Summary
Gene duplication produces the material that fuels evolutionary innovation. The “out-of-testis” hypothesis suggests that sperm competition creates selective pressure encouraging the emergence of new genes in male germline, but the somatic expression and function of the newly evolved genes are not well understood. We systematically mapped the expression of young duplicate genes throughout development in Caenorhabditis elegans using both whole-organism and single-cell transcriptomic data. Based on the expression dynamics across developmental stages, young duplicate genes fall into three clusters that are preferentially expressed in early embryos, mid-stage embryos, and late-stage larvae. Early embryonic genes are involved in protein degradation and develop essentiality comparable to the genomic average. In mid-to-late embryos and L4-stage larvae, young genes are enriched in intestine, epidermal cells, coelomocytes, and amphid chemosensory neurons. Their molecular functions and inducible expression indicate potential roles in innate immune response and chemosensory perceptions, which may contribute to adaptation outside of the sperm.
Keywords: gene duplication, young genes, Caenorhabditis elegans, developmental stage, cell type, somatic expression, adaptation, evolution
Graphical abstract

Highlights
-
•
Identifying gene-duplication events in eleven Caenorhabditis species
-
•
Three clusters of young duplicates based on developmental expression dynamics
-
•
High expression heterogeneity for young duplicates among lineages and cell types
-
•
Their weak constraints, high polymorphism, and potential roles in immune response
Ma et al. systematically mapped the somatic expression of recently duplicated genes in nematodes across developmental stages using both whole-organism and single-cell transcriptomic data. Young duplicate genes are enriched in early- and middle-stage embryos and late-stage larvae in specific lineages and cell types. They may contribute to innate immune response.
Introduction
Gene duplication is a mechanism to produce new genetic material, and the duplicate genes can undergo subfunctionalization, neofunctionalization, and degeneration.1,2,3,4 The evolutionary mechanism that drives the preservation of duplicate genes is under intense research. One prominent idea is the “out-of-testis” hypothesis, which posits that the strong selective pressure in the male germline encourages the evolution of new genes, which may increase reproductive fitness and thus be positively selected.5 This idea is supported by the observation that new genes showed exclusive or biased expression in the testis of both Drosophila6,7 and mammals.8 After the retention through positive selection, these testis-specific genes may eventually acquire expression and functions in other tissues and be incorporated into other biological processes. However, to our knowledge, a genome-wide analysis of the somatic expression of young duplicate genes throughout development and at single-cell resolution is still lacking. Moreover, the somatic function of the young duplicate genes and the biological pathways in which they are involved are largely unknown.
The studies of the fate of young duplicate genes are also complicated by a controversy regarding the essentiality of the newly arisen genes. Based on RNAi results, Chen et al. found that 30% of young genes (95% of which were derived from duplication within the last 35 million years) in Drosophila were essential for viability, and this proportion of essentiality was similar to that (35%) of old genes (which originated over 40 million years ago).9 However, in a later study Kondo et al. disputed these results by examining loss-of-function mutants of the young genes and found that most of them were not lethal.7 Interestingly, a more recent study examined a large collection of knockdown phenotypes of 11,354 Drosophila genes and still revealed a high proportion (32.2%) of essential genes among the new genes that arose <40 million years ago.10 The authors attributed the discrepancy between the RNAi and knockout results to the potential transcriptional compensation effects of the knockout (i.e., knockout mutants often generate aberrant mRNAs that trigger the upregulation of paralogous genes). We reason that analyzing both the RNAi and knockout phenotypes of young duplicate genes from another organism may help resolve the controversy.
In this study, we analyzed the somatic expression and function of young duplicate genes, as well as their essentiality, in Caenorhabditis elegans. Previous spatial transcriptomic studies of C. elegans and Pristionchus pacificus identified enriched expression of novel genes in the sperm-related regions, providing the first evidence to support the “out-of-testis” hypothesis in nematodes.11 This finding is somewhat surprising because both C. elegans and P. pacificus are androdioecious species and their self-fertilizing hermaphrodites produce limited numbers of sperms during reproduction. One potential explanation is that many of the recent gene-duplication events may predate the evolution of hermaphroditism in C. elegans and P. pacificus (since their sister species are dioecious), so the creation of new duplicate genes could still be driven by sperm competition. On the other hand, weaker sperm competition in these androdioecious species compared to gonochoristic species might encourage somatic expression of the young duplicate genes.
Focusing on C. elegans, we first analyzed the genomes of 11 Caenorhabditis species and identified C. elegans genes duplicated at different times in evolution and compared their expression patterns using both whole-organism and single-cell transcriptomic data. We found that young duplicate genes showed more dynamic expression throughout development than older genes and single-copy genes but were more restricted to specific cell types among the differentiated tissues. We identified three groups of young genes with peak expression at early embryonic, mid-embryonic, and late-larval stages, respectively. Essentiality analysis found that although young genes overall had much lower percentage of essential genes than older genes and single-copy genes, the group of young genes that had biased expression during early embryogenesis indeed develop essentiality at a level similar to that of the genomic average. In larvae and adults, young genes showed enriched expression not only in the sperms but also in the intestine, epidermis, and chemosensory neurons and may contribute to innate immunity and sensory perception. Overall, our studies provided a comprehensive analysis of the expression of young duplicate genes across developmental stages and tissue types and proposed potential somatic functions of the young genes out of the male germline.
Results
Gene duplication shapes genomic evolution in eleven Caenorhabditis species
To analyze gene duplication in the Caenorhabditis genus, we collected the protein sequences from 11 Caenorhabditis species that have high-quality genomic data (Table S1) and constructed orthogroups (OGs) among the homologous genes using Orthofinder2.12 Most (>90%) of the genes were included in OGs for each species (Figure 1A and Table S2). By comparing the gene tree of each OG with the species tree under a duplication-loss-coalescence (DLC) model,13 we identified duplication events and genes derived from duplications at each branch of the phylogenetic tree (Table S3). We grouped the duplicated genes by their origin from recent, old, or ancient duplications (see STAR Methods for estimated time of duplication). Considerably more genes were duplicated in recent times than in earlier and ancient times for all species, ranging from 14% to 34% of the genomes (Figure 1A).
Figure 1.
Duplication characteristics among 11 Caenorhabditis species
(A) Phylogenetic tree of the 11 Caenorhabditis species used to identify duplication events in a DLC model. The number of OGs, genes in OGs, and duplicates generated in different time periods are indicated for each species. The number of significantly expanded (red) and contracted (green) OGs on the branches (both terminal and internal) are shown.
(B) The number of genes from the top three gene families of recently duplicated genes for each species, and the number of genes gained or lost in the significantly expanded or contracted OGs, respectively, defined by the difference between the number of genes in one species and the average of the other ten species. Only the top three affected gene families that gained ≥30 genes or lost ≥10 genes are shown.
(C) The number of expanded (red) and contracted (green) OGs with specific domains or functions in the 11 species. See Table S5 for details.
(D) Enriched gene ontology terms for the 2,251 genes in the C. elegans-expanded and -specific OGs.
Using PfamScan,14 we searched for domain information of the recently duplicated genes and found that several domains were common among these genes (Table S3), including G-protein-coupled receptors (GPCRs), F-box domain (FBP), BTB domain, DUF3557 domain, and others, suggesting that similar families of genes were duplicated in different Caenorhabditis lineages (Figure 1B). Interestingly, lineages leading to C. inopinata had significant duplications in genes involved in DNA transposition (DDE_3 domain) and retrotransposition (RVT_1 domain), which did not occur in the sister species C. elegans. These findings are consistent with the expansion of transposable elements in C. inopinata.15,16
Focusing on C. elegans, we generated a list of 4,962 recently duplicated genes that include the 67 genes duplicated at the N4 branch, 4,496 genes duplicated at the C. elegans terminal branch (Figure 1A), and 399 genes found in C. elegans-specific OGs that only contained two or three genes and were thus excluded from the DLC model. These 4,962 N4/Cel genes are the main set of young duplicate genes analyzed in this study (Table S4). In addition, we also identified 256 and 1,406 genes that originated from ancient (at N0 branch) and old (at N1 and N3 branches) duplications, respectively, as well as 4,736 single-copy genes from the OGs that had only one ortholog from each species (Table S4). For genomic localizations, recently duplicated genes were enriched on the arms of chromosomes I, II, and V, whereas single-copy genes showed a more even distribution along the genome with a slight concentration on the chromosomal centers (Figure S1). This localization bias is consistent with previous observations.17,18 Most duplications were segmental or tandem, at least for the top three duplicate gene families, and the duplication of genomic blocks within chromosomes V and II and between them were the major driving forces for gene duplications (Figures S2A and S2B).
Expansion of specific gene families through duplications
We next identified the gene families that underwent significant expansion through duplication by applying the computational analysis of gene family evolution (CAFE) algorithm, which uses a stochastic birth and death process to model the evolutionary change of gene family sizes.19 This analysis uncovered 922 and 299 OGs that were significantly expanded and contracted, respectively, at the species level (Figure 1A). C. nigoni had the largest number of expanded OGs, whereas its sister species C. briggsae had the largest number of contracted OGs, which explains the differences in their total gene numbers (Figure 1A). This difference may be related to the evolution of distinct reproductive modes since gene loss may be a consequence of genomic adaptation to self-fertilization in C. briggsae.20,21 Nevertheless, the gene-number difference is not obvious between C. elegans and its outcrossing sister species C. inopinata.
The significantly expanded or contracted OGs share certain molecular structure or biological functions (Figure 1B and Table S5). For example, GPCRs, FBP genes, and BTB genes were commonly found in both the expanded and contracted OGs. GPCRs detect extracellular cues and transduce signals across the membrane; both FBP and BTB proteins act as substrate-specific adaptors for E3 ubiquitin ligase and function in protein degradation. Previous studies also found adaptive evolution of the FBP and BTB gene families.22 Interestingly, different species appear to expand and contract OGs with distinct domains. C. nigoni and C. remanei significantly expanded the FBP gene families, while the most expanded OGs in C. becei, C. briggsae, C. elegans, and C. panamensis contained GPCRs (Figure 1B). We also observed the previously reported contraction of GPCR gene families in C. inopinata.15 Genes involved in DNA transposition, retrotransposition, metabolism, and signaling, as well as transcription factors, helicases, and peptidases were also found in OGs with species-level expansion or contraction (Figure 1C).
Next, we focused on the 1,128 genes in the 71 C. elegans expanded OGs and the 1,123 genes in 264 C. elegans-specific OGs, which were excluded from the above CAFE analysis because they only contained genes from one species (Table S6). Ninety-five percent of these 2,251 genes in the combined C. elegans-expanded and -specific OGs were found in the 4,962 recently duplicated genes (Figure S2C), confirming that duplication contributes to gene family expansion. GPCR, FBP, LEC (Lectin_C domain), NHR (nuclear hormone receptor), IRLD (Recep_L_domain), MATH, BTB, histones, DUF3557, OAC (Acyl_transf_3 domain), and other families were highly enriched in these OGs (Table S6). Gene ontology analysis found the enrichment of these genes in functional terms consistent with domain characterization (Figure 1D). The expansion of gene families involved in sensory perception, innate immunity, and other pathways might facilitate the adaptation to environmental changes during the evolution of C. elegans.
Expression of recently duplicated genes at specific developmental stages
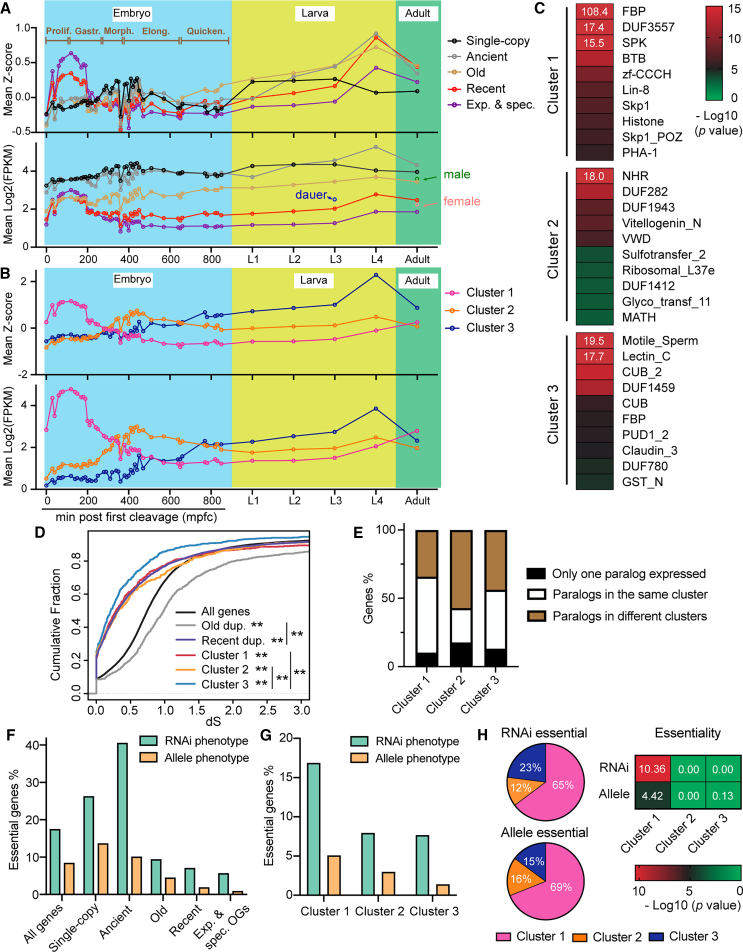
To understand when the recently duplicated genes are expressed throughout development, we used whole-embryo RNA-sequencing (RNA-seq) data with high temporal resolution23 and aggregate larval and adult-stage expression data (see STAR Methods). Although the recently duplicated genes generally had lower expression than older duplicate genes and single-copy genes, their expression showed strong enrichment at early embryonic stage and the late-larval stage (Figure 2A). Genes from the C. elegans-expanded and -specific OGs are the youngest among the recently duplicated genes and showed even stronger enrichment in early embryos. Older genes and single-copy genes showed much less dynamics throughout development. We next classified the young duplicate genes based on the Z-score profile across developmental stages and identified three clusters with stage-specific expression. Cluster 1 genes (1,000) had peak expression in early embryos (30–190 min post first cleavage [mpfc]), cluster 2 genes (402) had enriched expression at mid-embryonic stage (∼400 mpfc), and cluster 3 genes (781) had peak expression at L4 stage (Figures 2B and S3). Genes (2,779 or 56% of the young duplicates) that had very low expression (maximum expression across stages <10 fragments per kilobase of transcript per million mapped reads [FPKM]) were excluded from the clustering.
Figure 2.
Expression dynamics of duplicate genes throughout development in C. elegans
(A) Mean expression or standardized Z score for single-copy genes, genes duplicated at the N0 (ancient), N1/N3 (old), and N4/Cel (recent) branches, and genes in the C. elegans-expanded and -specific OGs across developmental stages. The Z score is calculated by normalizing expression values to the mean of all developmental time points. The average expression of recently duplicated genes in male and fog-2(−) female adults and dauers are also shown.
(B) Mean expression or Z score for the three clusters of recent duplicated genes throughout development.
(C) Top ten enriched domains in each cluster. The −log10 transformed p values from a hypergeometric test are shown, with values outside the color range denoted by white numbers.
(D) Cumulative plots for the dS of indicated genes. Double asterisks indicate p < 0.01 in a two-sample Kolmogorov-Smirnov test for comparison with all genes or between indicated pairs.
(E) Percentage of genes that are the only paralog in the OG with expression above threshold (FPKM > 10) or from the OGs in which expressed paralogs belong to the same or different clusters.
(F) Percentage of genes that were found to be essential based on RNAi or mutant allele phenotypes in each group.
(G) Percentage of essential genes in the three clusters.
(H) Distribution and enrichment of essential genes in each cluster, with the p value indicating the significance of enrichment in one cluster relative to others.
Different clusters showed the enrichment of different families of genes. Cluster 1 had mostly FBP and BTB genes, as well as DUF3557 and SPK genes; cluster 2 had enrichment of NHR, DUF282, and other genes; cluster 3 was enriched with genes encoding motile sperm domain-containing proteins, lectin, CUB-domain proteins, and others (Figure 2C). The molecular functions of the young genes appear to match their stage-specific expression. For example, the FBP and BTB genes expressed in early embryos may help degrade maternal proteins to promote the maternal-to-zygotic transition; the motile sperm genes may contribute to spermatogenesis and LEC and CUB genes for innate immune response in larvae and adults.
Since the duplicate genes in the three clusters almost all originated in C. elegans after the separation from C. inopinata, we used synonymous substitution rate (dS) to further estimate their ages.24 We found that cluster 3 genes had smaller dS and may be younger than the other two clusters (Figure 2D), suggesting that young embryonic genes may be derived from relatively earlier duplications than the young genes expressed in larval tissues. In addition, in clusters 1 and 3, ∼10% of genes are the only expressed paralog in the OG, ∼50% of the genes are from OGs in which all paralogs belong to the same cluster, and the rest are from OGs associated with more than one cluster. Cluster 2 genes were more likely to be mixed with other cluster genes in the same OG (Figure 2E). These findings may provide insights into the potential sub- and neofunctionalization of the duplicate genes if we interpret the different timing of expression as the sign of functional divergence.
Young genes expressed in early embryos develop essentiality
We next examined the essentiality of the young duplicate genes. To account for the potential discrepancy between RNAi and mutant phenotypes, we compiled two lists of RNAi-lethal and allele-lethal genes based on the curated phenotypes in WormBase (WS279). Both the knockdown and mutant data indicated lower percentages of essential genes in the recently duplicated genes compared to genes duplicated earlier and single-copy genes (Figure 2F). Interestingly, the old duplicates that originated at N1 and N3 branches (Figure 1A) had a level of essentiality below the genomic average, whereas the ancient duplicates that originated at N0 branch had a high percentage of essential genes comparable to single-copy genes. These results suggest that it may take a long time for the duplicate genes to develop essentiality. Although more genes appear to be essential based on the RNAi data compared to the mutant data (which could be due to the unavailability of knockout phenotypes for some genes or the real difference between RNAi and mutant phenotypes), young genes are much less likely to become essential than old genes.
Interestingly, within the recently duplicated genes, cluster 1 genes, which are expressed in early embryos, showed the highest level of essentiality. Based on the RNAi phenotype, 15.9% of cluster 1 genes were essential, similar to the genomic average (17.6%). In contrast, less than 8% of cluster 2 and cluster 3 genes were essential (Figure 2G). In fact, over 65% of the essential genes among the three clusters are from cluster 1, showing strong enrichment (Figure 2H). These results suggest that although young genes in general are rarely essential, a subset of them could acquire expression in early embryos and quickly develop essential functions by integrating into vital pathways of early embryogenesis. Thus, our studies in C. elegans may help reconcile the controversy about the essentiality of young genes in Drosophila.7,9,10
Young duplicate genes are enriched in specific cell types at the single-cell level
Using available single-cell transcriptomic data of embryos and larvae at L2 and L4 stages, we studied the expression pattern of the recently duplicated genes across cell types throughout development. For early embryos, cluster 1 genes showed the strongest expression in the AB lineage descendants at the 16-cell stage (∼70 mpfc), matching the time of their peak expression found in the bulk RNA-seq data (Figure 3A). The AB lineage gives rise to ectoderm, mesoderm, and the pharynx. Significant expression was also seen in the EMS blastomere, its daughter cells (MS and E), and granddaughter cells, as well as the C blastomere. MS lineage gives rise to mesoderm and pharynx, E for endoderm, and C for mesoderm and ectoderm. Nevertheless, the expression in the lineage leading to the germline (P1–P4 lineage) was much weaker compared to other lineages. The expression of cluster 1 genes among the embryonic cells was reduced in general from the 64-cell stage (∼130 mpfc) onward. In contrast, we did not see such dynamics and lineage specificity for the single-copy genes (Figure 3A). A total of 357 genes (35.7%) in cluster 1 had maternally deposited mRNAs in the 1-cell embryo according to a published list of maternal genes,25 suggesting that the early embryonic expression of cluster 1 genes may be at least partly derived from maternal deposition of mRNAs.
Figure 3.
Lineage-resolved expression dynamics of the three clusters of recently duplicated genes
(A) Average expression of cluster 1 genes and single-copy genes from zygote (P0) to embryonic cells at the 64-cell stage based on single-cell transcriptomic data. Early blastomeres are labeled in white, and the cells at the 64-cell stage are listed in black.
(B) The top 25 cell types that showed the highest mean Z scores for cluster 2 and cluster 3 genes among all cell types at 440 mpfc and L4 stage, respectively. Their average expression in these cell types is also shown. Values outside of the range are labeled.
(C) Average expression of cluster 2 and cluster 3 genes in the cells derived from AB, C, D, E, MS, and P4 lineages across developmental stages. The number of cells for each cell type at a given time point was used as the weight when calculating the average expression.
For cluster 2 genes which showed peak expression at ∼440 mpfc, we found enriched expression in the developing epidermal cells (e.g., seam cells, hyp7, hyp4–6, tail hypodermis), intestine, and body wall muscle (BWM). Cluster 3 genes which had peak expression at L4 stage showed enriched expression in sperm, differentiated intestine, epidermis, sensory neurons (e.g., ASK, ASJ, and ADF), and other tissues (Figure 3B). Thus, recently duplicated genes are not only expressed at specific developmental stages but also preferentially expressed in specific cell types. Interestingly, the same tissue types (such as epidermis, intestines, coelomocytes, and rectal glands) showed enriched expression of different sets of young genes (clusters 2 and 3) at different developmental time points. The expression of these young genes might help the developmental innovation or functional adaptation of these tissues.
The strongest enrichment of cluster 3 genes in the sperm of C. elegans hermaphrodites is consistent with the male-biased expression of the recently duplicated genes in the bulk RNA-seq data (e.g., higher expression in adult males than hermaphrodites and fog-2(−) females [Figure 2A] and stronger enrichment in genes upregulated in males [Figure S4A]). Thus, our results support previous studies11 of the “out-of-testis” hypothesis in C. elegans.
The single-cell resolution of the expression pattern also provided opportunities to assess expression divergence among the duplicates. Using cluster 3 genes as an example, we calculated the expression correlation of all paralogous pairs across the 164 cell types at the L4 stage and found that ∼40% of the pairs show highly correlated expression (correlation coefficient is ∼1), while another ∼25% of the pairs had almost no correlation (coefficient is ∼0; Figures S4B and S4C). The divergence in expression patterns may indicate functional divergence.
The three cluster genes (2,183 in total) represent about half of the 4,962 recently duplicated genes that had high enough expression to be detected in the whole-organism transcriptomic data. The vast majority (2,371 or 85%) of the other half that had no or low expression in the bulk RNA-seq data showed expression (transcripts per million [TPM] > 10) in at least one cell type in the single-cell transcriptomic data. Strikingly, these genes also showed similar enrichment among the cell types as the three cluster genes (Figures S5A–S5D). Thus, both highly and lowly expressed young genes showed the same cell-type specificity.
Furthermore, we compared the expression of cluster 2 and cluster 3 genes across the six lineages throughout development (Figure 3C). In general, cluster 2 genes had the peak expression in mid-to-late embryos except for the P lineage that gives rise to the germline (sperms and oocytes); cluster 3 genes had the peak expression in the larval stages. Cluster 2 genes also had higher embryonic expression than cluster 3 genes. The E lineage that gives rise to the endoderm (i.e., intestine) had stronger expression of both cluster 2 and cluster 3 genes in late embryos (e.g., 500 mpfc onward) than the other lineages. The strong enrichment of the young genes in the sperms drove the sharp increase of expression in the P lineage at the L4 stage (Figure 3C).
Different cell lineages show distinct expression dynamics for young genes
By tracing the lineage precursors of the terminally differentiated cells, we further mapped the expression of cluster 2 and cluster 3 genes throughout lineage progression at the single-cell level and uncovered heterogeneity of their expression dynamics across tissue types throughout development.
For cluster 2 genes, intestine and BWM lineages had the typical expression pattern with the peak expression occurring in mid-to-late embryos, whereas hypodermal lineages showed strong expression at both mid-embryonic and L4 stages (Figure 4A). Among other non-neuronal lineages, the arcade cells (interfacial epithelial cells) stood out because their cluster 2 gene expression peaked at L4 (Figure S6A). Similar variations in expression dynamics were also observed among the neuronal lineages, many of which had significant expression of cluster 2 genes at L4 stage (Figure 4A). This variation may be expected, as C. elegans neurons are generated non-clonally from many different lineages.26 Moreover, since most embryonic lineages complete the last round of cell division before 400 mpfc, the peak expression of cluster 2 genes at around 400 mpfc likely occurred shortly after the generation of the terminal cells and at the time when the post-mitotic cells start to differentiate in mid-to-late embryos.
Figure 4.
Heterogeneity of expression dynamics among cell lineages for young duplicate genes
(A and B) Average Z score for cluster 2 and cluster 3 genes in individual lineages that give rise to hypodermis, intestine, BWM, and neurons. Hypodermis and BWM cells are derived from multiple lineages. Each line represents the developmental expression dynamics during the progression of a single lineage that generates a terminal cell.
(C and D) Z-score profiles of some cluster 2 and cluster 3 genes throughout the lineages that produced two non-neuronal cells (BWM and hypodermis) and two neurons (ASEL and ASG). Lineage precursors are labeled with arrows. Numbers in parentheses indicate the number of genes.
For cluster 3 genes, although most cell lineages followed the whole-organism expression profile with enriched expression at larval stages (especially L4), we still found a few exceptions (Figures 4B and S6B). For example, the BWM lineages had prominent expression of cluster 3 genes in the embryos that were comparable if not higher than their larval expression, and the lineages that led to the generation of several neurons (e.g., ASE, IL1, and OLQ) also showed peak expression in mid-stage embryos instead of L4 (Figure 4B). These results suggest that the overall developmental dynamics of gene expression does not necessarily reflect the dynamics of individual lineages. The contribution of young genes to development depends on the tissue type.
Which gene families may show different expression profiles among different cell lineages? We found that NHR, GPCR, FBP, and DUF282 genes in cluster 2 showed the typical peak expression in the mid-stage embryos for a BWM lineage but had uncommon peak expression at the L4 stage for a hypodermal lineage (Figure 4C). Similar disparity was found between the lineages that generate chemosensory neurons, ASEL and ASG (Figure 4C). Moreover, FBP, LEC, motile sperm, CUB, and collagen genes in cluster 3 also showed distinct expression dynamics among individual lineages (Figure 4D). Thus, the single-cell transcriptomic analysis provides a granular view of the possible site of action for the young genes throughout development.
Expression of young duplicate genes is highly restricted among differentiated cell types
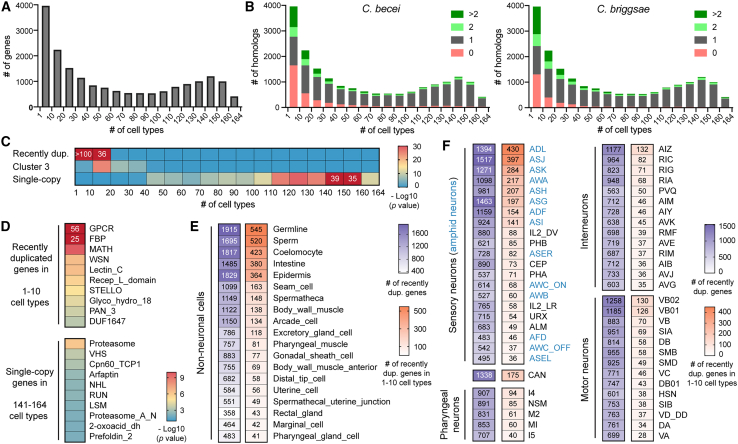
Compared to older genes and single-copy genes, the expression pattern of recently duplicated genes is not only more dynamic during development but also more restricted among differentiated cells. Using the transcriptomes of the 164 terminally differentiated cell types at the L4 stage,27 we found that among all genes in the genome, 3,963 and 2,238 were expressed in only 1–10 or 11–20 types of cells. Interestingly, the distribution of genes based on the number of cell types in which they are expressed appeared to be bimodal with preferences for genes to be either highly specific or highly ubiquitous, and more genes had specific expression than ubiquitous expression (Figures 5A and S7A). Another surprising finding was that a much larger fraction of C. elegans genes that had tissue-specific expression (i.e., expressed in ≤10 cell types) was absent or gained additional orthologs in related Caenorhabditis species than the more broadly expressed C. elegans genes (Figures 5B and S7B). This result hinted that these cell-specific genes were likely derived from gene duplications and underwent rapid evolution to be lost or further duplicated in the related species.
Figure 5.
Expression of recently duplicated genes is enriched in a few cell types
(A) The number of genes expressed in the indicated number of cell types at L4 stage according to single-cell transcriptomic data (genes with maximum TPM < 10 were removed). The number of cell types were binned into 17 groups.
(B) The number of C. elegans genes that have 0, 1, 2, or >2 homologs in C. becei and C. briggsae (according to the OGs in Table S2) for each of the 17 groups based on the number of cell types expressing the gene. Chi-squared test found statistical significance (p < 0.01) for the comparison between the cell-specific (i.e., ≤10 cell types) and ubiquitous (≥141 cell types) genes.
(C) Enrichment of recently duplicated genes, cluster 3 genes, and single-copy genes in the 17 groups, with p values calculated in a hypergeometric test.
(D) Domain enrichment for recently duplicated genes expressed in ≤10 cell types and single-copy genes expressed in ≥141 cell types.
(E and F) The top non-neuronal cell types (E) and neurons that showed the expression of the young duplicate genes and cell-specific young duplicates (F). The numbers of expressed young genes are shown. Neuronal classification was based on Cook et al.51; CAN neurons have unknown functions; amphid sensory neurons are in blue.
Supporting this hypothesis, we found that recently duplicated genes were highly enriched among the genes expressed in 1–10 and 11–20 cell types (Figure 5C). In fact, more than half of the genes expressed in 1–10 cell types were recently duplicated genes. Cluster 3 genes, which had the strongest expression at L4 stage, appeared to be enriched among the genes expressed in 11–20, 20–30, and 30–40 cell types, suggesting that young duplicate genes with higher overall expression tend to be expressed in more cell types. As a comparison, single-copy genes were much more likely to have ubiquitous expression across cell types (Figure 5C).
In terms of gene families, GPCR, FBP, MATH, WSN, LEC, and IRLD genes were the top enriched families among the highly cell-specific young duplicate genes, whereas genes involved in proteasome, chaperone function (Cpn60_TCP1), vesicle trafficking (VHS, arfaptin), protein-protein interactions (NHL, RUN), and RNA splicing (LSM) were enriched among the single-copy genes with ubiquitous expression (Figure 5D). Among tissue types, cell type-specific young genes were mostly expressed in the germ cells (including germline that produces oocytes at the L4 stage and the sperms), coelomocytes, intestine, and epidermis (Figure 5E). In the nervous system, young genes showed specific expression in the amphid gustatory and olfactory neuron (e.g., ADL, ASJ, ASK, and AWA) (Figure 5F). As expected, a large portion (∼60%) of these young genes were GPCRs, which function as receptors for various chemical cues. The expression and function of recently duplicated genes in selective cell types may contribute to adaptation.
Young duplicate genes are subjected to weak purifying selection and show a high degree of intraspecific polymorphism
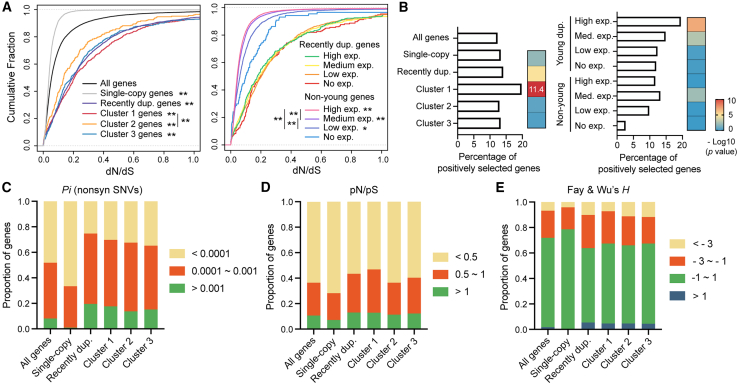
Next, we assessed the selective pressure on recently duplicated genes using the ratio of non-synonymous to synonymous substitution rates (dN/dS) and found that they generally had a higher dN/dS ratio than the genomic average, suggesting relaxed evolutionary constraints (Figures 6A and S8A; Table S4). In contrast, single-copy genes had lower than average dN/dS ratio, indicating strong purifying selection. Among the three clusters, cluster 1 genes had slightly higher dN/dS ratio than cluster 2 genes. We also compared the selective pressure on genes with different expression levels. All young duplicate genes showed similarly high dN/dS ratio regardless of expression levels, but for older genes higher expression correlated with lower dN/dS ratio, reflecting stronger purifying selection on highly expressed genes (Figure 6A).
Figure 6.
Young duplicate genes are subjected to weak evolutionary constraints and have large intraspecific variations
(A) Cumulative plots showing the dN/dS ratio of different groups of genes. Genes with a dN or dS value smaller than 0.0005 or equal to 999 were removed. Low expression means that the maximum FPKM across all developmental time points in the bulk RNA-seq data is between 0 and 10, medium expression for 10–100, and high expression for >100. Single and double asterisks indicate p < 0.05 and p < 0.01, respectively, in a two-sample Kolmogorov-Smirnov test in comparison with the genomic average or genes with no expression or between specific pairs.
(B) Percentage of positively selected genes identified by the branch-site model in CodeML.28 Enrichment p value was calculated from a hypergeometric test.
(C) Distribution of non-synonymous single-nucleotide polymorphisms for the indicated gene sets among 773 wild strains of C. elegans.
(D) Distribution of the ratio between the rates of non-synonymous and synonymous polymorphisms.
(E) Distribution of Fay and Wu’s H for high-frequency-derived non-synonymous SNVs using the highly divergent wild strain XZ1516 as the outgroup.
Because the above analyses were done using orthologs from the 11 genetically divergent Caenorhabditis species, one concern is that selection may need time to purge deleterious variants, but the length of the 11 terminal branches of the phylogenetic tree are not the same, leading to potential overestimation of dN in some cases. Another concern is that dS may be smaller for genes in OGs with C. elegans-specific duplications compared to OGs in which the closest ortholog of a C. elegans gene is from its sister species. To address these biases, we recalculated the dN/dS ratios using only the orthologs between C. elegans and C. inopinata, as the comparisons among genes would be roughly at the same evolutionary distance. The results supported that the young duplicate genes have higher dN/dS ratios or evolutionary rates than the genomic average and single-copy genes (Figure S8B).
Higher dN/dS ratio could indicate not only relaxed evolutionary constraints but also potential positive selection. To formally test for positive selection, we used the branch-site method,28,29 which is based on a likelihood ratio test, to detect positive selection along the C. elegans lineage of the tree. We found that the recently duplicated genes, especially cluster 1 genes and genes with high expression, were indeed enriched among the positively selected genes (Figure 6B and Table S4), supporting potential positive selection on the young duplicates, especially the ones expressed in early embryos.
We also examined the intraspecific variations of the recently duplicated genes and found that they carried higher polymorphism of non-synonymous single-nucleotide variants (SNVs) than an average gene or the single-copy genes (Figure 6C and Table S7). Their ratio of non-synonymous to synonymous polymorphisms (pN/pS) was also greater, suggesting weak purifying selection and rapid evolution within the species (Figure 6D). We also found that a smaller percentage of the young duplicate genes had Fay and Wu’s H value close to 0 (i.e., neutrality) compared to the single-copy genes (Figure 6E), while higher percentages of genes had either an excess (H < −1) or deficit (H > 1) of high-frequency derived non-synonymous SNVs due to potential positive or balancing selection, respectively. Previous work identified genomic regions that are hyperdivergent among the wild isolates.30 We found that 37% of the young duplicate genes are located in the hyperdivergent region (compared to 5% of single-copy genes; Figure S8C), supporting a high degree of polymorphism among young genes. More importantly, when removing these genomic localization biases and only focusing on the genes outside of the hyperdivergent region, we still observed a higher degree of polymorphism, a higher pN/pS ratio, and more negative H values for the recently duplicated genes compared to genomic average (Figures S8D–S8F), suggesting that the intraspecific variation on the young duplicate genes were largely independent of their locations. Interestingly, fewer genes had H > 1 after removing the hyperdivergent regions, supporting the previous hypothesis that the hyperdivergent haplotypes are maintained by balancing selection.30
Cluster 3 duplicate genes are induced by pathogenic infections
Given that many of the recently duplicated genes, such as FBP, MATH, and LEC family genes, are thought to be involved in innate immunity and stress response,31 we hypothesized that some young duplicates may function in the response toward pathogenic infection, thus contributing to the fitness of the animal. To test this hypothesis, we examined the transcriptomic data for gene-expression changes upon exposure to nine pathogens (Table S8) and found that recently duplicated genes, especially the cluster 3 genes, were enriched among the genes significantly upregulated by at least one pathogen (Figures 7A and 7B). Genes whose basal expression level was low or medium appeared to be much more enriched in the inducible genes than the ones with no or high expression. Moreover, the fold change of the recently duplicated genes was bigger than that of the single-copy genes (Figure 7C), and the young duplicates also appeared to respond to more than one infectious condition, whereas the single-copy genes responded specifically to only one condition on average (Figure 7D). These results support the findings in yeast that duplicate genes have higher plasticity and larger dynamic range in conditionally responsive expression.32 Nonetheless, in yeast, duplicate genes respond to specific stress and single-copy genes respond more generally to many stress conditions33; we found the opposite in C. elegans.
Figure 7.
Cluster 3 genes are induced by microbial infection
(A) Volcano plot showing significant upregulation (fold change >3) of genes by at least one pathogen. Green, FBP genes; blue, MATH genes; red, LEC genes.
(B) Enrichment of the 12 types of genes among the genes significantly upregulated by at least one pathogen, with p value from a hypergeometric test.
(C) Box-and-whisker plots showing the fold changes of recently duplicated genes with low and medium expression, cluster 3 genes, and single-copy genes among all upregulated genes (fold change >0). “+” indicates the mean value. Double asterisks indicate p < 0.01 in ANOVA and Dunnett’s test.
(D) Box-and-whisker plots showing the number of upregulations per gene among all significant upregulation events (fold change >3).
(E) The top cell types with the strongest expression of significantly upregulated cluster 3 genes, presented as the average of Z scores across cell types based on the single-cell transcriptomic data at L4 stage. Amphid sensory neurons are in blue. The top cell types for all cluster 3 genes are shown as a comparison.
(F and G) The top gene families among the pathogen-inducible recently duplicated genes and cluster 3 genes.
Using single-cell RNA-seq data, we found that the pathogen-inducible young duplicate genes showed the highest expression in the intestine (Figure 7E), which is a major tissue for innate immune response against bacterial infection in C. elegans.34 Domain analysis found that the top families of young genes that respond to pathogenic infections included FBP, LEC, MATH, CUB, collagen, Shk, and GPCR genes (Figures 7A, 7F, and 7G), all of which have known functions in innate immunity.35,36,37 In addition, the pals genes, which encode proteins with the ALS2CR12 domain, were previously found to respond to intracellular pathogens and may be under balancing selection38; we found that 29 (out of 39) pals genes belonged to the recently duplicated genes and that 24 were in the significantly upregulated gene list. Moreover, we found the enrichment of the pathogen-induced genes in amphid chemosensory neurons, including ADL and ASH neurons (Figure 7E), which were previously found to mediate the avoidance of toxin-producing Streptomyces through GPCRs.39 Thus, young genes might function not only in intestine and epidermis but also in sensory neurons to enhance immune response. Compared to infectious stresses, the young duplicates were less induced by non-infectious stresses, such as heat shock, cold stress, and hypoxia (Table S8 and Figure S9), suggesting that they might be more involved in innate immunity rather than general stress response. This finding also hints that the response to abiotic stress may be ancient, while the host response to its pathogens is more recently evolved and is continuously co-evolving with the pathogens.
Discussion
In this study, we systematically mapped the expression of young duplicate genes in C. elegans throughout development at single-cell resolution and found that they had enriched expression at three specific developmental stages and in specific somatic cell types.
One set (cluster 1) of the young duplicates had strong expression at early embryonic stages from proliferation to early gastrulation and were preferentially expressed in the blastomeres that give rise to somatic tissues rather than the germline. Many of them are involved in protein ubiquitination and degradation and may regulate early embryogenesis by inducing the degradation of maternal proteins, which is critical for maternal-to-zygotic transition.40 In fact, we found that this set of young genes had an essentiality level comparable to that of the genomic average, suggesting that a significant portion of them have been quickly integrated into vital pathways of embryonic development. In addition to acquiring essential functions, these young genes may also be the source of evolutionary innovation in early embryonic development, given that large morphologic and temporal variations were observed among the nematode species at the early embryonic stages.41,42 Supporting this idea, cluster 1 genes had the highest dN/dS ratios and the strongest enrichment of genes with positively selected sites among the young duplicate genes, suggesting potential positive selection on early embryonic development.
Another set (cluster 2) had the strongest expression in mid-to-late embryos, when most embryonic cells exit the cell cycle and begin to acquire their terminal cell fates. The enrichment of NHR family transcription factors in these young genes is interesting, since several NHR genes were known to regulate cell fate specification.43 These newly evolved genes might become cell fate regulators to facilitate differentiation or to help generate new cell types. Alternatively, NHR genes were also known to control metabolic networks,43 which might contribute to the morphogenesis of differentiating cells. In fact, other families of metabolic genes were also enriched in cluster 2.
The third set (cluster 3) showed the peak expression at late-larval stage. As expected, these genes were heavily enriched in sperms, supporting the “out-of-testis” theory. It is worth noting that the sperm genes are largely the same in hermaphrodites and males,44 so we would expect these young genes to be similarly enriched in male sperms. Among the somatic tissues, these young duplicate genes showed enriched expression in intestines, epidermis, and coelomocyte (the three major tissues that contact foreign pathogens and evoke innate immune response) as well as amphid chemosensory neurons. The molecular functions of the cluster 3 genes in innate immunity and chemosensory perception are consistent with their site of expression. We hypothesize that the young genes may enhance the immunity against pathogens and the sensing of environmental cues, which may contribute to adaptation.
Moreover, distinct sets (clusters 2 and 3) of young genes were enriched in the same cell types (e.g., intestine, hypodermis, seam cells, coelomocytes, chemosensory neurons) at different stages of development, suggesting that these tissues may be the major somatic targets of evolutionary innovation in C. elegans or nematodes in general. The intestine-biased expression of young genes is particularly interesting because C. elegans is a bacterivore and interacts with hundreds of bacterial species (both dietary and pathogenic) in their natural habitat.45 The complex bacteria-host interaction may drive the selection of young genes in the intestine. This observation in worms appears to be strikingly similar to the liver-specific somatic expression of young genes in mammals.46 Like the intestine in C. elegans, liver is both a digestive organ and a frontline innate immune organ in mammals.47 Therefore, young genes expressed in liver may drive dietary adaptation and enhance innate immunity in mammals, similar to the intestine-specific genes in C. elegans. This deep conservation in both sperm and somatic expression of young genes points to a potentially universal mechanism underlying genomic innovation.
How do the young genes acquire somatic expression? One possible mechanism is that young duplicate genes are first expressed in the sperms and are positively selected based on their contributions to sperm competitiveness. Once actively maintained, they can then acquire somatic expression by evolving cis-regulatory enhancers that are active in somatic tissues. In fact, only 29 (<1%) recently duplicated genes were expressed exclusively in the sperm, suggesting that vast majority of the young genes would broaden their expression into somatic cells. If the somatic function of the young gene becomes essential or is itself positively selected, the original sperm expression and function, in theory, may become redundant or even be lost. In fact, among the 4,168 recently duplicated genes that have expression in at least one somatic cell type, only 1,666 (∼40%) showed sperm expression, suggesting that a large fraction of the young duplicate genes may have indeed lost sperm expression and moved entirely outside of the testis.
Alternatively, some young duplicates might have originated directly from somatic tissues and were never expressed in the male germline. The finding that different evolutionary forces control the emergence of young testis- and liver-specific genes in mammals supports this hypothesis.46 In C. elegans, the numbers of recently duplicated genes that show expression in sperms (1,695), intestine (1,485), epidermis (1,829), and coelomocytes (1,817) are comparable, suggesting the possibility of multiple origins of new genes.
Regardless of the mechanisms, gene duplication plays an important role in the evolution and adaptation of C. elegans. For example, the FBP gene fog-2, which is responsible for the evolution of hermaphroditism in C. elegans,48 was created through a dramatic OG expansion that generated 80 new FBP genes after separation from C. inopinata. Neofunctionalization of one of them gave rise to self-fertilization in C. elegans. Interestingly, it was proposed that this reproductive mode transition from gonochorism to androdioecy might be driven by natural selection to achieve reproductive assurance because the requirement of mating may limit reproduction.49 As another example, srg-36 and srg-37 were duplicate pheromone receptor GPCR genes that promote dauer formation, and a natural variation in srg-37 reduced the dauer pheromone sensitivity in some wild populations.50 Thus, duplication creates additional gene copies that can be modified for the purpose of niche-specific adaptation.
Finally, the young duplicate genes count for more than half of the cell type-specific genes (expressed in ≤10 cell types) and thus contribute significantly to the molecular differences among cell types and may play important roles in their morphological and functional specification. Therefore, gene duplication may fuel the generation of the extraordinary cell type diversity in multicellular organisms.
Limitations of the study
Given the limited availability of high-quality genome assemblies in the Caenorhabditis genus, some species used in our phylogenetic reconstruction may be too distant to allow the identification of duplication events with high temporal resolution. For example, C. inopinata is the closest known sister species of C. elegans, but they are still separated at a relatively early evolutionary time point (much earlier than the separation of C. briggsae and C. nigoni). This may lead to overestimation of species-specific duplicates in C. elegans. If future sampling efforts can yield a closer sister species of C. elegans, the estimation of duplication time will be more accurate. Moreover, in this study we identified duplicated genes based on protein sequence similarity. It will be interesting to find out whether more duplication events can be identified by combining sequence orthology with genomic synteny in future studies.
The finding of sperm-enriched expression of young duplicates in the androdiecious nematodes is somewhat surprising because of the presumably weak sperm competition, which, on the other hand, may encourage somatic expression of the young genes. Thus, it will be of great importance to confirm our results of the somatic expression pattern of the young duplicates in gonochoric nematode species as well as in species from other phyla.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Genome assemblies of Caenorhabditis species | WormBase Parasite52 | https://parasite.wormbase.org/index.html |
| Pfam-A | Mistry et al.14 | https://doi.org/10.1093/nar/gkaa913 |
| Whole-embryo transcriptomic data | Hashimshony et al.23 | https://doi.org/10.1038/nature13996 |
| Whole-organism transcriptomic data at larval, adult, and dauer stages | WormBase | https://downloads.wormbase.org/releases |
| Male and female transcriptomic data at adult stage | Thomas et al.53 | https://doi.org/10.1016/j.cub.2012.09.038 |
| Single-cell transcriptomic data in early embryos | Tintori et al.54 | https://doi.org/10.1016/j.devcel.2016.07.025 |
| Single-cell transcriptomic data during embryogenesis | Packer et al.55 | https://doi.org/10.1126/science.aax1971 |
| Single-cell transcriptomic data at L2 larval stage | Cao et al.56 | https://doi.org/10.1126/science.aam8940 |
| Single-cell transcriptomic data at L4 larval stage | Taylor et al.27 | https://doi.org/10.1016/j.cell.2021.06.023 |
| Curated RNAi and mutant allele phenotype for essentiality | WormBase WormMine | http://intermine.wormbase.org/tools/wormmine/templates.do |
| Transcriptomic data for response to pathogenic infection and non-infectious stresses | See Table S8 | See Table S8 |
| Software and algorithms | ||
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com/ |
| R (v4.2.1) | R Project | https://www.r-project.org |
| Orthofinder (v2.5.4) | Emms et al.12 | https://doi.org/10.1186/s13059-019-1832-y |
| STRIDE | Emms et al.57 | https://doi.org/10.1093/molbev/msx259 |
| STAG | Emms et al.58 | https://doi.org/10.1101/267914 |
| TimeTree | Kumar et al.59 | http://timetree.org/ |
| Duplication-Loss Coalescence | Wu et al.13 | https://doi.org/10.1101/gr.161968.113 |
| r8s | Sanderson60 | https://doi.org/10.1093/bioinformatics/19.2.301 |
| CAFE (v4.2.1) | De Bie et al.19 | https://hahnlab.github.io/CAFE/manual.html |
| ClueGO | Bindea et al.61 | https://doi.org/10.1093/bioinformatics/btp101 |
| Hypergeometric test and Hierarchical clustering | R package - Stats | https://stat.ethz.ch/R-manual/R-devel/library/stats/html/00Index.html |
| PfamScan | Mistry et al.14 | https://doi.org/10.1093/nar/gkaa913 |
| edgeR | Robinson et al.62 | https://doi.org/10.1186/gb-2010-11-3-r25 |
| STAR (v2.7) | Dobin et al.63 | https://doi.org/10.1093/bioinformatics/bts635 |
| DESeq2 | Love et al.64 | https://doi.org/10.1186/s13059-014-0550-8 |
| MUSCLE (v3.8.31) | Edgar65 | https://doi.org/10.1093/nar/gkh340 |
| ClipKIT | Steenwyk et al.66 | https://doi.org/10.1371/journal.pbio.3001007 |
| MegaX | Kumar et al.67 | https://doi.org/10.1093/molbev/msy096 |
| PAML (v4.9j) | Yang28 | https://doi.org/10.1093/molbev/msm088 |
| PopGenome | Pfeifer et al.68 | https://doi.org/10.1093/molbev/msu136 |
Resource availability
Lead contact
Further information and requests for resources and methodologies should be directed to and will be fulfilled by the lead contact, Chaogu Zheng (cgzheng@hku.hk).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Data used in this study are all previously published datasets. The codes used to generate the results in this study can be found at https://github.com/Fuqiang-Ma/Cel_dup_development (https://doi.org/10.5281/zenodo.10104147).
Method details
Construction of orthogroups (OGs) and the identification of duplicate genes
From the genome assemblies of twenty-one Caenorhabditis species available in the WormBase ParaSite database (Table S1), we chose eleven species based on the assembly contiguity (N50 > 200,000) and the completeness of genome assembly and annotation (BUSCO assembly >90%). We did not include C. brenneri due to the reported high heterozygosity.69 The longest protein isoform encoded by each gene in the eleven species was extracted and used for constructing OGs using the OrthoFinder v2.5.412 with default parameters (Table S2). The species tree was inferred from all genes using the STAG software58 and then rooted using STRIDE.12,57 Duplication events at each branch in the species tree were identified using the Duplication-Loss-Coalescence algorithm13 implemented in OrthoFinder; duplication events with possibility lower than 0.5 were discarded. Genes with duplication events in more than one branch were assigned to the most recent one. Due to the lack of fossil records, accurate dating of the divergence time among the nematode species is difficult. Using genome rearrangement rate, neutral mutation rate, and pairwise ortholog comparison, previous works estimated the divergence times between C. elegans and C. briggsae to be in a range from 18.6 to 101.5 million years ago (mya)70,71,72 and the divergence time between C. nigoni and C. briggsae to be ∼3.5 mya.73 Using this information, we calibrated the species tree and estimated the origin of recently duplicated genes to be within 60 million years based on the median time of the estimated range, old duplication to be 60–130 mya, and ancient duplication to be > 130 mya.
Identification of C. elegans expanded and specific OGs
To identify significantly expanded and contracted OGs, we used the gene birth-death model implemented in CAFE v4.2.119 to analyze the evolution of gene family size on 24,422 OGs. Species tree constructed by OrthoFinder was used as the input for CAFE. A single birth and death parameter λ was estimated based on the estimated divergence time between C. briggsae and C. elegans. The rapidly evolving gene families at each branch on the tree were then identified as significantly expanded and contracted OGs and were used for further analysis. C. elegans specific genes refer to the genes in the OGs that only contained C. elegans genes. In total, we obtained 1,128 genes from 71 C elegans expanded OGs and 1,123 genes from 264 C elegans specific OGs.
Gene ontology analysis
To understand the molecular functions of duplicate genes, we used PfamScan to search for the potential domains in the proteins coded by each gene in the Pfam-A protein database.14 HMM file for specific domain was downloaded from the Pfam website (https://pfam.xfam.org/) and was used to search against the protein sequence files of the eleven species using hmmsearch. The resultant domain information for duplicate genes in the eleven Caenorhabditis species and for genes in the C. elegans expanded and specific OGs are in Table S3 and Table S6, respectively. One gene often carries more than one domain. Thus, we merged genes carrying the F box, FTH, FBA_2 and HTH_48 domains as F box proteins (FBPs) and merged all GPCR subfamilies as GPCRs (see Table S5). We also subjected the duplicate genes to gene ontology analysis using the ClueGO tools in Cytoscape to identify enriched functional modules.61
Whole-organism and single-cell transcriptomic data analysis
Time series of embryonic transcriptomic data were obtained from Hashimshony et al.23 Whole-animal transcriptomic data at the four larval and adult stages, as well as the dauer stage, were from the aggregated median expression on WormBase (WS279). The expression data for males and fog-2(−) females were from Thomas et al.53 Expression datasets used in this study are summarized in Table S8. All bulk RNA-seq data at different developmental stages were normalized to Fragments Per Kilobase of transcript per Million mapped reads (FPKM). We then performed hierarchical clustering (using the R Stats package) on the expression profiles of the recently duplicated genes across developmental stages. Genes with maximum FPKM <10 among all stages were excluded from the clustering. Based on the results of the clustering, we divided the recently duplicated genes into three clusters with peak expression in early embryos, mid-to-late embryos, and late larval stages, respectively.
We also obtained annotated single-cell transcriptomic data from four studies, which cover the developmental stages from embryogenesis to larval stages. The study by Tintori et al.54 covers the embryonic cells from zygote to 16-cell stage. Transcriptomes of other nonapoptotic embryonic lineage cells (including both precursors and terminal cells) were obtained from Packer et al.55 Data for differentiated individual cell type at L2 and L4 stages were obtained from the studies of Cao et al.56 and Taylor et al.,27 respectively. For analysis, we combined the four sets of scRNA-seq data and applied a pseudo-bulk approach to compute the expression level of every gene among the identified cell types across all developmental time points. Briefly, the counts of individual cells that belong to the same cell type were summed up in each batch of samples. Replicates with 5 or fewer cells were removed. After this filtering, we obtained the data for in total 1,208 cell types throughout development, including 531 embryonic lineage cells, 394 terminal cells at late embryonic stage, 117 cells at L2 stage, and 164 cell types at L4 stage. We then normalized the library size for each individual cell type by applying the trimmed mean of M-values (TMM) method implemented in edgeR.62 Gene expression values were calculated by averaging the expression across the pseudo-replicates for each cell type based on the effective library size and were presented as transcripts per million (TPM). Genes with maximum TPM <10 among all cell types were excluded from most analyses.
Gene essentiality analysis
Gene essentiality analysis was performed based on the curated RNAi and mutant phenotype data for lethality on WormBase (WS279). We used WormMine to extract genes whose RNAi or allele phenotypes contain “lethal”, “embryonic_lethal”, “adult_lethal”, “embryonic_terminal_arrest_variable_emb”, “embryonic_lethal_late_emb, larval_lethal”, “larval_arrest”, “late_larval_lethal”, “late_larval_arrest”, or “one_cell_arrest_early_emb”. We compiled 3,681 RNAi-lethal genes and 2,019 allele-lethal genes; 980 genes are common between the two lists. Essential genes in single-copy genes, C. elegans expanded & specific OGs, and duplicate genes at N0 (ancient), N1/N3 (old), and N4/Cel (recent) branches were then counted, and percentages were calculated.
Single-cell expression dynamics for individual cell lineages throughout development
To characterize the expression dynamics of cluster 2 and 3 young genes in specific lineages across development, we set up time points from 55 to 800 mpfc at 10-min intervals and added L2 and L4 stages as the last two time points. We then identified the cells that existed at a given time based on a timetable of birth and division time for every embryonic cell.74 For time points before 510 mpfc, we used the single-cell transcriptomic data for embryonic lineages and for time points beyond 510 mpfc, we used the data for terminal cell types. To compare the expression profiles of AB, C, D, E, MS, and P4 lineages during development, we identified the cells derived from the six lineages at a given time point and calculated the weighted average of gene expression within each lineage based on cell numbers.
For individual cell types, we identified all lineage precursors and the annotated terminal cells in the single-cell transcriptomic datasets and mapped these cells onto the developmental timeline based on their birth and division time to create time-resolved expression profiles for individual lineages throughout development. Using AVK neuron as an example, AVKL and AVKR cells are derived from ABplpapapa and ABprpapapa lineages, respectively. The single-cell transcriptomic datasets covered all six lineage cells from ABpxp to ABpxpapapa, as well as the terminal embryonic AVK neurons in different time windows (AVK:390_510, AVK:510_650, AVK:gt_650) and the differentiated AVK neurons at L2 and L4 stages. Since AVKL and AVKR are transcriptionally indistinguishable along development, we merged the two lineages into AVK lineage. Cells like the AVHL and AVHR neurons, whose lineage precursors have distinguishable transcriptomes (e.g., ABalapaaa for AVHL and ABalappap for AVHR), were analyzed separately as independent cell lineages. In total, we analyzed 171 lineages that generate 72 neurons and 99 non-neuronal cells. For each lineage, the expression profiles across developmental time points were created for individual genes and Z score across these time points were calculated. The average expression or Z score was then calculated for a given set of genes.
RNA-seq analysis for infectious and non-infectious stress-induced expression
We collected RNA-seq data for pathogenic infection or non-infectious stress-induced expression changes from previous studies (see Table S8 for references). The exact conditions of these experiments (all performed on the wild-type N2 strain) and the unique identifiers for the transcriptomic datasets can be found in Table S8. We obtained the raw reads and aligned them to the WBcel235 reference genome of C. elegans by STAR v2.7.63 Gene count was computed using STAR option --quantMode GeneCounts for RNA-seq data under different treatments. We then used DESeq264 to identify differentially expressed genes under each stress condition and combined them to generate a list of stress-upregulated and downregulated genes under infectious and non-infectious conditions for enrichment analysis. A gene qualifies as a stress-regulated gene if it is significantly changed by at least one condition. If a gene is regulated by multiple conditions, we consider the regulation as independent events and plotted all of them in Figures 7A and 7C.
Calculation of dN/dS ratios and identification of positively selected genes
To understand the evolutionary rate of different groups of C. elegans genes, protein sequences from the same OG were first aligned using MUSCLE v3.8.3165 and then aligned by codon alignment program in MegaX.67 Gaps were removed after the alignment. We then applied the CodeML program of PAML v4.9j28 to compute the evolutionary rate. The dN/dS values for each gene were computed using a free-ratio model which allows ω to vary along branches (NSsites = 0, model = 1). We calculated dN/dS values using either all the orthologs from the 11 terminal species (Figure 6A) or only the orthologs between C. elegans and C. inopinata (Figure S8B). Lack of synonymous or non-synonymous mutations would result in extreme value, and thus we excluded the dN or dS value smaller than 0.0005 or equal to 999 for ω analysis. The potential substitution saturation may also bias the estimation of evolutionary rate estimation. To reduce such confounding effects, we applied ClipKIT66 to trim the extremely divergent alignments and retain parsimony-informative sites. We also redid the analysis after filtering out the genes with dS > 2 as suggested by previous studies75 and obtained the same results (Figure S8A). To assess the effect of selective pressure on genes with different expression levels, we divided the genes into four categories based on their maximum expression levels across all developmental time points: no expression for maximum FPKM = 0, low for 0–10, medium for 10–100, and high for >100.
To identify positively selected genes, we run the branch site model in CodeML,28 which allows ω to vary among both sites in proteins and branches on the tree, on the orthologous sequences from the eleven species. This model detects positively selected amino acid sites in specific lineages of the phylogenetic tree. We tested the positive selection by comparing twice the log likelihood difference between null model (NSites = 2, model = 2, ω = 1) and alternative model (NSites = 2, model = 2, ω = 0) with a χ2-distribution in the likelihood ratio test (LRT). Positively selected genes were then identified based on the presence of positively selected sites that show statistical significance.
Calculation of population genetics statistics for C. elegans wild strains
We obtained the genotyped VCF file for the single-nucleotide variants (SNVs) of 773 wild isolates of C. elegans (CaeNDR 2020 release downloaded from https://caendr.org/data/data-release/c-elegans/20200815). To investigate the intraspecific evolution of recently duplicated genes among these wild isolates, we used PopGenome68 to calculate the polymorphism Pi76 and Fay and Wu’s H77 for each gene. When calculating H, we used a highly divergent strain, XZ1516, as the outgroup and excluded six other divergent strains (ECA1465, ECA1467, ECA1493, ECA1515, ECA701, and ECA702) similar to XZ1516. We also computed the ratio of non-synonymous (pN) to synonymous (pS) polymorphism using previous formula.78
Quantification and statistical analysis
Statistical analyses were performed using R (v4.2.1) and GraphPad Prism 8. Details of each test, sample sizes, and p values are described in the figure legends. In summary, hypergeometric tests were used throughout the study to test for the significance of enrichment of one set of genes in another set of genes. The test was applied in the essentiality analysis, the enrichment of particular gene families among the young duplicate genes, and the enrichment of gene expression in specific cell types. One-way ANOVA and Dunnett’s tests were used for multiple comparison of expression fold changes. Two-sample Kolmogorov-Smirnov tests were performed to compare different classes of genes for their evolutionary rate and synonymous substitution rate; p values were corrected by the number of pairwise comparisons.
Acknowledgments
We thank the WormBase ParaSite for sharing genomic data of Caenorhabditis species, and we thank the CeNGEN project; the Goldstein, Murray, Shendure, and Yanai labs; and others for sharing RNA-seq datasets. Some panels in the graphical abstract were prepared using BioRender. This study was supported by funds from the National Natural Science Foundation of China (Excellent Young Scientists Fund for Hong Kong and Macau, 32122002), the Research Grant Council of Hong Kong (GRF 17107021, GRF 17106322, and CRF C702620G), and the Health Bureau of Hong Kong (HMRF 09201426) to C.Z. Computational works were performed using research computing facilities offered by Information Technology Services at the University of Hong Kong.
Author contributions
F.M. and C.Z. designed the study, conducted most of the analyses, interpreted the data, prepared the figures, and wrote the manuscript. C.Y.L. performed some of the analyses. C.Z. conceived the project, supervised the work, and wrote the manuscript. All authors critically reviewed the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: December 18, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2023.100467.
Supplemental information
Genome assembly files and the information about the assembly quality were obtained for 21 Caenorhabditis species from the WormBase ParaSite website (https://parasite.wormbase.org/ftp.html). Eleven species (red) with high-quality genome assemblies were used in this study.
The longest transcript for each gene in the 11 species was used to construct OGs. In total, 24,422 OGs were constructed.
Domain information for the genes duplicated in different time periods were obtained by searching against the Pfam database using hmmsearch.
C. elegans genes generated by duplication events at different branches of the phylogenetic tree (including N0, N1/N3, and N4/Cel branches). C. elegans single-copy genes were also included for comparison. Positively selected genes and calculated dN/dS values for each gene can be found here.
OGs were grouped together based on the molecular or functional similarities of the domains identified in the OGs. Some OGs contained multiple domains, so the number of OGs in specific gene families may be smaller than the sum of the OGs with different domains.
The top gene families among the C. elegans-expanded or -specific OGs were listed, and their enrichment folds were calculated.
The polymorphism for non-synonymous SNVs (Pi), pN/pS ratios, and the Fay and Wu’s H of various types of genes.
Detailed information of all expression datasets used in this study including the references and the identifiers for the datasets.
References
- 1.Force A., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kondrashov F.A., Kondrashov A.S. Role of selection in fixation of gene duplications. J. Theor. Biol. 2006;239:141–151. doi: 10.1016/j.jtbi.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 3.Ohno S. Springer-Verlag; 1970. Evolution by Gene Duplication. [Google Scholar]
- 4.Taylor J.S., Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu. Rev. Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- 5.Kaessmann H. Origins, evolution, and phenotypic impact of new genes. Genome Res. 2010;20:1313–1326. doi: 10.1101/gr.101386.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haerty W., Jagadeeshan S., Kulathinal R.J., Wong A., Ravi Ram K., Sirot L.K., Levesque L., Artieri C.G., Wolfner M.F., Civetta A., Singh R.S. Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics. 2007;177:1321–1335. doi: 10.1534/genetics.107.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kondo S., Vedanayagam J., Mohammed J., Eizadshenass S., Kan L., Pang N., Aradhya R., Siepel A., Steinhauer J., Lai E.C. New genes often acquire male-specific functions but rarely become essential in Drosophila. Genes Dev. 2017;31:1841–1846. doi: 10.1101/gad.303131.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luis Villanueva-Cañas J., Ruiz-Orera J., Agea M.I., Gallo M., Andreu D., Albà M.M. New Genes and Functional Innovation in Mammals. Genome Biol. Evol. 2017;9:1886–1900. doi: 10.1093/gbe/evx136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S., Zhang Y.E., Long M. New genes in Drosophila quickly become essential. Science. 2010;330:1682–1685. doi: 10.1126/science.1196380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia S., VanKuren N.W., Chen C., Zhang L., Kemkemer C., Shao Y., Jia H., Lee U., Advani A.S., Gschwend A., et al. Genomic analyses of new genes and their phenotypic effects reveal rapid evolution of essential functions in Drosophila development. PLoS Genet. 2021;17 doi: 10.1371/journal.pgen.1009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rödelsperger C., Ebbing A., Sharma D.R., Okumura M., Sommer R.J., Korswagen H.C. Spatial Transcriptomics of Nematodes Identifies Sperm Cells as a Source of Genomic Novelty and Rapid Evolution. Mol. Biol. Evol. 2021;38:229–243. doi: 10.1093/molbev/msaa207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emms D.M., Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y.C., Rasmussen M.D., Bansal M.S., Kellis M. Most parsimonious reconciliation in the presence of gene duplication, loss, and deep coalescence using labeled coalescent trees. Genome Res. 2014;24:475–486. doi: 10.1101/gr.161968.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J., et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanzaki N., Tsai I.J., Tanaka R., Hunt V.L., Liu D., Tsuyama K., Maeda Y., Namai S., Kumagai R., Tracey A., et al. Biology and genome of a newly discovered sibling species of Caenorhabditis elegans. Nat. Commun. 2018;9:3216. doi: 10.1038/s41467-018-05712-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodruff G.C., Teterina A.A. Degradation of the Repetitive Genomic Landscape in a Close Relative of Caenorhabditis elegans. Mol. Biol. Evol. 2020;37:2549–2567. doi: 10.1093/molbev/msaa107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas J.H. Analysis of homologous gene clusters in Caenorhabditis elegans reveals striking regional cluster domains. Genetics. 2006;172:127–143. doi: 10.1534/genetics.104.040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.C elegans Sequencing Consortium Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 19.Han M.V., Thomas G.W., Lugo-Martinez J., Hahn M.W. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 2013;30:1987–1997. doi: 10.1093/molbev/mst100. [DOI] [PubMed] [Google Scholar]
- 20.Fierst J.L., Willis J.H., Thomas C.G., Wang W., Reynolds R.M., Ahearne T.E., Cutter A.D., Phillips P.C. Reproductive Mode and the Evolution of Genome Size and Structure in Caenorhabditis Nematodes. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin D., Schwarz E.M., Thomas C.G., Felde R.L., Korf I.F., Cutter A.D., Schartner C.M., Ralston E.J., Meyer B.J., Haag E.S. Rapid genome shrinkage in a self-fertile nematode reveals sperm competition proteins. Science. 2018;359:55–61. doi: 10.1126/science.aao0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas J.H. Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res. 2006;16:1017–1030. doi: 10.1101/gr.5089806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashimshony T., Feder M., Levin M., Hall B.K., Yanai I. Spatiotemporal transcriptomics reveals the evolutionary history of the endoderm germ layer. Nature. 2015;519:219–222. doi: 10.1038/nature13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katju V., Lynch M. The structure and early evolution of recently arisen gene duplicates in the Caenorhabditis elegans genome. Genetics. 2003;165:1793–1803. doi: 10.1093/genetics/165.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quarato P., Singh M., Cornes E., Li B., Bourdon L., Mueller F., Didier C., Cecere G. Germline inherited small RNAs facilitate the clearance of untranslated maternal mRNAs in C. elegans embryos. Nat. Commun. 2021;12:1441. doi: 10.1038/s41467-021-21691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hobert O. 2010. Neurogenesis in the Nematode Caenorhabditis elegans; pp. 1–24. WormBook. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor S.R., Santpere G., Weinreb A., Barrett A., Reilly M.B., Xu C., Varol E., Oikonomou P., Glenwinkel L., McWhirter R., et al. Molecular topography of an entire nervous system. Cell. 2021;184:4329–4347.e23. doi: 10.1016/j.cell.2021.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 29.Yang Z., Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- 30.Lee D., Zdraljevic S., Stevens L., Wang Y., Tanny R.E., Crombie T.A., Cook D.E., Webster A.K., Chirakar R., Baugh L.R., et al. Balancing selection maintains hyper-divergent haplotypes in Caenorhabditis elegans. Nat. Ecol. Evol. 2021;5:794–807. doi: 10.1038/s41559-021-01435-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakowski M.A., Desjardins C.A., Smelkinson M.G., Dunbar T.L., Lopez-Moyado I.F., Rifkin S.A., Cuomo C.A., Troemel E.R. Ubiquitin-mediated response to microsporidia and virus infection in C. elegans. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehner B. Conflict between noise and plasticity in yeast. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mattenberger F., Sabater-Muñoz B., Toft C., Fares M.A. The Phenotypic Plasticity of Duplicated Genes in Saccharomyces cerevisiae and the Origin of Adaptations. G3 (Bethesda) 2017;7:63–75. doi: 10.1534/g3.116.035329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulenburg H., Kurz C.L., Ewbank J.J. Evolution of the innate immune system: the worm perspective. Immunol. Rev. 2004;198:36–58. doi: 10.1111/j.0105-2896.2004.0125.x. [DOI] [PubMed] [Google Scholar]
- 35.Simonsen K.T., Gallego S.F., Færgeman N.J., Kallipolitis B.H. Strength in numbers: "Omics" studies of C. elegans innate immunity. Virulence. 2012;3:477–484. doi: 10.4161/viru.21906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reboul J., Ewbank J.J. GPCRs in invertebrate innate immunity. Biochem. Pharmacol. 2016;114:82–87. doi: 10.1016/j.bcp.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 37.Martineau C.N., Kirienko N.V., Pujol N. Innate immunity in C. elegans. Curr. Top. Dev. Biol. 2021;144:309–351. doi: 10.1016/bs.ctdb.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Sluijs L., Bosman K.J., Pankok F., Blokhina T., Wilten J.I.H.A., Te Molder D.M., Riksen J.A.G., Snoek B.L., Pijlman G.P., Kammenga J.E., Sterken M.G. Balancing Selection of the Intracellular Pathogen Response in Natural Caenorhabditis elegans Populations. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.758331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tran A., Tang A., O'Loughlin C.T., Balistreri A., Chang E., Coto Villa D., Li J., Varshney A., Jimenez V., Pyle J., et al. C. elegans avoids toxin-producing Streptomyces using a seven transmembrane domain chemosensory receptor. Elife. 2017;6 doi: 10.7554/eLife.23770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toralova T., Kinterova V., Chmelikova E., Kanka J. The neglected part of early embryonic development: maternal protein degradation. Cell. Mol. Life Sci. 2020;77:3177–3194. doi: 10.1007/s00018-020-03482-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brauchle M., Kiontke K., MacMenamin P., Fitch D.H.A., Piano F. Evolution of early embryogenesis in rhabditid nematodes. Dev. Biol. 2009;335:253–262. doi: 10.1016/j.ydbio.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schierenberg E. 2006. Embryological Variation during Nematode Development; pp. 1–13. WormBook. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antebi A. 2015. Nuclear receptor signal transduction in C. elegans; pp. 1–49. WormBook. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ebbing A., Vertesy A., Betist M.C., Spanjaard B., Junker J.P., Berezikov E., van Oudenaarden A., Korswagen H.C. Spatial Transcriptomics of C. elegans Males and Hermaphrodites Identifies Sex-Specific Differences in Gene Expression Patterns. Dev. Cell. 2018;47:801–813.e6. doi: 10.1016/j.devcel.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 45.Samuel B.S., Rowedder H., Braendle C., Félix M.A., Ruvkun G. Caenorhabditis elegans responses to bacteria from its natural habitats. Proc. Natl. Acad. Sci. USA. 2016;113:E3941–E3949. doi: 10.1073/pnas.1607183113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guschanski K., Warnefors M., Kaessmann H. The evolution of duplicate gene expression in mammalian organs. Genome Res. 2017;27:1461–1474. doi: 10.1101/gr.215566.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao B., Jeong W.I., Tian Z. Liver: An organ with predominant innate immunity. Hepatology. 2008;47:729–736. doi: 10.1002/hep.22034. [DOI] [PubMed] [Google Scholar]
- 48.Nayak S., Goree J., Schedl T. fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biol. 2005;3:e6. doi: 10.1371/journal.pbio.0030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cutter A.D., Morran L.T., Phillips P.C. Males, Outcrossing, and Sexual Selection in Caenorhabditis Nematodes. Genetics. 2019;213:27–57. doi: 10.1534/genetics.119.300244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee D., Zdraljevic S., Cook D.E., Frézal L., Hsu J.C., Sterken M.G., Riksen J.A.G., Wang J., Kammenga J.E., Braendle C., et al. Selection and gene flow shape niche-associated variation in pheromone response. Nat. Ecol. Evol. 2019;3:1455–1463. doi: 10.1038/s41559-019-0982-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cook S.J., Jarrell T.A., Brittin C.A., Wang Y., Bloniarz A.E., Yakovlev M.A., Nguyen K.C.Q., Tang L.T.H., Bayer E.A., Duerr J.S., et al. Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature. 2019;571:63–71. doi: 10.1038/s41586-019-1352-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Howe K.L., Bolt B.J., Shafie M., Kersey P., Berriman M. WormBase ParaSite - a comprehensive resource for helminth genomics. Mol. Biochem. Parasitol. 2017;215:2–10. doi: 10.1016/j.molbiopara.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas C.G., Li R., Smith H.E., Woodruff G.C., Oliver B., Haag E.S. Simplification and desexualization of gene expression in self-fertile nematodes. Curr. Biol. 2012;22:2167–2172. doi: 10.1016/j.cub.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tintori S.C., Osborne Nishimura E., Golden P., Lieb J.D., Goldstein B. A Transcriptional Lineage of the Early C. elegans Embryo. Dev. Cell. 2016;38:430–444. doi: 10.1016/j.devcel.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Packer J.S., Zhu Q., Huynh C., Sivaramakrishnan P., Preston E., Dueck H., Stefanik D., Tan K., Trapnell C., Kim J., et al. A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science. 2019;365 doi: 10.1126/science.aax1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao J., Packer J.S., Ramani V., Cusanovich D.A., Huynh C., Daza R., Qiu X., Lee C., Furlan S.N., Steemers F.J., et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017;357:661–667. doi: 10.1126/science.aam8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emms D.M., Kelly S. STRIDE: Species Tree Root Inference from Gene Duplication Events. Mol. Biol. Evol. 2017;34:3267–3278. doi: 10.1093/molbev/msx259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emms D.M., Kelly S. STAG: Species Tree Inference from All Genes. bioRxiv. 2018 Preprint at. [Google Scholar]
- 59.Kumar S., Stecher G., Suleski M., Hedges S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- 60.Sanderson M.J. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- 61.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steenwyk J.L., Buida T.J., 3rd, Li Y., Shen X.X., Rokas A. ClipKIT: A multiple sequence alignment trimming software for accurate phylogenomic inference. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pfeifer B., Wittelsbürger U., Ramos-Onsins S.E., Lercher M.J. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 2014;31:1929–1936. doi: 10.1093/molbev/msu136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barrière A., Yang S.P., Pekarek E., Thomas C.G., Haag E.S., Ruvinsky I. Detecting heterozygosity in shotgun genome assemblies: Lessons from obligately outcrossing nematodes. Genome Res. 2009;19:470–480. doi: 10.1101/gr.081851.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cutter A.D. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol. Biol. Evol. 2008;25:778–786. doi: 10.1093/molbev/msn024. [DOI] [PubMed] [Google Scholar]
- 71.Blair J.E., Shah P., Hedges S.B. Evolutionary sequence analysis of complete eukaryote genomes. BMC Bioinf. 2005;6:53. doi: 10.1186/1471-2105-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Coghlan A., Wolfe K.H. Fourfold faster rate of genome rearrangement in nematodes than in Drosophila. Genome Res. 2002;12:857–867. doi: 10.1101/gr.172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomas C.G., Wang W., Jovelin R., Ghosh R., Lomasko T., Trinh Q., Kruglyak L., Stein L.D., Cutter A.D. Full-genome evolutionary histories of selfing, splitting, and selection in Caenorhabditis. Genome Res. 2015;25:667–678. doi: 10.1101/gr.187237.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sulston J.E., Schierenberg E., White J.G., Thomson J.N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 75.Wu C., Zhang D., Kan M., Lv Z., Zhu A., Su Y., Zhou D., Zhang J., Zhang Z., Xu M., et al. The draft genome of the large yellow croaker reveals well-developed innate immunity. Nat. Commun. 2014;5:5227. doi: 10.1038/ncomms6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nei M., Li W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fay J.C., Wu C.I. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nei M., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genome assembly files and the information about the assembly quality were obtained for 21 Caenorhabditis species from the WormBase ParaSite website (https://parasite.wormbase.org/ftp.html). Eleven species (red) with high-quality genome assemblies were used in this study.
The longest transcript for each gene in the 11 species was used to construct OGs. In total, 24,422 OGs were constructed.
Domain information for the genes duplicated in different time periods were obtained by searching against the Pfam database using hmmsearch.
C. elegans genes generated by duplication events at different branches of the phylogenetic tree (including N0, N1/N3, and N4/Cel branches). C. elegans single-copy genes were also included for comparison. Positively selected genes and calculated dN/dS values for each gene can be found here.
OGs were grouped together based on the molecular or functional similarities of the domains identified in the OGs. Some OGs contained multiple domains, so the number of OGs in specific gene families may be smaller than the sum of the OGs with different domains.
The top gene families among the C. elegans-expanded or -specific OGs were listed, and their enrichment folds were calculated.
The polymorphism for non-synonymous SNVs (Pi), pN/pS ratios, and the Fay and Wu’s H of various types of genes.
Detailed information of all expression datasets used in this study including the references and the identifiers for the datasets.
Data Availability Statement
Data used in this study are all previously published datasets. The codes used to generate the results in this study can be found at https://github.com/Fuqiang-Ma/Cel_dup_development (https://doi.org/10.5281/zenodo.10104147).