, 20 s at optimised annealing temperature for each primer set, optimised length of extension at 72 °C; 2 min at 72 °C for final extension.
b Double concentration of Q5 polymerase.
c PCR protocol 2: 20 µL volume containing 10 µL H2O, 4 µL PrimeSTAR GXL Buffer (5X), 0.8 µL of each primer, 1.6 µL dNTP mixture, 0.4 µL PrimeSTAR GXL DNA Polymerase (1.25 U/μL) (TaKaRa Bio, Kusatsu, Shiga, Japan), and 3 µL of gDNA. Initial denaturation for 5 s at 98 °C; 35 cycles consisting of 10 s at 98 °C, 15 s at optimised annealing temperature, optimised length of extension at 68 °C; 5 min at 68 °C for final extension.
d PCR protocol 3: 20 µL volume containing 6.2 µL H2O, 10 µL OneTaq Hot Start Quick-Load 2X Master Mix with Standard Buffer (New England Biolabs, Ipswich, USA) 0.4 µL of each primer, and 3 µL of gDNA. Initial denaturation for 5 s at 98 °C; 35 cycles consisting of 30 s at 98 °C, 30 s at optimised annealing temperature, optimised length of extension at 72 °C; 7 min at 72 °C for final extension.
e Primers used exclusively for sequencing.
f Two primer combinations were used: ITS1/ITS4 or ITS6/ITS4.
PCR products were visualised by gel electrophoresis (300V; 5 min) using 2 % agarose gel stained by DNA Stain G (SERVA, Heidelberg, Germany). All amplicons were purified and sequenced in both directions by Eurofins Genomics GmbH (Cologne and Ebersberg, Germany) using the amplification primers except for the tigA and LSU amplicons which required two additional primers (Table 2).
Electropherograms were quality checked and forward and reverse reads were compiled using Geneious Prime® v. 2021.2.2 (Biomatters Ltd., Auckland, New Zealand). Pronounced double peaks were considered as heterozygous positions and labelled according to the IUPAC coding system. All sequences generated in this study were deposited in GenBank and accession numbers are given in Table 1.
Phylogenetic analysis
In the past decade LSU and/or ITS sequences of numerous unidentified Halophytophthora isolates and taxa have been submitted to GenBank. To provide an updated phylogenetic structure of the genus Halophytophthora s.str. and resolve the phylogenetic positions of the 10 new Halophytophthora taxa from Portugal within the genus, two datasets comprising all relevant and distinctive (i) LSU and (ii) ITS sequences available from GenBank and respective sequences from the new Portuguese taxa were analysed. In the LSU analysis representative species of related genera in the Peronosporaceae, Pythiaceae and Salisapiliaceae were included and Aphanomyces euteiches (CBS 156.73) used as outgroup taxon (dataset: 59 isolates and 1 289 characters). In the ITS analysis Phytophthora castanetorum (CBS 142.299) and P. × cambivora (CBS 141.218) were used as outgroup taxa (dataset: 63 isolates and 1 254 characters).
To study the relative phylogenetic positions of the nine new Halophytophthora taxa from Portugal for which living isolates were available and related Halophytophthora species, i.e., H. avicennae, H. fluviatilis and H. polymorphica, (iii) a full 9-partition dataset (9 loci: ITS-LSU-rpl10-Btub-hsp90-tigA-cox1-nadh1-rps10) was analysed with P. castanetorum (CBS 142.299) and P. × cambivora (CBS 141.218) as outgroup taxa (dataset: 60 isolates and 8 759 characters).
The sequences of the loci used in the analyses were aligned using the MAFFT v. 7 (Katoh & Standley 2013) plugin within the Geneious software by the E-INS-I strategy (ITS) or the G-INS-I strategy (all other loci). The ITS alignments in this study were manually edited and adjusted.
Bayesian Inference (BI) analysis was performed using MrBayes v. 3.2.7 (Ronquist & Huelsenbeck 2003) into partitions with the GTR Gamma + I nucleotide substitution model. Four Markov chains were run for 10 M generations, sampling every 1 000 steps, and with a burn in at 6 000 trees. Maximum-Likelihood (ML) analyses were carried out using raxmlGUI v. 2.0 (Edler et al. 2021) with a GTR Gamma + I nucleotide substitution model. There were 10 runs of the ML and bootstrap (thorough bootstrap) analyses with 1 000 replicates used to test the support of the branches.
Phylogenetic trees were visualized in TreeGraph2 v. 2.15.0-887 beta (Stöver & Müller 2010) and/or MEGA X v. 10.2.6 (Kumar et al. 2018) and edited in figure editor programs.
All datasets and original trees deriving from BI and ML analyses were deposited in the Dryad Digital Repository (https://datadryad.org; https://doi.org/10.5061/dryad.gf1vhhmr2).
Morphology of asexual and sexual structures
Characteristic morphological features of sporangia, oogonia, oospores, antheridia, chlamydospores, hyphal swellings and aggregations of the eight new Halophytophthora species and H. avicennae were recorded and compared with each other.
Sporangia formation and zoospore release were induced using a modified protocol of Nakagiri et al. (1994), Jesus et al. (2016) and Jung et al. (2017c). Two 12–15 mm square discs were cut from the growing edge of a 2–5-d-old V8A colony and submerged in a 90 mm diam Petri dish in 50 % non-sterile seawater. The Petri dishes were incubated at 20 °C in natural light and the water was changed after c. 6 h. Shape and special features of sporangia and the formation of hyphal swellings and aggregations were recorded after 18–48 h depending on the isolate. For each isolate 50 fully mature sporangia and 20 encysted zoospores and exit pores, chosen at random, were measured at × 400 using a compound microscope (Axio Imager.Z2), a digital camera (AxioCam ICc3) and a biometric software (ZEN) (all Zeiss, Jena, Germany).
Gametangia formation (oogonia and antheridia) and their cha-racteristic features were examined after 21–30 d growth at 20 °C in the dark on V8A. For each non-sterile isolate each 50 oogonia, oospores and antheridia chosen at random were measured under a compound microscope at × 400. The oospore wall index was calculated according to Dick (1990).
Colony morphology, growth rates and cardinal temperatures
Colony growth patterns of all eight Halophytophthora species were described from 7-d-old cultures grown at 20 °C in the dark in 90 mm plates on sCA, vegetable juice seawater agar (sV8A), and potato dextrose seawater agar (sPDA; Oxoid Ltd., UK) prepared like normal CA, V8A and PDA (Erwin & Ribeiro 1996, Jung & Burgess 2009, Jung et al. 2017c) but with 50 % seawater.
Temperature-growth relationships were determined by subculturing three to five representative isolates per Halophytophthora species onto 90 mm sV8A Petri dishes and incubating them for 24 h at 20 °C to stimulate onset of growth (Jung et al. 1999). Three replicate plates per isolate were subsequently incubated at 10, 15, 20, 25, 27.5, 30, 32.5 and 35 °C. Radial growth was recorded daily for 5 d or until the isolate almost reached the edge of the Petri dish, along two lines drawn to overlap in the centre of the inoculum plug at right angles. The mean growth rates (mm/d) were calculated and the cardinal temperatures established. Petri dishes showing no growth at 27.5, 30, 32.5 and 35 °C were re-incubated at 20 °C to check for viability and determine the lethal temperature (Jung et al. 2017d).
RESULTS
Phylogenetic analysis
For all three datasets the ML and BI analyses produced phylogenetic trees with similar topologies. Since the BI analyses provided higher support values than the ML analyses for the deeper nodes and partly also for the terminal nodes the BI trees are presented with both BI Posterior Probability values and ML bootstrap values included Fig. 2–4, Dryad dataset: (https://doi.org/10.5061/dryad.gf1vhhmr2). The LSU dataset included 22 isolates of nine new Halophytophthora taxa from Portugal and eight described Halophytophthora species, 21 unidentified Halophytophthora isolates and 15 isolates from representative species of other genera in the Peronosporaceae, Pythiaceae and Salisapiliaceae. In the LSU analysis Halophytophthora s.str. resided in sister position to Phytophthora and revealed a structure of eight phylogenetic clades which are designated here as Clades 1 to 8 (Fig. 2). Clade 1 comprised the ex-type isolate of H. batemanensis from New South Wales, Australia, 11 unidentified isolates from the Philippines which most likely belong to H. batemanensis (2–4 bp differences), three further isolates from the Philippines (DGPBL2, PSQNBL3 and PSQNYB2) belonging to an undescribed species, and a group of three isolates from Brazil (CCIBt4143 and CCIBt4146) and Florida (CBS 152.96) which were previously assigned to H. vesicula and constitute two or three new species. Clade 2 included two undescribed taxa from the Philippines and clustered in sister position to Clade 1 while Clades 3, 4, 5, 7 and 8 each contained only one species, H. insularis, H. avicennae, H. fluviatilis, H. souzae and the distinct undescribed Halophytophthora sp. PNGLYBE, respectively (Fig. 2). All nine new taxa from Portugal resided in Clade 6 together with the ex-type isolates of H. vesicula (CBS 393.81) and H. polymorphica (CBS 680.84), Halophytophthora sp. CLE33, an undescribed species from California related to H. vesicula and H. lateralis from Portugal, two new species related to H. polymorphica, Halophytophthora sp. PQ1YB from the Philippines and Halophytophthora ‘polymorphica’ CCIBt 4112 from Brazil, and two other potentially new species from Georgia, USA, Halophytophthora sp. 1 LT6430 and Halophytophthora sp. 2 LT6445, both related to H. brevisporangia and H. celeris from Portugal. The LSU sequences of H. vesicula and H. lateralis were identical. Both the deepest and second deepest nodes within Halophytophthora s.str. were highly supported. A polytomy at the second deepest node indicated either an ancient radiation or a character conflict. ‘Halophytophthora’ exoprolifera was basal to the Halophytophthora-Phytophthora-Nothophytophthora cluster (Fig. 2) and belongs to an undescribed genus.
Fig. 2.
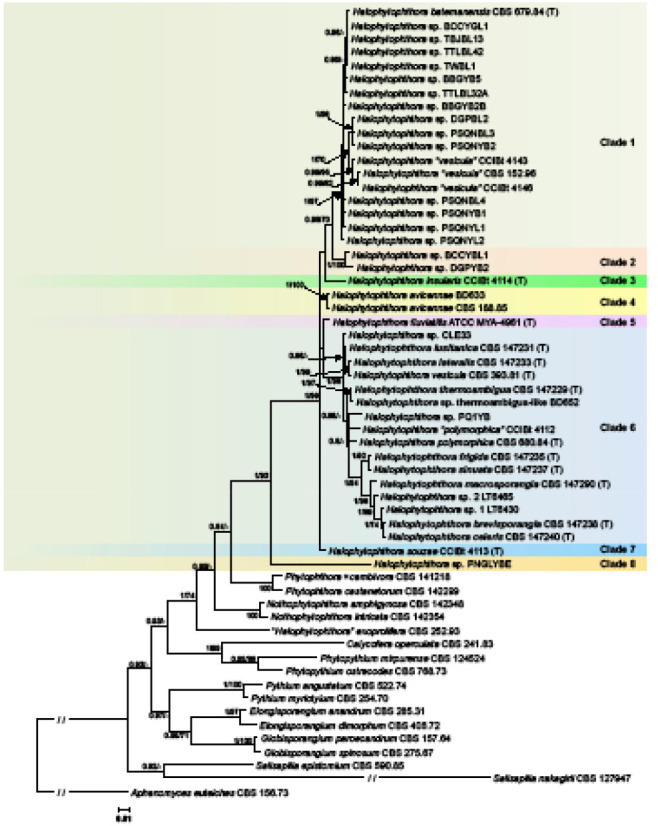
Fifty percent majority rule consensus phylogram derived from Bayesian inference analysis of an LSU dataset of Halophytophthora s.str. and representative species from related genera in the Peronosporaceae, Pythiaceae and Salisapiliaceae. Bayesian posterior probabilities and ML bootstrap values (in %) are indicated but not shown below 0.80 and 70 %, respectively. Aphanomyces euteiches was used as outgroup taxon. Scale bar indicates 0.01 expected changes per site per branch.
Fig. 4.
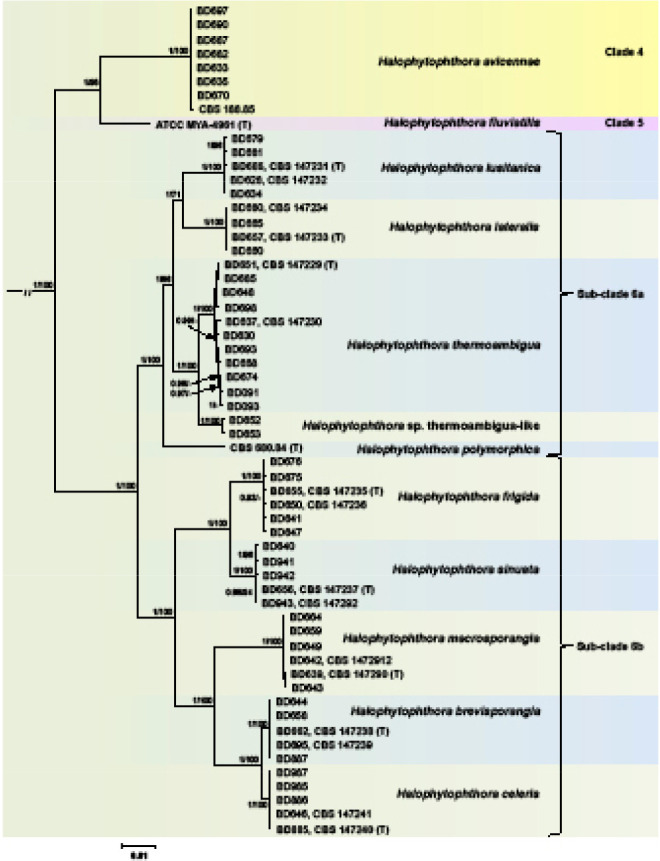
Fifty percent majority rule consensus phylogram derived from Bayesian inference analysis of a concatenated nine-locus (ITS-LSU-rpl10-Btub-hsp90-tigA-cox1-nadh1-rps10) dataset of Halophytophthora s.str. Clades 4, 5 and 6. Bayesian posterior probabilities and ML bootstrap values (in %) are indicated but not shown below 0.80 and 70 %, respectively. Phytophthora castanetorum (CBS 142.299) and P. × cambivora (CBS 141.218) were used as outgroup taxa (not shown). Scale bar indicates 0.01 expected changes per site per branch.
The BI and ML analyses of the ITS dataset comprising 61 Halophytophthora s.str. isolates confirmed the LSU clade structure of the genus expanding it by two further clades designated here as Clades 9 and 10 (Fig. 3). Clade 1 again contained the H. batemanensis ex-type, 11 isolates from the Philippines and Japan which should be assigned to H. batemanensis, isolate DGPBL2 from the Philippines belonging to an undescribed species, and three isolates from Brazil (CCIBt 4142 and CCIBt 4144) and Florida (CBS 152.96) previously assigned to H. vesicula and representing one or two new species (Fig. 3). Different from the LSU analysis the distinct Halophytophthora sp. PNGLYBE from Clade 8 resided in sister position to the latter cluster from Clade 1. Clades 1 and 2, the latter represented by an undescribed species comprising isolate BCCYBL1 from the Philippines and isolate IMB147 from Taiwan which was previously assigned to H. vesicula, constituted sister clades as in the LSU analysis. Halophytophthora avicennae (Clade 4) and H. fluviatilis (Clade 5) formed a polytomy together with the common ancestor of Clades 1 and 2 (Fig. 3). This four-clades cluster formed another polytomy together with Clades 3, 6, 7, 10 and the common ancestor of Clades 7 and 9. Clade 9 included the undescribed Halophytophthora sp. 2 KC-2014 from a stream in Washington State, USA, and resided in sister position to the Clade 7 species H. souzae from Brazil whereas Clade 10 contained the undescribed taxon Halophytophthora sp. Portugal_9 (Fig. 3). As in the LSU phylogeny, Clade 6 comprised nine new species from Portugal, the ex-types of H. vesicula and H. polymorphica, an undescribed species represented by the Brazilian isolate CCiBt 4112, previously assigned to H. polymorphica, and 11 unidentified isolates (Fig. 3). Halophytophthora sp. 1 KC-2014 from a stream in Washington State, USA, belongs to a new species related to H. lusitanica (15 bp differences = 1.2 %) while the undescribed Halophytophthora sp-4 (isolate PD6234625) from the Thau lagoon in Southern France resided in a basal position to this cluster. The informally designated Halophytophthora sp. Zostera (isolate CBS 140648) was identical to Halophytophthora sp-3 (isolate CBS 140657) from Z. marina in the North Sea and most likely belongs to H. lateralis (1 bp difference) which clustered in sister position to H. vesicula differing from the latter at 39 positions (= 3.1 %). This cluster together with a cluster comprising H. thermoambigua, H. sp. thermoambigua-like, H. polymorphica and Halophytophthora ‘polymorphica’ CCIBt 4112 constituted a distinct subclade designated here as Subclade 6a (Fig. 3). Halophytophthora thermoambigua and H. sp. thermoambigua-like are sister species differing in ITS at 7–9 positions. Subclade 6b contained a cluster comprising the sister species H. frigida and H. sinuata from Portugal and a larger cluster with an undescribed species from the German North Sea coast (Halophytophthora sp. 1 of Nigrelli & Thines 2013; isolates EMTD10 and EMTS19) residing in a basal position of a cluster containing H. macrosporangia, H. brevisporangia and H. celeris from Portugal and five unidentified isolates. Similar to the LSU analysis, H. sp. 2 LT6465 from Georgia, USA, constituted a new species basal to a cluster which included the two sister species H. brevisporangia and H. celeris from Portugal, H. sp. 1 LT6430 from Georgia and three isolates from the North Sea (CBS 140651, EMTD7 and EMTD12). With differences at 7–12 positions H. sp. 1 LT6430 from Georgia might belong to H. brevisporangia or to a closely related unknown species whereas the 5–6 polymorphisms differentiating isolates CBS 140651 (Halophytophthora sp-1), EMTD7 and EMTD12 (both Halophytophthora sp. 2 of Nigrelli & Thines 2013) from the Portuguese isolates of H. celeris are most likely within the variation of the latter species.
Fig. 3.
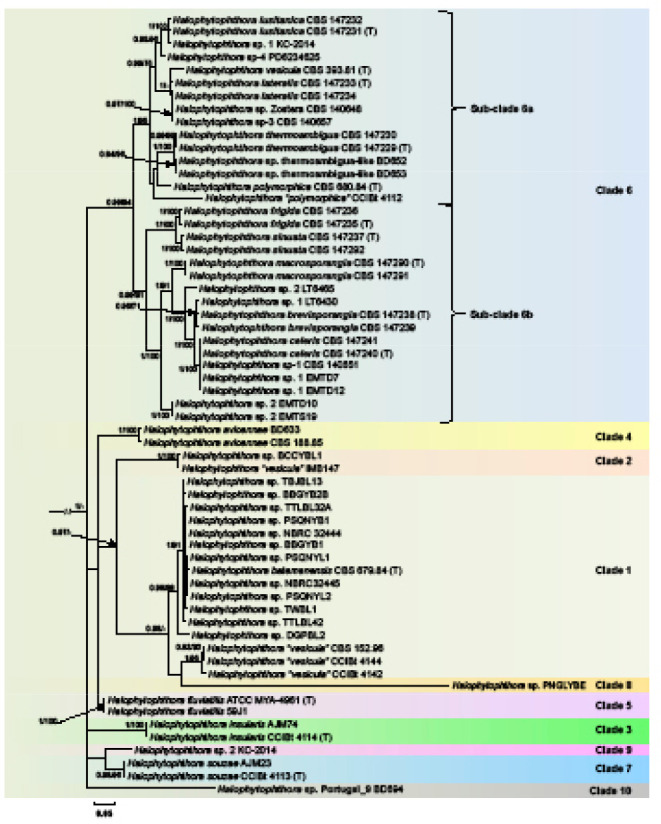
Fifty percent majority rule consensus phylogram derived from Bayesian inference analysis of an ITS dataset of Halophytophthora s.str.. Bayesian posterior probabilities and ML bootstrap values (in %) are indicated but not shown below 0.80 and 70 %, respectively. Phytophthora castanetorum (CBS 142.299) and P. × cambivora (CBS 141.218) were used as outgroup taxa (not shown). Scale bar indicates 0.05 expected changes per site per branch.
When the phylogeny of the nine new Halophytophthora taxa from Portugal within Clade 6 was analysed with the 9-partition dataset (ITS-LSU-rpl10-Btub-hsp90-tigA-cox1-nadh1-rps10), the Portuguese isolates formed nine fully supported distinct clades (Fig. 4). Within Subclade 6a, H. lateralis with H. lusitanica and H. thermoambigua with H. sp. thermoambigua-like clustered in sister position to each other with both clusters being well supported in both analyses and H. polymorphica residing in a basal position to them. Within Subclade 6b, H. frigida and H. sinuata clustered in sister position to each other while the sister species H. brevisporangia and H. celeris formed another cluster together with H. macrosporangia which resided in a basal position (Fig. 4). Across the 8 759 character multigene alignment there were 637 unique polymorphic sites (7.3 %) within Clade 6.
Halophytophthora brevisporangia, H. celeris, H. frigida, H. lateralis, H. lusitanica, H. macrosporangia, H. polymorphica, H. sinuata, H. thermoambigua and H. sp. thermoambigua-like had 55–61, 32–36, 50–51, 68–69, 53–71, 76–82, 61, 52–63, 23–70 and 29–32 unique polymorphisms, respectively, and differed from each other at 82–501 positions corresponding to sequence similarities of 94.3–99.1 %. The sister species H. lateralis with H. lusitanica, H. frigida with H. sinuata, H. brevisporangia with H. celeris, and H. thermoambigua with H. sp. thermoambigua-like shared 12, 56, 63 and 38–41 unique polymorphisms and differed from each other at 267–283, 170–178, 82–90 and 106–125 positions, respectively, corresponding to sequence similarities of 96.8–96.9, 98–98.1, 98.6–98.8 and 99–99.1 %, respectively. The ITS alignment contained several indels of up to 29 characters length which were partly shared between the Clade 6 species and also with H. fluviatilis (Clade 5) but were absent in H. avicennae (Clade 4). In hsp90 two isolates of H. sinuata (BD656 and BD943) had a 6 bp insertion at positions 433–438 whereas H. brevifolia, H. celeris and H. fluviatilis shared a 3 bp deletion at positions 497–499.
Heterozygous positions were present in all three nuclear single-copy genes, Btub, hsp90 and tigA, and in all Clade 6 species except of H. frigida. In the other nine species the frequencies of heterozygous sites varied considerably between and within species. While H. lateralis, H. sinuata, H. polymorphica, H. macro-sporangia, H. sp. thermoambigua-like, H. celeris and H. lusitanica had in total only 2, 2, 3, 5, 6 and 13 heterozygous sites across the 3 450 character alignment, H. brevisporangia and H. thermoambigua were heterozygous at 37 and 49 positions potentially indicating hybrid origin. Interestingly, the frequency of heterozygous sites varied considerably between individual isolates of H. thermoambigua with one isolate (BD637) being fully homozygous and the other eight isolates having between 1 and 25 heterozygous sites. In contrast to the nuclear genes, the mitochondrial cox1, nadh1 and rps10 genes contained no heterozygous sites.
Taxonomy
Morphological and physiological characters and morphometric data of the eight new Halophytophthora species and, for comparison, the seven known species of Halophytophthora s.str. are listed in Table 3, 4.
Table 3.
Morphological characters and dimensions (µm), cardinal temperatures (°C) and temperature-growth relations (mm/d) of eight new Halophytophthora species. Most discriminating characters are highlighted in bold.
| H. brevisporangia | H. celeris | H. frigida | H. lateralis | H. lusitanica | H. macrosporangia | H. sinuata | H. thermoambigua | |
|---|---|---|---|---|---|---|---|---|
| No. of isolates | 3a | 3a | 7a | 4a | 6a | 7a | 6a | 11a |
| Sporangia | ovoid 86 %, obpyriform, (distorded, ellipsoid, obturbinate) | ovoid 60 %, obpyriform, (subglobose, limoniform) | obpyriform 83 %, ovoid, (distorted, obturbinate, limoniform, ellipsoid) | ovoid 95% (sub-globose, ovoid-obpyri-form, distorted) | obpyriform 54 %, ovoid (ellipsoid, limoniform, obturbinate, distorted) | obpyriform 56 %, ovoid, (subglobose, distorted, peanut-shaped) | obpyriform 85 %, ovoid, (distorted, limoniform), often asymmetric | obpyriform 50 %, ovoid, (limoniform, ellipsoid, ampulliform, distorted) |
| l×b mean | 57.6±10.9 × 42.1±8.8 | 60.5±12.5 × 46.1±10.4 | 80.4±19.3 × 50.1±13.8 | 75.9±16.1 × 52.4±12.5 | 84.0±15.8 × 54.3±10.6 | 97.5±24.8 × 55.0±14.8 | 74.1±19.5 × 44.0±12.6 | 75.3±15.7 × 48.1±9.7 |
| range of isolate means | 54.7–59.7 × 39.9–45.9 | 56.2–64.6 × 43.9–47.3 | 72.7–85.7 × 44.5–57.1 | 67.7–89.5 × 45.4–63.6 | 76.2–95.4 × 47.1–60.2 | 76.8–118.9 × 42.9–67.4 | 56.9–82.1 × 32.5–51.4 | 69.2–83.2 × 40.3–57.1 |
| total range | 31.2–101.4 × 20.9–78.8 | 29.4–92.8 × 20.8–78.0 | 32.4–150.7 × 18.8–98.1 | 41.6–122 × 22.8–100.3 | 40.5–162.5 × 25.4–86.3 | 58–186.7 × 32.3–106.3 | 28.1–129.2 × 19.8–89.5 | 43.4–185.3 × 25.3–83.9 |
| l/b ratio | 1.4 ± 0.1 | 1.3 ± 0.1 | 1.6 ± 0.3 | 1.5 ± 0.2 | 1.6 ± 0.3 | 1.8 ± 0.4 | 1.7 ± 0.3 | 1.6 ± 0.3 |
| apexb | non-papillate, often pointed, 2 apices not observed | non-papillate, often pointed, 2 apices rare | non-papillate, pointed to protuberant, some curved or 2 apices | non-papillate or semi-papillate, sometimes pointed or 2 apices | non-papillate or rarely semipapillate, some-times 2 apices or curved | non-papillate, some-times pointed or 2 apices | non-papillate, often pointed, sometimes 2 apices | non-papillate, pointed to protuberant, some curved, 2 apices common |
| special features | basal plug frequent, lateral attachment occasionally | protruding basal plugs, sometimes intercalary or lateral attachment (13 %) | frequently intercalary or lateral attachment | lateral attachment 41.5 %, sometimes protruding basal plug | some with lateral attachment, hyphal beak or basal plug | sometimes lateral attachment, small vacuoles, swelling close to base | lateral attachment 31.4 %, often 1 or more vacuoles | some intercalary, lateral attachment 38.4 %, basal plug common |
| external proliferation | common, lax sympodia | rare | infrequent | infrequent | rare | infrequent | rare | infrequent |
| zoospore release | through semi-persistent elongated vesicle | through semi-persistent elongated vesicle | through semi-persistent elongated vesicle | through semi-persistent elongated vesicle | through semi-persistent elongated vesicle | through semi-persistent elongated vesicle | direct or through semi-persistent vesicle | through semi-persistent elongated vesicle |
| exitpores | 9.8 ± 1.7 | 10.5 ± 1.6 µm | 9.5 ± 1.6 | 12.6 ± 2.4 | 10.6 ± 2.1 | 10.3 ± 1.7 | 8.9 ± 1.5 | 10.3 ± 2.3 |
| zoospore cysts | 7.9 ± 0.7 | 8.8 ± 0.8 | 9.1 ± 2.0 | 7.9 ± 0.7 | 8.2 ± 0.9 | 8.2 ± 2.1 | 7.5 ± 1.1 | 8.6 ± 0.9 |
| Breeding system | sterile | sterile | homothallic | sterile | sterile | homothallic | homothallic | sterile |
| Oogonia | – | – | smooth-walled, some slightly ornamented | – | – | smooth-walled | wavy to slightly verru-cose goldenbrown wall | – |
| mean diam | 47.8 ± 4.0 | 48.2 ± 4.6 | 54.2 ± 3.8 | |||||
| range of isolate means | 43.0–49.9 | 46.7–49.2 | 51.6–55.6 | |||||
| total range | 32.0–58.6 | 18.4–58.7 | 39.4–64.3 | |||||
| Oospores | – | – | 99.3 % plerotic | – | – | 100 % plerotic | 95 % plerotic | – |
| mean diam | 45.3 ± 4.1 | 43.7 ± 4.2 | 47.5 ± 3.7 | |||||
| total range | 29.1–54.9 | 30.1–54.5 | 33.9–57.6 | |||||
| wall diam | 2.2 ± 0.5 | 1.7 ± 0.4 | 2.9 ± 0.6 | |||||
| oospore wall index | 0.26 ± 0.05 | 0.22 ± 0.04 | 0.32 ± 0.05 | |||||
| Abortion rate | 22.7 % (2–50 %) | 41.5 % (14–82 %) | 4.2 % (0–13 %) | |||||
| Antheridia | – | – | paragynous, mostly intricate stalks | – | – | paragynous intricate stalks occasionally | paragynous, very rarely intricate stalks | – |
| size | 18.3±3.6 × 10.3±2.7 | 15.2±3.1 × 7.8±1.7 | 14.9±2.8 × 11.6±1.8 | |||||
| Hyphal swellings | – | infrequent, triangular | rare | – | rare | infrequent | – | infrequent; 33.8 ± 9.5 |
| Hyphal aggregations | – | common | – | – | – | – | – | – |
| Colonies on sV8A | faintly radiate, limited cottony mycelium | uniform, limited cottony | faint radiate to petaloid, limited aerial or cottony | stellate, limited aerial mycelium | petaloid-faintly petaloid, limited aerial mycelium | faint stellate, limited aerial, submerged edge | uniform, limited aerial mycelium | stellate-radiate or petaloid, limited aerial |
| Colonies on sCA | uniform, limited cottony mycelium | uniform, limited cottony | similar to sV8A | faintly petaloid, limited aerial mycelium | petaloid-faintly petaloid, limited aerial mycelium | uniform, limited aerial, submerged edge | faintly radiate, limited aerial mycelium | stellate-radiate or petaloid, limited aerial |
| Colonies on sPDA | uniform or petaloid, dense-felty appressed | faint petaloid – petaloid, felty, cottony margin | faint petaloid, felty-cottony, submerged edge | dense-felty, uniform | felty, uniform, ring of collapsed aerial mycelium | stoloniferous-petaloid, appressed-submerged | faintly petaloid, cottony | felty, petaloid |
| Maximum temperature | 32.5 | 32.5 | 25 | 32.5 | 32.5 | 27.5 | 27.5 | 32.5 |
| Optimum temperature | 25 | 25 | 15 (20) | 25 | 25 (20, 27.5) | 15, (20), 25 | 25 | 25 (20, 27.5) |
| Growth rate sV8A 20 °C | 18.5 ± 1.3 | 20.4 ± 0.7 | 8.3 ± 2.3 | 10.3 ± 0.7 | 7.3 ± 0.6 | 4.2 ± 1.1 | 14.9 ± 0.3 | 5.2 ± 1.2 |
| Growth rate sCA 20 °C | 16.7 ± 0.5 | 17.0 ± 0.1 | 9.4 ± 0.8 | 12.0 ± 0.4 | 6.9 ± 0.8 | 5.1 ± 1.4 | 12.8 ± 0.1 | 5.2 ± 1.4 |
| Growth rate sPDA 20 °C | 13.3 ± 1.4 | 15.7 ± 0.3 | 5.7 ± 0.6 | 9.8 ± 0.3 | 6.7 ± 0.4 | 4.1 ± 1.1 | 13.3 ± 0.4 | 5.0 ± 2.1 |
a Numbers of isolates included in the growth tests: H. thermoambigua - 5; H. lusitanica - 5; H. lateralis - 4; H. frigida - 5; H. sinuata - 5; H. macrosporangia - 5; H. brevisporangia - 3; H. celeris - 3.
b Apex in all 8 new Halophylefththora species becoming pseudo-papillate during zoospore differentiation due to the shrinkage of the proleftlasm away from the apex.
– = character not observed; n.a. = not available.
Table 4.
Morphological characters and dimensions (µm), cardinal temperatures (°C) and temperature-growth relations (mm/d) of the seven known species of Halophytophthora s.str. Most discriminating characters are highlighted in bold
| H. avicennae | H. batemanensis | H. fluviatilis | H. insularis | H. polymorphica | H. souzae | H. vesicula | ||
|---|---|---|---|---|---|---|---|---|
| Source / no. of isolates | Gerrettson-Cornell & Simpson (1984) / n.a. | this study / 7 | Gerrettson-Cornell & Simpson (1984) ) / n.a. | Yang & Hong (2014) / 6 | Jesus et al. (2019) / 3 | Gerrettson-Cornell & Simpson (1984) / n.a. | Jesus et al. (2019) / 3 | Anastasiou & Churchland (1969) / n.a. |
| Sporangia> | ovoid, obpyriform, ob- clavate, botuliform, reniform, distorted) | obpyriform 56%, ovoid (limoniform, distorted, obturbinate) | ovoid, ellipsoid,limoniform | globose-ovoid, (limoni- form, obovoid, distorted) | limoniform, ovoid, obpyriform | ovoid, obpyriform, variable, asymetric, distorted | limoniform, ovoid, obpyriform | ovoid, obpyriform, (fusiform, distorted) |
| l×b mean > | 75 × 31 | 65.5±10.6 × 43.8±8.5 | 64 × 48 | 38.4±5.8 × 28.8±4.4 | 71.1 × 51.6 | 72 × 58 | 93.5 × 56.5 | 117 × 59 |
| range of isolate means > | n.a. | 56.1–71.3 × 36.9–50.9 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| total range > | 44–121 × 18–44 | 43.0–98.8 × 28.2–74.3 | 33–96 × 26–81 | 28.3–58.2 × 20.1–41.0 | 38.9–105.3 × 28.2–80.9 | 44–102 × 33–84 | 52.5–162.5 × 37.5–77.5 | 47–192 × 24–100 |
| l/b ratio > | 2.6 (1.6–4.8) | 1.5 ± 0.2 | 1.3 (1.1–1.6) | 1.3 | 1.4 | 1.3 (1.0–1.6) | 1.7 | 2.0 |
| apexa | non-papillate, some-times pointed or 2 apices | non-papillate, some-times pointed (9.2%) or 2 apices | non-papillate, some-times pointed or protuberant | non-papillate, often protuberant | non-papillate, pointed, sometimes 2 apices | non-papillate, sometimes protuberant or 2 apices | non-papillate, pointed, often 2 apices | papillate, sometimes bi-papillate |
| special features > | sometimes lateral attachment or 1–2 vacuoles | 4% lateral attachment, 0.8% with 1–2 vacuoles | sometimes lateral attachment | often basal plug, wide base, or rarely vacuoles | usually basal plug | often lateral attachment,hyphal beak, 1–3 vacuoles | usually basal plug, often curved | |
| external proliferation > | common | rare | rare | common, lax or compound sympodium | common | rare | infrequent | common, lax sympodia |
| zoospore release > | directly or through variable persistent vesicle; operculum-like structure not reportedb | through semi-persistent vesicle; operculum-like structure not observed | directly or through elongated persistent vesicle; operculum-like structure not reportedb | directly, no vesicle | through semipersistent globose or elongated vesicle with operculum- like structure | directly or through persistent vesicle; operculum-like structure not reportedb | through semipersistent globose or elongated vesicle with operculum- like structure | through semipersistent vesicle and dehiscence tube; operculum-like structure frequent |
| exitpores > | 10 (9–13) | 10.6 ± 1.5 | 9 (7–15) | n.a. | n.a. | 10 (7–12) | n.a. | n.a. |
| zoospore cysts > | c. 8 | 8.8 ± 1.0 | c. 8 | n.a. | 11.2 (10–12.5) | n.a. | 10.2 (8.7–12.5) | n.a. |
| Breeding system > | sterile | sterile | sterile | homothallic | sterile | sterile | homothallic | homothallic |
| Oogonia > | – | – | – | smooth-walled | – | – | smooth-walled | smooth-walled |
| mean diam > | 28.2±2.6 | 38.6 | 46.3 | |||||
| range of isolate means > | n.a. | n.a. | n.a. | |||||
| total range > | 23.4–35.1 | 25–45 | 32.1–59.7 | |||||
| Oospores > | – | – | – | plerotic | – | – | plerotic, yellow-brown | plerotic |
| mean diam > | 25.2±2.1 | 38.3 | 42.2 | |||||
| Total range > | 21.8–29.3 | 25–45 | 29.7–49.4 | |||||
| wall diam > | n.a. | 3 (2.5–5.0) | 2.5–5.0 | |||||
| oospore wall index > | n.a. | n.a. | n.a. | |||||
| Abortion rate > | n.a. | n.a. | n.a. | |||||
| Antheridia | – | – | – | paragynous, intricate | – | – | paragynous, some amphigynous? | paragynous, 1–3 |
| size > | < 5 | < 5 | (15–25) 20.5 × (8–13) 9 | |||||
| Hyphal swellings | – | – | – | common, limoniform | n.a. | – | n.a. | reniform or tuberous |
| Hyphal aggregations > | n.a. | – | n.a. | n.a. | n.a. | infrequent | n.a. | n.a. |
| Colonies on V8A > | petaloid, scanty aerial mycelium | petaloid-faintly petaloid, limited aerial mycelium | petaloid, submerged | faintly striate, limited aerial, submerged | n.a. | petaloid, scanty aerial mycelium | n.a. | n.a. |
| Colonies on CMA | Coralloid, scanty aerial to submerged | n.a. | petaloid, submerged | n.a. | n.a. | radiate, submerged | n.a. | rosaceous, submerged, limited aerial |
| Colonies on PDA > | no growth | n.a. | no growth | n.a. | n.a. | no growth | n.a. | n.a. |
| Maximum temperature | n.a. | 32.5 | n.a. | 29 | 35 | n.a. | c. 30 | n.a. |
| Optimum temperature | n.a. | 20 | n.a. | 25 | 25 | n.a. | 15 | n.a. |
| Growth rate V8A 25 °C | 2.8 (25 °C) | 4.1 ± 0.5 | 3.8 (25 °C) | 2.8 | n.a. | 4.3 (25 °C) | n.a. | n.a. |
| Growth rate CMA 25 °C > | 0.8 (25 °C) | n.a. | 0.9 (25 °C) | n.a. | n.a. | 2.8 (25 °C) | n.a. | 1–2 |
a Apex in all 7 known Halophytophthora species becoming pseudo-papillate during zoospore differentiation due to the shrinkage of the protoplasm away from the apex.
b Operculum-like structure reported by Jesus et al. (2019) but not mentioned in the original description by Gerrettson-Cornell & Simpson (1984).
– = character not observed; n.a. = not available.
Halophytophthora brevisporangiaT. Jung, C. Maia, G. Carella, M. Horta Jung, sp. nov. — MycoBank MB 838602; Fig. 5
Fig. 5.
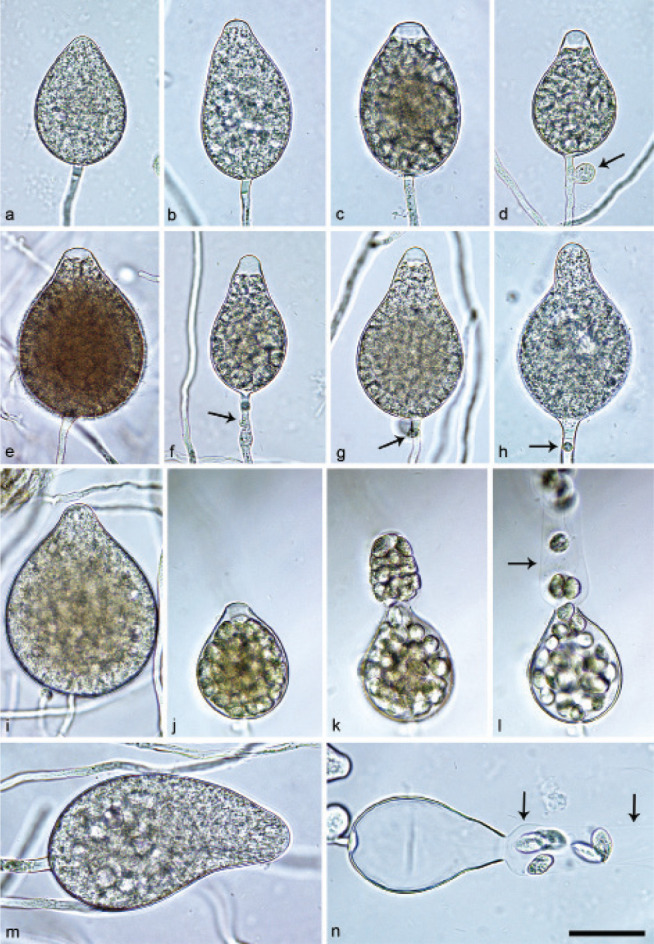
Sporangia of Halophytophthora brevisporangia formed on saltwater V8 agar (sV8A) flooded with nonsterile 50 % seawater. — a–b. Non-papillate, with a conspicuous basal plug; a. ovoid; b. obpyriform; c–g. pseudo-papillate apex due to shrinkage of protoplasm before zoospore release; c. ovoid, d. obpyriform, with external proliferation (arrow); e. broad-ovoid with conspicuous basal plug; f–h. obpyriform, with external proliferation (arrows) and conspicuous basal plug; h. non-papillate; i. broad-ovoid, with non-papillate pointed apex; j–l. ovoid, becoming pseudo-papillate and then releasing zoospores through an elongated semi-persistent vesicle (arrow); m. elongated-obpyriform with non-papillate curved apex and a conspicuous basal plug; n. obpyriform, with a conspicuous basal plug and external proliferation, releasing zoospores through an elongated semi-persistent vesicle (arrows). — Scale bar = 30 μm, applies to a–n.
Etymology. Name refers to the relatively short length of most sporangia.
Typus. PORTUGAL, Parque Natural da Ria Formosa, Quelfes, isolated from a tidal pond in a coastal saltmarsh, T. Jung, 2015 (CBS H-24574 holotype, dried culture on sV8A, Herbarium CBS-KNAW Fungal Biodiversity Centre, CBS 147238 = BD662, ex-type culture). ITS and cox1 sequences GenBank OK033641 and OK091206, respectively.
Sporangia, hyphal swellings and chlamydospores (Fig. 5) — Sporangia were not observed in solid agar but were abundantly produced in a mixture of distilled water and non-sterile seawater (1 : 1). They were produced terminally, mainly on unbranched sporangiophores (Fig. 5a–c, e, i, m) or in lax sympodia resulting from external proliferation (Fig. 5d, f–h, n). Sporangia were non-caducous and non-papillate becoming pseudo-papillate immediately before zoospore release due to shrinkage of proto-plasm near the apex (Fig. 5c–g, j), often with a pointed apex (34 %; Fig. 5f–i). Sporangial shapes were mostly ovoid (86.0 %; Fig. 5a,c, e, i–l) and less frequently obpyriform or elongated obpyriform (11.5 %; Fig. 5c–d, f–h, m), distorted (1.0 %), ellipsoid (1.0 %) and obturbinate (0.5 %). Sporangia with two apices were not observed. A conspicuous basal plug was frequently formed (Fig. 5a–b, d–e, h, m–n) while lateral attachment of the sporangiophore occurred only occasionally. Sporangial dimensions averaged 57.6 ± 10.9 × 42.1 ± 8.8 µm with an overall range of 31.2–101.4 × 20.9–78.8 µm and a range of isolate means of 54.7–59.7 × 39.9–45.9 µm. The l/b ratio was 1.4 ± 0.1. Release of zoospores occurred through an exit pore of 9.8 ± 1.7 µm width into a semi-persistent elongated vesicle (Fig. 5j–l, n). Zoospore cysts measured 7.9 ± 0.7 µm. Hyphal swellings or chlamydospores were not observed.
Oogonia, oospores and antheridia — All four isolates of H. brevisporangia were self-sterile.
Colony morphology, growth rates and cardinal temperatures (Fig. 13, 15) — All isolates formed limited cottony mycelium on sV8A and sCA and dense-felty appressed mycelium on sPDA. Colonies showed faintly radiate and uniform patterns on sV8A and sCA, respectively, and were uniform or petaloid on sPDA (Fig. 13). All isolates tested shared the same optimum and maximum temperatures of 25 and 32.5 °C, respectively (Fig. 15). All isolates were not growing at 35 °C but resumed growth after being re-incubated at 20 °C. Therefore, the lethal temperature is > 35 °C. All isolates were fast growing with radial growth rates on sV8A at 20 and 25 °C of 18.5 ± 1.3 and 20.2 ± 2.1 mm/d, respectively (Fig. 15). On sCA and sPDA radial growth at 20 °C was 16.7 ± 0.5 and 13.3 ± 1.4 mm/d, respectively.
Fig. 13.
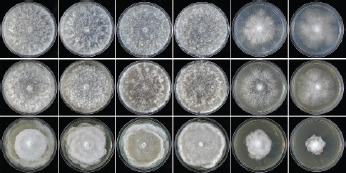
Colony morphology of Halophytophthora brevisporangia (BD662 and BD695), H. celeris (BD646 and BD885) and H. frigida (BD650 and BD655) (from left to right) after 7 d growth at 20 °C on sV8A, sCA and sPDA (from top to bottom).
Fig. 15.
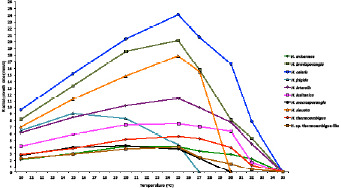
Mean radial growth rates of Halophytophthora avicennae (three isolates), H. brevisporangia (three isolates), H. celeris (three isolates), H. frigida (five isolates), H. lateralis (four isolates), H. lusitanica (five isolates), H. macrosporangia (five isolates), H. sinuata (five isolates), H. thermoambigua (five isolates) and H. sp. thermoambigua-like (two isolates) on sV8A at different temperatures.
Additional specimens examined. PORTUGAL, Sapal de Castro Marim / Rio Guadiana, Castro Marim, isolated from brackish water in the river estuary, G. Carella & M. Horta Jung, 2015; CBS 147239 = BD695; Parque Natural da Ria Formosa, Santa Luzia, isolated from a tidal channel in a coastal saltmarsh, T. Jung, 2015; BD644; Parque Natural da Ria Formosa, Quelfes, isolated from a tidal pond in a coastal saltmarsh, T. Jung, 2015; BD658.
Halophytophthora celerisT. Jung, C. Maia, G. Carella,
M. Horta Jung, sp. nov. — MycoBank MB 838603; Fig. 6
Fig. 6.
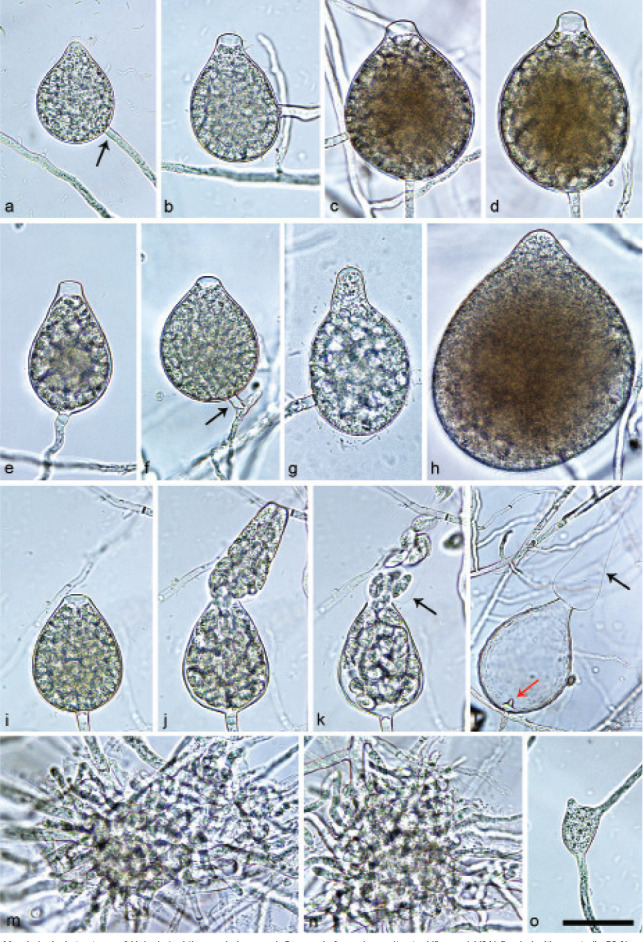
Morphological structures of Halophytophthora celeris. — a–l. Sporangia formed on saltwater V8 agar (sV8A) flooded with nonsterile 50 % seawater, most of them with a conspicuous basal plug (a, d–g, i–k); a. ovoid with non-papillate pointed apex, laterally attached, with conspicuous basal plug (arrow); b–f. pseudo-papillate apex due to shrinkage of protoplasm before zoospore release; b. ovoid, laterally attached; c. broad-ovoid, intercalary; d. broad-ovoid;e. ovoid to obpyriform; f. ovoid, on a short lateral hypha (arrow); g. obpyriform, non-papillate, laterally attached; h. broad-ovoid with non-papillate pointed apex; i–k. ovoid, becoming pseudo-papillate and then releasing zoospores through an elongated semi-persistent vesicle (arrow); l. ovoid, after zoospore release with an elongated semi-persistent vesicle (black arrow) and a conspicuous basal plug protruding into the empty sporangium (red arrow); m–n. hyphal aggregations; o. triangular hyphal swelling. — Scale bar = 30 μm, applies to a–o.
Etymology. Name refers to the fast growth in culture (celeris Latin = fast).
Typus. PORTUGAL, Parque Natural da Ria Formosa, Santa Luzia, isolated from a tidal channel in a coastal saltmarsh, T. Jung, 2015 (CBS H-24575 holotype, dried culture on sV8A, Herbarium CBS-KNAW Fungal Biodiversity Centre, CBS 147240 = BD885, ex-type culture). ITS and cox1 sequences GenBank OK033645 and OK091210, respectively.
Sporangia, hyphal swellings and chlamydospores (Fig. 6) — Sporangia were not observed in solid agar but were readily produced in a mixture of distilled water and non-sterile seawater (1 : 1). Sporangia were non-papillate, often with a pointed apex (20.7 %; Fig. 6a, h), becoming pseudo-papillate immediately before releasing zoospores (Fig. 6b–f, i), non-caducous and formed usually terminally (Fig. 6a–b, d–e, g–l) on unbranched sporangiophores or sometimes intercalary (Fig. 6c) or on short lateral hyphae (Fig. 6f). External proliferation was only rarely observed. Sporangial shapes varied mainly between ovoid to broad-ovoid (60 %; Fig. 6a–d, f, h–l), obpyriform (30.7 %; Fig. 6g) and ovoid to obpyriform (7.3 %; Fig. 6e) whereas subglobose (1.3 %) and limoniform (0.7 %) sporangia were rare. Lateral attachment of the sporangiophore (13.3 %; Fig. 6a–b, f–h) and a conspicuous basal plug (Fig. 6a,e–g,i) sometimes protruding into the empty sporangium (Fig. 6l) were common, small hyphal beaks infrequent. Size of the sporangia averaged 60.5 ± 12.5 × 46.1 ± 10.4 µm with a total range of 29.4–92.8 × 20.8–78.0 µm, a range of isolate means of 56.2–64.6 × 43.9–47.3 µm and a l/b ratio of 1.3 ± 0.1. Zoospores were released through an exit pore with a diameter of 10.5 ± 1.6 µm and a semi-persistent elongated vesicle (Fig. 6j–l). Zoospore cysts averaged 8.8 ± 0.8 µm. Hyphal aggregations (Fig. 6m–n) were common while globose or triangular hyphal swellings (Fig. 6o) were occasionally formed. Chlamydospores were not observed.
Oogonia, oospores and antheridia — All isolates of H. celeris were self-sterile.
Colony morphology, growth rates and cardinal temperatures (Fig. 13, 15) — All isolates formed uniform colonies with sparse cottony mycelium on sV8A and sCA and dense-felty colonies with cottony margins and faintly petaloid to petaloid patterns on sPDA (Fig. 13). All three isolates included in the growth tests had similar optimum, maximum and lethal temperatures of 25, 32.5 and 35 °C, respectively (Fig. 15) and showed fast growth. Radial growth rates at 20 and 25 °C on sV8A were 20.4 ± 0.7 and 24.0 ± 0.4 mm/d, respectively (Fig. 15). Radial growth on sCA and sPDA at 20 °C was 17.0 ± 0.1 and 15.7 ± 0.3 mm/d, respectively.
Additional specimens examined. PORTUGAL, Parque Natural da Ria Formosa, Santa Luzia, isolated from a tidal channel in a coastal saltmarsh, T. Jung, 2015; CBS 147241 = BD646, BD886.
Halophytophthora frigidaT. Jung, C. Maia, G. Carella,
M. Horta Jung, sp. nov. — MycoBank MB 838587; Fig. 7
Fig. 7.
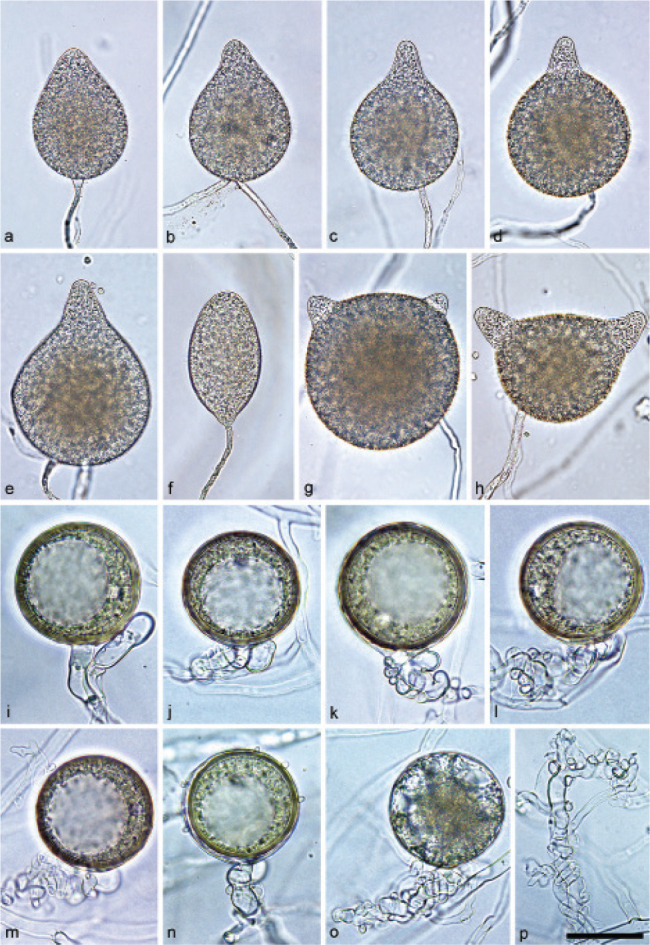
Morphological structures of Halophytophthora frigida. — a–h. Sporangia formed on saltwater V8 agar (sV8A) flooded with nonsterile 50 % seawater; a. ovoid with non-papillate pointed apex; b. obpyriform with non-papillate pointed apex, intercalary inserted; c–h. non-papillate with pointed protruding apices and lateral attachment of the sporangiophores; c–e. broad-obpyriform; f. elongated-obpyriform with curved apex; g. globose with two apices; h. distorted with two apices; i–n. mature globose oogonia formed in single culture in sV8A, containing thick-walled plerotic oospores with large lipid globules, and paragynous antheridia; i–m. smooth-walled; j–m. antheridial stalks entangling the oogonial stalks (intricate); n. ornamented oogonial wall; o. aborted oogonium with intricate antheridial stalk; p. vegetative hyphae entangling each other. — Scale bar = 40 μm in a–h and 30 μm in i–p.
Etymology. Name refers to the low optimum and maximum temperatures for growth.
Typus. PORTUGAL, Parque Natural da Ria Formosa, Santa Luzia, isolated from a tidal pond in a coastal saltmarsh, T. Jung, 2015 (CBS H-24571 holotype, dried culture on sV8A, Herbarium CBS-KNAW Fungal Biodiversity Centre, CBS 147235 = BD655, ex-type culture). ITS and cox1 sequences GenBank OK033652 and OK091217, respectively.
Sporangia, hyphal swellings and chlamydospores (Fig. 7a–h) — Sporangia were infrequently observed in solid agar but were abundantly produced in a mixture of distilled water and non-sterile seawater (1 : 1). They were usually formed terminally on mostly unbranched sporangiophores (Fig. 7a, c–h) or in lax sympodia of 2–3 sporangia although a few intercalary sporangia (Fig. 7b) could be observed in all isolates. Sporangia were non-papillate, usually with a pointed often protuberant apex (Fig. 7b–h), and non-caducous. Sporangial shapes were mainly obpyriform, broad-obpyriform and elongated obpyriform (83.1 %; Fig. 7b–f), ovoid (7.1 %; Fig. 7a) or globose to distorted with two apices (4.7 %; Fig. 7g–h) and, less frequently, obturbinate (1.8 %), limoniform (1.2 %), ellipsoid (0.6 %) or subglobose (0.3 %). Lateral attachment of the sporangiophore was common (15.7 %; Fig. 7d–h) and apices were occasionally curved (3.0 %; Fig. 7f). Sporangial dimensions of seven isolates averaged 80.4 ± 19.3 × 50.1 ± 13.8 µm, with an overall range of 32.4–150.7 × 18.8–98.1 µm and an isolate range of 72.7–85.7 × 44.5–57.1 µm. The l/b ratio was 1.6 ± 0.3. The zoospores, 9.1 ± 2.0 µm after encystment, were released through an exit pore of 9.5 ± 1.6 µm and a semi-persistent vesicle. Chlamydospores were not observed.
Oogonia, oospores and antheridia (Fig. 7i–o) — Gametangia were produced in single-culture in sV8A after 14–20 d. Mature oogonia were globose and mostly smooth-walled (Fig. 7i–m,o), although slightly ornamented oogonia were occasionally observed (Fig. 7n). Oogonial diameter was 47.8 ± 4.0 µm with an overall range of 32.0–58.6 µm and a range of isolate means of 43.0–49.9 µm. Oospores were plerotic (99.3 %) with a big reserve globule (ooplast), averaging 45.3 ± 4.1 µm (overall range 29.1–54.9 µm), with a wall diameter of 2.2 ± 0.5 µm and an oospore wall index of 0.26 ± 0.05. The abortion rate varied between isolates (2–50 %; Fig. 7o) with an average of 22.7 %. Antheridia were exclusively paragynous, measuring 18.3 ± 3.6 × 10.3 ± 2.7 µm, with intricate stalks commonly entangling the oogonial stalks (Fig. 7i–o). Vegetative hyphae were also frequently entangling each other (Fig. 7p).
Colony morphology, growth rates and cardinal temperatures (Fig. 13, 15) — There was variation in the colony morphology of the isolates tested. In one group the colonies on sV8A and sCA had a faintly radiate pattern with submerged margins and denser aerial mycelium in the centre or as a ring around the centre, while in the other group colonies on sV8A and sCA were faintly petaloid with limited aerial mycelium across the colonies. In both groups colonies on sPDA were faintly petaloid with irregular margins and felty to cottony aerial mycelium (Fig. 13). Both groups of colony patterns were present at the same sampling site. The optimum temperature for growth was generally low, 15 °C for four of the five isolates tested and 20 °C for the other isolate, while the maximum temperature was 25 °C for all isolates (Fig. 15). To verify the lethal temperature, all Petri dishes incubated for 1 wk at 27.5, 30, 32.5 and 35 °C were transferred to 20 °C. All Petri dishes from 27.5 and 30 °C resumed growth whereas those from 32.5 and 35 °C failed to grow, confirming 32.5 °C as the lethal temperature. Radial growth rates on sV8A at 15 and 20 °C were 9.0 ± 1.6 mm/d and 8.3 ± 2.3 mm/d, respectively (Fig. 15) whereas radial growth at 20 °C on sCA and sPDA was 9.4 ± 0.8 and 5.7 ± 0.6 mm/d, respectively.
Additional specimens examined. PORTUGAL, Parque Natural da Ria Formosa, Santa Luzia, isolated from a tidal pond in a coastal saltmarsh, T. Jung, 2015; CBS 147236 = BD650, BD647, BD654; Parque Natural da Ria Formosa, Santa Luzia, isolated from a tidal channel in a coastal saltmarsh, T. Jung, 2015; BD641; Ria de Alvor, Alvor, isolated from seawater in a lagoon, T. Jung & C. Maia, 2015; BD675, BD676.
Halophytophthora lateralisT. Jung, C. Maia, G. Carella, M. Horta Jung, sp. nov. — MycoBank MB 838586; Fig. 8
Fig. 8.
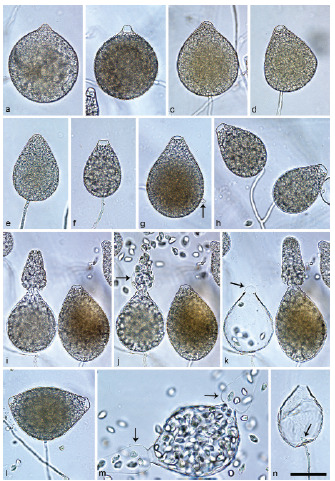
Sporangia of Halophytophthora lateralis formed on saltwater V8 agar (sV8A) flooded with nonsterile 50% seawater. — a–b. Subglobose, laterally attached; a. non-papillate pointed apex; b. pseudo-papillate apex due to shrinkage of protoplasm before zoospore release; c. broad-ovoid, shallow semi-papillate; d. broad-ovoid, with pseudo-papillate apex before zoospore release, laterally attached; e–f. ovoid; e. non-papillate, laterally attached, with conspicuous basal plug; f. with pseudo-papillate apex before zoospore release; g. ovoid to obpyriform, shallow semi-papillate, laterally attached, with hyphal projection (arrow);h. lax sympodium of two ovoid, laterally attached sporangia with pseudo-papillate apices before zoospore release; i–k. ovoid to obpyriform sporangia releasing zoospores through elongated semi-persistent vesicles (arrows); k. shrinking vesicle (arrow) closing exit pore; l. limoniform with two pseudo-papillate apices before zoospore release; m. limoniform with two apices, releasing zoospores through elongated semi-persistent vesicles (arrows); n. ovoid with a conspicuous basal plug (arrow) protruding into the empty sporangium after zoospore release. — Scale bar = 40 μm, applies to a–n.
Etymology. Name refers to the frequent lateral insertion of the sporangiophores to the sporangia.
Typus. PORTUGAL, Parque Natural da Ria Formosa, Quelfes, isolated from a tidal pond in a coastal saltmarsh, T. Jung, 2015 (CBS H-24570 holotype, dried culture on sV8A, Herbarium CBS-KNAW Fungal Biodiversity Centre, CBS 147233 = BD657, ex-type culture). ITS and cox1 sequences GenBank OK033655 and OK091220, respectively.
Sporangia, hyphal swellings and chlamydospores (Fig. 8) — Sporangia were not observed in solid agar but were abundantly produced in a mixture of distilled water and nonsterile seawater (1 : 1). Sporangia formed predominantly on unbranched sporangiophores or occasionally in lax sympodia (Fig. 8h) and were mostly ovoid (95.0 %, Fig. 8c–f, h–k, n) or rarely subglobose (2.0 %; Fig. 8a–b), limoniform or distorted with two apices (2.0 %; Fig. 8l–m) and ovoid to obpyriform (1.0 %; Fig. 8g). Sporangial apices were non-papillate (Fig. 8a, e) or shallow semipapillate (Fig. 8c, g), sometimes pointed (Fig. 8a), becoming pseudo-papillate due to the shrinkage of the protoplasm near the apex during zoospore differentiation (Fig. 8b, d, f, h– j, l). Lateral attachment of the sporangiophore was common (41.5 %, Fig. 8a – b, d, g–h) and short hyphal projections (Fig. 8g) and a conspicuous basal plug, protruding into the sporangium (Fig. 8n), were occasionally formed. Sporangial dimensions of four isolates averaged 75.9 ± 16.1 × 52.4 ± 12.5 µm, with an overall range of 41.6–122.0 × 22.8–100.3 µm, a range of isolate means of 67.7–89.5 × 45.4–63.6 µm and a l/b ratio of 1.5 ± 0.2. Limoniform to reniform zoospores were released through a wide exit pore (12.6 ± 2.4 µm) into a semi-persistent elongated vesicle (Fig. 8i– k, m) which after zoospore release shrinks relatively fast sometimes closing the empty sporangium again (Fig. 8k). In sporangia with two apices zoospores are often released through both apices via exit pores and vesicles (Fig. 8m). Zoospore cysts averaged 7.9 ± 0.7 µm. Hyphal swellings & chlamydospores were not observed.
Oogonia, oospores and antheridia — All isolates of H. lateralis were self-sterile.
Colony morphology, growth rates and cardinal temperatures (Fig. 14, 15) — Colonies on sV8A and sCA were stellate and faintly petaloid, respectively, with limited aerial mycelium whereas colonies on sPDA were dense-felty and uniform (Fig. 14). All four isolates included in the temperature tests had the same optimum and maximum growth temperature of 25 °C and 32.5 °C, respectively. The isolates did not grow at 35 °C and did not resume growth when transferred to 20 °C. Radial growth on sV8A at 20 and 25 °C was 10.3 ± 0.7 and 11.4 ± 0.9 mm/d, respectively (Fig. 15). On sCA and sPDA radial growth rates at 20 °C were, 12.0 ± 0.4 and 9.8 ± 0.3 mm/d, respectively.
Fig. 14.
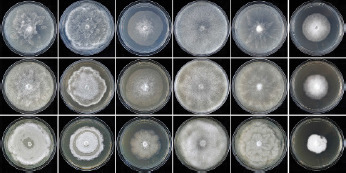
Colony morphology of Halophytophthora lateralis (BD680), H. lusitanica (BD628), H. macrosporangia (BD642), H. sinuata (isolate BD656), H. thermoambigua (isolate BD637) and H. sp. thermoambigua-like (isolate BD652) (from left to right) after 7 d growth at 20 °C on sV8A, sCA and sPDA (from top to bottom).
Additional specimens examined. PORTUGAL, Parque Natural da Ria Formosa, Almancil, isolated from a tidal pond in a coastal saltmarsh, T. Jung, 2015; CBS 147234 = BD680; Parque Natural da Ria Formosa, Quelfes, isolated from a tidal pond in a coastal saltmarsh, T. Jung, 2015; BD660, BD665.
Halophytophthora lusitanicaT. Jung, C. Maia, G. Carella, M. Horta Jung, sp. nov. — MycoBank MB 838585; Fig. 9
Fig. 9.
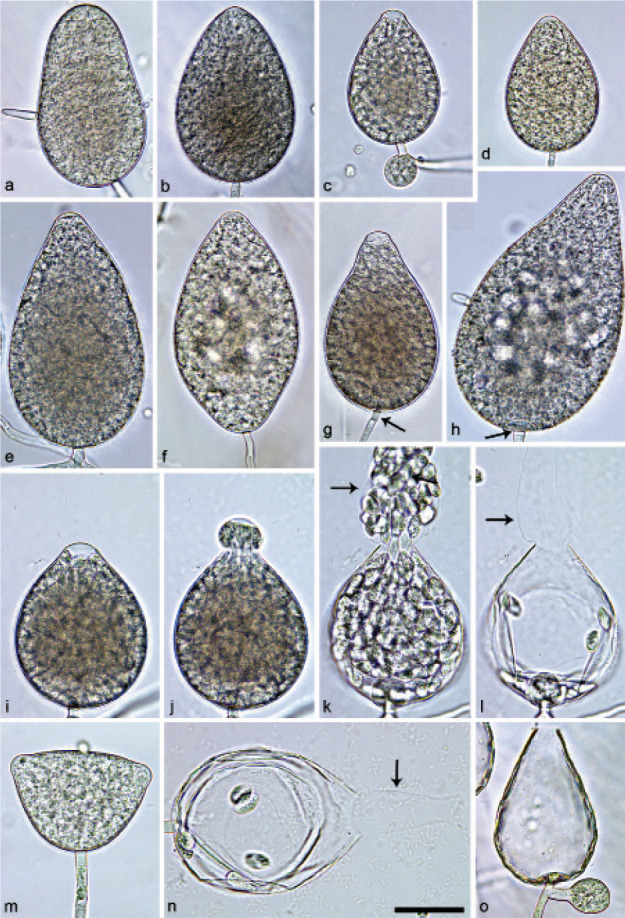
Sporangia of Halophytophthora lusitanica formed on saltwater V8 agar (sV8A) flooded with nonsterile 50 % seawater. — a. Ellipsoid, non-papillate, laterally attached, with short hyphal projection; b. ovoid, non-papillate; c. ovoid, with pseudo-papillate apex due to shrinkage of protoplasm before zoospore release, and globose swelling close to sporangial base; d. ovoid with pointed apex; e. elongated ovoid, on a short lateral hypha with non-papillate to shallow semi-papillate apex; f. non-papillate, limoniform; g. obpyriform, with pseudo-papillate apex before zoospore release, and conspicuous basal plug (arrow);h. elongated obpyriform, non-papillate, laterally attached, with short hyphal projection; i–l. broad-ovoid, becoming pseudo-papillate and then releasing zoospores through an elongated semi-persistent vesicle (arrows); m. distorted with two non-papillate apices; n. broad-ovoid after release of most zoospores through an elongated semi-persistent vesicle (arrow); o. ovoid to obpyriform, after zoospore release, with conspicuous basal plug and external proliferation. — Scale bar = 30 μm, applies to a–o.
Etymology. Name refers to the origin of the first isolates in Portugal (lusitanica Latin = from Lusitania which is the old Roman name for Portugal).
Typus. PORTUGAL, Parque Natural da Ria Formosa, Almancil, isolated from a tidal pond in a coastal saltmarsh, T. Jung, 2015 (CBS H-24569 holotype, dried culture on sV8A, Herbarium CBS-KNAW Fungal Biodiversity Centre, CBS 147231 = BD686, ex-type culture). ITS and cox1 sequences GenBank OK033663 and OK091228, respectively.
Sporangia, hyphal swellings and chlamydospores (Fig. 9) — Sporangia were not observed in solid agar but were abundantly produced in a mixture of distilled water and nonsterile seawater (1 : 1). Sporangial apices were mostly non-papillate or rarely shallow semipapillate (Fig. 9e) becoming pseudo-papillate due to the shrinkage of the protoplasm near the apex during zoospore differentiation (Fig. 9c, g, i). Sporangia were non-caducous and formed mostly terminally on unbranched sporangiophores or occasionally on short lateral hyphae (Fig. 9e). External proliferation close to the sporangial base was only rarely observed (Fig. 9o). Special features like lateral attachment of the sporangiophore (33.3 % of sporangia; Fig. 9a, h), short hyphal projections (Fig. 9e, h), a conspicuous basal plug (Fig. 9g–h, o) and a curved apex occurred in all isolates. Sporangial shapes were variable, ranging from obpyriform (54.0 %; Fig. 9g–h), ovoid to obpyriform (12.0 %; Fig. 9o) and ovoid, broad-ovoid or elongated ovoid (27.3 %, Fig. 9b–e, i–l, n) to ellipsoid (3.0 %; Fig. 9a), limoniform (1.0 %; Fig. 9f), obturbinate (1.0 %), ampulliform (0.7 %) and distorted shapes with two apices (1.0 %; Fig. 9m). Sporangia of six isolates averaged 84.0 ± 15.8 × 54.3 ± 10.6 µm, with an overall range of 40.5–162.5 × 25.4–86.3 µm, and a range of isolate means of 76.2–95.4 × 47.1–60.2 µm. The l/b ratio was 1.6 ± 0.3. Limoniform to reniform zoospores were released through an exit pore of 10.6 ± ٢.1 µm width into a semi-persistent elongated vesicle (Fig. 9i–l, n); zoospores measuring 8.2 ± 0.9 µm after encystment. Hyphal swellings were sometimes formed close to the base of the sporangium (Fig. 9c). Chlamydospores were not observed.
Oogonia, oospores and antheridia — All six isolates of H. lusi-tanica were self-sterile.
Colony morphology, growth rates and cardinal temperatures (Fig. 14, 15) — All isolates formed petaloid to faintly petaloid colonies with limited aerial mycelium on sV8A and sCA and uniform felty colonies with an appressed ring of collapsed aerial mycelium on sPDA (Fig. 14). The optimum temperature varied between the isolates. Three of the five tested isolates had an optimum temperature on sV8A of 25 °C, while the remaining two isolates had their optimum at 20 and 27.5 °C, respectively, averaging at 25 °C. The maximum growth temperature of all isolates was 32.5 °C. No growth occurred after transferring isolates that were incubated for 1 wk at 35 °C to 20 °C. Radial growth rates on sV8A at 20 and 25 °C were 7.3 ± 0.6 and 7.5 ± 0.8 mm/d, respectively (Fig. 15). Radial growth on sCA and sPDA at 20 °C was 6.9 ± 0.8 and 6.7 ± 0.4 mm/d, respectively.
Additional specimens examined. PORTUGAL, Rio Séqua, Tavira, isolated from brackish river water in the tidal zone near the estuary, T. Jung, 2015; CBS 147232 = BD628; BD632, BD634; Parque Natural da Ria Formosa, Almancil, isolated from a tidal pond in a coastal saltmarsh, T. Jung, 2015; BD679, BD681.
Halophytophthora macrosporangiaT. Jung, C. Maia, G. Carella, M. Horta Jung, sp. nov. — MycoBank MB 838597; Fig. 10
Fig. 10.
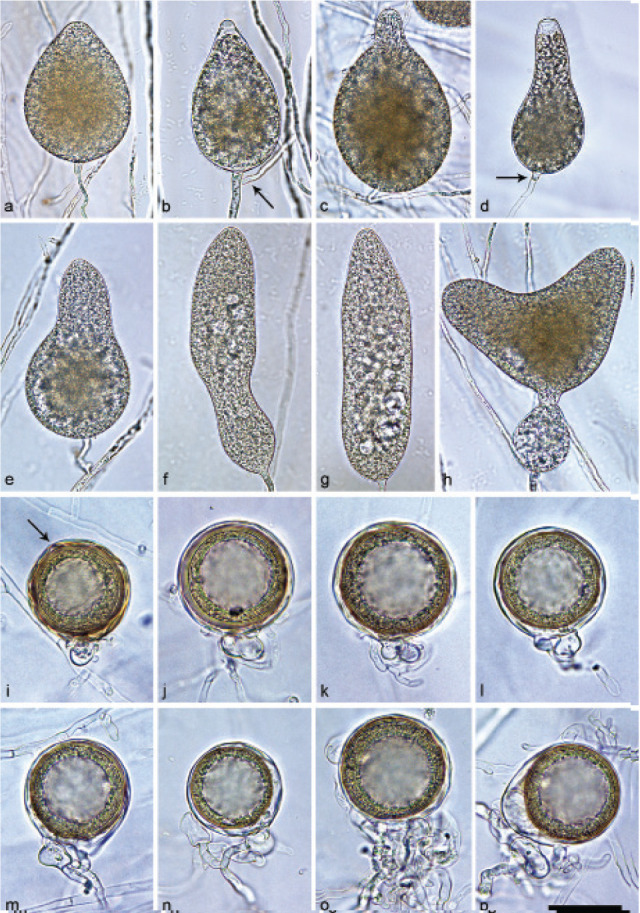
Morphological structures of Halophytophthora macrosporangia. — a–h. Sporangia formed on saltwater V8 agar (sV8A) flooded with nonsterile 50 % seawater; a. ovoid, non-papillate; b. ovoid to obpyriform, with pseudo-papillate apex due to shrinkage of protoplasm before zoospore release, and external proliferation (arrow); c–e. elongated-obpyriform; c. non-papillate pointed apex, laterally attached; d. with pseudo-papillate apex before zoospore release, and conspicuous basal plug (arrow); e. non-papillate; f. elongated peanut-shaped to ampulliform, non-papillate, with small lipid globules; g. elongated-ellipsoid, non-papillate, with small lipid globules; h. distorted with two non-papillate apices and a subglobose swelling close to the sporangial base; i–p. mature, globose golden-brown oogonia formed in single culture in sV8A with mostly thick-walled (a–m, o–p) or rarely thin-walled (n), plerotic to nearly plerotic oospores containing large lipid globules, and paragynous antheridia; i. globose with slightly ornamented wall (arrow); j–o. globose, smooth-walled; o. antheridial stalk entangling the oogonial stalk; p. excentric, smooth-walled. — Scale bar = 40 μm in a–h and 30 μm in i–p.
Etymology. Name refers to the large size of the sporangia.
Typus. PORTUGAL, Parque Natural da Ria Formosa, Santa Luzia, isolated from a tidal channel in a coastal saltmarsh, T. Jung, 2015 (CBS H-24573 holotype, dried culture on sV8A, Herbarium CBS-KNAW Fungal Biodiversity Centre, CBS 147290 = BD639, ex-type culture). ITS and cox1 sequences GenBank OK033664 and OK091229, respectively.
Sporangia, hyphal swellings and chlamydospores (Fig. 10a–h) — Sporangia were not observed in solid agar but were slowly produced in a mixture of distilled water and non-sterile seawater (1 : 1). Sporangia were formed terminally, mostly on unbranched sporangiophores (Fig. 10a–d, f–h), or less frequently on short lateral hyphae (Fig. 10e). External proliferation occurred infrequently (Fig. 10b). Sporangia were non-papillate, sometimes with a pointed apex (Fig. 10b–c), becoming pseudo-papillate during zoospore differentiation due to the shrinkage of the protoplasm near the apex (Fig. 10b, d), and non-caducous. Sporangial shapes were diverse ranging from obpyriform or elongated obpyriform (55.7 %; Fig. 10c–e), ovoid (29.3 %; Fig. 10a) and ovoid to obpyriform (6.0 %; Fig. 10b) to subglobose (3.7 %) and much less frequently distorted with often two apices (2.0 %; Fig. 10h), peanut-shaped to ampulliform (2.7 %; Fig. 10f), elongated-ellipsoid (0.3 %; Fig. 10g) and obturbinate (0.3 %). Special features like a slightly laterally displaced attachment of the sporangiophore (16 %; Fig. 10c), small vacuoles (Fig. 10g), a conspicuous basal plug (Fig. 10d) or a swelling close to the sporangial base (Fig. 10h) were observed in all isolates. Sporangial dimensions averaged 97.5 ± 24.8 × 55.0 ± 14.8 µm, with an overall range of 58.0–186.7 × 32.3–106.3 µm, a range of isolate means of 76.8–118.9 × 42.9–67.4 µm and a l/b ratio of 1.8 ± 0.4. Zoospores were released through an exit pore with a mean diameter of 10.3 ± 1.7 µm and a semipersistent vesicle. Zoospore cysts measured 8.2 ± 2.1 µm. Chlamydospores were not produced.
Oogonia, oospores and antheridia (Fig. 10i–p) — Gametangia were produced in single-culture in sV8A after 14–20 d. Oogonia were mostly globose (Fig. 10i–o) and less frequently, excentric (Fig. 10p), mostly with smooth, sometimes slightly wrinkled walls (Fig. 10j–p) or rarely slightly ornamented (Fig. 10i). Oogonia had an average diameter of 48.2 ± 4.6 µm, with an overall range of 18.4–58.7 µm and an isolate range of 46.7–49.2 µm. Oospores were plerotic or nearly plerotic with a large ooplast and an average diameter of 43.7 ± 4.2 µm, an overall range of 30.1–54.5 µm, a wall diameter of 1.7 ± 0.4 µm and an oospore wall index of 0.22 ± 0.04. The abortion rate varied between isolates from 14 to 82 % averaging 41.5 %. Antheridia were exclusively paragynous with an average size of 15.2 ± 3.1 × 7.8 ± 1.7 µm. Antheridial stalks entangled the oogonial stalks occasionally (Fig. 10o).
Colony morphology, growth rates and cardinal temperatures (Fig. 14, 15) — All isolates produced colonies with wide submerged margins and limited aerial mycelium in the centre on sV8A and sCA, faintly stellate on sV8A and uniform on sCA whereas colonies on sPDA were stoloniferous to petaloid and appressed with submerged margins (Fig. 14). Regarding optimum temperature for growth, the five isolates tested showed considerable variation. The optimum temperature was 15 °C for each one isolate from Santa Luzia and Quelfes, 20 °C for another isolate from Quelfes and 25 °C for two other isolates from Santa Luzia. While the maximum growth temperature was 27.5 °C in all isolates there was also variation in the lethal temperature, which was 32.5 °C in two isolates and 35 °C in three isolates. Radial growth rates on sV8A at 20 and 25 °C were 4.2 ± 1.1 and 3.7 ± 0.3 mm/d (Fig. 15). Radial growth on sCA and sPDA at 20 °C was 5.1 ± 1.4 and 4.1 ± 1.1 mm/d.
Additional specimens examined. PORTUGAL, Parque Natural da Ria Formosa, Santa Luzia, isolated from a tidal channel in a coastal saltmarsh, T. Jung, 2015; CBS 147291 = BD642, BD643, BD645; Parque Natural da Ria Formosa, Santa Luzia, isolated from a tidal pond in a coastal saltmarsh, T. Jung, 2015; BD649; Parque Natural da Ria Formosa, Quelfes, isolated from a tidal pond in a coastal saltmarsh, T. Jung, 2015; BD659, BD664.
Halophytophthora sinuataT. Jung, C. Maia, G. Carella, M. Horta Jung, sp. nov. — MycoBank MB 838590; Fig. 11
Fig. 11.
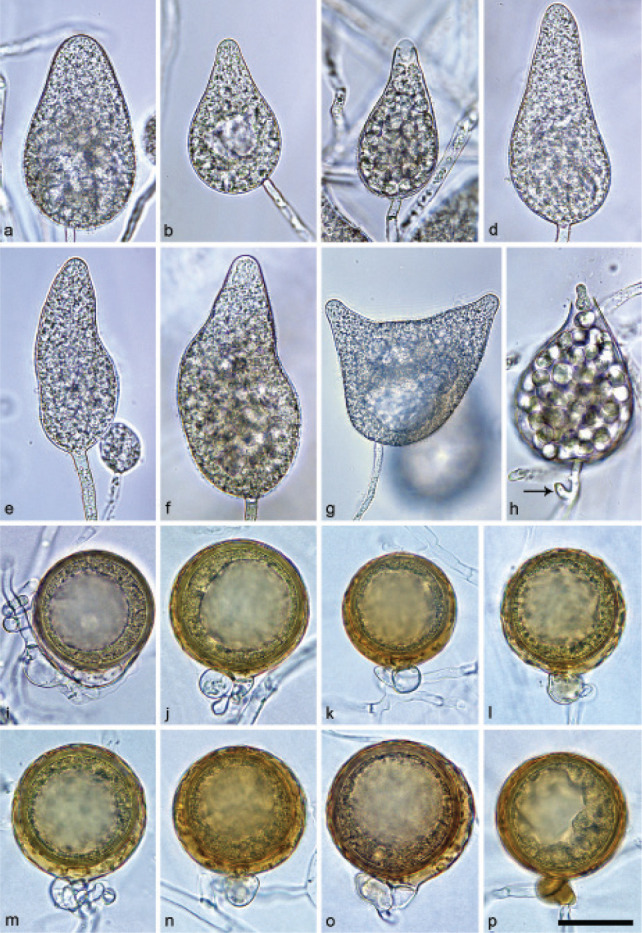
Morphological structures of Halophytophthora sinuata. — a–h. Sporangia formed on saltwater V8 agar (sV8A) flooded with nonsterile 50 % seawater; a. elongated-ovoid, non-papillate; b. obpyriform, with non-papillate pointed apex and lipid globule, laterally attached; c. obpyriform, with pseudo-papillate apex due to shrinkage of protoplasm before zoospore release; d–f. elongated-obpyriform, slightly excentric, non-papillate with pointed apex; d, e. with conspicuous basal plug; g. distorted with two pointed non-papillate apices and lipid globules; h. ovoid to obpyriform, releasing zoospores directly without vesicle, with beginning external proliferation (arrow); i–p. mature, globose golden-brown oogonia formed in single culture in sV8A containing thick-walled, plerotic to nearly plerotic oospores with large lipid globules, and paragynous antheridia; i–j. smooth-walled; k–p. with sinuate-wavy to wrinkled walls; p. with golden-brown antheridium. — Scale bar = 30 μm, applies to a–p.
Etymology. Name refers to the sinuate-waved oogonial walls.
Typus. PORTUGAL, Parque Natural da Ria Formosa, Santa Luzia, isolated from a tidal pond in a coastal saltmarsh, T. Jung, 2015 (CBS H-24572 holotype, dried culture on sV8A, Herbarium CBS-KNAW Fungal Biodiversity Centre, CBS 147237 = BD656, ex-type culture). ITS and cox1 sequences GenBank OK033671 and OK091236, respectively.
Sporangia, hyphal swellings and chlamydospores (Fig. 11a–h) — Sporangia were not observed in solid agar but were slowly produced in a mixture of distilled water and non-sterile seawater (1 : 1). They were formed terminally, almost exclusively on unbranched sporangiophores (Fig. 11a–h). External sporangial proliferation occurred very rarely (Fig. 11h). Sporangia had non-papillate apices which were often pointed (Fig. 11b–g), becoming pseudo-papillate due to the shrinkage of the protoplasm near the apex during zoospore differentiation (Fig. 11c), and were non-caducous with sporangial shapes ranging from mostly obpyriform or elongated and often asymmetrically obpyriform (84.7 %; Fig. 11b – f) to elongated ovoid (6.3 %; Fig. 11a), distorted with two apices (5.0 %; Fig. 11g), ovoid to obpyriform (3.3 %; Fig. 11h), limoniform (0.3 %), ampulliform (0.3 %) and obturbinate (0.3 %). Lateral attachment of the sporangiophore occurred in 31.3 % of the sporangia (Fig. 11b). One or more vacuoles were frequently observed (Fig. 11b, g). Sporangia averaged 74.1 ± 19.5 × 44.0 ± 12.6 µm, with an overall range of 28.1–129.2 × 19.8–89.5 µm and a range of isolate means of 56.9–82.1 × 32.5–51.4 µm. The l/b ratio was 1.7 ± 0.3. The zoospores were released through a narrow exit pore of 8.9 ± 1.5 µm width, either directly (Fig. 11h) or through a semipersistent vesicle. After encystment, zoospores had an average diameter of 7.5 ± 1.1 µm. Hyphal swellings & chlamydospores were not observed.
Oogonia, oospores and antheridia (Fig. 11i–p) — Gametangia were abundantly produced in sV8A after 14–20 d. Mature oogonia were globose with usually sinuate wavy to wrinkled (Fig. 11j–p) or, more rarely, smooth walls (Fig. 11i) and a golden-brown colour. Size of the oogonia averaged 54.2 ± 3.8 µm with an overall range of 39.4–64.3 µm and a range of isolate means of 51.6–55.6 µm. Oospores were mostly plerotic or nearly plerotic (95 %; Fig. 11i–p), with a big lipid globule, a diameter of 47.5 ± 3.7 µm (overall range 33.9–57.6 µm), a wall diameter of 2.9 ± 0.6 µm and an oospore wall index of 0.32 ± 0.05. With 4.2 % (isolate range 0–13 %) abortion rate was low. Antheridia were exclusively paragynous (Fig. 11i–p), sometimes golden-brown (Fig. 11p), averaging 14.9 ± 2.8 × 11.6 ± 1.8 µm. Antheridial stalks almost never entangled the oogonial stalks (Fig. 11i–p).
Colony morphology, growth rates and cardinal temperatures (Fig. 14, 15) — All isolates of H. sinuata had similar patterns on all three agar media. Colonies had limited aerial mycelium on sV8A and sCA and were cottony on sPDA. Colony patterns were uniform on sV8A, uniform to faintly radiate on sCA and uniform to faintly petaloid on sPDA (Fig. 14). All five isolates tested had the same optimum, maximum, and lethal temperatures, 25 °C, 27.5 °C and 32.5 °C, respectively. On sV8A radial growth at 20 and 25 °C was 14.9 ± 0.3 and 17.9 ± 0.6 mm/d (Fig. 15). Radial growth rates, on sCA and sPDA at 20 °C were 12.8 ± 0.1 and 13.3 ± 0.4 mm/d, respectively.
Additional specimens examined. PORTUGAL, Parque Natural da Ria Formosa, Santa Luzia, isolated from a tidal channel in a coastal saltmarsh, T. Jung, 2015; CBS 147292 = BD943, BD640, BD941, BD942, BD944.
Halophytophthora thermoambiguaT. Jung, C. Maia, G. Carella, M. Horta Jung, sp. nov. — MycoBank MB 838582; Fig. 12
Fig. 12.
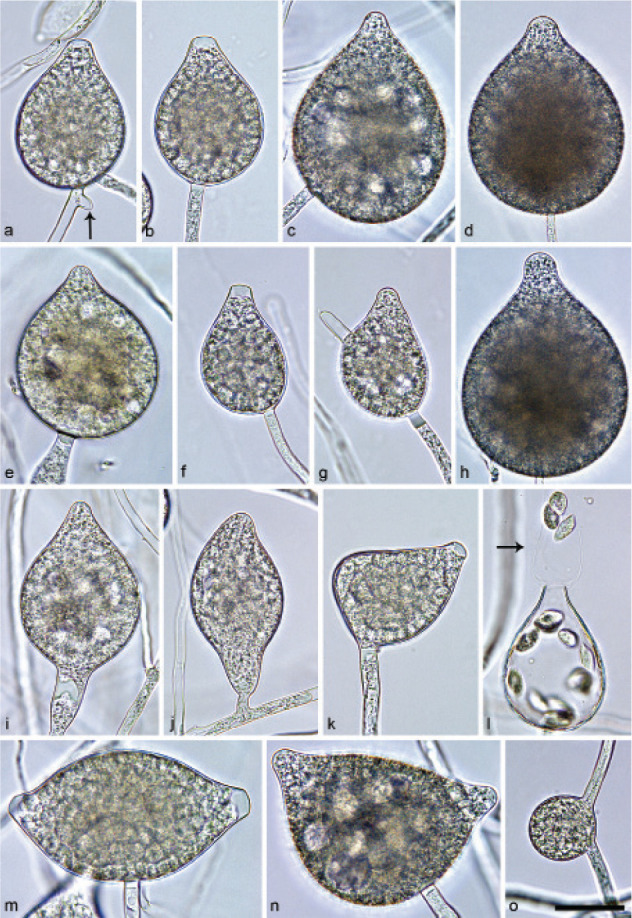
Morphological structures of Halophytophthora thermoambigua formed on saltwater V8 agar (sV8A) flooded with nonsterile 50 % seawater. — a–n. Sporangia; a–b. ovoid, pseudo-papillate apex due to shrinkage of protoplasm before zoospore release; a. intercalary with beginning external proliferation (arrow); b. conspicuous basal plug; c–e. broad-ovoid with non-papillate pointed apex; c. laterally attached; e. laterally attached with conspicuous basal plug; f. obpyriform, pseudo-papillate apex before zoospore release, laterally attached; g. non-papillate pointed with hyphal projection, laterally attached; h. broad-obpyriform with non-papillate pointed apex; i. ovoid to obpyriform with non-papillate pointed apex, widening sporangiophore and conspicuous basal plug; j. limoniform with non-papillate curved apex, on short lateral hypha; k. distorted, laterally attached with conspicuous basal plug and pseudo-papillate apex before zoospore release; l. ovoid to obpyriform, releasing zoospores through elongated semi-persistent vesicle (arrow); m. limoniform with two pseudo-papillate apices before zoospore release; n. limoniform with two non-papillate pointed apices; o. intercalary globose hyphal swelling. — Scale bar = 30 μm, applies to a–o.
Etymology. Name refers to the limited effect of temperature on growth rates and the variability of optimum temperatures between isolates.
Typus. PORTUGAL, Parque Natural da Ria Formosa, Santa Luzia, isolated from a tidal pond in a coastal saltmarsh, T. Jung, 2015 (CBS H-24568, holotype, dried culture on sV8A, Herbarium CBS-KNAW Fungal Biodiversity Centre, CBS 147229 = BD651, ex-type culture). ITS and cox1 sequences GenBank OK033680 and OK091244, respectively.
Sporangia, hyphal swellings and chlamydospores (Fig. 12) — Sporangia were infrequently observed on solid agar but were abundantly produced in a mixture of distilled water and nonsterile seawater (1 : 1). Sporangial apices were non-papillate, usually pointed and often protuberant (Fig. 12c–e, g–j, n), becoming pseudo-papillate due to the shrinkage of the protoplasm near the apex during zoospore differentiation (Fig. 12a–b, f, k, m); frequently two or rarely three apices (Fig. 12m–n). Sporangia were non-caducous and formed mostly terminally on unbranched sporangiophores (Fig. 12b–i, k, m –n) and less frequently intercalary (Fig. 12a) or on short lateral hypha (Fig. 12j). External proliferation was only infrequently observed (Fig. 12a). Lateral attachment of the sporangiophore (38.4 %, Fig. 12c, e, c, f– g, k, n) and a conspicuous basal plug were common (Fig. 12b, e, g, i, k, m–n) while other special features such as short hyphal projections (Fig. 12g), widening of the sporangiophore (Fig. 12i) and a curved apex (Fig. 12f, j – k) occurred occasionally. Shapes were mostly obpyriform (50.4 %; Fig. 12f – h), ovoid to broad-ovoid (33.1 %; Fig. 12a, d), ovoid to obpyriform (7.3 %; Fig. 12b– c, e, i, l) and less frequently limoniform to asymmetrically limoniform (Fig. 12j, m–n), ellipsoid, ampulliform and distorted (Fig. 12k).Sporangial dimensions of nine isolates of H. thermoambigua averaged 75.3 ± 15.7 × 48.1 ± 9.7 µm with an overall range of 43.4–185.3 × 25.3–83.9 µm and a range of isolate means of 69.2–83.2 × 40.3–57.1 µm and a length/breadth (l/b) ratio of 1.6 ± 0.3. Zoospores were differentiated inside the sporangium and released through an exit pore of 10.3 ± 2.3 µm width and a semi-persistent apically rupturing vesicle (Fig. 12l). Zoospore cysts averaging 8.6 ± 0.9 µm. Hyphal swellings (33.8 ± 9.5 µm) were infrequently produced (Fig. 12o). Chlamydospores were not observed.
Oogonia, oospores and antheridia — All nine isolates of H. thermoambigua were self-sterile.
Colony morphology, growth rates and cardinal temperatures (Fig. 14, 15) — All isolates produced stellate to radiate colonies with limited aerial mycelium on sV8A and sCA, respectively, and felty petaloid colonies on sPDA (Fig. 14). Halophytophthora thermoambigua showed a flat temperature-growth curve (Fig. 15). The five isolates had optimum growth temperatures of 20, 25 and 27.5 °C, respectively. The maximum growth temperature was 32.5 °C in all isolates. None of the isolates grew at 35 °C, and they did not resume growth after being re-incubated at 20 °C. Radial growth on sV8A at 20 and 25 °C was 5.2 ± 1.2 and 5.6 ± 1.0 mm/d, respectively (Fig. 15). Radial growth rates on sCA and sPDA at 20 °C were 5.2 ± 1.4 and 5.0 ± 2.1 mm/d, respectively.
Additional specimens examined. PORTUGAL, Rio Séqua, Tavira, isolated from brackish river water in the tidal zone near the estuary, T. Jung, 2015; CBS 147230 = BD637; BD631; Parque Natural da Ria Formosa, Santa Luzia, isolated from a tidal pond in a coastal saltmarsh, T. Jung, 2015; BD652, BD653; Ribeira de Odelouca, Silves, isolated from brackish river water in the tidal zone near the estuary, T. Jung, 2014; BD91, BD93; Ria de Alvor, Alvor, isolated from seawater in a lagoon, T. Jung & C. Maia, 2015; BD668, BD673, BD674; Sapal de Castro Marim / Rio Guadiana, Castro Marim, isolated from brackish water in the river estuary, G. Carella & M. Horta Jung, 2015; BD698.
Notes — Across the 8 759 character alignment of LSU, ITS, Btub, hsp90, rpl10, tigA, cox1, nadh1 and rps10 the Clade 6 species H. brevisporangia, H. celeris, H. frigida, H. lateralis, H. lusitanica, H. macrosporangia, H. sinuata, H. thermoambigua and H. sp. thermoambigua-like differ from each other and from the other Clade 6 species H. polymorphica at 82–501 positions and contain 55–61, 32–36, 50–51, 68–69, 53–71, 76–82, 52–63, 23–70 and 29–32 unique polymorphisms, respectively. Halophytophthora lateralis differs in ITS from the ex-type isolate of H. vesicula at 39 positions. In addition, unique combinations of morphological and physiological characters allow to distinguish the eight new species from each other and from the seven known Halophytophthora species. The most discriminating features are highlighted in bold in Table 3 and 4. For the known Halophytophthora species morphological and physiological information were taken from the original descriptions (Anastasiou & Churchland 1969, Gerrettson-Cornell & Simpson 1984, Yang & Hong 2014, Jesus et al. 2019). In addition, seven isolates of H. avicennae from Portugal were examined in this study. Having a homothallic breeding system H. frigida, H. macrosporangia and H. sinuata can easily be separated from the sterile H. avicennae, H. batemanensis, H. brevisporangia, H. celeris, H. insularis, H. lateralis, H. lusitanica, H. polymorphica and H. thermoambigua. Halophytophthora sinuata differs from the other five homothallic species, H. fluviatilis, H. frigida, H. macrosporangia, H. souzae and H. vesicula, by having on average the largest oogonia with mostly sinuate-wavy oogonial walls and a high proportion of asymmetric sporangia. Furthermore, it shows over the whole range between 10 and 27 °C much faster growth than H. frigida and H. macrosporangia (Fig. 14). Halophytophthora frigida is unique in forming both smooth-walled and slightly ornamented oogonia and mostly intricate antheridial stalks. It also differs from all other homothallic Halophytophthora species except of H. souzae by its low optimum temperature of 15 to 20 °C and from H. souzae by producing markedly larger oogonia. Halophytophthora macrosporangia is separated from all other homothallic Halophytophthora species except of H. souzae by its large sporangia and from the latter species by having on average larger oogonia and a lower maximum temperature for growth. Halophytophthora frigida, H. macrosporangia and H. sinuata are also distinguished from each other by having different colony morphologies on all three agar media tested (Fig. 12, 13). The sterile H. brevisporangia and H. celeris differ from the other new sterile species from Portugal, H. lateralis, H. lusitanica and H. thermoambigua, by having on average smaller sporangia with lower l/b ratios, different colony morphologies on all three agar media and faster growth between 10 and 27 °C. (Fig. 12 – 14). Halophytophthora celeris can be distinguished from H. brevisporangia by its faster growth between 10 and 32.5 °C and by forming hyphal aggregations (Fig. 14). Halophytophthora lateralis, H. lusitanica and H. thermoambigua have different colony morphologies on all three agar media (Fig. 12). Furthermore, unlike H. lateralis both H. thermoambigua and H. lusitanica have no clear optimum temperature for growth (Fig. 14). The latter two species can be separated by the slower growth of H. thermoambigua between 10 and 32.5 °C, and the high proportion of laterally attached sporangia and the predominance of ovoid sporangia in H. lateralis (Fig. 14). Finally, H. lateralis differs from the homothallic H. vesicula by being sterile.
Colony morphologies, growth rates, morphological characters and morphometric data from different studies can only be compared with caution due to differences in the preparation of agar media and the different methods of producing and measuring morphological structures. For instance, the l/b ratio of the sporangia from seven H. avicennae isolates from Portugal in the present study was 1.5 compared to 2.6 in the original description from Australia (Gerrettson-Cornell & Simpson 1984). Another example are the operculum-like structures which become visible after opening of the sporangial apex for zoospore release and were first reported for Calycofera operculata (previously Phytophthora operculata and Halophytophthora operculata) (Pegg & Alcorn 1982, Ho & Jong 1990). Jesus et al. (2019) observed operculum-like structures also in isolates of H. avicennae, H. batemanensis, H. polymorphica and H. vesicula although these were not reported in the original descriptions by Anastasiou & Churchland (1969) and Gerrettson-Cornell & Simpson (1984). Also in the present study, despite of studying high numbers of sporangia per isolate and species, operculum-like structures could not be observed in any of the eight new Halophytophthora species or the seven isolates of H. avicennae from Portugal.
Portuguese isolates of H. thermoambigua, H. lateralis and H. frigida, previously informally named Halophytophthora sp. 1, Halophytophthora sp. 3 and Halophytophthora sp. 4, respectively, were demonstrated to host a diverse assembly of eight putative novel mycoviruses from the order Bunyavirales (Botella et al. 2020). The virus-infected Halophytophthora isolates did not show obvious phenotypic differences compared to non-infected isolates.
Geographical distribution and diversity of Halophytoph- thora species
The distribution of the eight new Halophytophthora species, H. avicennae, Halophytophthora sp. thermoambigua-like, and Halophytophthora sp. Portugal_9 (of which the only isolate died during long-term storage), and the exact location of the seven sampled marine and brackish water sites along the Algarve coast in Portugal are given in Table 5. Halophytophthora thermoambigua was most widespread (6 sites) followed by H. avicennae (5 sites). Both species were isolated from all three ecosystem types, i.e., brackish river estuaries, brackish to marine tidal ponds or channels in saltmarshes and a marine lagoon. Halophytophthora brevisporangia and H. lusitanica were found at three sites in a river estuary and in saltmarshes while H. frigida occurred in the marine lagoon at Ria de Alvor and the saltmarsh in Santa Luzia. The remaining three new species, H. celeris, H. lateralis and H. sinuata, were each isolated from one or two saltmarsh sites. Halophytophthora sp. thermoambigua-like and Halophytophthora sp. Portugal_9 were recovered from each one site. Within-site diversity ranged from two Halophytophthora species in brackish water of the Ribeira de Odelouca estuary, 3–5 Halophytophthora taxa in tidal saltmarsh ponds in Quelfes and Almancil, the lagoon of Ria de Alvor and the Rio Séqua and Rio Guadiana estuaries, to six Halophytophthora species plus Halophytophthora sp. thermoambigua-like in a tidal pond and a neighbouring tidal channel in the saltmarsh at Santa Luzia (Table 5). Noteworthy, six Phytophthora species, i.e., P. condilina, P. gonapodyides, P. plurivora, P. inundata, P. pseudocryptogea and the undescribed P. sp. Clade06a New PT, were also occasionally isolated (Table 1, 5).
Table 5.
Geographical distribution of Halophytophthora spp. in marine and brackish-water ecosystems along the Algarve coast in Portugal.
| Sitea | Geographical coordinates | Ecosystem, water type | Halophytophthora spp.b |
|---|---|---|---|
| Ribeira de Odelouca, Silves | N37 12.284, W8 29.484 | Estuary, brackish | AVI, THE |
| Rio Séqua, Tavira | N37 7.930, W7 39.367 | Estuary, brackish | AVI, LUS, THEc |
| Rio Guadiana, Castro Marim | N37 14.476, W7 25.585 | Estuary, brackish | AVI, BRE, LUS, PT9, THEd |
| PN da Ria Formosa, Santa Luzia | N37 05.346, W7 40.261 | Tidal pond and channel in saltmarsh; marine | BRE, CEL, FRI, MAC, SIN, THE, THE-like |
| PN da Ria Formosa, Quelfes | N37 01.835, W7 49.104 | Tidal pond in saltmarsh; marine | BRE, LAT, MACe |
| PN da Ria Formosa, Almancil | N37 01.030, W7 59.619 | Tidal pond in saltmarsh; brackish | AVI, LAT, LUS, THEf |
| Ria de Alvor, Alvor | N37 08.538, W8 35.606 | Lagoon; marine | AVI, FRI, THEg |
a PN = Parque Natural.
b AVI = Halophytophthora avicennae, BRE = H. brevisporangia, CEL = H. celeris, FRI = H. frigida, LAT = H. lateralis, LUS = H. lusitanica, MAC = H. macrosporangia, PT9 = Halophytophthora sp. Portugal_9, SIN = H. sinuata, THE = H. thermoambigua
c Phytophthora plurivora and P. pseudocryptogea also isolated.
d P. plurivora also isolated.
e P. gonapodyides and P. condilina also isolated.
f P. inundata, P. pseudocryptogea and P. sp. Clade06a New PT also isolated.
g P. condilina and P. inundata also isolated.
DISCUSSION
To resolve the polyphyletic nature of the genus Halophytophthora (Lara & Belbahri 2011, Nigrelli & Thines 2013, Jung et al. 2017d) 10 previous Halophytophthora species were transferred to the genera Calycofera, Phytopythium, Salisapilia and Salispina (Hulvey et al. 2010, Thines 2014, Li et al. 2016, Bennett et al. 2017a, 2018, Bennet & Thines 2019). As a consequence of these reorganisations, seven described species, H. avicennae, H. batemanensis, H. fluviatilis, H. insularis, H. polymorphica, H. souzae and H. vesicula (Yang & Hong 2014, Jung et al. 2017d, Bennet & Thines 2019, Jesus et al. 2019) and the informally designated Halophytophthora sp. Zostera (Govers et al. 2016) and Halophytophthora sp. 1–4 (Nigrelli & Thines 2013, Man in ‘t Veld et al. 2019) remained in Halophytophthora s.str.
The oomycete survey performed with a single sampling date at seven sites in marine and brackish water of tidal ponds and channels in saltmarshes, lagoon ecosystems and two river estuaries distributed along an 80 km stretch of the Algarve coast in the South of Portugal unveiled an unprecedented bounty of 10 previously unknown Halophytophthora taxa plus H. avicennae, originally described from New South Wales and also reported from Brazil (Gerrettson-Cornell & Simpson 1984, Jesus et al. 2016), and six Phytophthora species. Based on differences in morphology and temperature-growth relations and a multigene phylogeny, eight new Halophytophthora species from Portugal are described here as H. brevisporangia, H. celeris, H. frigida, H. lateralis, H. lusitanica, H. macrosporangia, H. sinuata and H. thermoambigua thus doubling the number of described species in Halophytophthora s.str.
The phylogenetic analyses of separate LSU and ITS datasets, comprising all described Halophytophthora species, the 10 new Halophytophthora taxa from Portugal and all relevant and distinctive sequences available from GenBank, provided for the first time a comprehensive phylogeny of the genus Halophytophthora s.str. with a structure of ten clades which are designated as Clades 1–10. Except of Halophytophthora sp. Portugal_9 which constituted Clade 10, all new Halophytophthora taxa from the Algarve coast resided in Clade 6 together with H. polymorphica and H. vesicula. In the phylogenetic analyses of a concatenated nine-loci (ITS, LSU, Btub, hsp90, rpl10, tigA, cox1, nadh1, rps10) dataset H. brevisporangia, H. celeris, H. frigida, H. lateralis, H. lusitanica, H. macrosporangia, H. polymorphica, H. sinuata, H. thermoambigua and H. sp. thermoambigua-like formed two distinct strongly supported sub-clades 6a and 6b. In the LSU analysis Halophytophthora s.str. clustered in sister position to Phytophthora with Nothophytophthora residing in a basal position to this cluster. In contrast, a recent phylogenetic analysis of a comprehensive three-loci (LSU, ITS, cox1) dataset placed Halophytophthora s.str. in a basal position to the sister genera Nothophytophthora and Phytophthora reflecting better the closer similarities in morphology, lifestyles and habitats between Phytophthora and Nothophytophthora as compared to Halophytophthora (Jung et al. 2017d). ‘Halophytophthora’ exoprolifera belongs to an undescribed genus basal to the Halophytophthora-Nothophytophthora-Phytophthora cluster supporting a previous phylogeny (Jung et al. 2017d). For the cryptic species H. porrigovesica only a 18S ribosomal RNA gene sequence (GU994168) and an ITS sequence (GU258844) of isolate P15166 from Thailand could be retrieved from GenBank and an rps10 sequence of the Japanese ex-type isolate CBS 125230 from (http://oomycetedb.cgrb.oregonstate.edu) (Foster et al. 2021). BLAST ®searches for these three sequences showed that all closest hits belong to Phytophthora. ML and BI analyses using a 1 373 characters dataset including 69 ITS sequences of H. porrigovesica and representative species from all phylogenetic clades of Halophytophthora and Phytophthora, and other genera in the Peronosporaceae, Pythiaceae and Salisapiliaceae demonstrated that H. porrigovesica belongs to Phytophthora Clade 9 (see Appendix; alignment and trees deposited at Dryad: (https://doi.org/10.5061/dryad.gf1vhhmr2). Besides many freshwater and a few terrestrial species Phytophthora Clade 9 also includes several species from marine and brackish-water habitats, i.e., P. estuarina and P. rhizophorae (Li et al. 2016). Many ITS and LSU sequences retrieved from GenBank for isolates from marine and brackish-water ecosystems along the coasts of North America, Brazil, the Philippines and the North Sea could not be assigned to any of the known Halophytophthora species or the new Halophytophthora species from Portugal. Most likely, these isolates constitute multiple undescribed species in Halophytophthora Clades 1, 2, 6, 8 and 9.
Halophytophthora diversity at the individual marine sites in Portugal was high, ranging from two species in the brackish water upstream of the Ribeira de Odelouca estuary to seven species in the saltmarsh at Santa Luzia. With the two known species H. avicennae and H. polymorphica, the two new species H. insularis and H. souzae, and another unknown species, erroneously identified as H. vesicula, Halophytophthora diversity in tropical mangrove ecosystems along the Perequê river in Brazil was nearly as high as in Portugal (Jesus et al. 2016, 2019). In contrast, oomycete surveys in brackish and marine water ecosystems in British Columbia, the South-eastern USA and the Caribbean, New South Wales, Singapore, the Philippines and Japan showed lower diversity of Halophytophthora s.str. than in Portugal and Brazil (Fell & Master 1975, Nakagiri et al. 1994, 2001, Tan & Pek 1997, Nakagiri 2000, Hulvey et al. 2010, Bennett et al. 2018, Bennett & Thines 2019, 2020). However, differences between the various surveys might have been strongly biased by the use of different isolation and sampling methods. For instance, in the North Sea in total three Halophytophthora taxa, all of them unknown to science, were obtained during various oomycete surveys (Nigrelli & Thines 2013, Govers et al. 2016, Man in ‘t Veld et al. 2019). However, these surveys were focussed on direct isolations from necrotic Z. marina seeds and leaves and in situ baiting tests would most likely obtain a wider range of Halophytophthora species from the North Sea. Interestingly, in the south-eastern USA, the Caribbean, Southeast Asia and Japan the genera Salisapilia and Salispina seem to be more common than Halophytophthora s.str. (Fell & Master 1975, Nakagiri et al. 1994, 2001, Tan & Pek 1997, Nakagiri 2000, Hulvey et al. 2010, Bennett et al. 2018, Bennett & Thines 2019, 2020). In contrast, Salisapilia or Salispina species could not be isolated from any of the seven sites sampled in this study in Portugal or from the mangrove sites along the Perequê river in Brazil (Jesus et al. 2016, 2019). Also in Chesapeake Bay in Maryland and the North Sea Salisapilia species seem to be rare and there is only one record of S. sapolensis from a Zostera leaf at the German island of Sylt (Man in ‘t Veld et al. 2019).
Three of the eight new Halophytophthora species, H. frigida, H. macrosporangia and H. sinuata, have a homothallic breeding system with exclusively paragynous antheridia like H. fluviatilis, H. souzae and H. vesicula (Anastasiou & Churchland 1969, Yang & Hong 2014, Jesus et al. 2019). Since an amphigynous antheridial insertion, as observed in many Phytophthora species and in Nothophytophthora amphigynosa (Erwin & Ribeiro 1996, Jung et al. 2017a, c) has never been observed in any known species of the genera Halophytophthora and Phytopythium (De Cock et al. 2015) this morphological character most likely evolved for the first time in the common ancestor of Phytophthora and Nothophytophthora, Protophytophthora (Jung et al. 2017d). Homothallic species are mainly inbreeding rather than outcrossing (Erwin & Ribeiro 1996, Brasier et al. 2003, Jung et al. 2011). Consequently, the main evolutionary role of the oospores in H. frigida, H. macrosporangia and H. sinuata is most likely the survival of unsuitable environmental conditions rather than the creation of genetic diversity. Intriguingly, these three homothallic species have comparably low maximum temperatures for growth (25 – < 30 °C) and, hence, cannot spread and grow when shallow tidal ponds heat up in the sun during low tide. Therefore, they most likely survive low tides via oospores formed in plant debris or in infected living plant tissues, either submerged or temporarily exposed to the air and drying. In contrast, due to their relatively high maximum temperatures for growth (32 – < 35 °C) the five sterile new Halophytophthora species, H. brevisporangia, H. celeris, H. lateralis, H. lusitanica and H. thermoambigua, are better adapted to remain active in shallow warm tidal ponds and channels during low tides. The combination of high cardinal temperatures and a sterile or silent breeding system, enabling allocation of most resources to continuous asexual reproduction and spread via zoospores, is a typical feature of primarily aquatic Phytophthora species and considered as an adaptation to a mainly saprotrophic lifestyle as litter decomposers and opportunistic pathogens of riparian vegetation (Brasier et al. 2003, Jung et al. 2011).
In general, Halophytophthora species, like species from the related genera Salisapilia, Salispina, Phytopythium and Calycofera, play a key role as decomposers of plant litter in tropical and subtropical mangrove ecosystems and saltmarshes (Fell & Master 1975, Pegg & Alcorn 1982, Gerrettson-Cornell & Simpson 1984, Newell et al. 1987, Ho & Jong 1990, Ho et al. 1991, 1992, Newell & Fell 1992, 1995, Nakagiri et al. 1994, 2001, Raghukumar et al. 1995, Tan & Pek 1997, Leaño et al. 1998, 2000, Marano et al. 2016, Jesus et al. 2016, 2019, Bennett et al. 2017a, b, Bennett & Thines 2020, Su et al. 2020). Accordingly, also the eight new Halophytophthora species and H. avicennae most likely have, at least partially, a saprophytic lifestyle in the coastal ecosystems in Portugal. However, marine oomycetes are also known as serious plant pathogens. Species from early diverging oomycete genera, including Anisolpidium (Anisolpidiaceae, Anisolpidiales), Eurychasma (Eurychasmales, previously Saprolegniales), Olpidiopsis (Olpidiopsidaceae, Olpidiopsidales), Ectrogella s.lat. (Olpidiopsidales or Anisolpidiales), Pontisma (Pontismataceae, Lagenidiales) and Sirolpidium (Sirolpidiacae, Lagenidiales), are common pathogens of a wide range of algal groups (Sparrow 1960, West et al. 2006, Gachon et al. 2009, 2017, Badis et al. 2019, Buaya et al. 2019, 2021, Garvetto et al. 2020). Marine Halophytophthora and Phytophthora species are occasionally found associated with diseases of higher plants, and the low number of such reports most likely reflects the scarcity of marine phytopathological studies rather than the true importance of these genera as plant pathogens. In severely declining mangrove stands of Queensland an unidentified Halophytophthora species resembling H. vesicula was causing stem cankers and root rot of Avivennia marina trees (Pegg et al. 1980). In the early 1930s, a large-scale loss of up to 90 % of the marine foundation species Z. marina on both sides of the North Atlantic was attributed to the so-called ‘wasting disease’ associated with Labyrinthula zosterae (Labyrinthulomycetes, Straminipila). Later studies, however, have casted doubt on the primary role of Labyrinthula in the decline of eelgrass (Vergeer & Den Hartog 1994, Brakel et al. 2014, Martin et al. 2016). Recent isolations of Phytophthora and Halophytophthora species from necrotic seeds and leaves of declining eelgrass in the North Sea and at Chesapeake Bay on the US Atlantic coast raised the question whether Phytophthora and Halophytophthora species are involved as causal agents in the global seagrass decline (Man in ‘t Veld et al. 2011, 2019, Govers et al. 2016). Both Halophytophthora sp. Zostera, which is most likely identical to H. lateralis, and P. gemini were isolated from almost all Z. marina seeds collected at the island Sylt in the North Sea, and severely reduced seed germination rate in a pathogenicity trial (Govers et al. 2016). Interestingly, three of the eight new Halophytophthora species from Portugal, H. lusitanica, H. lateralis and H. thermoambigua, were recently isolated from necrotic Z. marina leaves at the Atlantic coast of Norway (T. Jung, I. Milenković, T. Corcobado & V. Talg ø unpubl. data). The potential involvement of these and other Halophytophthora and Phytophthora species in eelgrass decline is currently being investigated in Norway and Portugal. In the Netherlands, P. inundata was recently isolated from leaves of the halophytic Aster tripolium and Salicornia europaea (Man in ’t Veld et al. 2019). In the three saltmarshes sampled in Parque Natural da Ria Formosa in Portugal during the present study, scattered and patch mortality of halophytic plants, including Salicornia and Sarcocornia spp., has been observed. It remains unclear whether this mortality was caused by the harsh environmental conditions, characterised by periodical flooding and drying out and continuous, often strong winds, or by pathogen infections which might have been triggered by environmental stress. To clarify the potential involvement of the eight new Halophytophthora species in eelgrass decline and the mortality of halophytic saltmarsh plants in Portugal their pathogenicity to Z. marina, Salicornia ramomissima and Sarcocornia spp. is currently being tested. In addition, leaf litter decomposition trials with the seagrasses Cymodocea nodosa and Z. marina (Santschi et al. 2017, Aram & Rizzo 2019) are underway to get first insights into the ecological role and lifestyle of all new Halophytophthora species in the marine and brackish-water ecosystems along the Algarve coast.
The co-occurrence of 10 Halophytophthora taxa in these ecosystems raises the question whether some of them had been introduced from overseas, e.g., with water, sediments, and biofilm discharged from big ships’ ballast water tanks which were demonstrated to contain high concentrations of bacteria and viruses (Ruiz et al. 2000a, b, Drake et al. 2007) and most likely also provide suitable habitats for the transport of oomycetes. More oomycete surveys in yet under-surveyed regions of the world and population genetic or phylogenomic analyses of global populations are needed to unveil the true diversity of oomycetes in marine ecosystems and clarify the origin of these Halophytophthora species as has been done recently for the widespread plant pathogens P. cinnamomi and P. ramorum (Jung et al. 2021, Shakya et al. 2021).
Acknowledgments
The authors are grateful to the Portuguese Science and Technology Foundation (FCT) for co-financing with Portuguese national funds the European BiodivERsA project ‘RESIPATH: Responses of European Forests and Society to Invasive Pathogens’ (BIODIVERSA/0002/2012), and to the Czech Ministry for Education, Youth and Sports and the European Regional Development Fund for financing the Project ‘Phytophthora Research Centre’ (Reg. No. CZ.02.1.01/0.0/0.0/15_003/0000453). Cristiana Maia acknowledges FCT and the European Social Fund (FSE) for her Ph.D. grant (SFRH/BD/136277/2018). We also thank the Institute for the Conservation of Nature and Forestry (ICNF) for logistic support during the fieldwork in Portugal. DNA sequencing was partly supported by the Italian Ministry of Education, University and Research (MIUR) funded project ‘Metagenomic strategies to assess genetic diversity in soil-borne Phytophthora species’. Aneta Bačová, Henrieta Ďatková and Milica Raco (all Mendel University in Brno) are acknowledged for technical support.
Declaration on conflict of interest The authors declare that there is no conflict of interest.
Appendix
Fifty percent majority rule consensus phylogram derived from Bayesian inference analysis of an ITS dataset of ‘Halophytophthora’ porrigovesica isolate WPC P15166 from Thailand, representative species from all phylogenetic Clades of Phytophthora and Halophytophthora s.str., and representative species from related genera in the Peronosporaceae, Pythiaceae and Salisapiliaceae. Bayesian posterior probabilities and ML bootstrap values (in %) are indicated but not shown below 0.70 and 70 %, respectively. Aphanomyces euteiches was used as outgroup taxon. Scale bar indicates 0.1 expected changes per site per branch.
REFERENCES
- Anastasiou CJ, Churchland LM. 1969. Fungi on decaying leaves in marine habitats. Canadian Journal of Botany 47: 251–257. [Google Scholar]
- Aram K, Rizzo DM.2019. Phytophthora ramorum and Phytophthora gonapodyides differently colonize and contribute to the decomposition of green and senesced Umbellularia californica leaves in a simulated stream environment. Forests 10: 434. [Google Scholar]
- Badis Y, Klochkova TA, Strittmatter M. et al. 2019. Novel species of the oomycete Olpidiopsis potentially threaten European red algal cultivation. Journal of Applied Phycology 31: 1239–1250. [Google Scholar]
- Bennett RM, Cock AWAM, Lévesque A. et al. 2017a. Calycofera gen. nov., an estuarine sister taxon to Phytopythium, Peronosporaceae. Mycological Progress 16: 947–954. [Google Scholar]
- Bennett RM, Devanadera MK, Dedeles GR. et al. 2018. A revision of Salispina, its placement in a new family, Salispinaceae (Rhipidiales), and de-scription of a fourth species, S. hoi sp. nov. IMA Fungus 9: 259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RM, Nam B, Dedeles GR. et al. 2017b. Phytopythium leanoi sp. nov. and Phytopythium dogmae sp. nov., Phytopythium species associated with mangrove leaf litter from the Philippines. Acta Mycologia 52: 1103. [Google Scholar]
- Bennett RM. Thines M. 2019. Revisiting Salisapiliaceae. Fungal Systematics and Evolution 3: 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RM, Thines M. 2020. An overview on Philippine estuarine oomycetes. Philippine Journal of Systematic Biology 14: 1–14. [Google Scholar]
- Blair JE, Coffey MD, Park S-Y. et al. 2008. A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genetics and Biology 45: 266–277. [DOI] [PubMed] [Google Scholar]
- Botella L, Janoušek J, Maia C. et al. 2020. Marine oomycetes of the genus Halophytophthora harbor viruses related to Bunyaviruses. Frontiers in Microbiology 11: 1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella L, Jung T. 2021. Multiple viral infections detected in Phytophthora condilina by total and small RNA sequencing. Viruses 13: 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakel J, Werner FJ, Tams V. et al. 2014. Current European Labyrinthula zosterae are not virulent and modulate seagrass (Zostera marina) defense gene expression. PLoS One 9: e92448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasier CM, Cooke DEL, Duncan JM. et al. 2003. Multiple new phenotypic taxa from trees and riparian ecosystems in Phytophthora gonapodyides – P. megasperma ITS Clade 6, which tend to be high-temperature tolerant and either inbreeding or sterile. Mycological Research 107: 277–290. [DOI] [PubMed] [Google Scholar]
- Buaya AT, Ploch S, Inaba S. et al. 2019. Holocarpic oomycetes parasitoids of red algae are not Olpidiopsis. Fungal Systematics and Evolution 4: 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buaya AT, Scholz B, Thines M. 2021. Sirolpidium bryopsidis, a parasite of green algae, is probably conspecific with Pontisma lagenidioides, a parasite of red algae. Fungal Systematics and Evolution 7: 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballol M, Štraus D, Macia H. et al. 2021. Halophytophthora fluviatilis pathogenicity and distribution along a Mediterranean-subalpine gradient. Journal of Fungi 7: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke DEL, Drenth A, Duncan JM. et al. 2000. A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genetics and Biology 30: 17–32. [DOI] [PubMed] [Google Scholar]
- De Cock AWAM, Lodhi AM, Rintoul TL. et al. 2015. Phytopythium: molecular phylogeny and systematics. Persoonia 34: 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick MW. 1990. Keys to Pythium. University of Reading Press, Reading, UK. [Google Scholar]
- Drake LA, Doblin MA, Dobbs FC. 2007. Potential microbial bioinvasions via ships’ ballast water, sediment, and biofilm. Marine Pollution Bulletin 55: 333–341. [DOI] [PubMed] [Google Scholar]
- Edler D, Klein J, Antonelli A. et al. 2021. raxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods in Ecology and Evolution 12: 373–377. [Google Scholar]
- Erwin DC, Ribeiro OK. 1996. Phytophthora diseases worldwide. APS Press St. Paul, Minnesota. [Google Scholar]
- Fell JW, Master IM. 1975. Phycomycetes (Phytophthora spp. nov. and Pythium sp. nov.) associated with mangrove (Rhizophora mangle) leaves. Canadian Journal of Botany 53: 2908–2922. [Google Scholar]
- Foster ZSL, Albornoz FE, Fieland VJ. et al. 2021. A new oomycete metabarcoding method using the rps10 gene. bioRxiv: 2021.09.22.460084 [Google Scholar]
- Gachon CMM, Strittmatter M, Badis Y. et al. 2017. Pathogens of brown algae: culture studies of Anisolpidium ectocarpii and A. rosenvingei reveal that the Anisolpidiales are uniflagellated oomycetes. European Journal of Phycology 52: 133–148. [Google Scholar]
- Gachon CMM, Strittmatter M, Müller DG. et al. 2009. Detection of differential host susceptibility to the marine oomycete pathogen Eurychasma dicksonii by real-time PCR: not all algae are equal. Applied and Environmental Microbiology 75: 322–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbelotto MM, Lee HK, Slaughter G. et al. 1997. Heterokaryosis is not required for virulence of Heterobasidion annosum. Mycologia 89: 92–102. [Google Scholar]
- Garvetto A, Perrineau M-M, Dressler-Allame M. et al. 2020. ‘Ectrogella’ parasitoids of the diatom Licmophora sp. are polyphyletic. Journal of Eukaryotic Microbiology 67: 18–27. [DOI] [PubMed] [Google Scholar]
- Gerrettson-Cornell L, Simpson J. 1984. Three new marine Phytophthora species from New South Wales. Mycotaxon 19: 453–470. [Google Scholar]
- Govers LL, Man in ´t Veld WA, Meffert JP. et al. 2016. Marine Phytophthora species can hamper conservation and restoration of vegetated coastal ecosystems. Proceedings of the Royal Society B: Biological Sciences 283: 20160812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho HH, Chang HS, Hsieh SY. 1991. Halophytophthora kandeliae, a new marine fungus from Taiwan. Mycologia 83: 419–424. [Google Scholar]
- Ho HH, Chang HS, Huang SH. 2003. Halophytophthora elongata, a new marine species from Taiwan. Mycotaxon 85: 417–422. [Google Scholar]
- Ho HH, Jong SC. 1990. Halophytophthora, gen nov., a new member of the family Pythiaceae. Mycotaxon 36: 377–382. [Google Scholar]
- Ho HH, Nakagiri A, Newell SY. 1992. A new species of Halophytophthora from Atlantic and Pacific subtropical islands. Mycologia 84: 548–554. [Google Scholar]
- Hopple JS, Vilgalys R. 1994. Phylogenetic relationships among coprinoid taxa and allies based on data from restriction site mapping of nuclear rDNA. Mycologia 86: 96–107. [Google Scholar]
- Hüberli D, Hardy GEStJ, White D. et al. 2013. Fishing for Phytophthora from Western Australia’s waterways: A distribution and diversity survey. Australasian Plant Pathology 42: 251–260. [Google Scholar]
- Hulvey J, Telle S, Nigrelli L. et al. 2010. Salisapiliaceae – a new family of oomycetes from marsh grass litter of southeastern North America. Persoonia 25: 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesus AL, Marano AV, Gonçalve DR. et al. 2019. Two new species of Halophytophthora from Brazil. Mycological Progress 18: 1411–1421. [Google Scholar]
- Jesus AL, Marano AV, Jerônimo GH. et al. 2016. The genus Halophytophthora (Peronosporales, Straminipila) in Brazil: first descriptions of species. Brazilian Journal of Botany 39: 729–739. [Google Scholar]
- Jung T, Blaschke H, Neumann P. 1996. Isolation, identification and pathogenicity of Phytophthora species from declining oak stands. European Journal of Forest Pathology 26: 253–272. [Google Scholar]
- Jung T, Burgess TI.. 2009. Re-evaluation of Phytophthora citricola isolates from multiple woody hosts in Europe and North America reveals a new species Phytophthora plurivora sp nov. Persoonia 22: 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung T, Chang TT, Bakonyi J. et al. 2017a. Diversity of Phytophthora species in natural ecosystems of Taiwan and association with disease symptoms. Plant Pathology 66: 194–211. [Google Scholar]
- Jung T, Cooke DEL, Blaschke H. et al. 1999. Phytophthora quercina sp. nov., causing root rot of European oaks. . Mycological Research 103: 785–798. [Google Scholar]
- Jung T, Horta Jung M, Cacciola SO. et al. 2017b. Multiple new cryptic pathogenic Phytophthora species from Fagaceae forests in Austria, Italy and Portugal. IMA Fungus 8: 219–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung T, Horta Jung M, Scanu B. et al. 2017c. Six new Phytophthora species from ITS Clade 7a including two sexually functional heterothallic hybrid species detected in natural ecosystems in Taiwan. Persoonia 38: 100–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung T, Horta Jung M, Webber JF.et al. 2021. The destructive tree pathogen Phytophthora ramorum originates from the Laurosilva forests of East Asia. Journal of Fungi 7: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung T, Pérez-Sierra A, Durán A. et al. 2018. Canker and decline diseases caused by soil- and airborne Phytophthora species in forests and woodlands. Persoonia 40: 182–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung T, Scanu B, Bakonyi J. et al. 2017d. Nothophytophthora gen. nov., a new sister genus of Phytophthora from natural and semi-natural ecosystems. Persoonia 39: 143–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung T, Stukely MJC, Hardy GEStJ. et al. 2011. Multiple new Phytophthora species from ITS Clade 6 associated with natural ecosystems in Australia: evolutionary and ecological implications. Persoonia 26: 13–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon LPNM, Bakker FT, Van den Bosch GBM, et al. 2004. Phylogenetic analysis of Phytophthora species based on mitochondrial and nuclear DNA sequences. Fungal Genetics and Biology 41: 766–782. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M. et al. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamour K. (ed). 2013. Phytophthora: A global perspective. CABI, Wallingford, UK. [Google Scholar]
- Lamour KH, Finley L. 2006. A strategy for recovering high quality genomic DNA from a large number of Phytophthora isolates. Mycologia 98: 514–517. [DOI] [PubMed] [Google Scholar]
- Lara E, Belbahri L. 2011. SSU rRNA reveals major trends in oomycete evolution. Fungal Diversity 49: 93–100. [Google Scholar]
- Leaño EM, Jones EBG, Vrijmoed LLP. 2000. Why are Halophytophthora species well adapted to mangrove habitats? In: Hyde KD, Ho WH, Pointing SB. (eds), Aquatic mycology across the Fungal Diversity 5: 131–151. [Google Scholar]
- Leaño EM, Vrijmoed LLP, Jones ERG. 1998. Physiological studies on Halophytophthora vesicula (straminipilous fungi) isolated from fallen mangrove leaves from Mai Po, Hong Kong. Botanica Marina 41: 411–419. [Google Scholar]
- Li GJ, Hyde KD, Zhao RL.et al. 2016. Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 78: 1–237. [Google Scholar]
- Man in ‘t Veld WA, Rosendahl KC, Brouwer H, et al. 2011. Phytophthora gemini sp. nov., a new species isolated from the halophilic plant Zostera marina in the Netherlands. Fungal Biology 115: 724–732. [DOI] [PubMed] [Google Scholar]
- Man in ‘t Veld WA, Rosendahl KCHM, Van Rijswick PCJ. et al. 2019. Multiple Halophytophthora spp. and Phytophthora spp. including P. gemini, P. inundata and P. chesapeakensis sp. nov.isolated from the seagrass Zostera marina in the Northern hemisphere. European Journal of Plant Pathology 153: 341–357. [Google Scholar]
- Marano AV, Jesus AL, De Souza JI. et al. 2016. Ecological roles of saprotrophic Peronosporales (Oomycetes, Straminipila) in natural environments. Fungal Ecology 19: 77–88. [Google Scholar]
- Martin DL, Chiari Y, Boone E. et al. 2016. Functional, phylogenetic and host-geographic signatures of Labyrinthula spp. provide for putative species delimitation and a global-scale view of seagrass wasting disease. Estuaries and Coasts 39: 1403–1421. [Google Scholar]
- Martin FN, Tooley PW. 2003. Phylogenetic relationships among Phytophthora species inferred from sequence analysis of mitochondrially encoded cytochrome oxidase I and II genes. Mycologia 95: 269–284. [PubMed] [Google Scholar]
- Muehlstein LK, Porter D, Short FT. 1988. Labyrinthula sp., a marine slime mold producing symptoms of wasting disease in eelgrass, Zostera marina. Marine Biology 99: 465–472. [Google Scholar]
- Nakagiri A. 2000. Ecology and biodiversity of Halophytophthora species. In: Hyde KD, Ho WH, Pointing SB. (eds), Aquatic mycology across the Millennium. Fungal Diversity 5: 153–164. [Google Scholar]
- Nakagiri A, Ito T, Manoch L. et al. 2001. A new Halophytophthora species, H. porrigovesica, from subtropical and tropical mangroves. Mycoscience 42: 33–41. [Google Scholar]
- Nakagiri A, Newell SY, Ito T. 1994. Two new Halophytophthora species, H. tar-tarea and H. masteri, from intertidal decomposing leaves in saltmarsh and mangrove regions. Mycoscience 35: 223–232. [Google Scholar]
- Newell SY, Fell JW. 1992. Distribution and experimental responses to substrate of marine oomycetes (Halophytophthora spp.) in mangrove ecosystems. Mycological Research 96: 851–856. [Google Scholar]
- Newell SY, Fell JW. 1995. Do halophytophthoras (marine Pythiaceae) rapidly occupy fallen leaves by intraleaf mycelial growth? Canadian Journal of Botany 73:761–765. [Google Scholar]
- Newell SY, Miller JD, Fell JW. 1987. Rapid and pervasive occupation of fallen mangrove leaves by a marine zoosporic fungus. Applied and Environmental Microbiology 53: 2464–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigrelli L, Thines M. 2013. Tropical oomycetes in the German Bight – Climate warming or overlooked diversity? Fungal Ecology 6: 152–160. [Google Scholar]
- Pegg KG, Alcorn JL. 1982. Phytophthora operculata sp. nov. a new marine fungus. Mycotaxon 16: 99–102. [Google Scholar]
- Pegg KG, Gillespie NC, Forsberg LI. 1980. Phytophthora sp. associated with mangrove death in Central Coastal Queensland. Australasian Plant Pathology 9: 6–7. [Google Scholar]
- Raghukumar S, Sathe-Pathak V, Sharma S, et al. 1995. Thraustochytrid and fungal component of marine detritus 111. Field studies on decomposition of leaves of the mangrove Rhizophora apiculata. Aquatic Microbial Ecology 9: 117–125. [Google Scholar]
- Rahman MZ, Uematsu S, Coffey MD. et al. 2014. Re-evaluation of Japanese Phytophthora isolates based on molecular phylogenetic analyses. Mycoscience 55: 314–327. [Google Scholar]
- Reeser PW, Sutton W, Hansen EM. et al. 2011. Phytophthora species in forest streams in Oregon and Alaska. Mycologia 103: 22–35. [DOI] [PubMed] [Google Scholar]
- Robideau GP, De Cock AW, Coffey MD. et al. 2011. DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Molecular Ecology Resources 11: 1002–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Ruiz GM, Fofonoff PW, Carlton JT. et al. 2000b. Invasion of coastal marine communities in North America: Apparent patterns, processes, and biases. Annual Review of Ecology and Systematics 31: 481–531. [Google Scholar]
- Ruiz GM, Rawlings TK, Dobbs FC. et al. 2000a. Global spread of microorganisms by ships. Nature 408: 49–50. [DOI] [PubMed] [Google Scholar]
- Santschi F, Gounand I, Harvey E. et al. 2017. Leaf litter diversity and structure of microbial decomposer communities modulate litter decomposition in aquatic systems. Functional Ecology 32: 522–532. [Google Scholar]
- Shakya SK, Grünwald NJ, Fieland VJ. et al. 2021. Phylogeography of the wide-host range panglobal plant pathogen Phytophthora cinnamomi. Molecular Ecology 30: 5164–5178. [DOI] [PubMed] [Google Scholar]
- Sparrow FK. 1960. Aquatic Phycomycetes. The University of Michigan Press, Ann Arbor, USA. [Google Scholar]
- Stöver BC, Müller KF. 2010. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su CJ, Hsieh SY, Chiang MWL. et al. 2020. Salinity, pH and temperature growth ranges of Halophytophthora isolates suggest their physiological adaptations to mangrove environments. Mycology 11: 256–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BK, Trevathan-Tackett SM, Neuhauser S. et al. 2018. Host-pathogen dynamics of seagrass diseases under future global change. Marine Pollution Bulletin 134: 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TK, Pek CL. 1997. Tropical mangrove leaf litter fungi in Singapore with an emphasis on Halophytophthora. Mycological Research 101: 165–168. [Google Scholar]
- Thines M. 2014. Phylogeny and evolution of plant pathogenic oomycetes – a global overview. European Journal of Plant Pathology 38: 431–447. [Google Scholar]
- Vergeer LHT, Den Hartog C. 1994. Omnipresence of Labyrinthulaceae in seagrasses. Aquatic Botany 48: 1–20. [Google Scholar]
- West JA, Klochkova TA, Kim GH. et al. 2006. Olpidiopsis sp., an oomycete from Madagascar that infects Bostrychia and other red algae: Host species susceptibility. Phycological Research 54: 72–85. [Google Scholar]
- White TJ, Bruns T, Lee S. et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Gelfand DH, Sninsky JJ, Innes MA, et al. (eds), PCR protocols: a guide to methods and applications: 315– 322. Academic Press, San Diego, California, USA. [Google Scholar]
- Yang X, Hong C. 2014. Halophytophthora fluviatilis sp.nov. from freshwater in Virginia. FEMS Microbiology Letters 352: 230–237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fifty percent majority rule consensus phylogram derived from Bayesian inference analysis of an ITS dataset of ‘Halophytophthora’ porrigovesica isolate WPC P15166 from Thailand, representative species from all phylogenetic Clades of Phytophthora and Halophytophthora s.str., and representative species from related genera in the Peronosporaceae, Pythiaceae and Salisapiliaceae. Bayesian posterior probabilities and ML bootstrap values (in %) are indicated but not shown below 0.70 and 70 %, respectively. Aphanomyces euteiches was used as outgroup taxon. Scale bar indicates 0.1 expected changes per site per branch.


