Graphical abstract
Summary of the main findings of our study on the effects of EDS on the activity promoter of genes important for Leydig cell (R2C and MA-10) endocrine function (Star and Insl3) and response to toxic agents (Gsta3), as well as EDS-responsive region in the Star gene promoter.

Highlights
-
•
Ethylene dimethanesulfonate (EDS) is a molecule with known selective toxicity on Leydig cells.
-
•
EDS can impair the transcriptional level and affect gene promoter activity in immortalized Leydig cells derived from rat (R2C) and mouse (MA-10).
-
•
Leydig cells derived from rat (R2C) are more sensitive to EDS effects than mouse Leydig cells (MA-10).
-
•
An EDS-responsive region in the Star gene promoter was identified.
Abstract
Ethylene dimethanesulfonate (EDS) is a molecule with known selective cytotoxicity on adult Leydig cells. A single intraperitoneal injection in rats but not mice, leads to male androgen deprivation and infertility. In vitro studies using rat and mouse immortalized Leydig cell lines, showed similar effects of cell death promoted by EDS in rat cells as seen in vivo, and suggest that EDS affects gene transcription, which could firstly compromise steroidogenesis before the apoptosis process. Using gene reporter assay, this study aimed to investigate EDS effects on the promoter activity of genes important for endocrine function (Star, Insl3) and response to toxic agents (Gsta3) in immortalized Leydig cell lines (rat R2C and mouse MA-10 cells), as well as identify possible EDS-responsive elements in the Star gene promoter. EDS exposure of R2C and MA-10 Leydig cells increased Gsta3 promoter activity after 4 h of treatment and decreased Insl3 promoter activity only in R2C cells after 24 h of treatment. EDS also decreased Star promoter activity in both Leydig cell lines. Using R2C cells, the EDS-responsive region in the Star promoter was located between −400 and −195 bp. This suggests that this region and the associated transcription factors, which include MEF2, might be targeted by EDS. Additional somatic gonadal cell lines expressing Star were used and EDS did not affect Star promoter activity in DC3 granulosa cells while Star promoter activity was increased in MSC-1 Sertoli cells after 24 h of treatment. This study contributes to the knowledge regarding the mechanism of EDS action in Leydig cells, and in other gonadal cell lineages, and brings new light regarding the rats and mice differential susceptibility to EDS effects, in addition to providing new avenues for experimental approaches to better understand Leydig cell function and dynamics in different rodent species.
1. Introduction
Leydig cells are a cell population that resides in the testicular interstitium and are the main source of testosterone and insulin-like 3 (INSL3), two hormones that regulate male reproductive development and function (Tremblay, 2015, de Mattos et al., 2022, Zirkin and Papadopoulos, 2018). In certain species including the rat, Leydig cells are targeted by ethylene (or ethane) dimethanesulfonate (EDS), an alkylating agent with a structural analogy to the chemotherapeutic busulfan (Klinefelter et al., 1990, Teerds and Rijntjes, 2007). In adult rats, a single intraperitoneal EDS injection causes Leydig cell death within a few days leading to sterility in about seven days (Jackson, 1973). Although EDS can target Leydig cells of different vertebrates, its effects seem to be species-dependent, especially in rodents, with the rat being highly sensitive while the mouse being more resistant to EDS action (Teerds and Rijntjes, 2007, Rommerts et al., 2004, Kerr et al., 1987).
At the physiological level, EDS leads to male sterility due to androgen depletion in the testicles, since this alkylating agent acts as a cytotoxic agent selectively on Leydig cells (Jackson, 1973, Molenaar et al., 1985, Klinefelter et al., 1991). The sharp reduction in testosterone levels causes weight reduction of androgen-dependent organs, such as the prostate, the seminal vesicle, and the epididymis (Molenaar et al., 1985). In addition, studies conducted by Klinefelter et al. (Klinefelter et al., 1990, Klinefelter et al., 1991, Klinefelter et al., 1997) revealed that EDS is also efficient at reducing male fertility by direct effects on the epididymal function, compromising the sperm maturation process. Interestingly, EDS effects are reversible since the population of Leydig cells affected by the compound is reestablished about one month after exposure, and with the resumption of testosterone production, fertility is restored (O’Leary et al., 1986). Thus, studies using EDS are very promising to better understand androgen-dependent reproductive processes (Klinefelter et al., 1991).
Furthermore, recent studies have used EDS-treated rats to study the dynamics of Leydig cell depletion, which is followed by induction of mitosis of the resident stem Leydig cells, ultimately leading to the process of Leydig cell regeneration (Mo et al., 2019). This represents a powerful model to study the molecular factors, as well as the morphological and physiological features involved during Leydig cell differentiation (Mo et al., 2019, Duan et al., 2019, Li et al., 2019, Ni et al., 2019). Also, the process of Leydig cell regeneration that occurs following EDS-induced depletion seems to be very similar to the development of this cell population during puberty, which makes this model relevant to study testicular events during male puberty even in adult rats (Mo et al., 2019).
In addition to being efficient in whole animal models, in vitro approaches using EDS on different Leydig cell lines have revealed that EDS is similarly effective as in vivo approaches. Although cell death was triggered in EDS-treated rat (H540) and mouse (MA-10 and TM3) Leydig cell lines, the differential sensitivity to EDS was maintained, with rat Leydig cell lines being more sensitive to EDS than the mouse cell lines (Rommerts et al., 2004, King et al., 1998, Lee et al., 2012, Li et al., 1822). These studies also highlighted some of the pathways that are affected by EDS treatment causing Leydig cell death. These include a decrease in steroidogenic function, mitochondrial damage, increased oxidative stress, and ultimately apoptosis.
Although the various immortalized Leydig cell lines from rat (H540, R2C) and mouse (MA-10, MLTC-1, TM3) originate from Leydig cell tumors, they have nonetheless retained the steroidogenic machinery and ability to respond to gonadotrophin/hormone stimulation with increased steroidogenic output, which makes these cell lines valuable models for studying Leydig cell function and regulation in culture (Zirkin and Papadopoulos, 2018, Ascoli et al., 2007). The steroidogenic process in Leydig cell is stimulated mainly by LH, which binds to its G protein-coupled receptor causing an increase in cAMP production leading to the activation of several pathways and downstream transcription factors ultimately increasing the expression of several genes required for steroidogenesis (Tremblay, 2015, Ascoli et al., 2007, de Mattos et al., 2023).
As revealed by in vivo approaches and studies using Leydig cell lines, EDS invariably leads to Leydig cell death mainly by apoptosis involving the caspase signaling pathway (Teerds and Rijntjes, 2007, Rommerts et al., 2004, King et al., 1998). There is also evidence that oxidative stress might trigger these events leading to apoptosis (Lee et al., 2012). However, before apoptosis is triggered, there is a significant decrease in steroidogenesis and loss of steroidogenic function. This is mainly attributed to an important reduction in the expression of the Star gene, which codes for the steroidogenic acute regulatory protein (STAR), a key protein for steroidogenesis (Teerds and Rijntjes, 2007, King et al., 1998, Lee et al., 2012). In addition, EDS was found to activate the transcription factor NF-κB, which binds to and activates the Ndrg2 promoter leading to increased levels of NDRG2, a pro-apoptotic protein (Li et al., 1822). However, it remains unclear whether EDS effects on Leydig cell endocrine function are due to impairment in transcriptional, post-transcriptional, and/or translational processes. Thus, it is necessary to investigate if EDS affects gene expression at the transcriptional level.
The aim of this study was to determine whether EDS affects Leydig cell endocrine function by targeting transcriptional processes. This was achieved by investigating the effects of EDS on the promoter activity of genes important for endocrine function (Star, Insl3) and response to toxic agents (Gsta3) in immortalized rat and mouse Leydig cell lines.
2. Material and methods
2.1. Chemicals
Ethylene dimethanesulfonate (EDS) was obtained from Toronto Research Chemicals (C4H10O6S2; CAS #4672-49-5; Lot number 4-OBI-74-1; 98 % purity; Toronto, Canada). 8-bromo-cyclicAMP (8Br-cAMP), dimethyl sulfoxide (DMSO), and polyethyleneimine hydrochloride (PEI) were obtained from Sigma-Aldrich Canada (Oakville, Ontario, Canada). EDS was initially diluted at a concentration of 1 M in pure DMSO. Then, serial dilutions were made in distilled water and 0.5 % DMSO to the working concentrations.
2.2. Cell lines
Rat and mouse immortalized Leydig cell lines, R2C (ATCC, Manassas, Virginia, USA) and MA-10 (ATCC, Manassas, Virginia, USA), respectively, were used for this study from passages 40 to 52 (R2C) or 08 to 31 (MA-10). The study also included rat immortalized granulosa DC3 cells (passages 8 to 16, provided by Dr. Riaz Farookhi, from McGill University, Montreal, ON, Canada) and mouse immortalized MSC-1 Sertoli cells (passages 6 to 11, provided by Dr. Michael Griswold, from the Washington State University, Pullman, WA, USA). All cell lines were validated by morphology, and Leydig cell lines were also validated by steroid hormone production, as previously described (Abdou et al., 2014). Cells were cultured in HAMs-F10 supplemented with 15 % horse serum and 2.5 % fetal bovine serum (for R2C); Dulbecco’s Modified Eagle Medium-F12 (DMEM-F12 for MA-10) supplemented with 15 % horse serum; or DMEM supplemented with 10 % fetal bovine serum (for DC3 and MSC-1). Additionally, cell culture media was supplemented with penicillin–streptomycin, and cells were incubated at 37 °C, in 5 % CO2, in a humidified incubator.
2.3. Experimental design
For this study, the Leydig cell lines R2C and MA-10 were seeded in a 24-well plate (50,000 cells per well). Both cell lines produce steroids either constitutively (R2C) or in response to hormone stimulation (MA-10). The fact that they are derived from different species (R2C from rat and MA-10 from mouse), allows for the study of EDS differential effects and potential mechanisms of action between these two species. The next day the media was replaced, and the cells were treated, in duplicate, with increasing concentrations of EDS ranging from 1 to 50 mM (1, 2, 5, 10, 20, and 50 mM), to determine the lowest dose to cause adverse effects for each cell line (LOAEL). Control duplicate received only vehicle (distilled water or 0.5 % DMSO). Once the LOAEL was defined, lower concentrations (0.01, 0.1, and 1 mM) were chosen to study EDS effects on the activity of gene promoters by performing a pilot dose–response study, through luciferase assays following cell transfections, after 24 h of EDS exposure. The promoter constructs used in this study are related to Leydig cell endocrine function (Star, Cyp17a1, and Insl3) and defense against toxic agents (Gsta3). Cell counts were also performed in an automated cell counter (TC-20TM, Bio-Rad) to assess if EDS at 1 or 2 mM was affecting Leydig cell proliferation after 4 and 24 h of EDS exposure.
Based on the results from preliminary experiments, improvements were made to the EDS concentrations (1 mM for rat R2C cells and 2 mM for mouse MA-10 cells) and treatment periods (4 h for short-term effects and 24 h for long-term effects). Additional Star gene promoter constructs were also included in order to locate the EDS-responsive region(s) in the Leydig cell line shown to be more sensitive to EDS effects. Cell lines representing other gonadal lineages (Sertoli and granulosa) were included in the study to determine whether EDS affects Star gene promoter activity in gonadal somatic cells other than Leydig cells and to understand if the mechanisms of action would be exclusive or not to Leydig cells. DC3 granulosa cells were exposed to 1 mM EDS, and Sertoli MSC-1 cells to 2 mM EDS, similarly to concentrations used for each rat- and mouse-derived Leydig cell lines, respectively.
2.4. Plasmids, Transfections, and luciferase reporter assays
The following gene promoters were subcloned upstream of the Firefly luciferase reporter gene, generating reporter plasmids, and used for this study: mouse Star with −980 (full-length), −400, −195, and −43 (minimal promoter) to + 16 base pairs (bp) (Martin et al., 2008, Tremblay and Viger, 2001); −447 to + 36 bp rat Cyp17a1 (Tremblay and Viger, 2003); −1087 (full-length) and −79 (minimal) to + 5 bp mouse Insl3 (Robert et al., 2006, Laguë and Tremblay, 2008, Mendoza-Villarroel et al., 2014); −2062 (full-length) and −72 (minimal) to + 38 bp mouse Gsta3 (Di-Luoffo et al., 2015). For transfections, cells were plated in 24-well plates at 50,000 cells per well. On the next day, cells were transiently transfected with 500 ng of reporter plasmid using polyethyleneimine hydrochloride (1 μg/μL; PEI: Plasmid = 2:1) diluted in Opti-MEM media (GIBCO by Life Technologies, Burlington, ON, Canada). The transfection mix was prepared 20 min before cell transfection. Additionally, transfections in R2C and MA-10 were made with a plasmid containing the green fluorescent protein (GFP) gene, as a control to assess transfection efficiency. After sixteen hours, the media was replaced, and cells were allowed to grow for additional 32 h. After replacing the media, DMSO (control) or EDS was added for 4 or 24 h before cell harvesting. For cells transfected with Star promoter constructs, the cells were stimulated or not (basal) for an additional 4 h with 0.5 mM 8Br-cAMP (dilution in the standard cell culture media of each cell line, prepared immediately before the stimulation). Cells were then lysed, the lysates were collected, and placed in a 96-well plate, and the luciferase measurements (counts/second of the lights derived from luciferin oxidation by luciferase) were performed using a Tecan Spark 10 M multimode plate reader (Tecan, Morrisville, NC, USA). Each experiment was performed at least 3 to 5 times, in triplicate, and the number of replicates is indicated in each figure legend. After each experiment, intra-assays were normalized to obtain the fold activation related to the mean of the control triplicate.
2.5. Statistical analysis
Data are present as the mean ± standard error of the mean or as the median (interquartile ranges 25 and 75). Data were analyzed using Shapiro‐Wilk’s normality test in the intragroup values, then results were compared with the control group by one‐way analysis of variance (ANOVA) followed by Dunnett’s test (more than two groups) or by Welch’s t-test (for two groups), for parametric variables, or by Kruskal‐Wallis followed by Dunn’s test, for nonparametric variables. Differences were considered statistically significant when p < 0.05. Statistical analyses were performed using the software GraphPad Prism (Version 9.0).
3. Results
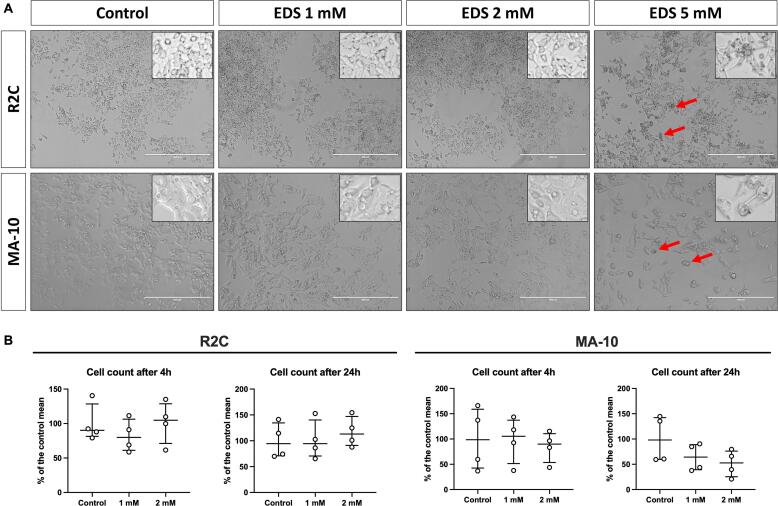
Exposure of rat R2C and mouse MA-10 Leydig cells to increasing EDS concentrations showed that 5 mM (Fig. 1A) or higher concentrations (data not shown) affects cell confluency and morphology, leading to cell death after 24 h of treatment. Concentrations of 1 and 2 mM did not affect Leydig cell confluency and number 24 h post-EDS exposure (Fig. 1B).
Fig. 1.
(A) Representative aspect of cells after 24 h of exposure to ethylene dimethanesulfonate (EDS) of rat R2C and mouse MA-10 Leydig cells. Note that at 5 mM, the cell confluency and morphological characteristic seem qualitatively impaired (red arrows; insert), indicating the cytotoxicity caused by EDS. (B) Cell count of R2C and MA-10 cells after 4 and 24 h of EDS exposure at concentrations of 1 or 2 mM (n = 4, each in duplicate). Values are expressed as median (interquartile ranges). Kruskal-Wallis followed by Dunn’s test (p > 0.05).
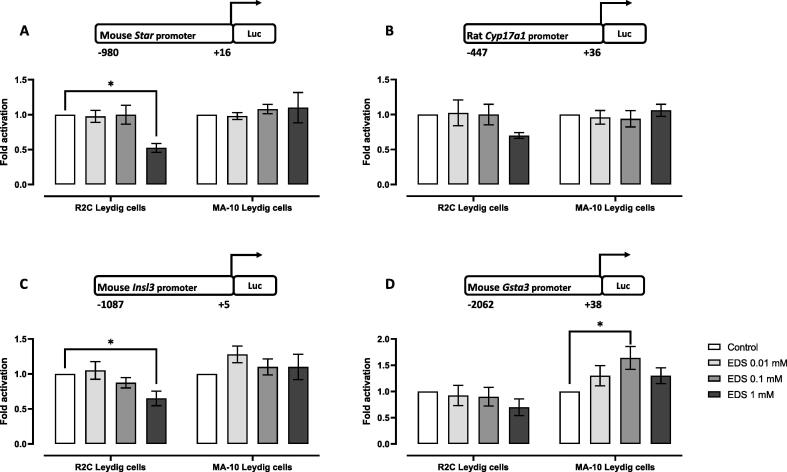
Next, the effect of EDS on the activity of selected Leydig cell gene promoters was assessed. To determine the optimal EDS concentration, rat R2C and mouse MA-10 Leydig cells were transiently transfected with various promoter constructs (Star, Cyp17a1, Insl3, Gsta3) and treated with increasing EDS concentrations. As shown in Fig. 2, increasing EDS concentration led to a statistically significant decrease in Star and Insl3 promoter activity in rat R2C but not in mouse MA-10 Leydig cells (Fig. 2A and 2C). The activity of the Cyp17a1 and Gsta3 promoter was reduced in rat R2C Leydig cells, but it did not reach statistical significance (Fig. 2B and 2D). Interestingly, in mouse MA-10 Leydig cells, a statistically significant increase in Gsta3 promoter activity was observed (Fig. 2D). Based on these experiments, EDS concentrations of 1 mM and 2 mM were chosen for experiments performed in rat R2C and mouse MA-10 Leydig cells, respectively.
Fig. 2.
A dose–response study with reporter luciferase assays to determine the effects of EDS on (A) Star, (B) Cyp17a1, (C) Insl3, and (D) Gsta3 promoter activity. Rat R2C (n = 4, each in triplicate) and mouse MA-10 (n = 5, each in triplicate) Leydig cells were transfected with different promoter constructs and treated for 24 h with increasing concentrations of EDS as indicated. Luciferase reporter (Luc). Values are expressed as mean ± S.E.M. ANOVA followed by Dunnett’s test (*p < 0.05 compared to the control group).
Despite rat R2C Leydig cells showing a lower transfection efficiency compared to mouse MA-10 Leydig cells (Supplemental Fig. 1), after 24 h of exposure to EDS (1 mM), a decrease in Insl3 promoter activity was observed in rat R2C Leydig cells, while having no effect in mouse MA-10 Leydig cells treated with 2 mM (Fig. 3). Short-term (4 h) EDS exposure had no effect on Insl3 promoter activity in both Leydig cell lines (Fig. 3). Regarding Gsta3 promoter activity, no change was observed after 24 h EDS exposure in both Leydig cell lines. However, EDS at 1 and 2 mM was found to increase Gsta3 promoter activity after 4 h of treatment in both R2C and MA-10 Leydig cells (Fig. 3).
Fig. 3.
EDS effects on the activity of the Insl3 and Gsta3 gene promoter after 4 and 24 h of exposure to EDS of rat R2C (1 mM of EDS, n = 4–5, each in triplicate) and mouse MA-10 (2 mM of EDS, n = 5, each in triplicate) Leydig cells. Luciferase reporter (Luc). Values are expressed as mean ± S.E.M. Welch’s t-test (*p < 0.05).
The effects of EDS on the activity of the hormone-inducible Star promoter were next assessed. As shown in Fig. 4, a 4 h EDS exposure of both rat R2C and mouse MA-10 Leydig cell lines (stimulated with 8Br-cAMP or not) had no effect on Star promoter activity. However, 24 h treatment with EDS led to a decrease in basal Star promoter activity in rat R2C cells, and following stimulation with 8Br-cAMP in both rat R2C and mouse MA-10 Leydig cell lines (Fig. 4). To confirm that the EDS-mediated reduction in 8Br-cAMP-stimulated Star promoter activity was specific to Leydig cells, transfections were performed in two non-Leydig cell lines: DC3 granulosa cells and MSC-1 Sertoli cells, both of which also endogenously express the Star gene, albeit at a lower level than Leydig cells. Exposure to 1 and 2 mM of EDS for 24 h did not affect the confluency or morphology of DC3 and MSC-1 cells (Supplemental Fig. 2). In unstimulated or 8Br-cAMP-stimulated DC3 granulosa cells, Star promoter activity was not affected after 24 h of treatment with 1 mM EDS (Fig. 4). However, in MSC-1 Sertoli cells, 24 h exposure with 2 mM EDS increased Star promoter activity after 8Br-cAMP stimulation (Fig. 4). In all transfections, the activity of the various minimal promoter constructs was not affected by EDS indicating that the EDS-mediated effects are specific. Altogether, these data show that rat- and mouse-derived Leydig cell lines respond differently to a given EDS concentration and that rat Leydig cells are more sensitive to EDS than mouse Leydig cells. In addition, these data indicate that EDS does modulate promoter activity, and therefore gene transcription, in Leydig cells.
Fig. 4.
EDS effects on Star promoter activity after 4 h of exposure to EDS of rat R2C (1 mM of EDS, n = 5, each in triplicate) and mouse MA-10 (2 mM of EDS, n = 5, each in triplicate) Leydig cells. EDS effects on Star promoter activity after 24 h of exposure to EDS of rat R2C (1 mM of EDS, n = 3, each in triplicate) and mouse MA-10 (2 mM of EDS, n = 5, each in triplicate) Leydig cells, DC3 granulosa cells (1 mM of EDS, n = 5, each in triplicate), and MSC-1 Sertoli cells (2 mM of EDS, n = 5, each in triplicate). EDS effects were assessed in basal and stimulated (0.5 mM 8Br-cAMP) conditions in R2C Leydig cells. Luciferase reporter (Luc). Values are expressed as mean ± S.E.M. Welch’s t-test (*p < 0.05).
To locate the EDS-responsive element in the Star promoter and therefore better define the molecular mechanism of EDS action on gene transcription, transient transfections were next performed in R2C Leydig cells (more sensitive to EDS effects) using 5′ progressive deletion constructs of the Star promoter (-400 and −195 to + 16 bp). As expected, the activity of the −980 bp Star reporter was inhibited by EDS (1 mM, 24 h exposure) in unstimulated and 8Br-cAMP-stimulated R2C cells (Fig. 5). Interestingly, with a −400 bp Star promoter construct, only basal activity was inhibited by EDS (Fig. 5). Further Star promoter deletion to −195 bp completely eliminated the EDS repressive effects, similar to the minimal Star promoter construct (-43 to + 16 bp) (Fig. 5). These data indicate that the EDS-responsive region is located between −400 and −195 bp in the Star promoter, a region that contains the binding element for the MEF2 transcription factor (Fig. 5).
Fig. 5.
The EDS-responsive region is located between −400 and −195 bp of the Star promoter. Progressive 5′ deletion constructs of Star gene promoter were transfected in rat R2C Leydig cells and treated with 1 mM EDS for 24 h (n = 3–5, each in triplicate) in the absence or presence of 0.5 mM 8Br-cAMP for 4 h. Luciferase reporter (Luc). Values are expressed as mean ± S.E.M. Welch’s t-test (*p < 0.05).
4. Discussion
EDS is a classical compound with known selective cytotoxicity on rodent Leydig cells. This study was designed to investigate the mechanisms of EDS action in rat R2C and mouse MA-10 Leydig cell lines and more specifically to determine whether EDS affects promoter activity of genes related to endocrine function and defense against toxic agents in Leydig cells.
Different studies of EDS exposure of immortalized cell lines showed that cell death of rat-derived cells and mouse-derived cells require different EDS concentrations (Rommerts et al., 2004, King et al., 1998, Lee et al., 2012, Li et al., 1822). In the present study, 5 mM of EDS was sufficient to impair cell growth and morphology in both Leydig cell lines tested, reaching cell death after 24 h of incubation with EDS. However, even at lower EDS concentrations, which did not affect cell number, EDS-induced effects between the two Leydig cell lines were both similar and different depending on the parameter assessed. This provides clues about the differential susceptibility of rats and mice to EDS.
The Gsta3 gene codes for the glutathione S-transferase alpha 3 (GSTA3) enzyme. The main functions of GSTA3 are to protect the cell against toxic agents and to contribute to steroidogenesis by catalyzing obligatory double-bond isomerization in the biosynthesis of progesterone and testosterone (Lindström et al., 2018, Mannervik et al., 2021). In the present work, Gsta3 promoter activity was increased after 4 h of EDS treatment in both Leydig cell lines tested. An increase in GSTA3 synthesis in response to EDS is consistent with a role for glutathione in protecting Leydig cells from the EDS cytotoxic effects, even at very low concentrations, as seen exclusively in mouse MA-10 Leydig cells treated with 0.01 mM of EDS. This indicates that mouse Leydig cells more efficiently respond by activating their defense mechanism when exposed to a very low dose of EDS. This may explain, at least in part, the resistance of mouse cells to EDS compared to rat cells. Our findings may explain the results reported in a previous study with isolated rat adult Leydig cells exposed for 4 h to EDS, which shows a decrease in intracellular levels of glutathione (Kelce and Zirkin, 1993), and this could be due an increase in GSTA3 enzyme, that catalyzes the conjugation of glutathione with the xenobiotic, generating the glutathione-S-conjugates in order to facilitate elimination of toxic agents from the cells (Jd et al., 2005, Cooper and Hanigan, 2018).
In addition to steroid hormones, Leydig cells also produce a peptide hormone, INSL3, which is responsible for the testis descent during fetal life and exerts anabolic effects on bone and skeletal muscle in adult life (Facondo et al., 2020, Ivell et al., 2013). In this study, EDS was found to decrease Insl3 promoter activity only in rat R2C Leydig cells, which is an additional indication that rat cells are more sensitive to EDS than mouse cells.
The STAR protein regulates the rate-limiting step in steroidogenesis by shuttling the cholesterol (substrate for steroid hormone synthesis) from the outer to the inner mitochondrial membrane where steroidogenesis is initiated (Tugaeva and Sluchanko, 2019). From previous studies, EDS was shown to first affect steroidogenesis by decreasing STAR protein levels, before leading to Leydig cell death (King et al., 1998). Herein, EDS was found to affect Star gene expression at the transcriptional level, by inhibiting Star promoter activity in both rat R2C and mouse MA-10 Leydig cells. Progressive deletions of the Star promoter revealed that the EDS-responsive element(s) are located between −400 and −195 bp, a region that includes a binding site for the MEF2 (DNA binding element sequence: 5′-CTATATATAC-3′) transcription factor (de Mattos et al., 2022, Daems et al., 2015, Di-Luoffo et al., 2016). Additional experiments, such as transfection of Leydig cells with a Star promoter construct harboring a mutation in the MEF2 binding site, are needed to better understand EDS mechanism of action.
The MEF2 family of transcription factors includes four members (MEF2A-D) that can bind to AT-rich sequences in regulatory regions of target genes leading to their activation. MEF2 factors were also found to act in cooperation with other transcription factors, such as GATA4 and COUP-TFII, in order to further increase the activity of various promoters including Nur77 (a transcription factor known to activate Insl3 and Star expression), Star, and Gsta (Tremblay, 2015, de Mattos et al., 2022, Di-Luoffo et al., 2015, Daems et al., 2015, Di-Luoffo et al., 2016, Abdou et al., 2016). Interestingly, target genes for MEF2 also include genes related to cell survival and other functions (McKinsey et al., 2002, Pon and Marra, 2015, Potthoff and Olson, 2007), which provides additional clues regarding the mechanism of EDS action in triggering cell death. Further investigations regarding the effects of EDS on MEF2 transcription factors, as well as on upstream elements are warranted.
It is interesting to note that the effects of EDS on the Star promoter in two non-Leydig somatic gonadal cell types were different, which supports the concept that EDS, by mechanisms that remain to be fully deciphered, affects specifically Leydig cells. While no effect was found in DC3 granulosa cells, in Sertoli MSC-1 cells EDS increased Star promoter activity. This is consistent with the lack of reported deleterious effects of EDS on granulosa and Sertoli cells, two lineages that are embryologically related (Piprek et al., 2016). However, a study on the effects of EDS in rat Sertoli cells revealed that exposure to EDS decreased STAR protein levels (King et al., 1998). This could be a consequence of the high EDS concentration used in that study, a concentration that also caused Sertoli cell apoptosis (King et al., 1998). Furthermore, MSC-1 is a mouse-derived Sertoli cell line, and mice are more resistant to EDS. Finally, although Star is expressed in Sertoli cells at low levels, its function in these cells remains unknown and STAR knockout mice have no Sertoli cell defects (Caron et al., 1997, Hasegawa et al., 2000).
The data herein provide additional evidence of the selectivity of EDS effects on somatic male gonadal cells, with Leydig cells from rat origin being the most sensitive. However, it is still not clear whether EDS targets mouse Leydig cells in the same manner as it does rat Leydig cells, or if EDS has different targets in the mouse testis (Rommerts et al., 2004, Kerr et al., 1987, Tarka-Leeds et al., 2003). EDS may affect both transcription and translation processes, and additional work including assessing mRNA and protein levels, is needed to thoroughly address these possibilities. Furthermore, our study functionally corroborates the concept that the mechanisms of EDS action include effects on gene promoter activity and finds that EDS impairs both rat and mouse Leydig cell lines, although higher EDS concentrations are needed to affect the mouse cell line.
5. Conclusion
This study assessed the effects of EDS on the activity of promoters of genes important for Leydig cell endocrine function and response to toxic agents in various cell lines from mouse and rat. We found that EDS impairs the activity of the Star and Insl3 promoters in rat Leydig cells, which reinforces the idea of Leydig cell endocrine function impairment prior to cell death. Furthermore, our results using rat and mouse cell lines support the concept that rats are more sensitive to EDS than mice, since higher concentrations of EDS are needed to lead to similar effects in mouse cells. Finally, we found that the EDS-responsive region in the Star promoter lies between −400 and −195 bp, a region that contains binding sites for various transcription factors, including MEF2. All these transcription factors therefore constitute possible EDS targets. Our findings provide new information about the molecular mechanism of EDS action in Leydig cells, and provides new avenues for experimental approaches to better understand Leydig cell function and dynamics in different rodent species.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank Dr. Riaz Farookhi (McGill University, Montreal, ON, Canada) and Dr. Michael Griswold (Washington State University, Pullman, WA, USA), who kindly provided the DC3 and MSC-1 cells, respectively. The authors thank the São Paulo Research Foundation (FAPESP) for the Ph.D. Scholarship (Grant numbers #2021/09882-2 and #2022/16531-4) provided to Jorge W. F. de Barros, graduate student from the General and Applied Biology Program, Institute of Biosciences, São Paulo State University (UNESP). Kenley Joule Pierre is the recipient of a studentship from the Canadian Institutes of Health Research (CIHR, funding reference number FBD-189317). The authors also thank the Canadian Institutes of Health Research (CIHR) for the funds provided to Dr. Jacques J. Tremblay (funding reference number MOP-81387).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crtox.2023.100147.
Contributor Information
Jorge W.F. de Barros, Email: barros.jwf@gmail.com.
Wilma De G. Kempinas, Email: wilma.kempinas@unesp.br.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Green fluorescent protein (GFP) was used as a control to assess transfection efficiency.
(A) Representative aspect of cells after 24 h of exposure to 1 mM of EDS of DC3 granulosa cells. (B) Representative aspect of cells after 24 h of exposure to 2 mM EDS of MSC-1 Sertoli cells.
Data availability
Data will be made available on request.
References
- Abdou H.S., Bergeron F., Tremblay J.J. A Cell-Autonomous Molecular Cascade Initiated by AMP-Activated Protein Kinase Represses Steroidogenesis. Mol. Cell Biol. 2014;34:4257–4271. doi: 10.1128/MCB.00734-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdou H.S., Robert N.M., Tremblay J.J. Calcium-dependent Nr4a1 expression in mouse Leydig cells requires distinct AP1/CRE and MEF2 elements. J. Mol. Endocrinol. 2016;56:151–161. doi: 10.1530/JME-15-0202. [DOI] [PubMed] [Google Scholar]
- Ascoli M. In: The Leydig Cell in Health and Disease. Payne A.H., Hardy M.P., editors. Humana Press; Totowa, NJ: 2007. Immortalized Leydig Cell Lines as Models for Studying Leydig Cell Physiology; pp. 373–381. [DOI] [Google Scholar]
- Caron K.M., Soo S.C., Wetsel W.C., Stocco D.M., Clark B.J., Parker K.L. Targeted disruption of the mouse gene encoding steroidogenic acute regulatory protein provides insights into congenital lipoid adrenal hyperplasia. PNAS. 1997;94:11540–11545. doi: 10.1073/pnas.94.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A.J.L., Hanigan M.H. Metabolism of Glutathione S-Conjugates: Multiple Pathways, Comprehensive. Toxicology. 2018:363. doi: 10.1016/B978-0-12-801238-3.01973-5. [DOI] [Google Scholar]
- Daems C., Di-Luoffo M., Paradis É., Tremblay J.J. MEF2 Cooperates With Forskolin/cAMP and GATA4 to Regulate Star Gene Expression in Mouse MA-10 Leydig Cells. Endocrinology. 2015;156:2693–2703. doi: 10.1210/en.2014-1964. [DOI] [PubMed] [Google Scholar]
- de Mattos K., Viger R.S., Tremblay J.J. Transcription Factors in the Regulation of Leydig Cell Gene Expression and Function. Front Endocrinol (lausanne). 2022;13 doi: 10.3389/fendo.2022.881309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mattos K., Pierre K.J., Tremblay J.J. Hormones and Signaling Pathways Involved in the Stimulation of Leydig Cell Steroidogenesis. Endocrines. 2023;4:573–594. doi: 10.3390/endocrines4030041. [DOI] [Google Scholar]
- Di-Luoffo M., Brousseau C., Bergeron F., Tremblay J.J. The Transcription Factor MEF2 Is a Novel Regulator of Gsta Gene Class in Mouse MA-10 Leydig Cells. Endocrinology. 2015;156:4695–4706. doi: 10.1210/en.2015-1500. [DOI] [PubMed] [Google Scholar]
- Di-Luoffo M., Brousseau C., Tremblay J.J. MEF2 and NR2F2 cooperate to regulate Akr1c14 gene expression in mouse MA-10 Leydig cells. Andrology. 2016;4:335–344. doi: 10.1111/andr.12150. [DOI] [PubMed] [Google Scholar]
- Duan Y., Wang Y., Li X., Mo J., Guo X., Li C., Tu M., Ge F., Zheng W., Lin J., Ge R. Fibroblast growth factor 16 stimulates proliferation but blocks differentiation of rat stem Leydig cells during regeneration. J. Cell Mol. Med. 2019;23:2632–2644. doi: 10.1111/jcmm.14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facondo P., Delbarba A., Maffezzoni F., Cappelli C., Ferlin A. INSL3: A Marker of Leydig Cell Function and Testis-Bone-Skeletal Muscle Network. Protein Pept. Lett. 2020;27:1246–1252. doi: 10.2174/0929866527666200925105739. [DOI] [PubMed] [Google Scholar]
- Hasegawa T., Zhao L., Caron K.M., Majdic G., Suzuki T., Shizawa S., Sasano H., Parker K.L. Developmental roles of the steroidogenic acute regulatory protein (StAR) as revealed by StAR knockout mice. Mol. Endocrinol. 2000;14:1462–1471. doi: 10.1210/mend.14.9.0515. [DOI] [PubMed] [Google Scholar]
- Ivell R., Wade J.D., Anand-Ivell R. INSL3 as a biomarker of Leydig cell functionality. Biol. Reprod. 2013;88:147. doi: 10.1095/biolreprod.113.108969. [DOI] [PubMed] [Google Scholar]
- Jackson H. Chemical Methods of Male Contraception: Treatment at different stages in the spermatogenic process by steroids and other chemicals yields significant data for future fertility control. Am. Sci. 1973;61:188–193. [PubMed] [Google Scholar]
- Jd H., Ju F., Ir J. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005;45 doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- Kelce W.R., Zirkin B.R. Mechanism by Which Ethane Dimethanesulfonate Kills Adult Rat Leydig Cells: Involvement of Intracellular Glutathione. Toxicol. Appl. Pharmacol. 1993;120:80–88. doi: 10.1006/taap.1993.1089. [DOI] [PubMed] [Google Scholar]
- Kerr J.B., Knell C.M., Abbott M., Donachie K. Ultrastructural analysis of the effect of ethane dimethanesulphonate on the testis of the rat, guinea pig, hamster and mouse. Cell Tissue Res. 1987;249 doi: 10.1007/BF00215530. [DOI] [PubMed] [Google Scholar]
- King S.R., Rommerts F.F., Ford S.L., Hutson J.C., Orly J., Stocco D.M. Ethane dimethane sulfonate and NNN’N’-tetrakis-(2-pyridylmethyl)ethylenediamine inhibit steroidogenic acute regulatory (StAR) protein expression in MA-10 Leydig cells and rat Sertoli cells. Endocr. Res. 1998;24:469–478. doi: 10.3109/07435809809032635. [DOI] [PubMed] [Google Scholar]
- Klinefelter G.R., Laskey J.W., Roberts N.R., Slott V., Suarez J.D. Multiple effects of ethane dimethanesulfonate on the epididymis of adult rats. Toxicol. Appl. Pharmacol. 1990;105:271–287. doi: 10.1016/0041-008x(90)90189-2. [DOI] [PubMed] [Google Scholar]
- Klinefelter G.R., Laskey J.W., Roberts N.L. In vitro/in vivo effects of ethane dimethanesulfonate on Leydig cells of adult rats. Toxicol. Appl. Pharmacol. 1991;107:460–471. doi: 10.1016/0041-008x(91)90309-3. [DOI] [PubMed] [Google Scholar]
- Klinefelter G.R., Laskey J.W., Ferrell J., Suarez J.D., Roberts N.L. Discriminant analysis indicates a single sperm protein (SP22) is predictive of fertility following exposure to epididymal toxicants. J. Androl. 1997;18:139–150. [PubMed] [Google Scholar]
- Laguë E., Tremblay J.J. Antagonistic effects of testosterone and the endocrine disruptor mono-(2-ethylhexyl) phthalate on INSL3 transcription in Leydig cells. Endocrinology. 2008;149:4688–4694. doi: 10.1210/en.2008-0310. [DOI] [PubMed] [Google Scholar]
- Lee E.-H., Oh J.-H., Lee Y.-S., Park H.-J., Choi M.-S., Park S.-M., Kang S.-J., Yoon S. Gene expression analysis of toxicological pathways in TM3 leydig cell lines treated with Ethane dimethanesulfonate. J. Biochem. Mol. Toxicol. 2012;26:213–223. doi: 10.1002/jbt.21409. [DOI] [PubMed] [Google Scholar]
- Li T., Hu J., He G.-H., Li Y., Zhu C.-C., Hou W.-G., Zhang S., Li W., Zhang J.-S., Wang Z., Liu X.-P., Yao L.-B., Zhang Y.-Q. Up-regulation of NDRG2 through nuclear factor-kappa B is required for Leydig cell apoptosis in both human and murine infertile testes. BBA. 1822;2012:301–313. doi: 10.1016/j.bbadis.2011.11.013. [DOI] [PubMed] [Google Scholar]
- Li H., Zhu Q., Wang S., Huang T., Li X., Ni C., Fang Y., Li L., Lian Q., Ge R.-S. Paraquat exposure delays stem/progenitor Leydig cell regeneration in the adult rat testis. Chemosphere. 2019;231:60–71. doi: 10.1016/j.chemosphere.2019.05.104. [DOI] [PubMed] [Google Scholar]
- Lindström H., Peer S.M., Ing N.H., Mannervik B. Characterization of equine GST A3–3 as a steroid isomerase. J. Steroid Biochem. Mol. Biol. 2018;178:117–126. doi: 10.1016/j.jsbmb.2017.11.011. [DOI] [PubMed] [Google Scholar]
- Mannervik B., Ismail A., Lindström H., Sjödin B., Ing N.H. Glutathione Transferases as Efficient Ketosteroid Isomerases. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.765970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L.J., Boucher N., Brousseau C., Tremblay J.J. The orphan nuclear receptor NUR77 regulates hormone-induced StAR transcription in Leydig cells through cooperation with Ca2+/calmodulin-dependent protein kinase I. Mol. Endocrinol. 2008;22:2021–2037. doi: 10.1210/me.2007-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey T.A., Zhang C.L., Olson E.N. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 2002;27:40–47. doi: 10.1016/S0968-0004(01)02031-X. [DOI] [PubMed] [Google Scholar]
- Mendoza-Villarroel R.E., Di-Luoffo M., Camiré E., Giner X.C., Brousseau C., Tremblay J.J. The INSL3 gene is a direct target for the orphan nuclear receptor, COUP-TFII, in Leydig cells. J. Mol. Endocrinol. 2014;53:43–55. doi: 10.1530/JME-13-0290. [DOI] [PubMed] [Google Scholar]
- Mo J., Chen X., Ni C., Wu K., Li X., Zhu Q., Ma L., Chen Y., Zhang S., Wang Y., Lian Q., Ge R.-S. Fibroblast growth factor homologous factor 1 stimulates Leydig cell regeneration from stem cells in male rats. J. Cell Mol. Med. 2019;23:5618–5631. doi: 10.1111/jcmm.14461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar R., de Rooij D.G., Rommerts F.F., Reuvers P.J., van der Molen H.J. Specific destruction of Leydig cells in mature rats after in vivo administration of ethane dimethyl sulfonate. Biol. Reprod. 1985;33:1213–1222. doi: 10.1095/biolreprod33.5.1213. [DOI] [PubMed] [Google Scholar]
- Ni C., Fang Y., Chen X., Wu K., Li H., Wang Y., Zhenkun L., Lian Q., Ge R.-S. Stem Leydig cell regeneration in the adult rat testis is inhibited after a short-term triphenyltin exposure. Toxicol. Lett. 2019;306:80–89. doi: 10.1016/j.toxlet.2019.02.010. [DOI] [PubMed] [Google Scholar]
- O’Leary P., Jackson A.E., Averill S., de Kretser D.M. The effects of ethane dimethane sulphonate (EDS) on bilaterally cryptorchid rat testes. Mol. Cell. Endocrinol. 1986;45:183–190. doi: 10.1016/0303-7207(86)90146-2. [DOI] [PubMed] [Google Scholar]
- Piprek R.P., Kloc M., Kubiak J.Z. In: Molecular Mechanisms of Cell Differentiation in Gonad Development. Piprek R.P., editor. Springer International Publishing; Cham: 2016. Early Development of the Gonads: Origin and Differentiation of the Somatic Cells of the Genital Ridges; pp. 1–22. [DOI] [PubMed] [Google Scholar]
- Pon J.R., Marra M.A. MEF2 transcription factors: developmental regulators and emerging cancer genes. Oncotarget. 2015;7:2297–2312. doi: 10.18632/oncotarget.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff M.J., Olson E.N. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- Robert N.M., Martin L.J., Tremblay J.J. The orphan nuclear receptor NR4A1 regulates insulin-like 3 gene transcription in Leydig cells. Biol. Reprod. 2006;74:322–330. doi: 10.1095/biolreprod.105.044560. [DOI] [PubMed] [Google Scholar]
- Rommerts F.F.G., Kühne L., van Cappellen G.W.A., Stocco D.M., King S.R., Jankowska A. Specific dose-dependent effects of ethane 1,2-dimethanesulfonate in rat and mouse Leydig cells and non-steroidogenic cells on programmed cell death. J. Endocrinol. 2004;181:169–178. doi: 10.1677/joe.0.1810169. [DOI] [PubMed] [Google Scholar]
- Tarka-Leeds D.K., Suarez J.D., Roberts N.L., Rogers J.M., Hardy M.P., Klinefelter G.R. Gestational Exposure to Ethane Dimethanesulfonate Permanently Alters Reproductive Competence in the CD-1 Mouse1. Biol. Reprod. 2003;69:959–967. doi: 10.1095/biolreprod.103.017343. [DOI] [PubMed] [Google Scholar]
- Teerds, K., Rijntjes, E., 2007. Dynamics of Leydig Cell Regeneration After EDS, in: A.H. Payne, M.P. Hardy (Eds.), The Leydig Cell in Health and Disease, Humana Press, Totowa, NJ, 2007: pp. 91–116. https://doi.org/10.1007/978-1-59745-453-7_6.
- Tremblay J.J. Molecular regulation of steroidogenesis in endocrine Leydig cells. Steroids. 2015;103:3–10. doi: 10.1016/j.steroids.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Tremblay J.J., Viger R.S. GATA factors differentially activate multiple gonadal promoters through conserved GATA regulatory elements. Endocrinology. 2001;142:977–986. doi: 10.1210/endo.142.3.7995. [DOI] [PubMed] [Google Scholar]
- Tremblay J.J., Viger R.S. Transcription Factor GATA-4 Is Activated by Phosphorylation of Serine 261 via the cAMP/Protein Kinase A Signaling Pathway in Gonadal Cells*. J. Biol. Chem. 2003;278:22128–22135. doi: 10.1074/jbc.M213149200. [DOI] [PubMed] [Google Scholar]
- Tugaeva K.V., Sluchanko N.N. Steroidogenic Acute Regulatory Protein: Structure, Functioning, and Regulation. Biochemistry (Mosc.) 2019;84:S233–S253. doi: 10.1134/S0006297919140141. [DOI] [PubMed] [Google Scholar]
- Zirkin B.R., Papadopoulos V. Leydig cells: formation, function, and regulation. Biol. Reprod. 2018;99:101–111. doi: 10.1093/biolre/ioy059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Green fluorescent protein (GFP) was used as a control to assess transfection efficiency.
(A) Representative aspect of cells after 24 h of exposure to 1 mM of EDS of DC3 granulosa cells. (B) Representative aspect of cells after 24 h of exposure to 2 mM EDS of MSC-1 Sertoli cells.
Data Availability Statement
Data will be made available on request.