Abstract
Background
Newborn screening (NBS) aims to detect congenital anomalies, and next‐generation sequencing (NGS) has shown promise in this aspect. However, the NBS strategy for monogenic inherited diseases in China remains insufficient.
Methods
We developed a NeoEXOME panel comprising 601 genes that are relevant to the Chinese population found through extensive research on available databases. An interpretation system to grade the results into positive (high‐risk, moderate‐risk, and low‐risk genotypes), negative, and carrier according to the American College of Medical Genetics (ACMG) guidelines was also developed. We validated the panel to evaluate its efficacy by using data from the “1000 Genomes Project” and conducted a pilot multicenter study involving 3423 neonates.
Results
The NGS positive rate in the 1000 Genomes Project was 7.6% (23/301), whereas the rate was 12.0% in the multicenter study, including 3249 recruited neonates. Notably, in 200 neonates, positive per conventional NBS, 58.5% (69/118) showed results consistent with NGS. In the remaining 3049 neonates showing negative results in conventional NBS, 271 (8.9%) were positive per NGS, and nine of them were clinically diagnosed with diseases in the follow‐up.
Conclusion
We successfully designed a NeoEXOME panel for targeted sequencing of monogenic inherited diseases in NBS. The panel demonstrated high performance in the Chinese population, particularly for the early detection of diseases with no biochemical markers.
Keywords: monogenic inherited diseases, NeoEXOME panel performance, newborn screening, pilot study, targeted sequencing
In this study, we designed a gene–disease association list (NeoEXOME panel) for the Chinese population with 601 genes based on databases. Then, we evaluated the application of our panel in the 1000 Genomes Project and a pilot multicenter study. Our study designed a personal NBS targeted‐sequencing NeoEXOME panel of monogenic inherited diseases for the Chinese, which has shown acceptable performance.
1. INTRODUCTION
Congenital anomalies are significantly responsible for neonatal mortality globally. The Ministry of Public Health of China in 2012 reported that approximately 5.6% of births in China have congenital anomalies, resulting in approximately 900,000 new cases each year (Ministry of Health of the People's Republic of China, 2012). To address this, the World Health Organization (WHO) has developed a three‐level prevention strategy based on the causes and epidemiology of congenital anomalies. The strategy includes preconception screening to increase the likelihood of a healthy birth, peri‐conception screening to help predict the risk of abnormalities and neonatal screening to reduce mortality and morbidity due to congenital disorders (World Health Organization, 2010). Newborn screening (NBS), the third step in the prevention of congenital anomalies, aims to identify serious diseases during the neonatal period to facilitate earlier referral and the initiation of medical or surgical treatment, thus potentially reducing child mortality and morbidity (Fabie et al., 2019; Zhou & Zhao, 2021).
NBS was initiated in the 1960s when Professor Guthrie first applied the method of bacterial inhibition to screen for phenylketonuria (PKU) in dried blood spots (DBS; Guthrie & Susi, 1963). Since then, NBS has advanced under the several screening guidelines (Rajabi, 2018; Wilson & Jungner, 1968). In 2006, the American College of Medical Genetics (ACMG) published a Recommended Uniform Screening Panel for NBS, which has undergone continuous updates (Watson et al., 2006). In China, NBS began in 1981 and focused mainly on screening for PKU and congenital hypothyroidism (CH; Zhan et al., 2009). Tandem mass spectrometry (MS) is the main conventional NBS method that has been widely used in clinical practice for the last 20 years (Ombrone et al., 2016). Although MS accelerated the development of NBS, it is associated with a false‐positive/negative ratio (Fabie et al., 2019; Mendell et al., 2012; Slaughter et al., 2010). In adddition, MS and other conventional NBS methods have a limited coverage of diseases that can be detected. New methods are thus urgently needed to address the limitations of NBS.
Recently, the high‐throughput sequencing (NGS) technology has been widely used in tumor‐targeted gene testing (Rizvi et al., 2018), pathogen detection (Han et al., 2019), as well as in three‐level screening for congenital anomalies (Swedish Council on Health Technology A, 2016). NGS has been successfully applied for NBS and has been shown to be useful for second‐tier confirmation of some diseases (Lin et al., 2021; Parad et al., 2020). In addition, the ability of NGS to detect disease‐genes in patients in the neonatal intensive care units (NICU) has also been uncovered (Meng et al., 2017; Zhu et al., 2020). However, there are challenges associated with the use of NGS in NBS, including a large number of variants with uncertain significance (VUS) and the lack of recognized and recommended list of gene–disease association conditions.
To address these challenges, researchers have been working to establish an appropriate criterion for the gene–disease association list and report strategies in NGS for NBS (Ceyhan‐Birsoy et al., 2017; Milko et al., 2019). However, the high incidence of monogenic disorders in the Chinese population is quite different from that of other nations due to diverse ethnicities and geographical areas. Therefore, the gene–disease association list and reporting strategy cannot be fully implemented. To address the unique genetic landscape of the Chinese population, we designed a severe and actionable inherited gene–disease list called NeoEXOME panel for NBS. We then conducted a multicenter study including 3423 neonates to evaluate the feasibility of the NeoEXOME panel and compare it with other NGS panels for NBS.
2. METHODS
2.1. Establishment of the NGS panel
2.1.1. Generation of gene–disease association list
We researched monogenic inherited diseases that are severe and actionable (outline in the result section) and screened for heritable genes corresponding to the diseases using the Online Mendelian Inheritance in Man (OMIM) database (https://www.omim.org/). Genes with a “phenotype mapping key” of “3,” which indicates that the molecular basis of the disease is known, were included.
2.1.2. Age of onset
We manually sorted diseases records in the OMIM database to identify the known minimum onset age of diseases and categorized them into six groups based on the age of onset: <1 year of age (infants), 1–3 years of age (toddlers), 3–6 years of age (preschoolers), 6–12 years of age (middle childhood), 12–18 years of age (young teens and teenagers), >18 years of age (adulthood).
2.2. Ethical compliance
This study was approved by the Ethics Committee of Children's Hospital of Chongqing Medical University (2019‐R‐171‐1). Guardians of neonates provided written and signed informed consent.
2.3. Participants
The study has been registered in ClinicalTrials.gov (Number: NCT03984266). The inclusion criteria were meeting both of the following: all newborns (including hospitalized infants and infants with abnormal results of conventional NBS) and the guardian's agreement to sign the informed consent and participate. The exclusion criteria were meeting one or more of the following criteria: other similar clinical studies are underway and receiving transfusion of allogeneic blood products in the past 2 weeks. Rejection criteria: Specimen cannot be tested due to improper collection or storage; samples that failed quality control; participants with no follow‐up data; guardian's request to withdraw.
From October 2019 to September 2021, a total of 3423 neonatal subjects were enrolled from five hospitals: Children's Hospital of Chongqing Medical University, Inner Mongolia Maternal and Child Health Hospital, Northwest Women and Children's Medical Center, Dalian Maternal and Child Health Hospital, and Xuzhou Maternal and Child Health Care Hospital.
2.4. Sample preparation
The heel blood of the participants was collected into a special filter paper and allowed to dry naturally at room temperature forming DBS. After conventional NBS, the DBS was stored at 4°C. Genomic DNA was extracted from DBS using QIAamp DNA Mini Kit (Qiagen, German) according to the operation manual. In brief, a punched‐out circle from a DBS was placed into a 1.5 mL microtube with Buffer ATL to dissolve DNA and incubated at 85°C for 10 min. Protein K was added to digest proteins. After incubation at 70°C and the addition of ethanol, the buffer was transferred into a Mini spin column. Finally, the DNA solution was eluted using the elution buffer after a series of centrifugation. The quality and quantity of the DNA were assessed by Qubit® 3.0 Fluorometer (Thermo Fisher Scientific Inc., USA) according to the manufacturer's instructions.
2.5. Library construction and sequencing
A library was constructed according to the standard procedures of Illumina (Illumina, Inc., USA), including terminal repair, adaptor connection, and polymerase chain reaction enrichment. The Neonatal Gene Capture Kit NeoExome™ (MyGenostics GenCap® Enrichment technologies, China) was designed based on the targeted genes in our study and used for hybridization capture of relevant target regions. The final library was determined using the Qubit® 3.0 Fluorometer and Agilent 2100 Bioanalyzer system (Agilent Technologies, USA) and sequenced using the HiSeq X Ten System (Illumina, Inc., USA) for 150‐bp double‐terminal sequencing (Zhu et al., 2020). After sequencing, we compared the reads to the UCSC HG19 reference genome and identified variants using a bioinformatics process set by MyGenostics (based on GATK, The Genome Analysis Toolkit). Variations were annotated using the ANNOVAR databases. Variants with high population frequency (>1/1000) were filtered out using the dbSNP 1381000 Genome Project, ESP6500SI, and ExAC (the Exome Aggregation Consortium) browser.
2.6. Probe design description
The design of the liquid‐phase hybridization capture probe was based on the GenCap technology of MyGenostics (Yu et al., 2020). In addition to designing probes that cover the exon region of related genes (covering the exon region, exon flanks ~50 bp, and non‐coding disease‐causing regions reported by the Human Gene Mutation Database, HGMD), encryption designing probes were designed for 25 CNV high‐risk genes (increasing the coverage of 300 bp intron region by designing probes on the exon flanks of related genes). We also designed probes that included full‐length coverage of HBA1/HBA2/HBB/SMN1/SMN2 genes (coding region and intron region of coverage gene). The probes covered the hot spot variations of the mitochondrial genes, MT‐RNR1, MT‐TL1, MT‐ND3, and MT‐ATP6. We covered some regional genes of Prader‐William/Angleman, DiGeorge, and Williams in the microdeletion syndrome and encrypted the probe for the analysis of copy number variation in related regions. The length, density, and position of the probe were adjusted based on the Probe design software BaitDesigner of MyGenostics to improve the capture efficiency of the probe according to the GC content, region size, Tm value, and other parameters of the target region.
2.7. Interpretation of gene variants
Based on the ACMG guidelines, the pathogenicity of the variation was classified into pathogenic, likely pathogenic, unknown, likely benign, and benign. The gene results were graded to positive (high‐, moderate‐, and low‐risk genotypes), negative, and carrier according to the pathogenicity of the variation and the genetic inheritance pattern (Figure S1).
2.7.1. High risk
High‐risk genotypes were defined as one pathogenic or likely pathogenic (P/LP) variant in the autosomal dominant or Y‐linked genes; two P/LP variants in the autosomal recessive genes; one P/LP variant in females in X‐linked dominant genes, or two P/LP variants in X‐linked recessive genes; and one P/LP variant in males in X‐linked genes.
2.7.2. Moderate risk
Moderate risk genotypes were defined as one P/LP variant, and one VUS variant with Bayesian point >3 in autosomal recessive genes; one P/LP variant, and one VUS variant with Bayesian point >3 in females in X‐linked recessive genes.
2.7.3. Low risk
Low‐risk genotypes were defined as one P/LP/VUS variant, and one VUS variant with Bayesian point <3 in the autosomal recessive genes; one VUS variant with Bayesian point <3 in the autosomal dominant or Y‐linked genes; one P/LP/VUS variant, and one VUS variant with Bayesian point <3 in the X‐linked recessive genes in females.
2.7.4. Carrier
Carrier genotypes were defined as one P/LP variant in autosomal recessive genes; one P/LP variant in females in X‐linked recessive genes.
2.8. Venny analysis
The Venny 2.1 software was used to compare different NBS NGS panels with our panel through an interactive tool (Oliveros, 2007; https://bioinfogp.cnb.csic.es/tools/venny/). Multivariate Venn's Diagrams were drawn using the R version 4.0.5 software (https://www.r‐project.org/).
3. RESULTS
3.1. Generation and features of the NeoExome panel
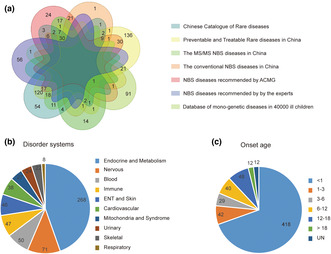
To design an NGS panel for NBS that covers severe and actionable monogenic diseases for the Chinese population, we combined the categorization of diseases based on the China National Catalog of Rare Diseases (The People's Republic of China, 2018), Preventable and Treatable Rare Diseases in China (Shanghai Municipal Health Commission, 2016), conventional MS/MS NBS project in China (Interlaboratory Quality Evaluation Committee of Neonatal Genetic and Metabolic Disease Screening CTC, Ministry of Health, 2019), conventional NBS project in China (National Health Commission of the People's Republic of China, 2010), ACMG recommendations (Fabie et al., 2019), Chinese NBS Laboratory Committee recommendations, and the MyGenostics database of monogenic diseases in 40,000 ill newborns. We collected the diseases list from each database, and then the disease panel was created. The OMIM database was used to standardize and screen out the diseases‐related genes and the disease‐genes panel (named NeoEXOME) was created (Figure 1a). Our NeoEXOME panel comprised of 601 genes and 542 diseases (Table S1).
FIGURE 1.
Characteristics of 601 gene‐diseases associations. (a) 7 categorizations of disease were collected and included to generate the gene panel. The disease system (b) and onset age (c) for those gene–disease associations were demonstrated. <1, <1 year of age (Infants); 1–3, 1–3 years of age (Toddlers); 3–6, 3–6 years of age (Preschoolers); 6–12, 6–12 years of age (Middle Childhood); 12–18, 12–18 years of age (Young Teens and Teenagers); >18, >18 years of age (Adulthood); UN, unknown.
The panel covered diseases with several systems, including skeletal, respiratory, urinary, immune system, nervous system, cardiovascular system, blood, endocrine system, metabolism, and mitochondria‐related diseases. Among them, metabolic system diseases accounted for the highest proportion (Figure 1b). In addition, most of the 601 gene‐diseases have an onset age in childhood (96%), with 418 having onset before 1 year and 577 having onset before 18 years (Figure 1c).
3.2. Performance of NeoEXOME
3.2.1. Validation results of “1000 Genomes project”
Firstly, we evaluated the performance of NeoEXOME by validating it with the data of unrelated 301 Chinese from the “1000 Genomes Project”. Of those individuals, 72 were negative, 223 were carriers, and 23 were positive, with a positivity rate of 7.6%. The variants of FLG and GJB2 account for the most frequently affected genes, at 47.8% (11/23) and 21.7% (5/23), respectively (Table S2).
3.2.2. A pilot study of the performance of NeoEXOME for NBS in China
We conducted a pilot, multicenter clinical trial to evaluate the performance of NeoEXOME. From October 2019 to September 2021, 3423 neonates were enrolled from five centers (Figure 2). DBS was collected within 3 days following birth, and NGS was performed based on our NeoEXOME panel.
FIGURE 2.
The overall flowchart of pilot multicenter study. 3423 neonates meet the inclusion criteria were enrolled from five institutions. CQ, Children's Hospital of Chongqing Medical University; XZ, Xuzhou Maternal and Child Health Care Hospital; NM, Inner Mongolia Maternal and Child Health Hospital; XB, Northwest Women and Children's Medical Center; DL, Dalian Maternal and Child Health Hospital.
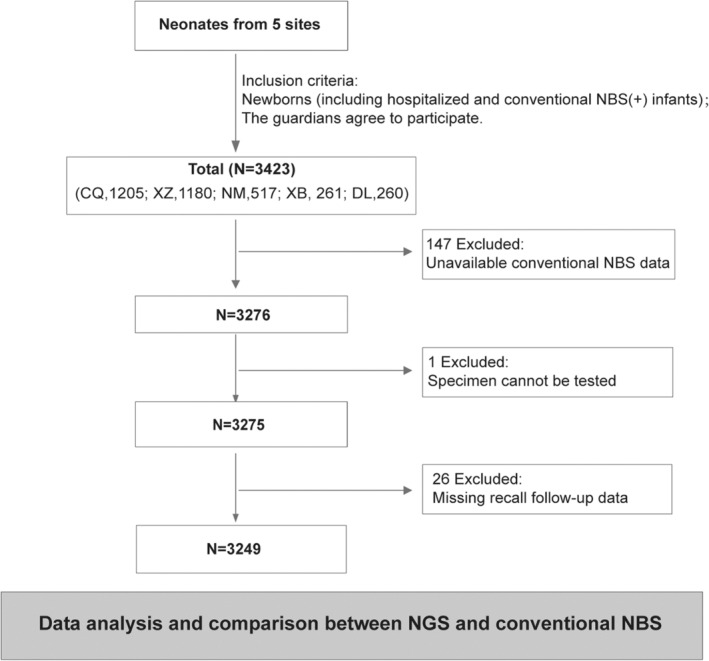
Among the 3249 neonates eligible for the analysis, 934 were NGS negative, 389 were NGS positive (244 high risk, 4 moderate risk, 141 low risk), and 1926 were carriers. The NGS positivity rate was 12.0% with most neonates genetically susceptible to monogenic‐related diseases (Figure 3a,b). Autosomal recessive inheritance was the most common inheritance pattern (Figure 3a,b). Endocrine and metabolism system disorders accounted for the highest proportion among neonates (Figure 3c). Most of the diseases detected were predicted to develop within the first year of life (Figure 3d). The variants of DUOX2 accounted for the most frequent changed genes (18.3%, 131/716), followed by UGT1A1, PAH, GJB2, FLG (Figure 3e). The top related disorders were thyroid dyshormonogenesis, hyperbilirubinemia, phenylketonuria, deafness, hyperbilirubinemia, and ichthyosis vulgaris, with the frequency of variant 5.7%, 5.4%, 5.0%, 4.1%, respectively. Of the 1926 infants that were tested as carriers, 3462 variants were found, including 1224 pathogenic variants and 2238 likely pathogenic variants (Figure 4a). The variants of GJB2 were the most common among all the centers, followed by UGT1A1, DUOX2, FLG, and SLC25A13 (Figure 4b).
FIGURE 3.
NeoEXOME detection results of positive cases. (a) Gene variants interpretation (risk grade and inheritance patterns) of neonates with mono‐gene change. (b) Gene variants interpretation (risk grade and genetic patterns) of neonates with multi‐genes change. (c) Disease system distribution of neonates that were NGS positive. (d) Gene–disease associations' onset age of neonates that were NGS positive. (e) Top 20 genes distribution of neonates with NGS positive cases. LR, Low risk; MR, Moderate risk; HR, High risk; AD, Autosomal dominant; AR, Autosomal recessive; XLD, X‐linked dominant; Mito, Mitochondrial; Syns, Syndrome; ENT, ear, nose, and throat; Endo, Endocrine; Meta, metabolism.
FIGURE 4.
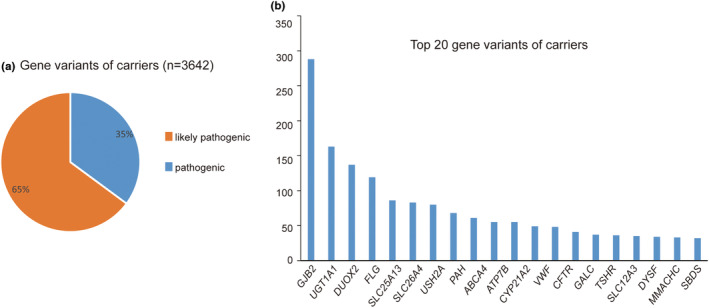
NGS detection results of carrier. (a) Gene variants characteristics of neonates that were carrier. (b) Top 20 genes distribution of neonates that were carrier.
To further validate the use of NGS in NBS, we collected conventional NBS results to compare the consistency of both approaches. Conventional NBS results of CH, PKU, hearing screening (HS), congenital adrenal hyperplasia (CAH), glucose‐6‐phosphate dehydrogenase (G6PD), and tandem MS detection were collected from the five centers (Table S3). However, CAH and G6PD detection were detected only in the Children's Hospital of Chongqing Medical University and Xuzhou Maternal and Child Health Care Hospital. In the 200 neonates that had positive results of conventional NBS, 118 were NGS positive (64 high risk, 2 moderate risk, 52 low risk), 14 were NGS negative, and 68 were NGS carriers (Table 1). Of the 118 neonates that were positive for both conventional NBS and NGS, 69 (58.5%) had consistent results with conventional NBS and NGS (Table S4). In 3049 neonates that had negative results of conventional NBS, 271 (8.9%) were NGS positive (180 high risk, 2 moderate risk, 89 low risk), 920 were NGS negative, and 1858 were NGS carriers (Table 1). Among the 271 neonates that had positive results of NGS, and negative results of conventional NBS, 168 were clinically followed up, and 9 of them (including genes of DUXO2, PAH, MUT, WAS, and SLC22A5) were clinically diagnosed with diseases.
TABLE 1.
Comparison between NGS and conventional newborn screening.
| NeoEXOME+ | Carriers | NeoEXOME‐ | Total | |||
|---|---|---|---|---|---|---|
| High risk | Moderate risk | Low risk | ||||
| Conventional NBS+ | 64 | 2 | 52 | 68 | 14 | 200 |
| Conventional NBS‐ | 180 | 2 | 89 | 1858 | 920 | 3049 |
| Total | 244 | 4 | 141 | 1926 | 934 | 3249 |
Abbreviation: NBS, newborn screening; NGS, next‐generation sequencing.
3.3. Comparison of NeoEXOME with other NBS panels
Finally, we compared our NeoEXOME panel with other NGS panels and whole genome sequencing (WGS) gene–disease lists globally in terms of gene–disease system and the disease onset age. The gene–disease information from “BabySeq” (Ceyhan‐Birsoy et al., 2017), “NC NEXUS” (Milko et al., 2019), and “NESTS” (Luo et al., 2020) projects were collected. There were 414 common genes in BabySeq and our panel (Figure 5a), most of which were endocrine and metabolism system disease‐genes. Nervous (SLC12A6 and GFAP) system, ear, nose, throat (ENT) (AMELX and DFNA5) system, and syndromes (CUL7, ERCC6) disease‐genes were higher in the BabySeq panel. Also, the BabySeq panel included gene–disease pairs of the digestive system and tumors (such as SLC26A3, APC, MSH2, SMAD4, PTEN, and TP53), which was not covered in our panel. There were 268 common genes in the NC NEXUS project and our panel (Figure 5b). ENT (USH1E, EPS8L2) system, respiratory (ABCA3, SFTPB) system, and syndromes (ALMS1, CACNB2) disease‐genes were higher in the NC NEXUS panel. Similar to the BabySeq panel, NC NEXUS also covered gene–disease pairs of the digestive system and tumor (such as CDH1, GIF, POLE, and PDX1). We compared our panel with the “NESTS” panel for Chinese newborns recently reported by Dr. Li. There were 191 common genes in the two panels (Figure 5c). Deafness‐related genes (KCNQ4, PTPRQ) dominated the gene list of the NESTS panel. Genes related to the respiratory system (RSPH4A and DNAAF1) were higher in the NESTS panel. Two genes MUTYH and APC that were related to multiple colorectal adenomas and adenomatous polyposis coli were also included in the NESTS panel. However, the NESTS panel did not include genes of DUOX2, UGT1A1, FLG, and ATP7B, which showed a high positive rate in our NeoEXOME panel. In general, the gene–disease association list designed by different projects vary in terms of focus and characteristics.
FIGURE 5.
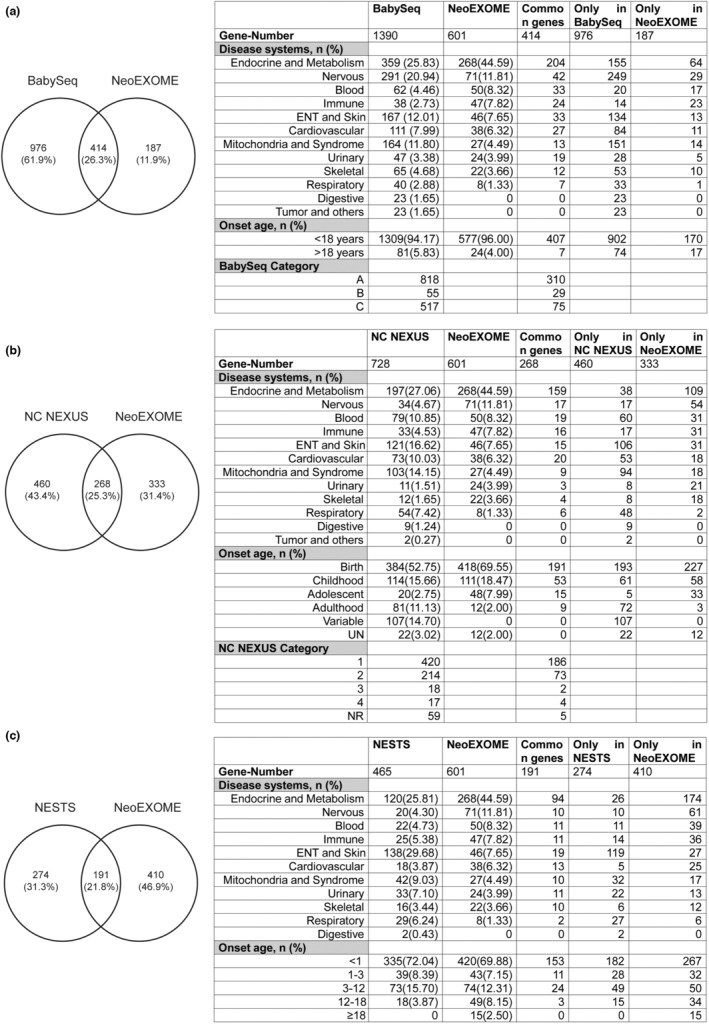
Comparison of NeoEXOME with other NBS panel. (a) NeoEXOME and BabySeq. (b) NeoEXOME and NC NEXUS. (c) NeoEXOME and NESTS.
4. DISCUSSION
NGS may play an important role in the detection of congenital anomalies. Carrier screening is a useful tool for detecting severe genetic defects. It involves the screening of both prospective parents to determine whether they carry any harmful genetic mutations that could be transmitted to their offspring. NGS‐based carrier screening allows to detect multiple variants simultaneously, providing a more comprehensive screening approach (Gregg et al., 2021). As the second‐step prevention of congenital anomalies, peri‐conception screening using NGS can detect chromosomal abnormalities in fetal free DNA of the maternal blood. The chromosomal abnormalities mainly involve Trisomy 21, 18, 13 syndromes (Breveglieri et al., 2019). The peri‐conception screening offers a non‐invasive and reliable alternative to invasive prenatal diagnostic testing, such as amniocentesis or chorionic villus sampling, reducing the risk of miscarriage. NBS using NGS is a critical tool for detecting severe, actionable, and early‐onset monogenic inherited diseases in newborns (Zhou & Zhao, 2021). In our study, we aimed to provide suitable technology, a set of rational diseases, and a gene–disease association list for NBS. We designed a NeoEXOME panel covering diseases that are severe and actionable for the Chinese population and verified it through a multicenter neonate study. We also compared our NeoEXOME panel with other NGS‐based NBS panels to illustrate the respective characteristics.
The gene–disease list is critical in the design of the NGS panel. The BabySeq project conducted a study to curate a catalog of gene–disease pairs based on the ACMG, the ClinGen clinical validity classification framework criteria, penetrance, and age of onset. After screening, they identified 954 genes that met their inclusion criteria (Ceyhan‐Birsoy et al., 2017). The NC NEXUS project developed an age‐based framework to assess the gene–disease list and categorized 822 gene–disease pairs into four groups (Milko et al., 2019). In China, several NGS screening studies for NBS have been conducted, including the panel designed by Dr. Yu (573 genes) (Luo et al., 2020), Dr. Li (465 genes) (Hao et al., 2022), Dr. Zhao(134 genes) (Huang et al., 2022), and Dr. Xu(164 genes; Tong et al., 2022; Wang et al., 2022). In our study, we integrated the diseases catalog of Rare Diseases in China, routine NBS diseases in China, ACMG and mainland experts' recommendation, and the database of genetic diseases in 40,000 ill‐children to generate a comprehensive list of 601 genes for the NeoEXOME panel for NBS.
The positivity rate of our NGS panel in the “1000 Genomes Project” was 7.6%, higher than that of previous studies (Zhang et al., 2015). The most frequently changed genes in our analysis were FLG and GJB2. FLG is a pathogenic gene for ichthyosis vulgaris, which encodes filaggrin and plays a key role in epidermal terminal differentiation and skin barrier formation. The proportion of FLG gene variation in ichthyosis vulgaris was reported to be 55.6%, and patients having ichthyosis vulgaris with FLG gene variant experienced more severe diseases (Qian et al., 2015). However, FLG also has high variation in the normal population, Palmer and colleagues carried out FLG analysis with 1008 people of the European origin and found that the functional deletion variants of the FLG gene were approximately 9% (Palmer et al., 2006). We believed that the FLG gene should not be included in our panel. Therefore, we recalculate NGS positivity rate excluding the FLG variants, 376 infants were NGS positive (11.57%) among the 3249 neonates eligible for the analysis. In addition, GJB2 gene variation is generally considered to be a common cause of non‐syndromic deafness (Koohiyan et al., 2020). Our analysis identified 5 GJB2 variants, four of which were c.109G>A variant (Table S2), a pathogenic variant with incomplete penetrance and having a high carrier rate among Asians (Shen et al., 2019). Our study highlights the importance of genetic counseling for such genetic variations.
Moreover, it is also necessary to consider whether genes linked to adult‐onset diseases should be detected during the neonatal stages. In the previous study conducted by the BabySeq project, a BRCA2 pathogenic variant was discovered in an infant, which increases the risk of breast cancer by 45%, and ovarian cancer by 11% in women (Ceyhan‐Birsoy et al., 2019). The researchers faced moral distress and proposed returning the adult‐onset genetic variants (Holm et al., 2019). However, this sparked a heated ethical debate: Lainie FR published an article arguing that researchers should avoid identifying adult‐onset genetic variants as it could have a psychological impact on the child and their parents and deprive the child of the right to an open future (Ross & Clayton, 2019). Our current panel included 12 adult‐onset genes and 12 genes with unknown onset age which we plan to exclude in the next round of study.
Our study has several limitations. The primary disadvantage is the lack of complete follow‐up data for some participants in our validation cohort. Our study indicated low concordance between NGS and conventional NBS (Table 1), which was also reported in the BabySeq project (Wojcik et al., 2021). One of the reasons for this is the incomplete conventional NBS data collected due to the varying detection indexes in different regions. Additionally, sequencing cannot fully represent phenotypes, and some participants may be gene‐positive, but negative for the clinical phenotype (Hao et al., 2022). This could be due to the differences in the time of disease onset or the low penetrance of the gene variant. Once the penetrance of a gene variant is low, the individual with this variant has a low chance to be symptomatic (Seaby & Ennis, 2020). This is also one limitation in our study. We did not include penetrance in our report interpretation category. Our report category needs modification, and we must strengthen our follow‐up in the subsequent study.
Although studies have shown that NGS may play a role as second‐tier screening in NBS and cannot replace MS, we believe that sequencing can be performed simultaneously with conventional NBS if the cost is reasonable. Sequencing could enable the early detection of diseases that have no biochemical markers, such as spinal muscular atrophy (SMA), Duchenne's muscular dystrophy (DMD), and cardiovascular diseases. Therefore, we proposed that the NeoEXOME panel may have the following applications in the clinical practice: (1) as a first‐tier NBS for genetic diseases with no biochemical markers; (2) as an adjunct diagnostic tool for monogenic inherited diseases with obvious abnormal phenotype; (3) as a second‐tier NBS in combination with conventional NBS to enhance diagnostic accuracy. The application of sequencing in healthy infants with negative conventional NBS results can help to determine the gene variants in the early stage of their lifetime to prevent mortality; (4) as an exclusion (quasi‐first‐line) screening for infants with low phenotypic specificity (jaundice, etc.). Further clinical trials are needed to validate the use of NeoEXOME in clinical practice.
In conclusion, we designed a promising targeted‐sequencing panel for NBS in China and evaluated this approach in a multicenter pilot study. As the technology continues to evolve, NGS is expected to increasingly contribute to improving our ability to prevent and manage congenital anomalies.
AUTHOR CONTRIBUTIONS
Conceptualization, LZ, GJY, and JW.; Investigation, ZYC, XYH, DJW, MSG, FS, RQ, RXZ, CRS, XHW BZ, DHC, HHY, YPQ, GSS, JW, and PPW; Validation, ZYC, JXW, HYZ, ZJY, and LZ; Writing—Original Draft Preparation, ZYC and LZ; Writing—Review & Editing, ZYC and LZ; Visualization, ZYC and PPW; Supervision, LZ, and GJY; Funding acquisition, LZ, and GJY; All authors have read and agreed to the published version of the manuscript.
FUNDING INFORMATION
This work is partially supported by the Chongqing Science and Technology Bureau (cstc2019jscx‐msxm0189), Shanghai Hospital Development Center (SHDC2020CR1047B), and the Natural Science Foundation of China (82070167, 82270160).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Supporting information
Table S1.
Table S2.
Table S3.
Table S4.
Figure S1.
ACKNOWLEDGMENTS
We thank all the participants and their guardians in the research.
Cao, Z. , He, X. , Wang, D. , Gu, M. , Suo, F. , Qiang, R. , Zhang, R. , Song, C. , Wang, X. , Zhu, B. , Cao, D. , Yu, H. , Qu, Y. , Shen, G. , Wu, J. , Wang, P. , Wang, J. , Zhang, H. , Yan, Z. … Zou, L. (2024). Targeted exome sequencing strategy (NeoEXOME) for Chinese newborns using a pilot study with 3423 neonates. Molecular Genetics & Genomic Medicine, 12, e2357. 10.1002/mgg3.2357
Contributor Information
Guangjun Yu, Email: gjyu@shchildren.com.cn.
Lin Zou, Email: zoulin@shchildren.com.cn, Email: zoulin74@126.com.
DATA AVAILABILITY STATEMENT
All data are available in the main text or the Supplementary Materials.
REFERENCES
- Breveglieri, G. , D'Aversa, E. , Finotti, A. , & Borgatti, M. (2019). Non‐invasive prenatal testing using fetal DNA. Molecular Diagnosis & Therapy, 23(2), 291–299. [DOI] [PubMed] [Google Scholar]
- Ceyhan‐Birsoy, O. , Machini, K. , Lebo, M. S. , Yu, T. W. , Agrawal, P. B. , Parad, R. B. , Holm, I. A. , McGuire, A. , Green, R. C. , Beggs, A. H. , & Rehm, H. L. (2017). A curated gene list for reporting results of newborn genomic sequencing. Genetics in Medicine: Official Journal of the American College of Medical Genetics, 19(7), 809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceyhan‐Birsoy, O. , Murry, J. B. , Machini, K. , Lebo, M. S. , Yu, T. W. , Fayer, S. , Genetti, C. A. , Schwartz, T. S. , Agrawal, P. B. , Parad, R. B. , Holm, I. A. , McGuire, A. L. , Green, R. C. , Rehm, H. L. , Beggs, A. H. , Agrawal, P. B. , Beggs, A. H. , Betting, W. N. , Ceyhan‐Birsoy, O. , … Yu, T. W. (2019). Interpretation of genomic sequencing results in healthy and ill newborns: Results from the BabySeq Project. American Journal of Human Genetics, 104(1), 76–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabie, N. A. V. , Pappas, K. B. , & Feldman, G. L. (2019). The current state of newborn screening in the United States. Pediatric Clinics of North America, 66(2), 369–386. [DOI] [PubMed] [Google Scholar]
- Gregg, A. R. , Aarabi, M. , Klugman, S. , Leach, N. T. , Bashford, M. T. , Goldwaser, T. , Chen, E. , Sparks, T. N. , Reddi, H. V. , Rajkovic, A. , Dungan, J. S. , & ACMG Professional Practice and Guidelines Committee . (2021). Screening for autosomal recessive and X‐linked conditions during pregnancy and preconception: A practice resource of the American College of Medical Genetics and Genomics (ACMG). Genetics in Medicine: Official Journal of the American College of Medical Genetics, 23(10), 1793–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, R. , & Susi, A. (1963). A simple phenylalanine method for detecting phenylketonuria in large populations of newborn infants. Pediatrics, 32, 338–343. [PubMed] [Google Scholar]
- Han, D. , Li, Z. , Li, R. , Tan, P. , Zhang, R. , & Li, J. (2019). mNGS in clinical microbiology laboratories: On the road to maturity. Critical Reviews in Microbiology, 45(5–6), 668–685. [DOI] [PubMed] [Google Scholar]
- Hao, C. , Guo, R. , Hu, X. , Qi, Z. , Guo, Q. , Liu, X. , Liu, Y. , Sun, Y. , Zhang, X. , Jin, F. , Wu, X. , Cai, R. , Zeng, D. , Hu, X. , Wang, X. , Ji, X. , Li, W. , Xing, Q. , Mu, L. , … Li, W. (2022). Newborn screening with targeted sequencing: A multicenter investigation and a pilot clinical study in China. Journal of Genetics and Genomics, 49(1), 13–19. 10.1016/j.jgg.2021.08.008 [DOI] [PubMed] [Google Scholar]
- Holm, I. A. , McGuire, A. , Pereira, S. , Rehm, H. , Green, R. C. , & Beggs, A. H. (2019). Returning a genomic result for an adult‐onset condition to the parents of a newborn: Insights from the BabySeq Project. Pediatrics, 143(Suppl 1), S37–S43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Wu, D. , Zhu, L. , Wang, W. , Yang, R. , Yang, J. , He, Q. , Zhu, B. , You, Y. , Xiao, R. , & Zhao, Z. (2022). Application of a next‐generation sequencing (NGS) panel in newborn screening efficiently identifies inborn disorders of neonates. Orphanet Journal of Rare Diseases, 17(1), 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Interlaboratory Quality Evaluation Committee of Neonatal Genetic and Metabolic Disease Screening CTC, Ministry of Health . (2019). Expert consensus on tandem mass spectrometry for neonatal disease screening. Chinese Journal of Laboratory Medicine, 42(42), 89–97. [Google Scholar]
- Koohiyan, M. , Koohian, F. , & Azadegan‐Dehkordi, F. (2020). GJB2‐related hearing loss in central Iran: Review of the spectrum and frequency of gene mutations. Annals of Human Genetics, 84(2), 107–113. [DOI] [PubMed] [Google Scholar]
- Lin, Y. , Zhang, W. , Huang, C. , Lin, C. , Lin, W. , Peng, W. , Fu, Q. , & Chen, D. (2021). Increased detection of primary carnitine deficiency through second‐tier newborn genetic screening. Orphanet Journal of Rare Diseases, 16(1), 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, X. , Sun, Y. , Xu, F. , Guo, J. , Li, L. , Lin, Z. , Ye, J. , Gu, X. , & Yu, Y. (2020). A pilot study of expanded newborn screening for 573 genes related to severe inherited disorders in China: Results from 1,127 newborns. Annals of Translational Medicine, 8(17), 1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell, J. R. , Shilling, C. , Leslie, N. D. , Flanigan, K. M. , al‐Dahhak, R. , Gastier‐Foster, J. , Kneile, K. , Dunn, D. M. , Duval, B. , Aoyagi, A. , Hamil, C. , Mahmoud, M. , Roush, K. , Bird, L. , Rankin, C. , Lilly, H. , Street, N. , Chandrasekar, R. , & Weiss, R. B. (2012). Evidence‐based path to newborn screening for Duchenne muscular dystrophy. Annals of Neurology, 71(3), 304–313. [DOI] [PubMed] [Google Scholar]
- Meng, L. , Pammi, M. , Saronwala, A. , Magoulas, P. , Ghazi, A. R. , Vetrini, F. , Zhang, J. , He, W. , Dharmadhikari, A. V. , Qu, C. , Ward, P. , Braxton, A. , Narayanan, S. , Ge, X. , Tokita, M. J. , Santiago‐Sim, T. , Dai, H. , Chiang, T. , Smith, H. , … Lalani, S. R. (2017). Use of exome sequencing for infants in intensive care units: Ascertainment of severe single‐gene disorders and effect on medical management. JAMA Pediatrics, 171(12), e173438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milko, L. V. , O'Daniel, J. M. , DeCristo, D. M. , Crowley, S. B. , Foreman, A. K. M. , Wallace, K. E. , Mollison, L. F. , Strande, N. T. , Girnary, Z. S. , Boshe, L. J. , Aylsworth, A. S. , Gucsavas‐Calikoglu, M. , Frazier, D. M. , Vora, N. L. , Roche, M. I. , Powell, B. C. , Powell, C. M. , & Berg, J. S. (2019). An age‐based framework for evaluating genome‐scale sequencing results in newborn screening. The Journal of Pediatrics, 209, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ministry of Health of the People's Republic of China . (2012). The report on prevention and treatment of birth defects‐ Ministry of Public Health of China. http://www.gov.cn/gzdt/2012‐09/12/content_2223371.htm
- National Health Commission of the People's Republic of China . (2010). Technical specification for screening of neonatal diseases. http://www.nhc.gov.cn/wjw/gfxwj/201304/e6215cd2b1c541c6914aefeb542e3467.shtml
- Oliveros, J. C. (2007. ‐2015). Venny. An interactive tool for comparing lists with Venn's diagrams. https://bioinfogp.cnb.csic.es/tools/venny/index.html
- Ombrone, D. , Giocaliere, E. , Forni, G. , Malvagia, S. , & la Marca, G. (2016). Expanded newborn screening by mass spectrometry: New tests, future perspectives. Mass Spectrometry Reviews, 35(1), 71–84. [DOI] [PubMed] [Google Scholar]
- Palmer, C. N. , Irvine, A. D. , Terron‐Kwiatkowski, A. , Zhao, Y. , Liao, H. , Lee, S. P. , Goudie, D. R. , Sandilands, A. , Campbell, L. E. , Smith, F. J. , O'Regan, G. M. , Watson, R. M. , Cecil, J. E. , Bale, S. J. , Compton, J. G. , DiGiovanna, J. J. , Fleckman, P. , Lewis‐Jones, S. , Arseculeratne, G. , … McLean, W. H. (2006). Common loss‐of‐function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nature Genetics, 38(4), 441–446. [DOI] [PubMed] [Google Scholar]
- Parad, R. B. , Kaler, S. G. , Mauceli, E. , Sokolsky, T. , Yi, L. , & Bhattacharjee, A. (2020). Targeted next generation sequencing for newborn screening of Menkes disease. Molecular Genetics and Metabolism Reports, 24, 100625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, Q. , Cheng, R. , Li, M. , Guo, Y. , Yu, X. , Zhang, Z. , Yu, H. , Qi, H. , & Yao, Z. (2015). Prevalence of atopic dermatitis, ichthyosis and filaggrin mutations in adolescents in a middle school in Shanghai. Chinese Journal of Dermatology, 48, 629–632. [Google Scholar]
- Rajabi, F. (2018). Updates in newborn. Screening, 47(5), e187–e190. [DOI] [PubMed] [Google Scholar]
- Rizvi, H. , Sanchez‐Vega, F. , La, K. , Chatila, W. K. , Jonsson, P. , Halpenny, D. , Plodkowski, A. J. , Long, N. M. , Sauter, J. L. , Rekhtman, N. , Hollmann, T. J. , Schalper, K. A. , Gainor, J. F. , Shen, R. , Ni, A. , Arbour, K. C. , Merghoub, T. , Wolchok, J. D. , Snyder, A. , … Hellmann, M. D. (2018). Molecular determinants of response to anti‐programmed cell death (PD)‐1 and anti‐programmed death‐ligand 1 (PD‐L1) blockade in patients with non‐small‐cell lung cancer profiled with targeted next‐generation sequencing. Journal of Clinical Oncology, 36(7), 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, L. F. , & Clayton, E. W. (2019). Ethical issues in newborn sequencing research: The case study of BabySeq. Pediatrics, 144(6), e20191031. 10.1542/peds.2019-1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaby, E. G. , & Ennis, S. (2020). Challenges in the diagnosis and discovery of rare genetic disorders using contemporary sequencing technologies. Briefings in Functional Genomics, 19(4), 243–258. [DOI] [PubMed] [Google Scholar]
- Shanghai Municipal Health Commission . (2016). List of major rare diseases in Shanghai. https://wsjkw.sh.gov.cn/fybj2/20180815/0012‐59645.html
- Shen, J. , Oza, A. M. , Del Castillo, I. , Duzkale, H. , Matsunaga, T. , Pandya, A. , Kang, H. P. , Mar‐Heyming, R. , Guha, S. , Moyer, K. , Lo, C. , Kenna, M. , Alexander, J. J. , Zhang, Y. , Hirsch, Y. , Luo, M. , Cao, Y. , Wai Choy, K. , Cheng, Y. F. , … ClinGen Hearing Loss Working Group . (2019). Consensus interpretation of the p.Met34Thr and p.Val37Ile variants in GJB2 by the ClinGen hearing loss expert panel. Genetics in Medicine: Official Journal of the American College of Medical Genetics, 21(11), 2442–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter, J. L. , Meinzen‐Derr, J. , Rose, S. R. , Leslie, N. D. , Chandrasekar, R. , Linard, S. M. , & Akinbi, H. T. (2010). The effects of gestational age and birth weight on false‐positive newborn‐screening rates. Pediatrics, 126(5), 910–916. [DOI] [PubMed] [Google Scholar]
- Swedish Council on Health Technology A . (2016). SBU systematic review summaries. In Prenatal diagnosis through next generation sequencing (NGS). Swedish Council on Health Technology Assessment (SBU) Copyright © 2016 by the Swedish Council on Health Technology Assessment. [PubMed] [Google Scholar]
- The People's Republic of China . (2018). Notice on publication of the list of Rare disease. https://www.gov.cn/zhengce/zhengceku/2018‐12/31/content_5435167.htm
- Tong, F. , Wang, J. , Xiao, R. , Wu, B. B. , Zou, C. C. , Wu, D. W. , Wang, H. , Zou, H. , Han, L. S. , Yang, L. , Zou, L. , Hei, M. Y. , Yang, R. L. , Yuan, T. M. , Wen, W. , Huang, X. W. , Gu, X. F. , Yang, Y. L. , Huang, Y. L. , … Zhao, Z. Y. (2022). Application of next generation sequencing in the screening of monogenic diseases in China, 2021: A consensus among Chinese newborn screening experts. World Journal of Pediatrics: WJP, 18(4), 235–242. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Wang, Y. Y. , Hong, D. Y. , Zhang, Z. L. , Li, Y. H. , Yang, P. Y. , Sun, Y. , Jiang, T. , & Xu, Z. F. (2022). Combined genetic screening and traditional biochemical screening to optimize newborn screening systems. Clinica Chimica Acta, 528, 44–51. [DOI] [PubMed] [Google Scholar]
- Watson, M. S. , Mann, M. Y. , Lloyd‐Puryear, M. A. , Rinaldo, P. , Rodney Howell, R. , & American College of Medical Genetics Newborn Screening Expert Group . (2006). Newborn screening: toward a uniform screening panel and system—executive summary. Pediatrics, 117(5 Pt 2), S296–S307. [DOI] [PubMed] [Google Scholar]
- Wilson, J. M. , & Jungner, Y. G. (1968). Principles and practice of mass screening for disease. Boletín de la Oficina Sanitaria Panamericana, 65(4), 281–393. [PubMed] [Google Scholar]
- Wojcik, M. H. , Zhang, T. , Ceyhan‐Birsoy, O. , Genetti, C. A. , Lebo, M. S. , Yu, T. W. , Parad, R. B. , Holm, I. A. , Rehm, H. L. , Beggs, A. H. , Green, R. C. , Agrawal, P. B. , Agrawal, P. B. , Beggs, A. H. , Betting, W. N. , Ceyhan‐Birsoy, O. , Christensen, K. D. , Dukhovny, D. , Fayer, S. , … Yu, T. W. (2021). Discordant results between conventional newborn screening and genomic sequencing in the BabySeq project. Genetics in Medicine: Official Journal of the American College of Medical Genetics, 23(7), 1372–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . (2010). Sixty‐third World Health Assembly, Birth defects.
- Yu, X. , Lin, Y. , & Wu, H. (2020). Targeted next‐generation sequencing identifies separate causes of hearing loss in one deaf family and variable clinical manifestations for the p.R161C mutation in SOX10. Neural Plasticity, 2020, 8860837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, J. Y. , Qin, Y. F. , & Zhao, Z. Y. (2009). Neonatal screening for congenital hypothyroidism and phenylketonuria in China. World Journal of Pediatrics: WJP, 5(2), 136–139. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Walsh, M. F. , Wu, G. , Edmonson, M. N. , Gruber, T. A. , Easton, J. , Hedges, D. , Ma, X. , Zhou, X. , Yergeau, D. A. , Wilkinson, M. R. , Vadodaria, B. , Chen, X. , McGee, R. B. , Hines‐Dowell, S. , Nuccio, R. , Quinn, E. , Shurtleff, S. A. , Rusch, M. , … Downing, J. R. (2015). Germline mutations in predisposition genes in pediatric cancer. The New England Journal of Medicine, 373(24), 2336–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, W. H. , & Zhao, Z. Y. (2021). Genomic newborn screening: Opportunities and challenges. Zhonghua Er Ke Za Zhi, 59(7), 541–544. [DOI] [PubMed] [Google Scholar]
- Zhu, T. , Gong, X. , Bei, F. , Ma, L. , Chen, Y. , Zhang, Y. , Wang, X. , Sun, J. , Wang, J. , Qiu, G. , Sun, J. , Sun, Y. , & Zhang, Y. (2020). Application of next‐generation sequencing for genetic diagnosis in neonatal intensive care units: Results of a multicenter study in China. Frontiers in Genetics, 11, 565078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Table S2.
Table S3.
Table S4.
Figure S1.
Data Availability Statement
All data are available in the main text or the Supplementary Materials.


