Figure 4. Exceptional gene regulation of cranial motor neuron master regulator Isl1.
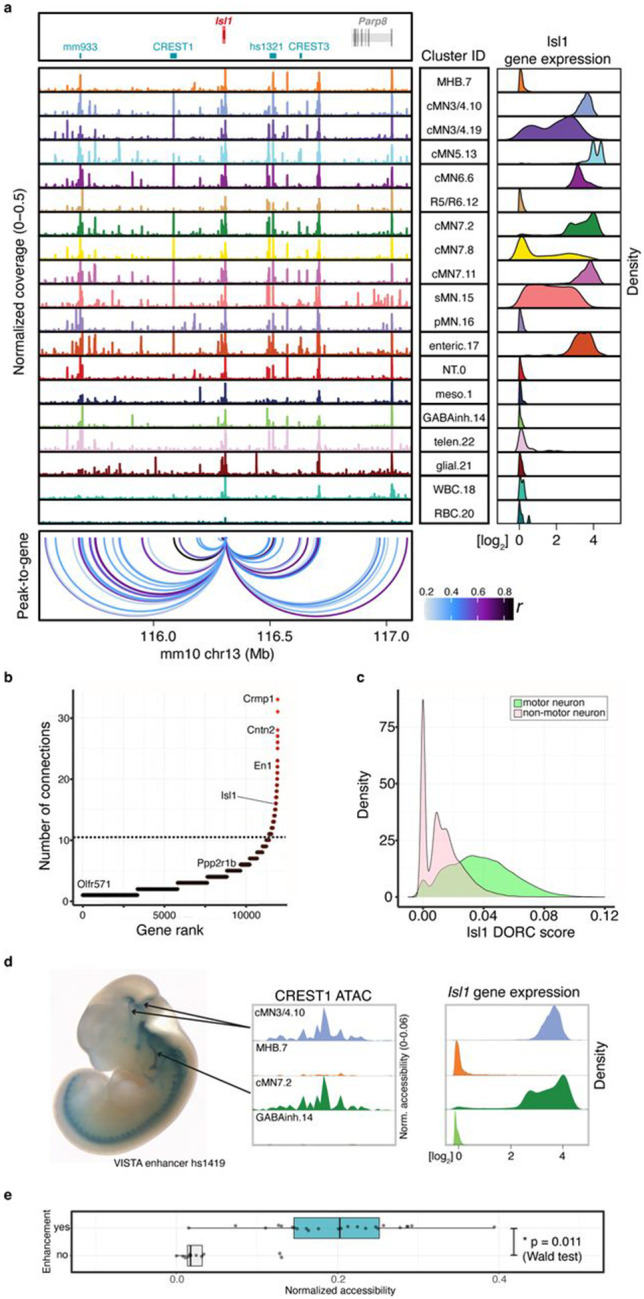
a. Pseudobulked chromatin accessibility profiles for all annotated clusters over a 1.5 Mb window centered about Isl1. Imputed gene expression profiles for each cluster are shown to the right. Isl1 is located within a gene desert with the nearest up- and downstream flanking genes 1.2 and 0.7 Mb away, respectively. Peak-to-gene predictions match known Isl1 enhancers (CREST1 in motor neurons and CREST3 in sensory neurons75; mm933 in multiple cranial motor nerves, dorsal root ganglion, and nose; hs1321 in multiple cranial motor nerves and forebrain) and identify additional putative enhancers surrounding Isl1.
b. The number of normalized regulatory connections for each rank ordered gene. Isl1 ranks in the top 1% of all genes with at least one regulatory connection. The inflection point of the plotted function is demarcated with a dotted line.
c. Per-cell Domain of Regulatory Chromatin (DORC) scores for Isl1 gene. DORC scores are significantly higher for cells from motor neuron clusters relative to non-motor neuron clusters (p-value < 1 x 10−15, ANOVA).
d. (Left) Lateral whole mount In vivo reporter assay testing CREST1 (VISTA enhancer hs1419) enhancer activity. CREST1 drives expression in cranial nerves 3, 4, and 7 (black lines; there is also expression in trigeminal motor nerve). (Right) Single cell ATAC profiles and imputed gene expression for a subset of corresponding clusters. CREST1 accessibility and Isl1 gene expression are positively correlated with in vivo expression patterns.
e. Boxplot depicting normalized accessibility levels for enhancers CREST1, CREST3, mm933, and hs1321 within nine scATAC clusters corresponding to distinct anatomic regions. Manually scored enhancer activity (“enhancement”) is significantly correlated with normalized accessibility (p-value = 0.011, Wald test). Center line: median; box limits: upper and lower quartiles; whiskers – 1.5 x interquartile range.
