Abstract
Ascosphaera (Eurotiomycetes: Onygenales) is a diverse genus of fungi that is exclusively found in association with bee nests and comprises both saprophytic and entomopathogenic species. To date, most genomic analyses have been focused on the honeybee pathogen A. apis, and we lack a genomic understanding of how pathogenesis evolved more broadly in the genus. To address this gap we sequenced the genomes of the leaf-cutting bee pathogen A. aggregata as well as three commensal species: A. pollenicola, A. atra and A. acerosa. De novo annotation and comparison of the assembled genomes was carried out, including the previously published genome of A. apis. To identify candidate virulence genes in the pathogenic species, we performed secondary metabolite-oriented analyses and clustering of biosynthetic gene clusters (BGCs). Additionally, we captured single copy orthologs to infer their phylogeny and created codon-aware alignments to determine orthologs under selective pressure in our pathogenic species. Our results show several shared BGCs between A. apis, A. aggregata and A. pollenicola, with antifungal resistance related genes present in the bee pathogens and commensals. Genes involved in metabolism and protein processing exhibit signatures of enrichment and positive selection under a fitted branch-site model. Additional known virulence genes in A. pollenicola, A. acerosa and A. atra are identified, supporting previous hypotheses that these commensals may be opportunistic pathogens. Finally, we discuss the importance of such genes in other fungal pathogens, suggesting a common route to evolution of pathogenicity in Ascosphaera.
Keywords: Eurotiomycetes, biosynthetic gene clusters, functional enrichment, positive selection, Chalkbrood, evolutionary genomics
Introduction
Ascosphaera is a diverse genus of fungi that only associates with bee nests and comprises species that can range from commensals to pathogens (Anderson et al. 1998). Its type species, A. apis, causes chalkbrood disease in Apis mellifera honey bees, while another known pathogen in the genus, A. aggregata, infects the alfalfa leaf-cutting bee Megachile rotundata. The infection starts after the ingestion of ascospores and subsequent germination in the insect midgut. The pathogen then penetrates the gut epithelium and invades the hemocoel with subsequent systemic mycosis (Aronstein and Murray 2010). However, the mechanisms of pathogenicity are yet to be fully elucidated (Boomsma et al. 2014). Genomic and transcriptomic studies have heavily emphasized A. apis, showing the role of secondary metabolites, transcription factors, mating loci and even basal metabolism genes in its virulence (Cornman et al. 2012; Getachew et al. 2020, 2018), but little-to-no genomic information exists for other Ascosphaera species. Nonetheless, it has been proposed that some these species originally thought to occupy a commensal niche may be considered opportunistic pathogens (Klinger et al. 2013), although the majority of Ascosphaera species have been reported in saprophytic associations (Aronstein and Murray 2010; Pitts-Singer and Cane 2011; Bissett et al. 1996; James and Skinner 2005).
Generally, genes emphasized in pathogenicity for A. apis are related to sexual reproduction (Aronstein and Colby 2015) or penetration of the pathogen through the peritrophic membrane in the bees’ midgut. However, once the fungus has reached the hemocoel, overcoming the host’s innate immune system is also key to a successful infection (Aronstein and Holloway 2013; Valero-Jiménez et al. 2016). These mechanisms of avoiding or suppressing immune responses are well characterized in several fungal pathogens and represent important virulence factors for entomopathogenic fungi (Avulova and Rosengaus 2011; Xu et al. 2017; Zhong et al. 2017). Other fundamental factors for the infection rely on how the pathogen’s metabolism interacts with the host to acquire nutrients and growth factors necessary for its development (DaFu et al. 2017). For instance, reactive oxygen species (ROS) defense responses and modulation of the oxidative stress in the midgut of infected bees seem to play a major role in the pathogenesis of A. apis to honey bees (Li et al. 2020). However, virulence factors in A. aggregata remain largely unclear and the role the commensal species play in bee health is opaque. Therefore, a comparative genomics approach is important to understanding the transitions and intermediate forms along the symbiotic spectrum from commensal to pathogen.
Previous phylogenies of Ascosphaera species were built on analysis of only a few gene regions (Klinger et al. 2013). While this multi locus approach improved the understanding of Ascosphaera evolution in comparison to prior analyses (Anderson et al. 1998) that included only ITS regions, we wanted to test for robustness of the phylogenetic relationships with genomic scale data. Therefore, in this study, we aimed to unravel the genomic diversity underlying Ascosphaera species with different lifestyles and characterize potential genes related to the evolution of pathogenicity. This study represents the first effort to sequence the whole genomes of four Ascosphaera species. We then annotated and compared the genomes of five species of Ascosphaera, including two species that are known pathogens (A. apis, A. aggregata) and three that are considered saprophytic (A. pollinicola, A. atra, A. acerosa). We expected that the evolution of pathogenicity would leave signatures of enrichment and natural selection in genes that contribute to virulence. Specifically, we predicted that biosynthetic and secondary metabolite genes will be present or altered in only the pathogenic species and not the commensal species.
Results
Genome assembly
Genome quality statistics and BUSCOs completeness scores are summarized in depth in Table 1. In brief, our genome completeness ranged from 24.4%-73.8%, N50 from 2,504 - 482,601, and L50 from 13 - 3,523 (Table 1). The coverage ranged from 57.9-99x, the GC content from 47.12-61.64, the heterozygosity from 0.18-3.17 and the repeat content from 3.90-32.20 (Table 1).
Table 1:
Genome assembly and contig statistics
|
Ascosphaera Species |
acerosa | aggregata | pollenicola | atra | apis |
|---|---|---|---|---|---|
| Accession Numbers | JAGYHY000000000 | JAGYHZ000000000 | JAGYIB000000000 | JAGYIA000000000 | GCA_001636715.1 |
| Depth of Coverage | 97.8X | 60.1X | 99X | 57.9X | - |
| Scaffold count | 5020 | 4503 | 5538 | 12891 | 82 |
| Total Length | 18611625 | 19806921 | 19988041 | 29072572 | 20313079 |
| Min | 500 | 500 | 500 | 996 | 1075 |
| Max length (bp) | 38,273 | 41,254 | 31,524 | 19,378 | 159,2427 |
| Median contig length (bp) | 2089 | 2780 | 2153 | 1741 | 129110 |
| Mean contig length (bp) | 3707.5 | 4398.61 | 3609.25 | 2255.26 | 247720.48 |
| L50 | 822 | 797 | 925 | 3523 | 13 |
| L90 | 3123 | 2735 | 3265 | 10268 | 37 |
| N50 | 6542 | 7511 | 6319 | 2504 | 482601 |
| N90 | 1529 | 2048 | 1624 | 1228 | 168129 |
| GC content | 61.64% | 47.12% | 50.22% | 51.41% | 47.66% |
| Heterozygosity | 0.41% | 0.19% | 0.18% | 3.17% | - |
| Repeat content | 8.01% | 5.59% | 3.55% | 9.26% | - |
| BUSCO Completeness Score | 44.6% | 59.4% | 53.5% | 24.4% | 73.8% |
| Single Copy | 1782 (44%) | 2401 (59.3%) | 2139 (52.9%) | 885 (21.9%) | 2981 (73.7%) |
| Duplicated | 23 (0.6%) | 5 (0.1%) | 26 (0.6%) | 100 (2.5%) | 5 (0.1%) |
A gene set enrichment analysis of MEROPS and CAZYme families did not reveal any significant hits.
Phylogeny and single copy orthologs under positive selection for pathogens
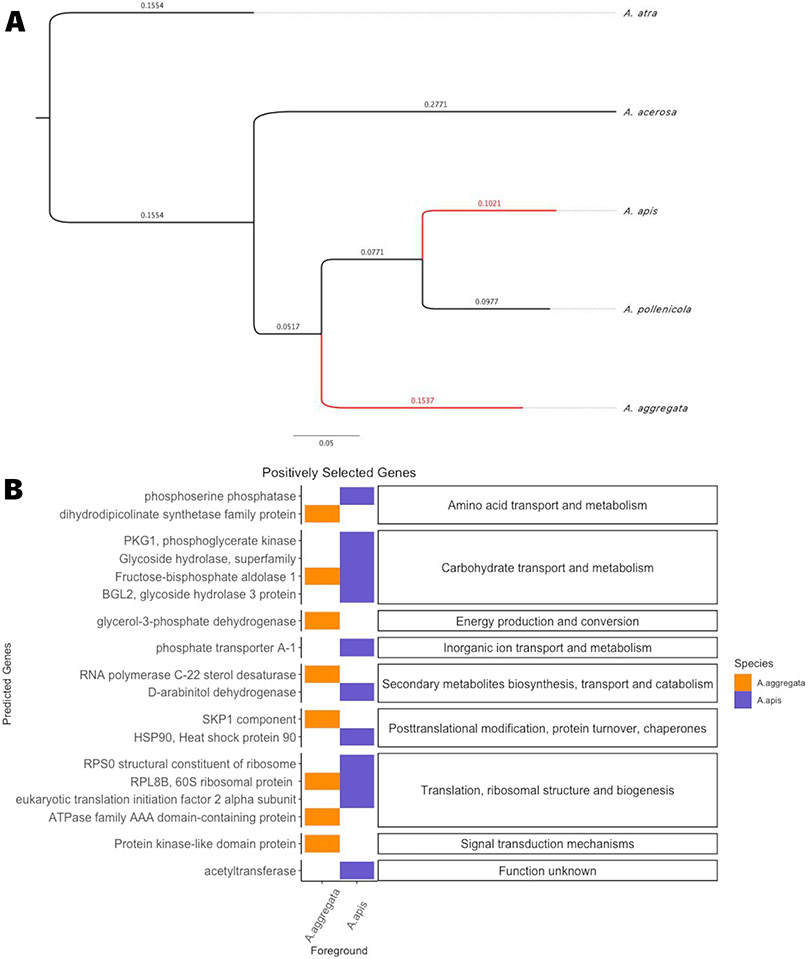
To further characterize genes within these families and how they are tied to the evolutionary history of the genus Ascosphaera, we assessed the selective pressure in different branches of their phylogeny. Across all five species, we detected 1,602 single copy orthologs. When we excluded A. atra we detected 3,058 single copy orthologs. Our resulting species tree revealed that A. apis and A. pollenicola were most closely related to each other, forming a pair of sister taxa (Fig. 1A). Ascosphaera aggregata was sister to the A. apis and A. pollenicola pair, followed by A. acerosa and most distantly A. atra (Fig. 1A). After intensive quality control measures about 20% (n = 279 for all 5 species, n= 616 when A. atra excluded) of the single copy orthologs analyzed were converted to nucleotide alignments and used in CodeML analyses to detect signatures of positive selection in the pathogens. After manually inspecting each result from CodeML and removing cryptic paralogs to eliminate false positives, we had a total of 18 genes with significantly elevated DN/DS ratios in the pathogens (FDR < 5%, Fig. 1B, supplementary table S1). The two most abundant classes of genes were related to primary metabolism and protein processing based on gene ontology (GO) terms. Broadly, 7 of these genes relate to amino acid, carbohydrate and inorganic ion transport and metabolism, 6 are involved in RNA processing, translation and post translational modification, 2 in secondary metabolite biosynthesis, transport and catabolism and 2 in energy production and signaling (Fig. 1B, supplementary table S1).
Figure 1:
Natural selection on carbohydrate metabolism and transport may be the most important process underlying evolution of pathogenicity in the genus Ascosphaera. A) Species tree generated in Orthofinder (v 2.3.12) by pulling single copy orthologs (Emms D.M. & Kelly S. 2019; Emms D.M. & Kelly S. 2017; Emms D.M. & Kelly S. 2018) and visualized with FigTreev1.4.4. Red lines indicate the known pathogen lineages. Bootstrap supports 100% for all nodes, branch lengths represent nucleotide evolution. B) Out of 1,602 single copy orthologs for the 5 species analysis and 3,058 when excluding A. atra, 128 showed signatures of significant positive selection (FDRP < 0.055%). The x-axis represents which species were marked as the foreground for CodeML analysis before running branch-site models. The group “5 species” represents the analysis including A. apis, A. pollenicola, A. aggregata, A. atra, and A. acerosa. The “4 species” is the same but excludes A. atra. The facet on the right side represents the GO terms for the predicted genes on the left.
Shared and unique biosynthetic gene clusters between species
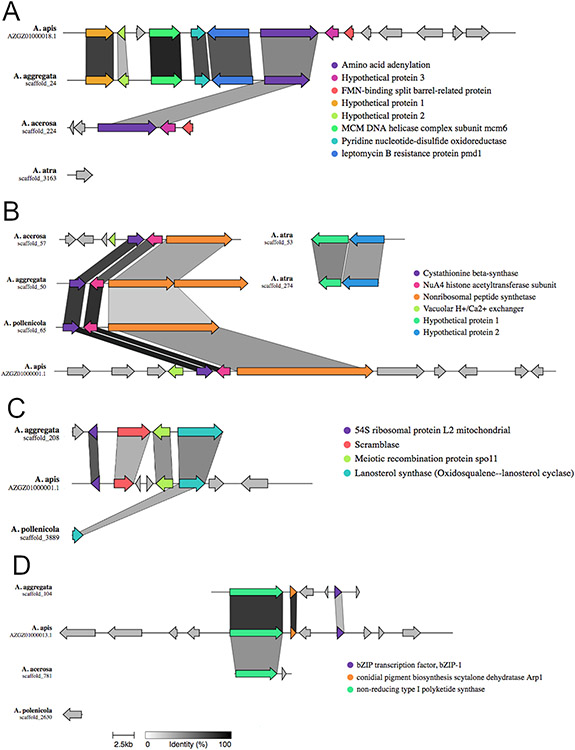
In order to investigate what positively selected genes may be involved in biosynthetic pathways, we screened for BGCs that would provide clues to the virulence of the pathogenic Ascosphaera species. For this, we predicted and compared secondary metabolite genes in the newly assembled genomes to our new annotation of the A. apis genome (Fig. 2). Ascosphaera aggregata displayed the most BGCs in common with the honeybee pathogen (A. apis). Similarities included the terpene oxidosqualene lanosterol cyclase (87.64% identity with A. apis from BLASTp). In A. apis, this cluster included the translation initiation factor 4B, SGT1 and CS domain protein, and 54S ribosomal protein L2 mitochondrial genes. Ascosphaera aggregata displayed genes for DNA damage repair protein Rad9 and 60S ribosomal protein L2 with 66.77% and 71.76% identity to A. apis, respectively (supplementary table S2). The meiotic recombination protein spo11 is also present in the cluster of both species (Fig. 2C). Another cluster shared by the two pathogen species was a non-reducing type 1 polyketide synthase (T1PKS), classified by SMCOG as a beta-ketoacyl synthase (Score: 206.9; E-value: 8.9e-63 in A. apis, fig. 2D). The cluster has genes encoding for conidial pigment biosynthesis scytalone dehydratase Arp1, Zn(2)-C6 fungal-type DNA-binding domain protein and bZIP-1 transcription factor in both pathogens. Unlike the pathogen species, A. pollenicola exhibited 100% similarity in the T1PKS cluster with the 1,3,6,8-tetrahydroxynaphthalene biosynthetic gene cluster from Nodulisporium sp. ATCC74245 (supplementary table S2). A nonribosomal polyketide synthase cluster containing genes encoding nucleotide oxidoreductases and leptomycin resistance proteins were also shared among the two pathogens (Fig. 2A). This cluster’s primary biosynthetic enzyme, which encodes an amino acid adenylation domain, is similar to a copy found in A. acerosa, with approximately 50% of identity (Fig. 2A). Ascosphaera apis was the only species to contain a fungal ribosomally synthesized and post-translationally modified peptides (F-RiPP) cluster (supplementary table S2).
Figure 2:
Alignment and synteny of homologous Biosynthetic Gene Clusters (BGCs) in Ascosphaera spp. The shading of the bars connecting aligned regions represents the percent identity between shared genes as indicated in the scale bar. A) Non-ribosomal peptide synthase cluster 1, B) Non-ribosomal peptide synthase cluster 2, C) Terpene cluster, D) Type 1 polyketide synthase cluster.
No clusters shared by or exclusive to the commensal species were found, although A. acerosa displayed one exclusive cluster containing a AMP-dependent synthetase and ligase, with 55.75% identity to Hydroxamate-type ferrichrome siderophore peptide synthetase from Penicillium digitatum Pd1 (supplementary table S2). Additionally, all of the species presented one Nonribosomal peptides (NRPS) cluster with an AMP-dependent synthetase and ligase gene (Fig. 2B) that varied between 40% and 60% identity with D-alanine-poly(phosphoribitol) ligase subunit 1 from Paracoccidioides lutzii Pb01. The exception was A. acerosa, to which the BLASTp search returned N-(5-amino-5-carboxypentanoyl)-L-cysteinyl-D-valine synthase from Coccidioides immitis with 81% identity. A. aggregata harbored, in this cluster, another AMP-dependent synthetase and ligase gene that returned 44% identity with enterobactin synthetase component F from Coccidioides immitis RMSCC 2394, but did not align with the other genes (supplementary table S2). Another gene found in this cluster for all species was a cysteine synthase that showed 83.68% identity with a cystathionine beta-synthase gene from A. apis. Finally, and also present in all species, an NRPS-like cluster containing AMP-dependent synthetase and ligase as its main gene, returned matches from BLASTp ranging from 58% to 100% identity with Male sterility, NAD-binding protein from A. apis. A second copy of this NRPS-like cluster existed in all species, with the same BLASTp hit in A. apis and A. polenicola for the primary biosynthetic gene, but with different secondary genes (supplementary table S2).
Discussion
Natural selection on carbohydrate metabolism and protein processing genes may be the most important process underlying evolution of pathogenicity in the genus Ascosphaera. In this paper, we describe the genomes of four Ascosphaera species and provide, for the first time, a thorough gene content analysis and comparative genomics of these species along with the previously published A. apis genome. Assessment of the quality of the genome assemblies indicate they were not as complete as preferred for comparative genomics. This could be due to the repeat content because our GC content, heterozygosity and DNA extractions do not appear problematic (Table 1, supplementary figure S1). Nevertheless, this makes our results more conservative because they represent only a subset of the possible genes. Although BUSCO scores indicate that some genes may be absent due to the incomplete nature of the genomes, our analysis identifies important metabolic and virulence genes for the new genomes of A. aggregata, A. acerosa, A pollenicola and A. atra.
Glycoside Hydrolases may represent a novel potential for characterization of commensalism in these fungi. The GH2 family includes β-galactosidases, β-glucuronidases, β-mannosidases, which are mainly involved in the hydrolysis of sucrose, maltose and trehalose and have been associated with saprophytic niches of pollen colonization (Gilliam et al., 1989). Such enzymes were also reported in Ascosphaera, being proposed as potential markers for the identification of A. apis causing the chalkbrood disease in bee colonies (Gilliam and Lorenz, 1993). This, coupled with the fact that Glycoside Hydrolases are observed in this study being under positive selection in A. apis, suggests that this gene likely derived from those homologs present in commensal species and that its functions are advantageous to the pathogenic niche, thereby being fixated in its populations. Whether this is due to paralogy and exactly what functions are involved in these different niches, however, still needs to be elucidated by further studies.
Primary metabolic flexibility has also been shown to play a key role in virulence for several pathogenic fungi (Ene et al. 2014). Many of the orthologous genes that have signatures of positive selection in our study perform primary metabolic functions with an emphasis on carbohydrate transport and metabolism. This is consistent with other studies where the highest percentage of enriched pathways in A. apis during infection had to do with metabolism related genes (Getachew et al. 2020). Similarly, metabolic flexibility has been demonstrated in the human pathogenic yeast Candida albicans in its ability to express glycolytic, gluconeogenic and glyoxylate cycle enzymes simultaneously allowing for assimilation of several carbon sources at once (Sandai et al. 2012). Furthermore, virulence was reduced when any of the three mentioned cycles were disrupted in Ca. albicans as well as in Cryptococcus neoformans mutants with glycolytic defects (Barelle et al. 2006; Price et al. 2011). In fact, in Cr. neoformans the intimate role of metabolism regulation in virulence is well documented (Kronstad et al. 2012). Still, researchers were surprised to find that A. apis could grow well on several different tested substrates besides cellulose indicating that metabolic flexibility may be key to its success as well (Shang et al. 2016). These findings are consistent with our results showing that the abundance of genes coding for carbohydrate active enzymes seem to be driving the evolution pathogenicity in A. apis and A. aggregata. One of the specific enzymes under positive selection in both pathogens was fructose-bisphosphate aldolase 1 (FBA1), which has been shown to be better at hexose synthesis through the gluconeogenic pathway (Marsh and Lebherz 1992). Hence, it might prove important for the Ascosphaera pathogens to utilize different sugar metabolic pathways. This specific enzyme, FBA1, is a key virulence factor in Mycobacterium tuberculosis, Toxoplasma gondii, and Francisella novicida (Puckett et al. 2014; Blume et al. 2015; Ziveri et al. 2017). It is, therefore, tempting to speculate that such enzymes may play a similar role in utilizing alternative carbon sources in entomopathogenic Ascosphaera species. Another carbohydrate metabolism enzyme showing signatures of positive selection in our Ascosphaera pathogens was D-arabitinol dehydrogenase. It was first described in the context of pathogenicity in plants (Hallborn et al. 1995), then subsequently found in the plant pathogen Uromyces fabae with high titers of D-arabitinol dehydrogenase in the haustorium of the fungus, one of its pathogenic structures, used to penetrate the cell wall and infect the plant (Link et al. 2005). Additionally, this enzyme has a detoxifying activity for reactive oxygen species (ROS) produced by the host as a defense mechanism against the fungus (Link et al. 2005). Furthermore, D-arabitinol dehydrogenase was found to be the most highly down-regulated gene in a transcriptomic analysis of A. apis in vivo-versus in vitro (Cornman et al. 2012). These two lines of evidence - transcription and positive selection - strongly suggest that D-arabitinol dehydrogenase is an important gene for Ascosphaera pathogenicity. This has interesting implications for the role this enzyme might play not only for the metabolism, but also for the pathogenicity of entomopathogenic fungi in general, especially considering studies that have demonstrated that filamentous endophytic insect pathogenic fungi such as Metarhizium spp. and Beauveria spp., use very similar mechanisms to infect their hosts. In fact, the same gene appears to be involved in the processes of virulence in insects and in plant colonization (Branine et al. 2019).
Secondary metabolite genes also seem to be involved in pathogenicity and are in general observed more in the clade comprising both bee pathogens and A. pollenicola. For example, A. apis presented an exclusive fungal ribosomally synthesized and post-translationally modified peptides (F-RiPP) biosynthetic cluster, comprising the cytochrome P450 gene. Molecular characterization of this enzyme family in the entomopathogenic fungus B. bassiana revealed that cytochrome P450 is involved in the degradation of hydrocarbons in the outermost layer of insects’ epicuticle (Pedrini et al. 2010). As previously mentioned, most filamentous entomopathogenic fungi penetrate the exoskeleton to infect insect hosts. However, Ascosphaera spp. spores infect bees after being ingested, and thus need to overcome the peritrophic membrane (PM) barrier to cause disease. Although hydrocarbons haven’t yet been described in the PM, its composition is mostly of chitin, glycosaminoglycans, as well as proteins and proteoglycans (Hegedus et al. 2009). N-acetyltransferase O1 and gamma-glutamyl transpeptidase, present in the F-RiPP biosynthetic cluster may be involved in the adhesion and degradation of the PM by this pathogen. A class of acetyltransferase also showed signatures of significant positive selection in A. apis, once again highlighting the potential arsenal of carbohydrate active enzymes on the capacity of the genus to infect its hosts. Similar mechanisms of chitin degradation enabling active PM invasion have been reported for other bee pathogens, such as that of the bacteria Paenibacillus larvae (Garcia-Gonzalez and Genersch 2013).
The presence of this gene cluster is especially interesting to consider in light of our other positively selected genes in the pathogen clade being implicated in post translational modifications, as they could be potentially playing ubiquitin-like roles (Getachew et al. 2020). Post translational modification might be especially useful inside a host or between different lifestyles as in opportunistic pathogens (Lorenz 2013). In Cr. neoformans, upon temperature shifts associated with entry into a host, the mRNA encoding ribosomal proteins are rapidly degraded (Bloom et al. 2019). This transcriptional and translational rewiring has been suggested to be a key mechanism for Cryptococci to withstand stress and evade the innate immune system (Bloom et al. 2019). Similarly, our positively selected gene dihydrodipicolinate synthase, present in a T1PKS biosynthetic cluster in A. apis and A. aggregata, was shown to be important in stress tolerance, vegetative growth and pathogenesis in Fusarium asiaticum (Ren et al. 2018).
Infections caused by A. apis and A. aggregata are known to be temperature sensitive (Bailey 1968, Xu and James 2012). Interestingly, Apis mellifera (A. apis host) can recognize A. apis infections and induce a “social fever” to heat up the brood cells and kill the fungi before infection progresses too far (Starks et al. 2000). In the alfalfa leafcutting bee (Megachile rotundata), both high and low temperatures appear to induce an immune response before infection can take hold (Xu and James 2012). Temperature stress on Ascosphaera is especially interesting because heat shock proteins present in biosynthetic clusters for A. apis and A. pollenicola show signatures of positive selection in A. apis. Such proteins have been implicated in virulence in several other pathogenic fungi. For instance in Metarhizium robertsii (a widely distributed insect pathogen used for insect pest control) when genes responsible for heat tolerance were knocked out, the expression of five different heat shock proteins were eliminated and virulence was highly reduced in the Galleria mellonella host (Xie et al. 2019). Similarly, Aspergillus fumigatus demonstrated an inability to grow in vitro and a loss of virulence in a murine model when HSP90 was repressed (Lamoth et al. 2014), demonstrating the importance of chaperone-like functions in the virulence of other Eurotiales and possibly in the genus Ascosphaera.
In regard to the ribosomal proteins under positive selection, there are several “extra ribosomal” functions notably implicated in immune signaling and disease (Zhou et al. 2015). For instance, some of these ribosomal proteins are found in the terpene lanosterol synthase (oxidosqualene cyclase) cluster, showing their involvement in the biosynthesis of terpenes. This BGC is present only in the bee pathogens A. apis and A. aggregata and in A. pollenicola. Lanosterol synthase catalyzes the conversion of (3S)-2,3-oxidosqualene to lanosterol in the biosynthetic pathway of sterols and triterpenes (Abe 2007; Shang et al. 2010). The lanosterol backbone can be, then, modified to a variety of different structures. A similar lanosterol cyclase cluster has been proven to be upregulated in Metarhizium anisopliae during the early stages of infection in the tick Rhipicephalus microplus, which indicates an involvement with the pathogenic process in this fungus (Sbaraini et al. 2016) and quite possibly in the pathogenesis to both honeybees and leaf-cutting bees as well.
We demonstrate that several genes playing classic entomopathogenic roles in ascomycete fungi are also present in Ascosphaera species previously thought to be commensal. For example, A. acerosa presented an exclusive BGC containing only a hydroxamate-type ferrichrome siderophore peptide synthetase gene, which is a common type of siderophore produced by hypocrealean fungi (Liu et al. 2017) suggesting a possible virulent activity for the species. Additionally, a biosynthetic cluster similar to D-alanine-poly (phosphoribitol) ligase was found in all species, except in A. acerosa. This cluster displayed high similarity to N-(5-amino-5-carboxypentanoyl)-L-cysteinyl-D-valine synthase. To date, there is no comprehensive characterization of this ligase in fungi, but there is very robust evidence that it is necessary for the avoidance of insect cationic antimicrobial peptides by the modification of teichoic acids in the cell wall. In the entomopathogenic bacteria Bacillus thurigiensis and B. cereus (Wu et al. 2019), the mechanism involves positively charging the cell surface thus repelling these AMPs and resisting the host’s immune response. Ascosphaera aggregata displayed a second gene in this biosynthetic cluster with high amino acid identity to the enterobactin synthetase component F, a homolog of the catecholate-like siderophore produced by Enterobacteriaceae and other Eurotiomycetes (Haas et al. 2003). Furthermore, an NRPS cluster in A. apis, A. aggregata and A. acerosa contained a pmd1 gene, responsible for resistance to the potent antifungal Leptomycin B. This is, to our knowledge, the first report of antifungal resistance genes in Ascosphaera spp. The biosynthetic cluster has highly similar genes to the siderophore dimethylcoprogen produced by the citrus pathogen Alternaria alternata (Chen et al. 2013), which has also been previously identified in entomopathogenic fungi (Krasnoff et al. 2020; Molnár et al. 2010). Taken together, our findings agree with the previous hypothesis that additional Ascosphaera species besides A. apis and A. aggregata, have the potential to be opportunistic pathogens of bees (Bissett et al. 1996; Klinger et al. 2013; Skou and Hackett 1979), implicating that entomopathogenicity is a characteristic that most likely evolved from a saprophyte or commensal niche.
Concluding remarks
Overall, our study provides a genomic and evolutionary landscape for the under-studied virulence factors of bee infections in the fungal genus Ascosphaera. Ribosomal genes that play a role in both primary and secondary metabolism may be important in these interactions by regulating genes involved in sensing temperature, nutrient availability, and hypoxia. Additionally, secondary metabolism genes involved in fungicide resistance, iron scavenging and polyketide synthases seem promising as candidate virulence factors. More experimental work is needed to understand the function of such factors and how/if they relate directly to virulence in the bee hosts by entomopathogenic and opportunistic Ascosphaera. Co-genomic approaches that take into account the host genome are needed to understand whether the genes under positive selection in our study are a consequence of coevolution or other environmental pressures. Lastly, additional work should be directed toward understanding where each species falls on the symbiotic spectrum, because we demonstrate that species thought to be ‘commensal’ may be beneficial or opportunistic pathogens. This information is necessary before we can further understand the evolution of pathogenicity in the group.
Materials and Methods
Fungal strains and growth conditions
Cultures for A. pollenicola (strain ATCC 62712) and A. acerosa (strain ATCC 201316) were obtained from American Type Culture Collection (ATCC; Manassas, VA). The culture for A. atra (ARSEF 5147) was obtained from the ARS Collection of Entomopathogenic Fungal Cultures (ARSEF; Ithaca NY). These fungi were plated on aseptic Sabouraud Dextrose Agar (SDA) and Potato Dextrose Agar (PDA), both formulations obtained from Difco (BD companies, Franklin Lakes, NJ) and the resultant growth was used for DNA extraction. Ascosphaera aggregata (strain wild2) culture was grown from a single spore isolated from a diseased Megachile rotundata cadaver collected in Logan, UT by the Pollinating Insect Biology Management and Systematics Unit (USDA-ARS PIMBSR, Logan, Utah). The cadaver was confirmed as infected with a single A. aggregata infection through the use of the species-specific forward (5’-GCACTCCCACCCTTGTCTA-3’) and reverse (5’-CTCGTCGAGGGTCTTTTCC-3’) primers modified from James and Skinner (2005) for qPCR (Klinger et al. 2015). Spore isolation and resulting A. aggregata hyphal growth used for DNA extraction were performed on V-8 media (James and Buckner 2004). All strain and growth condition information is summarized in Table 2.
Table 2:
Fungal isolate source table. Sources, culture media and extraction methods for fungal species used in this study.
|
Ascosphaera species |
Straina | Isolation Sourceb | DNA Extraction method |
|---|---|---|---|
| aggregata | USDA-ARS PIBMSR wild2 | Single spore isolate from Megachile rotundata: Logan, UT; Cultured in lab on V-8 media | PureGene Salty |
| atra | ARSEF 5147 | Isolated from honey in Apis mellifera colony: Waroona, Western Australia; Cultured in lab on SDA | PureGene Salty |
| pollenicola | ATCC 62712 | Isolated from pollen stores of Megachile rotundata: Western Canada; Cultured in lab on PDA | PureGene Salty |
| acerosa | ATCC 201316 | Isolated from Megachile rotundata: Lethbridge, Alberta; Cultured in lab on SDA | MoBio Ultra Clean Plant DNA Kit |
ARSEF: Agricultural Research Service Collection of Entomopathogenic Fungi, Ithaca, New York; ATCC: American Type Culture Collection, Manassas, Virginia; USDA-ARS PIBMSR: Pollinating Insect-Biology, Management, Systematics Research: Logan, Utah.
SDA: Saborard Dextrose Agar; PDA: Potato Dextrose Agar; V8: Modified V8 agar (James and Buckner 2004)
DNA extraction and Sequencing
DNA was extracted for all species, using the CTAB DNA extraction procedure outlined in Carter-House et al. 2020. Libraries were prepared at the Genomics Core Facility in the Institute for Integrative Genome Biology of the University of California, Riverside, using the SeqOnce RhinoSeq protocol (SeqOnce Biosciences, Pasadena, CA). This protocol includes random enzymatic fragmentation resulting in fragments of 100-1000 base pairs. After adapter ligation and PCR amplification, these libraries were then dual size-selected for 200-600 base pair reads using AMPure XP beads. These were then sent to UC San Francisco and sequenced on the Novaseq 6000 using the NovaSeq 6000 S4 Reagent Kit v1.5 (300 cycles) with 2x150 paired-end reads.
Genome Assembly and Annotation
The heterozygosity of the genomes was inferred in GenomeScope (Vurture et al., 2017) using forward and reverse raw reads, after counting k-mers of length 21, using a hash size of 100M and exporting its histogram in jellyfish (Marcais and Kingsford, 2011). Genome assembly was performed by the Automatic Assembly For The Fungi - AAFTF v 0.2.4 (Stajich and Palmer 2018). This tool combines Trimmomatic v0.4 (Bolger et al. 2014) to clip Illumina adapters and low quality sequences (Q < 3) as well as SPAdes (Prjibelski et al. 2020) and BBTools (Bushnell B., BBMap, sourceforge.net/projects/bbmap/, May 20, 2020) for assembling the reads, NCBI-BLAST to search and trim vector and contamination, and polish contigs for accurate base pair identification with Pilon (Walker et al. 2014).
Genome annotations for A. acerosa, A. aggregata, A. atra, A. pollenicola as well as for the complete A. apis genome (accession: GCA_001636715.1) (Qin et al. 2006; Shang et al. 2016) were performed in the Funannotate v1.7.2 pipeline (Palmer and Stajich 2019). We used RepeatModeler v1.0.11 (Hubley et al. 2016) to build a species-specific library of repetitive elements followed by RepeatMasker v4.0.6 (Smit et al. 2015) to find and mask transposable elements, interspersed repeats and low complexity DNA sequences. The masked genome was used for ab initio prediction of the gene models in Augustus v3.3.3 (Stanke and Morgenstern 2005) with default parameters. It was trained based on initial Aspergillus nidulans seed species, along with protein evidence provided by the UniProt/SwissProt database (v2019_11) to EVidence Modeler (Haas et al. 2008) and GeneMark-ES (Borodovsky and Lomsadze 2011) using Funannotate default evidence weights along with ‘--optimize_augustus’ and ‘--keep_no_stops’ arguments to refine training and avoid losing valid models, with a minimum number of 150 models for the training. Evidence was also provided by BUSCO 2.0 (Simão et al. 2015), using default parameters, based on the conservation of 4,046 universal single-copy orthologs in the eurotiomycetes_odb9 dataset (creation date: 02/13/2016, https://busco-archive.ezlab.org/v2/), which was also used to assess the completeness of the genomes.
Functional annotations for the predicted proteins were obtained using Diamond (Buchfink et al. 2015) to search the UniProt/SwissProt protein database (v2019_11) using all default parameters provided by the pipeline. Putative protein function was assigned by sequence similarity to InterProScan v5.48-83.0 (Jones et al. 2014), EggNog v1.0.3 (Huerta-Cepas et al. 2019), dbCAN and CAZyme 9.0 (Lombard et al. 2014), as well as Pfam (Mistry et al. 2020) and MEROPS v12.0 (Rawlings et al. 2014) databases. Gene ontology terms were assigned by InterPro, using default parameters. The secretome was predicted using SignalP v5.0 (Nielsen 2017; Almagro Armenteros et al. 2019) and Phobius v1.01 (Käll et al. 2007), identifying proteins carrying a signal peptide. Assembly statistics were generated in AAFTF v0.2.4 and QUAST v4.6.3 (Gurevich et al., 2013). A gene-set enrichment analysis was performed on the putative CAZYme and MEROPS peptidase content using the Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa et al., 2016) collection of the Molecular Signatures Database (MSigDB; Liberzon et al., 2011) in the software GSEA (Subramanian et al., 2005), with adjusted settings for limited number of gene sets, as follows, and otherwise default parameters: Number of permutations = 1000, Collapse to gene symbols = No_collapse, Permutation type = Gene_set, Metric for ranking genes = Ratio_of_classes, Max size = 500, Min size = 0.
Prediction of Secondary Metabolites
To mine the secondary metabolite genes in Ascosphaera spp., we employed the antibiotics and Secondary Metabolites Analyses Shell (antiSMASH) (Blin et al. 2019) pipeline v.5.1.1 for biosynthetic gene cluster (BGCs) predictions, using relaxed strictness parameters of profile hidden Markov Models (pHMM). All non-annotated genes of the new assemblies were submitted to BLASTp searches against the Non-Redundant (NR) NCBI database. The details are displayed in supplementary table S2, along with the most significant hits and the SMCOGS references when applicable. In order to investigate the relationships between BGCs of different species, we summarized antiSMASH results in a distance metric fashion with the Biosynthetic Genes Similarity Clustering and Prospecting Engine (BiG-SCAPE), which uses a combination of the Jaccard of domain types, Domain Sequence Similarity and Adjacency indices (Navarro-Muñoz et al. 2020). Clusters grouped together in BiG-SCAPE that displayed similar BLASTp hits were submitted to further global alignments and synteny comparisons using the Clinker pipeline and visualized in clustermap.js (Gilchrist and Chooi 2020).
Detection of single copy orthologs under positive selection
We used Orthofinder (v 2.3.12) to identify single copy orthologs and infer a species tree (Emms and Kelly 2019), using predicted amino acid sequences for the species. We specified multiple sequence alignment and BLAST as the sequence search program. The species tree was generated from concatenated alignments of the single copy orthogroups generated with STAG (Emms and Kelly 2018). We used FastTree as the tree inference program, using a maximum likelihood model (Emms and Kelly 2017). We performed codon aware alignments with the nucleotide and amino acid sequences corresponding to the identified single copy orthologs (SCOs) with ete3 (v 2.0.3) mixed mode alignment (Huerta-Cepas et al, 2010). We used the “ete3 build” command to convert amino acids to nucleotides if the average protein similarity was higher than 90%.
To detect signatures of positive selection, we used the “ete3 (v 2.0.3) evol” function to run a branch-site model on nucleotide alignments of the SCOs (Huerta-Cepas et al, 2010). We marked the tree by setting each pathogen alone as the foreground against the other four species as the background to determine genes specific to each pathogen under positive selection. We repeated these steps after excluding A. atra (a commensal) due to the relative incompleteness of its genome compared to others. This allowed us to pull even more high-quality single copy orthologs under significant positive selection between the two pathogens. The results from the five species versus four species (no A. atra) comparison are delineated in Supplemental Table 1.
Quality Control
After only pulling out single copy orthologs that showed signatures of significant positive selection (p < 0.05) for each model, we then went through each alignment by hand to rule out false positives due to missing pieces or misalignments. To account for multiple tests we used the qvalue package (version 2.22.0) in R (version 4.0.3) to convert p-values from the CodeML output to q-values (Dabney et al. 2010). We removed any genes with false discovery rates (FDR) greater than 5% (or q-value > 0.05). To remove cryptic paralogs, we used Notung which is a gene-tree species tree reconciliation software (Chen et al. 2000). We generated pairwise predictions for orthology versus paralogy in each gene across all 5 species. Genes were only kept if no paralogs were present in the resulting gene homology table. We then designated any alignments with average Ks values above three as saturated and removed them (De La Torre et al. 2017). For the branch-site model, saturation does not increase the rate of false positives, rather a high dS is more of a concern for a loss of power (false negatives) (Gharib and Robinson-Rechavi 2013). With that in mind, we provide conservative estimates of genes under positive selection.
Supplementary Material
Significance Statement.
Identification of the genes involved in pathogenicity is a necessary initial step for therapeutic development and obviating the evolution of novel pathogens. We found genomic signatures that suggest that protein processing and metabolism are important processes for the evolution of pathogenicity in bee-associated fungi in the genus Ascosphaera. By using whole genomes to differentiate pathogens from non-pathogens at a gene level we found that rapid, efficient replication may be the most important characteristic of pathogenic Ascosphaera species.
Acknowledgements
This work was supported in part by USDA Agriculture Experimental Station at the University of California, Riverside and NIFA Hatch projects [CA-R-PPA-5062-H to J.E.S and CA-R-ENT-5109-H to Q.S.M]. JES is a Fellow in the CIFAR program Fungal Kingdom: Threats and Opportunities. Additional support came from the National Institutes of Health (NIH) National Institute of General Medical Science Grant [5R01GM122060-02]. Library preparation and sequencing was facilitated by the UC Riverside Genomics Core facility. Analyses were performed on the high-performance computing resources of the Institute for Integrative Genome Biology (IIGB) at UC Riverside [National Science Foundation DBI-1429826 and NIH S10-OD016290]. We would lastly like to thank Derreck Carter-House for all of his help with DNA extraction.
Data Availability Statement
These Whole Genome Shotgun project assemblies and annotation have been deposited at DDBJ/ENA/GenBank under the accessions JAGYHY000000000, JAGYHZ000000000, JAGYIA000000000 and JAGYIB000000000. The versions described in this paper are versions JAGYHY010000000, JAGYHZ010000000, JAGYIA010000000 and JAGYIB010000000. The Illumina sequence data are associated with the BioProjects PRJNA725040, PRJNA725042, PRJNA725043 and PRJNA725050.
References
- 1.Abe I. 2007. Enzymatic synthesis of cyclic triterpenes. Nat. Prod. Rep 24: 1311–1331. 10.1039/b616857b [DOI] [PubMed] [Google Scholar]
- 2.Almagro Armenteros JJ et al. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nature Biotechnology. Springer US, 37(4):420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 3.Anderson DL, Gibbs AJ, Gibson NL. 1998. Identification and phylogeny of spore-cyst fungi (Ascosphaera spp.) using ribosomal DNA sequences. Mycol. Res 102: 541–547. 10.1017/S0953756297005261 [DOI] [Google Scholar]
- 4.Aronstein K, Colby D. 2015. A multiplex PCR assay for determination of mating type in isolates of the honey bee fungal pathogen, Ascosphaera apis. J. Apic. Res 54:105–107. 10.1080/00218839.2015.1109917 [DOI] [Google Scholar]
- 5.Aronstein K, Holloway B. 2013. Honey bee fungal pathogen, Ascosphaera apis; current understanding of host-pathogen interactions and host mechanisms of resistance. In: Mendez-Vilas A, editor. Microbial pathogens and strategies for combating them: science, technology and education. Spain: Formatex Research Centre. 402–410. [Google Scholar]
- 6.Aronstein KA, Murray KD. 2010. Chalkbrood Disease in Honey Bees. Journal of Invertebrate Pathology. 103 Suppl 1 (January): S20–29. [DOI] [PubMed] [Google Scholar]
- 7.Avulova S, Rosengaus RB. 2011. Losing the battle against fungal infection: Suppression of termite immune defenses during mycosis. J. Insect Physiol 57: 966–971. 10.1016/j.jinsphys.2011.04.009 [DOI] [PubMed] [Google Scholar]
- 8.Bailey L. 1968. Honey bee pathology. Annu. Rev. Entomol 13(1):191–212. [Google Scholar]
- 9.Barelle CJ et al. 2006. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol. 8: 961–971. doi: 10.1111/j.1462-5822.2005.00676.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bissett J, Duke G, Goettel M. 1996. Ascosphaera acerosa sp. nov. isolated from the alfalfa leafcutting bee, with a key to the species of Ascosphaera. Mycologia. 88, 797–803. 10.1080/00275514.1996.12026717 [DOI] [Google Scholar]
- 11.Blin K et al. 2019. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47: W81–W87. 10.1093/nar/gkz310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bloom ALM et al. 2019. Thermotolerance in the pathogen Cryptococcus neoformans is linked to antigen masking via mRNA decay-dependent reprogramming. Nat. Commun, 10(1): 4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blume M. et al. 2015. A Toxoplasma gondii Gluconeogenic Enzyme Contributes to Robust Central Carbon Metabolism and Is Essential for Replication and Virulence. Cell Host Microbe. 18:210–220. doi: 10.1016/j.chom.2015.07.008 [DOI] [PubMed] [Google Scholar]
- 14.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England), 30(15): 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boomsma JJ, Jensen AB, Meyling NV, Eilenberg J. 2014. Evolutionary Interaction Networks of Insect Pathogenic Fungi. Annu. Rev. Entomol 59:467–485. 10.1146/annurev-ento-011613-162054 [DOI] [PubMed] [Google Scholar]
- 16.Borodovsky M, Lomsadze A. 2011. Eukaryotic Gene Prediction Using GeneMark.hmm-E and GeneMark-ES. Curr. Protoc. Bioinforma Ed. Board Andreas Baxevanis Al; CHAPTER, Unit-4.610. 10.1002/0471250953.bi0406s35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Branine M, Bazzicalupo A, Branco S. 2019. Biology and applications of endophytic insect-pathogenic fungi. PLoS Pathog. 15. 10.1371/journal.ppat.1007831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12:59–60. 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- 19.Chen K, Durand D, and Farach-Colton M. 2000. “NOTUNG: A Program for Dating Gene Duplications and Optimizing Gene Family Trees.” Journal of Computational Biology: A Journal of Computational Molecular Cell Biology 7 (3-4): 429–47. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Lin C, Chung K. 2013. A nonribosomal peptide synthetase mediates siderophore production and virulence in the citrus fungal pathogen Alternaria alternata. Mol. Plant Pathol 14: 497–505. 10.1111/mpp.12021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cornman RS et al. 2012. Transcriptome analysis of the honey bee fungal pathogen, Ascosphaera apis: implications for host pathogenesis. BMC Genomics. 13:285. 10.1186/1471-2164-13-285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dabney Alan, Storey John D., and Warnes GR. 2010. “Qvalue: Q-Value Estimation for False Discovery Rate Control.” R Package Version 1 (0). [Google Scholar]
- 23.DaFu C et al. 2017. Transcriptomic analysis of Ascosphaera apis stressing larval gut of Apis mellifera ligustica (Hyemenoptera: Apidae). Acta Entomol. Sin 60:401–411. [Google Scholar]
- 24.De La Torre AR, Li Z, Van de Peer Y, Ingvarsson PK. 2017. Contrasting Rates of Molecular Evolution and Patterns of Selection among Gymnosperms and Flowering Plants. Molecular Biology and Evolution 34(6):1363–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Vries S, de Vries J. 2020. A Global Survey of Carbohydrate Esterase Families 1 and 10 in Oomycetes. Front. Genet 11. 10.3389/fgene.2020.00756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emms DM, Kelly S. 2019. OrthoFinder: Phylogenetic Orthology Inference for Comparative Genomics. Genome Biology. 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emms DM, Kelly S. 2018. STAG: Species Tree Inference from All Genes. Cold Spring Harbor Laboratory. 10.1101/267914. [DOI] [Google Scholar]
- 28.Emms DM, Kelly S. 2017. STRIDE: Species Tree Root Inference from Gene Duplication Events. Mol Biol Evol. 34(12): 3267–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ene IV, Brunke S, Brown AJP, Hube B. 2014. Metabolism in Fungal Pathogenesis. Cold Spring Harbor Perspectives in Medicine. 4(12): a019695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Gonzalez E, Genersch E. 2013. Honey bee larval peritrophic matrix degradation during infection with Paenibacillus larvae, the aetiological agent of American foulbrood of honey bees, is a key step in pathogenesis. Environ. Microbiol 15:2894–2901. 10.1111/1462-2920.12167 [DOI] [PubMed] [Google Scholar]
- 31.Getachew A et al. 2020. Transcriptome profiling reveals insertional mutagenesis suppressed the expression of candidate pathogenicity genes in honeybee fungal pathogen, Ascosphaera apis. Sci. Rep 10:7532. 10.1038/s41598-020-64022-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Getachew A et al. 2018. Molecular identification of pathogenicity associated genes in honeybee fungal pathogen, Ascosphaera apis, by Restricted Enzyme-Mediated Integration (REMI) constructed mutants. Int. J. Agric. Biol 20: 2879–2890. [Google Scholar]
- 33.Gharib WH, Robinson-Rechavi M. 2013. The Branch-Site Test of Positive Selection Is Surprisingly Robust but Lacks Power under Synonymous Substitution Saturation and Variation in GC. Molecular Biology and Evolution 30(7): 1675–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilchrist CLM, Chooi YH. 2020. clinker & clustermap.js: Automatic generation of gene cluster comparison figures (preprint). Bioinformatics. 10.1101/2020.11.08.370650 [DOI] [PubMed] [Google Scholar]
- 35.Gilliam M, Lorenz BJ. 1993. Enzymatic activity of strains of Ascosphaera apis, an entomopathogenic fungus of the honey bee, Apis mellifera. Apidologie. 24:1, 19–23. 10.1051/apido:19930102 [DOI] [Google Scholar]
- 36.Gilliam M, Prest DB, Lorenz BJ. 1989. Microbiology of pollen and bee bread: taxonomy and enzymology of molds. Apidology. 20, 53–68. 10.1051/apido:19890106 [DOI] [Google Scholar]
- 37.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies, Bioinformatics. 29:8, 1072–1075. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haas BJ et al. 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 9, R7. 10.1186/gb-2008-9-1-r7mmml,m;m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haas H et al. 2003. Characterization of the Aspergillus nidulans transporters for the siderophores enterobactin and triacetylfusarinine C. Biochem. J 371:505–513. 10.1042/BJ20021685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hallborn J, Walfridsson M, Penttilä M, Keränen S, Hahn-hägerdal B. 1995. A short-chain dehydrogenase gene from Pichia stipitis having D-arabinitol dehydrogenase activity. Yeast. 11(9):839–847. [DOI] [PubMed] [Google Scholar]
- 41.Hegedus D, Erlandson M, Gillott C, Toprak U. 2009. New Insights into Peritrophic Matrix Synthesis, Architecture, and Function. Annu. Rev. Entomol 54:285–302. 10.1146/annurev.ento.54.110807.090559 [DOI] [PubMed] [Google Scholar]
- 42.Hubley R et al. 2016. The Dfam database of repetitive DNA families. Nucleic Acids Research. 44:81–89. doi: 10.1093/nar/gkv1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huerta-Cepas J et al. 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Research, 47(D1):D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huerta-Cepas J, Dopazo J, Gabaldón T. 2010. ETE: a python Environment for Tree Exploration. BMC Bioinformatics. 11:1. doi: 10.1186/1471-2105-11-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.James RR, Skinner JS. 2005. PCR diagnostic methods for Ascosphaera infections in bees. J. Invertebr. Pathol 90:98–103. 10.1016/j.jip.2005.08.004 [DOI] [PubMed] [Google Scholar]
- 46.James RR, Buckner JS. 2004. Lipids stimulate spore germination in the entomopathogenic ascomycete Ascosphaera aggregata. Mycopathologia.158(3):293–302. [DOI] [PubMed] [Google Scholar]
- 47.Jones P et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics. 30:1236–1240. 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Käll L, Krogh A, Sonnhammer ELL. 2007. Advantages of combined transmembrane topology and signal peptide prediction-the Phobius web server. Nucleic Acids Res. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Research. 44:D1, D457–D462. 10.1093/nar/gkv1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keller A et al. 2018. Wild Bees and Their Nests Host Paenibacillus Bacteria with Functional Potential of Avail. Microbiome. 6(1): 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klinger EG, James RR, Youssef NN, Welker DL. 2013. A multi-gene phylogeny provides additional insight into the relationships between several Ascosphaera species. J. Invertebr. Pathol 112:41–48. 10.1016/j.jip.2012.10.011 [DOI] [PubMed] [Google Scholar]
- 52.Klinger EG, Vojvodic S, DeGrandi-Hoffman G, Welker DL, James RR. Mixed infections reveal virulence differences between host-specific bee pathogens. J. Invertebr. pathol 2015. Jul 1:129:28–35. [DOI] [PubMed] [Google Scholar]
- 53.Krasnoff SB, Howe KJ, Heck ML, Donzelli BGG. 2020. Siderophores from the Entomopathogenic Fungus Beauveria bassiana. J. Nat. Prod 83:296–304. 10.1021/acs.jnatprod.9b00698 [DOI] [PubMed] [Google Scholar]
- 54.Kronstad J et al. 2012. Adaptation of Cryptococcus Neoformans to Mammalian Hosts: Integrated Regulation of Metabolism and Virulence. Eukaryotic Cell 11(2): 109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lamoth F, Juvvadi PR, Gehrke C, Asfaw YG, Steinbach WJ. 2014. Transcriptional Activation of Heat Shock Protein 90 Mediated via a Proximal Promoter Region as Trigger of Caspofungin Resistance in Aspergillus Fumigatus. The Journal of Infectious Diseases. 209(3):473–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Z et al. 2020. Changes in Antioxidant Enzymes Activity and Metabolomic Profiles in the Guts of Honey Bee (Apis mellifera) Larvae Infected with Ascosphaera apis. Insects. 11: 419. 10.3390/insects11070419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. 2011. Molecular signatures database (MSigDB) 3.0, Bioinformatics. 27:12, 1739–1740. 10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Link T et al. 2005. Characterization of a novel NADP+-dependent D-arabitol dehydrogenase from the plant pathogen Uromyces fabae. Biochem. J 389(2):289–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu H et al. 2017. The Stress-Responsive and Host-Oriented Role of Nonribosomal Peptide Synthetases in an Entomopathogenic Fungus, Beauveria bassiana. J. Microbiol. Biotechnol 27:439–449. 10.4014/jmb.1606.06056 [DOI] [PubMed] [Google Scholar]
- 60.Lombar V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. 2013. The carbohydrate-active enzymes database (CAZy). Nucleic Acids Res. 42(D1):D490–D495, 10.1093/nar/gkt117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lorenz MC. 2013. Carbon catabolite control in Candida albicans: new wrinkles in metabolism. MBio. 4(1): e00034--13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marcais G and Kingsford C. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 27:6, 764–770. doi: 10.1093/bioinformatics/btr011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marsh JJ, Lebherz HG. 1992. Fructose-bisphosphate aldolases: an evolutionary history. Trends Biochem. Sci 17:110–113. doi: 10.1016/0968-0004(92)90247-7 [DOI] [PubMed] [Google Scholar]
- 64.Mistry J et al. 2021. Pfam: The protein families database in 2021, Nucleic Acids Res. 49(D1):D412–D419. 10.1093/nar/gkaa913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Molnár I, Gibson DM, Krasnoff SB. 2010. Secondary metabolites from entomopathogenic Hypocrealean fungi. Nat. Prod. Rep 27:1241–1275. 10.1039/C001459C [DOI] [PubMed] [Google Scholar]
- 66.Navarro-Muñoz JC et al. 2020. A computational framework to explore large-scale biosynthetic diversity. Nat Chem Biol. 16:60–68. 10.1038/s41589-019-0400-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nielsen H. 2107. Predicting Secretory Proteins with SignalP. Methods Mol Biol. 1611:59–73. doi: 10.1007/978-1-4939-7015-5_6. [DOI] [PubMed] [Google Scholar]
- 68.Palmer MJ, Stajich JE. 2019. Funannotate. doi: 10.5281/zenodo.3679386. [DOI] [Google Scholar]
- 69.Pedrini N, Zhang S, Juárez MP, Keyhani NO. 2010. Molecular characterization and expression analysis of a suite of cytochrome P450 enzymes implicated in insect hydrocarbon degradation in the entomopathogenic fungus Beauveria bassiana. Microbiology. 156:2549–2557. 10.1099/mic.0.039735-0 [DOI] [PubMed] [Google Scholar]
- 70.Pitts-Singer TL, Cane JH. 2011. The Alfalfa Leafcutting Bee, Megachile Rotundata: The World’s Most Intensively Managed Solitary Bee. Annual Review of Entomology 56: 221–37. [DOI] [PubMed] [Google Scholar]
- 71.Price MS et al. 2011. Cryptococcus neoformans requires a functional glycolytic pathway for disease but not persistence in the host. mBio. 2:e00103–11. doi: 10.1128/mBio.00103-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. 2020. Using SPAdes De Novo Assembler. Current Protocols in Bioinformatics. 70 (1): e102. [DOI] [PubMed] [Google Scholar]
- 73.Puckett S et al. 2014. Inactivation of fructose-1,6-bisphosphate aldolase prevents optimal co-catabolism of glycolytic and gluconeogenic carbon substrates in Mycobacterium tuberculosis. PLoS Pathog. 10:e1004144. doi: 10.1371/journal.ppat.1004144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qin X, Evans JD, Aronstein KA, Murray KD, Weinstock GM. 2006. Genome Sequences of the Honey Bee Pathogens Paenibacillus Larvae and Ascosphaera Apis. Insect Molecular Biology. 15(5): 715–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rawlings ND, Waller M, Barrett AJ, Bateman A. 2013. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 42(DI):D503–9. doi: 10.1093/nar/gkt953. Epub 2013 Oct 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ren W, Tao J, Shi D, Chen W, Chen C. 2018. Involvement of a dihydrodipicolinate synthase gene FaDHDPS1 in fungal development, pathogenesis and stress responses in Fusarium asiaticum. BMC Microbiol. 18(1):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carter-House D, Stajich J, Unruh S, Kurbessoian T. 2020. Fungal CTAB DNA Extraction. Protocols.io. dx.doi.org/ 10.17504/protocols.io.bhx8j7rw [DOI] [Google Scholar]
- 78.Sandai et al. 2012. The Evolutionary Rewiring of Ubiquitination Targets Has Reprogrammed the Regulation of Carbon Assimilation in the Pathogenic Yeast Candida Albicans. mBio. 3(6). 10.1128/mBio.00495-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sbaraini N et al. 2016. Secondary metabolite gene clusters in the entomopathogen fungus Metarhizium anisopliae: genome identification and patterns of expression in a cuticle infection model. BMC Genomics. 17:736. 10.1186/s12864-016-3067-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shang CH, Shi L, Ren A, Qin L, Zhao MW. 2010. Molecular Cloning, Characterization, and Differential Expression of a Lanosterol Synthase Gene from Ganoderma lucidum. Biosci. Biotechnol. Biochem 74:974–978. 10.1271/bbb.90833 [DOI] [PubMed] [Google Scholar]
- 81.Shang Y et al. Divergent and Convergent Evolution of Fungal Pathogenicity. Genome Biology and Evolution. 8(5):1374–1387, 10.1093/gbe/evw082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- 83.Skou JP, Hackett K. 1979. A New, Homothallic Species of Ascosphaera. Friesia. 11: 265–271. [Google Scholar]
- 84.Smit A, Hubley R, Green P. 2015. RepeatMasker. Open-4.0 Available at: http://www.repeatmasker.org. [Google Scholar]
- 85.Stajich JE, Palmer J. 2018. AAFTF: Automated Assembly for the Fungi, v0.2.1. doi: 10.5281/zenodo.1658103. [DOI] [Google Scholar]
- 86.Starks PT, Blackie CA, Seeley TD. 2000. Fever in honeybee colonies. Naturwissenschaften. 87(5):229–231. [DOI] [PubMed] [Google Scholar]
- 87.Stanke M, Morgenstern B. 2005. AUGUSTUS : a web server for gene prediction in eukaryotes that allows user-defined constraints. 33:465–467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences. 102:43, 15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tatusov RL et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Valero-Jiménez CA, Wiegers H, Zwaan BJ, Koenraadt CJM, van Kan JAL. 2016. Genes involved in virulence of the entomopathogenic fungus Beauveria bassiana. J. Invertebr. Pathol 133:41–49. 10.1016/j.jip.2015.11.011 [DOI] [PubMed] [Google Scholar]
- 91.Vuong HQ, McFrederick QS. 2019. Comparative Genomics of Wild Bee and Flower Isolated Lactobacillus Reveals Potential Adaptation to the Bee Host. Genome Biology and Evolution 11(8): 2151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vurture GW, Sedlazeck FJ, Nattestad M, Underwood CJ, Fang H, Gurtowski J, Schatz MC. 2017. GenomeScope: fast reference-free genome profiling from short reads, Bioinformatics. 33:14, 2202–2204. 10.1093/bioinformatics/btx153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Walker BJ et al. 2014. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PloS One. 9 (11): e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Williams MK, Tripodi AD, Szalanski AL. 2019. Molecular survey for the honey bee (Apis mellifera L.) trypanosome parasites Crithidia mellificae and Lotmaria passim. Journal of Apicultural Research. 58(4):553–8. [Google Scholar]
- 95.Wu H et al. 2019. The dlt operon in Bacillus thuringiensis confers resistance to cationic antimicrobial peptides and virulence to insect. [Google Scholar]
- 96.Xie T et al. 2019. MrSVP, a Secreted Virulence-Associated Protein, Contributes to Thermotolerance and Virulence of the Entomopathogenic Fungus Metarhizium Robertsii. BMC Microbiology. 19:25. 10.1186/s12866-019-1396-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu J et al. 2017. The Entomopathogenic Fungi Isaria fumosorosea Plays a Vital Role in Suppressing the Immune System of Plutella xylostella: RNA-Seq and DGE Analysis of Immunity-Related Genes. Front. Microbiol 8. 10.3389/fmicb.2017.01421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xu J, James RR. 2012. Temperature stress affects the expression of immune response genes in the alfalfa leafcutting bee, Megachile rotundata. Insect Mol. Biol 21(2):269–280. [DOI] [PubMed] [Google Scholar]
- 99.Zhong K, Liu ZC, Wang JL, Liu XS. 2017. The entomopathogenic fungus Nomuraea rileyi impairs cellular immunity of its host Helicoverpa armigera. Arch. Insect Biochem. Physiol 96:e21402. 10.1002/arch.21402 [DOI] [PubMed] [Google Scholar]
- 100.Zhou X, Liao WJ, Liao JM, Liao P, Lu H. 2015. Ribosomal proteins: functions beyond the ribosome. J. Mol. Cell Biol 7(2):92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ziveri J et al. 2017. The metabolic enzyme fructose-1,6-bisphosphate aldolase acts as a transcriptional regulator in pathogenic Francisella. Nat. Commun 6:853. doi: 10.1038/s41467-017-00889-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
These Whole Genome Shotgun project assemblies and annotation have been deposited at DDBJ/ENA/GenBank under the accessions JAGYHY000000000, JAGYHZ000000000, JAGYIA000000000 and JAGYIB000000000. The versions described in this paper are versions JAGYHY010000000, JAGYHZ010000000, JAGYIA010000000 and JAGYIB010000000. The Illumina sequence data are associated with the BioProjects PRJNA725040, PRJNA725042, PRJNA725043 and PRJNA725050.