Abstract
Chronic inflammation rather than invasion is characteristic of some forms of superficial candidiasis such as denture stomatitis. We hypothesized that Candida albicans may play a critical role in the pathogenesis of inflammatory lesions observed in chronic candidiasis by activating the proinflammatory cytokine interleukin-1β (IL-1β) from epithelial stores of the precursor. The aim of this study was therefore to demonstrate the proteolytic cleavage and activation of the inactive precursor of IL-1β (pro-IL-1β) by C. albicans. After incubation of either blastospores or hyphae with the inactive precursor, proteolytic cleavage was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis Western immunoblotting analysis, and the biological activity of the cleavage products was tested in a bioassay. We report here that late-stationary-growth-phase blastospores as well as hyphae of C. albicans, but not exponentially growing cells, can efficiently cleave pro-IL-1β to yield fragments of molecular masses compatible with mature biologically active IL-1β (17 to 19 kDa). Assays conducted in the presence of selected proteinase inhibitors suggest that the cleavage of pro-IL-1β involves the participation of one or more aspartyl proteinases. Cleavage products showed a dose-dependent IL-1β-like activity in a thymocyte proliferation bioassay, which was inhibited by anti-IL-1β neutralizing antibodies. The present data thus suggest a role for C. albicans proteinases in the activation and maintenance of the inflammatory response at epithelial surfaces.
Candida-associated denture stomatitis is the most common Candida infection and is estimated to affect about 50% of complete-denture wearers (4, 6). A characteristic feature of this form of candidiasis is a chronic inflammation of the palatal mucosa in contact with the prosthesis, and a diagnostic criterion is the topographical relationship between the inflamed mucosa and Candida albicans growth on the fitting surface of the denture (5). The carriage rate of Candida among denture wearers is very high, both in stomatitis patients (93%) and in denture wearers with clinically healthy mucosa (86%), compared to the general population (30 to 40%) (6). The mechanism by which C. albicans, a common oral commensal, may occasionally change to a pathogenic form is not clear. One hypothesis is that infective strains of serotype A are more virulent than others, although no mechanisms have been proposed (reviewed in references 4 and 34). Budtz-Jorgensen and Bertram have suggested that both an increase in yeast counts and the unique microbial ecology of denture plaque may contribute to the emergence of the pathogenic potential of C. albicans (reviewed in reference 5). We propose here that the proteolytic activity of C. albicans may participate in the pathogenesis of the inflammatory lesions observed in Candida-associated stomatitis via the enzymatic activation of the proinflammatory cytokine interleukin-1β (IL-1β) from epithelial stores of the precursor.
IL-1 is thought to play a central role in the induction, progression, and maintenance of an inflammatory response (reviewed in reference 17). While inflammation is obviously critical to the mediation of host responses to injury and infection, inappropriate or prolonged inflammatory responses can produce a variety of pathological conditions including inflammatory dermatoses (16, 30). Keratinocytes are the primary source of IL-1 in both normal and diseased epithelia (21, 22, 58). IL-1β is produced in large quantities in keratinocytes and stored in the superficial layers of the epithelium as an inactive, 31- to 33-kDa precursor (pro-IL-1β) (36). While monocytes produce a cysteine proteinase that cleaves pro-IL-1β to produce the 17- to 18-kDa active form (28), keratinocytes do not produce an IL-1β convertase enzyme (ICE) (36). Consequently, IL-1β activity is not observed with normal keratinocytes, despite high precursor content.
Although ICE is exquisitely specific in cleaving pro-IL-1β into a biologically active 17- to 18-kDa fragment, it is not the only proteinase that can activate pro-IL-1β. Serine proteinases such as neutrophil elastase and cathepsin G, mast cell chymase, and granzyme A can all generate active fragments from pro-IL-1β (2, 24, 37). It has been postulated that this property may contribute to the pathology of rheumatoid arthritis and other forms of acute or chronic inflammation (16). The possibility that microbial enzymes display such convertase-like activity was tested with Streptococcus pyogenes, and it was found that exotoxin B, a cysteine proteinase, possessed ICE-like activity (27). The ability of mucosal pathogens to cleave and activate epithelial stores of pro-IL-1β has not yet been investigated. Generating proinflammatory cytokines by enzymatic cleavage may represent a novel pathogenic mechanism for proteolytic microbes, especially if the inflammatory reaction is inefficient in clearing the microorganisms. We therefore hypothesized that, given the strategic storage of pro-IL-1β in superficial epithelial cells of squamous epithelia (15, 21), membrane-bound and secreted proteinases may act as virulence factors in chronic candidiases of the skin and mucosae by triggering a damaging inflammation cascade. The aim of this study was to test this hypothesis by demonstrating the proteolytic cleavage and activation of pro-IL-1β by clinical isolates of C. albicans.
MATERIALS AND METHODS
C. albicans strains and culture conditions.
C. albicans isolates 2 through 6 from Candida-associated stomatitis patients were cultured in modified Sabouraud medium (20 g of glucose and 10 g of tryptose per liter) in shaking flasks (100 rpm) at 25°C. Blastospores were harvested at various times during growth (between 8 h [exponential growth phase] and 120 h [stationary growth phase]), washed three times in sterile RPM1-F12 (Gibco BRL, Burlington, Ontario, Canada), pH 6, and resuspended at 1.5 × 109 cells/ml in RPMI-F12 culture medium unless otherwise indicated. In selected experiments, age-matched culture supernatants of the blastospores were also collected for analysis. Mycelium formation was induced by incubation of washed, stationary-growth-phase (96-h) blastospores in Iscove’s modified Dulbecco medium (Sigma Chemical Co., St. Louis, Mo.) for 4 h at 37°C as previously described (14). Blastospores, mycelial cells, and culture supernatants prepared as described above were used to determine their potential to degrade pro-IL-1β.
Fluorometric assay for ICE-like activity.
The fluorometric assay for the determination of ICE-like activity was adapted from that of Moncla et al. (38). The fluorogenic substrate acetyl-Tyr-Val-Ala-Asp-7-amido-4-methylcoumarin was obtained from Peptides International (Louisville, Ky.). A 10 mM stock solution was prepared in dimethyl formamide and kept at −80°C. Prior to the assay, the substrate was diluted to 50 μM in 0.1 M Tris HCl (pH 8.0), and 30-μl drops were applied to a cellulose paper strip (6 by 66 mm; Whatman Inc., Fairfield, N.J.). After drying at room temperature, C. albicans culture supernatants (20 μl), or 3 × 108 blastospores in 20 μl, were applied to the spots. Hydrolysis was monitored by UV illumination (366 nm) after a 2-h incubation at 37°C in a humidified atmosphere. Treponema denticola ATCC 35405 cells were used as a positive control (1).
SDS-PAGE Western immunoblotting analysis of pro-IL-1β cleavage.
Recombinant pro-IL-1β (9 ng) (33 kDa; Cistron Biotechnology, Pine Brook, N.J.) and 1.9 × 107 Candida cells in RPMI-F12 medium were incubated in 24-μl volumes at 37°C for 8 h unless otherwise indicated. When culture supernatants of C. albicans were tested, a 12-μl aliquot was mixed with an equal volume of RPMI-F12 medium containing 24 ng of pro-IL-1β. Following the incubation, sodium dodecyl sulfate (SDS) sample buffer was added, and the mixture was boiled for 5 min. Polyacrylamide gel electrophoresis (PAGE) was performed with a minigel system (Bio-Rad Laboratories, Mississauga, Ontario, Canada). Samples (32 μl) were loaded on a 12% polyacrylamide gel, and the electrophoresis was carried out at 100 V for 90 min (31). Proteins were electrophoretically transferred to a nitrocellulose membrane (Gibco BRL) for 60 min at 100 V (54). The membrane was blocked in 20 mM Tris–500 mM NaCl buffer (Tris-buffered saline) containing 18% glucose, 10% glycerol, 0.6% Tween 20, and 2.5% bovine serum albumin (blocking buffer) and stained by a three-layer immunoenzymatic technique. Membranes were sequentially incubated with anti-IL-1β goat polyclonal antibodies (R & D Systems, Minneapolis, Minn.) diluted to 2 μg/ml in Tris-buffered saline–0.5% bovine serum albumin, biotin-labeled donkey anti-goat immunoglobulin G (Jackson ImmunoResearch Laboratories Inc., West Grove, Pa.) diluted to 1.2 μg/ml in blocking buffer, and streptavidin-alkaline phosphatase (Cedarlane Laboratories Ltd., Hornby, Ontario, Canada) diluted 1/1,000 in blocking buffer. Membranes were developed in a solution containing nitroblue tetrazolium chloride (1.65 mg) and 5-bromo-4-chloro-3-indolylphosphate p-toluidine salt (0.8 mg) in 5 ml of 100 mM Tris-HCl (pH 9.5) containing 100 mM NaCl and 50 mM MgCl2. Positive controls, recombinant pro-IL-1β, and recombinant mature IL-1β (Cistron Biotechnology) were included in each gel. Pancreatic α-chymotrypsin (12 μg/ml; Sigma) was also used as a positive control for the generation of a 18- to 19-kDa fragment from recombinant pro-IL-1β, as already reported (2, 36). Negative controls included omitting each reagent in turn.
Effect of heat treatment and proteinase inhibitors on pro-IL-1β cleavage.
A preliminary characterization of the enzymatic activity involved heating suspensions of C. albicans for 30 min at temperatures ranging from 56 to 100°C prior to adding the pro-IL-1β substrate. In other assays, the following proteinase inhibitors, at a final concentration of 5 mM, were added to 1.9 × 107 Candida cells in 0.1 M citrate buffer (pH 6.0) 15 min before adding the pro-IL-1β: EDTA (Sigma) and 1,10-phenanthroline (Sigma) for metalloproteinases, iodoacetamide (Sigma) for cysteine proteinases, 4-(2-aminoethyl)benzensulfonyl fluoride (AEBSF; ICN Biochemical Inc., Aurora, Ohio) for serine proteinases, Nα-p-tosyl-l-lysyl chloromethyl ketone (TLCK; Sigma) for serine and cysteine proteinases, and pepstatin A (Sigma) for aspartyl proteinases.
Effect of pH on pro-IL-1β cleavage.
The enzymatic cleavage of pro-IL-1β by Candida cells was tested by using 0.1 M buffers at various pHs: citrate buffer at pH 4.0, 5.0, and 6.0; phosphate buffer at pH 7.0 and 8.0; Tris HCl buffer at pH 9.0; and carbonate buffer at pH 10.0. The mixtures were incubated at 37°C for 8 h, and postincubation pHs were verified with a microelectrode. Controls included omitting C. albicans cells to detect spontaneous hydrolysis of pro-IL-1β.
Bioassay for determining IL-1β activity.
The assay mixtures analyzed by SDS-PAGE Western immunoblotting were also analyzed, after filtration, by using a murine thymocyte proliferation assay adapted from that of Gery and Waksman (20). The bioassay measured the comitogenic effect of mature IL-1β, or biologically active fragments derived from pro-IL-1β, on concanavalin A (ConA; Flow Laboratories, Mississauga, Ontario, Canada) submitogenic stimulation of thymocyte proliferation. Thymocyte cultures were grown in RPM1-F12 supplemented with 25 mM HEPES, gentamicin (50 μg/ml), penicillin (100 U/ml), streptomycin (100 μg/ml), 5% decomplemented fetal calf serum (FetalClone; HyClone Laboratories Inc., Logan, Utah), 50 μM β-mercaptoethanol, and 3 mM glutamine. ConA was added at 0.75 or 2.0 μg/ml, as indicated, to 1.5 × 106 thymocytes/well in flat-bottom Nunclon 96-well culture plates (Nunc InterMed, Roskilde, Denmark). Standard curves were determined by simultaneously adding ConA and graded concentrations (0 and 1,000 pg/ml) of recombinant mature IL-1β. To titrate the biological activity of the Candida supernatants incubated (8 h in RPMI-F12 medium, pH 6.0) in the presence of 24 ng of pro-IL-1β, standard curves were constructed in the presence of control Candida supernatants (i.e., supernatants of Candida incubation in RPMI-F12 medium without pro-IL-1β.) Such standard curves were required because of an intrinsic inhibitory activity of Candida soluble products in supernatants (see below). An MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] colorimetric assay adapted from that of Mosmann (42) was used to measure cell proliferation after a 72-h incubation at 37°C (13). Specificity was confirmed with anti-human IL-1β neutralizing antibodies used at 1 μg/well. The means ± standard deviations of data obtained from duplicate cultures and from three similar experiments were calculated.
RESULTS
Cleavage of pro-IL-1β by C. albicans.
Aliquots from a C. albicans culture were recovered at various times (up to 120 h) to test the capacity of blastospores and culture supernatants to hydrolyze a fluorogenic substrate that closely matches the amino-terminal sequence of the cleavage site of monocyte ICE. No hydrolysis was detected with C. albicans, whereas T. denticola ATCC 35405, which was used as a positive control (1), caused an intense, fluorescent reaction.
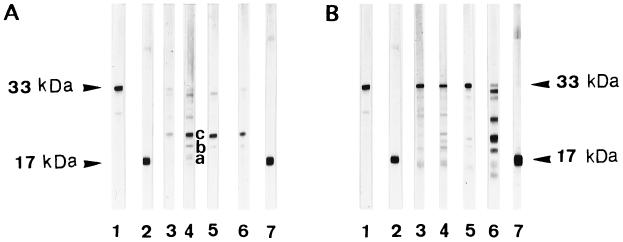
The ability of blastospores and culture supernatants to cleave the natural pro-IL-1β substrate was then analyzed by SDS-PAGE Western immunoblotting. Weak activity was detected with exponentially growing cells (<18 h), whereas more efficient cleavage was observed with stationary-growth-phase (≥72-h) blastospores (Fig. 1A). The major digestion product derived from pro-IL-1β had an apparent molecular mass of approximately 21 kDa (band c). Additional bands with molecular masses in the 17- to 19-kDa range were also detected (bands a and b). Fragments with molecular masses greater than 21 kDa were also produced but were not studied further because their sizes were not compatible with mature, biologically active IL-1β (17 to 19 kDa) (2, 24, 37). In addition, mycelial cells recovered after a 4-h induction at 37°C showed a pattern of cleavage similar to that of the 96-h blastospores from which they were generated (data not shown). When blastospores culture supernatants (cultured for the same time periods) were tested for hydrolytic activity against pro-IL-1β, efficient cleavage also occurred, generating fragments a, b, and c (Fig. 1B). However, it took 120 h for culture supernatants to produce the major 21-kDa band, which is generated by 72-h stationary-growth-phase blastospores (Fig. 1B, lane 6, versus Fig. 1A, lane 4).
FIG. 1.
SDS-PAGE Western immunoblotting analysis of the cleavage of pro-IL-1β by C. albicans as a function of growth phase. Recombinant pro-IL-1β (24 ng) was incubated with cells or supernatants at 37°C for 8 h in RPMI-F12 medium (pH 6.0). (A) C. albicans (isolate 2) blastospores in exponential (8 h; lane 3) and late stationary (72, 96, and 120 h; lanes 4 through 5, 6) growth phases. Purified recombinant pro-IL-1β (33 kDa; lane 1) and mature IL-1β (17 kDa; lanes 2 and 7) are shown for reference. (B) Culture supernatants corresponding to the C. albicans blastospores used for panel A. Supernatants were harvested during the exponential (8 h; lane 3) and late stationary (72, 96, and 120 h; lanes 4 through 6) growth phases. Purified recombinant pro-IL-1β (33 kDa; lane 1) and mature IL-1β (17 kDa; lanes 2 and 7) are shown for reference. Bands a, b, and c are approximately 17, 18, and 21 kDa, respectively.
The fact that a significant decrease in the 33-kDa recombinant pro-IL-1β band with the production of few digestion products was observed with exponential-growth-phase blastospores suggests that pro-IL-1β binds to the cell surface. This was confirmed by analyzing whole blastospores by SDS-PAGE Western immunoblotting following an 8-h incubation with pro-IL-1β and two washes in phosphate-buffered saline. About half of the pro-IL-1β was recovered, apparently unaffected, from the washed blastospores (data not shown).
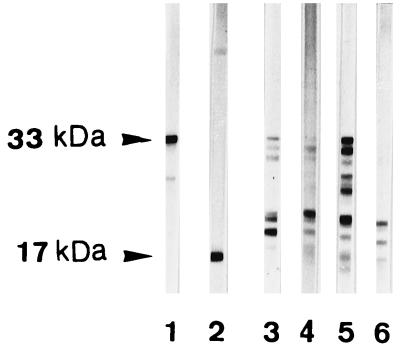
Four other C. albicans isolates obtained from four different patients with denture stomatitis were grown to the stationary phase and tested for the ability to cleave pro-IL-1β at pH 6. These isolates were all found to generate a fragment profile compatible with mature, biologically active IL-1β (17 to 19 kDa) (2, 24, 37), although there was some heterogeneity in the molecular masses and intensities of the various fragments (Fig. 2). Clinical isolate 2 was further investigated for its capacity to cleave pro-IL-1β and generate biologically active fragments.
FIG. 2.
SDS-PAGE Western immunoblotting analysis of the cleavage of pro-IL-1β by different clinical isolates of C. albicans. Blastospores from C. albicans isolates 3 through 6 (lanes 3 through 6) from four different patients with denture stomatitis were harvested in the stationary growth phase (96 h) and incubated with pro-IL-1β for 8 h at pH 6.0 in RPMI-F12. Purified recombinant pro-IL-1β (33 kDa) and mature IL-1β (17 kDa) (lanes 1 and 2) are shown for reference.
The patterns shown in Fig. 1 were obtained with an 8-h incubation at pH 6.0 in RPM1-F12 medium. We used SDS-PAGE Western immunoblotting to determine whether changing the reaction conditions would modify pro-IL-1β cleavage by stationary-growth-phase blastospores. The data obtained are summarized in Table 1. Pro-IL-1β cleavage by C. albicans blastospores did not occur at pH 4.0. At pH 5.0, two pro-IL-1β cleavage fragments, one of approximately 21 kDa (band c) and a second of approximately 19 kDa (band b), were generated. Reactions at pH 6.0 appeared to be optimal for the generation of these two fragments as well as a 17-kDa fragment (band a). In general, raising the pH from 6.0 to 7.0 produced a more heterogeneous pattern of digestion and decreased the yield of low-molecular-mass (17- to 19-kDa) fragments. Virtually no digestion occurred at pH 8.0. Control assays at different pHs, without Candida cells or culture supernatants, indicated that no significant spontaneous degradation of pro-IL-1β occurs under these conditions (data not shown). Measurements at the end of the incubation period revealed that pHs remained constant throughout the enzymatic assay.
TABLE 1.
Generation of pro-IL-1β cleavage fragments by C. albicans blastospores under various assay conditions
| Assay conditions | Pro-IL-1β fragmentsa
|
||
|---|---|---|---|
| a | b | c | |
| Controls | |||
| Recombinant mature IL-1β | + | − | − |
| α-Chymotrypsin cleavage | − | + | − |
| Incubation period (h) at pH 6.0 | |||
| 3 | − | + | + |
| 8 | + | + | + |
| 18 | + | + | − |
| Reactional pH for an 8-h assay | |||
| 4 | − | − | − |
| 5 | − | + | + |
| 6 | + | + | + |
| 7 | − | − | + |
| 8 | − | − | − |
| Proteinase inhibitor at pH 6 for an 8-h assay | |||
| EDTA | + | + | + |
| AEBSF | + | + | + |
| TLCK | + | + | + |
| 1,10-Phenanthroline | + | + | + |
| Iodoacetamide | + | + | + |
| Pepstatin A | − | − | − |
Approximate molecular masses of fragments as in Fig. 1: a, ≈17 kDa; b, ≈19 kDa; c, ≈21 kDa.
Pro-IL-1β cleavage by C. albicans cells was assayed after various incubation times (Table 1). Cleavage was observed after a 3-h incubation at pH 6.0 but was much more evident after 8 h. Following an 18-h incubation, the pattern became sketchy because the high-molecular-mass (≥21-kDa) fragments were further degraded or unstable under these conditions. Moreover, the substrate (pro-IL-1β) became undetectable in the assay medium when the enzymatic reaction was allowed to continue for 18 h.
We used the optimal conditions (pH 6.0, 8-h incubation) to evaluate the effect of class-specific proteinase inhibitors on the cleavage of pro-IL-1β by C. albicans (Table 1). Pepstatin A, an inhibitor of aspartyl proteinases, was the only inhibitor tested that prevented the cleavage of pro-IL-1β into low-molecular-mass (17- to 21-kDa) fragments. Cleavage was also completely abolished by prior heating of C. albicans for 30 min at 70 or 100°C (data not shown).
Production of biologically active fragments from pro-IL-1β by C. albicans.
Supernatants from C. albicans blastospores incubated with and without pro-IL-1β in RPM1-F12 medium (pH 6.0, 8 h) were tested in the IL-1β bioassay. Since the bioassay is based on costimulation by ConA and mature IL-1β (or eventually biologically active fragments derived from pro-IL-1β), it was expected that neither ConA (at suboptimal mitogenic concentrations) nor mature recombinant IL-1β alone would produce significant proliferation. Assaying the costimulatory activity of pro-IL-1β cleavage products on ConA-stimulated thymocytes was, however, hampered by the inhibitory activity of soluble material released from Candida blastospores during the 8-h incubation in RPMI-F12 medium on ConA stimulation. Control supernatants of Candida blastospores incubated in RPMI-F12 medium without pro-IL-1β inhibit in a dose-dependent manner the proliferative response to ConA and to ConA plus 1,000 pg of mature recombinant IL-1β per ml (data not shown). This inhibition starts weakening when the control supernatant is diluted 1/100 and is completely lifted at a dilution of 1/1,000. However, such high dilutions precluded the detection of IL-1β-like activity in similarly diluted assay supernatants (Candida cells incubated with pro-IL-1β).
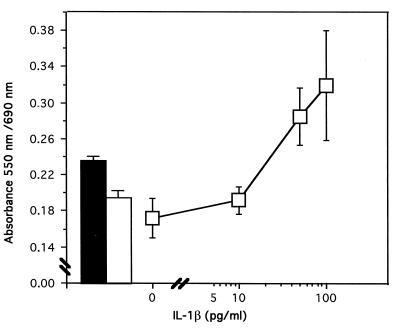
We hypothesized that the inhibition could be due to ConA sequestration by soluble Candida mannan or mannoproteins (7) and that ConA signalization would be somewhat limited when the proliferative assay was carried out in the presence of these soluble Candida products. We thus increased the ConA concentration to 2.0 μg/ml to overcome this inhibition. Figure 3 shows that under these conditions, the thymocytes proliferated following a normal dose-response curve with graded concentrations of recombinant mature IL-1β notwithstanding the presence of control Candida supernatant (Candida cells incubated in RPMI-F12 medium without pro-IL-1β) diluted 1/10. These conditions thus enabled us to assess the biological activity of assay supernatants (supernatants of Candida cells incubated with pro-IL-1β). At a 1/10 dilution, assay supernatants containing pro-IL-1β cleavage products can elicit a proliferative response corresponding to the costimulatory activity of 20 to 30 pg of mature recombinant IL-1β per ml. This IL-1β-like costimulatory activity was neutralized in the presence of anti-IL-1β antibodies, and the proliferative response decreased to the level of ConA stimulation alone, indicating an almost complete loss of comitogenic activity.
FIG. 3.
IL-1β-like costimulatory activity from proteolytic cleavage of pro-IL-1β by C. albicans. A dose-response curve of mature recombinant IL-1β was generated by using the bioassay described in Materials and Methods. To titrate the biological activity of the supernatants of Candida incubated in the presence of 24 ng of pro-IL-1β, this standard curve was constructed in the presence of control supernatants (i.e., supernatants of C. albicans incubated in RPMI-F12 medium for 8 h at 37°C without pro-IL-1β). These adapted standard curves were required because of the inhibitory effect of Candida supernatants on this bioassay. The ConA concentration was increased to 2.0 μg/ml to overcome this inhibitory activity. Control supernatants were diluted (1/10) and added simultaneously with graded concentrations of recombinant mature IL-1β and ConA. The closed bar shows the proliferative response when the assay is conducted in the presence of pro-IL-1β cleavage products (assay supernatants) instead of mature recombinant IL-1β. At this dilution (1/10), Candida supernatants containing cleavage products generated from pro-IL-1β generate a proliferative response corresponding to the costimulatory activity of 20 to 30 pg of mature IL-1β per ml. This biological activity was neutralized by antibodies directed against mature IL-1β (open bar). Means ± standard deviations from three experiments are shown.
DISCUSSION
C. albicans commonly colonizes various stratified squamous epithelia, including those of the oral cavity, the pharynx, the eosophagus, the vagina, and the epidermis. Because it possesses several attributes that are critical for colonizing these sites, it is considered not only a successful commensal but also an opportunistic pathogen since the expression of C. albicans virulence is often signalled by a breakdown in surveillance by innate and acquired defense mechanisms, for example, in AIDS. However, defense mechanisms may also participate in the pathogenesis of superficial candidiases in situations where C. albicans is allowed to sustain a chronic inflammatory reaction. The inflammatory lesions observed in mucosal candidiases are thought to be triggered by Candida aggressins (44, 45), and although several authors (9, 35) report that a number of C. albicans constituents can act as inflammatory aggressins in vivo, no mechanism has been proposed to explain the transition from asymptomatic carriage to proinflammatory potential. Candida proteinases are suspected to act as virulence factors, possibly by facilitating fungal access and adherence to epithelial cells, invasion of host tissues, and interference with host defense mechanisms (reviewed in references 18, 35, and 49). We report here that C. albicans proteinase(s) can cleave pro-IL-1β to produce biologically active fragments. This is the first time to our knowledge that Candida proteinases have been proposed as proinflammatory agents at mucosal surfaces.
C. albicans proteinases can apparently cause limited proteolysis of the Hageman factor, leading to activation of the kallikrein-kinin system, which in turn generates bradykinin and causes increased vascular permeability (26). The release of inflammatory mediators through cell wall-induced complement activation via the alternative cascade or direct production of a neutrophil chemotactic attractant by the fungus may also lead to inflammatory reactions (10, 11, 29). However, neither complement nor Hageman factor is available at the skin surface or in the upper layers of stratified mucosal epithelia (53). Therefore, direct activation of pro-IL-1β reserves would be an alternate but powerful inflammatory mechanism. One may argue that inflammation would benefit the host rather than the fungus. However, in chronic inflammatory diseases such as denture stomatitis, the inflammatory reaction not only is inefficient but accounts for much of the etiopathology.
The extracellular proteolytic activity of C. albicans has been extensively studied and results mainly from secreted aspartyl proteinases (Sap) whose activity is restricted to the acidic pH range (reviewed in reference 11). Sap isoenzymes are not specific to a single substrate and can break down a number of host proteins found in the oral cavity, including salivary proteins, immunoglobulin A, mucin, and epithelial keratin (8, 25, 43, 48, 50). Sap isoenzymes are now known to be the products of a family of genes that are expressed and regulated differentially (33, 39, 49, 51, 56). The expression of the SAP1 and SAP3 genes is regulated during phenotypic switching from the white to the opaque form of strain WO-1 (41). Sap2 is the main enzyme secreted in vitro by the yeast form of many strains of C. albicans, including white and opaque cells of strain WO-1 (23, 56, 57). The expression of the SAP4, SAP5, and SAP6 genes has been detected at neutral pH during serum-induced yeast-to-hypha transition, whereas the expression of the SAP7 gene has not yet been detected under any in vitro conditions (23, 56). Among the products of the eight SAP genes identified so far, only the products of genes SAP1 to -3 and a putative Sap8 protein have been isolated and characterized (40, 55). While it is becoming clear that the pattern of Sap production is environmentally regulated in C. albicans, at least in vitro, the attribution of a particular proteolytic pattern to commensal versus pathogenic strains of C. albicans remains speculative. Moreover, as the entire substrate specificity or spectrum of C. albicans proteinases has not yet been defined, a more complete characterization of proteolytic enzymes as putative virulence factors is clearly needed to clarify these issues.
In mucosal infections, proteolytic enzymes with mucinolytic and keratinolytic activities may enable proinflammatory enzymes to gain access to stratum corneum stores of pro-IL-1β and thus participate in the cleavage and activation of this precursor. Our study of this activity suggests that it is not due to a typical convertase-like enzyme because the classic fluorogenic ICE substrate is not hydrolyzed by Candida and because pro-IL-1β is cleaved into multiple fragments rather than a predominant, low-molecular-mass (17- to 18-kDa) fragment as is the case for monocyte ICE (28). We do not know whether the high-molecular-mass (25- to 30-kDa) fragments are intermediates in pro-IL-1β processing to smaller fragments with biological activity (17 to 19 kDa) or whether the cleavage pattern results from a number of concurrently acting proteinases. However, the pepstatin sensitivity of pro-IL-1β cleavage and the increase in enzymatic activity during the stationary growth phase that follows the decrease in the pH of the culture medium suggest a similarity between Saps and the pro-IL-1β-cleaving enzyme(s) (55). In addition, data presented here do not exclude that a pro-IL-1β-cleaving enzyme(s) may be associated with the Candida cell surface, a feature already demonstrated for Saps on C. albicans cells that adhere to human, nonkeratinized, buccal epithelia (3). The fact that Saps function optimally at pH 4 whereas pro-IL-1β cleavage occurs mainly at pH 6 under our conditions suggests that a putative neutral proteinase (32) may also be at work, possibly with Saps, to cleave the precursor.
Generally, proteolytic processing of pro-IL-1β by ICE or other convertase-like enzymes generates 17- to 19-kDa fragments resistant to further proteolytic degradation. In our study, we observed that the 17- to 19-kDa fragments are also stable end products of C. albicans proteolytic activity. In fact, after 18 h of incubation, the 17- to 19-kDa fragments increase in concentration and are the only fragments detectable. The five C. albicans isolates studied demonstrated an ability to generate 17- to 19-kDa fragments from pro-IL-1β, although each strain showed a particular cleavage pattern. This finding may be related to different panels of proteinases or to the amount of enzyme released, which may vary by 2 orders of magnitude (12).
As the expression of proteinases by C. albicans has been reported for human skin mycoses and the initial stages of mucosal candidiases (3, 46, 47), we suggest a role for pro-IL-1β cleavage and activation by Candida proteinases in the pathogenesis of these inflammatory lesions. The mobilization of keratinocyte pro-IL-1β into biologically active cleavage products may have immediate effects on contiguous keratinocytes, fibroblasts, and endothelial cells. As a proinflammatory cytokine, IL-1β initiates and perpetuates the deleterious inflammation cascade by mediating neutrophil emigration and releasing secondary lipid-derived mediators and chemotactic cytokines, which in turn participate in the expression of vascular endothelial adhesion molecules and lead to an amplified recruitment of inflammatory cells (reviewed in references 19 and 52). Such an immunopathologic sequence would persist only in situations where host defense mechanisms are deficient or inefficient.
The transition of C. albicans from an innocuous commensal to an opportunistic pathogen thus likely depends on cumulative adaptive attributes that may be controlled by rapid phenotypic variability, which in turn is regulated by microenvironmental stimuli. According to the current, integrated models of composite virulence phenotypes proposed by Cutler (11) and Odds (45), the C. albicans virulence profile would vary depending on the site and stage of infection and the nature of the host response. Adherence to and invasion of epithelia, together with the ability to evade local host defenses, may be selected in mucosal microniches, and there is evidence that this selection may be linked to the secretion (and the secretion rate) of proteinases (11, 45). We proposed here that C. albicans proteinases may contribute to the inflammatory nature of mucosal candidiasis by the activation of an epithelial, proinflammatory cytokine. That epithelial pro-IL-1β may be mobilized for in situ activation by Candida enzymes is presently being tested in assays human keratinocytes cultured as stratified dermal equivalents. A precise characterization of the pro-IL-1β-cleaving enzymes and of their regulation and activity at mucosal surfaces will further clarify how C. albicans may occasionally convert to a harmful pathogen and how Saps may act as pathogenic determinants in infections of the stratified epithelia.
ACKNOWLEDGMENTS
This study was supported by the Medical Research Council of Canada, the Fonds de la Recherche en Santé du Québec, and the Fonds Emile-Beaulieu. A.B. was supported by a studentship from the Fonds pour la Formation de Chercheurs et l’Aide à la Recherche.
We thank Gene Bougeau (Anglocom) for editorial assistance.
REFERENCES
- 1.Beauséjour A, Deslauriers N, Grenier D. Activation of the interleukin-1β precursor by Treponema denticola: a potential role in chronic inflammatory periodontal disease. Infect Immun. 1997;65:3199–3202. doi: 10.1128/iai.65.8.3199-3202.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black R A, Kronheim S R, Cantrell M, Deeley M C, March C J, Prickett K S, Wignall J, Conlon P J, Cosman D, Hopp T P, Mochizuki D Y. Generation of biologically active interleukin-1β by proteolytic cleavage of the inactive precursor. J Biol Chem. 1988;263:9437–9442. [PubMed] [Google Scholar]
- 3.Borg M, Rüchel R. Expression of extracellular acid proteinase by proteolytic Candida spp. during experimental infection of oral mucosa. Infect Immun. 1988;56:626–631. doi: 10.1128/iai.56.3.626-631.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Budtz-Jorgensen E. Candida associated denture stomatitis and angular cheilitis. In: Samaranyake L P, MacFarlane T W, editors. Oral candidosis. London, England: Wright; 1990. pp. 156–183. [Google Scholar]
- 5.Budtz-Jorgensen E, Bertram U. Denture stomatitis. I. The etiology in relation to trauma and infection. Acta Odontol Scand. 1970;28:71–92. doi: 10.3109/00016357009033133. [DOI] [PubMed] [Google Scholar]
- 6.Budtz-Jorgensen E, Stenderup A, Grabowski M. An epidemiologic study of yeast in elderly denture wearers. Community Dent Oral Epidemiol. 1975;3:115–119. doi: 10.1111/j.1600-0528.1975.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 7.Cassone A, Mattia E, Boldrini L. Agglutination of blastospores of Candida albicans by concanavalin A and its relationship with the distribution of mannan polymers and the ultrastructure of the cell wall. J Gen Microbiol. 1978;105:263–273. doi: 10.1099/00221287-105-2-263. [DOI] [PubMed] [Google Scholar]
- 8.Colina A R, Aumont F, Deslauriers N, Belhumeur P, De Repentigny L. Evidence for degradation of gastrointestinal mucin by Candida albicans secretory aspartyl proteinase. Infect Immun. 1996;64:4514–4519. doi: 10.1128/iai.64.11.4514-4519.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cutler J E, Friedman L, Milner K C. Biological and chemical characterization of toxic substances from Candida albicans. Infect Immun. 1972;6:616–627. doi: 10.1128/iai.6.4.616-627.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cutler J E. Chemotactic factor produced by Candida albicans. Infect Immun. 1977;18:568–573. doi: 10.1128/iai.18.3.568-573.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cutler J E. Putative virulence factors of Candida albicans. Annu Rev Microbiol. 1991;45:187–218. doi: 10.1146/annurev.mi.45.100191.001155. [DOI] [PubMed] [Google Scholar]
- 12.De Bernardis F, Chiani P, Ciccozzi M, Pellegrini G, Ceddia T, D’Offizzi G, Quinti I, Sullivan P A, Cassone A. Elevated aspartyl proteinase secretion and experimental pathogenicity of Candida albicans isolates from oral cavities of subjects infected with human immunodeficiency virus. Infect Immun. 1996;64:466–471. doi: 10.1128/iai.64.2.466-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deslauriers N, Côte L, Montplaisir S, De Repentigny L. Oral carriage of Candida albicans in murine acquired immunodeficiency syndrome. Infect Immun. 1997;65:661–667. doi: 10.1128/iai.65.2.661-667.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deslauriers N, Michaud J, Carré B, Léveillé C. Dynamic expression of cell-surface antigens probed with Candida albicans-specific monoclonal antibodies. Microbiology. 1996;142:1239–1248. doi: 10.1099/13500872-142-5-1239. [DOI] [PubMed] [Google Scholar]
- 15.Didierjean L, Salomon D, Merot Y, Siegenthaler G, Shaw A, Dayer J M, Saurat J H. Localization and characterization of the interleukin-1 immunoreactive pool (IL-1 α and β forms) in normal human epidermis. J Invest Dermatol. 1985;92:809–816. doi: 10.1111/1523-1747.ep12696825. [DOI] [PubMed] [Google Scholar]
- 16.Dinarello C A, Wolff S M. The role of interleukin-1 in disease. N Engl J Med. 1993;328:106–108. doi: 10.1056/NEJM199301143280207. [DOI] [PubMed] [Google Scholar]
- 17.Dinarello C A. The biological properties of interleukin-1. Eur Cytokine Netw. 1994;5:517–531. [PubMed] [Google Scholar]
- 18.Douglas L J. Candida proteinases and candidosis. Crit Rev Biotechnol. 1988;8:121–129. doi: 10.3109/07388558809150541. [DOI] [PubMed] [Google Scholar]
- 19.Downey G P. Mechanisms of leukocyte motility and chemotaxis. Curr Opin Immunol. 1994;6:113–124. doi: 10.1016/0952-7915(94)90042-6. [DOI] [PubMed] [Google Scholar]
- 20.Gery I, Waksman B. Potentiation of the T-lymphocyte response to mitogens II. The cellular source of potentiating mediators. J Exp Med. 1972;136:143–155. doi: 10.1084/jem.136.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hauser C, Saurat J H, Schmitt A, Jaunin F, Dayer J M. Interleukin-1 is present in normal human epidemis. J Immunol. 1986;136:3317–3323. [PubMed] [Google Scholar]
- 22.Hazuda D J, Strickler J, Kueppers F, Simon P L, Young P R. Processing of precursor interleukin-1β and inflammatory disease. J Biol Chem. 1990;265:6318–6322. [PubMed] [Google Scholar]
- 23.Hube B, Monod M, Schofield D A, Brown A J P, Gow N A R. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol Microbiol. 1994;14:87–99. doi: 10.1111/j.1365-2958.1994.tb01269.x. [DOI] [PubMed] [Google Scholar]
- 24.Irmler M, Hertig S, Robson MacDonald H, Sadoul R, Becherer J D, Proudfoot A, Solari R, Tschopp J. Granzyme A is an interleukin-1β converting enzyme. J Exp Med. 1995;181:1917–1922. doi: 10.1084/jem.181.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaminishi H, Hagihara Y, Hayashi S, Cho T. Isolation and characterization of collagenolytic enzyme produced by Candida albicans. Infect Immun. 1986;53:312–316. doi: 10.1128/iai.53.2.312-316.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaminishi H, Tanaka M, Cho T, Maeda H, Hagihara Y. Activation of the plasma kallikrein-kinin system by Candida albicans proteinase. Infect Immun. 1990;58:2139–2143. doi: 10.1128/iai.58.7.2139-2143.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapur V, Majesky M W, Li L L, Black R A, Musser J M. Cleavage of interleukin-1β (IL-1β) precursor to produce active IL-1β by a conserved extracellular cysteine protease from Streptococcus pyogenes. Proc Natl Acad Sci USA. 1993;90:7676–7680. doi: 10.1073/pnas.90.16.7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kostura M J, Tocci M J, Limjuco G, Chin J, Cameron P, Hillman A G, Chartrain N A, Schmidt J A. Identification of a monocyte specific prointerleukin-1β convertase activity. Proc Natl Acad Sci USA. 1989;86:5227–5231. doi: 10.1073/pnas.86.14.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozel T R, Brown R R, Pfrommer G S. Activation and binding of C3 by Candida albicans. Infect Immun. 1987;55:1890–1894. doi: 10.1128/iai.55.8.1890-1894.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kupper T S, Groves R W. The interleukin-1 axis and cutaneous inflammation. J Invest Dermatol. 1995;105:62S–66S. doi: 10.1111/1523-1747.ep12316087. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli U K. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Logan D A. Neutral proteinase activity of Candida albicans. Exp Mycol. 1986;10:157–162. [Google Scholar]
- 33.Magee B B, Hube B, Wright R J, Sullivan P A, Magee P T. The genes encoding the secreted aspartyl proteinases of Candida albicans constitute a family with at least three members. Infect Immun. 1993;61:3240–3243. doi: 10.1128/iai.61.8.3240-3243.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin M V, Lamb D J. Frequency of Candida albicans serotypes in patients with denture-induced stomatitis and in normal denture wearers. J Clin Pathol. 1982;35:888–891. doi: 10.1136/jcp.35.8.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCullough M J, Ross B C, Reade P C. Candida albicans: a review of its history, taxonomy, epidemiology, virulence attributes and methods of strain differentiation. J Oral Maxillofac Surg. 1996;25:136–144. doi: 10.1016/s0901-5027(96)80060-9. [DOI] [PubMed] [Google Scholar]
- 36.Mizutani H, Black R, Kupper T S. Human keratinocytes produce but do not process pro-interleukin-1 (IL-1) beta. J Clin Invest. 1991;87:1066–1071. doi: 10.1172/JCI115067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizutani H, Schechter N, Lazarus G, Black R A, Kupper T S. Rapid and specific conversion of precursor interleukin-1β (IL-1β) to an active IL-1 species by human mast cell chymase. J Exp Med. 1991;174:821–825. doi: 10.1084/jem.174.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moncla B J, Braham P, Rabe L K, Hillier S L. Rapid presumptive identification of black-pigmented gram-negative anaerobic bacteria using 4-methylumbelliferin derivatives. J Clin Microbiol. 1991;29:1955–1958. doi: 10.1128/jcm.29.9.1955-1958.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monod M, Togni G, Hube B, Sanglerd D. Multiplicity of genes encoding secreted aspartic proteinases in Candida species. Mol Microbiol. 1994;13:357–368. doi: 10.1111/j.1365-2958.1994.tb00429.x. [DOI] [PubMed] [Google Scholar]
- 40.Morrison C J, Hurst S F, Bragg S L, Kuykendall R J, Diaz H, Pohl J, Reiss E. Heterogeneity of the purified extracellular aspartyl proteinase from Candida albicans: characterization with monoclonal antibodies and N-terminus amino acid sequence analysis. Infect Immun. 1993;61:2030–2036. doi: 10.1128/iai.61.5.2030-2036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morrow B, Srikantha T, Soll D R. Transcription of the gene for a pepsinogen, PEP1, is regulated by white-opaque switching in Candida albicans. Mol Cell Biol. 1992;12:2997–3005. doi: 10.1128/mcb.12.7.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mosmann T. Rapid colorimetric assay for cellular grow and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 43.Negi M, Tsuboi R, Matsui T, Ogawa H. Isolation and characterization of proteinase from Candida albicans: substrate specificity. J Invest Dermatol. 1984;83:32–36. doi: 10.1111/1523-1747.ep12261656. [DOI] [PubMed] [Google Scholar]
- 44.Odds F C. Candidosis of the oropharynx. In: Odds F C, editor. Candida and candidosis. London, England: Baillière Tindall; 1988. pp. 117–123. [Google Scholar]
- 45.Odds F C. Candida species and virulence. ASM News. 1994;60:313–318. [Google Scholar]
- 46.Ray T L, Wuepper K D. Experimental cutaneous candidiasis in rodents. II. Role of the stratum corneum barrier and serum complement as mediator of a proteolytic enzyme from Candida albicans. Arch Dermatol. 1978;114:539–543. doi: 10.1001/archderm.114.4.539. [DOI] [PubMed] [Google Scholar]
- 47.Rüchel R. On the role of proteinases from Candida albicans in the pathogenesis of acronecrosis. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt I Orig Reihe A. 1983;255:524–536. [PubMed] [Google Scholar]
- 48.Rüchel R. Cleavage of immunoglobulins by pathogenic yeasts of the genus Candida. Microbiol Sci. 1986;3:316–319. [PubMed] [Google Scholar]
- 49.Rüchel R, De Bernardis F, Ray T L, Sullivan P A, Cole G T. Candida acid proteinases. J Med Vet Mycol. 1992;30:123–132. [PubMed] [Google Scholar]
- 50.Samaranayake Y H, MacFarlane T W, Samaranayake L P, Aitchison T. The in vitro proteolytic and saccharolytic activity of Candida species cultured in human saliva. Oral Microbiol Immunol. 1994;9:229–235. doi: 10.1111/j.1399-302x.1994.tb00063.x. [DOI] [PubMed] [Google Scholar]
- 51.Smolensky G, Sullivan P A, Cutfield S M, Cutfield J F. Analysis of secreted aspartic proteinases from Candida albicans: purification and characterization of individual Sap1, Sap2 and Sap3 isoenzymes. Microbiology. 1997;143:349–356. doi: 10.1099/00221287-143-2-349. [DOI] [PubMed] [Google Scholar]
- 52.Springer T A. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multi-step paradigm. Cell. 1994;76:301–304. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 53.Squier C A, Johnson N W, Hackeman M. Structure and function of normal human oral mucosa. In: Dolby A E, editor. Oral mucosa in health and disease. Oxford, England: Blackwell; 1975. pp. 1–95. [Google Scholar]
- 54.Towbin H, Staehellin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White T C, Miyasaki S H, Agibian N. Three distinct secreted aspartyl proteinases in Candida albicans. J Bacteriol. 1993;175:2997–3005. doi: 10.1128/jb.175.19.6126-6133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White T C, Agabian N. Candida albicans secreted aspartyl proteinases: isoenzyme pattern is determined by cell type, and levels are determined by environmental factors. J Bacteriol. 1995;177:5215–5221. doi: 10.1128/jb.177.18.5215-5221.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wright R J, Carne A, Hieber A D, Lamont I L, Emerson G E, Sullivan P A. A second gene for a secreted aspartyl proteinase in Candida albicans. J Bacteriol. 1992;174:7848–7853. doi: 10.1128/jb.174.23.7848-7853.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamamoto T, Osaki T. Characteristic cytokines generated by keratinocytes and mononuclear infiltrates in oral lichen planus. J Invest Dermatol. 1995;104:784–788. doi: 10.1111/1523-1747.ep12606990. [DOI] [PubMed] [Google Scholar]