Abstract
Background
Cryptic translocations can be identified via genetic analysis of aborted tissues or malformed infants, but it is difficult to deduce the parental origins of the translocations. In the absence of such information, it is not easy to distinguish translocations from normal embryos during pre‐implantation genetic testing, that seeks to block familial transmission of translocations.
Methods
Here, we present a new method that detects cryptic translocations and blocks familial transmission thereof. Whole‐genome, low‐coverage mate‐pair sequencing (WGLMPS) revealed chromosome breakpoint sequences, and preimplantation genetic haplotyping (PGH) was then used to discard embryos with cryptic translocations.
Results
Cryptic translocations were found in all four families, and familial transmission was successfully blocked in one family.
Conclusion
Whole‐genome, low‐coverage mate‐pair sequencing combined with preimplantation genetic haplotyping methods powerfully and practically identify cryptic translocations and block familial transmissions.
Keywords: breakpoint sequencing, cryptic translocation, PGH, WGLMPS
Previously, it was difficult to identify and block familial transmission of cryptic translocations, especially when parental origins of the translocations are absence. In this work, we report a WGLMPS (whole‐genome, low‐coverage mate‐pair sequencing) method that can effectively detect chromosome breakpoint. Using this method, we identified cryptic translocations in four families and successfully blocked the familial transmission in one family.
1. INTRODUCTION
Reciprocal translocation is one of the most common chromosomal structural abnormalities (0.074%–0.152% of all newborns) (Jacobs et al., 1992; Maeda et al., 1991). Carriers with balanced translocation are typically phenotypically normal. However, such translocations trigger infertility, repeat miscarriages, and birth defects, because gamete production is unbalanced (Fiorentino et al., 2011; Zhang et al., 2014). In vitro fertilization (IVF) centers encounter many patients who have undergone translocations. The prevalence rate is 0.6% in infertile males (Mau‐Holzmann, 2005), and 2.7% and 2.4% in couples experiencing repeat implantation failures and miscarriages, respectively (Clifford et al., 1994; Stern et al., 1999).
Most translocations can be detected by traditional G‐band karyotyping of peripheral blood, but cryptic translocations are difficult to discern. There are three main types of such translocations: (1) the translocation segments exhibit similar bands, (2) the translocations occur in shallow zones, or (3) the translocated fragments are smaller than the resolution of the analysis (Dong et al., 2019).
Although it is difficult to identify cryptic translocations during conventional peripheral blood karyotyping, unbalanced outcomes that reflect translocations can be detected via amniocentesis or analysis of chorionic villi or embryos (Chau et al., 2019; Wang et al., 2021); meiotic segregation of each type of translocation leads to characteristic outcomes (Zhang et al., 2018). Of all the possible gametes, only two (the results of alternate segregation) yielded normal (balanced) chromosomes. The others are genetically unbalanced, with adjacent‐1, adjacent‐2, 3:1, or 4:0 disjunctions (Snider et al., 2021).
Thus, it is possible to detect cryptic translocation in one couple. Several cytogenetic and molecular methods [high‐resolution G‐band karyotyping (HRGBK), fluorescence in situ hybridization (FISH), and whole‐genome sequencing (WGS)], can be used to study balanced translocations (Aristidou et al., 2017). However, HRGBK does not detect shallow zones, FISH does not precisely identify translocation chromosomal breakpoints (and is applicable to only a few chromosomes), and WGS is time‐consuming and costly.
In recent years, a robust global detection method of balanced chromosomal rearrangements (whole‐genome low‐coverage mate‐pair sequencing [WGLMPS]) has been developed (Dong et al., 2014) (exploiting CNV‐seq and link technologies) to detect simultaneously fragment abnormalities and yield breakpoint information (Ou et al., 2020). This technique identifies almost all cryptic chromosomal abnormalities and breakpoint sequences that greatly aid preimplantation genetic testing (PGT).
After deducing the parental origin of a cryptic translocation, PGT distinguishes translocations from normal embryos; familial translocation transmission is blocked. Preimplantation genetic haplotyping (PGH) using a single‐nucleotide polymorphism (SNP) chip is efficient (Handyside et al., 2010; Li et al., 2021; Zhang et al., 2017). The haplotypes of all chromosomes involved in translocation and the corresponding normal homologous chromosomes were established using informative SNP markers. Finally, the predicted PGH results of the transferred embryos were validated via second trimester amniotic fluid breakpoint sequencing.
2. MATERIALS AND METHODS
2.1. Patients
We included four families with cryptic translocations. All IVF cycles featured either a long protocol or gonadotropin‐releasing hormone (GnRH) antagonist protocol to control ovarian hyperstimulation. Embryo culture and biopsy were performed as described previously (Ou et al., 2015). All couples received genetic counseling. Amniocentesis (prenatal diagnosis) after PGT and embryo transfer.
2.2. Whole‐genome low‐coverage mate‐pair sequencing
All couples underwent the WGLMPS. Genomic DNA was extracted from peripheral blood using a Qiagen DNA extraction kit and used to construct a non‐size‐selected mate‐pair library (Luo et al., 2018) that was subjected to BGISeq‐500, 50‐bp‐end multiplex sequencing. High‐quality paired‐end reads were aligned with the NCBI human reference genome (hg19, GRCh37.1) using SOAP2 (Ou et al., 2020). Only uniquely mapped reads were analyzed (Li et al., 2014). The breakpoints of the translocations were validated by junction‐spanning PCR (Aristidou et al., 2017).
2.3. Blastocyst biopsy and whole‐genome amplification
Five to ten cells were removed from the trophectoderm at the blastocyst stage, rinsed three times with G‐MOPS (Vitrolife) medium, and transferred to RNAse‐/DNAse‐free PCR tubes (Axygen) in minimal medium (Ou et al., 2015). Whole‐genome amplification (WGA) was performed using the multiple displacement amplification (MDA) method. Isothermal DNA amplification with phi 29 DNA polymerase was performed (Repli‐g single‐cell kit, QIAGEN GmbH, Hilden, Germany) according to the manufacturer's protocol. Amplification was performed at 30°C for 8 h, and the reaction was stopped by incubation at 65°C for 3 min. To avoid contamination, all steps were performed in a biosafety cabinet (Ou et al., 2015).
2.4. Preimplantation genetic haplotyping
We used the Illumina Human Karyomap‐12 V1.0 (Handyside et al., 2010) SNP microarray to perform genome‐wide PGH analysis of the embryos of two families. The flanking and breakpoint regions identified using WGLMPS were assessed by establishing their haplotypes. This information was used to identify balanced and normal embryos. Molecular karyotyping and haplotype linkage analyses were performed using Bluefuse‐multi software (Illumina Inc., San Diego, CA, USA 17).
3. RESULTS
Case 1
A 29‐year‐old woman experienced four failed pregnancies. In 2016, a fetus died at 32 weeks of gestation and labor was induced in utero. In 2018, biochemical pregnancy (only) occurred. In 2019, embryo development ceased at 11 weeks of gestation, and chromosomal microarray analysis (CMA) of villi revealed a 1p36.33p36.23 deletion and 19p13.3 duplication. In 2020, an embryo died at 2 months of gestation, and next‐generation sequencing (NGS) of the villi revealed the deletion and duplication mentioned above. Conventional karyotyping of the husband's peripheral blood revealed no anomalies (Figure 1a). However, chromosome 1 and 19 translocations in the husband were confirmed by WGLMPS breakpoint sequencing and junction‐spanning PCR (Figure 1b). The breakpoint of chromosome 19 was at chr19:4530606, and no gene was affected. The breakpoint of chromosome 1 was at chr1:9068476; thus, it was in the SLC2A7 gene, which is not associated with disease. This translocation was confirmed via limited FISH probing of chromosome 1 subtelomeres (Figure 1c).
Case 2
A 28‐year‐old woman experienced two spontaneous abortions. In 2018, embryonic development ceased at 2 months of gestation, but no chromosomal examination was performed. In 2019, embryo development ceased at two months of gestation. The villus revealed a 6q25q27 deletion and an 8q24.13q27.3 duplication. Karyotyping of the couple's peripheral blood revealed no abnormalities, but breakpoint sequencing confirmed the reciprocal translocation of chromosomes 6 and 8 in females (Figure 1d). The breakpoint of chromosome 8 was at chr8:125492064, and thus, in the RNF139 gene. The OMIM database does not include any diseases associated with this gene. The breakpoint of chromosome 6 was at chr6:149194749, and thus, in the UST gene. The OMIM database does not include any diseases associated with this gene. In addition, a chromosome 6 deletion (17‐kb) at the breakpoint of the reciprocal translocation, which also involved the UST gene. However, no related CNV cases in this region have been reported in the literature or in any database.
Case 3
A 30‐year‐old woman had not been pregnant in the previous 2 years and had undergone IVF because of fallopian tube problems. She did not become pregnant after the first IVF transplantation in 2018 but gave birth to a child after a second IVF transplantation in the same year. However, the patient had multiple malformations. Whole‐exome sequencing revealed a 1p36.33p36.32 duplication and a 6q26q27 deletion. Karyotyping of the couple's peripheral blood revealed no abnormalities, but breakpoint sequencing confirmed the reciprocal translocation of chromosomes 1 and 6 in females (Figure 1e). The breakpoint of chromosome 6 is at chr6:162492303; thus, in the PARK2 gene, which is associated with autosomal recessive juvenile Parkinson's disease‐2. The breakpoint of chromosome 1 was at chr1:4005764 and was absent in any of the genes.
Case 4
A 28‐year‐old woman experienced two arrested embryonic developments in early pregnancy, but the chorionic villi were not examined. In 2019, chorionic villi were assessed for neck cystic hygroma. CMA revealed a 6p25.3p25.1 deletion and an 11q24.1q25 duplication. Karyotyping of the couple's peripheral blood revealed no abnormalities, but breakpoint sequencing confirmed the reciprocal translocation of chromosomes 6 and 11 in females (Figure 1f). The breakpoint of chromosome 11 was at chr11:123441622, thus, in the GRAMD1B gene. The OMIM database does not include any diseases associated with this gene. The breakpoint of chromosome 6 was at chr6:5216702 and was not present in any protein‐coding gene. In addition, a 626‐kb paracentric inversion of chromosome 13q14.3 was detected. The breakpoints were located near chr13:52924035 and chr13:53550510, respectively. No breakpoint was found in the protein‐coding gene. A 626‐kb duplication of chromosome 13 was also.3, involving SUGT1, PCDH8, THSD1, and six other protein‐coding genes. The OMIM database indicates that intracranial berry aneurysm type 12 is associated with THSD1; OMIM does not list a phenotype for any of the other genes. CNV pathogenicity has not been reported in the literature or in any databases. A duplication of 114‐kb in q14.3 of chromosome 13 was also detected, involving the OLFM4 gene. However, the OMIM database does not list any phenotypes associated with this gene. CNV pathogenicity has not been reported in the literature or in any databases.
FIGURE 1.
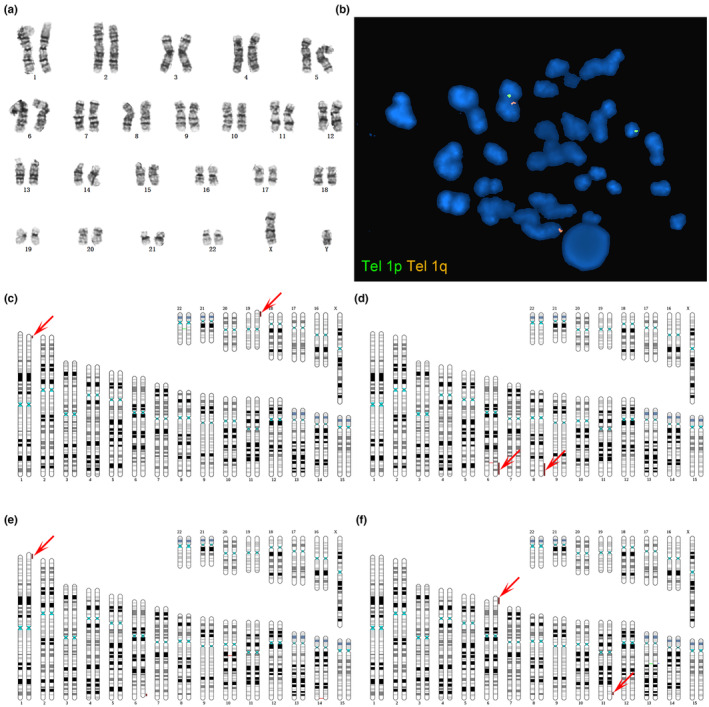
Breakpoint sequencing and FISH probing confirmed cryptic translocation of chromosomes 1 and 19. (a): Conventional karyotyping of the male's peripheral blood. (b). Breakpoint sequencing confirming a cryptic translocation of chromosomes 1 and 19 (indicated by arrows). (c): Limited FISH probing of the chromosome 1 subtelomeres confirmed the translocation. (d): Breakpoint sequencing confirmed reciprocal translocation of chromosomes 6 and 8 (red arrows). (e): Breakpoint sequencing confirmed reciprocal translocation of chromosomes 1 and 6 (red arrows). (f): Breakpoint sequencing confirmed the reciprocal translocation of chromosomes 6 and 11 (red arrows).
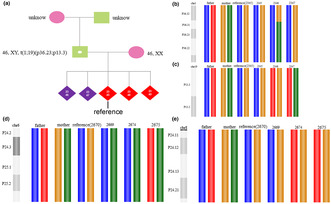
In Case 1, PGH was performed. Breakpoint sequencing confirmed reciprocal translocation of chromosomes 1 and 19 in males. However, given the high cost of such sequencing and the difficulties associated with drawing blood in older adults, the chromosomes of the male parents were not checked. Unbalanced embryo 2345 served as a reference when constructing the haplotype (Figure 2a). This embryo exhibited adjacent‐1 segregation, resulting in one normal chromosome 1 and one derivative chromosome 19. Linkage analysis (PGH) revealed that embryos 2346 and 2347 were normal (Figure 2b,c).
FIGURE 2.
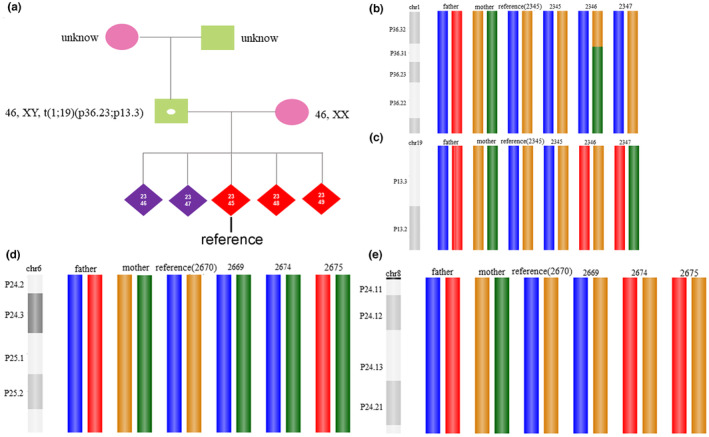
Haplotyping results of each embryo. (a): The unbalanced embryo 2345 served as the reference when constructing the haplotype. (b): The haplotypes of chromosome 1 of each embryo (blue: normal, red: derivative). (c): The haplotypes of chromosome 19 of each embryo (red: normal, blue: derivative). (d): Haplotyping of chromosome 6 of each embryo (yellow: normal, green: derivative). (e): Haplotyping of chromosome 8 of each embryo (green: normal, yellow: derivative).
In Case 2, PGH was performed. After breakpoint sequencing confirmed the reciprocal translocation of chromosomes 6 and 8 in females, the haplotype was constructed using unbalanced embryo 2670 (adjacent‐1 segregation) as the reference. Linkage analysis revealed that embryos 2669, 2674, and 2675 carried the translocation (Figure 2d,e). Embryo 2669 did not yield a pregnancy after embryo transfer, whereas 2674 did. Breakpoint sequencing of the second trimester amniotic fluid revealed translocation.
4. DISCUSSION
We report four carriers of cryptic balanced translocations revealed by breakpoint sequencing that had evaded detection on G‐band karyotyping (which often fails to detect aberrations close to telomeres because most terminal bands are G‐negative) (Yi Ning et al., 1996). Cryptic translocations occurred in shallow zones, and the sizes of translocated fragments were below the karyotyping resolution in Cases 1, 3, and 4. Although the translocation fragment was large in Case 2, it was missed by G‐band karyotyping because the other bands were similar. Although it is difficult to find cryptic translocations during conventional peripheral blood karyotyping, we found translocation‐associated unbalanced outcomes (adjacent‐1) in the chorionic villi of three cases and in the newborn in one case using CMA or NGS. Below, we summarize the meiotic segregation models of translocations, as revealed by the embryonic NGS results: (1) Alternate segregation is similar to that of a balanced beam: (2) Adjacent segregation is similar to a seesaw; and 3. 3:1 segregation is like a double stick (Figure 3).
FIGURE 3.

The meiotic segregation models of translocation as revealed by embryonic NGS. (a) Alternate segregation can trigger translocations or the genome may remain completely normal. As genomic deletion and duplication are lacking, the points of the chromosomes (except the sex chromosomes) all lie the same plane, and the graph is suggestive of a balanced beam. (b) Adjacent segregation is more complex, featuring both duplication and deletion, just as a seesaw can go high or low. However, the adjacent segregations differ. Adjacent‐1 segregation involves the segregation of homologous centromeres; the translocation involves chromosome duplication and deletion (generally of small segments) inside the arm (thus a “small seesaw”). (c) Adjacent‐2 segregation does not involve segregation of homocentromeres. The duplications and deletions of chromosomes involved in translocation extend beyond one arm, generally involving large segments (thus a “big seesaw”). (d) Adjacent‐2 recombination can produce two identical (normal) chromosomes; the chromosome involved in translocation is simultaneously duplicated and deleted (thus, a “very big seesaw”). (e) Adjacent‐2 recombination can also produce two identical chromosomes, triggering two duplications and deletions of/in each chromosome involved in translocation (thus, a “double seesaws”). (f) 3:1 segregation resembles a double stick, which can be combined into one stick or remain divided. The “two in one” scenario involves the duplication or deletion of one (entire) chromosome of the two chromosomes involved in translocation. (g) The “division into two” scenario is associated with the synchronous duplication or deletion of both chromosomes involved in translocation, thus a “double stick raised.” (h) The stick can also be laid down.
We inferred cryptic translocations in four families when the villi and newborns exhibited imbalances. We used WGLMPS to detect precise breakpoints. WGLMPS detects all CNVs and structural variations (SVs). Thus, other chromosomal abnormalities (not translocations) were found in Case 4, similar to the corresponding breakpoints (to accuracies of 1 kb) (Peng et al., 2021). SLC2A7, RNF139, UST, PARK2, and GRAMD1B (and a microdeletion) are located in the breakpoint regions; some of these genes are associated with disease. After identifying the parental origin of a cryptic translocation, we blocked the familial transmission of this translocation by selecting only normal embryos during PGT. Haplotype linkage analysis was used to distinguish between the two possible forms of alternate segregation. Currently, SNPs or short tandem repeats (STRs) are widely used to construct haplotypes (Handyside et al., 2010; Renwick et al., 2006). SNP frequencies exceed 1%, SNPs are widely distributed and genetically stable, and analytical automation is simple.
PGH is based on an SNP chip that compares DNAs from embryos, parents, and close relatives using genome‐wide SNP genotyping of subjects with balanced translocations. Next, genome‐wide haplotyping of the carrier family was performed using linkage analysis. Finally, the chromosomal status of embryos was determined. Embryos with translocational haplotypes can be distinguished from normal embryos. To ensure successful haplotyping, we preferred MDA over DOP‐PCR when performing single‐cell amplification. MDA affords much better genomic coverage and fewer duplications than DOP‐PCR (Hou et al., 2015). Haplotypes may be constructed in two ways, of which the first is the “three tube blood model.” Hereditary translocations are identified by analysis of the two peripheral bloods of the couple and that of one of the carrier's parents. This optimally identifies the balanced translocation carriers. The second is the “el” two‐tube blood model. A new familial translocation (the inheritance of which is unknown) is detected via assay of the blood of the couple only, and haplotypes are constructed using unbalanced embryos as a reference.
In such cases, adjacent‐1 segregation is the first choice. As this is homocentric, such segregation generally yields more genetic information than other patterns. Adjacent‐2 segregation generally lacks most of the information on one chromosome. The 3:1 segregation also lacks such information, but paradoxically may contain excessive information on other regions. Adjacent‐2 recombination may render haplotype construction impossible because recombination boundaries are uncertain.
5. CONCLUSIONS
Accurate breakpoint mapping is a key process in identifying cryptic translocations. In this study, we used the robust WGLMPS method to study four families with cryptic translocations and performed PGH on two families to distinguish translocated embryos from normal embryos. Amniotic fluid breakpoint sequencing verified the PGH results. Thus, WGLMPS combined with PGH is both powerful and practical for identifying the source of cryptic translocation and blocking familial transmission.
AUTHOR CONTRIBUTIONS
Hong Li, Ling‐Yin Kong, and Bo Liang provided the conceptualization, funding acquisition, and project administration. Jian Ou, Jian Sun, Chuan‐Chun Yang, Meng‐Xia Ni, Qin‐Yan Zou, Shi‐Yu Xing, Lin Chun‐Hua Lin provided the data curation, methodology, and project administration. Qing‐Xia Meng, Jie Ding, Ai‐Yan Zheng, and Yan Zhang analyzed the data. Jian Ou wrote the original draft. All the authors have written, reviewed, and edited the final manuscript.
FUNDING INFORMATION
This work was funded by grants from Jiangsu Province ‘13th Five‐Year’ Youth Medical Talent Program (QNRC2016246), Suzhou Clinical Medicine Talent Program (SZYJTD201708) and Jiangsu Provincial Medical Key Discipline (Laboratory) (JSDW202214).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
The study was approved by the Institutional Review Board. All the procedures adhered to the principles of the Declaration of Helsinki.
INFORMED CONSENT
Written informed consent was obtained from all the couples.
ACKNOWLEDGEMENTS
The authors thank all the staffs who have contributed to this study and the patients who participated in this study.
Ou, J. , Sun, J. , Yang, C.‐C. , Ni, M.‐X. , Zou, Q.‐Y. , Xing, S.‐Y. , Lin, C.‐H. , Meng, Q.‐X. , Ding, J. , Zheng, A.‐Y. , Zhang, Y. , Kong, L.‐Y. , Liang, B. , & Li, H. (2024). Identification and interruption of inheritance of familial cryptic translocations: A case report. Molecular Genetics & Genomic Medicine, 12, e2356. 10.1002/mgg3.2356
Jian Ou, Jian Sun, and Chuan‐Chun Yang contributed equally to this work.
DATA AVAILABILITY STATEMENT
The data underlying this article is available on request to the corresponding author upon reasonable request.
REFERENCES
- Aristidou, C. , Koufaris, C. , Theodosiou, A. , Bak, M. , Mehrjouy, M. M. , Behjati, F. , Tanteles, G. , Christophidou‐Anastasiadou, V. , Tommerup, N. , & Sismani, C. (2017). Accurate breakpoint mapping in apparently balanced translocation families with discordant phenotypes using whole genome mate‐pair sequencing. PLoS ONE, 12(1), e0169935. 10.1371/journal.pone.0169935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau, M. H. K. , Cao, Y. , Kwok, Y. K. Y. , Chan, S. , Chan, Y. M. , Wang, H. , Yang, Z. , Wong, H. K. , Leung, T. Y. , & Choy, K. W. (2019). Characteristics and mode of inheritance of pathogenic copy number variants in prenatal diagnosis. American Journal of Obstetrics and Gynecology, 221(5), 493.e1–493.e11. 10.1016/j.ajog.2019.06.007 [DOI] [PubMed] [Google Scholar]
- Clifford, K. , Rai, R. , Watson, H. , & Regan, L. (1994). An informative protocol for the investigation of recurrent miscarriage: Preliminary experience of 500 consecutive cases. Human Reproduction, 9(7), 1328–1332. 10.1093/oxfordjournals.humrep.a138703 [DOI] [PubMed] [Google Scholar]
- Dong, Z. , Jiang, L. , Yang, C. , Hu, H. , Wang, X. , Chen, H. , Choy, K. W. , Hu, H. , Dong, Y. , Hu, B. , Xu, J. , Long, Y. , Cao, S. , Chen, H. , Wang, W.‐J. , Jiang, H. , Xu, F. , Yao, H. , Xu, X. , & Liang, Z. (2014). A robust approach for blind detection of balanced chromosomal rearrangements with whole‐genome low‐coverage sequencing. Human Mutation, 35(5), 625–636. 10.1002/humu.22541 [DOI] [PubMed] [Google Scholar]
- Dong, Z. , Yan, J. , Xu, F. , Yuan, J. , Jiang, H. , Wang, H. , Chen, H. , Zhang, L. , Ye, L. , Xu, J. , Shi, Y. , Yang, Z. , Cao, Y. , Chen, L. , Li, Q. , Zhao, X. , Li, J. , Chen, A. , Zhang, W. , … Chen, Z. J. (2019). Genome sequencing explores complexity of chromosomal abnormalities in recurrent miscarriage. American Journal of Human Genetics, 105(6), 1102–1111. 10.1016/j.ajhg.2019.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentino, F. , Spizzichino, L. , Bono, S. , Biricik, A. , Kokkali, G. , Rienzi, L. , Ubaldi, F. M. , Iammarrone, E. , Gordon, A. , & Pantos, K. (2011). PGD for reciprocal and Robertsonian translocations using array comparative genomic hybridization. Human Reproduction, 26(7), 1925–1935. 10.1093/humrep/der082 [DOI] [PubMed] [Google Scholar]
- Handyside, A. H. , Harton, G. L. , Mariani, B. , Thornhill, A. R. , Affara, N. , Shaw, M. A. , & Griffin, D. K. (2010). Karyomapping: A universal method for genome wide analysis of genetic disease based on mapping crossovers between parental haplotypes. Journal of Medical Genetics, 47(10), 651–658. 10.1136/jmg.2009.069971 [DOI] [PubMed] [Google Scholar]
- Hou, Y. , Wu, K. , Shi, X. , Li, F. , Song, L. , Wu, H. , Dean, M. , Li, G. , Tsang, S. , Jiang, R. , Zhang, X. , Li, B. , Liu, G. , Bedekar, N. , Lu, N. , Xie, G. , Liang, H. , Chang, L. , Wang, T. , … Wang, J. (2015). Comparison of variations detection between whole‐genome amplification methods used in single‐cell resequencing. GigaScience, 4, 37. 10.1186/s13742-015-0068-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, P. A. , Browne, C. , Gregson, N. , Joyce, C. , & White, H. (1992). Estimates of the frequency of chromosome abnormalities detectable in unselected newborns using moderate levels of banding. Journal of Medical Genetics, 29(2), 103–108. 10.1136/jmg.29.2.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Chen, H. , Yin, C. , Yang, C. , Wang, B. , Zheng, S. , Zhang, J. , & Fan, W. (2014). Mapping breakpoints of a familial chromosome insertion (18,7) (q22.1; q36.2q21.11) to DPP6 and CACNA2D1 genes in an azoospermic male. Gene, 547(1), 43–49. 10.1016/j.gene.2014.06.007 [DOI] [PubMed] [Google Scholar]
- Li, R. , Wang, J. , Gu, A. , Xu, Y. , Guo, J. , Pan, J. , Zeng, Y. , Ma, Y. , Zhou, C. , & Xu, Y. (2021). Feasibility study of using unbalanced embryos as a reference to distinguish euploid carrier from noncarrier embryos by single nucleotide polymorphism array for reciprocal translocations. Prenatal Diagnosis, 41(6), 681–689. 10.1002/pd.5897 [DOI] [PubMed] [Google Scholar]
- Luo, A. , Cheng, D. , Yuan, S. , Li, H. , Du, J. , Zhang, Y. , Yang, C. , Lin, G. , Zhang, W. , & Tan, Y. Q. (2018). Maternal interchromosomal insertional translocation leading to 1q43‐q44 deletion and duplication in two siblings. Molecular Cytogenetics, 11, 24. 10.1186/s13039-018-0371-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, T. , Ohno, M. , Matsunobu, A. , Yoshihara, K. , & Yabe, N. (1991). A cytogenetic survey of 14,835 consecutive liveborns. Jinrui Idengaku Zasshi, 36(1), 117–129. 10.1007/bf01876812 [DOI] [PubMed] [Google Scholar]
- Mau‐Holzmann, U. A. (2005). Somatic chromosomal abnormalities in infertile men and women. Cytogenetic and Genome Research, 111(3–4), 317–336. 10.1159/000086906 [DOI] [PubMed] [Google Scholar]
- Ou, J. , Wang, W. , Feng, T. , Liao, L. , Meng, Q. , Zou, Q. , Ding, J. , Zheng, A. , Duan, C. , Li, P. , Liu, Q. , Lin, C. , & Li, H. (2015). Identification of small segmental translocations in patients with repeated implantation failure and recurrent miscarriage using next generation sequencing after in vitro fertilization/intracytoplasmic sperm injection. Molecular Cytogenetics, 8, 105. 10.1186/s13039-015-0207-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, J. , Yang, C. , Cui, X. , Chen, C. , Ye, S. , Zhang, C. , Wang, K. , Chen, J. , Zhang, Q. , Qian, C. , Fang, G. , & Zhang, W. (2020). Successful pregnancy after prenatal diagnosis by NGS for a carrier of complex chromosome rearrangements. Reproductive Biology and Endocrinology, 18(1), 15. 10.1186/s12958-020-00572-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y. , Yang, S. , Xi, H. , Hu, J. , Jia, Z. , Pang, J. , Liu, J. , Yu, W. , Tang, C. , & Wang, H. (2021). Whole genome sequencing reveals translocation breakpoints disrupting TP63 gene underlying split hand/foot malformation in a Chinese family. Molecular Genetics & Genomic Medicine, 9(3), e1604. 10.1002/mgg3.1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renwick, P. J. , Trussler, J. , Ostad‐Saffari, E. , Fassihi, H. , Black, C. , Braude, P. , Ogilvie, C. M. , & Abbs, S. (2006). Proof of principle and first cases using preimplantation genetic haplotyping—A paradigm shift for embryo diagnosis. Reproductive Biomedicine Online, 13(1), 110–119. 10.1016/s1472-6483(10)62024-x [DOI] [PubMed] [Google Scholar]
- Snider, A. C. , Darvin, T. , Spor, L. , Akinwole, A. , Cinnioglu, C. , & Kayali, R. (2021). Criteria to evaluate patterns of segmental and complete aneuploidies in preimplantation genetic testing for aneuploidy results suggestive of an inherited balanced translocation or inversion. F&S Reports, 2(1), 72–79. 10.1016/j.xfre.2020.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, C. , Pertile, M. , Norris, H. , Hale, L. , & Baker, H. W. (1999). Chromosome translocations in couples with in‐vitro fertilization implantation failure. Human Reproduction, 14(8), 2097–2101. 10.1093/humrep/14.8.2097 [DOI] [PubMed] [Google Scholar]
- Wang, X. , Wu, C. , Hao, D. , Zhang, J. , Tan, C. , Cheng, D. H. , Fei, J. , & Yu, Y. (2021). One healthy live birth after preimplantation genetic testing of a cryptic balanced translocation (9;13) in a family with cerebral palsy and glaucoma: A case report. BMC Medical Genomics, 14(1), 82. 10.1186/s12920-021-00938-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Ning, A. R. , Smith, A. C. M. , Macha, M. , Precht, K. , Riethman, H. , Ledbetter, D. H. , Flint, J. , Horsley, S. , Regan, R. , Kearney, L. , Knight, S. , Kvaloy, K. , & Brown, W. R. A. (1996). A complete set of human telomeric probes and their clinical application. National Institutes of Health and Institute of Molecular Medicine collaboration. Nature Genetics, 14(1), 86–89. 10.1038/ng0996-86 [DOI] [PubMed] [Google Scholar]
- Zhang, S. , Lei, C. , Wu, J. , Sun, H. , Zhou, J. , Zhu, S. , Wu, J. , Fu, J. , Sun, Y. , Lu, D. , Sun, X. , & Zhang, Y. (2018). Analysis of segregation patterns of quadrivalent structures and the effect on genome stability during meiosis in reciprocal translocation carriers. Human Reproduction, 33(4), 757–767. 10.1093/humrep/dey036 [DOI] [PubMed] [Google Scholar]
- Zhang, S. , Lei, C. , Wu, J. , Zhou, J. , Sun, H. , Fu, J. , Sun, Y. , Sun, X. , Lu, D. , & Zhang, Y. (2017). The establishment and application of preimplantation genetic haplotyping in embryo diagnosis for reciprocal and Robertsonian translocation carriers. BMC Medical Genomics, 10(1), 60. 10.1186/s12920-017-0294-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Zhu, S. , Wu, J. , Liu, S. , & Sun, X. (2014). Quadrivalent asymmetry in reciprocal translocation carriers predicts meiotic segregation patterns in cleavage stage embryos. Reproductive Biomedicine Online, 29(4), 490–498. 10.1016/j.rbmo.2014.06.010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article is available on request to the corresponding author upon reasonable request.


