Abstract
The tick-borne relapsing fever spirochete Borrelia hermsii evades the mammalian immune system by periodically switching expression among members of two multigene families that encode immunogenic, antigenically distinct outer surface proteins. The type strain, B. hermsii HS1, has at least 40 complete genes and pseudogenes that participate in this multiphasic antigenic variation. Originally termed vmp (for variable major protein) genes, they have been reclassified as vsp (for variable small protein) and vlp (for variable large protein) genes, based on size and amino acid sequence similarities. To date, antigenic variation in B. hermsii has been studied only in the type strain, HS1. Nucleotide sequence comparisons of 23 B. hermsii HS1 genes revealed five distinct groups, the vsp gene family and four subfamilies of vlp genes. We used PCR with family- and subfamily-specific primers, followed by restriction fragment length polymorphism analysis, to compare the vsp and vlp repertoires of HS1 and seven other B. hermsii isolates from Washington, Idaho, and California. This analysis, together with pulsed-field gel electrophoresis genome profiles, revealed that the eight isolates formed three distinct groups, which likely represent clonal lineages. Members of the three groups coexisted in the same geographic area, but they could also be isolated across large geographical distances. This population structure may result from immune selection by the host, as has been proposed for other pathogens with polymorphic antigens.
Relapsing fever is the medical term for a disease characterized by a cyclic rise and fall in body temperature caused by arthropod-transmitted spirochetes of the genus Borrelia. Louse-borne or epidemic relapsing fever is solely a human disease, whereas tick-borne relapsing fever, with one exception, is a zoonosis that can be transmitted to humans. Several tick-borne relapsing fever Borrelia species, each associated with a different tick vector and mammalian hosts, occur within discrete geographical areas throughout the world (6). Borrelia hermsii, for example, persists in natural cycles that involve the soft tick Ornithodoros hermsi, chipmunks, and tree squirrels in high-altitude forests of California, Arizona, Nevada, Colorado, Oregon, Idaho, Washington, and British Columbia.
After injection into a mammal by an infected, feeding tick, B. hermsii multiplies in the blood stream, achieving numbers that can exceed 10 million per ml of peripheral blood (48, 53). Fever occurs as these organisms are cleared by an immune response directed against a prominent lipoprotein that coats the outer surfaces of spirochetes. Spirochetes virtually disappear from the blood, and the fever subsides; however, spirochetemia and fever recur 4 to 7 days later. This second population consists of spirochetes that are coated with a new lipoprotein which is antigenically different from the one expressed by the original infecting bacteria. This antigenic variation is multiphasic; relapse and recovery can repeat for several cycles, and each relapse population of spirochetes expresses an antigenically distinct surface lipoprotein that is encoded by a separate gene (4). At least 40 distinct serotypes have previously been identified in the progeny of a single cell of B. hermsii HS1, and most of the corresponding genes have been cloned and sequenced (7, 41). These genes were originally named variable major protein genes (vmp1, vmp2, vmp3, etc.). A recent comparison of their amino acid sequences indicated that the vmp genes comprise two distinct multigene families; therefore, they have been redesignated vsp and vlp genes (for variable small and variable large proteins, respectively) (2, 15).
Genes of both families are found in two locations on linear plasmids in B. hermsii HS1. In one locus, the expression site, a single vsp or vlp gene is positioned immediately downstream of a promoter near one end (or telomere) of a 28- to 30-kb linear plasmid and only that gene is expressed (27). A second copy of the expressed gene and all other genes of the two gene families exist in nonexpressed or silent form at nontelomeric locations on the same linear plasmid or on different linear plasmids of about the same size. Antigenic variation results from inter- or intraplasmid DNA rearrangements that replace the gene at the telomeric expression site with a different vsp or vlp gene, augmented in some cases by postswitch mutations of this previously silent gene (5, 27, 36, 40–42). Any silent vsp or vlp gene can replace any expressed vsp or vlp gene, although with different probabilities (3, 8, 53).
Our understanding of the genetic mechanisms of antigenic variation of relapsing fever Borrelia spp. in the vertebrate host has emerged from studies of a single isolate of B. hermsii, the type strain HS1. Here we begin an analysis at the population level of the genes involved in B. hermsii antigenic variation. We compared the vsp and vlp repertoire of the type strain HS1 to those of seven other isolates of B. hermsii from different enzootic foci in the western United States. Parasite genes that encode surface antigens often seem to be subject to a faster molecular evolutionary clock than do genes for nonimmunogenic proteins, probably because allelic (genetic) polymorphism arises through immune selection of escape variants in previously exposed hosts (12, 57). We reasoned, therefore, that comparisons of the vsp and vlp genes would reveal finer phylogenetic differences than would comparisons of conserved, presumably less variable markers, such as 16S DNA coding for rRNA or flagellin genes. The results showed extensive variation in the vsp and vlp genes of different isolates. The variation was not continuous, however. The eight isolates examined exhibited one of three basic vsp and vlp profiles, suggesting a population composed of clones or families of closely related clones.
A second motive for this study arose from our interest in the interaction of B. hermsii with its tick vector. Originally isolated in 1968 (56), present stocks of the type strain HS1 have decreased infectivity for ticks, probably as a result of laboratory passage in vitro and in mice (6). To investigate the role of variable surface lipoproteins in the invertebrate host, the tick O. hermsi, we utilized specific Vsp and Vlp antibodies and gene sequences derived from analysis of the HS1 strain. The present study identified a new isolate, B. hermsii DAH, that appears to be identical to B. hermsii HS1 in its vsp and vlp repertoire and that is infectious for both invertebrate and vertebrate hosts.
MATERIALS AND METHODS
Borrelia strains.
All Borrelia strains were from the culture collection of the Rocky Mountain Laboratories. The eight isolates of B. hermsii used in this study, their geographic origins, and dates of isolation are shown in Fig. 1 and Table 1. The histories, identifications, and characterizations of some of these isolates have been described previously (47). A clone of B. hermsii HAN was obtained by limiting dilution in BSK medium; B. hermsii HS1 has also been previously cloned (53). Other tick-borne relapsing fever species used were B. turicatae and B. parkeri, (from western North America), B. crocidurae (from Africa and the Middle East), and B. anserina, the agent of avian borreliosis (found worldwide). B. coriaceae is the putative cause of tick-borne epizootic bovine abortion (29), and B. burgdorferi B31 is the type strain of the Lyme disease agent (13).
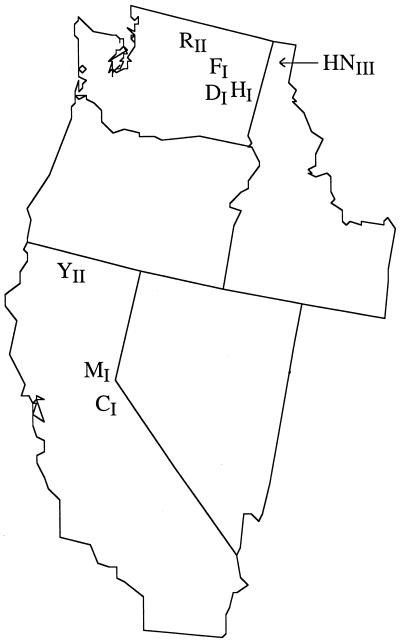
FIG. 1.
Map of western United States showing the geographic origins of the eight B. hermsii isolates used in this study. A Roman numeral subscript indicates the group to which an isolate belongs, based on the similarity of the vsp and vlp gene repertoire. C, CON; D, DAH; F, FRO; H, HS1; HN, HAN; M, MAN; R, REN; Y, YOR.
TABLE 1.
B. hermsii isolates used in this study
| Isolate | Geographic origin | Source | Yr of isola- tion | Refer- ence |
|---|---|---|---|---|
| HS1 | Spokane, Wash. | O. hermsi tick | 1968 | 56 |
| DAH | Northwest of Cheney, Wash. | Human blood | 1991 | 47 |
| FRO | Eastern Washington | Human blood | 1987 | 46 |
| MAN | Sierra Nevada Mountains, Calif. | Human blood | 1960s | 46 |
| CON | Sierra Nevada Mountains, Calif. | Human blood | 1960s | 46 |
| HAN | Bonner’s Ferry, Idaho | Human blood | 1990 | 47 |
| YOR | Siskiyou Mountains, Calif. | Human blood | 1964 | 46 |
| REN | Winthrop, Wash. | Human blood | 1992 | 47 |
B. hermsii HS1 vsp and vlp gene sequence comparisons.
The nucleotide sequences of 23 vsp and vlp genes previously cloned from B. hermsii HS1 (7, 14, 41, 43) were compared by using the multiple-sequence analysis programs PILEUP and PRETTY of UNIX (version 7; Genetics Computer Group, Inc., Madison, Wis.). The individual genes and their GenBank/EMBL accession numbers are as follows: vsp1, L33870; vsp2, L33897; vsp3, L04789; vsp6, L33898; vsp8, L33899; vsp11, L33900; vsp13, L33901; vsp22, L33902; vsp24, L04786; vlp4, U51926; vlp5, U52035; vlp7, X53926; vlp9, U52036; vlp10, U52037; vlp12, U52038; vlp14, U52148; vlp15, U52039; vlp17, L04788; vlp18, U52149; vlp19, U52040; vlp21, M57256; vlp23, U52041; and vlp25, L04787.
PCR analysis of the vsp and vlp genes.
Genomic DNA was isolated by a standard method from Borrelia cultures in BSK II medium (36). PCR (30 cycles) was performed with 50 ng of Borrelia DNA by using a thermal cycler (Perkin-Elmer, Foster City, Calif.) and vsp family- and vlp subfamily-specific primer sets designed from B. hermsii HS1 sequences (Table 2). A 659-bp fragment of the flagellin gene was also amplified from Borrelia species with a generic primer set homologous to conserved regions of the gene (39). A portion of each PCR was electrophoresed on 4% polyacrylamide gels in TBE buffer (90 mM Tris [pH 8.0], 90 mM borate, 2 mM EDTA).
TABLE 2.
PCR primers
| Target | Sequencea | Annealing temp (°C) | References |
|---|---|---|---|
| vsp family | 5′-AAGTCTGACGGAACAGTACT-3′ | 50 | 7, this study |
| 5′-GTTATTTTGAGAAGGTTTTTC-3′ | |||
| vlp family | |||
| α subfamily | 5′-CTAATGATAGGWTGTGGACAAC-3′ | 55 | 7, this study |
| 5′-CTCAAGAACATTCTTTACTGTC-3′ | |||
| β subfamily | 5′-GTGATGCATTAGGATTTAATGC-3′ | 55 | 7, this study |
| 5′-CCTAATACCTTATTTACWGCAC-3′ | |||
| γ subfamily | 5′-CTAGTGACCCAATTGCTAATGT-3′ | 55 | 7, this study |
| 5′-TCTAGTGCTTTAGTAATTGCAC-3′ | |||
| δ subfamily | 5′-ATACTAAGAAAAGTGATATAGG-3′ | 50 | 7, this study |
| 5′-CCATTGCTCGCAGTGCAATGCC-3′ | |||
| Flagellin gene | 5′-ACATATTCAGATGCAGACAGAGGT-3′ | 60 | 39, this study |
| 5′-GCAATCATTGCCATTGCAGATTGT-3′ |
W, A or T.
RFLP analysis of the vsp and vlp genes.
PCR-amplified DNA fragments were ethanol precipitated, washed with 70% ethanol, and resuspended in TE (10 mM Tris [pH 8], 1 mM EDTA). DNAs from vsp family- and vlp subfamily-specific PCRs were digested in separate reactions with the following restriction endonucleases: DdeI and RsaI (vsp family); HindIII and PstI (vlpα); Sau3AI, ScaI, and SspI (vlpβ); BglII, DdeI, and a PvuII-HaeIII double digest (vlpγ); and Sau3AI and SspI (vlpδ). DNAs from flagellin gene PCRs were digested with AluI, PvuII, RsaI, and Sau3AI. Digests were electrophoresed on 3% SeaPlaque GTG agarose (FMC BioProducts, Rockland, Maine) or 12% polyacrylamide gels with TBE buffer. Restriction fragment length polymorphism (RFLP) patterns of DNA amplified from each B. hermsii isolate were individually compared with that of the HS1 strain to determine a similarity coefficient (S), the proportion of shared restriction fragments, as a measure of genetic distance. Each DNA band was treated as a separate character, and bands of the same electrophoretic mobility were considered to be shared, regardless of differences in staining intensity. We used the formula of Nei and Li (37), S(x,y) = 2nxy/(nx + ny), in which nxy is the number of fragments shared by the two isolates and nx and ny are the total numbers of fragments produced from isolates x and y, respectively.
Pulsed-field gel electrophoresis (PFGE) and Southern blotting.
Borrelia cells from BSK II cultures were centrifuged, washed twice with TN (50 mM Tris [pH 8], 150 mM NaCl), and resuspended in TN to a concentration of approximately 109 per ml. An equal volume of molten (37°C) 1% InCert low-melting-temperature agarose (FMC) was mixed with the cell suspension, which was aliquoted to a mold and allowed to gel in the form of agarose blocks. Cells were lysed in situ by incubating agarose blocks for 16 h at 45°C in 50 mM Tris (pH 8)–50 mM EDTA–1% sodium dodecyl sulfate (SDS) containing 1 mg of proteinase K per ml (19). Blocks were washed four times, 1 h each, with TE. Intact genomic DNA from blocks was separated on a 1% agarose gel by transverse alternating field electrophoresis by using the Geneline II system (Beckman Instruments, Palo Alto, Calif.) and a previously described protocol (33). After transverse alternating field electrophoresis, the DNA on the gel was stained with ethidium bromide, photographed, and then transferred from the gel to a Hybond-N membrane (Amersham, Arlington Heights, Ill.) by vacuum blotting (VacuGene; Pharmacia Biotech, Piscataway, N.J.).
For use as probes on Southern blots, vsp and vlp-specific PCRs were diluted 1:1,000 and reamplified. Reamplified DNA fragments were purified by using Sephacryl S-400 microspin columns (Pharmacia) and labelled with [α-32P]dATP by means of a random primer labelling kit (Boeringher Mannheim, Indianapolis, Ind.). Southern hybridization was performed at 37°C for 16 h in 50% formamide–6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–5× Denhardt’s solution–0.5% SDS–0.1 mg of denatured salmon sperm DNA per ml. Final washes were done with 0.1× SSC–0.1% SDS–0.1 mM EDTA at 63°C.
RESULTS
Four subfamilies of the vlp gene family of B. hermsii HS1.
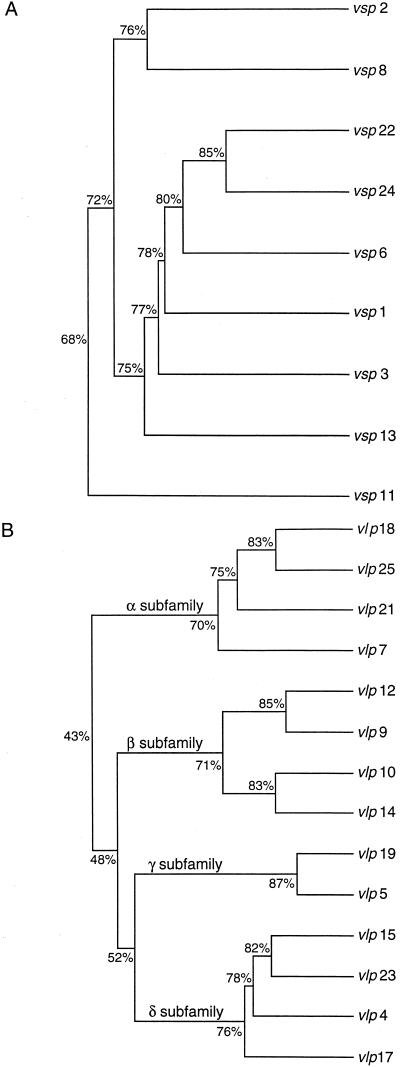
The nucleotide sequences of 9 vsp genes (vsp1, -2, -3, -6, -8, -11, -13, -22, and -24) and 14 vlp genes (vlp4, -5, -7, -9, -10, -12, -14, -15, -17, -18, -19, -21, -23, and -25) of B. hermsii HS1 (2, 7, 43) were compared by using multiple-sequence analysis programs of the Genetics Computer Group. As previously described (43), the nine vsp genes comprised one family of genes that ranged from 632 to 654 bp, with 68 to 85% nucleotide sequence similarity. The 14 vlp genes examined ranged from 1,023 to 1,110 bp and formed four distinct groups, labelled subfamilies α to δ (Fig. 2). The branchpoints that separate the four vlp subfamilies occur much deeper in the dendrogram than do the branchpoints that separate individual gene sequences within each subfamily. This is a reflection of the 70 to 87% similarity between members of each particular vlp subfamily, compared to the 39 to 51% nucleotide sequence similarities of vlp genes from different subfamilies.
FIG. 2.
Dendrograms comparing the genetic relatedness of 9 vsp (A) and 14 vlp (B) gene sequences of B. hermsii HS1. The vlp genes are subdivided into four subfamilies, labelled α to δ. The percentage of nucleotide sequence similarity between genes or gene clusters is indicated at each branchpoint.
Polymorphism of vsp and vlp genes among B. hermsii isolates.
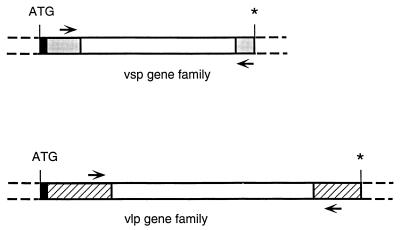
A schematic comparison of vsp and vlp genes is shown in Fig. 3. Probably because of common transport, lipidation, and membrane-anchoring mechanisms of these surface lipoproteins (14), the 5′ ends of all of the vsp and vlp genes examined are conserved. All 23 genes are identical for the first 26 bp, and all, except for vlp subfamily α genes, are identical for the first 77 bp. Downstream from this constant leader sequence, the 5′ and 3′ portions of the genes of the vsp family or a particular vlp subfamily are additionally conserved (7, 43). Nucleotide sequence alignments confirmed that most of the variation between individual vsp family and vlp subfamily members occurs in a central region of the genes. From the conserved 5′ and 3′ regions, sequences were identified for use as vsp family- and vlp subfamily-specific PCR primers. Each of the five primer sets was predicted to amplify the central variable segment of every member of the vsp family or a particular vlp subfamily of the HS1 strain (except for vsp family primers, which amplify all vsp genes except for vsp11) but not those of genes outside of the subfamily. Because the primer sequences occurred within genes, both silent and expressed forms were amplified.
FIG. 3.
Nucleotide sequence features of vsp and vlp genes of B. hermsii HS1. The central, most variable segment of these genes is flanked by conserved regions (shaded areas for vsp and hatched areas for vlp) that contain the vsp family- or vlp subfamily-specific PCR primer binding sites (arrows). The short 5′ leader sequence shared by genes of both families is indicated by a black box, and asterisks denote stop codons.
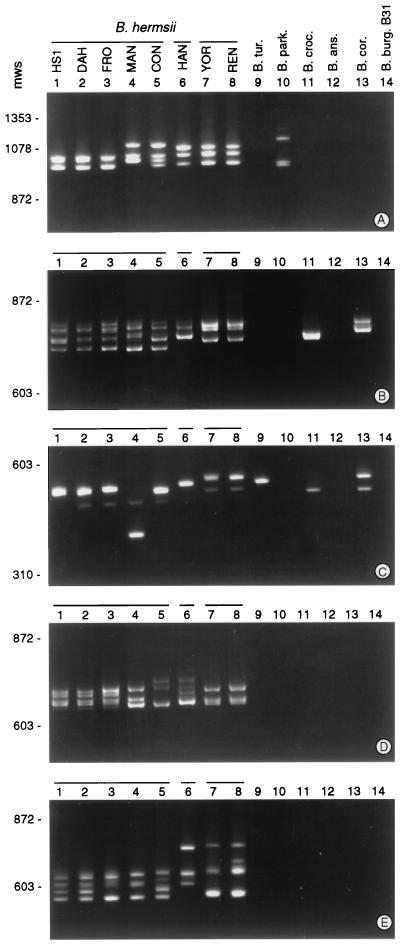
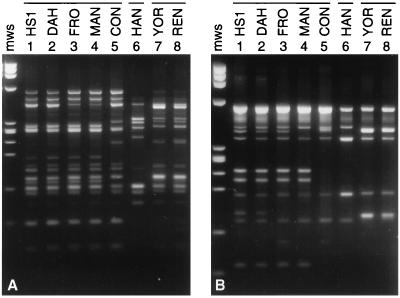
To detect genetic variability of the vsp and vlp repertoire among isolates of B. hermsii, we performed PCR with each of the five primer sets and genomic DNAs isolated from B. hermsii HS1 and seven other B. hermsii isolates (Table 1). Because the collection of vsp or vlp products generated in each PCR differed in size by 5% or less, they usually migrated as a single band on agarose gels (data not shown). Greater resolution was achieved on polyacrylamide gels, on which amplified vsp and vlp fragments yielded family or subfamily profiles (Fig. 4). This analysis revealed polymorphism in the vsp and vlp genes among isolates of this species. B. hermsii DAH and FRO displayed profiles similar to that of the type strain HS1, but the five other B. hermsii isolates generated different patterns. The profiles of B. hermsii YOR and REN appeared to be identical; the remaining three isolates yielded unique vsp and vlp profiles. Some of the polymorphism evident in Fig. 4 could be due to heteroduplex formation or to amplification of intergenic sequences if adjacent silent genes were arrayed in opposite orientations. Nevertheless, identical profiles resulted from each of three independent PCR experiments.
FIG. 4.
PCR profiles of vsp and vlp genes of relapsing fever Borrelia species. PCRs with primer sets specific for B. hermsii HS1 vlp subfamilies α through δ (A through D, respectively) and vsp family (E) were performed with DNAs isolated from the indicated Borrelia species. PCRs were analyzed on 4% polyacrylamide gels. Lanes 1 through 8, the indicated isolates of B. hermsii; lanes 9 through 11, relapsing fever agents B. turicatae (B. tur.), B. parkeri (B. park.), and B. crocidurae (B. croc.), respectively; lanes 12, B. anserina (B. ans.); lanes 13, B. coriaceae (B. cor.); lanes 14, from B. burgdorferi (B. burg.) B31. Horizontal bars designate the three groups of related B. hermsii isolates. The sizes (in base pairs) of selected HaeIII-digested φX170 DNA molecular weight standards (mws) are shown on the left.
Two other relapsing fever Borrelia species from the western United States were tested (Fig. 4). B. parkeri yielded a product only with the vlpα subfamily primers, and B. turicatae yielded a product only with the vlpγ subfamily primers. B. crocidurae, a relapsing fever agent from the Mediterranean region, and B. coriaceae, the putative agent of epizootic bovine abortion isolated from California, generated products from both the vlpβ and vlpγ subfamily primer sets. B. burgdorferi, the agent of Lyme borreliosis, and B. anserina, the agent of avian borreliosis, were negative for all five PCRs.
The PCR analysis discussed above utilized conserved 5′ and 3′ sequences to amplify the central variable regions of all the vsp and vlp genes. Many of the individual products were predicted to be nearly the same size and not resolvable, even by polyacrylamide gel electrophoresis. Thus, the observed number of PCR fragments generated from the HS1 strain (Fig. 4) was less than the number of known genes (Fig. 2). To further compare the vsp and vlp repertoires of isolates, portions of the PCRs of Fig. 4 were digested with restriction endonucleases prior to gel electrophoresis. Because the most variable regions of these genes were amplified, we expected this subsequent RFLP analysis to increase observable differences among isolates. Restriction enzymes with known sites in HS1 sequences were chosen. The RFLP profiles for the vsp family are shown in Fig. 5; the results for the vlp subfamilies are not shown. The RFLP patterns generated from the type strain HS1 included all of the fragments predicted from the known sequences of the 23 genes analyzed and a few fragments presumably derived from other vsp and vlp genes. The RFLP patterns of the seven previously uncharacterized B. hermsii isolates were compared to that of HS1 by calculating an estimate of genetic distance, S, the proportion of restriction fragments shared by the pair of isolates (37). S values range from 1.0 (complete identity) to 0 (total dissimilarity). For comparison, RFLP analysis was also conducted with a 659-bp fragment amplified from the flagellin gene of each of the eight B. hermsii isolates. RFLP analyses of two separate sets of PCRs were identical.
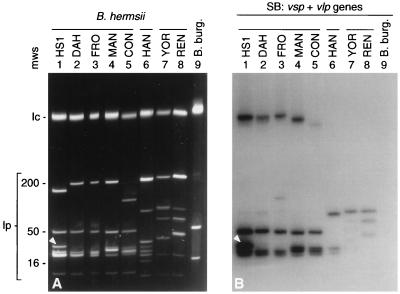
FIG. 5.
vsp gene family RFLP profiles of eight B. hermsii isolates. Gene fragments amplified by PCR with vsp family-specific primers were digested with DdeI (A) or RsaI (B) and analyzed on ethidium bromide-stained 3% agarose gels. Lanes 1 through 8, the indicated B. hermsii isolates (horizontal bars designate the three groups of related isolate); lanes mws, HaeIII-digested φX170 DNA molecular weight standards.
Several points can be made from the results (Fig. 5; Table 3). B. hermsii DAH was identical and B. hermsii FRO was nearly identical to the type strain HS1 across the entire vsp and vlp repertoire. B. hermsii MAN and CON were related to HS1 (S = 0.83 and 0.77, respectively, for the entire repertoire). These related isolates originated from eastern Washington state, except for the central California isolates MAN and CON (Fig. 1). B. hermsii YOR (from northern California) and REN (from northcentral Washington) were identical to each other but were not closely related to HS1. B. hermsii HAN (from northern Idaho) was not similar to any of the other isolates and yielded fewer total restriction digest products. The greatest heterogeneity among all these isolates was seen in genes of the smallest subfamily, vlpγ. Genetic variability among vsp and vlp genes appeared to be much greater than that among the flagellin genes of the eight isolates. B. hermsii HS1, DAH, FRO, MAN and CON were identical in their flagellin RFLP profiles for all four restriction enzymes used. B. hermsii HAN, YOR, and REN were identical and differed from the others by a single RsaI site. The nucleotide sequence of a 220-bp portion of the flagellin gene of B. hermsii YOR has previously been determined; it differs from the HS1 flagellin gene sequence at five nucleotide positions (2.3% base substitution) (39).
TABLE 3.
RFLP S values of seven B. hermsii isolates compared to the type strain HS1
| Genetic locus |
S value fora:
|
||||||
|---|---|---|---|---|---|---|---|
| DAH | FRO | MAN | CON | HAN | YOR | REN | |
| vsp family | 1.0 (27) | 0.96 (27) | 0.96 (25) | 0.67 (24) | 0.56 (16) | 0.55 (24) | 0.55 (24) |
| vlp family | |||||||
| α subfamily | 1.0 (15) | 1.0 (15) | 0.73 (15) | 0.71 (13) | 0.40 (8) | 0.71 (16) | 0.71 (16) |
| β subfamily | 1.0 (36) | 0.94 (32) | 0.83 (34) | 0.89 (38) | 0.57 (27) | 0.54 (26) | 0.54 (26) |
| γ subfamily | 1.0 (11) | 0.91 (12) | 0.57 (10) | 0.91 (12) | 0.30 (9) | 0.07 (17) | 0.07 (17) |
| δ subfamily | 1.0 (23) | 1.0 (23) | 0.86 (21) | 0.65 (11) | 0.61 (13) | 0.62 (16) | 0.62 (16) |
| Total | 1.0 (112) | 0.96 (109) | 0.83 (105) | 0.77 (98) | 0.54 (73) | 0.53 (99) | 0.53 (99) |
| Flagellin gene | 1.0 (23) | 1.0 (23) | 1.0 (23) | 1.0 (23) | 0.93 (22) | 0.93 (22) | 0.93 (22) |
Data are the proportions of restriction fragments shared by the indicated isolate and HS1. Values range from 1.0 (identical to HS1) to 0 (totally dissimilar to HS1). See Materials and Methods for details. Parenthetical data are the total numbers of different restriction fragments from each isolate. The numbers of fragments for HS1 were the same as those for the DAH isolate.
Linear plasmid location of the vsp and vlp gene families.
Silent and expressed vsp and vlp genes of B. hermsii HS1 have previously been mapped to linear plasmids (27, 40). To see whether this is true for the other isolates, PFGE was performed with genomic DNA from each of the eight isolates, followed by Southern hybridization with HS1 vsp and vlp gene probes. The ethidium bromide-stained gel showed that the linear chromosome and linear plasmid profiles of the eight isolates correlated with the classification described above for the vsp and vlp gene family repertoire. Isolates HS1, DAH, FRO, MAN and CON formed one group, YOR and REN formed a second group, and the unique HAN isolate formed a third group (Fig. 6A). Among the HS1 group, the largest linear plasmid (180 to 200 kb) of the CON isolate was noticeably smaller than those of the other four isolates. Strain HS1 had a linear plasmid of about 40 kb that was not present in the others (Fig. 6). This difference, however, was due to a size increase of the HS1 expression plasmid from 30 to 40 kb, which occurred during in vitro passage, probably as a result of intragenic plasmid recombination (2).
FIG. 6.
(A) PFGE gel showing linear-chromosome (lc) and linear-plasmid (lp) profiles of eight B. hermsii isolates (lanes 1 through 8) and high-passage B. burgdorferi (B. burg.) B31 (lane 9). Arrowheads indicate the HS1 expression plasmid. During in vitro passage after isolation, the size of this plasmid increased by about 10 kb. (B) Southern blot (SB) probed with the total collection of PCR-amplified B. hermsii HS1 vsp and vlp gene fragments (vsp + vlp genes). The sizes (in kilobases) of selected lambda DNA concatemers and the 16-kb linear plasmid of B. burgdorferi B31, used as molecular weight standards (mws), are shown on the left. Horizontal bars designate the three groups of related B. hermsii isolates.
PCR-amplified gene fragments of the HS1 vsp family and each of four vlp subfamilies hybridized to two to four linear plasmids of approximately 30 to 50 kb in the five HS1-related isolates and more weakly to linear plasmids of 30 to 90 kb in the three isolates more distantly related to HS1 (23). Thus, genes of the vsp family and of each vlp subfamily are apportioned to different linear plasmids; in other words, individual linear plasmids typically contain a mixture of vsp and vlp genes. Although not all of these plasmids contained representatives of every group, even the smallest vlp subfamily, vlpγ (Fig. 2B), mapped to at least two linear plasmids. A composite in which the hybridization probe was a mixture of the products of all five vsp and vlp PCRs from HS1 is shown in Fig. 6B. The hybridization patterns and linear-plasmid profiles are consistent with and thus reinforce the grouping of isolates based on vsp and vlp polymorphisms. Individual vlp subfamily probes (but not the vsp family probe) also hybridized weakly to the chromosomes of the five related strains or to a DNA species that comigrated with the chromosome on PFGE gels. Whether this was due to the occurrence of vlp-related sequences on the chromosome or minor amplification of an unrelated chromosomal segment during PCR generation of the probes is not known.
DISCUSSION
The antigenic variation of B. hermsii, revealed by many years of studying the type strain HS1, is due to frequent gene conversions in which an expressed gene at a recombinogenic telomeric expression locus is replaced by a different, previously silent gene (27, 40). The 23 vsp and vlp genes of B. hermsii HS1 that we compared compose five distinct groups, based on nucleotide sequence similarity (Fig. 2). In an earlier study, the order of vsp and vlp genes expressed during relapsing fever did not follow a preordained program nor was it completely random (3, 8, 53). The three most common first-relapse serotypes, however, resulted from switches to vlp7 (α subfamily), vsp2, or vlp17 (δ subfamily). Since Vsp proteins or Vlp proteins of the same subfamily can share epitopes (9), switching to an antigenically unrelated gene of another family or subfamily may be important in prolonging infection.
All previous work with the vsp and vlp genes of B. hermsii has been done with HS1, the type strain. This study is the first investigation at the population level of these two B. hermsii gene families that underlie the outer surface protein antigenic variation which occurs during relapsing fever. Allelic polymorphism of immunogenic outer surface protein genes at the population level has previously been observed in viruses (1, 44, 45), protozoa (16), and bacteria (12), including the circular plasmid-borne gene encoding outer surface lipoprotein OspC of the Lyme disease spirochete B. burgdorferi. Like the Vsp and Vlp lipoproteins of B. hermsii, OspC appears to be the predominant outer surface lipoprotein during initial infection of a mammal (20, 38, 49), and different isolates of B. burgdorferi exhibit extensive allelic polymorphism of this gene (24, 31, 52, 54, 55, 58). To extend the comparison further, ospC of B. burgdorferi has previously been shown to be similar in sequence to vsp33 of B. hermsii HS1 (17, 34).
We compared the vsp and vlp repertoires of seven isolates of B. hermsii from California, Idaho, and Washington (47) to that of the HS1 type strain, originally isolated in 1968 near Spokane, Wash. (56) (Fig. 1). This comparison revealed another type of genetic variation of vsp and vlp genes, polymorphism at the population level. The polymorphism was not continuous but discrete; the eight isolates appeared to represent three groups. The HS1-related group contained DAH, FRO, MAN, CON, and the type strain. These five had not only 77% or greater RFLP similarities in vsp and vlp profiles but also identical flagellin gene RFLP profiles (Fig. 5; Table 3) and similar genome profiles, with vsp and vlp genes located on linear plasmids of similar sizes (Fig. 6). Of these five isolates, B. hermsii DAH was identical to HS1 in its vsp and vlp profile. For unknown reasons which do not involve apparent changes in vsp or vlp gene repertoire, laboratory passage of B. hermsii HS1 (as well as Lyme disease Borrelia spp.) can result in loss of infectivity for mammals and serial passage in the vertebrate host can result in loss of infectivity for ticks (6, 46). Thus, low-passage, fully infectious B. hermsii DAH should be useful in studying relapsing fever pathogenesis and transmission, because the vsp- and vlp-specific antisera and sequence data accrued from many years of work with the type strain can potentially be utilized. The type strain-related group also included B. hermsii FRO, which was 96% identical to HS1 in its vsp and vlp profile, and the MAN and CON isolates, which were 83 and 77% identical to HS1, respectively. A second group was composed of B. hermsii YOR and REN. These two isolates had identical vsp and vlp profiles, which were only 53% similar to the HS1 profile, and identical genome profiles. The third group was represented by a single isolate, B. hermsii HAN, which had a vsp and vlp profile that was only 54% similar to that of HS1 and had a unique genome profile.
There are interesting parallels between the antigenic variation of this spirochete at the clonal level and that at the population level. A single B. hermsii cell contains distinct groups of outer surface antigen genes (Fig. 2), and the total population appears to be made up of distinct groups of isolates with different allelic repertoires of these genes. Just as the ability of a single spirochete to switch expression among antigenically distinct vsp and vlp genes allows escape from an individual host’s immune response, allelic polymorphism or genetic variability of vsp and vlp genes within the total spirochete population may help to evade herd immunity (21, 22). This may be particularly important for a parasite, such as B. hermsii, that is transmitted within discrete enzootic foci by a nest-dwelling tick.
A population composed of independent strains with nonoverlapping repertoires of polymorphic antigenic determinants is characteristic of several pathogens. African trypanosomes, which exhibit multiphasic antigenic variation of variable surface glycoproteins (Vsg proteins) during infection in a manner strikingly similar to that of B. hermsii (11, 27), provide one example. Populations of Trypanosoma species are made up of different serodemes, or strains with distinct repertoires of vsg alleles (35), which are analogous to the three groups of B. hermsii delineated in this study. A stable collection of strains with allelic differences in immunogenic surface antigen genes also typifies populations of Plasmodium falciparum (variable antigen types) and the bacterium Neisseria meningitidis (polymorphic epitopes of the outer membrane protein PorA) (21). Immune selection by the host has previously been proposed as the driving force that organizes a parasite population into independently transmitted strains that do not share alleles. According to this model, exposure to one strain leads to complete or partial cross-protection against all members of the same strain but the host remains susceptible to other strains circulating in the herd (21). A second model is suggested by the fact that the three species of North American relapsing fever borreliae have complete specificity for three species of Ornithodoros ticks (6). Likewise, the groups of B. hermsii detected here could have resulted from coevolution with distinct, reproductively isolated strains of the O. hermsi vector. Although little is known about the population genetics of O. hermsi, the coexistence of all three B. hermsii groups within the same enzootic focus (Fig. 1) appears to be inconsistent with this hypothesis.
Because the population structure described here has previously been observed for both sexual (frequently recombining) and clonal (rarely recombining) pathogens, the B. hermsii groups we detected cannot be assumed to consist of independent clonal lineages. Nevertheless, our results suggest that B. hermsii, like B. burgdorferi (10, 18, 30), has a clonal population structure. Clonality of bacteria at the population level refers to infrequent genetic exchange and recombination between different cell lines (50, 51). One criterion of a clonal population is linkage disequilibrium, the nonrandom association of genetic markers. Although only eight isolates were examined and their individual vsp and vlp gene sequences were not compared, evidence of linkage disequilibrium was detected. Three B. hermsii groups were apparent by RFLP analysis of the vsp and vlp genes, and this conformed with the classification based on other markers, including RFLP patterns of the flagellin gene, PFGE genome profiles, and hybridization patterns of the two multigene families (Fig. 6). In addition, previous characterizations of plasmid and total protein profiles, Western and Southern blot reactivities, genomic DNA RFLP profiles, and PCRs of vlp7 and vlp21 genes of the eight isolates were consistent with the classification based on vsp and vlp polymorphisms (46, 47). Further evidence for or against linkage disequilibrium and a clonal population structure will come from comparing phylogenetic trees of the vsp and vlp gene sequences with those of other genetic markers, such as 16S DNA coding for rRNA and flagellin genes, as well as from other population genetics analyses, such as multilocus enzyme electrophoresis. A second characteristic of clonal populations is the ability to recover isolates of identical genotypes over large geographic areas and long periods (50). Perhaps most telling are the allopatric northern Californian YOR and Washington state REN isolates, which appeared to be identical in vsp and vlp genes even though they were separated by over 400 miles (Fig. 1). B. hermsii is not spread rapidly by birds or other migratory hosts, suggesting that this dispersal required many years and generations. The apparent genetic identity of the sympatric B. hermsii HS1 and DAH isolates, which were isolated from the Spokane, Wash., area 24 years apart, is also evidence of infrequent gene flow.
Although the eight isolates, except for HS1 and HAN, have not been cloned by limiting dilution, it is unlikely that the vsp and vlp polymorphism seen is due to the isolates being composed of a mixture of clones. Rather, the results indicate that the human blood samples from which the new isolates were cultured contained clonal populations, as evidenced by the identity of independent, geographically and temporally distant isolates. The isolation procedure itself likely entails cloning. Greater than 99% of the population in a spirochetemia express the same lipoprotein gene (41), indicating they are the progeny of a single cell. During isolation, spirochetemic human blood was first used to infect mice and spirochetemic mouse blood was then inoculated into BSK II medium (47). As evidence of this, clones of B. hermsii HAN were identical to the parent isolate in our analyses.
Simple genetic drift might be expected to affect all the vsp and vlp genes equally, with the consequence that the estimated genetic distance between isolates would be equivalent for each multigene family and subfamily. In fact, marked differences were seen. For example, within the HS1-related group, the vsp genes of the MAN isolate had an S value of 0.96 by RFLP to the HS1 vsp genes; however, the S value for their vlpγ genes was only 0.57 (Table 3). Conversely, the vsp genes of the CON isolate had an S value of only 0.67 to the HS1 vsp genes, whereas the S value for their vlpγ genes was 0.91. A greater disparity was evident in the vlpα and vlpγ genes of the group composed of the identical YOR and CON isolates, compared to those of HS1. Their vlpα genes had an S value of 0.71, but the S value for their vlpγ genes was only 0.07. The reason for the apparent inconsistency in genetic relatedness across the vsp family and vlp subfamilies is unclear at this level of analysis. One possibility is that genetic exchange can occur between isolates. Evidence for possible horizontal transfer of plasmid-borne outer surface protein gene sequences of B. burgdorferi has previously been reported (18, 25, 31, 32). Alternatively, intra- and interplasmid recombination events (28, 41, 42) that differentially affect the multigene families in clones may result in different mutation rates.
The following relapsing fever Borrelia species contained sequences that were PCR amplified with the B. hermsii HS1 vlp subfamily-specific primer sets indicated (Fig. 4): B. turicatae (γ subfamily) and B. parkeri (α subfamily), which (like B. hermsii) are from the western United States; and B. crocidurae (β and γ subfamilies), which is from the Mediterranean region. The suspected agent of epizootic bovine abortion in the western United States, B. coriaceae, also yielded a product with the vlpβ and vlpγ subfamily PCR primers. In a previous study, HS1 vlp7 and vlp21 gene probes (α subfamily) hybridized weakly to B. turicatae and B. coriaceae DNAs (47). Our results support the conclusion (47) that B. coriaceae is more closely related to B. hermsii than is B. anserina, the cause of avian borreliosis, or B. burgdorferi, neither of which were positive in any of the PCRs. RFLP analysis of the PCR products derived from the other Borrelia species examined revealed that they were not highly similar to the analogous B. hermsii products (23).
Finally, an ongoing controversy in molecular evolution is the degree to which genetic variation, such as vsp and vlp polymorphism, is selected, in this case by the host’s immune system (positive or Darwinian selection), or results from random fixation via genetic drift (neutral theory of evolution) (26). Evidence has previously been found for positive selection of B. burgdorferi ospC variants (54). An ideal test case of the positive-selection hypothesis may be provided by the vsp and vlp genes, because relapsing fever spirochetes rely on them to thwart the mammalian immune response and thus to prolong infection and increase the likelihood of transmission. Answers to these and other questions raised by this study await nucleotide sequence analysis of these two multigene families in multiple B. hermsii isolates.
ACKNOWLEDGMENTS
Donald Anderson, Sacred Heart Medical Center, Spokane, Wash., provided infected human blood samples from which three of the B. hermsii isolates used in this study were cultured. We are grateful to Robert Karstens for technical assistance; to Robert Belland, Patricia Rosa, and John Swanson for reviewing the manuscript; and to Robert Evans and Gary Hettrick for help with preparing figures.
This work was supported in part by NIH grant AI24424 to A.G.B.
REFERENCES
- 1.Balanant J, Guillot S, Candrea A, Delpeyroux F, Crainic R. The natural genomic variability of poliovirus analyzed by a restriction fragment length polymorphism assay. Virology. 1991;184:645–654. doi: 10.1016/0042-6822(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 2.Barbour, A. G. Unpublished data.
- 3.Barbour A G. Clonal polymorphism of surface antigens in a relapsing fever Borrelia species. In: Jackson G G, Thomas H, editors. Bayer-Symposium VIII: the pathogensis of bacterial infections. Berlin, Germany: Springer-Verlag; 1985. pp. 235–245. [Google Scholar]
- 4.Barbour A G. Antigenic variation of a relapsing fever Borrelia species. Annu Rev Microbiol. 1990;44:155–171. doi: 10.1146/annurev.mi.44.100190.001103. [DOI] [PubMed] [Google Scholar]
- 5.Barbour A G, Burman N, Carter C J, Kitten T, Bergström S. Variable antigen genes of the relapsing fever agent Borrelia hermsii are activated by promoter addition. Mol Microbiol. 1991;5:489–493. doi: 10.1111/j.1365-2958.1991.tb02132.x. [DOI] [PubMed] [Google Scholar]
- 6.Barbour A G, Hayes S F. Biology of Borrelia species. Microbiol Rev. 1986;50:381–400. doi: 10.1128/mr.50.4.381-400.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbour, A. G., and B. I. Restrepo. Unpublished data.
- 8.Barbour A G, Stoenner H G. Antigenic variation of Borrelia hermsii. UCLA Symp Mol Cell Biol. 1985;20:123–135. [Google Scholar]
- 9.Barstad P A, Coligan J E, Raum M G, Barbour A G. Variable major proteins of Borrelia hermsii. Epitope mapping and partial sequence analysis of CNBr peptides. J Exp Med. 1985;161:1302–1314. doi: 10.1084/jem.161.6.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boerlin P, Peter O, Bretz A-G, Postic D, Baranton G, Piffaretti J-C. Population genetic analysis of Borrelia burgdorferi isolates by multilocus enzyme electrophoresis. Infect Immun. 1992;60:1677–1683. doi: 10.1128/iai.60.4.1677-1683.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borst P, Greaves D R. Programmed gene rearrangements altering gene expression. Science. 1987;235:658–667. doi: 10.1126/science.3544215. [DOI] [PubMed] [Google Scholar]
- 12.Brunham R C, Plummer F A, Stephens R S. Bacterial antigenic variation, host immune response, and pathogen-host coevolution. Infect Immun. 1993;61:2273–2276. doi: 10.1128/iai.61.6.2273-2276.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burgdorfer W, Barbour A G, Hayes S F, Benach J L, Grunwaldt E, Davis J P. Lyme disease—a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 14.Burman N, Bergström S, Restrepo B I, Barbour A G. The variable antigens Vmp7 and Vmp21 of the relapsing fever bacterium Borrelia hermsii are structurally analogous to the VSG proteins of the African trypanosome. Mol Microbiol. 1990;4:1715–1726. doi: 10.1111/j.1365-2958.1990.tb00549.x. [DOI] [PubMed] [Google Scholar]
- 15.Cadavid D, Pennington P M, Kerentseva T A, Bergström S, Barbour A G. Immunologic and genetic analyses of VmpA of a neurotropic strain of Borrelia turicatae. Infect Immun. 1997;65:3352–3360. doi: 10.1128/iai.65.8.3352-3360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carcy B, Bonnefoy S, Schrevel J, Mercereau-Puijalon O. Plasmodium falciparum: typing of malaria parasites based on polymorphism of a novel multigene family. Exp Parasitol. 1995;80:463–472. doi: 10.1006/expr.1995.1058. [DOI] [PubMed] [Google Scholar]
- 17.Carter C J, Bergström S, Norris S J, Barbour A G. A family of surface-exposed proteins of 20 kilodaltons in the genus Borrelia. Infect Immun. 1994;62:2792–2799. doi: 10.1128/iai.62.7.2792-2799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dykhuizen D E, Polin D S, Dunn J, Wilske B, Preac-Mursic V, Dattwyler R J, Luft B J. Borrelia burgdorferi is clonal: implications for taxonomy and vaccine development. Proc Natl Acad Sci USA. 1993;90:10163–10167. doi: 10.1073/pnas.90.21.10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferdows M S, Barbour A G. Megabase-sized linear DNA in the bacterium Borrelia burgdorferi, the Lyme disease agent. Proc Natl Acad Sci USA. 1989;86:5969–5973. doi: 10.1073/pnas.86.15.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fung B P, McHugh G L, Leong J M, Steere A C. Humoral immune response to outer surface protein C of Borrelia burgdorferi in Lyme disease: role of the immunoglobulin M response in the serodiagnosis of early infection. Infect Immun. 1994;62:3213–3221. doi: 10.1128/iai.62.8.3213-3221.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta S, Maiden M C J, Feavers I M, Nee S, May R M, Anderson R M. The maintenance of strain structure in populations of recombining infectious agents. Nat Med. 1996;2:437–442. doi: 10.1038/nm0496-437. [DOI] [PubMed] [Google Scholar]
- 22.Gupta S, Trenholme K, Anderson R M, Day K P. Antigenic diversity and the transmission dynamics of Plasmodium falciparum. Science. 1994;263:961–963. doi: 10.1126/science.8310293. [DOI] [PubMed] [Google Scholar]
- 23.Hinnebusch, B. J. Unpublished data.
- 24.Jauris-Heipke S, Fuchs R, Motz M, Preac-Mursic V, Schwab E, Soutschek E, Will G, Wilske B. Genetic heterogeneity of the genes coding for the outer surface protein C (OspC) and the flagellin of Borrelia burgdorferi. Med Microbiol Immunol (Berlin) 1993;182:37–50. doi: 10.1007/BF00195949. [DOI] [PubMed] [Google Scholar]
- 25.Jauris-Heipke S, Liegl G, Preac-Mursic V, Robler D, Schwab E, Soutschek E, Will G, Wilske B. Molecular analysis of genes encoding outer surface protein C (OspC) of Borrelia burgdorferi sensu lato: relationship to ospA genotype and evidence of lateral gene exchange of ospC. J Clin Microbiol. 1995;33:1860–1866. doi: 10.1128/jcm.33.7.1860-1866.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura M. The neutral theory of molecular evolution. Cambridge, United Kingdom: Cambridge University Press; 1983. [Google Scholar]
- 27.Kitten T, Barbour A G. Juxtaposition of expressed variable antigen genes with a conserved telomere in the bacterium Borrelia hermsii. Proc Natl Acad Sci USA. 1990;87:6077–6081. doi: 10.1073/pnas.87.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitten T, Barrera A V, Barbour A G. Intragenic recombination and a chimeric outer membrane protein in the relapsing fever agent Borrelia hermsii. J Bacteriol. 1993;175:2516–2522. doi: 10.1128/jb.175.9.2516-2522.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lane R S, Burgdorfer W, Hayes S F, Barbour A G. Isolation of a spirochete from the soft tick, Ornithodoros coriaceus: a possible agent of epizootic bovine abortion. Science. 1985;230:85–87. doi: 10.1126/science.3898367. [DOI] [PubMed] [Google Scholar]
- 30.Lebech A-M, Hansen K, Wilske B, Theisen M. Taxonomic classification of 29 Borrelia burgdorferi strains isolated from patients with Lyme borreliosis: a comparison of five different phenotypic and genotypic typing schemes. Med Microbiol Immunol (Berlin) 1994;183:325–341. doi: 10.1007/BF00196683. [DOI] [PubMed] [Google Scholar]
- 31.Livey I, Gibbs C P, Schuster R, Dorner F. Evidence for lateral transfer and recombination in OspC variation in Lyme disease Borrelia. Mol Microbiol. 1995;18:257–269. doi: 10.1111/j.1365-2958.1995.mmi_18020257.x. [DOI] [PubMed] [Google Scholar]
- 32.Marconi R T, Samuels D S, Landry R K, Garon C F. Analysis of the distribution and molecular heterogeneity of the ospD gene among the Lyme disease spirochetes: evidence for lateral gene exchange. J Bacteriol. 1994;176:4572–4582. doi: 10.1128/jb.176.15.4572-4582.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marconi R T, Samuels D S, Schwan T G, Garon C T. Identification of a protein in several species of Borrelia related to OspC of the Lyme disease spirochetes. J Clin Microbiol. 1993;31:2577–2583. doi: 10.1128/jcm.31.10.2577-2583.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margolis N, Hogan D, Cieplak W, Jr, Schwan T G, Rosa P A. Homology between Borrelia burgdorferi OspC and members of the family of Borrelia hermsii variable major proteins. Gene. 1994;143:105–110. doi: 10.1016/0378-1119(94)90613-0. [DOI] [PubMed] [Google Scholar]
- 35.Masake R A, Nantulya V M, Musoke A J, Moloo S K, Nguli K. Characterization of Trypanosoma congolense serodemes in stocks isolated from cattle introduced onto a ranch in Kilifi, Kenya. Parasitology. 1987;94:349–357. doi: 10.1017/s0031182000054007. [DOI] [PubMed] [Google Scholar]
- 36.Meier J T, Simon M I, Barbour A G. Antigenic variation is associated with DNA rearrangements in a relapsing fever Borrelia. Cell. 1985;41:403–409. doi: 10.1016/s0092-8674(85)80013-1. [DOI] [PubMed] [Google Scholar]
- 37.Nei M, Li W-H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Padula S J, Sampieri A, Dias F, Szczepanski A, Ryan R W. Molecular characterization and expression of p23 (OspC) from a North American strain of Borrelia burgdorferi. Infect Immun. 1993;61:5097–5105. doi: 10.1128/iai.61.12.5097-5105.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Picken R N. Polymerase chain reaction primers and probes derived from flagellin gene sequences for specific detection of the agents of Lyme disease and North American relapsing fever. J Clin Microbiol. 1992;30:99–114. doi: 10.1128/jcm.30.1.99-114.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plasterk R H A, Simon M I, Barbour A G. Transposition of structural genes to an expression sequence on a linear plasmid causes antigenic variation in the bacterium Borrelia hermsii. Nature. 1985;318:257–263. doi: 10.1038/318257a0. [DOI] [PubMed] [Google Scholar]
- 41.Restrepo B I, Barbour A G. Antigen diversity in the bacterium B. hermsii through “somatic” mutations in rearranged vmp genes. Cell. 1994;78:867–876. doi: 10.1016/s0092-8674(94)90642-4. [DOI] [PubMed] [Google Scholar]
- 42.Restrepo B I, Carter C J, Barbour A G. Activation of a vmp pseudogene in Borrelia hermsii: an alternate mechanism of antigenic variation during relapsing fever. Mol Microbiol. 1994;13:287–299. doi: 10.1111/j.1365-2958.1994.tb00423.x. [DOI] [PubMed] [Google Scholar]
- 43.Restrepo B I, Kitten T, Carter C J, Infante D, Barbour A G. Subtelomeric expression regions of Borrelia hermsii linear plasmids are highly polymorphic. Mol Microbiol. 1992;6:3299–3311. doi: 10.1111/j.1365-2958.1992.tb02198.x. [DOI] [PubMed] [Google Scholar]
- 44.Robertson D L, Sharp P M, McCautchan F E, Hahn B H. Recombinations in HIV-1. Nature. 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 45.Saitou N, Nei M. Polymorphism and evolution of influenza A virus genes. Mol Biol Evol. 1986;3:57–74. doi: 10.1093/oxfordjournals.molbev.a040381. [DOI] [PubMed] [Google Scholar]
- 46.Schwan, T. G. Unpublished data.
- 47.Schwan T G, Gage K L, Hinnebusch J. Analysis of relapsing fever spirochetes from the western United States. J Spirochetal Tick-borne Dis. 1995;2:3–8. [Google Scholar]
- 48.Schwan, T. G., and B. J. Hinnebusch. Unpublished data.
- 49.Schwan T G, Piesman J, Golde W T, Dolan M C, Rosa P A. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci USA. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selander R K, Musser J M. Population genetics of bacterial pathogens. In: Iglewski B H, Clark V L, editors. Molecular basis of bacterial pathogenesis. London, United Kingdom: Academic Press; 1990. pp. 11–36. [Google Scholar]
- 51.Smith J M, Smith N H, O’Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stevenson B, Barthold S W. Expression and sequence of outer surface protein C among North American isolates of Borrelia burgdorferi. FEMS Microbiol Lett. 1994;124:367–372. doi: 10.1111/j.1574-6968.1994.tb07310.x. [DOI] [PubMed] [Google Scholar]
- 53.Stoenner H G, Dodd T, Larsen C. Antigenic variation in B. hermsii. J Exp Med. 1982;156:1297–1311. doi: 10.1084/jem.156.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Theisen M, Borre M, Mathiesen M J, Mikkelson B, Lebech A-M, Hansen K. Evolution of the Borrelia burgdorferi outer surface protein OspC. J Bacteriol. 1995;177:3036–3044. doi: 10.1128/jb.177.11.3036-3044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Theisen M, Frederiksen B, Lebech A-M, Vuust J, Hansen K. Polymorphism in ospC gene of Borrelia burgdorferi and immunoreactivity of OspC protein: implications for taxonomy and for use of OspC protein as a diagnostic antigen. J Clin Microbiol. 1993;31:2570–2576. doi: 10.1128/jcm.31.10.2570-2576.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson R S, Burgdorfer W, Russell R, Francis B J. Outbreak of tick-borne relapsing fever in Spokane County, Washington. JAMA. 1969;210:1045–1050. [PubMed] [Google Scholar]
- 57.Tibayrenc M. Population genetics of parasitic protozoa and other microorganisms. Adv Parasitol. 1995;36:48–115. doi: 10.1016/s0065-308x(08)60490-x. [DOI] [PubMed] [Google Scholar]
- 58.Wilske B, Preac-Mursic V, Jauris S, Hofmann A, Pradel I, Soutschek E, Schwab E, Will G, Wanner G. Immunological and molecular polymorphisms of OspC, an immunodominant major outer surface protein of Borrelia burgdorferi. Infect Immun. 1993;61:2182–2191. doi: 10.1128/iai.61.5.2182-2191.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]