Abstract
The fyuA-irp gene cluster contributes to the virulence of highly pathogenic Yersinia (Yersinia pestis, Yersinia pseudotuberculosis, and Yersinia enterocolitica 1B). The cluster encodes an iron uptake system mediated by the siderophore yersiniabactin and reveals features of a pathogenicity island. Two evolutionary lineages of this “high pathogenicity island” (HPI) can be distinguished on the basis of DNA sequence comparison: a Y. pestis group and a Y. enterocolitica group. In this study we demonstrate that the HPI of the Y. pestis evolutionary group is disseminated among species of the family Enterobacteriaceae which are pathogenic to humans. It prevails in enteroaggregative Escherichia coli and in E. coli blood culture isolates (93 and 80%, respectively), but is rarely found in enteropathogenic E. coli, enteroinvasive E. coli, and enterotoxigenic E. coli isolates. In contrast, the HPI was absent from enterohemorrhagic E. coli, Shigella, and Salmonella enterica strains investigated. Polypeptides encoded by the fyuA, irp1, and irp2 genes located on the HPI could be detected in E. coli strains pathogenic to humans. However, these E. coli strains showed a reduced sensitivity to the bacteriocin pesticin, whose uptake is mediated by the FyuA receptor. Escherichia strains do not possess the hms gene locus thought to be a part of the HPI of Y. pestis. Deletions of the fyuA-irp gene cluster affecting solely the fyuA part of the HPI were identified in 3% of the E. coli strains tested. These results suggest horizontal transfer of the HPI between Y. pestis and some pathogenic E. coli strains.
The pathogenicity of Yersinia pestis, Yersinia pseudotuberculosis, and Yersinia enterocolitica is determined by a 70-kb virulence plasmid, pYV (11, 38). Y. pestis carries two additional plasmids which are required for full virulence: a 100-kb plasmid encoding capsular antigen fraction 1 and mouse exotoxin and a 9.5-kb plasmid coding for the bacteriocin pesticin and a plasminogen activator (35, 36). Y. pestis, Y. pseudotuberculosis serotype O1, and Y. enterocolitica biotype 1B strains possess a chromosomal determinant that is involved in iron uptake mediated by the siderophore yersiniabactin (7, 22, 23). This iron acquisition determinant, recently designated the “high-pathogenicity island” (HPI), is absolutely necessary for expression of the trait of virulence to mice (9, 16).
In Y. pestis, the HPI is known as the pgm locus, which comprises about 102 kb of chromosomal DNA and includes the following genes involved in iron storage and uptake: (i) the hemin storage (hms) locus (37); (ii) the irp1 and irp2 genes coding for the iron-repressible high-molecular-weight proteins HMWP1 and HMWP2, which presumably are involved in the production of yersiniabactin (28); and (iii) the fyuA or psn gene (for ferric yersiniabactin uptake or pesticin sensitivity) coding for the yersiniabactin receptor FyuA that also serves as a receptor for pesticin (9, 10, 23, 40). The 102-kb HPI of Y. pestis is flanked by single copies of the IS100 insertion element. Homologous recombination between these two IS100 copies may be responsible for the deletion of the intervening 102-kb DNA segment. In contrast to Y. pestis, highly pathogenic strains of Y. enterocolitica and almost all Y. pseudotuberculosis strains lack the hms locus and therefore are pigmentation negative but do carry a 45-kb stretch of chromosomal DNA comprising the irp1-irp2 and fyuA genes (fyuA-irp gene cluster [Fig. 1]) (4, 8, 34).
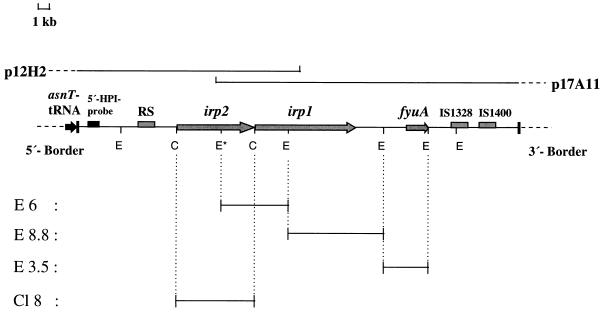
FIG. 1.
Physical map of the HPI of Y. enterocolitica O8 WA-C with the presumed 5′ and 3′ borders of the HPI. Horizontal lines below the restriction map represent different subcloned DNA fragments (E6, E8.8, E3.5, and Cl8) which were used as probes in hybridization assays. Arrows and bars within the restriction map indicate the locations of the identified genes and repeated sequence (RS) elements, respectively. E, EcoRI; C, ClaI; p12H2 and p17A11, cosmid clones. The locations of the 5′-HPI probe and the adjacent asnT tRNA are indicated. The asterisk indicates an additional internal EcoRI site found only in the irp2 genes of Y. enterocolitica biotype 1B strains.
The irp1-irp2-fyuA fragment is unstable in Y. enterocolitica and Y. pseudotuberculosis. In Y. enterocolitica, this instability may be due to the IS1328 insertion element or to other insertion elements adjacent to fyuA which have been recognized recently (8, 41). DNA sequencing of irp2 and fyuA revealed a G+C content of about 60 and 56 mol%, respectively, much higher than the G+C content of the whole Yersinia chromosome (46 to 50 mol%). These data suggest that pathogenic Yersinia spp. have acquired their HPIs from an organism with a higher G+C content by horizontal transfer.
Recently, we have shown that some colicin-producing E. coli strains carry genes with sequences nearly identical to those of the irp2 and fyuA genes of Yersinia spp. (41). This observation prompted us to investigate the possible dissemination of the HPI of Yersinia spp. (Yersinia HPI) among species of Enterobacteriaceae pathogenic to humans. In this study, we used PCR and Southern hybridization assays to screen for the presence of irp2 and fyuA sequences within a panel of five established pathotypes of E. coli (enterotoxigenic E. coli [ETEC], enteroaggregative E. coli [EAEC], enteropathogenic E. coli [EPEC], enterohemorrhagic E. coli [EHEC], and enteroinvasive E. coli [EIEC]) as well as Shigella species and certain serotypes of Salmonella enterica. Additionally, we screened E. coli strains isolated from blood cultures of inpatients and stool samples of healthy individuals.
Surprisingly, the irp2-fyuA gene cluster was detected in more than 93% of EAEC strains, about 27% of EIEC strains, and 5% of ETEC and EPEC strains but in none of the EHEC, Salmonella, or Shigella strains studied.
MATERIALS AND METHODS
Bacterial strains.
We analyzed 220 E. coli strains, 24 S. enterica strains, and 14 Shigella strains of different species (Table 1). The E. coli strains included 135 strains of the five established enteric pathotypes, EPEC (serotypes O111, O119, O127, and O157), EIEC (O124, O143, and O145), ETEC (O6, O25, O128, and O148), EHEC (O157:H7, O26, O55, and O111), and EAEC (O44, O86, O119, and O125), which were obtained either from E. coli strain collections of the national reference centers of Salmonella and other enteric bacteria (Robert Koch Institut, Berlin, and Hygienisches Institut, Hamburg, Germany) or from children with diarrhea from different countries, partially listed in reference 44. The enteric E. coli pathotypes were confirmed by PCR amplification with specific primers for virulence genes as described previously (17, 18, 27, 33, 43). Additionally, all EAEC strains were proved to be enteroaggregative by the HEp-2 cell assay (12, 45, 47). EAEC 17-2 has been described previously (29, 31, 48) and was kindly provided by J. P. Nataro (Center for Vaccine Development, Baltimore, Md.). The toxin production of ETEC and EHEC strains was tested by enzyme-linked immunosorbent assay, and the invasive phenotype of EIEC strains was confirmed by invasion assays (5).
TABLE 1.
Presence of fyuA and irp2 genes in different pathogenic species of Enterobacteriaceae isolated from humans
| Species and pathotype or serotype | Total no. | No. with:a
|
|
|---|---|---|---|
| irp2 | fyuA | ||
| E. coli | |||
| EAEC | 60 | 56 | 55 |
| EIEC | 15 | 3 | 2 |
| ETEC | 20 | 1 | 1 |
| EPEC | 20 | 1 | 1 |
| EHEC | 20 | 0 | 0 |
| S. enterica | |||
| Enteritidis | 10 | 0 | 0 |
| Typhimurium | 10 | 0 | 0 |
| Typhi | 4 | 0 | 0 |
| Shigella flexneri | 3 | 0 | 0 |
| Shigella boydii | 3 | 0 | 0 |
| Shigella sonnei | 5 | 0 | 0 |
| Shigella dysenteriae | 3 | 0 | 0 |
| E. coli from blood cultures | 60 | 50 | 48 |
| E. coli from stool samples (healthy individuals) | 25 | 8 | 7 |
The presence of the fyuA and irp2 genes was determined by PCR with appropriate primers and confirmed by DNA hybridization.
In addition, 60 E. coli strains were isolated from blood cultures of inpatients at the university hospitals of Würzburg and Munich, Germany (serotypes of blood culture isolates were O2, O4, O6, O11, O25, O40, O54, and O125). Twenty-five E. coli strains were isolated from stool samples of healthy individuals. It was shown by PCR that the E. coli isolates from both inpatient blood cultures and stool samples of healthy individuals did not belong to any of the established E. coli enteric pathotypes.
Isolation of plasmid and genomic DNAs.
Plasmid DNA for large- and small-scale preparations was extracted by the alkaline lysis method (1). Large-scale plasmid preparations were further purified with anion-exchange resin columns (Macherey-Nagel, Düren, Germany). For EAEC strains, it turned out to be advantageous to wash the bacterial cell pellet twice in phosphate-buffered saline before starting the alkaline lysis. Bacterial genomic DNA was isolated by a lysozyme-sodium dodecyl sulfate (SDS)-proteinase K method (1) and further purified by phenol and chloroform extractions.
Hybridization and DNA probes.
Restriction enzyme-digested genomic and plasmid DNA fragments were resolved through 0.8% agarose gels, and DNA was transferred to Zeta-Probe BT blotting membranes (Bio-Rad Laboratories) with a vacuum blotter (Pharmacia, Freiburg, Germany), modifying the method of Southern (46). In order to generate DNA probes of the Yersinia HPI, two overlapping cosmids of a genomic library from Y. enterocolitica O8 WA-C (40) covering the entire Yersinia HPI (Fig. 1) were subjected to endonuclease digestion. Three EcoRI fragments and one ClaI fragment were subcloned into the pBluescript KS vector, resulting in plasmids E3.5, E8.8, E6, and Cl8, respectively (Fig. 1). These cloned fragments of the Yersinia HPI were used as the HPI probes in the hybridization assays. Digoxigenin labeling of the probes and hybridization were performed with a DNA labeling and detection kit (Boehringer, Mannheim, Germany). After prehybridization at 68°C for 2 h and addition of heat-denatured probe, the blots were incubated overnight at 68°C in the absence of formamide. The detection was performed according to the manufacturer’s instructions.
PCR.
Different sets of primers used for PCR amplification were synthesized by Carl Roth (Karlsruhe, Germany). PCR amplifications were performed in an automated thermal cycler (TRIO Thermoblock; Biometra, Göttingen, Germany) as described by Saiki et al. (42) with TaqI polymerase and different pairs of oligonucleotide primers. The initial denaturation step (94°C, 5 min) was followed by 35 cycles of denaturation (94°C, 1 min), annealing (at the annealing temperature [Tm], 1 min), and extension (72°C, 1 min), with one final extension step (72°C, 8 min). The sequences of the forward (FP) and reverse (RP) primers used for PCR reactions, the size of the amplified fragment (S), and Tm were as follows: (i) irp2 (FP), 5′-AAGGATTCGCTGTTACCGGAC-3′; irp2 (RP), 5′-TCGTCGGGCAGCGTTTCTTCT-3′ (S, 280 bp; Tm, 61°C); (ii) irp2-P242 (FP), 5′-AAGGATTCGCTGTTACCGGAC-3′; irp2-P505 (RP), 5′-TCGTCGGGCAGCGTTTCTTCT-3′ (S, 264 bp; Tm, 58°C); (iii) fyuA (FP), 5′-GCGAC GGGAAGCGATTTA-3′; fyuA (RP), 5′-CGCAGTAGGCACGATGTTGTA-3′ (S, 780 bp; Tm, 60°C); (iv) hmsR (FP) 5′-TAAAGAAAGACCCCACCAATC-3′, hmsR (RP), 5′-ATCATCGGCATCAAGCAAATC-3′ (S, 730 bp; Tm, 56°C); entF (FP), 5′-TATCAGCGTTATCACCATTTG-3′, entF (RP), 5′-CCAGTTCCGGCAGCGTTTCTT-3′ (S, 511 bp; Tm, 55°C); and Ec-chrom (FP), 5′-TTTATTCCGTTGCGTGAGGTT-3′, HPI-5end (RP), 5′-TAGGATACCTTCACGCTGCTGTCGCGC-3′ (S, 900 bp; Tm, 52°C).
Sequence analysis of the fyuA gene of EAEC 17-2.
In order to sequence the fyuA genes of E. coli strains, overlapping PCR fragments were generated with primers which were derived from the Y. enterocolitica fyuA gene as described previously (41). The PCR products were purified with anion-exchange resin columns (Quiagen, Hilden, Germany). A total of 100 ng of purified PCR product was subjected to Taq cycle-sequencing reactions with the Prism Ready Reaction dye dideoxy terminator cycle-sequencing kit (Applied Biosystems, Darmstadt, Germany) according to the manufacturer’s instructions. Electrophoresis of sequencing products was performed on 7% polyacrylamide gels with an automated sequencer (model 373A; Applied Biosystems). The nucleotide sequences were analyzed with the ANALYSIS program version 1.2 (Applied Biosystems) and the DNASIS program version 2.0 (Hitachi Software Engineering Co., Tokyo, Japan).
Detection of FyuA, HMWP2, and HMWP1.
Whole cells of Yersinia and E. coli strains growing under iron-deficient conditions were incubated for 1 h with 35S-labeled amino acids (Amersham, Braunschweig, Germany) (15). Labeled proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE). Dried gels were exposed to Kodak BioMax MR film at room temperature.
For immunoblotting, ultrasonicated bacterial cell pellets were treated with 2% Triton X-100 and the insoluble membrane material was purified and subjected to SDS-PAGE as described previously (20, 23). After electrotransfer to polyvinylidene difluoride membranes (Millipore, Eschborn, Germany), FyuA was detected by antiserum (anti-FyuA) raised against FyuA of Y. enterocolitica O8 Y1852 in rabbits (23). HMWP1 and HMWP2 were detected with an antiserum raised against purified HMWP1 and HMWP2 of Y. enterocolitica O8 Ye 8081 in mice (7). Goat anti-rabbit and goat anti-mouse antibodies conjugated to alkaline phosphatase (Sigma Chemical Co., St. Louis, Mo.) were employed as second antibodies, and the reaction was developed with Nitro Blue Tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (21).
Pesticin sensitivity test.
The pesticin assay was performed as described elsewhere (13, 23). Briefly, the pesticin-producing strain Y. pestis EV76 was grown overnight at 26°C, and pesticin production was induced by an additional incubation of 16 h in the presence of 0.3 μg of mitomycin C ml−1. The cells were collected by centrifugation, and the supernatant was used as a crude pesticin preparation after sterilization with 0.1% chloroform (25). Sensitivity to pesticin was monitored by serial dilutions (1:2) of crude pesticin preparations on mid-log-phase bacterial cultures by a double-layer technique. The plates were incubated at 37°C for 16 to 18 h. Y. enterocolitica Y8081 and Y. pseudotuberculosis Y-P-I were used as reference pesticin-sensitive strains (25).
RESULTS
Survey for the presence of irp2 and fyuA genes among E. coli strains pathogenic to humans.
The results of the survey with PCR to detect the irp2 and fyuA genes in E. coli strains pathogenic to humans are shown in Table 1. The irp2 gene was detected in 93% and the fyuA gene was detected in about 92% of EAEC strains. Among EIEC strains, 20% yielded an amplified irp2 fragment and 13% yielded a fyuA fragment. Five percent of EPEC and ETEC strains harbored the irp2 and fyuA genes. In contrast, no irp2 or fyuA gene was detectable in EHEC, Salmonella, or Shigella strains tested in this study. Additionally, E. coli strains isolated from blood cultures of inpatients were subjected to the same PCR assays. Surprisingly, in 83% of the isolates from blood cultures the irp2 gene was detectable by PCR (Table 1). In contrast, the irp2 and fyuA genes were detectable in only 32 and 28%, respectively, of E. coli strains isolated from stool samples of healthy individuals (n = 25).
DNA hybridization assays were subsequently performed with chromosomal DNA of E. coli strains in which a fragment of irp2 was amplified by PCR.
To determine the location of the E. coli fyuA-irp gene cluster, both plasmid and chromosomal DNAs of EAEC, ETEC, EIEC, and EPEC strains were purified, digested with EcoRI endonuclease, and used for Southern hybridization with probes E3.5 and Cl8 (Fig. 1). Corresponding DNA restriction fragments were detectable only in E. coli chromosomal DNA preparations.
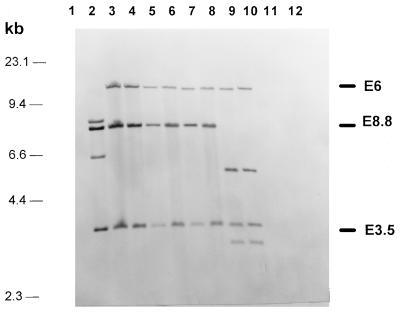
Probes E6, E8.8, and E3.5, covering the central part of the Yersinia HPI (Fig. 1), were used to evaluate the degree of conservation of the fyuA-irp gene cluster among different E. coli strains. Forty-seven of 57 E. coli strains which were positive for irp2 and fyuA by PCR displayed Southern hybridization patterns identical to those of Y. pestis and Y. pseudotuberculosis but different from that of Y. enterocolitica O8 (WA-314, biotype 1B). This difference is due to an additional internal EcoRI restriction site present in the irp2 gene of Y. enterocolitica biotype 1B strains but absent from other irp2-positive Yersinia strains (Fig. 1). Ten E. coli strains positive for irp2 and fyuA by PCR showed a slightly different hybridization pattern, which is apparently due to an additional EcoRI restriction site in the DNA fragment corresponding to E8.8 (Fig. 1 and 2). Interestingly, this hybridization pattern was shared by E. coli strains which belonged to different pathotypes.
FIG. 2.
Southern hybridizations of EcoRI-digested genomic DNA of Y. enterocolitica and different E. coli strains with four DNA fragments, E6, E8.8, E3.5, and Cl8 (as shown in Fig. 1), representing the central part of the Yersinia HPI. Lane 1, E. coli DH5α; lane 2, Y. enterocolitica O8 WA-C; lane 3, Y. pseudotuberculosis O1; lane 4, EAEC 17-2 (HPI positive); lane 5, EPEC 12-1; (HPI positive); lane 6, EIEC E12860 (HPI positive); lane 7, ETEC H488/84 (HPI positive); lane 8, E. coli 14094/96 from blood culture; (HPI positive); lane 9, EAEC DMI 155 (HPI positive); lane 10, EIEC H823/88 (HPI positive); lane 11, EHEC 1249/87 (HPI negative); and lane 12, EAEC 4827/94; (HPI negative).
Five E. coli strains which have been shown to be irp2 positive and fyuA negative by PCR gave no hybridization signal with E3.5 and E8.8 probes located in the fyuA region (i.e., the 3′ part of the fyuA-irp gene cluster) but revealed a hybridization signal with an E6 probe, indicating that all of the deletions affected the fyuA region. To examine possible variations of the irp2 region (i.e., the 5′ region) of the fyuA-irp gene cluster, a hybridization analysis was performed with a probe derived from a 0.9-kb BglII-SacII DNA fragment which was previously shown to be located at the 5′ border of the Yersinia HPI in the Y. enterocolitica O8 strain Ye 8081 (8) (Fig. 1). This probe hybridized to a single EcoRI chromosomal fragment in Y. enterocolitica WA-C and all irp2-positive E. coli strains used in this study (data not shown). The sizes of these fragments varied from 4 to 12 kb, suggesting a different position of the EcoRI restriction site located outside the fyuA-irp gene cluster. No signal was detectable in irp2-negative E. coli strains used as controls.
As mentioned above, the size of the HPI of Y. pestis differs from those of other Yersinia spp., and this HPI is thought to be composed of two parts, the fyuA-irp gene cluster and the hemin storage (hms) locus, respectively. In order to determine whether irp2-positive E. coli strains possess genes homologous to hms of Y. pestis, hybridizations with a probe derived from the hmsR gene of Y. pestis were performed. This probe did not hybridize either with the irp2-positive E. coli strains or with Y. enterocolitica O8 WA-C but recognized the corresponding fragment in Y. pestis (data not shown).
Sequence analysis of the fyuA-irp gene cluster in E. coli.
In order to determine the nucleotide sequence conservation of the fyuA gene of E. coli, PCR-amplified fyuA DNA fragments of EAEC 17-2 were sequenced and compared with Yersinia fyuA sequences. The fyuA gene of strain 17-2 has 99.6, 99.8, and 98.8% identity with those of Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica, respectively. Comparing the nucleotide substitutions of the fyuA genes from EAEC and Y. pestis, only five of nine nucleotide substitutions caused amino acid mismatches. The G+C content of the fyuA gene was 56.2 mol%, which is much higher than the overall G+C content of E. coli (48 to 52 mol%) or Y. enterocolitica (46 to 50 mol%) chromosomes.
To investigate the conservation of the irp2 gene, a small fragment which, in Y. enterocolitica, is located at the beginning of the irp2 gene (bp 242 to 526) was sequenced in EAEC 17-2 and EIEC E12860. The sequences of the EIEC E12860 and EAEC 17-2 irp2 fragments were identical and had 99.7, 99.7, and 98.6% identity with those of Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica, respectively.
In addition, the linkage of the Yersinia HPI to asnT tRNA in E. coli was demonstrated by PCR analysis. We have generated a PCR primer derived from the E. coli chromosome adjacent to the asnT tRNA gene. This FP (Ec-chrom) was used for PCR in combination with a RP (HPI-5end) located at the 5′ end of the Yersinia HPI. This RP was derived from the 0.9-kb BglII-SacII DNA fragment which was previously used as a 5′-border probe. By means of this 5′-border PCR, we screened a panel of 10 HPI-positive E. coli strains. All of the strains revealed single PCR products of the same size. We have sequenced these PCR fragments and found the Yersinia HPI to be directly linked to the asnT tRNA gene in these E. coli strains (data not shown).
Presence of HMWP1, HMWP2, and FyuA proteins in E. coli strains.
By immunoblot analysis with anti-FyuA rabbit serum, FyuA was detectable in the outer membrane fraction of fyuA-positive E. coli strains (Table 1 and Fig. 3). Since FyuA has been shown to function as a pesticin receptor (23), the sensitivity of FyuA-positive E. coli strains to pesticin was tested. A moderate degree of pesticin sensitivity was detected in some FyuA-positive E. coli strains. However, the diameter of the inhibition zone was smaller and the halo appeared more turbid than those obtained with the pesticin-sensitive strain Y. pseudotuberculosis Y-P-I. The majority of FyuA-positive and all fyuA-negative E. coli strains turned out to be insensitive to pesticin under the test conditions.
FIG. 3.
Immunoblot of outer membrane proteins probed with anti-FyuA rabbit serum. Lane 1, Y. enterocolitica O8 WA-C; lane 2, E. coli 14094/96 from blood culture (HPI positive); lane 3, EAEC 17-2 (HPI positive); and lane 4, EAEC 4827/94 (HPI negative).
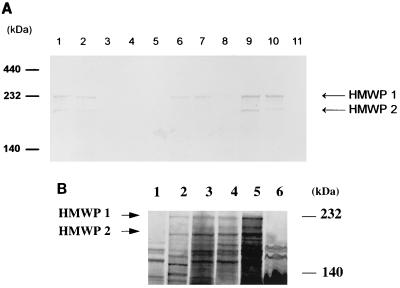
Additionally, cell lysates of EAEC cultivated under iron-restricted conditions were analyzed by SDS-PAGE and immunoblotting with mouse anti-HMWP1 and anti-HMWP2 antiserum (7). E. coli strains harboring irp1 and irp2 genes were shown to express the corresponding iron-repressible HMWP1 and HMWP2 (Fig. 4). However, in some E. coli strains, HMWP1 and HMWP2 were not detected, even though the irp1 and irp2 genes were detected by PCR and DNA hybridization.
FIG. 4.
(A) Immunoblot of outer membrane proteins probed with anti-HMWP1 and anti-HMWP2 mouse serum. Lanes: 1, Y. enterocolitica O8 WA-C; 2, Y. pseudotuberculosis O1; 3, E. coli DH5α; 4, EAEC 4827/94 (HPI negative); 5, EPEC 2403/85 (HPI negative); 6, EAEC 17-2 (HPI positive); 7, EPEC 12-1 (HPI positive); 8, EIEC E12860 (HPI positive); 9, ETEC H488/84 (HPI positive); 10, E. coli 14094/96 from blood culture (HPI positive); and 11, EHEC 1249/87 (HPI negative). (B) Fluorographs of a polyacrylamide gel containing 35S-labeled proteins from E. coli DH5α (lane 1), Y. enterocolitica O8 WA-C (lane 2), EPEC 2403/85 (HPI positive) (lane 3), EIEC E12860 (HPI positive) (lane 4), EAEC 17-2 (HPI positive) (lane 5), and EAEC 4827/94 (HPI negative) (lane 6).
DISCUSSION
The fyuA-irp gene cluster identified primarily in yersiniae meets the basic criteria of so-called pathogenicity islands, such as (i) atypical G+C content of the 45-kb chromosomal DNA region, (ii) requiring a cluster of genes for virulence, (iii) the presence of “mobility” genes (e.g., insertion elements) as well as association with a tRNA gene at one boundary, and (iv) instability (8, 14, 39). Moreover, we recently demonstrated that the fyuA-irp gene cluster comprises two evolutionary lineages, one assigned to Y. enterocolitica biotype 1B strains and the other to Y. pestis and Y. pseudotuberculosis. Surprisingly, the fyuA-irp gene cluster of the latter lineage could also be detected in four pesticin-sensitive E. coli strains, indicating that it might be transmissible (41). Therefore, we investigated the distribution of the fyuA-irp gene cluster among some pathogenic species of the family Enterobacteriaceae.
We demonstrated that the fyuA-irp gene cluster is absent in Shigella, S. enterica serotypes Enteritidis and Typhimurium, and Shiga toxin-producing E. coli (EHEC). The gene cluster is infrequently detected in E. coli strains of EPEC, ETEC, and EIEC pathotypes. Strikingly, the EAEC pathotype, which causes acute and chronic diarrhea in infants (26), frequently harbors a chromosomal fyuA-irp gene cluster. Nearly 93% of the 60 EAEC strains studied were irp2 positive. This result prompted us to examine the prevalence of the fyuA-irp gene cluster in clinical E. coli isolates from blood cultures of hospitalized patients. Most of the E. coli blood culture isolates (80%) carried the fyuA-irp gene cluster. In contrast, less than one-third of E. coli strains isolated from stool samples of healthy individuals possessed the fyuA and irp genes.
This high prevalence of the fyuA-irp gene cluster among isolates of E. coli from humans raises the question of why, so far, pesticin-sensitive E. coli strains have been considered rare. By immunoblotting, we were able to demonstrate that irp1, irp2, and fyuA are expressed by almost all E. coli strains carrying the fyuA-irp gene cluster. However, only some fyuA-positive strains showed low-level sensitivity to pesticin, while the majority of FyuA-HMWP-positive E. coli strains were insensitive to pesticin. This may be due to point mutations of the fyuA gene or other, e.g., posttranslational, modifications of the FyuA protein in these E. coli strains. On the other hand, the pesticin receptor FyuA may be not accessible to the 40-kDa pesticin polypeptide due to masking of the receptor by surface components, e.g., the capsule or pili. Nevertheless, one can assume that E. coli strains carrying the fyuA-irp gene cluster are able to produce the siderophore yersiniabactin, which might contribute to the virulence of these E. coli isolates. This attractive hypothesis has to be verified by comparing the virulence of the parental strain and of the isogenic fyuA and irp mutants in a suitable infection model.
Sequencing of the fyuA gene of EAEC 17-2 and internal fragments of irp2 of various E. coli strains indicated that the highest homology was found with the fyuA and irp2 genes of the Y. pestis-Y. pseudotuberculosis lineage group. This observation was supported by Southern hybridization of EcoRI-digested chromosomal DNA from E. coli strains of different pathotypes, using probes covering the entire fyuA-irp gene cluster (Fig. 2). Moreover, we could demonstrate that, as with Yersinia, the HPI in E. coli is linked to the asnT tRNA gene. From this, we conclude that the fyuA-irp gene cluster might be a mobile genetic element which has spread by horizontal transfer through different genera of the family Enterobacteriaceae but could be only maintained in certain pathotypes, such as Yersinia species virulent in mice and enteroaggregative and septicemic E. coli isolates, which probably colonized the intestines of these patients.
As with Y. enterocolitica, we could not detect the hms locus of Y. pestis in fyuA-irp-positive E. coli isolates. This is not surprising, since the hms locus was shown to be important for Y. pestis to block the flea proventriculus (24). Therefore, one possible explanation would be that the original HPI of Y. pestis, composed of the hms locus and the fyuA-irp gene cluster, spread by horizontal transfer into the family Enterobacteriaceae, followed by a deletion of the hms region. However, a more likely hypothesis is that the fyuA-irp gene cluster forms a mobile element that can spread independently of the hms region. The different G+C contents of the two loci (47 mol% for hms versus 56 to 59 mol% for fyuA-irp) and the distinct codon usage argue for the latter hypothesis (16).
Certain E. coli strains (up to 3% of irp2-positive strains) carry a truncated fyuA-irp gene cluster, with deletions proceeding from fyuA or both fyuA and irp1 to the 3′ end of the HPI. Such spontaneous deletions involve only the 3′ part of the fyuA-irp gene cluster and abrogate expression of the yersiniabactin receptor. These E. coli isolates may have lost the receptor after adaptation to a new environment. All E. coli strains used in this study possess the highly efficient enterobactin siderophore system (detected by PCR [data not shown]) (3, 32). Therefore, it seems unlikely that iron acquisition is the primary reason for the wide dissemination of yersinia genes among E. coli strains pathogenic to humans. It is rather tempting to suggest that other effects of gene products of the fyuA-irp gene cluster are of benefit for certain E. coli pathotypes. The possible cytotoxic effect on T cells of the siderophore desferrioxamine B (2) could be an attractive alternative to the yersiniabactin function. Interestingly, it has been shown that the yersiniabactin-like siderophore pyochelin of Pseudomonas aeruginosa promotes damage to endothelial cells by formation of free radicals (6). On the other hand, the irp2 gene product, HMWP2, has extensive similarity to a superfamily of adenylate-forming enzymes involved in the nonribosomal peptide synthesis of not only siderophores but also peptide antibiotics (19). Therefore, it is also conceivable that other products of the putative enzyme HMWP2 might be of benefit to the E. coli strains that carry the fyuA-irp gene cluster.
It remains unclear whether multiple insertion elements located within the fyuA-irp gene cluster are responsible for the observed deletions. However, genetic instability of the HPI is well known in Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica, causing spontaneous loss of iron acquisition in vivo and consequently virulence attenuation. Thus, strong selective pressure is necessary for the maintenance of HPIs. By analogy with yersiniae, we can suggest that the fyuA-irp gene cluster may also contribute to the virulence of certain clinically important E. coli pathotypes, such as EAEC and septicemia-causing E. coli strains. The reason for the high-frequency distribution of the fyuA-irp gene cluster among EAEC strains remains unknown. The genes located on this cluster are apparently not involved in the EAEC phenotype, since we could detect EAEC strains that exhibit the aggregative adherent (AA) phenotype in HEp-2 cell assay and lack the fyuA-irp gene cluster. Moreover, Nataro et al. (30) have shown that this phenotype is closely associated with the presence of a 60-MDa plasmid which encodes bundle-forming fimbriae. It has been shown that cloning of two distinct parts of this plasmid is sufficient to express the AA phenotype in an E. coli HB101 strain that lacks the fyuA-irp gene cluster.
Taken together, these results indicate that (i) the fyuA-irp gene cluster of Yersinia spp. is largely conserved among different HPI-positive E. coli strains even though some distinct changes of the hybridization patterns exist; (ii) deletions of the fyuA-irp gene cluster in E. coli are of different sizes and involve the right-hand side of the HPI, including the fyuA locus; and (iii) the fyuA-irp gene cluster of E. coli strains is more closely related to the HPIs of Y. pestis and Y. pseudotuberculosis than to the HPI of Y. enterocolitica serotype O8. However, the exact function of the fyuA-irp gene cluster in E. coli strains still remains unclear, and our future work will focus on the effect of irp2 and fyuA mutations on the virulence of pathogenic E. coli strains.
ACKNOWLEDGMENTS
We thank Daniela Fischer for excellent technical assistance.
This study was supported by a grant from the Deutsche Forschungsgemeinschaft to J.H. (HE 1297/2-3).
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1989. [Google Scholar]
- 2.Autenrieth I B, Hantke K, Heesemann J. Immunosuppression of the host and delivery of iron to the pathogen: a possible dual role of siderophores in the pathogenesis of microbial infections? Med Microbiol Immunol. 1991;180:135–141. doi: 10.1007/BF00206117. [DOI] [PubMed] [Google Scholar]
- 3.Bagg A, Neilands J B. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol Rev. 1987;51:509–518. doi: 10.1128/mr.51.4.509-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bearden S W, Fetherston J D, Perry R D. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect Immun. 1997;65:1659–1668. doi: 10.1128/iai.65.5.1659-1668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bettelheim K A, Hanna N, Smith D L, Dwyer B W. Evaluation of the Phadebact ETEC-LT test for the heat-labile enterotoxin of Escherichia coli. Int J Med Microbiol. 1989;271:70–76. doi: 10.1016/s0934-8840(89)80055-6. [DOI] [PubMed] [Google Scholar]
- 6.Britigan B E, Rasmussen G T, Cox C D. Pseudomonas siderophore pyochelin enhances neutrophil-mediated endothelial cell injury. Am J Physiol. 1994;266:L192–L198. doi: 10.1152/ajplung.1994.266.2.L192. [DOI] [PubMed] [Google Scholar]
- 7.Carniel E, Antoine J-C, Guiyoule A, Guiso N, Mollaret H H. Purification, location, and immunological characterization of the iron-regulated high-molecular-weight proteins of the highly pathogenic yersiniae. Infect Immun. 1989;57:540–545. doi: 10.1128/iai.57.2.540-545.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carniel E, Guilvout I, Prentice M. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J Bacteriol. 1996;178:6743–6751. doi: 10.1128/jb.178.23.6743-6751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carniel E, Guiyoule A, Guilvout L, Mercereau-Puijalon O. Molecular cloning, iron-regulation and mutagenesis of the irp2 gene encoding HMWP2, a protein specific for the highly pathogenic Yersinia. Mol Microbiol. 1992;6:379–388. doi: 10.1111/j.1365-2958.1992.tb01481.x. [DOI] [PubMed] [Google Scholar]
- 10.Carniel E, Mercereau-Puijalon O, Bonnefoy S. The gene coding for the 190,000-dalton iron-regulated protein of Yersinia species is present only in the highly pathogenic strains. Infect Immun. 1989;57:1211–1217. doi: 10.1128/iai.57.4.1211-1217.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornelis G, Laroche Y, Balligand G, Sory M P, Wauters G. Yersinia enterocolitica, a primary model for bacterial invasiveness. Rev Infect Dis. 1987;9:64–87. doi: 10.1093/clinids/9.1.64. [DOI] [PubMed] [Google Scholar]
- 12.Cravioto A, Gross R J, Scotland S, Rowe B. An adhesive factor found in strains of Escherichia coli belonging to the traditional enteropathogenic serotypes. Curr Microbiol. 1979;3:95–99. [Google Scholar]
- 13.Davies J K, Reeves P. Genetics of resistance to colicins in Escherichia coli K-12: cross-resistance among colicins of group B. J Bacteriol. 1975;123:96–101. doi: 10.1128/jb.123.1.96-101.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Almeida A M, Guiyoule A, Guilvout I, Iteman I, Baranton G, Carniel E. Chromosomal irp2 gene in Yersinia: distribution, expression, deletion and impact on virulence. Microb Pathog. 1993;14:9–21. doi: 10.1006/mpat.1993.1002. [DOI] [PubMed] [Google Scholar]
- 15.Fetherston J D, Perry R D. The pigmentation locus of Yersinia pestis KIM6+ is flanked by an insertion sequence and includes the structural genes for pesticin sensitivity and HMWP2. Mol Microbiol. 1994;13:697–708. doi: 10.1111/j.1365-2958.1994.tb00463.x. [DOI] [PubMed] [Google Scholar]
- 16.Fetherston J D, Schuetze P, Perry R D. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol Microbiol. 1992;6:2693–2704. doi: 10.1111/j.1365-2958.1992.tb01446.x. [DOI] [PubMed] [Google Scholar]
- 17.Franke J, Franke S, Schmidt H, Schwarzkopf A, Wieler L H, Baijer G, Beutin L, Karch H. Nucleotide sequence analysis of enteropathogenic Escherichia coli (EPEC) adherence factor probe and development of PCR for rapid detection of EPEC harboring virulence plasmids. J Clin Microbiol. 1994;32:2460–2463. doi: 10.1128/jcm.32.10.2460-2463.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frankel G, Riley L, Giron J A, Valmassoi J, Friedmann A, Strockbine N, Falkow S, Schoolnik G K. Detection of Shigella in feces using DNA amplification. J Infect Dis. 1990;161:1252–1256. doi: 10.1093/infdis/161.6.1252. [DOI] [PubMed] [Google Scholar]
- 19.Guilvout I, Mercereau-Puijalon O, Bonnefoy S, Pugsley A P, Carniel E. High-molecular-weight protein 2 of Yersinia enterocolitica is homologous to AngR of Vibrio anguillarum and belongs to a family of proteins involved in nonribosomal peptide synthesis. J Bacteriol. 1993;175:5488–5504. doi: 10.1128/jb.175.17.5488-5504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hames B D. Introduction to polyacrylamide gel electrophoresis. In: Hames B D, Rickwood D, editors. Gel electrophoresis of proteins: a practical approach. Washington, D.C: IRL Press; 1987. pp. 1–91. [Google Scholar]
- 21.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 22.Heesemann J. Chromosomal-encoded siderophores are required for mouse virulence of enteropathogenic Yersinia species. FEMS Microbiol Lett. 1987;48:229–233. [Google Scholar]
- 23.Heesemann J, Hantke K, Vocke T, Saken E, Rakin A, Stojiljkovic I, Berner R. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane protein of 65000 Da and pesticin sensitivity. Mol Microbiol. 1993;8:397–408. doi: 10.1111/j.1365-2958.1993.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 24.Hinnebusch B J, Perry R D, Schwan T G. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science. 1996;273:367–370. doi: 10.1126/science.273.5273.367. [DOI] [PubMed] [Google Scholar]
- 25.Hu P C, Yang G C, Brubaker R R. Specificity, induction, and absorption of pesticin. J Bacteriol. 1972;112:212–219. doi: 10.1128/jb.112.1.212-219.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huppertz H, Rutkowski S, Aleksic S, Karch H. Acute and chronic diarrhoea and abdominal colic associated with enteroaggregative Escherichia coli in young children living in western Europe. Lancet. 1997;349:1660–1662. doi: 10.1016/S0140-6736(96)12485-5. [DOI] [PubMed] [Google Scholar]
- 27.Karch H, Meyer T. Single primer pair for amplifying segments of distinct Shiga-like-toxin genes by polymerase chain reaction. J Clin Microbiol. 1989;27:2751–2757. doi: 10.1128/jcm.27.12.2751-2757.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucier T S, Fetherston J D, Brubaker R R, Perry R D. Iron uptake and iron-repressible polypeptides in Yersinia pestis. Infect Immun. 1996;64:3023–3031. doi: 10.1128/iai.64.8.3023-3031.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nataro J P. Enteroaggregative and diffusely adherent Escherichia coli. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press; 1995. pp. 727–737. [Google Scholar]
- 30.Nataro J P, Deng Y, Maneval D R, German A L, Martin W C, Levine M M. Aggregative adherence fimbriae I of enteroaggregative Escherichia coli mediate adherence to HEp-2 cells and hemagglutination of human erythrocytes. Infect Immun. 1992;60:2297–2304. doi: 10.1128/iai.60.6.2297-2304.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nataro J P, Yikang D, Giron J A, Savarino S J, Kothary M H, Hall R. Aggregative adherence fimbria I expression in enteroaggregative Escherichia coli requires two unlinked plasmid regions. Infect Immun. 1993;61:1126–1131. doi: 10.1128/iai.61.3.1126-1131.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neilands J B. Siderophores of bacteria and fungi. Microbiol Sci. 1984;1:9–14. [PubMed] [Google Scholar]
- 33.Olive D M. Detection of enterotoxigenic Escherichia coli after polymerase chain reaction amplification with a thermostable DNA polymerase. J Clin Microbiol. 1989;27:261–265. doi: 10.1128/jcm.27.2.261-265.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pelludat, C., A. Rakin, C. A. Jacobi, S. Schubert, and J. Heesemann. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 35.Pendrak M L, Perry R D. Proteins essential for expression of the Hms+ phenotype of Yersinia pestis. Mol Microbiol. 1993;8:857–864. doi: 10.1111/j.1365-2958.1993.tb01632.x. [DOI] [PubMed] [Google Scholar]
- 36.Perry R D, Lucier T S, Sikkema D J, Brubaker R R. Storage reservoirs of hemin and inorganic iron in Yersinia pestis. Infect Immun. 1993;61:32–39. doi: 10.1128/iai.61.1.32-39.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perry R D, Pendrak M L, Schuetze P. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J Bacteriol. 1990;172:5929–5937. doi: 10.1128/jb.172.10.5929-5937.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Portnoy D A, Martinez R J. Role of a plasmid in the pathogenicity of Yersinia species. Curr Top Microbiol Immunol. 1985;118:29–51. doi: 10.1007/978-3-642-70586-1_3. [DOI] [PubMed] [Google Scholar]
- 39.Rakin A, Heesemann J. Virulence-associated fyuA/irp2 gene cluster of Yersinia enterocolitica biotype 1B carries a novel insertion sequence IS1328. FEMS Microbiol Lett. 1995;129:287–292. doi: 10.1111/j.1574-6968.1995.tb07594.x. [DOI] [PubMed] [Google Scholar]
- 40.Rakin A, Saken E, Harmsen D, Heesemann J. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol Microbiol. 1994;13:253–263. doi: 10.1111/j.1365-2958.1994.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 41.Rakin A, Urbitsch P, Heesemann J. Evidence for two evolutionary lineages of highly pathogenic Yersinia species. J Bacteriol. 1995;177:2292–2298. doi: 10.1128/jb.177.9.2292-2298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt H, Knop C, Franke S, Aleksic S, Heesemann J, Karch H. Development of PCR for screening of enteroaggregative Escherichia coli. J Clin Microbiol. 1995;33:701–705. doi: 10.1128/jcm.33.3.701-705.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt H, Russmann H, Schwarzkopf A, Aleksic S, Heesemann J, Karch H. Prevalence of attaching and effacing Escherichia coli in stool samples from patients and controls. Int J Med Microbiol Virol Parasitol Infect Dis. 1994;281:201–213. doi: 10.1016/s0934-8840(11)80571-2. [DOI] [PubMed] [Google Scholar]
- 45.Scotland S M, Smith H R, Said B, Willshaw G A, Cheasty T, Rowe B. Identification of enteropathogenic Escherichia coli isolated in Britain as enteroaggregative or as members of a subclass of attaching-and-effacing E. coli not hybridizing with the EPEC adherence-factor probe. J Med Microbiol. 1991;35:278–283. doi: 10.1099/00222615-35-5-278. [DOI] [PubMed] [Google Scholar]
- 46.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 47.Vial P A, Mathewson J J, Dupont H L, Guers L, Levine M M. Comparison of two assay methods for patterns of adherence to HEp-2 cells of Escherichia coli from patients with diarrhea. J Clin Microbiol. 1990;28:882–885. doi: 10.1128/jcm.28.5.882-885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vial P A, Robins Browne R, Lior H, Prado V, Kaper J B, Nataro J P, Maneval D, Elsayed A, Levine M M. Characterization of enteroadherent-aggregative Escherichia coli, a putative agent of diarrheal disease. J Infect Dis. 1988;158:70–79. doi: 10.1093/infdis/158.1.70. [DOI] [PubMed] [Google Scholar]