Abstract
Many bioinformatics tools are available for the quantitative analysis of proteomics experiments. Most of these tools use a dedicated statistical model to derive absolute quantitative protein values from mass spectrometry (MS) data. Here, we present iSanXoT, a standalone application that processes relative abundances between MS signals and then integrates them sequentially to upper levels using the previously published Generic Integration Algorithm (GIA). iSanXoT offers unique capabilities that complement conventional quantitative software applications, including statistical weighting and independent modeling of error distributions in each integration, aggregation of technical or biological replicates, quantification of posttranslational modifications, and analysis of coordinated protein behavior. iSanXoT is a standalone, user-friendly application that accepts output from popular proteomics pipelines and enables unrestricted creation of quantification workflows and fully customizable reports that can be reused across projects or shared among users. Numerous publications attest the successful application of diverse integrative workflows constructed using the GIA for the analysis of high-throughput quantitative proteomics experiments. iSanXoT has been tested with the main operating systems. Download links for the corresponding distributions are available at https://github.com/CNIC-Proteomics/iSanXoT/releases.
Keywords: Mass spectrometry, Quantitative proteomics, Proteomics pipeline, Generic integration algorithm, WSPP model, Protein coordination
Graphical Abstract
1. Introduction
There are many bioinformatics packages available for the quantitative analysis of MS-based proteomics data, including MaxQuant [1], FragPipe [2], and Trans Proteomics Pipeline [3]. Most of these packages process raw data (the MS signal) to derive a quantitative value for each protein in each sample. The SanXoT package [4] was designed to process relative abundances between MS signals, which are sequentially integrated to derive weighted averages of protein ratios through the iterative application of the Generic Integration Algorithm (GIA) [5]. This approach exploits the stability of intensity ratios within the same scan (spectrum) in stable isotope-labeling experiments, enabling separate and accurate modeling of error sources at the scan, peptide, and protein levels. Originally conceived for analyzing data from 18O labeling [6], this concept was subsequently generalized to other approaches in the form of the WSPP model [7]. The SanXoT suite also facilitates various types of quantitative analysis through direct applications of the GIA. Examples include the integration of protein values from experiments conducted using different techniques [7], the analysis of coordinated protein behavior using the Systems Biology Triangle model (SBT model) [5], and the quantitative analysis of post-translational modifications (PTMs). The use of peptide to protein integration for detecting PTM changes was initially introduced for analyzing oxidized Cys-containing peptides in redox proteomics [8]. This approach was later generalized using the GIA for the quantitative analysis of any type of PTM identified employing open-search methods [9]. SanXoT has found application in over 60 published quantitative proteomics studies (refer to, for instance, [10], [11], [12], [13], [14], [15], [16], [17], [18], [19]).
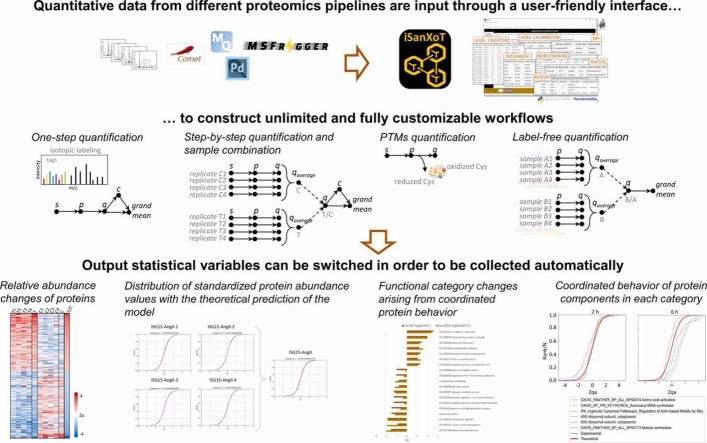
The original SanXoT package was designed for use by bioinformaticians and required the modular assembly of a series of scripts to construct quantitative workflows, which once arranged could not be easily reused for other experiments. Moreover, SanXoT lacked tools for preparing result tables or adapting data from other software packages. Therefore, despite SanXoT’s strengths, these limitations have hindered the widespread uptake of this package by the proteomics community. Here, we present iSanXoT, an open-source application that access SanXoT modules through a user-friendly interface without compromising its adaptability to the construction of unlimited, fully customizable user-designed quantitative workflows. Once arranged, workflows can be stored and easily reused for the analysis of new datasets. iSanXoT includes a reporting module that allows users to extract the quantitative information and generate user-customizable result tables that compile the desired quantitative data from the specified levels of analysis. iSanXoT also includes a general adaptor for processing quantitative data obtained with widely used software tools, such as MaxQuant, Fragpipe, and Proteome Discoverer (Fig. 1).
Fig. 1.
iSanXoT modular design. iSanXoT accepts quantitative data from widely used proteomics pipelines. The set of modules needed to run the quantitative workflows is invoked within a user-friendly interface based on task tables, allowing the construction of fully customizable user-designed quantitative workflows.
2. Materials and methods
iSanXoT uses the GIA [5] to sequentially integrate quantitative information (relative abundance) into higher levels, allowing the construction of fully customizable workflows that can be adapted to a wide range of analyses. iSanXoT houses several modules that build upon the programs included in SanXoT. In general, the workflows start by adapting the output from other software packages for use with iSanXoT. This is followed by the sequential integration of the quantitative information into higher levels and the generation of reports containing the desired information, specified by the user through a user-friendly task table interface. This interface contains several basic modules that perform one-step tasks, as well as compound modules featuring ready-for-use general purpose workflows. iSanXoT also allows newly created workflows to be reused by the same or other users. In the present study, we include four representative workflows that illustrate the flexibility of the package and that can be reused or adapted by any user. Detailed information on the use of these workflows and the processing of quantitative data obtained with some widely used proteomics pipelines is presented in the Supplementary Material.
2.1. Input data adapter
iSanXoT requires a TSV (tab-separated values) file containing, at a minimum, the quantified features together with their quantitative values (ID-q.tsv). The required information is extracted from the ID-q file to create the relation tables and to integrate the quantified features into higher levels. Most proteomics software tools generate tables that can be easily adapted for this purpose (see Table S1 in the Supplementary Material). The Input Data Adapter module converts external TSV files into ID-q files that can be directly used by iSanXoT.
2.2. Relation tables module
This module automatically relates lower-level elements to higher-level elements, allowing the user to create the relation tables needed to perform the integrations specified in the Basic and Compound modules. For instance, the relation table peptide2protein.tsv indicates which peptides belong to each protein. The module displays a task table in which the user indicates the information from the ID-q file (or any other TSV file) to be used to create the relation table.
2.3. Basic modules
The LEVEL CREATOR module is used to define how the ratios between quantitative values are to be calculated from the ID-q table and to assign a name to each ratio.
The LEVEL CALIBRATOR module calibrates the statistical weights (defined as the inverse of variances) associated with each quantitative value at the lowermost level by fitting a model that describes the relation between signal intensity and variance. The statistical weights are used to control the propagation of errors to higher levels and to calculate weighted averages at each integration step.
The INTEGRATE module contains a task table in which the user indicates the integrations from lower to higher levels to be performed with the GIA [5]. This module also generates a plot per integration that can be used to check the fit of the standardized variable distribution to the expected normal distribution.
The NORCOMBINE module integrates technical or biological replicates into a single value using the GIA [5]. As well as calculating weighted averages, GIA integrations take account of error propagation and estimate a global variance across replicates. As above, the accuracy of the integration model can be checked in the normality plots.
The RATIOS module calculates the log2-ratio between the elements of two samples. This step is useful for the direct comparison of results between two conditions.
The SBT module applies the Systems Biology Triangle algorithm [5], which integrates data from a lower level, an intermediate level, and a higher level. This module is best suited to the detection of functional category changes produced by the coordinated behavior of the component proteins. In this case, the module uses the protein (lower), category (intermediate), and grand mean (higher) levels.
2.4. Compound modules
The WSPP-SBT module automatically constructs the relation tables and performs the integrations needed to calculate quantitative protein values using the WSPP model [7] and the SBT algorithm [5]. The WSPP model initially calibrates the quantitative ratios at the scan level and then integrates these first into peptide values and then into protein values. The SBT algorithm is used to detect changes in functional categories due to protein coordination, as described above. Since this is one of the most common analyses in quantitative proteomics, we found it useful to include a compound module combining all the required tasks.
The three remaining compound modules are extensions of the WSPP-SBT module. The WSPPG-SBT module includes an intermediate protein-to-gene integration, after which the SBT algorithm is applied at the gene level to reveal coordinated gene behavior. Analysis at the gene level can be advantageous in some situations because functional classification is more developed at the gene than at the protein level. In addition, protein-to-gene integration avoids dilution of quantitative values due to the presence of isoforms or peptide misassignment and at the same time may detect protein isoforms whose quantitative behavior deviates from that of other isoforms generated from the same gene. The WPP-SBT and WPPG-SBT modules are similar to the other compound modules, except that they perform the initial calibration at the peptide level. These modules are useful in label-free experiments quantified at the peptide level.
2.5. Report modules
The REPORT module generates TSV files displaying the quantitative results at the levels and from the samples selected by the user. This highly configurable module allows data to be gathered from different levels and the inclusion of information from other TSV tables, such as whole protein descriptions or functional classifications.
The SANSON module helps to resolve functional category redundancy and depicts the global category changes detected by the SBT model. This module generates a similarity graph showing relationships between functional categories on the basis of the protein elements they share.
3. Results and discussion
To illustrate some applications of iSanXoT, we analyzed three proteomics datasets obtained by isobaric labeling and previously explored by integrative analysis with SanXoT [5], [20], [21] plus a dataset from a label-free experiment [22]. Using these datasets as case studies, we describe four representative workflows that exemplify the versatility of iSanXoT for a variety of quantitative analyses beyond mere protein abundance. The workflows are described in detail in the Supplementary Material and can be downloaded, together with the necessary input data, from https://cnic-proteomics.github.io/iSanXoT.
4. Workflow 1 (one-step quantification in a labeled experiment)
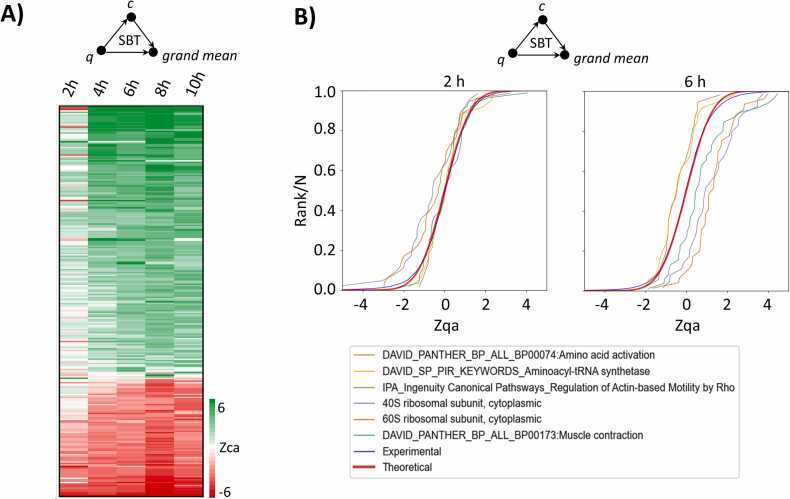
This workflow illustrates the use of the iSanXoT compound modules through an analysis of quantitative data from García-Marqués et al. [5]. This study characterized molecular alterations in vascular smooth muscle cells (VSMCs) stimulated with angiotensin-II (AngII) for 0, 2, 4, 6, 8, and 10 h. Quantitative proteomics was performed by isobaric iTRAQ 8-plex labeling. Here, we used the WSPP-SBT module in iSanXoT to integrate the quantitative information at the scan, peptide, and protein levels (WSPP model) [23] and to perform a functional category analysis (SBT model) [5] (Fig. 2A). The WSPP-SBT module conducts a fully automated analysis of each sample indicated in the task table. The module calculates relative protein abundance changes (Supplementary Figure S6), producing normally-distributed, standardized quantitative Z values while allowing full control of error sources at each integration level (scan, peptide, and protein; see below). The module also applies the SBT algorithm to detect functional protein alterations produced from coordinated protein behavior [5] (Fig. 3). The triangular integration in the SBT model improves the estimation of protein variance from the protein to category integration [5]. By considering protein variability within functional categories, the SBT model reduces the influence of protein changes in the biological model on the protein variance estimation [5].
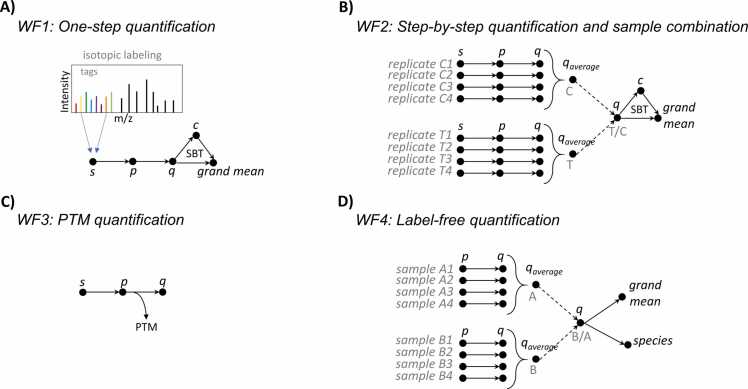
Fig. 2.
Scheme of the four representative workflows described in this study. These panels illustrate how iSanXoT integrates quantitative information at higher levels or combines it across sample types in each workflow (WF). Note that whereas WF1 (A) uses a compound module that automatically performs all required steps, the other three workflows (B-D) were constructed by indicating the tasks to be performed step-by-step via the interface. The rationale for the workflows is described in Results and Discussion. More specific details about how to use the interface are provided in the Supplementary Material file. All four workflows can be reused or adapted to other sample configurations according to user needs. The letters s, p, q and c refer to integration levels: scan, peptide, protein and category, respectively. PTM: post-translational modification; SBT: Systems Biology Triangle.
Fig. 3.
Representative results obtained with workflow 1. A. Heatmap depicting functional category changes produced by coordinated protein behavior over time. Each row refers to a functional protein category. Category changes are expressed in terms of the standardized variable (i.e., log2(fold-change) of the category in units of standard deviation). The subscript ca means that the Z values are calculated from the category (c) to grand mean (a) integration (see Fig. 2A). B. Examples of functional category changes in relation to the null hypothesis after 2 h and 6 h incubation with angiotensin-II. The null hypothesis (thick red line) corresponds to the cumulative standard normal distribution function ; the other lines are the cumulative distribution functions of the protein changes in each one of the indicated categories, expressed as standardized variables (i.e., log2(fold-changes) of proteins in units of standard deviation, determined from the protein (q) to grand mean (a) integration, as indicated in Fig. 2A).
5. Workflow 2 (step-by-step quantification and sample combination in a labeled experiment)
In this case study, we used the data from González-Amor et al. [21] to illustrate the step-by-step construction of a quantitative workflow from the full list of Basic modules in iSanXoT. In this case, the workflow is designed to calculate protein values in several samples by integrating the quantitative data at the scan, peptide, and protein levels in each sample (Fig. 2B). The workflow then integrates the quantitative information from technical or biological replicates within user-defined sample groups and analyzes protein abundance changes across these groups (Fig. 2B). Finally, the workflow captures category abundance changes produced by coordinated protein behavior (Fig. 2B). As reported previously [21], the workflow revealed a coordinated alteration of proteins implicated in cardiovascular function, extracellular matrix and remodeling, and vascular redox state in aortic tissue from AngII-infused ISG15-KO mice (Figure S20A). The coordinated protein behavior in some of the altered categories can be easily analyzed in the sigmoidal plots (Figure S20B).
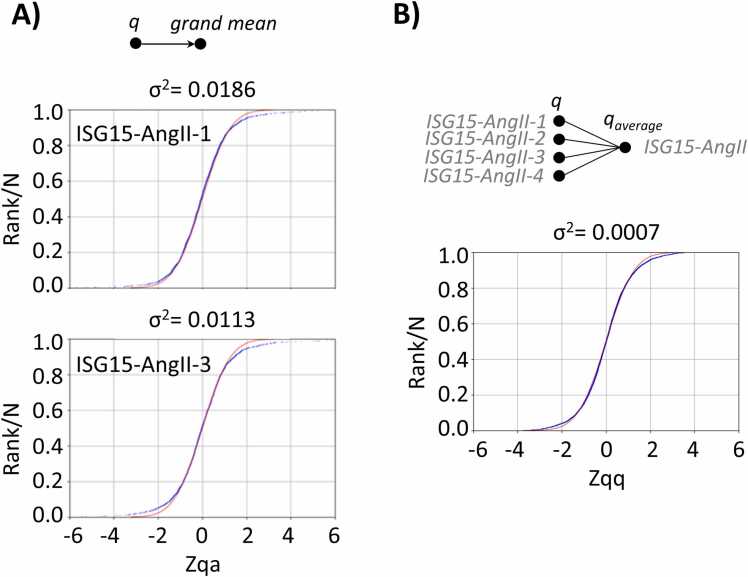
This workflow also illustrates two key features of the statistical approach used in iSanXoT. The first is the automated calibration step (performed by the LEVEL CALIBRATOR module), in which a statistical weight is assigned to each quantitative measure at the initial (scan) level. The statistical weight is the inverse of the variance of each quantification and is estimated from the signal intensity by fitting a global model. iSanXoT automatically generates plots that can be used to test the accuracy of the model (Supplementary Figure S12). This initial calibration step allows subsequent integrations to be performed using weighted averages and with control of error sources at all levels. The second key feature is the ability of iSanXoT to integrate protein values from multiple samples (NORCOMBINE module) by applying the GIA, which calculates a weighted average and accurately models the dispersion of protein values around this average, taking account of error propagation and allowing full control of outliers [7] (Fig. 4). This unique design allows the integration of protein values obtained in non-balanced samples, in different experiments or mass spectrometers, and even with different labeling techniques, as we showed previously [7].
Fig. 4.
Representative results obtained with workflow 2. A. Distribution of standardized protein abundance values, showing the good alignment with the theoretical prediction of the model for ISG15-AngII-1 and ISG15-AngII-3 samples from WF 2. B. Distribution of the standardized variable at the protein level around the average during replicate integration from the ISG15-AngII sample group, demonstrating the accuracy of the GIA.
6. Workflow 3 (quantification of posttranslationally modified peptides in a labeled experiment)
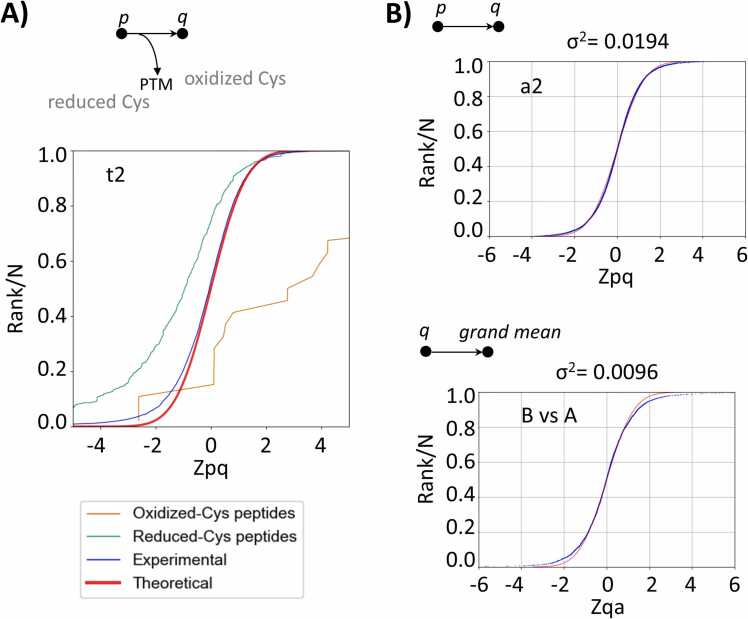
This workflow was designed to analyze quantitative data on PTMs detected in the study by Bonzon-Kulichenko et al. [20], which used the redox proteomics technique FASILOX to analyze reversible oxidation on Cys residues in mouse embryonic fibroblast preparations subjected to chemical oxidation. FASILOX labels reduced and oxidized Cys residues with distinct alkylating reagents, allowing these two groups of Cys sites to be quantified separately [20]. Workflow 3 carries out the conventional integrations from the scan-to-peptide and peptide-to-protein levels and uses the GIA output to model the dispersion of peptide values when they are integrated to their proteins, thereby detecting peptidoforms that deviate significantly from the expected behavior, independently of protein abundance changes (Fig. 2C). The standardized variable used by the model to describe the quantitative behavior of all peptidoforms in the experiment accurately follows the expected normal distribution (Fig. 5A, red and blue lines). This makes this workflow very useful for the detection of statistically significant abundance changes in posttranslationally modified peptides, decoupling them from their parent proteins on the basis of a robust and validated statistical framework [8], [9] (Fig. 5A, brown and green lines). For the sake of simplicity, here we show a simple application to redox proteomics [8], but the same workflow can be used to analyze high-throughput PTM data, including data obtained from open searches, as we have demonstrated previously [9].
Fig. 5.
Representative results obtained with workflows 3 and 4. A. Workflow 3: quantification of posttranslationally modified peptides in a labeled experiment. Distribution of the standardized variable at the peptide level around the protein averages for all peptidoforms in the proteome (blue), peptidoforms of oxidized Cys-containing peptides (brown), and peptidoforms of reduced-Cys containing peptides (green) in a FASILOX experiment sample. Note that the model accurately captures the distribution of peptide quantifications and how the oxidized and reduced forms tend to increase or decrease, respectively, deviating from the theoretical distribution (red). B. Workflow 4: label-free quantification. Distribution of the standardized variable at the peptide level (top) and protein level (bottom) in a sample from a label-free experiment, illustrating the accurate description of quantitative variability.
7. Workflow 4 (label-free quantification)
In this case study, we utilized a dataset derived from the analysis of samples prepared according to Navarro et al. [22] to demonstrate the application of iSanXoT in analyzing quantitative data obtained through label-free approaches. The study involved a comparison of two mixtures containing human, yeast, and Escherichia coli proteins in different proportions. The MS data were processed to the peptide level using MaxQuant, and the resulting output was adapted for use by iSanXoT with the Input Data Adapter module.
Workflow 4 was designed to assess changes in protein abundance between the two sample groups. This involved transforming the quantitative peptide data into relative abundances at the peptide level, followed by subsequent integrations as outlined in Workflow 2 (refer to Fig. 2D). Notably, our approach diverges from the majority of existing software, which typically performs statistical analysis on absolute protein abundances. While a comprehensive discussion on the choice between relative or absolute abundances from label-free data to feed quantitative statistical models is beyond the scope of this work, certain distinctive features of iSanXoT analysis are worth highlighting. The built-in GIA performs variance calibration, normalization, standardization, and statistical weighting at each integration step, all through a user-friendly interface and in a fully automated manner, eliminating the need for adjustable parameters. Additionally, iSanXoT provides complete control and inspection of all analysis steps. This includes direct inspection of variance calibration (refer to the example from Workflow 2 in Figure S12) and assessment of the normality of standardized quantitative variables (illustrated in Fig. 5B at both the peptide and protein levels; see also Figure S37). These steps are fundamental for accurately estimating the statistical significance of observed changes.
Moreover, iSanXoT obviates the need for quality control, data filtering, or pre-processing, as the data is weighted according to quantitative accuracy. This often renders missing value imputation unnecessary, even in the presence of highly unbalanced data. The GIA can integrate data from different experiments, generating a unique quantitative variable usable for statistical inference (Fig. 5B, bottom).
iSanXoT also supports selective normalization based on a data subset, such as a group of proteins from the same species (human proteins in Supplementary Figure S32; see the normalized values in Figure S38). Furthermore, the iSanXoT interface makes it easy the introduction of intermediate levels of quantitative integration, including protein-to-species integration (Fig. 2D), which models the dispersion of protein values within each species.
This integration can be performed in each sample separately (Figure S40), or after replicates have been integrated within each sample (Figure S41), ensuring that all data are centered around the null hypothesis (Figure S43). This analysis allows the determination of the quantitative accuracy with which each protein is measured relative to the other proteins of the same species. A benchmark summarizing iSanXoT performance for various tests conducted in this label-free experiment is also presented in the Supplementary Material (Tables S2-S4).
8. Conclusions
iSanXoT harnesses the unique statistical characteristics of the GIA to expand the capabilities of conventional packages employed in the analysis of quantitative proteomics data. Because each GIA integration models and standardizes the integrated data independently, the quantitative information can be extended without limit to higher levels or integrated across samples within a given level, while maintaining full control of error sources at each step. By performing serial GIA integrations, this approach naturally allows the creation of quantitative workflows adaptable to a large variety of user needs.
For this study, we selected four case studies that illustrate how iSanXoT can be used to create workflows designed to address different specific biological questions. Workflow 1 exemplifies the use of iSanXoT for the automated analysis of functional categories that change in response to coordinated protein behavior. This analysis requires a statistical model able to capture the underlying protein variance within the functional categories used for protein classification, and in iSanXoT this task is performed by the dedicated WSPP-SBT module. Workflow 2 illustrates the use of the GIA to integrate and compare protein values across sample types, whereas Workflow 3 presents the use of the GIA for PTM analysis, demonstrating the statistical model’s highly accurate description of quantitative variance at the peptide level. High-throughput quantitative PTM analysis is attracting increasing interest, particularly with the ongoing development of open-search approaches. Finally, Workflow 4 presents the application of the GIA to the analysis of label-free data; this workflow has advantages over conventional approaches in some situations, for example when the data are highly unbalanced or when the analysis requires careful control of variance at the peptide level.
iSanXoT is a standalone application with a user-friendly interface, enabling versatile and highly automatable creation of integrative quantification workflows and report tables. Once workflows are prepared, their export and reuse in multiple projects is straightforward. The current version of iSanXoT accepts the direct output from several popular proteomics pipelines, making the application readily available for the proteomics community.
CRediT authorship contribution statement
Jose Manuel Rodríguez: Conceptualization, Methodology, Software, Data Curation, Formal analysis, Validation, Writing - Original Draft, Writing - Review and Editing, Visualization. Emilio Camafeita: Validation, Data Curation, Writing - Review and Editing. Inmaculada Jorge: Validation, Writing - Review and Editing. Rafael Barrero-Rodríguez and Ricardo Magni: Software. Ana Martínez del Val, Estefanía Núñez, Andrea Laguillo, Cristina A. Devesa and Juan A. López: Validation, Data Curation. Jesús Vázquez: Conceptualization, Supervision, Funding acquisition, Writing – Review and Editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could influence the work reported in this paper.
Acknowledgments
This study was supported by competitive grants from the Spanish Ministry of Science, Innovation and Universities (PGC2018–097019-B-I00, PID2021–122348NB-I00, PLEC2022–009235 and PLEC2022–009298), the Instituto de Salud Carlos III (Fondo de Investigación Sanitaria grant PRB3 (PT17/0019/0003- ISCIII-SGEFI / ERDF, ProteoRed), Comunidad de Madrid (IMMUNO-VAR, P2022/BMD-7333), the Spanish Ministry of Education, Culture and Sports (FPU18/03882, FPU20/03365 and PRE2019-090019), and the “la Caixa” Banking Foundation (project codes HR17–00247 and HR22–00253). The CNIC is supported by the Instituto de Salud Carlos III (ISCIII), the Ministerio de Ciencia e Innovación (MCIN), and the Pro CNIC Foundation) and is a Severo Ochoa Center of Excellence (grant CEX2020–001041-S funded by MICIN/AEI/10.13039/501100011033).
Author contributions
JMR conceived the project, developed the methodology and software, curated the data, conducted formal analysis, validated the results, wrote the original draft, and contributed to text review and editing. EC contributed to data validation, managed data curation, and contributed to text review and editing. IJ contributed to data validation and text editing. RBR and RM contributed to program development. AMV, EN, AL, CAD, and JAL contributed to results validation and data curation. JV contributed to project conceptualization, acquired funding, provided supervision, contributed to writing and review, and participated in editing.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.12.034.
Contributor Information
Emilio Camafeita, Email: ecamafeita@cnic.es.
Jesús Vázquez, Email: jvazquez@cnic.es.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11(12):2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 2.Kong A.T., Leprevost F.V., Avtonomov D.M., Mellacheruvu D., Nesvizhskii A.I. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat Methods. 2017;14(5):513–520. doi: 10.1038/nmeth.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deutsch E.W., Mendoza L., Shteynberg D.D., Hoopmann M.R., Sun Z., et al. Trans-Proteomic Pipeline: Robust Mass Spectrometry-Based Proteomics Data Analysis Suite. J Proteome Res. 2023;22(2):615–624. doi: 10.1021/acs.jproteome.2c00624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trevisan-Herraz M., Bagwan N., Garcia-Marques F., Rodriguez J.M., Jorge I., et al. SanXoT: a modular and versatile package for the quantitative analysis of high-throughput proteomics experiments. Bioinformatics. 2019;35(9):1594–1596. doi: 10.1093/bioinformatics/bty815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Marques F., Trevisan-Herraz M., Martinez-Martinez S., Camafeita E., Jorge I., et al. A Novel Systems-Biology Algorithm for the Analysis of Coordinated Protein Responses Using Quantitative Proteomics. Mol Cell Prote: MCP. 2016;15(5):1740–1760. doi: 10.1074/mcp.M115.055905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jorge I., Navarro P., Martinez-Acedo P., Nunez E., Serrano H., et al. Statistical model to analyze quantitative proteomics data obtained by 18O/16O labeling and linear ion trap mass spectrometry: application to the study of vascular endothelial growth factor-induced angiogenesis in endothelial cells. Mol Cell Prote: MCP. 2009;8(5):1130–1149. doi: 10.1074/mcp.M800260-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Navarro P., Trevisan-Herraz M., Bonzon-Kulichenko E., Nunez E., Martinez-Acedo P., et al. General statistical framework for quantitative proteomics by stable isotope labeling. J Proteome Res. 2014;13(3):1234–1247. doi: 10.1021/pr4006958. [DOI] [PubMed] [Google Scholar]
- 8.Martinez-Acedo P., Nunez E., Gomez F.J., Moreno M., Ramos E., et al. A novel strategy for global analysis of the dynamic thiol redox proteome. Mol Cell Prote: MCP. 2012;11(9):800–813. doi: 10.1074/mcp.M111.016469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bagwan N., Bonzon-Kulichenko E., Calvo E., Lechuga-Vieco A.V., Michalakopoulos S., et al. Comprehensive Quantification of the Modified Proteome Reveals Oxidative Heart Damage in Mitochondrial Heteroplasmy. Cell Rep. 2018;23(12):3685–3697. doi: 10.1016/j.celrep.2018.05.080. e4. [DOI] [PubMed] [Google Scholar]
- 10.Nunez E., Fuster V., Gomez-Serrano M., Valdivielso J.M., Fernandez-Alvira J.M., et al. Unbiased plasma proteomics discovery of biomarkers for improved detection of subclinical atherosclerosis. EBioMedicine. 2022;76 doi: 10.1016/j.ebiom.2022.103874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Garcia M., Sanchez-Perales S., Jarabo P., Calvo E., Huyton T., et al. Mechanical control of nuclear import by Importin-7 is regulated by its dominant cargo YAP. Nat Commun. 2022;13(1) doi: 10.1038/s41467-022-28693-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez-Lopez A., Espinos-Estevez C., Gonzalez-Gomez C., Gonzalo P., Andres-Manzano M.J., et al. Cardiovascular Progerin Suppression and Lamin A Restoration Rescue Hutchinson-Gilford Progeria Syndrome. Circulation. 2021;144(22):1777–1794. doi: 10.1161/CIRCULATIONAHA.121.055313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorenzo C., Delgado P., Busse C.E., Sanz-Bravo A., Martos-Folgado I., et al. ALDH4A1 is an atherosclerosis auto-antigen targeted by protective antibodies. Nature. 2021;589(7841):287–292. doi: 10.1038/s41586-020-2993-2. [DOI] [PubMed] [Google Scholar]
- 14.de la Fuente-Alonso A., Toral M., Alfayate A., Ruiz-Rodriguez M.J., Bonzon-Kulichenko E., et al. Aortic disease in Marfan syndrome is caused by overactivation of sGC-PRKG signaling by NO. Nat Commun. 2021;12(1) doi: 10.1038/s41467-021-22933-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Mashhadi R.H., Al-Mashhadi A.L., Nasr Z.P., Mortensen M.B., Lewis E.A., et al. Local Pressure Drives Low-Density Lipoprotein Accumulation and Coronary Atherosclerosis in Hypertensive Minipigs. J Am Coll Cardiol. 2021;77(5):575–589. doi: 10.1016/j.jacc.2020.11.059. [DOI] [PubMed] [Google Scholar]
- 16.Nicolas-Avila J.A., Lechuga-Vieco A.V., Esteban-Martinez L., Sanchez-Diaz M., Diaz-Garcia E., et al. A Network of Macrophages Supports Mitochondrial Homeostasis in the Heart. Cell. 2020;183(1):94–109. doi: 10.1016/j.cell.2020.08.031. e23. [DOI] [PubMed] [Google Scholar]
- 17.Martinez-Lopez D., Roldan-Montero R., Garcia-Marques F., Nunez E., Jorge I., et al. Complement C5 Protein as a Marker of Subclinical Atherosclerosis. J Am Coll Cardiol. 2020;75(16):1926–1941. doi: 10.1016/j.jacc.2020.02.058. (JV and JLMV are co-corresponding authors) [DOI] [PubMed] [Google Scholar]
- 18.Martin-Cofreces N.B., Chichon F.J., Calvo E., Torralba D., Bustos-Moran E., et al. The chaperonin CCT controls T cell receptor-driven 3D configuration of centrioles. Sci Adv. 2020;6(49) doi: 10.1126/sciadv.abb7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forte D., Garcia-Fernandez M., Sanchez-Aguilera A., Stavropoulou V., Fielding C., et al. Bone Marrow Mesenchymal Stem Cells Support Acute Myeloid Leukemia Bioenergetics and Enhance Antioxidant Defense and Escape from Chemotherapy. Cell Metab. 2020;32(5):829–843. doi: 10.1016/j.cmet.2020.09.001. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonzon-Kulichenko E., Camafeita E., Lopez J.A., Gomez-Serrano M., Jorge I., et al. Improved integrative analysis of the thiol redox proteome using filter-aided sample preparation. J Proteom. 2020;214 doi: 10.1016/j.jprot.2019.103624. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez-Amor M., Garcia-Redondo A.B., Jorge I., Zalba G., Becares M., et al. Interferon stimulated gene 15 pathway is a novel mediator of endothelial dysfunction and aneurysms development in angiotensin II infused mice through increased oxidative stress. Cardiovasc Res. 2021 doi: 10.1093/cvr/cvab321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navarro P., Kuharev J., Gillet L.C., Bernhardt O.M., MacLean B., et al. A multicenter study benchmarks software tools for label-free proteome quantification. Nat Biotechnol. 2016;34(11):1130–1136. doi: 10.1038/nbt.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonzon-Kulichenko E., Perez-Hernandez D., Nunez E., Martinez-Acedo P., Navarro P., et al. A robust method for quantitative high-throughput analysis of proteomes by 18O labeling. Mol Cell Proteom. 2011;10(1) doi: 10.1074/mcp.M110.003335. M110 003335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material