Abstract
Human lactoferrin (hLf), a glycoprotein released from neutrophil granules during inflammation, and the lipopolysaccharide (LPS)-binding protein (LBP), an acute-phase serum protein, are known to bind to the lipid A of LPS. The LPS-binding sites are located in the N-terminal regions of both proteins, at amino acid residues 28 to 34 of hLf and 91 to 108 of LBP. Both of these proteins modulate endotoxin activities, but they possess biologically antagonistic properties. In this study, we have investigated the competition between hLf and recombinant human LBP (rhLBP) for the binding of Escherichia coli 055:B5 LPS to the differentiated monocytic THP-1 cell line. Our studies revealed that hLf prevented the rhLBP-mediated binding of LPS to the CD14 receptor on cells. Maximal inhibition of LPS-cell interactions by hLf was raised when both hLf and rhLBP were simultaneously added to LPS or when hLf and LPS were mixed with cells 30 min prior to the incubation with rhLBP. However, when hLf was added 30 min after the interaction of rhLBP with LPS, the binding of the rhLPS-LBP complex to CD14 could not be reversed. These observations indicate that hLf competes with rhLBP for the LPS binding and therefore interferes with the interaction of LPS with CD14. Furthermore, experiments involving competitive binding of the rhLBP-LPS complex to cells with two recombinant mutated hLfs show that in addition to residues 28 to 34, another basic cluster which contains residues 1 to 5 of hLf competes for the binding to LPS. Basic sequences homologous to residues 28 to 34 of hLf were evidenced on LPS-binding proteins such as LBP, bactericidal/permeability-increasing protein, and Limulus anti-LPS factor.
Bacterial lipopolysaccharides (LPS) are potent activators of the immune system. They stimulate host cells, mainly monocytes/macrophages and neutrophils, to produce endogeneous mediators such as cytokines (24, 48). The presence of large amounts of LPS leads to excessive release of these mediators, resulting in septic shock (34).
By their ability to interact with anionic LPS, a variety of serum cationic proteins were shown to modulate the LPS-mediated activation of cells (41). One of these proteins, a 60-kDa acute-phase protein named LPS-binding protein (LBP), is present in normal plasma (37, 42). LBP binds to the lipid A portion of LPS (43) and mediates the transfer of LPS to CD14, a glycosylphosphatidylinositol-anchored membrane protein present on myeloid cells (26, 46, 51). The recognition of the LBP-LPS complex by the CD14 receptor leads to the activation of monocytes and macrophages (47).
Other cationic molecules, such as bactericidal/permeability-increasing protein (BPI) (12, 13), polymyxin B (31), lysozyme (33), Limulus anti-LPS factor (LALF) (15, 32), and lactoferrin (Lf) (3, 11), were found to bind LPS and to inhibit endotoxin activity. Human Lf (hLf) is an iron-binding glycoprotein (30) found in exocrine secretions of mammals and released from granules of neutrophils during inflammatory responses (25). hLf is associated with host defense through its antibacterial properties (4, 36) and immunological activities (5, 53). This glycoprotein inhibits in vitro the release of tumor necrosis factor alpha (TNF-α), interleukin-1 (IL-1) (7), and IL-6 (27) from LPS-stimulated monocytes and decreases the priming of neutrophils by LPS (6, 50). In vivo, hLf also regulates the release of TNF-α and protects mice against a lethal dose of Escherichia coli (22, 52). Since many immunomodulatory activities of hLf may be relevant to the interactions of hLf with LPS, several studies focused on the molecular basis of such interactions. First, hLf was shown to bind to the lipid A region of LPS with a high affinity (3). Second, site-directed mutagenesis experiments with hLf demonstrated that the loop region of amino acids 28 to 34 of hLf interacts with the E. coli 055:B5 LPS (11). This region also plays important roles in the binding of hLf to its specific receptor on activated lymphocytes (9, 10, 18, 28) and in the antimicrobial activity of human and bovine Lfs (4, 44). Furthermore, another cationic region involving amino acid residues 2 to 5, located in the vicinity of amino acids 28 to 34, has been identified as a recognition site for anionic molecules such as heparin (23).
Since hLf interacts with LPS, it might be assumed that the protein competes with LBP in serum for the binding of LPS, thereby preventing the binding of the LPS-LBP complex to the CD14 receptor. To check this hypothesis, we have studied the binding of the E. coli 055:B5 LPS to the differentiated monocytic cell line THP-1, mediated either by LBP-containing human serum or by purified recombinant human LBP (rhLBP). These experiments were performed in the presence of various concentrations of hLf. A further insight in the role of hLf domain N-I in the competition for LPS binding with rhLBP was gained with mutated recombinant hLfs. For this purpose, two hLf variants were produced by site-directed mutagenesis and assayed in competitive experiments: EGS-rhLf, in which residues 28 to 34 were replaced by a loop of the C-terminal lobe counterpart of hLf, and G4R-rhLf, which lacked residues 1 to 5 of rhLf. Finally, a comparison of the primary and secondary structures of the potential LPS-binding sites located in hLf, LBP, BPI, and LALF suggested the involvement of a conserved consensus sequence for LPS binding.
MATERIALS AND METHODS
Materials.
Bovine serum albumin (BSA), Dulbecco’s phosphate-buffered saline (PBS), E. coli 055:B5 LPS labeled with fluorescein isothiocyanate (FITC), and FITC-conjugated goat anti-mouse immunoglobulin G (IgG) were purchased from Sigma Chemical Co. (St. Louis, Mo.). Anti-CD14 monoclonal antibody IOM2 and isotype control mouse IgG2a were obtained from Immunotech (Marseille, France), RPMI 1640 medium and gentamicin were from Gibco BRL (Eragny, France), and fetal calf serum was from Techgen International (Les Ullis, France). 1,25-Dihydroxy-vitamin D3 was obtained from Calbiochem (La Jolla, Calif.), and glutamine was obtained from Eurobio (Les Ullis, France). Human serum was purchased from a local blood transfusion center and heated at 56°C for 30 min.
Proteins.
hLf was purified from pooled human lactoserum by ion-exchange chromatography and iron saturated, as previously described (29, 38). Homogeneity of the protein was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (17). hLf samples were passed through a Detoxi-Gel column (Pierce Chemicals Co., Rockford, Ill.) before use. Pyrogen-free water was used to prepare all buffer solutions. LPS contamination of hLf and buffer solutions was estimated by Limulus amoebocyte lysate assays (QCL1000; BioWhitaker, Walkersville, Md.). rhLBP was purified by the method of Theofan et al. (40) from supernatants of CHO cells transfected with the cDNA of hLBP.
Expression and purification of recombinant hLfs.
A full-length 2.3-kbp cDNA coding for hLf was obtained from a human mammary gland cDNA library (Clontech, Palo Alto, Calif.) (19). Nonmodified rhLf has been produced in a baculovirus expression system and purified as previously described (35). Two rhLf variants were obtained by site-directed mutagenesis of the cDNA coding sequence of hLf by using the Sculptor in vitro mutagenesis system kit (Amersham International, Amersham, United Kingdom). G4R-rhLf, a mutated rhLf in which the residues 1GRRRR5 were deleted, was obtained as previously reported (20). EGS-rhLf corresponds to rhLf in which the sequence 28RKVRGPP34 was replaced by the sequence 365EGS367 (11). For this purpose, a mutagenic oligonucleotide, 5′TGGCAAAGGAATATGGAAGGTTCTGT3′, was synthesized by Eurogentec (Seraing, Belgium). The template for the mutagenesis was the phage M13-mp11, containing a 346-bp EcoRI-AccI fragment of the coding sequence cloned into the pBluescript SK plasmid (19). After mutagenesis, the deletion was confirmed by DNA sequence analysis and the mutated EcoRI-AccI fragment was ligated back into pBluescript SK with the 3′ complementary part of the full-length cDNA of hLf as described previously (19). Finally, the mutated cDNA was subcloned into pVL1392 which was previously digested with EcoRI and dephosphorylated with calf intestine alkaline phosphatase (Stratagene, La Jolla, Calif.) to yield the pVL1392-EGS-rhLf construct. EGS-rhLf was produced in the baculovirus expression system and purified on an SP-Sepharose fast-flow column, as previously described (35). The purity of the rhLf mutant was checked by sodium dodecyl sulfate–7.5% polyacrylamide gel electrophoresis. N-terminal amino acid sequence analysis of the mutant protein was performed by the Edman degradation procedure, using an Applied Biosystems 477 protein sequencer.
Cell culture.
Human promonocytic THP-1 cells (ECACC no. 88081201) were grown in RPMI 1640 supplemented with 10% fetal calf serum, 2 mM l-glutamine, and 2 × 10−5 M β-mercaptoethanol in a 5% CO2–air humidified atmosphere at 37°C. THP-1 cells were induced to express CD14 by treatment for 48 h with 50 nM 1,25-dihydroxy-vitamin D3 (49) in the presence of 100 U of gamma interferon per ml. Viability was over 96% as determined by trypan blue dye exclusion.
CD14 expression at the cell surface.
CD14 expression was detected on the surface of the differentiated THP-1 cells by flow cytometry. Cells (300,000) were incubated at 4°C for 30 min with anti-CD14 monoclonal antibody IOM2 in RPMI containing 0.2% BSA and 0.04% NaN3. An isotype control IgG2a was used as a negative control. The cells were washed twice with PBS and stained with FITC-conjugated goat anti-mouse IgG for 30 min at 4°C. After two washes, cells were analyzed with a Becton Dickinson FACScan flow cytometer. Cells were gated for forward- and side-angle light scatters, and 10,000 particles of the gated population were analyzed. The fluorescence channels were set on a logarithmic scale, and the mean fluorescence intensity was determined.
Binding of FITC-labeled LPS to cells.
Differentiated THP-1 cells were adjusted to 300,000 cells in 200 μl of RPMI and 0.04% NaN3. Cells were incubated with 1 μg of FITC-labeled E. coli 055:B5 LPS per ml in the presence of 10% human serum or 1.5 μg of purified rhLBP per ml. For a negative control, 0.4% BSA was added with the FITC-labeled LPS. After 1 h at 4°C, cells were centrifuged at 400 × g for 5 min, washed twice with PBS, and analyzed by flow cytometry.
To inhibit the binding of LPS to the CD14 receptor, cells diluted in RPMI supplemented with 0.04% NaN3 and 0.2% BSA were pretreated for 30 min at 4°C with anti-CD14 antibody at 10 μg/ml. An isotype mouse IgG2a was used as a negative control. After three washes, cells were incubated with FITC-LPS in presence of 10% human serum or 1.5 μg of rhLBP per ml, as described above.
Effect of hLf on the binding of FITC-labeled LPS to differentiated THP-1 cells.
Inhibition of the binding of FITC-labeled LPS to differentiated THP-1 cells was performed as described above but in the presence of hLf concentrations ranging from 5 to 40 μg/ml. Briefly, cells were incubated with 1 μg of FITC-labeled E. coli 055:B5 LPS per ml and 1.5 μg of rhLBP per ml in RPMI–0.04% NaN3. hLf samples were added to cells at the same time as rhLBP or 30 min before or after rhLBP addition. Similar experiments were performed with 10% human serum instead of rhLBP. After 1 h at 4°C, cells were centrifuged at 400 × g for 3 min and the supernatant was removed. Cells resuspended in PBS were analyzed by flow cytometry and the fluorescence was detected, as described above. The mean of the fluorescence intensity obtained with rhLBP or serum defined the total LPS binding to cells. The results obtained in the presence of hLf were expressed as percentages of the total LPS binding.
Statistical analysis.
Data are presented as the mean ± standard error for the indicated number of independent experiments. Statistical significance was analyzed with a Student’s t test for unpaired data. Values with P < 0.05 were considered to be significant.
RESULTS
Effects of hLf on serum-mediated binding of LPS to differentiated THP-1 cells.
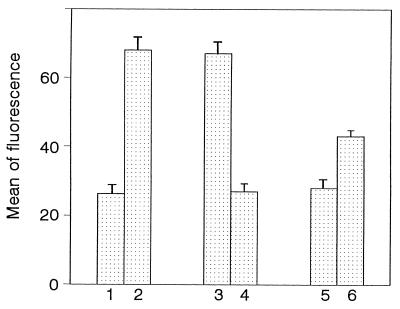
It is well known that LBP present in serum is responsible for the LPS binding to the CD14 receptor (16). As shown in Fig. 1, differentiated THP-1 cells, incubated with 1 μg of FITC-labeled E. coli 055:B5 LPS per ml and BSA, showed only a low fluorescence intensity (27.2 ± 2.8). In contrast, when 10% human serum was added to cells in the presence of LPS, the mean fluorescence intensity was increased (68.1 ± 3.8). This result suggests that the binding of E. coli 055:B5 LPS to cells was mediated by LBP present in serum. Anti-CD14 antibodies were then used to confirm that the CD14 receptor was responsible for the LPS binding. As illustrated in Fig. 1, preincubation of cells with anti-CD14 antibodies suppressed the serum-mediated LPS binding (28.1 ± 2.7).
FIG. 1.
LPS binding to differentiated THP-1 cells in the presence of human serum. As described in Materials and Methods, cells were incubated with 1 μg of FITC-labeled E. coli 055:B5 LPS per ml in the presence of 0.4% BSA (bar 1), 10% human serum (bar 2), 10% human serum after preincubation of cells with isotype control IgG2a (bar 3), 10% human serum after previous incubation of cells with anti-CD14 monoclonal antibody (bar 4), 20 μg of hLf per ml (bar 5), or 10% human serum and 20 μg of hLf per ml added at the same time (bar 6). The mean fluorescence intensity was determined by flow cytometry. The results (means ± standard errors) were calculated from five separate experiments.
We investigated the effect of hLf on the serum-dependent binding of FITC-labeled E. coli 055:B5 LPS to differentiated THP-1 cells. The addition of hLf to cells, at a concentration of 20 μg/ml, decreased the fluorescence intensity (44.1 ± 1.8). This result indicates that smaller amounts of LPS were bound to monocytes in the presence of hLf and suggests that the hLf-LPS complex interferes with the serum-mediated binding of LPS through the CD14 receptor.
A control experiment performed with hLf and LPS, without serum, gave results similar to those obtained with BSA (29.0 ± 2.7) (Fig. 1). This experiment confirmed that hLf did not promote the binding of LPS to the CD14 receptor but rather inhibited the serum-mediated interaction of LPS with cells.
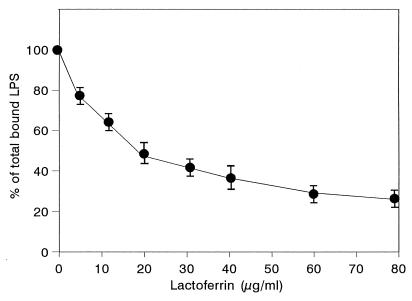
The concentration of hLf required for maximal inhibition of binding of 1 μg of LPS per ml was determined (Fig. 2). hLf blocked LPS binding to cells in a concentration-dependent manner. Up to 75% ± 5% inhibition was obtained in the presence of 80 μg of hLf per ml.
FIG. 2.
Inhibition of LPS binding to differentiated THP-1 cells by hLf in the presence of human serum. Cells were incubated with 1 μg of FITC-labeled LPS per ml and 10% human serum in the presence of increasing concentrations of hLf, as described in Materials and Methods. The results are expressed as percentages of the total LPS bound to cells with 10% serum alone. Each point represents the mean ± standard error from four experiments.
Effects of hLf on rhLBP-mediated binding of LPS to differentiated THP-1 cells.
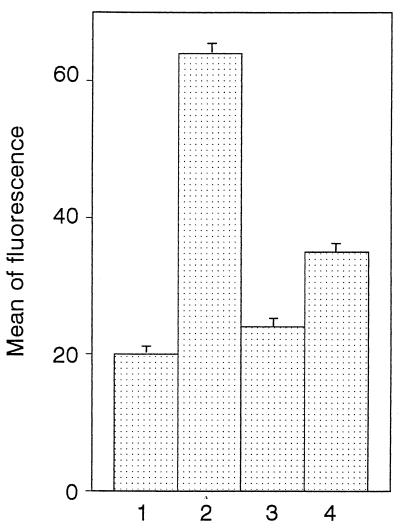
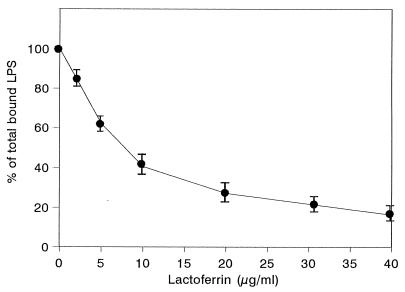
We investigated whether hLf could interfere directly with rhLBP for the LPS binding to differentiated THP-1 cells. As illustrated in Fig. 3, cells showed a brighter fluorescence in the presence of 1.5 μg of rhLBP per ml (64.3 ± 1.6) than with BSA (control) (20.3 ± 1.3). This increased fluorescence intensity was similar to that detected with 10% human serum (68.1 ± 3.8) (Fig. 1). rhLBP promotes the LPS binding to differentiated THP-1 cells expressing CD14, since the binding was inhibited by preincubation of cells with anti-CD14 monoclonal antibody (24.3 ± 1.2) (Fig. 3). As seen in the experiments with serum, 20 μg of hLf per ml inhibited the fluorescence caused by binding of rhLBP-LPS complex (35.3 ± 1.9). When increasing concentrations of hLf were added to cells simultaneously with both rhLBP and FITC-labeled LPS, the rhLBP-mediated binding of LPS to THP1 cells was decreased (Fig. 4). Only 5 μg of hLf per ml was sufficient to provide 38% ± 5% inhibition. Maximal inhibition (78% ± 4%) was obtained at an hLf concentration of 40 μg/ml. These findings indicate that hLf and rhLBP may compete for the same LPS-binding sites, blocking the binding of rhLBP-LPS complexes to cells.
FIG. 3.
LPS binding to differentiated THP-1 cells in the presence of rhLBP. As described in Materials and Methods, cells were incubated with 1 μg of FITC-labeled LPS per ml in the presence of 0.4% BSA (bar 1), 1.5 μg of rhLBP per ml (bar 2), 1.5 μg of rhLBP per ml after preincubation of cells with anti-CD14 monoclonal antibody (bar 3), or 1.5 μg of rhLBP per ml and 20 μg of hLf per ml added at the same time (bar 4). The mean fluorescence intensity was determined. The results (means ± standard errors) were calculated from four separate experiments.
FIG. 4.
Inhibition of LPS binding to differentiated THP-1 cells by hLf in the presence of rhLBP. The binding of FITC-labeled LPS (1 μg/ml) to cells was performed in the presence of 1.5 μg of rhLBP per ml and increasing concentrations of hLf, added simultaneously. The results are expressed as percentages of the total LPS bound to cells with rhLBP alone. Each point represents the mean ± standard error from four experiments.
Time-dependent inhibition by hLf of rhLBP-mediated binding of LPS to differentiated THP-1 cells.
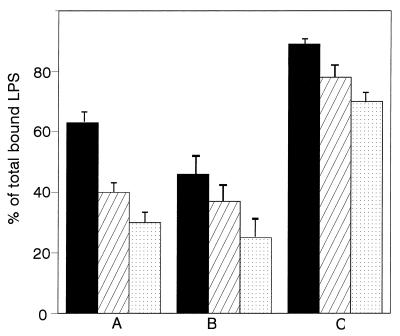
Three concentrations of Lf were added to cells at various times relative to the preincubation of LPS with rhLBP. When LPS, rhLBP, and hLf were simultaneously incubated with differentiated THP-1 cells, hLf was able to inhibit the LPS binding to cells (Fig. 5A). About 35% ± 4% inhibition was obtained with 5 μg of hLf per ml, and a maximum of 68% ± 5% was obtained with 20 μg/ml. When LPS was first preincubated with hLf and cells, 30 min before addition of rhLBP, the inhibitory effect of hLf was enhanced (Fig. 5B). Indeed, under these experimental conditions, 48% ± 6% inhibition was then detected with only 5 μg of hLf per ml. However, a low inhibition (28% ± 3%) was measured when 20 μg of hLf per ml was added to cells previously incubated with both LPS and rhLBP for 30 min (Fig. 5C).
FIG. 5.
Time-dependent inhibition of rhLBP-mediated binding of LPS to differentiated THP-1 cells in the presence of hLf. The binding of FITC-labeled LPS was studied, as described in Materials and Methods, in the presence of rhLBP (1.5 μg/ml) and three concentrations of hLf added at various intervals: 5 μg/ml (filled bars), 10 μg/ml (hatched bars), and 20 μg/ml (stippled bars). hLf was added to LPS at the same time as rhLBP (A), 30 min before rhLBP (B), or 30 min after rhLBP (C). Results are percentages of the total LPS bound to cells in the presence of rhLBP alone. Data are expressed as the means ± standard errors from three replicates.
Inhibition of binding of LPS to differentiated THP-1 cells by rhLf and mutated rhLfs.
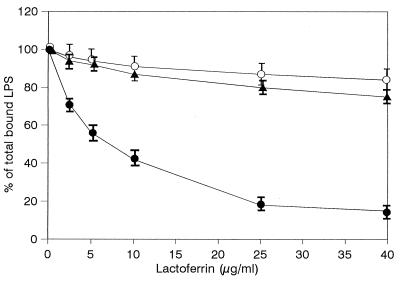
Amino acid residues 28 to 34 are involved in the interactions of hLf with anionic molecules such as LPS (11) or heparin (23). Additionally, residues 2 to 5 of hLf may interact with heparin (23). These two N-terminal basic clusters of the protein have been mutated. The rhLf mutants were produced in insect Sf9 cells infected by baculovirus, purified, and assayed in competitive experiments with rhLBP. As illustrated in Fig. 6, the rhLBP-mediated binding of LPS was inhibited by increasing concentrations of native rhLf. Maximal inhibition (78% ± 4%) was obtained with 25 μg of rhLf per ml used as a control. In contrast, neither EGS-rhLf nor G4R-rhLf was able to significantly prevent the binding of LPS to cells. Indeed, no more than 22% ± 5% or 15% ± 6% inhibition was detected with 40 μg of protein per ml, respectively.
FIG. 6.
Inhibition of LPS binding to differentiated THP-1 cells by rhLf and mutated rhLfs in the presence of rhLBP. Cells were incubated for 1 h at 4°C simultaneously with 1 μg of FITC-labeled LPS per ml, 1.5 μg of rhLBP per ml, and increasing concentrations of rhLf (•), EGS-rhLf (▴), or G4R-rhLf (○). Results are percentages of total LPS bound to cells in the presence of rhLBP alone. Each point represents the mean ± standard error from four experiments.
DISCUSSION
The ability of hLf to form complexes with LPS (3, 11) and thus to inhibit the LPS-induced release of cytokines by mononuclear phagocytes (7, 27) makes it a potentially important molecule in the inflammatory response. In contrast to hLf, LBP, another LPS-binding protein present in serum, enhances the LPS-induced cell activation in mediating the transfer of LPS to CD14 on monocytes or macrophages (26, 46, 51). Since both LBP (43) and hLf (3) are known to bind to the lipid A moiety of LPS, we investigated whether the antagonistic effects of these two basic proteins are due to a competition between hLf and rhLBP for binding to LPS.
First, flow cytometry experiments demonstrated that E. coli 055:B5 LPS bound specifically to CD14 on differentiated monocytic THP-1 cells, in the presence of either human serum or rhLBP. In contrast to the case for rhLBP, our results showed that hLf did not promote the interaction of endotoxin with CD14 but even prevented the rhLBP-mediated binding of LPS to CD14. This effect appeared to be dependent on the hLf concentration, although the inhibition was not total even at 80 or 40 μg of hLf per ml in the presence of serum or rhLBP, respectively. The binding of LPS to cells was efficiently inhibited when hLf interacted with LPS prior to rhLBP. However, once the rhLBP-LPS complex was formed, hLf was unable to block its binding to cells.
Our results indicate that both rhLBP and hLf compete for the binding to E. coli LPS, therefore interfering with the rhLBP-mediated interaction of LPS with CD14. The close affinities of both hLf and LBP for LPS (11, 26) and the fact that both cationic proteins bind to lipid A of LPS (3, 43), probably at or near the same epitope, strongly support these results. Concerning the LPS-binding site present in hLf, it has been recently demonstrated that the loop region containing amino acids 28 to 34 located in the N-I domain of hLf is involved in the high-affinity interaction with LPS (11). In this study, experiments performed with EGS-rhLf, a protein in which residues 28 to 34 were replaced by the loop of the C-terminal lobe counterpart, indicated that residues 28 to 34 are essential to inhibit the rhLBP-mediated binding of LPS to CD14. This region is also present in lactoferricin, a bactericidal pepsin-derived fragment of Lf (4, 44) which in vitro suppresses the release of IL-6 from monocytic THP-1 cells stimulated by LPS (27).
Furthermore, we investigated the role of another cationic region, involving residues 1GRRRR5 of hLf, in the competition with rhLBP for LPS binding. As reported from the crystallographic structure analysis of hLf (2), this sequence is located in the vicinity of residues 28 to 34. The recombinant hLf lacking residues 1 to 5 (G4R-rhLf) did not inhibit the rhLBP-mediated binding of LPS to cells. Based on these observations, we can postulate that residues 1 to 5 of hLf may interact synergically with residues 28 to 34 as a cationic cradle to bind LPS. A similar interaction between hLf and heparin, another anionic molecule, has been previously suggested by Mann et al. (23).
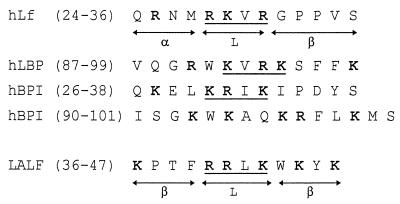
The comparison of the LPS-binding sites of both hLf and LBP should explain how and why these two cationic proteins compete for endotoxin interactions. From the analysis of the properties of truncated forms of LBP, the LPS-binding region of LBP has been located between amino acid residues 1 and 197 (14) and, more precisely, between residues 91 and 108 (39). Another LPS-binding protein, BPI, which is found in neutrophil granules (13) and has 44% sequence homology with LBP, also inhibits some biological activities of LPS, such as polymorphonuclear cell priming and cytokine production by monocytes (8). The antagonistic properties of both BPI and LBP can be explained by competitive effects for the binding to LPS. Three regions of the N-terminal domain of BPI, amino acid residues 17 to 45, 65 to 99, and 142 to 169, interact with LPS and exhibit a heparin-binding capacity (1, 21). Interestingly, the alignment of the N-terminal sequences of hLf, BPI, and LBP shows similarities between amino acid residues 24 to 36, 26 to 38, and 87 to 99, respectively, of the proteins (Fig. 7). It is indeed worth noting that a pattern of three basic amino acids separated by one hydrophobic amino acid is present in hLf (28RKVR31), BPI (30KRIK33), and LBP (92KVRK95). Another LPS-binding protein, LALF, possesses a similar structural motif (40RRLK43) (32). Amino acids 40 to 43 of LALF, whose three-dimensional structure has been defined (15), form a solvent-accessible loop at the protein surface, exactly as do amino acids 28 to 31 of hLf (18) (Fig. 7).
FIG. 7.
Alignment of amino acid residues 24 to 36 of hLf, 87 to 99 of hLBP, 26 to 38 of hBPI, and 36 to 47 of LALF. Homologous residues are in boldface. The positions of sheets (β), helices (α), and loops (L) are indicated for hLf and LALF.
Moreover, hLf, LBP, and BPI also bind to heparin (1, 21, 23). Interestingly, a consensus sequence (XBBXBX, where X is any hydrophobic amino acid and B is any basic amino acid) has been identified in various heparin-binding proteins (21). Based on this observation, it can be suggested that the structural motif (BBXB) is a binding site for both LPS and heparin. Additionally, our results suggest that a cluster of four consecutive arginine residues in hLf (residues 2 to 5), involved in the binding with heparin (23), is also essential for LPS interactions. However, this basic cluster is not encountered in the sequences of all of the different LPS-binding proteins. It should be interesting to determine if heparin could affect the interaction of LPS with hLf or with LBP and BPI.
Thus, this paper demonstrates the ability of hLf to inhibit, in vitro, the rhLBP-mediated binding of endotoxin to differentiated monocytic THP-1 cells. This property of hLf may be explained by a competition between rhLBP and hLf to bind LPS. Amino acids 28 to 34 and 1 to 5 of hLf are involved in the competition for the LPS binding. Residues 28 to 31 of hLf exhibit homologies with the LPS-binding sites located in other cationic proteins, such as LBP, BPI, and LALF.
The ability of Lf to limit, in vitro, the binding of LPS to CD14 indicates that Lf might modulate the inflammatory processes in vivo. This hypothesis is supported by a previous study reporting the protective function of Lf against sublethal doses of LPS in mice (52). Indeed, injection of bovine Lf into mice prior to LPS challenge decreases the release of TNF-α, a major inducer of inflammatory process (22, 52). Moreover, it has been demonstrated that the repeated bacterial infections in neutropenic patients can be reduced by Lf treatment (45). Nevertheless, the LPS-neutralizing activity of Lf may depend on the presence and concentration of other LPS-binding proteins. In contrast to the case for LBP, the physiological concentration of Lf in serum is low but drastically increases during infection. Following the LBP-mediated stimulation of the immune system, Lf released from neutrophilic granules could neutralize the excess of LPS at the site of inflammation and protect the host against the excessive release of cytokines. Although the minimal concentration of endotoxin which can be bound to Lf has not been investigated, the effect of Lf on the cytokine release induced by LPS was detected even with 10 ng of LPS per ml (27). This suggests that due to its high affinity for LPS, Lf could, in vivo, absorb small amounts of LPS. Further in vivo studies are needed to investigate whether Lf could directly overcome the LBP-mediated activation of cells in the host and modulate the CD14-independent LPS signalling pathways.
ACKNOWLEDGMENTS
This work was supported in part by the Université des Sciences et Technologies de Lille I, the Centre National de la Recherche Scientifique (U.M.R. du CNRS no. 111 [director, A. Verbert]), and the Programme International de Coopération Scientifique of the CNRS with Rumania (contract no. 232).
We are grateful to M. Masson and M. C. Slomianny for their skillful technical assistance and to C. Motas and J. Montreuil for their involvement in the initiation of contract no. 232. We thank S. Krag (Department of Biochemistry, Johns Hopkins University, Baltimore, Md.) for reviewing the manuscript.
REFERENCES
- 1.Abrahamson S L, Wu H S, Williams R E, Der K, Ottah N, Little R, Gazzano-Santoro H, Theofan G, Bauer R, Leigh S, Orme A, Horwitz A H, Caroll S F, Dedrick R L. Biochemical characterization of recombinant fusions of LPS-binding protein and bactericidal/permeability-increasing protein. J Biol Chem. 1997;272:2149–2155. doi: 10.1074/jbc.272.4.2149. [DOI] [PubMed] [Google Scholar]
- 2.Anderson B F, Baker H M, Norris G E, Rice D W, Baker E N. Structure of human lactoferrin: crystallographic structure analysis and refinement at 2.8 Å resolution. J Mol Biol. 1989;209:711–734. doi: 10.1016/0022-2836(89)90602-5. [DOI] [PubMed] [Google Scholar]
- 3.Appelmelk B J, An Y Q, Geerts M, Thijs B G, De Boer H A, MacLaren D M, De Graaff J, Nuijens J H. Lactoferrin is a lipid A binding protein. Infect Immun. 1994;62:2628–2632. doi: 10.1128/iai.62.6.2628-2632.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellamy W, Takase M, Yamauchi K, Wakabayashi H, Kawase K, Tomita M. Identification of the bactericidal domain of lactoferrin. Biochim Biophys Acta. 1992;1121:130–136. doi: 10.1016/0167-4838(92)90346-f. [DOI] [PubMed] [Google Scholar]
- 5.Brock J. Iron and cells of the immune system. In: de Souza M, Brock J, editors. Iron in immunity, cancer and inflammation. Chichester, England: John Wiley and Sons; 1986. pp. 81–98. [Google Scholar]
- 6.Cohen M S, Mao J, Rasmusen G T, Serody J S, Britigan B E. Interaction of lactoferrin and lipopolysaccharide: effects on the antioxidant property of lactoferrin and the ability of lipopolysaccharide to prime human neutrophils for enhanced superoxide anion. J Infect Dis. 1992;166:1375–1378. doi: 10.1093/infdis/166.6.1375. [DOI] [PubMed] [Google Scholar]
- 7.Crouch S M, Slater K J, Fletcher J. Regulation of cytokine release from mononuclear cells by the iron-binding protein lactoferrin. Blood. 1992;80:235–240. [PubMed] [Google Scholar]
- 8.Dentener M A, von Asmuth E J U, Fraucot G J M, Marra M N, Buurman W A. Antagonistic effects of lipopolysaccharide binding protein and bactericidal/increasing permeability protein on LPS induced cytokine release by mononuclear phagocytes. J Immunol. 1993;151:4258–4265. [PubMed] [Google Scholar]
- 9.Elass A, Vergoten G, Legrand D, Mazurier J, Elass-Rochard E, Spik G. Processes underlying interactions of human lactoferrin with the Jurkat human lymphoblastic T-cell line receptor. I. Quantitative structure affinity relationships studies. Quant Struct Act Relat. 1996;15:95–101. [Google Scholar]
- 10.Elass A, Vergoten G, Legrand D, Mazurier J, Elass-Rochard E, Spik G. Processes underlying interactions of human lactoferrin with the Jurkat human lymphoblastic T-cell line receptor. II. Comparative molecular field analysis. Quant Struct Act Relat. 1996;15:102–107. [Google Scholar]
- 11.Elass-Rochard E, Roseanu A, Legrand D, Trif M, Salmon V, Motas C, Montreuil J, Spik G. Lactoferrin-LPS interactions: involvement of the 28–34 loop region of human lactoferrin in the high-affinity binding to Escherichia coli 055:B5 lipopolysaccharides. Biochem J. 1995;312:839–845. doi: 10.1042/bj3120839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gazzano-Santoro H, Parent J B, Conlon P J, Kasler H G, Tsai C M, Lill-Elghanian D A, Hollingsworth R I. Characterization of the structural elements in lipid A required for binding of a recombinant fragment of bactericidal/permeability-increasing protein. Infect Immun. 1995;63:2201–2205. doi: 10.1128/iai.63.6.2201-2205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray P W, Flaggs G, Leong S R, Gumina R J, Weiss J, Ooi C E, Elsbach P. Cloning of the cDNA of a human neutrophil bactericidal protein-structure and functional correlations. J Biol Chem. 1989;264:9505–9509. [PubMed] [Google Scholar]
- 14.Han J, Mathison J, Ulevitch R, Tobias P S. Lipopolysaccharide binding protein truncated at Ile-197 binds LPS but does not transfer LPS to CD14. J Biol Chem. 1994;269:8172–8177. [PubMed] [Google Scholar]
- 15.Hoess A, Watson S, Siber G R, Liddington R. Crystal structure of an endotoxin-neutralizing protein from the horseshoe crab, Limulus anti-LPS factor, at 1.5Å resolution. EMBO J. 1993;12:3351–3356. doi: 10.1002/j.1460-2075.1993.tb06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirkland T N, Finley F, Leturcq D, Moriarty A, Lee J D, Ulevitch R J, Tobias P S. Analysis of lipopolysaccharide binding to CD14. J Biol Chem. 1993;268:24818–24823. [PubMed] [Google Scholar]
- 17.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Legrand D, Mazurier J, Elass A, Rochard E, Vergoten G, Maes P, Montreuil J, Spik G. Molecular interactions between human lactoferrin and the PHA-activated human lymphocyte lactoferrin receptor lie in the two loop containing regions of the N-terminal domain I of human lactoferrin. Biochemistry. 1992;31:9243–9251. doi: 10.1021/bi00153a018. [DOI] [PubMed] [Google Scholar]
- 19.Legrand D, Salmon V, Coddeville B, Benaïssa M, Plancke Y, Spik G. Structural determination of two N-linked glycans isolated from recombinant human lactoferrin expressed in BHK cells. FEBS Lett. 1995;365:57–60. doi: 10.1016/0014-5793(95)00441-b. [DOI] [PubMed] [Google Scholar]
- 20.Legrand D L, van Berkel P H C, Salmon V, van Veen H A, Slomianny M C, Nuijens J H, Spik G. The N-terminal Arg2, Arg3 and Arg4 of human lactoferrin interact with sulfated molecules but not with the receptor expressed in the Jurkat human lymphoblastic T-cells. Biochem J. 1997;327:841–846. doi: 10.1042/bj3270841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Little R G, Kelner D N, Lim E, Burke D J, Colon P J. Functional domains of recombinant bactericidal/increasing permeability protein (rBPI23) J Biol Chem. 1994;269:1865–1872. [PubMed] [Google Scholar]
- 22.Machnicki M, Zimecki M, Zagulski T. Lactoferrin regulates the release of tumor necrosis factor α, Il-6 in vivo. J Exp Pathol. 1993;74:433–439. [PMC free article] [PubMed] [Google Scholar]
- 23.Mann D M, Romm E, Migliorini M. Delineation of the glycosaminoglycan-binding site in the human inflammatory response protein lactoferrin. J Biol Chem. 1994;269:23661–23667. [PubMed] [Google Scholar]
- 24.Martin T R, Mathison J C, Tobias P S, Leturcq D J, Moriarty A M, Maunder R J, Ulevitch R J. Lipopolysaccharide-binding protein enhances the responsiveness of alveolar macrophages to bacterial lipopolysaccharide: implications for cytokine production in normal and injured lungs. J Clin Invest. 1992;90:2209–2219. doi: 10.1172/JCI116106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masson P L, Heremans J F, Schönne E. Lactoferrin an iron binding protein in neutrophilic leukocytes. J Exp Med. 1969;130:643–656. doi: 10.1084/jem.130.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathison J C, Tobias P S, Wolfson E, Ulevitch R J. Plasma LPS binding protein, a key component in macrophage recognition of gram (−) LPS. J Immunol. 1992;194:200–206. [PubMed] [Google Scholar]
- 27.Mattsby-Baltzer I, Roseanu A, Motas C, Elverfors J, Engberg I, Hanson L A. Lactoferrin or a fragment thereof inhibits the endotoxin-induced interleukin 6 response in human monocytic cells. Pediatr Res. 1996;40:257–261. doi: 10.1203/00006450-199608000-00011. [DOI] [PubMed] [Google Scholar]
- 28.Mazurier J, Legrand D, Hu W L, Montreuil J, Spik G. Expression of human lactoferrin receptors in PHA-stimulated human peripheral blood lymphocytes: isolation of the receptors by anti-ligand affinity chromatography. Eur J Biochem. 1989;179:481–487. doi: 10.1111/j.1432-1033.1989.tb14578.x. [DOI] [PubMed] [Google Scholar]
- 29.Mazurier J, Spik G. Comparative study of the iron-binding properties of human transferrins. Complete and sequential iron saturation and desaturation of the lactotransferrin. Biochim Biophys Acta. 1980;629:399–408. doi: 10.1016/0304-4165(80)90112-9. [DOI] [PubMed] [Google Scholar]
- 30.Montreuil J, Tonnelat J, Mullet S. Préparation et propriétés de la lactotransferrine du lait de femme. Biochim Biophys Acta. 1960;45:413–421. doi: 10.1016/0006-3002(60)91478-5. [DOI] [PubMed] [Google Scholar]
- 31.Morrison D C, Jacobs D M. Binding of polymyxin B to the lipid A portion of bacterial lipopolysaccharide. Immunochemistry. 1976;13:813–818. doi: 10.1016/0019-2791(76)90181-6. [DOI] [PubMed] [Google Scholar]
- 32.Muta T, Miyata T, Tokunaga F, Nakamura T, Iwanaga S. Primary structure of anti-LPS factor from American horseshoe crab, Limulus polyphenus. J Biochem. 1987;101:1321–1330. doi: 10.1093/oxfordjournals.jbchem.a121999. [DOI] [PubMed] [Google Scholar]
- 33.Ohno N, Morrison D C. Lipopolysaccharide interaction with lysozyme. J Biol Chem. 1989;264:4434–4441. [PubMed] [Google Scholar]
- 34.Parillo J E. Pathogenetic mechanisms of septic shock. N Engl J Med. 1993;328:1471–1477. doi: 10.1056/NEJM199305203282008. [DOI] [PubMed] [Google Scholar]
- 35.Salmon V, Legrand D, Georges B, Slomianny M C, Coddeville B, Spik G. Characterization of human lactoferrin produced in the baculovirus expression system. Protein Express Purif. 1996;9:203–210. doi: 10.1006/prep.1996.0687. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez L, Calvo M, Brock J H. Biological role of lactoferrin. Arch Dis Child. 1992;67:657–661. doi: 10.1136/adc.67.5.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schumann R R, Leong S R, Flaggs G W, Gray P W, Wright S D, Mathison J C, Tobias P S, Ulevitch R J. Structure and function of LPS-binding protein. Science. 1990;249:1429–1433. doi: 10.1126/science.2402637. [DOI] [PubMed] [Google Scholar]
- 38.Spik G, Strecker G, Fournet B, Bouquelet S, Montreuil J, Dorland L, van Halbeek H, Vliegenthart J F G. Primary structure of the glycans from human lactotransferrin. Eur J Biochem. 1982;121:413–419. doi: 10.1111/j.1432-1033.1982.tb05803.x. [DOI] [PubMed] [Google Scholar]
- 39.Taylor A H, Heauner G, Nedelman M, Sherris D, Brunt E, Knight D, Ghrayeb J. Lipopolysaccharide neutralizing peptides reveal a lipid A binding site of LPS binding protein. J Biol Chem. 1995;270:17934–17938. doi: 10.1074/jbc.270.30.17934. [DOI] [PubMed] [Google Scholar]
- 40.Theofan G, Horwitz A H, Williams R E, Liu P S, Chan I, Birr C, Carroll S F, Meszaros K, Parent J B, Kasler H, Aberle S, Trown P W, Gazzano-Santoro H. An amino-terminal fragment of human LPS-binding protein retains lipid A binding but not CD14-stimulatory activity. J Immunol. 1994;152:3623–3629. [PubMed] [Google Scholar]
- 41.Tobias P S, Mathison J C, Ulevitch R J. A family of LPS-binding proteins involved in responses to gram (−) sepsis. J Biol Chem. 1988;263:13479–13481. [PubMed] [Google Scholar]
- 42.Tobias P S, Soldau K, Ulevitch R J. Isolation of a LPS binding acute phase reactant from rabbit serum. J Exp Med. 1986;164:777–793. doi: 10.1084/jem.164.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tobias P S, Soldau K, Ulevitch R J. Identification of a lipid A binding site in the acute phase reactant LPS binding protein. J Biol Chem. 1989;264:10867–10871. [PubMed] [Google Scholar]
- 44.Tomita M, Takase M, Bellamy W, Shimamura S. A review: the active peptide of lactoferrin. Acta Paed Japonica. 1994;36:585–591. doi: 10.1111/j.1442-200x.1994.tb03250.x. [DOI] [PubMed] [Google Scholar]
- 45.Trumpler U P, Straub W, Rosenmund A. Antibacterial prophylaxis with lactoferrin in neutropaenic patients. Eur J Clin Microbiol Infect Dis. 1989;8:310–318. doi: 10.1007/BF01963459. [DOI] [PubMed] [Google Scholar]
- 46.Ulevitch R J. Recognition of bacterial endotoxins by receptor-dependent mechanisms. Adv Immunol. 1993;53:267–289. doi: 10.1016/s0065-2776(08)60502-7. [DOI] [PubMed] [Google Scholar]
- 47.Ulevitch R J, Tobias P S. Recognition of endotoxin by cells leading to transmembrane signalling. Curr Opin Immunol. 1994;6:125–130. doi: 10.1016/0952-7915(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 48.Ulevitch R J, Tobias P S. Receptor-dependent mechanisms of cell stimulation by bacterial endotoxin. Annu Rev Immunol. 1995;13:437–457. doi: 10.1146/annurev.iy.13.040195.002253. [DOI] [PubMed] [Google Scholar]
- 49.Vey E, Zhang D E, Dayer J M. IFN γ and 1,25 (OH)2VitD3 induce on THP-1 cells distinct patterns of cell surface antigen expression. Cytokine production and responsiveness to contact with activated T cells. J Immunol. 1992;149:2040–2046. [PubMed] [Google Scholar]
- 50.Wang D, Pabst K M, Aida Y, Pabst M J. Lipopolysaccharide-inactivating activity of neutrophils is due to lactoferrin. J Leukocyte Biol. 1995;57:865–874. doi: 10.1002/jlb.57.6.865. [DOI] [PubMed] [Google Scholar]
- 51.Wright S D, Ramos R A, Tobias P S, Ulevitch R J, Mathison J C. CD14 serves as the cellular receptor for complexes of LPS with LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 52.Zagulski T, Lipinski P, Zagulska A, Broniek S, Jarzabek Z. Lactoferrin can protect mice against a lethal dose of Escherichia coli in experimental infection in vivo. Br J Exp Pathol. 1989;70:697–704. [PMC free article] [PubMed] [Google Scholar]
- 53.Zimecki M, Mazurier J, Machnicki M, Wieczorek Z, Montreuil J, Spik G. Immunostimulatory activity of lactoferrin and maturation of CD4(−)-CD8(−) murine thymocytes. Immunol Lett. 1991;30:377–382. doi: 10.1016/0165-2478(91)90099-v. [DOI] [PubMed] [Google Scholar]