Abstract
Carbon dot (CD) nanoparticles offer tremendous advantages as fluorescent probes in bioimaging and biosensing; however, they lack specific affinity for biomolecules, limiting their practical applications in selective targeting. Nanoparticles with intrinsic affinity for a target have applications in imaging, cytometry, therapeutics, etc. Toward that end, we report the transferrin receptor (CD71) targeting CDs, synthesized for the first time. The formation of these particles is truly groundbreaking, as direct tuning of nanoparticle affinity was achieved by simple and careful precursor selection of a protein, which has the targeting characteristic of interest. We hypothesized that the retention of the original protein’s peptides on the nanoparticle surface provides the CDs with some of the function of the precursor protein, enabling selective binding to the protein’s receptor. This was confirmed with FTIR (Fourier transform infrared) data and subsequent affinity-based cell assays. These transferrin (Tf)-derived CDs have been shown to possess an affinity for CD71, a cancer biomarker that is ubiquitously expressed in nearly every cancer cell line due to its central role mediating the uptake of cellular iron. The CDs were tested using the human leukemia cell line HL60 and demonstrated the selective targeting of CD71 and specific triggering of transferrin-mediated endocytosis via clathrin-coated pits. The particle characterization results reflect a carbon-based nanoparticle with bright violet fluorescence and 7.9% quantum yield in aqueous solution. These unpresented CDs proved to retain the functional properties of the precursor protein. Indicating that this process can be repeated for other disease biomarkers for applications ranging from biosensing and diagnostic bioimaging to targeted therapeutics.
Introduction
Traditional methods of affinity-based sensing and imaging frequently utilize multicomponent probes with a biorecognition element to detect the analyte of interest. Typically, the recognition molecules of choice are antibodies, other proteins, or aptamers.1−3 The resulting affinity biomoieties are most commonly comprised of antigen–antibody, DNA-oligonucleotides, or conjugated protein–protein complexes.4 Affinity biosensors must be selective and stable in order to form an analyte–receptor complex where a measurable signal can be detected.3,4 Antibodies (∼150 kDa) are currently the gold standard for clinical-assay-based detection methods. However, antibodies are limited by their stability, bulky size, and inability to interact with intracellular targets. These factors result in a subsequent reduction of sensitivity for the target biomolecule. Antibodies and aptamers are subject to enzymatic cleavage and degrade swiftly in biological systems when in contact with proteases and nucleases, respectively.5,6 Antibodies are also sensitive to changes in pH, salinity, temperature, glycosylation, light exposure, and agitation.7 Stability within a biological environment is especially an issue when conjugating the antibody to a nanoparticle or drug, where studies indicate that antibody conjugates are even less stable than the unchanged antibody protein.8 The main benefit of antibodies is their near unparalleled target specificity; however, once bound the antibody–antigen complex is near irreversible, excluding high sensitivity.9 This is by design; antibodies have evolved to tag antigens to alert specialized immune cells. As such, antibodies are difficult to use for the continuous measurement of real samples.
Most protein–protein interactions of interest occur intracellularly. Antibodies have been used to visualize protein interactions and targets for cancer diagnostics and research. However, the cell membrane impermeability of antibodies limits the feasibility of intracellularly targeted therapeutics and diagnostics. Therefore, antibody-based sensing is almost exclusively limited to cell surface proteins, which accounts for less than 1% of protein–protein interactions.10 While certain cell processes, such as endocytosis, can be exploited to get antibodies into a cell, once there the protease-rich environment results in degradation of the antibody, or antibody-complex linkage.6,11 Moreover, conjugating drugs or nanomaterial to an antibody can affect the stability and affinity of the recognition molecule.8,12 The coupling of an antibody or aptamer to a nanoparticle results in a physical space being inserted between the fluorophore and the targeted biomolecule. Affinity biomolecules such as antibodies can range from 10 nm to over 40 nm in size, which negatively impacts the resulting resolution of super-resolution fluorescence microscopy images captured using these types of probes.13
A cutting-edge solution for such technical hindrances will require a biosensing and luminescent nanoparticle that possesses innate selectivity for a target biomolecule without the restrictions of conventional conjugation-based sensing techniques. One such nanoparticle is the carbon dot (CD), which is a type of quantum dot typically 1–10 nm in diameter and has been shown to possess surface chemistry and chemical function similar to that of its carbon precursor.14,15 Fluorescent CDs derived from proteins have also been shown to have sensing capabilities.16 Recently, our lab has developed and reported a novel synthesis process where fibronectin-derived photoluminescent CDs selectively bound to breast cancer cells’ extracellular matrix where the fibronectin receptor, integrin α5β1, resides.17 The success of the fibronectin-derived affinity CDs indicates that more protein based affinity CDs could be synthesized using our method to produce particles that can target a plethora of biomarkers of interest.
Transferrin-mediated endocytosis is a cellular uptake pathway of interest due to its ability to internalize material and the high expression of the transferrin receptor (CD71) in most cancer cell lines.18 Transferrin is an 80 kDa glycoprotein whose primary function is to bind ferric iron circulating in the body and then transport and deliver the iron into cells. Iron is an essential metal cofactor used by numerous proteins that mediate important cell processes such as cellular respiration, DNA synthesis, and proliferation.19 Transferrin selectively binds to CD71, triggering transferrin-mediated endocytosis (Figure 1). This is the primary mechanism of iron uptake for cells. Due to iron’s important functions in cell growth, cancer cells have an increased demand for iron. As such, higher expression of CD71 is seen in malignant cells, and CD71 levels have been correlated to level of malignancy.18,20 Benign and cancerous cells have differing levels of CD71 expression with some cancer cell lines exhibiting several hundred-fold higher levels of CD71 expression.19
Figure 1.
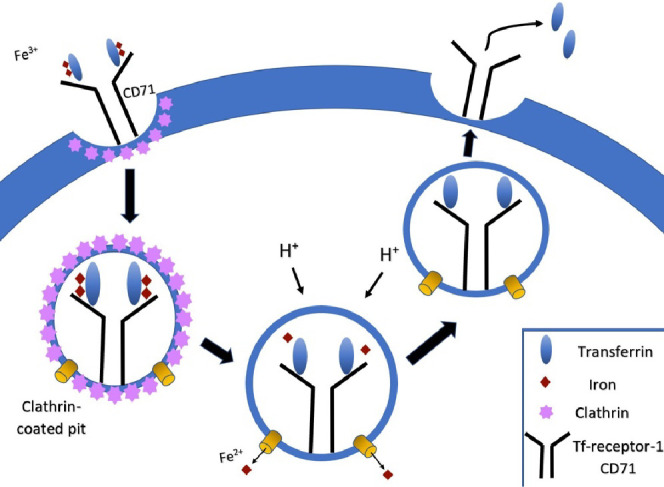
Process of transferrin-mediated endocytosis. Each transferrin protein binds two ferric ions and then binds to CD71. This triggers cell membrane invagination and formation of clathrin-coated pits. The vesicles coated in the clathrin protein then pinch off the membrane and move the transferrin-CD71 complex into the cell. The clathrin coat in the vesicle is degraded, leaving the complex in an endosome. A reduction in pH to 5.5 causes transferrin to lose its affinity for the ferric ions, which are then released into the cell, and the transferrin either is recycled back out of the cell or is sent along the transferrin degradation pathway. The transferrin-CD71 recycling pathway is depicted here, as it is the most common route. This is the primary mechanism of iron uptake into cells.
In the past few years, several publications have begun to report using proteins and other biomass carbon precursors during the synthesis. There are dozens of biological based reported precursors ranging from proteins such as bovine serum albumin (BSA) to plants such as Tinospora cordifolia to food-based products such as milk.21−23 In overwhelming majority of cases, the biomass carbon source was used in large part due to the environmentally friendly nature of the carbon precursor for bioimaging applications.24 In several studies, it was noticed that by varying the carbon precursor, the resulting CD selectivity was subsequently changed. Studies done by Liu et al. have reported that by varying the biological sources, CDs were synthesized with intrinsic selectivity for various metal ions.25 Charkaborty et al. reported using hemoglobin-derived CDs to sense H2O2.26 Oftentimes, the rationale for selecting a precursor is not given.
There is, therefore, a largely unexplored area of rational design used in CD research where precursors are carefully selected based on known affinity. Several researchers are beginning to explore this new area of predesigned CD affinity where Li et al. reported using amino acids found known to be antimicrobial in order to develop antimicrobial CDs.27 There is a need to bridge the understanding that CDs can retain the original precursor functionality and the thousands of cancer biomarkers that can be targeting by utilizing ligands with specific affinity for the biomarker of interest. While understanding which biological carbon precursors facilitate affinity CDs of interest is certainly a gap in the knowledge that requires deep exploration, the focus of this publication is to bring the knowledge gained in metal and chemical sensing CD research to the benefit of targeted cancer biosensing and ideally targeted therapeutics.
We have designed and developed a simple yet innovative affinity CD that possesses intrinsic selectivity and specificity for CD71 by synthesizing for the first time a nanoparticle with transferrin protein as the carbon precursor. Here, we examine the properties and CD71 selectivity of transferrin-derived carbon dots along with their potential to be used in biosensing applications. First, we synthesized the nitrogen-doped nanoparticles out of transferrin protein, and then we purified the particles and characterized the chemical and physical properties of the resulting nanomaterial. Finally, we confirmed receptor affinity via two different cell-based assays using the HL60 human leukemia cell line.
The affinity of the Tf-derived CDs is believed to be due to the retention of transferrin amino acid sequence functional groups, enabling active selective binding to the CD71 receptor and triggering CD71-mediated endocytosis. Previous literature supports the hypothesis that carbon precursor properties are retained by the subsequent CD formation due to incomplete carbonization enabling inherent selectivity of the CD.28−30 Since only 8.2 mg of protein is used in the synthesis, and the amount of carbon precursor can vary based on total sample volume needs, this synthesis method is consistent with other cost-effective and economical nanoparticle synthesis methods.31−33 We foresee this new type of nanoparticle to be an attractive alternative for traditional biosensing recognition molecules, such as antibodies or conjugated nanoparticles. These protein-derived affinity CDs offer nearly limitless customization as the physical and chemical properties of the particles such as affinity, size, doped heteroatoms, fluorescence intensity, and emission wavelength can all be directly tuned for whatever application required of the CD.
Results and Discussion
Protein-Derived Carbon Dot Size and Morphology
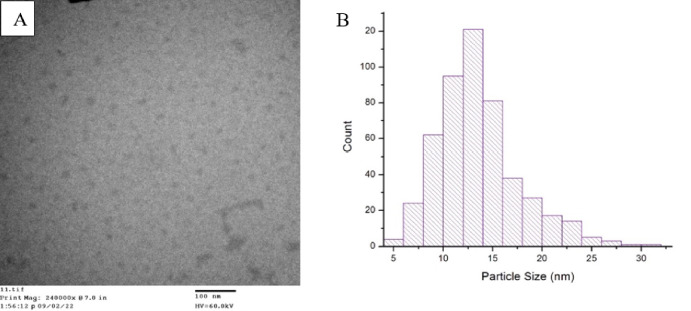
Transmission electron microscopy (TEM) was conducted to confirm the CD formation. The TEM images showed abundant material with sizes and morphology consistent with CDs.34 The average size of the CDs is 13 nm, with a size range of 5–32 nm (Figure 2). These particles range in size from qualifying as a carbon dot (<20 nm) to entering the size range of a small nanoparticle (>20 nm).15 The larger size of these protein-derived nanoparticles, compared to those of other CDs reported, might be due to the size of the carbon precursor. We implemented the “bottom-up” synthesis method for manufacturing affinity carbon dots. The “bottom-up” technique utilizes smaller molecules that are fragmented then chemically assembled into carbon nanostructures resulting in a high-yield of particles.35 However, the size of more commonly used organic small molecules for carbon dot synthesis, such as citric acid (192.124 g/mol) or glucose (180.156 g/mol), is much less than other precursors such has proteins like transferrin (80 kDa).36−38 Therefore, the larger carbon precursor could result in a larger average particle diameter due to there being more initial material, in terms of mass, during the synthesis process. We hypothesize that some amount of the large size we observe in the TEM is due to the presence of the original protein’s peptides retained in the structure and on the surface of the CDs. Nanoparticles conjugated to peptides have been known to drastically increase in size, indicating that peptide surface chemistry can impact overall particle diameter.39,40
Figure 2.
(A) TEM image of transferrin-derived carbon dots on a 3 nm carbon substrate. Size and morphology consistent with CD formation indicating successful synthesis of carbon nanoparticles. (B) Histogram of transferrin protein-derived carbon dots’ size range. The diameters vary in size from 5 to 32 nm, with the average diameter being 13 nm.
The larger diameter of these protein-derived affinity CDs may have an additional benefit over other quantum dots or small molecules when targeting CD71 because receptor-mediated endocytosis requires a membrane wrapping process triggered by an affinity ligand binding to the receptor. However, thermodynamics limit the size of the particle that can efficiently prompt membrane wrapping processes.41−43 For spherical nanoparticles, this enthalpic limit is about 30 nm, where particles smaller than this cannot efficiently drive the membrane wrapping processes of endocytosis. Additionally, nanoparticles larger in diameter than 60 nm are hindered from effective cellular uptake due to binding to a vast quantity of receptors and prohibiting multiple nanoparticles binding and triggering receptor-mediated endocytosis at the receptor of interest.44 In general, in vitro cell assays indicate that optimal receptor-mediated particle internalization occurs within the 10–60 nm range.42,45−47 Thus, small molecules and quantum dots below 10 nm in size are unable to trigger most cell uptake pathways.42,44 The CD71 affinity CDs described here are well within this optimal cell uptake size range and should, therefore, more efficiently bind to CD71 and trigger receptor-mediated endocytosis than small molecules, peptides, or quantum dots.
Chemical Composition of Transferrin Carbon Dots
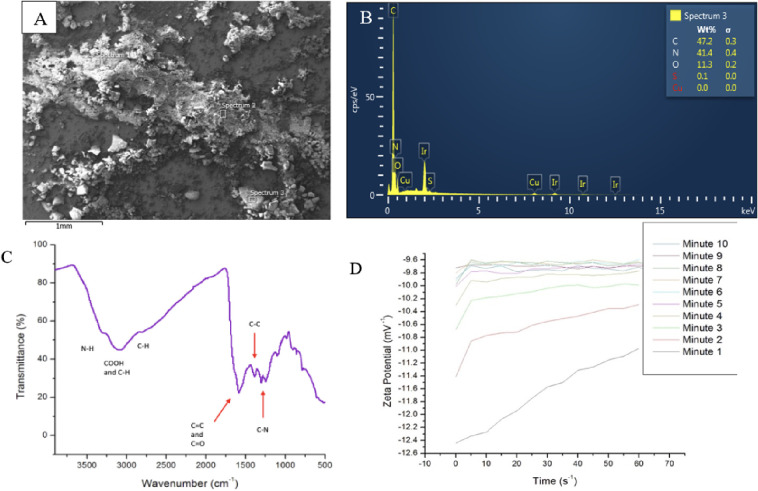
Nitrogen doping of the nanomaterial was confirmed with an SEM equipped with EDS capabilities. We see strong peaks confirming the presence of carbon and nitrogen, with a moderate peak indicating some oxygen present in the sample. The nitrogen content in the sample nearly rivals that of carbon in weight percent. This is a much higher proportion of nitrogen into the carbon skeleton than seen in other protein-derived CD samples manufactured via 700 W of microwave radiation.17 Our hypothesis is that there is an energy barrier that the nitrogen must overcome in order to be inserted into the carbon core. The higher temperature of the oven overcame this energy barrier and made the process of heteroatom doping into protein CDs more efficient. The SEM images in Figure S2 show homogeneous distribution of all the carbon, oxygen, and nitrogen species. Some regions of the sample also yielded trace elements, such as iron and sulfur (Figure S2). The trace levels of sulfur are thought to be from the disulfide bonds found in the transferrin protein.
FTIR-ATR was conducted on the transferrin-derived carbon dots (Figure 3C). An intense peak at 1600–1585 cm–1 designates the aromatic C–C bend, and 1680–1640 cm–1 labels the aromatic C=C bond. Both peaks confirm a graphene lattice core. A broad peak at 2800–3500 cm–1 indicates the presence of an O–H stretch found in carboxylic acids. Moderate peaks show primary amine N–H stretches at 3400–3300 cm–1 and 3330–3250 cm–1. The presence of primary amines and carboxylic acids suggests that amino acids might be present on the nanoparticles as each amino acid sequence begins and is terminated by the primary amine and carboxylic acid. Additional peaks such as the C–N aromatic amine stretch at 1340–1260 cm–1, the C=O stretch at 1650–1850 cm–1, the C–H aromatic stretch at 3030 cm–1, and the C–H stretch at 2800 cm–1 are all found in various amino acid structures.
Figure 3.
(A) SEM electron image of bulk transferrin-derived nanoparticles. (B) EDS spectrum of a spot location of the transferrin-derived nanomaterial. The majority of the sample is made of carbon, then nitrogen, and then oxygen with trace amounts of sulfur. The sample was sputter coated with iridium to reduce surface charging. (C) ATR-FTIR spectrum of CD71 affinity CDs. Strong peaks at 1600–1585 cm–1 and 1680–1640 cm–1 indicate a graphene CD core. Additional peaks indicate amino acid functional groups present in the nanoparticles. (D) Zeta potentials of 39 μM transferrin carbon dots in DI water. After about 5 min, the zeta potential of the nanoparticles stabilized to indicate a zeta potential of −9.7 mV–1.
The zeta potential of the CDs suspended in DI water was determined to be about −9.7 mV–1 after 10 min of measurement. It can be seen in Figure 3D that the zeta potential of the nanoparticles started around −12.5 mV–1 and over the next 10 min evened out to a −9.7 mV–1. The current hypothesis for the small zeta potential is that the number of primary amines far exceeds the number of carboxylic acids found on the surface of the carbon dot. The EDS data indicate that a much larger weight % of the bulk material is made up of nitrogen rather than oxygen, leading to a small negative charge on the particle instead of a larger negative charge (Figure 3A,B). Zeta potentials can be used to determine the stability of particles in a solution. High zeta potentials, either positive or negative, result in a stable mixture due to sufficient repulsion forces. However, small zeta potentials can indicate inferior physical stability due to interparticle attraction and aggregation.48,49
Fluorescent Properties of CD71 Affinity Carbon Dots
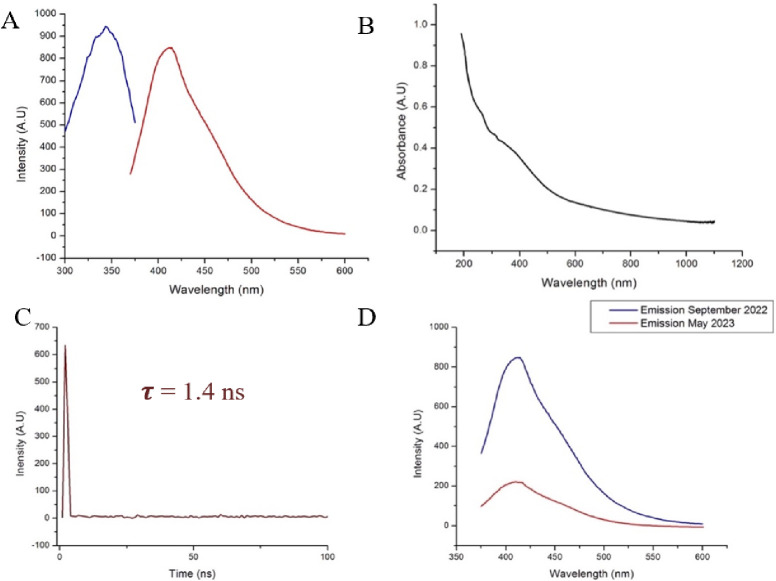
CDs are intrinsically capable of fluorescence emission and are distinct in their fluorescence capabilities in that the band gap fluorescence emission of CDs can be modulated.15 The conjugated π domain of the carbon core is believed to be the nanoparticles fluorescence center.50,51 The band gap between valence band and conduction band of the particle increases with decreasing particle size, resulting in enhanced quantum yield and fluorescent emission due to the band gap transition in the ultraviolet region.52 It can be seen in Figure 4A that the fluorescence emission of the transferrin CDs is 413 nm under 343 nm UV excitation. This indicates successful formation of luminescent CDs from the transferrin protein.
Figure 4.
(A) Fluorescence excitation (blue) and emission (red) spectra of 0.39 μM transferrin-derived affinity carbon dots in DI water. The peak excitation wavelength is 343 nm. At that excitation wavelength, the resulting peak emission wavelength is 413 nm. (B) The UV–visible spectrum was 0.39 μM. A slight peak is seen in the curve at from 300 to 400 nm. This is in the same region as the nanoparticles have peak absorption, as can be seen in the fluorescence emission spectrum. (C) Lifetime fluorescence emission decay of affinity CDs using time-correlated single photon spectroscopy. Equation 2 was used to calculate the lifetime decay and found to be 1.4 ns. (D) Fluorescence emission profile of transferrin nanoparticles. Maximum emission was observed at 413 nm under 343 nm excitation.
The affinity transferrin-derived CDs described here are larger than typical CDs with an average diameter of over 10 nm. The quantum yield of the transferrin CDs suspended in DI water was calculated to be 7.94%, where the standard used was 1 ppm of quinine sulfate in 0.05 M H2SO4. The quantum yield of quinine sulfate was 0.54. The area under the photoluminescent curve (I) of quinine sulfate was 15 760, and the absorbance intensity of quinine sulfate at 343 nm was 0.01459 A.U. (Figure S2). Previous protein-derived carbon dots manufactured in this lab had an average quantum yield (eq 1) of 1.417 ± 0.0085%.17 This indicates that the combined synergistic effect of codoping heteroatoms resulted in an enhanced quantum yield despite both emission wavelengths being similar. This is especially impressive as DI water does not seem to be a solvent that yields high photoemission intensity compared to other organic solvents such as ethanol.53,54 The fluorescence emission lifetime of the nanoparticles was calculated (eq2) via time-correlated single-photon counting spectroscopy (Figure 4) and was found to be 1.4 ns. The lifetime decay of the affinity carbon dots is, thus, on the same scale as traditional fluorophores used in bioimaging.55
A way to improve quantum yield in CDs would be doping in heteroatoms such as nitrogen into the carbon skeleton to redshift the particles emission as well as enhance quantum yield.56 Nitrogen is of similar radius size to that of carbon and can be easily integrated within the carbon structure forming new surface state energy levels lowering the band gap energy often resulting in red-shifted emission wavelengths and improved quantum yield.52 Formamide and ethylenediamine were the organic solvents used in conjunction with the transferrin protein. Both solvents are rich in nitrogen to aid in the incorporation of nitrogen into the carbon dot during particle nucleation and subsequent carbonization. Transferrin itself is a protein with multiple elements composing the main structure including carbon, oxygen, nitrogen, sulfur, and iron in the case of holo-transferrin.57 Nitrogen and sulfur codoped carbon dots both can aid in modulating the emission wavelength and improving quantum yield.58 Co-doping with multiple heteroatoms can have a synergistic effect where additional band gaps are formed resulting in a higher quantum yield and longer emission wavelengths.52
The small negative zeta potential of the transferrin-derived carbon dots stipulates the low physical stability of the nanoparticles in an aqueous solution. This is confirmed in Figure 4D, where the emission intensity of the CDs was tracked over a period of 9 months. At the end of the 9 months, the fluorescence emission intensity of the CDs had dropped 74.11%. However, even at the end of 9 months of storage and exposure to ambient temperatures and overhead lights, the same sample of CDs was still fluorescent. This is due to the conduction band electron bandgap transitions of conjugated π bond domains that make up the fluorescence mechanism of the CDs. This allows CDs to resist complete photobleaching for long periods of time.15,50,52
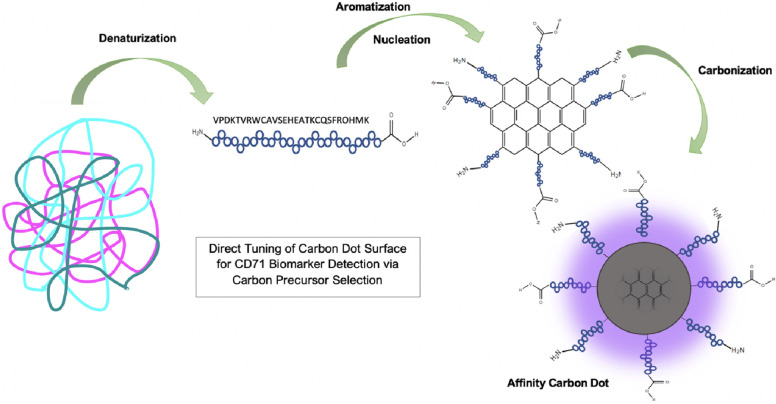
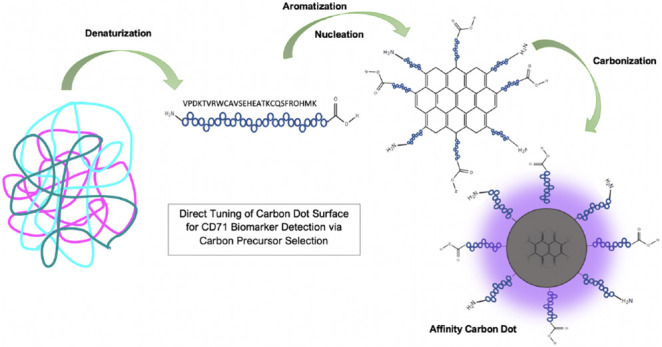
Using the data from Figures 3 and 4, the proposed mechanism of affinity CD synthesis is outlined in Figure 5. We hypothesize that the proteins are denatured to the amino acid peptide sequence when dissolved in solution and during initial heating. Upon further heating, a critical energy point is reached where the peptides begin to form aromatic C=C bonds and nucleate. Carbonization ensues as new C=C bonds appear at the surface of the particle using carbon and nitrogen supplied in the protein amino acid structure. A layered graphene-like core begins to build into a quasi-spherical particle. If the carbonization processes are ended before all the amino acids are carbonized into the graphene core, the unreacted amino acid peptides are left on the surface of the carbon nanoparticle (Figure 5).14,16 In this way, the CD retains the original proteins affinity for its receptor. This process provides an intrinsically fluorescent and selective CD, without the need for further modification.
Figure 5.
Proposed mechanism of affinity CD formation. Proteins are denatured, then form carbon aromatic rings, and then carbonize until a CD is formed.
CD71 Affinity Biosensing Assays
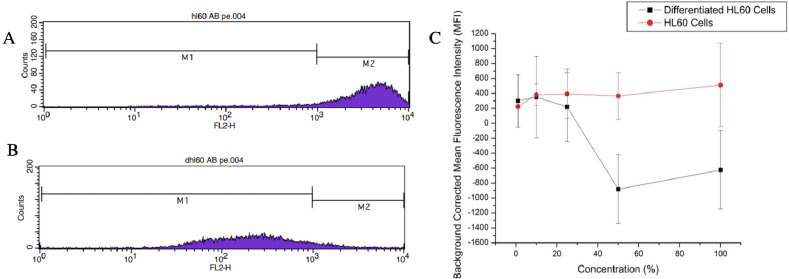
To evaluate the transferrin-derived CDs affinity for the transferrin receptor CD71, a differentiated cell assay was conducted. The HL60 cell line was cultured in 1.5% DMSO and differentiated to suppress the CD71 expression. The differentiation of the HL60 cells was confirmed with flow cytometry using Mouse Anti-Human CD71 antibody fluorescently tagged with PE (Figure 6A,B). Both differentiated and undifferentiated cell populations were then stained with varying concentrations of the transferrin-derived carbon dots ranging from 1% (0.39 μM) to 100% (39.25 μM). The cells were then imaged on a fluorescence microscope, the mean fluorescence intensity (MFI) of each stained cell population was calculated, and Student’s t-test was used to determine any statistical difference in the cells MFI (Figure 6C). The lower staining concentrations did not result in a statistically different MFI until the concentration reached 25% (9.75 μM), from there until 100% (39.25 μM) the differentiated and undifferentiated HL60 cells possessed statistically different MFI. Thus, at 9.75 μM staining concentrations and higher, we can determine nanoparticle affinity for the receptor.
Figure 6.
(A) Flow cytometry data showing undifferentiated HL60 cells incubated with the PE tagged CD71 antibody. (B) Flow cytometry data of differentiated HL60 cells incubated with the PE tagged CD71 antibody. The fluorescence intensity of the differentiated cells shifted to the left indicating the antibody had less transferrin receptors to bind to. Thus, CD71 expression was suppressed (Table S1). (C) Background corrected MFI of the differentiated and undifferentiated HL60 cells stained with transferrin-derived nanoparticles. Statistical difference indicates nanoparticle affinity for CD71 receptor.
In Figure 6, the differentiated cells decreased in fluorescence down into the negatives. This is because each image set measured the intensity of the cells and background corrected for the fluorescence in the solution the cells were imaged in. The differentiated cells have less intensity than the background due to the nanoparticles not entering the cell and staying in solution. This led to a “negative” background corrected MFI. The continuous increase of the undifferentiated cells MFI compared to the lowering of the differentiated cells MFI indicates nanoparticle affinity for transferrin receptor CD71. The HL60 cells expressing high levels of CD71 are internalizing more affinity CDs resulting in a measurable increase in cells’ fluorescence intensity. The HL60 cells expressing low levels of CD71 have a much lower uptake of the affinity carbon dots resulting in a lower fluorescence intensity. This indicates that the CDs are entering the cell through transferrin-mediated endocytosis and the CDs are selective for CD71, meaning we have successfully synthesized CD71 affinity CDs.
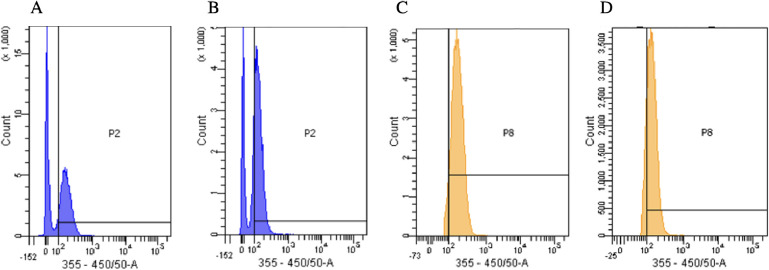
A competitive inhibition assay (Figure 7) was then conducted to confirm the results in Figure 6 and Table S1. A portion of the HL60 cells were incubated with biotinylated CD71 antibodies, and another were incubated in PBS only. The binding of the CD71 antibody inhibits internalization of transferrin as the antibody competes for the binding site and then irreversibly blocks transferrin uptake.59Figure 7 shows that the MFI of the cells stained with 3.90 μM affinity CDs was 187. However, the cells incubated with the CD71 antibody and then stained with the same concentration of affinity CDs had a lower MFI of 149 AU. Furthermore, a population closer between the autofluorescence of the cell and the fluorescence of the affinity CDs had a significant shift in fluorescence intensity. The MFI of the cells stained with the affinity CDs had an MFI of 186. The MFI of the cells shifted to 147 when they were incubated with the untagged CD71 antibody.
Figure 7.
(A) Flow cytometry data of HL60 cells stained with 3.90 μM affinity CDs. Population 2 (P2) indicated the fluorescence of the affinity CDs and expressed an MFI of 187. (B) HL60 cells incubated in biotinylated CD71 antibody and then stained with 3.0 μM CD71 affinity CDs. Population MFI of 149. The decrease in MFI indicates that the nanoparticles have affinity for CD71. (C) Another population of cells, named population 8 (P8) adjacent to P2, showed a similar shift. The cells stained with 3.90 μM affinity CDs yielded an MFI of 186. (D) However, the cells incubated with the CD71 antibody and then stained with 3.90 μM affinity CDs resulted in a lower MFI of 147.
Both populations have decreased fluorescence intensity when the CD71 antibody blocks the CD71 receptor, preventing selective binding of the affinity CDs and resulting in less internalization of the nanoparticles. These results show that even at the relatively low concentration of 3.90 μM affinity CDs, there is a measurable difference in uptake of CDs between the control and test cell samples whereas a concentration of 9.75 μM was the cutoff for statistical difference in MFI for the cell differentiation experiment seen in Figure 6. Literature works previously report CDs formed using ethylenediamine have a carbon source.60 In order to prove that the selectivity of the CD71 receptor was due to the inclusion of a transferrin precursor, CDs synthesized from formamide ethylenediamine were tested for selectivity to transferrin. The final product had a different consistency and color than the original Tf-derived nanoparticles. We then performed a competitive inhibition assay with HL60 cells under the exact same conditions. Cells were analyzed via flow cytometry, as previously described. The cells stained with the ethylenediamine-formamide derived CD had mean fluorescence intensities of 184 and 180 in the two population gates. The cells first incubated with the CD71 antibody and then stained with the ethylenediamine-formamide derived particles had a mean fluorescence intensity of 192 and 187. Since the CD71 inhibited cells did not show decreased fluorescence, this indicates that the nanoparticles are not entering the cell through the CD71-mediated pathway and are not selective. Therefore, the Tf precursor provides selectivity via retention of functional groups. The data show a slight increase in MFI when inhibited but this could be explained by the slight MFI increase attributed to autofluorescence of the biotinylated antibody added to the autofluorescence of the cells (Figures S5 and S7). This flow cytometry data confirms the differentiation experiment conclusion that transferrin-derived CDs possess an affinity for CD71.
Biocompatibility of CD71 Affinity Carbon Dots
Biocompatibility tests were performed to determine the toxicity effects of the affinity CDs on HL60 cells (Figure 8). The experiment was conducted using flow cytometry to determine rate of HL60 cell death when stained with 3.9 μM of affinity CDs. The cells were costained with PI to identify occurrence of cell death as PI cannot permeate the intact cell membrane of a living cell. Additionally, PI emits in the red region of the electromagnetic spectrum and can be excited by a 488 nm laser and its emission detected by a 610/20 bandpass. This enables minimal overlap with the CDs, which emit in the violet region of the spectrum and are excited with a 355 nm laser.
Figure 8.
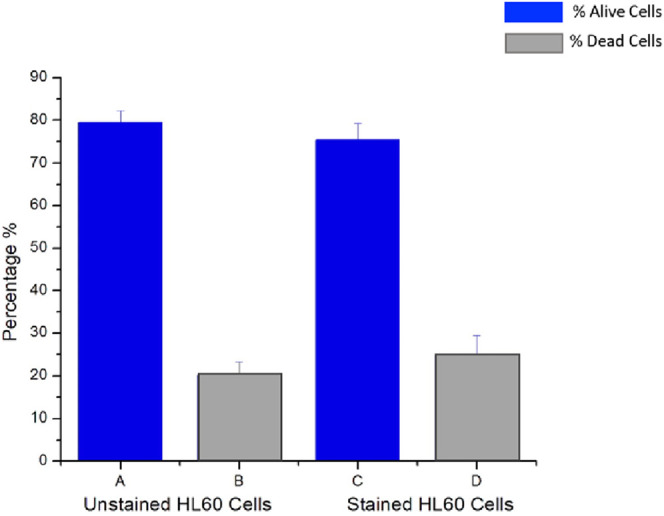
Flow cytometry was used to determine the biocompatibility of HL60 cells stained with 3.9 μM affinity carbon dots. The p-value is 0.218747; at 95% CI indicate no statistical increase in cell death.
Student’s t test was done to determine if there was a statistical difference in cell death. A p value of 0.218747 shows that the results were not statistically different (with a critical value of 0.05). Thus, there was no statistical difference in cell death between the control sample and the cells stained with affinity CDs. This indicates a high level of biocompatibility between affinity CDs and cells. Higher concentrations of CDs were unable to be analyzed in the same manner as the solution became so opaque that the detector was unable to detect the emission of the PI.
Cell Imaging Via CD71 Affinity Nanoparticles
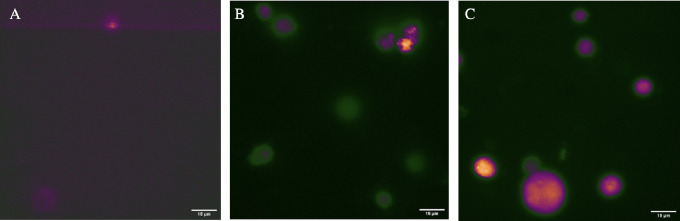
Fluorescence microscope images were taken of the HL60 cells stained with CD71 affinity CDs with concentrations ranging from 0.39 to 5.85 μM. A UV filter cube was used to isolate the proper excitation and emission wavelengths for these CDs. It can be seen in Figure 9 that the nanoparticles were internalized into the cell and had minimal accumulation into small organelles. The distribution of affinity nanoparticles appears to be homogeneous across the cell. This distribution is to be expected as CD71 receptors densely populate the entire cell membrane, constantly internalizing transferrin that is then shuttled into the cell and then either degraded or recycled back out of the cell.61 Comparing the control sample of cells with no affinity CDs added to the sample, the autofluorescence of the HL60 leukemia cells is significantly less than the images of cells stained with 0.39 and 5.85 μM affinity CDs. Therefore, even at the lowest concentration (0.39 μM) and 5 months after CDs synthesis, the nanoparticles still have a suitable fluorescence emission intensity for bioimaging based experiments.
Figure 9.
Fluorescence microscopy image of HL60 cells under UV excitation using an oil immersion 100× objective. (A) HL60 cells with no CD71 affinity CDs. (B) HL60 cells stained with 0.39 μM CD71 affinity CDs and (C) 5.85 μM CD71 affinity CDs.
Some bright points in the cells in Figure 9B,C could possibly be an endosome loaded with nanoparticles or loaded into a lysosome if the transferrin-derived particle is following the degradation pathway. Additionally, in Figure 9B, some of the cells appear not to have as much internalization of the affinity nanoparticles within the cell, hence, the lower fluorescence intensity. However, in Figure S6, the white light images show that some of the cells were not within the plane of focus on the microscope, explaining the lack of bright internal fluorescence. Higher resolution images, such as from super resolution microscopy, could be done to have a better understanding of the particle’s intracellular movement. While it is clear from Figures 6 and 7 that the affinity CDs are entering cells via the transferrin-mediated endocytosis iron uptake pathway, the exact effect of the affinity nanoparticles are unknown. It is also unknown what becomes of the affinity CDs once inside the cell. Future work will focus on understanding the role these affinity CDs have within cancer cells.
Conclusion
Traditional biosensing techniques rely on affinity biomolecules, such as antibodies or aptamers, to target various receptors and proteins in biological environments. These standard recognition molecules are not without limits including an inability to enter a cell, large physical size, and instability.9,11,13,62 Here, we describe an affinity CD that is fundamentally fluorescent and selective for the target of interest without the need for any post synthesis modification. The CDs were synthesized out of holotransferrin and were found to have an average diameter of 13 nm. The CDs were nitrogen doped using formamide and ethylenediamine. The affinity CDs appear to retain transferrin’s affinity for CD71 as differentiation assays and competitive binding cell assays were conducted on the leukemia HL60 cell line to determine nanoparticle affinity for CD71. The nanoparticles were determined to be selective for CD71, and this is believed to be due to the incomplete carbonization of the proteins polypeptides resulting in a layered graphene like core with a transferrin amino acid functionalized surface chemistry.
The retained amino acid functional groups of the transferrin are then thought to interact with CD71 enabling binding and subsequent triggering of transferrin-mediated endocytosis. The direct binding capabilities of these nanoparticles to transferrin enable it to be internalized into the cell and used for bioimaging applications. These affinity CDs remove the physical distance between probe and target caused by linker molecules or bulky biomolecules and enable direct and precise labeling of the target, ensuring accurate and high-resolution final images. The lack of linked or conjugated fluorophores or recognition molecules also removes added instabilities where the cleave of any of those linkage sites results in loss of signal or function of the biosensing affinity complex.8,12 While these affinity CDs offer a new approach to affinity biosensing and bioimaging, much is unknown about their interactions with at the cellular level. Future work must be done to evaluate the effect affinity CDs have on biological systems, such as how they specifically trigger receptor-mediated endocytosis and their subsequent rate of uptake into the cell. Future studies will also need to be conducted to test the efficacy of affinity carbon dots compared to more established techniques. Ultimately, affinity CDs are a new platform in nanomaterial science with cross-cutting applications across varying fields with potential uses in cancer research, disease diagnostics, and potentially even targeted therapeutics.
Materials and Methods
Materials
Holo-Transferrin derived from human blood plasma was obtained from R&D systems. Ethylenediamine and formamide were acquired from Fisher chemical. Acetone was obtained from Macron fine chemicals, and the phosphate-buffered saline (PBS) solution was secured from Mediatech, Inc. NaCl was obtained from Sigma, and the propidium iodide (PI) was acquired from Molecular Probes Invitrogen Detection Technologies. PE (R-phycoerythrin) Mouse Anti-Human CD71 and biotinylated mouse Antihuman CD71 was procured from BD Pharmigen. TEM support films Carbon Type B 400 mesh Cu were obtained from TED Pella, Inc. Human leukemia cell line (HL60) procured from the American Type Culture Collection (ATCC), was cultured in RPMI 1640 medium (Hyclone) containing 1% penicillin-streptomycin (Sigma-Aldrich) and 10% fetal bovine serum (FBS, Hyclone), and was incubated at 37 °C with 5% CO2.
General Synthesis Procedure for Transferrin-Derived Carbon Dots
8.2 mg of transferrin was suspended in 20 mL of formamide and vortexed until completely dissolved. Then 0.525 mL of ethylenediamine was incorporated into the solution and vortexed until thoroughly mixed. The homogeneous solution was then transferred to an acid digestion vessel (Parr Acid Digestion Vessel 4748 125 mL Large Capacity Vessel) and placed into an oven (Thermo Scientific PR305225G Precision Compact Gravity Oven). The oven was set to the maximum temperature (an average of 214 °C with a heating cycle range of 206–222 °C), and then the vessel was heated for 6 h. Upon completion of the heating phase, the sample vessel was removed from the oven and left to cool back down to room temperature. Once cooled, the solution was filtered with a 0.20 μm pore Nylon disk filter. The nanoparticles were then precipitated from the filtered solution with 50 mL of acetone. The precipitated nanoparticle solution was then transferred into Teflon centrifuge tubes (Thermo Scientific Oak ridge FEP 50 mL). The solution was then centrifuged at 10,000 rpm where the supernatant was removed leaving the nanoparticle precipitate. The nanoparticle pellet was then washed twice by a 1:1 (v/v) acetone and ethanol mixture. After the final wash, the nanoparticle precipitate was suspended in wash solution, transferred to a Petri dish, and left in a fume hood to evaporate off the organic solvents, leaving behind the CDs. Once completely dried, the CD nanoparticles can be suspended in any desired solution of any volume. For these experiments, the nanoparticles were suspended in 50 mL of ultrapure water or left as a solid, depending on the requirements of the characterization method. All raw data are available upon request.
Characterization of Transferrin-Derived Carbon Dots
Nanoparticle fluorescence spectra were acquired with the Agilent Cary eclipse Fluorescence Spectrometer. Ultraviolet–visible (UV–vis) absorption data were obtained with the Agilent 8453 UV–vis Spectrometer. Quantum yield was calculated using the following equation:
| 1 |
where QQS is the quantum yield of quinine sulfate standard, QCD is the quantum yield of the CDs sample, and ηx is the refractive index of the CDs sample or quinine sulfate standard. Refractive index for sample and reference: water = 1.33. ICD is the area under the photoluminescent curve (I) of the sample, IQS is area under the photoluminescent curve (I) of the reference, ACD is absorbance intensity of the sample, and AQS is absorbance intensity of the reference. Lifetime decay of the transferrin-derived carbon dots was calculated using the lifetime decay equation:
| 2 |
The lifetime data were acquired using a Horiba Johbin Yvon single Photon Counting Controller Fluorohub with a MicroHR Horiba Jobin Yvon spectrometer. The zeta potential of the nanoparticle solution was obtained via a Microtrac Stabino particle charge mapping system. Transmission electron microscopy (TEM) of the nanomaterial sample was conducted on a Hitachi H-7650. Scanning electron microscopy (SEM) was conducted on a Hitachi H-4300 SEM instrument rigged with an EDAX Energy Dispersive X-ray (EDS) spectrometer. A Thermo Scientific Nicolet iS10 Smart ITR FTIR instrument with an ATR crystal was used to acquire the FTIR spectrum of the nanoparticles. For flow cytometry data, a BD FACSAria cell sorter as well as a BD FACSCalibur flow cytometer were utilized. Epifluorescence microscopy images were taken using a Nikon Eclipse Ti2 inverted microscope fitted with an sCMOS PCO edge camera and a high-powered LED light source.
Differentiated HL60 Cell Line CD71 Affinity Assay of Transferrin Carbon Dots
Two flasks of HL60 cells were cultured, where one flask was cultured with RPMI 1640 medium (undifferentiated HL60 cell line) and the other flask was cultured in RPMI 1640 medium with 1.5% (v/v) dimethyl sulfoxide (DMSO), which suppresses CD71 expression (differentiated cell line).63 After sufficient population growth, cells were incubated in affinity carbon dot solutions for 1 h at 1%, 10%, 25%, 50%, and 100% concentrations diluted in PBS, along with a control with no nanoparticle staining. Additionally, small aliquots of differentiated and undifferentiated cells were stained with a fluorescently labeled CD71 antibody to confirm successful differentiation via flow cytometry and fluorescence microscopy. After the incubation period ended, the stained cells were then imaged on the microscope using a 20× objective, with an exposure time of 10 ms and under UV excitation via a DAPI filter cube with a 378/52 excitation bandpass filter and a 447/60 emission bandpass filter. The number of cells and the background subtracted mean fluorescence intensity of each cell was calculated using ImageJ software.
Anti-CD71 Competitive Inhibition CD71 Affinity Assay of Transferrin Carbon Dots
HL60 cells were cultured for 1 week to maximize cell population and then were split into two equal aliquots. One aliquot of cells was washed with PBS and then incubated with 100 μL of the untagged biotinylated CD71 antibody, and the other was washed with PBS and then incubated without the CD71 antibody. Afterward the cells were incubated for 30 min in a 10% affinity carbon dot diluted in PBS and washed in PBS twice. The mean fluorescence intensity of each aliquot of cells was then calculated using the FACS Aria flow cytometer with a 355 nm excitation laser to excite the CDs.
Biocompatibility Experiment of HL60 Cells and CD71 Affinity Carbon Dots
HL60 cells were cultured for a week and then washed with PBS to remove the used medium. The nanoparticles were then resuspended in a 10% affinity carbon dots solution diluted in PBS in 3 vials for triplicate measurements and left to incubate for 30 min. An additional 3 vials of cells were washed with PBS and incubated with no carbon dots added to serve as the control. Twenty microliters of PI was added to each vial of cells for 5 min and was then immediately analyzed on the flow cytometer. A flow cytometer equipped with a 488 nm laser was used to excite PI, which can enter a cell and bind to the cellular nuclear content only if the membrane is compromised. The number of dead and alive cells was then calculated based on differences in cellular fluorescence intensity by PI staining.
HL60 Cell Staining with CD71 Affinity Carbon Dots
HL60 cells were cultured for at least 2 days and then placed into vials and washed with PBS. The cells were then incubated for 30 min in 1%, 5%, and 15% nanoparticle solutions suspended in PBS. An additional vial was left as the control, and no nanoparticles are added to the cells. Then the cells were washed with PBS twice more and transferred to a glass coverslip. Fluorescence microscopy was conducted to visualize CDs-stained and control cells. Images were taken using a 100× oil immersion objective with a camera exposure time of 100 ms. A DAPI filter cube, which excites in the UV (378 nm/52 nm) and emits violet (447 nm/60 nm), was used to excite the cells stained with nanoparticles under UV light and collect the violet fluoresce emission from the cell sample. The Blue Orange ICG lookup table was applied in ImageJ software for visualization of images (raw images are available upon request).
Acknowledgments
The authors would like to thank the College of Arts and Sciences Microscopy (CASM) of Texas Tech University for the use of their facilities and the expertise of their research scientists B. Zhao and S. Lizarraga.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c07744.
SEM images of precipitated out transferrin-derived nanomaterial and EDS spectra at various locations. Elemental mapping of nanomaterial determining elemental composition made of EDS data points. Tabular data of HL60 differentiation experiment determining statistically significant differences in cellular fluorescence with affinity CDs. Flow cytometry data of competitive inhibition of CD71 receptor on HL60 cells with anti-CD71 antibody (PDF)
Author Contributions
The manuscript was written through contributions of all authors. S.S. designed experimental protocols and methodology, performed the experiments, analyzed the data, and wrote the paper. D.P. designed experimental methodology and edited the paper. M.J. edited the paper and contributed to data collection. L.F. and L.H. contributed to data collection and analysis. Raw data available upon request from the authors.
This work was supported by the National Science Foundation (no. CBET 1849063).
The authors declare no competing financial interest.
Supplementary Material
References
- Tan G.; Kantner K.; Zhang Q.; Soliman M. G.; Del Pino P.; Parak W. J.; Onur M. A.; Valdeperez D.; Rejman J.; Pelaz B. Conjugation of Polymer-Coated Gold Nanoparticles with Antibodies-Synthesis and Characterization. Nanomaterials 2015, 5 (3), 1297–1316. 10.3390/nano5031297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobysh M.; Ramanaviciene A.; Viter R.; Ramanavicius A. Affinity Sensors for the Diagnosis of COVID-19. Micromachines 2021, 12 (4), 390. 10.3390/mi12040390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzin L.; Shamsipur M. Recent advances in design of electrochemical affinity biosensors for low level detection of cancer protein biomarkers using nanomaterial-assisted signal enhancement strategies. J. Pharm. Biomed. Anal. 2018, 147, 185–210. 10.1016/j.jpba.2017.07.042. [DOI] [PubMed] [Google Scholar]
- Arugula M. A.; Simonian A. L. Novel trends in affinity biosensors: Current challenges and perspectives. Meas. Sci. Technol. 2014, 25, 032001. 10.1088/0957-0233/25/3/032001. [DOI] [Google Scholar]
- Kubiak R. J.; Lee N.; Zhu Y.; Franch W. R.; Levitskaya S. V.; Krishnan S. R.; Abraham V.; Akufongwe P. F.; Larkin C. J.; White W. I. Storage Conditions of Conjugated Reagents Can Impact Results of Immunogenicity Assays. J. Immunol Res. 2016, 2016, 1485615. 10.1155/2016/1485615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry T.; Hough R.; Rogers S.; Rechsteiner M. Intracellular distribution and degradation of immunoglobulin G and immunoglobulin G fragments injected into HeLa cells. J. Cell Biol. 1983, 96 (2), 338–46. 10.1083/jcb.96.2.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H.; Ó’Fágáin C.; O’Kennedy R. Antibody stability: A key to performance - Analysis, influences and improvement. Biochimie 2020, 177, 213–225. 10.1016/j.biochi.2020.08.019. [DOI] [PubMed] [Google Scholar]
- Acchione M.; Kwon H.; Jochheim C. M.; Atkins W. M. Impact of linker and conjugation chemistry on antigen binding, Fc receptor binding and thermal stability of model antibody-drug conjugates. MAbs 2012, 4 (3), 362–372. 10.4161/mabs.19449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock B. Antibodies for immunosensors a review. Anal. Chim. Acta 1997, 347 (1), 177–186. 10.1016/S0003-2670(97)00167-0. [DOI] [Google Scholar]
- Slastnikova T. A.; Ulasov A. V.; Rosenkranz A. A.; Sobolev A. S. Targeted Intracellular Delivery of Antibodies: The State of the Art. Front. Pharmacol. 2018, 9, 1208. 10.3389/fphar.2018.01208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoof M.; Corr S. J.; Kaluarachchi W. D.; Massey K. L.; Briggs K.; Zhu C.; Cheney M. A.; Wilson L. J.; Curley S. A. Stability of antibody-conjugated gold nanoparticles in the endolysosomal nanoenvironment: Implications for noninvasive radiofrequency-based cancer therapy. Nanomedicine 2012, 8 (7), 1096–1105. 10.1016/j.nano.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorywalska M.; Strop P.; Melton-Witt J. A.; Hasa-Moreno A.; Farias S. E.; Galindo Casas M.; Delaria K.; Lui V.; Poulsen K.; Loo C.; Krimm S.; Bolton G.; Moine L.; Dushin R.; Tran T.-T.; Liu S.-H.; Rickert M.; Foletti D.; Shelton D. L.; Pons J.; Rajpal A. Effect of Attachment Site on Stability of Cleavable Antibody Drug Conjugates. Bioconjugate Chem. 2015, 26 (4), 650–659. 10.1021/bc5005747. [DOI] [PubMed] [Google Scholar]
- Shaw A.; Hoffecker I. T.; Smyrlaki I.; Rosa J.; Grevys A.; Bratlie D.; Sandlie I.; Michaelsen T. E.; Andersen J. T.; Högberg B. Binding to nanopatterned antigens is dominated by the spatial tolerance of antibodies. Nat. Nanotechnol. 2019, 14 (2), 184–190. 10.1038/s41565-018-0336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigodanza F.; Burian M.; Arcudi F.; Đorđević L.; Amenitsch H.; Prato M. Snapshots into carbon dots formation through a combined spectroscopic approach. Nat. Commun. 2021, 12 (1), 2640. 10.1038/s41467-021-22902-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Li R.; Yang B. Carbon Dots: A New Type of Carbon-Based Nanomaterial with Wide Applications. ACS Cent. Sci. 2020, 6 (12), 2179–2195. 10.1021/acscentsci.0c01306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai L.; Chen J.; Chang Q.; Shi W.; Chen Q.; Huang L. Protein-derived carbon nanodots with an ethylenediamine-modulated structure as sensitive fluorescent probes for Cu2+ detection. RSC Adv. 2017, 7 (27), 16608–16615. 10.1039/C7RA01441D. [DOI] [Google Scholar]
- Strickland S.; Jorns M.; Heyd L.; Pappas D. Novel synthesis of fibronectin derived photoluminescent carbon dots for bioimaging applications. RSC Adv. 2022, 12 (47), 30487–30494. 10.1039/D2RA05137K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels T. R.; Bernabeu E.; Rodríguez J. A.; Patel S.; Kozman M.; Chiappetta D. A.; Holler E.; Ljubimova J. Y.; Helguera G.; Penichet M. L. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim. Biophys. Acta 2012, 1820 (3), 291–317. 10.1016/j.bbagen.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorella S.; Karagiannis T. C. Transferrin receptor-mediated endocytosis: A useful target for cancer therapy. J. Membr. Biol. 2014, 247 (4), 291–307. 10.1007/s00232-014-9637-0. [DOI] [PubMed] [Google Scholar]
- Ponka P.; Lok C. N. The transferrin receptor: Role in health and disease. Int. J. Biochem. Cell Biol. 1999, 31 (10), 1111–37. 10.1016/S1357-2725(99)00070-9. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Hao J.; Zhang J.; Zhang B.; Tang J. Protein as the source for synthesizing fluorescent carbon dots by a one-pot hydrothermal route. RSC Adv. 2012, 2 (23), 8599–8601. 10.1039/c2ra21217j. [DOI] [Google Scholar]
- Wang D.; Zhu L.; McCleese C.; Burda C.; Chen J.-F.; Dai L. Fluorescent carbon dots from milk by microwave cooking. RSC Adv. 2016, 6 (47), 41516–41521. 10.1039/C6RA06120F. [DOI] [Google Scholar]
- Mohapatra D.; Pratap R.; Pandey V.; Dubey P. K.; Agrawal A. K.; Parmar A. S.; Sahu A. N. Tinospora cordifolia Leaves Derived Carbon dots for Cancer Cell Bioimaging, Free radical Scavenging, and Fe3+ Sensing Applications. J. Fluoresc. 2022, 32 (1), 275–292. 10.1007/s10895-021-02846-6. [DOI] [PubMed] [Google Scholar]
- Naik K.; Chaudhary S.; Ye L.; Parmar A. S. A Strategic Review on Carbon Quantum Dots for Cancer-Diagnostics and Treatment. Front. Bioeng. Biotechnol. 2022, 10, 882100. 10.3389/fbioe.2022.882100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Li T.; Hou Y.; Wu Q.; Yi J.; Zhang G. Microwave synthesis of carbon dots with multi-response using denatured proteins as carbon source. RSC Adv. 2016, 6 (14), 11711–11718. 10.1039/C5RA23081K. [DOI] [Google Scholar]
- Chakraborty D.; Sarkar S.; Das P. K. Blood Dots: Hemoglobin-Derived Carbon Dots as Hydrogen Peroxide Sensors and Pro-Drug Activators. ACS Sustainable Chem. Eng. 2018, 6 (4), 4661–4670. 10.1021/acssuschemeng.7b03691. [DOI] [Google Scholar]
- Li P.; Han F.; Cao W.; Zhang G.; Li J.; Zhou J.; Gong X.; Turnbull G.; Shu W.; Xia L.; Fang B.; Xing X.; Li B. Carbon quantum dots derived from lysine and arginine simultaneously scavenge bacteria and promote tissue repair. Appl. Mater. Today 2020, 19, 100601. 10.1016/j.apmt.2020.100601. [DOI] [Google Scholar]
- Liu H.; Li Z.; Sun Y.; Geng X.; Hu Y.; Meng H.; Ge J.; Qu L. Synthesis of Luminescent Carbon Dots with Ultrahigh Quantum Yield and Inherent Folate Receptor-Positive Cancer Cell Targetability. Sci. Rep. 2018, 8 (1), 1086. 10.1038/s41598-018-19373-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M.; Ruan S.; Liu S.; Sun T.; Qu D.; Zhao H.; Xie Z.; Gao H.; Jing X.; Sun Z. Self-Targeting Fluorescent Carbon Dots for Diagnosis of Brain Cancer Cells. ACS Nano 2015, 9 (11), 11455–11461. 10.1021/acsnano.5b05575. [DOI] [PubMed] [Google Scholar]
- Yao H.; Li J.; Song Y.; Zhao H.; Wei Z.; Li X.; Jin Y.; Yang B.; Jiang J. Synthesis of ginsenoside Re-based carbon dots applied for bioimaging and effective inhibition of cancer cells. Int. J. Nanomed. 2018, 13, 6249–6264. 10.2147/IJN.S176176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khairol Anuar N. K.; Tan H. L.; Lim Y. P.; So’aib M. S.; Abu Bakar N. F. A Review on Multifunctional Carbon-Dots Synthesized From Biomass Waste: Design/ Fabrication, Characterization and Applications. Frontiers in Energy Research 2021, 9, 67. 10.3389/fenrg.2021.626549. [DOI] [Google Scholar]
- Juan A.; Cimas F. J.; Bravo I.; Pandiella A.; Ocaña A.; Alonso-Moreno C. An Overview of Antibody Conjugated Polymeric Nanoparticles for Breast Cancer Therapy. Pharmaceutics 2020, 12, 9. 10.3390/pharmaceutics12090802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z.; Al Zaki A.; Hui J. Z.; Muzykantov V. R.; Tsourkas A. Multifunctional nanoparticles: Cost versus benefit of adding targeting and imaging capabilities. Science 2012, 338 (6109), 903–910. 10.1126/science.1226338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cailotto S.; Amadio E.; Facchin M.; Selva M.; Pontoglio E.; Rizzolio F.; Riello P.; Toffoli G.; Benedetti A.; Perosa A. Carbon Dots from Sugars and Ascorbic Acid: Role of the Precursors on Morphology, Properties, Toxicity, and Drug Uptake. ACS Med. Chem. Lett. 2018, 9 (8), 832–837. 10.1021/acsmedchemlett.8b00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi B.; Cui Y.; Wang S.; Frank B. P.; Williams D. N.; Brown R. P.; Melby E. S.; Hamers R. J.; Rosenzweig Z.; Fairbrother D. H.; Orr G.; Haynes C. L. Malic Acid Carbon Dots: From Super-resolution Live-Cell Imaging to Highly Efficient Separation. ACS Nano 2018, 12 (6), 5741–5752. 10.1021/acsnano.8b01619. [DOI] [PubMed] [Google Scholar]
- Chahal S.; Yousefi N.; Tufenkji N. Green Synthesis of High Quantum Yield Carbon Dots from Phenylalanine and Citric Acid: Role of Stoichiometry and Nitrogen Doping. ACS Sustainable Chem. Eng. 2020, 8 (14), 5566–5575. 10.1021/acssuschemeng.9b07463. [DOI] [Google Scholar]
- Javed M.; Saqib A. N. S.; Ata-ur-Rehman; Ali B.; Faizan M.; Anang D. A.; Iqbal Z.; Abbas S. M. Carbon quantum dots from glucose oxidation as a highly competent anode material for lithium and sodium-ion batteries. Electrochim. Acta 2019, 297, 250–257. 10.1016/j.electacta.2018.11.167. [DOI] [Google Scholar]
- Shen Y.; Li X.; Dong D.; Zhang B.; Xue Y.; Shang P. Transferrin receptor 1 in cancer: a new sight for cancer therapy. Am. J. Cancer Res. 2018, 8 (6), 916–931. [PMC free article] [PubMed] [Google Scholar]
- Ribeiro M. C.; Corrêa V. L. R.; da Silva F. K. L.; de Oliveira Neto J. R.; Casas A. A.; de Menezes L. B.; Amaral A. C. Improving peptide quantification in chitosan nanoparticles. Int. J. Biol. Macromol. 2018, 119, 32–36. 10.1016/j.ijbiomac.2018.07.119. [DOI] [PubMed] [Google Scholar]
- Kozlowski R.; Ragupathi A.; Dyer R. B. Characterizing the Surface Coverage of Protein-Gold Nanoparticle Bioconjugates. Bioconjug Chem. 2018, 29 (8), 2691–2700. 10.1021/acs.bioconjchem.8b00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias D. R.; Poloukhtine A.; Popik V.; Tsourkas A. Effect of ligand density, receptor density, and nanoparticle size on cell targeting. Nanomedicine 2013, 9 (2), 194–201. 10.1016/j.nano.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshyar N.; Gray S.; Han H.; Bao G. The effect of nanoparticle size on in vivo pharmacokinetics and cellular interaction. Nanomedicine 2016, 11 (6), 673–692. 10.2217/nnm.16.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanese A.; Tang P. S.; Chan W. C. W. The Effect of Nanoparticle Size, Shape, and Surface Chemistry on Biological Systems. Annu. Rev. Biomed. Eng. 2012, 14 (1), 1–16. 10.1146/annurev-bioeng-071811-150124. [DOI] [PubMed] [Google Scholar]
- Yuan H.; Li J.; Bao G.; Zhang S. Variable Nanoparticle-Cell Adhesion Strength Regulates Cellular Uptake. Phys. Rev. Lett. 2010, 105 (13), 138101. 10.1103/PhysRevLett.105.138101. [DOI] [PubMed] [Google Scholar]
- Liu X.; Huang N.; Li H.; Jin Q.; Ji J. Surface and Size Effects on Cell Interaction of Gold Nanoparticles with Both Phagocytic and Nonphagocytic Cells. Langmuir 2013, 29 (29), 9138–9148. 10.1021/la401556k. [DOI] [PubMed] [Google Scholar]
- Huang J.; Bu L.; Xie J.; Chen K.; Cheng Z.; Li X.; Chen X. Effects of Nanoparticle Size on Cellular Uptake and Liver MRI with Polyvinylpyrrolidone-Coated Iron Oxide Nanoparticles. ACS Nano 2010, 4 (12), 7151–7160. 10.1021/nn101643u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.-H.; Lee C.-W.; Chiou A.; Wei P.-K. Size-dependent endocytosis of gold nanoparticles studied by three-dimensional mapping of plasmonic scattering images. J. Nanobiotechnol. 2010, 8 (1), 33. 10.1186/1477-3155-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samimi S.; Maghsoudnia N.; Eftekhari R. B.; Dorkoosh F.. Chapter 3 - Lipid-Based Nanoparticles for Drug Delivery Systems. In Characterization and Biology of Nanomaterials for Drug Delivery; Mohapatra S. S.; Ranjan S.; Dasgupta N.; Mishra R. K.; Thomas S., Eds.; Elsevier, 2019; pp 47–76. [Google Scholar]
- Joseph E.; Singhvi G.. Chapter 4 - Multifunctional nanocrystals for cancer therapy: A potential nanocarrier. In Nanomaterials for Drug Delivery and Therapy; Grumezescu A. M., Ed.; William Andrew Publishing, 2019; pp 91–116. [Google Scholar]
- Sk M. A.; Ananthanarayanan A.; Huang L.; Lim K. H.; Chen P. Revealing the tunable photoluminescence properties of graphene quantum dots. J. Mater. Chem. C 2014, 2 (34), 6954–6960. 10.1039/C4TC01191K. [DOI] [Google Scholar]
- Kwon W.; Lee G.; Do S.; Joo T.; Rhee S.-W. Size-controlled soft-template synthesis of carbon nanodots toward versatile photoactive materials. Small 2014, 10 (3), 506–513. 10.1002/smll.201301770. [DOI] [PubMed] [Google Scholar]
- Yan F.; Sun Z.; Zhang H.; Sun X.; Jiang Y.; Bai Z. The fluorescence mechanism of carbon dots, and methods for tuning their emission color: A review. Mikrochim. Acta 2019, 186 (8), 583. 10.1007/s00604-019-3688-y. [DOI] [PubMed] [Google Scholar]
- Zhao X.; Zhang M.; Zhang Z. Ultra-fast microwave-assisted synthesis of photoluminescent carbon dots with an ultra-high quantum yield for H2O2 detection. J. Environ. Chem. Eng. 2022, 10 (6), 109008. 10.1016/j.jece.2022.109008. [DOI] [Google Scholar]
- Tao S.; Song Y.; Zhu S.; Shao J.; Yang B. A new type of polymer carbon dots with high quantum yield: From synthesis to investigation on fluorescence mechanism. Polymer 2017, 116, 472–478. 10.1016/j.polymer.2017.02.039. [DOI] [Google Scholar]
- Chang C. W.; Sud D.; Mycek M. A.. Fluorescence Lifetime Imaging Microscopy. In Methods in Cell Biology; Academic Press, 2007; Vol. 81; pp 495–524. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Chen Y.; Liang M.; Xu L.; Qi S.; Chen H.; Chen X. Solid-phase synthesis of highly fluorescent nitrogen-doped carbon dots for sensitive and selective probing ferric ions in living cells. Anal. Chem. 2014, 86 (19), 9846–9852. 10.1021/ac502446m. [DOI] [PubMed] [Google Scholar]
- MacGillivray R. T.; Mendez E.; Sinha S. K.; Sutton M. R.; Lineback-Zins J.; Brew K. The complete amino acid sequence of human serum transferrin. Proc. Natl. Acad. Sci. U. S. A. 1982, 79 (8), 2504–8508. 10.1073/pnas.79.8.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y.; Pang H.; Yang H. B.; Guo C.; Shao J.; Chi Y.; Li C. M.; Yu T. Carbon-based dots co-doped with nitrogen and sulfur for high quantum yield and excitation-independent emission. Angew. Chem., Int. Ed. Engl. 2013, 52 (30), 7800–7804. 10.1002/anie.201301114. [DOI] [PubMed] [Google Scholar]
- Candelaria P. V.; Leoh L. S.; Penichet M. L.; Daniels-Wells T. R. Antibodies Targeting the Transferrin Receptor 1 (TfR1) as Direct Anti-cancer Agents. Front. Immunol. 2021, 12, 607692. 10.3389/fimmu.2021.607692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan P.; Zhi B.; Coburn L.; Haynes C. L.; Schmidt-Rohr K. A molecular fluorophore in citric acid/ethylenediamine carbon dots identified and quantified by multinuclear solid-state nuclear magnetic resonance. Magn. Reson. Chem. 2020, 58 (11), 1130–1138. 10.1002/mrc.4985. [DOI] [PubMed] [Google Scholar]
- Mayle K. M.; Le A. M.; Kamei D. T. The intracellular trafficking pathway of transferrin. Biochim. Biophys. Acta 2012, 1820 (3), 264–281. 10.1016/j.bbagen.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhin A. V.; Tarantul V. Z.; Gening L. V. Aptamers: Problems, solutions and prospects. Acta Nat. 2013, 5 (4), 34–43. 10.32607/20758251-2013-5-4-34-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons V. J.; Pappas D. Affinity separation and subsequent terminal differentiation of acute myeloid leukemia cells using the human transferrin receptor (CD71) as a capture target. Analyst 2019, 144 (10), 3369–3380. 10.1039/C8AN02357C. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.