Abstract
Acute thymic atrophy occurs following type 1 inflammatory conditions such as viral infection and sepsis, resulting in cell death and disruption of T cell development. However, the impact type 1 immunity has on thymic-resident innate lymphoid cells (ILCs) remains unclear. Single-cell RNA sequencing revealed neonatal thymic–resident type 1 ILCs (ILC1s) as a unique and immature subset compared to ILC1s in other primary lymphoid organs. Culturing murine neonatal thymic lobes with the type 1 cytokines interleukin-12 (IL-12) and IL-18 resulted in a rapid expansion and thymic egress of KLRG1+CXCR6+ cytotoxic ILC1s. Live imaging showed the subcapsular thymic localization and exit of ILC1s following IL-12 + IL-18 stimulation. Similarly, murine cytomegalovirus infection in neonates resulted in thymic atrophy and subcapsular localization of thymic-resident ILC1s. Neonatal thymic grafting revealed that type 1 inflammation enhances the homing of cytokine-producing thymus-derived ILC1s to the liver and peritoneal cavity. Together, we show that type 1 immunity promotes the expansion and peripheral homing of thymic-derived ILC1s.
Type 1 immunity results in expansion and peripheral homing of neonatal thymic–derived ILC1s.
INTRODUCTION
The type 1 immunity–inducing cytokines interleukin-12 (IL-12) and IL-18 are critical in viral infection (1, 2) and sepsis (3, 4). Human and murine cytomegalovirus (CMV) infections are associated with increased production of IL-12 and IL-18 (2, 5, 6), while congenital and perinatal human CMV infections can result in morbidity or life-long neurological complications (7, 8). Although neonatal CMV infections rarely result in long-term complications, preterm infants can develop serious sepsis-like syndrome (8, 9). During viral infection and sepsis, the thymus undergoes acute thymic atrophy (10, 11), where T cell development is disrupted but can recover when the inflammation is resolved (12, 13). Acute thymic atrophy differs from age-related thymic involution, where the thymic tissue is slowly replaced with fibroblasts and adipose tissue (14). Although infection-induced thymic atrophy is evolutionarily conserved in vertebrates (11, 15), the underlying causality and selective advantage for the host remains unclear.
Group-1 innate lymphoid cells (ILCs) comprise ILC1s and conventional natural killer (cNK) cells. ILC1s primarily reside in the tissues and mediate an early response against viral infections and cancer, whereas cNK cells circulate and act throughout the immune response (16–18). ILC1s are seeded in the body during several distinct perinatal waves (19, 20) and are the dominant population of group-1 ILCs across tissues at birth (19). On the other hand, cNK cells originate from the bone marrow and develop postnatally to become the primary group-1 ILC population in the spleen and bone marrow within the first 2 weeks of life (19). However, contrary to cNK cells, the ontogeny of the different perinatal ILC1 waves remains elusive (19, 21, 22). In the thymus, group-1 ILCs are recognized as a rare population, distinct from other peripheral group-1 ILCs (23, 24), and can develop from early thymic progenitors (ETPs) both from humans and mice (25–28). Group-1 ILCs derived from embryonic ETPs in mice have been shown to comprise both ILC1s and NK cells and derive from bipotent T/ILC-restricted progenitors from the fetal liver (29). Unlike cNK cells, thymic group-1 ILC development depends on IL-7 and the transcription factor GATA3 (23), similar to ILC1s, ILC2s, and ILC3s (30). However, whether thymic-derived ILC1s and cNK cells contribute to the peripheral compartments remains unclear (22). ILC1 numbers increase in all examined tissues during perinatal development in mice from embryonic day 14.5 (E14.5) until postnatal day 3 (P3) (19). The only exception is the thymus, where ILC1 cell numbers reach maximum levels around birth and decrease between P1 and P3 (19). Whether this thymic-specific decrease in postnatal ILC1s signifies cell death or egress to peripheral tissues remains to be determined.
ILC1s and cNK cells express classical pan-NK cell markers, such as NKp46, CD122, and NK1.1, while being negatively delineated for T and B cell lineage markers (19, 30, 31). During viral infection and in cancer models in adult mice, cNK cells acquire “ILC1-like” features, such as tissue residency and enhanced cytokine production (32–34). Despite their phenotypical overlap, ILC1s and cNK cells are considered separate lineages with no developmental interconversion (19, 31, 35–37). ILC1s that display a high capacity for cytokine production and low levels of granzyme have been coined “helper-like” ILC1s based on the parallels with CD4 helper T cells (31, 38). Moreover, IL-7R+ helper–like ILC1s represent a less mature state of ILC1s that can differentiate into Granzyme B+IL-7R− “cytotoxic” ILC1s (38). This observation challenges the classical delineation between ILC1s and cNK cells based on their respective capacity for cytokine production and cytotoxic killing (19, 38).
To examine the effect of type 1 immunity on ILC1s in the thymus, neonates were infected with murine CMV (MCMV) and by sterile inflammation via administering combined IL-12 and IL-18. We show that both MCMV infection and IL-12 + IL-18 injections result in acute thymic atrophy and thymic ILC1 activation. Single-cell RNA sequencing (scRNA-seq) reveals that neonatal thymic–resident ILC1s have a unique and immature phenotype compared to other primary lymphoid organs. Using neonatal thymic organ cultures (NTOCs), we show that IL-12 + IL-18 stimulation results in the expansion and activation of thymic-resident ILC1s exiting from the outer capsule region of the thymus. Subcapsular kidney grafting showed that ILC1s can egress the thymus in vivo in response to type 1 cytokines and home to the liver and peritoneal cavity. Together, our results indicate that type 1 immunity promotes the neonatal thymus to produce peripheral homing ILC1s.
RESULTS
Type 1 inflammation induces expansion of thymus-exiting ILC1s during NTOC
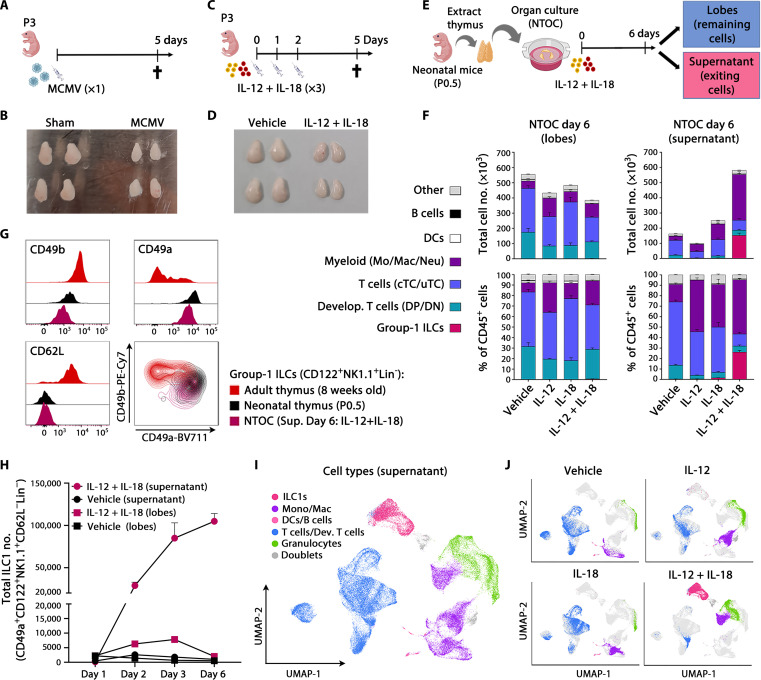
Infecting neonatal mice with MCMV results in acute thymus atrophy within 5 days (Fig. 1, A and B). Since MCMV infection rapidly induces high levels of the type-1 cytokines IL-12 and IL-18 (2), we investigated whether the injection of IL-12 and IL-18 could cause thymic atrophy during sterile inflammation. Similar to MCMV infection (Fig. 1, A and B), intraperitoneal injection of IL-12 and IL-18 in both neonatal and adult mice for three consecutive days resulted in severe acute thymus atrophy, here shown on day 5 (Fig. 1, C and D, and fig. S1, A and B). To investigate the direct effect of IL-12 + IL-18 on the thymus, we cultured neonatal thymic lobes in an NTOC in the presence of the two cytokines (Fig. 1E). IL-12 + IL-18 administration resulted in cell loss compared to the vehicle condition (Fig. 1F and fig. S1D), similar as observed in vivo. Unexpectedly, we detected a large population of group-1 ILCs (either ILC1s or cNK cells) exiting into the supernatant of the organ culture within 6 days and comprising ~25% of the thymus-exiting cells. Furthermore, various myeloid subsets were also observed exiting from the thymus into the supernatant under all four conditions (Fig. 1F and fig. S1C). The group-1 ILCs expressed CD122+IL-18Rα+NK-1.1+ and were lineage-negative (Lin−: TCRβ−TCRγδ−CD3e−CD4−CD8β−Ly-6G−CD19−CD115−CD11c−) (fig. S1C). As ILC1s express CD49b at birth, it is not an exclusive cNK cell marker (19); however, ILC1s are CD49a+CD62L−, and cNK cells are defined as CD49b+CD49a− (19, 21, 30). Thus, when comparing the NTOC-exiting group-1 ILCs with 8-week-old adults and P0.5 neonatal thymic group-1 ILCs (CD122+NK1.1+Lin− cells), we confirmed them as ILC1s based on their CD49a+CD62L− phenotype. The neonatal thymus ILC1s had a similar CD49a+CD62L− phenotype as the NTOC-derived ILC1 population. Only a minority of the adult thymic group-1 ILCs had the CD49a+CD62L− ILC1 phenotype observed in neonatal mice (Fig. 1G). Although Eomes and CD49a are coexpressed in some group-1 ILCs, such as in the salivary gland, hepatic ILC1s and NK cells have a distinct CD49a+Eomes− and CD49a−Eomes+ phenotype, respectively (21, 38). Thus, to uniformly compare ILC1s and cNK cells across different tissues in neonates and adult mice, we gated on CD122+NK1.1+Lin− cells and defined ILC1s as CD49a+CD62L− and cNK cells as CD49b+CD49a−, confirming this definition based on intracellular Eomes expression (fig. S1, E and F). Apart from ILC1s and cNK cells, a subset of NK cells in the adult thymus could be defined as CD49b+CD49a+CD62L+ and displayed high Eomes expression (fig. S1, E and F), as previously described (24). Contrary to adult thymus ILC1s, both the neonatal thymus ILC1s and NTOC-derived ILC1s had a low expression of Eomes (fig. S1E). Kinetic measurements revealed that most of the initial exit and expansion of thymic-derived ILC1s in the supernatant of IL-12 + IL-18–treated NTOC occurred rapidly in the first 3 days after the organ culture was set up (Fig. 1H). Cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) was performed on viable cells from the supernatant after 6 days of NTOC (fig. S1G). Twenty-three distinct cell populations were identified from the four conditions (vehicle, IL-12, IL-18, and IL-12 + IL-18) (fig. S2, A to C). On the basis of the top log-fold differentially expressed genes and manually curated genes (fig. S2, A and B), we identified the overall cell type of each of the thymus-derived populations (Fig. 1I and fig. S2C). The lack of Cd3e, Trdc, Trac, and Bcl11b expression further confirmed the expanded NTOC population as ILC1s rather than T cells. The expression of Ncr1, Tbx21, Cxcr6, Il2rb, and the absence of Sell (encoding CD62L) corroborated their ILC1 identity (fig. S2B). The single-cell data showed a large population of thymic-derived ILC1s exiting the NTOC in response to IL-12 + IL-18 stimulation (Fig. 1J). Along with the ILC1s, several large populations of granulocytes, monocytes, and macrophages were also observed to exit the NTOC lobes (Fig. 1I), confirming the corresponding findings by flow cytometry (Fig. 1, F and J, and fig. S2B). In addition, CITE-seq antibody labeling revealed the group-1 ILC differentiation marker KLRG1 (39) to be highly expressed on the thymic-exiting ILC1s following IL-12 + IL-18 stimulation (fig. S2D). In summary, type 1 immunity results in neonatal thymic atrophy, and the cytokines IL-12 and IL-18 synergize in the production of thymus-exiting ILC1s during NTOC.
Fig. 1. Type 1 inflammation induces expansion of thymus-exiting ILC1s during NTOC.
(A) Schematic of MCMV infections in neonates. (B) Picture of thymic lobes 5 days after infection with MCMV or sham, representative of five independent experiments, as shown in (A). (C) Schematic of IL-12 + IL-18 injections in neonates. (D) Picture of thymic lobes 5 days after the first injection with IL-12 + IL-18 or vehicle, representative of four independent experiments, as shown in (C). (E) Schematic of NTOC strategy for cellular analysis; dividing each well into lobes and supernatant. (F) Stacked bar plots of NTOC performed as illustrated in (E), showing the total number (top) and percentage (bottom) of CD45+ cell types in thymic lobes (left) and supernatant (right) (gating shown in fig. S1C). Data are representative of five independent experiments and shown for n = 3 biological replicates per condition with error bars displaying SEM. DCs, dendritic cells; DN, double negative; DP, double positive; cTC, conventional T cells; uTC, unconventional T cells; Mo, monocytes; Mac, macrophages; Neu, neutrophils. (G) Flow cytometry comparison of ILC1 and cNK markers on group-1 ILCs (CD122+NK1.1+Lin−) from the thymus in adults (8 weeks old), neonates (P0.5), and IL-12 + IL-18–stimulated NTOC supernatant (day 6). Histograms and density plots are representative of n = 6 biological replicates from two independent experiments. (H) Kinetic analysis of ILC1 expansion in NTOC (lobes and supernatant) as shown in (E). Data are representative of three independent experiments and shown for n = 3 biological replicates per condition. The y axis is divided into two different segments. Error bars display mean with SEM. (I) Uniform Manifold Approximation and Projection (UMAP) plot of scRNA-seq data on day 6 NTOC supernatant cells as shown in (E), from four conditions: (i) vehicle, (ii) IL-18, (iii) IL-12, and (iv) IL-12 + IL-18. The major cell types are divided by color. (J) Split UMAPs of the four conditions as in (I); Lin−, Lineage negative, defined as TCRβ−TCRγδ−CD3e−CD4−CD8β−Ter-119−Ly-6G−CD19−CD11c−F4/80−.
NTOC-derived ILC1s are cytokine-producing and have cytotoxic functionality
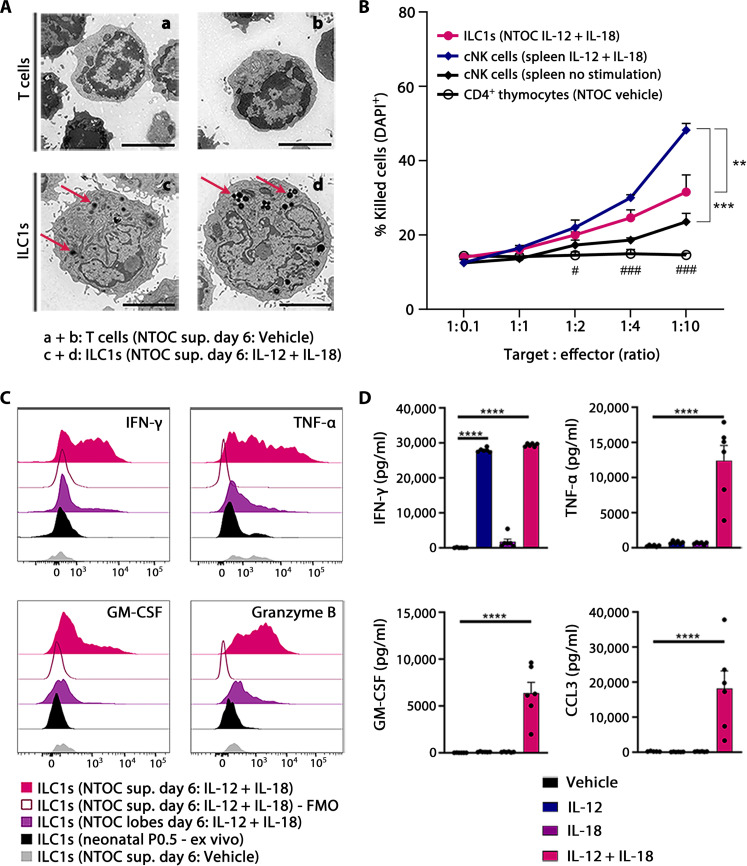
Electron microscopy on sorted ILC1s and T cells from NTOC supernatant revealed the presence of dense cytotoxic–like granules (Fig. 2A) (40). We then sorted ILC1s from IL-12 + IL-18–treated NTOCs, which exhibited a dose-dependent killing of YAC-1 target cells, and confirmed the thymic-derived ILC1s as being cytotoxic (Fig. 2B). The NTOC-derived ILC1s were less cytotoxic compared to splenic cNK cells activated for 18 hours with IL-12 + IL-18. Intracellular flow cytometry verified the NTOC-derived ILC1s expression of Granzyme B, along with enhanced and active production of the cytokines interferon-γ (IFN-γ), tumor necrosis factor–α (TNF-α), and granulocyte-macrophage colony-stimulating factor (GM-CSF) following IL-12 + IL-18 stimulation (Fig. 2C). Protein measurement on the NTOC supernatant after 6 days revealed high levels of secreted IFN-γ, TNF-α, CCL3, and GM-CSF following IL-12 + IL-18 stimulation (Fig. 2D). Combined, these results show that the thymic-derived ILC1s are functional and capable of both cytotoxic killing and cytokine production.
Fig. 2. NTOC-derived ILC1s are cytokine-producing and have cytotoxic functionality.
(A) Transmission electron microscopy pictures of sorted NTOC-derived CD4 T cells (a + b; vehicle supernatant) or ILC1s (c + d; IL-12 + IL-18 supernatant); representative cells are shown from two independent experiments. Red arrows in ILC1 pictures indicate black organelles identified as dense cytotoxic–like granules. Scale bars, 2 μm. (B) YAC-1 killing assay, showing dose-dependent killing of YAC-1 target cells following increasing cell numbers of four different types of sorted effector cells: (i) ILC1s (NTOC IL-12 + IL-18), (ii) cNK cells (adult spleen IL-12 + IL-18), (iii) cNK cells (adult spleen), and (iv) noncytotoxic CD4+ thymocytes (NTOC vehicle). Killing was shown as 4′,6-diamidino-2-phenylindole–positive (DAPI+) YAC-1 cells after 4 hours of coculture. Data are pooled from three independent experiments showing n = 4 to 6 biological replicates per condition. Error bars display mean with SEM. (C) Histograms of intracellular flow cytometry, visualizing active expression of IFN-γ, TNF-α, GM-CSF, and Granzyme B in different NTOC-derived ILC1s compared to ex vivo P0.5 ILC1s. Expression is shown 4 hours after brefeldin A and monensin block and no further stimulation. Histograms are representative of n = 6 to 9 biological replicates per condition from two to three independent experiments. FMO, fluorescence minus one. (D) Bar plot of protein measurements of cytokines in the supernatant after 6 days of NTOC, measured by multiplex enzyme-linked immunosorbent assay. Data are pooled from three independent experiments showing n = 6 biological replicates per condition. Each dot represents a replicate, and error bars show mean with SEM. Statistical significance was calculated by (B) one-way analysis of variance (ANOVA) between cell types; (D) one-way ANOVA (TNF-α and CCL3). Nonparametric Kruskal-Wallis test (IFN-γ and GM-CSF). **P < 0.01, ***P < 0.001, and ****P < 0.0001. (B) One-way ANOVA between NTOC-derived ILC1 and noncytotoxic CD4+ thymocytes: #P < 0.05 and ###P < 0.001.
KLRG1+ ILC1s differentiate and expand from immature thymic ILC1s
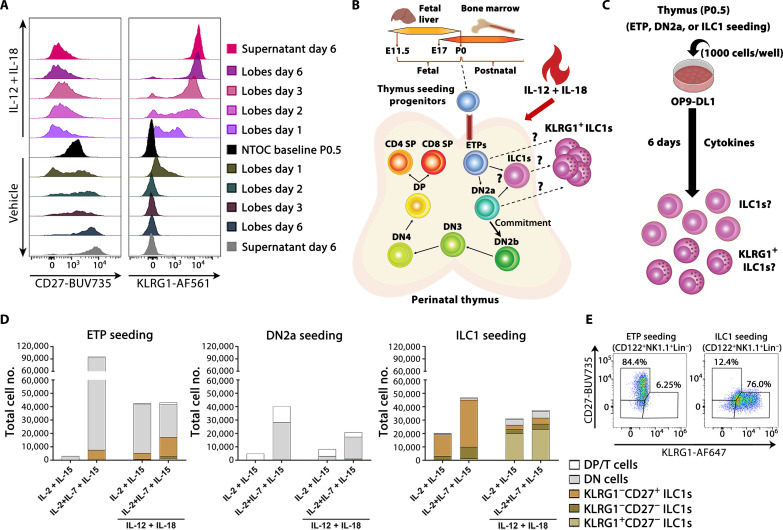
Neonatal thymic lobes from Rag2−/−OTI T cell receptor (TCR)–transgene mice still showed NTOC expansion of ILC1s (fig. S3A), thereby confirming the ontogenesis of thymic-derived ILC1s as independent from T cell development (25). Furthermore, we confirmed that the differentiation marker KLRG1 (39) gradually increased on the ILC1s inside the thymic lobes following IL-12 + IL-18 stimulation. This high KLRG1 expression on ILC1s inside the lobes corresponded to the equally high KLRG1 expression on ILC1s in the supernatant. Conversely, the vehicle control and ex vivo extracted ILC1s displayed an immature CD27+/low phenotype and only expressed low levels of KLRG1 on day 1 of the NTOC (Fig. 3A). Because thymus-seeding ETPs have multipotent potential (41), we hypothesize that the KLRG1high ILC1s differentiate directly either from ETPs, from uncommitted double negative 2a (DN2a) cells, or from immature thymic ILC1s (Fig. 3B). To investigate this, we sorted ETPs, DN2a, and ILC1s from ex vivo neonatal (P0.5) thymus lobes. Each of the three populations was cocultured on OP9-DL1 cells with the common γ-chain cytokines (IL-2, IL-7, and IL-15) in the presence or absence of IL-12 + IL-18 to reveal the precursor of the KLRG1high expanding ILC1s (Fig. 3C). We found that ETPs could develop into group-1 ILCs in a highly IL-7–dependent manner (Fig. 3D), confirming previous findings (23, 25, 27). Their CD122+CD49a+CD62L− phenotype verified their ILC1 identity, and neonatal DN2a-seeded cells consistently only produced few ILC1s (Fig. 3D and fig. S3B), indicating a notable loss of ILC1-potential compared to ETPs. ETP-derived ILC1s primarily displayed a KLRG1−CD27+ phenotype, both in the presence and in the absence of IL-12 + IL-18 (Fig. 3, D and E). Last, seeding neonatal thymic ILC1s followed by 6 days of OP9-DL1 culture with IL-2 + IL-7 + IL-15 mainly yielded an expansion of the immature KLRG1−CD27+ ILC1s, while adding IL-12 + IL-18 resulted in the differentiation of KLRG1 + CD27- ILC1s (Fig. 3, D and E). These results indicate that most of the observed NTOC-exiting KLRG1high ILC1s are likely differentiating and expanding from immature KLRG1− thymic ILC1s rather than ETPs.
Fig. 3. KLRG1+ ILC1s differentiate and expand from immature thymic ILC1s.
(A) Flow cytometry comparison of neonatal thymic ILC1 expression of the maturation markers CD27 and KLRG1 inside the lobes during NTOC, from baseline (P0.5) and days 1, 2, 3, and 6 compared with day 6 in the supernatant. Histograms are representative of n = 9 biological replicates per condition from three independent experiments. (B) Schematic of thymic development, with T cell commitment at the DN2b stage, visualizing hypothesized precursors for the KLRG1+ ILC1s. (C) Schematic of OP9-DL1 organ culture setup to test the hypothesis illustrated in (B), where 1000 cell-sorted progenitor cells were added to each 96-well on top of OP9-DL1 cells to determine whether the rapidly expanded KLRG1+ ILC1s derived from neonatal (P0.5) ETPs or immature ILC1s. DL1 notch ligand–expressing OP9 cells were used to simulate T cell promoting thymic environment to make sure T cell development remained possible. (D) Stacked bar plots showing developing T cells or ILC1s based on their expression of KLRG1 and CD27 following 6 days of OP9-DL1 coculture for ETPs [CD117hiCD44+CD25−CD122−Lin−], DN2a [CD117hiCD44+CD25+CD122−Lin−], or ILC1s (CD122+Lin−) from the neonatal thymus, stimulated with IL-2, IL-7, and IL-15 with or without IL-12 + IL-18. Populations are defined as follows: DP/T cells (CD3e+/CD4+/CD8b+), DN (CD122−Lin−), and ILC1s (CD122+NK1.1+Lin−). The y axes are divided into two segments. Results are shown as the mean of two replicates and representative of n = 4 to 6 biological replicates per condition from two to three independent experiments. (E) Gating on KLRG1 and CD27 expression in CD122+NK1.1+Lin−-gated ILC1s as shown in (D) following 6 days of culture with IL-12 + IL-18 + (IL-2 + IL-7 + IL-15) under ETP and ILC1-seeded conditions. SP, single positive.
Neonatal thymic ILC1s shift expression from CX3CR1 to CXCR6 upon IL-12 + IL-18 stimulation
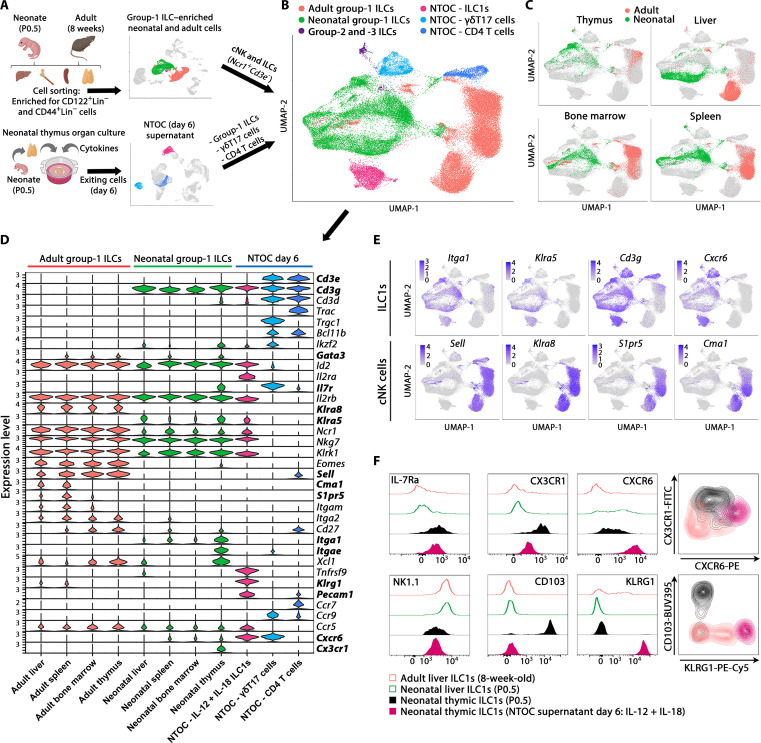
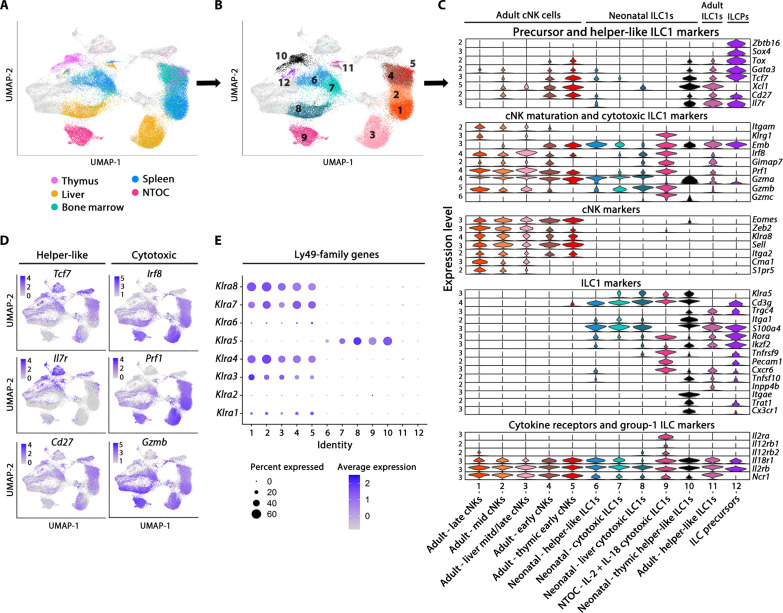
To compare the NTOC-derived ILC1s with other group-1 ILCs, we performed scRNA-seq on group-1 ILC–enriched cells from the liver, spleen, bone marrow, and thymus from P0.5 neonates and 8-week-old adult mice, where group-1 ILCs were identified on the basis of their Ncr1+Cd3e− phenotype. All cells colocalizing with the group-1 ILCs (ILC precursors, ILC2s, and ILC3s) were included in the downstream comparison. ILC1s from NTOC supernatant were incorporated along with two internal control clusters (CD4 T cells and yδT17 cells) (Fig. 4A) to avoid proliferation genes confounding the clustering; only nonproliferating cells were included from the NTOC supernatant. After integration, Uniform Manifold Approximation and Projection (UMAP) and marker gene expression showed six major groups of cells as indicated by color (Fig. 4, B and C, and fig. S4A).
Fig. 4. Thymic ILC1s shift expression from CX3CR1 to CXCR6 upon IL-12 + IL-18 stimulation.
(A) Schematic of scRNA-seq strategy to extract the liver, bone marrow, spleen, and thymus from P0.5 neonates and 8-week-old adult mice. Group-1 ILC cell enrichment was performed, and CD122+Lin− cells and CD44+Lin− cells were sorted for the enrichment of group-1 ILCs (ILC1s and cNK cells) and progenitor cells from each organ. The resulting eight scRNA-seq datasets were combined, and from these, the clusters primarily containing ILC1s and cNK cells (identified by Ncr1+Cd3e− expression) were selected for downstream analysis. In addition, three clusters from the scRNA-seq data in Fig. 1 were selected from the combined four conditions of NTOC supernatant for integration with the new single-cell data: (i) ILC1s (IL-12 + IL-18), (ii) γδT17 cells, and (iii) CD4 T cells. The two T cell clusters were chosen as internal controls. (B) UMAP of Harmony-integrated data with the same colors as in (A), showing the six major groupings divided on the basis of sample origin and gene expression in the case of group-2 and group-3 ILCs. (C) UMAP visualization of cells derived from the four tissues green (neonates) and red (adults). (D) Violin plots showing gene expression of curated genes with colors based on (B), showing the group-1 ILCs and the eight different tissues of origin and the three NTOC clusters. (E) Feature plots of ILC1 markers (top row) and cNK cell markers (bottom row). (F) Flow cytometry comparison of ILC1s across tissues (CD122+NK1.1+CD49a+CD62L−Lin−) from adult liver (8 weeks old), neonatal liver and thymus (P0.5), and IL-12 + IL-18–stimulated NTOC supernatant (day 6), displayed as histograms with thymus-derived ILC1s (closed) and liver ILC1s (open) or density plots (gating shown in fig. S4C). Histograms and density plots are representative of n = 6 biological replicates per condition from two independent experiments. FITC, fluorescein isothiocyanate; PE, phycoerythrin.
All group-1 ILCs (ILC1s and cNK cells) were further divided on the basis of their organ of origin and compared to the three NTOC clusters identified in the cells from the supernatant at day 6 (ILC1s, γδT17 cells, and CD4+ T cells). The group-1 ILCs from all organs shared expression of Id2, IL2rb, Ncr1, Nkg7, and Klrk1 (Fig. 4D). Notably, there was a clear dichotomy in the expression profile between adult and neonatal-derived group-1 ILCs. The neonatal and NTOC group-1 ILCs expressed the ILC1-specific (19, 38) markers Itga1, Klra5, Cd3g, and Cxcr6. However, they did not express the cNK cell–specific (38) markers Sell, Klra8, S1pr5, or Cma1, thus reaffirming that neonatal group-1 ILCs primarily consist of ILC1s (Fig. 4, D and E). Neonatal thymic ILC1s showed high expression of several markers that were not correspondingly expressed in the other tissue-derived group-1 ILCs, such as Il7r, Itgae, and Cx3cr1. In contrast, IL-12 + IL-18–activated NTOC ILC1s displayed high expression of Il2ra, Klrg1, Tnfrsf9, Pecam1, Cxcr6, Ifng, Prf1, Gzmb, and Gzmc (Fig. 4E and fig. S4B). Flow cytometry validation showed that the majority of neonatal thymic ILC1s could be defined as CX3CR1+CD103+CD122+Lin−. Conversely, following stimulation with IL-12 + IL-18, NTOC-derived ILC1s showed no CD103 expression, reduced CX3CR1 and NK1.1 expression, and shifted to high expression of CXCR6 (Fig. 4F and fig. S4C). The shift of thymic ILC1s going from high in situ CX3CR1 expression to enhanced CXCR6 expression in thymic-exiting ILC1s may indicate a changed homing capacity in response to type 1 cytokine activation.
Steady-state neonatal thymic ILC1s display a unique and highly immature phenotype
Following the Harmony integration, the UMAP of adult, neonatal, and NTOC-derived cells were divided into 20 clusters containing ILCs, NK cells, T cells, and ILC progenitors (fig. S5A). Several group-1 ILC populations clustered according to their cell state rather than cell type or tissue imprinting (fig. S5, B and C). To focus the downstream analysis, we selected the 11 group-1 ILC clusters and 1 cluster of ILC precursors (numbered 1 to 12), which clustered according to cell type– and tissue-specific imprinting (Fig. 5, A and B). Of the cell state–clustering group-1 ILC, cluster 13 contains dying cells (fig. S5D), cluster 14 displays a high expression of interferon-stimulated genes (fig. S5C), and clusters 15 and 16 comprise proliferating cells (fig. S5C). A small population were identified as adult ILC1s (cluster 11) with higher gene expression similarity to neonatal ILC1s than adult cNK cells. On the basis of differentially expressed genes, ILC1 and cNK cell–specific markers were determined across tissues in neonates and adult mice (Fig. 5C and fig. S5E). In addition, to further parse out the impact of tissue imprinting on the ILC1 differentiation state, we evaluated Helper-like and cytotoxic ILC1 genes (Fig. 5D). As shown in the perinatal liver (19, 38), the neonatal liver group-1 ILCs (cluster 8) mainly comprise cytotoxic ILC1s based on their high Irf8, Prf1, and Gzmb expression, similar as observed in NTOC-derived ILC1s (Fig. 5, C and D). The neonatal thymic ILC1s (cluster 10) displayed an immature helper-like phenotype and expressed higher levels of associated markers Tcf7, Cd27, and Il7r (19, 38) compared to other neonatal ILC1 clusters (Fig. 5, C and D). Analysis of group-1 ILC transcription factors among neonatal ILC1 populations revealed that the thymic ILC1s expressed the highest levels of ILC precursor markers, such as Tcf7, Gata3, Batf3, and Tox (Fig. 5C and fig. S5F) (38, 42). The vast majority of the adult group-1 ILCs was identified as cNK cells. On the basis of previous single-cell studies (19), the genes Tcf7, Cd27, and Emb decreased according to cNK maturation level, while the genes Itgam, Klrg1, Prf1, Gzmb, Zeb2, Cma1, and S1pr5 increased according to cNK maturation level. These genes were, therefore, used to divide cNK cells into early, mid, and late developmental stages (Fig. 5C and fig. S5E). The Ly49 family genes (Klra1 to Klra8) allowed the separation of the adult group-1 ILCs and neonatal ILC1s (Fig. 5E), confirming previous results showing Klra5 as a specific embryonic-wave ILC1 marker (19, 20). In conclusion, the steady-state thymus primarily contains embryonic-wave ILC1s and displays a unique ILC1 phenotype with higher levels of helper-like ILC1 genes than other neonatal ILC1 clusters.
Fig. 5. Steady-state neonatal thymic ILC1s display a unique and immature phenotype.
(A) Tissue overview showing a UMAP of Harmony-integrated scRNA-seq comparison of group-1 ILCs in 8-week-old adult and P0.5 neonatal mice across tissues from (Fig. 4A), showing an overview of the different tissues (thymus, liver, spleen, and bone marrow) and NTOC clusters. As visualized in (B), only cells from the 12 selected clusters are shown, representing tissue-specific group-1 ILCs and ILC precursors (ILCPs) (neonatal and adult tissues are shown with the same color). (B) UMAP overview of the 12 selected clusters in Harmony-integrated scRNA-seq comparison. (C) Violin plot showing gene expression level of curated cell type and functional genes in selected clusters. Cluster order is based on the similarity in expression of selected genes between clusters. (D) Feature plots showing gene signatures associated with the two ILC1 maturation stages; Helper-like ILC1s (left column) and cytotoxic ILC1s genes (right column). (E) Expression of Ly49 family genes in 12 selected clusters.
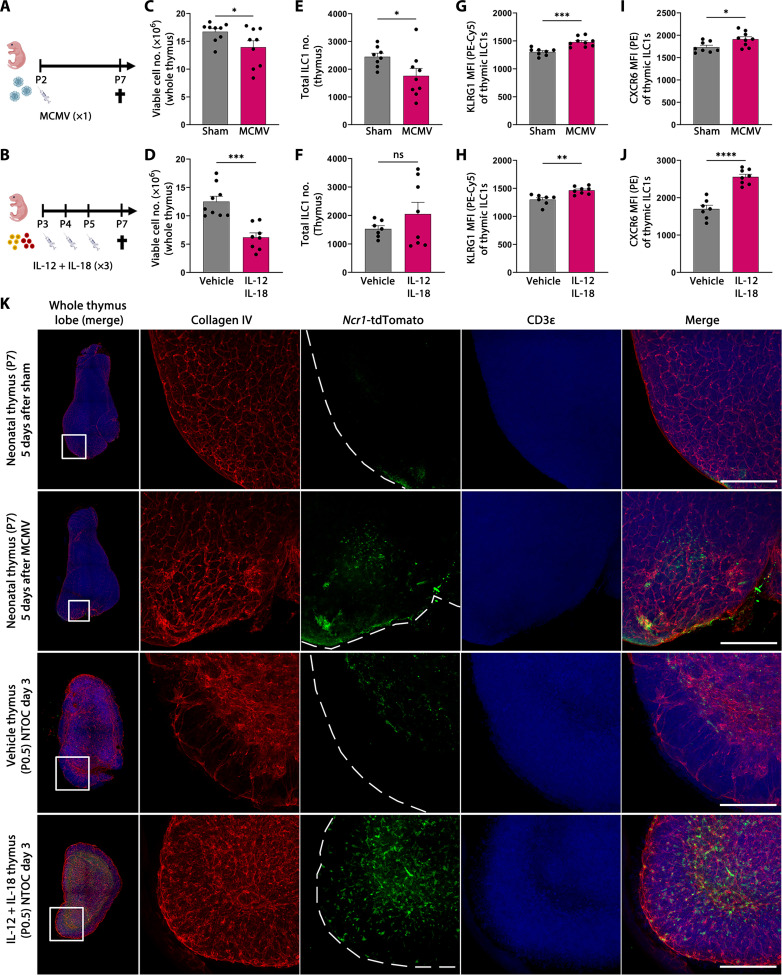
Thymic ILC1s display enhanced CXCR6 and KLRG1 expression during MCMV infection
To investigate the in vivo effect on the thymus and the thymic ILC1 compartment following type 1 inflammation, we infected neonatal mice with MCMV (Fig. 6A) and compared these with neonatal mice receiving three injections with IL-12 + IL-18 (Fig. 6B). In both models, the thymic cellularity was reduced compared to control mice, revealing thymic atrophy (Fig. 6, C and D). In the MCMV-infected neonates, the number of thymic ILC1s was reduced compared to sham control mice (Fig. 6E). However, no difference in ILC1 numbers was observed between IL-12 + IL-18–injected mice and vehicle control (Fig. 6F), which may reflect different kinetics of the two models. Nevertheless, similar as observed in the NTOCs (Fig. 4F), the thymic ILC1s expressed higher levels of KLRG1 and CXCR6 in both models compared to controls (Fig. 6, G and J, and fig. S6, B to E), indicating enhanced activation of the thymic ILC1s. We further investigated whether the thymic group-1 ILCs expressed the NK receptor Ly49H, a lectin-like receptor recognizing MCMV-infected cells (43). However, only 1 to 3% of the CD122+NK1.1+Lin−–expressing thymic group-1 ILCs expressed Ly49H, and no differences were observed between MCMV-infected mice and sham controls (fig. S5F) or between vehicle and IL-12 + IL-18–injected neonates (fig. S6G). Therefore, the observed thymic effects are most likely a result of the type 1 immune response rather than the direct MCMV interaction with the Ly49H receptor (43). In addition, the similarity between the thymic cellularity and ILC1 activation in the two models reveals high similarity of MCMV infection and IL-12 + IL-18 administration.
Fig. 6. Thymic ILC1s display enhanced CXCR6 and KLRG1 expression following MCMV infection.
(A) Schematic of neonates infected with MCMV or sham for 5 days. (B) Schematic of neonates injected three times with IL-12 + IL-18 or vehicle. (C and D) Thymic cellularity at P7 in (C) MCMV-infected and (D) IL-12 + IL-18–injected neonates. (E and F) Flow cytometry–based ILC1 numbers in the thymus at P7 in (E) MCMV-infected and (F) IL-12 + IL-18–injected neonates (gating shown in fig. S6A). (G and H) Flow cytometry–based mean fluorescence intensity of KLRG1 expression on thymic ILC1s in (G) MCMV-infected and (H) IL-12 + IL-18–injected neonates. (I and J) Flow cytometry–based mean fluorescence intensity of CXCR6 expression on thymic ILC1s in (I) MCMV-infected and (J) IL-12 + IL-18–injected neonates. (C) to (J) Data are shown for n = 8 to 9 (MCMV) and n = 7 to 9 (IL-12 + IL-18) neonates and representative of two independent experiments. Bar graphs indicate individual mice (symbol), and error bars display means with SEM. (K) (Column 1) Confocal 3D imaging of neonatal thymic lobes, showing the whole thymic lobe and the zoomed-in section used in the other four columns. (Columns 2 to 4) Single colors and (columns 1 and 5) merged overlay of all colors are shown as maximum projection intensity: red (collagen IV), green (Ncr1-tdTomato), and blue (CD3). (Rows 1 and 2) Neonatal thymus from sham or MCMV-infected mice. (Rows 3 and 4) Thymic lobes from day 3 vehicle or IL-12 + IL-18–stimulated NTOC. Scale bars, 200 μm. Images are representative of n = 3 to 5 mice or biological replicates per condition from two independent experiments. Statistical significance was calculated by unpaired t test: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. ns, not significant.
To further evaluate the effects of MCMV infection on the thymic ILC1 compartment, we used “Ncr1-tdTomato” mice used for group-1 ILCs fate mapping (44), enabling the visualization of the ILC1s in the neonatal thymus. We performed confocal three-dimensional (3D) imaging of the entire thymic lobe. Following MCMV infection on P2, Ncr1-tdTomato+ cells were observed in the cortex and capsular region of the thymus 5 days after (P7). This altered localization of Ncr1-tdTomato+ cells was not observed in the Sham control thymus (Fig. 6K, rows 1 and 2). Similarly, NTOCs using Ncr1-tdTomato thymic lobes showed enhanced subcapsular localization of Ncr1+ cells on day 3 following IL-12 + IL-18 stimulation compared to the vehicle condition (Fig. 6K, rows 3 and 4). To further substantiate these images, we performed live imaging of vehicle and IL-12 + IL-18–stimulated NTOC lobes, and along with the subcapsular localization of thymic ILC1s, it was possible to observe individual ILC1s exiting into the medium (movie S1). Together, these data reveal neonatal thymic ILC1 activation and thymic exit during type 1 immunity.
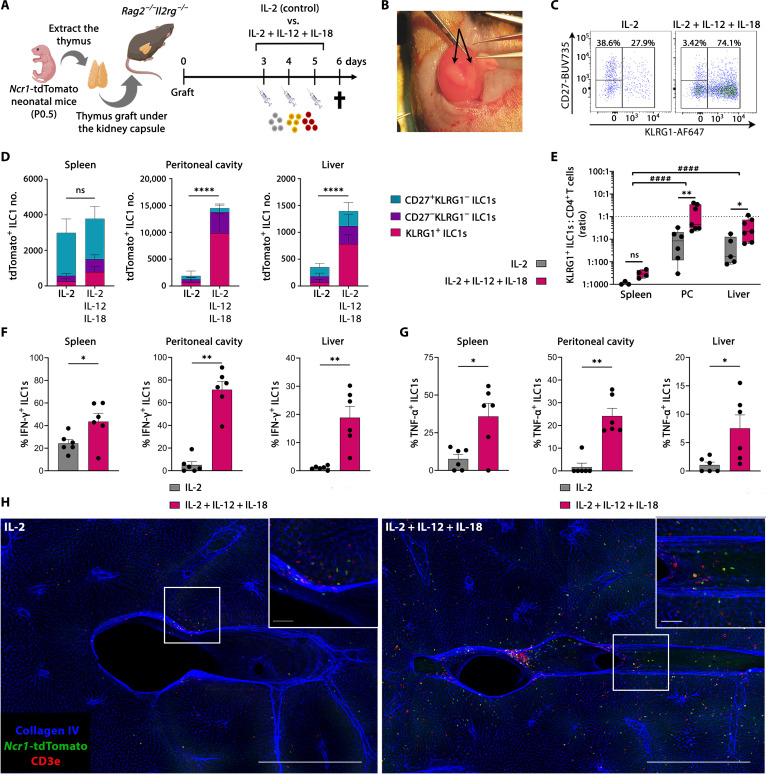
KLRG1+ thymic ILC1s are homing to the liver and the peritoneal cavity
The NTOC cultures revealed thymic ILC1 exit and expansion combined with a high CXCR6 expression, which may indicate a capacity for liver homing (38, 45). Thus, we established a thymic kidney capsule implantation model (46) where P0.5 neonatal thymic lobes from Ncr1-tdTomato mice were grafted under the kidney capsule of Rag2−/−Il2rg−/− mice, which lack T cells, B cells, and ILCs. In this model, all T cells and tdTomato+ ILC1s observed in the transplanted mice are derived from the neonatal thymus graft. IL-2 is critical for the IL-12 + IL-18 synergy (47, 48), and because of their lack of T cells, Rag2−/−Il2rg−/− mice have severely impaired IL-2 production (49). Therefore, IL-2 was either administered alone, as a control, or in combination with IL-12 + IL-18, as illustrated (Fig. 7, A and B). We examined the thymus graft, spleen, liver, and peritoneal cavity by flow cytometry for the presence of thymic graft–derived T cells and tdTomato+ ILC1s (Fig. 7, C and D, and fig. S7A) and confirmed the successful grafting by repopulating the spleen with CD4+ and CD8+ T cells (fig. S7B). A substantial influx of neonatal tdTomato+ ILC1s into the liver, peritoneal cavity, and spleen was observed after administration of IL-2. The administration of IL-12 + IL-18 enhanced the total homing of ILC1s to the liver and peritoneum compared to IL-2–injected mice, but not to the spleen (Fig. 7D). This shows that thymic-derived ILC1s develop and are exiting in the absence of type 1–induced inflammation. Moreover, under both conditions, only low numbers of ILC1s remained in the thymic graft, further indicative of thymic-derived ILC1s’ capacity for peripheral expansion (fig. S7C). To determine the activation status of the thymic-derived ILC1s, we evaluated their CD27 and KLRG1 expression (Fig. 7, C and D). Although the CD27+KLRG1− ILC1s did not differ between treatments (fig. S7D), the thymic-derived KLRG1+ ILC1s displayed enhanced homing to the spleen, liver, and peritoneal cavity when receiving IL-12 + IL-18 (Fig. 7D and fig. S7E). Moreover, the homing of thymus graft–derived CD4 T cells was not significantly changed by the treatment (fig. S7F). To evaluate the homing efficiency in 6 days following the grafting, we determined the ratios of ILC1s to CD4 T cells in each organ. These ratios revealed a preferential homing of ILC1s to the liver and peritoneal cavity (fig. S7G), with KLRG1+ ILC1s and CD4 T cells entering the peritoneal cavity and the liver approximately in a 1:1 ratio and a 1:2 ratio, respectively, in the IL-12 + IL-18 containing treatment (Fig. 7E). Thus, the KLRG1+ ILC1s display a high preferential ILC1 homing to these two compartments during type 1 immunity, indicating that the IL-12 + IL-18–induced KLRG1+ ILC1 differentiation is accompanied by an increased capacity for tissue homing for ILC1s from the neonatal thymus. Thymus-derived ILC1s in the spleen, peritoneal cavity, and liver exhibit enhanced and active production of both IFN-γ and TNF-α in mice receiving IL-12 + IL-18 (Fig. 7, F and G, and fig. S7, H to J). These results demonstrate that the thymic-derived ILC1s are functional and capable of cytokine production after entering other tissues. Last, to visualize the thymus-derived Ncr1-tdTomato exit and homing to the periphery, we performed 3D confocal imaging on the liver of these mice. Similar to the flow cytometry results, we found a homing and tissue localization of Ncr1-tdTomato ILC1s in the liver parenchyma, which was exacerbated in mice receiving IL-12 + IL-18 (Fig. 7H). In summary, thymic-derived ILC1s are exiting the neonatal thymus to peripheral tissues both in the presence and in the absence of type 1 cytokines. However, following IL-12 + IL-18 activation, thymic-derived ILC1s display preferential homing to the liver and peritoneal cavity compared to the spleen and produce IFN-γ and TNF-α upon entering the new tissues.
Fig. 7. KLRG1+ thymic ILC1s are homing to the liver and the peritoneal cavity.
(A) Schematic of neonatal Ncr1-tdTomato thymus graft experiments. (B) Picture of a kidney with two recently grafted neonatal thymic lobes (arrows). (C) Representative flow plots of tdTomato+ ILC1s from the peritoneal cavity. (D) Stacked bar plots showing total thymic graft–derived ILC1s divided on the basis of CD27 and KLRG1 expression in the indicated organs [gating shown in (C) and fig. S7A]. Error bars are shown as SEM. (E) Count ratio (log10 scale) between thymus graft–derived KLRG1+ ILC1s and CD4 T cells for determining differential homing in indicated tissues. Data are shown as box-and-whiskers plots displaying median, minimum, and maximum. (C) and (E) Data are pooled from two independent experiments from n = 3 to 4 (spleen), n = 6 to 7 (peritoneal cavity), and n = 5 to 7 (liver) and representative of four independent experiments. (F and G) Intracellular cytokine expression showing the percentage of thymus graft–derived ILC1 population actively expressing (F) IFN-γ or (G) TNF-α in indicated organs (gating shown in fig. S7H). Data are pooled from two independent experiments with n = 6 mice per condition. Error bars are shown as SEM. (E) to (G) Each symbol represents an individual mouse. (H) Confocal 3D imaging of 500-μm-thick liver samples from thymus-grafted mice. Representative pictures are shown as maximum projection intensity. Blue, collagen IV; green, Ncr1-tdTomato; red, CD3e. Scale bars, 500 μm (bottom right corner) and 50 μm (zoomed-in section). Images are representative of n = 3 to 4 from two independent experiments. Statistical significance was calculated by (D) unpaired t test (total cell count), (E) two-way ANOVA between treatments and tissues, [(F) and (G); except spleen IFN-γ] Kruskal-Wallis test, and [(F); spleen IFN-γ] unpaired t test, *P < 0.05, **P < 0.01, and ****P < 0.0001. (E) Two-way ANOVA between tissues, ####P < 0.0001.
DISCUSSION
Group-1 ILCs can develop from ETPs in mice and humans (25–28). Nevertheless, the relationship between thymic group-1 ILCs and peripheral group-1 ILCs remains unresolved (24). To illuminate unexplored areas of group-1 ILC ontogeny, Sparano et al. (19) used fate mapping and scRNA-seq by describing different embryonic and neonatal waves of ILC1s and cNK cells in the liver and bone marrow. Here, we show that neonatal thymic ILC1s can migrate to peripheral organs and rapidly expand in response to IL-12 and IL-18. On the basis of scRNA-seq analysis, we confirmed that at birth, the vast majority of group-1 ILCs is ILC1s with embryonic-wave characteristics (Klra5 expression) (Fig. 5E) (19, 20). The immune response of ILC1s and cNK cells exhibit different temporal kinetics during viral infection and metastatic cancer (16, 18). Specifically, tissue-resident ILC1s display early functionality in controlling MCMV infection (18). Moreover, ILC1s are critical for preventing metastatic colonization in the liver, while recruited cNK cells inhibit metastatic outgrowth and progression (16). However, in the absence of ILC1-specific genetic ablation models, it remains challenging to disentangle their tissue-specific functions compared to cNK cells and other lymphocytes. Accordingly, our cross-tissue comparison of group-1 ILCs in neonates and adults using scRNA-seq may help address the functional redundancy between ILC1s and cNK cells by determining the most appropriate genes to target.
In this study, we identified neonatal thymic ILC1s as a distinct subset as compared to neonatal ILC1s from other primary lymphoid organs with their unique expression of Itgae, Trat1, and Cx3cr1 (Fig. 5C and fig. S4B). Furthermore, neonatal thymic ILC1s express high levels of helper-like and ILC precursor markers compared to other neonatal ILC1 populations, such as Tcf7, Il7r, Tox, Batf3, and Cd27+ (Fig. 5C and fig. S5, E and F) (38, 42). As many aspects of the layered ILC1 ontogeny remain unknown, this finding opens the possibility for some subsets of embryonic wave–seeded ILC1s to originate from the perinatal thymus.
In parallel to ILC1s, ILC2s can also develop from progenitors in the thymus (50, 51). A study on the intrathymic transcriptional checkpoints revealed that Rora expression in embryonic thymic progenitors repressed T cell fate and promoted thymic ILC2 commitment (50). Although Rora expression was highly increased in the thymus-exiting ILC1 population (Fig. 5C), it remains unknown whether thymic ILC1s have an equivalent Rora dependency as thymic ILC2. Similar to previous findings (25, 27), we show that perinatal ETPs have a substantial group-1 ILC potential, even when cultured under T cell–skewing OP9-DL1 conditions (Fig. 3D). While the neonatal ETP-derived group-1 ILCs were identified as ILC1s (Fig. 3D and fig. S3B), these ILC1s lack the differentiation marker KLRG1 following 6 days of culture, despite the presence of IL-12 + IL-18. Only after seeding immature neonatal thymic–derived ILC1s and stimulating with IL-12 + IL-18, did the ILC1s acquire the terminal differentiation marker KLRG1 (39), similar to the NTOC-exiting ILC1s (Fig. 3, A and D). These data indicate that the NTOC-expanding ILC1s mainly derive from the immature ILC1s already present in the neonatal thymus rather than inducing their de novo development from the ETPs. Although group-1 ILCs can develop from human thymic progenitors following in vitro culture, direct in situ group-1 ILC development in the human thymus has yet to be confirmed (26, 28). Whether infection or other inflammatory triggers are required to perturb steady-state thymic development to allow in situ group-1 ILC development remains to be determined.
Following MCMV infection and injection of IL-12 + IL-18 into neonates, the thymic ILC1s up-regulated CXCR6 and KLRG1 (Fig. 6, G to J), similar to what was observed upon IL-12 + IL-18 stimulation in the NTOC (Fig. 4F). Thus, the enhanced CXCR6 and KLRG1 expression observed on thymic ILC1s is likely a direct effect of the type 1 immune response. As CXCR6 expression on ILC1s is associated with liver homing (38, 45), the enhanced CXCR6 expression could be related to establishing tissue residency when exiting the thymus. In addition, enhanced KLRG1 expression is associated with differentiation (39) and likely reflects a changed activation state of the thymic ILC1s. Performing live imaging and 3D confocal imaging of the NTOC-cultured thymic lobes, we show that the thymic ILC1s localize inside the thymic parenchyma. Upon IL-12 + IL-18 stimulation, they localize in the outer capsule region of the thymus, from where the ILC1s can be observed exiting into the culture medium (Fig. 6K and movie S1). Following MCMV infection in neonates, a similar enhanced ILC1 localization was observed in the subcapsular region and thymus parenchyma (Fig. 6K). In the NTOC, although the thymic ILC1s display the same KLRG1+ phenotype inside the lobes as in the supernatant following IL-12 + IL-18 stimulation (Fig. 3A), the ILC1 numbers inside the thymic lobes remained low compared with ILC1s found in the supernatant (Fig. 1H). Therefore, it is plausible that the thymic microenvironment restricts the amount of thymic-resident ILC1s and that the thymus primarily functions as a repository or staging ground for their exit. To further evaluate the capability of thymic ILC1s to exit the thymus and enter other organs, we used thymic kidney grafts to track ILC1s from the P0.5 neonatal thymus. Despite ILC1s being a rare subset in the thymus, a substantial population of thymic-derived ILC1 was observed migrating to the spleen, peritoneal cavity, and liver 6 days after grafting, independent of treatment (Fig. 7C), which further implicates the thymus as a source for peripheral homing ILC1s. In addition, thymic-derived ILC1 homing to the liver and the peritoneal cavity increased substantially in response to IL-2 + IL-12 + IL-18 compared to the IL-2–injected control mice consisting primarily of differentiated KLRG1+ ILC1s. Unexpectedly, we found that the number of thymic graft–derived ILC1s and CD4+ T cells in the liver and peritoneal cavity is relatively similar in both the liver and peritoneal cavity in the IL-12 + IL-18–treated mice; however, the amount of thymic-derived ILC1s homing to the spleen was similar under both conditions and 75 to 120 times lower compared to CD4 T cells (Fig. 7E and fig. S7G). These results and the proportional rarity of thymic-resident ILC1s indicate a preferential homing of thymic ILC1s toward the liver and the peritoneal cavity during type 1 immunity. Furthermore, we confirmed that after entering the spleen, peritoneal cavity, and liver, the thymic-derived ILC1s were functional and responded to IL-12 + IL-18 activation with increased levels of intracellular IFN-γ and TNF-α. Since liver-resident ILC1s previously have been shown to limit MCMV infection in an IFN-γ–dependent manner (18), the capacity for liver homing and IFN-γ production by thymic-derived ILC1s indicate a possible function during viral responses in early life. TCRα-knockout thymus grafts were previously used to show group-1 ILC homing to the spleen (23), although wild-type thymus grafts were not known to produce peripheral homing ILC1s. The peripheral homing of thymic-derived ILC1s, combined with high expression of ILC precursor markers on neonatal thymic ILC1s, indicates a thymic ontogeny for some peripherally seeded ILC1 subsets.
Although the underlying causality for infection-induced thymus atrophy is poorly understood (11), infection with murine roseolovirus has been shown to disrupt central tolerance in the thymus (52). Here, we show that acute thymus atrophy not only is a process where developing T cells die but can be accompanied by a subcapsular ILC1 localization during type 1 immunity (Fig. 6 and movie S1) and greater peripheral homing of thymic ILC1s (Fig. 7, C, E, and H). Our results contribute to existing evidence that the thymus not only is a T cell–producing organ but also can produce peripheral homing ILC1s, which is augmented during acute type 1 inflammation. A limitation of this study is that it does not definitively show whether specific perinatal waves of ILC1s derive from the thymus. However, this study shows that some subsets of perinatally seeded ILC1s are likely to have a thymic origin.
Collectively, our work uses a multitude of approaches to studying the impact of type 1 immunity on the thymus. It identifies neonatal thymic ILC1s as a subset with ILC precursor characteristics, distinct from other embryonic-wave ILC1s from primary lymphoid organs. Furthermore, we demonstrate that thymic-derived ILC1s can rapidly contribute to peripheral ILC1 compartments, indicative of tissue-resident ILC1 subsets with thymic ontogeny.
MATERIALS AND METHODS
Experimental design
The aim of this study was to elucidate the impact of type 1 inflammatory conditions on the neonatal thymus, with a particular focus on the expanding ILC1s observed exiting the thymic lobes. For this purpose, we used scRNA-seq, functional assays, and flow cytometry to validate their ILC1 identity and functionality. Additional scRNA-seq was used to compare them with group-1 ILCs from other tissues. Last, we used genetic fate mapping and confocal imaging to determine their in situ thymic expansion and peripheral homing capacity. Each experiment was performed independently at least two times; the number of replicates and repetitions is indicated in the figure legend. In all mouse experiments, the mice were randomly assigned a group. GPower3.1 software was used to predetermine sample sizes in mouse models based on mean variances of previous experiments. In case of no prior experience with the animal model, a minimum of three biological replicates were sampled. For all experiments, male and female mice were used.
Mice and ethical considerations
C57BL/6N mice were used in all experiments except for the transgene mice and their wild-type controls, which were on a C57BL/6J background. Transgene mice: Ncr1iCre mice [Ncr1tm1.1(icre)Viv] were provided by Vivier and coworkers (44) and crossed with ROSA26–flox-stop-flox-tdTomato mice [B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J] to create Ncr1-tdTomato mice, Rag2−/−OTI [B6.129S6-Rag2tm1Fwa Tg(TcraTcrb)1100Mjb], and Rag2−/−Il2rg−/− mice (C;129S4-Rag2tm1.1Flv Il2rgtm1.1Flv/J). Purchased time-mated pregnant females were bought from Janvier. All transgene mice were bred and kept in-house at the VIB Center for Inflammation Research under specific pathogen-free conditions. All mice were kept in individually ventilated GM500 cages (Techniplast) with a 14/10-hour light/dark cycle in a temperature-controlled (21°C) room. Mice were provided water and food ad libitum. All mouse experiments were conducted according to institutional, national, and European animal regulations. Ghent University’s Ethics Committee approved all experimental animal procedures. Experiments were conducted in agreement with the European Parliament’s Directive 2010/63/EU and the 22-09-2010 Council on the protection of animals used for scientific purposes.
Genotyping
Genotyping of the different transgene mouse lines was performed by polymerase chain reaction (PCR). Ncr1-tdTomato mice: tdTomatofl/fl mice were genotyped using the PCR primers: wild-type, CAGCTTCTTGTAATCGGGGA (forward) and GCGAGGAGGTCATCAAAGAG (reverse); transgene, TGGCTTCTGAGGACCGCCCTGGGC (forward) and CACCTGTTCAATTCCCCTGCA (reverse). Yielding a 160–base pair (bp) wild-type DNA band is at 160 bp, and a transgene DNA band is at 245 bp. Ncr1iCre mice were genotyped using the PCR primers: forward-1, ACCGCCTGATCTATGGTGCC; forward-2, GGGTGGGTGTAGCCTCTATC; reverse, CACTCCTACCCCTTCATTTCTGA, yielding a 350-bp wild-type DNA band and a transgene DNA band of 280 to 300 bp. Rag2−/−OTI mice [B6.129S6-Rag2tm1Fwa Tg(TcraTcrb)1100Mjb] were genotyped using the PCR primers: Rag2−/−, CAGGGTCGCTCGGTGTTC (forward-1), CTTGCCAGGAGGAATCTCTG (forward-2), and GTTTCCCATGTTGCTTCCA (reverse), yielding a 200-bp wild-type DNA fragment and a 388-bp knockout DNA fragment. OTI: OTI TCR, CAGCAGCAGGTGAGACAAAGT (forward) and GGCTTTATAATTAGCTTGGTCC (reverse); OT Int Ctrl, CAAATGTTGCTTGTCTGGTG (forward) and GTCAGTCGAGTGCACAGTTT (reverse), yielding a 200-bp wild-type DNA band and a 300-bp transgene DNA band. Rag2−/−Il2rg−/− mice were genotyped using the same Rag2−/− primers described above and Il2rg−/−: GTGGGTAGCCAGCTCTTCAG (common forward), CCTGGAGCTGGACAACAAAT (wild-type reverse), and GCCAGAGGCCACTTGTGTAG (knockout reverse), yielding a 269-bp wild-type DNA fragment and a 349-bp knockout DNA fragment.
Cytokines
The following cytokines were used: recombinant murine IL-2 (protein core VIB, Ghent, Belgium), recombinant murine IL-7 (catalog no. 217-17, PeproTech, Rocky Hill, NJ), recombinant murine IL-12 (catalog no. 210-12, PeproTech, Rocky Hill, NJ), recombinant murine IL-15 (catalog no. 210-15, PeproTech), and recombinant murine IL-18 (catalog no. P70380, R&D Systems, Minneapolis, MN). All lyophilized cytokines were reconstituted in phosphate-buffered saline (PBS) with 0.1% bovine serum albumin before storing at −80°C. The concentration of bovine serum albumin under all vehicle control conditions corresponded to the highest concentration within the specific experiment.
Neonatal thymic organ cultures
Neonates were euthanized by decapitation at P0.5, and thymic lobes were extracted and cleaned before setup. Three neonatal thymus lobes were cultured per well unless otherwise stated. Only intact neonatal thymic lobes were cultured on top of a 0.8-μm Isopore Membrane Filter (catalog no. ATTP01300, Merck Millipore), which was suspended on top of 2 ml of NTOC medium in 12-well plates and cultured at 37°C with 5% CO2. Cytokines were used at the following concentrations: IL-12 (4 ng/ml) and IL-18 (40 ng/ml). The NTOC medium was composed of Dulbecco’s modified Eagle’s medium (DMEM) (catalog no. 31330-038, Gibco) supplemented with 20% fetal bovine serum (FBS) (lot nos. 71133 and 90439, brand: Tico Europe and Greiner Bio-One), 20 μM l-glutamine, 50 μM 2-mercaptoethanol, 0.8% penicillin-streptomycin (catalog no. P4333, Sigma-Aldrich), 0.4 mM Na pyruvate (catalog no. S-8636, Sigma-Aldrich), and 1× nonessential amino acids (catalog no. M7145, Sigma-Aldrich). Different types of FBS were tested for optimal cellular output of the NTOC to ensure that it supported ILC1 development.
OP9-DL1 cocultures
A total of 1000 OP9-DL1–engineered bone marrow stromal cells were plated in 96-well plates 24 hours before coculture. We sorted the population of interest: ETPs (CD44+CD25−c-KithighLin−), DN2a (CD44+CD25+c-KithighLin−), and immature ILCs (CD44+CD122+Lin−) in a FACSymphony A5 sorter (BD Biosciences). Cocultures were initiated by seeding 1000 cell-sorted precursors/immature ILC1s, which were seeded and treated with cytokines. Cytokines were used at the following concentrations: IL-2 (20 ng/ml), IL-7 (20 ng/ml), IL-15 (20 ng/ml), IL-12 (2 ng/ml), and IL-18 (20 ng/ml) in a final volume of 200 μl on the day of the setup. On day 3 of the coculture, 50 μl of medium-diluted cytokines IL-2, IL-7, and IL-15 were supplemented in the same amount as on setup day to the respective conditions. The OP9-DL1 cell line and cocultures were maintained at 37°C with 5% CO2, in α–minimum essential medium (catalog no. 22571-020, Gibco) containing 20% FBS (Tico Europe), 20 μM l-glutamine, 50 μM 2-mercaptoethanol, 0.8% penicillin-streptomycin (catalog no. P4333, Sigma-Aldrich), 0.,4 mM Na pyruvate (catalog no. S-8636, Sigma-Aldrich), and 1× nonessential amino acids (catalog no. M7145, Sigma-Aldrich). On day 6, the coculture was analyzed by flow cytometry on a FACSymphony A5 (BD Biosciences).
YAC-1 killing assay
YAC-1 target cells were stained with eFluor 670 dye (catalog no. 65-0840-85, Thermo Fisher Scientific), and 5000 stained YAC-1 cells were seeded in each well of a 96-well U-bottomed plate. Four different populations of effector cells were sorted and cultured with YAC-1 target cells: (i) ILC1s (CD122+CD62L−Lin−) from the supernatant of day 6 NTOC stimulated with IL-12 + IL-18; (ii) noncytotoxic CD4+ thymocytes (CD4+CD122−CD19−Ter119−CD11c−F4/80−) from day 6 vehicle NTOC lobes and supernatant; (iii) unstimulated cNK cells (CD122+CD62L+Lin−) from freshly isolated 8-week-old adult C57BL/6 spleens; and (iv) IL-12 + IL-18–stimulated cNK cells (CD122+CD62L+Lin−) from 8-week-old adult C57BL/6 spleens following 18 hours of culture of 200,000 splenocytes per well in 96-well U-bottomed plates with the cytokines IL-12 (4 ng/ml) and IL-18 (40 ng/ml). Lin− was defined as CD3−TCRb−TCRgd−CD19−CD4−CD8b−Ter119−CD11c−F4/80−. An increasing number of effector cells were added to the wells with the freshly seeded YAC-1 cells to create different target:effector ratios with a constant amount of target cells. Cells were centrifuged [300g for 2 min at room temperature (RT)] immediately after both target and effector cells were added and incubated at 37°C with 5% CO2. The dose-dependent killing was determined by flow cytometry following 4 hours of coculture, by 4′,6-diamidino-2-phenylindole (DAPI) staining (0.1 μg/ml) to determine the percentage of dying DAPI+ cells from the eFluor 670+ YAC-1 cells. YAC-1 and cocultured cells were maintained in RPMI 1640 medium (catalog no. R1780-500ML, Sigma-Aldrich) containing 10% FBS (Greiner Bio-One), 50 μM 2-mercaptoethanol, and 0.8% penicillin-streptomycin (catalog no. P4333, Sigma-Aldrich) and split at least two times before seeding.
Electron microscopy
Thymic CD4+ T cells (TCRb+CD4+CD8b−) and ILC1s (CD122+CD4−CD8b−TCRb−TCRgd−CD19−Ly-6G−MHCII−CD11c−) were sorted from NTOC supernatant day 6 treated with vehicle and IL-12 + IL-18, respectively. Sorted cells were fixed in 4% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M Na cacodylate buffer (pH 7.2) and centrifuged at 1500 rpm. Low–melting point agarose was used to keep the cells concentrated for further processing. Cells were fixed for 4 hours at RT, followed by fixation overnight at 4°C after replacement with fresh fixative. After washing in buffer, they were postfixed in 1% OsO4 with 1.5% K3Fe(CN)6 in 0.1 M Na cacodylate buffer at RT for 1 hour. After washing, cells were subsequently dehydrated through a graded ethanol series, including a bulk staining with 1% uranyl acetate at the 50% ethanol step, followed by embedding in Spurr’s resin. Ultrathin sections of a gold interference color were cut using an ultramicrotome (Leica EM UC6), followed by a poststaining in a Leica EM AC20 for 40 min in uranyl acetate at 20°C and for 10 min in the lead stain at 20°C. Sections were collected on Formvar-coated copper slot grids. Grids were viewed with a JEM-1400Plus transmission electron microscope (JEOL, Tokyo, Japan) operating at 80 kV.
Multiplex enzyme-linked immunosorbent assay
The customizable multiplex U-PLEX enzyme-linked immunosorbent assay (MSD, Rockville, MD) was used to measure protein concentrations of cytokines in the supernatant of NTOC culture. Murine cytokines were measured: IFN-γ, TNF-α, GM-CSF, and CCL3. NTOC supernatant was centrifuged at 10,000g to remove cells and cell debris from samples before assay measurements.
Neonatal thymus grafts
The neonatal thymic grafts were performed as previously described (46). Adult Rag2−/−Il-2rγ−/− male mice were provided analgesia in drinking water the day before the grafting procedure. Mice were anesthetized in an isoflurane box and placed in an isoflurane gas mask on a 37° heat plate. The mice were shaven on the left dorsal side at the height of the kidney and disinfected with 70% ethanol, followed by isobetadine and another round of 70% ethanol. A 1.5-cm longitudinal incision in the skin and muscle was made at the height of the right kidney, and the kidney was exposed. A small horizontal incision was made in the kidney capsule on the caudal kidney pole with a 18-gauge needle. A subcapsular space was created underneath the kidney capsule by gently moving a blunt forceps underneath the capsule in the presence of sterile PBS. Subsequently, two thymus lobes from neonatal Ncr1-tdTomato mice were inserted in the subcapsular space and gently pushed to the cranial pole of the kidney. The grafted kidney was placed back in its original location, the muscles were sutured, and the skin was closed with clips and disinfected with isobetadine. The mice recovered, and the grafted thymus was allowed to vascularize to the ample kidney vasculature for 3 days. The thymus-grafted mice then received three intraperitoneal injections with a 24-hour interval of cytokines as described in the “Cytokine injections” section. The mice were euthanized 6 days after grafting.
Cytokine injections
Adult and neonatal mice were randomly assigned to their group. All mice received three intraperitoneal injections of cytokines or vehicle with a 24-hour interval. Neonates were injected intraperitoneally with cytokines diluted in DMEM in a total volume of 20 μl using a 29-gauge needle. The needle opening was facing toward the animal upon injection to avoid abdominal fluid exiting during the procedure. In neonates, the cytokine concentrations per injection were as follows: 10 ng of [IL-12] and 100 ng of [IL-18]. The cytokine injection in adult C57BL/6N mice was modified from a systemic inflammatory IL-12 + IL-18 model in BALB/c mice (53). In the adult thymus-grafted Rag2−/−Il2rg−/− mice, the model was adapted to an IL-2 + IL-12 + IL-18 model, considering the lacking IL-2 production in Rag2−/−Il2rg−/− mice (49). For all adult mice, the cytokines were diluted in PBS, and each mouse was injected intraperitoneally with a total volume of 0.2 ml. In adult mice, the cytokine concentrations per injection were as follows: 100,000 IU of [IL-2], 150 ng of [IL-12], and 750 ng of [IL-18].
Neonatal MCMV infection
C57BL/6N and Ncr1-tdTomato mice neonates were injected once intraperitoneally with 20 μl of MCMV diluted in DMEM (catalog no. 31330-038, Gibco) using 0.5 ml of 29-gauge BD Microfine (catalog no. 324824, BD Biosciences). The needle opening was facing toward the animal upon injection to avoid abdominal fluid exiting during the procedure. The MCMV batch was obtained by infecting MCMV-susceptible BALB/c mice and harvesting salivary glands as described (54). All mice were either sham-injected (DMEM) or infected with a sublethal MCMV dose of 300 plaque-forming units/μl. Sham and MCMV-infected mice were kept in separate cages in a BL2 animal facility at the VIB-IRC, and all handling of the virus and tissues was performed in a BL2 facility.
Confocal 3D imaging
Confocal imaging on the liver and thymic lobes was performed using the Ce3D clearing method (55). Briefly, adult Ncr1-tdTomato thymus-grafted mice were perfused with PBS with 1% heparin, and the liver, more specifically the lobus sinister medialis hepatis, was fixed overnight at 4°C in 4% paraformaldehyde. In addition, Ncr1-tdTomato thymic lobes from MCMV-infected neonates, controls, and NTOC lobes were fixed overnight at 4°C with PLP buffer as previously described (55). All organs were incubated at RT under gentle shaking (150 to 220 rpm) with primary and secondary antibodies for 5 days each, followed by 3 days of Ce3D clearing solution, and then mounted on glass slides, and 2 × 2 or 3 × 3 tiles were imaged using a Zeiss LSM 880 AiryScan (Zaventem, Belgium) on Fast Airy mode with a Plan-Apochromat 10×/0.45, at a z-interval between 7 and 15 μm. Images were processed using ZEN software. Montages were made with Fiji [National Institutes of Health (NIH), Bethesda, Maryland, USA], and cell quantifications in thymic lobes were performed using Arivis (Rostock, Germany). Antibodies used for 3D imaging can be found in table S1.
Live cell imaging
NTOC lobes were prestained at RT for 30 min in NTOC medium derived from the individual conditions (vehicle day 4) and (IL-12 + IL-18 day 4). Antibodies (table S1) were present in the medium with Hoechst (catalog no. H21492, Thermo Fisher Scientific) throughout the experiment to stain thymus exiting cells. Each NTOC lobe was placed and imaged in a μ-Slide 15-well chamber (ibidi, Germany) containing the same staining medium with Hoechst and antibodies. Cells were imaged with the respective laser lines: 405 (Hoechst), 488 (CD11b), 561 (tdTomato-Ncr1), and 639 (TCRβ, TCRγδ, and CD3). Only laser lines 405, 561, and 639 are shown. Live cell imaging was performed on an Axio Observer.Z1 (Zeiss, Germany) equipped with a CSU-X1 Yokogawa spinning-disk head (Yokogawa Corporation of America) and a Photometrics Prime 95B (Teledyne Photometrics), with an EC Plan-Neofluar 10×/0.30 dry objective, and equipped with temperature and humidity controls (37°C with 5% CO2, respectively). Z-stacks were acquired every 30 min for a period of 10 hours. Image reconstruction was performed with ImageJ (NIH). The resulting movie is shown for one lobe and representative of 10 biological replicates per group from two independent experiments (movie S1).
Tissue preparation for flow cytometry and cell sorting
NTOC supernatant cells were harvested by repeated pipetting of the NTOC well and filtered after the lobes were removed from the well. Preparation of the thymic lobes and spleen for single-cell suspension used for flow cytometry and cell sorting (scRNA-seq analysis and OP9-DL1 coculture) were obtained by smashing organs through a 70-μm cell strainer. The strainers were then washed with 2% FBS in PBS. Perfusion was performed to remove erythrocytes from adult liver samples. The mice were anesthetized (ketamine/xylazine), and blood was extracted by retro-orbital punction, followed by cardiac perfusion with 15 ml of PBS with 1% heparin, using a perfusion pump. Leucocytes were extracted from adult and neonatal livers by mincing using a gentleMACS Dissociator (catalog no. 130-093-235, Miltenyi Biotec). This was followed by collagenase treatment with prewarmed RPMI containing collagenase A (1 mg/ml; catalog no. 11088793001, Merck Millipore). The resulting tissue homogenate was centrifuged on top of a 5 ml of 37.5% Percoll (catalog no. P1644-1 L, Merck Millipore) gradient (700g for 10 min at 4°C, break 1/acceleration 4). Adult bone marrow cells were acquired by cutting the knee-end of tibia and fibula, placing the open-end downward in the tube and using centrifugal force to extract cells from bones (1900g for 1 min at RT). Neonatal bone marrow was acquired by isolating femurs and humeri from neonates, crushing the cleaned bones with mortar and pestle, and filtering cells through a 70-μm cell strainer. To further enrich for lymphocytes, the red blood cells were lysed using ACK buffer (catalog no. 10-548E, Lonza) for 2 min at RT before antibody staining. Subsequent flow cytometry or fluorescence-activated cell sorting prep was performed on ice. Antibody staining was performed 30 min in the dark. For intracellular staining of transcription factors, cells were fixed after extracellular staining using eBioscience Foxp3/Transcription Factor Staining Buffer Set (catalog no. 00-5523-00, Thermo Fisher Scientific). For intracellular cytokine measurements, cells were first extracted from each tissue as mentioned above and incubated with 1.5 μM brefeldin A (catalog no. B6542-25MG, Sigma-Aldrich) and Monensin solution (1000×; catalog no. 420701, BioLegend) for 4 hours (with no further stimulation, to evaluate the cytokines being produced at the time of cell isolation). For the staining procedure, the cells were first stained extracellularly, then fixed, and permeabilized using BD Cytofix/Cytoperm (catalog no. 554714, BD Biosciences), followed by intracellular staining. Cell counts (Figs. 3 and 7) were derived by flow cytometry by adding Precision count beads to the sample after single-cell suspension according to the manufacturer’s instructions (catalog no. 424902, BioLegend), and to acquire absolute NTOC and neonatal thymus cell counts (Figs. 1 and 6), they were counted on a FACSVerse using volume metrics and counting of viable (DAPI) and death cells for 15 s immediately following single-cell suspension. Fixable Viability Dye eFluor 780 (FVD-eF780; catalog no. 65-0865-14, Thermo Fisher Scientific) was used to exclude dead cells in all flow cytometry and cell sorting experiments unless otherwise stated. All antibodies used for flow cytometry and cell sorting are shown in table S1. All flow cytometry antibodies were validated by the supplier. Sorting of viable NTOC cells (DAPI negative) for CITE-seq and electron microscopy was performed on a BD FACSAria III cell sorter. Sorting of cell types ex vivo for scRNA-seq (tissues) and for OP9-DL1 cultures was performed on the BD FACSymphony S6 cell sorter. All flow cytometry was performed on a FACSymphony A5 cell analyzer. Data were analyzed with FlowJo software, version 10.8.
scRNA-seq and CITE-seq
For the first CITE-seq (Fig. 1), all viable (DAPI negative) cells from day 6 NTOC supernatant (Figs. 1, 4, and 5) treated with (i) vehicle, (ii) IL-12, (iii) IL-18, and (iv) IL-12 + IL-18 were sorted (Aria III, BD Biosciences) and pelleted by centrifugation at 400g for 5 min at 4°C. When CITE-seq was to be performed, cells were stained with CD16/CD32 monoclonal antibody (catalog no. 553142, BD Biosciences) to block nonspecific Fc receptor binding of CITE-seq antibodies for 20 min at 4°C, before being washed in excess PBS with 2% FBS used directly in downstream CITE-seq analysis. For the second CITE-seq (Figs. 4 and 5) experiment comparing tissue group-1 ILCs from 8-week-old adult and P0.5 neonatal mice from ex vivo extracted the thymus, liver, spleen, and bone marrow, the following samples were used: adult mice: 12 thymus, 4 spleens, 12 perfused livers, and bone marrow collected from two hind legs (tibia and fibia) of four mice. For all adult organs, an equal number of male and female mice were used. Neonatal mice: 90 thymus, 45 spleen, 45 bone marrow samples (arm and leg bones from 45 neonates), and 30 livers. Following single-cell suspension as described in the previous paragraph, type 1 ILCs were enriched by negative selection using magnetic beads (MagniSort Mouse NK cell Enrichment Kit, catalog no. 8804-6828-74, Thermo Fisher Scientific). The samples were then sorted to enrich for viable group-1 ILCs (CD122+Lin−, 50%), DN1-DN2 cells (CD44+Lin−, 25%), and DN3-DN4 cells (CD44−Lin−, 25%) for parallel comparison of group-1 ILCs and progenitor cells across tissues (Lin−, CD4−CD8a−CD5−TCRb−TCRgd−Ter-119−CD11c−F4/80−Ly-6G/Ly-6C−). Following sorting, both the NTOC supernatant cells and ex vivo tissue comparison cells were stained with mouse cell surface protein TotalSeq-A antibodies panels containing 9 isotype controls and 77 (NTOC) or 174 (tissue) oligo-conjugated antibodies (TotalSeq-A, BioLegend) (tables S2 and S3). The sorted single-cell suspensions were resuspended at a final estimated concentration of 1500 and 1100 cells/μl for NTOC and tissues, respectively. The cells were loaded on a Chromium GemCode Single Cell Instrument (10x Genomics) to create single-cell gel beads-in emulsion (GEM). The scRNA-seq libraries were prepared using GemCode Single Cell 3′ Gel Bead and Library kit, version 3 (NTOC) and version NextGEM 3.1 (tissue) (10x Genomics) according to the manufacturer’s instructions with the addition of amplification primer (3 nM, 5’CCTTGGCACCCGAGAATT*C*C) during cDNA amplification to enrich the TotalSeq-A cell surface protein oligos. Additional information about the processing, quality control, UMAP clustering, visualizations, and differential gene expression analyses can be found in Supplementary Materials and Methods.
Statistical analysis
The differences in variances between treatment groups were tested with an F test, and when the F test showed differences between conditions, statistics were performed on log2-transformed values. In case the variances remained different on log2-transformed data, a nonparametric test was used as indicated. The test used to evaluate differences between condition groups is specified in the figure legend. Statistical analyses were carried out using GraphPad Prism version 9.4.1 (GraphPad Software Inc., La Jolla, CA).
Acknowledgments
We thank the VIB Single Cell Core, VIB Nucleomics Core, VIB Flow Core Ghent, and VIB Bioimaging Core for support and access to the instrument park (vib.be/core-facilities). The research unit of P.V. is part of Cancer Research Institute Ghent (CRIG) and Ghent Gut Inflammation Group (GGIG) consortia. We want to thank S. Skov (UCPH-SUND), J. Maelfait (VIB-UGent), K. Ravichandran (VIB-UGent), A. Brown (VIB-UGent), and B. Lambrecht (VIB-UGent) for critical input to the project. We are grateful to E. Vivier (CIML) for providing us with the Ncr1-iCre mouse line and B. Lambrecht for giving us the Rag2−/− OTI and the Rag2−/−Il2rg−/− mouse lines. We thank B. Maes (VIB-UGent) for feedback on the manuscript. We thank statistician M. Vuylsteke (VIB-UGent) for input on the statistics shown in the paper. BioRender was used to create some figures. Last, we thank the VIB-UGent animal house staff.
Funding: This work was supported by FWO Research Grants G.0B96.20N (to P.V. and P.T.), G.0C76.18N (to P.V.), G.0B71.18N (to P.V.), G.0A93.22N (to P.V.), EOS MODEL-IDI Grant (30826052) (to P.V.), and EOS CD-INFLADIS (40007512) (to P.V.), Special Research Fund UGent [Methusalem grant BOF16/MET_V/007 (to P.V.) BOF22/MET_V/007 (to P.V.), and iBOF ATLANTIS grant 20/IBF/039 (to P.V.)], Foundation against Cancer (F/2016/865 and F/2020/1505) (to P.V.), and VIB Flanders Institute for Biotechnology (to P.V.).
Author contributions: Conceptualization: P.T., P.V., and M.R.P. Investigation: P.T., M.R.P., W.S., and T.B.B. Methodology: P.T., M.R.P., W.S., J.H., T.D., and J.V. Critical data interpretation: T.T., N.T., S.J., and G.L. Visualization (imaging): A.G. Visualization (scRNA-seq): B.V., R.R., and T.B.B. Supervision: P.T., P.V., S.J., and G.L. Writing (original draft): P.T. Writing (review and editing): P.T., P.V., A.G., M.R.P., W.S., T.T., T.B.B., and G.L. All of the authors have read and provided comments on the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The single-cell sequencing has been deposited in the Gene Expression Omnibus under the ID: GSE229632. The Ncr1-iCre mice were provided by Eric Vivier Marseille-Luminy Immunology Center (CIML), pending scientific review, and a completed material transfer agreement. Requests for the Ncr1-iCre mice should be submitted to E. Vivier (vivier@ciml.univ-mrs.fr). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All mouse lines, reagents, and software used are listed in the paper or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Materials and Methods
Figs. S1 to S7
Tables S1 to S28
Legend for movie S1
Legend for data file S1
References
Other Supplementary Material for this manuscript includes the following:
Movie S1
Data file S1
REFERENCES AND NOTES
- 1.Gherardi M. M., Ramírez J. C., Esteban M., IL-12 and IL-18 act in synergy to clear vaccinia virus infection: Involvement of innate and adaptive components of the immune system. J. Gen. Virol. 84 (Pt. 8), 1961–1972 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Pien G. C., Satoskar A. R., Takeda K., Akira S., Biron C. A., Cutting edge: Selective IL-18 requirements for induction of compartmental IFN-γ responses during viral infection. J. Immunol. 165, 4787–4791 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Vanden Berghe T., Demon D., Bogaert P., Vandendriessche B., Goethals A., Depuydt B., Vuylsteke M., Roelandt R., Van Wonterghem E., Vandenbroecke J., Choi S. M., Meyer E., Krautwald S., Declercq W., Takahashi N., Cauwels A., Vandenabeele P., Simultaneous targeting of IL-1 and IL-18 is required for protection against inflammatory and septic shock. Am. J. Respir. Crit. Care Med. 189, 282–291 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Mera S., Tatulescu D., Cismaru C., Bondor C., Slavcovici A., Zanc V., Carstina D., Oltean M., Multiplex cytokine profiling in patients with sepsis. APMIS. 119, 155–163 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Bourgon N., Fitzgerald W., Aschard H., Magny J. F., Guilleminot T., Stirnemann J., Romero R., Ville Y., Margolis L., Leruez-Ville M., Cytokine profiling of amniotic fluid from congenital cytomegalovirus infection. Viruses 14, 2145 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Renneson J., Dutta B., Goriely S., Danis B., Lecomte S., Laes J. F., Tabi Z., Goldman M., Marchant A., IL-12 and type I IFN response of neonatal myeloid DC to human CMV infection. Eur. J. Immunol. 39, 2789–2799 (2009). [DOI] [PubMed] [Google Scholar]
- 7.U. S. Akpan, L. S. Pillarisetty, Congenital cytomegalovirus infection, StatPearls, 1–3 (2022); www.ncbi.nlm.nih.gov/books/NBK541003/. [PubMed]
- 8.Osterholm E. A., Schleiss M. R., Impact of breast milk-acquired cytomegalovirus infection in premature infants: Pathogenesis, prevention, and clinical consequences? Rev. Med. Virol. 30, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Namba F., Nakagawa R., Haga M., Yoshimoto S., Tomobe Y., Okazaki K., Nakamura K., Seki Y., Kitamura S., Shimokaze T., Ikegami H., Nishida K., Mori S., Tamai K., Ozawa J., Tanaka K., Miyahara N., Cytomegalovirus-related sepsis-like syndrome in very premature infants in Japan. Pediatr. Int. 64, e14994 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Kong Y., Li Y., Zhang W., Yuan S., Winkler R., Kröhnert U., Han J., Lin T., Zhou Y., Miao P., Wang B., Zhang J., Yu Z., Zhang Y., Kosan C., Zeng H., Sepsis-induced thymic atrophy is associated with defects in early lymphopoiesis. Stem Cells 34, 2902–2915 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Luo M., Xu L., Qian Z., Sun X., Infection-associated thymic atrophy. Front. Immunol. 12, 652538 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ansari A. R., Liu H., Acute thymic involution and mechanisms for recovery. Arch. Immunol. Ther. Exp. 65, 401–420 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Lee E. N., Park J. K., Lee J.-R., Oh S.-O., Baek S.-Y., Kim B.-S., Yoon S., Characterization of the expression of cytokeratins 5, 8, and 14 in mouse thymic epithelial cells during thymus regeneration following acute thymic involution. Anat. Cell Biol. 44, 14–24 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearse G., Normal structure, function and histology of the thymus. Toxicol. Pathol. 34, 504–514 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Berthault C., Larcher T., Härtle S., Vautherot J. F., Trapp-Fragnet L., Denesvre C., Atrophy of primary lymphoid organs induced by Marek’s disease virus during early infection is associated with increased apoptosis, inhibition of cell proliferation and a severe B-lymphopenia. Vet. Res. 49, 31 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ducimetière L., Lucchiari G., Litscher G., Nater M., Heeb L., Nuñez N. G., Wyss L., Burri D., Vermeer M., Gschwend J., Moor A. E., Becher B., van den Broek M., Tugues S., Conventional NK cells and tissue-resident ILC1s join forces to control liver metastasis. Proc. Natl. Acad. Sci. U.S.A. 118, e2026271118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon J. P., Vrba S. M., Reynoso G. V., Wynne-Jones E., Kamenyeva O., Malo C. S., Cherry C. R., McManus D. T., Hickman H. D., Group 1 innate lymphoid-cell-derived interferon-γ maintains anti-viral vigilance in the mucosal epithelium. Immunity 54, 276–290.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El Weizman O., Adams N. M., Schuster I. S., Krishna C., Pritykin Y., Lau C., Degli-Esposti M. A., Leslie C. S., Sun J. C., O’Sullivan T. E., ILC1 confer early host protection at initial sites of viral infection. Cell 171, 795–808.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sparano C., Solís-Sayago D., Vijaykumar A., Rickenbach C., Vermeer M., Ingelfinger F., Litscher G., Fonseca A., Mussak C., Mayoux M., Friedrich C., Nombela-Arrieta C., Gasteiger G., Becher B., Tugues S., Embryonic and neonatal waves generate distinct populations of hepatic ILC1s. Sci. Immunol. 7, eabo6641 (2022). [DOI] [PubMed] [Google Scholar]
- 20.Chen Y., Wang X., Hao X., Li B., Tao W., Zhu S., Qu K., Wei H., Sun R., Peng H., Tian Z., Ly49E separates liver ILC1s into embryo-derived and postnatal subsets with different functions. J. Exp. Med. 219, e20211805 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopes N., Galluso J., Escalière B., Carpentier S., Kerdiles Y. M., Vivier E., Tissue-specific transcriptional profiles and heterogeneity of natural killer cells and group 1 innate lymphoid cells. Cell Rep. Med. 3, 100812 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kogame T., Egawa G., Nomura T., Kabashima K., Waves of layered immunity over innate lymphoid cells. Front. Immunol. 13, 957711 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vosshenrich C. A. J. J., García-Ojeda M. E., Samson-Villéger S. I., Pasqualetto V., Enault L., Le Goff O. R.-L., Corcuff E., Guy-Grand D., Rocha B., Cumano A., Rogge L., Ezine S., Di Santo J. P., A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat. Immunol. 7, 1217–1224 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Gabrielli S., Sun M., Bell A., Zook E. C., de Pooter R. F., Zamai L., Kee B. L., Murine thymic NK cells are distinct from ILC1s and have unique transcription factor requirements. Eur. J. Immunol. 47, 800–805 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vargas C. L., Poursine-Laurent J., Yang L., Yokoyama W. M., Development of thymic NK cells from double negative 1 thymocyte precursors. Blood 118, 3570–3578 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cordes M., Canté-Barrett K., van den Akker E. B., Moretti F. A., Kiełbasa S. M., Vloemans S. A., Garcia-Perez L., Teodosio C., van Dongen J. J. M., Pike-Overzet K., Reinders M. J. T., Staal F. J. T., Single-cell immune profiling reveals thymus-seeding populations, T cell commitment, and multilineage development in the human thymus. Sci. Immunol. 7, eade0182 (2022). [DOI] [PubMed] [Google Scholar]
- 27.Schmitt T. M., Ciofani M., Petrie H. T., Zúñiga-Pflücker J. C., Carlos Zúñiga-Pflücker J., Maintenance of T cell specification and differentiation requires recurrent Notch receptor-ligand interactions. J. Exp. Med. 200, 469–479 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavaert M., Liang K. L., Vandamme N., Park J. E., Roels J., Kowalczyk M. S., Li B., Ashenberg O., Tabaka M., Dionne D., Tickle T. L., Slyper M., Rozenblatt-Rosen O., Vandekerckhove B., Leclercq G., Regev A., Van Vlierberghe P., Guilliams M., Teichmann S. A., Saeys Y., Taghon T., Integrated scRNA-seq identifies human postnatal thymus seeding progenitors and regulatory dynamics of differentiating immature thymocytes. Immunity 52, 1088–1104.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Elsaid R., Meunier S., Burlen-Defranoux O., Soares-da-Silva F., Perchet T., Iturri L., Freyer L., Vieira P., Pereira P., Golub R., Bandeira A., Perdiguero E. G., Cumano A., A wave of bipotent T/ILC-restricted progenitors shapes the embryonic thymus microenvironment in a time-dependent manner. Blood 137, 1024–1036 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stokic-Trtica V., Diefenbach A., Klose C. S. N., NK cell development in times of innate lymphoid cell diversity. Front. Immunol. 11, 813 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klose C. S. N., Flach M., Möhle L., Rogell L., Hoyler T., Ebert K., Fabiunke C., Pfeifer D., Sexl V., Fonseca-Pereira D., Domingues R. G., Veiga-Fernandes H., Arnold S. J., Busslinger M., Dunay I. R., Tanriver Y., Diefenbach A., Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell 157, 340–356 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Gao Y., Souza-Fonseca-Guimaraes F., Bald T., Ng S. S., Young A., Ngiow S. F., Rautela J., Straube J., Waddell N., Blake S. J., Yan J., Bartholin L., Lee J. S., Vivier E., Takeda K., Messaoudene M., Zitvogel L., Teng M. W. L., Belz G. T., Engwerda C. R., Huntington N. D., Nakamura K., Hölzel M., Smyth M. J., Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat. Immunol. 18, 1004–1015 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Flommersfeld S., Böttcher J. P., Ersching J., Flossdorf M., Meiser P., Pachmayr L. O., Leube J., Hensel I., Jarosch S., Zhang Q., Chaudhry M. Z., Andrae I., Schiemann M., Busch D. H., Cicin-Sain L., Sun J. C., Gasteiger G., Victora G. D., Höfer T., Buchholz V. R., Grassmann S., Fate mapping of single NK cells identifies a type 1 innate lymphoid-like lineage that bridges innate and adaptive recognition of viral infection. Immunity 54, 2288–2304.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park E., Patel S., Wang Q., Andhey P., Zaitsev K., Porter S., Hershey M., Bern M., Plougastel-Douglas B., Collins P., Colonna M., Murphy K. M., Oltz E., Artyomov M., Sibley L. D., Yokoyama W. M., Toxoplasma gondii infection drives conversion of NK cells into ILC1-like cells. eLife 8, e47605 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Constantinides M. G., McDonald B. D., Verhoef P. A., Bendelac A., A committed precursor to innate lymphoid cells. Nature 508, 397–401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daussy C., Faure F., Mayol K., Viel S., Gasteiger G., Charrier E., Bienvenu J., Henry T., Debien E., Hasan U. A., Marvel J., Yoh K., Takahashi S., Prinz I., De Bernard S., Buffat L., Walzer T., T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J. Exp. Med. 211, 563–577 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gordon S. M., Chaix J., Rupp L. J., Wu J., Madera S., Sun J. C., Lindsten T., Reiner S. L., The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity 36, 55–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedrich C., Taggenbrock R. L. R. E., Doucet-Ladevèze R., Golda G., Moenius R., Arampatzi P., Kragten N. A. M., Kreymborg K., Gomez de Agüero M., Kastenmüller W., Saliba A. E., Grün D., van Gisbergen K. P. J. M., Gasteiger G., Effector differentiation downstream of lineage commitment in ILC1s is driven by Hobit across tissues. Nat. Immunol. 22, 1256–1267 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malaisé M., Rovira J., Renner P., Eggenhofer E., Sabet-Baktach M., Lantow M., Lang S. A., Koehl G. E., Farkas S. A., Loss M., Agha A., Campistol J. M., Schlitt H. J., Geissler E. K., Kroemer A., KLRG1+ NK cells protect T-bet-deficient mice from pulmonary metastatic colorectal carcinoma. J. Immunol. 192, 1954–1961 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Nakatani K., Kaneda K., Seki S., Nakajima Y., Pit cells as liver-associated natural killer cells: Morphology and function. Med. Electron Microsc. 37, 29–36 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Shin S. B., McNagny K. M., ILC-you in the thymus: A fresh look at innate lymphoid cell development. Front. Immunol. 12, 681110 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiong J., Zhao Y., Lin Y., Chen L., Weng Q., Shi C., Liu X., Geng Y., Liu L., Wang J., Zhang M., Identification and characterization of innate lymphoid cells generated from pluripotent stem cells. Cell Rep. 41, 111569 (2022). [DOI] [PubMed] [Google Scholar]
- 43.Orr M. T., Sun J. C., Hesslein D. G. T., Arase H., Phillips J. H., Takai T., Lanier L. L., Ly49H signaling through DAP10 is essential for optimal natural killer cell responses to mouse cytomegalovirus infection. J. Exp. Med. 206, 807–817 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narni-Mancinelli E., Chaix J., Fenis A., Kerdiles Y. M., Yessaad N., Reynders A., Gregoire C., Luche H., Ugolini S., Tomasello E., Walzer T., Vivier E., Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc. Natl. Acad. Sci. U.S.A. 108, 18324–18329 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ran G. H., Lin Y. Q., Tian L., Zhang T., Yan D. M., Yu J. H., Deng Y. C., Natural killer cell homing and trafficking in tissues and tumors: From biology to application. Signal Transduct. Target. Ther. 7, 205 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morillon Y. M., Manzoor F., Wang B., Tisch R., Isolation and transplantation of different aged murine thymic grafts. J. Vis. Exp., e52709 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao W., Lin J. X., Wang L., Li P., Leonard W. J., Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat. Immunol. 12, 551–559 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshimoto T., Takeda K., Tanaka T., Ohkusu K., Kashiwamura S., Okamura H., Akira S., Nakanishi K., IL-12 up-regulates IL-18 receptor expression on T cells, Th1 cells, and B cells: Synergism with IL-18 for IFN-γ production. J. Immunol. 161, 3400–3407 (1998). [PubMed] [Google Scholar]
- 49.Zhao Y., Liu P., Xin Z., Shi C., Bai Y., Sun X., Zhao Y., Wang X., Liu L., Zhao X., Chen Z., Zhang H., Biological characteristics of severe combined immunodeficient mice produced by CRISPR/Cas9-mediated Rag2 and IL2rg mutation. Front. Genet. 10, 401 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferreira A. C. F., Szeto A. C. H., Heycock M. W. D., Clark P. A., Walker J. A., Crisp A., Barlow J. L., Kitching S., Lim A., Gogoi M., Berks R., Daly M., Jolin H. E., McKenzie A. N. J., RORα is a critical checkpoint for T cell and ILC2 commitment in the embryonic thymus. Nat. Immunol. 22, 166–178 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qian L., Bajana S., Georgescu C., Peng V., Wang H. C., Adrianto I., Colonna M., Alberola-Ila J., Wren J. D., Sun X. H., Suppression of ILC2 differentiation from committed T cell precursors by E protein transcription factors. J. Exp. Med. 216, 884–899 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bigley T. M., Yang L., Kang L. I., Saenz J. B., Victorino F., Yokoyama W. M., Disruption of thymic central tolerance by infection with murine roseolovirus induces autoimmune gastritis. J. Exp. Med. 219, e20211403 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tougaard P., Martinsen L. O. L. O., Zachariassen L. F. L. F., Krych L., Nielsen D. S. D. S., Buus T. B. T. B., Pedersen A. E. A. E., Hansen A. K. A. K., Skov S., Hansen C. H. F. C. H. F., TL1A aggravates cytokine-induced acute gut inflammation and potentiates infiltration of intraepithelial natural killer cells in mice. Inflamm. Bowel Dis. 25, 510–523 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Kamimura Y., Lanier L. L., Rapid and sequential quantitation of salivary gland-associated mouse cytomegalovirus in oral lavage. J. Virol. Methods 205, 53–56 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li W., Germain R. N., Gerner M. Y., High-dimensional cell-level analysis of tissues with Ce3D multiplex volume imaging. Nat. Protoc. 14, 1708–1733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hao Y., Hao S., Andersen-Nissen E., Mauck W. M., Zheng S., Butler A., Lee M. J., Wilk A. J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E. P., Jain J., Srivastava A., Stuart T., Fleming L. M., Yeung B., Rogers A. J., McElrath J. M., Blish C. A., Gottardo R., Smibert P., Satija R., Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korsunsky I., Millard N., Fan J., Slowikowsk K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P. R., Raychaudhuri S., Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Figs. S1 to S7
Tables S1 to S28
Legend for movie S1
Legend for data file S1
References
Movie S1
Data file S1