Abstract
The Bordetella pertussis proteins P.69 (also designated pertactin) and pertussis toxin are important virulence factors and have been shown to confer protective immunity in animals and humans. Both proteins are used in the new generation of acellular pertussis vaccines (ACVs), and it is therefore important to study the degree of antigenic variation in these proteins. Sequence analysis of the genes for P.69 and the pertussis toxin S1 subunit, using strains collected from Dutch patients in the period 1949 to 1996, revealed three P.69 and three S1 variants which show differences in amino acid sequence. Polymorphism in P.69 was confined to a region comprised of repeats and located proximal to the RGD motif involved in adherence to host tissues. Variation in S1 was observed in two regions previously identified as T-cell epitopes. P.69 and S1 variants, identical to those included in the Dutch whole-cell pertussis vaccine (WCV), were found in 100% of the strains from the 1950s, the period when the WCV was introduced in The Netherlands. However, nonvaccine types of P.69 and S1 gradually replaced the vaccine types in later years and were found in ∼90% strains from 1990 to 1996. These results suggest that vaccination has selected for strains which are antigenically distinct from vaccine strains. Analysis of strains from vaccinated and nonvaccinated individuals indicated that the WCV protects better against strains with the vaccine type P.69 than against strains with non-vaccine types (P = 0.024). ACVs contain P.69 and S1 types which are found in only 10% of recent Dutch B. pertussis isolates, implying that they do not have an optimal composition. Our findings cast a new light on the reemergence of pertussis in highly vaccinated populations and may have major implications for the long-term efficacy of both WCVs and ACVs.
In the prevaccination era, pertussis was a major cause of child morbidity and mortality (35). In the 1950s, many countries, including The Netherlands, introduced whole-cell pertussis vaccines (WCVs) which greatly reduced the pertussis burden, and in such countries the disease more or less disappeared. In the 1970s, interest in pertussis revived due to side effects caused by pertussis vaccination (10, 35). This has resulted in the development of acellular pertussis vaccines (ACVs), which are now being introduced in various countries (14, 30, 31). In recent years, interest in pertussis has increased because in a number of countries which use WCVs, such as Australia, Canada, the United States, and The Netherlands, there is evidence that the incidence of pertussis is increasing despite high vaccination coverage (1, 4, 5, 12). This is exemplified by the pertussis epidemic in The Netherlands in 1996, which showed an incidence which was fivefold higher than in previous epidemics (11). The resurgence of pertussis may be caused by several factors (4, 11, 25) such as waning vaccine-induced immunity, a decrease in vaccine quality (e.g., due to changes in production processes), or a decrease in vaccine coverage. Further, improved surveillance and changes in case definition may result in seemingly higher incidences. Another cause for the reemergence of pertussis may be the expansion of strains which are antigenically distinct from vaccine strains (35). Using DNA fingerprinting, we have shown that the population structure of Bordetella pertussis in The Netherlands has changed over time, and we suggested that these changes may have been driven by vaccination (34). Here we investigate this hypothesis further by analyzing antigenic shifts in the Dutch B. pertussis population. We focused on two B. pertussis proteins, P.69 (also designated P.69/pertactin) and pertussis toxin, which are part of most ACVs and have been shown to confer protective immunity in animals and humans (16, 27, 31).
P.69 is produced as a large (910-amino-acid) precursor molecule. It is proteolytically processed at its N and C termini to produce P.69 and P.30, which are located at the cell surface and in the outer membrane, respectively (7, 8). P.69 contains the amino acid triplet arginine-glycine-aspartic acid (RGD), a sequence motif which functions as a cell-binding site in a number of mammalian proteins, and it has been shown that the P.69 RGD sequence is also involved in adherence to host cells (17). Pertussis toxin is composed of five subunits (S1 to S5); the toxic, catalytic functions are located in the S1 subunit, which comprises 235 amino acids (20, 24). Like P.69, pertussis toxin is excreted and may be found loosely associated with the outer membrane. Pertussis toxin has numerous biological activities and probably plays a role in hampering the host immune response (32).
Both P.69 and pertussis toxin are part of most ACVs, and it is therefore important to study the degree of antigenic variation in these proteins. Variation in the S1 pertussis toxin subunit was observed previously with a limited number of strains (2). Here we extend these observations by using a well-defined strain collection. Further, we show, for the first time, that B. pertussis strains show variation in P.69. Temporal trends in the frequencies of P.69 and S1 variants suggest that the emergence of novel variants has been driven by vaccination. Our findings cast new light on the reemergence of pertussis in highly vaccinated populations and may have major implications for the long-term efficacy of both WCVs and ACVs.
MATERIALS AND METHODS
Strains.
B. pertussis strains were collected in the years 1949 to 1996. The distribution of the strains over different periods was as follows: 1949 to 1980, 35; 1981, 17; 1982, 11; 1983, 6; 1984, 6; 1985, 9; 1986, 7; 1987, 12; 1988, 12; 1989; 27; 1990, 21; 1991, 0; 1992, 16; 1993, 21; 1994, 45; 1995, 18; and 1996, 49. Most strains were sent to the National Institute for Public Health and the Environment (RIVM) by regional laboratories for serotyping or confirmation of identification. Detailed information, such as the region where the strain was isolated and patient age, was available for strains isolated in 1988 and later. B. pertussis strains were grown on Bordet-Gengou agar (Difco Catalog no. 0048-15-7) supplemented with 1% glycerol and 15% sheep blood or in Verwey medium (33) at 35°C for 3 days.
DNA sequencing.
DNA was isolated essentially as described in reference 3. DNA sequencing was performed by PCR amplification of chromosomal DNA followed by direct sequencing of the PCR products. Conditions for amplification of the prn and s1 genes were as follows. The prn gene was amplified in 50 μl containing 1 mM Tris-HCl (pH 8), 2 mM MgCl2, 10% dimethyl sulfoxide, 200 μM each deoxynucleotide, 10 pmol of each primer, and 0.75 U of AmpliTaq polymerase (Perkin-Elmer). The reaction mixtures were preheated at 95°C for 3 min, and 30 amplification cycles were performed in a Hybaid OmniGene incubator, using the following program: 20 s at 95°C, 30 s at 55°C, and 1 min at 72°C. The last cycle was concluded with reaction for 7 min at 72°C. The s1 gene was amplified in 50 μl containing 10 mM Tris-HCl (pH 9), 1.5 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, 0.01% gelatin, 5% dimethyl sulfoxide, 200 μM each deoxynucleotide, 10 pmol of each primer, and 0.75 U of AmpliTaq polymerase (Perkin-Elmer). The reaction mixtures were preheated at 95°C for 3 min, and 30 amplification cycles were performed in a Hybaid OmniGene incubator, using the following program: 15 s at 95°C, 15 s at 59°C, and 1 min at 72°C. The last cycle was concluded with reaction for 10 min at 72°C. PCR fragments were purified with a Qiaquick (Qiagen) PCR purification kit and sequenced on both strands with the primers used for amplification in combination with internal primers (see below). Sequence reactions were carried out with an ABI prism dye terminator Cycle Sequencing Ready reaction kit, and the products were analyzed on a model 373 or 377 ABI DNA sequencer (Perkin-Elmer–Applied Biosystems).
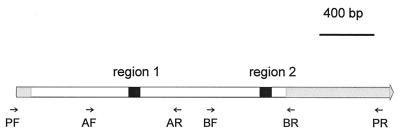
The sequences and positions of the primers used to amplify and sequence the prn and s1 genes are indicated in Table 1 and Fig. 1. A region coding for P.69 was amplified with primers PF and PR. The P.69-encoding region from six strains, isolated in the years 1954 (n = 2), 1990 (n = 1), and 1994 (n = 3), was sequenced completely, using a combination of the primers depicted in Fig. 1. Two polymorphic regions of the prn gene, designated regions 1 and 2 (Fig. 1), were sequenced in a larger number of strains. A DNA fragment containing regions 1 and 2 was amplified with primers AF and BR. Subsequently, region 1 was sequenced by using primers AF and AR, while region 2 was sequenced with primers BF and BR (Fig. 1). Region 1 was sequenced in all 312 strains used in this study. Region 2 was sequenced in 126 strains. The s1 gene was sequenced completely, using three primers (Table 1). Forty-nine strains were analyzed, isolated in three periods spanning approximately 7 years: 1949 to 1954 (n = 12), 1978 to 1985 (n = 15), and 1990 to 1996 (n = 17).
TABLE 1.
Primers used in this studya
| Primer | Sequence (5′-3′) | Gene | Position |
|---|---|---|---|
| PF | TGTCTCTGTCACGCATTGTC | prn | 152–171 |
| AF | GCCAATGTCACGGTCCAA | prn | 649–666 |
| AR | GCAAGGTGATCGACAGGG | prn | 1234–1217 |
| BR | CGGATTCAGGCGCAACTC | prn | 2076–2059 |
| BF | AGCTGGGCGGTTCAAGGT | prn | 1542–1559 |
| PR | ATGCCGTTGGTGTGTACCGT | prn | 2714–2695 |
| S1F | TAGGCACCATCAAAACGCAG | s1 | 474–493 |
| S1FM | ACAATGCCGGCCGTATCCTC | s1 | 946–965 |
| S1R | TCAATTACCGGAGTTGGGCG | s1 | 1350–1330 |
FIG. 1.
Structure of the prn gene, coding for the P.69 precursor. The regions coding for the N- and C-terminal parts of the P.69 precursor, which are removed, are indicated in gray. The central white region codes for P.69, which is exposed at the cell surface. Regions 1 and 2, which code for the repeats GGxxP and PQP, respectively, are indicated in black. The small arrows show the approximate positions of primers used for PCR and sequencing. The figure is based on data in reference 8.
Statistical analysis.
P values (one sided) were calculated by using the Mantel-Haenszel chi-square test for trend analysis (29).
RESULTS
Polymorphism in P.69.
The gene for the B. pertussis P.69 precursor (prn) has been sequenced (8), and its gene structure suggested that polymorphism may occur in two regions, designated regions 1 and 2 (Fig. 1), that are comprised of repeats coding for the amino acid sequences GGxxP and PQP, respectively. Region 1 is near the RGD motif, which is involved in adherence to host receptors (17). DNA regions with repeated sequences show a high mutation frequency due to recombination between repeats (28). That polymorphism may occur in these two regions was also suggested by a comparison of the B. pertussis P.69 with the homologous proteins derived from the closely related species B. bronchiseptica and B. parapertussis. Although amino acid substitutions are found over the whole length of the molecule, the largest differences are found in regions 1 and 2 (19). Further, the differences in regions 1 and 2 can be explained by the addition or removal of the repeat unit, which is in accordance with the proposed mechanism underlying polymorphism in repeated regions.
To identify polymorphic regions in the B. pertussis P.69 molecule, we sequenced the prn genes from six B. pertussis strains isolated in the years 1950, 1990, and 1994. The region sequenced comprised more than 90% of the prn gene and completely encompassed the DNA coding for the cell surface-exposed P.69 protein (Fig. 1). All differences observed between the six sequences were confined to region 1. A more extensive analysis, focused on regions 1 and 2 and involving 126 strains from different periods, also revealed variation in region 1 only, and subsequent analyses were confined to this region (total of 312 strains).
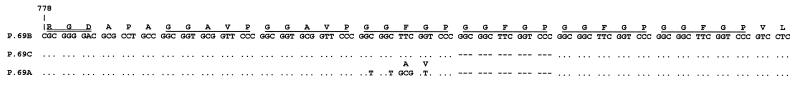
An analysis of Dutch strains isolated in the years 1949 to 1996 revealed three P.69 types (designated P.69A, -B, and -C), which differed in two amino acid substitutions and/or the number of GGxxP repeats (Fig. 2). P.69B contained six repeats, while P.69A and -C contained five. P.69A was identical to the previously characterized P.69 molecule (8). The two strains used for the Dutch WCV were found to produce P.69A.
FIG. 2.
Variants of P.69 observed in the Dutch B. pertussis population. Depicted are the DNA sequence and corresponding amino acid sequence of the single polymorphic region observed. In P.69C and P.69A, only differences with respect to P.69B are indicated. Dots and dashes indicate identical bases and gaps, respectively. GGxxP repeats are underlined. The RGD sequence, which is involved in attachment to mammalian cells, is indicated by double underlining. The number 778 refers to the position of the first base of the indicated sequence in the prn gene.
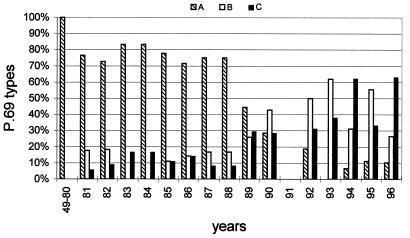
When the percentages of the three P.69 types in different years were determined, interesting trends were observed (Fig. 3). In strains isolated in the period 1949 to 1980, only P.69A was found. In 1981, two novel types (P.69B and P.69C) appeared, and the percentages of these types remained more or less constant, comprising 20 to 30% of all isolates, until 1988. From 1989 on, an increase in strains with P.69B and P.69C was observed until they comprised ∼90% of the B. pertussis population. In 1993 to 1996, the predominance of P.69B and P.69C alternated in successive years.
FIG. 3.
Temporal trends in the frequency of P.69 variants in the Dutch B. pertussis population. The percentage of strains harboring distinct P.69 variants was determined in Dutch strains collected between 1949 and 1996 for each period indicated on the x axis. Strains collected between 1949 and 1980 contained P.69A only. Insufficient strains from 1991 were available. The Dutch WCV contains P.69A.
Distribution of P.69 types in groups with different levels of vaccine-induced immunity.
The two strains which comprise the Dutch WCV both produce P.69A, and it is conceivable that the displacement of strains carrying this type has been driven by vaccination. This hypothesis predicts that the percentage of P.69A strains isolated should be lower in vaccinated individuals than in nonvaccinated individuals. In The Netherlands, children are vaccinated with the WCV at the ages of 3, 4, 5, and 11 months. Further, it has been well established that vaccine-induced immunity wanes after a few years, and in older age groups vaccine-induced immunity is supplemented and possibly eventually overshadowed by immunity acquired by infection (23). Thus, we presumed that the age groups 0 to 3, 4 to 11, 12 to 48, and >48 months were comprised of individuals with no, partial, optimal, and waning vaccine-induced immunity, respectively. It should be noted that no data were available about the vaccination status of the individuals from which the strains were isolated. However, in the period 1993 to 1996, approximately 85% of the culture-confirmed pertussis patients older than 12 months were vaccinated (11). There was a correlation between the percentage of P.69A strains isolated from an age group and the degree of vaccine-induced immunity (Table 2). The percentages of P.69A strains found in the categories with no, partial, and optimal vaccine-induced immunity were 22, 11, and 8%, respectively. This downward trend was found to be significant (P = 0.024). When the groups with waning and optimal vaccine-induced immunity were compared, again a downward trend (from 18 to 8%) was observed. This trend was marginally significant (P = 0.066). These observations are consistent with the notion that vaccine-induced immunity against P.69A strains is stronger than that against P.69B and P.69C strains.
TABLE 2.
Percentage of P.69 types isolated from different age groupsa
| Age group (mo) | No. of strains | Vaccine-induced immunityb | % of P.69 types isolatedc
|
|
|---|---|---|---|---|
| Vaccine type (P.69A) | Non-vaccine types (P.69B and P.69C) | |||
| 0–3 | 55 | No | 22 | 78 |
| 4–11 | 35 | Partial | 11 | 89 |
| 12–48 | 40 | Optimal | 8 | 93 |
| >48 | 67 | Waning | 18 | 82 |
| Total | 197 | 16 | 84 | |
Data are from the years 1989 to 1996.
Estimation of the level of vaccine-induced immunity was based on the fact that in The Netherlands, children are vaccinated at 3, 4, 5, and 11 months of age. Further, vaccine-induced immunity is known to wane after a few years.
Percentages were calculated within each age group. The downward trend observed in the percentage of vaccine type P.69 in the categories with no, partial, and optimal vaccine-induced immunity was significant (P = 0.024), and the upward trend observed in the categories with optimal and waning immunity was marginally significant (P = 0.066).
Polymorphism in the S1 subunit of pertussis toxin.
Approximately 15 strains from each of three periods 1949 to 1954, 1978 to 1985, and 1990 to 1996 were selected to study polymorphism in the s1 gene. Three S1 variants, S1A, S1B, and S1D, were found in the Dutch B. pertussis population (S1C was observed in a non-Dutch strain) (Fig. 4). Interestingly, all mutations observed in the s1 gene resulted in amino acid substitutions (i.e., were nonsilent). Further, the mutations were found in two regions identified as T-cell epitopes (26). The s1 genes from a number of B. pertussis strains have been sequenced (2, 20, 21, 24), and small differences in sequence were observed. A comparison of the S1 sequences revealed that the previously published sequences correspond to S1A, S1B, and S1D. Thus, S1C represents a novel S1 type. The two Dutch vaccine strains produced S1B and S1D.
FIG. 4.
Variants of the pertussis toxin S1 subunit. Depicted are the DNA and corresponding amino acid sequences of the two polymorphic regions observed. In S1B and S1D, only differences with respect to S1A are indicated. Dots indicate identical bases. The numbers refer to positions in the s1 gene.
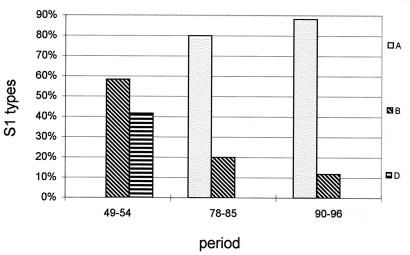
A comparison of the three different periods analyzed (Fig. 5) revealed that two S1 types, S1B and S1D, were found in the period 1949 to 1954. The frequency of S1B decreased in time and was found in 58, 20, and 12% of the strains in the periods 1949 to 1954, 1978 to 1985, and 1990 to 1996, respectively. The other S1 type observed in the period 1949 to 1954, S1D, was not found in later periods. In the period 1978 to 1985, a novel S1 type, S1A, was observed in 80% of the isolates. The frequency of the S1A type increased in time and was found in 88% of the isolates from the most recent period analyzed, 1990 to 1996.
FIG. 5.
Temporal trends in the frequencies of the pertussis toxin S1 subunit in the Dutch B. pertussis population. The percentages of each variant in three periods were determined. The strains used for the Dutch WCV produce S1B and S1D.
DISCUSSION
To our knowledge, this is the first study on the effect of long-term (∼44 years) vaccination on antigenic shifts in a bacterial population. We found that two B. pertussis antigens, P.69 and the S1 subunit of pertussis toxin, are polymorphic and that the frequencies of their variants show shifts in time. Molecular and immunological arguments suggest that variation in P.69 and S1 has been driven by immune selection. First, although a large number of s1 genes were sequenced, all observed mutations were nonsilent. In the prn gene, two of the four codons affected by point mutations contained nonsilent mutations (Fig. 2). Without selective pressure, the ratio of silent to nonsilent mutations is predicted to be 20 (25); thus, it seems likely that the observed changes in S1 and P.69 were fixed in the population due to positive selection. Second, variation in P.69 occurred in a region comprised of repeats, and such regions have been implicated in evasion of the host immune response (22, 28). Third, there is evidence that protective antibodies induced by P.69 are directed against variable regions. B. pertussis P.69 does not cross-protect against B. parapertussis infection in a mouse model, although the two molecules show 93% sequence identity (15). However, this lack of cross-protection cannot be attributed to a particular region of P.69, since differences between the two species are found in both region 1 and region 2, and amino acid substitutions are also found outside these regions (19). Finally, in P.69 and S1, the mutations were confined to regions of the molecule which have been defined as B-cell and T-cell epitopes, respectively (9, 26).
Epidemiological data also provided evidence that the observed variation in P.69 and S1 was driven by immune selection. Temporal trends in the frequencies of P.69 and S1 variants indicated that divergence had occurred between vaccine strains and clinical isolates with respect to these two antigens. The WCV was introduced in The Netherlands in the early 1950s, and in this period all isolates had the same P.69 and S1 types as the two vaccine strains (i.e., P.69A, S1B, and S1D). This is not unexpected since the vaccine strains were clinical isolates obtained in this period. In later periods, strains with the vaccine type P.69 or S1 were replaced by strains carrying novel P.69 and S1 variants (i.e., P.69B, P.69C, and S1A), and such strains comprised ∼90% of the B. pertussis population in the period 1990 to 1996 (Fig. 3 and 5). These observation suggest that the observed shifts in P.69 and S1 have been driven by vaccination with the WCV. Further, we found that in recent years, P.69B and P.69C alternate in successive years, a phenomenon which probably reflects frequency-dependent selection (18) due to waning and increasing naturally acquired immunity against a particular P.69 type.
If the displacement of strains carrying the vaccine-type P.69 (i.e., P.69A) by strains with non-vaccine-type P.69 has been driven by vaccination, one would expect an inverse relationship between the percentage of P.69A strains isolated from individuals and their degree of vaccine-induced immunity. This is exactly what we observed: the percentages of P.69A strains isolated from groups with no, partial, or optimal vaccine-induced immunity were 22, 11, and 8%, respectively (Table 2). This downward trend was significant (P = 0.024). A similar, marginally significant (P = 0.066), downward trend was observed when groups with waning and optimal vaccine-induced immunity were compared (with 18 and 8% P.69A types, respectively). These observations are consistent with the notion that vaccine-induced immunity against P.69A strains is stronger than that against P.69B and P.69C strains. Since the vaccination status is a function of age, we cannot exclude age-related effects other than vaccine-induced immunity on the distribution of P.69 variants. However, the oldest (>48 months) and youngest (0 to 3 months) groups that we studied contained approximately the same percentage of P.69A strains (22 and 18%, respectively), indicating that the distribution of P.69 variants over different categories was not determined simply by age. A similar analysis for S1 was not possible due to the limited number of strains typed.
It is conceivable that the expansion of particular P.69 and S1 types is not a consequence of variation in these molecules only but is also due to other bacterial antigens which affect strain fitness and which may be linked to certain P.69 and S1 types. To investigate this, we are analyzing polymorphism in other surface molecules of B. pertussis.
Both region 1 and region 2 of P.69 are comprised of repeats, are part of immunodominant B-cell epitopes, and show variation within the genus Bordetella (9, 19). Nevertheless, we observed variation only in region 1. Region 1 is located adjacent to the RGD sequence, which has been implicated in the biological function of P.69, i.e., adherence to host cells (17). The X-ray structure of P.69 revealed that the RGD sequence is located in a loop which protrudes from the molecule (13). Thus, antibodies directed against this section of the molecule may effectively block the function of P.69, and consequently variation in this region may benefit the bacteria more than variation in region 2.
There is evidence that the incidence of pertussis is increasing in populations vaccinated with WCVs (1, 4, 5, 11, 12), and our results suggest that one of the factors which has contributed to this phenomenon may be the decline of vaccine efficacy due to antigenic shifts in the B. pertussis population. Our findings also may have implications for the efficacy of ACVs, many of which contain both P.69 and pertussis toxin (35). Published sequences (6, 8, 20, 21, 24) suggest that a number of ACVs contain P.69A and S1B or S1D pertussis toxin subunits, i.e., variants which are found in only ∼10% of recent Dutch B. pertussis isolates. Clearly these vaccines do not have an optimal composition for the Dutch situation. This is not to say that these ACVs will not be effective in The Netherlands, but that inclusion of other P.69 and pertussis toxin variants could make these vaccines more effective. This may also be true for other countries. The inclusion of several P.69 and pertussis toxin types should be considered to prevent the expansion of strains expressing variant antigens which are not included in vaccines. Such a vaccine-driven expansion, documented by us here for a WCV, may occur more rapidly with ACVs than with WCVs and also have a more pronounced effect on vaccine efficacy, since ACVs induce a narrower spectrum of antibody specificities than WCVs. Our results may have implications for the interpretation of the outcome of field trials with ACVs, since the efficacy of a vaccine may differ in different regions due to differences in the population structure of B. pertussis (i.e., the frequency of antigenic variants). The field trials with ACVs were generally held in unvaccinated populations, and our data presented here and previously (34) suggest that the population structure of B. pertussis may be distinct in vaccinated and unvaccinated populations. In collaboration with others, we are currently performing a Europe-wide study of the population structure of B. pertussis in vaccinated and unvaccinated populations.
ACKNOWLEDGMENTS
We are grateful to Nico Nagelkerke and Han de Neeling for advice in statistical analyses and to Joop Schellekens and Hester de Melker for helpful discussions.
Part of this work was supported by grant 25-2545 from the Praeventiefonds.
REFERENCES
- 1.Andrews R, Herceq A, Roberts C. Pertussis notifications in Australia. Commun Dis Intell. 1997;21:145–148. doi: 10.33321/cdi.1997.21.30. [DOI] [PubMed] [Google Scholar]
- 2.Arico B, Gross R, Smida J, Rappuoli R. Evolutionary relationships in the genus Bordetella. Mol Microbiol. 1987;1:301–308. doi: 10.1111/j.1365-2958.1987.tb01936.x. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Brooklyn, N.Y: Green Publishing Associates; 1989. pp. 2.4.1–2.4.2. [Google Scholar]
- 4.Bass J W, Stephenson S R. The return of pertussis. Pediatr Infect Dis J. 1987;6:141–144. doi: 10.1097/00006454-198702000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Bass J W, Wittler R R. Return of epidemic pertussis in the United States. Pediatr Infect Dis J. 1994;13:343–345. doi: 10.1097/00006454-199405000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Capiau C, Carr S A, Hemling M E, et al. Purification, characterization and immunological evaluation of the 69-kDa outer membrane protein of Bordetella pertussis. In: Manclark C R, editor. Proceedings of the Sixth International Symposium on Pertussis. Publication no. (FDA) 90-1164. Bethesda, Md: Department of Health and Human Services; 1990. pp. 75–85. [Google Scholar]
- 7.Charles I, Fairweather N, Pickard D, Beesley J, Anderson R, Dougan G, Roberts M. Expression of the Bordetella pertussis P.69 pertactin adhesin in Escherichia coli: fate of the carboxy-terminal domain. Microbiology. 1994;140:3301–3308. doi: 10.1099/13500872-140-12-3301. [DOI] [PubMed] [Google Scholar]
- 8.Charles I G, Dougan G, Pickard D, Chatfield S, Smith M, Novotny P, Morrissey P, Fairweather N F. Molecular cloning and characterization of protective outer membrane protein P.69 from Bordetella pertussis. Proc Natl Acad Sci USA. 1989;86:3554–3558. doi: 10.1073/pnas.86.10.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charles I G, Li J L, Roberts M, Beesley K, et al. Identification and characterization of a protective immunodominant B cell epitope of pertactin (P.69) from Bordetella pertussis. Eur J Immunol. 1991;21:1147–1153. doi: 10.1002/eji.1830210509. [DOI] [PubMed] [Google Scholar]
- 10.Cherry, J. D. 1996. Historical review of pertussis and the classical vaccine. J. Infect. Dis. 174(Suppl. 3):S259–S263. [DOI] [PubMed]
- 11.deMelker H E, Conyn-van Spaendock M A E, Rümke H C, van Wijngaarden J K, Mooi F R, Schellekens J F P. Pertussis in the Netherlands: an outbreak despite high levels of immunization with whole cell vaccine. Emerg Infect Dis. 1997;3:175–178. doi: 10.3201/eid0302.970211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeSerres G, Boulianne N, Douville-Fradet M, Duval B. Pertussis in Quebec: ongoing epidemic since the late 1980s. Can Commun Dis Rep. 1995;15:45–48. [PubMed] [Google Scholar]
- 13.Emsley P, Charles I G, Fairweather N F, Isaacs N W. Structure of Bordetella pertussis virulence factor P.69 pertactin. Nature. 1966;381:90–92. doi: 10.1038/381090a0. [DOI] [PubMed] [Google Scholar]
- 14.Greco D, Salmaso S, Mastrantonio P. A controlled trial of two acellular vaccines and one whole-cell vaccine against pertussis. N Engl J Med. 1996;334:341–348. doi: 10.1056/NEJM199602083340601. [DOI] [PubMed] [Google Scholar]
- 15.Khelef N, Danve B, Quentin-Millet M J, Guiso N. Bordetella pertussis and Bordetella parapertussis: two immunologically distinct species. Infect Immun. 1993;61:486–490. doi: 10.1128/iai.61.2.486-490.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobisch M, Novotny P. Identification of a 68-kilodalton outer membrane protein as the major protective antigen of Bordetella bronchiseptica by using specific-pathogen-free piglets. Infect Immun. 1990;58:352–357. doi: 10.1128/iai.58.2.352-357.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leininger E, Ewanowich C A, Bhargava A, Peppler M S, Kenimer J G, Brennan M J. Comparative roles of the Arg-Gly-Asp sequence present in the Bordetella pertussis adhesins pertactin and filamentous hemagglutinin. Infect Immun. 1992;60:2380–2385. doi: 10.1128/iai.60.6.2380-2385.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levin B R. Frequency-dependent selection in bacterial populations. Philos Trans R Soc Lond Ser B. 1988;319:459–472. doi: 10.1098/rstb.1988.0059. [DOI] [PubMed] [Google Scholar]
- 19.Li L J, Fairweather N F, Novotny P, Dougan G, Charles I G. Cloning, nucleotide sequence and heterologous expression of the protective outer-membrane protein P.68 pertactin from Bordetella bronchiseptica. J Gen Microbiol. 1992;138:1697–1705. doi: 10.1099/00221287-138-8-1697. [DOI] [PubMed] [Google Scholar]
- 20.Locht C, Keith J M. Pertussis toxin gene: nucleotide sequence and genetic organization. Science. 1986;232:1258–1264. doi: 10.1126/science.3704651. [DOI] [PubMed] [Google Scholar]
- 21.Loosmore S, Cunningham J, Bradley W, Yao F, Dekaban G, Klein M. A unique sequence of the Bordetella pertussis toxin operon. Nucleic Acids Res. 1989;17:8365. doi: 10.1093/nar/17.20.8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madoff L C, Michel J L, Gong E W, Kling D E, Kasper D L. Group B streptococci escape host immunity by deletion of tandem repeat elements of the alpha C protein. Proc Natl Acad Sci USA. 1996;93:4131–4136. doi: 10.1073/pnas.93.9.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nennig M E, Shinefield H R, Edwards K M, Black S B, Fireman B H. Prevalence and incidence of adult pertussis in an urban population. JAMA. 1996;275:1672–1674. [PubMed] [Google Scholar]
- 24.Nicosia A, Perugini M, Franqini C, et al. Cloning and sequencing of the pertussis toxin genes: operon structure and gene duplication. Proc Natl Acad Sci USA. 1986;83:4631–4635. doi: 10.1073/pnas.83.13.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochman H, Wilson A C. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J Mol Evol. 1987;26:74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- 26.Peppoloni S, Pizza M, De Magistris M T, Bartoloni A, Rappuoli R. Acellular pertussis vaccine composed of genetically inactivated pertussis toxin. Physiol Chem Phys Med NMR. 1995;27:355–361. [PubMed] [Google Scholar]
- 27.Roberts M, Tite J, Fairweather N, Dougan G, Charles I. Recombinant P69/pertactin: immunogenicity and protection of mice against Bordetella pertussis infection. Vaccine. 1992;10:43–52. doi: 10.1016/0264-410x(92)90418-j. [DOI] [PubMed] [Google Scholar]
- 28.Robertson B D, Meyer T F. Genetic variation in pathogenic bacteria. Trends Genet. 1992;8:422–427. doi: 10.1016/0168-9525(92)90325-x. [DOI] [PubMed] [Google Scholar]
- 29.SAS Institute Inc. SAS/STAT user’s guide, version 6. 4th ed. Vol. 2. Cary, N.C: SAS Institute Inc.; 1989. [Google Scholar]
- 30.Schmitt H J, Vonkonig C H W, Neiss A. Efficacy of acellular pertussis vaccine in early childhood after household exposure. JAMA. 1996;275:37–41. [PubMed] [Google Scholar]
- 31.Trollfors B, Taranger J, Lagergard T, Lind L, Sundh V, Zackrisson G, Lowe C U, Blackwelder W, Robbins J B. A placebo-controlled trial of a pertussis-toxoid vaccine. N Engl J Med. 1995;333:1045–1050. doi: 10.1056/NEJM199510193331604. [DOI] [PubMed] [Google Scholar]
- 32.Ui M. The multiple biological activities of pertussis toxin. In: Wardlaw A C, Parton R, editors. Pathogenesis and immunity in pertussis. New York, N.Y: John Wiley & Sons; 1988. pp. 39–74. [Google Scholar]
- 33.Verwey W F, Thiele E H, Sage D N, Suchardt L T. A simple liquid culture medium for the growth of Haemophilus pertussis. J Bacteriol. 1949;58:127–134. [PMC free article] [PubMed] [Google Scholar]
- 34.VanderZee A, Vernooij S, Peeters M, VanEmbden J D A, Mooi F R. Dynamics of the population structure of Bordetella pertussis as measured by IS1002-associated RFLP: comparison of pre- and postvaccination strains and global distribution. Microbiology. 1996;42:3479–3485. doi: 10.1099/13500872-142-12-3479. [DOI] [PubMed] [Google Scholar]
- 35.Willems R J L, Mooi F R. From whole cell to acellular vaccines. Rev Med Microbiol. 1996;7:13–21. [Google Scholar]