Abstract
Background
Dilated cardiomyopathy with ataxia (DCMA) is an autosomal recessive disorder arising from truncating mutations in DNAJC19, which encodes an inner mitochondrial membrane protein. Clinical features include an early onset, often life-threatening, cardiomyopathy associated with other metabolic features. Here, we aim to understand the metabolic and pathophysiological mechanisms of mutant DNAJC19 for the development of cardiomyopathy.
Methods
We generated induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) of two affected siblings with DCMA and a gene-edited truncation variant (tv) of DNAJC19 which all lack the conserved DnaJ interaction domain. The mutant iPSC-CMs and their respective control cells were subjected to various analyses, including assessments of morphology, metabolic function, and physiological consequences such as Ca2+ kinetics, contractility, and arrhythmic potential. Validation of respiration analysis was done in a gene-edited HeLa cell line (DNAJC19tvHeLa).
Results
Structural analyses revealed mitochondrial fragmentation and abnormal cristae formation associated with an overall reduced mitochondrial protein expression in mutant iPSC-CMs. Morphological alterations were associated with higher oxygen consumption rates (OCRs) in all three mutant iPSC-CMs, indicating higher electron transport chain activity to meet cellular ATP demands. Additionally, increased extracellular acidification rates suggested an increase in overall metabolic flux, while radioactive tracer uptake studies revealed decreased fatty acid uptake and utilization of glucose. Mutant iPSC-CMs also showed increased reactive oxygen species (ROS) and an elevated mitochondrial membrane potential. Increased mitochondrial respiration with pyruvate and malate as substrates was observed in mutant DNAJC19tv HeLa cells in addition to an upregulation of respiratory chain complexes, while cellular ATP-levels remain the same. Moreover, mitochondrial alterations were associated with increased beating frequencies, elevated diastolic Ca2+ concentrations, reduced sarcomere shortening and an increased beat-to-beat rate variability in mutant cell lines in response to β-adrenergic stimulation.
Conclusions
Loss of the DnaJ domain disturbs cardiac mitochondrial structure with abnormal cristae formation and leads to mitochondrial dysfunction, suggesting that DNAJC19 plays an essential role in mitochondrial morphogenesis and biogenesis. Moreover, increased mitochondrial respiration, altered substrate utilization, increased ROS production and abnormal Ca2+ kinetics provide insights into the pathogenesis of DCMA-related cardiomyopathy.
Keywords: Dilated cardiomyopathy with ataxia, Genetics, Metabolism, Mitochondria, OXPHOS, ROS, Contractility
Highlights
-
•
Loss of DNAJC19's DnaJ domain disrupts cardiac mitochondrial structure, leading to abnormal cristae formation in iPSC-CMs.
-
•
Impaired mitochondrial structures lead to an increased mitochondrial respiration, ROS and an elevated membrane potential.
-
•
Mutant iPSC-CMs show sarcomere dysfunction and a trend to more arrhythmias, resembling DCMA-associated cardiomyopathy.
1. Introduction
DCMA (dilated cardiomyopathy with ataxia syndrome; MIM, #610198) is an autosomal recessive mitochondrial disorder arising from homozygous mutations in the DNAJC19 gene. The major clinical features include cerebellar ataxia, early onset dilated cardiomyopathy (DCM), prolonged QT-interval, 3-methylglutaconic aciduria (3-MGA), as well as variable clinical features including failure to thrive, developmental delay and abnormalities of male genitalia. Early studies on DCMA revealed a genetic association of DNAJC19 encoding the DnaJ Heat Shock Protein Family (Hsp40) Member C19 with DCMA and identified a single splice site mutation (NM_145261.4):c.130-1G > C resulting in a loss of the full-length transcript [1,2]. Additional families with biallelic truncating mutations in DNAJC19 have been reported since then, confirming it as a disease gene for DCMA and suggesting a loss of function mechanism [[3], [4], [5]]. The early, often life-threatening cardiac phenotype is characterized by great variability and remains one of the most important challenges in the clinical management of these patients [2,6].
DNAJC19 is integrated into the inner mitochondrial membrane (IMM) via its NH2-terminal transmembrane domain, while a conserved DnaJ domain at the COOH-terminus faces the matrix side [7]. DnaJ proteins act as molecular co-chaperones for Hsp70 by aiding the folding of newly imported proteins [8,9]. DNAJC19 shows similarities to Pam18/Tim14 in yeast, a constituent of the TIM23 import machinery [10]. Therefore, it has been suggested to be part of the motor complex, which associates with the core TIM23 translocon and mediates the import of precursor proteins destinated for the matrix in an ATP- and membrane potential dependent manner [11]. By interacting with prohibitins (PHB1+2), DNAJC19 is also involved in the formation of prohibitin scaffolds, which form large ring structures in the IMM and thereby controls cristae morphology and the functional integrity of mitochondria [12]. Prohibitins segregate specific membrane domains and restrict phospholipids, including cardiolipin (CL), into specific membrane domains [13,14]. CLs are involved in shaping mitochondrial morphology [15], interact with complexes of oxidative phosphorylation (OXPHOS) and enzymes of the Krebs cycle (TCA) as well as with carrier proteins and the mitochondrial calcium uniporter (MCU) [[16], [17], [18]].
Changes in CL species occur in Barth Syndrome (BTHS), an inherited cardiomyopathy caused by mutations of the transacylase tafazzin (TAZ) [7,19]. However, a knockdown of DNAJC19 in HEK293T cells showed normal monolysocardiolipin levels, but a shift of CLs to longer and more unsaturated acyl chains [12]. Furthermore, the depletion of PHB2 or DNAJC19 resulted in misbalanced processing of the CL dependent mitochondrial fusion protein OPA1. The exact processing of the mitochondrial membrane protein OPA1 [12] is an essential regulator of mitochondrial morphology. In line with this, imbalanced OPA1 processing has been observed in induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) from patients with DCMA [20]. Interestingly, CL profiles were neither significantly altered in fibroblasts nor in iPSC-CMs [20,21] suggesting a more complex interdependence of OPA1 processing and PHB [11,13,22]. Taken together, the structural and functional consequences of mutant DNAJC19 on cardiac mitochondria and how disturbed CL remodeling and an altered mitochondrial biogenesis cause severe heart failure and arrhythmias remain unknown.
Mitochondria are the primary source of energy in the heart as they provide adenosine triphosphate (ATP) required for excitation-contraction coupling. To meet energy demands, the cardiac muscle has a very high mitochondrial density, occupying about 35% of the cardiomyocyte volume [23,24]. Mitochondrial bioenergetics and ROS production are tightly regulated, and impaired mitochondrial function in the heart can have many pathological downstream effects subsequently leading to heart failure, to which mutations in the mitochondrial DNAJC19 may contribute. Therefore, we used a personalized in vitro model system of human iPSC-CMs from DCMA patients and genome edited iPSCs with a truncating variant (tv) of DNAJC19 to unravel pathological mechanisms of heart disease due to mutations in DNAJC19 [25].
In this study, we report that the loss of the DnaJ interaction domain in mutant DNAJC19 cells leads to loss of full-length DNAJC19, fragmented, mitochondria with altered cristae structures and a disturbed mitochondrial network. Functionally, we observed upregulation of respiratory chain (RC) complexes, an excess in oxygen consumption rates (OCRs) and mitochondrial respiration, which indicate a higher electron transport chain activity to meet ATP demands. An increase in extracellular acidification rates (ECARs) suggested alterations in substrate metabolism, which was confirmed by a decreased fatty acid uptake and utilization of alternative fuels such as glucose. Finally, we could show downstream effects on mutant DNAJC19 iPSC-CMs with respect to their electrophysiological properties and altered calcium homeostasis.
2. Methods
Detailed methods are provided in Appendix A.
2.1. Study approval
The study was approved by the Conjoint Health Research Ethics Board of the University of Calgary (Ethics ID: 20729; 23515). Written informed consent was obtained from all participants or their legal representatives before participation in the study. The study conformed to the principles outlined in the Declaration of Helsinki. Medical history of participating individuals was obtained from medical records at the time of inclusion.
2.2. Generation of iPSCs and stem cell maintenance
Dermal fibroblasts obtained from the patients DCMAP1 (LIBUCi001-A) and DCMAP2 (LIBUCi002-A) were reprogrammed into iPSC lines and validated for stem cell like characteristics as previously described [25]. Additionally, a control (CTRL1) line (JMUi001-A) was used to generate a DNAJC19 truncation variant (DNAJC19tv) in an isogenic background by genome editing via CRISPR/Cas9 technology as reported [25]. CTRL2 was separately reprogrammed from fibroblasts (Promocell, #C-12302) of a 47-year-old Caucasian female according to standard protocols (Invitrogen™) and validated as described in the Supplemental Data and shown in Figure S1.
iPSCs were maintained in feeder-free conditions using mTeSR™1 medium (STEMCELL Technologies Inc.) on Corning® Matrigel™-hESC-qualified Matrix (Corning Inc.) coated culture plates. Passaging of cells was performed at 70–80 % confluency every 2–5 days using Accutase® and Dulbecco's phosphate-buffered saline (dPBS). Cells were resuspended in mTeSR™1 medium supplemented with RHO/ROCK pathway inhibitor (RI) Y27632 (10 μM, Miltenyi Biotec GmbH) and reseeded. After passage, the culture medium was replaced every day.
2.3. Differentiation, purification and maturation of iPSC-CMs
For cardiac differentiation the modified protocol of Kadari et al. was implemented using iPSCs at 80–90 % confluency [26]. Differentiation was induced and further processed with supplemented CBM medium (CBM; RPMI 1640 medium, B-27™ supplement) and small molecules: CHIR99021, BMP4 and IWR1. After 8–10 days beating of iPSC-CMs was visually recognized and glucose-depleted and lactate-enriched medium, termed cardiac enrichment medium (CEM; RPMI 1640 medium without glucose, sodium l-lactate) was used for 3–7 days with daily medium change. Thereafter, iPSC-CMs were cultured between 60 and 120 days depending on the experiment. Further purification was done by the PSC-derived Cardiomyocyte Isolation Kit, (Magnetic-activated cell sorting, Miltenyi Biotec). Finally, iPSC-CMs were replated on Corning® hESC-Matrigel™-coated culture dishes or for enhanced maturation stages on matrigel-mattresses (Corning Inc.) according to Feaster et al. on corresponding time points required for the individual experiments [27].
2.4. Generation and maintenance of HeLa cell lines
Like genome editing in iPSCs, CRISPR/Cas9 technology was used to introduce the DNAJC19 truncation variant (DNAJC19tvHeLa) into the genetic background of a HeLa control (CTRLHeLa) cell line (ECACC). The same strategy as reported in [25] was used. HeLa cells were maintained in Dulbecco's modified Eagle medium supplemented with 10 % fetal bovine serum (FBS) and were passaged with 0.025 % trypsin (ThermoFischer) and dPBS after reaching 80–90% confluency.
2.5. Molecular, structural and functional analyses
Procedures for immunofluorescent staining and life cell imaging, transmission electron microscopy, real-time PCR, western blot analyses (from cell lysates or isolated HeLa mitochondria), the Mito Stress Test Assay, radioactive tracer uptake studies, live cell analyses of calcium transients, cell shortening and arrhythmia detection using the IonOptix system as well as respiratory analyses on the Oroboros Oxygraph-2K and assessment of cellular ATP concentrations in HeLa cells are described in the Supplementary Data. Primer sequences are listed in Table S1. Primary and secondary antibodies are listed in Table S2.
2.6. Statistical analysis
Data are presented as mean ± standard error of mean (SEM) in line charts or bar graphs with single data points superimposed. Statistical tests were performed using GraphPad Prism version 6.01 and 9.3.1 (GraphPad Software, San Diego, California USA). For comparisons, normally distributed data was analyzed by paired or unpaired two-tailed Student's t-test (two-group analysis), or two-way ANOVA analysis/mixed-effects analysis followed by Bonferroni post hoc test (multiple-group analysis). The normality of data was examined by the D'Agostino-Pearson test before parametric or non-parametric testing. Whenever normality was not present in the data set a non-parametric alternative was performed. Statistical significance was considered when ∗ p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
3. Results
3.1. Generation of patient-specific and CRISPR/Cas9 gene-edited iPSC-derived cardiomyocytes
DCMA is a complex metabolic disorder with an early onset cardiac phenotype. To investigate the molecular and metabolic alterations affecting cardiomyocytes, we generated two patient-specific iPSC lines, DCMAP1 (LIBUCi001-A) and DCMAP2 (LIBUCi002-A), and one CRISPR/Cas9 gene-edited iPSC line, DNAJC19tv (JMUi001-A-1) [25]. Both patients were clinically diagnosed with DCMA (DCMAP1, DCMAP2) and carry a homozygous splice acceptor site mutation (c.130-1G > C) in the DNAJC19 (NM_145261.4) gene, whereas the gene-edited mutation describes two allele-specific genetic variants, both leading to a premature stop-codon that predicts the loss of full-length DNAJC19 (c.[131_140del];[137_138insAGTATAATTGCC]). The latter was generated within the isogenic control iPSC line (CTRL1, JMUi001-A) of a six-year-old Caucasian male [25]. Additionally, a second control iPSC line was generated and validated (shown in Figure S1) from fibroblasts of a 47-year-old Caucasian female serving as a second control (CTRL2) for most experiments.
Both patient-specific iPSC lines originating from fibroblasts of a sib-pair - an 8-year-old boy (DCMAP1) and his 10-year-old sister (DCMAP2). Parents were second cousins and from the Canadian Dariusleut Hutterite population. The boy was born at term and diagnosed with deteriorating heart failure at 2 weeks of age leading to heart transplantation at 20 months of age. Cardiac exam at an age of 8 years revealed normal cardiac function despite a slightly prolonged corrected QT-interval (465 ms). In contrast, his sister was also born at term and first diagnosed with DCMA at an age of 18 months due to a developmental delay. Her cardiac exam showed a preserved left ventricular systolic function, but ECGs demonstrated prolonged QT-intervals (between 560 and 590 ms). Despite betablocker therapy, at an age of 6-years an internal cardioverter defibrillator (ICD) was implanted after an unexplained syncopal event. However, ever since no arrhythmic events have been documented. Her cardiac function remained stable. Both siblings demonstrated additional clinical features of DCMA including developmental and growth delay, cerebellar ataxia with frequent falls and 3-methylglutaconic aciduria (3-MGA).
For further characterization of the cardiac phenotype, all five iPSC lines (DCMAP1, DCMAP2, DNAJC19tv, CTRL1, CTRL2) were differentiated within 7–10 days into a 2D monolayer of contracting iPSC-CMs and cultivated for at least 60 days and characterized to ensure purity and a mature adult-like stage (Figure S2).
The DNAJC19 gene consists of three isoforms, with isoform 1 representing the full-length transcript of 525 nucleotides (nt), isoform 2 lacking the transmembrane domain due to an alternative start codon, and isoform 3 lacking the DnaJ domain caused by exon 4 deletion (Figure 1A). The homozygous c.130-1G > C mutation is predicted to affect splicing of isoform 1 and 2 resulting in deletion of exon 4 and an exclusive 445 nt alternative spliced transcript corresponding to isoform 3. DNAJC19 specific RT-PCR was performed on cDNA of mutant and control iPSC-CMs. In both CTRLs, the full-length transcript was the predominately observed product, whereas the two mutation carriers expressed only the shorter 445 bp product as previously reported in adult leukocytes of patients with the disease [1], (Figure 1B). RT-PCR from cardiac cDNA of the DNAJC19tv mutation revealed ∼537 bp and ∼515 bp stable fragments corresponding to the biallelic mutation c.[131_140del]; [137_138insAGTATAATTGCC] generated by CRISPR/Cas9 leading to a frameshift and premature stop codon. Next, we performed western blot analysis, which demonstrated the loss of the full-length DNAJC19 protein in all mutants compared to both CTRLs showing stable expression of the 12.5 kDa protein (Figure 1C).
Figure 1.
Loss of full-length DNAJC19 in patient-derived and gene-edited DNAJC19 mutants. (A) Schematic representation of DNAJC19 and corresponding RNA and protein isoforms. The transcription of DNAJC19 generates one full-length and one transcript variant lacking exon 4 (Δexon4). Both transcripts result in the expression of three DNAJC19 isoforms that comprise a transmembrane domain and/or a DnaJ domain. The Δexon4 transcript, the only existing transcript variant in DCMA mutants, results in isoform 3, consisting of the transmembrane domain only. (B) RT-PCR analysis of DNAJC19 with gene-specific primers (RTF & RTR) on cDNA of mutant DNAJC19 iPSC-CMs (DCMAP1, DCMAP2; DNAJC19tv) and controls (CTRL1, CTRL2). Note the strongly expressed full-length 525 bp transcript and a weaker 445 bp transcript (Δex4) in CTRLs, whereas DCMAP1 and DCMAP2 iPSC-CMs displayed only a strong 445 bp transcript (Δex4 transcript). Expression analysis of DNAJC19tv revealed the expected biallelic mutation introduced by CRISPR/Cas9 resulting in two different transcripts close to the 525 bp transcript (515 bp; 537 bp). Quantification was normalized to CTRL1 and normalized to GAPDH. Data are shown as mean ± SEM together with the biological replicates. (C) Loss of full-length DNAJC19 in all mutant iPSC-CMs via immunoblotting. Both controls displayed a strong expression of the 12.5 kDa DNAJC19 protein, whereas all mutants share the loss of the full-length DNAJC19. Data are shown as mean ± SEM with the biological replicates superimposed. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns p > 0.05 using unpaired t-test, calculated versus CTRL1 after normalization to GAPDH signal. bp = base pairs; kDa = kilo Dalton; GAPDH = glyceraldehyde 3-phosphate dehydrogenase; DNAJC19 = DnaJ heat shock protein family (Hsp40) member C19; DNAJC19tv = gene-edited DNAJC19 truncation variant in CTRL1; DCMAP1 = dilated cardiomyopathy with ataxia patient 1; DCMAP2 = dilated cardiomyopathy with ataxia patient 2, CTRL1 = normal control 1; CTRL2 = normal control 2.
Overall, patient-derived iPSC-CMs and gene-edited iPSCs behaved similarly, with minor line-specific differences with regards to differentiation and maturation. Mutant iPSC-CMs displayed loss of full-length DNAJC19 protein, suggesting at least a partial loss of function affecting the DnaJ domain.
3.2. Decreased mitochondrial protein expression and abnormal cristae formation of mutant DNAJC19 iPSC-derived CMs
Since DNAJC19 is an integral membrane protein in the IMM, we assessed different subcellular locations of iPSC-derived CMs at 60 days of age by immunofluorescent co-staining of DNAJC19 with MitoTracker for mitochondrial structures, DAPI for nuclear regions and cardiac TNT (cTNT) indicating the cytoplasmic localization. Control iPSC-CM displayed elongated mitochondrial fiber structures with dense regions in the perinuclear area and in some cases, mitochondrial aggregations close to the cell-cell-contact sites, which mainly co-localize with the green labeled DNAJC19 (Figure 2A, upper two panels). However, all mutant iPSC-CMs showed an enhanced aggregation and fragmentation of mitochondrial network structures distributed over the whole cell body and almost no signal of DNAJC19 at mitochondrial structures (Figure 2A, three lower panels), whereas some mis-localized DNAJC19 appeared in the nuclei. Mainly cardiomyocytes of DCMAP1 also displayed rare remaining signals of DNAJC19 at the cardiomyocyte edges, which was not seen in DNAJC19tv and DCMAP2 (Figure 2A). Overall, iPSC-CMs of all three mutants appeared to be smaller in size than CTRLs (Figure 2B; p = 0.042). To evaluate mitochondrial protein content of mutant iPSC-CMs compared to CTRLs, we performed immunoblot analysis of CM lysates collected at day 60 and quantified VDAC expression, which represents the mitochondrial content compared to α-actinin expression as a marker for cardiomyocytes. Interestingly, all DNAJC19 mutant iPSC-CMs unraveled significantly reduced mitochondrial expression compared to CTRLs (Figure 2C,D; p = 0.0406) suggesting reduced mitochondrial content in mutant CMs.
Figure 2.
Impact of mutant DNAJC19 on cell and mitochondrial morphology and expression. (A) Immunofluorescent co-staining of iPSC-CMs using cardiac troponin T (cTnT, red), DAPI (blue), MitoTracker-based mitochondrial membrane labelling (violet) and DNAJC19 (green). Control iPSC-CMs (CTRL1, CTRL2) displayed DNAJC19 localization adjacent to mitochondrial structures. DNAJC19 mutants showed absence of the signal of DNAJC19 expression and higher mitochondrial fragmentation (white arrows, DNAJC19tv, DCMAP1, DCMAP2). (B) CM areas in [μm2] were quantified from maximum intensity projections of z-stacks from at least 5–55 iPSC-CMs per biological replicate of each control (CTRL) and mutant DNAJC19 (DNAJC19mut) iPSC-CM cell line. DNAJC19mut iPSC-CMs demonstrated reduced areas compared to CTRL. (C) Immunoblotting showed reduced expression levels of the mitochondrial marker VDAC in mutant iPSC-CMs. VDAC expression of at least two technical replicates was normalized to α-Actinin (CM marker) and plotted as fold-change of CTRL expression. iPSC-CMs of both controls (CTRL1, CTRL2) and three mutants (DNAJC19tv, DCMAP1, DCMAP2) were analyzed together in groups. Data are shown as mean ± SEM or fold-change of CTRL ± SEM with the biological replicates superimposed. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns p > 0.05 using unpaired t-test or Mann–Whitney test whenever the data did not follow a Gaussian distribution. Scale bars = 50 μm; DAPI = 4′,6-diamidino-2-phenylindole.
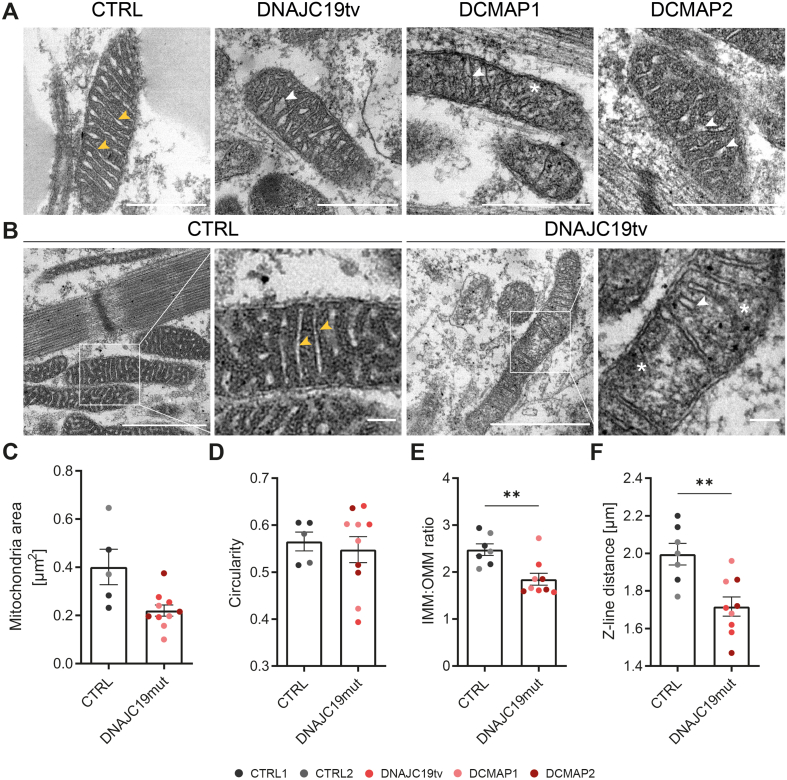
To further evaluate mitochondrial structure, we performed transmission electron microscopy (TEM) on mitochondria and analyzed the images in a blinded manner. Control iPSC-CMs demonstrated intermyofibrillar mitochondria with a clearly visible cristae lumen and densely packed mitochondrial matrix that complemented to a regular cristae pattern (Figure 3A,B). In contrast, mutant iPSC-CMs displayed smaller mitochondria with abnormal invaginations and disturbed mitochondrial structures, brighter matrix areas and local mitochondrial swelling. In addition, we observed reduced numbers and disarrayed cristae structures (Figure 3A,B). Size assessment of mitochondria showed a trend towards reduced areas in all three mutants, whereas the overall mitochondrial shape did not change compared to the controls (Figure 3C,D). As an additional parameter, we determined the ratio of the IMM to the outer mitochondrial membrane (OMM), which was significantly reduced in mutant DNAJC19tv compared to controls, suggesting reduced IMM content (Figure 3E; p = 0.0047). Finally, we assessed sarcomere structures with normal visual alignment, but interestingly reduced Z-line distances, suggesting overall shorter sarcomeres (Figure 3F; p = 0.0028).
Figure 3.
Abnormal cristae structures in DNAJC19 mutants. (A) Representative transmission electron microscopy (TEM) images and (B) higher magnifications are depicted for one control (CTRL2) and all three DCMA mutants (DNAJC19tv, DCMAP1 and DCMAP2). Control iPSC-CMs demonstrate normal inner mitochondrial membrane (IMM) invaginations as well as a regular cristae pattern (yellow arrows). DNAJC19 mutants display abnormal invaginations of the IMM, a reduced number of cristae (white arrows) and dissolved IMM structures (∗). (C–F) Quantification via ImageJ of (C) mitochondrial areas in [μm2], (D) mitochondrial shapes represented by circularity [0–1], (E) IMM to outer mitochondrial membrane (OMM) ratio and (F) Z-line distance in [μm] from at least 100 mitochondria per biological replicate. Note the trend towards smaller mitochondria with a similar shape, whereas IMM to OMM ratios and Z-line distances were significantly reduced in DNAJC19 mutants. iPSC-CMs of both controls (CTRL1, CTRL2) and three mutants (DNAJC19tv, DCMAP1, DCMAP2) were analyzed together in groups. Data are shown as mean ± SEM together with each biological replicate. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns p > 0.05 using unpaired t-test or Mann–Whitney test whenever the data did not follow a Gaussian distribution. (A) scale bars = 0.5 μm; (B) scale bars = 1 μm.
In summary, we observed fundamental structural differences of mutant mitochondria with loss of the mitochondrial network and disturbed cristae formation.
3.3. Elevated oxygen consumption, increased extracellular acidification and ROS production in mutant DNAJC19 iPSC-derived CMs
To assess mitochondrial function, we performed respiration analyses via measuring cellular oxygen consumption (OXPHOS) and proton excretion (as an index for glycolysis) on MACS-purified iPSC-CMs of CTRLs and mutant DNAJC19 iPSC-CMs. We simultaneously examined oxygen consumption rates (OCRs) corresponding to OXPHOS activities and extracellular acidification rates (ECARs) representing the rate of glycolysis through medium acidification such as lactate production (Figure S3A). Afterwards crystal violet absorbance was measured for normalization (Appendix A).
Compared to control iPSC-CMs, all three mutant DNAJC19 cell lines demonstrated significantly elevated OCRs (p < 0.0001; Figure 4A, Figure S3B). We observed an increase in basal-, maximal and ATP-linked respiration, indicating higher electron transport chain (ETC) activity as a potential compensatory mechanism to meet cellular ATP demands (p < 0.0001; Figure 4B, C, Figure S3C).
Figure 4.
Elevated oxygen consumption and increased extracellular acidification in DNAJC19 mutants. (A) Displayed are oxygen consumption rates (OCR), associated (B) respiratory changes, the accompanied impact on the (C) extracellular acidification rate (ECAR) and (D) acidification related categories of CTRL and DNAJC19mut iPSC-CMs performed with the Mito Stress Test Assay (1 μM FCCP, 1 μM Oligomycin and 0.5 μM Rotenone/Antimycin A) using the Seahorse XF96 system. iPSC-CMs of both controls (CTRL1, CTRL2) and three mutants (DNAJC19tv, DCMAP1, DCMAP2) were analyzed together in groups. (A) Mutant iPSC-CMs displayed an overall higher OCR compared to controls. (B) All categories for respiratory measurements demonstrated increased oxygen consumption. (C) ECARs were increased over time in mutant iPSC-CMs, which can be also seen in (D) showing that basal ECARs, oligomycin-induced and FCCP-mediated ECARs have higher flux traces of H+ in mutant DNAJC19 iPSC-CMs. The Mito Stress Test Assay was performed at three time points per condition from 4 to 5 independent experiments and 16–23 technical replicates. Data are shown as mean ± SEM twith the biological replicates superimposed (accordingly color-coded to control or DNAJC19-associated genotypes). ####p < 0.0001, ###p < 0.001, ##p < 0.01, #p < 0.05, ns p > 0.05 using regular two-way ANOVA calculated versus healthy CTRL CMs (A, C) and ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns p > 0.05 using unpaired t-test, calculated versus CTRL (B, D). OCR = oxygen consumption rate; ECAR = extracellular acidification rate; FCCP = Carbonyl cyanide 4-(trifluoromethoxy)-phenylhydrazone; OLIGO = Oligomycin; ROT & AA = Rotenone/Antimycin A.
Under normal conditions, energy generation of iPSC-CMs mainly relies on mitochondrial OXPHOS, whereas under stress conditions (in particular, hypoxia), a switch to non-oxidative glycolysis occurs, where pyruvate is converted to lactate instead of being taken up into mitochondria. In fact, the rates of glycolysis (as assessed by ECAR) were increased in mutant DNAJC19 compared to control cell lines, already at baseline conditions (Figure 4C,D). Furthermore, glycolysis was also elevated in mutant lines after interrupting mitochondrial ATP production and thereby, electron flux along the ETC with oligomycin, a blocker of the F1Fo-ATP synthase (Figure 4C,D). Also, when dissipating the proton gradient across the IMM with FCCP, which maximally accelerates electron flux along the ETC (Figure 4A), or after blockade of electron flux along the ETC by the addition of rotenone and antimycin A (which inhibit complexes I and III or the ETC, respectively), glycolytic flux was consistently higher in mutant DNAJC19 compared to control cell lines (Figure 4D; Figure S3C).
As a proof-of-concept, we interrogated whether the impairment of OXPHOS increases mitochondrial reactive oxygen species (ROS), and whether there are consequences on the mitochondrial membrane potential. The extent of MitoSOX or tetramethylrhodamine (TMRM) accumulation normalized to the MitoTracker signal was used to measure ROS or the mitochondrial membrane potential, respectively, via life cell imaging. As supposed, in DNAJC19tv iPSC-CMs compared to the isogenic control iPSC-CMs (CTRL1), MitoSOX was significantly increased, indicating increased ROS (p = 0.0079; Figure 5A). However, we also observed an elevated TMRM signal, suggesting an increased mitochondrial membrane potential, which might be indicative of an increased reduction of pyridine nucleotide by the TCA cycle (p = 0.0286; Figure 5B). In summary, these data show a clear link towards an increased metabolism and increased ROS production in mutant DNAJC19 iPSC-CMs.
Figure 5.
Increased reactive oxygen species (ROS) and elevated inner mitochondrial membrane potential (Δψm) in mutant DNAJC19tv. Mutant DNAJC19tv CMs displayed elevated MitoSOX/MitoTracker (A) and TMRM/MitoTracker levels (B) compared to CTRL1 iPSC-CMs, indicating elevated ROS levels and increased mitochondrial membrane potential, respectively. Representative confocal live cell images show co-localization of MitoSOX or TMRM with MitoTracker. DNAJC19 iPSC-CMs demonstrated a spotty distribution of mitochondria, most dense around the nucleus while CTRL1 CMs displayed an elongated string-like, more equally distributed structure. Data are shown as mean ± SEM for each biological replicate, with at least 50 iPSC-CMs. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns p > 0.05 using unpaired t-test calculated versus the normalized ratio of CTRL1 iPSC-CMs. Scale bars = 50 μm. TMRM = tetramethylrhodamine.
3.4. Upregulation of respiratory chain (RC) complex expression and increased mitochondrial respiration in DNAJC19tv HeLa cells
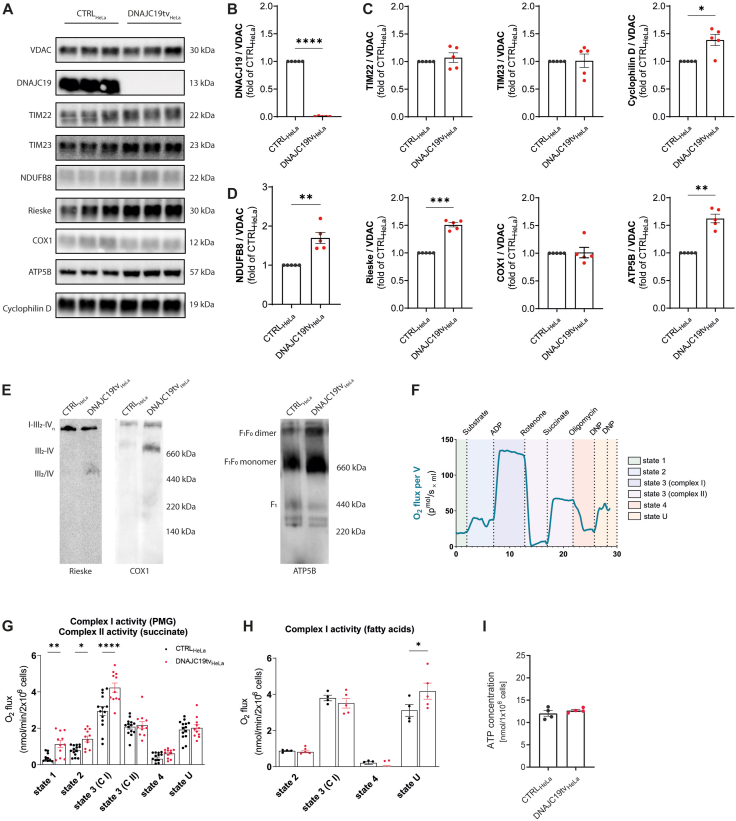
To investigate mitochondrial respiration in a second cell system, we introduced a homozygous truncation variant in DNAJC19 by CRISPR/Cas9 in a HeLa cell line. The genomic variation consists of a 60 bp deletion covering the intron-exon boundary of exon 4 resulting in a deletion of exon 4 at the cDNA level and causing a frameshift with premature stop codon (Figure S4A and B; DNAJC19tvHeLa). Western blot analysis confirmed markedly reduced DNAJC19 protein (p = 0.0031; Figure S4C and D) similar to what was observed in mutant iPSC-CMs. We quantified the amount of VDAC in relation to β-actin, which served as a housekeeping protein and observed no difference in mutant and control HeLa lysates suggesting equal amounts of mitochondria in both cell lines (p = 0.9003; Figure S4C and D). Next, we used isolated mitochondria and confirmed the loss of DNAJC19 in DNAJC19tv cells (p < 0.0001; Figure 6A,B). DNAJC19 has been suggested to be associated with TIM23 mediating the import of protein precursors [28]. Therefore, we investigated the abundance of the central proteins of the translocase of inner membrane (TIM) 23 and 22, respectively. Interestingly, both proteins TIM23 and TIM22 showed equal protein levels in mutant vs. control cells (p = 0.4558 and p = 0.9197, respectively), whereas cyclophilin D, which regulates the mitochondrial permeability transition pore was increased (p = 0.0187; Figure 6A,C).
Figure 6.
Upregulation of respiratory chain complex expression and increased mitochondrial respiration in DNAJC19tv HeLa cells. (A) Immunoblots of isolated mitochondria from CTRLHeLa and DNAJC19tvHeLa normalized to VDAC. Note the loss of mitochondrial import protein DNAJC19 (B) and equal expression of TIM22, TIM23 in DNAJC19tvHeLa cells. Cyclophilin D levels, regulating the mitochondrial permeability transition pore were increased DNAJC19tvHeLa cells (C). RC complexes I, III, V were upregulated in DNAJC19tvHeLa cells, whereas cytochrome C oxidase (COX1), a mediator between complex III and IV, remains unchanged (D). Paired t-test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, mean ± SEM. Blue-Native (BN) PAGE analysis of Digitonin-solubilized membranes and western blotting for complex III complex using antibody against RIESKE and complex IV using antibody against COX1. Assembly states of respiratory chain complexes are indicated with III2-IV and III2/IV, supercomplexes are indicated by I-III-IV. Monomeric and dimeric forms of complex V were detected using antibodies against ATP5B. (E). Schematic presentation of the protocol (F) for respiration analysis of DNAJC19tvHeLa compared to CTRLHeLa cells with either pyruvate, malate and glutamate (PMG) (G) or a mixture of long-chain fatty acids as substrates (H). Increased respiration becomes evident under basal respiration (state 1) and after PMG (state 2). State 3 respiration was measured for complex I and II separately and increased for complex I in (G). Measurement of residual respiration was induced by inhibition of the electron transport chain (state 4). Uncoupling of the respiratory chain enabled measurement of maximum electron transfer capacity by titration of DNP (state U). Uncoupled respiration showed increased electron transfer capacity in DNAJC19tvHeLa cells in (H). Mixed-effects analysis with Bonferroni's post-test for multiple comparisons, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, mean ± SEM. In an ATP assay, intracellular ATP concentrations of CTRLHeLa and DNAJC19tvHeLa cells were measured and showed equal ATP levels in both cell lines (I), unpaired t-test, ∗p < 0.05.
To validate previous findings of elevated oxygen consumption rates in mutant iPSC-CMs we examined the expression of RC complexes and performed a mitochondrial respiration analysis in DNAJC19tv compared to CTRL HeLa cells. Protein expression levels of the RC complexes I (NDUFB8; p = 0.0078), III (Rieske; p = 0.0002), and V (ATP5B; p = 0.0013) were all significantly upregulated in mitochondria of mutant DNAJC19tv cells compared to controls, except for the cytochrome c oxidase which demonstrated equal protein abundance (p = 0.9068; Figure 6A,D). We investigated if the elevated levels of complex I and III constituents are reflected in a stable or even greater abundance of respiratory chain complexes. We therefore solubilized isolated mitochondria in the mild detergent digitonin, which preserves respiratory chain supercomplexes (respirasomes). Separation of solubilized mitochondrial membrane proteins by blue native gel electrophoresis (BN-PAGE; [29]) and subsequent western blotting using antibodies against complex I (NDUFB8) and III (RIESKE) revealed in a greater abundance of individual complexes (III2/IV and III2-IV) and a stable presence of supercomplexes (I-III2-IV). Western blotting analysis for ATP5B demonstrated elevated amounts of the monomeric and dimeric form of complex V (Figure 6E).
To confirm that increased RC protein abundance is linked to higher mitochondrial respiration, we performed mitochondrial respiration analysis using mutant DNAJC19tv and CTRL HeLa permeabilized cells with the protocol shown in Figure 6F on the Oroboros Oxygraph-2K system. Basal respiration (state 1) on endogenous substrates was increased in DNAJC19tv cells (p = 0.001). Upon addition of pyruvate, malate and glutamate (PMG) as substrates (state 2) as well as application of ADP to convey a high energy demand to the mitochondria (state 3) respiration was consistently elevated in DNAJC19tvHeLa cells (p = 0.0267 and p < 0.0001, respectively) indicating increased maximum respiration to generate more NADH from substrates and possibly more ATP. With rotenone we have blocked complex I respiration and added succinate to measure the activity of complex II, which was not changed compared to the CTRL cells (p > 0.999). To selectively inhibit the F1FO-ATPase and thus stop the proton flux towards the mitochondrial matrix, oligomycin was added (state 4). However, the extent of proton leak between mutant and control cells appeared to be comparable (p > 0.999), suggesting that mutant mitochondria are generally well coupled. Next, we applied the uncoupling agent DNP so that the proton gradient collapsed and the complex II-coupled ETC operated at its maximum capacity to maintain the gradient (state U). There was also no difference between both conditions indicating that uncoupled complex II respiration remains the same (p > 0.999; Figure 6G). In a second approach, we used long-chain fatty acids as a substrate and measured complex I activity. However, in contrast to PMG leading to increased mitochondrial respiration, long-chain fatty acids did not cause an increase, instead respiration remained the same in DNAJC19tv cells (p > 0.999). Finally, when DNP was added to uncouple the ETC, mutant cells appeared to have a higher ETC capacity than CTRL cells (p = 0.0269; Figure 6H).
In summary, mitochondrial respiration was increased at baseline and accelerated with carbohydrates as substrate in DNAJC19tv cells, which was not seen with fatty acids which may indicate substrate preference. Overall ETC capacity was increased, which would suggest higher ATP levels. Therefore, we performed an ATP-assay to measure the ATP concentration in the cells, but unexpectedly we observed similar ATP levels in DNAJC19tv and CTRL cells (p = 0.6957; Figure 6I). This may suggest a higher ATP consumption or demand of cellular processes in mutant cells.
3.5. Alterations in cardiac metabolism in mutant DNAJC19 iPSC-derived CMs
To further assess substrate utilization, we performed radioactive tracer uptake studies on 60-days-old iPSC-CMs to simultaneously measure fatty acid and glucose uptake using 125I-BMIPP (125I-β-methyl-iodo-phenyl-pentadecanoic acid) and 18F-FDG (18F-2-fluoro-2-deoxy-d-glucose), respectively. In addition, we measured the activity of the cysteine/glutamate transporter by quantification of the cellular uptake of 18F-FASu (5-18F-l-aminosuberic acid; Figure 7A). The cellular uptake of each radioactively labelled tracer was normalized to the uptake of CTRL1. The utilization of fatty acids represented by 125I-BMIPP uptake was significantly decreased in all three mutant DNAJC19 iPSC-CMs, with the most significant decrease in DCMAP2 (p < 0.0001) and to a lower extent in DCMAP1 (p = 0.0299) and DNAJC19tv (p = 0.001), respectively (Figure 7B). Simultaneous assessment of 18F-FDG uptake revealed a significantly increased uptake of DCMAP1 compared to CTRL1 (p = 0.0197), suggesting higher glucose utilization. In contrast, DCMAP2 (n.s.) and DNAJC19tv (p = 0.0046) showed decreased 18F-FDG uptakes, suggesting a shift towards utilization of other substrates (Figure 7C). Finally, we measured 18F-FASu uptake to address the cellular response to oxidative stress via the cysteine/glutamate transporter in CTRLs and DNAJC19tv. Interestingly, the DNAJC19tv iPSC-CMs showed an increased cellular response (p = 0.0385), which suggests a potential defense mechanism against increased ROS (Figure 7D).
Figure 7.
Radioactive tracer uptakes reveal metabolic alterations in DNAJC19 mutants. (A) Schematic description of radioactive tracer experiments of iPSC-CMs with 125I-BMIPP and 18F-FDG or 18F-FASu to examine the cellular uptake of radioactive tracers to evaluate fatty acid and glucose utilization or the functionality of the cystine/glutamate transporter, respectively. (B) Mutant DNAJC19 iPSC-CMs displayed a reduced uptake of 125I-BMIPP to different degrees. (C) Simultaneous assessment of 18F-FDG uptake demonstrated decreased uptake in DNAJC19tv and DCMAP2, whereas uptake of DCMAP1 was increased. (D) Radioactive tracer uptake of 18F-FASu was quantified to assess the cellular response to oxidative stress via the cysteine/glutamate transporter. DNAJC19tv displayed increased cellular response to oxidative stress and the potential to switch to l-glutamate. Data for each biological replicate were examined by calculating of the uptake index after normalization to protein content using BCA Protein Assay and normalization to the uptake of CTRL1. Each biological replicate consisted of 2–4 technical replicates that were examined based on 4–6 independent cultures. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns p > 0.05 using paired t-test calculated versus the normalized uptake index of CTRL1 iPSC-CMs; CPM = counts per minute; 18F-FDG = 18F-2-fluoro-2-deoxy-d-glucose; 125I-BMIPP = 125I-β-methyl-iodo-phenyl-pentadecanoic acid; 18F-FASu = 5-18F-l-aminosuberic acid (incorporated characteristics of both natural substrates: l-cystine and l-glutamate). The schematic illustration in (A) was created with Biorender.com.
Overall, mutant iPSC-CMs displayed a decreased fatty acid uptake and utilization of other substrates, which confirms changes in metabolic features in mutant DNAJC19 iPSC-CMs.
3.6. Altered calcium homeostasis, contractility and arrhythmic events detected in mutant DNAJC19 iPSC-CMs
Since the processes of excitation-contraction (EC) coupling demand large amounts of ATP from mitochondria, we determined the impact of DNAJC19 mutations on cytosolic Ca2+ handling, cell shortening and the occurrence of arrhythmic events in 120-days-old iPSC-CM in the absence and presence of β-adrenergic stimulation with increasing concentrations of isoproterenol (10–100 nM). IPSC-CMs were all exposed to electrical field stimulation at 0.5 Hz. Since control iPSC-CMs had a slow spontaneous beating rate of <0.5 Hz, they largely followed the external electrical pacing at 0.5 Hz in the absence, but also the presence of β-adrenergic stimulation (Figure 8A). In contrast, DCMAP2 and DNAJC19tv iPSC-CMs had either a spontaneous beating rate that was substantially higher than 0.5 Hz (p < 0.0001), or more delayed- and early after depolarizations (DAD/EADs) that lead to arrhythmia, and this was further increased by isoproterenol (Figure 8A). Conversely, DCMAP1 iPSC-CMs had a much lower spontaneous beating rate or arrhythmia affinity that was only marginally higher compared to paced control iPSC-CMs (p = 0.037; Figure 8A). Systolic cell shortening was reduced in all three DCMA mutations compared to control iPSC-CM (p = 0.0097 or less; Figure 8B). This difference was unlikely related to the higher beating rates, since the DCMAP1 mutant had the lowest cell shortening amplitude (Figure 8B) despite a largely comparable beating rate close to 0.5 Hz (Figure 8A). Interestingly, when monitoring intracellular Ca2+ concentrations ([Ca2+]c) using Indo-1 AM, we observed substantially elevated diastolic [Ca2+]c in all three DNAJC19 mutants (p < 0.0001; Figure 8C), which was in line with shortened diastolic sarcomere (Z-disc) lengths (Figure 3F), while systolic [Ca2+]c were comparable between the four different cell lines.
Figure 8.
DNAJC19 mutants display alterations in calcium homeostasis and arrhythmic events. (A)–(D) Indo-1-AM based calcium transients and edge-detection based cell contraction displayed increased (A) frequency, decreased (B) cell shortenings; elevated (C) diastolic calcium concentrations and (D) prolonged time of recovery in all mutant iPSC-CMs. Sin exp tau was frequency corrected by using Bazett's formula. (E) Arrhythmic events were quantified under basal conditions (NT) by subjective assignment to defined groups based on the occurrence of individual characteristics: breaks, delayed after depolarization (DAD)/early after depolarization (EADs) and fibrillation-like events (threshold >1 Hz), when present in at least 4 events using Indo-1-AM based visualization of calcium transients. Arrhythmic events are displayed in percent as mean ± SEM. with ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns p > 0.05 using unpaired t-test calculated versus CTRL. Each biological replicate consisted of 2–8 technical replicates (iPSC-CMs) that were examined based on 3–6 independent experiments with at least n = 13–35 iPSC-CMs per group. (F) Beat-to-beat rate variability was calculated using cell contractions identified by edge detection according to the following formula: . Each biological replicate consisted of 3–6 independent cultures with at least n = 14–23 iPSC-CMs per group. Note, the beat-to-beat rate variability of DNAJC19tv was increased at baseline and in all three mutant lines upon raised isoprenaline (Iso) concentrations to discordant levels. Statistical analysis was performed using regular two-way ANOVA with ####p < 0.0001, ###p < 0.001, ##p < 0.01, #p < 0.05, ns p > 0.05 with ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns p > 0.05 Bonferroni post-tests calculated versus CTRL. At least 120 days in vitro aged mattress-cultured iPSC-CMs of CTRL and DNAJC19mut lines were investigated towards alterations in calcium homeostasis and contractility using Indo-1-AM staining and edge-detection via the IonOptix system. Single-cell iPSC-CMs were electrically stimulated with a frequency of 0.5 Hz and a stress protocol was performed including N-Tyrode (NT) and Iso concentrations 10 nM, 30 nM, 100 nM.
The main mechanism of cytosolic Ca2+ removal during diastole is the sarcoplasmic reticulum (SR) Ca2+ ATPase (SERCA), and the rate of SERCA is reflected best by the time constant of cytosolic Ca2+ decay (τ). This value, which was corrected to the respective beating rate using the Bazett's formula, was substantially slower in the three mutant DCMA compared to the control iPSC-CMs at baseline, and even in response to isoproterenol (p < 0.0001; Figure 8D), indicating slower SERCA activity as a likely explanation for elevated diastolic [Ca2+]c.
Since elevated diastolic [Ca2+]c may cause spontaneous SR Ca2+ release events, we monitored arrhythmic events by visually determining pauses, DAD/EADs and a fibrillation-like phenotype over a time frame of 120 s for each treatment condition in spontaneously beating iPSC-CM (Figure S5A, S5B, S5C and S5D). Overall, DNAJC19tv and DCMAP1 demonstrated a significantly higher rate of arrhythmic events compared to CTRLs (p = 0.0056 and p = 0.0311, respectively), whereas DCMAP2 did not (Figure 8E). However, when categorizing the events, DCMAP1 revealed a higher rate of DAD/EADs, whereas all other events were more equally distributed (Figure S5E). Additionally, we calculated the beat-to-beat rate variability index displaying the variability in time between each Ca2+ transient (difference between beat-to-beat rate). Here, all three mutant lines demonstrated a significantly higher beat-to-beat rate variability with increasing β-adrenergic stimulation compared to CTRLs (p < 0.0001; Figure 8F).
Taken together, loss-of-function mutations in DNAJC19 and the associated mitochondrial and metabolic remodeling observed in human iPSC-CMs were associated with disturbed EC coupling, where slowed SERCA activity (which is particularly sensitive to drops in cellular ATP/ADP ratios) [30,31] accounts for elevated diastolic [Ca2+]c, which likely underlies shorter diastolic sarcomere length reflecting diastolic dysfunction, and potentially also cellular arrhythmias, which is aggravated during β-adrenergic stimulation.
4. Discussion
To unravel the pathogenesis of mutant DNAJC19 in cardiac disease, we investigated iPSC-derived CMs from two patients with discordant phenotypes and a gene-edited truncation variant of DNAJC19 in an isogenic background cell line, thereby providing for the first time a powerful platform for modeling DCMA related cardiac features in a comprehensive set-up of mature (>60 days old), purified CMs in a 2D culture system. We found fragmented and reduced amounts of mitochondria with disturbed cristae morphology, density and distribution in mutant DNAJC19 iPSC-CMs. Unexpectedly, we observed higher oxygen consumption rates and an upregulated mitochondrial respiration in mutant DNAJC19 cells. Elevated mitochondrial respiration was not accompanied by increased fatty acid transport and oxidation, indicating substantial shifts in the substrate choice and potential elevation of alternative substrates. In addition, on single CM level we discovered decreased contractility, altered cytosolic Ca2+ homeostasis and a trend towards cellular arrhythmias.
DCMA associated cardiomyopathy is caused by truncation mutations in DNAJC19 suggesting a partial loss of function mechanism. Therefore, we assessed the remaining DNAJC19 protein in iPSC-CMs carrying the homozygous DNAJC19 (NM_145261.4):c.130-1G > C mutation, which is predicted to lead to a C-terminal truncated protein with a loss of the DnaJ domain. Despite the absence of full-length DNAJC19 proteins in all our three mutant cell lines shown by western blot analysis (Figure 1C), we observed remaining protein expression by immunofluorescent staining with a signal mainly restricted in the nucleus (Figure 2A). This might be surprising as the predicted N-terminal transmembrane domain anchoring DNAJC19 in the IMM remains intact while the matrix facing DnaJ domain is absent [1,12]. Interestingly, PHB1 as well as PHB2 are also localized in both mitochondria and nuclei and influence transcription, modulate proteins involved in apoptosis and affect the cell cycle [32,33]. However further studies are required to validate a potential role of DNAJC19 in the nucleus.
The exact function of DNAJC19 is currently unknown and different hypotheses predict a role in protein import into mitochondria or a role, related to its interaction with prohibitins and their ability to control inner membrane organization. Regarding a potential function in protein import, we did not find evidence for deficient TIM23 translocase as protein levels of its central component Tim23 or of proteins with predicted presequences, like ATP5B were unaltered [34]. The mitochondria-associated phenotype of mitochondrial fragmentation was detected in co-localization studies of mutant CMs in addition to a significantly decreased mitochondrial content in mutant DNAJC19 CMs (Figure 2). The analysis of intermyofibrillar mitochondria sizes and shapes revealed a trend towards smaller sized mitochondria with otherwise unchanged shapes. We also observed a loss of highly tubular mitochondria that were partly still associated with sarcomeres in mutant iPSC-CMs. The balance of mitochondrial to myofibrillar area is essential to ensure proper cardiac function and requires to be maintained throughout the whole development of the human heart. Comparing fetal embryonic stem cell-derived CMs' mitochondrial content with adult CMs' content, the mitochondrial mass increases over prolonged cultivation times [35,36]. The assessment of 60-day-old mutated iPSC-CMs unraveled decreased mitochondrial contents with shared fiber densities, suggesting a pathogenic relevance of DNAJC19 in mitochondrial biosynthesis. It is known that early CMs present higher mitochondrial synthesis rates as well as faster dynamics than CMs after postnatal development [37,38]. Interestingly, fragmentation of mitochondria and decreased perimeters have been previously observed in more immature, 21-day- old iPSC-derived CMs of two other DCMA patients with the same mutation, suggesting that defects in mitochondrial dynamics might be essential in the early disease process [20]. The same authors also observed a shift in the ratio between long (L-) and short (S-) OPA1 forms with implications for mitochondrial fusion [20,39,40]. In line, we found that loss of full-length DNAJC19 caused alterations in mitochondrial cristae formation such as an irregular, less dense pattern. The IMM invaginations seemed to be shorter, did not reach deep into the matrix and either had a hardly visible or a bulky cristae lumen when analyzed on the ultrastructural level. In addition, the ratio of IMM to OMM was significantly decreased in mitochondria of mutant iPSC-CMs (Figure 3). The same observations of abnormal cristae morphologies were also reported in the study of Richter-Dennerlein et al. upon siRNA-mediated knock-down of DNAJC19 in HEK293T cells [12]. In this report, defects in cristae formation was shown to been independent of OPA1 processing. This may suggest that not OPA1 regulation but rather alterations in CL affect mitochondrial morphology in DCMA [12,41].
We assessed the contribution of both energetic pathways, OXPHOS and glycolysis by measuring OCRs and ECARs. Elevated basal respiration, ATP linked respiration and maximal respiration indicate an elevated oxygen turnover (Figure 4). In particular, the elevated ATP-linked respiration indicated that higher ETC activities are required to establish the mitochondrial membrane potential and the corresponding proton motive force to meet cellular ATP demands. FCCP, which leads to the collapse of the proton gradient and allows direct diffusion of protons through the IMM into the matrix resulted in full activation of the ETC to re-establish the mitochondrial membrane potential. There are multiple causes of increased OCR levels which such as proton leakage due to disintegrated IMM, mismatch of ATP supply and demands, mispositioning of the ETC complexes, abnormal supercomplex formation and loss of the proton trap function. To assess mutant DNAJC19 proteins in this context, the expression of RC proteins and mitochondrial respiration in mutant DNAJC19 HeLa cells. In line with the iPSC CM data, succinate and fatty acid driven respiration were unchanged and pyruvate-malate driven respiration were even increased in DNAJC19 HeLa cells, compared to control (Figure 6G, H). Increased respiration was associated with increased protein levels of key proteins of the respiratory chain (NDUFB8, complex I, RIESKE, complex III and ATP5B, complex V; Figure 6A–D). Moreover, increased levels of respiratory chain subunits resulted in an elevated provision of respiratory chain complexes to form supercomplexes (respirasomes) and dimeric complex V (Figure 6E). It can be speculated that the assembly of respiratory chain supercomplexes may reach a limit due to structural limitations of HeLa cell line mitochondria. Increased respiration may serve to maintain similar levels of ATP (Figure 6I).
Few reports exist, where material from single patients with DNAJC19 defects has been analyzed without providing a comprehensive data set. In one patient in alterations in the RC activity has been demonstrated by reduced complex IV activity in muscle tissue [5]. Biochemical analysis of skeletal muscle tissue from another DCMA patient revealed slightly reduced ATP production as well as partially reduced complex I, complex II and complex IV activities and stable respiratory chain complexes in BN-PAGE analysis [3]. The report of Al Teneiji et al. used a more specified analysis and found a particular reduction of complex I and I + III activities, while complex II + III and complex IV activities were rather increased in muscle biopsies and dermal fibroblasts from DCMA patients [4]. Together with our analysis, these publications underline DCMA-specific alterations in mitochondrial energy metabolism. Mitochondrial dysfunction due to DNAJC19 deficiency results in a compensatory increase in respiratory activity to cover energy demands.
Dysfunction of OXPHOS and uncoupling of mitochondrial ETC are a major source of ROS production causing oxidative stress on CMs with potentially subsequent effects on signaling pathways and contractile function [42]. Indeed, we found increased mitochondrial ROS measured by MitoSOX in DNAJC19tv iPSC-CMs compared to their isogenic control (Figure 5) suggesting that ROS may contribute to heart failure in DCMA related cardiomyopathy. Notably, similar observations were seen in BTHS iPSC-CMs showing sarcomeric disarray and reduced contractility, which could be reversed by scavenging of ROS via MitoTEMPO, which may also be beneficial in DCMA cardiomyopathy [43].
Generation of ATP in healthy human CMs relies mainly on fatty acid metabolism, but changes in energy metabolism in many disease states have demonstrated metabolic flexibility, allowing a switch between different carbon sources such as glucose [44,45]. Glucose utilization was indirectly assessed by ECARs, which were increased basally, indicating an increase in overall metabolic flux in mutant DNAJC19 iPSC-CMs. Next, we investigated substrate utilization by radiotracer uptake studies. It is important to note that the metabolic switch towards maximal fatty acid utilization in iPSC-CMs requires about 56 days of culture, which stresses the importance of using at least 60-days-old iPSC-CMs for such investigations [46]. Uptake studies of 125I-BMIPP consistently showed the decrease of fatty acid uptakes in all mutant CMs. However, the simultaneous investigation of glucose uptake via 18F-FDG demonstrated an increase only in DCMAP1, whereas the two other mutant iPSC-CMs indicated an opposing trend (Figure 7). Notably, in BTHS similar metabolic alterations have been reported – a reduced long chain fatty acid utilization with enhanced glucose uptake in mutant TAZ iPSC-CMs. 18F-FASu measurements indicated increased cystine uptake in DCMA iPSC-CM. Also in BTHS, elevated cystine uptake was considered to fuel glutathione biosynthesis to protect from oxidative damage [47].
Early onset of heart failure due to cardiomyopathy is the most common cause of death in DCMA. Therefore, we investigated for the first time in isolated iPSC-CMs the downstream effects on cardiac contractility, calcium homeostasis and arrhythmias. Despite a high variability between the three different mutant iPSC-CM lines, it became clear that spontaneous beating frequencies are increased with more beat-to-beat variability. Here, we speculate that those may contribute to an increased energy demand and subsequently an increased respiration. In addition, we observed increased diastolic [Ca2+]c and reduced cell shortening as a measure for contractility in mutant isolated CMs (Figure 8). Based on the ultra-structurally intact sarcomeres, albeit shorter, we assume that the main cause of CM injury is not the sarcomeres themselves, but the underlying mitochondrial dysfunction and increased ROS production which may impinge particularly on SERCA activity, which can be reduced by either energetic deficit and/or ROS [30,31]. However, our observations are limited to single CMs from a 2D culture, and more detailed investigations of 3D microtissue simulating myocardial tissue or even animal models are required to characterize DCMA related cardiomyopathy in more detail and provide the link of mechano-energetic coupling and altered mitochondrial function [43]. In this sense, a recent study of BTHS related cardiomyopathy in the Taz-KD mouse needs to be noted showing that the disruption of mechano-energetic uncoupling rather than previously suggested mitochondrial respiration and oxidative stress are the primary problem of BTHS-cardiomyopathy [18].
Finally, DCMA is a multisystemic disease with a heterogenous clinical presentation and a high mortality within the first year of life where patients are mainly suffering from heart failure. A recent cohort study of DNAJC19 mutation carriers from the Canadian Hutterite population showed that half demonstrated dilated cardiomyopathy and more than 80% a prolonged QT interval [6]. Prospectively, it would be interesting to get further insight in the cause of the prolonged QT-interval by studying ion channel currents and the action potential in more detail.
Despite being siblings with a similar genetic background, both investigated patients displayed different phenotypes as DCMAP1 required a heart transplant early after birth, where DCMAP2 showed only minor signs of cardiomyopathy underlying intrafamilial variability. Our investigations of patient-derived iPSC-CMs and the gene-edited DNAJ19tv showed many similarities with gradual differences with regards to structural changes of mitochondria and changes in OXPHOS indicating robust upstream processes primarily associated to the loss of the DnaJ domain of DNAJC19. Substrate utilization,Ca2+ handling and arrhythmias appear to be more variable between the different mutant conditions possibly also due to modifying factors such as the genetic background, but also culture conditions and the limitations of the in vitro model itself [48,49].
5. Conclusion
In conclusion, we provide for the first time a comprehensive in vitro model of iPSC-derived CMs from two patients and a gene-edited truncated DNAJC19 line in an isogenic control background which underlines DCMA as a mitochondrial disease with substantial structural alterations of mitochondria and an increased OXPHOS activity. In line, mutant DNAJC19 HeLa cells displayed upregulation of respiratory chain complexes and an increased coupled mitochondrial respiration. However, underlying mechanisms explaining those novel and unexpected observations remain unknown. But downstream, mutant CMs demonstrated disturbed Ca2+ homeostasis, decreased contractility and arrhythmias mimicking the DCMA associated cardiomyopathy.
Funding
This work was supported by the Federal Ministry of Education and Research (BMBF; grant no: 01EO1504) and by the German Research Foundation (DFG; SFB 1525; project number: 453989101; to B. Gerull, T. Higuchi, K. Lorenz, J. Dudek and C. Maack). B. Gerull and P. Wörsdörfer received funding from the IZKF at the University of Würzburg (project E-D410). The Transmission Electron Microscope JEOL JEM-1400Flash was funded by the DFG grant GEPRIS number 426173797 (INST 93/1003-1 FUGG) and the Transmission Electron Microscope JEOL JEM-2100 was funded by DFG grant GEPRIS number 218894163. J. Dudek (DU1839/2-1) and C. Maack (MA 2528/7-1, 8-1) are supported by the DFG.
CRediT authorship contribution statement
Anna Janz: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. Katharina Walz: Data curation, Formal analysis, Investigation, Validation, Visualization, Writing – review & editing. Alexandra Cirnu: Data curation, Formal analysis, Investigation, Validation, Writing – review & editing. Jessica Surjanto: Data curation, Investigation, Visualization. Daniela Urlaub: Data curation, Investigation, Methodology, Validation. Miriam Leskien: Data curation, Formal analysis, Investigation, Methodology. Michael Kohlhaas: Formal analysis, Investigation, Methodology, Writing – review & editing. Alexander Nickel: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. Theresa Brand: Methodology, Resources, Validation, Writing – review & editing. Naoko Nose: Data curation, Formal analysis, Validation. Philipp Wörsdörfer: Data curation, Formal analysis, Resources, Visualization. Nicole Wagner: Data curation, Formal analysis, Methodology, Visualization. Takahiro Higuchi: Resources, Supervision, Validation, Writing – review & editing. Christoph Maack: Methodology, Resources, Supervision, Validation, Writing – review & editing. Jan Dudek: Methodology, Resources, Validation, Writing – original draft, Writing – review & editing. Kristina Lorenz: Methodology, Resources, Supervision, Writing – review & editing. Eva Klopocki: Methodology, Resources, Writing – review & editing. Süleyman Ergün: Resources, Supervision, Writing – review & editing. Henry J. Duff: Conceptualization, Resources, Supervision, Writing – review & editing. Brenda Gerull: Conceptualization, Data curation, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Brenda Gerull reports a relationship with Chiesi Pharmaceuticals Inc that includes: speaking and lecture fees and travel reimbursement.
Acknowledgments
We are grateful to the patients participating in this study and to the genetic counsellors of the Alberta Children's Hospital Genetics Clinic for supporting this research. We would like to thank Martina Regensburger (posthum), Anne Kilinc, Susanne Schraut, Anja Sauer for their excellent technical help. Furthermore, we thank Saskia Mühlig and Lars Mayer for assisting with the tracer uptake studies. We would also like to thank Christian Stigloher, Daniela Bunsen and Claudia Gehrig of the Imaging Core Facility (Biocenter, Julius-Maximilians-University Würzburg, Würzburg, Germany) for their excellent support with the electron microscopy analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2023.101859.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Davey K.M., Parboosingh J.S., McLeod D.R., Chan A., Casey R., Ferreira P., et al. Mutation of DNAJC19, a human homologue of yeast inner mitochondrial membrane co-chaperones, causes DCMA syndrome, a novel autosomal recessive Barth syndrome-like condition. J Med Genet. 2006;43 doi: 10.1136/jmg.2005.036657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sparkes R., Patton D., Bernier F. Cardiac features of a novel autosomal recessive dilated cardiomyopathic syndrome due to defective importation of mitochondrial protein. Cardiol Young. 2007;17 doi: 10.1017/S1047951107000042. [DOI] [PubMed] [Google Scholar]
- 3.Ojala T., Polinati P., Manninen T., Hiippala A., Rajantie J., Karikoski R., et al. New mutation of mitochondrial DNAJC19 causing dilated and noncompaction cardiomyopathy, anemia, ataxia, and male genital anomalies. Pediatr Res. 2012;72 doi: 10.1038/pr.2012.92. [DOI] [PubMed] [Google Scholar]
- 4.Al Teneiji A., Siriwardena K., George K., Mital S., Mercimek-Mahmutoglu S. Progressive cerebellar atrophy and a novel homozygous pathogenic DNAJC19 variant as a cause of dilated cardiomyopathy ataxia syndrome. Pediatr Neurol. Sep 2016;62:58–61. doi: 10.1016/j.pediatrneurol.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 5.Ucar S.K., Mayr J.A., Feichtinger R.G., Canda E., Coker M., Wortmann S.B. Previously unreported biallelic mutation in DNAJC19: are sensorineural hearing loss and basal Ganglia Lesions additional features of dilated cardiomyopathy and ataxia (DCMA) syndrome? JIMD Rep. 2017;35:39–45. doi: 10.1007/8904_2016_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Machiraju P., Degtiarev V., Patel D., Hazari H., Lowry R.B., Bedard T., et al. Phenotype and pathology of the dilated cardiomyopathy with ataxia syndrome in children. J Inherit Metab Dis. Mar 2022;45:366–376. doi: 10.1002/jimd.12441. [DOI] [PubMed] [Google Scholar]
- 7.Lu Y.W., Claypool S.M. Disorders of phospholipid metabolism: an emerging class of mitochondrial disease due to defects in nuclear genes. Front Genet. 2015;6:3. doi: 10.3389/fgene.2015.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohtsuka K., Hata M. Mammalian HSP40/DNAJ homologs: cloning of novel cDNAs and a proposal for their classification and nomenclature. Cell Stress Chaperones. Apr 2000;5:98–112. doi: 10.1379/1466-1268(2000)005<0098:mhdhco>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayer M.P., Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. Mar 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mokranjac D., Sichting M., Neupert W., Hell K. Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J. Oct 1 2003;22:4945–4956. doi: 10.1093/emboj/cdg485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Laan M., Meinecke M., Dudek J., Hutu D.P., Lind M., Perschil I., et al. Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nat Cell Biol. Oct 2007;9:1152–1159. doi: 10.1038/ncb1635. [DOI] [PubMed] [Google Scholar]
- 12.Richter-Dennerlein R., Korwitz A., Haag M., Tatsuta T., Dargazanli S., Baker M., et al. DNAJC19, a mitochondrial cochaperone associated with cardiomyopathy, forms a complex with prohibitins to regulate cardiolipin remodeling. Cell Metabol. Jul 1 2014;20:158–171. doi: 10.1016/j.cmet.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 13.Merkwirth C., Langer T. Prohibitin function within mitochondria: essential roles for cell proliferation and cristae morphogenesis. Biochim Biophys Acta. Jan 2009;1793:27–32. doi: 10.1016/j.bbamcr.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 14.Osman C., Merkwirth C., Langer T. Prohibitins and the functional compartmentalization of mitochondrial membranes. J Cell Sci. Nov 1 2009;122:3823–3830. doi: 10.1242/jcs.037655. [DOI] [PubMed] [Google Scholar]
- 15.Acehan D., Malhotra A., Xu Y., Ren M., Stokes D.L., Schlame M. Cardiolipin affects the supramolecular organization of ATP synthase in mitochondria. Biophys J. 2011;100 doi: 10.1016/j.bpj.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chatzispyrou I.A., Guerrero-Castillo S., Held N.M., Ruiter J.P.N., Denis S.W., L I.J., et al. Barth syndrome cells display widespread remodeling of mitochondrial complexes without affecting metabolic flux distribution. Biochim Biophys Acta, Mol Basis Dis. Nov 2018;1864:3650–3658. doi: 10.1016/j.bbadis.2018.08.041. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh S., Basu Ball W., Madaris T.R., Srikantan S., Madesh M., Mootha V.K., et al. An essential role for cardiolipin in the stability and function of the mitochondrial calcium uniporter. Proc Natl Acad Sci U S A. Jul 14 2020;117:16383–16390. doi: 10.1073/pnas.2000640117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertero E., Nickel A., Kohlhaas M., Hohl M., Sequeira V., Brune C., et al. Loss of mitochondrial Ca(2+) uniporter limits inotropic reserve and provides trigger and substrate for arrhythmias in Barth syndrome cardiomyopathy. Circulation. Nov 23 2021;144:1694–1713. doi: 10.1161/CIRCULATIONAHA.121.053755. [DOI] [PubMed] [Google Scholar]
- 19.Dudek J., Maack C. Barth syndrome cardiomyopathy. Cardiovasc Res. Mar 15 2017;113:399–410. doi: 10.1093/cvr/cvx014. [DOI] [PubMed] [Google Scholar]
- 20.Rohani L., Machiraju P., Sabouny R., Meng G., Liu S., Zhao T., et al. Reversible mitochondrial fragmentation in iPSC-derived cardiomyocytes from children with DCMA, a mitochondrial cardiomyopathy. Can J Cardiol. Apr 2020;36:554–563. doi: 10.1016/j.cjca.2019.09.021. [DOI] [PubMed] [Google Scholar]
- 21.Machiraju P., Wang X., Sabouny R., Huang J., Zhao T., Iqbal F., et al. SS-31 peptide reverses the mitochondrial fragmentation present in fibroblasts from patients with DCMA, a mitochondrial cardiomyopathy. Front Cardiovasc Med. 2019;6:167. doi: 10.3389/fcvm.2019.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson C.J., Kahl A., Fruitman H., Qian L., Zhou P., Manfredi G., et al. Prohibitin levels regulate OMA1 activity and turnover in neurons. Cell Death Differ. Jun 2020;27:1896–1906. doi: 10.1038/s41418-019-0469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabbah H.N. Targeting the mitochondria in heart failure: a translational perspective. JACC Basic Transl Sci. Jan 2020;5:88–106. doi: 10.1016/j.jacbts.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bers D.M., Perez-Reyes E. Ca channels in cardiac myocytes: structure and function in Ca influx and intracellular Ca release. Cardiovasc Res. May 1999;42:339–360. doi: 10.1016/s0008-6363(99)00038-3. [DOI] [PubMed] [Google Scholar]
- 25.Janz A., Chen R., Regensburger M., Ueda Y., Rost S., Klopocki E., et al. Generation of two patient-derived iPSC lines from siblings (LIBUCi001-A and LIBUCi002-A) and a genetically modified iPSC line (JMUi001-A-1) to mimic dilated cardiomyopathy with ataxia (DCMA) caused by a homozygous DNAJC19 mutation. Stem Cell Res. Jul 2020;46 doi: 10.1016/j.scr.2020.101856. [DOI] [PubMed] [Google Scholar]
- 26.Kadari A., Mekala S., Wagner N., Malan D., Koth J., Doll K., et al. Robust generation of cardiomyocytes from human iPS cells requires precise modulation of BMP and WNT signaling. Stem Cell Rev Rep. Aug 2015;11:560–569. doi: 10.1007/s12015-014-9564-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feaster T.K., Cadar A.G., Wang L., Williams C.H., Chun Y.W., Hempel J.E., et al. Matrigel mattress: a method for the generation of single contracting human-induced pluripotent stem cell-derived cardiomyocytes. Circ Res. Dec 4 2015;117:995–1000. doi: 10.1161/CIRCRESAHA.115.307580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinha D., Srivastava S., D'Silva P. Functional diversity of human mitochondrial J-proteins is independent of their association with the inner membrane presequence translocase. J Biol Chem. Aug 12 2016;291:17345–17359. doi: 10.1074/jbc.M116.738146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wittig I., Braun H.P., Schägger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 30.Tian R., Nascimben L., Ingwall J.S., Lorell B.H. Failure to maintain a low ADP concentration impairs diastolic function in hypertrophied rat hearts. Circulation. Aug 19 1997;96:1313–1319. doi: 10.1161/01.cir.96.4.1313. [DOI] [PubMed] [Google Scholar]
- 31.Tian R., Ingwall J.S. Energetic basis for reduced contractile reserve in isolated rat hearts. Am J Physiol. Apr 1996;270:H1207–H1216. doi: 10.1152/ajpheart.1996.270.4.H1207. [DOI] [PubMed] [Google Scholar]
- 32.Signorile A., Sgaramella G., Bellomo F., De Rasmo D. Prohibitins: a critical role in mitochondrial functions and implication in diseases. Cells. Jan 18 2019;8 doi: 10.3390/cells8010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatsuta T., Model K., Langer T. Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol Biol Cell. Jan 2005;16:248–259. doi: 10.1091/mbc.E04-09-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohta S., Tomura H., Matsuda K., Kagawa Y. Gene structure of the human mitochondrial adenosine triphosphate synthase beta subunit. J Biol Chem. 1988/08/15/1988;263:11257–11262. [PubMed] [Google Scholar]
- 35.Cao F., Wagner R.A., Wilson K.D., Xie X., Fu J.D., Drukker M., et al. Transcriptional and functional profiling of human embryonic stem cell-derived cardiomyocytes. PLoS One. 2008;3:e3474. doi: 10.1371/journal.pone.0003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu X.Q., Soo S.Y., Sun W., Zweigerdt R. Global expression profile of highly enriched cardiomyocytes derived from human embryonic stem cells. Stem Cell. Sep 2009;27:2163–2174. doi: 10.1002/stem.166. [DOI] [PubMed] [Google Scholar]
- 37.Piquereau J., Caffin F., Novotova M., Lemaire C., Veksler V., Garnier A., et al. Mitochondrial dynamics in the adult cardiomyocytes: which roles for a highly specialized cell? Front Physiol. 2013;4:102. doi: 10.3389/fphys.2013.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y., Liu Y., Dorn G.W. Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circ Res. Dec 9 2011;109:1327–1331. doi: 10.1161/CIRCRESAHA.111.258723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishihara N., Fujita Y., Oka T., Mihara K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. Jul 12 2006;25:2966–2977. doi: 10.1038/sj.emboj.7601184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anand R., Wai T., Baker M.J., Kladt N., Schauss A.C., Rugarli E., et al. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J Cell Biol. Mar 17 2014;204:919–929. doi: 10.1083/jcb.201308006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khalifat N., Puff N., Bonneau S., Fournier J.B., Angelova M.I. Membrane deformation under local pH gradient: mimicking mitochondrial cristae dynamics. Biophys J. Nov 15 2008;95:4924–4933. doi: 10.1529/biophysj.108.136077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinberg S.F. Oxidative stress and sarcomeric proteins. Circ Res. Jan 18 2013;112:393–405. doi: 10.1161/CIRCRESAHA.111.300496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang G., McCain M.L., Yang L., He A., Pasqualini F.S., Agarwal A., et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med. Jun 2014;20:616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanley W.C., Recchia F.A., Lopaschuk G.D. Myocardial substrate metabolism in the normal and failing heart. Physiol Rev. Jul 2005;85:1093–1129. doi: 10.1152/physrev.00006.2004. [DOI] [PubMed] [Google Scholar]
- 45.Chabowski A., Górski J., Glatz J.F., JJ P.L., Bonen A. Protein-mediated fatty acid uptake in the heart. Curr Cardiol Rev. Feb 2008;4:12–21. doi: 10.2174/157340308783565429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nose N., Werner R.A., Ueda Y., Gunther K., Lapa C., Javadi M.S., et al. Metabolic substrate shift in human induced pluripotent stem cells during cardiac differentiation: functional assessment using in vitro radionuclide uptake assay. Int J Cardiol. Oct 15 2018;269:229–234. doi: 10.1016/j.ijcard.2018.06.089. [DOI] [PubMed] [Google Scholar]
- 47.Fatica E.M., DeLeonibus G.A., House A., Kodger J.V., Pearce R.W., Shah R.R., et al. Barth syndrome: exploring cardiac metabolism with induced pluripotent stem cell-derived cardiomyocytes. Metabolites. Dec 17 2019;9 doi: 10.3390/metabo9120306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rouhani F., Kumasaka N., de Brito M.C., Bradley A., Vallier L., Gaffney D. Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet. Jun 2014;10 doi: 10.1371/journal.pgen.1004432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bassett A.R. Editing the genome of hiPSC with CRISPR/Cas9: disease models. Mamm Genome. Aug 2017;28:348–364. doi: 10.1007/s00335-017-9684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.