Highlights
-
•
UV–Vis and SERR spectroelectrochemistry for determination of the E°' of solution/immobilized DyPs.
-
•
Simultaneous monitoring of spectroscopic and electrochemical properties of heme proteins.
-
•
Assessment of E°' of co-existing heme spin populations by SERR spectroelectrochemistry.
Keywords: Heme proteins, Redox potential, Spectroelectrochemistry, Dye decolorizing peroxidases
Abstract
Dye-decolorizing peroxidases (DyPs) are heme-containing enzymes that are structurally unrelated to other peroxidases. Some DyPs show high potential for applications in biotechnology, which critically depends on the stability and redox potential (E°') of the enzyme. Here we provide a comparative analysis of UV–Vis- and surface-enhanced resonance Raman-based spectroelectrochemical methods for determination of the E°' of DyPs from two different organisms, and their variants generated targeting E°' upshift. We show that substituting the highly conserved Arginine in the distal side of the heme pocket by hydrophobic amino acid residues impacts the heme architecture and redox potential of DyPs from the two organisms in a very distinct manner. We demonstrate the advantages and drawbacks of the used spectroelectrochemical approaches, which is relevant for other heme proteins that contain multiple heme centers or spin populations.
Graphical abstract
1. Introduction
Heme proteins perform fascinatingly diverse cellular functions that include electron transfer (ET), oxygen transport and storage, catalysis, oxidative phosphorylation and signal transduction, among others [[1], [2], [3]]. The reactivity of the heme moiety and its redox potential (E°') are governed by the properties of the cofactor and of the surrounding protein, which together ensure the range of chemical characteristics needed for these diverse functions. The E°' of the ferric/ferrous iron (Fe(III)/Fe(II)) redox couple span from +450 to −550 mV (vs. SHE) in heme proteins [4]. Such a large variation has been attributed to an interplay of several interconnected factors that include the immediate molecular heme environment, such as the nature of axial ligands, the spin state of the heme iron, the heme geometry and its exposure to the solvent. For instance, in human myoglobin and cytochromes, mutations of axial ligands can alter the redox potential by more than 400 mV [5,6].
The E°' of the Fe(III)/Fe(II) redox couple of heme peroxidases, enzymes that catalyse the oxidation of structurally diverse organic compounds with concomitant reduction of hydrogen peroxide, also cover a wide range, comparable to that of other heme proteins [4]. The E°' defines the range of oxidizable substrates and is thus linked to peroxidase specificities for distinct physiological substrates [7]. The reaction mechanism of peroxidases does not involve the Fe(III)/Fe(II) redox couple, however the value of E°' Fe(III)/Fe(II) is a good indicator of the stability and availability of the resting state ferric enzyme (Fe (III)) for the catalytic reaction, as well as of the catalytically relevant redox couples E°' Fe(III)/Compound I [Fe(IV)=O]+•, Compound I/Compound II [Fe(IV)=O]+ and Compound II/Fe(III) [4]. Moreover, measurement of the intrinsically very positive redox potential of the catalytic intermediates, which typically ranges between +0.8 and +1.2 V, is often a challenge due to their instability and extreme reactivity [[8], [9], [10]].
Determination of E°' Fe(III)/Fe(II) of heme proteins can be performed by i) chemical redox titrations (sometimes referred to as potentiometric titrations in the literature), in which stepwise chemical reduction or oxidation of the protein in solution is monitored by spectroscopic changes of the heme group; the most commonly used methods are UV–Visible [11,12], electron paramagnetic resonance (EPR) [13], nuclear magnetic resonance (NMR) [[14], [15], [16]], resonance Raman (RR) [17], Magnetic Circular Dichroism (MCD) [18], and to a lower extent Infrared (IR) spectroscopies [19]; ii) electrochemical methods, such as cyclic voltammetry (CV), in which the current changes due to protein oxidation / reduction are monitored as the electrode potential is swept upward or downward across the protein's E°' [20,21]; and iii) spectroelectrochemical methods, in which spectroscopic changes (most commonly UV–Vis, IR and surface-enhanced RR (SERR)) of the protein are monitored along an electrochemical redox titration. The spectroelectrochemical methods enable the identification of electroactive species of interest and simultaneously provide information about their molecular structure in situ [[22], [23], [24], [25], [26], [27], [28], [29], [30]].
(Spectro)electrochemistry can be performed employing the heme protein either in solution or attached to solid electrodes, thus relying on mediated or direct electron transfer (i.e. MET and DET) to the heme active site, respectively. It is noteworthy that in the case of biotechnological applications, immobilized enzymes offer a distinct advantage over enzymes in solution, due to physical separation of the biocatalyst from the reactants / products and its re-usability.
Solution measurements of E°', both by chemical and (spectro)electrochemical oxidation / reduction, typically rely on the presence of redox mediators to facilitate the ET between protein and electrode, i.e. MET, and significant amounts of protein. These approaches often suffer from poor reversibility, which results from the use of mediators that can potentially cause chemical instability, side reactions (e.g. with residual O2 present in solution), baseline drifting and appearance of additional absorption bands in the UV−Vis spectra overlapping with the protein [31]. On the other hand, (spectro)electrochemical strategies that rely on protein immobilization and DET are not necessarily straightforward either. In some cases, suitable immobilization platforms that ensure attachment of the protein to the electrode cannot be found or the interactions with the electrode can induce conformational changes and/or denaturation [32,33]. Despite the large spectrum of immobilization strategies that include physical adsorption, encapsulation, chemical binding and biocompatible electrodes, it is not uncommon that the immobilization results in the formation of non-native protein with altered redox potential [34] and loss of catalytic activity [35]. To that end, electrochemical methods provide limited or no molecular information about the electroactive species. Besides, not all proteins can be electrically wired to electrodes [36]; the heme cofactors are frequently deeply buried within the polypeptide chain, or they cannot be properly orientated towards the electrode, thereby hampering ET and measurement of CV signals. Hence, it is of crucial importance to consider all the above methodological constrains and the intrinsic properties of the heme protein, i.e. stability, surface charge distribution, heme exposure and heme cavity electrostatics and flexibility, when selecting the experimental tool for determination of E°'. We highlight SERR spectroelectrochemistry, in which the heme protein is immobilized on plasmonic Ag substrates that simultaneously provide surface enhancement of the RR signals and serve as electrodes [[37], [38]]. (SE)RR spectra obtained upon excitation into the Soret electronic transition band of the porphyrin display core-size marker bands (e.g. ν4, ν3, ν2) originating mainly from complex stretching coordinates of the porphyrin. These marker bands are sensitive to the redox state of the heme iron (ferric or ferrous), its spin (e.g. high-spin (HS) and low-spin (LS)), and coordination pattern (number and nature of the axial ligands) [[38], [39]]. By comparison of the RR spectra of the ferric (or ferrous) protein in solution with the SERR spectra of the protein immobilized on a metal electrode poised at a potential that ensures the protein is oxidized (or reduced), the eventual immobilization induced non-native population(s) can be identified [[37], [40]]. Furthermore, changes of SERR bands attributed to different electroactive species can be monitored along an electrochemical titration, which allows determining the E°' of distinct populations. Therefore, SERR spectroscopy has a superior capacity to i) detect immobilization induced structural changes and ii) provide the E°' of different spin / coordination state sub-populations, if present [[34], [41]].
Dye-decolorizing peroxidases (DyPs) have been attracting a lot of interest for the development of biotechnological applications due to their intrinsic capacity to degrade lignin and environmentally harmful molecules, including anthraquinone-based dyes that possess high redox potentials [42,43]. For that purpose, it is essential to understand their mechanistic properties and the factors that control their E°'. Despite comparable proximal and distal heme cavity architectures, DyPs from different sources show remarkable differences in their specific activities and hence distinct potential for applications [44]. DyPs have a unique GXXDG motif at the distal heme side, which lacks the highly conserved His that acts as an acid-base catalyst in the catalytic reaction in peroxidase-catalase and peroxidase-cyclooxygenase super-families [[43], [45]]. Instead, all known DyP structures possess an Asp(Glu) / Arg couple at the distal side [[46], [47], [48], [49]]. Studies of the role of the distal residues in the catalytic reaction of a number of DyPs indicated that either Asp or Arg can take up the role of the distal His [[9], [48], [49], [50], [51], [52]]. Moreover, the distal mutations of the wild-type (WT) enzymes that modify the H-bonding network and the polarity of the heme cavity, can change the coordination state of the resting Fe(III) and consequently influence the E°' Fe(III)/Fe(II) [9]. For example, previous studies of the effect of Arg substitution on the E°' of DyPs, which employed a variety of methodologies, including UV–Vis chemical and spectroelectrochemical titrations and CV, showed case-dependent effects. A negligible ΔE°' was observed for Arg/Leu substitution in the DyP from Klebsiella pneumoniae (KpDyP) [49], while negatively shifted E°’ were measured for the Arg/Leu variant of Bacillus subtilis DyP (BsDyP) [48] and the Arg/Ala variant of Thermomonospora curvata enzyme (TcDyP) [50].
Here we employed different spectroelectrochemical approaches to study the redox properties of DyPs from Cellulomonas bogoriensis (CboDyP) and Pseudomonas putida (PpDyP), which show remarkable activity in solution towards multiple anthraquinone and azo dyes, as well as phenols and metal ions in the case of PpDyP [46,53]. Enzyme variants, in which the hydrophilic positively charged distal Arg was substituted by hydrophobic residues: non-polar Ile or amphipathic Trp were generated by direct mutagenesis aiming at rationally altering their E°'. The effect of the distal substitutions on the heme cavity and redox properties of CboDyP and PpDyP was probed by RR spectroscopy, UV–Vis- and SERR-based spectroelectrochemistry.
2. Materials and methods
2.1. Reagents and chemicals
8-Amino-1-octanethiol hydrochloride (AOT), 6-mercapto-1-hexanol (MOH) and 1-undecanethiol (1-UDT) were purchased from Sigma-Aldrich, 11-amino-1-undecanethiol hydrochloride (AUT) was acquired from Dojindo. All the other chemicals were purchased from Sigma-Aldrich and were of the highest purity available. Solutions were prepared using deionized water form a Milli-Q® Water Purification System (Merck Millipore).
2.2. Construction, overexpression, and purification of cboDyP and ppDyP enzyme variants
Single amino acid replacements were created using the QuikChange site-directed mutagenesis protocol. For CboDyP variants, plasmid pBAD-histag-SUMO-CboDyP (containing the WT cboDyP sequence) was used as the template [46] and the primers were designed using the AA scan software [54]. For PpDyP variants, the plasmid of WT ppDyP, cloned in pET15b, was used as the template [9]. The primers, forward PpDyP_Opt_R214W_Fw 5′ CGGAAGCGTTTATGGTGTGGCGTAGCGTTAGCTGGG 3′ and reverse PpDyP_Opt_R214W_Rv 5′ CCCAGCTAACGCTACGCCACACCATAAACGCTTCCG 3′ were used to create the R214W mutation; forward PpDyP_Opt_R214I_Fw 5′ CGGAAGCGTTTATGGTGATTCGTAGCGTTAGCTGGG 3′ and PpDyP_Opt_R214I_Rv 5′ CCCAGCTAACGCTACGAATCACCATAAACGCTTCCG 3′ were used to create the R214I mutation. DNA sequence analysis was used to confirm the presence of the desired mutation in the resulting plasmids and the absence of unwanted mutations in other regions of the insert. The plasmids containing the cboDyP and ppDyP genes were transformed into NEB 10 β cells and Escherichia coli Tuner strains, respectively. The recombinant PpDyP variants were produced under the control of the T7lac promoter.
The proteins were expressed and purified as previously described [[9], [46], [55], [56]]. The heme content of the DyPs was determined by the pyridine ferrohemochrome method using an extinction coefficient of ε (R–O 556 nm) = 28.32 mM−1 cm−1 [57]. The UV–Vis spectra of the purified enzymes were recorded on a UV/Visible spectrophotometer Perkin Elmer Lambda 650. The enzymes were stored at –20 °C until use.
2.3. Resonance Raman (RR) and surface-enhanced RR (SERR) spectroscopy measurements
RR and SERR spectra were acquired with a Raman spectrometer (Jobin Yvon LabRam 800 HR) with a back-illuminated CCD detector cooled by liquid nitrogen; an Olympus 20x objective was used for laser focusing on the sample and light collection in the backscattering geometry. A 405 nm diode laser was used as excitation source (Toptica Photonics AG).
RR spectra of the enzymes were measured at room temperature using a rotating quartz cuvette (Hellma) to prevent prolonged sample exposure to laser, as previously described [38]. Sample concentration was 20–80 μM in 12.5 mM potassium phosphate buffer and 12.5 mM K2SO4 at pH 7.0 (buffer A).
The Ag ring electrodes used in SERR experiments were electrochemically roughened in 0.1 M KCl to create a plasmonic-active surface. Following an initial potential step at +2.0 V (40 s), the electrodes were treated with three repetitive oxidation/reduction cycles at +0.3/−0.3 V (1st cycle 60 s; 2nd 30 s; and 3rd 30 s and 300 s for oxidation and reduction steps, respectively). After the roughening process, the SER-active Ag electrodes were immediately immersed in a 1 mM solution of alkanethiol self-assembled monolayer (SAM) prepared in ethanol. The SAM was allowed to form overnight. The following mixed SAMs were used: 1:3 AOT/MOH for WT PpDyP and variants and 1:2 1-UDT/AUT for WT CboDyP and variants. Prior to SERR experiments the Ag coated electrodes were immersed for 30 min in 0.5 µM enzyme solution prepared in 10 mL buffer A. SERR measurements were carried as previously described [[35], [55]], in argon-purged supporting electrolyte (buffer A) to avoid formation of O2 reduction products that could interact with the immobilized enzymes. The electrode was kept at constant rotation (2600 rpm) to avoid prolonged exposure of individual enzyme molecules to laser irradiation.
The laser power was 3 mW and 1.4 mW in RR and SERR experiments, respectively. The accumulation times were 20–30 s; to improve the signal to noise (S/N) ratio 4–12 spectra were co-added in each experiment. After polynomial baseline subtraction all spectra were subjected to component analysis, as described previously [58].
2.4. Determination of redox potentials
2.4.1. UV–Vis spectroelectrochemistry
UV–Vis-based spectroelectrochemical titrations of the enzymes in solution were carried out using a Pine Research Instrumentation spectroelectrochemistry kit (DRP10208). The enzyme solution (15–25 μM) in 50 mM phosphate buffer, 75 mM NaCl, pH 7.0 (buffer B) was purged with argon and kept under argon atmosphere throughout the experiment. The following mediators were added in a ratio of 1:1 to the enzyme solution: trimethylhydroquinone (+115 mV), 1,4-naphthoquinone (+60 mV), duroquinone (+5 mV), indigo trisulphonate (−70 mV), indigo disulphonate (−125 mV), anthraquinone-2-sulphonate (−225 mV), safranine O (−284 mV), benzyl viologen (−345 mV), and methyl viologen (−440 mV). A gold screen-printed electrode card composed of a perforated honeycomb working and solid counter electrodes (Pine Research Instrumentation) was placed inside a quartz cell (optical path 1.7 mm) containing the enzyme/mediator solution; a cylindrical Ag/AgCl electrode (3 M KCl, World Precision Instruments, WPI) was used as reference. The electrodes were connected to a Princeton Applied Research 263A potentiostat that controlled the potential applied to the cell. The UV–Vis spectra were recorded using a Sarspec STD spectrophotometer. Enzyme reduction and oxidation were monitored by the evolution of the ferrous Soret absorption band at 430 nm. Preliminary assays performed with low mediator concentration revealed that the reduction and oxidation Nernst curves were not superimposable, with gaps of up to 80 mV observed between reductive and oxidative titrations for some enzymes (Fig. S1A). To minimize this hysteresis effect, the mediators were used in equimolar concentration compared to the enzyme, which allowed obtaining reversible redox titrations (Fig. S1B). In these experimental conditions, mediator absorption influenced the measurement of Soret band intensities, therefore the latter were corrected by subtraction of the absorbance of control spectra attained from titrations performed with mediators only. The E°' values were obtained by fitting the Nernst equation (Eq. (1)),
| (1) |
in which, R is the universal gas constant, T is the temperature, n is the number of electrons involved in the reaction, F is the Faraday constant and [Ox] and [Red] represent the concentrations of oxidized and reduced species, respectively, to the potential dependent corrected and normalized absorption at 430 nm.
2.4.2. SERR spectroelectrochemistry
SERR-based spectroelectrochemistry experiments were performed as previously described, with the enzymes immobilized on SAM modified Ag electrodes [[35], [55]]. Briefly, a home-built spectroelectrochemical cell with a three-electrode arrangement was used: the SAM/enzyme modified Ag working electrode, an Ag/AgCl (3 M KCl) reference electrode (WPI) and a platinum wire counter electrode (Goodfellow). A Princeton Applied Research 263A potentiostat was used to control the electrode potentials. The experiments were carried out as described in Section 2.3. Determination of E°' for the different spin species was based on the relative SERR intensities of a single oxidation state marker band, obtained from the component analysis (ν3 mode was used for PpDyP R214W and, due to the lower spectral quality, the most intense ν4 mode was used for PpDyP R214I). The E°' were obtained by fitting the Nernst equation (Eq. (1)) to the potential dependent relative SERR intensities of each species [40].
3. Results and discussion
3.1. Heme configuration in DyP variants
The distal cavity of DyPs is lined by conserved amino acid residues, namely Asp (Glu in CboDyP) and Arg, which are involved in the formation of the catalytic intermediate Compound I, upon H2O2 binding to the heme at the onset of catalysis [[59], [60]]. Herein we investigate the effects of substituting the conserved Arg residue, R307 in CboDyP and R214 in PpDyP, by amino acids with hydrophobic side chains (Trp or Ile) on the heme configuration and the E°'.
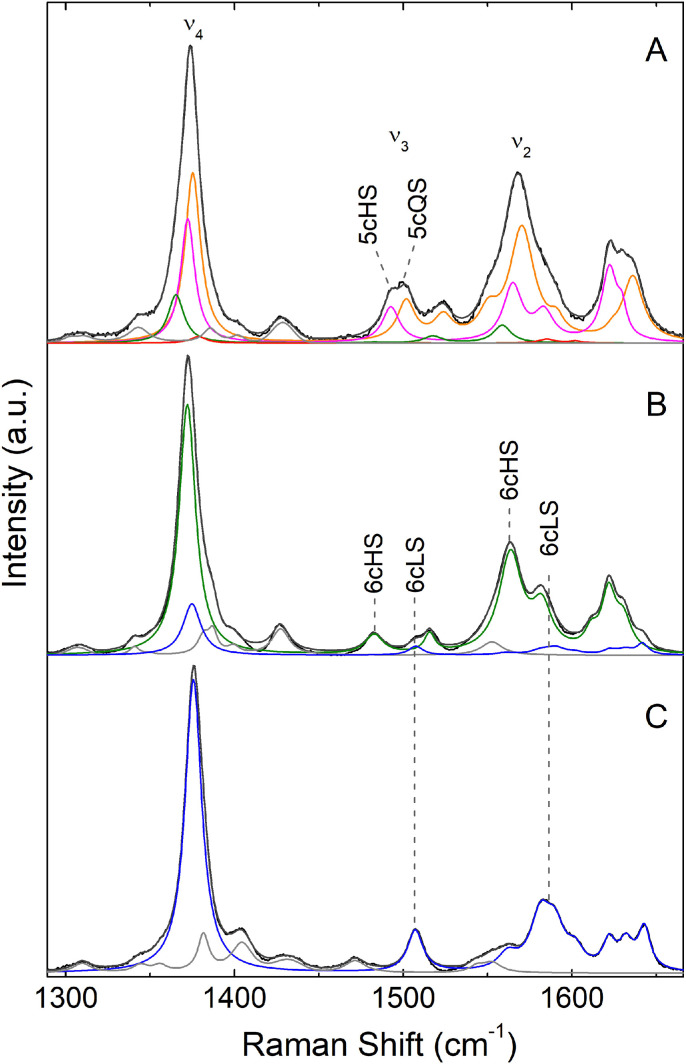
The UV–Vis spectra of R307W and R307I CboDyP variants are identical to those of the WT CboDyP (Fig. S2A-C). RR spectroscopy, which provides a more detailed description of the heme pocket configuration, indicates that R307W (Fig. 1B) indeed reveals a uniform six-coordinated high-spin (6cHS) population, with oxidation state marker ν4 band centered at 1373 cm−1, and oxidation/spin state marker bands, ν3 and ν2, at 1483 and 1563 cm−1, respectively, as was previously observed for the WT enzyme (Fig. 1A, Table S1) [35]. Component analysis of the RR spectrum of the R307I variant indicates, besides a predominant 6cHS species, a presence of an additional six-coordinated low-spin (6cLS) population (ν4, ν3 and ν2 centered at 1375, 1508 and 1582 cm−1) with ca. 25 % relative abundance (Fig. 1C, Table S1).
Fig. 1.
Experimental and component RR spectra of ferric WT CboDyP and variants. (A) WT; (B) R307W and (C) R307I variants. The component spectra representing 6cHS (green) and 6cLS (blue) heme species and non-assigned bands (light grey) are fitted to the experimental spectra (black); the overall component spectra are depicted in dark grey. The spectra of 20–80 µM proteins were measured with 405 nm excitation and 3 mW laser power. (For interpretation of the references to color in this figure, the reader is referred to the web version of this article).
UV–Vis spectra of PpDyP variants are indicative of strongly affected heme structure by the R214 substitution (Fig. S2D-F). The Soret band red-shifts 8–14 nm in the variant's spectra in comparison with WT enzyme, and the Q-bands by ca. 30 nm [9]. The absence of charge transfer (CT) bands in PpDyP R214I spectrum suggests that the high-spin heme species identified in WT enzyme are not present. The RR spectra of PpDyP R214W and R214I reveal similar scenario: the two major species in the RR spectrum of WT PpDyP (Fig. 2A), the five-coordinated high-spin (5cHS) and five-coordinated quantum mechanically mixed-spin (5cQS) populations are absent in the spectra of the variants (Fig. 2B, C). RR spectra of R214W show the presence of 6cHS and 6cLS populations (ν4, ν3 and ν2 centered at 1372, 1483 and 1564 cm−1 and at 1375, 1507 and 1582 cm−1, respectively), while R214I variant reveals a uniform 6cLS population (ν4, ν3 and ν2 centered at 1376, 1507 and 1581 cm−1), cf. Table S1. These species represent only minor contributions (< 10 %) in the RR spectra of WT PpDyP [40], which is consistent with drastic structural alterations of the distal heme pocket of PpDyP variants.
Fig. 2.
Experimental and component RR spectra of ferric WT PpDyP and variants. (A) WT; (B) R214W and (C) R214I variants. The component spectra representing 6cHS (green), 6cLS (blue), 5cHS (magenta) and 5cQS (orange) heme species, and non-assigned bands (light grey) are fitted to the experimental spectra (black); the overall component spectra are depicted in dark grey. The spectra of 20–80 µM proteins were measured with 405 nm excitation and 3 mW laser power. (For interpretation of the references to color in this figure, the reader is referred to the web version of this article).
Replacement of the distal Arg in PpDyP variants (and to lower extent also in CboDyP) leads to the formation of hexacoordinated heme, carrying a thus far unknow distal ligand, since neither Ile or Trp are likely to coordinate the heme iron. This is consistent with the highly flexible heme pocket observed in resting peroxidases, which is capable of adopting different configurations. They most commonly result in a 5cHS heme iron that has a vacant sixth axial position for hydrogen peroxide binding and/or a 6cHS configuration in which the heme carries a loosely bound axial ligand [61]. Owing to its importance for catalysis, Arg variants of DyPs from different organisms have been previously studied. The substitution of the Arg residue is shown to have a variable effect on the heme cavity structure, ranging from moderate to complete attenuation of CT bands in the UV–Vis spectra of Arg/Leu substituted PpDyP and DyPB from Rhodococcus jostii RHA1 [9,52] to very subtle or no spectral differences in the case of Arg/Ala variants of BsDyP from B. subtilis, KpDyP from K. pneumoniae, TcDyP from T. curvata and ElDyP from Enterobacter lignolyticus [[48], [49], [50], [62]].
3.2. Determination of DyP redox potentials
In the next step, we set to determine the E°’ of the studied DyPs employing UV–Vis spectroelectrochemistry, in which heme reduction/oxidation is monitored via the changes in the Soret bands and as such has limited ability to discriminate between distinct heme populations. The individual E°’ values of structurally different heme species were addressed by SERR spectroelectrochemistry, which has a capacity to provide information on redox properties of co-existing spin population, as demonstrated on proteins containing multiple heme groups and/or spin populations, including PpDyP [[17], [34], [40], [63]].
UV–Vis spectroelectrochemistry: The E°’ of the Fe(III)/Fe(II) redox couple of the CboDyPs and PpDyPs was probed by UV–Vis spectroelectrochemistry in solution, employing a cell equipped with a gold electrode as the electron donor/acceptor, in the presence of a mixture of redox mediators, which cover a range of 550 mV (cf. Section 2.4.1). The UV–Vis spectra were acquired at a series of poised potentials, after equilibrium was reached, i.e. no spectral variations were detected (ca. 5 min). The potential of the working electrode was varied in successive steps between ca. +300 and −500 mV for reductive titrations and backwards for oxidative titrations (Fig. 3A). The E°’ value was determined from fits of the Nernst equation (Eq. (1)) to plots of normalized absorbance as a function of poised potential (cf. Fig. 3B for CboDyP R307I variant). Owing to the strong absorption of redox mediators in the Q and the ferric Soret band regions, we monitored the ferrous Soret band.
Fig. 3.
(A) UV–Vis spectral changes along the oxidative titration of CboDyP variant R307I performed from −395 mV (green line) to +305 mV (black line) poised potential. (B) Relative absorption of the ferrous protein at 430 nm plotted as a function of the cell potential. The solid line represents the fit of the Nernst equation to the experimental data, yielding E°’ = −155 ± 20 mV. (For interpretation of the references to color in this figure, the reader is referred to the web version of this article).
CboDyP variants show comparable E°’ (−160 ± 12 mV for R307W and −155 ± 20 mV for R307I), which are significantly upshifted in respect to that of the WT enzyme, E°’ = −320 ± 10 mV, Table S2. The value reported herein for the WT CboDyP is identical to the one previously determined by UV–Vis-based chemical titrations with dithionite/ferricyanide as reducing/oxidizing agents [35]. Based on pure electrostatic effects, the removal of the positively charged Arg should contribute to the stabilization of the Fe(III) form, thereby decreasing the E°' of the variants. We propose that the upshifted E°’ is a consequence of decreased hydration of the heme environment due to the introduction of the hydrophobic Trp and Ile residues in the heme cavity. According to the Kassner relation, the increase in hydrophobicity can account for up to 200 mV upshifts of E°' in heme proteins [64].
The E°’ values of the two PpDyP variants do not show significant differences (−270 ± 10 mV and −260 ± 10 mV for R214W and R214I, respectively) and are comparable to the E°’ of the WT PpDyP (−290 ± 5 mV), determined herein by UV–Vis spectroelectrochemistry (Table S2) and previously by UV–Vis chemical and SERR-based spectroelectrochemical titrations [40]. The Nernst curves obtained for all probed enzymes, including those that exist as a mixture of spin populations, reveal single redox transitions. This suggests that the E°’ values of the different populations are very close, but it may also indicate that UV–Vis spectroscopy is not sufficiently sensitive to distinguish the redox properties of the different heme species. In the next step, we employed SERR spectroelectrochemistry to test the above hypothesis.
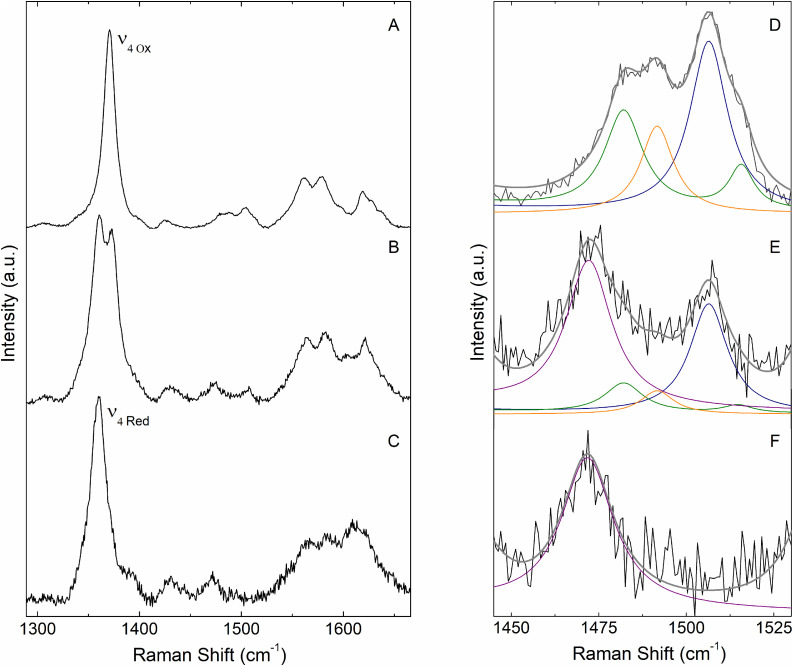
SERR spectroelectrochemistry: PpDyP R214W variant, which in solution presents a mixture of 6cHS and 6cLS states, was attached to Ag electrodes coated with SAMs composed of mixed polar and positively charged alkanethiols, which were previously shown to ensure a biocompatible interface for the WT enzyme adsorption. It was demonstrated that the structural integrity of the heme pocket in WT PpDyP was preserved under these conditions [40]. The SERR spectra of the immobilized PpDyP R214W, which were shown to be reversible upon varying electrode potentials (Fig. 4A–C), were first compared with the enzyme's RR spectra in solution. Component analysis of the SERR spectrum of R214W in the ferric state (obtained at +310 mV poised potential at the electrode) reveals similar frequencies and bandwidths for the 6cHS and 6cLS heme populations identified in the RR spectrum of the ferric enzyme. This suggests that R214W heme architecture is largely preserved in the immobilized enzyme. Still, an additional 5cHS species with ν3 at 1492 cm−1 (Figs. 4D and S3) could be identified in the spectrum (ca. 30 % abundance), which is assigned to a non-native heme state formed upon adsorption onto the electrode [65].
Fig. 4.
Left panel: SERR spectra of immobilized PpDyP variant R214W in the (A) ferric (at +310 mV poised potential), (B) semi-reduced (−240 mV poised potential) and (C) ferrous (−390 mV poised potential) state. Right panel: (D–F) Component analysis of the ν3 region of the respective SERR spectra shown on the left panel. The component spectra represent the ν3 band of ferric populations: 6cHS at 1483 cm−1 (green) and 6cLS at 1506 cm−1 (blue), ferrous species: 5cHS at 1472 cm−1 (purple), together with the non-native ferric population: 5cHS at 1492 cm−1 (orange). The experimental spectra are depicted in black and the overall component spectra in dark grey. Measurements were performed with 405 nm excitation and 1.4 mW laser power. (For interpretation of the references to color in this figure, the reader is referred to the web version of this article).
The E°’ of individual, native and non-native spin populations that are simultaneously present at the electrode surface were determined by deconvolution of potential dependent SERR spectra, in which the individual component bands that represent certain species were fit to experimental spectra recorded at each poised potential [[17], [40]]. Spectral parameters (band frequencies and widths) of each spin population were kept constant, thus the only variables for a SERR spectrum obtained at a given potential were the amplitudes of the individual component spectra, which are proportional to the species concentration. The Nernst fits to the relative spectral contributions of the native ferric species of PpDyP R214W variant as a function of electrode potential, Fig. S4, yield E°’ of −280 ± 9 mV and −260 ± 5 mV for the 6cHS and 6cLS populations, respectively. These values indicate that the two spin species have intrinsically similar E°’. On the other hand, the non-native 5cHS ferric species reveals upshifted E°’ (−70 mV). We speculate that immobilization-induced structural changes increase the hydrophobicity of the heme cavity, resulting in a denatured 5cHS form of the enzyme, which has lost axial ligands and has a higher E°’.
In the next step we probed the redox potentials of PpDyP R214I, and CboDyP R307W and R307I variants, for which only one main spin configuration is identified in solution. The SERR spectrum of ferric PpDyP R214I variant, acquired at +300 mV, indicates large immobilization induced alterations of the active site, since the newly formed 5cHS species (ν4 and ν3 centered at 1370 and 1492 cm−1, respectively), which is not present in the solution RR spectrum (Fig. S3C, D), became the major population in the spectrum of the immobilized enzyme. The E°’ of −230 ± 10 mV determined for the native 6cLS population is equivalent to the value determined by UV–Vis spectroelectrochemistry, while the E°’ of the non-native species, which could not be detected by the UV-Vis experiments, is −125 ± 18 mV. Similar to the R214W variant, we hypothesize that the non-native 5cHS population with upshifted E°’ is a result of immobilization induced changes in R214I's heme cavity. The two variants appear to be more prone to conformational alterations upon attachment to electrodes than the WT PpDyP. This is likely an outcome of destabilization of the heme cavity caused by the Arg substitution. The finding that the E°' of the native population in PpDyP variants is not affected by the substitution of the distal Arg by either Trp or Ile (cf. Section 3.2 UV–Vis spectroelectrochemistry) is nevertheless surprising, considering that the spin populations (in solution) underwent major alterations from 5cQS and 5cHS in WT enzyme to 6cLS (R214I) and a mixture of 6cLS and 6cHS (R214W) species. We attribute the small ΔE°’ to a complex combination of competing factors that can act on the heme upon substitution of the distal Arg. As previously proposed for other heme proteins, the removal of the positive charge is expected to lower the E°’ as the electrostatic repulsion with Fe(III) is diminished. On the other hand, the introduction of non-polar residues can reduce the polarity in the distal cavity and disrupt the H-bonding network linking the distal and proximal sides of the heme, leading to a loss of imidazolate character of the proximal His, which tends to stabilize the ferrous heme and increase the E°' [4,9]. The two opposite effects seem to counterbalance in PpDyP variants, resulting in a negligible influence of the Arg substitution on the overall E°’. A possible re-arrangement of the solvent molecules and H-bonding network around the distal cavity likely contributes to the maintenance of the polarity of the heme environment, thereby favoring the stabilization of the ferric iron and explaining the low E°’ of the R214W and R214I variants.
CboDyP R307W and R307I variants undergo major alterations of the active site upon immobilization (Fig. S5). The SERR spectra of the two enzymes, which in solution mainly adopt a 6cHS heme configuration, reveal heterogeneous heme coordination with two co-existing non-native populations assigned to 5cHS and 6cLS species (ν3 centered at 1492 and 1503 cm−1, respectively for R307W variant and at 1491 and 1504 cm−1 for R307I variant). This is in contrast with the behavior of WT CboDyP, for which it was demonstrated that more than half of the population retains its native structure upon attachment of the enzyme to the electrode [35]. The difference in behavior could be due to a lower stability of the variants. Since under the employed experimental conditions, immobilization induces significant structural changes, SERR spectroelectrochemistry could not be used for the characterization of the CboDyP variants.
4. Concluding remarks
Herein we discuss different approaches for the determination of the E°’ of DyP peroxidases and heme proteins in general. We highlight the potentials and drawbacks of UV–Vis- and SERR-based spectroelectrochemical methods, which rely on MET and DET respectively, and which have distinct intrinsic capacities to distinguish between E°’ of multiple heme populations. Although it could be used for the determination of the E°’ of all studied DyPs, UV–Vis spectroelectrochemistry requires thorough control of the ET efficiency between the electrode and the enzyme in order to ensure reversibility. The experiments often require high concentration of redox mediators that facilitate the electron exchange process, which in the case of the two studied DyPs reached equimolar enzyme/mediator ratio to minimize the hysteresis effects observed in UV–Vis spectroelectrochemical titrations. The use of high amounts of mediators, which can mask subtle changes of the protein's spectral features, is a potential disadvantage of this approach, together with the lack of sensitivity for simultaneous evaluation of the E°’ of multiple heme species. SERR spectroelectrochemistry, unlike electrochemical methods for determination of E°’, such as protein film voltammetry, enables the straightforward identification of native and non-native heme configurations, it uses low amounts of enzyme for electrode preparation (ca. 0.5 µM) and does not require redox mediators. However, SERR spectroelectrochemistry critically relies on preservation of the heme active site upon immobilization of the enzyme. This requires systematic search for proper SAM composition, i.e. immobilization conditions that ensure biocompatibility, which is not always easy or possible to achieve, as demonstrated here for CboDyP variants, and previously for e.g. cytochrome cd1 nitrite reductase, the DyPs from Thermobifida fusca and Streptomyces coelicolor (TfuDyP and ScoDyP), and cytochrome P450 [[17], [34], [35]]. On the other hand, there are numerous examples of proteins and enzymes that were shown to maintain their native structure and catalytic activity when immobilized on SAM coated electrodes, including DyPs (e.g. PpDyP), cytochrome PccH, aa3 oxygen reductase, cbb3 oxygen reductase and cytochrome c nitrite reductase [[40], [63], [66], [67], [68]]. Since it simultaneously offers information on E°ʼ of the immobilized species, ET efficiency between enzyme and electrode, enzyme's structure, stability and electrocatalytic activity, SERR spectroelectrochemistry is also a valuable tool for evaluating the performance of bioelectronic devices based on immobilized heme enzymes, such as 3rd generation electrochemical biosensors [55]. It allows for the rational design of bioelectronic devices, providing in situ information on enzyme structural properties upon attachment to electrodes, the range of substrates that can be explored, and the working conditions in which the devices can operate.
CRediT authorship contribution statement
Catarina Barbosa: Formal analysis, Investigation, Validation, Visualization, Writing – original draft. Carolina F. Rodrigues: Investigation. Nikola Lončar: Funding acquisition, Writing – review & editing. Lígia O. Martins: Funding acquisition, Supervision, Writing – review & editing. Smilja Todorovic: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – original draft, Writing – review & editing. Célia M. Silveira: Conceptualization, Formal analysis, Investigation, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank Dr. Caterina Martin, from GECCO Biotech B.V., for the help in the construction of CboDyP variants and Prof. Marco W. Fraaije for helpful discussions. This work was supported by FCT - Fundação para a Ciência e a Tecnologia, I.P., through MOSTMICRO-ITQB R&D Unit (UIDB/04612/2020, UIDP/04612/2020) and LS4FUTURE Associated Laboratory (LA/P/0087/2020). The authors acknowledge the support from the European Union's Horizon 2020 Research and Innovation Program, through B-LigZymes project (grant agreement number 824017) and from COST Action CA21115, supported by COST (European Cooperation in Science and Technology). CB and CRF acknowledge FCT Ph.D. studentships 2020.05017.BD and UI/BD/153388/2022, respectively.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.bbadva.2023.100112.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- 1.Poulos T.L. Heme enzyme structure and function. Chem. Rev. 2014;114(7):3919–3962. doi: 10.1021/cr400415k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spiro T.G., Jarzecki A.A. Heme-based sensors: theoretical modeling of heme-ligand–protein interactions. Curr. Opin. Chem. Biol. 2001;5(6):715–723. doi: 10.1016/S1367-5931(01)00271-X. [DOI] [PubMed] [Google Scholar]
- 3.Sykes A.G., Mauk G. Vol. 51. Academic Press; 2000. (Heme-Fe Proteins). [Google Scholar]
- 4.Battistuzzi G., Bellei M., Bortolotti C.A., Sola M. Redox properties of heme peroxidases. Arch. Biochem. Biophys. 2010;500(1):21–36. doi: 10.1016/j.abb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Adachi S., Nagano S., Watanabe Y., Ishimori K., Morishima I. Alteration of human myoglobin proximal histidine to cysteine or tyrosine by site-directed mutagenesis: characterization and their catalytic activities. Biochem. Biophys. Res. Commun. 1991;180(1):138–144. doi: 10.1016/S0006-291X(05)81266-5. [DOI] [PubMed] [Google Scholar]
- 6.Mauk A.G., Moore G.R. Control of metalloprotein redox potentials: what does site-directed mutagenesis of hemoproteins tell us? J. Biol. Inorg. Chem. 1997;2(1):119–125. doi: 10.1007/s007750050115. [DOI] [Google Scholar]
- 7.Ayala M., Roman R., Vazquez-Duhalt R. A catalytic approach to estimate the redox potential of heme-peroxidases. Biochem. Biophys. Res. Commun. 2007;357(3):804–808. doi: 10.1016/j.bbrc.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 8.Liers C., Aranda E., Strittmatter E., Piontek K., Plattner D.A., Zorn H., Ullrich R., Hofrichter M. Phenol oxidation by DyP-type peroxidases in comparison to fungal and plant peroxidases. J. Mol. Catal. B Enzym. 2014;103:41–46. doi: 10.1016/j.molcatb.2013.09.025. [DOI] [Google Scholar]
- 9.Mendes S., Brissos V., Gabriel A., Catarino T., Turner D.L., Todorovic S., Martins L.O. An integrated view of redox and catalytic properties of B-type PpDyP from Pseudomonas putida MET94 and its distal variants. Arch. Biochem. Biophys. 2015;574:99–107. doi: 10.1016/j.abb.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 10.Mondal M.S., Fuller H.A., Armstrong F.A. Direct measurement of the reduction potential of catalytically active cytochrome c peroxidase compound I: voltammetric detection of a reversible, cooperative two-electron transfer reaction. J. Am. Chem. Soc. 1996;118(1):263–264. doi: 10.1021/ja952489f. [DOI] [Google Scholar]
- 11.Alves M.N., Fernandes A.P., Salgueiro C.A., Paquete C.M. Unraveling the electron transfer processes of a nanowire protein from Geobacter sulfurreducens. Biochim. Biophys. Acta (BBA) Bioenerg. 2016;1857(1):7–13. doi: 10.1016/j.bbabio.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Guerrero F., Sedoud A., Kirilovsky D., Rutherford A.W., Ortega J.M., Roncel M. A high redox potential form of cytochrome c550 in photosystem II from Thermosynechococcus elongatus. J. Biol. Chem. 2011;286(8):5985–5994. doi: 10.1074/jbc.M110.170126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novak D., Mojovic M., Pavicevic A., Zatloukalova M., Hernychova L., Bartosik M., Vacek J. Electrochemistry and electron paramagnetic resonance spectroscopy of cytochrome c and its heme-disrupted analogs. Bioelectrochemistry. 2018;119:136–141. doi: 10.1016/j.bioelechem.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 14.Paquete C.M., Turner D.L., Louro R.O., Xavier A.V., Catarino T. Thermodynamic and kinetic characterisation of individual haems in multicentre cytochromes c3. Biochim. Biophys. Acta (BBA) Bioenerg. 2007;1767(9):1169–1179. doi: 10.1016/j.bbabio.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Pessanha M., Rothery E.L., Miles C.S., Reid G.A., Chapman S.K., Louro R.O., Turner D.L., Salgueiro C.A., Xavier A.V. Tuning of functional heme reduction potentials in Shewanella fumarate reductases. Biochim. Biophys. Acta (BBA) Bioenerg. 2009;1787(2):113–120. doi: 10.1016/j.bbabio.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Santos H., Moura J.J.G., Moura I., LeGall J., Xavier A.V. NMR studies of electron transfer mechanisms in a protein with interacting redox centres: desulfovibrio gigas cytochrome c3. Eur. J. Biochem. 1984;141(2):283–296. doi: 10.1111/j.1432-1033.1984.tb08190.x. [DOI] [PubMed] [Google Scholar]
- 17.Silveira C.M., Quintas P.O., Moura I., Moura J.J.G., Hildebrandt P., Almeida M.G., Todorovic S. SERR spectroelectrochemical study of cytochrome cd1 nitrite reductase Co-immobilized with physiological redox partner cytochrome c552 on biocompatible metal electrodes. PLOS One. 2015;10(6) doi: 10.1371/journal.pone.0129940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marritt S.J., van Wonderen J.H., Cheesman M.R., Butt J.N. Magnetic circular dichroism of hemoproteins with in situ control of electrochemical potential: “MOTTLE”. Anal. Biochem. 2006;359(1):79–83. doi: 10.1016/j.ab.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 19.Melin F., Hellwig P. Reference Module in Chemistry, Molecular Sciences and Chemical Engineering. Elsevier; 2015. Combining electrochemistry and infrared spectroscopy for the study of proteins. [DOI] [Google Scholar]
- 20.Borsari M., Benini S., Marchesi D., Ciurli S. Cyclic voltammetry and spectroelectrochemistry of cytochrome c8 from Rubrivivax gelatinosus. Implications in photosynthetic electron transfer. Inorg. Chim. Acta. 1997;263(1–2):379–384. doi: 10.1016/S0020-1693(97)05667-3. [DOI] [Google Scholar]
- 21.Howes B.D., Brissett N.C., Doyle W.A., Smith A.T., Smulevich G. Spectroscopic and kinetic properties of the horseradish peroxidase mutant T171S. Evidence for selective effects on the reduced state of the enzyme. FEBS J. 2005;272(21):5514–5521. doi: 10.1111/j.1742-4658.2005.04943.x. [DOI] [PubMed] [Google Scholar]
- 22.Ash P.A., Vincent K.A. Spectroscopic analysis of immobilised redox enzymes under direct electrochemical control. Chem. Commun. 2012;48(10):1400–1409. doi: 10.1039/c1cc15871f. [DOI] [PubMed] [Google Scholar]
- 23.Cerda J.F., Guzman C.X., Zhang H., Amendola E.J., Castorino J.D., Millet N., Fritz A.L., Houchins D.N., Roeder M.H. Spectroelectrochemical measurements of redox proteins by using a simple UV/visible cell. Electrochem. Commun. 2013;33:76–79. doi: 10.1016/j.elecom.2013.05.004. [DOI] [Google Scholar]
- 24.Kaim W., Fiedler J. Spectroelectrochemistry: the best of two worlds. Chem. Soc. Rev. 2009;38(12):3373–3382. doi: 10.1039/b504286k. [DOI] [PubMed] [Google Scholar]
- 25.Kornienko N., Ly K.H., Robinson W.E., Heidary N., Zhang J.Z., Reisner E. Advancing techniques for investigating the enzyme-electrode interface. Acc. Chem. Res. 2019;52(5):1439–1448. doi: 10.1021/acs.accounts.9b00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortimer R.J. 3rd. Academic Press; 2017. Spectroelectrochemistry, Applications. In Encyclopedia of Spectroscopy and Spectrometry; pp. 160–171. [DOI] [Google Scholar]
- 27.Reuillard B., Ly K.H., Hildebrandt P., Jeuken L.J.C., Butt J.N., Reisner E. High performance reduction of H2O2 with an electron transport decaheme cytochrome on a porous ITO electrode. J. Am. Chem. Soc. 2017;139(9):3324–3327. doi: 10.1021/jacs.6b12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilgus H., Ranweiler J.S., Wilson G.S., Stellwagen E. Spectral and electrochemical studies of cytochrome c peptide complexes. J. Biol. Chem. 1977;253(9):3265–3272. [PubMed] [Google Scholar]
- 29.Wilson G.S. Spectroelectrochemistry of proteins. Electroanalysis. 2022;34(12):1834–1841. doi: 10.1002/elan.202100535. [DOI] [Google Scholar]
- 30.Zhong F., Pletneva E.V. Ligation and reactivity of methionine-oxidized cytochrome c. Inorg. Chem. 2018;57(10):5754–5766. doi: 10.1021/acs.inorgchem.8b00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dutton P.L. Vol. 54. 1978. Redox potentiometry: determination of midpoint potentials of oxidation-reduction components of biological electron-transfer systems; pp. 411–435. (Methods Enzymol.). VolIssue. [DOI] [PubMed] [Google Scholar]
- 32.Oellerich S., Wackerbarth H., Hildebrandt P. Spectroscopic characterization of nonnative conformational states of cytochrome c. J. Phys. Chem. B. 2002;106(25):6566–6580. doi: 10.1021/jp013841g. [DOI] [Google Scholar]
- 33.Wackerbarth H., Klar U., Günther W., Hildebrandt P. Novel time-resolved surface-enhanced (resonance) Raman spectroscopic technique for studying the dynamics of interfacial processes: application to the electron transfer reaction of cytochrome c at a silver electrode. Appl. Spectrosc. 1999;53(3):283–291. doi: 10.1366/0003702991946668. [DOI] [Google Scholar]
- 34.Todorovic S., Jung C., Hildebrandt P., Murgida D.H. Conformational transitions and redox potential shifts of cytochrome P450 induced by immobilization. J. Biol. Inorg. Chem. 2006;11(1):119–127. doi: 10.1007/s00775-005-0054-9. [DOI] [PubMed] [Google Scholar]
- 35.Zuccarello L., Barbosa C., Galdino E., Lončar N., Silveira C.M., Fraaije M.W., Todorovic S. SERR spectroelectrochemistry as a guide for rational design of DyP-based bioelectronics devices. Int. J. Mol. Sci. 2021;22(15):7998. doi: 10.3390/ijms22157998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nöll T., Nöll G. Strategies for “wiring” redox-active proteins to electrodes and applications in biosensors, biofuel cells, and nanotechnology. Chem. Soc. Rev. 2011;40(7):3564–3576. doi: 10.1039/c1cs15030h. [DOI] [PubMed] [Google Scholar]
- 37.Sezer M., Millo D., Weidinger I.M., Zebger I., Hildebrandt P. Analyzing the catalytic processes of immobilized redox enzymes by vibrational spectroscopies. IUBMB Life. 2012;64(6):455–464. doi: 10.1002/iub.1020. [DOI] [PubMed] [Google Scholar]
- 38.Siebert F., Hildebrandt P. Wiley-VCH Verlag; 2008. Vibrational Spectroscopy in Life Science. [Google Scholar]
- 39.Hildebrandt P. Encyclopedia of Biophysics. Springer; Berlin Heidelberg: 2018. Resonance Raman spectroscopy of protein–cofactor complexes; pp. 1–10. [DOI] [Google Scholar]
- 40.Sezer M., Genebra T., Mendes S., Martins L.O., Todorovic S. A DyP-type peroxidase at a bio-compatible interface: structural and mechanistic insights. Soft Matter. 2012;8(40):10314–10321. doi: 10.1039/c2sm26310f. [DOI] [Google Scholar]
- 41.Murgida D.H., Hildebrandt P., Todorovic S. Immobilized redox proteins: mimicking basic features of physiological membranes and interfaces. Biomim. Learn. Nat. 2010:21–48. doi: 10.5772/8778. [DOI] [Google Scholar]
- 42.Colpa D.I., Fraaije M.W., van Bloois E. DyP-type peroxidases: a promising and versatile class of enzymes. J. Ind. Microbiol. Biotechnol. 2014;41(1):1–7. doi: 10.1007/s10295-013-1371-6. [DOI] [PubMed] [Google Scholar]
- 43.Sugano Y. DyP-type peroxidases comprise a novel heme peroxidase family. Cell. Mol. Life Sci. 2009;66(8):1387–1403. doi: 10.1007/s00018-008-8651-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshida T., Sugano Y. A structural and functional perspective of DyP-type peroxidase family. Arch. Biochem. Biophys. 2015;574:49–55. doi: 10.1016/j.abb.2015.01.022. [DOI] [PubMed] [Google Scholar]
- 45.Singh R., Eltis L.D. The multihued palette of dye-decolorizing peroxidases. Arch. Biochem. Biophys. 2015;574:56–65. doi: 10.1016/j.abb.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 46.Habib M.H., Rozeboom H.J., Fraaije M.W. Characterization of a New DyP-peroxidase from the alkaliphilic cellulomonad, cellulomonas bogoriensis. Molecules. 2019;24(7):1208. doi: 10.3390/molecules24071208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lučić M., Wilson M.T., Svistunenko D.A., Owen R.L., Hough M.A., Worrall J.A.R. Aspartate or arginine? Validated redox state X-ray structures elucidate mechanistic subtleties of FeIV = O formation in bacterial dye-decolorizing peroxidases. J. Biol. Inorg. Chem. 2021;26(7):743–761. doi: 10.1007/s00775-021-01896-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mendes S., Catarino T., Silveira C., Todorovic S., Martins L.O. The catalytic mechanism of A-type dye-decolourising peroxidase BsDyP: neither aspartate nor arginine is individually essential for peroxidase activity. Catal. Sci. Technol. 2015;5(12):5196–5207. doi: 10.1039/C5CY00478K. [DOI] [Google Scholar]
- 49.Pfanzagl V., Nys K., Bellei M., Michlits H., Mlynek G., Battistuzzi G., Djinovic-Carugo K., Van Doorslaer S., Furtmüller P.G., Hofbauer S., Obinger C. Roles of distal aspartate and arginine of B-class dye-decolorizing peroxidase in heterolytic hydrogen peroxide cleavage. J. Biol. Chem. 2018;293(38):14823–14838. doi: 10.1074/jbc.RA118.004773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C., Shrestha R., Jia K., Gao P.F., Geisbrecht B.V., Bossmann S.H., Shi J., Li P. Characterization of dye-decolorizing peroxidase (DyP) from Thermomonospora curvata reveals unique catalytic properties of A-type DyPs. J. Biol. Chem. 2015;290(38):23447–23463. doi: 10.1074/jbc.M115.658807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfanzagl V., Bellei M., Hofbauer S., Laurent C.V.F.P., Furtmüller P.G., Oostenbrink C., Battistuzzi G., Obinger C. Redox thermodynamics of B-class dye-decolorizing peroxidases. J. Inorg. Biochem. 2019;199 doi: 10.1016/j.jinorgbio.2019.110761. [DOI] [PubMed] [Google Scholar]
- 52.Shrestha R., Jia K., Khadka S., Eltis L.D., Li P. Mechanistic insights into DyPB from Rhodococcus jostii RHA1 via kinetic characterization. ACS Catal. 2021;11(9):5486–5495. doi: 10.1021/acscatal.1c00703. [DOI] [Google Scholar]
- 53.Santos A., Mendes S., Brissos V., Martins L.O. New dye-decolorizing peroxidases from Bacillus subtilis and Pseudomonas putida MET94: towards biotechnological applications. Appl. Microbiol. Biotechnol. 2014;98(5):2053–2065. doi: 10.1007/s00253-013-5041-4. [DOI] [PubMed] [Google Scholar]
- 54.Sun D., Ostermaier M.K., Heydenreich F.M., Mayer D., Jaussi R., Standfuss J., Veprintsev D.B. AAscan, PCRdesign and MutantChecker: a suite of programs for primer design and sequence analysis for high-throughput scanning mutagenesis. PLOS One. 2013;8(10):e78878. doi: 10.1371/journal.pone.0078878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barbosa C., Silveira C.M., Silva D., Brissos V., Hildebrandt P., Martins L.O., Todorovic S. Immobilized dye-decolorizing peroxidase (DyP) and directed evolution variants for hydrogen peroxide biosensing. Biosens. Bioelectron. 2020;153 doi: 10.1016/j.bios.2020.112055. [DOI] [PubMed] [Google Scholar]
- 56.Brissos V., Tavares D., Sousa A.C., Robalo M.P., Martins L.O. Engineering a bacterial DyP-type peroxidase for enhanced oxidation of lignin-related phenolics at alkaline pH. ACS Catal. 2017;5:3454–3465. doi: 10.1021/acscatal.6b03331. [DOI] [Google Scholar]
- 57.Berry E.A., Trumpower B.L. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal. Biochem. 1987;161(1):1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 58.Döpner S., Hildebrandt P., Mauk A.G., Lenk H., Stempfle W. Analysis of vibrational spectra of multicomponent systems. Application to pH-dependent resonance Raman spectra of ferricytochrome c. Spectrochim. Acta Part A. 1996;51:573–584. [Google Scholar]
- 59.Yoshida T., Tsuge H., Konno H., Hisabori T., Sugano Y. The catalytic mechanism of dye-decolorizing peroxidase DyP may require the swinging movement of an aspartic acid residue. FEBS J. 2011;278(13):2387–2394. doi: 10.1111/j.1742-4658.2011.08161.x. [DOI] [PubMed] [Google Scholar]
- 60.Jones P., Dunford H.B. The mechanism of compound I formation revisited. J. Inorg. Biochem. 2005;99(12):2292–2298. doi: 10.1016/j.jinorgbio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 61.Smulevich G., Feis A., Howes B.D. Fifteen years of Raman spectroscopy of engineered heme containing peroxidases: what have we learned? Acc. Chem. Res. 2005;38(5):433–440. doi: 10.1021/ar020112q. [DOI] [PubMed] [Google Scholar]
- 62.Shrestha R., Huang G., Meekins D.A., Geisbrecht B.V., Li P. Mechanistic insights into dye-decolorizing peroxidase revealed by solvent isotope and viscosity effects. ACS Catal. 2017;7(9):6352–6364. doi: 10.1021/acscatal.7b01861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Todorovic S., Verissimo A., Wisitruangsakul N., Zebger I., Hildebrandt P., Pereira M.M., Teixeira M., Murgida D.H. SERR-Spectroelectrochemical study of a cbb3 oxygen reductase in a biomimetic construct. J. Phys. Chem. B. 2008;112(51):16952–16959. doi: 10.1021/jp807862m. [DOI] [PubMed] [Google Scholar]
- 64.Kassner R.J. Theoretical model for the effects of local nonpolar heme environments on the redox potentials in cytochromes. J. Am. Chem. Soc. 1973;95(8):2674–2677. doi: 10.1021/ja00789a044. [DOI] [PubMed] [Google Scholar]
- 65.Wackerbarth H., Hildebrandt P. Redox and conformational equilibria and dynamics of cytochrome c at high electric fields. ChemPhysChem. 2003;4(7):714–724. doi: 10.1002/cphc.200200618. [DOI] [PubMed] [Google Scholar]
- 66.Silveira C.M., Castro M.A., Dantas J.M., Salgueiro C., Murgida D.H., Todorovic S. Structure, electrocatalysis and dynamics of immobilized cytochrome PccH and its microperoxidase. Phys. Chem. Chem. Phys. 2017;19(13):8908–8918. doi: 10.1039/c6cp08361g. [DOI] [PubMed] [Google Scholar]
- 67.Todorovic S., Pereira M.M., Bandeiras T.M., Teixeira M., Hildebrandt P., Murgida D.H. Midpoint potentials of hemes a and a3 in the quinol oxidase from Acidianus ambivalens are inverted. J. Am. Chem. Soc. 2005;127(39):13561–13566. doi: 10.1021/ja052921l. [DOI] [PubMed] [Google Scholar]
- 68.Todorovic S., Rodrigues M.L., Matos D., Pereira I.A.C. Redox properties of lysine- and methionine-coordinated hemes ensure downhill electron transfer in NrfH2A4 nitrite reductase. J. Phys. Chem. B. 2012;116(19):5637–5643. doi: 10.1021/jp301356m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.