Abstract

Mycobacterium tuberculosis is responsible for tuberculosis (TB) all over the world. Despite tremendous advancements in biomedical research, new treatment approaches, and preventive measures, TB incidence rates continue to ascend. The herbaceous plant Acalypha indica, also known as Indian Nettle, belongs to the Euphorbiaceae family and is known as one of the most important sources of medicines and pharmaceuticals for the medical therapy for a range of ailments. However, the precise molecular mechanism of its therapeutic action is still unknown. In this study, an integrated network pharmacology approach was employed to explore the potential mechanism of A. indica phytochemicals against TB. The active chemical components of A. indica were collected from two independent databases and published sources, whereas SwissTargetPrediction was used to identify the target genes of these phytochemicals. GeneCards and DisGeNET databases were employed to retrieve tuberculosis-related genes and variants. Following the evaluation of overlapped genes, gene enrichment analysis and PPI network analysis were performed using the DAVID and STRING databases, respectively. Later, to identify the potential target(s) for the disease, molecular docking was performed. A. indica revealed 9 active components with 259 potential therapeutic targets; TB attributed 694 intersecting genes from the two data sets; and both TB and A. indica overlapped 44 potential targets. The in-depth analysis based on the degree revealed that AKT1 and EGFR formed the foundation of the PPI network. Moreover, docking analysis followed by molecular dynamics simulations revealed that phytosterol and stigmasterol have higher binding affinities to AKT1 and EGFR to suppress tuberculosis. This study provides a convincing proof that A. indica can be exploited to target TB after experimental endorsement; further, it lays the framework for more experimental research on A. indica’s anti-TB activity.
1. Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), is a major public health problem, which is a leading cause of death globally and causes substantial health problems. TB is a potentially dangerous infectious disease that mostly affects the lungs. People contract TB from one another by coughing and sneezing, which release microscopic droplets of germs into the air. Mtb infections cause a couple of million deaths each year, and this higher mortality has been linked to poor diagnostic methods and the rapid evolution of multidrug-resistant TB (MDR-TB).1,2 10.4 million TB cases were reported in 2016 and 1.7 million deaths were reported annually, according to the World Health Organization (WHO). India alone reported 26% of the total number of TB deaths worldwide in the same year.3
The current TB treatment options are effective only against active TB and do not address any side effects. The researchers’ main goal is to find and create novel drugs that can boost safety, modify the treatment course, and treat both infections and side effects.4 In the absence of efficient therapy, medications for TB, hope is placed in plant-based natural products due to their chemical diversity and crucial significance as phyto-drugs.5 Medicinal plants are an everlasting gift of nature that have been utilized to heal a variety of ailments in humans since time immemorial. According to WHO estimates, traditional medicines are used by 80% of the population in poor countries for basic healthcare.6 The utilization of therapeutic plant extracts and phytochemicals is extremely important in the treatment of a variety of disorders, including TB. Plants including Acalypha indica, Allium sativum, Adhatoda vasica, Cassia sophera, and Andrographis paniculata have shown considerable antimycobacterial efficacy against TB under vitro conditions.7
A. indica Linn. (Family Euphorbiaceae) is a common annual shrub cultivated as a weed throughout India. It is an erect, herbaceous plant that can reach a height of 100 cm. It has traditionally been used as a diuretic, an anthelmintic, and a respiratory aid and in the treatment of rheumatoid arthritis, scabies, and other skin infections.8 Previous research has found that A. indica has antibacterial,9 anti-inflammatory,10 hepatoprotective,11 antifungal,12 antifertility,13 anthelmintic,14 antiulcer,15 antioxidant, and anticancer activities.16 Individuals in India have used A. indica to cure any disease associated with the respiratory system.17 Gupta et al. used an aqueous extract of A. indica to conduct an experiment with different tuberculosis bacteria. As a result, the aqueous extract of A. indica inhibited the growth of Mtb H37Rv and two multidrug-resistant Mtb.18
AKT1 (also known as protein kinase B or PKB) and EGFR (epidermal growth factor receptor) are both cell surface receptors that play an important role in cell signaling cascades. They are significant in the context of cancer and have been examined as potential biomarkers in a variety of diseases, including TB. Network pharmacology is a cross-disciplinary field that analyzes the network of biological systems and selects key signal nodes for drug molecular design by combining the core ideas and research methodologies of bioinformatics, network science, mathematics, and computer science.19,20 As a novel holistic, multidisciplinary, integrative area, it analyzes drug–target interactions through the prism of systems-based techniques, providing a new paradigm for breakthrough drug development.21,22 Molecular docking is a method of drug design that involves the interaction of drug molecules and receptors. It has been widely used in drug research and development in recent years.23,24 Hence, we employed network pharmacology in combination with molecular docking, followed by molecular dynamics simulation to investigate the potential mechanism of A. indica against tuberculosis. To the best of our knowledge, this is the very first study to explore the efficacy and mechanistic actions of A. indica phytocompounds against TB as well as offer theoretical support and a scientific basis for experimental validation of our current findings.
2. Materials and Methods
2.1. Screening of Bioactive Chemicals of A. indica
The chemical information was acquired from published literature and by using Dr. Duke’s Phytochemical and Ethnobotanical Database (https://phytochem.nal.usda.gov/phytochem/search)25 and Indian Medicinal Plants, Phytochemistry and Therapeutics (IMPPAT) (https://cb.imsc.res.in/imppat/),26 two independent, publicly accessible databases of phytochemicals. The bioactive chemical components were virtually filtered out through Traditional Chinese Medicine System Pharmacology (TCMSP) (https://tcmsp-e.com/tcmsp.php)27 based on pharmacological indexes of OB and DL greater than 30% and 0.18, respectively. OB is for the oral bioavailability of a potential therapeutic component, whereas DL represents drug-component affinity, which may indicate a potential drug. Both DL and OB are more significant characteristics of a drug.
2.2. Target Profiling of A. indica and Tuberculosis
The target profiling is the preliminary step to highlight the interaction frameworks of herbs used to treat a range of ailments. SwissTargetPrediction (http://www.swisstargetprediction.ch/) was accessed to retrieve the target genes of corresponding bioactive chemical based on the simplified molecular input line entry system (SMILES), confined to “Homo sapiens.”28 Only targets with a combined score of ≥0.7 were selected for subsequent analysis. The duplicated genes were eliminated by aligning the UNIPROT IDs by using UniProtKB (https://www.uniprot.org/help/uniprotkb). The target genes and their variants related to tuberculosis were acquired from DisGeNET (https://www.disgenet.org/) and GeneCards (https://www.genecards.org/) databases using the terms “TB and tuberculosis.” Moreover, these databases provide a summary of genomic data sets coupled with physiological annotations. The records of target genes acquired from these databases were overlapped to predict more unique target genes involved in disease pathways. The target genes for A. indica in TB were mapped using a Venn diagram to identify putative targets. The researches were advanced by considering these mapped genes.
2.3. Compound–Target Network
To visualize the molecular processes of A. indica in tuberculosis, Cytoscape was used to build the interaction network between active molecules and probable targets.29 The network’s nodes signify bioactive chemicals and potential targets. The lines indicate the interaction of nodes known as edges. Later, a Cytoscape plugin, Network Analyzer, was used to reveal the topological characteristics of the network. The network was evaluated on the basis of the degree of connectivity.
2.4. Protein–Protein Interaction Network
Protein–protein interaction30 network of potential targets was built with a confidence score of >0.4 by using STRING database. The isolated points found in the network were deleted, and the network is ready to import into Cytoscape. Cytoscape was used to optimize the PPI network on the basis of degree of connectivity, and the nodes with higher degree in the network were bigger. To find out the key targets, a cytoscape plugin, CytoHubba, was employed. These key potential targets may serve as potential therapeutic targets that might be involved in disease pathways.
2.5. Gene Ontology and Pathway Enrichment Analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and gene enrichment analysis were carried out employing a database of functional annotations, DAVID (https://david.ncifcrf.gov/). The gene function of potential targets was categorized into biological processes (BP), cellular components,30 and molecular functions (MFs). The pathway enrichment analysis was carried out through KEGG database. These ontologies and pathways were screened out based on the probability value of <0.05. Later on, top 10 enriched ontologies and pathways were displayed in bubble maps in the R language using the ggplot2 package. To find out the interactions of key hub, targets to top 10 enriched pathways were visualized in Cytoscape (https://cytoscape.org/).
2.6. Compound–Target–Pathway Network
The compound–target and pathway–target networks were merged to build the compound–target–pathway network by using Cytoscape. The nodes in the network represent the active chemicals, potential targets, and signaling pathways. The edges depict the relationship between these three types of nodes and how they interact with each other.
2.7. Molecular Docking
Protein–ligand molecular docking is employed to anticipate the binding sites of bioactive constituents and potential targets identified via network pharmacology. Molecular docking predicts interactions that bind ligands to paired receptors in a molecular system. Binding energy is the primary characteristic of the analysis evaluation. The 3D conformations of ligands were retrieved from PubChem database.31 The 3D structures of receptor proteins were retrieved from Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSBPDB) (https://www.rcsb.org/).32 To preprocess and minimize the receptor protein, a chimera was used. Autodock Vina features a user-friendly interface that has been used to dock the receptor protein to ligands.33 Afterward, chimera was used to visualize the interactions between the receptors and ligands.34
2.8. Molecular Dynamics Simulation
The complexes were MD simulated at 100 ns using the Schrodinger suite’s Desmond software.35 In the simulation, the results of molecular docking were used to analyze the dynamic interactions of the protein–ligand complex. Using the OPLS_2005 force field, the complexes were preprocessed, optimized, and minimized.36 In an orthorhombic box with dimensions of 10 × 10 × 10 Å3, the system builder tool was used to add a transferable intermolecular potential with the 3-point TIP3P solvent model.37 Counter ions were introduced when needed to establish neutrality, and a concentration of 0.15 M NaCl was included to imitate a physiological state. The NPT ensemble was used to simulate the conditions of 1 atm pressure and 300 K temperature. Before starting, the complex was relaxed, and simulated trajectories were saved every 50 ps for analysis.
3. Results
3.1. Screening of Bioactive Chemicals of A. indica
The phytochemical information on A. indica was collected from different independent databases and literature mining.38−43 A total of 134 chemicals of the leaves of A. indica were collected. Among these chemicals, 12 chemicals (Ar–curcumene, aurantiamide, β-sitosterol, campesterol, catechin, flavoxanthin, clionasterol, kaempherol, squalene, stigmasterol, diisooctyl phthalate, and 2-monoolein) were screened based on pharmacokinetic indices of DL and OB. These 12 chemicals with their chemical properties are presented, which may serve as therapeutic drugs against the disease (Table 1).
Table 1. Twelve Bioactive Phytochemicals of A. indica, Their Properties, Pharmacokinetic Attributes, and 2D Structures.
3.2. Target Profiling of A. indica and Tuberculosis
A total of 259 unique target genes were collected for the 9 bioactive chemicals of A. indica leaves’ extract with a probability score of greater than zero. As there was no target gene mapped to Ar–curcumene and catechin, these chemicals were dropped from further investigations. Another chemical, flavoxanthin, was also removed since it did not meet the criteria of probability larger than zero. Following the identification of the most potential pharmacological targets, 694 unique disease-related genes were revealed. Subsequently, a Venn diagram was plotted to figure out the potential target genes of A. indica and tuberculosis. A total of 44 target genes were identified as potential targets and further used for subsequent investigations (Figure 1).
Figure 1.

Venn diagram represents the potential genes between the herb (A. indica) and TB.
3.3. Compound–Target Network
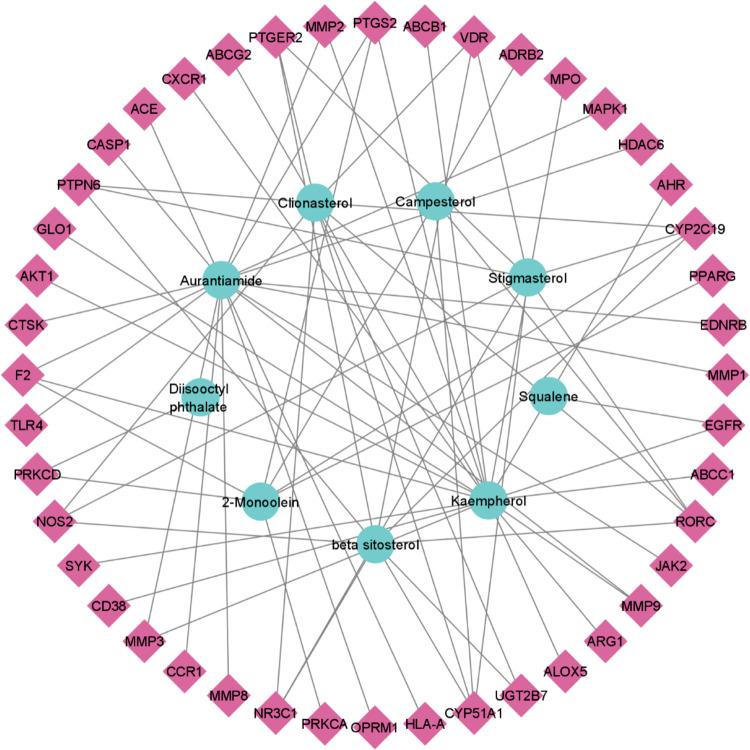
The active phytochemicals and potential target genes were evaluated, as the mode of function remains a riddle. To uncover the molecular mechanism, a network of 9 bioactive chemicals and 44 potential target genes was constructed. The network contained 53 nodes and 73 edges. The topological analysis of the network revealed that it consists of 0.053 density, 1.310 heterogeneity, 0.305 centralization, and 3.771 average shortest path length (Figure 2).
Figure 2.
Interaction network of 9 compounds and 44 potential target genes. The circular nodes in the inner circle represent the bioactive chemicals, diamond nodes in the outer circle represent potential target genes, and lines between them represent the interaction between nodes.
The chemical aurantiamide (18), kaempherol (18), β-sitosterol (9), clionasterol (9), and stigmasterol (8) exhibited higher degree of connectivity in the network, indicating that these chemicals may serve as therapeutic compounds (Table 2).
Table 2. Degree of Connectivity of Chemicals Explored via Cytoscape.
| sl. no. | chemical name | class | degree |
|---|---|---|---|
| 01 | aurantiamide | peptides | 18 |
| 02 | kaempherol | flavonoid | 18 |
| 03 | β-sitosterol | phytosterol | 9 |
| 04 | clionasterol | phytosterol | 9 |
| 05 | stigmasterol | phytosterol | 8 |
| 06 | 2-monoolein | glyceride | 7 |
| 07 | campesterol | phytosterol | 2 |
| 08 | squalene | terpenoid | 1 |
| 09 | diisooctyl phthalate | phthalic acid | 1 |
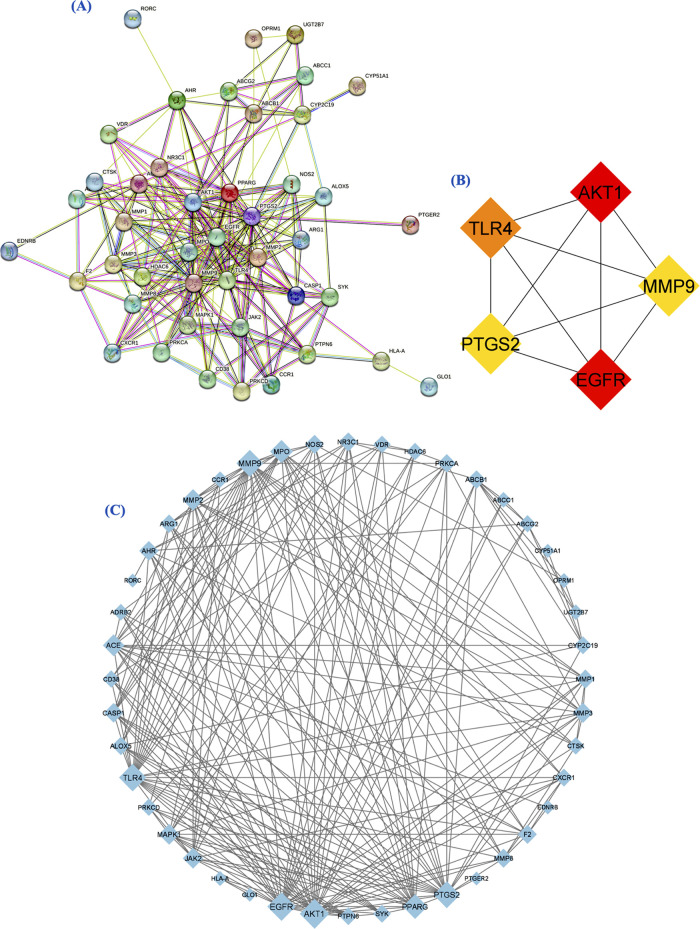
3.4. Protein–Protein Interaction Network
The signaling pathways of proteins are often mediated by protein interactions. A PPI network with a confidence score of 0.4 was built to comprehend the functional mechanism of 44 potential target genes. The network contained 44 nodes and 223 edges greater than the expected number of edges, which are 57. The target genes were closely related to each other, and no isolated point was found in the network (Figure 3A). The network was found to be significant. The network was optimized in Cytoscape, and the larger nodes such as AKT1 (29) and EGFR (27) showed higher degree of connectivity. As the degree of connectivity decreases, the size of the nodes gradually decreases in the network. Target proteins such as GLO1 (1) and PTGER2 (1) with less degree of connectivity showed a smaller sized node in the network (Figure 3B). Later, overlapped genes were analyzed, including AKT1, EGFR, TLR4, MMP9, and PTGS2. These targets were highly connected in the network (Figure 3C).
Figure 3.
(A) Interaction analysis of potential 44 target genes (protein) constructed through the STRING database. (B) The PPI network was optimized through Cytoscape. (C) Candidate potential targets evaluated through CytoHubba based on degree.
3.5. Gene Ontology and Pathway Enrichment Analysis
To gain a more comprehensive knowledge of the bioactivity of the targets addressed by the beneficial drugs, a gene function annotation analysis was carried out. The potential targets were involved in 155 BPs, 29 CCs, 44 MFs, and 73 signaling pathways with an ease of <0.05 and a gene count of >2. The top 10 enriched annotations were taken because the targeted genes with their best ligands were presented in top 10 enriched ontologies and 20 pathways with least p value plotted in bubble charts (Figure 4).
Figure 4.
Bubble map visualization of enriched gene functional annotation and signaling pathways. (A) Biological processes, (B) cellular components, (C) molecular function, and (D) KEGG pathways.
The interactions of candidate potential targets to the top 20 pathways were built through Cytoscape. Earlier, it has been disclosed that the key target genes were mainly involved in estrogen signaling pathway, relaxin signaling pathway, C-type lectin receptor signaling pathway, HIF-1 signaling pathway, and tuberculosis (Figure 5).
Figure 5.
Interaction framework of candidate potential targets to enriched signaling pathways. The elliptical nodes represent the signaling pathway, diamond nodes represent the potential targets, and lines between nodes represent the pattern of their interactions.
3.6. Compound–Target–Pathway Network
To visualize the interactions underlying candidate potential targets, bioactive chemical components, and disease pathways, compound–target networks and pathway–target networks were combined. Each active constituent is interconnected to multiple targets that are correspondents to signaling pathways. It may impose synergistic effect to the disease treatment (Figure 6).
Figure 6.
A. indica-induced pathways. The circular nodes represent active chemicals in the inner circle, diamond nodes in the middle circle represent potential targets, and hexagons in the outer circle represent signaling pathways.
3.7. Molecular Docking
Molecular docking was performed to verify candidate target genes for chemicals that can inhibit tuberculosis. The bioactive chemicals found in A. indica (aurantiamide, β-sitosterol, campesterol, clionasterol, kaempherol, squalene, stigmasterol, diisooctyl phthalate, and 2-monoolein) and candidate targets, namely, AKT1 and EGFR. Molecular docking was used to evaluate candidate bioactive target genes to reduce the possibility of tuberculosis prevalence. The significantly docked complexes were screened on the basis of binding affinity. The binding affinity of docking complexes ranges from −4.5 to −9.2 kcal/mol (Table 3).
Table 3. Binding Affinities of Docking Complexes and Their Interacting Residues.
| binding
affinities (kcal/mol) |
|||
|---|---|---|---|
| sl. no. | ligands | AKT1 | EGFR |
| 01 | aurantiamide | –7.8 | –6.9 |
| 02 | β-sitosterol | –8.5 | –8.6 |
| 03 | campesterol | –8.7 | –8.7 |
| 04 | clionasterol | –8.3 | –8.6 |
| 05 | kaempherol | –7.6 | –7.8 |
| 06 | squalene | –5.9 | –4.5 |
| 07 | stigmasterol | –9 | –9.2 |
| 08 | diisooctyl phthalate | –6.3 | –5.9 |
| 09 | 2-monoolein | –5.4 | –5.2 |
All of the 9 ligands successfully bind to the target proteins AKT1 and EGFR (Figures 7 and 8). But stigmasterol was the most prominently complexed with AKT1 and EGFR with −9 and −9.2 kcal/mol, respectively.
Figure 7.
Docking complexes of AKT1 with 9 active chemicals.
Figure 8.
EGFR binding site residues to the chemical components of A. indica.
3.8. Molecular Dynamics Simulation
3.8.1. Root-Mean-Square Deviation
To assess the stability of the protein–ligand complex, the root-mean-square deviation (RMSD) of carbon α atoms in protein and ligand trajectories was computed. Both complexes were found to be equilibrated after 10 ns (Figure 9). After equilibration, the RMSD of AKT1-complex maintained a RMSD value of ∼2–2.5 Å until 40 ns. During 40–60 ns, RMSD deviated in the range of ∼1.5–3 Å. After 60 ns, the RMSD value gradually decreased to ∼2 Å at 70 ns and then attained the stability in the range of ∼2–3 Å until the end of simulation. The RMSD of the ligand (stigmasterol) remained in the range of ∼1–2 Å throughout the simulation. The RMSD plot of the EGFR-complex showed that the RMSD remained in the range ∼1.5–1.75 Å until 80 ns and then displayed some deviations during 80–90 ns. The RMSD attained the previous values in the last part of simulation. Similarly, the RMSD of stigmasterol in this complex remained in the range of ∼1–1.5 Å throughout the simulation.
Figure 9.
RMSD plots of AKT1 and EGFR complexes during the 100 ns simulation. Blue plot shows the RMSD of AKT1 protein, cyan plot shows the RMSD of EGFR, and green plot shows the RMSD of stigmasterol.
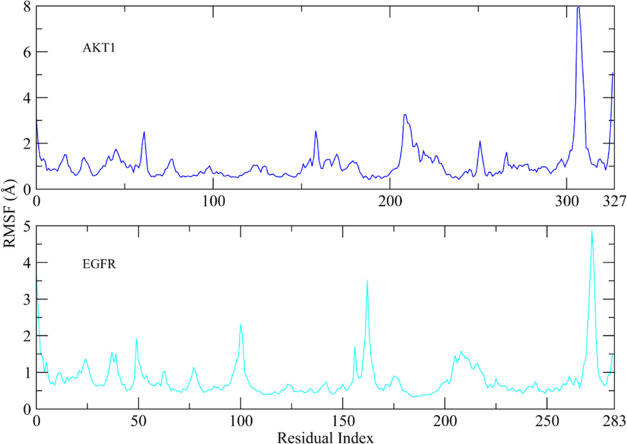
3.8.2. Root-Mean-Square Fluctuations
To examine the dynamic behavior of protein restudies when bound to the ligand, root-mean-square fluctuation (RMSF) values were computed. Except for the loop areas, the RMSF values of protein residues fluctuate less than 1 Å during the simulation period (Figure 10). The RMSF figure demonstrated that the protein residues were stiff and did not display significant variations during simulation, implying that the protein–ligand combination was stable. The RMSF values of loop residues reached a maximum of ∼8 Å in the AKT1-complex, while the greatest RMSF of loop regions in the EGFR-complex was ∼5 Å.
Figure 10.
Fluctuations of protein residual during simulations as determined by RSMF values. Blue plot shows the RMSF plot of the AKT1-complex, while cyan plot shows that of the EGFR-complex.
3.8.3. Protein–Ligand Contacts
MD simulations revealed that hydrogen bonds and hydrophobic interactions were the most critical interactions between the protein and its ligands (Figure 11). The residues involved in hydrogen bonding in the AKT-complex were Val185, Lys189, and Glu191, while Lys716 and Lys728 made hydrogen bonds with stigmasterol in the EGFR-complex.
Figure 11.
Interaction of protein–ligand interactions during MDS. (A) The protein–ligand contacts of AKT1-complex. (B) The protein–ligand contacts of EGFR-complex. The interacting residues are shown are shown as large, stacked bars.
3.8.4. Principal Component Analysis
The dynamic behavior of the protein in both complexes was analyzed by the principal component analysis, which helps to identify the collective motions of the MD trajectories. The PCA plots of both complexes are shown in Figure 12A,B. The proportion of the variance was plotted against the eigenvalue, which represents the dynamic motions in hyperspace. Only three PCs were plotted as covering the major fluctuations. In the AKT1-complex, it can be observed that PC1 showed the highest variation of 35.37% than others. PC2 and PC3 depicted the variability of 14.22 and 7.83%, respectively (Figure 12A). While in the EGFR-complex, the highest variation was 13.51% in PC1. Other variations in PC2 and PC3 were 10.73 and 8.12%, respectively (Figure 12B). The PCA analysis indicated conformational changes in all clusters through simple clustering in the PC subspace; blue regions exhibited the most significant movement, white regions showed intermediate movement, and red regions showed less flexibility movement.
Figure 12.
Principal component analysis of AKT1 and EGFR complexes. (A) The PCA plot of AKT1-complex with overall flexibility of 57.42%. (B) The PCA plot of EGFR-complex with overall flexibility of 32.36% in three hyper spaces.
3.8.5. Cross Correlation
The correlation among the protein residues was investigated by a cross-correlation matrix, Figure 13A,B. The cyan color showed positively correlated residues, while magenta color represented anticorrelated residues. It can be observed that most of the residues had positive correlation, while the anticorrelated residues were too short to observe. The diagonal lines showed the positive correlation of topological proximate residues. This analysis suggested that the protein residues were highly correlated to each other when bound to the respective compounds during simulation.
Figure 13.
Dynamic cross-correlation matrix of the protein complexes to find the positive correlation in protein residues. (A) The DCCM of AKT1-complex. (B) The DCCM of EGFR-complex.
3.8.6. MM/GBSA
Molecular mechanics generalized Born surface area (MM/GBSA) technique was adopted to compute the total binding free energy (ΔGtotal) for both complexes. ΔGtotal value is generally utilized for estimating the stability of the complex of protein–ligand. The lower values of ΔGtotal indicate that the complex is more stable and vice versa. It was estimated as an addition of the protein–ligand complex and the difference of the AKT1 and EGFR proteins and ligands free energies. The total binding free energy calculated using the MM/GBSA model is the resultant of the participation of several protein–ligand interactions including van der Waals energy (ΔEvdW), electrostatic energy (ΔEele), and ΔGGB (electrostatic contribution to solvation free energy by generalized Born). The total binding free energies are provided in Table 4.
Table 4. Binding Free Energies of the Complexes Calculated by Implying the MM/GBSA Module.
| energy components | AKT1 | EGFR |
|---|---|---|
| ΔEvdW | –57.44 ± 0.47 | –58.29 ± 0.37 |
| ΔEele | 1.01 ± 0.20 | 0.23 ± 0.38 |
| ΔEGB | 19.12 ± 0.22 | 16.92 ± 0.40 |
| ΔEsurf | –6.06 ± 0.03 | –5.87 ± 0.02 |
| ΔGgas | –56.43 ± 0.49 | –58.05 ± 0.48 |
| ΔGsolv | 13.05 ± 0.22 | 11.05 ± 0.40 |
| ΔGtotal | –43.37 ± 0.52 | –47.00 ± 0.35 |
The ΔEvdW contribution of the EGFR-complex was slightly higher than that of the AKT1-complex. The electrostatic contribution was negligible in both complexes. The GB contribution showed that the AKT1-complex has higher GB value than the EGFR-complex. The total binding free energy of both complexes was −43.37 ± 0.52 for AKT1-complex and −47.00 ± 0.35 for EGFR-complex.
4. Discussion
Plant-derived product investigations have gained great attention in recent years.44 An in silico drug discovery technique, network pharmacology facilitates comprehension of the complex correlations within drugs and their potential target genes as well as their underlying action mechanisms.45 Furthermore, the variety of innovative drugs derived from plants brings systematic complications.46 Natural substances, primarily derived from herbs, are frequently used as potential therapies, such as taxol.30 Tuberculosis is indeed a persistent, contagious, and lethal disorder.47 Antibiotics such as pyrazinamide and rifampicin are being used as first-line therapies for tuberculosis.48,49 Unfortunately, a major challenge is the development of pathogenic antibiotic resistance to such contemporary medications.50 In the previous 46 years, only a few drugs, including pretomanid, delamanid, and bedaquiline, have been approved to relieve pathogens-resistant tuberculosis. However, using herbal remedies with antipathogenic extracts will not let the pathogens become resistant to them. A. indica has significant clinical value in terms of boosting human health. A. indica leaf extracts offer hope for the evolution of medicines, as it possesses antituberculosis, antimalarial, analgesic, antiulcer, antioxidant, anti-inflammatory, and antivenom properties. A. indica is a primary source of metabolites, including 134 chemical components, including peptides, flavonoids, phytosterols, glycerides, terpenoids, and phthalic acids. This step establishes a protocol for the investigation of biologically active components of A. indica as well as a unique pharmacological notion to investigate the molecular mechanism of A. indica for tuberculosis treatment.
In this investigation, a variety of potential target genes have mediated numerous disease-related signaling pathways. By targeting these genes that can control disease-related pathways, disease progression can be minimized. The PPI interaction analysis revealed that two potential target genes, AKT1 and EGFR, were closely linked to other potential targets. AKT1 is a serine/threonine protein kinase that belongs to the Akt kinase family (Akt1, Akt2, and Akt3).51 Mtb’s subcellular proliferation is controlled by the serine/threonine kinase AKT1.52 In mice macrophages, the AKT1 inhibitor H-89 was reported to minimize pathogen growth and AKT1 and AKT2 knockdown in human THP-1 cells decreased the intracellular proliferation of pathogens.53,54 AKT1 signaling can also influence the adaptive immune response, which includes T and B cells. T-cell activation and function are critical for TB infection control, and AKT1 may play a role in modulating T-cell responses. The epidermal growth factor receptor (EGFR), also known as ErbB1 or HER1, is a cell surface receptor that regulates cell development and differentiation in response to EGF and similar ligands. EGFR inhibition reported to regulate subcellular mycobacterial proliferation by promoting autophagy, leading to increased elimination of pathogen.55 EGFR is highly expressed on the surface of epithelial cells, especially those lining the respiratory system. In TB, the first point of interaction with Mtb is frequently in the lungs. EGFR activation can affect the function of lung epithelial cells and their response to Mtb infection. Although the EGFR can influence several aspects of the host response to tuberculosis, its role in tuberculosis pathogenesis is complex and context-dependent. More researches are needed to completely understand the mechanisms involved in the relationship between EGFR and tuberculosis infection.
Gene ontology (GO) enrichment analysis is critical in understanding the complex biological mechanisms underlying diseases, such as tuberculosis. Researchers acquire important insights into disease pathogenesis by categorizing genes into biological processes (BPs), cellular components (CCs), and molecular functions (MFs). BPs emphasize processes such as immunological response and apoptosis, offering light on how the host combats Mtb infection. CCs can identify the cellular sites where TB-related genes are active, which is important for understanding Mtb’s intracellular survival strategies, such as those identified in phagosomes or cytokine receptor complexes. MFs highlight the specific roles that genes perform, such as identifying kinases that modulate immunological signaling or transcription factors that control tuberculosis-related gene expression. The potential targets were mainly involved in serine-type peptidase activity, ligand-activated transcription binding activity, ABC-type xenobiotic transporter activity, and efflux transmembrane transporter activity. The pathway enrichment analysis revealed that potential targets are primarily involved in key signaling pathways related to tuberculosis and the HIF-1 signaling pathway. Hypoxia-inducible factor 1 (HIF-1) activation in the HIF-1 signaling pathway regulates mycobacteria infection through enhanced phagocytosis in infected cells.56
The interaction analysis of bioactive compounds, potential target genes, and signaling pathways highlighted the strong interrelation of aurantiamide, kaempherol, β-sitosterol, clionasterol, and stigmasterol in the compound–target–pathway network, indicating that they possess antituberculosis action. Moreover, the findings of docking studies confirmed the existence of a significant binding affinity between both the bioactive components and the receptor proteins. Of note, stigmasterol was prominently bound to the candidate targets AKT1 and EGFR with higher binding affinity. In accordance with subsequent network analysis, A. indica inhibits the primary target genes and has therapeutic benefits against tuberculosis. In light of network pharmacology, the current study elaborates on the active chemicals, their potential targets, and related pathways to treat tuberculosis (TB), providing a conceptual structure for more scientific investigation.
There were various limitations to our study. To begin, more testing is warranted to endorse our findings. Second, to improve the accuracy of network pharmacology analysis results, a larger library of traditional medicine target genes is necessary. Third, even after combining the findings of network pharmacology and molecular docking, we failed to fully comprehend A. indica’s particular therapeutic mechanism. Despite the fact that we have shown some basic encouraging results, in-depth research and clinical trials are needed to evaluate the potential of A. indica targeting TB and validate its medical applications.
5. Conclusions
The current study establishes a new scientific foundation for determining the efficacy of multitherapeutic components, multiple potential targets, and the evaluation of novel targets against targets against tuberculosis. Our findings reveal that active compounds from A. indica, such as aurantiamide, kaempherol, β-sitosterol, clionasterol, and stigmasterol, have a high affinity for AKT1 and EGFR. The current findings were corroborated by molecular docking and simulation studies. These findings will serve as a solid basis for future research on A. indica’s method of action against TB. However, the current study has some constraints since further chemical constituents and therapeutic investigation are required to validate our findings.
Acknowledgments
This project was funded by the Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, under Grant No. G: 660-141-1439. The authors, therefore, gratefully acknowledge with thanks DSR for technical and financial support.
Special Issue
Published as part of ACS Omegavirtual special issue “Phytochemistry.”
The authors declare no competing financial interest.
Special Issue
Published as part of ACS Omegavirtual special issue “Phytochemistry”.
References
- Jordao L.; Vieira O. V. Tuberculosis: new aspects of an old disease. Int. J. Cell Biol. 2011, 2011, 403623 10.1155/2011/403623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaiss W. M.; Thaiss C. C.; Thaiss C. A. Recent developments in the epidemiology and management of tuberculosis–new solutions to old problems?. Infect. Drug Resist. 2012, 5, 1–8. 10.2147/IDR.S27604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C. W. M.; et al. Epidemic and pandemic viral infections: impact on tuberculosis and the lung: A consensus by the World Association for Infectious Diseases and Immunological Disorders (WAidid), Global Tuberculosis Network (GTN), and members of the European Society of Clinical Microbiology and Infectious Diseases Study Group for Mycobacterial Infections (ESGMYC). Eur. Respir. J. 2020, 56 (4), 2001727 10.1183/13993003.01727-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntutela S.; Smith P.; Matika L.; et al. Efficacy of Artemisia afra phytotherapy in experimental tuberculosis. Tuberculosis 2009, 89, S33–S40. 10.1016/S1472-9792(09)70009-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan B.; Vaidya A. D.; Chorghade M. Ayurveda and natural products drug discovery. Curr. Sci. 2004, 10, 789–799. [Google Scholar]
- Ekor M. The growing use of herbal medicines: issues relating to adverse reactions and challenges in monitoring safety. Front. Pharmacol. 2014, 4, 177. 10.3389/fphar.2013.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani R.; Narayanasamy M.; Feussner K.-D. Plant-derived antimicrobials to fight against multi-drug-resistant human pathogens. 3 Biotech 2017, 7, 172. 10.1007/s13205-017-0848-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaiah D.; et al. Studies on phytochemical constituents of six Malaysian medicinal plants. J. Med. Plants Res. 2009, 3 (2), 67–72. [Google Scholar]
- Jayakumari M.; et al. Antibacterial potential of Acalypha Indica against human pathogens. Int. J. Current Res. 2010, 1, 001–004. [Google Scholar]
- Muzammil M. S.; et al. Anti-inflammatory studies on Acalypha indica L. leaves by membrane stabilization. Indian J. Nat. Prod. Resour. 2014, 5, 195–197. [Google Scholar]
- Kumar S. S.; Kumar C. V.; Vardhan A. V. Hepatoprotective activity of Acalypha indica linn against thioacetamide induced toxicity. Int. J. Pharm. Pharm. Sci. 2013, 5, 3–6. [Google Scholar]
- Kurandawad J. M.; Lakshman H. Diversity of the endophytic fungi isolated from Acalypha Indica Linn–A Promising medicinal plant. Int. J. Sci. Res. Publ. 2014, 4, 2250–3153. [Google Scholar]
- Hiremath S. P.; Rudresh K.; Badami S.; et al. Post-coital antifertility activity of Acalypha indica L. J. Ethnopharmacol. 1999, 67 (3), 253–258. 10.1016/S0378-8741(98)00213-X. [DOI] [PubMed] [Google Scholar]
- Ranju G.; et al. In vitro anthelmintic activity of Acalypha indica leaves extracts. Int. J. Res. Ayurveda Pharmacol. 2011, 2 (1), 247–249. [Google Scholar]
- Manjulatha K. Comparitive Study of Leaves and Roots Ethanolic Extracts of Acalypha Indica on Peptic Ulcers Induced by Physical and Chemical Agents in Rodents. VRI Phytomed. 2013, 1 (1), 19–25. [Google Scholar]
- Sanseera D.; et al. Antioxidant and anticancer activities from aerial parts of Acalypha indica Linn. Chiang Mai Univ. J. Nat. Sci. 2012, 11, 157–168. [Google Scholar]
- Jayaprakasam R.; Ravi T. Evaluation of anti arthritic activity of the root extract of Acalypha indica Linn. using in vitro techniques. Int. J. Phyto Pharm. 2012, 2, 169–173. [Google Scholar]
- Gupta R.; Thakur B.; Singh P.; et al. Anti-tuberculosis activity of selected medicinal plants against multi-drug resistant Mycobacterium tuberculosis isolates. Indian J. Med. Res. 2010, 131 (6), 809–813. [PubMed] [Google Scholar]
- Hopkins A. L. Network pharmacology. Nat. Biotechnol. 2007, 25 (10), 1110–1111. 10.1038/nbt1007-1110. [DOI] [PubMed] [Google Scholar]
- Song X.; Zhang Y.; Dai E.; et al. Mechanism of action of celastrol against rheumatoid arthritis: A network pharmacology analysis. Int. Immunopharmacol. 2019, 74, 105725 10.1016/j.intimp.2019.105725. [DOI] [PubMed] [Google Scholar]
- Hashemi D.; Ma X.; Ansari R.; et al. Design principles for the energy level tuning in donor/acceptor conjugated polymers. Phys. Chem. Chem. Phys. 2019, 21 (2), 789–799. 10.1039/C8CP03341B. [DOI] [PubMed] [Google Scholar]
- Luo T.-t.; Lu Y.; Yan S. k.; et al. Network pharmacology in research of Chinese medicine formula: methodology, application and prospective. Chin. J. Integr. Med. 2020, 26 (1), 72–80. 10.1007/s11655-019-3064-0. [DOI] [PubMed] [Google Scholar]
- Dong D.; Xu Z.; Zhong W.; et al. Parallelization of molecular docking: a review. Curr. Top. Med. Chem. 2018, 18 (12), 1015–1028. 10.2174/1568026618666180821145215. [DOI] [PubMed] [Google Scholar]
- Pinzi L.; Rastelli G. Molecular docking: shifting paradigms in drug discovery. Int. J. Mol. Sci. 2019, 20 (18), 4331. 10.3390/ijms20184331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lans C.; van Asseldonk T. J. M.. Dr. Duke’s Phytochemical and Ethnobotanical Databases, a Cornerstone in the Validation of Ethnoveterinary Medicinal Plants, as Demonstrated by Data on Pets in British Columbia; Springer: Cham, 2020; pp 219–246. [Google Scholar]
- Mohanraj K.; et al. IMPPAT: A curated database of Indian Medicinal Plants, Phytochemistry And Therapeutics. Sci. Rep. 2018, 8 (1), 4329 10.1038/s41598-018-22631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru J.; et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014, 6, 13 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller D.; et al. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014, 42 (W1), W32–W38. 10.1093/nar/gku293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl M.; Wiese S.; Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol. Biol. 2011, 291–303. 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- Nobili S.; et al. Natural compounds for cancer treatment and prevention. Pharmacol Res. 2009, 59 (6), 365–378. 10.1016/j.phrs.2009.01.017. [DOI] [PubMed] [Google Scholar]
- Burley S. K.; et al. Protein Data Bank (PDB): the single global macromolecular structure archive. Methods Mol. Biol. 2017, 1607, 627–641. 10.1007/978-1-4939-7000-1_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S. K.; Bhikadiya C.; Bi C.; et al. RCSB Protein Data Bank (RCSB. org): delivery of experimentally-determined PDB structures alongside one million computed structure models of proteins from artificial intelligence/machine learning. Nucleic Acids Res. 2023, 51 (D1), D488–D508. 10.1093/nar/gkac1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt J.; Santos-Martins D.; Tillack A. F.; et al. AutoDock Vina 1.2. 0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 2021, 61 (8), 3891–3898. 10.1021/acs.jcim.1c00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25 (13), 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Bowers K. J.; Chow D. E.; Xu H.. et al. In Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters, Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, 2006.
- Shivakumar D.; et al. Improving the prediction of absolute solvation free energies using the next generation OPLS force field. J. Chem. Theory Comput. 2012, 8 (8), 2553–2558. 10.1021/ct300203w. [DOI] [PubMed] [Google Scholar]
- Price D. J.; Brooks C. L. III A modified TIP3P water potential for simulation with Ewald summation. J. Chem. Phys. 2004, 121 (20), 10096–10103. 10.1063/1.1808117. [DOI] [PubMed] [Google Scholar]
- Sinha T.; Bandyopadhaya A. Ethno-pharmacological importance and valuable phytochemicals of Acalypha indica (L.) a review. J. Ethnopharmacol. 2012, 3, 360–368. [Google Scholar]
- Chekuri S.; et al. Acalypha indica L.-an important medicinal plant: a brief review of its pharmacological properties and restorative potential. Eur. J. Med. Plants 2020, 31 (11), 1–10. 10.9734/ejmp/2020/v31i1130294. [DOI] [Google Scholar]
- Ravi S.; et al. Identification of food preservative, stress relief compounds by GC–MS and HR-LC/Q-TOF/MS; evaluation of antioxidant activity of Acalypha indica leaves methanolic extract (in vitro) and polyphenolic fraction (in vivo). J. Food Sci. Technol. 2017, 54, 1585–1596. 10.1007/s13197-017-2590-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivalingam A. S. GC–MS Analysis of Bioactive Compounds Present In Ethanolic Leaf Extract Acalypha indica. Asian J. Adv. Res. 2021, 11, 16–22. [Google Scholar]
- Rajkumar P.; et al. GC-MS, Phytochemical Analysis and In Silico Approaches of a Medicinal Plant Acalypha indica. J. Sci. Res. 2022, 14 (2), 671–684. 10.3329/jsr.v14i2.56648. [DOI] [Google Scholar]
- Ravi S.; et al. A Comprehensive Review on Traditional Knowledge, Phytochemistry and Pharmacological Properties of Acalypha indica L. Pharmacogn. Rev. 2021, 15 (30), 134–185. 10.5530/phrev.2021.15.16. [DOI] [Google Scholar]
- Hiebl V.; et al. Natural products as modulators of the nuclear receptors and metabolic sensors LXR, FXR and RXR. Biotechnol. Adv. 2018, 36 (6), 1657–1698. 10.1016/j.biotechadv.2018.03.003. [DOI] [PubMed] [Google Scholar]
- Berger S. I.; Iyengar R. Network analyses in systems pharmacology. Bioinformatics 2009, 25 (19), 2466–2472. 10.1093/bioinformatics/btp465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai F.-F.; et al. Yinchenhao decoction suppresses rat liver fibrosis involved in an apoptosis regulation mechanism based on network pharmacology and transcriptomic analysis. Biomed. Pharmacother. 2019, 114, 108863 10.1016/j.biopha.2019.108863. [DOI] [PubMed] [Google Scholar]
- Combrink M.; du Preez I.; et al. Metabolomics describes previously unknown toxicity mechanisms of isoniazid and rifampicin. Toxicol. Lett. 2020, 322, 104–110. 10.1016/j.toxlet.2020.01.018. [DOI] [PubMed] [Google Scholar]
- Türktaş H.; et al. Hepatotoxicity of antituberculosis therapy (rifampicin, isoniazid and pyrazinamide) or viral hepatitis. Tubercle Lung Dis. 1994, 75 (1), 58–60. 10.1016/0962-8479(94)90104-X. [DOI] [PubMed] [Google Scholar]
- Halsey N. A.; et al. Randomised trial of isoniazid versus rifampicin and pyrazinamide for prevention of tuberculosis in HIV-1 infection. Lancet 1998, 351 (9105), 786–792. 10.1016/S0140-6736(97)06532-X. [DOI] [PubMed] [Google Scholar]
- Lynch N.; Berry D. Differences in perceived risks and benefits of herbal, over-the-counter conventional, and prescribed conventional, medicines, and the implications of this for the safe and effective use of herbal products. Complement. Ther. Med. 2007, 15 (2), 84–91. 10.1016/j.ctim.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Cohen M. M. Jr. AKT genes and their roles in various disorders. Am. J. Med. Genet. A 2013, 161 (12), 2931–2937. 10.1002/ajmg.a.36101. [DOI] [PubMed] [Google Scholar]
- Wang X.; et al. AKT1 polymorphisms are associated with tuberculosis in the Chinese population. Int. J. Immunogenet. 2010, 37 (2), 97–101. 10.1111/j.1744-313X.2010.00897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijl C.; et al. Intracellular bacterial growth is controlled by a kinase network around PKB/AKT1. Nature 2007, 450 (7170), 725–730. 10.1038/nature06345. [DOI] [PubMed] [Google Scholar]
- Jayaswal S.; et al. Identification of host-dependent survival factors for intracellular Mycobacterium tuberculosis through an siRNA screen. PLoS Pathog. 2010, 6 (4), e1000839 10.1371/journal.ppat.1000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley S. A.; et al. Identification of host-targeted small molecules that restrict intracellular Mycobacterium tuberculosis growth. PLoS Pathog. 2014, 10 (2), e1003946 10.1371/journal.ppat.1003946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; et al. Activation of hypoxia-inducible factor 1 (Hif-1) enhanced bactericidal effects of macrophages to Mycobacterium tuberculosis. Tuberculosis 2021, 126, 102044 10.1016/j.tube.2020.102044. [DOI] [PubMed] [Google Scholar]