ABSTRACT
We describe the genome of a lytic phage EKq1 isolated on Klebsiella quasipneumoniae, with activity against Klebsiella pneumoniae. EKq1 is an unclassified representative of the class Caudoviricetes, similar to Klebsiella phages VLCpiS8c, phiKp_7-2, and vB_KleS-HSE3. The 48,244-bp genome has a GC content of 56.43% and 63 predicted protein-coding genes.
KEYWORDS: Klebsiella quasipneumoniae, Klebsiella pneumoniae, phage EKq1, complete genome sequence, class Caudoviricetes, unclassified Caudoviricetes, lytic phage, therapeutic candidate
ANNOUNCEMENT
Phages are used as alternative or adjunct antibacterials against multidrug-resistant (MDR) Klebsiella pneumoniae infections (1 – 3). K. pneumoniae phages usually have limited host ranges (4), but phages with broader activity can be isolated on near-neighbor species (5, 6). Here, we describe the genome of phage EKq1 isolated on Klebsiella quasipneumoniae and capable of lysing some MDR K. pneumoniae clinical isolates.
EKq1 was isolated from sewage collected on 16 March 2022, in Montgomery County, Maryland, using a human blood isolate, K. quasipneumoniae MRSN 829456, for phage enrichment. The enrichment was performed as described (7), in broth with shaking at 37°C, and phage plaques were detected on double-layer agar plates. The phage was purified by three single plaque isolations, and its DNA was extracted from lysate with the QIAamp DNA Mini Kit (Qiagen, Germantown, MD), per the manufacturer’s protocol. A library was constructed using the KAPA HyperPlus Kit (Roche Diagnostics, Indianapolis, IN) and sequenced on an Illumina MiSeq (Illumina, San Diego, CA) with a 600-cycle MiSeq Reagent Kit v3 that produced 300-bp paired-end reads. Paired-end sequences (1,864,294 reads total) were assessed for quality using FastQC 0.11.9 (8) and trimmed with Trimmomatic (9) v0.39. Phage EKq1 genome was assembled de novo using Unicycler 0.4.8 (10), its termini and DNA packaging mechanism were determined using PhageTerm (11), and lifestyle was predicted using BACPHLIP (12). Protein-coding sequences (CDSs) were annotated using the Pharokka pipeline (13 – 23). Amino acid sequence similarity searches were performed in Diamond (24, 25) against the nr database downloaded in January 2023. All tools were run with default parameters.
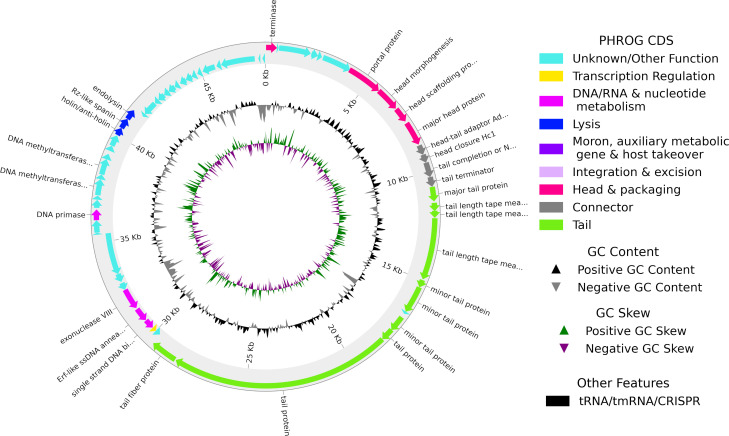
The average read coverage was 977×; EKq1 genome was 48,244 bp long, with G + C content of 56.43%, contained 63 predicted CDSs (Fig. 1) and direct terminal repeats of 8,798 bp. Mash alignment (23) to the INPHARED database (22) placed EKq1 among Klebsiella siphophages currently classified as class Caudoviricetes, with highest DNA identity, >98%, to VLCpiS8c (GenBank ON602734), phiKp_7-2 (LC768468), and vB_KleS-HSE3 (26; MT075871). Phage vB_KleS-HSE3 was lytic against one of four tested MDR K. pneumoniae strains (26). Nucleotide identity higher than 95% indicates that phages EKq1, VLCpiS8c, phiKp_7-2, and vB_KleS-HSE3 belong to the same species (27). These phages contain a putative Cas4-like exonuclease. A similar exonuclease in Campylobacter phages stimulated acquisition of host-derived spacers by the bacterial CRISPR-Cas system that might be a decoy to prevent phage DNA acquisition and, therefore, an anti-CRISPR measure (28).
Fig 1.
Genome organization of EKq1.
BACPHLIP scored EKq1 genome at 89%, while the threshold for high-confidence lytic lifestyle is 95% (12). Additionally, nucleotide BLAST search (29) against the nr database found a region of EKq1 DNA with 78%–89% identity to bacterial chromosomes, within prophage genes (e.g., MKK01_09025 and MKK01_09030 in Klebsiella variicola, GenBank CP092632). However, these genes encode a carbohydrate-binding domain protein and a tail fiber protein and have no relation to lysogenicity. EKq1 putative proteins showed no homology to products related to lysogenic lifestyle, gene transfer, and bacterial proteins including antibiotic resistance determinants (18) and virulence factors (19). Thus, EKq1 appears to be a lytic phage and a candidate for therapeutic use.
ACKNOWLEDGMENTS
We thank Yunxiu He and Caleb Jones for their technical assistance.
Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the authors and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense.
This study was supported by the Military Infectious Diseases Research Program, grant MI210045. The Multidrug-Resistant Organism Repository and Surveillance Network (MRSN) at the Walter Reed Army Institute of Research provided the strain K. quasipneumoniae MRSN 829456 used for phage isolation and K. pneumoniae clinical isolates for phage host range testing.
Contributor Information
Andrey A. Filippov, Email: andrey.a.filippov.ctr@health.mil.
Simon Roux, DOE Joint Genome Institute, Berkeley, California, USA .
DATA AVAILABILITY
The EKq1 genome BioProject, BioSample, GenBank, and the NCBI Sequence Read Archive accession numbers are PRJNA1016341, SAMN37380043, OR555718, and SRR26063729, respectively.
REFERENCES
- 1. Herridge WP, Shibu P, O’Shea J, Brook TC, Hoyles L. 2020. Bacteriophages of Klebsiella spp., their diversity and potential therapeutic uses. J Med Microbiol 69:176–194. doi: 10.1099/jmm.0.001141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eskenazi A, Lood C, Wubbolts J, Hites M, Balarjishvili N, Leshkasheli L, Askilashvili L, Kvachadze L, van Noort V, Wagemans J, Jayankura M, Chanishvili N, de Boer M, Nibbering P, Kutateladze M, Lavigne R, Merabishvili M, Pirnay J-P. 2022. Combination of pre-adapted bacteriophage therapy and antibiotics for treatment of fracture-related infection due to pandrug-resistant Klebsiella pneumoniae. Nat Commun 13:302. doi: 10.1038/s41467-021-27656-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Doub JB, Shishido A, Srikumaran U, Haskoor J, Tran-Nguyen P, Lee M, Würstle S, Lee A, Kortright K, Chan BK. 2022. Salphage: salvage bacteriophage therapy for a recalcitrant Klebsiella pneumoniae prosthetic shoulder infection - a case report. Acta Orthop 93:756–759. doi: 10.2340/17453674.2022.4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kęsik-Szeloch A, Drulis-Kawa Z, Weber-Dąbrowska B, Kassner J, Majkowska-Skrobek G, Augustyniak D, Lusiak-Szelachowska M, Zaczek M, Górski A, Kropinski AM. 2013. Characterising the biology of novel lytic bacteriophages infecting multidrug resistant Klebsiella pneumoniae. Virol J 10:100. doi: 10.1186/1743-422X-10-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jensen EC, Schrader HS, Rieland B, Thompson TL, Lee KW, Nickerson KW, Kokjohn TA. 1998. Prevalence of broad-host-range lytic bacteriophages of Sphaerotilus natans, Escherichia coli, and Pseudomonas aeruginosa. Appl Environ Microbiol 64:575–580. doi: 10.1128/AEM.64.2.575-580.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu L-T, Chang S-Y, Yen M-R, Yang T-C, Tseng Y-H. 2007. Characterization of extended-host-range pseudo-T-even bacteriophage Kpp95 isolated on Klebsiella pneumoniae. Appl Environ Microbiol 73:2532–2540. doi: 10.1128/AEM.02113-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mencke JL, He Y, Filippov AA, Nikolich MP, Belew AT, Fouts DE, McGann PT, Swierczewski BE, Getnet D, Ellison DW, Margulieux KR. 2022. Identification and characterization of vB_PreP_Epr2, a lytic bacteriophage of pan-drug resistant Providencia rettgeri. Viruses 14:708. doi: 10.3390/v14040708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- 9. Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garneau JR, Depardieu F, Fortier L-C, Bikard D, Monot M. 2017. PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci Rep 7:8292. doi: 10.1038/s41598-017-07910-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hockenberry AJ, Wilke CO. 2021. BACPHLIP: predicting bacteriophage lifestyle from conserved protein domains. PeerJ 9:e11396. doi: 10.7717/peerj.11396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bouras G, Nepal R, Houtak G, Psaltis AJ, Wormald P-J, Vreugde S. 2023. Pharokka: a fast scalable bacteriophage annotation tool. Bioinformatics 39:btac776. doi: 10.1093/bioinformatics/btac776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16. doi: 10.1093/nar/gkh152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, Hugenholtz P. 2007. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 8:209. doi: 10.1186/1471-2105-8-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Steinegger M, Söding J. 2017. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol 35:1026–1028. doi: 10.1038/nbt.3988 [DOI] [PubMed] [Google Scholar]
- 17. McNair K, Zhou C, Dinsdale EA, Souza B, Edwards RA. 2019. PHANOTATE: a novel approach to gene identification in phage genomes. Bioinformatics 35:4537–4542. doi: 10.1093/bioinformatics/btz265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen A-L, Cheng AA, Liu S, et al. 2020. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48:D517–D525. doi: 10.1093/nar/gkz935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, Jin Q. 2005. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 33:D325–8. doi: 10.1093/nar/gki008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chan PP, Lin BY, Mak AJ, Lowe TM. 2021. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res 49:9077–9096. doi: 10.1093/nar/gkab688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Terzian P, Olo Ndela E, Galiez C, Lossouarn J, Pérez Bucio RE, Mom R, Toussaint A, Petit M-A, Enault F. 2021. PHROG: families of prokaryotic virus proteins clustered using remote homology. NAR Genom Bioinform 3:lqab067. doi: 10.1093/nargab/lqab067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cook R, Brown N, Redgwell T, Rihtman B, Barnes M, Clokie M, Stekel DJ, Hobman J, Jones MA, Millard A. 2021. INfrastructure for a PHAge REFerence database: identification of large-scale biases in the current collection of cultured phage genomes. Phage (New Rochelle) 2:214–223. doi: 10.1089/phage.2021.0007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17:132. doi: 10.1186/s13059-016-0997-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- 25. Buchfink B, Reuter K, Drost H-G. 2021. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods 18:366–368. doi: 10.1038/s41592-021-01101-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peng Q, Fang M, Liu X, Zhang C, Liu Y, Yuan Y. 2020. Isolation and characterization of a novel phage for controlling multidrug-resistant Klebsiella pneumoniae. Microorganisms 8:542. doi: 10.3390/microorganisms8040542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adriaenssens E, Brister JR. 2017. How to name and classify your phage: an informal guide. Viruses 9:70. doi: 10.3390/v9040070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hooton SPT, Connerton IF. 2014. Campylobacter jejuni acquire new host-derived CRISPR spacers when in association with bacteriophages harboring a CRISPR-like Cas4 protein. Front Microbiol 5:744. doi: 10.3389/fmicb.2014.00744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The EKq1 genome BioProject, BioSample, GenBank, and the NCBI Sequence Read Archive accession numbers are PRJNA1016341, SAMN37380043, OR555718, and SRR26063729, respectively.