Abstract
Many of the world’s most important vegetables and medicinal crops, including carrot, celery, coriander, fennel, and cumin, belong to the Apiaceae family. In this review, we summarize the complex origins of Apiaceae and the current state of research on the family, including traditional and molecular breeding practices, bioactive compounds, medicinal applications, nanotechnology, and omics research. Numerous molecular markers, regulatory factors, and functional genes have been discovered, studied, and applied to improve vegetable and medicinal crops in Apiaceae. In addition, current trends in Apiaceae application and research are also briefly described, including mining new functional genes and metabolites using omics research, identifying new genetic variants associated with important agronomic traits by population genetics analysis and GWAS, applying genetic transformation, the CRISPR-Cas9 gene editing system, and nanotechnology. This review provides a reference for basic and applied research on Apiaceae vegetable and medicinal plants.
Introduction
Apiaceae contains 434 genera and nearly 3780 species, including many important vegetables, such as carrot (Daucus carota), coriander (Coriandrum sativum), and celery (Apium graveolens) [1], which are mainly distributed in northern temperate regions [2]. Apiaceae also contains important medicinal plants, including Angelica sinensis, Peucedanum praeruptorum, and Angelica dahurica, which are aromatic herbs with alternating feathered leaves that are sheathed at the base of a shortened stem [3]. The flowers of Apiaceae plants are usually bisexual and include five sepals and petals, as well as an enlarged disk at the base of the style, and form a conspicuous flat-topped umbel [4, 5]. The cremocarp consists of two parts that split open in Apiaceae seeds [6].
Previous studies have revealed that the Apiaceae family is rich in secondary metabolites that have medicinal value [7, 8]. The Apiaceae family includes many vegetable crops that are rich in flavonoids, carotenoids, coumarin, coumarin derivatives, vitamins, and minerals [8], such as celery, carrot, parsley (Petroselinum crispum), and fennel (Foeniculum vulgare) [9]. Apiaceae plants are also used as herbs and spices, including dill (Anethum graveolens), coriander (Coriandrum sativum), caraway (Carum carvi), and cumin (Cuminum cyminum) [10, 11]. Moreover, some species were used as herbal folk remedies in ancient times, including gum ammoniac (Dorema ammoniacum), goutweed (Aegopodium podagraria), Peucedanum luxurians, and Seseli devenyense [12–15]. Some Apiaceae species are grown as ornamental flowering plants, such as masterwort (Astrantia) [16], blue lace flower (Trachymene caerulea) [17], and sea holly (Eryngium maritimum) [18]. In addition, the Apiaceae family includes many toxic perennial plants, such as poison hemlock (Conium maculatum), which contains the toxin coniine [19, 20], water hemlock (Cicuta maculata), which contains the toxin cicutoxin [21], and fool’s parsley (Aethusa cynapium), which contains the toxin coniine [22, 23]. The major Apiaceae vegetable species and medicinal species are summarized in Table 1.
Table 1.
Medicinal applications of major compounds in Apiaceae vegetable and medicinal species.
| Common name | Latin name | Edible parts | Main compounds | Use |
|---|---|---|---|---|
| Anise | Pimpinella anisum | Seed | Trans-anethole, p-anisaldehyde, estragole, farnesol, limonene, 4′-methoxypropiophenone [24, 25] | Edible |
| Asafoetida | Ferula assafoetida | Root | Quercetin, gallic acid, phenol, arsine triethyl, 8-acetoxy-5-S-hydroxyumbelliprenin, asadisulfide, vanillin, β-sitosterol [26, 27] | Antifungal, antidiabetic, anti-inflammatory, antimutagenic, antiviral [28] |
| Bei Chaihu | Bupleurum chinense | Root | Saikosaponin-D, 1-O-caffeoylglycerol, esculetin, scopoletin, α-spinasterol [29, 30] | Antioxidant, hepatoprotective, anti-inflammatory, antipyretic, analgesic, immunomodulatory [31, 32] |
| Caraway Carrot | Carum carvi Daucus carota | Seed, root | Limonene, carvacrol, carvone, carvenone, linalool, p-hydroxybenzoic acid, kaempferol, naringenin [33, 34], β-carotene, quercetin, luteolin, kaempferol, myricetin [35, 36] | Antispasmodic, carminative, astringent [33], edible |
| Celery | Apium graveliens | Petiole, leaves | Apigenin, luteolin, kaempferol, caffeic and ferulic acids [37, 38] | Edible |
| Chinese angelica | Angelica sinensis | Root | Ferulic acid, Z-ligustilide, Z-butylidenephthalide, N-butylidenephthalide, E-ligustilide, p-hydroxybenzoic acid [39–41] | Anti-inflammatory, immunostimulatory, anticancer, neuroprotective, antihepatotoxic, antioxidative,anticardiovascular [42] |
| Chuanminshen | Chuanminshen violaceum | Root | Bergapten, ficusin, 2,3-dihydro-3,5-dihydroxy-6-methyl-4H-pyran-4-one, falcarinol [43] | Antioxidant, immunomodulatory, anti-inflammatory, antitussive [44, 45] |
| Cnidium | Cnidium monnieri | Fruit | Osthol, osthenol, imperatorin, isopimpinellin, bergapten, xanthotoxol, isoporalen, isopimpinelline [46, 47] | Anthelmintic, anti-allergic, anti-atherosclerosis, analgesic, antibacterial [48] |
| Coral vegetable | Glehnia littoralis | Root | α-Pinene, limonene, β-phellandrene, germacrene B, spathulenol [49] | Antioxidant, antitumor, anti-amnesic, immunomodulatory, antimicrobial, allelopathic [50, 51] |
| Coriander | Coriandrum sativum | Stem, leaves | β-Carotene, β-cryptoxanthin epoxide, lutein-5,6-epoxide, violaxanthin, neoxanthin [52] | Edible |
| Cumin | Cuminum cyminum | Seed | p-Cymene, thymoquinone, α-thujene, gallic and vanillic acids, luteolin, catechin, coumarin, eugenol [53, 54] | Edible |
| Dill | Anethum graveolens | Seed | Carvone, trans-dihydrocarvone, dill ether, α-phellandrene, limonene [55] | Edible |
| Dwarf pennywort | Hydrocotyle sibthorpioides | All | Quercetin, isorhamnetin, 6-caffeoylgalactoside, stigmasterol, daucosterol [56] | Antihyperglycemic, antioxidant, antitumor [57, 58] |
| Fennel | Foeniculum vulgare | Stem, leaves, seed | Trans-anethole, estragole, fenchone, limonene, rosmarinic acid [59] | Edible |
| Ferula | Ferula sinkiangensis | Stem, leaves | Coumarins, sesquiterpenes, sesquiterpene lactones, sesquiterpene coumarins, glucuronic acid [60] | Antineuroinflammatory, antibacterial, antimicrobial, anti-inflammatory, anticancer, antioxidant, antileishmanial [61] |
| Gotu kola | Centella asiatica | All | Chlorogenic acid, madecassoside, asiaticoside, madecassic acid, asiatic acid [62] | Antidiabetic, wound-healing, antimicrobial, memory-enhancing, antioxidant, neuroprotecting [63] |
| Japanese parsley | Cryptotaenia japonica | Stem, leaves | Luteolin, apigenin, p-coumaric acid, caffeic, ferulic acid [64] | Antioxidant, antibacterial, anti-inflammatory [64] |
| Lovage | Levisticum officinale | Leaves | Falcarinol, (Z)-ligustilide, (Z)-3-butylidenephthalide, trans-β-farnesene, β-phellandrene [65] | Edible |
| Notopterygium | Notopterygium incisum | Stem, root | Notopterol, bergapten, imperatorin, isoimperatorin, cnidilin, pabulenol, alaschanioside C [66, 67] | Analgesic, antioxidant, anti-inflammatory, antiviral, anti-arrhythmic, immunosuppressive [66] |
| Parsley | Petroselinum crispum | Petiole, root, stem, leaves | Apigenin, phenylpropanoids apiol, oleanolic acid, furanocoumarins, isoimperatorin, oxypeucedanin [68, 69] | Antioxidant, hepatoprotective, brain protective, antidiabetic, analgesic, spasmolytic, immunosuppressant, antiplatelet [70] |
| Parsnip | Pastinaca sativa | Leaves, root | Xanthotoxin, bergapten, isopimpinellin, imperatorin [71] | Edible |
| Radix changii | Changium smyrnioides | Root | 4-Methoxycinnamic acid, 7-hydroxy coumarin, cafeic acid, α-terpinene, β-patchoulene [72] | Antitussive, eliminating phlegm, anti-asthmatic, immunoregulatory, antioxidant, antitumor, antifatigue, antihypoxia, anti-atherosclerotic [72] |
| Slender celery | Apium leptophyllum | Seed | β-Sitosterol, apigenin, quercetin, luteolin, kaempferol, isorhamnetin, β-selinene, p-cresyl iso-valerate [73–75] | Edible |
| Water Dropwort | Oenanthe javanica | Petiole, leaves | Persicarin, apigenin, isorhamnetin, quercetin, hyperoside, azelaic acid, myristic acid, catechol, 3,5-dihydroxybenzoic acid [76, 77] | Edible |
| Water dropwort | Ostericum sieboldii | Petiole, leaves | Myristicin, α-terpineol, α-cadinol, β-farnesene, linalool [78] | Analgesic, anti-inflammatory [78] |
| Wild chervil | Anthriscus sylvestris | Stem, leaves, root | β-Phellandrene, Z-β-ocimene, α-pinene, (−)-deoxypodophyllotoxin, chlorogenic acid, luteolin-7- O-glucoside, isoflavone, picropodophyllotoxin, falcarindiol [79–83] | Antitumor, antimicrobial, anti-inflammatory, antioxidant [80] |
In recent years, Apiaceae plants have been studied with respect to bioactive compounds, medicinal applications, omics, and traditional or modern separation techniques for rare compounds. Many important substances and mechanisms of Apiaceae plants have been fully revealed, and summarizing these research advances can further promote the application of Apiaceae plants. In this review, we summarize the complex origins of Apiaceae and the current state of research on the family, including traditional and molecular breeding practices, bioactive compounds, medicinal applications, omics research, molecular markers, regulatory factors, functional genes, genomics research, functional gene mining, and molecular breeding, and discuss future perspectives.
Apiaceae origin
The complex origins of vegetables and medicinal plants in Apiaceae
Although >70% of Apiaceae family genera are distributed in the Northern Hemisphere [84], biogeographical and molecular phylogenetic studies demonstrated that the Apiaceae family originated in the Southern Hemisphere [84, 85]. Furthermore, Australasia was estimated to be the place of origin of crown Apiaceae plants during the early Paleogene [86].
The Apiaceae family has been mainly divided into four subfamilies: Azorella, Centella, Apioideae, and Eryngium [87, 88]. Apioideae subfamilies include several important vegetable crops: celery, carrot, parsley, water dropwort, and coriander [89–91]. However, each of these Apiaceae species has distinct origins. Carrot and celery originated in Middle Asia around Afghanistan [8, 75, 92], and slowly spread into the Mediterranean area [93]. The earliest recorded carrots were mainly purple or yellow, with some white or black species, instead of orange [94]. Parsley originated in the late third century BC on the Mediterranean coast [95], where it was used for decoration and seasoning [96]. Water dropwort originated in Europe and the Mediterranean region, whereas coriander originated in the Middle East region [97, 98]. The Apiaceae family also contains many important Chinese herbal plants [99]; the origins of many of these plants remain unclear. For example, recent studies revealed that the Angelica group has been cultivated for food and medicine since at least 800 AD [100], and originated in the Middle East [101], possibly Syria, or northern European countries [102]. Although the Apiaceae family contains many species, most of the members of this family have not been comprehensively investigated, especially vegetables and medicinal species.
Bioactive compounds in vegetables and medicinal plants in Apiaceae
All vegetables and medicinal species in Apiaceae have effective secretory systems involving different organs, including roots, stems, leaves, flowers, and fruit [103–105]. According to previous studies, the biologically active compounds of Apiaceae plants can be divided into two groups: nutrients and nutraceuticals [106]. Nutrients are important plant growth regulators that mainly include minerals, proteins, fiber, carbohydrates, and lipids [107]. In contrast, nutraceuticals, a portmanteau word derived from ‘nutrition’ and ‘pharmaceutics’, are non-nutritive plant compounds with high antioxidant activity [108–110]. Nutraceuticals, which mainly include polyphenolic compounds, polyacetylenes, and terpenoids [106], are thought to promote health and are used in the food processing and pharmaceutical industries [111–113].
Phenolic compounds
Phenolic compounds, such as phenolic acids, simple phenols, flavonoids, and hydroxycinnamic acid derivatives [114], are responsible for the flavor, color, and sensory properties of plant-derived foods and beverages [112, 115], and they also contribute to the nutritional qualities of vegetables and medicinal plants [116]. Several studies have pointed out the value of phenolic compounds in some Apiaceae plants [117–119]. The phenolic compounds, such as flavonoids, phenolic acids, coumarin and tannins in fennel, apiin and malonylapiin in parsley, and apiin in celery, are responsible for organoleptic characteristics, such as bitterness, astringency, color, flavor, and odor [117]. The antioxidant activity of many Apiaceae plants has also been attributed primarily to phenolic compounds [120, 121]. Celery contains the flavonoids apigenin, luteolin, kaempferol, isorhamnetin, and quercetin, and extracts of celery have antibacterial, anti-inflammatory, antioxidation, antitumor, and cardiovascular protective activities [75]. The luteolin-7-O-β-d-glucoside (cynaroside) from Anthriscus sylvestris displays biological activity, especially against Gram-negative bacteria, exhibits antimutagenic activity, suppresses biofilm formation of Pseudomonas aeruginosa and Staphylococcus aureus, and increases the frequency of mutations leading to ciprofloxacin resistance in Salmonella typhimurium [122]. Moreover, phenolic compounds can be used to extend the shelf life of foods, delay the oxidation of inclusions [123], and reduce the risk of cancer and cardiovascular, cerebrovascular, and nervous system diseases [124]. Ferulic acid from Angelica sinensis and Ferula teterrima exhibited a therapeutic effect on membranous nephropathy-induced proteinuria and breast cancer [125, 126]. In addition, the content of various bioactive substances in plants is regulated by many factors, including environment, cultivation techniques, varieties, and harvest time [127–129].
Polyacetylenes
Recent studies demonstrated that several polyacetylenes isolated from Apiaceae plants have high toxicity to bacteria, fungi, and mammalian cells [130–132], as well as neurotoxicity, an inhibitory effect on platelet aggregation, and the potential to cause allergic skin reactions [131]. In mammalian experiments, polyacetylenes inhibited tumor formation [131, 133], indicating that these compounds may have clinical applications. A group of aliphatic C17 polyacetylenes in carrot, celery, parsley, and parsnip have revealed interesting antitumor (namely antileukemic), anti-inflammatory and antiplatelet aggregatory effects in mammals [134]. However, polyacetylenes have a negative impact on the taste of the roots of Apiaceae vegetables and medicinal plants, such as parsnip, celeriac, parsley, carrot, and fennel bulbs, because they increase bitterness [135]. In addition, some conjugated polyacetylenes (cicutoxin, oenanthotoxin, virol A, virol B, and virol C) produced by species of the Oenanthe (O. crocata) and Cicuta genera (C. virosa, C. maculata, and C. douglasii), have been identified amongst the strongest plant neurotoxins [134].
Terpenoids
Terpenoids, such as aromatic and essential oils, are the largest group of specialized metabolites in plants [136]. Some terpenoids are specifically distributed in the Apiaceae plants, such as carotol in carrot, trans-anethole in anise and fennel, and carvone in caraway and dill. These species are commonly used as food supplements for their aromatic qualities, which can enhance the smell and taste of foods [137, 138]. Terpenoids possess antioxidant and antimicrobial activities and are the main components of essential oils [139]. The antioxidant effects of terpenoids have led them to be used to improve and treat some diseases, such as cancer, cirrhosis, rheumatoid arthritis, and arteriosclerosis [140]. Asiaticoside in Centella asiatica suppressed the viability of colorectal cancer and increased cell apoptosis by inhibiting the activation of the NF-κB signaling pathway by downregulating IκBα phosphorylation [141]. Moreover, the antimicrobial activities of terpenoids have led them to be used to make efficient antibiotics and antimycotic agents [142, 143].
Applications of edible organs from representative Apiaceae species
The purpose of this review is to investigate edible organs containing nutraceuticals and having medicinal value in representative plants of the Apiaceae family. Nutraceuticals and medicinal value in the Apiaceae family play an important role in food safety and have health benefits [144, 145]. The seed is the characteristic reproductive body of both angiosperms and gymnosperms. The seeds of Apiaceae plants, which are actually dried fruit, are used as natural food additives for spices and seasonings [146]. Acimovic and Milic [103] have summarized the types and uses of nutraceuticals in 12 Apiaceae plants in detail. For example, the dried fruit of Apium graveolens, Carum sativum, and Foeniculum vulgare are used in salads, cakes, sausages, curries, soups, vegetables, and other foods [103]. Moreover, essential oil may be extracted from the seeds of some plants of the Apiaceae family [118]. For example, Carum carvi, Petroselinum crispum, Cuminum cyminum, and Daucus carota may be used to produce essential oils for use in food processing [8], and as additives in candy, chewing gum, soft drinks, and beer [147]. Parsnip has a sweet taste similar to nutmeg and cinnamon, as well as a unique aromatic character [148]. Previous studies have revealed that the seeds of other vegetables and medicinal plants in Apiaceae, such as dill, coriander, and fennel, also contain many different types of nutraceuticals with important medicinal value at varying concentrations [149]. For example, dill seeds are used to relieve colic pain and treat diarrhea, asthma, neuralgia, diabetes, cardiovascular diseases, gallbladder disease, and other conditions [150, 151]. Cumin seeds have been used widely in traditional Chinese medicinal practices to treat toothaches, diarrhea, epilepsy, dyspepsia, and jaundice [152]. Carrot seeds were shown to improve memory when administered to Alzheimer’s patients and were found to have hypoglycemic and hypolipidemic properties [153].
The leaves, stems, and roots of Apiaceae family plants are important vegetative organs that are used for pickling, as well as being consumed fresh. The fresh leaves of dill, coriander, parsley, and celery are used in many countries as garnishes and to flavor salads, dips, snacks, and soups [103, 154]. Petioles, such as celery, are used for the preparation of salads, juices, soups, stews, and sauces [155]. The roots of Apiaceae members are used as food and medicine. For example, Angelica and lovage roots are used to flavor meat and canned vegetables, but they can also be used as raw material for the production of herbal liqueurs and bitter spirits [106, 156]. The fresh roots of carrot and parsnip are the most widely consumed Apiaceae root vegetables, and they are primarily eaten raw in juices or salads, or for pickling, soups, and cakes. More importantly, carrot taproots contain effective anti-inflammatory and anticancer compounds, as well as constituents with hypoglycemic and hypolipidemic properties [157].
Genetic breeding
Male sterile breeding
Genetic male sterility and cytoplasmic–nuclear male sterility (CMS) have been utilized for the production of hybrid cultivars in Apiaceae [158]. A male-sterile line was also used in the mechanization of hybrid seed production, which simplified the procedure and reduced its cost [159, 160]. The difficulty of obtaining F1 hybrid seeds in vegetables and medicinal plants in Apiaceae is due to a lack of emasculation methods. Thus, male-sterility breeding is used for Apiaceae crop breeding, including carrot, celery, coriander, and others [161]. Male sterility can be genetically or/and cytoplasmically determined [162, 163]. Currently used cytoplasmic male sterility (CMS) systems include the ‘brown anther’ type and ‘petaloid’ type [164]. The progress of carrot male sterile breeding research were summarized by many researchers in various years [165–167].
F1 hybrid celery seeds are difficult to obtain, because celery flowers are small, numerous, and easily self-pollinate [163]. The first sterile male celery line was the Iranian accession P1229526, in which sterility is conferred by a single recessive ms-1 gene [168]. Furthermore, Quiros et al. [168] found unstable celery CMS in an unidentified wild celery plant, and Gao et al. [169] identified a sterile male celery plant (01-3A) from the inbred line 01-3.
Disease resistance breeding
Three main methods, (i) selecting disease-resistant varieties, (ii) strengthening cultivation management, and (iii) applying fungicides, are commonly used to prevent and control the occurrence and spread of plant diseases [170]. Selection of disease-resistant germplasm resources has been the most effective method of reducing the occurrence of diseases in vegetables and other food crops [171].
Numerous studies have shown that powdery mildew infects a wide range of Apiaceae plants, including carrot, parsnip, celery, dill, and fennel [172]. Powdery mildew (Blumeria graminis f. sp. hordei) mainly occurs in leaves and petioles, and can cause fatal damage to Apiaceae vegetable crops. The first report of powdery mildew in Apiaceae vegetables was a report about carrot and parsley crops in the state of Washington in the USA [173]. In addition, Alternaria radicina, a seed-borne fungal disease, can decrease seed quality [174].
Early blight, caused by Cercospora apii, is a highly transmissible disease of Apium [175]. The celery ‘Floribelle M9’ cultivar with superior resistance to early blight was developed in the 1990s and used to develop early blight-resistant cultivars, such as ‘FBL 5-2 M’ [169]. Late blight, caused by Septoria apiicola, is an important leaf disease that infects celery, celeriac, and carrot [176]. In addition, two Septoria-resistant celery species (Apium chilense and A. panul) have been crossed to generate plants with enhanced disease resistance [177].
Fusarium oxysporum is a soil-borne fungus that causes fusarium yellows disease in celeriac, celery and carrot [169]. UC1 is a fusarium yellow disease-resistant celery breeding line that has been backcrossed with elite varieties to create the resistant lines UC8-1, UC10-1, and UC26-1 [178]. Somaclonal variation has been used to select Fusarium-resistant celery plants, such as the MSU-SHK5 line, during regeneration from cell suspensions [179]. In 2017, three potentially resistant celeriac accessions from Turkey and an additional resistant accession from China were identified as sources of F. oxysporum resistance [180].
Leaf blight, caused by Alternaria dauci, is a fungal leaf disease that negatively impacts carrot and coriander cultivation [174, 181]. Gugino and colleagues [182] research identified five carrot cultivars (‘Bolero’, ‘Carson’, ‘Calgary’, ‘Ithaca’, and ‘Fullback’) with relatively low susceptibility to A. dauci, as well as three cultivars (‘Bolero’, ‘Carson’, and ‘Bergen’) that showed relatively low susceptibility to Cercospora carotae. However, carrot cultivar ‘Fontana’ was found to be highly susceptible to these two diseases [182]. Infection of coriander plants becomes apparent when they bloom, the flowers turn yellow and are generally taller than those of uninfected plants [183]. Moreover, infection of coriander plants becomes apparent when they bloom, the flowers turn yellow, and the plants are generally taller than uninfected plants [184].
Sclerotinia disease, caused by Sclerotinia sclerotiorum, S. minor, and S. trifoliorum, can cause severe damage to stored Apiaceae vegetables, especially carrot [185, 186]. Jensen et al. [186] revealed that Daucus carota, as a susceptible host to Sclerotinia sclerotiorum, can obtain disease-resistance genes from disease-resistant cultivated species during flowering to produce resistant offspring. Although sclerotinia disease also occurs in celery and parsley, the impact on these species is minimal. Aster yellows, caused by a bacterium-like organism called a phytoplasma, is a common destructive disease worldwide [184].
Celery mosaic virus (CeMV) is transmitted by aphids and is the most common viral disease in celery [187]. A single recessive locus and markers linked to CeMV resistance genes were identified in 2001 [188]. Using post-transcriptional gene silencing technology, previous studies attempted to produce celery and carrot plants with resistance to CeMV and carrot virus Y (CarVY), but resistant celery plants were not obtained [169].
Root-knot nematodes (RKNs, Meloidogyne spp.) are major pathogens that affect carrot [189] and other Apiaceae species, including celery [190] and parsnip [191]. The roots of carrot plants infected by RKNs displayed malformed, stubby, hairy roots with tough galls and thick skin. Moreover, the aerial parts of infected plants become yellow and display inhibited growth and development [192]. At the same time, wounds produced by RKNs on carrots increase the probability of infection by diseases and other pests. RKNs show strong adaptability and can adapt to complex and variable environments. The best carrot RKN-resistant varieties obtained so far include ‘Brasilia’ and ‘Tropical’, and two resistance genes (Mj-1 and Mj-2) have been identified [193–196]. Another resistance locus was identified in the ‘PI652188’ cultivar in 2014 and mapped to a different position in chromosome 8 [189]. Furthermore, RKNs also affect production of fennel; infection increases the size of root galls, decreases plant vigor, and causes a yellow phenotype [197]. Other methods of reducing nematode populations in the soil include solarizing the soil and crop rotation [198].
Breeding for insect pest resistance
The carrot fly (Chamaepsila rosae), a small black-bodied fly, affects many members of the Apiaceae family, including celery, parsnip, parsley, carrot, and other carrot-family herbs [199]. Plant roots attacked by carrot fly larvae are destroyed, causing fatal damage to affected plants. Bacterial diseases that infect plants through wounds (soft rot or parsnip blight) [200] are the main reason why wounded roots are difficult to store, especially in carrot production [201]. Previous studies revealed that breeding resistant varieties is an effective method of mitigating the effects of carrot fly infestation [202, 203].
The carrot weevil (Listronotus oregonensis) is a pest of parsley, carrots, and celery. When carrots are attacked by the carrot weevil, only the ribs of the leaves and stalks are left [140]. This pest causes significant damage to agricultural production and cannot be effectively controlled. At present, no resistant varieties are available [204].
Carrot willow aphid (Cavariella aegopodii) is a widespread temperate species that feeds on members of the Apiaceae family [205]. The carrot willow aphid causes direct and indirect damage to plants. Direct damage is mainly caused when the aphid draws juice from plant leaves [206], and indirect damage is caused by the transmission of viral diseases, such as Carrot red leaf virus (CRLV), Parsnip mosaic virus (PMV), and Parsnip yellow fleck virus (PYFV). The main method of controlling carrot willow aphid infestation in agricultural production is the application of pesticides.
Celery fly (Euleia heraclei) is a small brown-winged, green-eyed European fly, whose larvae are leaf miners that attack celery and parsnips [207]. These pests burrow inside and destroy the leaves of celery and parsnip, and infested plants show large yellow or brown blotches that are approached by a short gallery [208]. Removing the affected leaves or plants is an effective way of controlling celery fly infestation.
Aphids (Aphidoidea), armyworm (Mythimna separata), and cutworms (Agrotis spp.) affect many Apiaceae plants, especially fennel plants. Aphids are soft-bodied insects that cause discoloration of leaves, necrotic spots, and stunted growth. The use of resistant varieties and insecticides can effectively control the spread of aphids [209]. The application of Bacillus thuringiensis efficiently blocked the spread of armyworm [210]. Cutworms mainly attack the roots of plants, cutting off the transport of water and nutrients between the roots and the aboveground parts. Field management is the main measure used to prevent and control the occurrence of cutworms. These three pests also harm parsley [211].
Beet armyworm (Spodoptera exigua) is a pest that is difficult to control and affects celery and celeriac [212]. However, celery cultivars ‘K-26[1]’, ‘K-I08[3]2’, ‘K-I28’, ‘F-128[3]1’, and ‘F-128[4]’ with resistance against beet armyworm have been identified [213]. Moreover, plants with resistance to fusarium yellows displayed a significant increase in beet armyworm resistance [214]. Beet armyworm-resistant cultivars were obtained from 13 cultivars of varieties rapaceum, dulce, and secalinum in 1991 [215].
Late-bolting breeding
Early bolting significantly decreases the quality and yield of Apiaceae vegetables [216], such as carrot, celery, and parsley [217]. The demand of Apium species for a cold period is affected by their environments and genetics [207]. Wohlfeiler et al. [218] revealed that the vernalization requirement of carrot was controlled by a multiallelic digene. Previous studies found that annual and biennial celery cultivars bolt easily, and cultivars with strong bolting resistance are rare [169, 219]. A single locus, Hb, was identified from F2 hybrids and found to control the bolting time of celery [220]. Slow-bolting celery cultivars ‘Florida Sloblot M68’ [221] and ‘Juventus’ [222] were generated by single selection and crossing, respectively.
Molecular marker-assisted breeding
Modern molecular markers include amplified fragment length polymorphisms (AFLPs), simple sequence repeats (SSRs), PCR-based markers, and inter-simple sequence repeat (ISSRs) [223]. These molecular markers have been widely used in breeding members of the Apiaceae family. Que et al. [8] summarized the application of molecular markers (polymerase chain reaction (RAPD), AFLPs, quantitative trait locus (QTL) and SSRs) in carrot research, including genetic diversity, population structure, and identification of the difference between CMS and fertile carrots. In celery, RAPD markers were used to explore the genetic diversity of 23 celery cultivars and classify 40 celery varieties from the major regions of China, which showed that celery may be divided into four groups, 12 varieties, and three cultivated types (salad, turnip, and cutting celery) [224, 225]. AFLP technology was used to identify 245 polymorphic sites in 24 celery cultivars using eight AFLP primers [75]. Moreover, five ISSR primers were used to study the genetic diversity of 105 celery accessions, which were classified into five groups [75]. A study of the linkage relationships of 34 markers in celery showed that they were distributed in eight linkage groups, including 21 restriction fragment length polymorphisms (RFLPs), 11 isozymes, and 2 morphological traits, and the total covered length was 318 centimorgans (cM) [226]. In 1995, F2 population genetic linkage maps of two celery varieties were constructed; these maps contained 29 RFLPs and 100 RAPDs, and they covered a total length of 803 cM [227]. Expressed sequence tags (EST)-SSR fingerprinting, including eight SSR markers, was used to explore the genetic diversity of 11 celery varieties [228]. RNA-seq technology was used to identify 1939 and 2004 SSRs in the ‘Ventura’ and ‘Jinnan Shiqin’ varieties, respectively [229, 230]. In coriander research, many molecular makers, including RAPDs, ISSRs, and SSRs, were also used alone or in combination to explore the genetic diversity of coriander varieties [231–233]. Transcriptome analysis of different tissues of coriander identified 9746 SSRs [234]. In addition, 120 primers were randomly selected to verify 14 coriander accessions in India [234]. Apart from the three plants mentioned above, molecular markers were also widely used to study other Apiaceae species. For example, SSRs and AFLPs were used to investigate the genetic diversity of Eryngium alpinum [235, 236]. Single-nucleotide polymorphism (SNP) was used to investigate the genetic diversity and population structure of 78 Western type open-pollinated carrot cultivars [237]. Transcriptome sequencing for high-throughput SNPs revealed that Western carrots may originate from Eastern carrots. The reduction in genetic diversity in Western cultivars due to domestication bottleneck/selection may have been offset by introgression from wild carrot [238]. In addition, ISSR markers were used to determine the phylogenetic relationships among the taxa of Johrenia [239–241].
Transgenic breeding
Agricultural biotechnologies use different techniques to modify the genetic structure of plants to produce genetically modified plants [242]. Transgenic technology can be used to improve plant traits (yield and quality) and solve agricultural problems (biotic and abiotic stresses) [243]. Transgenic systems have been established for only a few Apiaceae vegetables, including carrot and celery. Permyakova et al. [244] established transgenic carrot lines overexpressing the cfp10, esat6, and dIFN genes (encoding deltaferon) from Mycobacterium tuberculosis, which produce CFP10-ESAT6-dIFN protein in the roots of transgenic carrots, by Agrobacterium-mediated transformation. It is most important to emphasize that this genetically modified carrot does not induce immune responses in mice and has no side effects [244]. In addition, transgenic carrot plants expressing human interferon α-2b have been generated [245, 246]. Moreover, in carrot, combined expression of lipid transfer protein (ltp) and chitinase (chi-2) genes enhanced resistance to foliar fungal pathogens [247–249]. Tan [250] revealed that overexpression of the AgFNS gene from purple celery increased apigenin content and decreased anthocyanin content in transgenic celery. Ding et al. [251] found that AgZDS, a gene encoding ζ-carotene desaturase, increases lutein and β-carotene contents in transgenic Arabidopsis and celery. Wang et al. [252] reported that AgMYB12, a novel R2R3-MYB transcription factor, regulates apigenin biosynthesis by interacting with the AgFNS gene in celery. Overall, the application of genetically modified Apiaceae species will accelerate the breeding of Apiaceae vegetables.
Genome editing in Apiaceae vegetables
The CRISPR/Cas9 system has been used for targeted mutagenesis in plants, including gene knockout, multiplex gene editing, and insertion and deletion of large fragments [253–255]. A previous study knocked out the carrot gene encoding flavanone 3-hydroxylase (F3H), a critical gene for anthocyanin biosynthesis, by genome editing [256]. The results showed that the purple callus in which CRISPR/Cas9 vectors targeted the F3H gene became discolored [256, 257]. This gene editing system was also used to knock out other Apiaceae vegetable genes, including carrot GGred (geranylgeranyl diphosphate reductase), LCYE (lycopene ε-cyclase), CENH3 (centromeric histone H3), and DcCCD4 (carotenoid cleavage dioxygenases) [256, 258, 259]. Xu et al. [260] also established a stable gene-editing system in carrot, and the system could be used for generating stable gene-edited carrot plants.
Nanoparticles in Apiaceae plants
Based on previous studies of nanoparticles, it has become evident that nanotechnology can play a vital role in agricultural production, especially regarding gene modification and pest control [261, 262]. Although fertilizers are very important to vegetable crops at all stages, most fertilizers are wasted due to leaching and degradation by various factors. Thus, it is necessary to reduce nutrient waste and increase crop yield through the use of nanomaterials [263]. Nanofertilizers could be more effective than conventional fertilizers because they are capable of releasing nutrients to plants on demand when necessary [264, 265]. At present, the application of nanotechnology is still its infancy in vegetables and medicinal plants of Apiaceae. However, a recent study found that nano-enhanced ammonium bicarbonate increased celery yield and reduced fertilizer requirements [266, 267]. This area of research also provides a new way to perform gene manipulation and expression regulation in plant cells or tissues [268, 269]. In comparison with the widely used Agrobacterium-mediated transformation method, nanotechnology can be used to deliver chemicals, proteins, and nucleotides to confer targeted traits on non-genetically modified plants [270].
Omics research in vegetables and medicinal plants of Apiaceae
Genomics
Omics research attempts to comprehensively understand the biological molecules in an organism at a particular functional level, such as the genome, transcriptome, or proteome [271–273]. Apiaceae is a large angiosperm family that includes many medicinal, edible, and spice species, which play important roles in daily life around the world [272]. A gene-editing system for carrots was established and used to determine the inheritance of anthocyanin sites in carrots, providing new ideas and methods for transgenic carrot breeding [260, 274].
Although the members of Apiaceae have a wide geographical distribution and rich nutritional and medicinal value, little research has been performed on the genomes of Apiaceae species [275]. Here, we summarize genomic information for five representative species that have been sequenced, assembled, and annotated well, including coriander (2n = 2x = 22), celery (2n = 2x = 22), and carrot (2n = 2x = 18). In 2014, Xiong’s group established CarrotDB, a genomic and transcriptomic database for carrot [276, 277]. In 2016, Simon’s group published a high-quality carrot genome sequence assembly (421.5 Mb) with the N50 scaffold length of 64.5 kb [278]. In 2018, Feng et al. [279] established CeleryDB, a celery genome database. Later, Li et al. [280] published the genome sequence of celery and identified important functional genes. More recently, a high-quality celery genome sequence, with N50 scaffold length of 289.78 Mb, was made available [281]. The coriander genome sequence was published in 2020. The total assembled coriander genome size is 2.13 Gb, which is divided over 6186 scaffolds with an N50 scaffold length of 160.99 Mb [282].
In addition to celery, carrot, and coriander, the genomes of two other Apiaceae plants have been sequenced. Oenanthe javanica (Blume) DC., a Chinese herbal medicine, belongs to the Apiaceae family [283]. The O. javanica genome was published in 2021 [284]. The assembled O. javanica genome contains 149 923 scaffolds, the size of the assembled genome is 1.28 Gb, and the N50 scaffold length is 13.093 Mb. Fennel, belonging to the genus Foeniculum in Apiaceae, is a Chinese herbal plant used to treat various diseases [285]. The assembled genome of fennel consists of 300 377 scaffolds, the total length of the genome is 1010.97 Mb, and the N50 scaffold length is 18.88 Mb. Many studies have revealed that plant genomes contain abundant repeat sequences. Genomic sequences and annotation have provided important information that has contributed to studies of the functions of genes involved in regulating the yield and quality traits of horticultural crops [286]. The further study of important gene functions and breeding, as well as comparative genomic analysis of Apiaceae, will provide new methods for genetic and breeding research using Apiaceae vegetable crops and medicinal plants. Genome information on five Apiaceae plants is shown in Table 2.
Table 2.
Genome information on five sequenced Apiaceae plants.
| Species | Source | Genus | Gene size (Gb) | Number of genes | Website link |
|---|---|---|---|---|---|
| Coriandrum sativum | Bio2RDF | Coriandrum | 2.13 | 40 747 | http://cgdb.bio2db.com/databases.html# |
| Daucus carota | NCBI | Daucus | 0.41 | 37 099 | https://www.ncbi.nlm.nih.gov/genome/?term=Daucus+carota |
| Foeniculum vulgare | NCBI | Foeniculum | 0.99 | 43 936 | https://www.ncbi.nlm.nih.gov/genome/?term=Foeniculum+vulgare |
| Oenanthe javanica | NCBI | Oenanthe | 1.28 | 42 270 | https://www.ncbi.nlm.nih.gov/genome/?term=Oenanthe+javanica |
| Apium graveolens | NCBI | Apium | 3.25 | 31 326 | https://www.ncbi.nlm.nih.gov/genome/11000 |
Transcriptomics
Transcriptome data are widely used in gene expression analysis, gene function discovery, and molecular marker development [287, 288]. Although the Apiaceae family has a large number of members, some Apiaceae vegetable crops have undergone transcriptome analysis [8, 277–284, 289–290]. Besides, transcriptome technology has also been applied in research on stress response [291], root development [292], and lignin biosynthesis in carrot [293].
Moreover, transcriptome analysis has been used widely in celery research. Jia et al. [294] revealed the mechanism of formation of lignin and hormones based on transcriptome profiles of celery at different developmental stages. Through transcriptome analysis, Liu et al. [295] found that multiple genes controlling hormone synthesis in celery were associated with leaf development. Li et al. [296] demonstrated the relationship between related gene expression profiling and accumulation of β-carotene in celery leaves and petioles using transcriptome analysis. Jiang et al. [297] identified the response genes of Oenanthe javanica under abiotic stress through transcriptome assembly and gene annotation. Tan and colleagues [298] analyzed temperature stress response genes by de novo assembly and transcriptome characterization in Cryptotaenia japonica. Li et al. [299] also identified abiotic stress-related AP2/ERF transcription factors by transcriptome sequencing and analysis of parsley.
Transcriptomics have also been applied to study Apiaceae plants used in Chinese herbal medicine. For example, transcriptome analysis of different tissues from Ferula assa-foetida revealed candidate genes for terpene and phenylpropyl metabolism [300]. In conclusion, the application of transcriptomics allows researchers to explore the phenotypic characteristics of vegetables and medicinal plants in Apiaceae and the physiological functions of Apiaceae genes.
MicroRNAs
MicroRNAs (miRNAs) are endogenous small RNAs that play important roles in regulating plant growth and development [301, 302]. In the process of plant development, miRNAs play key roles at every major stage [302–304]. Drikvand et al. [305] identified three miRNAs (csa-miR162, csa-miR169, and csa-miR399) in coriander and found that the target genes of these miRNAs displayed differential expression in seed and leaf samples. A total of 431 and 346 miRNAs were identified in celery varieties ‘Ventura’ and ‘Jinnan Shiqin’, respectively, and 6 of these miRNAs were found to be involved in responses to cold and heat stresses [229]. Najafabadi et al. [306] identified the top five miRNAs (2919, 5251, 838, 5021, and 5658) involved in the biosynthesis and regulation of terpenes in Ferula gummosa. Jia et al. [307] identified 344 conserved miRNAs associated with leaf development in celery. Jiang and colleagues also identified microRNAs affected by abiotic stress in celery [308]. Bhan et al. (2019) surveyed the miRNAs in two carrot variants with different colors (orange-red and purple) using RNA-seq, leading to the validation of 2 novel miRNAs and 11 known miRNAs [309]. Recently, the responses to water stress were investigated using integrative genome, transcriptome, miRNA and degradome analysis in O. javanica [284].
Proteomics
Proteomics is now considered one of the most important ‘post-genomic’ approaches to help us understand the function of genes. In fact, some genomics companies have launched large-scale proteomics projects [310]. Proteomics, of course, is widely used to study the Apiaceae plants. Huang et al. [311] performed proteomic analysis of temperature stress-responsive proteins in celery leaves and identified 71 temperature-responsive proteins. Khodadadi et al. [312] elucidated the response mechanism in drought-sensitive and -tolerant genotypes of fennel leaf using a gel-free/label-free proteomic technique, and further analysis revealed that drought stress may limit photorespiration by reducing the activity of cobalamin-independent methionine synthase in drought-sensitive genotypes. Bai et al. [313] reported the precise mechanism by which asafoetida extract influenced the growth of Pleurotus ferulae mycelium using comparative proteomic analysis, and the results showed that asafoetida extracts significantly affected the growth and metabolism of P. ferulae [313]. Comparative proteomic analysis also provides new insights into gene mining in carrot plants [314, 315].
Metabolomics
Metabolomics encompasses all chemical reactions occurring in cells. GC–MS technology has been used for metabolite profiling since the early 1990s [316]. Plant metabolites have been used as chemical markers to distinguish differences among vegetables and medicinal plants of the Apiaceae family [317]. In carrot research, metabolomics analysis revealed that wild and cultivated carrots showed differences in metabolites [318] that were consistent with their genotypes. Identification of the WtDcTPS1 gene, which is involved in the synthesis of geraniol in wild carrot, was achieved by metabolomics analysis [319].
NMR-based metabolomics has been used to discriminate celery from different geographical origins [320, 321]. Based on UHPLC–QTOF–MS/MS metabolomics analysis, nine chemical markers were used to distinguish Radix Angelica sinensis samples from different regions [322, 323]. Radix bupleuri is one of the most popular traditional Chinese herbal drugs [324–326]. Studies have shown that R. bupleuri protects the liver by interacting with various metabolic processes [327–329]. DG (Danggui, A. sinensis) products were found to significantly relieve blood stasis syndrome in rats, and Jiu Danggui was the most effective type [330]. In addition, plant metabolites are involved in the color, taste, and scent of fruits and flowers, and they also contribute to the regulation of various resistance and stress responses [331].
In recent years, environmental scientists have developed practical applications for metabolomics. In carrot research, Koutouan et al. [181] reported a link between leaf secondary metabolites and resistance to Alternaria dauci. Another recent study identified genes and metabolites in important biological pathways that may regulate selenium tolerance in celery [332]. Many plants of the Apiaceae family are used as condiments or vegetables, and some of them have medicinal properties that may be related to secondary metabolites [333]. In summary, metabolomics analysis is an important method for the in-depth study of the physiological and biochemical processes of vegetables and medicinal plants in Apiaceae, and could provide new possibilities for human use.
Functional genes involved in the synthesis of nutraceuticals in Apiaceae vegetables and medicinal plants
Vegetables and medicinal plants in the Apiaceae family are good sources of many secondary metabolites, such as carotenoids, anthocyanins, terpenes, and dietary fiber [118, 334]. Information on identified functional genes in some Apiaceae plants is shown in Table 3.
Table 3.
Information on identified functional genes in some Apiaceae plants.
| Species | Gene name | Gene expression status | GenBank | Function |
|---|---|---|---|---|
| Daucus carota | PSY | Overexpression | NM_001329177.1 | Increased content of carotenoids [340] |
| PDS | Overexpression | NM_001329175.1 | Produced β-carotene and α-carotene [341] | |
| ZISO | Overexpression | XM_017363269.1 | Produced β-carotene and α-carotene [341] | |
| ZDS | Overexpression | NM_001329165.1 | Produced β-carotene and α-carotene [341] | |
| CRTISO | Overexpression | XM_017392673.1 | Produced β-carotene and α-carotene [341] | |
| LCYB | Overexpression | NM_001329160.1 | Produced β-carotene and α-carotene [342] | |
| LCYE | Overexpression | NM_001329163.1 | Produced β-carotene and α-carotene [342] | |
| CYP97A3 | Overexpression | JQ655297.1 | Decreased content of α-carotene in roots [345] | |
| F3H | Overexpression | XM_017385173.1 | Regulated biosynthesis of anthocyanins [256] | |
| UCGalT1 | Overexpression | KP319022.1 | Regulated biosynthesis of anthocyanins [371] | |
| MYB6 | Overexpression | XM_017379690.1 | Regulated biosynthesis of anthocyanins [368] | |
| MYB7 | Overexpression | XM_017385289.1 | Regulated biosynthesis of anthocyanins [274] | |
| MYB113 | transcriptome | XM_017383803.1 | Regulated biosynthesis of anthocyanins [271] | |
| USAGT1 | Overexpression | KT595241.1 | Regulated biosynthesis of anthocyanins [370] | |
| bHLH | Overexpression | QEA09235.1 | Colored with carrot taproot anthocyanin [372] | |
| GST | Overexpression | XM_017389912.1 | Colored with carrot taproot anthocyanin [372] | |
| TPS04 | Overexpression | XM_017390437.1 | Produced α-terpineol, sabinene, β-limonene, β-pinene, myrcene [388] | |
| TPS26 | Recombinant protein expression in Escherichia coli | XM_017390438.1 | Regulated monoterpene production [289–291] | |
| TPS27 | Recombinant protein expression in Escherichia coli | KZM99345.1 | Regulated monoterpene production [289–291] | |
| TPS54 | Recombinant protein expression in Escherichia coli | KZM99341.1 | Formed sabinene [388] | |
| TPS55 | Recombinant protein expression in Escherichia coli | KZM99344.1 | Regulated monoterpene production [387–389] | |
| TPS1 | Recombinant protein expression in E. coli | DcTPS58617 | Synthesized (E)-β-caryophyllene α-humulene [390] | |
| TPS2 | Recombinant protein expression in Escherichia coli | XM_017389213.1 | Synthesized monoterpene synthase with geraniol [390] | |
| atp6 | Overexpression | JQ248574.1 | Associated with carrot male sterility [160] | |
| atp9 | Overexpression | AJ009982.1 | Associated with carrot male sterility [165] | |
| DFR2 | Overexpression | AF184272_1 | Involved in anthocyanin synthesis [8] | |
| UFGT | Overexpression | XM_017392428.1 | Involved in anthocyanin synthesis [8] | |
| FLS1 | Overexpression | XM_017372509.1 | Involved in anthocyanin synthesis [8] | |
| LDOX2 | Overexpression | AF184274.1 | Involved in anthocyanin synthesis [8] | |
| MYB2 | Overexpression | Participated in anthocyanin synthesis regulation in purple celery [373] | ||
| Apium graveolens | γTRPS | Recombinant protein expression in Escherichia coli | KF700699.1 | Catalyzed the conversion of geranyl diphosphate [392] |
| FNS | Overexpression | AY817676.1 | Increased content of apigenin, decreased content of anthocyanin in petiole of transgenic celery [252] | |
| Coriandrum sativum | LINS | Recombinant protein expression in Escherichia coli | KF700700.1 | Catalyzed conversion of geranyl diphosphate [392] |
Carotenoids
Carotenoids are natural pigments that are widely distributed in photosynthetic organisms and may provide health benefits [335, 336]. The first committed step in carotenoid biosynthesis is catalyzed by phytoene synthase (PSY) [337–339]. Overexpression of PSY increased the content of carotenoids in transgenic plants [340]. The PSY product, 15-cis-phytoene, is further desaturated and isomerized to form all-trans-lycopene by phytoene desaturase (PDS), 15-cis-ζ-carotene isomerase (Z-ISO), ζ-carotene desaturase (ZDS), and cis-trans-isomerization (CrtISO) [341]. Next, lycopene cyclases, including lycopene cyclase β (LCYB) and lycopene cyclase ε (LCYE), are involved in producing β-carotene and α-carotene [342]. β-Carotene, known as the orange-red pigment in carrots, has been shown to account for 80% of the total carotene content in this vegetable [343]. Moreno et al. [344] revealed that DcLcyb1 plays an essential role in the accumulation of β-carotene in carrot plants. Arango et al. [345] found that overexpression of CYP97A3 in orange carrots significantly decreased the content of α-carotene in roots without significantly changing the content of α-carotene in leaves. Analysis of domesticated varieties and wild carrot accessions revealed a significant genomic region that contains the Or (Orange) gene, which is a candidate for carotenoid presence in carrot [346]. Coe et al. [347] have revealed that Or and CH are likely involved in controlling the accumulation of β-carotene and modulated carotenoid flux in carrot. Wang et al. [348] found that the expression profiles of the genes related to carotenoid biosynthesis were closely related to carotenoid content in carrots with different colors. Zhang et al. [349] found that the carotenoid contents and expression of related genes were affected by drought stress in carrot taproots. Then, Li et al. [350] reported that Arabidopsis plants hosting the DcBCH1 gene, encoding non-heme carotene hydroxylase (BCH), improves tolerance to drought in transgenic plants. BCH is a key regulatory enzyme in the β-branch of the carotenoid biosynthesis pathway. In addition, multiple paralogs of carotenoid pathway genes have been identified in carrot, suggesting that different paralogs are involved in the precise temporal regulation of carotene synthesis in different tissues, developmental stages, and environmental conditions [351].
The expression of carotenoid pathway genes also increases the accumulation of other pigments in Apiaceae vegetables [352–356]. In celery, the relative expression levels of AgPSY1 and AgLCYE in the ‘Ventura’ cultivar were significantly higher than those in the ‘Liuhe Yellow Heart Celery’ cultivar [357]. Furthermore, transcriptome profiling of biosynthesis genes and β-carotene content in the leaf blades and petioles of celery demonstrated that AgPSY1, AgCRTISO2, and AgBCH1 may play important roles in the accumulation of β-carotene [296]. Similar results have been reported in carrot [290, 358, 359]. Ding et al. [360] found that the expression levels of AgLCYB and AgPSY2 genes were significantly correlated with lutein and β-carotene contents in yellow celery. Yin et al. [361] and Ding et al. [360] demonstrated that overexpression of the genes AgLCY-ε and AgZDS, encoding lycopene epsilon cyclase and ζ-carotene desaturase, increased lutein and β-carotene accumulation in transgenic Arabidopsis.
Anthocyanins
Anthocyanins are phenolic compounds that are synthesized via the phenylpropanoid pathway and add pigmentation to several organs and tissues of many plant species [362]. Anthocyanins protect plants from UV radiation, contribute to plant adaptation to different abiotic and biotic stresses, and delay plant senescence [363, 364]. In addition, anthocyanins promote various health benefits due to their antioxidant effects and anti-inflammatory properties [365]. Research on anthocyanins in Apiaceae has mainly focused on a few species, including carrot and purple celery.
In carrot research, previous studies have provided many sources of information on anthocyanins in carrot, such as the content of anthocyanins of different varieties (purple, yellow and orange carrots), structural genes encoding key enzymes, and transcription factors regulating anthocyanin biosynthesis [274, 366–371]. Furthermore, Chialva et al. [253] identified long non-coding RNAs (lncRNAs) involved in regulating anthocyanin biosynthesis in taproots. A transcriptome analysis strongly suggested that transcription factors bHLH and GST are involved in anthocyanin pigmentation in carrot roots [372].
Recent studies found that transcription factors AgMYB1/AgMYB2 and OjMYB1 are involved in the regulation of anthocyanin biosynthesis in purple celery (Apium graveolens) and Oenanthe javanica, respectively [373–376]. AgMYB12, a R2R3-MYB transcription factor, regulates apigenin biosynthesis in transgenic celery. Overexpression of AgMYB12 in celery improved the accumulation of apigenin by interacting with the AgFNS [252, 377]. Feng et al. [378] demonstrated that the gene AgUCGalT1, encoding galactosyltransferase, regulated anthocyanin galactosylation in purple celery.
Terpenes
Terpenes are an important group of secondary metabolites that affect taste and flavor [379]. Terpene synthases (TPSs) are catalysts responsible for the formation of sesquiterpenes, monoterpenes, and diterpenes [380–382], which are widely distributed in many plants [382–385]. In carrot research, Keilwagen et al. [386] identified 65 putative TPS family genes. A previous study identified a carrot TPS gene cluster on chromosome 4 that was found to be related to monoterpene production, including DcTPS04, DcTPS26, DcTPS27, DcTPS54, and DcTPS55 [387–389]. In vitro enzyme assays of DcTPS54 and DcTPS04 showed that DcTPS54 is responsible for the formation of sabinene, whereas DcTPS04 is involved in producing the major products α-terpineol, sabinene, β-limonene, β-pinene, and myrcene [388]. Analysis by Yahyaa et al. [390] revealed the function of two TPSs, the sesquiterpene synthase DcTPS1 and the monoterpene synthase DcTPS2. DcTPS1 is responsible for the synthesis of (E)-β-caryophyllene and α-humulene in carrot [391]. In Coriandrum sativum, two TPSs, the recombinant proteins CsγTRPS and CsLINS, were found to catalyze the conversion of geranyl diphosphate [392]. Song et al. [282] first systematically identified TPS family genes in C. sativum.
Dietary fiber
Dietary fiber in plants is classified as soluble or insoluble. Soluble fiber is found in many plants, including carrots, broccoli, onions, barley, bananas, berries, apples, and pears. Insoluble fiber is found in whole grain, wheat, bran, nuts, seeds, and some fruits and vegetables [393, 394]. Dietary fiber plays an important role in moderating the postprandial insulin response and reducing cholesterol and the incidence of heart disease, among other beneficial effects [395]. The plant cell wall, including the primary and secondary wall, which contain lignin and cellulose, is the source of most of the dietary fiber in plants [396].
Hormones play important roles in lignin biosynthesis in celery [397–399] and carrot [400–403]. Transcription factors were important regulators of lignin biosynthesis in celery and carrot [404, 405]. Hypoxia, caused by elevated CO2 concentration also affected lignin content in celery and carrot [315, 406, 407]. The chemical molecular structures of the main bioactive compounds in Apiaceae plants are shown in Fig. 1.
Figure 1.
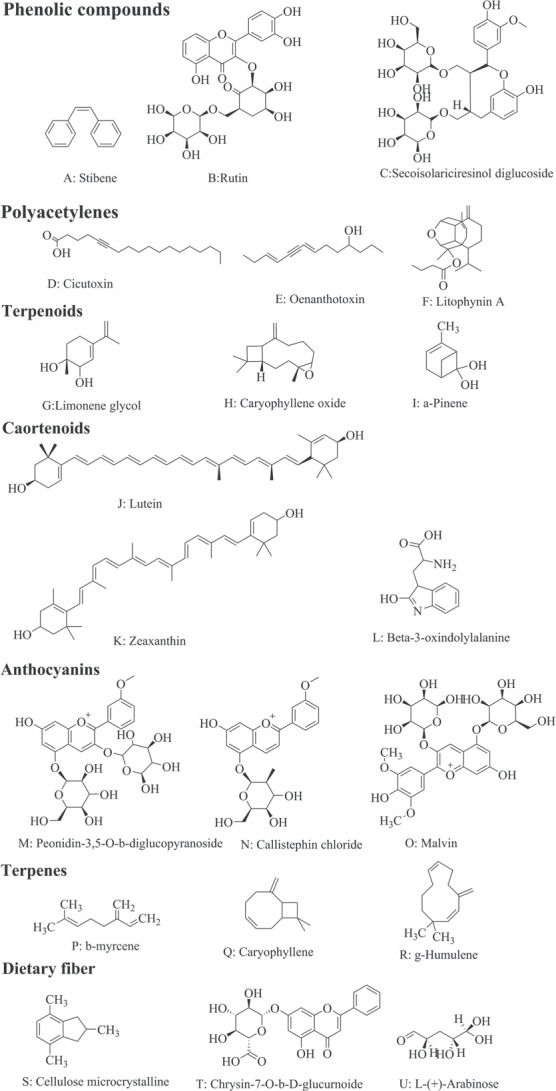
Molecular structure of main bioactive compounds of Apiaceae plants.
Conclusions and future perspectives
Vegetables and medicinal plants are essential foods in human diets and health care, and can provide various necessary nutrients and nutraceuticals. With the strengthening of people’s health consciousness, the diversification, quality, nutritional value, and medicinal value of vegetables and medicinal plants are increasing. Vegetable and medicinal plant research has become increasingly important. In this review, we summarize the origin of Apiaceae plants, common vegetables, and medicinal plants of Apiaceae, bioactive compounds, medicinal applications, traditional and molecular breeding, functional genes, omics analysis, and other aspects. Although a number of Apiaceae plants have been discovered, few members have been utilized for the specific compounds they contain. In the future we should collect Apiaceae germplasm. Omic analysis (transcriptome, genome, and metabolome) is used to explore gene information and bioactive substances in Apiaceae plants. Integration of DNA molecular markers and genome-wide association analysis (GWAS) explores the relationship between genotypes and phenotypes and mine the variation in genomic loci associated with the important agronomic traits. Molecular breeding, including genetic transformation and the CRISPR-Cas9 gene editing system, will be widely used in Apiaceae plant breeding.
Improving breeding level
In crop breeding, excellent varieties have been selected and planted for quality, size, and biotic and abiotic tolerances. Although a few members of the Apiaceae, such as carrot, coriander, and celery, are the most widely grown vegetable crops in the world, their cultivars are insufficient to meet the demand from health-conscious consumers looking for more vegetables and medicinal plants among the Apiaceae. China is rich in wild germplasm resources of Apiaceae. In the future, the purpose of research is to collect and domesticate wild germplasm resources, and increase the exploration and utilization of wild germplasm resources to create more cultivated varieties. In addition, more effective breeding platforms and technology fully combine traditional breeding programs with modern molecular technologies should be established. Molecular markers, GWAS, genetic modification (usually using CRISPR-Cas9 technology to create non-transgenic mutant plants), and nanotechnology should be widely used to guide traditional breeding or molecular breeding.
Mining functional genes
Plant genomes and transcriptomes have been used to explore gene information. Combined transcriptome and metabolome analysis has explored bioactive compounds, functional genes, and transcription factors. The yeast one-hybrid and yeast two-hybrid systems are widely recognized as valuable and straightforward techniques to study interactions between transcription factors and between DNA and transcription factors. Integration of DNA molecular markers and GWAS explore the relationship between genotypes and phenotypes, and mine the variation in genomic loci associated with the important agronomic traits and detected key genes.
Extraction and utilization of bioactive ingredients
Vegetables and medicinal plants in Apiaceae are an excellent source of secondary metabolites, which specifically modulate health-maintaining processes. However, the sample extraction techniques severely block the isolation and extraction of individual secondary metabolites in Apiaceae plants, which severely restricts the development of traditional Chinese medicine. In addition, the pharmacological mechanisms of active ingredients in many vegetables and medicinal plants of Apiaceae are still unclear due to the lack of the animal studies and clinical trials. With the innovations of new technology and the development of molecular biology, research on bioactive ingredients mainly focuses on their isolation and extraction, structure analysis, metabolic pathway analysis, and molecular mechanisms.
Omics of vegetables and medicinal plants in Apiaceae
The family Apiaceae is in the major group flowering plants, and contains >3700 species in 434 genera. However, there are only a few species with available genomes. Innovations in sequencing technology and reduction of sequencing costs provide a great opportunity for studying Apiaceae plant genomes. High-quality genomes of Apiaceae plants contribute to faster and more accurate understanding of genome structures, functional gene information, and other sequences. Moreover, comparative genomics research is commonly used to explore the origin and evolution history of vegetables and medicinal plants in Apiaceae. The applications of GWAS help us identify SNPs and InDels among the different varieties of Apiaceae crops. In addition, comprehensive transcriptome, proteome, and metabolome analysis promotes discoveries in expression patterns and gene function and structure, as well as metabolite components in vegetables and medicinal plants in Apiaceae.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31872098; 32072563); the Guizhou Science and Technology Support Project [Qiankehe Support (2019) 2257]; the Jiangsu seed industry revitalization project [JBGS (2021) 068]; the Guizhou Academy of Agricultural Sciences Support Project (Qian Nongkeyuan Support [2021] No. 05); Construction of Modern Agricultural Technology System in Guizhou Province [GZCYTX (2011-0101)]; the Guizhou Academy of Agricultural Sciences Support Project [Qian Nongkeyuan Science and Technology Innovation (2022) No. 07], and the Priority Academic Program Development of Jiangsu Higher Education Institutions Project (PAPD).
Author contributions
X.J.W., G.F.T., and A.S.X. conceived the outline of the manuscript; X.J.W., G.F.T., and A.S.X. wrote the manuscript; Q.L., T.L., P.H.M., Y.T.P., J.X.L., J.Z., and H.L. provided revisions. All authors read and approved the final manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1. McCormick N. RNA-mediated virus resistance for carrot (Daucus carota var. sativum) and celery (Apium graveolens var. dulce). Ph.D. Thesis, University of Melbourne, 2006.
- 2.Affolter JM. A monograph of the genus Lilaeopsis (Umbelliferae). Syst Bot Monogr. 1985;6:1–140. [Google Scholar]
- 3.Sahebkar A, Iranshahi M. Biological activities of essential oils from the genus Ferula (Apiaceae). Asian Biomed. 2010;4:835–47. [Google Scholar]
- 4.Ajani Y, Bull-Herenu K, Claßen-Bockhoff R. Patterns of flower development in Apiaceae–Apioideae. Flora. 2016;221:38–45. [Google Scholar]
- 5.Ellis PR, Hardman JA. Pests of umbelliferous crops. In: McKinlay RG, ed. Vegetable Crop Pests. Palgrave Macmillan: London, 1992, 327–78. [Google Scholar]
- 6.Qiu YX, Hong DY, Fu CX et al. Genetic variation in the endangered and endemic species Changium smyrnioides (Apiaceae). Biochem Syst Ecol. 2004;32:583–96. [Google Scholar]
- 7.Pollastro F, Gaeta S. Apiaceae, a family of species rich in secondary metabolites: aromatic compounds and medicinal attributes. In: Geoffriau E, Simon PW, eds. Carrots and Related Apiaceae Crops, 2nd ed. CABI Publishing: London, UK, 2020, 35–46. [Google Scholar]
- 8.Que F, Hou XL, Wang GL et al. Advances in research on the carrot, an important root vegetable in the Apiaceae family. Hortic Res. 2019;6:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan J, Yu L, Xu S et al. Assay and evaluation of flavonoid content in Chinese celery. Agr Sci Technol. 2014;15:1200–4. [Google Scholar]
- 10.Saleem F, Eid AH, Shetty K. Potato–herb synergies as food designs for hyperglycemia and hypertension management. In: Paliyath G, Bakovic M, Shetty K, Functional Foods, Nutraceuticals, and Degenerative Disease Prevention. John Wiley & Sons Inc. (NYSE:JW.A): Hoboken, USA, 2011, 325–40. [Google Scholar]
- 11.Ferrie AMR, Bethune TD, Waterer GCA. Field evaluation of doubled haploid plants in the Apiaceae: dill (Anethum graveolens L.), caraway (Carum carvi L.), and fennel (Foeniculum vulgare mill.). Plant Cell Tissue Organ Cult. 2011;104:407–13. [Google Scholar]
- 12.Sepanlou MG, Ardakani MM, Mannan H et al. Ethnobotanical and traditional uses, phytochemical constituents and biological activities of Eryngium species growing in Iran. Tradit Med Res. 2019;4:148–59. [Google Scholar]
- 13.Tovchiga O. Metabolic effects of goutweed (Aegopodium podagraria L.) tincture and metformin in dexamethasone. J Dis Med Plants. 2016;2:117–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Widelski J, Luca SV, Skiba A et al. Isolation and antimicrobial activity of coumarin derivatives from fruits of Peucedanum luxurians Tamamsch. Molecules. 2018;23:1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Widelski J, Luca SV, Skiba A et al. Coumarins from Seseli devenyense Simonk.: isolation by liquid–liquid chromatography and potential anxiolytic activity using an in vivo zebrafish larvae model. Int J Mol Sci. 2021;22:e1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borgen L, Guldahl AS. Great-granny's garden: a living archive and a sensory garden. Biodivers Conserv. 2011;20:441–9. [Google Scholar]
- 17.Wick RL, Rane KK, Sutton DP. Two new ornamental hosts for Phytophthora cactorum: Trachymene caerulea and Gypsophila paniculata. Plant Dis. 1987;71:281–4. [Google Scholar]
- 18.Wei Z, Chunfeng S, Baocheng W et al. Wild ornamental plant resources of Apiaceae in China and their application to landscaping. Chin Wild Plant Resour. 2017;36:68–70. [Google Scholar]
- 19.Hotti H, Seppänen-Laakso T, Arvas M et al. Polyketide synthases from poison hemlock (Conium maculatum L.). FEBS J. 2015;282:4141–56. [DOI] [PubMed] [Google Scholar]
- 20.Radulović N, Dorđević N, Denić M et al. A novel toxic alkaloid from poison hemlock (Conium maculatum L., Apiaceae): identification, synthesis and antinociceptive activity. Food Chem Toxicol. 2012;50:274–9. [DOI] [PubMed] [Google Scholar]
- 21.Stonecipher CA, Welch KD, Lee ST et al. Geographical and seasonal variation in water hemlock (Cicuta maculata) toxins. Biochem Syst Ecol. 2020;89:104012. [Google Scholar]
- 22.Burrows ND, Ward B, Cranfield R. Short-term impacts of logging on understorey vegetation in a jarrah forest. Aust For. 2002;65:47–58. [Google Scholar]
- 23.James LF, Ralphs MH, Nielsen DB. The Ecology and Economic Impact of Poisonous Plants on Livestock Production. CRC Press: Florida, USA, 2019. [Google Scholar]
- 24.Kosalec I, Pepeljnjak S, Kustrak D. Antifungal activity of fluid extract and essential oil from anise fruits (Pimpinella anisum L., Apiaceae). Acta Pharma. 2005;55:377–85. [PubMed] [Google Scholar]
- 25.Shahrajabian MH, Sun W, Cheng Q. Chinese star anise and anise, magic herbs in traditional Chinese medicine and modern pharmaceutical science. Asian J Med Biol Res. 2019;5:162–79. [Google Scholar]
- 26.Dehpour AA, Ebrahimzadeh MA, Fazel NS. et al. Antioxidant activity of the methanol extract of Ferula assafoetida and its essential oil composition. Grasas Aceites 2009;60:405–12. [Google Scholar]
- 27.Amalraj A, Gopi S. Biological activities and medicinal properties of asafoetida: a review. J Tradit Complement Med. 2017;7:347–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iranshahy M, Iranshahi M. Traditional uses, phytochemistry and pharmacology of asafoetida (Ferula assa-foetida oleo-gum-resin) – a review. J Ethnopharmacol. 2011;134:1–10. [DOI] [PubMed] [Google Scholar]
- 29.Hu SCS, Lee IT, Yen MH et al. Anti-melanoma activity of Bupleurum chinense, Bupleurum kaoi and nanoparticle formulation of their major bioactive compound saikosaponin-d. J Ethnopharmacol. 2016;179:432–42. [DOI] [PubMed] [Google Scholar]
- 30.Wang M, Liu P, Feng X et al. Studies on the chemical constituents of the aerial part of Bupleurum chinense (II). J Chinese Med Mat. 2009;32:367–9. [PubMed] [Google Scholar]
- 31.Zhao W, Li JJ, Yue SQ et al. Antioxidant activity and hepatoprotective effect of a polysaccharide from bei chaihu (Bupleurum chinense DC). Carbohydr Polym. 2012;89:448–52. [DOI] [PubMed] [Google Scholar]
- 32.Xie JY, Di HY, Li H et al. Bupleurum chinense DC polysaccharides attenuates lipopolysaccharide-induced acute lung injury in mice. Phytomedicine. 2012;19:130–7. [DOI] [PubMed] [Google Scholar]
- 33.Agrahari P, Singh DK. A review on the pharmacological aspects of Carum carvi. J Biol Earth Sci. 2014;4:M1–13. [Google Scholar]
- 34.Vallverdú-Queralt A, Regueiro J, Alvarenga JFR et al. Characterization of the phenolic and antioxidant profiles of selected culinary herbs and spices: caraway, turmeric, dill, marjoram and nutmeg. Food Sci Technol. 2015;35:189–95. [Google Scholar]
- 35.Bystrická J, Kavalcová P, Musilová J et al. Carrot (Daucus carota L. ssp. sativus (Hoffm.) Arcang.) as source of antioxidants. Acta Agric Slov. 2015;105:303–11. [Google Scholar]
- 36.Poulin MJ, Bel-Rhlid R, Piché Y et al. Flavonoids released by carrot (Daucus carota) seedlings stimulate hyphal development of vesicular-arbuscular mycorrhizal fungi in the presence of optimal CO2 enrichment. J Chem Ecol. 1993;19:2317–27. [DOI] [PubMed] [Google Scholar]
- 37.Shin JY, Che DN, Cho BO et al. Anti-inflammatory effect of hydrolyzed celery leaves extract in murine primary splenocyte. J Food Biochem. 2019;43:e12970. [DOI] [PubMed] [Google Scholar]
- 38.Yao Y, Ren G. Effect of thermal treatment on phenolic composition and antioxidant activities of two celery cultivars. LWT – Food Sci Technol. 2011;44:181–5. [DOI] [PubMed] [Google Scholar]
- 39.Lao SC, Li SP, Kan KKW et al. Identification and quantification of 13 components in Angelica sinensis (Danggui) by gas chromatography–mass spectrometry coupled with pressurized liquid extraction. Anal Chim Acta. 2004;526:131–7. [Google Scholar]
- 40.Li P, Li SP, Lao SC et al. Optimization of pressurized liquid extraction for Z-ligustilide, Z-butylidenephthalide and ferulic acid in Angelica sinensis. J Pharm Biomed Anal. 2006;40:1073–9. [DOI] [PubMed] [Google Scholar]
- 41.Zhao C, Jia Y, Lu F. Angelica stem: a potential low-cost source of bioactive phthalides and phytosterols. Molecules. 2018;23:3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chao WW, Lin BF. Bioactivities of major constituents isolated from Angelica sinensis (Danggui). Chin Med. 2011;6:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin S, Li HY, Wang ZY et al. Analysis of methanolic extracts and crude polysaccharides from the leaves of Chuanminshen violaceum and their antioxidant activities. Antioxidants (Basel). 2019;8:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin S, Li HY, Yuan Q et al. Structural characterization, antioxidant activity, and immunomodulatory activity of non-starch polysaccharides from Chuanminshen violaceum collected from different regions. Int J Biol Macromol. 2020;143:902–12. [DOI] [PubMed] [Google Scholar]
- 45.Dong H, Zhang Q, Li L et al. Antioxidant activity and chemical compositions of essential oil and ethanol extract of Chuanminshen violaceum. Ind Crop Prod. 2015;76:290–7. [Google Scholar]
- 46.Lee TH, Chen YC, Hwang TL et al. New coumarins and anti-inflammatory constituents from the fruits of Cnidium monnieri. Int J Mol Sci. 2014;15:9566–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song G, Zhou L, Sheng N et al. Simultaneous quantification of 16 bioactive constituents in common Cnidium fruit by liquid chromatography–electrospray ionization-mass spectrometry. J Pharm Biomed. 2015;107:304–10. [DOI] [PubMed] [Google Scholar]
- 48.Sun Y, Yang AWH, Lenon GB. Phytochemistry, ethnopharmacology, pharmacokinetics and toxicology of Cnidium monnieri (L.) Cusson. Int J Mol Sci. 2020;21:e1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyazawa M, Kurose K, Itoh A et al. Comparison of the essential oils of Glehnia littoralis from northern and southern Japan. J Agric Food Chem. 2001;49:5433–6. [DOI] [PubMed] [Google Scholar]
- 50.Huang GJ, Deng JS, Liao JC et al. Inducible nitric oxide synthase and cyclooxygenase-2 participate in anti-inflammatory activity of imperatorin from Glehnia littoralis. J Agric Food Chem. 2012;60:1673–81. [DOI] [PubMed] [Google Scholar]
- 51.Yoon TS, Choo BK, Cheon MS et al. Pharmacological activities of Glehnia littoralis. Korean J Orient Med. 2008;14:123–8. [Google Scholar]
- 52.Guerra NB, Almeida Melo E, Mancini Filho J. Antioxidant compounds from coriander (Coriandrum sativum L.) etheric extract. J Food Compos Anal. 2005;18:193–9. [Google Scholar]
- 53.Hassanien MF, Assiri AM, Alzohairy AM et al. Health-promoting value and food applications of black cumin essential oil: an overview. J Food Sci Technol. 2015;52:6136–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mnif S, Aifa S. Cumin (Cuminum cyminum L.) from traditional uses to potential biomedical applications. Chem Biodivers. 2015;12:733–42. [DOI] [PubMed] [Google Scholar]
- 55.Sharopov FS, Wink M, Gulmurodov IS et al. Composition and bioactivity of the essential oil of Anethum graveolens L. from Tajikistan. Int J Med Arom Plants. 2013;3:125–30. [Google Scholar]
- 56.Swargiary A, Daimari M. GC–MS analysis of phytocompounds and antihyperglycemic property of Hydrocotyle sibthorpioides Lam. SN Appl Sci. 2021;3:1–11. [Google Scholar]
- 57.Kumari S, Elancheran R, Kotoky J et al. Rapid screening and identification of phenolic antioxidants in Hydrocotyle sibthorpioides Lam. by UPLC–ESI-MS/MS. Food Chem. 2016;203:521–9. [DOI] [PubMed] [Google Scholar]
- 58.Yu F, Yu F, McGuire PM et al. Effects of Hydrocotyle sibthorpioides extract on transplanted tumors and immune function in mice. Phytomedicine. 2017;14:166–71. [DOI] [PubMed] [Google Scholar]
- 59.Ahmed AF, Shi M, Liu C et al. Comparative analysis of antioxidant activities of essential oils and extracts of fennel (Foeniculum vulgare Mill.) seeds from Egypt and China. Food Sci Hum Wellness. 2019;8:67–72. [Google Scholar]
- 60.Iranshahi M, Rezaee R, Najafi MN et al. Cytotoxic activity of the genus Ferula (Apiaceae) and its bioactive constituents. Avicenna J Phytomed. 2018;8:296–312. [PMC free article] [PubMed] [Google Scholar]
- 61.Mohammadhosseini M, Venditti A, Sarker SD et al. The genus Ferula: ethnobotany, phytochemistry and bioactivities – a review. Ind Crop Prod. 2019;129:350–94. [Google Scholar]
- 62.Sabaragamuwa R, Perera CO, Fedrizzi B. Ultrasound assisted extraction and quantification of targeted bioactive compounds of Centella asiatica (gotu kola) by UHPLC-MS/MS MRM tandem mass spectroscopy. Food Chem. 2022;371:131187. [DOI] [PubMed] [Google Scholar]
- 63.Chandrika UG, Kumara PAP. Gotu kola (Centella asiatica): nutritional properties and plausible health benefits. Adv Food Nutr Res. 2015;76:125–57. [DOI] [PubMed] [Google Scholar]
- 64.Lu J, Fu X, Liu T et al. Phenolic composition, antioxidant, antibacterial and anti-inflammatory activities of leaf and stem extracts from Cryptotaenia japonica Hassk. Ind Crop Prod. 2018;122:522–32. [Google Scholar]
- 65.Santos PA, Figueiredo AC, Oliveira MM et al. Growth and essential oil composition of hairy root cultures of Levisticum officinale WDJ Koch (lovage). Plant Sci. 2005;168:1089–96. [Google Scholar]
- 66.Azietaku JT, Ma H, Yu XA et al. A review of the ethnopharmacology, phytochemistry and pharmacology of Notopterygium incisum. J Ethnopharmacol. 2017;202:241–55. [DOI] [PubMed] [Google Scholar]
- 67.Xu K, Jiang S, Sun H et al. New alkaloids from the seeds of Notopterygium incisum. Nat Prod Res. 2012;26:1898–903. [DOI] [PubMed] [Google Scholar]
- 68.Meyer H, Bolarinwa A, Wolfram G et al. Bioavailability of apigenin from apiin-rich parsley in humans. Ann Nutr Metab. 2006;50:167–72. [DOI] [PubMed] [Google Scholar]
- 69.Sbai H, Saad I, Ghezal N et al. Bioactive compounds isolated from Petroselinum crispum L. leaves using bioguided fractionation. Ind Crop Prod. 2016;89:207–14. [Google Scholar]
- 70.Farzaei MH, Abbasabadi Z, Ardekani MRS et al. Parsley: a review of ethnopharmacology, phytochemistry and biological activities. J Tradit Chin Med. 2013;33:815–26. [DOI] [PubMed] [Google Scholar]
- 71.Stegelmeier BL, Colegate SM, Knoppel EL et al. Wild parsnip (Pastinaca sativa) induced photosensitization. Toxicon. 2019;167:60–6. [DOI] [PubMed] [Google Scholar]
- 72.Lei LJ, Wang WL, Wang J et al. A review of the ethnopharmacology, phytochemistry, and pharmacology of Changium smyrnioides Wolff. J Nat Med. 2019;73:1–10. [DOI] [PubMed] [Google Scholar]
- 73.Helal IE, Galala AA, HEA S et al. Bioactive constituents from Apium leptophyllum fruits. J Pharm Res Int. 2016;14:1–8. [Google Scholar]
- 74.Sahoo HB, Patro SK, Sagar Ret al. Mutagenic evaluation and spectroscopic characterization of flavonoidal fraction of Apium leptophyllum (Pers.) fruit. Int J Nut Pharmacol Neurol Dis 2015;5:82–88. [Google Scholar]
- 75.Li MY, Hou XL, Wang F et al. Advances in the research of celery, an important Apiaceae vegetable crop. Crit Rev Biotechnol. 2017;38:172–83. [DOI] [PubMed] [Google Scholar]
- 76.Kumar S, Huang X, Li G et al. Comparative transcriptomic analysis provides novel insights into the blanched stem of Oenanthe javanica. Plants (Basel). 2021;10:2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun L, Chang Y, Han S et al. Metabolomics analysis of bioactive compounds in culture water planting floating bed water dropwort in different development stages. Res Environ Sci. 2021;34:1860–75. [Google Scholar]
- 78.Liu ZL, Chu SS, Jiang GH. Insecticidal activity and composition of essential oil of Ostericum sieboldii (Apiaceae) against Sitophilus zeamais and Tribolium castaneum. Rec Nat Prod. 2011;5:74–81. [Google Scholar]
- 79.Bos R, Koulman A, Woerdenbag HJ et al. Volatile components from Anthriscus sylvestris (L.) Hoffm. J Chromatogr. 2002;966:233–8. [DOI] [PubMed] [Google Scholar]
- 80.Olaru OT, Niţulescu GM, Orţan A et al. Ethnomedicinal, phytochemical and pharmacological profile of Anthriscus sylvestris as an alternative source for anticancer lignans. Molecules. 2015;20:15003–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Velescu BS, Anuţa V, Niţulescu GM et al. Pharmaceutical assessment of Romanian crops of Anthriscus sylvestris (Apiaceae). Farmacia. 2017;65:824–31. [Google Scholar]
- 82.Jeong GS, Kwon O-K, Park B-Y et al. Lignans and coumarins from the roots of Anthriscus sylvestris and their increase of caspase-3 activity in hl-60 cells. Biol Pharm Bull. 2007;30:1340–3. [DOI] [PubMed] [Google Scholar]
- 83.Vyas A, Shukla SS, Pandey R et al. Chervil: a multifunctional miraculous nutritional herb. Asian J Plant Sci. 2012;11:163–71. [Google Scholar]
- 84.Calvino CI, Teruel FE, Downie SR. The role of the Southern Hemisphere in the evolutionary history of Apiaceae, a mostly north temperate plant family. J Biogeogr. 2016;43:398–409. [Google Scholar]
- 85.Calvino CI, Downie SR. Circumscription and phylogeny of Apiaceae subfamily Saniculoideae based on chloroplast DNA sequences. Mol Phylogenet Evol. 2007;44:175–91. [DOI] [PubMed] [Google Scholar]
- 86.Banasiak L, Piwczyński M, Uliński T et al. Dispersal patterns in space and time: a case study of Apiaceae subfamily Apioideae. J Biogeogr. 2013;40:1324–35. [Google Scholar]
- 87.Nicolas AN, Plunkett GM. The demise of subfamily Hydrocotyloideae (Apiaceae) and the re-alignment of its genera across the entire order Apiales. Mol Phylogenet Evol. 2009;53:134–51. [DOI] [PubMed] [Google Scholar]
- 88.Downie SR, Katz-Downie DS, Cho KJ. Phylogenetic analysis of Apiaceae subfamily Apioideae using nucleotide sequences from the chloroplast rpoC1 intron. Mol Phylogenet Evol. 1996;6:1–18. [DOI] [PubMed] [Google Scholar]
- 89.Calvino CI, Martinez SG, Downie SR. Morphology and biogeography of Apiaceae subfamily Saniculoideae as inferred by phylogenetic analysis of molecular data. Am J Bot. 2008;95:196–214. [DOI] [PubMed] [Google Scholar]
- 90.Downie S, Katz-Downie D, Watson M. A phylogeny of the flowering plant family Apiaceae based on chloroplast DNA rpl16 and rpoC1 intron sequences: towards a suprageneric classification of subfamily Apioideae. Am J Bot. 2000;87:273–92. [PubMed] [Google Scholar]
- 91.Plunkett GM, Downie SR. Major lineages within Apiaceae subfamily Apioideae: a comparison of chloroplast restriction site and DNA sequence data. Am J Bot. 1999;86:1014–26. [PubMed] [Google Scholar]
- 92.Degtjareva GV, Pimenov MG, Samigullin TH. Molecular data allow to elucidate the taxonomic placement of some Umbelliferae from middle Asia and Afghanistan (Pinacantha, Ladyginia, Peucedanum mogoltavicum). Phytotaxa. 2018;350:42–50. [Google Scholar]
- 93.Gallie DR. Transgenic carrot (Daucus carota L.). In: Bajal YPS (ed.), Transgenic Crops II. Springer: Berlin, 2001,147–58. [Google Scholar]
- 94.Simon PW, Wolff XY. Carotenes in typical and dark orange carrots. J Agric Food Chem. 1987;35:1017–22. [Google Scholar]
- 95.Mahmood S, Hussain S, Malik F. Critique of medicinal conspicuousness of parsley (Petroselinum crispum): a culinary herb of Mediterranean region. Pak J Pharm Sci. 2014;27:193–202. [PubMed] [Google Scholar]
- 96.Chomchalow N. Production of herbs in Asia: an overview. AU J Technol. 2002;6:95–108. [Google Scholar]
- 97.Gunn D. Water dropwort. In Practice. 1992;14:409–10. [Google Scholar]
- 98.Uchibayashi M. The coriander story. Yakushigaku Zasshi. 2001;36:56–7. [PubMed] [Google Scholar]
- 99.Wei J, Gao YZ, Zhou J et al. Collection and sorting of medicinal plants in Chinese Apiaceae (Umbelliferae). China J Chin Mat Med. 2019;44:5329–35. [DOI] [PubMed] [Google Scholar]
- 100.Rautio AM, Axelsson W, Östlund LL. They followed the power of the plant: historical sami harvest and traditional ecological knowledge (Tek) of Angelica archangelica in northern Fennoscandia. J Ethnobiol. 2016;36:617–36. [Google Scholar]
- 101.Liao CY, Downie SR, Yu Y et al. Historical biogeography of the Angelica group (Apiaceae tribe Selineae) inferred from analyses of nrDNA and cpDNA sequences. J Syst Evol. 2012;50:206–17. [Google Scholar]
- 102.Teixidor-Toneu I, Kjesrud K, Kool A. Sweetness beyond desserts: the cultural, symbolic, and botanical history of Angelica (Angelica archangelica) in the Nordic region. J Ethnobiol. 2020;40:289–304. [Google Scholar]
- 103.Acimovic MG, Milic NB. Perspectives of the Apiaceae hepatoprotective effects- a review. Nat Prod Commun. 2017;12:309–17. [PubMed] [Google Scholar]
- 104.Nebija F, Stefkov G, Karapandzova M et al. Morphological and anatomical characteristics of the root and herb from Eryngium campestre L. (Apiaceae). Maced Pharm Bull. 2006;52:57–64. [Google Scholar]
- 105.Kotina EL, Wyk BV, Tilney PM et al. The systematic significance of bark structure in southern African genera of tribe Heteromorpheae (Apiaceae). Bot J Linn Soc. 2012;169:677–91. [Google Scholar]
- 106.Aćimović MG. Nutraceutical potential of Apiaceae. In: Mérillon JM, Ramawat K, eds. Bioactive Molecules in Food . Reference Series in Phytochemistry. Springer: Cham, 2017, 1–31. [Google Scholar]
- 107.Anderson JW, Chen W. Plant fiber. Carbohydrate and lipid metabolism. Am J Clin Nutr. 1979;32:346–63. [DOI] [PubMed] [Google Scholar]
- 108.Szerszunowicz I, Kobukowski J. Characteristics of potential protein nutraceuticals of plant origin with antioxidant activity. Molecules. 2020;25:e1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ibe C, Jacobs CC, Imo C et al. Evaluation of the antioxidant activities of Psidium guajava and Aloe vera. J Pharm Res Int. 2014;4:397–406. [Google Scholar]
- 110.Cornelli U. Antioxidant use in nutraceuticals. Clin Dermatol. 2009;27:175–94. [DOI] [PubMed] [Google Scholar]
- 111.Pandey M, Verma RK, Saraf SA. Nutraceuticals: new era of medicine and health. Asian J Pharm Clin Res. 2010;3:11–5. [Google Scholar]
- 112.Shahidi F. Nutraceuticals, functional foods and dietary supplements in health and disease. J Food Drug Anal. 2012;20:226–30. [Google Scholar]
- 113.Gilsing V, Nooteboom B. Exploration and exploitation in innovation systems: the case of pharmaceutical biotechnology. Res Policy. 2006;35:1–23. [Google Scholar]
- 114.Nenadis N, Wang LF, Tsimidou M et al. Estimation of scavenging activity of phenolic compounds using the ABTS(*+) assay. J Agric Food Chem. 2004;52:4669–74. [DOI] [PubMed] [Google Scholar]
- 115.Soares S, Kohl S, Thalmann S et al. Different phenolic compounds activate distinct human bitter taste receptors. J Agric Food Chem. 2013;61:1525–33. [DOI] [PubMed] [Google Scholar]
- 116.Cheynier V. Phenolic compounds: from plants to foods. Phytochem Rev. 2012;11:153–77. [Google Scholar]
- 117.Thiviya P, Gamage A, Piumali D et al. Apiaceae as an important source of antioxidants and their applications. Cosmetics. 2021;8:e111. [Google Scholar]
- 118.Sayed B, Talou T, Saad Z et al. The Apiaceae: ethnomedicinal family as source for industrial uses. Ind Crop Prod. 2017;109:661–71. [Google Scholar]
- 119.Christova-Bagdassarian VL, Bagdassarian KS, Atanassova MS. Phenolic profile: antioxidant and antibacterial activities from the Apiaceae family (dry seeds). Mintage J Pharm Med Sci. 2013;2:26–31. [Google Scholar]
- 120.Materska M, Perucka I. Antioxidant activity of the main phenolic compounds isolated from hot pepper fruit (Capsicum annuum L.). J Agric Food Chem. 2005;53:1750–6. [DOI] [PubMed] [Google Scholar]
- 121.Kähkönen MP, Hopia AI, Vuorela HJ et al. Antioxidant activity of plant extracts containing phenolic compounds. J Agric Food Chem. 1999;47:3954–62. [DOI] [PubMed] [Google Scholar]
- 122.Žemlička L, Fodran P, Lukeš V et al. Physicochemical and biological properties of luteolin-7-O-β-d-glucoside (cynaroside) isolated from Anthriscus sylvestris (L.) Hoffm. Monatsh Chem - Chemical Monthly. 2014;145:1307–18. [Google Scholar]
- 123.Xie HK, Zhou D-Y, Yin F-W et al. Mechanism of antioxidant action of natural phenolics on scallop (Argopecten irradians) adductor muscle during drying process. Food Chem. 2019;281:251–60. [DOI] [PubMed] [Google Scholar]
- 124.Patricia RR, Varela-López A, Forbes-Hernández TY et al. Phenolic compounds isolated from olive oil as nutraceutical tools for the prevention and management of cancer and cardiovascular diseases. Int J Mol Sci. 2018;19:e2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cheng CW, Chang WL, Chang LC et al. Ferulic acid, an Angelica sinensis-derived polyphenol, slows the progression of membranous nephropathy in a mouse model. Evid Based Complement Alternat Med. 2012;3:161235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang X, Lin D, Jiang R et al. Ferulic acid exerts antitumor activity and inhibits metastasis in breast cancer cells by regulating epithelial to mesenchymal transition. Oncol Rep. 2016;36:271–8. [DOI] [PubMed] [Google Scholar]
- 127.Josuttis M, Krüger E, Dietrich H. Influence of cultivation techniques on health beneficial components in strawberry. Acta Hortic. 2009;11:205–10. [Google Scholar]
- 128.Romo-Hualde A, Sáiz-Abajo MJ, Yetano-Cunchillos AI et al. Characterization of bioactive substances in various artichoke varieties. Acta Hortic. 2011;58:401–6. [Google Scholar]
- 129.Juhaimi F, Ghafoor K, Uslu N et al. The effect of harvest times on bioactive properties and fatty acid compositions of prickly pear (Opuntia ficus-barbarica A. Berger) fruits. Food Chem. 2020;303:125387. [DOI] [PubMed] [Google Scholar]
- 130.Negri R. Polyacetylenes from terrestrial plants and fungi: recent phytochemical and biological advances. Fitoterapia. 2015;106:92–109. [DOI] [PubMed] [Google Scholar]
- 131.Christensen LP, Brandt K. Bioactive polyacetylenes in food plants of the Apiaceae family: occurrence, bioactivity and analysis. J Pharm Biomed Anal. 2006;41:683–93. [DOI] [PubMed] [Google Scholar]
- 132.Bijttebier S, D'Hondt E, Apers S et al. Identification and quantification of bioactive polyacetylenes in supermarket products and food processing waste. Planta Med. 2013;79:3–5. [Google Scholar]
- 133.Metzger BT, Barnes DM, Reed JD. Purple carrot (Daucus carota L.) polyacetylenes decrease lipopolysaccharide-induced expression of inflammatory proteins in macrophage and endothelial cells. J Agric Food Chem. 2008;56:3554–60. [DOI] [PubMed] [Google Scholar]
- 134.Sousa RMO, Cunha AC, Fernandes-Ferreira M. The potential of Apiaceae species as sources of singular phytochemicals and plant-based pesticides. Phytochemistry. 2021;187:e112714. [DOI] [PubMed] [Google Scholar]
- 135.Rawson A, Hossain MB, Patras A et al. Effect of boiling and roasting on the polyacetylene and polyphenol content of fennel (Foeniculum vulgare) bulb. Food Res Int. 2013;50:513–8. [Google Scholar]
- 136.Yazaki K, Arimura G-I, Ohnishi T. 'Hidden' terpenoids in plants: their biosynthesis, localization and ecological roles. Plant Cell Physiol. 2017;58:1615–21. [DOI] [PubMed] [Google Scholar]
- 137.Singh RP, Gangadharappa HV, Mruthunjaya K. Cuminum cyminum – a popular spice: an updated review. Pharmacogn J. 2017;9:292–301. [Google Scholar]
- 138.Vidal M. Fennel: a choice aromatic vegetable for those who like the taste of aniseed. Jardins De France. 1982;3:116–8. [Google Scholar]
- 139.Verdeguer M, Sánchez-Moreiras AM, Araniti F. Phytotoxic effects and mechanism of action of essential oils and terpenoids. Plants (Basel). 2020;9:1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Grassmann J. Terpenoids as plant antioxidants. Vitam Horm. 2005;72:505–35. [DOI] [PubMed] [Google Scholar]
- 141.Zhou X, Ke C, Lv Y et al. Asiaticoside suppresses cell proliferation by inhibiting the NF-κB signaling pathway in colorectal cancer. Int J Mol Med. 2020;46:1525–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vazquez-Muoz R, Meza-Villezcas A, Fournier PGJ et al. Enhancement of antibiotics antimicrobial activity due to the silver nanoparticles impact on the cell membrane. PLoS One. 2019;14:e0224904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tapwal P, Pradhan S, Chandra S et al. Antimycotic activity and phytochemical screening of fungal endophytes associated with Santalum album. Nusantara Biosci. 2016;8:14–7. [Google Scholar]
- 144.Dutta S, Ali KM, Dash SK et al. Role of nutraceuticals on health promotion and disease prevention: a review. J Drug Deliv Ther. 2018;8:42–7. [Google Scholar]
- 145.Dillard CJ, German JB. Phytochemicals: nutraceuticals and human health. J Sci Food Agric. 2000;80:1744–56. [Google Scholar]
- 146.Mann C. Super cilantro, mann, chris. Better Nutr. 2008;81:252–8. [Google Scholar]
- 147.Costa AG, Bertolucci SKV, Chagas JH et al. Biomass production, yield and chemical composition of peppermint essential oil using different organic fertilizer sources. Cienc Agrotecnol. 2013;37:202–10. [Google Scholar]
- 148.Andruţa ME, Vlaic R, Muresan V et al. Development and characterization of a biologically active white sauce based on horseradish, onion, parsley and parsnip. Hop Med Plants. 2017;27:139–48. [Google Scholar]
- 149.Srinivasan K. Role of spices beyond food flavoring: nutraceuticals with multiple health effects. Food Rev Int. 2005;21:167–88. [Google Scholar]
- 150.Tahereh S, Seyedeh M, Hekmatzadeh F et al. Evaluating the effects of dill (Anethum graveolens) seed on the duration of active phase and intensity of labour pain. J Herb Med. 2015;5:26–9. [Google Scholar]
- 151.Mansouri M, Nayebi N, Keshtkar A et al. The effect of 12 weeks Anethum graveolens (dill) on metabolic markers in patients with metabolic syndrome; a randomized double blind controlled trial. Daru. 2012;20:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Vishnu PR. Studies on bioactive properties of coriander (Coriandrum sativum L.) foliage. Indian Bee J. 2015;31:67–82. [Google Scholar]
- 153.Mani V, Parle M, Ramasamy K et al. Anti-dementia potential of Daucus carota seed extract in rats. Pharmacologyonline. 2010;1:552–65. [Google Scholar]
- 154.Athar M, Bokhari TZ. Ethnobotany and production constraints of traditional and commonly used vegetables of Pakistan. J Veg Sci. 2008;12:27–38. [Google Scholar]
- 155.Greathouse WV, Shearer CE. Celery product and method for its production. United States Patent US20020114865, 2002.
- 156.Casselman B. Angelica: the angels' herb and Diaghilev's surname. Vocab Rev. 2006;17:58–67. [Google Scholar]
- 157.Dias JCS. Nutritional and health benefits of carrots and their seed extracts. Food Nutr Sci. 2014;5:2147–56. [Google Scholar]
- 158.Mogie M. The Evolution of Asexual Reproduction in Plants. London: Chapman and Hall; 1992. [Google Scholar]
- 159.Zeng Q, Deng L, Hu W et al. Verification of glyphosate resistance, lepidopteran resistance and wide compatibility of male sterile line E1C4008S in rice. Rice Sci. 2020;27:215–26. [Google Scholar]
- 160.Tan GF, Wang F, Zhang XY et al. Different lengths, copies and expression levels of the mitochondrial atp6 gene in male sterile and fertile lines of carrot (Daucus carota L). Mitochondrial DNA A DNA Mapp Seq Anal. 2018;29:446–54. [DOI] [PubMed] [Google Scholar]
- 161.Nothnagel T, Straka P, Linke B. Male sterility in populations of Daucus and the development of alloplasmic male-sterile lines of carrot. Plant Breed. 2010;119:145–52. [Google Scholar]
- 162.Schnable PS, Wise RP. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 1998;3:175–80. [Google Scholar]
- 163.Colombo N, Galmarini CR, Havey M. The use of genetic, manual and chemical methods to control pollination in vegetable hybrid seed production: a review. Plant Breed. 2017;136:287–99. [Google Scholar]
- 164.Kozik E, Nowak R, Nowakowska M et al. Level of sterility and morphological flowers differentiation of petaloid male-sterile plants of carrot. J Agric Sci. 2012;4:187–94. [Google Scholar]
- 165.Kalia P, Mangal M, Singh S et al. Morphological and molecular changes on cytoplasmic male sterility (CMS) introgression in Asiatic carrot (Daucus carota L.). Planta. 2019;250:507–18. [DOI] [PubMed] [Google Scholar]
- 166.Yamamoto T, Nakajima Y, Oeda K. Morphological changes in homeotic cytoplasmic male-sterile carrots combined with fertile cytoplasm by asymmetrical cell fusion. Plant Cell Rep. 2000;19:363–70. [DOI] [PubMed] [Google Scholar]
- 167.Timin NI, Vasilevsky VA. Genetic peculiarities of carrot (Daucus carota L.). J Appl Genet. 1997;38:232–6. [Google Scholar]
- 168.Quiros CF, Rugama A, Dong YY, Orton TJ. Cytological and genetical studies of a male sterile celery. Euphytica. 1986;35:867–75. [Google Scholar]
- 169.Gao GX, Jin LZ, Lu ZM, Gu ZH. Discovery and botanical characters of celery male sterile material. Tianjin Agric Sci. 2006;12:9–11. [Google Scholar]
- 170.Russell PE. Fungicide resistance: occurrence and management. J Agric Sci. 1995;124:317–8. [Google Scholar]
- 171.Nelson R, Wiesner-Hanks T, Wisser R et al. Navigating complexity to breed disease-resistant crops. Nat Rev Genet. 2018;19:21–33. [DOI] [PubMed] [Google Scholar]
- 172.Watson A. Managing carrot powdery mildew. Australas Plant Pathol. 2016;45:29–35. [Google Scholar]
- 173.Glawe DA, Pelter GQ, Toit LJD. First report of powdery mildew of carrot and parsley caused by Erysiphe heraclei in Washington state. Plant Health Prog. 2005;114:21–3. [Google Scholar]
- 174.Scott DJ, Wenham HT. Occurrence of two seed-borne pathogens, Alternaria radicina and Alternaria dauci, on imported carrot seed in New Zealand. N Z J Agric Res. 1973;16:247–50. [Google Scholar]
- 175.Milosavljević A, Pfaf-Dolovac E, Mitrović M et al. First report of Cercospora apii, causal agent of Cercospora early blight of celery, in Serbia. Plant Dis. 2014;98:1157. [DOI] [PubMed] [Google Scholar]
- 176.Trueman CL, McDonald MR, Gossen BD et al. Evaluation of disease forecasting programs for management of septoria late blight (Septoria apiicola) on celery. Can J Plant Pathol. 2007;29:330–9. [Google Scholar]
- 177.Ochoa O, Quiros CF. Apium wild species: novel sources for resistance to late blight in celery. Plant Breed. 1989;102:317–21. [Google Scholar]
- 178.Quiros CF, D'Antonio V, Greathead AS et al. UC8-1, UC10-1, and UC26-1: three celery lines resistant to fusarium yellows. HortScience. 1993;28:351–2. [Google Scholar]
- 179.Lacy ML, Grumet R, Toth KF et al. MSU-SHK5: a somaclonally derived fusarium yellows-resistant celery line. HortScience. 1996;31:289–90. [Google Scholar]
- 180.Epstein L, Kaur S, Chang PL et al. Races of the celery pathogen Fusarium oxysporum f. sp. apii are polyphyletic. Phytopathology. 2017;107:463–73. [DOI] [PubMed] [Google Scholar]
- 181.Koutouan C, Le V, Raymonde C et al. Link between carrot leaf secondary metabolites and resistance to Alternaria dauci. Sci Rep. 2018;8:e13746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gugino BK, Carroll JE, Widmer TL et al. Field evaluation of carrot cultivars for susceptibility to fungal leaf blight diseases in New York. Crop Prot. 2007;26:709–14. [Google Scholar]
- 183.Mansur PS, Silva AL, Salcedo SS et al. Alternaria dauci causes leaf spots and leaf blight of coriander (Coriandrum sativum) in Brazil. Australas Plant Dis Notes. 2020;15:1–5. [Google Scholar]
- 184.Lee I. ‘Candidatus Phytoplasma asteris’, a novel phytoplasma taxon associated with aster yellows and related diseases. Int J Syst Evol Microbiol. 2004;54:1037–48. [DOI] [PubMed] [Google Scholar]
- 185.Saharan GS, Mehta N. Introduction. In: Saharan GS, Mehta N. Sclerotinia Diseases of Crop Plants: Biology, Ecology and Disease Management. New York: Springer, 2008, 1–11. [Google Scholar]
- 186.Jensen BD, Finckh MR, Munk L et al. Susceptibility of wild carrot (Daucus carota ssp. carota) to Sclerotinia sclerotiorum. Eur J Plant Pathol. 2008;122:359–67. [Google Scholar]
- 187.Khoshkhatti N, Koohi Habibi M, Mosahebi G. Characterization of celery mosaic virus from celery in Tehran province. Iranian. Iran J Virol. 2011;5:10–4. [Google Scholar]
- 188.Ruiz J, Pico B, Li G et al. Identification of markers linked to a celery mosaic virus resistance gene in celery. J Am Soc Hortic Sci. 2001;126:432–5. [Google Scholar]
- 189.Parsons J, Matthews W, Iorizzo M et al. Meloidogyne incognita nematode resistance QTL in carrot. Mol Breed. 2015;35:e114. [Google Scholar]
- 190.Vovlas N, Lucarelli G, Sasanelli N et al. Pathogenicity and host-parasite relationships of the root-knot nematode Meloidogyne incognita on celery. Plant Pathol. 2008;57:981–7. [Google Scholar]
- 191.Rady GH. Evaluation of some root-knot nematode management strategies in sugarbeet fields at West Nubaryia district. Ann Agric Sci Moshtohor. 2021;59:617–30. [Google Scholar]
- 192.Karssen G, Moens M. Root-knot nematodes. Plant Nematol. 2013;73:74–108. [Google Scholar]
- 193.Huang SP, Vecchia PT, Ferreira PE. Varietal response and estimates of heritability of resistance to Meloidogyne javanica in carrots. J Nematol. 1986;18:496–501. [PMC free article] [PubMed] [Google Scholar]
- 194.Boiteux L, Belter J, Roberts P et al. RAPD linkage map of the genomic region encompassing the root-knot nematode (Meloidogyne javanica) resistance locus in carrot. Theor Appl Genet. 2000;100:439–46. [Google Scholar]
- 195.Simon P, Matthews W, Roberts P. Evidence for simply inherited dominant resistance to Meloidogyne javanica in carrot. Theor Appl Genet. 2000;100:735–42. [Google Scholar]
- 196.Ali A, Matthews WC, Cavagnaro PF et al. Inheritance and mapping of Mj-2, a new source of root-knot nematode (Meloidogyne javanica) resistance in carrot. J Hered. 2013;105:288–91. [DOI] [PubMed] [Google Scholar]
- 197.Lazarova S, Coyne D, Rodríguez GM et al. Functional diversity of soil nematodes in relation to the impact of agriculture – a review. Diversity. 2021;13:e64. [Google Scholar]
- 198.Candido V, Miccolis V, Basile M et al. Soil solarization for the control of Meloidogyne javanica on eggplant in southern Italy. Acta Hortic. 2005;698:195–200. [Google Scholar]
- 199.Selby MJ. Chemical ecology of the carrot fly, Psila rosae (F.): laboratory and field studies. Ph.D. Thesis, University of Nottingham, 2004.
- 200.Hardman JA, Ellis PR, Saw PL. Further investigations of the host range of the carrot fly, Psila rosae (F.). Ann Appl Biol. 1992;117:495–506. [Google Scholar]
- 201.Sunley R. EPPO workshop on carrot fly (Psila rosae): integrated approaches for pest control. EPPO Bull. 2009;39:113–5. [Google Scholar]
- 202.Guerin PM, Ryan MF. Relationship between root volatiles of some carrot cultivars and their resistance to the carrot fly, Psila rosae. Entomol Exp Appl. 1984;36:217–24. [Google Scholar]
- 203.Ellis P, Hardman J. The consistency of the resistance of eight carrot cultivars to carrot fly attack at several centres in Europe. Ann Appl Biol. 1981;98:491–7. [Google Scholar]
- 204.Boivin G. Integrated management for carrot weevil. Integr Pest Manag Rev. 1999;4:21–37. [Google Scholar]
- 205.Gratwick M. Willow-carrot aphid. In: Gratwick M (ed.), Crop Pests in the UK. Dordrecht: Springer, 1992, 73–7. [Google Scholar]
- 206.Hoffmann H, Botha J. Aphids, mealybugs and scales; common sapsuckers in the home garden. Department of Agriculture and Food, Government of Western Australia. 2011;Note no. 499. [Google Scholar]
- 207.Gratwick M. In: Gratwick M, ed. Celery fly, Crop Pests in the UK. Springer: Dordrecht, 1992,252–4. [Google Scholar]
- 208.Krivosheina MG, Ozerova NA. To the biology of celery fly Euleia heraclei (Linnaeus, 1758) (Diptera: Tephritidae) – pest of alien Apiaceae species in Moscow region. Russian Entomol J. 2016;25:209–13. [Google Scholar]
- 209.Vanlerberghe-Masutti F, Guillemaud T. Aphid resistance to insecticides. Biofutur. 2007;279:27–30. [Google Scholar]
- 210.Sanchis V, Bourguet D. Bacillus thuringiensis: applications in agriculture and insect resistance management. A review. Agron Sustain Dev. 2008;28:11–20. [Google Scholar]
- 211.Webb SE. Insect management for celery and parsley. ENY 463, Entomology and Nematology Department, Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida, 2017. [Google Scholar]
- 212.Griswold MJ, Trumble JT. Consumption and utilization of celery, Apium graveolens, by the beet armyworm Spodoptera exigua. Entomol Exp Appl. 2011;38:73–9. [Google Scholar]
- 213.Diawara M, Trumble J, Lacy M et al. Potential of somaclonal celeries for use in integrated pest management. J Econ Entomol. 1996;89:218–23. [Google Scholar]
- 214.Eigenbrode S, Trumble JT, Jones RA. Resistance to beet armyworm, hemipterans, and Liriomyza spp. in Lycopersicon accessions. J Amer Soc Hortic Sci. 1993;118:525–30. [Google Scholar]
- 215.Meade T, Hare JD. Differential performance of beet armyworm and cabbage looper (Lepidoptera: Noctuidae) larvae on selected Apium graveolens cultivars. Environ Entomol. 1991;6:1636–44. [Google Scholar]
- 216.Yang X, Yu YJ, Zhang FL et al. Linkage map construction and quantitative trait loci analysis for bolting based on a double haploid population of Brassica rapa. J Integr Plant Biol. 2007;49:664–71. [Google Scholar]
- 217.Ramin AA, Atherton JG. Manipulation of bolting and flowering in celery (Apium graveolens L. var. dulce). J Hortic Sci. 1994;69:861–8. [Google Scholar]
- 218.Wohlfeiler J, Alessandro MS, Cavagnaro PF et al. Multiallelic digenic control of vernalization requirement in carrot (Daucus carota L.). Euphytica. 2019;215:e37. [Google Scholar]
- 219.Jenni S, Gamache I, Côté JC et al. Early field detection of bolting in celery. Hortic Technol. 2005;15:843–5. [Google Scholar]
- 220.Quiros CF, Douches D, D'Antonio V. Inheritance of annual habit in celery: cosegregation with isozyme and anthocyanin markers. Theor Appl Genet. 1987;74:203–8. [DOI] [PubMed] [Google Scholar]
- 221.Wolf EA, White JM, Stubblefield RS et al. ‘Florida Slobolt M68’: a spring celery cultivar for Florida. HortScience. 1993;28:754–5. [Google Scholar]
- 222.Wang W, Wu F, Gao GX et al. A new slow-bolting celery cultivar 'Juventus'. Acta Hortic Sin. 2012;39:1007–8. [Google Scholar]
- 223.Landjeva S, Korzun V, Börner A. Molecular markers: actual and potential contributions to wheat genome characterization and breeding. Euphytica. 2007;156:271–96. [Google Scholar]
- 224.Liu PY, Zhang BH, Zhao H et al. The study of based on RAPD molecular markers in celery cultivars relationship. Sci Discov. 2017;5:312–6. [Google Scholar]
- 225.Yang X, Quiros C. Identification and classification of celery cultivars with RAPD markers. Theor Appl Genet. 1993;86:205–12. [DOI] [PubMed] [Google Scholar]
- 226.Huestis GM, McGrath JM, Quiros CF. Development of genetic markers in celery based on restriction fragment length polymorphisms. Theor Appl Genet. 1993;85:889–96. [DOI] [PubMed] [Google Scholar]
- 227.Yang X, Quiros CF. Construction of a genetic linkage map in celery using DNA-based markers. Genome. 1995;38:36–44. [DOI] [PubMed] [Google Scholar]
- 228.Fu N, Wang PY, Liu XD et al. Use of EST-SSR markers for evaluating genetic diversity and fingerprinting celery (Apium graveolens L.) cultivars. Molecules. 2014;19:1939–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Li MY, Wang F, Xu ZS et al. High throughput sequencing of two celery varieties small RNAs identifies microRNAs involved in temperature stress response. BMC Genomics. 2014;15:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Li MY, Wang F, Jiang Q et al. Identification of SSRs and differentially expressed genes in two cultivars of celery (Apium graveolens L.) by deep transcriptome sequencing. Hortic Res. 2014;1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Choudhary S, Sharma R, Jethra G et al. Molecular diversity in coriander (Coriandrum sativum) using RAPD and ISSR markers. Indian J Agric Sci. 2019;89:193–8. [Google Scholar]
- 232.Alp FM, Gebologlu MD. Two different molecular markers (SSR & IPBS) assessment on Coriandrum sativum L. with capillary electrophoresis. Fresenius Environ Bull. 2017;26:4568–73. [Google Scholar]
- 233.Singh RK, Verma SS, Meena RS et al. Characterization of coriander (Coriandrum sativum L.) varieties using SDS-PAGE and RAPD markers. Afr J Biotechnol. 2013;12:1189–95. [Google Scholar]
- 234.Tulsani NJ, Hamid R, Jacob F et al. Transcriptome landscaping for gene mining and SSR marker development in coriander (Coriandrum sativum L.). Genomics. 2019;112:1545–53. [DOI] [PubMed] [Google Scholar]
- 235.Gaudeul M, Till-Bottraud I, Barjon F et al. Genetic diversity and differentiation in Eryngium alpinum L. (Apiaceae): comparison of AFLP and microsatellite markers. Heredity. 2004;92:508–18. [DOI] [PubMed] [Google Scholar]
- 236.Gaudeul M, Taberlet P, Till-Bottraud I. Genetic diversity in an endangered alpine plant, Eryngium alpinum L. (Apiaceae), inferred from amplified fragment length polymorphism markers. Mol Ecol. 2000;9:1625–37. [DOI] [PubMed] [Google Scholar]
- 237.Stelmach K, Macko-Podgórni A, Allender C et al. Genetic diversity structure of western-type carrots. BMC Plant Biol. 2021;21:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Rong J, Lammers Y, Strasburg JL et al. New insights into domestication of carrot from root transcriptome analyses. BMC Genomics. 2014;15:895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Bekir D, Ahmet D, Yavuz B et al. Phylogenetic relationships among the taxa of the genus Johrenia DC. (Apiaceae) from Turkey based on molecular method. Bangladesh J Plant Taxon. 2010;17:113–20. [Google Scholar]
- 240.Duran A, Behçet L, Öztürk M. Diplotaenia bingolensis (Apiaceae), new species from East Anatolia, Turkey. Plant Syst Evol. 2015;301:467–78. [Google Scholar]
- 241.Rowland LJ, Alkharouf N, Darwish O et al. Generation and analysis of blueberry transcriptome sequences from leaves, developing fruit, and flower buds from cold acclimation through deacclimation. BMC Plant Biol. 2012;12:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Persidis A. Agricultural biotechnology. Nat Biotechnol. 1996;2:91–3. [DOI] [PubMed] [Google Scholar]
- 243.Newell CA. Plant transformation technology. Developments and applications. Mol Biotechnol. 2000;16:53–65. [DOI] [PubMed] [Google Scholar]
- 244.Permyakova NV, Zagorskaya AA, Belavin PA et al. Transgenic carrot expressing fusion protein comprising M. tuberculosis antigens induces immune response in mice. Biomed Res Int. 2015;2015:e417565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Rosales-Mendoza S, Tello-Olea MA. Carrot cells: a pioneering platform for biopharmaceuticals production. Mol Biotechnol. 2015;57:219–32. [DOI] [PubMed] [Google Scholar]
- 246.Luchakivskaya Y, Kishchenko O, Gerasymenko I et al. High-level expression of human interferon alpha-2b in transgenic carrot (Daucus carota L.) plants. Plant Cell Rep. 2011;30:407–15. [DOI] [PubMed] [Google Scholar]
- 247.Jayaraj J, Punja ZK. Combined expression of chitinase and lipid transfer protein genes in transgenic carrot plants enhances resistance to foliar fungal pathogens. Plant Cell Rep. 2007;26:1539–46. [DOI] [PubMed] [Google Scholar]
- 248.Huang Y, Liu H, Jia Z et al. Combined expression of antimicrobial genes (Bbchit1 and LJAMP2) in transgenic poplar enhances resistance to fungal pathogens. Tree Physiol. 2012;32:1313–20. [DOI] [PubMed] [Google Scholar]
- 249.Wally O, Jayaraj J, Punja ZK. Broad-spectrum disease resistance to necrotrophic and biotrophic pathogens in transgenic carrots (Daucus carota L.) expressing an Arabidopsis NPR1 gene. Planta. 2009;231:131–41. [DOI] [PubMed] [Google Scholar]
- 250.Tan GF. Study on the mechanism of anthocyanin and apigenin metabolization in celery, Doctoral dissertation. Nanjing Agricultural University, Nanjing, 2017. [Google Scholar]
- 251.Ding X, Liu JX, Li T et al. AgZDS, a gene encoding ζ-carotene desaturase, increases lutein and β-carotene contents in transgenic Arabidopsis and celery. Plant Sci. 2021;312:e111043. [DOI] [PubMed] [Google Scholar]
- 252.Wang H, Liu JX, Feng K et al. AgMYB12, a novel R2R3-MYB transcription factor, regulates apigenin biosynthesis by interacting with the AgFNS gene in celery. Plant Cell Rep. 2021;3:1–13. [DOI] [PubMed] [Google Scholar]
- 253.Chialva C, Blein T, Crespi M et al. Identification and functional prediction of anthocyanin biosynthesis regulatory long non-coding RNAs (lncRNAs) in carrot. BioRxiv. 2020;10:e356964. [Google Scholar]
- 254.Razzaq A, Saleem F, Kanwal M et al. Modern trends in plant genome editing: an inclusive review of the CRISPR/Cas9 toolbox. Int J Mol Sci. 2019;20:e4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Leena A, Alka N. Gene editing and crop improvement using CRISPR-Cas9 system. Front Plant Sci. 2017;8:e1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Klimek-Chodack M, Oleszkiewicz T, Qi Y et al. Carrot genome editing using CRISPR-based systems. Acta Hortic. 2019;1264:53–65. [Google Scholar]
- 257.Klimek-Chodacka M, Oleszkiewicz T, Lowder LG et al. Efficient CRISPR/Cas9-based genome editing in carrot cells. Plant Cell Rep. 2018;37:575–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Li T, Deng YJ, Liu JX et al. DcCCD4 catalyzes the degradation of α-carotene and β-carotene to affect carotenoid accumulation and taproot color in carrot. Plant J. 2021;108:1116–30. [DOI] [PubMed] [Google Scholar]
- 259.Dunemann F, Unkel K, Sprink T. Using CRISPR/Cas9 to produce haploid inducers of carrot through targeted mutations of centromeric histone H3 (CENH3). Acta Hortic. 2019;1264:211–20. [Google Scholar]
- 260.Xu ZS, Feng K, Xiong AS. CRISPR/Cas9-mediated multiply targeted mutagenesis in orange and purple carrot plants. Mol Biotechnol. 2019;61:191–9. [DOI] [PubMed] [Google Scholar]
- 261.Athanassiou CG, Kavallieratos NG, Benelli G et al. Nanoparticles for pest control: current status and future perspectives. J Pest Sci. 2018;91:1–15. [Google Scholar]
- 262.Wang P, Lombi E, Zhao FJ et al. Nanotechnology: a new opportunity in plant sciences. Trends Plant Sci. 2016;21:699–71. [DOI] [PubMed] [Google Scholar]
- 263.Servin A, Elmer W, Mukherjee A et al. A review of the use of engineered nanomaterials to suppress plant disease and enhance crop yield. J Nanopart Res. 2015;17:e92. [Google Scholar]
- 264.Mani P, Mondal S. Agri-nanotechniques for plant availability of nutrients. In: Kole C, Kumar D, Khodakovskaya M, eds. Plant Nanotechnology. Springer: Cham, 2016,263–303. [Google Scholar]
- 265.Kottegoda N, Munaweera I, Adassooriya N et al. A green slow-release fertilizer composition based on urea-modified hydroxyapatite nanoparticles encapsulated wood. Curr Sci. 2011;101:73–8. [Google Scholar]
- 266.Huang S, Wang L, Liu L et al. Nanotechnology in agriculture, livestock, and aquaculture in China. A review. Agron Sustain Dev. 2014;35:369–400. [Google Scholar]
- 267.Torney FO, Trewyn BG, Lin SY et al. Mesoporous silica nanoparticles deliver DNA and chemicals into plants. Nat Nanotechnol. 2007;2:295–300. [DOI] [PubMed] [Google Scholar]
- 268.Siddiqui DMH, Al-Whaibi M, Mohammad F. Nanotechnology and Plant Sciences. Cham: Springer; 2015. [Google Scholar]
- 269.Pramanik A, Datta AK, Das D et al. Assessment of nanotoxicity (cadmium sulphide and copper oxide) using cytogenetical parameters in Coriandrum sativum L. (Apiaceae). Cytol Genet. 2018;52:299–308. [Google Scholar]
- 270.Sekhon BS. Nanotechnology in agri-food production: an overview. Nanotechnol Sci Appl. 2014;7:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Veenstra TD. Omics in systems biology: current progress and future outlook. Proteomics. 2021;21:e2000235. [DOI] [PubMed] [Google Scholar]
- 272.Liu J, Shi L, Han J et al. Identification of species in the angiosperm family Apiaceae using DNA barcodes. Mol Ecol Resour. 2015;14:1231–8. [DOI] [PubMed] [Google Scholar]
- 273.Zhuang J, Zhang J, Hou XL et al. Transcriptomic, proteomic, metabolomic and functional genomic approaches for the study of abiotic stress in vegetable crops. Crit Rev Plant Sci. 2014;33:225–37. [Google Scholar]
- 274.Xu ZS, Yang QQ, Feng K et al. Changing carrot color: insertions in DcMYB7 alter the regulation of anthocyanin biosynthesis and modification. Plant Physiol. 2019;181:195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.McKeon TA, Douglas G, Hayes DF, Hildebrand RJW. Chapter 1-introduction to industrial oil crops. In: McKeon TA, Hayes DG, Hildebrand DF et al., eds. Industrial Oil Crops. Amsterdam, Holland: AOCS press, 2016, 359–78. [Google Scholar]
- 276.Freeman R. Hybrid carrot variety Nun 85190. USA Patent, 2014;14/186028.
- 277.Xu ZS, Tan HW, Wang F et al. CarrotDB: a genomic and transcriptomic database for carrot. Database (Oxford). 2014;2014:bau096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Iorizzo M, Ellison S, Senalik D et al. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat Genet. 2016;48:657–66. [DOI] [PubMed] [Google Scholar]
- 279.Feng K, Hou XL, Li MY et al. CeleryDB: a genomic database for celery. Database (Oxford). 2018;2018:e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Li MY, Feng K, Hou XL et al. The genome sequence of celery (Apium graveolens L.), an important leaf vegetable crop rich in apigenin in the Apiaceae family. Hortic Res. 2020;7:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Song X, Sun P, Yuan J et al. The celery genome sequence reveals sequential paleo-polyploidizations, karyotype evolution, and resistance gene reduction in Apiales. Plant Biotechnol J. 2020;19:731–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Song X, Wang J, Li N et al. Deciphering the high-quality genome sequence of coriander that causes controversial feelings. Plant Biotechnol J. 2020;18:1444–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Lu CL, Li XF. A review of Oenanthe javanica (Blume) DC. as traditional medicinal plant and its therapeutic potential. Evid Based Complement Alternat Med. 2019;2019:e6495819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Liu JX, Jiang Q, Tao JP et al. Integrative genome, transcriptome, microRNA, and degradome analysis of water dropwort (Oenanthe javanica) in response to water stress. Hortic Res. 2021;8:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Rather MA, Dar BA, Sofi SN et al. Foeniculum vulgare: a comprehensive review of its traditional use, phytochemistry, pharmacology, and safety. Arab J Chem. 2012;9:1574–83. [Google Scholar]
- 286.Sonah H, Deshmukh RK, Singh VP et al. Genomic resources in horticultural crops: status, utility and challenges. Biotechnol Adv. 2011;29:199–209. [DOI] [PubMed] [Google Scholar]
- 287.Xie C, Li B, Xu Y et al. Characterization of the global transcriptome for Pyropia haitanensis (Bangiales, Rhodophyta) and development of cSSR markers. BMC Genomics. 2013;14:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Verma M, Kumar V, Patel RK et al. CTDB: an integrated chickpea transcriptome database for functional and applied genomics. PLoS One. 2015;10:e0136880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Iorizzo M, Senalik DA, Grzebelus D et al. De novo assembly and characterization of the carrot transcriptome reveals novel genes, new markers, and genetic diversity. BMC Genomics. 2011;12:e389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Ma J, Li J, Xu Z et al. Transcriptome profiling of genes involving in carotenoid biosynthesis and accumulation between leaf and root of carrot (Daucus carota L.). Acta Biochim Biophys Sin. 2018;50:481–90. [DOI] [PubMed] [Google Scholar]
- 291.Hao JN, Wang YH, Duan AQ et al. NAC family transcription factors in carrot: genomic and transcriptomic analysis and responses to abiotic stresses. DNA Cell Biol. 2020;39:816–27. [DOI] [PubMed] [Google Scholar]
- 292.Wang GL, Jia XL, Xu ZS et al. Sequencing, assembly, annotation, and gene expression: novel insights into the hormonal control of carrot root development revealed by a high-throughput transcriptome. Mol Gen Genomics. 2015;290:1379–91. [DOI] [PubMed] [Google Scholar]
- 293.Wang GL, Huang Y, Zhang XY et al. Transcriptome-based identification of genes revealed differential expression profiles and lignin accumulation during root development in cultivated and wild carrots. Plant Cell Rep. 2016;35:1743–55. [DOI] [PubMed] [Google Scholar]
- 294.Jia XL, Wang GL, Xiong F et al. De novo assembly, transcriptome characterization, lignin accumulation, and anatomic characteristics: novel insights into lignin biosynthesis during celery leaf development. Sci Rep. 2015;5:e8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Liu J, Feng K, Hou X et al. Transcriptome profiling reveals the association of multiple genes and pathways contributing to hormonal control in celery leaves. Acta Biochim Biophys Sin. 2019;51:524–34. [DOI] [PubMed] [Google Scholar]
- 296.Liu J, Ma J, Feng K et al. Transcriptome profiling of β-carotene biosynthesis genes and β-carotene accumulation in leaf blades and petioles of celery cv. Jinnanshiqin. Acta Biochim Biophys Sin. 2019;51:116–9. [DOI] [PubMed] [Google Scholar]
- 297.Jiang Q, Wang F, Tan HW et al. De novo transcriptome assembly, gene annotation, marker development, and miRNA potential target genes validation under abiotic stresses in Oenanthe javanica. Mol Gen Genomics. 2015;290:671–83. [DOI] [PubMed] [Google Scholar]
- 298.Tan GF, Wang F, Li MY et al. De novo assembly and transcriptome characterization: novel insights into the temperature stress in Cryptotaenia japonica Hassk. Acta Physiol Plant. 2015;37:1–12. [Google Scholar]
- 299.Li MY, Tan HW, Wang F et al. De novo transcriptome sequence assembly and identification of AP2/ERF transcription factor related to abiotic stress in parsley (Petroselinum crispum). PLoS One. 2014;9:e108977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Amini H, Naghavi MR, Shen T et al. Tissue-specific transcriptome analysis reveals candidate genes for terpenoid and phenylpropanoid metabolism in the medicinal plant Ferula assafoetida. G3 (Bethesda). 2019;9:807–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Sunkar R, Li YF, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17:196–203. [DOI] [PubMed] [Google Scholar]
- 302.Megha B, Basu U, Kav N. Regulation of low temperature stress in plants by microRNAs. Plant Cell Environ. 2018;41:1–15. [DOI] [PubMed] [Google Scholar]
- 303.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. [DOI] [PubMed] [Google Scholar]
- 304.Zhang B-H, Pan XP, Cobb GP et al. Plant microRNA: a small regulatory molecule with big impact. Dev Biol. 2006;289:3–16. [DOI] [PubMed] [Google Scholar]
- 305.Drikvand RM, Sohrabi SS, Sohrabi SM et al. Identification and characterization of conserved miRNAs of Coriandrum sativum L. using next-generation sequencing data. J Crop Biotechnol. 2019;8:59–74. [Google Scholar]
- 306.Najafabadi AS, Naghavi MR. Mining Ferula gummosa transcriptome to identify miRNAs involved in the regulation and biosynthesis of terpenes. Gene. 2018;645:41–7. [DOI] [PubMed] [Google Scholar]
- 307.Jia XL, Li MY, Jiang Q et al. High-throughput sequencing of small RNAs and anatomical characteristics associated with leaf development in celery. Sci Rep. 2015;5:e11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Jiang Q, Wang F, Li MY et al. High-throughput analysis of small RNAs and characterization of novel microRNAs affected by abiotic stress in a local celery cultivar. Sci Hortic. 2014;169:36–43. [Google Scholar]
- 309.Bhan B, Koul A, Sharma D et al. Identification and expression profiling of miRNAs in two color variants of carrot (Daucus carota L.) using deep sequencing. PLoS One. 2019;14:e0212746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Huber LA. Is proteomics heading in the wrong direction? Nat Rev Mol Cell Biol. 2003;4:74–80. [DOI] [PubMed] [Google Scholar]
- 311.Huang W, Ma HY, Huang Y et al. Comparative proteomic analysis provides novel insights into chlorophyll biosynthesis in celery under temperature stress. Physiol Plant. 2017;161:468–85. [DOI] [PubMed] [Google Scholar]
- 312.Khodadadi E, Fakheri BA, Aharizad S et al. Leaf proteomics of drought-sensitive and-tolerant genotypes of fennel. Biochim Biophys Acta, Proteins Proteom. 2017;1865:1433–44. [DOI] [PubMed] [Google Scholar]
- 313.Bai Y, Huang W, Tao Y et al. Differential protein expression profiling in Pleurotus ferulae mycelium caused by asafoetida extracts using a proteomics approach. J Korean Soc Appl Biol Chem. 2014;57:97–103. [Google Scholar]
- 314.Louarn S, Nawrocki A, Thorup-Kristensen K et al. Proteomic changes and endophytic micromycota during storage of organically and conventionally grown carrots. Postharvest Biol Technol. 2013;76:26–33. [Google Scholar]
- 315.Wang YH, Wu XJ, Sun S et al. DcC4H and DcPER are important in dynamic changes of lignin content in carrot roots under elevated carbon dioxide stress. J Agric Food Chem. 2018;66:8209–20. [DOI] [PubMed] [Google Scholar]
- 316.Nassar AF, Wu T, Nassar SF et al. UPLC–MS for metabolomics: a giant step forward in support of pharmaceutical research. Drug Discov Today. 2017;22:463–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Hammami S, Snene A, Sirignano Cet al. Conference: 7th edition of international conference and exhibition on separation techniques, theme: launching the innovative ideas and technologies of separation techniques. Berlin, Germany, 2014, 5–7. [Google Scholar]
- 318.Grebenstein C, Choi YH, Rong J et al. Metabolic fingerprinting reveals differences between shoots of wild and cultivated carrot (Daucus carota L.) and suggests maternal inheritance or wild trait dominance in hybrids. Phytochemistry. 2011;72:1341–7. [DOI] [PubMed] [Google Scholar]
- 319.Yahyaa M, Ibdah M, Marzouk S et al. Profiling of terpene metabolome in carrot fruits of wild (Daucus carota L. ssp. carota) accessions and characterization of a geraniol synthase. J Agric Food Chem. 2016;66:2378–86. [DOI] [PubMed] [Google Scholar]
- 320.Lau H, Laserna AKC, Li SFY. 1H NMR-based metabolomics for the discrimination of celery (Apium graveolens L. var. dulce) from different geographical origins. Food Chem. 2020;332:e127424. [DOI] [PubMed] [Google Scholar]
- 321.Farhadi F, Asili J, Iranshahy M et al. NMR-based metabolomic study of asafoetida. Fitoterapia. 2019;139:104361. [DOI] [PubMed] [Google Scholar]
- 322.Zhang K, Yan M, Han S et al. Identification of chemical markers for the discrimination of radix Angelica sinensis grown in geoherb and non-geoherb regions using UHPLC-QTOF-MS/MS based metabolomics. Molecules. 2019;24:e3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Bai YJ, Xu JD, Kong M et al. Discovery of characteristic chemical markers for inspecting sulfur-fumigated radix Angelicae sinensis by ultra-high performance liquid chromatography–quadrupole/time-of-flight mass spectrometry based metabolomics and chemical profiling approach. Food Res Int. 2015;76:387–94. [DOI] [PubMed] [Google Scholar]
- 324.Xing J, Sun HM, Jia JP et al. Integrative hepatoprotective efficacy comparison of raw and vinegar-baked radix Bupleuri using nuclear magnetic resonance-based metabolomics. J Pharm Biomed. 2017;138:215–22. [DOI] [PubMed] [Google Scholar]
- 325.Jie X, Sun HM, Li ZY et al. Comparison of volatile components between raw and vinegar baked radix Bupleuri by GC-MS based metabolic fingerprinting approach. Evid Based Complement Alternat Med. 2015;2015:e653791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Chen Y, Wang J, Yuan L et al. Interaction of the main components from the traditional Chinese drug pair chaihu-shaoyao based on rat intestinal absorption. Molecules. 2011;16:9600–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Zhao R, Chen YJ, Cai JX. International Conference on Bioinformatics and Biomedicine Workshops. Atlanta, GA, USA; 2011:740–5. [Google Scholar]
- 328.Zhao R, Liu S, Mao S et al. Study on liver targeting effect of vinegar-baked Radix Bupleuri on resveratrol in mice. J Ethnopharmacol. 2009;126:415–20. [DOI] [PubMed] [Google Scholar]
- 329.Zhao R, Lijuan L, Yinjie W et al. Vinegar-baked Radix Bupleuri modulates the cell membrane constituents and inhibits the P-gp activity in rat hepatocytes. BMC Complement Altern Med. 2014;14:e357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Yuan Z, Zhong L, Hua Y et al. Metabolomics study on promoting blood circulation and ameliorating blood stasis: investigating the mechanism of Angelica sinensis and its processed products. Biomed Chromatogr. 2019;33:e4457. [DOI] [PubMed] [Google Scholar]
- 331.Hounsome N, Hounsome B, Tomos D et al. Plant metabolites and nutritional quality of vegetables. J Food Sci. 2008;73:48–65. [DOI] [PubMed] [Google Scholar]
- 332.Zhang C, Xu B, Zhao CR et al. Comparative de novo transcriptomics and untargeted metabolomic analyses elucidate complicated mechanisms regulating celery (Apium graveolens L.) responses to selenium stimuli. PLoS One. 2019;14:e0226752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Roman M, Baranski R, Baranska M. Nondestructive Raman analysis of polyacetylenes in Apiaceae vegetables. J Agric Food Chem. 2011;59:7647–53. [DOI] [PubMed] [Google Scholar]
- 334.Bagci E, Dogan G. Composition of the essential oils of two Umbelliferae herbs (Artedia squamata and Malabaila secacul) growing wild in Turkey. J Essent Oil Bear Plants. 2015;18:44–51. [Google Scholar]
- 335.Maoka T. Carotenoids as natural functional pigments. J Nat Med. 2020;74:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Rodriguez-Concepcion M, Avalos J, Bonet ML et al. A global perspective on carotenoids: metabolism, biotechnology, and benefits for nutrition and health. Prog Lipid Res. 2018;70:62–93. [DOI] [PubMed] [Google Scholar]
- 337.Fanciullino AL, Dhuique-Mayer C, Luro F et al. Carotenoid biosynthetic pathway in the Citrus genus: number of copies and phylogenetic diversity of seven genes. J Agric Food Chem. 2007;55:7405–17. [DOI] [PubMed] [Google Scholar]
- 338.Botella-Pavía P, Rodríguez-Concepción M. Carotenoid biotechnology in plants for nutritionally improved foods. Physiol Plant. 2006;126:369–81. [Google Scholar]
- 339.Cordero BF, Couso I, León R et al. Enhancement of carotenoids biosynthesis in Chlamydomonas reinhardtii by nuclear transformation using a phytoene synthase gene isolated from Chlorella zofingiensis. Appl Microbiol Biotechnol. 2011;91:341–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Shewmaker CK, Sheehy JA, Daley M et al. Seed-specific overexpression of phytoene synthase: increase in carotenoids and other metabolic effects. Plant J. 2010;20:401–12. [DOI] [PubMed] [Google Scholar]
- 341.Chen Y, Li F, Wurtzel ET. Isolation and characterization of the Z-ISO gene encoding a missing component of carotenoid biosynthesis in plants. Plant Physiol. 2010;153:66–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Takemura M, Maoka T, Misawa N. Carotenoid analysis of a liverwort Marchantia polymorpha and functional identification of its lycopene β-and ε-cyclase genes. Plant Cell Physiol. 2014;55:194–200. [DOI] [PubMed] [Google Scholar]
- 343.Kaur K. Kinetics of extraction of β-carotene from tray dried carrots by using supercritical fluid extraction technique. Food Nutr Sci. 2012;3:591–5. [Google Scholar]
- 344.Moreno JC, Cerda A, Simpson K et al. Increased Nicotiana tabacum fitness through positive regulation of carotenoid, gibberellin and chlorophyll pathways promoted by Daucus carota lycopene β-cyclase (Dclcyb1) expression. J Exp Bot. 2016;67:2325–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Arango J, Jourdan M, Geoffriau E et al. Carotene hydroxylase activity determines the levels of both α-carotene and total carotenoids in orange carrots. Plant Cell. 2014;26:2223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Ellison SL, Luby CH, Corak KE et al. Carotenoid presence is associated with the or gene in domesticated carrot. Genetics. 2018;210:1497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Coe KM, Ellison S, Senalik D et al. The influence of the or and carotene hydroxylase genes on carotenoid accumulation in orange carrots [Daucus carota (L.)]. Theor Appl Genet. 2021;134:3351–62. [DOI] [PubMed] [Google Scholar]
- 348.Wang YH, Li T, Zhang RR et al. Transcript profiling of genes involved in carotenoid biosynthesis among three carrot cultivars with various taproot colors. Protoplasma. 2020;257:949–63. [DOI] [PubMed] [Google Scholar]
- 349.Zhang RR, Wang YH, Li T et al. Effects of simulated drought stress on carotenoid contents and expression of related genes in carrot taproots. Protoplasma. 2021;258:379–90. [DOI] [PubMed] [Google Scholar]
- 350.Li T, Liu JX, Deng YJ et al. Overexpression of a carrot BCH gene, DcBCH1, improves tolerance to drought in Arabidopsis thaliana. BMC Plant Biol. 2021;21:475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Rodriguez-Concepcion M, Stange C. Biosynthesis of carotenoids in carrot: an underground story comes to light. Arch Biochem Biophys. 2013;539:110–6. [DOI] [PubMed] [Google Scholar]
- 352.Bowman MJ, Willis DK, Simon PW. Transcript abundance of phytoene synthase 1 and phytoene synthase 2 is associated with natural variation of storage root carotenoid pigmentation in carrot. J Am Soc Hortic Sci. 2014;139:63–8. [Google Scholar]
- 353.Clotault J, Peltier D, Berruyer R et al. Expression of carotenoid biosynthesis genes during carrot root development. J Exp Bot. 2008;59:3563–73. [DOI] [PubMed] [Google Scholar]
- 354.Perrin F, Brahem M, Dubois-Laurent C et al. Differential pigment accumulation in carrot leaves and roots during two growing periods. J Agric Food Chem. 2016;64:906–12. [DOI] [PubMed] [Google Scholar]
- 355.Perrin F, Geoffriau E, Peltier D et al. A focus on the regulation of carotenoid accumulation in carrot root. Acta Hortic. 2017;1153:101–8. [Google Scholar]
- 356.Wang H, Ou CG, Zhuang FY et al. The dual role of phytoene synthase genes in carotenogenesis in carrot roots and leaves. Mol Breed. 2014;34:2065–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Li JW, Ma J, Feng K et al. Carotenoid accumulation and distinct transcript profiling of structural genes involved in carotenoid biosynthesis in celery. Plant Mol Biol Rep. 2018;6:663–74. [Google Scholar]
- 358.Surles RL, Weng N, Simon PW et al. Carotenoid profiles and consumer sensory evaluation of specialty carrots (Daucus carota, L.) of various colors. J Agric Food Chem. 2004;52:3417–21. [DOI] [PubMed] [Google Scholar]
- 359.Wang R, Wang Y, Guo W et al. Stability and bioactivity of carotenoids from Synechococcus sp. PCC 7002 in zein/NaCas/gum arabic composite nanoparticles fabricated by pH adjustment and heat treatment antisolvent precipitation. Food Hydrocoll. 2021;117:e106663. [Google Scholar]
- 360.Ding X, Jia LL, Xing GM et al. The accumulation of lutein and β-carotene and transcript profiling of genes related to carotenoids biosynthesis in yellow celery. Mol Biotechnol. 2021;63:638–49. [DOI] [PubMed] [Google Scholar]
- 361.Yin L, Liu JX, Tao JP et al. The gene encoding lycopene epsilon cyclase of celery enhanced lutein and β-carotene contents and confers increased salt tolerance in Arabidopsis. Plant Physiol Biochem. 2020;157:339–47. [DOI] [PubMed] [Google Scholar]
- 362.Clifford MN. Anthocyanins – nature, occurrence and dietary burden. J Sci Food Agric. 2000;80:1118–25. [Google Scholar]
- 363.Landi T, Tattini M, Gould KS. Multiple functional roles of anthocyanins in plant-environment interactions. Environ Exp Bot. 2015;119:4–17. [Google Scholar]
- 364.Steyn WJ, Wand SJE, Holcroft DM et al. Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytol. 2002;155:349–61. [DOI] [PubMed] [Google Scholar]
- 365.Palungwachira P, Tancharoen S, Phruksaniyom C et al. Antioxidant and anti-inflammatory properties of anthocyanins extracted from Oryza sativa L. in primary dermal fibroblasts. Oxid Med Cell Longev. 2019;2019:2089817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Zhan L, Xu B, Wu T et al. Transcriptomic profiling of two Pak Choi varieties with contrasting anthocyanin contents provides an insight into structural and regulatory genes in anthocyanin biosynthetic pathway. BMC Genomics. 2017;18:e288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Yildiz M, Willis DK, Cavagnaro PF et al. Expression and mapping of anthocyanin biosynthesis genes in carrot. Theor Appl Genet. 2013;126:1689–702. [DOI] [PubMed] [Google Scholar]
- 368.Xu ZS, Feng K, Que F et al. A MYB transcription factor, DcMYB6, is involved in regulating anthocyanin biosynthesis in purple carrot taproots. Sci Rep. 2017;7:e45324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Xu ZS, Yang QQ, Feng K et al. DcMYB113, a root-specific R2R3-MYB, conditions anthocyanin biosynthesis and modification in carrot. Plant Biotechnol J. 2020;18:1585–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Chen YY, Xu ZS, Xiong AS. Identification and characterization of DcUSAGT1, a UDP-glucose:sinapic acid glucosyltransferase from purple carrot taproots. PLoS One. 2016;11:e0154938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Xu ZS, Ma J, Wang F et al. Identification and characterization of DcUCGalT1, a galactosyltransferase responsible for anthocyanin galactosylation in purple carrot (Daucus carota L.) taproots. Sci Rep. 2016;6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Meng G, Clausen SK, Rasmussen SK. Transcriptome analysis reveals candidate genes related to anthocyanin biosynthesis in different carrot genotypes and tissues. Plants (Basel). 2020;9:e344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Feng K, Liu JX, Duan AQ et al. AgMYB2 transcription factor is involved in the regulation of anthocyanin biosynthesis in purple celery (Apium graveolens L.). Planta. 2018;248:1249–61. [DOI] [PubMed] [Google Scholar]
- 374.Feng K, Xu ZS, Que F et al. An R2R3-MYB transcription factor, OjMYB1, functions in anthocyanin biosynthesis in Oenanthe javanica. Planta. 2018;247:301–15. [DOI] [PubMed] [Google Scholar]
- 375.Feng K, Xing GM, Liu JX et al. AgMYB1, an R2R3-MYB factor, plays a role in anthocyanin production and enhancement of antioxidant capacity in celery. Veg Res. 2021;1:e2. [Google Scholar]
- 376.Liu B, Ning Z, Gao J et al. Preparing apigenin from leaves of Adinandra nitida. Food Technol Biotechnol. 2008;46:111–5. [Google Scholar]
- 377.Tan GF, Ma J, Zhang XY et al. AgFNS overexpression increase apigenin and decrease anthocyanins in petioles of transgenic celery. Plant Sci. 2017;263:31–8. [DOI] [PubMed] [Google Scholar]
- 378.Feng K, Xu ZS, Liu JX et al. Isolation, purification, and characterization of AgUCGalT1, a galactosyltransferase involved in anthocyanin galactosylation in purple celery (Apium graveolens L.). Planta. 2018;247:1363–75. [DOI] [PubMed] [Google Scholar]
- 379.Mele M, Kang HM, Lee YT et al. Grape terpenoids: flavor importance, genetic regulation, and future potential. Crit Rev Food Sci Nutr. 2020;61:1429–47. [DOI] [PubMed] [Google Scholar]
- 380.Cheng AX, Lou YG, Mao YB et al. Plant terpenoids: biosynthesis and ecological functions. J Integr Plant Biol. 2010;49:179–86. [Google Scholar]
- 381.Keszei A, Brubaker CL, Foley WJ. A molecular perspective on terpene variation in Australian Myrtaceae. Aust J Bot. 2008;56:197–213. [Google Scholar]
- 382.Bohlmann J, Meyer-Gauen G, Croteau R. Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA. 1998;95:4126–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Nieuwenhuizen NJ, Green SA, Chen X et al. Functional genomics reveals that a compact terpene synthase gene family can account for terpene volatile production in apple. Plant Physiol. 2013;161:787–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Aubourg S, Lecharny A, Bohlmann J. Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Gen Genomics. 2002;267:730–45. [DOI] [PubMed] [Google Scholar]
- 385.Külheim C, Padovan A, Hefer CA et al. The eucalyptus terpene synthase gene family. BMC Genomics. 2015;16:e450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Keilwagen J, Lehnert H, Berner T et al. The terpene synthase gene family of carrot (Daucus carota L.): identification of QTLs and candidate genes associated with terpenoid volatile compounds. Front Plant Sci. 2017;8:e1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Muchlinski A, Ibdah M, Ellison S et al. Diversity and function of terpene synthases in the production of carrot aroma and flavor compounds. Sci Rep. 2020;10:e9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Reichardt S, Budahn H, Lamprecht D et al. The carrot monoterpene synthase gene cluster on chromosome 4 harbours genes encoding flavour-associated sabinene synthases. Hortic Res. 2020;7:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Ibdah M, Muchlinski A, Yahyaa M, et al. Carrot volatile terpene metabolism: terpene diversity and biosynthetic genes. In: Simon P, Iorizzo M, Grzebelus D et al. (eds), The Carrot Genome. Cham: Springer, 279–93. [Google Scholar]
- 390.Yahyaa M, Tholl D, Cormier G et al. Identification and characterization of terpene synthases potentially involved in the formation of volatile terpenes in carrot (Daucus carota L.) roots. J Agric Food Chem. 2015;63:4870–8. [DOI] [PubMed] [Google Scholar]
- 391.Yahyaa M, Matsuba Y, Brandt W et al. Identification, functional characterization, and evolution of terpene synthases from a basal dicot. Plant Physiol. 2015;169:1683–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Galata M, Sarker LS, Mahmoud SS. Transcriptome profiling, and cloning and characterization of the main monoterpene synthases of Coriandrum sativum L. Phytochemistry. 2014;102:64–73. [DOI] [PubMed] [Google Scholar]
- 393.Ortiz J, Romero N, Robert P et al. Dietary fiber, amino acid, fatty acid and tocopherol contents of the edible seaweeds Ulva lactuca and Durvillaea antarctica. Food Chem. 2006;99:98–104. [Google Scholar]
- 394.Soliman GA. Dietary fiber, atherosclerosis, and cardiovascular disease. Nutrients. 2019;11:e1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Anderson JW, Baird P, Davis RH Jr et al. Health benefits of dietary fiber. Nutr Rev. 2010;67:188–205. [DOI] [PubMed] [Google Scholar]
- 396.Selvendran RR. The plant cell wall as a source of dietary fiber: chemistry and structure. Am J Clin Nutr. 1984;39:320–37. [DOI] [PubMed] [Google Scholar]
- 397.Ding X, Liu JX, Xing GM et al. The accumulation of ascorbic acid and lignin, and differential expression of ascorbic acid and lignin related-genes in yellow celery. J Hortic Sci Biotechnol. 2020;95:722–33. [Google Scholar]
- 398.Yin L, Xing GM, Sun S et al. Comparison of ascorbic acid and lignin accumulation in four white celery varieties and transcriptional profiling of genes related to the metabolic pathways. Biotechnol Biotechnol Equip. 2020;34:532–41. [Google Scholar]
- 399.Duan AQ, Feng K, Wang GL et al. Elevated gibberellin enhances lignin accumulation in celery (Apium graveolens L.) leaves. Protoplasma. 2019;256:777–88. [DOI] [PubMed] [Google Scholar]
- 400.Khadr A, Wang YH, Zhang RR et al. Cytokinin (6-benzylaminopurine) elevates lignification and the expression of genes involved in lignin biosynthesis of carrot. Protoplasma. 2020;257:1507–17. [DOI] [PubMed] [Google Scholar]
- 401.Khadr A, Wang Y, Que F et al. Exogenous abscisic acid suppresses the lignification and changes the growth, root anatomical structure and related gene profiles of carrot. Acta Biochim Biophys Sin. 2020;52:97–100. [DOI] [PubMed] [Google Scholar]
- 402.Khadr A, Wang GL, Wang YH et al. Effects of auxin (indole-3-butyric acid) on growth characteristics, lignification, and expression profiles of genes involved in lignin biosynthesis in carrot taproot. PeerJ. 2020;8:e10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Que F, Wang YH, Xu ZS et al. DcBAS1, a carrot brassinosteroid catabolism gene, modulates cellulose synthesis. J Agric Food Chem. 2019;67:13526–33. [DOI] [PubMed] [Google Scholar]
- 404.Duan AQ, Tao JP, Jia LL et al. AgNAC1, a celery transcription factor, related to regulation on lignin biosynthesis and salt tolerance. Genomics. 2020;112:5254–64. [DOI] [PubMed] [Google Scholar]
- 405.Li T, Huang Y, Khadr A et al. DcDREB1A, a DREB-binding transcription factor from Daucus carota, enhances drought tolerance in transgenic Arabidopsis thaliana and modulates lignin levels by regulating lignin-biosynthesis-related genes. Environ Exp Bot. 2020;169:e103896. [Google Scholar]
- 406.Liu JX, Feng K, Wang GL et al. Elevated CO2 induces alteration in lignin accumulation in celery (Apium graveolens L.). Plant Physiol Biochem. 2018;127:310–9. [DOI] [PubMed] [Google Scholar]
- 407.Que F, Wang GL, Feng K et al. Hypoxia enhances lignification and affects the anatomical structure in hydroponic cultivation of carrot taproot. Plant Cell Rep. 2018;37:1021–32. [DOI] [PubMed] [Google Scholar]


