Abstract
Objective
Human functional genomics has proven powerful in discovering drug targets for common metabolic disorders. Through this approach, we investigated the involvement of the purinergic receptor P2RY1 in type 2 diabetes (T2D).
Methods
P2RY1 was sequenced in 9,266 participants including 4,177 patients with T2D. In vitro analyses were then performed to assess the functional effect of each variant. Expression quantitative trait loci (eQTL) analysis was performed in pancreatic islets from 103 pancreatectomized individuals. The effect of P2RY1 on glucose-stimulated insulin secretion was finally assessed in human pancreatic beta cells (EndoCβH5), and RNA sequencing was performed on these cells.
Results
Sequencing P2YR1 in 9,266 participants revealed 22 rare variants, seven of which were loss-of-function according to our in vitro analyses. Carriers, except one, exhibited impaired glucose control. Our eQTL analysis of human islets identified P2RY1 variants, in a beta-cell enhancer, linked to increased P2RY1 expression and reduced T2D risk, contrasting with variants located in a silent region associated with decreased P2RY1 expression and increased T2D risk. Additionally, a P2RY1-specific agonist increased insulin secretion upon glucose stimulation, while the antagonist led to decreased insulin secretion. RNA-seq highlighted TXNIP as one of the main transcriptomic markers of insulin secretion triggered by P2RY1 agonist.
Conclusion
Our findings suggest that P2RY1 inherited or acquired dysfunction increases T2D risk and that P2RY1 activation stimulates insulin secretion. Selective P2RY1 agonists, impermeable to the blood–brain barrier, could serve as potential insulin secretagogues.
Highlights
-
•
Loss-of-function P2RY1 variants were highly penetrant for type 2 diabetes.
-
•
eQTL analysis in human islets identified P2RY1 variants linked to reduced P2RY1 expression and increased T2D risk.
-
•
P2RY1 activation in human beta cells increases insulin secretion upon glucose stimulation.
-
•
TXNIP is a significant marker for insulin secretion triggered by P2RY1 agonist.
-
•
Impermeable-to-brain P2RY1 agonists could serve as potential insulin secretagogues.
1. Introduction
Type 2 diabetes (T2D) is a complex metabolic disorder that affects millions of people worldwide. T2D that is a multifactorial disease with a strong genetic component, arises from a dysfunction in the regulation of blood glucose levels, leading to chronic elevation of blood sugar. One crucial aspect of glucose homeostasis is insulin secretion from pancreatic beta cells [1]. These cells release insulin in response to elevated blood glucose levels, facilitating the uptake of glucose by body cells for energy and promoting the storage of excess glucose. This regulatory process helps keep blood glucose within a narrow and essential range, crucial for the overall functioning of the body.
P2RY1 has emerged as an intriguing putative candidate gene implicated in beta-cell function. It encodes a G protein-coupled receptor (GPCR) coupled to the Gq family of G protein alpha subunits [2,3]. P2RY1 belongs to the purinergic receptor family, which is involved in cellular responses to extracellular nucleotides, such as adenosine diphosphate (ADP), and, to a lesser extent, adenosine triphosphate (ATP). Upon activation, P2RY1 triggers an increase in intracellular calcium concentration by stimulating phospholipase C (PLC) [2,4,5]. P2RY1 is expressed in various human tissues, but particularly in pancreatic beta cells [5]. Here, following glucose uptake and glycolysis, an elevated ATP:ADP ratio activates the ATP-dependent potassium channels. This triggers membrane depolarization, opening of voltage-gated calcium channels, and influx of calcium ions. Elevated intracellular calcium levels eventually stimulate insulin-containing vesicle fusion with the cell membrane, releasing insulin into the bloodstream [6,7]. Genome-wide association studies have demonstrated a significant association between a block of common variants at the P2RY1 locus and the risk of T2D (adjusted or not for body mass index), as well as with random glucose and glycated hemoglobin A1c levels, according to the T2D Knowledge portal [8]. However, the potential involvement of P2RY1 in human beta-cell function and T2D has received limited exploration [9,10], in contrast to the thorough examination of P2RY1's role in platelet aggregation and smooth muscle contraction [[11], [12], [13]].
Here, we conducted human functional genomics to investigate the potential role of P2RY1 in insulin secretion and T2D.
2. Results
2.1. Contribution of rare loss-of-function P2RY1 variants to T2D
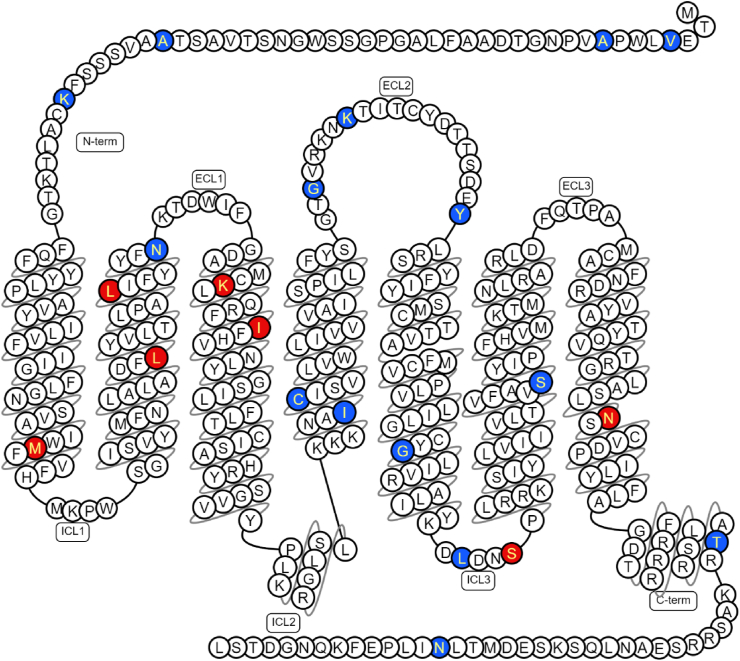
P2RY1 (NM_002563.5) was sequenced by next-generation sequencing in 9,266 adults, including 4,177 participants with T2D (Table S1). We identified 22 rare non-synonymous variants, including 11 novel variants (Table S2). Each variant was carried by one, two or three participants (Table S2). These variants were distributed throughout the receptor (Figure 1).
Figure 1.
Location of the variants found in P2RY1.
Schematic representation of the human form of the P2RY1 receptor (encoded by NM_002563.5) according to Swiss-Prot (via the GPCR database [https://gpcrdb.org/]). Red represents variant inducing amino acid changes linked to loss-of-function variants, while blue represents variant inducing amino acid changes linked to neutral variants according to our in vitro assays. C-term, Carboxyl-Terminus; ECL, Extra Cellular Loop; ICL, Intra Cellular Loop; N-term, Amino-Terminus. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Each mutant or wild-type P2RY1 was expressed in human embryonic kidney 293 cells (HEK293luc), which stably expressed the luciferase gene under the regulation of a promoter incorporating multiple Nuclear Factor of Activated T-cell response elements (NFAT-REs) that are activated by Gq-protein-coupled receptors [14,15]. Among the 22 P2RY1 variants tested, we observed a consistent reduction in basal activity compared to the wild-type receptor for five variants encoding p.M75I, p.L99W, p.L107V, p.K125E, and p.N316S (Fig. S1A). This reduction suggested that these specific variants negatively impact the receptor's ability to initiate downstream signaling pathways.
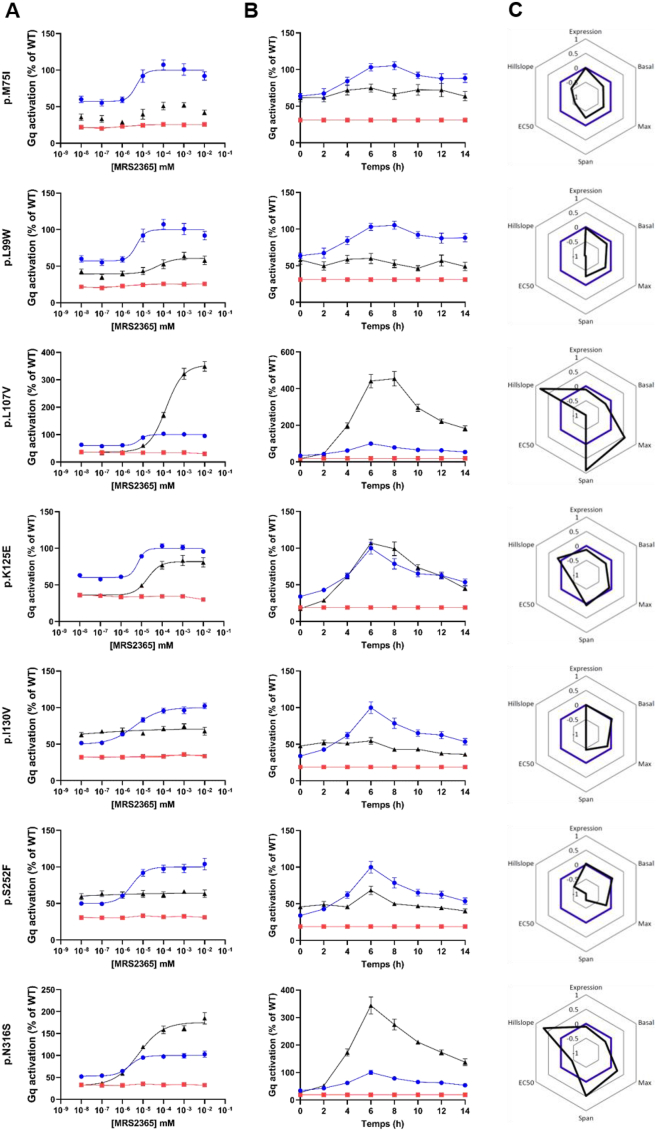
We then conducted a dose–response analysis to evaluate the luciferase activity of the 22 rare P2RY1 variants in response to an increasing dose of the P2RY1-specific agonist MRS2365 over a period of 6 h. Among the 22 P2RY1 variants tested, three variants encoding p.L107V, p.K125E, and p.N316S exhibited a shift in the EC50 value compared to the wild-type receptor (Figure 2A). This result indicates that these three variants displayed reduced ability to bind with MRS2365. The variant encoding p.K125E also displayed reduced activity throughout the dose–response analysis while the variants encoding p.L107V and p.N316S exhibited reduced basal activity and augmented maximum effect (Figure 2A). Furthermore, the variants encoding p.M75I, p.L99W, p.I130V, and p.S252F showed minimal to no response upon stimulation with the P2RY1-specific agonist MRS2365 (Figure 2A), suggesting a potential loss of receptor functionality and an impaired activation of downstream signaling pathways in these variants. The remaining 15 variants demonstrated similar activity to the wild-type P2RY1 (Fig. S1B).
Figure 2.
In vitro analyses for characterizing the activity of loss-of-function P2RY1 variants in the NFAT-RE pathway.
NFAT-RE luciferase activation was shown following (A) dose–response (during a 6-hour period) and (B) time course (at 10 uM concentration) experiments for each P2RY1 loss-of-function variant. Each mutant (depicted by black triangles) was compared to the activity of wild-type P2RY1 (positive control shown as blue dots) and a P2RY1 mutant harboring an early stop-codon variant (negative control represented by a red square). Data are presented as percentages of wild-type maximum activity. The results represent the means ± SEM of 4 independent experiments, each performed in technical quadruplicate.
(C) Radar charts represent signaling signature of the seven loss-of-function variants (represented in black) and wild-type P2RY1 (shown in blue). Parameters measured include protein expression, basal activity, maximum effect upon agonist stimulation (Max), gap between minimum and maximum effects (Span), half maximal effective concentration (EC50), and activity over time (Hillslope). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Furthermore, in a time-course luciferase assay experiment in response to 10 uM of the P2RY1-specific agonist MRS2365, we examined the activity of the seven loss-of-function P2RY1 variants identified in the previous experiments (Figure 2B). Upon stimulation with MRS2365, these variants exhibited equal (p.K125E) or significantly higher (p.L107V and p.N316S) activity response over time compared to the wild-type receptor (Figure 2B). This indicates that despite the reduced basal activity, these specific variants enhance the receptor's responsiveness to a very high dose of the P2RY1-specific agonist. On the other hand, the variants encoding p.M75I, p.L99W, p.I130V, and p.S252F demonstrated minimal to no response upon stimulation with MRS2365.
We then investigated the expression patterns of the seven loss-of-function P2RY1 mutants at the cell membrane through immunofluorescence experiments. Through quantification experiments, we observed a significantly reduced expression levels of three variants encoding p.L107V, p.K125E, and p.N316S, compared to the wild-type receptor (Figure 2C and Fig. S1C). This result suggests that these specific variants may have an impact on the stability or trafficking of the receptor, leading to lower overall expression levels at the cell surface. On the other hand, the variants encoding p.M75I, p.L99W, p.I130V, and p.S252F showed no significant change in expression compared to the wild-type control, indicating that these variants do not grossly affect the overall expression or localization of the receptor. However, all loss-of-function P2RY1 mutants were observed to be localized to the cell periphery, suggesting that these variants did not prevent the targeting of the receptor to the cell surface (Fig. S2).
In summary, the variants encoding p.L107V, p.K125E, and p.N316S resulted in reduced basal activity, expression, and altered EC50 values of the P2RY1 receptor. Furthermore, the variants encoding p.M75I, p.L99W, p.I130V, and p.S252F were associated with either a lack or a decreased response to MRS2365. All of these loss-of-function P2RY1 variants were situated within transmembrane regions, except for the variant encoding p.S252F, which was positioned within the third intracellular loop and was identified as a phosphorylation site (Figure 1) [16].
Of note, we found a significant association (P = 1.3 × 10−3) between the predicted in silico score of variants via REVEL [17] and the deleteriousness of P2RY1 variants based on our in vitro analyses (Table S2). Importantly, except for one variant (encoding p.S252F), all loss-of-function mutations of P2RY1 were novel, as not listed in the GnomAD browser (Table S2).
Among eight carriers of a loss-of-function P2RY1 variant, all exhibited glucose intolerance (i.e. one individual with pre-diabetes and six with overt T2D), except for a 51-year-old adult with normal weight. The carriers of a loss-of-function P2RY1 variant who had pre-diabetes or T2D were overweight (i.e. with a body mass index between 25 and 35 kg/m2), and were treated for T2D with metformin (one of whom was also on insulin). Therefore, the carriers of a loss-of-function P2RY1 variant had a common form of T2D.
In the TOPMed study from the AMP T2D knowledge portal [8], we found that the null P2RY1 variants (i.e. either frameshift or stop gain) were significantly associated with a higher risk of T2D in 44,083 participants (P = 0.043 with an odds ratio of 10.9).
2.2. Negative association between P2RY1 expression levels in human pancreatic islets and T2D risk alleles
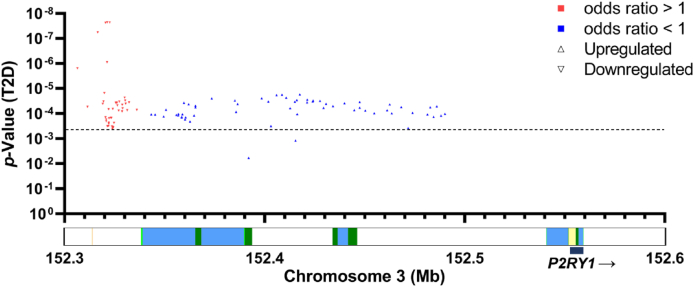
We then conducted an expression quantitative trait loci (eQTL) analysis in 103 pancreatectomized individuals [18,19]. We specifically analyzed the effect of single nucleotide polymorphisms (SNPs) at the P2RY1 locus, known to be associated with T2D [8], on P2RY1 expression in pancreatic islet samples from these living donors. Our eQTL analysis uncovered a negative association between P2RY1 expression levels and T2D risk alleles (Figure 3). Notably, all SNPs that contributed to an elevation in P2RY1 expression in pancreatic islets were consistently associated with a protective effect on T2D risk (Figure 3). In contrast, every SNP linked to a decrease in P2RY1 expression was found to be associated with increased T2D risk (Figure 3). This observation indicates that P2RY1 expression levels in human pancreatic islets play a crucial role in modulating the susceptibility to T2D. Of note, the SNPs associated with increased P2RY1 expression levels and reduced T2D risk were located within an enhancer region that is active in the human pancreatic beta-cell line EndocβH1, while the SNPs associated with decreased P2RY1 expression levels were situated in a silent chromatin region upstream of P2RY1 gene (Figure 3).
Figure 3.
Negative association between P2RY1 expression levels in human pancreatic islets and T2D risk alleles.
On the X-axis, the localization of expression quantitative trait loci (eQTLs) at the P2RY1 locus is represented, while the Y-axis represents the p-value of their association with T2D risk. Blue up-pointing triangles represent SNPs significantly associated with increased P2RY1 expression in human pancreatic islets and reduced T2D risk (with a p-value <0.05), while red down-pointing triangles represent SNPs significantly associated with decreased P2RY1 expression in islets and increased T2D risk (with a p-value <0.05). The dotted line indicates the multiple testing threshold.
Below the X-axis, a chromatin map from human pancreatic beta-cell line EndoCβH1 is represented. The chromatin elements are color-coded as follows: light green for weak enhancers, dark green for active enhancers, yellow for active transcription start sites (TSS), and blue for transcription sites. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.3. Regulation of insulin secretion from human pancreatic beta cells by P2RY1
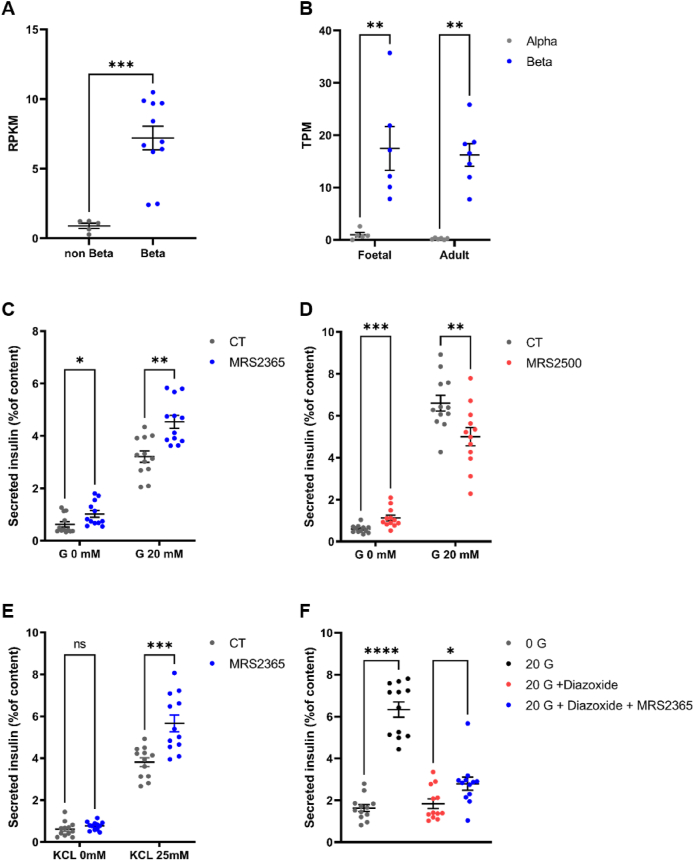
We next aimed to investigate the impact of P2RY1 on insulin secretion. For this purpose, we first assessed the expression of P2RY1 in human pancreatic beta cells. In addition, based on RNA sequencing (RNA-seq) data [20], we found that the expression of P2RY1 was significantly enriched in human pancreatic beta cells when compared to other cell types within pancreatic islets (Figure 4A). Upon examining RNA-Seq data from both fetal and adult human pancreatic beta cells [21], we observed that the expression of P2RY1 remained stable and unchanged during these developmental stages (Figure 4B). Using human pancreatic beta cells EndoCβH5 [22], we found that the stimulation of P2RY1 with its specific agonist MRS2365 for 40 min resulted in a 41 % increase in insulin secretion under high glucose conditions (Figure 4C), while insulin content remained stable (Fig. S4A). In contrast, the inhibition of P2RY1 with its specific antagonist MRS2500 resulted in a 24 % decrease in insulin secretion under high glucose conditions (Figure 4D), while insulin content remained stable (Fig. S4B).
Figure 4.
P2RY1 expression in human pancreatic beta cells and P2RY1 effect on glucose-stimulated insulin secretion upon its stimulation or inhibition.
(A) Expression of P2RY1 in human purified pancreatic beta cells (n = 11) and non-beta cell (remaining islet preparation) (n = 5) assessed via [20]. Data are mean ± SEM of RPKM (Reads Per Kilobase Million).
(B) Expression of P2RY1 in Fetal and Adult human beta and alpha cells (between 6 and 7 samples per condition) assessed via [21]. Data are mean ± SEM of TPM (Transcripts Per Million).
Glucose-stimulated insulin secretion (glucose [G] 0 mM versus [G] 20 mM) from EndoCβH5 treated with (C) ± 1 μM of P2RY1-specific agonist MRS2365; or (D) ± 1 μM of P2RY1-specific antagonist MRS2500.
(E) Potassium-chloride stimulated insulin secretion ([KCl] 0 mM versus [KCl] 25 mM) from EndoCβH5 treated with ±1 μM of P2RY1-specific agonist MRS2365.
(F) Glucose stimulated insulin secretion (glucose [G] 0 mM versus [G] 20 mM) from EndoCβH5 treated with ±1 μM Diazoxide ± 1 μM of P2RY1-specific agonist MRS2365.
Data are the means ± SEM from three independent experiments, each conducted in technical quadruplicate. All experiments were analyzed using a Mann–Whitney t-test: ∗P < 0.01; ∗∗P < 0.01; ∗∗∗P < 0.001. CT, control condition.
We conducted experiments using potassium chloride (KCl) instead of glucose to explore the influence of P2RY1 on insulin secretion independent of glucose metabolism. Notably, the P2RY1 agonist led to a significantly increased insulin secretion in response to 25 mM of KCl, when compared to the control condition (Figure 4E), while insulin content remained stable (Fig. S4C). This suggests that P2RY1 activation induces insulin secretion by directly mobilizing calcium ions.
On the other hand, we conducted experiments involving the hyperpolarization of the cell membrane with 250 μM of diazoxide. Although diazoxide inhibited insulin secretion under high glucose conditions, the P2RY1 agonist led to a significantly increased insulin secretion (Figure 4F), while insulin content remained stable (Fig. S4D). This underscores the notion that P2RY1 activation can enhance insulin secretion independently of glucose metabolism, emphasizing a direct role in calcium mobilization.
Upon treating EndoCβH5 cells with the P2RY1-specific agonist under high glucose conditions, RNA-seq analysis revealed the upregulation of 532 genes (with a Transcripts Per Million [TPM] greater than 1) and 296 downregulated genes (with a TPM greater than 1) in response to P2RY1 agonist (Fig. S3). Notably, we found enrichment of significantly upregulated genes in several pathways in response to P2RY1 agonist: nutrient levels (P = 3.3 × 10−12), extracellular stimulus (P = 3.7 × 10−12), starvation (P = 6.9 × 10−8), glucose starvation (P = 1.3 × 10−4), but also GPCR downstream signaling (P = 1.5 × 10−4) and more specifically NFAT pathway (P = 2.2 × 10−4) (Table S3). Of note, using the P2RY1 agonist alone (i.e. without glucose stimulation) only significantly induced an upregulation of genes linked to GPCR signaling pathway (data not shown).
Among the differentially regulated genes in response to P2RY1-specific agonist, TXNIP (encoding Thioredoxin Interacting Protein) emerged as the most significantly downregulated gene (Log2 Fold Change = −0.60; Padj = 3.0 × 10−44; Fig. S3). This is of particular interest, given the known role of TXNIP in T2D development. Indeed, elevated TXNIP expression has been linked to increased oxidative stress and impaired glucose uptake in pancreatic beta cells [23,24]. This, in turn, was suggested to lead to impaired insulin secretion and to contribute to diabetes development [25,26]. TXNIP has also been associated with the activation of inflammasomes, leading to increased production of pro-inflammatory cytokines, further promoting beta cell dysfunction and apoptosis [27]. The dysregulation of TXNIP in response to P2RY1 agonist stimulation may, therefore, represent a critical link between P2RY1 signaling and the development of T2D.
Of note, RNA-seq sequence reads did not reveal any coding P2RY1 coding mutations in EndoCβH5 cells.
3. Discussion
Based on complementary functional genomics tools, our study highlights a novel link between P2RY1 dysfunction (in terms of activity and/or expression) and increased T2D risk in humans. Using luciferase assays monitoring the activation of NFAT pathway upon P2RY1 stimulation, we indeed found that rare loss-of-function P2RY1 variants were mostly carried by individuals with typical T2D or prediabetes associated with overweight. Of note, some of these variants caused decreased P2RY1 expression, but most of them did not appear to grossly alter receptor expression. We confirmed in the large TOPMed cohort study that rare deleterious P2RY1 variants (focusing on null variants) are associated with T2D risk in human general populations.
Drawing from our eQTL analyses conducted on human pancreatic islet samples from living donors, coupled with in vitro experiments performed on human pancreatic beta cells, we posit that the connection between P2RY1 and T2D in humans involves P2RY1 function in modulating insulin secretion. Our present results are in line with previous data that showing that purinergic receptors may influence glucose homeostasis through insulin secretion, not only in rodents [[28], [29], [30]], but also in humans [9,10,30].
Our current study is constrained by our exclusive examination of the impact of rare P2RY1 variants on NFAT-RE luciferase activation and P2RY1 expression, leaving room for the exploration of various additional physiological outcomes. Our emphasis was indeed directed towards investigating the canonical pathway upon P2RY1 activation.
P2RY1 being a GPCR, it should be considered as a privileged drug target [31], and our findings may pave the way for innovative therapeutic strategies for T2D.
4. Methods
4.1. DNA sequencing
P2YR1 (NM_002563.5) was accurately sequenced in 9,266 French or Belgian adults including 4,177 participants with T2D (Table S1). A total of 7,268 samples are derived from the RaDiO study, and their sequencing has been comprehensively detailed in a separate publication [32]. A total of 1,998 samples were derived from the PreciDIAG study including patients diagnosed with T2D. The recruitment of patients started in 2019, in the Department of Diabetology of Liège University Hospital (Belgium). The large majority of these patients have been prospectively followed in this department. Clinical data in particular anthropometric and metabolic data are available for each patient at recruitment. Exclusion criteria included latent autoimmune diabetes of adulthood (LADA), type 1 diabetes (based on the presence of autoantibodies that are systematically assessed), and gestational diabetes. All included patients were more than 18 years old. DNA samples from PreciDIAG study were sequenced through whole-exome sequencing. Briefly, we used KAPA HyperExome probes (Roche, Pleasanton, CA, USA) and Illumina (San Diego, CA, USA) sequencing (NovaSeq6000 system, 150bp paired end reads mode). Sequence reads were aligned to the Human Genome Reference Sequence (GRCh38/hg38). We achieved an average sequencing depth of 80 × in the participants.
All cohort studies followed ethical principles defined in the Helsinki declaration (revised in 1996), and they were approved by local ethical committees from Corbeil-Essonnes hospital (France), Comité Consultatif de Protection des Personnes se prêtant à des Recherches Biomédicales (CCPPRB) of Lille - Lille Hospital (Lille, France), Hotel-Dieu hospital (France), Bicêtre hospital (France), and Liège (Belgium). All participants signed an informed consent form.
Prediabetes was defined as fasting plasma glucose levels ≥5.6 mmol/l and <7 mmol/l; and T2D was defined as fasting plasma glucose levels ≥7 mmol/l, glycated hemoglobin A1c higher than 6.5 % and/or use of drug therapy for hyperglycemia [33].
4.2. Plasmid generation
The human P2RY1 (NM_002563.5) plasmid was purchased from Origene (Rockville, USA). This plasmid includes the P2RY1 gene fused with MYc-DDK tag (at C-terminus). All plasmid including each variant was generated using the QuickChange II XL site-directed mutagenesis kit (Agilent Technologies, Santa Clara, USA). Each plasmid was verified by Sanger sequencing using 3730 Series Genetic Analyzers (Thermo Fisher Scientific, Waltham, USA) and then amplified in E. coli (Thermo Fisher Scientific, Waltham, USA) and extracted using NucleoBond Xtra Maxi Columns for transfection-grade plasmid DNA (Macherey–Nagel, Duren, Germany) according to the manufacturer's protocol.
4.3. HEK293luc cells culture
HEK293luc cells (E8510) (Promega, Madison, USA) were cultured in Dulbecco's Modified Eagle's Medium (Gibco, Waltham, USA) supplemented with 10 % fetal bovine serum and 50 μg/ml hygromycin B (Thermo Fisher Scientific, Waltham, USA) at 37 °C, 5 % CO2. HEK293luc cells represent a stable transfection cell line containing a plasmid including the luciferase gene regulated by a minimal TATA promoter that incorporates multiple Nuclear Factor of Activated T-cell response elements (NFAT-REs).
4.4. Plasmid electroporation
One million HEK293luc cells were electroporated with 5 μg of P2RY1 wild-type or variant plasmid (at a concentration of 0.5 μg/μl) using the Cell Line Nucleofector Kit V (Lonza, Basel, Switzerland) according to the manufacturer's protocol. Electroporation was performed using the Nucleofector 2 b device (Lonza, Basel, Switzerland) with the Q-001 program. The cells were then resuspended in Dulbecco's Modified Eagle's Medium (Gibco, Waltham, USA) supplemented with 10 % fetal bovine serum, 1 % penicillin/streptomycin (Gibco, Waltham, USA), and seeded in white opaque 96-well plates coated with Poly-d-lysine at a concentration of 20,000 cells/well (100 μl/well) and cultured for 48 h.
4.5. Luciferase assays
Electroporated HEK293luc cells (Promega, Madison, USA) were treated for 6 h with increasing doses of MRS2365 (Tocris Bioscience, Bristol, UK) ranging from 10−8 to 10−2 M diluted in Dulbecco's Modified Eagle's Medium (Gibco, Waltham, USA), supplemented with 1 % fetal bovine serum for 6 h for dose–response experiment, or with 10−2 M of MRS2365 diluted in Dulbecco's Modified Eagle's Medium (Gibco, Waltham, USA) supplemented with 1 % fetal bovine serum for 14 h with 2-hour intervals for time-course experiment. The cells were then lysed in 40 μl of passive lysis buffer (Promega) for 30 min. Luciferase activity was measured by adding 25 μl of luciferase assay system reagent (Promega) directly to the plate. Luminescence was read using a GloMax Luminometer (Promega). Each experiment was independently conducted four times and in technical quadruplicate.
4.6. Immunofluorescence assays
Electroporated HEK293luc cells (Promega, Madison, USA) were seeded onto poly-d-lysine-coated coverslips in a 12-well plate at a concentration of 100,000 cells/well (1000 μl/well) and cultured for 48 h. Two days after electroporation, cells were fixed in 4 % paraformaldehyde (Thermo Fisher Scientific, Waltham, USA) for 1 h, followed by three washes with phosphate-buffered saline (PBS). Subsequently, cells were permeabilized with 0.1 % Triton X-100 in PBS for 15 min. The unoccupied binding sites were blocked using a blocking buffer, Background Reducing (Agilent, Santa Clara, USA) for 1 h. Overnight incubation at 4 °C was performed with the primary antibody anti-DDK (TA-50011) (Origene, Rockville, USA) and anti-αTubulin (PA1-38814) (Thermo Fisher Scientific, Waltham, USA); both diluted at 1/1000 in the blocking buffer. After three washes with PBS containing 0.1 % Tween, cells were incubated with a fluorescent secondary antibody (A-11029 and A11012) (Sigma, Saint Louis, USA); both diluted at 1/1000 in blocking buffer, and nuclei were stained with DAPI (D9542) diluted at 1/1000 (Thermo Fisher Scientific, Waltham, USA) for 1 h in the dark. Following three washes with PBS containing 0.1 % Tween, slides were mounted using Prolong Gold Antifade Mountant (Thermo Fisher Scientific, Waltham, USA). Cells were observed using a Cell Observer NanoZoomer S20MD Slide scanner system (Hamamatsu, Shizuoka, Japan). This experiment was conducted three times independently. To quantify P2RY1 expression, DDK intensity was measured (using ImageJ Fiji) and normalized relative to DAPI intensity. This experiment was carried out across six regions on three separate slides.
4.7. Radar chart
Radar charts were generated using data from dose–response and time-course experiments, basal activity, and quantification of expression. In these graphs, each value was plotted using the logarithm base 10 (log10) transformation, which enabled a comprehensive representation of the data, emphasizing relative differences on a logarithmic scale. The activity of the wild-type P2RY1 was set as zero, and the scale for all radar chart ranges from −1 to +1. Variants with enhanced properties are depicted within the range of 0 to +1, while variants with impaired properties are depicted within the range of 0 to −1.
The term “Expression" represents the expression level of each P2RY1 mutant quantified from immunofluorescence assays. The “Basal" value represents the receptor activity under basal conditions (i.e., without agonist stimulation). The “Max" value represents the maximum effect, “Span" indicates the gap between the minimum and maximum effects, and “EC50″ represents the dose required to achieve 50 % of the maximum effect from the dose–response experiment. All these parameters were calculated using GraphPad Prism (version 9), employing a nonlinear regression (curve fit, variable slope, 4 parameters) for their determination. The “Hillslope" value was also calculated using GraphPad Prism 9, utilizing a simple linear regression from the time-course experiment data.
4.8. EndoCβH5 cell culture
EndoCβH5 cells were cultured in Optiβ1 medium (Univercell Biosolutions, Toulouse, France) at 37 °C with 5 % CO2. The cells were seeded at a density of 100,000 cells per well in a 96-well plate coated with βcoat (Univercell Biosolutions), following the manufacturer's protocol.
4.9. Insulin secretion assays
Six days after seeding, the cells were incubated in a starving medium for 24 h, followed by treatment with βKrebs Buffer (Univercell Biosolutions) for 60 min at 37 °C. Subsequently, the cells were treated with βKrebs Buffer containing ±20 mM glucose ±1 μM of MRS2365 or 1 μM of MRS2500 for 40 min. Subsequently, the cells were treated with βKrebs Buffer containing ±20 mM glucose; ±25 mM KCl; ±1 μM MRS2365 (Tocris Bioscience, Bristol, UK); ±1 μM MRS2500 (Tocris) and/or 250 μM diazoxide (Tocris) for 40 min. The supernatant was collected, and the cells were lysed in TETG buffer. Insulin concentrations in the supernatants and lysates were assessed using the ELISA Human Insulin kit (Mercodia, Uppsala, Sweden) according to the provided protocol. Prior to analysis, the samples were diluted 1:50 in water for the supernatants and 1:4000 for the lysates. Each experiment was conducted three times independently and in technical quadruplicate.
4.10. RNA sequencing
RNA was extracted from 0.5 M EndoCβH5 cells using the NucleoSpin RNA Mini kit for RNA purification (Macherey–Nagel, Duren, Germany). The quality of the RNA samples was verified using RNA 6000 nanochips on the Agilent 2100 Bioanalyzer. Purified RNA (500 ng) was utilized for library preparation. Briefly, RNA libraries were generated using the NextFlex poly(A) Beads 2.0 and NextFlex Rapid Directional RNA-seq Kit 2.0 (PerkinElmer, Waltham, MA, USA) following the manufacturer's instructions. The libraries were sequenced on the Illumina NovaSeq6000 system. Four biological replicates per condition were sequenced. Raw data were demultiplexed using bcl2fastq (Illumina; version v2.19.1.403). An adapter trimming step was done using trimmomatic (version 0.39). Mapping of reads on the human genome (Hg38) was performed using STAR (version 2.7.3a). On average, 85 million reads accurately mapped against the human genome. Raw and normalized counting steps were done using RSEM (version v1.3.1). Gene name annotations were done using a GTF from Encode (version 42), and Ensembl (version 108). Differential analysis was performed using DESeq2 focusing on genes with an average Transcripts Per Million (TPM) exceeding one (version 1.38.3). Volcano plot was generated using shinyapp tool (http://www.graphbio1.com/en/). Gene-set enrichment analysis was done using Metascape (accessed in July 2023) [34]. In this analysis, we included only genes that were significantly upregulated (Padj < 0.05). The sequencing data can be accessed with a GEO accession number that will be provided after acceptation. RNA-seq sequence reads at P2RY1 locus were analyzed through Integrative Genomics Viewer [35].
4.11. eQTL analysis
Blood and pancreatic islet samples were collected from a total of 103 pancreatectomized patients as previously described [18]. SNPs at the P2RY1 locus were identified through DNA microarray analysis (Illumina Omni2.5 M Beadchip array), while P2RY1 (NM_002563.5) expression was assessed using RNA sequencing [18,19]. The eQTL analysis was performed using FastQTL software [36], with sex, BMI, age and RNA batch effect as covariates. The eQTL analysis was focused on common variants at P2RY1 locus known to be genome-wide or nominally associated with T2D risk [8]. The chromatin map was generated using the shinyapp tool (https://shinyapps.jax.org/endoc-islet-multi-omics/) based on the analysis of chromatin states (ChromHMM) done in the human pancreatic beta-cell line EndoCβH1 [37].
Funding
We thank “France Génomique” consortium (ANR-10-INBS-009). This work was supported by grants from the French National Research Agency (ANR-10-LABX-46 [European Genomics Institute for Diabetes] and ANR-10-EQPX-07-01 [LIGAN-PM]), from the European Research Council (ERC OπO – 101,043,671, to AB), and from the National Center for Precision Diabetic Medicine – PreciDIAB, which is jointly supported by the French National Agency for Research (ANR-18-IBHU-0001), by the European Union (FEDER), by the Hauts-de-France Regional Council and by the European Metropolis of Lille (MEL).
CRediT authorship contribution statement
Mathilde Boissel: Formal analysis, Writing – review & editing. Souhila Amanzougarene: Formal analysis, Writing – review & editing. Guillaume Charpentier: Resources, Writing – review & editing. Martine Vaxillaire: Resources, Writing – review & editing. Hélène Loiselle: Investigation, Writing – review & editing. Beverley Balkau: Resources, Writing – review & editing. Raphaël Boutry: Investigation, Writing – review & editing. Morgane Baron: Supervision, Writing – review & editing. Marjorie Fadeur: Resources, Writing – review & editing. Bénédicte Toussaint: Investigation, Methodology, Writing – review & editing. Nicolas Paquot: Resources, Writing – review & editing. Emmanuel Vaillant: Investigation, Methodology, Writing – review & editing. Philippe Froguel: Funding acquisition, Supervision, Writing – review & editing. Michel Marre: Resources, Writing – review & editing. Arnaud Dance: Conceptualization, Formal analysis, Methodology, Writing – original draft, Investigation. Amelie Bonnefond: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Resources, Writing – original draft, Writing – review & editing. Marie Gernay: Resources, Writing – review & editing. Justine Fernandes: Investigation, Methodology, Writing – review & editing. Mehdi Derhourhi: Formal analysis, Supervision, Writing – review & editing. Sylvia Franc: Resources, Writing – review & editing. Amna Khamis: Resources, Writing – review & editing. Mark Ibberson: Resources, Writing – review & editing
Declaration of competing interest
Authors declare that they have no competing interests.
Acknowledgments
We are grateful to all individuals included in the different cohort studies. We thank Frédéric Allegaert and Timothée Beke. We thank Mickaël Canouil for his assistance in statistical analyses.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2023.101867.
Contributor Information
Philippe Froguel, Email: philippe.froguel@cnrs.fr.
Amélie Bonnefond, Email: amelie.bonnefond@inserm.fr.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.American Diabetes Association Professional Practice Committee 2. Classification and diagnosis of diabetes: standards of medical care in diabetes-2022. Diabetes Care. 2022;45(Suppl 1):S17–S38. doi: 10.2337/dc22-S002. [DOI] [PubMed] [Google Scholar]
- 2.Jin J., Daniel J.L., Kunapuli S.P. Molecular basis for ADP-induced platelet activation. II. The P2Y1 receptor mediates ADP-induced intracellular calcium mobilization and shape change in platelets. J Biol Chem. 1998;273(4):2030–2034. doi: 10.1074/jbc.273.4.2030. [DOI] [PubMed] [Google Scholar]
- 3.Murugappa S., Kunapuli S.P. The role of ADP receptors in platelet function. Front Biosci: J Vis Literacy. 2006;11:1977. doi: 10.2741/1939. 86. [DOI] [PubMed] [Google Scholar]
- 4.Novak I. Purinergic receptors in the endocrine and exocrine pancreas. Purinergic Signal. 2008;4(3):237–253. doi: 10.1007/s11302-007-9087-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balasubramanian R., de Azua I.R., Wess J., Jacobson K.A. Activation of distinct P2Y receptor subtypes stimulates insulin secretion in MIN6 mouse pancreatic β cells. Biochem Pharmacol. 2010;79(9):1317–1326. doi: 10.1016/j.bcp.2009.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gloyn A.L., Pearson E.R., Antcliff J.F., Proks P., Bruining G.J., Slingerland A.S., et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med. 2004;350(18):1838–1849. doi: 10.1056/NEJMoa032922. [DOI] [PubMed] [Google Scholar]
- 7.Bonfanti D.H., Alcazar L.P., Arakaki P.A., Martins L.T., Agustini B.C., de Moraes Rego F.G., et al. ATP-dependent potassium channels and type 2 diabetes mellitus. Clin Biochem. 2015;48(7–8):476–482. doi: 10.1016/j.clinbiochem.2014.12.026. [DOI] [PubMed] [Google Scholar]
- 8.Costanzo M.C., von Grotthuss M., Massung J., Jang D., Caulkins L., Koesterer R., et al. The Type 2 Diabetes Knowledge Portal: an open access genetic resource dedicated to type 2 diabetes and related traits. Cell Metabol. 2023;35(4):695–710.e6. doi: 10.1016/j.cmet.2023.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcheva, B., Weidemann, B.J., Taguchi, A., Perelis, M., Ramsey, K.M., Newman, M.V., et al., n.d. P2Y1 purinergic receptor identified as a diabetes target in a small-molecule screen to reverse circadian β-cell failure. Elife 11: e75132, Doi: 10.7554/eLife.75132. [DOI] [PMC free article] [PubMed]
- 10.Khan S., Ferdaoussi M., Bautista A., Bergeron V., Smith N., Poitout V., et al. A role for PKD1 in insulin secretion downstream of P2Y 1 receptor activation in mouse and human islets. Physiological Reports. 2019;7(19) doi: 10.14814/phy2.14250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smyth S.S., Woulfe D.S., Weitz J.I., Gachet C., Conley P.B., Goodman S.G., et al. G-protein-coupled receptors as signaling targets for antiplatelet therapy. Arterioscler Thromb Vasc Biol. 2009;29(4):449–457. doi: 10.1161/ATVBAHA.108.176388. [DOI] [PubMed] [Google Scholar]
- 12.Hechler B., Cattaneo M., Gachet C. The P2 receptors in platelet function. Semin Thromb Hemost. 2005;31(2):150–161. doi: 10.1055/s-2005-869520. [DOI] [PubMed] [Google Scholar]
- 13.Voss A.A. Extracellular ATP inhibits chloride channels in mature mammalian skeletal muscle by activating P2Y1 receptors. J Physiol. 2009;587(Pt 23):5739–5752. doi: 10.1113/jphysiol.2009.179275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbott K.L., Loss J.R., Robida A.M., Murphy T.J. Evidence that Galpha(q)-coupled receptor-induced interleukin-6 mRNA in vascular smooth muscle cells involves the nuclear factor of activated T cells. Mol Pharmacol. 2000;58(5):946–953. doi: 10.1124/mol.58.5.946. [DOI] [PubMed] [Google Scholar]
- 15.Kawano S., Otsu K., Kuruma A., Shoji S., Yanagida E., Muto Y., et al. ATP autocrine/paracrine signaling induces calcium oscillations and NFAT activation in human mesenchymal stem cells. Cell Calcium. 2006;39(4):313–324. doi: 10.1016/j.ceca.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Kim S.-A., Choi H.S., Ahn S.-G. Pin1 induces the ADP-induced migration of human dental pulp cells through P2Y1 stabilization. Oncotarget. 2016;7(51):85381–85392. doi: 10.18632/oncotarget.13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99(4):877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khamis A., Canouil M., Siddiq A., Crouch H., Falchi M., Bulow M. von., et al. Laser capture microdissection of human pancreatic islets reveals novel eQTLs associated with type 2 diabetes. Mol Metabol. 2019;24:98–107. doi: 10.1016/j.molmet.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wigger L., Barovic M., Brunner A.-D., Marzetta F., Schöniger E., Mehl F., et al. Multi-omics profiling of living human pancreatic islet donors reveals heterogeneous beta cell trajectories towards type 2 diabetes. Nat Metab. 2021;3(7):1017–1031. doi: 10.1038/s42255-021-00420-9. [DOI] [PubMed] [Google Scholar]
- 20.Nica A.C., Ongen H., Irminger J.-C., Bosco D., Berney T., Antonarakis S.E., et al. Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome. Genome Res. 2013;23(9):1554–1562. doi: 10.1101/gr.150706.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blodgett D.M., Nowosielska A., Afik S., Pechhold S., Cura A.J., Kennedy N.J., et al. Novel observations from next-generation RNA sequencing of highly purified human adult and fetal islet cell subsets. Diabetes. 2015;64(9):3172–3181. doi: 10.2337/db15-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanchi B., Taurand M., Colace C., Thomaidou S., Audeoud C., Fantuzzi F., et al. EndoC-βH5 cells are storable and ready-to-use human pancreatic beta cells with physiological insulin secretion. Mol Metabol. 2023;76 doi: 10.1016/j.molmet.2023.101772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Junn E., Han S.H., Im J.Y., Yang Y., Cho E.W., Um H.D., et al. Vitamin D3 up-regulated protein 1 mediates oxidative stress via suppressing the thioredoxin function. J Immunol. 2000;164(12):6287–6295. doi: 10.4049/jimmunol.164.12.6287. [DOI] [PubMed] [Google Scholar]
- 24.J Y., Y M., H T., H M., T I. Anti-inflammatory thioredoxin family proteins for medicare, healthcare and aging care. Nutrients. 2017;9(10) doi: 10.3390/nu9101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou R., Tardivel A., Thorens B., Choi I., Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol. 2010;11(2):136–140. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- 26.Ma M., O B., Cr K. Metabolic syndrome: is Nlrp3 inflammasome a trigger or a target of insulin resistance? Circ Res. 2011;108(10) doi: 10.1161/RES.0b013e318220b57b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J., Hui S.T., Couto F.M., Mungrue I.N., Davis D.B., Attie A.D., et al. Thioredoxin-interacting protein deficiency induces Akt/Bcl-xL signaling and pancreatic beta-cell mass and protects against diabetes. FASEB (Fed Am Soc Exp Biol) J: Official Publication of the Federation of American Societies for Experimental Biology. 2008;22(10):3581–3594. doi: 10.1096/fj.08-111690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Léon C., Freund M., Latchoumanin O., Farret A., Petit P., Cazenave J.-P., et al. The P2Y1 receptor is involved in the maintenance of glucose homeostasis and in insulin secretion in mice. Purinergic Signal. 2005;1(2):145–151. doi: 10.1007/s11302-005-6209-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mesto N., Bailbe D., Eskandar M., Pommier G., Gil S., Tolu S., et al. Involvement of P2Y signaling in the restoration of glucose-induced insulin exocytosis in pancreatic β cells exposed to glucotoxicity. J Cell Physiol. 2022;237(1):881–896. doi: 10.1002/jcp.30564. [DOI] [PubMed] [Google Scholar]
- 30.Todd J.N., Poon W., Lyssenko V., Groop L., Nichols B., Wilmot M., et al. Variation in glucose homeostasis traits associated with P2RX7 polymorphisms in mice and humans. J Clin Endocrinol Metabol. 2015;100(5):E688–E696. doi: 10.1210/jc.2014-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang D., Zhou Q., Labroska V., Qin S., Darbalaei S., Wu Y., et al. G protein-coupled receptors: structure- and function-based drug discovery. Signal Transduct Targeted Ther. 2021;6(1):7. doi: 10.1038/s41392-020-00435-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Folon L., Baron M., Toussaint B., Vaillant E., Boissel M., Scherrer V., et al. Contribution of heterozygous PCSK1 variants to obesity and implications for precision medicine: a case-control study. The Lancet. Diabetes & Endocrinology. 2023;11(3):182–190. doi: 10.1016/S2213-8587(22)00392-8. [DOI] [PubMed] [Google Scholar]
- 33.American Diabetes Association 2. Classification and diagnosis of diabetes: standards of medical care in diabetes-2019. Diabetes Care. 2019;42(Suppl 1):S13–S28. doi: 10.2337/dc19-S002. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ongen H., Buil A., Brown A.A., Dermitzakis E.T., Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics. 2016;32(10):1479–1485. doi: 10.1093/bioinformatics/btv722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawlor N., Márquez E.J., Orchard P., Narisu N., Shamim M.S., Thibodeau A., et al. Multiomic profiling identifies cis-regulatory networks underlying human pancreatic β cell identity and function. Cell Rep. 2019;26(3):788–801.e6. doi: 10.1016/j.celrep.2018.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.