Abstract
Kawasaki disease (KD) is a pediatric vasculitis caused by an unknown trigger in genetically susceptible children. The incidence varies widely across genetically diverse populations. Several associations with HLA Class I alleles have been reported in single cohort studies. Using a genetic approach, from the nine single nucleotide variants (SNVs) associated with KD susceptibility in children of European descent, we identified SNVs near the HLA-C (rs6906846) and HLA-B genes (rs2254556) whose association was replicated in a Japanese descent cohort (rs6906846 p=0.01, rs2254556 p=0.005). The risk allele (A at rs6906846) was also associated with HLA-C*07:02 and HLA-C*04:01 in both US multi-ethnic and Japanese cohorts and HLA-C*12:02 only in the Japanese cohort. The risk A-allele was associated with eight non-conservative amino acid substitutions (amino acid positions); Asp or Ser (9), Arg (14), Ala (49), Ala (73), Ala (90), Arg (97), Phe or Ser (99), and Phe or Ser(116) in the HLA-C peptide binding groove that binds peptides for presentation to cytotoxic T cells (CTL). This raises the possibility of increased affinity to a “KD peptide” that contributes to the vasculitis of KD in genetically susceptible children.
Keywords: Kawasaki disease, HLA-C, antigen presentation, amino acid substitution, cytotoxic T cells
Introduction
HLA class I molecules present peptide to CD8+ cytotoxic T-cells (CTL) that screen damaged cells and play a central role in vascular pathology in Kawasaki disease (KD), an acute pediatric vasculitis [1, 2]. Each HLA type has specific amino acid sequences in the peptide binding groove and single amino acid substitutions at critical positions for peptides and/or T cell receptors are known to influence disease susceptibility and response to antigens [3–5]. CTL infiltrate the arterial wall in KD vasculitis and may recognize peptides derived from the disease “trigger”. Therefore, finding specific HLA types that are associated with KD susceptibility and that share single amino acid substitutions at critical positions in the peptide-binding groove may contribute to defining the trigger for KD.
Although association of single nucleotide variants (SNVs) in the HLA region with KD susceptibility has been reported [6–12], only a SNV in the HLA class II region (rs2857151) discovered by a Japanese genome-wide association study (GWAS) [9] was successfully replicated in both Chinese and European descent cohorts [10, 13]. We previously performed a pathway analysis using European descent GWAS data and reported several genetic variants associated with KD susceptibility in the HLA class I region [14]. In the current study, we focused on the subset of KD risk-associated SNVs that were also validated in a Japanese cohort. We postulated that risk-associated SNVs are associated with certain HLA types that share amino acid substitutions in the peptide binding domain resulting in preferential peptide binding to the antigen trigger of KD.
Materials and Methods
Patient enrollment and sample collection
The patient cohorts are described in the study flow diagram (Fig.1). Diagnosis of KD was made according to American Heart Association criteria as previously reported [15]. Detailed information of the subjects for European descent Cohort for GWAS was previously described [16]. Multi-ethnic Cohort 1 (n=78, enrolled during 2002–2014) was enrolled at UCSD and enriched for carriers of the risk allele at rs6906846 (HLA-C) and/or rs2254556 (HLA-B) in order to analyze the relationship between SNVs, high-resolution HLA typing results, and HLA amino acid variation. Cohort 2 contained Japanese KD subjects (n=275) who were a subset of the published Japanese GWAS [9] and were typed for HLA Class I at high resolution. Demographic, clinical, and laboratory data were summarized for Cohort 1 (Supplementary Table 1) but were not available for Japanese Cohort 2. The Institutional Review Boards of the participating centers (UCSD and RIKEN) reviewed and approved this study and parental consent and assent as appropriate were obtained from parents and participants.
Fig 1. Study design.
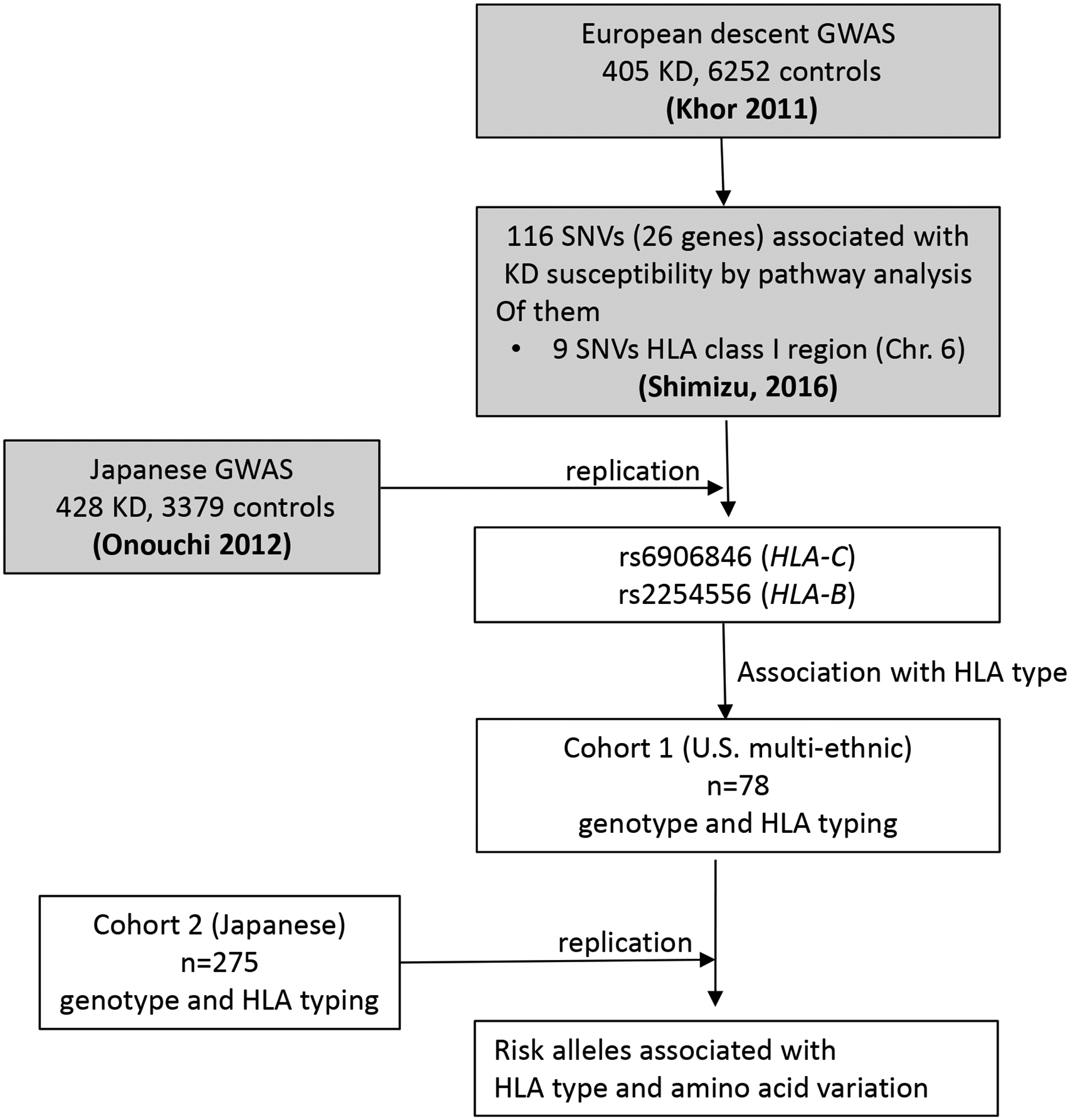
Summary of workflow including GWAS (Khor 2011 and Onouchi 2012), pathway analysis, gene stability selection (Shimizu 2016), and high-resolution HLA typing. GWAS: Genome-wide association study, SNV: single nucleotide variant, HLA: Human leukocyte antigen, Shadowed box: previously published analysis, Open box: analysis in this study.
Genotyping
For Cohort 1, DNA was collected from either whole blood or mouthwash samples as previously described [16] and genotyping for rs6906846 (HLA-C) and rs2254556 (HLA-B) was performed using TaqMan SNP genotype assays following the manufacturer’s instructions (Life Technologies, USA).
Pathway analysis and gene stability selection
The pathway analysis followed by gene stability selection to find the responsible SNVs/genes driving the pathway association using European descent GWAS data (405 KD subjects and 6252 controls) [16] was previously reported [14]. From this analysis, 116 SNVs in 26 genes were selected as drivers of the top 100 pathways associated with KD (Fig 1). In the current study, we focused on the nine SNVs in the HLA class I region. Since no other European descent GWAS dataset was available for replication, each SNV was evaluated for association with KD susceptibility (t-test, p<0.05) in a published Japanese GWAS dataset (428 KD subjects and 3379 controls) [9]. Meta-analysis was performed with PLINK.
High-resolution HLA typing and evaluation of amino acid variation
High-resolution typing for HLA-B and C was performed on Cohort 1 (mixed ethnicity). The sequence-specific oligonucleotide probe (SSO) method, sequence-specific primer method, and/or Sanger sequence-based typing methods were used. HLA-B and HLA-C high resolution typing for Japanese Cohort 2 was conducted by the SSO method using the WAKFlow (R) HLA Typing kit (Wakunaga Pharmaceutical) according to the manufacturer’s instructions.
Linkage between SNVs and HLA high-resolution types or amino acid sequence was identified in KD subjects who were homozygous for each SNV. The amino acid sequence encoded by HLA class I genes was aligned using the IMGT/HLA database (http://www.ebi.ac.uk/ipd/imgt/hla/).
Association between SNVs (rs6906846 (HLA-C) and rs2254556 (HLA-B)) and high resolution HLA types and amino acid sequence
Linkage between SNVs and HLA high-resolution types or amino acid sequences was restricted to KD subjects who were homozygous for each SNV since we were unable to assign SNV alleles and HLA sequences to the same chromosome. The amino acid sequence encoded by HLA class I genes was aligned using the IMGT/HLA database (http://www.ebi.ac.uk/ipd/imgt/hla/). For Cohorts 1 and 2, association between alleles and each HLA-type or amino acid sequence was calculated using the Chi-square test.
Statistical analysis:
Association between genetic variants and clinical parameters was performed using non-parametric tests due to the non-normal distribution. P-values were calculated by Mann–Whitney U test for continuous variables and Chi square test or Fisher’s exact test for categorical variables. For the comparison of ≥ 3 variants, the Kruskal–Wallis test was used.
Results
Identification of KD-associated SNVs in the HLA region
Of the 116 SNVs in 26 genes recently published as potential drivers of the top 100 pathways associated with KD susceptibility in a European GWAS dataset [14] (Fig 1), nine SNVs were located within a 100 kb stretch in the HLA class I region (6p21.33) (association p<0.05, Supplementary Fig 1). Five SNVs clustered upstream or downstream of the HLA-C gene, while the other four SNVs clustered upstream of the HLA-B gene. Since there was no European GWAS data with which to validate these results, we used a previously published Japanese GWAS dataset [9] to test the association with KD susceptibility. Association of two of the nine SNVs (rs6906846 and rs2254556, Linkage disequilibrium (LD): r2<0.2) was replicated (428 KD subjects and 3,379 controls; p=1.1xE-02 for rs6906846 and p=4.9xE-03 for rs2254556) (Table 1 and Supplementary Table 2). Meta-analysis confirmed significant association for these SNVs (p=7.5xE-05 and OR 1.2 for rs6906846; p=3.0xE-04 and OR1.3 for rs2254556). Risk allele frequencies of these SNVs in different populations were as follows: rs6906846 (A allele): Hispanic 0.32, Asian 0.26, and European 0.31 and rs2254556 (T allele): Hispanic 0.08, Asian 0.07, and European 0.16. Although two additional alleles (rs2596551 and rs2523535) were significantly associated in the meta-analysis, genotyping of the 78 individuals at these loci yielded only three homozygotes. Therefore, we confined our further analysis to the alleles that were individually associated in each cohort at p<0.05.
Table 1.
HLA class I SNVs discovered in a pathway/gene stability analysis on a European descent GWAS followed by validation in a Japanese GWAS dataset
| SNV | A1 | European descent GWAS (405 KD and 6252 controls) | Japanese GWAS (428 KD and 3379 controls) | Meta-analysis | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KD | Control | OR | p | KD | Control | OR | p | OR | p | ||||||||||
| Number of alleles | AF | Number of alleles | AF | Number of alleles | AF | Number of alleles | AF | ||||||||||||
| A1 | A2 | A1 | A2 | A1 | A2 | A1 | A2 | ||||||||||||
| rs2844623 | T | 107 | 701 | 0.13 | 2315 | 10065 | 0.19 | 0.7 | 1.0E-04 | 213 | 641 | 0.25 | 1519 | 5211 | 0.23 | 1.1 | 1.1E-01 | 0.9 | 2.4E-01 |
| rs2524099 | A | 350 | 460 | 0.43 | 5917 | 6517 | 0.48 | 0.8 | 1.6E-02 | 562 | 294 | 0.66 | 4226 | 2532 | 0.63 | 1.1 | 7.5E-02 | 1.0 | 6.1E-01 |
| rs2524070 | A | 111 | 699 | 0.14 | 2338 | 10096 | 0.19 | 0.7 | 2.9E-04 | 75 | 781 | 0.09 | 577 | 6173 | 0.09 | 1.0 | 8.4E-01 | 0.8 | 7.8E-03 |
| rs6906846 | A | 271 | 539 | 0.33 | 3536 | 8898 | 0.28 | 1.3 | 2.2E-03 | 280 | 576 | 0.33 | 1927 | 4831 | 0.29 | 1.2 | 1.1E-02 | 1.2 | 7.5E-05 |
| rs7382297 | T | 152 | 658 | 0.19 | 1936 | 10490 | 0.16 | 1.3 | 1.6E-02 | 112 | 744 | 0.13 | 816 | 5942 | 0.12 | 1.1 | 3.9E-01 | 1.2 | 1.8E-02 |
| rs2596551 | C | 116 | 694 | 0.14 | 2480 | 9954 | 0.20 | 0.7 | 9.3E-05 | 46 | 808 | 0.05 | 480 | 6268 | 0.07 | 0.7 | 6.2E-02 | 0.7 | 1.9E-05 |
| rs2844575 | C | 342 | 464 | 0.42 | 5728 | 6592 | 0.46 | 0.8 | 2.5E-02 | 438 | 418 | 0.51 | 3495 | 3253 | 0.52 | 1.0 | 7.3E-01 | 0.9 | 6.9E-02 |
| rs2523535 | G | 293 | 517 | 0.36 | 5322 | 7112 | 0.43 | 0.8 | 2.2E-04 | 98 | 758 | 0.11 | 926 | 5832 | 0.14 | 0.8 | 7.2E-02 | 0.8 | 4.5E-05 |
| rs2254556 | T | 172 | 636 | 0.21 | 2210 | 10202 | 0.18 | 1.2 | 1.3E-02 | 65 | 791 | 0.08 | 357 | 6401 | 0.05 | 1.5 | 4.9E-03 | 1.3 | 3.3E-04 |
p: nominal p-value, A1: Reported in forward orientation to genome, Bonferroni corrected p value threshold: 0.05/(9 SNVs in HLA-B and C region)=5.0E-03, Bold indicates SNVs with p<0.05 in both European descent and Japanese GWAS, AF: A1 allele frequencies, OR: odds ratio
Association of SNVs with HLA-B and C alleles
The antigen presenting site of the HLA class I molecule is on the α1 and α2 domains and is comprised of ~180 amino acids [17]. Amino acid variation in HLA molecules that affects peptide binding affinity influences the peptide repertoire presented to T cells and the immunogenicity of the HLA-peptide complex [4, 18, 19]. We postulated that the risk-associated SNVs (rs2254556 and rs690846) could influence the polymorphic class I HLA amino acid sequences in the peptide binding groove, which, in turn, affects CTL recognition. Therefore, we first studied the association between the two SNVs (rs6906846 and rs2254556) and high-resolution HLA-B and HLA-C types, using only homozygous subjects in U.S. Cohort 1 (rs6906846: AA n=24, GG n=29 and rs2254556: AA n=4, GG n=54) since we did not have the sequence for each chromatid.
The A-allele (risk) of rs6906846 was associated with certain HLA types, including HLA-C*07:02 (p=2.8E-10) and HLA-C*04:01 (p=8.6E-09) in the US multi-ethnic cohort (Table 2 and Supplementary Table 3). This same association was replicated using only homozygous subjects in the Japanese cohort (rs6906846: AA n=23, GG n=125). Again, HLA-C*07:02 (p=1.9E-17, US and Japanese combined p=2.8E-30) and HLA-C*04:01 (p=7.5E-05, US and Japanese combined p=1.1E-17) were associated with the A-allele (risk) of rs6906846 (Table 2 and Supplementary Table 3). HLA-C*12:02, which was not found in the US multi-ethnic Cohort 1, was strongly associated with the A-allele (risk) of rs6906846 in Japanese Cohort 2 (p=1.9E-18).
Table 2.
Association between rs6906846 and the three most significantly associated HLA-C types (See Supplementary Table 2)
| HLA-C type | US multi-ethnicity (Cohort 1) | Japanese (Cohort 2) | Combined | ||||
|---|---|---|---|---|---|---|---|
| rs6906846 | p | rs6906846 | p | pcombined | |||
| A allele | G allele | A allele | G allele | ||||
| n=48 | n=58 | n=46 | n=250 | ||||
| *04:01 | 20 | 0 | 8.6E-09 | 5 | 0 | 7.5E-05 | 1.1E-17 |
| *07:02 | 23 | 0 | 2.8E-10 | 20 | 1 | 1.9E-17 | 2.8E-30 |
| *12:02 | 0 | 0 | NA | 21 | 1 | 1.9E-18 | NA |
Bonferroni corrected p value threshold: 0.05/(50 HLA-B types + 26 HLA-C types)=6.6E-04, NA: not applicable, Only homozygous subjects of rs6906846 were included in the analysis.
The frequency of HLA-C*04:01, C*07:02, and C*12:02 (associated with A-allele at rs6906846) in all subjects in Cohort 2 (Japanese n=275, 550 chromatids) was compared with published Japanese data [20]. HLA-C*12:02 was over-represented in Japanese KD subjects compared to the Japanese population at large (HLA-C*12:02: 15.3% vs.11.2%, p= 3.5 E-03) (Table 3). This analysis could not be performed for multiethnic Cohort 1 that was intentionally enriched for subjects carrying the A-allele (rs6906846).
Table 3.
Frequencies of HLA-C*04:01, C*07:02 and C*12:02 in KD patients and the general Japanese population
| HLA-C type | Japanese | Odds ratio (CI) | p | |||
|---|---|---|---|---|---|---|
| KD (550 haplotypes) | General (19183 haplotypes) | |||||
| n | % | n | % | |||
| C*04:01 | 20 | 3.6 | 846 | 4.4 | 0.9 (0.6–1.4) | NS |
| C*07:02 | 69 | 12.5 | 2439 | 12.7 | 1.0 (0.8–1.3) | NS |
| C*12:02 | 84 | 15.3 | 2145 | 11.2 | 1.4 (1.1–1.8) | 3.5E-03 |
p: nominal p-value. CI: Confidence interval. Bonferroni corrected p value threshold: 0.05/3=0.0166.
We also explored the potential relationships between the risk alleles in the HLA-B region and high resolution HLA types. The T-allele (risk) of rs2254556 (HLA-B) was not associated with HLA-C high-resolution types (data not shown). For association with HLA-B high-resolution types, the A-allele of rs6906846 and the T-allele of rs2254556 showed only weak association with HLA-B*07:02 (p=8.1E-05 and p=3.5E-07, respectively).
Association of SNVs with HLA-B and C amino acid variants
Next, we analyzed the association between the SNVs and the amino acid sequence for 181 positions in the antigen presenting site of HLA-C and HLA-B using individuals homozygous for rs6906846 both in the U.S. (AA n=24, GG n=29; Cohort 1) and Japanese cohorts (AA n=23, GG n=125; Cohort 2). In both cohorts, we found highly significant associations (nominal p<1.0E-06) of certain amino acids with the risk allele of rs6906846 in eight positions in the peptide binding groove (α1 and α2 domain). The amino acids and their position in parentheses were as follows: Asp or Ser (9), Arg (14), Ala (49), Ala (73), Ala (90), Arg (97), Phe or Ser (99), and Phe or Ser (116) (Table 4). The association between these eight amino acid positions and rs6906846 was replicated in Japanese Cohort 2 with the strongest p value at amino acid position 73 (U.S. and Japanese combined p=7.7E-68) (Table 4).
Table 4.
Location and character of amino acids associated with the A and G alleles at rs6906846
| Location | Position | Amino acids | Amino acid characteristics | U.S. Cohort 1 | Japanese Cohort 2 | Combined | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Charge | Hydrophobia | rs6906846 allele | p | rs6906846 allele | p | pcombined | ||||||||
| Negative | Positive | Neutral | Hydrophobic | Hydrophilic | A | G | A | G | ||||||
| α1 domain | 9 | D | X | X | 24 | 14 | 4.0E-14 | 20 | 5 | 1.2E-16 | 6.6E-28 | |||
| S | X | X | 21 | 1 | 5 | 37 | ||||||||
| F | X | X | 0 | 13 | 0 | 79 | ||||||||
| Y | X | X | 3 | 30 | 21 | 129 | ||||||||
| 14 | R | X | X | 27 | 58 | 2.8E-09 | 41 | 250 | 7.5E-05 | 2.0E-18 | ||||
| W | X | X | 21 | 0 | 5 | 0 | ||||||||
| 49 | A | X | X | 27 | 58 | 2.8E-09 | 41 | 250 | 7.5E-05 | 2.0E-18 | ||||
| E | X | X | 21 | 0 | 5 | 0 | ||||||||
| 73 | A | X | X | 47 | 17 | 1.9E-14 | 46 | 6 | 9.4E-48 | 7.7E-68 | ||||
| T | X | X | 1 | 41 | 0 | 244 | ||||||||
| 90 | A | X | X | 1 | 44 | 2.8E-16 | 21 | 245 | 4.9E-19 | 1.3E-41 | ||||
| D | X | X | 47 | 14 | 25 | 5 | ||||||||
| α2 domain | 97 | R | X | X | 48 | 31 | 4.7E-09 | 46 | 132 | 5.7E-12 | 6.0E-22 | |||
| W | X | X | 0 | 26 | 0 | 118 | ||||||||
| S | X | X | 0 | 1 | 0 | 0 | ||||||||
| 99 | F | X | X | 22 | 1 | 5.9E-25 | 5 | 37 | 6.1E-20 | 6.1E-20 | ||||
| S | X | X | 23 | 0 | 20 | 1 | ||||||||
| C | X | X | 0 | 13 | 0 | 79 | ||||||||
| Y | X | X | 3 | 44 | 21 | 133 | ||||||||
| 116 | F | X | X | 23 | 9 | 1.7E-09 | 5 | 38 | 5.4E-23 | 9.3E-34 | ||||
| S | X | X | 25 | 20 | 41 | 43 | ||||||||
| Y | X | X | 0 | 27 | 0 | 157 | ||||||||
| L | X | X | 0 | 2 | 0 | 12 | ||||||||
Bonferroni corrected p value threshold: 0.05/181=2.8E-04, Underlined bold: Residues pointing toward the peptide binding site and can interact with the peptide (van Deutekom & Keşmir 2015), Characteristics of amino acids (Harper’s Illustrated Biochemistry (30th edition)
Based on crystallography [21], amino acid residues in five (position 9, 73, 97, 99 and 116) of the eight positions in the α−1 and 2 domains are predicted to point toward the peptide binding groove (Fig 2), and thus would be expected to interact with the peptide. None of the positions were at the sites that directly interact with the T-cell receptor.
Fig 2. HLA-C amino acid position in the peptide binding groove associated with rs6906846 A/G (risk/protective).
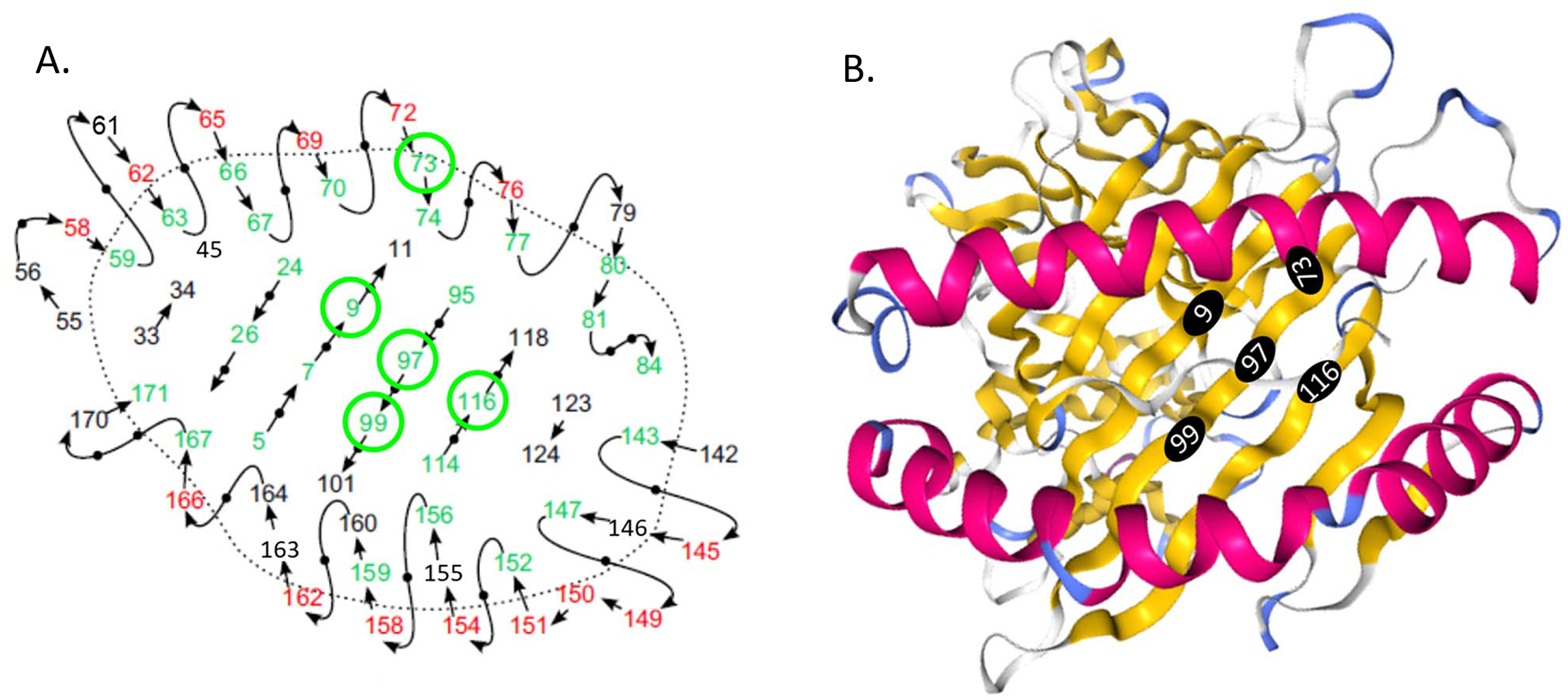
A schematic view of the peptide binding groove and amino acid positions (Adapted from van Deutekom & Keşmir 2015) (A). Residues pointing toward the peptide binding site can interact with the peptide and are colored in green. Residues pointing up from the peptide binding site can interact with the T cell receptor and are colored in red. The remaining residues are shown in black. The 3D structure of the HLA-C molecule (PDB id: 5VGE) and amino acid positions from the Protein Data Bank (PDB: http://www.rcsb.org/3d-view/5VGE) (B). Amino acid positions that are associated with rs6906846 A/G (risk/protective) and pointing towards the peptide binding groove are circled in green (A) and in black filled circles (B).
Association between rs2254556 and certain amino acids was evaluated using only homozygotes in U.S. Cohort 1 (TT n=4, CC n=54). The T-allele (risk) was not associated with amino acids in the antigen presenting site of HLA-B or HLA-C, although the number of subjects was limited. No association was detected between amino acids at position 77–80 (critical position for binding to NK cells) and the risk alelles at rs6906846 and rs2254556.
Discussion
Using a genetic approach, we identified two intergenic variants, rs6906846 (HLA-C) and rs2254556 (HLA-B) associated with KD susceptibility in both European descent and Japanese subjects. High-resolution HLA typing and amino acid sequence analysis in U.S. multi-ethnic and Japanese cohorts demonstrated that genetic variation at rs6906846 was associated with amino acids at position 9, 14, 49, 73, 90, 97, 99 and 116 in the peptide-binding groove of the subjects’ HLA-C molecule that defines the allele-specific peptide repertoire presented to CTL [22].
Other studies of the HLA-B and -C locus and KD pathogenesis have focused on allele associations in a Korean population, HLA/KIR relationships in an Italian KD population, and an analysis of the imputed HLA locus with HLA types in a European descent cohort. Our risk alleles were not in linkage disequilibrium with any of the risk alleles or associated with any HLA types reported in these studies [7, 11, 12, 23].
We propose a mechanism by which the risk alleles described here are linked to HLA-C types and amino acid substitutions that lead to preferential binding of KD antigen-derived peptides, which may lead to increased activation and expansion of CTL (Fig 3). As an alternative hypothesis, we need to consider that the amino acid substitutions associated with the rs6906846 (HLA-C) risk allele could have lower affinity for KD–associated peptides and therefore reduced presentation of relevant peptides to CTL, leading to reduced responses and delayed clearing of the inciting agent. Both hypotheses underscore the importance of antigen presentation to CTL in KD pathogenesis [1, 2].
Fig 3. Hypothesis for effect of risk alleles in HLA-C on KD pathogenesis.
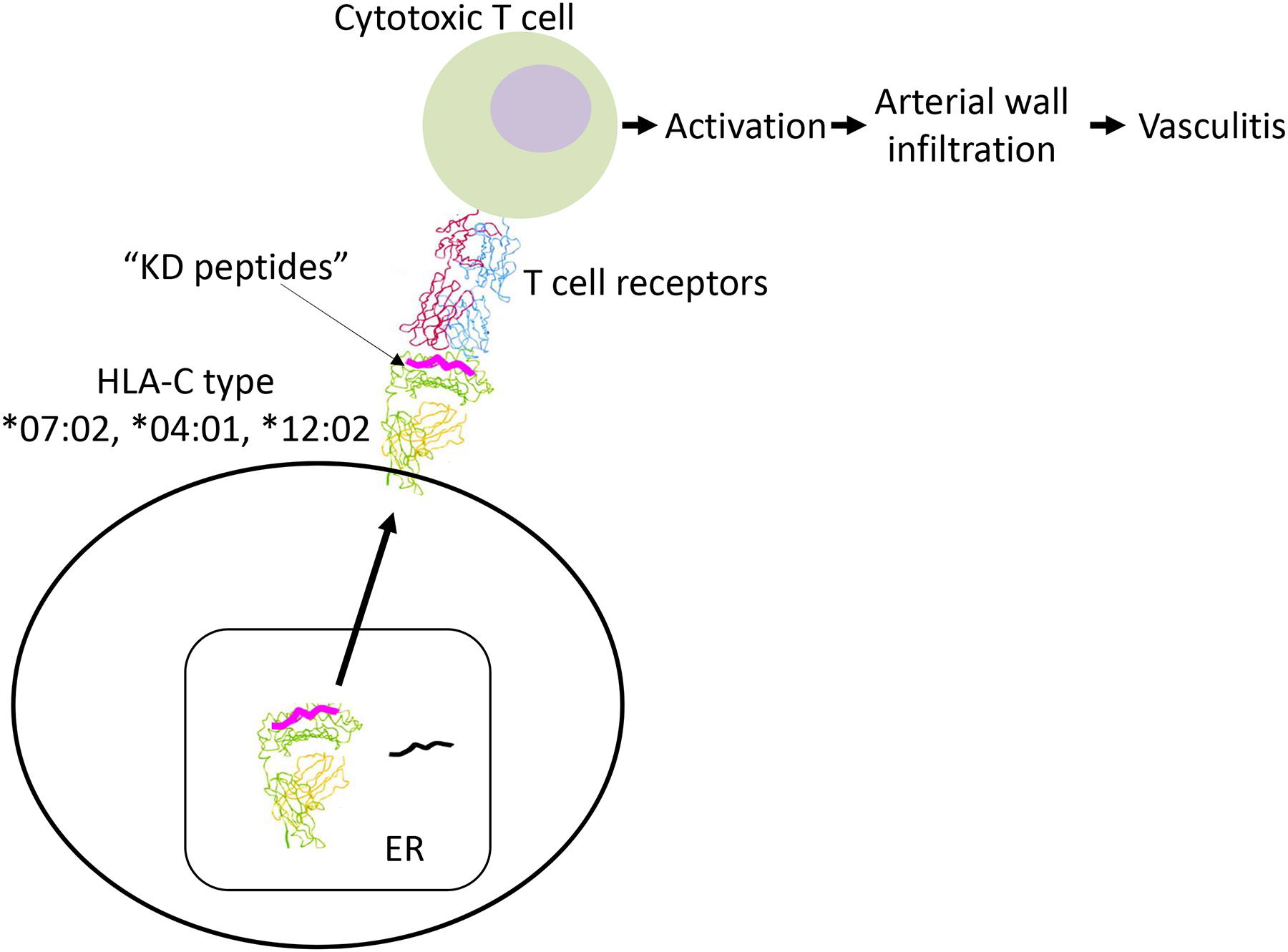
We propose distinct mechanisms by which the risk alleles described here may affect KD susceptibility due to preferential binding of KD pathogen-derived peptides in the HLA-C antigen binding groove. The amino acids associated with the HLA-C risk allele may preferentially bind to KD antigen-derived peptides leading to T cell activation and arterial wall infiltration of CD8+ cytotoxic T cells. ER: endoplasmic reticulum
Amino acid variation in HLA-C
Single amino acid substitutions at critical positions in the peptide-binding groove are known to influence disease susceptibility and response to pathogens [3–5]. As an example, of the five positions that were oriented toward the peptide binding site (position 9, 73, 97, 99 and 116, Fig 2), certain amino acids at positions 97, 99 and 116 have been linked to ankylosing spondylitis and graft versus host disease (GVHD) [24–26].
The strongest combined p-value was observed at position 73, which is located on the alpha 1-domain alpha helix at the edge of the antigen-binding groove. Amino acid changes that alter charge or hydrophobicity (Table 4) change the peptide-binding repertoire [4]. At position 73, the A allele (risk) of rs6906846 (HLA-C) is associated with the hydrophobic amino acid alanine (A) but the G allele (protective) is associated with the hydrophilic amino acid threonine (T) in both US and Japanese cohorts. At position 9, the A allele (risk) of rs6906846 (HLA-C) is associated with the negatively charged, hydrophilic amino acid, aspartic acid (D), but the G allele (protective) is associated with the neutral, hydrophobic amino acids phenylalanine (F) and tyrosine (Y) in both US and Japanese cohorts. These important amino acid changes would be expected to have a significant effect on peptide binding.
KD risk alleles and HLA-C peptide-binding specificities
The A (risk) allele at rs6906846 (HLA-C) was strongly linked to HLA-C*07:02, C*04:01, and C*12:02. The HLA type C*12:02 is not represented in US multi-ethnic populations and the frequency of this HLA type among the Japanese KD subjects was 15.3% compared to 11.2% among the Japanese population at large (p=3.5E-03). This is intriguing given the 12-fold higher rate of KD in children of Japanese descent compared to the rate in a mixed ethnic KD cohort from the U.S. [27]. Because the peptide repertoire is restricted by HLA type-specific amino acid variation in the peptide-binding grove, amino acid differences between the A (risk)-associated and G (protection)-associated HLA-C types may be helpful in identifying KD-specific antigens. Peptide binding affinity between candidate peptides (8–14-mers) and HLA-C types of interest can be calculated. (http://www.cbs.dtu.dk/services/NetMHC) [28] (Fig 4). In addition, a published analysis using the transmission disequilibrium test on 12 informative KD families showed over-transmission of HLA-C*15, which tagged with leucine in position 116 that is located in the middle of the peptide binding groove of the HLA-C molecule [11]. The environmental or infectious trigger for KD may contain peptides with configurations that are preferentially presented by these KD-associated HLA-C types.
Fig 4. Amino acid sequence of peptide nonamers that preferentially bind to HLA-C*04:01, C*07:02 and C*12:02.
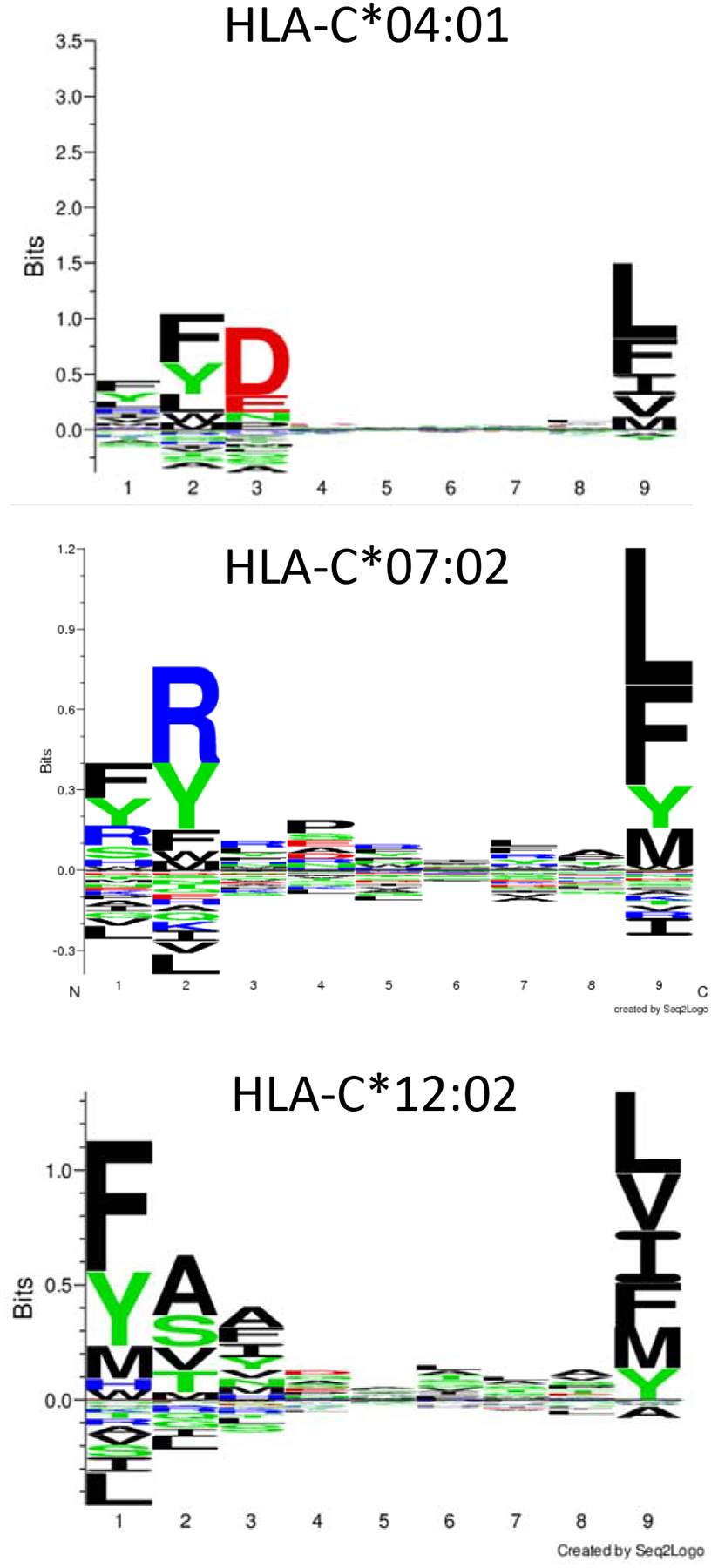
http://www.cbs.dtu.dk/services/NetMHC (4.0 server)
Strengths and limitations
A genetic pathway approach allowed us to discover HLA class I intergenic SNVs associated with KD susceptibility in both European descent and Japanese subjects. Because there were no available European GWAS datasets on which to perform replication, we used a Japanese GWAS to test for the association between SNVs and KD. Differences in LD structure between the two populations may have prevented us from replicating additional HLA variants identified through the pathway and gene stability analyses. Since association of these intergenic SNVs was discovered in ethnically diverse populations, we did not adjust for the population structure in subsequent analysis of tagged amino acids. Also, the minor allele frequency of rs2254556 was low, so it was difficult to assess whether the associations were lacking due to the small sample size, the low number of polymorphic alleles, or a true lack of association. Two SNVs (rs2596551 and 2523535) in HLA-B did not make the statistical cut-off of p<0.05 in Japanese cohort. However, there was a similar trend of association in both European descent cohort 1 and Japanese cohort 2. Future analysis of association between these SNVs and HLA-type is needed. An additional limitation of our study was the small sample size, which precluded a meaningful analysis of genotype vs. disease outcome (coronary artery status and response to treatment). High-resolution HLA typing was available for only a small number of our KD subjects. Enrichment of our mixed ethnicity Cohort 1 for individuals homozygous for the risk allele precluded a comparison to population frequencies of the associated HLA types in each individual ethnic group. Although interactions between HLA A, B and C and KIR genes have been reported in association with KD susceptibility, the KIR loci and HLA/KIR epistasis were not explored in the current study [23, 29].
Conclusion
Association of KD susceptibility with two SNVs in the HLA class I in European and Japanese GWAS datasets highlights the importance of HLA-C-mediated antigen presentation in KD pathogenesis. These SNVs were associated with specific high-resolution HLA-C types suggesting an influence on peptide binding. These risk SNVs may mediate the preferential binding of “KD-associated peptides” derived from the environmental trigger, which are presented to CTL and/or are involved in NK cell regulation. Future studies should focus on HLA class I pathway genes in existing KD GWAS databases and high-resolution class I HLA typing in KD populations.
Supplementary Material
Acknowledgements
We thank Joan Pancheri, RN for clinical sample collection, DeeAnna Scherrer for technical assistance, and Emelia Bainto and Nipha Sivilay for data base management.
International Kawasaki Disease Genetic Consortium: Annette L. Baker, David Burgner, Rolando Cimaz, Nagib Dahdah, Sonia Davila, Taco W Kuijpers, Jane W. Newburger, Anne H. Rowley, Stanford T. Shulman, Rae S. M. Yeung (alphabetical order)
Funding
This work supported in part by grants from the Gordon and Marilyn Macklin Foundation, the National Institutes of Health, National Heart, Lung, Blood Institute (HL69413) awarded to JCB and the NIH Roadmap for Medical Research, Grant U54HL108460 awarded to AHT.
Abbreviations:
- KD
Kawasaki disease
- SNV
Single nucleotide variant
- GWAS
genome-wide association study
Reference
- 1.Brown TJ, Crawford SE, Cornwall ML, Garcia F, Shulman ST, Rowley AH. CD8 T lymphocytes and macrophages infiltrate coronary artery aneurysms in acute Kawasaki disease. J Infect Dis. 2001;184(7):940–3. Epub 2001/08/31. doi: JID010422 [pii] 10.1086/323155. [DOI] [PubMed] [Google Scholar]
- 2.Rowley AH, Wylie KM, Kim KY, Pink AJ, Yang A, Reindel R, et al. The transcriptional profile of coronary arteritis in Kawasaki disease. BMC Genomics. 2015;16:1076. doi: 10.1186/s12864-015-2323-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao X, Nelson GW, Karacki P, Martin MP, Phair J, Kaslow R, et al. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N Engl J Med. 2001;344(22):1668–75. doi: 10.1056/NEJM200105313442203. [DOI] [PubMed] [Google Scholar]
- 4.Kloverpris HN, Harndahl M, Leslie AJ, Carlson JM, Ismail N, van der Stok M, et al. HIV control through a single nucleotide on the HLA-B locus. J Virol. 2012;86(21):11493–500. doi: 10.1128/JVI.01020-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strettell MD, Donaldson PT, Thomson LJ, Santrach PJ, Moore SB, Czaja AJ, et al. Allelic basis for HLA-encoded susceptibility to type 1 autoimmune hepatitis. Gastroenterology. 1997;112(6):2028–35. [DOI] [PubMed] [Google Scholar]
- 6.Kim JJ, Hong SJ, Hong YM, Kim S, Kang MJ, Kim KJ, et al. Genetic variants in the HLA-G region are associated with Kawasaki disease. Hum Immunol. 2008;69(12):867–71. doi: 10.1016/j.humimm.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Kim JJ, Yun SW, Yu JJ, Yoon KL, Lee KY, Kil HR, et al. A genome-wide association analysis identifies NMNAT2 and HCP5 as susceptibility loci for Kawasaki disease. J Hum Genet. 2017;62(12):1023–9. doi: 10.1038/jhg.2017.87. [DOI] [PubMed] [Google Scholar]
- 8.Lin YJ, Wan L, Wu JY, Sheu JJ, Lin CW, Lan YC, et al. HLA-E gene polymorphism associated with susceptibility to Kawasaki disease and formation of coronary artery aneurysms. Arthritis Rheum. 2009;60(2):604–10. doi: 10.1002/art.24261. [DOI] [PubMed] [Google Scholar]
- 9.Onouchi Y, Ozaki K, Burns JC, Shimizu C, Terai M, Hamada H, et al. A genome-wide association study identifies three new risk loci for Kawasaki disease. Nat Genet. 2012;44(5):517–21. Epub 2012/03/27. doi: 10.1038/ng.2220. [DOI] [PubMed] [Google Scholar]
- 10.Shendre A, Wiener HW, Zhi D, Vazquez AI, Portman MA, Shrestha S. High-density genotyping of immune loci in Kawasaki disease and IVIG treatment response in European-American case-parent trio study. Genes Immun. 2014;15(8):534–42. doi: 10.1038/gene.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shrestha S, Wiener HW, Aissani B, Shendre A, Tang J, Portman MA. Imputation of class I and II HLA loci using high-density SNPs from ImmunoChip and their associations with Kawasaki disease in family-based study. Int J Immunogenet. 2015;42(3):140–6. doi: 10.1111/iji.12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon YC, Sim BK, Yu JJ, Yun SW, Yoon KL, Lee KY, Kil HR, Kim GB, Han MK, Song MS, Lee HD, Jang GY, Hong YM, Kwon OJ, Oh HB, Lee JK HLA-B*54:01 is associated with susceptibility to Kawasaki Disease. Circ Genom Precis Med. 2019;(12):e002365. [DOI] [PubMed] [Google Scholar]
- 13.Yan Y, Ma Y, Liu Y, Hu H, Shen Y, Zhang S, et al. Combined analysis of genome-wide-linked susceptibility loci to Kawasaki disease in Han Chinese. Hum Genet. 2013;132(6):669–80. doi: 10.1007/s00439-013-1279-2. [DOI] [PubMed] [Google Scholar]
- 14.Shimizu C, Eleftherohorinou H, Wright VJ, Kim J, Alphonse MP, Perry JC, et al. Genetic Variation in the SLC8A1 Calcium Signaling Pathway Is Associated With Susceptibility to Kawasaki Disease and Coronary Artery Abnormalities. Circ Cardiovasc Genet. 2016;9(6):559–68. doi: 10.1161/CIRCGENETICS.116.001533. [DOI] [PubMed] [Google Scholar]
- 15.McCrindle BW, Rowley AH, Newburger JW, Burns JC, Bolger AF, Gewitz M, et al. Diagnosis, Treatment, and Long-Term Management of Kawasaki Disease: A Scientific Statement for Health Professionals From the American Heart Association. Circulation. 2017;135(17):e927–e99. doi: 10.1161/CIR.0000000000000484. [DOI] [PubMed] [Google Scholar]
- 16.Khor CC, Davila S, Breunis WB, Lee YC, Shimizu C, Wright VJ, et al. Genome-wide association study identifies FCGR2A as a susceptibility locus for Kawasaki disease. Nat Genet. 2011;43(12):1241–6. Epub 2011/11/15. doi: 10.1038/ng.981. [DOI] [PubMed] [Google Scholar]
- 17.Wieczorek M, Abualrous ET, Sticht J, Alvaro-Benito M, Stolzenberg S, Noe F, et al. Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Front Immunol. 2017;8:292. doi: 10.3389/fimmu.2017.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis DM, Mandelboim O, Luque I, Baba E, Boyson J, Strominger JL. The transmembrane sequence of human histocompatibility leukocyte antigen (HLA)-C as a determinant in inhibition of a subset of natural killer cells. J Exp Med. 1999;189(8):1265–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaefer MR, Williams M, Kulpa DA, Blakely PK, Yaffee AQ, Collins KL. A novel trafficking signal within the HLA-C cytoplasmic tail allows regulated expression upon differentiation of macrophages. J Immunol. 2008;180(12):7804–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikeda N, Kojima H, Nishikawa M, Hayashi K, Futagami T, Tsujino T, et al. Determination of HLA-A, -C, -B, -DRB1 allele and haplotype frequency in Japanese population based on family study. Tissue Antigens. 2015;85(4):252–9. doi: 10.1111/tan.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Deutekom HW, Kesmir C. Zooming into the binding groove of HLA molecules: which positions and which substitutions change peptide binding most? Immunogenetics. 2015;67(8):425–36. doi: 10.1007/s00251-015-0849-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsumura M, Fremont DH, Peterson PA, Wilson IA. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science. 1992;257(5072):927–34. [DOI] [PubMed] [Google Scholar]
- 23.Bossi G, Mannarino S, Pietrogrande MC, Salice P, Dellepiane RM, Cremaschi AL, et al. Genetic epistasis between killer immunoglobulin-like receptors and human leukocyte antigens in Kawasaki disease susceptibility. Genes Immun. 2015;16(7):481–7. doi: 10.1038/gene.2015.34. [DOI] [PubMed] [Google Scholar]
- 24.Ferrara GB, Bacigalupo A, Lamparelli T, Lanino E, Delfino L, Morabito A, et al. Bone marrow transplantation from unrelated donors: the impact of mismatches with substitutions at position 116 of the human leukocyte antigen class I heavy chain. Blood. 2001;98(10):3150–5. Epub 2001/11/08. [DOI] [PubMed] [Google Scholar]
- 25.Kim K, Bang SY, Lee S, Lee HS, Shim SC, Kang YM, et al. An HLA-C amino-acid variant in addition to HLA-B*27 confers risk for ankylosing spondylitis in the Korean population. Arthritis Res Ther. 2015;17:342. Epub 2015/11/29. doi: 10.1186/s13075-015-0855-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pidala J, Wang T, Haagenson M, Spellman SR, Askar M, Battiwalla M, et al. Amino acid substitution at peptide-binding pockets of HLA class I molecules increases risk of severe acute GVHD and mortality. Blood. 2013;122(22):3651–8. doi: 10.1182/blood-2013-05-501510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makino N, Nakamura Y, Yashiro M, Ae R, Tsuboi S, Aoyama Y, et al. Descriptive epidemiology of Kawasaki disease in Japan, 2011–2012: from the results of the 22nd nationwide survey. J Epidemiol. 2015;25(3):239–45. doi: 10.2188/jea.JE20140089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasmussen M, Harndahl M, Stryhn A, Boucherma R, Nielsen LL, Lemonnier FA, et al. Uncovering the peptide-binding specificities of HLA-C: a general strategy to determine the specificity of any MHC class I molecule. J Immunol. 2014;193(10):4790–802. Epub 2014/10/15. doi: 10.4049/jimmunol.1401689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Capittini C, Emmi G, Mannarino S, Bossi G, Dellepiane RM, Salice P, et al. An immune-molecular hypothesis supporting infectious aetiopathogenesis of Kawasaki disease in children. Eur J Immunol. 2018;48(3):543–5. doi: 10.1002/eji.201747226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


