Summary
Chronic kidney disease is a leading cause of death and disability globally and impacts individuals of African ancestry (AFR) or with ancestry in the Americas (AMS) who are under-represented in genome-wide association studies (GWASs) of kidney function. To address this bias, we conducted a large meta-analysis of GWASs of estimated glomerular filtration rate (eGFR) in 145,732 AFR and AMS individuals. We identified 41 loci at genome-wide significance (p < 5 × 10−8), of which two have not been previously reported in any ancestry group. We integrated fine-mapped loci with epigenomic and transcriptomic resources to highlight potential effector genes relevant to kidney physiology and disease, and reveal key regulatory elements and pathways involved in renal function and development. We demonstrate the varying but increased predictive power offered by a multi-ancestry polygenic score for eGFR and highlight the importance of population diversity in GWASs and multi-omics resources to enhance opportunities for clinical translation for all.
Keywords: kidney function, chronic kidney disease, genome-wide association study, multi-ancestry, admixed populations, eGFR, fine-mapping, expression quantitative trait locus, polygenic scores
Graphical abstract

Highlights
-
•
Meta-analysis of GWASs of AFR and AMS individuals identifies new loci for eGFR
-
•
There was no heterogeneity in allelic effects on eGFR due genetic ancestry or sex
-
•
Polygenic score performance into AFR/AMS studies depends on ancestry and sample size
Hughes et al. conducted a large genetic study of kidney function in individuals of African ancestry and ancestry in the Americas who are disproportionately impacted by chronic kidney disease (CKD). Their study highlighted the importance of studying these under-represented groups to understand CKD biology and improve prediction of kidney function.
Introduction
Chronic kidney disease (CKD) is a leading cause of death and disability globally,1,2 and incurs huge health care costs for its treatment, prescriptions, office visits, and hospitalizations.3 In the US, compared with European ancestry individuals, CKD more often impacts individuals of African ancestry (AFR) and with ancestry in the Americas (AMS), who also more often have advanced disease and complications.4,5,6 These differences in disease risk may be related to lifestyle and social determinants of health that are correlated with ancestry, in addition to genetic differences in the spectrum of causal alleles.7 There is still limited understanding of this relationship, primarily because of the bias of genome-wide association studies (GWASs) to individuals of European ancestry.8 Identification of genetic variants that contribute to disease risk in non-European ancestry populations is essential to advance understanding of disease biology that informs the development of therapeutics. Furthermore, polygenic scores developed from European ancestry GWASs have limited transferability into other populations, partially reflecting differences in allele frequencies, effect sizes, and linkage disequilibrium (LD) structure, which vary across ancestry groups.9,10
In clinical care, CKD diagnosis is based on abnormal levels of blood and urine biomarkers, which are used for estimated glomerular filtration rate (eGFR) and to assess kidney injury. Disease progression to end-stage kidney disease and clinical decision for kidney replacement therapy (dialysis) still rely mostly on eGFR. Mendelian randomization analyses have also highlighted a causal effect of eGFR on diastolic blood pressure and hypertension, a risk factor for CKD.11 The most recent GWAS of eGFR included 1.5 million participants, but only 7.3% were AFR or AMS individuals.12 To address this population bias, the Continental Origins and Genetic Epidemiology Network (COGENT) Kidney Consortium was established to expand the diversity of genetic ancestry in GWAS of kidney traits, enabling locus discovery in under-studied populations, improving methods for multi-ancestry analyses, and building resources for downstream functional studies.13 Prior multi-ancestry meta-analyses of eGFR from the COGENT-Kidney Consortium have highlighted the improvement in fine-mapping resolution afforded by non-European ancestry individuals and contributed knowledge on allelic effect heterogeneity across diverse populations at identified loci.11,14 These analyses highlighted candidate causal genes with cell-type-specific expression in the glomerulus, and in the proximal and distal nephron, and causal effects of eGFR on overall and cause-specific CKD, kidney stone formation, and diastolic blood pressure.
Here, we describe the results of our latest COGENT-Kidney GWAS meta-analysis in 145,732 AFR and AMS individuals from the Americas, Africa, and Europe. With these data, we demonstrate the value of analyses conducted in these under-represented and under-studied population groups to understand how eGFR-associated variants impact molecular processes underlying CKD, and to enhance trait prediction through development of multi-ancestry polygenic scores (Figure 1).
Figure 1.
Analytical pipeline
In step 1, we conducted multi-ancestry meta-analysis of eGFR GWAS in 145,732 AFR and AMS individuals from the COGENT Kidney Consortium and Million Veteran Program (MVP). In step 2, we performed downstream integration with functional genomics resources to understand the effector genes and molecular mechanisms through which eGFR association signals are mediated. These analyses included correlation with eQTL in kidney from the Human Kidney Tissue Resource and The Cancer Genome Atlas, and in blood from the Genes-Environments and Admixture in Latino Asthmatics study and the Study of African Americans, Asthma, Genes, and Environments, and the Multi-Ethnic Study of Atherosclerosis. In step 3, we assessed evidence of heterogeneity in allelic effects at eGFR association signals that is driven by sex and/or ancestry. In step 4, we constructed ancestry-specific and multi-ancestry polygenic scores to assess transferability into AFR and AMS individuals.
Results
Discovery of eGFR loci in AFR and AMS individuals
We aggregated newly generated sex-stratified eGFR association summary statistics from 22 GWASs in a total of 83,386 AFR and AMS individuals from the Americas, Africa, and Europe (Table S1, related to Figure 1), using standardized protocols (STAR Methods). Assigning appropriate genetic ancestry labels to the included GWAS is complex and consensus has not been reached on the best descriptors. For the purposes of this work, the AFR GWAS included African Americans from the US, West Africans from Nigeria and Ghana, and admixed Africans from the UK. The AMS GWAS included Hispanics/Latinos from the Americas and the UK, and American Indians from the US (the preferred descriptor by members of this community). Each GWAS was imputed to reference panels from the 1000 Genomes Project,15 Haplotype Reference Consortium,16 African Genome Resources,17,18 or Trans-Omics for Precision Medicine Project19 (Table S2, related to Figure 1). Within each GWAS, eGFR was derived from serum creatinine (mg/dL) using the 2009 Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) equation.20,21 Subsequent association analyses were conducted using inverse-rank normalized eGFR residuals and adjusted for population structure and relatedness (Table S2, related to Figure 1). Across studies, we performed multi-ancestry (AFR + AMS) meta-analysis, under a fixed-effects model with inverse-variance weighting of effect sizes (STAR Methods).
To increase power to detect eGFR association signals in AFR and AMS individuals, we conducted a combined meta-analysis by aggregating association summary statistics from the multi-ancestry (AFR + AMS) meta-analysis with those from two additional resources: (1) an African American GWAS from the US from the Million Veteran Program comprising 57,336 individuals22 and (2) a meta-analysis of non-overlapping Hispanic/Latino GWAS from the US and Mexico from the COGENT-Kidney Consortium11 comprising a total of 5,010 individuals (Table S3, related to Figure 1). In this combined meta-analysis of 145,732 individuals, we identified 41 loci attaining genome-wide significant (p < 5 × 10−8) evidence of eGFR association (Figure 2; Table S4), with the strongest signals mapping to GATM (rs1145085, p = 5.6 × 10−49), PRKAG2 (rs10265221, p = 1.0 × 10−20), and SLC22A2 (rs11753349, p = 1.3 × 10−19). Two of the 41 loci were not reported in the latest, predominantly European ancestry eGFR GWAS meta-analysis,12 or in recent multi-ancestry meta-analyses from the Million Veteran Program.22 The previously unreported loci mapped to/near GABBR2 (rs73490762, p = 6.3 × 10−9) and LCOR (rs12258469, p = 3.5 × 10−8). Furthermore, lead SNVs from our combined meta-analysis were independent of previously reported signals (AFR, AMS, and EUR r2 < 0.2) at six loci (Table S5, related to Figure 2): OR52H1 (rs73392143, p = 4.2 × 10−17), SLC47A1 (rs35790011, p = 3.3 × 10−16), APOL3 (rs2016708, p = 4.1 × 10−12), ARG1 (rs73544620, p = 8.2 × 10−10), OVOL1 (rs624307, p = 2.6 × 10−9), and ADGRV1 (rs148044830, p = 1.4 × 10−8). At seven of these previously unreported signals (one previously unreported locus and six independent signals at previously reported loci), the lead SNV was rare (minor allele frequency <1%) in European ancestry individuals, and more common in other ancestry groups, emphasizing the importance of increasing population diversity in complex trait GWAS.
Figure 2.
Manhattan plot and quantile-quantile (QQ) plot of genome-wide eGFR association from combined meta-analysis of up to 145,732 AFR and AMS individuals
In the Manhattan plot, each point represents an SNV passing quality control in the meta-analysis, plotted with their observed association p value (on a –log10 scale) as a function of genomic position (NCBI build 37). The genome-wide significance threshold (p < 5 × 10−8) is highlighted by the horizontal red line. The names and locations of novel loci are indicated. In the QQ plot, each point represents an SNV passing quality control in the meta-analysis, plotted with the observed association p value (on a –log10 scale) as a function of their expected association p value (on a –log10 scale).
We repeated our analyses in a subset of studies using the 2021 CKD-EPI equation,23 which has been developed for use without correction for race. At lead SNVs identified in our combined meta-analysis, we observed strong correlation in allelic effect sizes derived from analyses with the two equations (Figure S1). While the mean and variance of the eGFR distribution might vary between equations, we hypothesize that the relative ranks of individuals within the distributions were not substantially changed, and thus have little impact after inverse-rank normalization.
Multi-omics integration reveals regulatory elements, genes, and pathways involved in renal function and kidney development
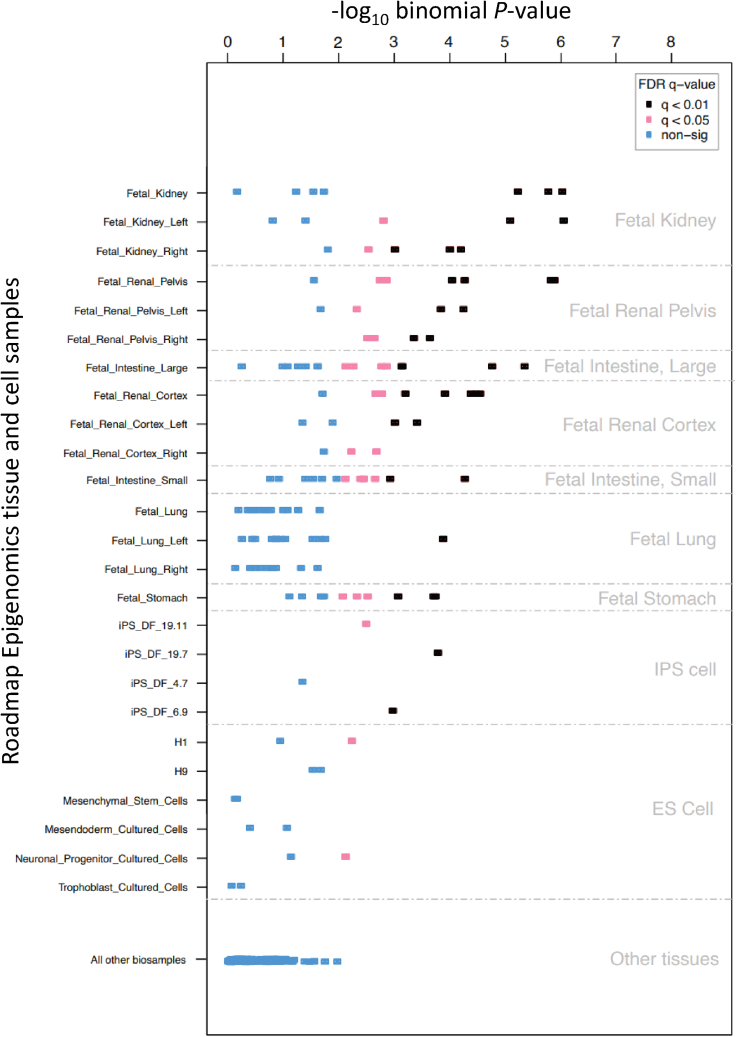
To gain insight into the key regulatory processes driving eGFR associations, and the genes and cell types through which their effects are mediated, we employed a series of complementary multi-omics analyses. We began by integrating tissue- and cell-type-specific regulatory elements across the 1,000 SNVs with the strongest eGFR association in our combined meta-analysis using FORGE224 (STAR Methods). Across unconsolidated Roadmap Epigenomics DNase I hypersensitive site data, eGFR associations were most significantly enriched for fetal kidney, fetal renal cortex, and fetal renal pelvis, with fetal intestine/stomach and lung also significantly enriched (Figure 3). The kidney tissue enrichment was replicated across more than 10 samples for each of these categories, and after extending analyses to include the 5,000 SNVs with the strongest eGFR association (Figure S2). To better understand the regulatory elements encoded by these kidney-specific DNase I hypersensitive sites, we then integrated transcription factor (TF) motif information from JASPAR, UniProt, Taipale, and Transfac databases with the same set of the 1,000 SNVs with the strongest eGFR association using FORGE2-TF25,26 (STAR Methods). Integration of these datasets revealed several key TFs with a role in renal function and kidney development (Table S6, related to Figure 3). In agreement with these findings, AmiGO2/PANTHER pathway analysis on significant TF genes (using a TF-specific background) revealed several kidney pathways including “renal system development” (p = 3.1 × 10−5), "regulation of metanephric glomerular mesangial cell proliferation" (p = 0.045), and "kidney development" (p = 2.3 × 10−5).
Figure 3.
Genomic variants associated with eGFR highlight kidney regulatory elements
Shown are the results of FORGE2 analysis for the top 1,000 eGFR SNVs. The horizontal axis shows FORGE2 enrichment (–log10 p value) of the eGFR SNV set with DNase I hotspots for a range of cell and tissue samples (vertical axis, significant samples in black). The top ranked sample set (highest black points) indicate the most significant association is for kidney samples (i.e., are highly ranked for the top 1,000 SNVs associated with eGFR).
To gain insight into genes and biological processes through which eGFR association signals are mediated, we began by conducting gene-based (MAGMA) expression analysis for 53 tissues from the Genotype Tissue Expression (GTEx) Project (version 8),27 implemented in FUMA.28,29 Using AFR and AMS LD reference panels from the 1000 Genomes Project, we observed significant enrichment (p < 0.00093, Bonferroni correction) of eGFR associations mediated through genes expressed in kidney medulla and kidney cortex, but not in any other tissues (Figure S3; Table S7, related to Figure 1). We then leveraged the diverse populations in our combined meta-analysis to fine-map causal variants driving eGFR associations at each of the 41 loci attaining genome-wide significance (STAR Methods). We assessed the evidence that 99% credible SNVs were significant cis-expression quantitative trait loci (eQTLs) in kidney tissue from 569 individuals of European ancestry from the Human Kidney Tissue Resource (HKTR) and The Cancer Genome Atlas (TCGA).30,31 We also considered overlap of our credible sets with significant cis-eQTLs in blood in 721 AFR and 610 AMS individuals from among African Americans, Puerto Ricans, and Mexican Americans from the Genes-Environments and Admixture in Latino Asthmatics (GALA II) study and the Study of African Americans, Asthma, Genes, and Environments (SAGE).32 Where 99% credible SNVs overlapped eQTLs, we considered the signals to be correlated when there was strong LD (AFR, AMS, and EUR r2 > 0.8) between the lead SNVs for the eGFR association and eQTL signal (Table S8, related to Figure 1).
We observed correlation of eGFR association signals with significant eQTLs in kidney (European ancestry individuals from HKTR/TCGA) at three loci: GBAP1 at the KRTCAP2 locus (rs2049805, p = 1.4 × 10−27), LARP4B at the LARP4B locus (rs80282103, p = 5.6 × 10−10), and GP2 at the PDILT locus (rs77924615, p = 7.2 × 10−9). LARP4B belongs to an evolutionarily conserved family of genes implicated in RNA metabolism and translation and we have previously shown differential expression of this gene in single-cell datasets for tubular epithelial cells in the distal nephron.11 At the KRTCAP2 locus, we observed correlation of the eGFR association signal with an eQTL in blood (AMS individuals from GALA II and SAGE) for GBAP1 (rs914615, p = 2.6 × 10−64), but also for TRIM46 (rs12411216, p = 7.0 × 10−7), which encodes a protein that interacts with Wnt/β-catenin signal pathways. In vitro studies suggest a role of the gene in hypoxia-induced kidney fibrosis.33
At the NFATC1 locus, we observed correlation of the eGFR association signal with significant eQTLs in blood for NFATC1 in both AFR and AMS individuals from GALA II and SAGE (rs8096658, AFR p = 2.6 × 10−41, AMS p = 6.7 × 10−36). The lead SNV (for both eGFR association and eQTL) was also a significant eQTL in blood in an additional 273 AFR individuals from the Multi-Ethnic Study of Atherosclerosis (p = 4.3 × 10−10) (STAR Methods). The same SNV failed quality control in European ancestry individuals from HKTR/TCGA and was therefore not tested in kidney. NFATC1 plays a central role in inducible gene transcription during immune response, and we have previously shown a role for this gene in salt sensitivity.14
Taken together, these results suggest that integration of large-scale epigenomic, TF motif, and transcriptomic data with our eGFR associations identified in under-represented population groups reveals key regulatory elements and pathways involved in renal function and kidney development, and highlights potential effector genes for eGFR signals in fine-mapped loci that play a role in kidney function and hypertension.
Portability of eGFR polygenic scores varies across AFR and AMS GWAS
Polygenic scores derived from GWAS undertaken in European ancestry populations have poor performance for prediction of complex traits and diseases when deployed in other population groups.9,10 This limited transferability can occur because European ancestry individuals are monomorphic for causal variants that are present in other ancestry groups, and/or because of differences in allele frequency, varying patterns of LD, and allelic effect heterogeneity between ancestries. For example, APOL1 variants have strong effects on kidney function and CKD in AFR individuals but are absent in individuals without AFR.34,35 Consequently, the inclusion of the APOL1 risk genotype in a polygenic score for CKD derived from GWAS in mostly European ancestry populations substantially improved discrimination in African Americans.36 Previous studies have also highlighted sex-differentiated effects on eGFR,37 which would impact the performance of polygenic scores derived from sex-combined meta-analyses for prediction in men and women.
We first assessed the evidence for differences in allelic effects on eGFR between GWASs due to genetic ancestry and/or sex. We used a meta-regression approach to partition heterogeneity in eGFR effects in the multi-ancestry (AFR + AMS) meta-analysis into three components.38 The first component captures heterogeneity that is correlated with genetic ancestry, represented by two axes of genetic variation derived from multidimensional scaling of a genetic distance matrix between GWASs (STAR Methods; Figure S4). The second component represents heterogeneity in allelic effects between males and females. The final component reflects residual heterogeneity due to differences in study design (for example, different sample characteristics, environmental exposures, or covariate adjustments between GWASs). We assessed the evidence of heterogeneity in allelic effects on eGFR across the 41 lead SNVs identified in the combined meta-analysis (Figure S5; Tables S9 and S10, related to Figure 1). None of the 41 lead SNVs demonstrated significant evidence of heterogeneity that was correlated with genetic ancestry or due to sex (Bonferroni-corrected statistical significance threshold, p < 0.0012). These results would suggest that polygenic score performance is not driven by differences in allelic effects between sexes or ancestry groups.
We next compared the performance of multi-ancestry (AFR + AMS) and ancestry-specific polygenic scores into AFR and AMS GWAS. We selected eight studies as “test GWASs.” The test GWASs were selected to represent the diversity of ancestry in our meta-analysis, including West Africans (AADM), African Americans (REGARDS, WHI-AA, BIOME-AA), Hispanics/Latinos (BIOME-HA, HCHS/SOL-MAIN, BAMBUI), and American Indians (SHS). For each test GWAS, we repeated multi-ancestry (AFR + AMS) and ancestry-specific meta-analyses, under a fixed-effects model, after excluding the test GWAS (STAR Methods). For comparison, we also considered two much larger published eGFR GWASs that were not matched for AFR/AMS ancestry: 143,658 individuals of East Asian ancestry from BioBank Japan39 and 1,004,040 individuals of European ancestry from the CKDGen Consortium.40 We then applied Polygenic Prediction via Bayesian Regression and Continuous Shrinkage Priors (PRS-CS)41 to derive five polygenic scores: multi-ancestry (AFR + AMS), AFR specific, AMR specific, East Asian ancestry specific, and European ancestry specific (Figure 4; Table S11, related to Figure 4).
Figure 4.
Transferability of multi-ancestry and ancestry-specific polygenic scores for eGFR into AFR and AMS test GWAS
Polygenic scores were constructed using PRS-CS, and their performance assessed in eight test GWASs. For each test GWAS, five polygenic scores were constructed: multi-ancestry (AFR + AMS), AFR specific, AMS specific, East Asian (EAS) specific from BioBank Japan, and European (EUR) specific from the CKDGen Consortium. Linkage disequilibrium (LD) was matched to the ancestry of the test GWAS. The eGFR variance explained by each polygenic score was estimated in each test GWAS. The relative performance of the polygenic scores varied across test GWAS. The multi-ancestry (AFR + AMS) and AFR-specific polygenic scores explained the highest proportion of eGFR variance in African American test GWAS (REGARDS, WHI-AA, BIOME-AA). In contrast, the EUR-specific polygenic score explained the highest proportion of eGFR variance in the AMS test GWAS (BIOME-HA, HCHS/SOL-MAIN, BAMBUI, SHS). All polygenic scores explained a low proportion of eGFR variance in West Africans from Nigeria and Ghana (AADM).
The multi-ancestry (AFR + AMS) and AFR-specific polygenic scores explained the highest proportion of eGFR variance in the African American test GWAS, despite substantially smaller sample sizes than the East Asian ancestry- and European ancestry-specific scores. In contrast, the European ancestry-specific polygenic score explained the highest proportion of eGFR variance in the AMS test GWAS (Hispanics/Latinos and American Indians). For three of the four AMS test GWASs, the AMS-specific polygenic score outperformed the AFR-specific score. However, interestingly, the AMS-specific polygenic score performed worse than the AFR-specific score in the Hispanic/Latino BIOME-HA test GWAS, which could reflect the fact that individuals in this study are more genetically similar to individuals in the AFR GWAS than in the AMR GWAS who have contributed to our meta-analysis (Figure S4). Finally, all polygenic scores explained a low proportion of eGFR variance in West Africans from Nigeria and Ghana.
Discussion
We have conducted a large meta-analysis of eGFR focused on AFR and AMS GWASs, bringing together a total of 105,607 individuals of AFR and 40,125 individuals with AMS. Our study contributes important insights into the genetic contribution of eGFR in these populations and provides resources for genetic prediction of kidney function. We have demonstrated important gains in discovery in meta-analyses of AFR and AMS GWASs, even for common SNVs. Identified loci in meta-analyses of AFR and AMS GWASs include genes relevant to kidney physiology and disease, and kidney development, consistent with reports in studies of predominantly European ancestry individuals.12,42 Using a comprehensive approach to query epigenetic data, we have shown that fine-mapped SNVs are in regulatory genomic regions in kidney tissue and cells, which are relevant to eGFR. These new GWAS findings support research focused on these populations to uncover the full spectrum of genetic variation related to disease susceptibility.
Interestingly, this study highlights the utility of transcriptome data generated from the same ancestry groups used in the GWAS discovery to identify potential effector genes for some association signals within identified loci. Differences in sample size between eQTL resources, as well as mismatch of LD patterns, allele frequencies, and imputation quality between ancestry groups will impact colocalization performance. Furthermore, methodology for formal colocalization of GWAS signals and molecular QTLs across admixed and mismatched ancestry groups is not well developed. The limited resources in kidney tissue in non-European ancestry populations are likely precluding discoveries for complex traits for which kidney-related pathways are of relevance.43 However, for some loci not driven by SNVs with allelic differences across ancestry groups, we were able to provide supporting evidence for effector genes using both blood and kidney tissue data (such as GBAP1 at the KRTCAP2 locus).
A major contribution of this study is the derivation of polygenic scores for eGFR in AFR and AMS individuals. The findings of our study indicate that the performance of polygenic scores depends on the sample size from which they are derived and the genetic distance from the test GWAS, consistent with findings in population biobanks.44,45 Our results indicate that these differences are not driven by ancestry-correlated heterogeneity in allelic effects. However, polygenic score performance will still vary between ancestry groups because of other factors that include differences in allele frequency and LD patterns. Our multi-ancestry (AFR + AMS)- and AFR-specific polygenic scores consistently outperformed European ancestry-specific scores for prediction into the African American test GWAS, despite the more than 10-fold difference in sample size. In contrast, the European ancestry-specific polygenic score consistently outperformed the better matched multi-ancestry (AFR + AMS)- and AMS-specific scores into AMS test GWAS. However, the relative performance of the AMS-specific and AFR-specific polygenic scores into the AMS test GWAS varied considerably. This is likely due to the complex and heterogeneous genetic background and admixture within AMS populations, suggesting that polygenic score prediction could be improved through alternative modeling approaches that are more representative of this population group. Our findings highlight the need for larger sample sizes in AFR and AMS GWAS, and the importance of development of polygenic score methodology that accounts for admixture to enhance the opportunities for risk prediction and patient stratification in under-studied and under-represented populations.
A strength of our study is that GWASs contributing to the COGENT-Kidney Consortium used standardized protocols for trait transformation and downstream statistical analysis. As is standard in large-scale GWAS meta-analyses, the genotyping arrays and imputation reference panels used varied between studies. We harmonized our choice of SNVs for inclusion across reference panels to minimize the bias introduced by varying imputation quality. Unfortunately, because the trait transformations were not consistent between the COGENT-Kidney Consortium and the Million Veteran Program, our combined meta-analysis across resources was conducted using Stouffer’s method, which does not provide combined estimates of allelic effect sizes or measures of heterogeneity.
Summary
Our large meta-analysis of AFR and AMS GWASs for eGFR contributes to the discovery of eGFR loci and provides insights into the utility of leveraging population-matched multi-omics resources in research of diverse populations, specifically for fine-mapping effector genes relevant to the trait. The study has also demonstrated the variable prediction performance of multi-ancestry (AFR + AMS)- and ancestry-specific polygenic scores into AFR and AMS individuals, dependent on the sample size and genetic distance from the discovery GWAS. These insights are essential for building relevant resources to enhance future opportunities for clinical translation of GWASs in these under-studied and under-represented populations and to reduce disparities in genomic research.
Limitations of the study
A potential limitation of our study was the use of the 2009 CKD-EPI equation20,21 to derive eGFR in the COGENT-Kidney Consortium (and Million Veteran Program). The more recent 2021 CKD-EPI equation,23 which has been developed for use without correction for race, has been shown to have less pronounced differences between Black and non-Black participants. However, when we compared allelic effect sizes at lead SNVs in a subset of AFR and AMS GWASs, we observed highly consistent results and strong correlation. This would indicate that, while the 2021 CKD-EPI equation might impact the distribution of eGFR within ancestry groups, the relative positions on individuals within the distribution does not vary substantially, and there is consequently high concordance after inverse rank normalization. Finally, while the transcriptomic data generation, processing, quality control, and analysis were not consistent across the kidney and whole-blood resources used in our analyses, the definition of significant cis-eQTLs was the same for both (FDR < 5%, within 1 Mb of the transcription start site).
Consortia
The following investigators of the Human Kidney Tissue Resource contributed to recruitment and/or phenotyping of human kidney gene expression studies: Wojciech Wystrychowski, Monika Szulinska, Andrzej Antczak, Maciej Glyda, Robert Król, Joanna Zywiec, Ewa Zukowska-Szczechowska, Pawel Bogdanski, and Bernard Keavney.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| GWAS meta-analysis summary statistics | This paper | GWAS Catalog: accession number GCST90295957 |
| Software and algorithms | ||
| MR-MEGA (version 0.2): | https://genomics.ut.ee/en/tools | |
| METAL (version 2011-03-25): | http://csg.sph.umich.edu/abecasis/Metal/index.html | |
| FORGE2 | https://forge2.altiusinstitute.org/ | |
| AmiGO2/PANTHER GO Ontology database https://doi.org/10.5281/zenodo.6399963, released 2022-03-22 | https://amigo.geneontology.org/amigo | |
| FUMA (version 1.5) | https://fuma.ctglab.nl/ | |
| LDlink (version 5.6.2) | https://ldlink.nih.gov/?tab=home | |
| PRS-CS (version 3 Nov 2022) | https://github.com/getian107/PRScs | |
Resource availability
Lead contact
Further information and requests for data availability should be directed to and will be fulfilled by the lead contact, Nora Franceschini (noraf@unc.edu).
Materials availability
No materials were generated in this study. GWAS meta-analysis summary statistics are available through the GWAS Catalog, accession number GCST90295957.
Data and code availability
-
•
This paper analyzes existing, publicly available genotype and phenotype cohort data. These accession numbers for the datasets are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
No experimental models were employed in this study.
Subjects included in multi-ancestry GWAS meta-analyses
We aggregated association summary statistics from 22 GWAS in a total of 83,386 AFR and AMS individuals (Table S1, related to Figure 1). We conducted a combined meta-analysis of these data with association summary statistics obtained from: (i) an African American GWAS from the Million Veteran Program comprising 57,336 individuals22; and (ii) a meta-analysis of Hispanic/Latino GWAS from the COGENT-Kidney Consortium11 comprising a total of 5,010 individuals (Table S3, related to Figure 1).
Subjects included in eQTL look-ups
We conducted a look-up of eQTLs derived from: (i) kidney tissue samples from 569 individuals of European ancestry from the Human Kidney Tissue Resource (HKTR) and The Cancer Genome Atlas (TCGA)30,31; (ii) whole-blood from 721 AFR and 610 AMS individuals from amongst African Americans, Puerto Ricans, and Mexican Americans from the Genes-Environments and Admixture in Latino Asthmatics (GALA II) study and the Study of African Americans, Asthma, Genes, and Environments (SAGE)32; and (iii) peripheral blood mononuclear cells (PBMCs) from 273 African American and 241 Hispanic/Latino individuals from the Multi-Ethnic Study of Atherosclerosis (MESA).
Method details
Study-level analyses
Individuals were assayed with a range of GWAS genotyping arrays or whole genome sequencing, with sample and SNV quality control (QC) undertaken within each study (Tables S1 and S2, related to Figure 1). All studies followed standardized protocols for phenotype definition and analytical pipelines. Within each study, individuals were first assigned to an ancestry group (AFR or AMS) using genetic ancestry, with population outliers excluded. Analyses were then conducted separately within each ancestry group (AFR or AMS). For each ancestry-specific GWAS not assayed via whole genome sequencing, individuals were pre-phased and imputed up to reference panels from the 1000 Genomes Project (phase 3, October 2014 release),15 Haplotype Reference Consortium,16 African Genome Resources,17,18 or Trans-Omics for Precision Medicine Project19 (Table S2, related to Figure 1). The COGENT Kidney Consortium analysis plan distributed to studies is provided below.
The 2009 CKD-EPI equation20,21 was used to calculate eGFR from serum creatinine (mg/dL) to be consistent with recently-published studies.12,22 Sex-stratified eGFR residuals were obtained after adjustment for age and other study-specific covariates (Table S2), and subsequently transformed using inverse rank normalization (IRN). Association with IRN eGFR was evaluated via linear regression, separately in males and females, under an additive model using allele dosage. Analyses accounted for structure (population stratification and/or familial relationships) by: (i) excluding related samples and adjusting for principal components derived from a genetic relatedness matrix (GRM) as additional covariates in the regression model; or (ii) incorporating a random effect for the GRM in a mixed model (Table S2). SNVs with poor imputation quality (info/r2 < 0.3) and/or minor allele count <5 were excluded. Sex-stratified study-level association summary statistics (p-values and standard error of allelic effects) were corrected for residual structure, not accounted for in the regression analysis, by means of genomic control46 if the inflation factor was >1 (Table S2). In a subset of studies, we repeated our analyses using the 2021 CKD-EPI equation23 and compared allelic effect sizes between the two equations.
COGENT Kidney Consortium analysis plan provided to studies
If your study includes multiple ancestry groups, please conduct analyses separately for each ancestry group. All analyses should be sex-stratified.
Phenotype
Estimated glomerular filtration rate: eGFR (ml/min/1.73m2). Use the 2009 CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) equation from serum creatinine measures, which is expressed as a single equation and calculated as follows:
If female and Scr ≤0.7 then eGFR = 144 x (SCr/0.7)−0.329 x 0.993Age [x 1.159 if black]
If female and Scr >0.7 then eGFR = 144 x (SCr/0.7)−1.209 x 0.993Age [x 1.159 if black]
If male and Scr ≤0.9 then eGFR = 141 x (SCr/0.9)−0.411 x 0.993Age [x 1.159 if black]
If male and Scr>0.9 then eGFR = 141 x (SCr/0.9)−1.209 x 0.993Age [x 1.159 if black]
SCr is serum creatinine (mg/dL). To convert SCr from μmol/L to mg/dl, divide by 88.4. Age is in years. Do not use the constant for black unless for African Americans. Creatinine calibration: if SCr was measured using a Jaffé assay before 2009, then multiply SCr mg/dl by 0.95 before including in the equation.
Genotypes
We recommend prephasing and imputation using the University of Michigan Imputation Server (https://imputationserver.readthedocs.io/en/latest/). For all analyses, use SNP dosage after imputing up to one of the following reference panels: 1000 Genomes Phase 3 (all ancestries); TOPMed; population-specific whole-genome sequence. Please do not filter SNPs on the basis of allele frequency or imputation quality before association analysis.
Association analysis
Within each sex, obtain eGFR residuals after adjustment for age and other study-specific covariates (but not principal components for population structure) and then perform inverse rank normalization. To test for association, under an additive genetic model, either use a linear model (unrelated individuals only) or a linear mixed model with a random effect for the genetic relationship matrix. Include principal components as covariates to account for population structure, as appropriate.
Preparing association summary statistics
To minimize the burden on analysts, we request upload of the output directly from the association analysis software (e.g., SNPTEST, BOLT-LMM) after removing monomorphic SNPs. Please provide filenames that include the following information: sex, ancestry, and analysis date. Please ensure that the following information is included (or can be derived) in the files: SNP ID (as per reference panel), chromosome and position; effect allele and other allele; beta (effect size of the effect allele) and corresponding standard error; p-value for association (please do not apply genomic control correction); effect allele frequency; imputation quality score (info or r2). If possible, please provide beta and standard error to at least five decimal places, and the association p-value to at least two-significant figures.
Multi-ancestry (AFR+AMS) GWAS meta-analysis
We considered autosomal biallelic SNVs that overlap the 1000 Genomes Project reference panel (phase 3, October 2014 release)15 and the Haplotype Reference Consortium reference panel.16 The Haplotype Reference Consortium panel includes re-sequenced samples from the 1000 Genomes Project reference panel. For these overlapping samples between reference panels, we compared their alternate allele frequency in the two panels. We then excluded SNVs that differed in allele frequency by >20% when comparing these two panels. We aggregated sex-stratified allelic effect estimates across GWAS via inverse-variance weighted fixed-effects meta-analysis using METAL.47 We corrected meta-analysis association summary statistics (p-values and standard error of allelic effects) for inflation due to residual structure between GWAS by genomic control adjustment.46
Combined meta-analysis
We performed a combined meta-analysis by aggregating association summary statistics from the multi-ancestry (AFR+AMS) GWAS meta-analysis with those from two additional resources: (i) an African American GWAS in 57,336 individuals from the Million Veteran Program22; and (ii) a meta-analysis of additional AMS GWAS in 5,010 individuals from a prior COGENT-Kidney Consortium publication11 (Table S3, related to Figure 1). We conducted fixed-effects meta-analysis using Stouffer’s method, implemented in METAL,47 because different approaches to eGFR transformation were used in the additional resources, and effect estimates were therefore not on the same scale. SNVs not reported in at least one of the additional resources were excluded to ensure signals were associated across multiple studies. Allelic effects were aligned to the eGFR decreasing allele. In total, the combined meta-analysis included 145,732 individuals.
African American GWAS from the Million Veteran Program
We obtained summary statistics of the Million Veteran Program eGFR GWAS from dbGap. Briefly, individuals were genotyped with a custom Affymetrix Axiom Biobank array, with sample and SNV QC previously described.22 Briefly, after pre-phasing with Eagle2,48 individuals were imputed to the 1000 Genomes Project reference panel (phase 3, October 2014 release)15 using minimac3.49 The 2009 CKD-EPI equation20,21 was used to calculate eGFR from serum creatinine (mg/dL). Individuals were stratified according to presence/absence of diabetes and hypertension. Within each stratum, association eGFR was evaluated via linear regression using SNPTEST50 under an additive model in the dosage of the minor allele, with adjustment for age, age,2 sex, body mass index, and ten principal components derived from the GRM to account for population structure. SNVs with poor imputation quality (info <0.4) were excluded. Allelic effect estimates for eGFR were aggregated across strata via inverse-variance weighted fixed-effects meta-analysis using METAL.47 Association summary statistics (p-values and standard error of allelic effects) were corrected for residual structure, not accounted for in the regression analysis, by means of genomic control.46
Meta-analysis of AMS GWAS from the COGENT-Kidney Consortium
Using GWAS data from our prior publication, we considered only those studies that did not overlap with those contributing to the current multi-ancestry GWAS meta-analysis. Briefly, individuals were assayed with a range of GWAS genotyping arrays, with sample and SNV QC undertaken within each study (Table S3, related to Figure 1). Individuals were then pre-phased and imputed up to the 1000 Genomes Project reference panel (phase 1, March 2012 release).51 The four variable Modification of Diet in Renal Disease (MDRD) equation52,53 was used to calculate eGFR from serum creatinine (mg/dL). Association with eGFR was evaluated via linear regression under an additive model in the dosage of the minor allele, with adjustment for study-specific covariates to account for population structure. SNVs with poor imputation quality (info <0.4, r2 < 0.3) were excluded. Association summary statistics (p-values and standard error of allelic effects) were corrected for residual structure, not accounted for in the regression analysis, by means of genomic control46 if the inflation factor was >1 (Table S3, related to Figure 1). Finally, allelic effect estimates for eGFR were aggregated across GWAS via inverse-variance weighted fixed-effects meta-analysis using METAL.47
Locus and lead SNV definition
We initially selected lead SNVs attaining genome-wide significant evidence of association (P < 5x10−8) in the combined meta-analysis that were separated by at least 500kb. Loci were first defined by the flanking genomic interval mapping 500kb up- and downstream of lead SNVs. Then, where lead SNVs were separated by less than 1Mb, the corresponding loci were aggregated as a single locus. The lead SNV for each locus was then selected as the SNV with minimum association p-value. A locus was considered novel if no previously reported lead SNVs for eGFR at genome-wide significance mapped within the locus boundaries. Effect allele frequencies for lead SNVs were obtained from the 1000 Genomes Project reference panel (phase 3, October 2014 release)15 using AFR, AMS, and European ancestry haplotypes. For each locus, LD between the lead SNVs from the combined meta-analysis and those reported in the largest published eGFR meta-analysis12 were obtained from the 1000 Genomes Project reference panel (phase 3, October 2014 release)15 using AFR and AMS haplotypes.
Integration of epigenomic data resources with eGFR associations
We used FORGE224 to perform functional overlap analysis of the 1,000 SNVs with the strongest eGFR associations (smallest p-values) from the combined meta-analysis across DNase I hotspots from the Roadmap Epigenomics Consortium.54 To evaluate whether the observed enrichment was consistent and robust, FORGE2 performed analysis across multiple replicate samples, obtaining a significant enrichment for at least 10 replicate samples in each kidney tissue. Analyses were repeated using the 5,000 SNVs with the strongest eGFR associations from the combined meta-analysis. We then performed TF motif analysis using data from JASPAR, UNIPROT, Taipale, and TRANSFAC databases for the SNVs underlying FORGE2 tissue-specific enrichment signal for kidney. To do this, we used the FORGE2-TF25,26 to identify the most important TF motifs associated with our DNase I hotspot enrichment. We then applied AmiGO2/PANTHER analysis to evaluate pathways associated with significant TF motifs.
Tissue expression analysis
To test the relationship between highly expressed genes in a specific tissue and eGFR associations from the combined meta-analysis, we conducted gene-property analysis using average expression of genes per tissue type as a gene covariate using FUMA.28,29 For 53 specific tissue types from GTEx version 8,27 gene expression values were log2 transformed per tissue type (winsorized at 50). MAGMA was performed using the results of gene-level analyses (gene-based p-values), and a one-sided test conducted with conditioning on average expression across all tissue types. We used the default gene annotation window size of 0kb upstream and downstream. We conducted separate analyses using LD from AFR and AMS haplotypes from the 1000 Genomes Project reference panel (phase 3, October 2014 release).15
Fine-mapping causal variants driving eGFR associations
For each locus attaining genome-wide significance in the combined meta-analysis, we localised causal variants driving the eGFR association through multi-ancestry fine-mapping. Within each locus, we approximated the Bayes’ factor, , in favor of eGFR association of the th SNV using summary statistics from the combined meta-analysis, given by
where is the association Z score and is the total sample size.55 The posterior probability for the th SNV was then given by . We derived a 99% credible set56 for the eGFR association signal by: (i) ranking all SNVs according to their posterior probability ; and (ii) including ranked SNVs until their cumulative posterior probability attains or exceeds 0.99.
Correlation of eGFR association signals with eQTLs in kidney and blood
We cross-referenced SNVs in the 99% credible set driving each eGFR association signal against significant eQTLs derived from: (i) kidney tissue samples from individuals of European ancestry from the Human Kidney Tissue Resource (HKTR) and The Cancer Genome Atlas (TCGA)30,31; and (ii) whole-blood from AFR and AMS individuals from the Genes-Environments and Admixture in Latino Asthmatics (GALA II) study and the Study of African Americans, Asthma, Genes, and Environments (SAGE).32.
The HKTR includes 478 kidney tissue samples from moleculAr analysis of the TRANScriptome of renaL human TissuE Study (TRANSLATE)57 and its extension (TRANSLATE-T)58 human kiDney-Manchester renal tIssue pRojEct (ADMIRE),59 moleculaR analysis of mEchanisms regulating gene exPression in post-ischAemic Injury to Renal allograft (REPAIR), and Renal gEne expreSsion and PredispOsition to cardiovascular and kidNey Disease (RESPOND) studies.30,59 In addition, 91 “control” kidney tissue samples from TCGA31 were included in the analysis. In brief, kidney specimens were secured from cancer-unaffected renal tissue after nephrectomy or from kidney biopsy preceding the transplantation, as reported previously.30 DNA and RNA were extracted and processed as reported in prior publications30,59 Genotype imputation into the Human Kidney Tissue Resource and TCGA were carried out separately on the Michigan Imputation Server49 using 1000 Genomes Project Phase 3 data15 as the reference panel applied to all genotyped variants that passed quality control. Minimac449 was used to perform imputations with the default phasing software Eagle v.2.4. We excluded variants with duplicate genomic locations, imputation score <0.40, MAF <1%, or HWE p < 10−6 at the post-imputation quality control level. The eQTL analysis was conducted using FastQTL.60 The normalised expression of each kidney gene was regressed against alternative allele dosage, age, sex, source of tissue indicator (nephrectomy/kidney biopsy), the top three principal components derived from genotyped autosomal variants, 120 hidden factors estimated using probabilistic estimation of expression residuals (PEER) factors61 and seven kidney cell-type proportions deconvolved from statistically normal kidney cells from the single cell dataset62 and the MuSiC R package.63 Only variants in cis (within 1Mb of the transcription start site of a gene) were included in the kidney eQTL analysis.
We considered whole-blood gene expression using whole-genome and RNA sequencing data from 2,733 African Americans, Puerto Ricans and Mexican Americans from GALA II and SAGE. Details of the data generation, processing, quality control, and eQTL analysis have been previously reported.32 We focused on cis-eQTLs (within 1Mb of the transcription start site of a gene) that identified in subsets of 721 participants with >50% AFR ancestry and 610 participants with >50% AMS ancestry. FastQTL60 was used to process raw gene counts and identify eQTLs, according to the GTEx v8 pipeline (https://github.com/broadinstitute/gtex-pipeline), adjusting for age, sex, asthma status, the first five genetic ancestry principal components and PEER factors61 as covariates.
For both resources, significant eQTLs were defined by FDR <5%. For each eGFR locus for which the credible set overlapped with a significant eQTL, we assessed LD between the lead eGFR SNV and the lead eQTL SNV using African, American and European ancestry haplotypes from the 1000 Genomes Project reference panel (phase 3, October 2014 release).15 We defined correlation between the eGFR association signal and eQTL only if the LD between lead SNVs was strong (r2 > 0.8) in all three ancestry groups.
Transcriptomic analyses and eQTL identification in the Multi-Ethnic Study of Atherosclerosis (MESA)
RNA was extracted using a Trizol protocol from cryopreserved PBMCs, which were isolated from baseline study visit (exam 1) blood samples. RNA sample quality was assessed using RNA Integrity Number (RIN, Agilent Bioanalyzer) prior to shipment to sequencing centers. All blood laboratory work was performed at the University of Vermont. The RNA was sequenced at the Broad Institute (n = 580) and at the Northwest Genomics Center (NWGC; n = 583) using harmonized protocols. RNA Quality Control was repeated at the sequencing centers by RIN analysis at the NWGC and by RNA Quality Score analysis (RQS, Caliper) at the Broad Institute. A minimum of 250ng RNA sample was required as input for library construction, performed using the Illumina TruSeqTM Stranded mRNA Sample Preparation Kit (polyA selection). RNA was sequenced as 2x101bp paired-end reads on the Illumina HiSeq 4000 according to the manufacturer’s protocols. Target coverage was of ≥40M reads.
Comprehensive information about the RNA-seq pipeline used for TOPMed can be found in https://github.com/broadinstitute/gtex-pipeline/blob/master/TOPMed_RNAseq_pipeline.md under MESA RNA-seq pilot. Briefly, reads were aligned using STAR64 and transcript-level expected counts quantified using RSEM v1.3.0.65 Additional QC checks were performed for sample swaps (RNA-seq vs. VCF fingerprinting) and expression-based sex checks (XIST and RPS4Y1 genes). Post-QC there were 461 (Broad) and 511 (UW) transcriptomes available for analysis. Mapping of eQTLs was performed using tensorQTL,66 separately in each ancestry group. The cis-gene mapping interval was set to +/−1Mb of the transcription start site (TSS) and variants with MAF ≥1% in MESA TOPMed samples were included. The MESA genotypes were taken from the main TOPMed whole genome sequencing program described elsewhere. To control for population stratification, TOPMed program genotype PCs 1–11, sequencing center, and PEER factors61 were included as covariates to control for both technical and biological variation. A correction for multiple testing used an empirical null association distribution derived from 10,000 permutations which was to calculate gene-level q-values67 with a fixed p-value interval for the estimation of pi_0 (the 'lambda' parameter was set to 0.85). Significant eQTLs were defined by FDR <5%.
Heterogeneity due to ancestry and sex
For studies contributing to the multi-ancestry (AFR+AMS) meta-analysis, we used meta-regression, implemented in MR-MEGA,38 to model allelic effect heterogeneity due to genetic ancestry and sex. We constructed a distance matrix of mean effect allele frequency differences between each pair of GWAS across a subset of 386,563 SNVs reported in all studies. We implemented multi-dimensional scaling of the distance matrix to obtain two principal components that defined axes of genetic variation to separate AFR and AMS GWAS. For each SNV, we modeled allelic effect estimates across GWAS via linear regression, weighted by the inverse of the variance of the effect estimates, incorporating the two axes of genetic variation and sex as covariates. We tested for heterogeneity in allelic effects on eGFR between GWAS that is: (i) correlated with genetic ancestry; and (ii) due to sex. We also tested for residual allelic effect heterogeneity between GWAS that was not accounted for by genetic ancestry or sex. For lead SNVs identified in the combined meta-analysis, we tested for evidence of enrichment in heterogeneity by means of a binomial test.
Ancestry-specific GWAS meta-analyses
We conducted AFR- and AMS-specific meta-analyses. For each ancestry, we aggregated sex-stratified allelic effect estimates across GWAS via inverse-variance weighted fixed-effects meta-analysis using METAL.47 We corrected meta-analysis association summary statistics (p-values and standard error of allelic effects) for inflation due to residual structure between GWAS by genomic control adjustment.46
Sex-specific GWAS meta-analyses
For each sex, we aggregated sex-specific allelic effect estimates across GWAS via inverse-variance weighted fixed-effects meta-analysis using METAL.47 We corrected meta-analysis association summary statistics (p-values and standard error of allelic effects) for inflation due to residual structure between GWAS by genomic control adjustment.46
Derivation and testing of eGFR polygenic scores across population groups
We selected eight studies as “test GWAS”: AADM, REGARDS, WHI-AA, BIOME-AA, BIOME-HA, HCHS/SOL-MAIN, BAMBUI, and SHS. For each test GWAS, we repeated multi-ancestry (AFR+AMS) and ancestry-specific meta-analyses, under a fixed-effects model, after excluding the test GWAS. We also obtained association summary statistics from published European and East Asian ancestry-specific eGFR GWAS meta-analyses. Within each test GWAS, we selected SNVs overlapping those reported in the multi-ancestry and ancestry-specific meta-analyses. We used PRS-CS,41 with LD reference aligned to the test GWAS and default settings, to derive LD-revised allelic effect estimates for each SNV to be used as weights in the polygenic score. LD references are provided by the PRS-CS software and are obtained from ancestry-specific haplotypes from the 1000 Genomes Project reference panel (phase 3, October 2014 release).15 For each test GWAS, we then regressed the observed allelic effect estimates at SNVs, weighted by their corresponding variances, on the weights, as implemented in grs.summary function of the gtx R package.68 We estimated the percentage of eGFR variance explained, measured by pseudo R2, and p-value for association with the polygenic score.
Acknowledgments
This study was supported by National Institutes of Health (NIH) grants R01-DK117445 and R01-MD012765 (to N.F. and A.P.M.), U01-DK130044, R01-HL163972 and K26-DK138425 (to N.F.), R01-HL142711, R01-HL127564, and U01-HG011719 (to P.N.), T32-HL125232 (to T.G.), K01-MH121659 (to E.G.A.), and R01-HL136666 (to M.R.I.). C.E.B. was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH. R.W.M. was supported by a Canadian Institutes of Health Research Fellowship during the completion of this work. M.T. and Human Kidney Tissue Resource are supported by grants from the British Heart Foundation (PG/17/35/33001, PG/19/16/34270, and PG/22/10957), Kidney Research UK (RP_017_20180302 and RP_013_20190305), and by the NIHR Manchester Biomedical Research Centre (NIHR203308). E.T.S. is supported by the Minas Gerais Research Agency (FAPEMIG) and the Brazilian National Research Council (CNPq). E.J.R. and E.J.P.-S. were supported by the Division of Intramural Research of the National Heart, Lung, and Blood Institute (NHLBI) and the National Institute on Minority Health and Health Disparities at the NIH. This work was supported in part by the Intramural Research Program of the National Human Genome Research Institute (NHGRI) (to A.R.B. and C.R.).
African American Diabetes Mellitus Study (AADM): the study was supported in part by the NIH Intramural Research Program in the Center for Research on Genomics and Global Health (CRGGH). The CRGGH is supported by the NHGRI, the National Institute of Diabetes and Digestive and Kidney Diseases, and the Office of the Director at the NIH (1ZIAHG200362). Support for participant recruitment and initial genetic studies of the AADM study was provided by NIH grant no. 3T37TW00041-03S2 from the Office of Research on Minority Health.
Atherosclerosis Risk in Communities Study (ARIC): the ARIC study has been funded in whole or in part with Federal funds from the NHLBI,NIH, Department of Health and Human Services, under contract numbers HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I, and HHSN268201700005I. The authors thank the staff and participants of the ARIC study for their important contributions. Funding for the GENEVA substudy was provided by NHGRI grant U01HG004402 (E. Boerwinkle). Access through dbGAP for ARIC (phs000280.v7).
Bambui (Brazil) Cohort Study of Ageing (BAMBUI): the EPIGEN-Brazil Initiative is funded by the Brazilian Ministry of Health (Department of Science and Technology from the Secretaria de Ciência, Tecnologia e Insumos Estratégicos) through Financiadora de Estudos e Projetos. This project was supported by the Intramural Research Program of the NHGRI, NIH, through the Center for Research on Genomics and Global Health (CRGGH).
BioME Biobank (BIOME): the Mount Sinai BioME Biobank is supported by The Andrea and Charles Bronfman Philanthropies.
Coronary Artery Risk Development in Young Adults Study (CARDIA): CARDIA is conducted and supported by the NHLBI in collaboration with the University of Alabama at Birmingham (HHSN268201800005I and HHSN268201800007I), Northwestern University (HHSN268201800003I), University of Minnesota (HHSN268201800006I), and Kaiser Foundation Research Institute (HHSN268201800004I). Genotyping was funded as part of the NHLBI Candidate-gene Association Resource (N01-HC-65226) and the NHGRI Gene Environment Association Studies (GENEVA) (U01-HG004729, U01-HG04424, and U01-HG004446).
Brazil Longitudinal Study of Adult Health (ELSA): the authors thank the staff and participants of the Elsa Study for their important contributions.
Hispanic Community Health Study and Study of Latinos (HCHS/SOL): HCHS/SOL was carried out as a collaborative study supported by contracts from the NHLBI to the University of North Carolina (N01-HC-65233), University of Miami (N01-HC-65234), Albert Einstein College of Medicine (N01-HC-65235), Northwestern University (N01-HC-65236), and San Diego State University (N01-HC-65237). The following contribute to the HCHS/SOL through a transfer of funds to the NHLBI: National Center on Minority Health and Health Disparities, the National Institute of Deafness and Other Communications Disorders, the National Institute of Dental and Craniofacial Research (NIDCR), the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Neurological Disorders and Stroke, and the Office of Dietary Supplements. The Genetic Analysis Center at the University of Washington was supported by NHLBI and NIDCR contracts (HHSN268201300005C AM03 and MOD03).
Howard University Family Study (HUFS): HUFS was supported by NIH grants S06GM008016-320107 to C.R. and S06GM008016-380111 to A.A. We thank the participants of the study, for which enrollment was carried out at the Howard University General Clinical Research Center, supported by NIH grant 2M01RR010284.
Jackson Heart Study (JHS): JHS is supported and conducted in collaboration with Jackson State University (HHSN268201800013I), Tougaloo College (HHSN268201800014I), the Mississippi State Department of Health (HHSN268201800015I), and the University of Mississippi Medical Center (HHSN268201800010I, HHSN268201800011I, and HHSN268201800012I) contracts from NHLBI and the National Institute on Minority Health and Health Disparities. The authors also wish to thank the staff and participants of the JHS. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of NHLBI, NIH; or the US Department of Health and Human Services.
Multi-Ethnic Study of Atherosclerosis (MESA): MESA and the MESA SHARe projects are conducted and supported by the NHLBI in collaboration with MESA investigators. Support for MESA is provided by contracts 75N92020D00001, HHSN268201500003I, N01-HC-95159, 75N92020D00005, N01-HC-95160, 75N92020D00002, N01-HC-95161, 75N92020D00003, N01-HC-95162, 75N92020D00006, N01-HC-95163, 75N92020D00004, N01-HC-95164, 75N92020D00007, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, and UL1-TR-001420, UL1TR001881, DK063491, and R01HL105756. Funding for SHARe genotyping was provided by NHLBI Contract N02-HL-64278. Genotyping was performed at Affymetrix (Santa Clara, CA) and the Broad Institute of Harvard and MIT (Boston, MA) using the Affymetrix Genome-Wide Human SNP Array 6.0. MESA Family is conducted and supported by the NHLBI in collaboration with MESA investigators. Support is provided by grants and contracts R01HL071051, R01HL071205, R01HL071250, R01HL071251, R01HL071258, R01HL071259, UL1TR001881, and DK063491, and by the National Center for Research Resources, grant UL1RR033176. The authors thank the other investigators, the staff, and the participants of the MESA study for their valuable contributions. A full list of participating MESA investigators and institutes can be found at http://www.mesa-nhlbi.org.
Sea Islands Genetic Network: Reasons for Geographic and Racial Differences in Stroke (REGARDS): the REGARDS study is supported by a cooperative agreement U01 NS041588 from the National Institute of Neurological Disorders and Stroke, NIH, U.S. Department of Health and Human Services. The authors thank the other investigators, the staff, and the participants of the REGARDS study for their valuable contributions. A full list of participating REGARDS investigators and institutions can be found at http://www.regardsstudy.org.
Strong Heart Study (SHS): SHS has been funded in whole or in part with federal funds from the NHLBI, NIH, Department of Health and Human Services, under contract nos. 75N92019D00027, 75N92019D00028, 75N92019D00029, and 75N92019D00030. The study was previously supported by research grants: R01HL109315, R01HL109301, R01HL109284, R01HL109282, and R01HL109319 and by cooperative agreements U01HL41642, U01HL41652, U01HL41654, U01HL65520, and U01HL65521. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Indian Health Service (IHS). The MEGA array was genotyped under the National Institute on Minority Health and Health Disparities, NIH R01-MD012765 to N.F.
UK Biobank (UKBB): access to the UK Biobank was provided through project 42633.
Women’s Health Initiative (WHI): the WHI program is funded by the NHLBI through contracts HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C. We thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: http://www.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Long%20List.pdf.
Genetics of Latinos Diabetic Retinopathy (GOLDR): GOLDR was supported in part by the Genetics of Latinos Diabetic Retinopathy (GOLDR) Study grant EY14684. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant, UL1TR001881, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Research Center.
Mexican American Hypertension and Insulin Resistance (HTNIR): this work was supported by the NHLBI (grant HL55005), the National Institutes of Health National Center for Research Resources, General Clinical Research Center (grants RR00425 to RR00430, M01-RR00865, M01-RR00043, M01-RR00425, and CA42710). The authors thank the families for their participation in this study.
Mexico City Studies (MC): in Mexico, this work was supported by the Fondo Sectorial de Investigación en Salud y Seguridad Social (SSA/IMSS/ISSSTE-CONACYT, project 150352), Temas Prioritarios de Salud Instituto Mexicano del Seguro Social (2014-FIS/IMSS/PROT/PRIO/14/34), and the Fundación IMSS. We thank Jorge Gutierrez Cuevas, Jaime Gómez Zamudio, and Araceli Méndez Padrón for technical support. In Canada, computations were performed on the GPC supercomputer at the SciNet HPC Consortium. SciNet is funded by: the Calgary Foundation for Innovation under the auspices of Compute Canada; the Government of Ontario, Ontario Research Fund – Research Excellence; and the University of Toronto. E.J.P. was supported by the Canadian Institutes of Health Research and the Banting and Best Diabetes Centre, University of Toronto.
Northern Manhattan Family Study (NOMAS): this work was supported by the NIH (R01-NS-065114 and R01-NS-29993).
Human Kidney Tissue Resource: the following investigators contributed to recruitment and/or phenotyping of human kidney gene expression studies: Wojciech Wystrychowski, Monika Szulinska, Andrzej Antczak, Maciej Glyda, Robert Król, Joanna Zywiec, Ewa Zukowska-Szczechowska, Pawel Bogdanski, and Bernard Keavney.
Author contributions
Central analysis group, O.H., A.R.B., C.E.B., A.P.M., and N.F.; study-level analyses, A.R.B., G.N., Q.S., B.M.L., T.G., M.C.M., J.D., L.M.R., H.K., R.W.M., A.C.P., M.H.G., E.G.A., A.V.-S., N.W.-R., N.D.D., X.G., and Y.H.; transcriptomic data generation and analysis, F.A., X.X., J.E., F.J.C., M.T., and J.C.M.; study-level genotyping, phenotyping, and additional analyses, A.A., L.G.B., J.C., G.C., A.D., M.O.G., E.I., M.R.I., M.J., A.C.J., C.K., E.M.L., M.B.L., J.P.L., R.J.F.L., V.T.H.M., J.P.-R., L.Q., L.J.R., S.S.R., E.J.R., E.T.-S., K.D.T., J.G.U., J.W., B.A.Y., Z.Y., and Y.Z.; study PI, Y.-D.I.C., T.R., J.I.R., M.C., M.F., M.F.L.-C., G.P., P.N., S.A.C., A.P.C., L.A.L., Y.L., E.J.P.-S., R.D., J.C.M., C.R., and N.F.; Consortium steering group, A.P.M. and N.F.; manuscript preparation, O.H., A.R.B., C.E.B., A.P.M., and N.F. All authors reviewed and approved final manuscript.
Declaration of interests
P.N. reports investigator-initiated research grants from Allelica, Apple, Amgen, Boston Scientific, Genentech/Roche, and Novartis, personal fees from Allelica, Apple, AstraZeneca, Blackstone Life Sciences, Eli Lilly & Co, Foresite Labs, Genentech/Roche, GV, HeartFlow, Magnet Biomedicine, and Novartis, scientific advisory board membership of Esperion Therapeutics, Preciseli, and TenSixteen Bio, scientific co-founder of TenSixteen Bio, equity in MyOme, Preciseli, and TenSixteen Bio, and spousal employment at Vertex Pharmaceuticals, all unrelated to the present work. M.B.L. has received speaker and advisory fees from Otsuka, Reata, Bayer, and Sanofi Genzyme. G.P. has received speaker and advisory fees from Amgen, Bayer, Novartis, and Sanofi, and has received research grants from Bayer and Sanofi. L.M.R. is a consultant for the TOPMed Administrative Coordinating Center (through Westat).
Published: December 22, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2023.100468.
Contributor Information
Andrew P. Morris, Email: andrew.morris-5@manchester.ac.uk.
Nora Franceschini, Email: noraf@unc.edu.
Supplemental information
References
- 1.GBD 2016 Causes of Death Collaborators Global, regional and national age-sex specific mortality for 264 causes of death. 1980-2016: a systematic analysis of the Global Burden of Disease Study 2016. Lancet. 2017;390:1151–1210. doi: 10.1016/S0140-6736(17)32152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.GBD 2016 Diseases and Injury Incidence and Prevalence Collaborators Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1211–1259. doi: 10.1016/S0140-6736(17)32154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johansen K.L., Chertow G.M., Gilbertson D.T., Herzog C.A., Ishani A., Israni A.K., Ku E., Li S., Li S., Liu J., et al. US Renal Data System 2021 Annual Data Report: Epidemiology of Kidney Disease in the United States. Am. J. Kidney Dis. 2022;79:A8–A12. doi: 10.1053/j.ajkd.2022.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xue J.L., Eggers P.W., Agodoa L.Y., Foley R.N., Collins A.J. Longitudinal study of racial and ethnic differences in developing end-stage renal disease among aged medicare beneficiaries. J. Am. Soc. Nephrol. 2007;18:1299–1306. doi: 10.1681/ASN.2006050524. [DOI] [PubMed] [Google Scholar]
- 5.Collins A.J., Foley R.N., Herzog C., Chavers B., Gilbertson D., Herzog C., Ishani A., Johansen K., Kasiske B., Kutner N., et al. US Renal Data System 2012 Annual Data Report. Am. J. Kidney Dis. 2013;61:2–7. doi: 10.1053/j.ajkd.2012.11.031. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Chronic Kidney Disease Basics. https://www.cdc.gov/kidneydisease/basics.html.
- 7.Borrell L.N., Elhawary J.R., Fuentes-Afflick E., Witonsky J., Bhakta N., Wu A.H.B., Bibbins-Domingo K., Rodríguez-Santana J.R., Lenoir M.A., Gavin J.R., et al. Race and Genetic Ancestry in Medicine - A Time for Reckoning with Racism. N. Engl. J. Med. 2021;384:474–480. doi: 10.1056/NEJMms2029562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sirugo G., Williams S.M., Tishkoff S.A. The Missing Diversity in Human Genetic Studies. Cell. 2019;177:26–31. doi: 10.1016/j.cell.2019.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin A.R., Gignoux C.R., Walters R.K., Wojcik G.L., Neale B.M., Gravel S., Daly M.J., Bustamante C.D., Kenny E.E. Human demographic history impacts genetic risk prediction across diverse populations. Am. J. Hum. Genet. 2017;100:635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morris A.P., Le T.H., Wu H., Akbarov A., van der Most P.J., Hemani G., Smith G.D., Mahajan A., Gaulton K.J., Nadkarni G.N., et al. Trans-ethnic kidney function association study reveals putative causal genes and effects on kidney-specific disease aetiologies. Nat. Commun. 2019;10:29. doi: 10.1038/s41467-018-07867-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H., Doke T., Guo D., Sheng X., Ma Z., Park J., Vy H.M.T., Nadkarni G.N., Abedini A., Miao Z., et al. Epigenomic and transcriptomic analyses define core cell types, genes and targetable mechanisms for kidney disease. Nat. Genet. 2022;54:950–962. doi: 10.1038/s41588-022-01097-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franceschini N., Morris A.P. Genetics of kidney traits in worldwide populations: the Continental Origins and Genetic Epidemiology Network (COGENT) Kidney Consortium. Kidney Int. 2020;98:35–41. doi: 10.1016/j.kint.2020.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahajan A., Rodan A.R., Le T.H., Gaulton K.J., Haessler J., Stilp A.M., Kamatani Y., Zhu G., Sofer T., Puri S., et al. Trans-ethnic Fine Mapping Highlights Kidney-Function Genes Linked to Salt Sensitivity. Am. J. Hum. Genet. 2016;99:636–646. doi: 10.1016/j.ajhg.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. The 1000 Genomes Project Consortium (2015). A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K., et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurdasani D., Carstensen T., Tekola-Ayele F., Pagani L., Tachmazidou I., Hatzikotoulas K., Karthikeyan S., Iles L., Pollard M.O., Choudhury A., et al. The African genome variation project shapes medical genetics in Africa. Nature. 2015;517:327–332. doi: 10.1038/nature13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurdasani D., Carstensen T., Fatumo S., Chen G., Franklin C.S., Prado-Martinez J., Bouman H., Abascal F., Haber M., Tachmazidou I., et al. Uganda genome resource enables insights into population history and genomic discovery in Africa. Cell. 2019;179:984–1002.e36. doi: 10.1016/j.cell.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levey A.S., Stevens L.A., Schmid C.H., Zhang Y.L., Castro A.F., Feldman H.I., Kusek J.W., Eggers P., van Lente F., Greene T., et al. A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levey A.S., Stevens L.A. Estimating GFR using the CKD Epidemiology Collaboration (CKD-EPI) creatinine equation: more accurate GFR estimates, lower CKD prevalence estimates, and better risk predictions. Am. J. Kidney Dis. 2010;55:622–627. doi: 10.1053/j.ajkd.2010.02.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hellwege J.N., Velez Edwards D.R., Giri A., Qiu C., Park J., Torstenson E.S., Keaton J.M., Wilson O.D., Robinson-Cohen C., Chung C.P., et al. Mapping eGFR loci to the renal transcriptome and phenome in the VA Million Veteran Program. Nat. Commun. 2019;10:3842. doi: 10.1038/s41467-019-11704-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inker L.A., Eneanya N.D., Coresh J., Tighiouart H., Wang D., Sang Y., Crews D.C., Doria A., Estrella M.M., Froissart M., et al. New Creatinine- and Cystatin C-Based Equations to Estimate GFR without. N. Engl. J. Med. 2021;385:1737–1749. doi: 10.1056/NEJMoa2102953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breeze C.E., Haugen E., Reynolds A., Teschendorff A., van Dongen J., Lan Q., Rothman N., Bourque G., Dunham I., Beck S., et al. Integrative analysis of 3604 GWAS reveals multiple novel cell type-specific regulatory associations. Genome Biol. 2022;23:13. doi: 10.1186/s13059-021-02560-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breeze C.E., Reynolds A.P., van Dongen J., Dunham I., Lazar J., Neph S., Vierstra J., Bourque G., Teschendorff A.E., Stamatoyannopoulos J.A., Beck S. eFORGE v2.0: updated analysis of cell type-specific signal in epigenomic data. Bioinformatics. 2019;35:4767–4769. doi: 10.1093/bioinformatics/btz456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breeze C.E. Cell type-specific signal analysis in epigenome-wide association studies. Methods Mol. Biol. 2022;2432:57–71. doi: 10.1007/978-1-0716-1994-0_5. [DOI] [PubMed] [Google Scholar]
- 27.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe K., Taskesen E., van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe K., Umićević Mirkov M., de Leeuw C.A., van den Heuvel M.P., Posthuma D. Genetic mapping of cell type specificity for complex traits. Nat. Commun. 2019;10:3222. doi: 10.1038/s41467-019-11181-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eales J.M., Jiang X., Xu X., Saluja S., Akbarov A., Cano-Gamez E., McNulty M.T., Finan C., Guo H., Wystrychowski W., et al. Uncovering genetic mechanisms of hypertension through multi-omic analysis of the kidney. Nat. Genet. 2021;53:630–637. doi: 10.1038/s41588-021-00835-w. [DOI] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research Network. Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R.M., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kachuri L., Mak A.C.Y., Hu D., Eng C., Huntsman S., Elhawary J.R., Gupta N., Gabriel S., Xiao S., Keys K.L., et al. Gene expression in African Americans, Puerto Ricans and Mexican Americans reveals ancestry-specific patterns of genetic architecture. Nat. Genet. 2023;55:952–963. doi: 10.1038/s41588-023-01377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao L., Duan L., Guo Y., Zhou B., Xu Q., Zhang C., Liu W., Liu W., Liu Z., Hu J., et al. TRIM46 upregulates Wnt/β-catenin signaling by inhibiting Axin1 to mediate hypoxia-induced epithelial-mesenchymal transition in HK2 cells. Mol. Cell. Biochem. 2022;477:2829–2839. doi: 10.1007/s11010-022-04467-4. [DOI] [PubMed] [Google Scholar]
- 34.Genovese G., Friedman D.J., Ross M.D., Lecordier L., Uzureau P., Freedman B.I., Bowden D.W., Langefeld C.D., Oleksyk T.K., Uscinski Knob A.L., et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parsa A., Kao W.H.L., Xie D., Astor B.C., Li M., Hsu C.y., Feldman H.I., Parekh R.S., Kusek J.W., Greene T.H., et al. APOL1 risk variants, race, and progression of chronic kidney disease. N. Engl. J. Med. 2013;369:2183–2196. doi: 10.1056/NEJMoa1310345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan A., Turchin M.C., Patki A., Srinivasasainagendra V., Shang N., Nadukuru R., Jones A.C., Malolepsza E., Dikilitas O., Kullo I.J., et al. Genome-wide polygenic score to predict chronic kidney disease across ancestries. Nat. Med. 2022;28:1412–1420. doi: 10.1038/s41591-022-01869-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graham S.E., Nielsen J.B., Zawistowski M., Zhou W., Fritsche L.G., Gabrielsen M.E., Skogholt A.H., Surakka I., Hornsby W.E., Fermin D., et al. Sex-specific and pleiotropic effects underlying kidney function identified from GWAS meta-analysis. Nat. Commun. 2019;10:1847. doi: 10.1038/s41467-019-09861-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mägi R., Horikoshi M., Sofer T., Mahajan A., Kitajima H., Franceschini N., McCarthy M.I., COGENT-Kidney Consortium T2D-GENES Consortium. Morris A.P., Morris A.P. Trans-ethnic meta-regression of genome-wide association studies accounting for ancestry increases power for discovery and improves fine-mapping resolution. Hum. Mol. Genet. 2017;26:3639–3650. doi: 10.1093/hmg/ddx280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanai M., Akiyama M., Takahashi A., Matoba N., Momozawa Y., Ikeda M., Iwata N., Ikegawa S., Hirata M., Matsuda K., et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. 2018;50:390–400. doi: 10.1038/s41588-018-0047-6. [DOI] [PubMed] [Google Scholar]
- 40.Stanzick K.J., Li Y., Schlosser P., Gorski M., Wuttke M., Thomas L.F., Rasheed H., Rowan B.X., Graham S.E., Vanderweff B.R., et al. Discovery and prioritization of variants and genes for kidney function in >1.2 million individuals. Nat. Commun. 2021;12:4350. doi: 10.1038/s41467-021-24491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ge T., Chen C.-Y., Ni Y., Feng Y.-C.A., Smoller J.W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 2019;10:1776. doi: 10.1038/s41467-019-09718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wuttke M., Li Y., Li M., Sieber K.B., Feitosa M.F., Gorski M., Tin A., Wang L., Chu A.Y., Hoppmann A., et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat. Genet. 2019;51:957–972. doi: 10.1038/s41588-019-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomaszewski M., Morris A.P., Howson J.M.M., Franceschini N., Eales J.M., Xu X., Dikalov S., Guzik T.J., Humphreys B.D., Harrap S., Charchar F.J. Kidney omics in hypertension: from statistical associations to biological mechanisms and clinical applications. Kidney Int. 2022;102:492–505. doi: 10.1016/j.kint.2022.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Privé F., Aschard H., Carmi S., Folkersen L., Hoggart C., O'Reilly P.F., Vilhjálmsson B.J. Portability of 245 polygenic scores when derived from the UK Biobank and applied to 9 ancestry groups from the same cohort. Am. J. Hum. Genet. 2022;109:12–23. doi: 10.1016/j.ajhg.2021.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding Y., Hou K., Xu Z., Pimplaskar A., Petter E., Boulier K., Privé F., Vilhjálmsson B.J., Olde Loohuis L.M., Pasaniuc B. Polygenic scoring accuracy varies across the genetic ancestry continuum. Nature. 2023;618:774–781. doi: 10.1038/s41586-023-06079-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 47.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genome-wide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loh P.-R., Danecek P., Palamara P.F., Fuchsberger C., A Reshef Y., K Finucane H., Schoenherr S., Forer L., McCarthy S., Abecasis G.R., et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M., et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 51.1000 Genomes Project Consortium. Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levey A.S., Bosch J.P., Lewis J.B., Greene T., Rogers N., Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Ann. Intern. Med. 1999;130:461–470. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- 53.Levey A.S., Coresh J., Greene T., Stevens L.A., Zhang Y.L., Hendriksen S., Kusek J.W., van Lente F., Chronic Kidney Disease Epidemiology Collaboration Using standardized serum creatinine values in the modification of diet in renal disease study equation for estimating glomerular filtration rate. Ann. Intern. Med. 2006;145:247–254. doi: 10.7326/0003-4819-145-4-200608150-00004. [DOI] [PubMed] [Google Scholar]
- 54.Roadmap Epigenomics Consortium. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kass R.E., Raftery A.E. Bayes factors. J. Am. Stat. Assoc. 1995;90:773–795. [Google Scholar]
- 56.Wellcome Trust Case Control Consortium. Maller J.B., McVean G., Byrnes J., Vukcevic D., Palin K., Su Z., Howson J.M.M., Auton A., Myers S., et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet. 2012;44:1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomaszewski M., Eales J., Denniff M., Myers S., Chew G.S., Nelson C.P., Christofidou P., Desai A., Büsst C., Wojnar L., et al. Renal Mechanisms of Association between Fibroblast Growth Factor 1 and Blood Pressure. J. Am. Soc. Nephrol. 2015;26:3151–3160. doi: 10.1681/ASN.2014121211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu X., Eales J.M., Akbarov A., Guo H., Becker L., Talavera D., Ashraf F., Nawaz J., Pramanik S., Bowes J., et al. Molecular insights into genome-wide association studies of chronic kidney disease-defining traits. Nat. Commun. 2018;9:4800. doi: 10.1038/s41467-018-07260-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang X., Eales J.M., Scannali D., Nazgiewicz A., Prestes P., Maier M., Denniff M., Xu X., Saluja S., Cano-Gamez E., et al. Hypertension and renin-angiotensin system blockers are not associated with expression of angiotensin-converting enzyme 2 (ACE2) in the kidney. Eur. Heart J. 2020;41:4580–4588. doi: 10.1093/eurheartj/ehaa794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ongen H., Buil A., Brown A.A., Dermitzakis E.T., Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics. 2016;32:1479–1485. doi: 10.1093/bioinformatics/btv722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stegle O., Parts L., Piipari M., Winn J., Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc. 2012;7:500–507. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Young M.D., Mitchell T.J., Vieira Braga F.A., Tran M.G.B., Stewart B.J., Ferdinand J.R., Collord G., Botting R.A., Popescu D.-M., Loudon K.W., et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science. 2018;361:594–599. doi: 10.1126/science.aat1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X., Park J., Susztak K., Zhang N.R., Li M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat. Commun. 2019;10:380. doi: 10.1038/s41467-018-08023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taylor-Weiner A., Aguet F., Haradhvala N.J., Gosai S., Anand S., Kim J., Ardlie K., van Allen E.M., Getz G. Scaling computational genomics to millions of individuals with GPUs. Genome Biol. 2019;20:228. doi: 10.1186/s13059-019-1836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dastani Z., Hivert M.-F., Timpson N., Perry J.R.B., Yuan X., Scott R.A., Henneman P., Heid I.M., Kizer J.R., Lyytikäinen L.P., et al. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper analyzes existing, publicly available genotype and phenotype cohort data. These accession numbers for the datasets are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.