Abstract
Purpose:
This study sought to identify β-catenin targets that regulate desmoid oncogenesis and determine whether external signaling pathways, particularly those inhibited by sorafenib (e.g., PDGFRβ), affect these targets to alter natural history or treatment response in patients.
Experimental Design:
In vitro experiments utilized primary desmoid cell lines to examine regulation of β-catenin targets. Relevance of results was assessed in vivo using Alliance trial A091105 correlative biopsies.
Results:
CTNNB1 knockdown inhibited hypoxia-regulated gene expression in vitro and reduced levels of HIF1α protein. ChIP-seq identified ABL1 as a β-catenin transcriptional target that modulated HIF1α and desmoid cell proliferation. Abrogation of either CTNNB1 or HIF1A inhibited desmoid cell–induced VEGFR2 phosphorylation and tube formation in endothelial cell co-cultures. Sorafenib inhibited this activity directly but also reduced HIF1α protein expression and c-Abl activity while inhibiting PDGFRβ signaling in desmoid cells. Conversely, c-Abl activity and desmoid cell proliferation were positively regulated by PDGF-BB. Reduction in PDGFRβ and c-Abl phosphorylation was commonly observed in biopsy samples from patients after treatment with sorafenib; markers of PDGFRβ/c-Abl pathway activation in baseline samples were associated with tumor progression in patients on the placebo arm and response to sorafenib in patients receiving treatment.
Conclusions:
The β-catenin transcriptional target ABL1 is necessary for proliferation and maintenance of HIF1α in desmoid cells. Regulation of c-Abl activity by PDGF signaling and targeted therapies modulates desmoid cell proliferation, thereby suggesting a reason for variable biologic behavior between tumors, a mechanism for sorafenib activity in desmoids, and markers predictive of outcome in patients.
Translational Relevance.
Desmoid-type fibromatosis is a mesenchymal neoplasm that can regress spontaneously or be locally aggressive, leading to significant morbidity. Desmoid tumors are universally associated with activation of β-catenin signaling, but secondary factors that modulate differences in tumor growth and affect response to drugs such as sorafenib have not been identified despite extensive genomic analyses. Here we demonstrate that PDGF receptor signaling can modulate desmoid cell proliferation, at least in part by promoting activity of the β-catenin transcriptional target c-Abl. We show that activity of the PDGFRβ/β-catenin/c-Abl signaling axis is associated with outcome both in desmoids managed with active observation and those treated with sorafenib by analyzing biopsies from Alliance A091105, a phase III, placebo-controlled trial evaluating sorafenib response in desmoid patients. This is the first study to provide mechanistic evidence leading to rational identification of predictive markers in desmoid patients.
Introduction
Desmoid-type fibromatosis (desmoids) is a mesenchymal neoplasm, which does not progress to high-grade disease or metastasize but can be locally aggressive. Surgery has been the standard treatment, but complete resection often requires morbid procedures and ∼30% of patients have local recurrence (1). Therefore, alternatives to surgery have been extensively studied. Large series of patients managed without medical or surgical intervention have shown that in approximately 50% of patients, the disease remains stable or regresses spontaneously without treatment (2, 3). For patients with progressive tumors at risk for significant symptoms such as limb contracture or life-threatening intestinal fistulas, a range of cytotoxic (e.g., doxorubicin-based regimens) and antihormonal therapies have been used (4). More recently, phase III randomized-controlled trials have shown significant responses to sorafenib or the γ-secretase inhibitor nirogacestat, although the mechanisms by which these drugs exert their effects are unclear (5, 6).
Activated β-catenin is associated with initiation of desmoids. Most desmoids have sporadic mutations in exon 3 of its encoding gene CTNNB1 (7, 8). These mutations prevent β-catenin phosphorylation, usually observed in the context of canonical Wnt activation. GSK3β phosphorylation of these sites (T41 or S45) leads to degradation of β-catenin when complexed with APC, Axin, and GSK3β. Exon 3 mutations therefore lead to accumulation of β-catenin in cancer cells (9). In a minority of cases, germline or sporadic loss-of-function mutations in the adenomatous polyposis coli (APC) gene prevent the turnover of β-catenin (10, 11). In the unphosphorylated state caused by CTNNB1 or APC mutation, β-catenin translocates into the nucleus, where it acts as a transcription factor for oncogenes such as CCND1 and MVC in most systems. These canonical targets do not appear to be upregulated in desmoid cells, and several genes upregulated in desmoids have not been linked to Wnt/β-catenin signaling (12). These suggest that alternative β-catenin transcriptional targets may be important for desmoid initiation.
Here, we sought to identify a subset of these targets to better understand how β-catenin may promote desmoid formation. Because secondary genomic events affecting desmoid progression and response to therapy have not been identified, we hypothesized that external signaling may modulate activity of the β-catenin targets and alter biologic behavior of tumors. Using an in vitro system, we examined whether one such pathway targeted by sorafenib, PDGFRβ signaling, could act in this manner. Our results identify a mechanism by which PDGF signaling affects oncogenic activity of ABL1, identified as a novel β-catenin transcriptional target, and HIF1α in desmoid cells. Analyses of pre- and post-treatment biopsies suggest that sorafenib may inhibit this pathway in desmoids and that activation of PDGF signaling may be a marker predictive of progression in untreated tumors.
Materials and Methods
Sample acquisition
Tumor and normal tissues were flash frozen at the time of surgery in patients who consented to Memorial Sloan Kettering Cancer Center (MSKCC) Institutional Review Board–approved protocol #02–060 (Supplementary Materials and Methods Table S1). Flash frozen, pre- and post-treatment biopsies from a subset of patients (n = 17) enrolled on Alliance for Clinical Trials in Oncology trial A091105 and who consented to A091105-ST1 were obtained by image guidance prior to treatment and between days 8 and 11 after initiation of sorafenib or placebo. Baseline FFPE specimens were available from a larger cohort (Supplementary Materials and Methods Tables S2 and S3; ref. 6). All patient procedures were performed in accordance with the Declaration of Helsinki and U.S. Common Rule; informed written consent was obtained from each subject or subject's guardian.
Cell culture
Desmoid cells were grown in a 1:1 mixture of DMEM high glucose and F12 medium with 2 mmol l-glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin and maintained in a 37°C incubator at 5% CO2. Puromycin (1 μg/mL), blasticidin (2 μg/mL), or G418 (1 mg/mL) was used for plasmid selection. Primary desmoid cell lines (DES9525, DES8163, and DES3726), derived from resected specimens by collagenase disaggregation, were grown in 50% FBS during initial passages and transitioned to 20% FBS for standard conditions; all three cell lines utilized in this manuscript were derived from lower extremity tumors. Desmoid cell lines were validated by Sanger sequencing (see below); only those demonstrating ∼50% variant allele frequency were utilized for long-term cultures and repeat sequencing was performed every 10 to 15 passages (Supplementary Materials and Methods Fig. S1). Desmoid cells were immortalized using Lenti-hTERT-Neo Virus and antibiotic selection between 5 and 8 passages after establishment (1 mg/mL neomycin; Applied Biological Materials) and tested for mycoplasma using MycoAlert-Plus (Lonza; August 2020). Effects of sorafenib (Selleckchem) and PDGF-BB (R&D Systems) were examined in the presence of 10% and 5% FBS, respectively.
HUVEC cells (Lonza) were cultured in Endothelial Cell Growth Media (Cedarlane Laboratories). Desmoid and HUVEC cells were co-cultured by seeding 18 × 103 desmoid cells onto 24-well inserts for 24 hours before transfer to 24-well plates coated with growth factor–reduced Matrigel (Corning) and containing 4 × 104 HUVEC cells in 400 μL desmoid medium with 1% FBS. Conditioned medium (CM) was collected after 48 hours of desmoid cell culture; the amount of CM used to treat HUVEC cells was adjusted on the basis of desmoid cell numbers at the time of collection.
Nucleic acid preparation and mutation, transcriptome, and transcription factor binding analysis
DNA and RNA were prepared using DNeasy and RNeasy Kits (Qiagen). CTNNB1 exon 3 was amplified from genomic DNA by PCR HotStart Taq DNA Polymerase (Qiagen) and primer sequences 5′-TGATGGAGTTGGACATGGCC-3′, 5′-CTCATACAGGACTTGGGAGG-3′. After purification (QiaQuick; Quiagen) PCR products were isolated by gel electrophoresis. Sanger sequencing was performed as reported previously (13). Tumor specimens were macrodissected from cryomolds and RNA samples prepared from specimens containing >90% tumor as determined by hematoxylin and eosin staining. RNA from tumor or cell line samples was isolated using RNeasy Kits (Qiagen).
Gene expression was analyzed by U133A 2.0 arrays (Affymetrix) to which cRNA (prepared as described previously) was hybridized (14). Array data were quantified and normalized using standard R/Bioconductor packages and differential expression was assessed by the LIMMA empirical Bayes method (15, 16). Libraries from cell-line RNA were prepared for sequencing on the Illumina HiSeq 4000 platform as described by Chi and colleagues (17). The raw count matrix generated by HTSeq was processed using the R/Bioconductor package DESeq, which both normalizes the full dataset and analyzes differential expression between sample groups (18, 19). Gene set enrichment analysis (GSEA) was performed using the Fast-GSEA (fgsea) package from R/Bioconductor (100,000,000 permutation setting; https://www.biorxiv.org/content/10.1101/060012v3). We used the bedtools coverage tool and RNAseq.bam files to calculate depth of coverage over the CTNNB1 locus for transcript ENST00000349496 (gencode version V19) and assess variant allele frequency; median read depth was ∼300 and 95% of bases were covered between ∼160 and 630× (20).
Nucleic acids for chromatin immunoprecipitation sequencing (ChIP-seq) were prepared from 1 × 106 cells using the SimpleChIP Enzymatic Kit (Cell Signaling Technology) with β-catenin antibody D10A8 (6 μg; Cell Signaling Technology) and amplified using Maxima SYBR Green Mix (Thermo Fisher Scientific). Immunoprecipitated DNA was quantified by PicoGreen, size evaluated by Agilent BioAnalyzer, and Illumina sequencing libraries prepared using the KAPA HTP Library Preparation Kit (Kapa Biosystems KK8234) per manufacturer's instructions with 0.1 to 16.8 ng input DNA and eight cycles of PCR. Barcoded libraries were run on the HiSeq 4000 in a PE100 run using the HiSeq 3000/4000 SBS Kit (Illumina; average of 30 million paired reads). Duplicate .fasta files from ChIP and input samples were concatenated and aligned to hg19 using bowtie2 with default settings (21). Peaks were annotated by MACS2 (broad peak calling, P-value = 10−7, data were normalized to the larger dataset and mfold parameter between 5 and 50; ref. 22). To generate bedgraph files for UCSC genome browser visualization, .sam files were processed using Homer software (final parameters of makeUCSCfile program, -fsize 5e7 -res 10; ref. 23). ChIP-qPCR used the same β-catenin antibody and the following primers for ABL1: F:TGCCATGTCCGAGAATCTTT and R: ACTGGATCTGTGCATGATTAACT.
In vitro analyses
Lentiviral infections, cell proliferation assays, and RT-PCR analyses were performed as described previously (14, 24, 25). Gene expression was inhibited using lentiviral pLKO.1 vectors (Horizon Discovery or MSKCC Gene Editing and Screening facility) carrying short hairpin RNA (shRNA) directed against relevant genes (Supplementary Materials and Methods Table S4) or a scramble control (#RHS6848; ref. 26). Overexpression of HIF1A utilized LentiORF [OHS5898–224626816; control RFP ORF (Horizon Discovery)]. Assays were performed 4 to 6 days following infection to allow for antibiotic selection. Cell proliferation was measured using the CyQuant Cell Proliferation Assay Kit (Invitrogen) with 103 cells seeded in 96-well plates. IC50 was calculated as the drug concentration reducing DNA content by 50% in cultures compared with untreated controls. For transcript analyses, cDNA was synthesized using cDNA Reverse Transcription Kit (Thermo Fisher Scientific) and amplified using Taqman Gene Expression Master Mix (Applied Biosystems) with gene-specific primers (ABL1 Taqman Probe Hs01104728_m1, HIF1a Hs00153153_m1, CTNNB1 Hs00355045_m1 Thermo Fisher Scientific Taqman Gene Expression Assays) on a ViiA 7 Real-Time PCR System (Thermo Fisher Scientific).
For luciferase reporter assays, cells were seeded on 48-well plates at 75% confluence before transfection with hypoxia-responsive luciferase and control Renilla constructs (Cigal HIF1 pathway reporter assay; Qiagen) using Lipofectamine 2000 (Invitrogen). Luciferase assays were performed 48 hours after transfection using the Dual-Glo Luciferase Assay System (Promega). Results were reported as the ratio of inducible to non-inducible signal. Endothelial tube formation was quantified manually in 6 to 10 representative fields of view using light microscopy 24 hours after co-culture of desmoid and HUVEC cells.
Immunoblotting, IHC, and antibodies
For immunoblots, cultured cells were lysed in 50 mmol/L Tris-HCl pH 7.4, 250 mmol/L NaCl, 5 mmol/L EDTA, and 0.5% NP-40. Supernatants were acquired from tissue preparations for immunoblot analysis after fresh-frozen biopsy specimens or macrodissected cryomolds were dissociated in resin/RIPA buffer (Cytiva Sample Grinding Kit) on ice. Equal quantities of protein (15–20 μg) were resolved using Mini-PROTEAN TGX Precast Protein Gels or Tetra Vertical Electrophoresis Cell (Bio-Rad). After transfer to 0.2 μmol/L polyvinylidene difluoride (PVDF) membrane (Bio-Rad) and blocking, membranes were incubated with antibodies in Starting Block T20 (Thermo Fisher Scientific) overnight, then horseradish peroxidase-conjugated mouse or rabbit IgG before development using an Enhanced Chemiluminescence (ECL) Kit (GE Healthcare), SuperSignal West Pico PLUS, or Femto Chemiluminescent Substrate (Thermo Fisher Scientific) according to the manufacturer's instructions. Primary antibodies against the following proteins and isoforms were used: HIF1α [Cell Signaling Technology (CST) 36169], β-catenin (CST 9562), pAkt (Ser473) (CST 9271), Akt (CST 9272), pPDGFRβ (Y751) (CST 3161), PDGFRβ (CST 3169), pVEGFR2 (Y1175) (CST 2478), c-Abl (CST 2862), pc-Abl Y412 (Millipore 07–788), pCrkl (CST 3181), pCrkII (CST 3491), β-actin (CST 8457), and vinculin (Abcam ab91459). Protein levels were quantified using ImageJ. Phospho-receptor tyrosine kinase array kits were evaluated per the manufacturer's instructions in the presence or absence of 40 μmol/L sorafenib (R&D Systems).
For IHC, paraffin-embedded tissue sections were cut at 5 μmol/L and heated at 58°C for 1 hour. Samples were loaded into Leica Bond RX and sections were dewaxed at 72°C before being pretreated with EDTA-based epitope retrieval ER2 solution (Leica, AR9640) for 20 minutes at 100°C. Slides were incubated with anti-FosB (0.5 μg/mL, CST 2251), anti-EGR1 (5 μg/mL, R&D Systems MAB2818), or anti-p21 (1: 150, EA10, Millipore OP64) for 60 minutes (30 minutes for p21), then with Leica Bond Polymer anti-rabbit HRP [included in Polymer Refine Detection Kit (Leica DS9800)] for 8 minutes, with anti-rat or mouse secondary antibody (Vector Laboratories, BA-4000) for 8 minutes, with Leica Bond Polymer anti-rabbit HRP, then mixed DAB reagent (Polymer Refine Detection Kit) for 10 minutes, followed by hematoxylin (Refine Detection Kit) counterstaining for 10 minutes. After staining, sample slides were washed in water, dehydrated using ethanol gradient (70%, 90%, 100%), washed three times in HistoClear II (National Diagnostics, HS-202), and mounted in Permount (Thermo Fisher Scientific, SP15).
Statistical analysis
Unless otherwise noted, in vitro experiments were performed at least three times. Figures show mean ± SE. For experiments performed using multiple cell lines and/or constructs, test results describe the range of means and least stringent P-value applicable to all cell lines and/or constructs examined. Statistical associations were assessed using the Fisher exact test for categorical variables, Welch t test for continuous variables and Kaplan–Meier curves, log-rank testing, and Cox regression for time-to-event variables. Best response was defined by RECIST v1.1 and progression-free survival (PFS) by time to clinical progression or >15% growth in size of the index lesion (as of May 4, 2023).
Data and resource availability
The data generated in this study are publicly available and accessible in Gene Bank Omnibus (GEO study GSE237692). Cell lines and lentiviral constructs are available on request. RRID for commercially available resources are noted in Supplementary Materials and Methods Table S5.
Results
β-Catenin regulates HIF1α-mediated paracrine signaling by desmoid cells
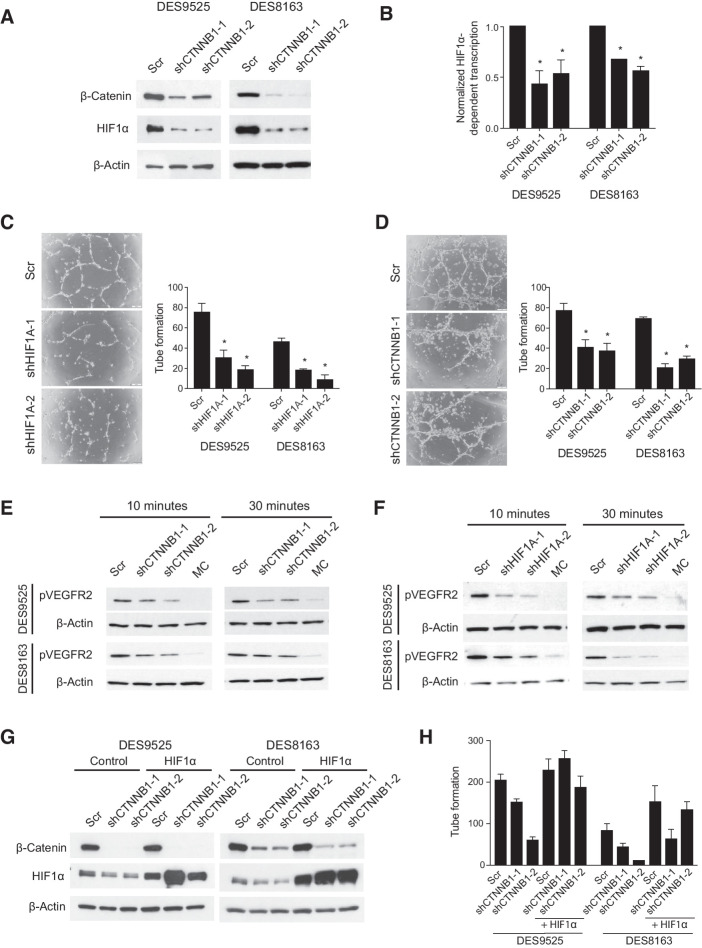
Assessment of transcriptomes of DES9525 cells in which CTNNB1 was knocked down (vs. scramble controls) using GSEA demonstrated dysregulation of multiple gene sets associated with hypoxia and HIF1 signaling (Supplementary Table S1). Supervised clustering of desmoid tumors (n = 65) and normal mesenchymal tissues (n = 8 skeletal muscle and 9 fat samples), analyzed by U133A 2.0 gene array, revealed that HIF1- and angiogenesis-associated genes separated tumors and normal tissue with 100% accuracy (Supplementary Table S2; Supplementary Fig. S1). HIF1α was also detectable in immunoblots of a small set of desmoids at levels proportional to β-catenin (Supplementary Fig. S2A). In vitro, HIF1α protein was detected in desmoid cell lines at levels higher than in control mesenchymal stem cells (Supplementary Fig. S2B). Assessment of these cells by immunoblot after CTNNB1 knockdown showed not only decreased β-catenin but also a decrease in HIF1α protein and HIF1 luciferase reporter activity compared with scramble controls (47%–57%, P ≤ 0.045 and 33%–44%, P ≤ 0.0029 reporter activity in DES9525 and 8163 cells, respectively; Fig. 1A and B).
Figure 1.
Desmoid cells promote endothelial cell tube formation by activating VEGFR2 in a HIF1A- and CTNNB1-dependent manner. A and B, Effect of CTNNB1 knockdown on (A) HIF1α protein and (B) mRNA expression assessed by dual luciferase assay. C and D, Effect of knockdown of (C) HIF1A or (D) CTNNB1 in DES9525 or DES8163 desmoid cells on co-culture–induced HUVEC tube formation. Representative photos show HUVECs following co-culture with DES9525 cells transduced as indicated; quantitated results average triplicate experiments. E and F, Effect of knockdown of (E) CTNNB1 or (F) HIF1A in desmoid cells on phosphorylation of VEGFR2 in HUVEC cells treated with desmoid-conditioned media for the indicated times; MC indicates nonconditioned media control. G and H, Effect of HIF1α overexpression on (G) HIF1α protein expression and (H) representative trial of co-culture–induced HUVEC tube formation in desmoid cells in which CTNNB1 is knocked down. Control for HIF1α overexpression is red fluorescent protein (RFP, Ctrl; *, P < 0.05).
Reduction in HIF1A using shRNA did not inhibit desmoid cell proliferation (Supplementary Fig. S2C and S2D), although knockdown of CTNNB1 reproducibly reduced cell proliferation by >90% (P ≤ 0.032; Supplementary Fig. S2E). Because HIF1α signaling was also regulated by CTNNB1 in desmoid cells, we examined desmoid cells co-cultured with HUVECs; a 3-fold increase in HUVEC tube formation was noted in co-cultures compared with HUVECs grown alone; conditioned media from desmoid cultures also induced VEGFR2 phosphorylation in HUVEC cells (Supplementary Fig. S3). Such paracrine effects appeared to depend on both HIF1α and β-catenin; HIF1A knockdown in desmoid cells led to decreased HUVEC tube formation: 61% to 77% in DES9525 cells (P ≤ 0.011) and 61% to 82% in DES8163 cells (P ≤ 0.0054; Fig. 1C). Abrogated CTNNB1 expression in desmoid cells similarly resulted in fewer HUVEC tubes when the endothelial cells were co-cultured with DES9525 (48%–52% reduction, P < 0.001) or DES8163 cells (56%–75%, P < 0.001; Fig. 1D). Conditioned media from both CTNNB1 and HIF1A knockdowns failed to induce VEGFR2 phosphorylation in HUVECs as robustly as media from control cultures and knockdown of CTNNB1 was associated with decreased VEGF secretion as detected by ELISA (Fig. 1E and F; Supplementary Fig. S4A). Enforced expression of HIF1A rescued compromised HUVEC tube formation and VEGF secretion caused by CTNNB1 knockdown, suggesting paracrine effects of desmoid cells on HUVECs were mediated by β-catenin in a HIF1-dependent manner (Fig. 1G and H; Supplementary Fig. S4B).
Sorafenib inhibits PDGFRβ on desmoid cells to modulate paracrine signaling and proliferation
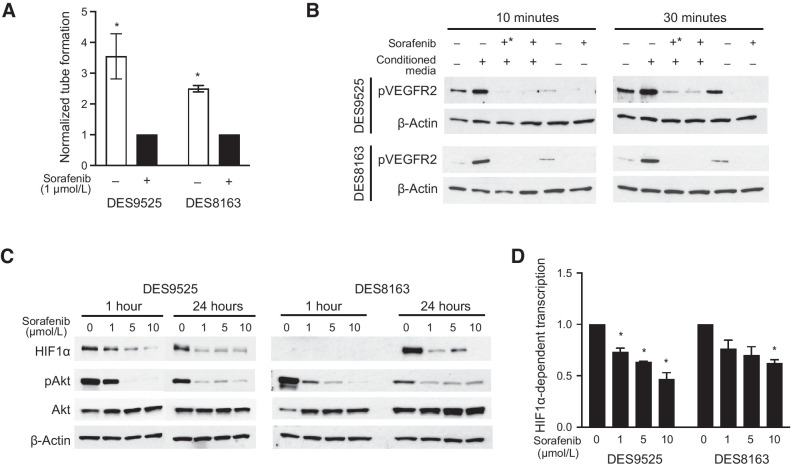
We examined whether sorafenib, a VEGFR2 inhibitor, prevented β-catenin/HIF1α paracrine signaling and proliferation. Relatively high concentrations were required to inhibit proliferation of desmoid cells grown under standard conditions (IC50 11 μmol/L and 13 μmol/L, respectively, for DES9525 and DES8163; Supplementary Fig. S5). Addition of sorafenib to desmoid/HUVEC co-cultures inhibited tube formation at significantly lower doses, however. Sorafenib (1 μmol/L) inhibited HUVEC tube formation by 72% in DES9525 co-cultures and 60% in DES8163 co-cultures (P ≤ 0.045; Fig. 2A). Similar concentrations of sorafenib were sufficient to suppress activation of VEGFR2 on HUVECs induced by desmoid-conditioned media when sorafenib was added either to desmoid cultures themselves or to endothelial cell cultures after treatment with conditioned media (Fig. 2B). Addition of sorafenib to desmoid cell cultures was associated with decreases in HIF1α protein expression at concentrations at least as low as 1 μmol/L and inhibited HIF1-mediated transcription as assessed by luciferase reporter assays (by 54% and 38% in DES9525 and DES8163, respectively, grown in 10 μmol/L, P ≤ 0.015; Fig. 2C and D).
Figure 2.
Sorafenib inhibits HIF1-mediated paracrine VEGF signaling. Effect of sorafenib on (A) the number of HUVEC tubes formed following co-culture with desmoid cells, (B) VEGFR2 phosphorylation in HUVEC cells treated with desmoid-conditioned media (+, desmoids grown in the presence of sorafenib; +*, sorafenib added to conditioned media after it was added to HUVEC cultures), (C) HIF1α expression and Akt phosphorylation, and (D) HIF1-mediated transcription as defined by a luciferase reporter assay (*, P < 0.05).
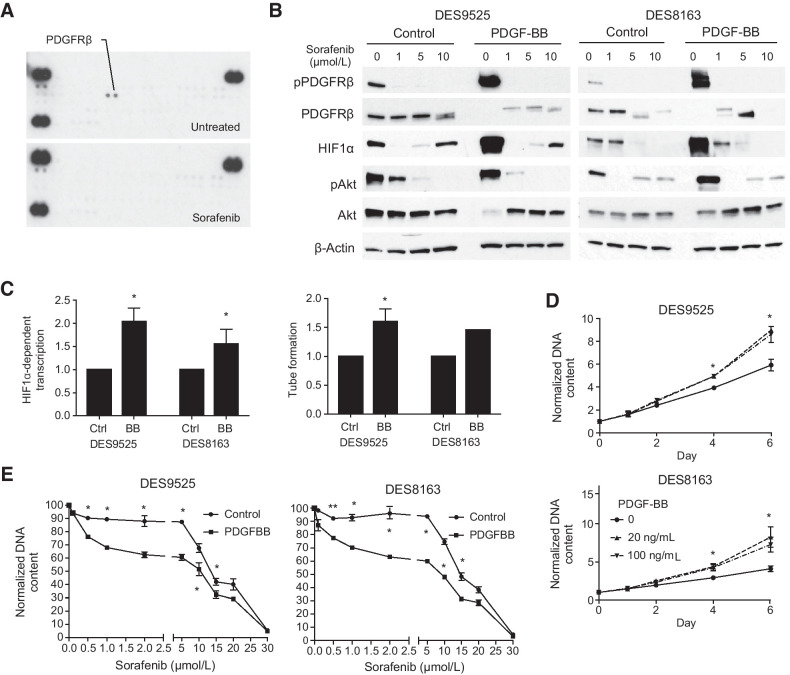
Sorafenib did not alter cellular levels of β-catenin itself (not shown) and given its ability to inhibit multiple tyrosine kinases, we examined which were inhibited by the drug in desmoid cells and whether such inhibition could modulate downstream effects of β-catenin. Sorafenib specifically reduced PDGFRβ phosphorylation in desmoid cells (Fig. 3A). Stimulation of PDGFRβ signaling by addition of PDGF-BB to cultures resulted in increased PDGFRβ and Akt phosphorylation and increased HIF1α and HIF1-mediated transcription (by 100% in DES9525 and 56% in DES8163, P = 0.027 and 0.096, respectively; Fig. 3B and C). The effect on PDGFR pathway activation was completely inhibited by sorafenib at concentrations as low as 1 μmol/L (Fig. 3B). Addition of PDGF-BB to desmoid and HUVEC co-cultures also increased HUVEC tube formation (by 34% in DES9525, P = 0.043 and 44% in a representative trial of DES8163; Fig. 3C). In addition to modulating paracrine effects, PDGF-BB was mitogenic, increasing desmoid proliferation by 51% and 50% in DES9525 and DES8163, respectively (P ≤ 0.034 in each cell line; Fig. 3D). This was associated with sensitization to sorafenib, with reduced IC50s observed in both DES9525 and DES8163 cell lines after addition of PDGF-BB to cell cultures (from 14 μmol/L to 10 μmol/L in DES9525 and 15 μmol/L to 10 μmol/L in DES8163 after addition of 10 ng PDGF-BB; Fig. 3E). Together, these data indicate PDGFRβ signaling supports desmoid paracrine signaling and proliferation and is inhibited by sorafenib.
Figure 3.
PDGF signaling is a sorafenib target that regulates HIF1α accumulation, enhances paracrine effects, and increases proliferation. A, Sorafenib (40 μmol/L) decreases phosphorylation of PDGFRβ as detected by receptor tyrosine kinase array. B, Immunoblot analysis of activation of PDGFRβ signaling pathway components and HIF1α expression in desmoid cells treated with sorafenib in the presence and absence of PDGF-BB. C–E, Effect of PDGF-BB on (C) relative HIF1α transcriptional activity assessed by luciferase reporter assay (20 ng/mL PDGF-BB), relative rate of HUVEC tube formation in desmoid co-cultures, and (D) desmoid cell proliferation as assessed by DNA quantification. E, Relative proliferation of desmoid cells grown in the presence or absence of PDGF-BB (20 ng/mL) at varying concentrations of sorafenib normalized to that in cultures without the drug at 72 hours after treatment. All quantitated results average triplicate experiments (*, P < 0.05; **, P < 0.005).
c-Abl is a transcriptional target of β-catenin and its activity is modulated by PDGFRβ signaling
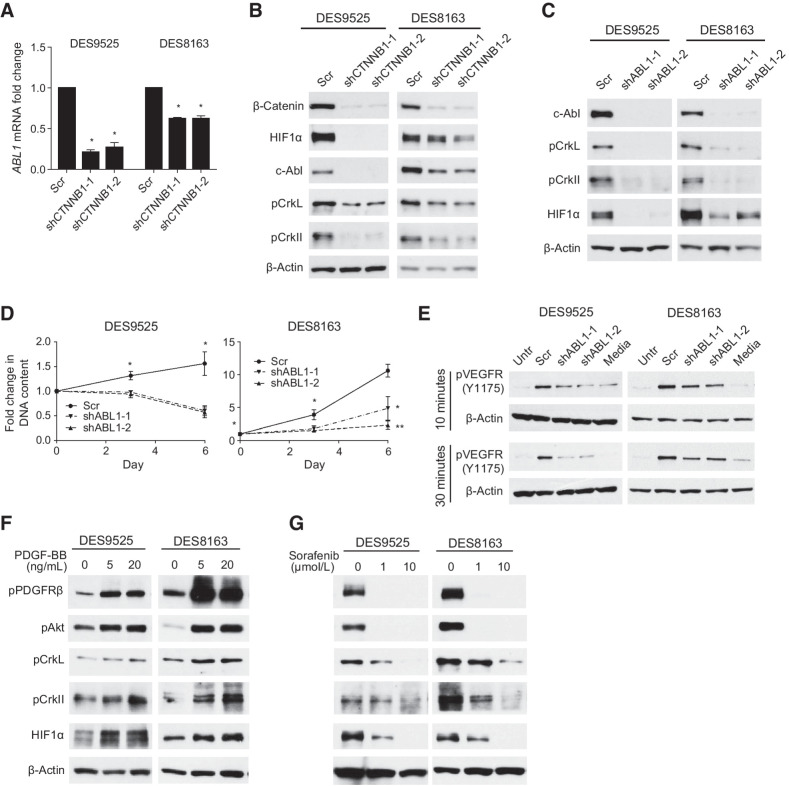
Knockdown of β-catenin had minimal effect on HIF1A transcription (not shown). ChIP-seq identified a potential β-catenin binding site in the region of ABL1, previously reported to regulate HIF1α translation (Supplementary Fig. S6A; ref. 27). This was confirmed by ChIP-qPCR and expression of the gene was 54% lower in CTNNB1 knockdown cells compared with scramble control based on RNA-seq (Supplementary Fig. S6B; Supplementary Table S3). CTNNB1 knockdown caused 73% to 79% downregulation of ABL1 mRNA expression by RT-PCR in DES9525 (P ≤ 0.0069) and 38% downregulation in DES8163 (P ≤ 0.0089; Fig. 4A). Immunoblotting confirmed this change was reflected at the protein level and demonstrated concordant decreases in phosphorylation of c-Abl targets Crkl and CrkII (Fig. 4B). HIF1α expression was also downregulated in ABL1-deficient cells (Fig. 4C).
Figure 4.
β-Catenin transcriptional target ABL1 is necessary for cell proliferation and HIF1α accumulation in desmoid cells. A and B, Effect of CTNNB1 knockdown on (A) relative transcript levels of ABL1 (detected by RT-PCR) and (B) expression of c-Abl and activation of its canonical phosphorylation targets. C–E, Effect of ABL1 knockdown on (C) activation of c-Abl phosphorylation targets, (D) proliferation (as measured by DNA content) of desmoid cells (average of triplicate experiment; *, P < 0.05; **, P < 0.005) and (E) VEGFR2 phosphorylation detected by immunoblot. F and G, Effect of (F) PDGF-BB and (G) sorafenib on activation of canonical PDGFRβ and c-ABL pathway components as measured by immunoblot.
Inhibition of ABL1 also completely suppressed proliferation in DES9525 and reduced proliferation by >50% in DES8163 cells (Fig. 4D). Significant death in ABL1 knockdowns complicated the ability to perform co-culture experiments to examine the role of c-Abl in regulating HUVEC tube formation. However, supporting a role for c-Abl/HIF1 in paracrine signaling to HUVEC cells, increased VEGFR2 phosphorylation was not observed when HUVEC cells were treated with conditioned media from desmoids in which ABL1 was knocked down (Fig. 4E). To determine whether c-Abl is regulated by PDGFRβ, we examined the effect of PDGF-BB on phosphorylation of canonical c-Abl targets; increased Crkl and CrkII phosphorylation were observed, supporting such an interaction (Fig. 4F). Sorafenib inhibited activation of these pathways (Fig. 4G). These results suggest c-Abl is a novel transcriptional target of β-catenin, is essential for desmoid survival, and regulates desmoid secretions via HIF1α.
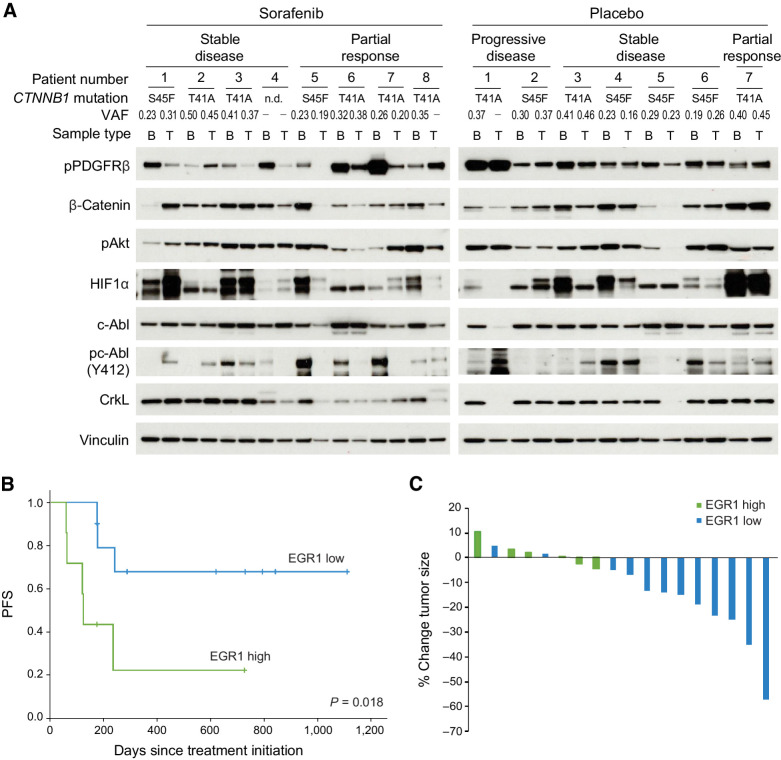
Analysis of specimens from alliance protocol A091105 suggests a role for PDGFRβ/c-Abl signaling in prediction of desmoid progression and response to sorafenib in patients
To determine whether PDGFRβ/β-catenin/c-Abl/HIF1α signaling was targeted by sorafenib in vivo, we examined biopsy samples from a double-blind, randomized phase III placebo-controlled clinical trial of sorafenib in desmoid patients (Alliance A091105, NCT02066181). Fresh-frozen samples from pre- and post-treatment biopsies were available in 8 patients treated with sorafenib (4 with partial response and 4 with stable disease) and 7 patients on the placebo arm (2 with progressive disease, 4 with stable disease, and 1 with partial response). Patients treated with sorafenib had a median difference in normalized pPDGRβ values of −0.66 (IQR: −3.77–0.00) in post-treatment biopsies compared with placebo patients (−0.29; IQR: 0.47–0.27; P = 0.30), and 6 of 8 (75%) sorafenib patients had a >50% decrease in pPDGFRβ compared with baseline compared with only 1 of 7 (14%) in patients receiving placebo (P = 0.041; Fig. 5A; Supplementary Fig. S7A). Decreased phosphorylation of c-Abl was also more common in post-treatment biopsies in patients treated with sorafenib compared with placebo [median change −0.49 (IQR: −1.51–0.13) vs. 0.09 (IQR: 0.00–0.44); P = 0.12; 62% vs. 14% of patients with >50% reduction; P = 0.12; Supplementary Fig. S7B and S7C]. Changes in HIF1α expression were not specifically observed in response to sorafenib (decreased in 38% of sorafenib-treated vs. 42% of placebo-treated tumors).
Figure 5.
Inhibition of PDGFRβ/c-Abl signaling may correlate with response to sorafenib in desmoid patients. A, Activation of PDGFRβ, c-Abl, and Akt and expression of HIF1α and β-catenin as detected by immunoblot in baseline (B) and post-treatment (T) biopsies in individual patients treated on the placebo and sorafenib arms of Alliance A091105. Best response (partial response, stable or progressive disease) as assayed by RECIST criteria or clinical progression, CTNNB1 mutations and their variant allele frequencies (VAF; identified by RNA-seq) are annotated. Samples with no mutations detected are annotated (-). B, PFS defined by clinical progression or radiographic increase in size (>15% above baseline) in patients and stratified according to high (≥50% of desmoid cells) or low (<50% of cells positive) levels of nuclear EGR1 staining on IHC of baseline samples obtained from patients in the placebo arm of the trial. C, Percent change in tumor size after four to five cycles of sorafenib in patients on Alliance A091105. Tumors with ≥90% cells staining for nuclear EGR1 are designated as EGR1 high as opposed to low.
Among patients receiving sorafenib, pronounced decrease in phosphorylation of c-Abl was more commonly observed in patients experiencing partial responses (3 of 4: 100% decrease) than in those with stable disease (2 of 4: 70% decrease, 100% decrease). This difference reflected higher baseline levels of pc-Abl in samples from patients who responded to treatment. In fact, pc-Abl was, on average, 5.3-fold higher in patients with response (after normalization to vinculin, P = 0.036) compared with those with stable disease (Supplementary Fig. S8A–S8C). Phospho-PDGFRβ levels averaged 2.6-fold higher in patients who responded versus those who did not, although this result was not statistically significant (P = 0.20). Interestingly, protein expression of β-catenin tended to decrease in patients treated with sorafenib who responded (3 of 4 vs. 1 of 4 patients with stable disease), although this did not correlate with a decrease in CTNNB1 mutant variant allele frequency as assessed by RNA-seq (Fig. 5A; Supplementary Table S4).
Neither CTNNB1 mutation site nor variant frequency was clearly associated with progression during active observation in the 7 patients with biopsies available. When normalized to vinculin, baseline pPDGFRβ tended to be higher in patients with progressive disease compared with those with stable disease in this cohort, but the small sample size prohibited definitive analysis (3.9-fold; Supplementary Fig. S9A and S9B). RNA-seq on DES9595 cells treated with PDGF-BB identified EGR1 as significantly upregulated after PDGFRβ stimulation and IHC on a desmoid TMA showed pronounced variability in in vivo expression (Supplementary Table S5; Supplementary Fig. S10). We examined EGR1 expression in biopsies from 17 patients treated on the placebo arm of Alliance A091105 (median follow-up 287 days). No patients with increasing tumor size in the cohort remained on trial until formal RECIST progression was noted. If progression was defined as clinical progression or >15% radiographic growth, however, those with EGR1 staining in ≥50% of tumor cells had shorter PFS compared with those with low EGR1 staining (median time to progression 123 days vs. not reached, P = 0.018; Fig. 5B). EGR1 staining appeared to be more predictive in the context of observation than did FosB and p21, markers of TERT-associated senescence, nominated by RNA-seq and GSEA on frozen baseline samples of two progressing and four stable tumors on the placebo arm of the trial as potentially prognostic (Supplementary Fig. S11; Supplementary Table S6). The association of EGR1 and progression while on sorafenib could not be assessed due to lack of events in the sorafenib arm, but EGR1 staining in ≥90% of tumor cells was associated with less tumor shrinkage after four cycles of drug (n = 6, average growth 1.1 ± 3.7%) compared with tumors with less EGR1 staining (n = 6, average shrinkage –17 ± 4.9%; P = 0.02; Fig. 5C). EGR1 staining thresholds used for stratification in the analyses were selected post hoc, but these results nominate PDGFR pathway activation as a biomarker of response to sorafenib in desmoid patients, and of progression in those under active observation.
Discussion
To understand the molecular biology underpinning variations in desmoid behavior and sensitivity to active therapies including sorafenib (7, 8), we investigated the oncogenic targets of β-catenin in desmoids and whether environmental signaling could alter their activity to produce changes in desmoid behavior. Because the genes most differentially regulated in desmoids (compared to normal mesenchymal tissues) do not include canonical β-catenin targets such as CCND1 or AXIN2 (28), we performed RNA-seq on desmoid cells to determine which genes β-catenin may regulate; many were associated with angiogenesis and HIF1 signaling. This was not due to a direct transcriptional effect on HIF1A or HIF1B per our ChIP-seq data; instead, we identified ABL1 as a β-catenin target that according to the literature translationally regulates HIF1 components (27). Both CTNNB1 and ABL1 are necessary for HIF1α accumulation in desmoid cells. We also found that c-Abl was necessary for both HIF1-associated paracrine activity and desmoid cell proliferation. Perhaps even more interestingly, the activity of c-Abl as assessed by phosphorylation of its canonical targets seems to be positively regulated by signaling through PDGFRβ, a protein implicated in desmoid pathogenesis (29). Taken together, these data present a novel model in which oncogenic effects of β-catenin are modulated not by secondary genomic events but instead by environmental signaling, offering an alternative explanation for desmoids’ variable behavior. We confirmed our in vitro results in two cell lines with CTNNB1 S45F mutations; select results were also confirmed in a cell line carrying the CTNNB1 T41A mutation because previous reports have suggested that different β-catenin mutations may affect cells differently (Supplementary Fig. S12; ref. 30). This cell line (DES3276), even after ectopic expression of TERT, has a prolonged doubling time, preventing complete analysis.
It is unclear whether the paracrine effects of HIF1α seen in vitro have clinical relevance. Although HIF1-associated gene expression differentiates desmoids from normal fat and skeletal muscle, whether these are the appropriate normal mesenchymal controls is open to debate. Also, CD34 staining for endothelial cells in banked specimens showed few blood vessels (not shown), and HIF1α protein levels were not altered by treatment in specimens obtained from patients on the sorafenib arm of Alliance trial A091105. Our identification of c-Abl as a potential dependency in desmoids may more accurately inform understanding of how tumors respond to current therapeutic regimens. Both sorafenib and imatinib (used to treat desmoids, particularly in Europe; ref. 31) inhibit PDGFRβ, which activates c-Abl by phosphorylation and can inhibit c-Abl directly. Preclinical data have also identified FAK inhibitors and dasatinib as potential therapies for in vivo validation (32). Both inhibit Src kinase, which, like PDGFRβ, can positively regulate c-Abl via posttranslational modification (Y412 phosphorylation). We show here that dasatinib, which inhibits PDGFRβ, c-Abl, and Src, inhibits proliferation at concentrations >10-fold lower than sorafenib, suggesting targeting such an axis at multiple levels may be even more efficient than targeting PDGFRβ and c-Abl alone (Supplementary Fig. S13). Whether these pathways represent the end targets of other desmoid therapies such as gamma secretase inhibitors is less clear. It is possible, however, as gamma secretase substrates such as CD44 and ephrin family members are highly expressed in desmoid cells (Supplementary Table S4) and each has been associated with regulation of c-Abl and/or Src activity (33–35).
The clinical relevance of a β-catenin/PDGFRβ/c-Abl pathway was interrogated using pre- and post-treatment biopsies from both the sorafenib and placebo arms of Alliance A091105. The limited number of samples prevents us from drawing definitive conclusions and effects of sampling error could be significant. The CTNNB1 mutation could not be detected in a subset of samples analyzed by immunoblot, which could reflect low variant allele frequency, alternate APC mutations that could not be detected in this study because the gene was poorly covered in our RNA-seq analysis (∼25–75×), or sampling error, which would affect the power of our analyses. Nonetheless, PDGFRβ phosphorylation was generally reduced in patients following treatment with sorafenib and baseline levels tended to be higher in patients who responded to the drug compared with those who had stable disease. This was consistent with in vitro findings demonstrating increased sensitivity to sorafenib in cell lines pretreated with PDGF-BB to stimulate PDGFRβ signaling. Perhaps even more convincing, phosphorylation of c-Abl was significantly higher in 3 of 4 patients with partial responses and available samples compared with those with stable disease; in all cases, kinase activation appeared to drop precipitously after treatment. c-Abl phosphorylation as a predictive marker of response may be more accurate than pPDGFRβ if additional pathways sensitive to sorafenib inhibition and upstream of c-Abl in parallel signaling cascades also play a role in desmoid pathogenesis (e.g., ephrins and MET are both inhibited by sorafenib and can phosphorylate c-Abl via Src; refs. 36–38). Interestingly, ephrin regulation of c-Abl is a targetable vulnerability in animal models of APC-driven intestinal tumorigenesis, suggesting interplay of this pathway with β-catenin signaling (39).
Surprisingly, c-Abl itself did not seem to predict progression during active observation, although again small sample size severely limits our ability to draw definitive conclusions. PDGFRβ phosphorylation did seem to be higher in the baseline specimen of one patient whose tumor later progressed. To evaluate the association with outcomes of variations in activity in this pathway in a larger cohort, we identified EGR1 as a gene significantly upregulated in desmoid cells after treatment with PDGF-BB. As EGR1 stains accurately by IHC (40), we utilized this as a surrogate for PDGFRβ stimulation in baseline biopsies from 17 patients treated with placebo in Alliance A091105. In patients with ≥50% of tumor cells staining with EGR1, PFS was significantly shorter than in patients with less robust EGR1 staining. This would be concordant with activation of PDGFRβ signaling predicting biologic behavior during periods of active observation, although the link may not be direct. EGR1 itself can bind to β-catenin to modulate its transcriptional activity (41). In addition, it has been shown in other systems to act as a downstream effector of Src (42). Future studies may assess whether the protein itself plays a functional role in regulation of desmoid oncogenesis.
Although analysis of PFS in patients treated with sorafenib was not feasible due to lack of events, analysis of radiographic growth suggested that high expression of EGR1 was associated with relative resistance to sorafenib. In hepatocellular carcinoma, a similar phenomenon has been noted and appears to reflect activation of parallel, upstream oncogenic pathways less sensitive to sorafenib (e.g., MET/Akt/ERK; ref. 43). We observed PDGFRβ inhibition in patients treated with sorafenib regardless of response. However, precipitous decreases in c-Abl phosphorylation from high baseline and, in fact, any decrease in Akt phosphorylation were observed only in patients with response. Decreases in activation of at least one of these proteins was observed in each of the patients with response (Fig. 5A). Post-treatment biopsies in patients with response had decreases in a median of four of the five markers total β-catenin, pAkt, pc-Abl, pCrkl, and HIF1α versus a median of one in patients with stable disease. These suggest that PDGFRβ inhibition's effects on downstream signaling pathways correlates with sorafenib's induction of significant tumor responses. This result, along with the ability of EGR1 to predict desmoid progression during observation, will need to be validated in secondary cohorts. Validations would be particularly important, since thresholds for defining desmoid subsets with high and low EGR1 staining were determined post hoc, introducing bias.
Together these data identify a mechanism by which external signaling pathways modify activity of β-catenin–mediated oncogenesis in desmoid tumors. In the absence of secondary molecular mutations that can lead to variable behavior during active observation or differences in drug response, environmental regulation may play a dominant role in determining tumor phenotype. Our molecular studies have nominated PDGFRβ signaling as a potential marker of aggressive and resistant disease. If validated, this finding may significantly improve tailoring of therapy for patients with desmoid-type fibromatosis.
Supplementary Material
Supplemental Methods Tables, Supplemental Methods Figures, Supplemental Figures
Gene set enrichment analysis of pathways altered in desmoid cells after CTNNB1 knockdown
Genes included in HIF1- and angiogenesis-related pathways and used for supervised clustering of desmoid tumors. Pathways in which each gene are included are annotated in the table.
Genes noted to be altered in DES9525 after CTNNB1 knockdown as identified by RNA-seq. Fold change is relative to expression in cells treated with scramble control.
CTNNB1 mutations detected in pre- and post-treatment biopsies from patients on ALLIANCE A091105.
Genes with altered expression in DES9525 cells after treatment with PDGF-BB as compared to untreated cells as determined by RNA-seq.
Gene set enrichment analysis performed on RNA-seq from pre-treatment biopsies taken from patients on the placebo arm of ALLIANCE A091105.
Acknowledgments
The work presented here was funded in part by NIH/NCI through U10 CA180821 (Alliance), U10 CA180882 (Alliance), the Cancer Center Support Grant P30 CA008748, SPORE in Soft Tissue Sarcoma P50 CA217694 (to M.R. Hameed, N.P. Agaram, N.D. Socci, L. Qin, S. Singer, A.M. Crago), Surgical Training Grant T32 CA009501 (to A.M. Villano, K. Prendergast), and R37 CA241856 (to A.M. Crago), by the FDA through R01 FD005105 (to M.M. Gounder, A.M. Crago), the American Cancer Society via Mentored Research Scientist Grant MSRG-15–064–01-TBG (to A.M. Crago), Bayer (Alliance), David and Alicia Pinkston (to A.M. Crago), and the Kristen Ann Carr Foundation (to A.M. Crago). ClinicalTrials.gov Identifier: NCT02066181. The authors would like to express appreciation to Jessica Moore for editorial support, Bhumika Jadeja for database support, Drs. Andrew Koff, Ping Chi, and Yueming Li for helpful discussions, Wenfei Kang and Marina Ahser for assistance with IHC, and Thomas Li for assistance with HIF1 experiments. We acknowledge the use of the MSKCC Integrated Genomics Operation Core. We would like to thank the patients who were willing to participate on Alliance trial A091105 and Alliance staff including Sherry Breaux, Sharmila Giri, Blake Waechter, Tiffany Shafer, and Sumithra Mandrekar (https://acknowledgments.alliancefound.org).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
A.M. Villano reports personal fees from Merck outside the submitted work. N.D. Socci reports grants from NIH during the conduct of the study. M.M. Gounder reports grants and personal fees from Bayer during the conduct of the study as well as grants and personal fees from Ayala, Epizyme, Rain Oncology, and Boehringer Ingelheim outside the submitted work; personal fees from Aadi Biosciences, Regeneron, and TYME; and grants from SpringWorks Therapeutics. A.M. Crago reports grants from NCI, FDA, American Cancer Society, and the Kristen Ann Carr Foundation during the conduct of the study and other support from SpringWorks Therapeutics outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
J. Hu: Conceptualization, data curation, formal analysis, supervision, investigation, visualization, methodology, writing–original draft, writing–review and editing. M.R. Hameed: Conceptualization, data curation, investigation, visualization, methodology, writing–review and editing. N.P. Agaram: Data curation, validation, investigation, methodology, writing–review and editing. K.A. Whiting: Data curation, formal analysis, validation, methodology, writing–review and editing. L.-X. Qin: Resources, formal analysis, supervision, validation, methodology, writing–review and editing. A.M. Villano: Resources, data curation, formal analysis, validation, investigation, methodology, writing–review and editing. R.B. O'Connor: Resources, formal analysis, supervision, validation, methodology, writing–review and editing. J.M. Rozenberg: Data curation, formal analysis, investigation, visualization, methodology, writing–review and editing. S. Cohen: Data curation, investigation, writing–review and editing. K. Prendergast: Data curation, writing–review and editing. S. Kryeziu: Data curation, writing–review and editing. R.L. White Jr: Resources, writing–review and editing. M.C. Posner: Resources, writing–review and editing. N.D. Socci: Formal analysis, investigation, methodology, writing–review and editing. M.M. Gounder: Resources, writing–review and editing. S. Singer: Conceptualization, resources, methodology, writing–review and editing. A.M. Crago: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, methodology, writing–original draft, project administration.
References
- 1. Crago AM, Denton B, Salas S, Dufresne A, Mezhir JJ, Hameed M, et al. A prognostic nomogram for prediction of recurrence in desmoid fibromatosis. Ann Surg 2013;258:347–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fiore M, Rimareix F, Mariani L, Domont J, Collini P, Le Pechoux C, et al. Desmoid-type fibromatosis: a front-line conservative approach to select patients for surgical treatment. Ann Surg Oncol 2009;16:2587–93. [DOI] [PubMed] [Google Scholar]
- 3. Colombo C, Fiore M, Grignani G, Tolomeo F, Merlini A, Palassini E, et al. A prospective observational study of active surveillance in primary desmoid fibromatosis. Clin Cancer Res 2022;28:4027–32. [DOI] [PubMed] [Google Scholar]
- 4. de Camargo VP, Keohan ML, D'Adamo DR, Antonescu CR, Brennan MF, Singer S, et al. Clinical outcomes of systemic therapy for patients with deep fibromatosis (desmoid tumor). Cancer 2010;116:2258–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gounder M, Ratan R, Alcindor T, Schoffski P, van der Graaf WT, Wilky BA, et al. Nirogacestat, a gamma-secretase inhibitor for desmoid tumors. N Engl J Med 2023;388:898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gounder MM, Mahoney MR, Van Tine BA, Ravi V, Attia S, Deshpande HA, et al. Sorafenib for advanced and refractory desmoid tumors. N Engl J Med 2018;379:2417–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crago AM, Chmielecki J, Rosenberg M, O'Connor R, Byrne C, Wilder FG, et al. Near universal detection of alterations in CTNNB1 and Wnt pathway regulators in desmoid-type fibromatosis by whole-exome sequencing and genomic analysis. Genes Chromosomes Cancer 2015;54:606–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alman BA, Li C, Pajerski ME, Diaz-Cano S, Wolfe HJ. Increased beta-catenin protein and somatic APC mutations in sporadic aggressive fibromatoses (desmoid tumors). Am J Pathol 1997;151:329–34. [PMC free article] [PubMed] [Google Scholar]
- 9. Macdonald BT, Semenov MV, He X. SnapShot: Wnt/beta-catenin signaling. Cell 2007;131:1204. [DOI] [PubMed] [Google Scholar]
- 10. Couture J, Mitri A, Lagace R, Smits R, Berk T, Bouchard HL, et al. A germline mutation at the extreme 3' end of the APC gene results in a severe desmoid phenotype and is associated with overexpression of beta-catenin in the desmoid tumor. Clin Genet 2000;57:205–12. [DOI] [PubMed] [Google Scholar]
- 11. Li C, Bapat B, Alman BA. Adenomatous polyposis coli gene mutation alters proliferation through its beta-catenin-regulatory function in aggressive fibromatosis (desmoid tumor). Am J Pathol 1998;153:709–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Skubitz KM, Skubitz AP. Gene expression in aggressive fibromatosis. J Lab Clin Med 2004;143:89–98. [DOI] [PubMed] [Google Scholar]
- 13. Pratilas CA, Hanrahan AJ, Halilovic E, Persaud Y, Soh J, Chitale D, et al. Genetic predictors of MEK dependence in non-small cell lung cancer. Cancer Res 2008;68:9375–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brill E, Gobble R, Angeles C, Lagos-Quintana M, Crago A, Laxa B, et al. ZIC1 overexpression is oncogenic in liposarcoma. Cancer Res 2010;70:6891–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003;4:249–64. [DOI] [PubMed] [Google Scholar]
- 16. Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004;3:Article3. [DOI] [PubMed] [Google Scholar]
- 17. Chi P, Chen Y, Zhang L, Guo X, Wongvipat J, Shamu T, et al. ETV1 is a lineage survival factor that cooperates with KIT in gastrointestinal stromal tumours. Nature 2010;467:849–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Putri GH, Anders S, Pyl PT, Pimanda JE, Zanini F. Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics 2022;38:2943–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010;26:841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu T. Use model-based Analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol Biol 2014;1150:81–95. [DOI] [PubMed] [Google Scholar]
- 23. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Singer S, Socci ND, Ambrosini G, Sambol E, Decarolis P, Wu Y, et al. Gene expression profiling of liposarcoma identifies distinct biological types/subtypes and potential therapeutic targets in well-differentiated and dedifferentiated liposarcoma. Cancer Res 2007;67:6626–36. [DOI] [PubMed] [Google Scholar]
- 25. Yuan JS, Reed A, Chen F, Stewart CN Jr. Statistical analysis of real-time PCR data. BMC Bioinf 2006;7:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep 2013;5:1704–13. [DOI] [PubMed] [Google Scholar]
- 27. Sourbier C, Ricketts CJ, Matsumoto S, Crooks DR, Liao PJ, Mannes PZ, et al. Targeting ABL1-mediated oxidative stress adaptation in fumarate hydratase-deficient cancer. Cancer Cell 2014;26:840–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 1999;398:422–6. [DOI] [PubMed] [Google Scholar]
- 29. Alman BA, Naber SP, Terek RM, Jiranek WA, Goldberg MJ, Wolfe HJ. Platelet-derived growth factor in fibrous musculoskeletal disorders: a study of pathologic tissue sections and in vitro primary cell cultures. J Orthop Res 1995;13:67–77. [DOI] [PubMed] [Google Scholar]
- 30. Lazar AJ, Tuvin D, Hajibashi S, Habeeb S, Bolshakov S, Mayordomo-Aranda E, et al. Specific mutations in the beta-catenin gene (CTNNB1) correlate with local recurrence in sporadic desmoid tumors. Am J Pathol 2008;173:1518–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kasper B, Gruenwald V, Reichardt P, Bauer S, Rauch G, Limprecht R, et al. Imatinib induces sustained progression arrest in RECIST progressive desmoid tumours: Final results of a phase II study of the German Interdisciplinary Sarcoma Group (GISG). Eur J Cancer 2017;76:60–7. [DOI] [PubMed] [Google Scholar]
- 32. Mercier KA, Al-Jazrawe M, Poon R, Acuff Z, Alman B. A metabolomics pilot study on desmoid tumors and novel drug candidates. Sci Rep 2018;8:584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bourguignon LY, Zhu H, Shao L, Chen YW. CD44 interaction with c-Src kinase promotes cortactin-mediated cytoskeleton function and hyaluronic acid-dependent ovarian tumor cell migration. J Biol Chem 2001;276:7327–36. [DOI] [PubMed] [Google Scholar]
- 34. Murakami D, Okamoto I, Nagano O, Kawano Y, Tomita T, Iwatsubo T, et al. Presenilin-dependent gamma-secretase activity mediates the intramembranous cleavage of CD44. Oncogene 2003;22:1511–6. [DOI] [PubMed] [Google Scholar]
- 35. Georgakopoulos A, Litterst C, Ghersi E, Baki L, Xu C, Serban G, et al. Metalloproteinase/Presenilin1 processing of ephrinB regulates EphB-induced Src phosphorylation and signaling. EMBO J 2006;25:1242–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol 2008;26:127–32. [DOI] [PubMed] [Google Scholar]
- 37. Mao W, Irby R, Coppola D, Fu L, Wloch M, Turner J, et al. Activation of c-Src by receptor tyrosine kinases in human colon cancer cells with high metastatic potential. Oncogene 1997;15:3083–90. [DOI] [PubMed] [Google Scholar]
- 38. Davy A, Robbins SM. Ephrin-A5 modulates cell adhesion and morphology in an integrin-dependent manner. EMBO J 2000;19:5396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kundu P, Genander M, Straat K, Classon J, Ridgway RA, Tan EH, et al. An EphB-Abl signaling pathway is associated with intestinal tumor initiation and growth. Sci Transl Med 2015;7:281ra44. [DOI] [PubMed] [Google Scholar]
- 40. Myung DS, Park YL, Kim N, Chung CY, Park HC, Kim JS, et al. Expression of early growth response-1 in colorectal cancer and its relation to tumor cell proliferation and apoptosis. Oncol Rep 2014;31:788–94. [DOI] [PubMed] [Google Scholar]
- 41. Lu D, Han C, Wu T. Microsomal prostaglandin E synthase-1 promotes hepatocarcinogenesis through activation of a novel EGR1/beta-catenin signaling axis. Oncogene 2012;31:842–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Qureshi SA, Cao XM, Sukhatme VP, Foster DA. v-Src activates mitogen-responsive transcription factor Egr-1 via serum response elements. J Biol Chem 1991;266:10802–6. [PubMed] [Google Scholar]
- 43. Chen HA, Kuo TC, Tseng CF, Ma JT, Yang ST, Yen CJ, et al. Angiopoietin-like protein 1 antagonizes MET receptor activity to repress sorafenib resistance and cancer stemness in hepatocellular carcinoma. Hepatology 2016;64:1637–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Methods Tables, Supplemental Methods Figures, Supplemental Figures
Gene set enrichment analysis of pathways altered in desmoid cells after CTNNB1 knockdown
Genes included in HIF1- and angiogenesis-related pathways and used for supervised clustering of desmoid tumors. Pathways in which each gene are included are annotated in the table.
Genes noted to be altered in DES9525 after CTNNB1 knockdown as identified by RNA-seq. Fold change is relative to expression in cells treated with scramble control.
CTNNB1 mutations detected in pre- and post-treatment biopsies from patients on ALLIANCE A091105.
Genes with altered expression in DES9525 cells after treatment with PDGF-BB as compared to untreated cells as determined by RNA-seq.
Gene set enrichment analysis performed on RNA-seq from pre-treatment biopsies taken from patients on the placebo arm of ALLIANCE A091105.