Extended Data Figure 9. Cell type-aware candidate variants alter reporter expression in vivo.
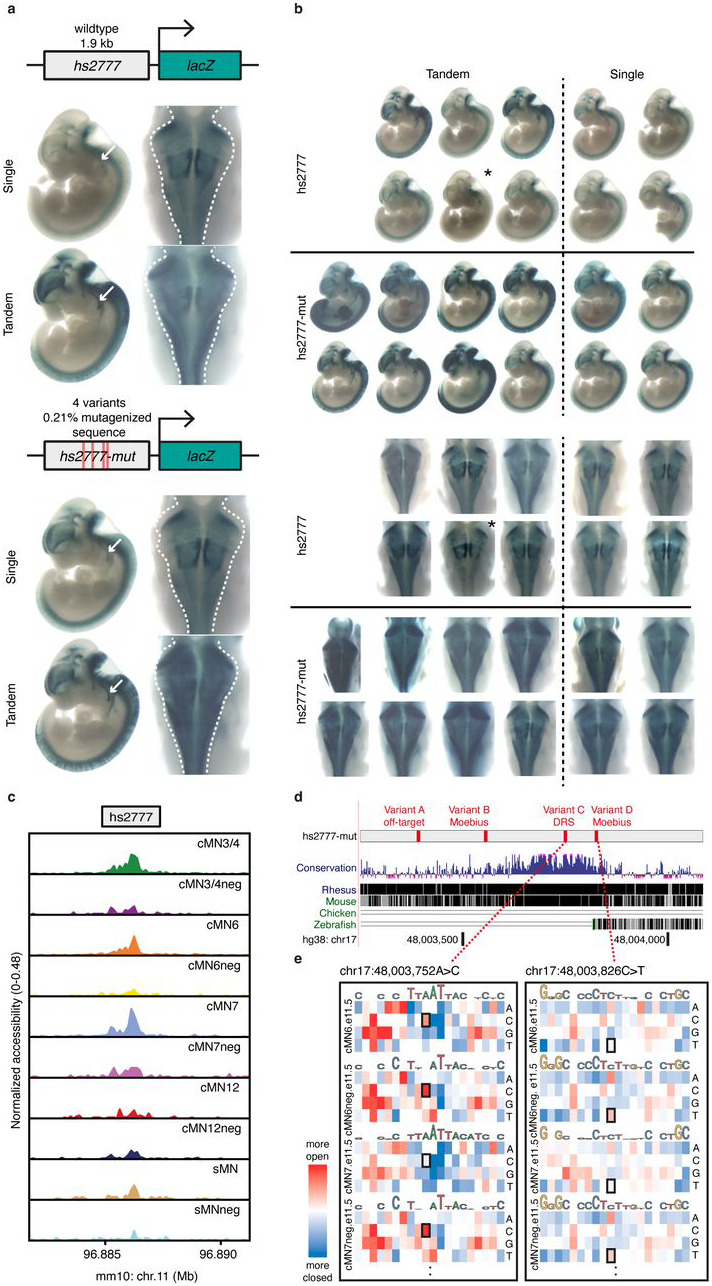
a. Representative whole mount in vivo enhancer reporter expression for (top) hs2777 wildtype and (bottom) hs2777-mut enhancer constructs. For each reporter insertion, dosage is labelled (“single”, “tandem”). Reporter expression views are shown as lateral (left) and dorsal through the 4th ventricle (right). Cranial nerve 7 (white arrows) and surrounding hindbrain tissue (dashed lines) show visible gain of reporter expression.
b. Additional replicates as in (a), matched by injection batch (top and bottom). hs2777-mut constructs reproducibly show increased reporter expression across midbrain, hindbrain, and neural tube. Random insertions are denoted by an asterisk.
c. hs2777 chromatin accessibility profiles in the cranial motor neurons and surrounding cell types. The wildtype element is accessible across multiple cMNs and surrounding cells.
d. UCSC screenshot depicting location of hs2777-mut variants: “Variant A” (chr17:48003393G>A, off-target), “Variant B” (chr17:48003557C>G, Moebius), “Variant C” (chr17:48003752A>C, DRS), and “Variant D” (chr17:48003826C>T, Moebius). hs2777-mut overlaps conserved non-coding sequence, particularly for Variants C and D.
e. Neural net-trained in silico saturation mutagenesis predictions for all possible nucleotide changes in hs2777 for selected samples cMN6 e11.5, cMN6neg e11.5, cMN7 e11.5, and cMN7neg e11.5. Predicted loss-of-function nucleotide changes are colored in blue and gain-of-function in red. Specific nucleotide changes corresponding to in vivo Variants C and D are boxed. Samples marked with “neg” are GFP-negative cells surrounding the motor neurons of interest. All other samples are GFP-positive motor neurons. Variants C and D are predicted to increase accessibility in relevant samples consistent with their corresponding phenotypes; DRS alters cMN6 but not cMN7 development (Variant C), while MBS alters both (Variant D).
