SUMMARY
COVID-19 has led to over 6.8 million deaths worldwide and continues to affect millions of people, primarily in low-income countries and communities with low vaccination coverage. Low-cost and rapid response technologies that enable accurate, frequent testing of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants are crucial for outbreak prevention and infectious disease control. Here we produce and characterize cellulose fibers naturally generated by the bacterium Gluconacetobacter hansenii as an alternative biodegradable substrate for manufacturing an eco-friendly diagnostic test for COVID-19. Using this green technology, we describe a novel and label-free potentiometric diagnostic test that can detect SARS-CoV-2 within 10 min and costs US$3.50 per unit. The test has bacterial cellulose (BC) as its substrate and a carbon-based electrode modified with graphene oxide and the human angiotensin-converting enzyme-2 (ACE2) as its receptor. Our device accurately and precisely detects emerging SARS-CoV-2 variants and demonstrates exceptional sensitivity, specificity, and accuracy for tested clinical nasopharyngeal/oropharyngeal (NP/OP) samples.
Graphical Abstract
de Lima et al. produce and characterize cellulose fibers naturally generated by the bacterium Gluconacetobacter hansenii as an alternative biodegradable substrate for manufacturing an eco-friendly biosensor test for COVID-19. This low-cost test (US$3.50 per test) is capable of detecting SARS-CoV-2 variants within 10 min.
INTRODUCTION
The COVID-19 pandemic, caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), has led to the death of over 6.8 million people worldwide and incalculable economic damage.1 Seeking to quell the pandemic, researchers have developed several diagnostic methods to meet the demand for reliable, fast tests designed for frequent use.2,3 Rapid tests have played an extremely important role in disease surveillance and control over the course of the pandemic, as they can quickly identify infected individuals and provide information that helps prevent viral spread.2 The reverse-transcription polymerase chain reaction (RT-PCR) is the gold standard for testing because of its high specificity and sensitivity for the detection of SARS-CoV-2 nucleic acids in nasopharyngeal sample fluids.4,5 However, testing with this technique takes a long time and requires expensive reagents, such as primers, and specialized labor.6 Thus, there is a need to find cheaper and more rapid tests.
Furthermore, several SARS-CoV-2 variants are already in circulation, and new variants are highly likely to emerge. The performance of diagnostic tests, mainly those based on the analysis of genetic material, may be affected by SARS-CoV-2 mutations, and these tests may consequently require recurrent updates and changes in the detection protocol to improve their sensitivity and avoid false-negative results.7,8 Despite the commitment to research on SARS-CoV-2 mutations, little is known about how these mutations may affect the antigenic phenotypes of the virus or interfere with vaccine effectiveness, as well as with the immunity acquired from natural infection.9,10
Biodegradability is also an important consideration in test development. Given the large quantities of materials being used to test for SARS-CoV-2 (as well as other pathogens and various clinical biomarkers), fabricating detection devices with recyclable or biodegradable materials would reduce their environmental impact.11 The most widely used substrate for manufacturing electrical circuits and consequently electrodes is the printed circuit board (PCB). PCBs contain Cu, Al, and Sn, consisting of nearly 28% metal. These metals in PCBs are more than 10 times purer than the metals in rich-content minerals.12 Because PCBs are used extensively and discarded afterward, the recycling of PCBs is not trivial. Moreover, the high percentage of non-metals in PCBs is around 70%, consisting mostly of thermoset resins and reinforcing materials; these materials pose a particularly challenging recycling problem. The network structure of thermoset resins hinders them from being remelted or reformed. Due to inorganic fillers such as glass fiber, with considerably lower fuel efficiency, incineration is not appropriate for treating nonmetals. Nonmetal components of PCBs are mostly disposed of in landfills, which can waste resources and produce significant secondary contamination.12 Other polymeric substrates, such as poly(ethylene terephthalate) (PET), polyurethane (PU), polylactide (PLA), polypropylene (PP), and polyethylene (PE), have been used as platforms in the sensing field, but their (bio)degradation in nature is slow and their recyclability is limited, contributing to increasing environmental pollution.13
With these limitations of existing materials in mind, we explored the use of bacterial cellulose (BC) as a substrate for the development of electrochemical devices. BC is an extracellular polymer synthesized by species of bacteria belonging to several genera: Agrobacterium, Gluconacetobacter, and Sarcina.14 Besides being biodegradable, BC is mechanically resistant, porous, crystalline, flexible, and biocompatible.14–17 BC is used in the wound care industry, in skin regenerative medicine, and point-of-care (POC) devices.15,18–21 As a material, BC is nontoxic and low cost22 and also exhibits several advantages over commercial paper, such as reduced fiber diameter, no use of chemical methods or processes in its manufacture, and high purity.
Here, we present an eco-friendly electrochemical biosensor to detect SARS-CoV-2 using a biodegradable BC substrate. The device does not require the use of a redox probe or additional reagents for transduction and diagnosis at the POC. Our technology uses carbon screen-printed electrodes modified with graphene oxide conjugated with polyethylene glycol (PEG), as G-PEG, for anchoring the angiotensin-converting enzyme-2 (ACE2) bioreceptor. This receptor is a remarkably selective bioelement that recognizes the SARS-CoV-2 spike protein (SP).3,4 Our device detected the SARS-CoV-2 virus in less than 10 min, displayed a limit of detection (LOD) of 4.26 × 10−18 g mL−1 SP or 0.05 copies μL−1, and presented excellent sensitivity, specificity, and accuracy when tested in 65 nasopharyngeal/oropharyngeal (NP/OP) clinical samples (25 negative samples and 40 positive samples confirmed by RT-PCR). However, increasing the number of clinical samples would likely affect the sensitivity, specificity, and accuracy profiles of our method. Nevertheless, we do think that the excellent reproducibility observed for the clinical samples tested is indicative of a robust biosensor for the detection of SARS-CoV-2 viral particles in complex samples.
RESULTS AND DISCUSSION
Fabrication and characterization of BC-based electrochemical biosensor
Gluconacetobacter hansenii was incubated in Hestrin-Schramm (HS) medium containing glucose for BC production. After 27 days, the BC material was collected and treated with 5 mmol L−1 NaOH at 80°C, which was subsequently washed abundantly with deionized water and, after drying, resulted in a clear sheet. The BC substrate was used as a flexible platform for the screen printing of the electrochemical systems using flexible carbon and Ag/AgCl conductive inks, which were cut to dimensions of 2.5 × 2.0 cm (Figure 1A).
Figure 1. Fabrication and characterization of the SARS-CoV-2 electrochemical biosensor using bacterially produced cellulose.
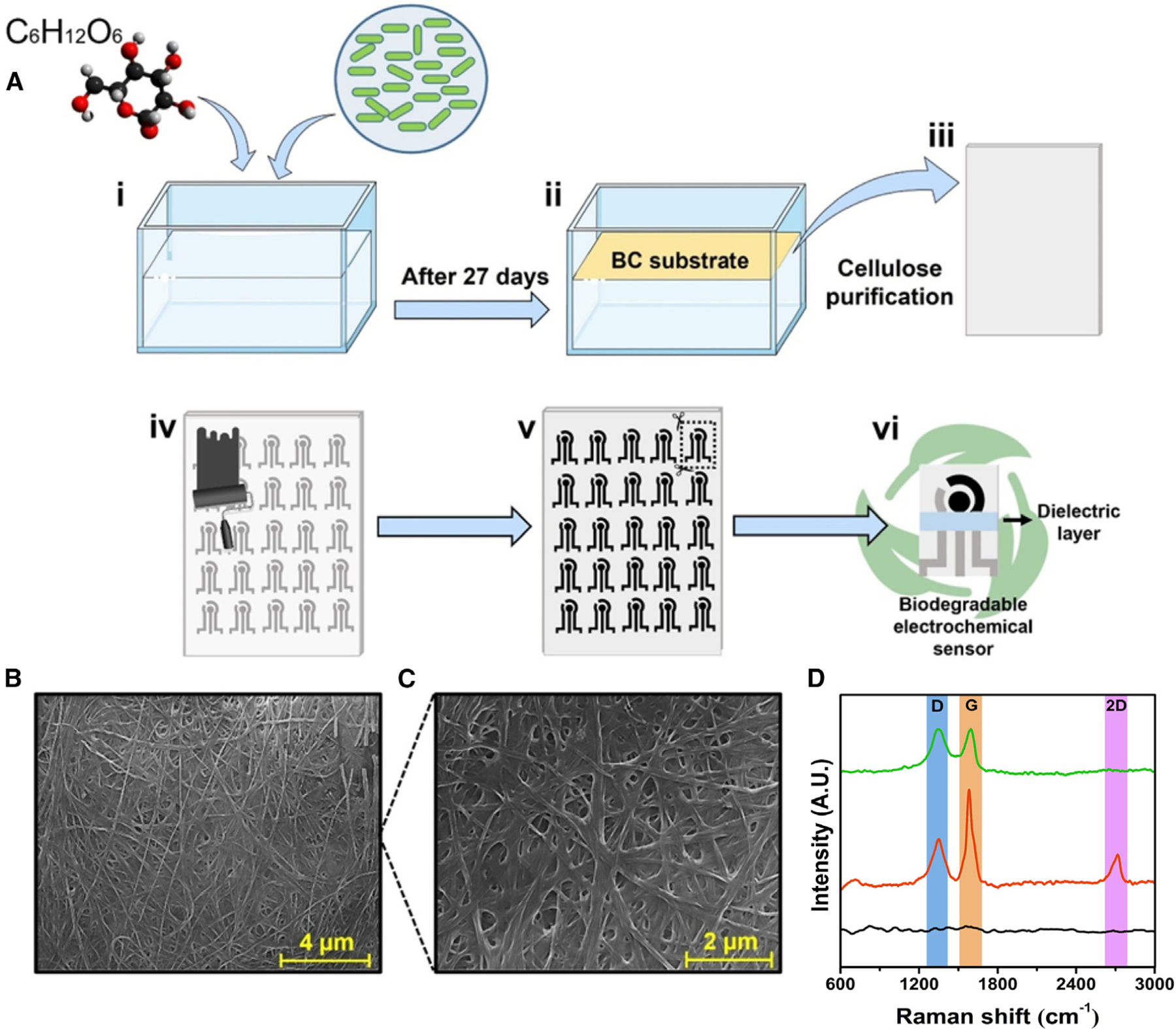
(A) Fabrication steps of the biodegradable BC substrate and the electrochemical devices. First, the bacterium Gluconacetobacter hansenii was incubated in HS medium with 20 g L−1 glucose (i); after 27 days, a BC substrate was collected and treated with 5 mmol L−1 NaOH at 80°C (ii), resulting in a clear sheet (iii). Next, the biodegradable BC substrate was screen printed with carbon and Ag/AgCl conductive ink (iv), resulting in a device with three electrodes (WE, CE, and RE), which were cut out using a scissor (v), yielding a portable, biodegradable, and inexpensive electrochemical sensor (vi).
(B and C) Micrograph of BC substrate at magnifications of (B) 13,000× (scale bar, 4 μm) and (C) 25,000× (scale bar, 2 μm).
(D) Raman spectra of the (●) BC substrate, (●) BC/carbon ink electrode, and (●) BC/carbon ink/G-PEG electrode. This figure was created in BioRender.com.
Morphological characterizations of the produced BC sheet revealed a typical pure mat of interwoven nanosized cellulose fibers (Figures 1B and 1C). The structural characterizations of BC and BC-based carbon screen-printed electrode with and without the superficial modification with G-PEG material (ACE2 receptor anchoring species) were performed by Raman spectroscopy (Figure 1D). The BC substrate presented low-intensity bands characteristics of the cellulosic material.23 It is worth noting the presence of the characteristic D band (~1,350 cm−1), G band (~1,580 cm−1), and the 2D band (~2,750 cm−1) related to graphene-based materials on the bare carbon electrode (Figure 1D).24 The G-PEG-modified electrode presented a suppression on the 2D band due to high structural defects of carbon rings in the graphene oxide flakes.24 The disorder degree level was calculated through the peak intensity ratio of the D and G bands (ID/IG). We obtained an ID/IG of 0.49 ± 0.13 and 0.99 ± 0.18 for BC-based carbon screen-printed electrode without and with the superficial modification with G-PEG material, respectively, demonstrating that the introduction of G-PEG enhances the disordered nature of the carbon electrode.25,26
Electrochemical characterization of the biosensor
To evaluate the electrochemical behavior and record measurements of each functionalization step of the biosensor, we used cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS) (Figures 2A–2C). A ferricyanide/ferrocyanide redox probe was used to compare the voltammetric and EIS behaviors between each functionalization step of the working electrode. However, this redox probe presents surface-sensitivity features, causing interactions with electrodic surface and inducing non-ideal behavior of an outer-sphere redox probe, which may lead to misinterpretations of the properties of carbon-based material and erroneous values for the calculated electroactive area.27–29 In addition, the cyclic voltammograms presented in Figure 2 were recorded using different electrodes freshly prepared at each functionalization step to avoid the influence of the previous measurement on the subsequent electrochemical response and in further modification steps of the sensor.
Figure 2. Electrochemical behavior of the eco-friendly bacterial cellulose-based biosensor.
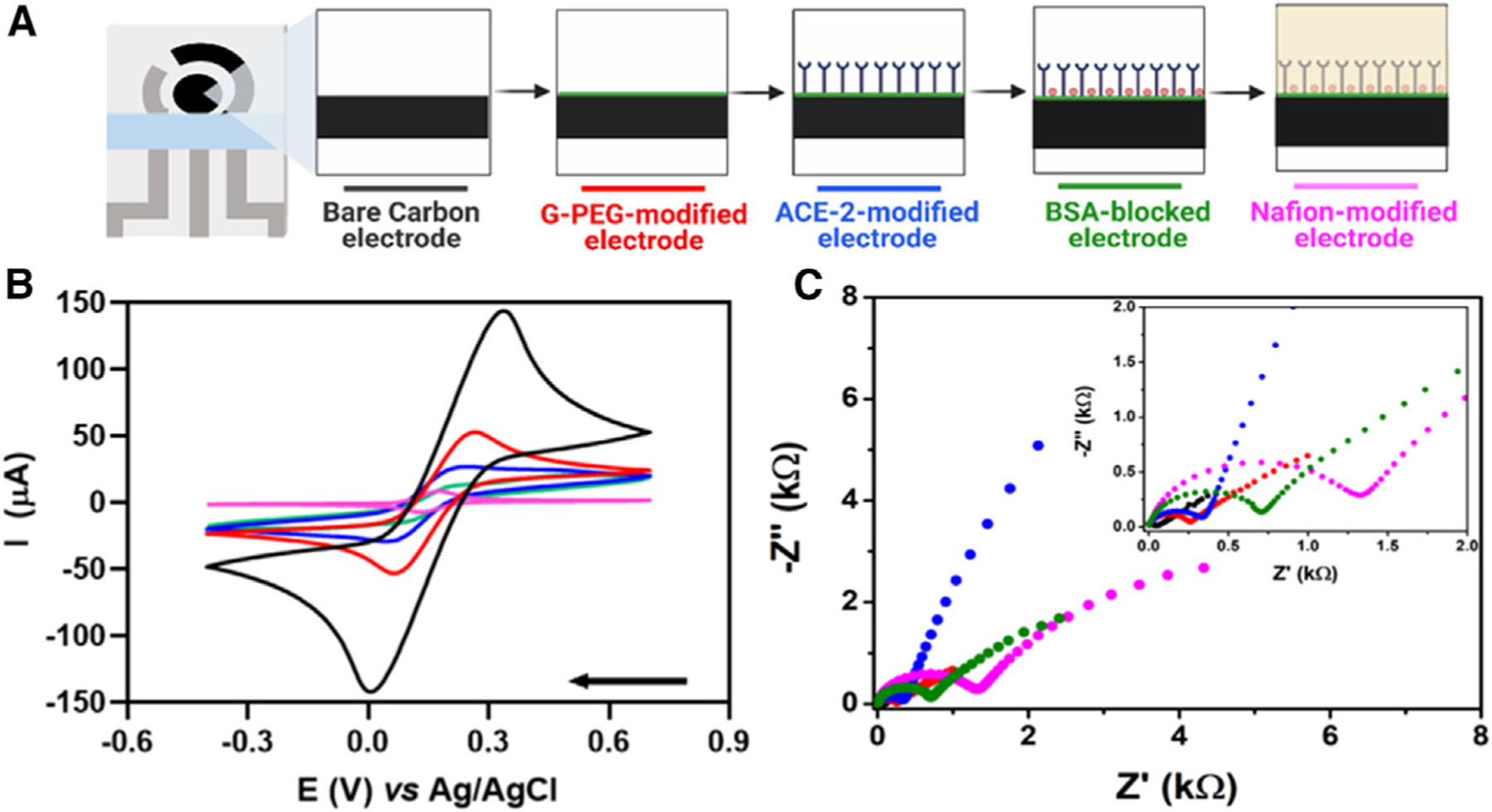
(A)Schematic representation of stepwise functionalization of the electrochemical biosensor.
(B) CVs recorded using different electrodes at each functionalization step of the electrochemical biosensor using 5.0 mmol L−1 [Fe(CN)6]3−/4− containing 0.1 mol L−1 KCl as supporting electrolyte in a potential window ranging from −0.4 to 0.7 V at a scan rate of 50 mV s−1.
(C) Nyquist plots were obtained in the same experimental conditions as used for (B). Inset shows a zoomed-in view of the plots at high-frequency regions. Conditions: frequency range from 1 × 105 Hz to 0.1 Hz and amplitude of 10 mV; measurements were performed at room temperature. The colors displayed in the CVs and Nyquist plots are related to each step modification illustrated in (A).
(A)Created in BioRender.com.
The bare carbon screen-printed electrode on the biodegradable BC substrate presented a defined redox process with peak currents (ip) of 148.5 μA and resistance to charge transfer (RCT) of 40.2 Ω (Figures 2B and 2C, respectively). After modifying the working electrode (WE) with G-PEG, the ip drastically decreased to 51.7 μA and the RCT increased to 312.6 Ω. These results were in line with the low electrical conductivity of graphene oxide; PEG contributes to a lower charge transfer.30,31 Next, ACE2 was covalently anchored to the WE surface by the N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (EDAC)-N-hydroxysuccinimide (NHS) approach. This step contributed to a decrease in the ip to 27.5 μA and increased the RCT value to 451.3 Ω. Subsequently, the remaining nonspecific sites of the electrode were blocked with 0.1% (m/v) bovine serum albumin (BSA), resulting in an ip of 14.0 μA and RCT of 824.7 Ω, which reflects the modification with a nonconductive layer on the WE surface. In the last step of biosensor fabrication, the WE surface was modified using a 1.0% (m/v) Nafion permeable membrane to enhance the robustness of the biosensor. Hence, the ip decreased to 7.9 μA and the RCT increased to 1,466 Ω.
Analytical performance of the biosensor
In this study, to achieve sensitive detection of SARS-CoV-2 directly at the point of need, we focused on an approach that does not require sophisticated instrumentation. Potentiometric measurements can be conducted using a low-cost and portable potentiometer that detects the difference in electrical potential between a stable reference electrode (RE) and a functional WE. The WE is selective toward the target analyte, which causes a charge change at its surface upon recognizing the target species, eliminating the need for a redox probe for analysis.
To obtain a highly selective biosensor, we modified the WE with ACE2, a natural receptor for entry of SARS-CoV-2 into the human body.32,33 Highly specific interactions between the SP and the ACE2-coated electrode induced a potential variation. In these interactions, the output voltage was logarithmically correlated with the concentration of the target species in solution, similar to traditional ion-selective electrodes (ISEs).34 However, our reagentless electroanalytical method is based on the affinity between the ACE2-functionalized biosensor and the target SP analyte. Therefore, it is a non-reversible sensor, which is a valuable diagnostic tool as a disposable miniaturized device,35 whose disposal is further facilitated by its eco-friendly features.
The potentiometric response used for diagnostic purposes emerges from differences between the open circuit potential (OCP) of the functional ACE2-modified electrode in PBS medium and the potential change induced by the selective binding of the SP to the ACE2-modified electrode. The selective accumulation of the charged SP species on the biosensor surface during the incubation period causes a potential change in the electrodic surface that can be used to monitor the presence of the virus.36 The potentiometric mechanism of detection of SP on our biosensor uses a similar rationale to previous studies that modified the carbon-based electrode surface with receptors to detect proteins and microorganisms.35,37,38
To obtain a selective and sensitive biosensor, we evaluated two approaches for anchoring the ACE2 receptor on the carbon screen-printed electrodes. First, the WE was modified with amine-functionalized G-PEG. PEG is a widely used method for the covalent attachment of biomolecules (PEGylation process) through their COOH-terminal groups.39,40 Second, the WE was modified with the conducting polymer polyethyleneimine (PEI), which also contains NH2 functional groups.41,42 G-PEG provided significant discrimination of the analytical signal at the low concentrations of SP analyzed (10−14–10−11 g mL−1) (Figure 3A), probably because the large surface area of the nanomaterial provided more bioconjugation sites, facilitating the interactions between ACE2 and SP.43 Thus, the G-PEG modification strategy was used throughout this study. All graphics were plotted as a potential difference, ΔE:
| (Equation 1) |
where is the potential measured in the presence of SARS-CoV-2 and is the potential obtained for the blank (i.e., 0.1 mol L−1 PBS at pH = 7.4). We expressed the analytical response as the difference between the sample and the blank (Equation 1) to minimize eventual fluctuation in potential response due to temperature differences at the testing sites, eliminating the need for recalibration of the biosensor for POC testing.
Figure 3. Optimization studies of the BC-based electrochemical biosensor.
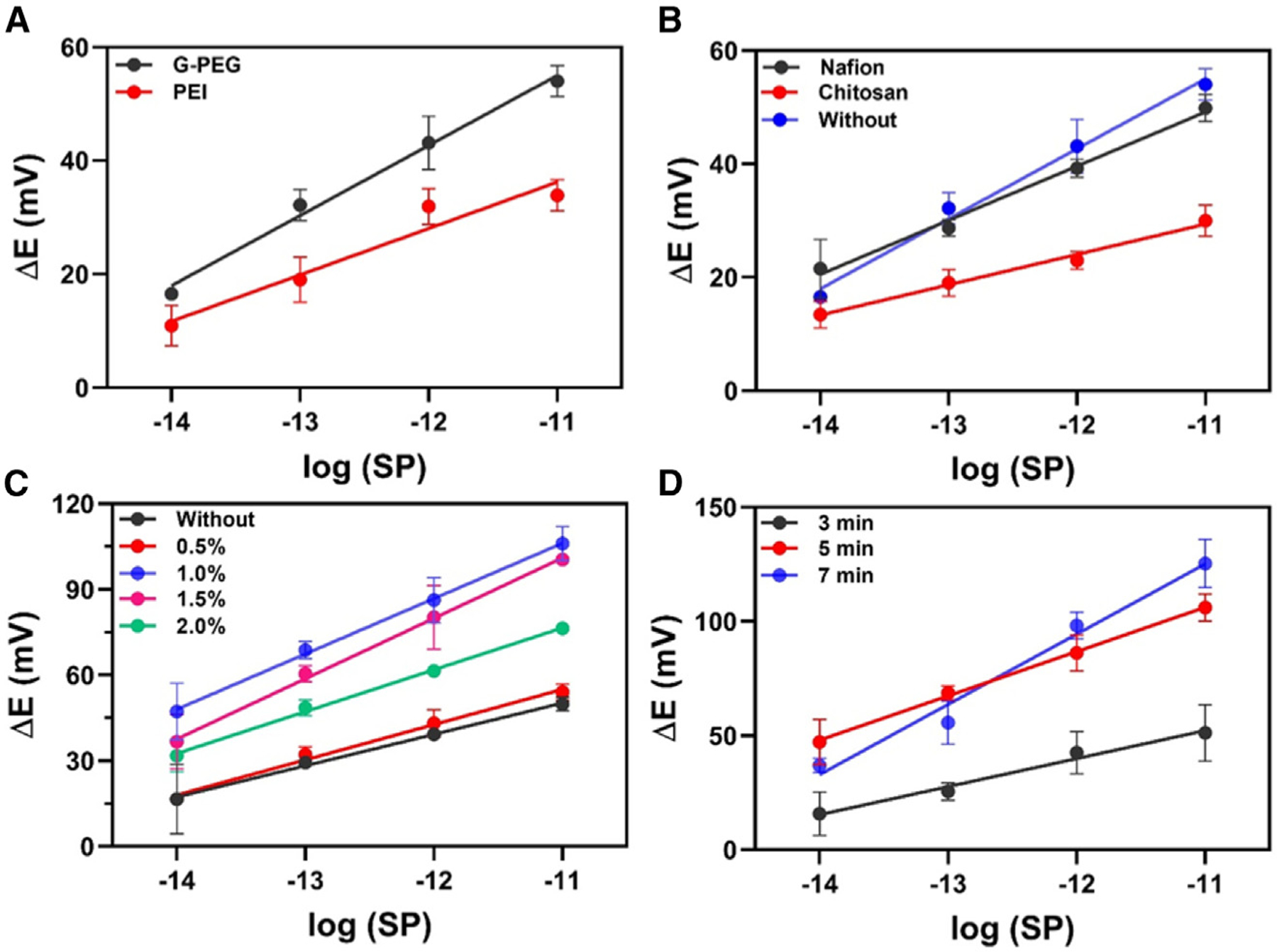
(A) Modification of the WE to anchor ACE2 using 2 mg mL−1 G-PEG (black circles) or1 mg mL−1 PEI (red circles).
(B) Performance of the electrochemical biosensor modified with 0.5% (m/v) Nafion (black circles), modified with 0.5% (m/v) chitosan (red circles), and without membrane layer (blue circles).
(C) Effect of Nafion concentration on the sensitivity of the method: 0.0% (black circles), 0.5% (m/v; red circles), 1.0% (m/v; blue circles), 1.5% (m/v; purple circles), and 2.0% (m/v; green circles).
(D) Incubation time study with time ranging from 3 to 7 min, using the BC-based biosensor. All experiments were carried out in triplicate (n = 3) using SP as a target in a concentration range from 10 fg mL−1 to 10 pg mL−1 at room temperature. The error bars correspond to the standard deviation. Potentiometric measurements were recorded in 0.1 mol L−1 PBS during 300 s of analysis.
The fabrication, modification, and functionalization steps were then optimized to obtain a more robust and sensitive biosensor for SARS-CoV-2 SP detection. The WE was modified with G-PEG using the drop-casting method and incubating for 60 min at 37°C to dry. This procedure introduces amine groups on the WE surface for bioconjugation. Next, the ACE2 receptor containing EDAC + NHS was dropped on the WE modified with G-PEG and kept for 30 min at 37°C. When the carboxyl groups of ACE2 were exposed to EDAC-NHS, they were activated to form a stable ester, which undergoes a nucleophilic addition with the amino groups present on the WE, resulting in a stable amide bond between the carbon WE/G-PEG and ACE2.31,41 The remaining unmodified sites of the WE surface were then blocked using a 1.0% (m/v) BSA solution.
Nafion and chitosan are polymeric membranes commonly used as coatings for biosensors to enhance the robustness of the electrochemical device. These membranes protect the electrode surface against biofouling when this surface is exposed to the sample’s complex matrix; these membranes can also provide superficial preconcentration of chemical species.44 For this study, analytical curves were made at concentrations ranging from 1 × 10−14 to 1 × 10−11 g mL−1 SP in 0.1 mol L−1 PBS (pH = 7.4) to compare three strategies of biosensor modification: (1) using 0.5% Nafion, (2) using 0.5% chitosan, and (3) without any permeable membrane. The Nafion layer resulted in the highest detectability of the biosensor (Figure 3B), because its anionic membrane allows small positively charged species to cross the biosensing surface and become preconcentrated close to this surface.45 Given the results presented in Figure 3B, we studied the effects of changing the proportion of Nafion on the modified biosensor since this proportion would directly affect membrane thickness. Figure 3C shows the performance of the biosensor at various Nafion concentrations; 0.5% Nafion did not present a significant difference in the detectability compared with the unmodified biosensor (i.e., in the absence of a Nafion layer). However, 1.0% Nafion exhibited the highest detectability, which is associated with adequate membrane thickness to enable the preconcentration of the SP and allow its binding to the ACE2-modified surface. When we increased the proportion of the semipermeable membrane (1.5 and 2.0% [m/v]) on the modified biosensor, the biosensor presented lower detectability and sensitivity compared with the use of 1.0% Nafion solution. This behavior was due to the higher thickness of the resulting membrane, which limited the mass transport of the target analyte (SPs or viral fragments containing SPs) to access the modified surface of the biosensor; i.e., a higher amount of Nafion membrane (>1.0% [m/v]) hindered the permeation of the target species and subsequent binding to the ACE2-modified WE surface, causing a loss in the sensitivity of the method. Collectively, our experiments revealed that 1.0% (m/v) Nafion provided the optimal detectability and was thus selected for our biosensor, in accordance with previous studies.36,43
We next evaluated which time of incubation of SP with the surface of the modified biosensor would yield the best analytical performance for SARS-CoV-2 detection. The experiment was carried out in triplicate with an interval concentration ranging from 10−14 to 10−11 g mL−1 of SARS-CoV-2 SP (Figure 3D). The results revealed that 5 min and 7 min of incubation time provided similar detectability. However, this optimization was based on the analytical sensitivity (slope) parameter obtained by analytical curves. Thus, we chose 7 min as the optimal incubation time, which provided high detectability and sensitivity. These results demonstrate the fast-binding kinetics between SP and the ACE2 receptor immobilized on the electrode surface, highlighting the efficiency of our biosensor architecture.
We then evaluated the electrochemical signal (potential difference) provided by our potentiometric biodegradable BC-based biosensor through dose-response curves with low concentrations of virus. All the potentiometric measures were carried out in triplicate using 0.1 mol L−1 PBS (pH = 7.4) for 300 s. The measurements were recorded by dropping 10 μL of SARS-CoV-2 SP or clinical samples onto the surface of the biosensor and incubating it for 7 min before each measurement. The biosensor required at least 30 s to provide a stable potential difference response in the presence of SARS-CoV-2 SP to stabilize the accumulated charge (Figure 4A). The electrical potential was sampled at 3 min for quantitative purposes to ensure a stable response. The signal for ΔE increased with the increase in the concentration of SARS-CoV-2 SP over the concentration range studied of 10.0 zg mL−1 to 1.0 μg mL−1 in 0.1 mol L−1 PBS at pH = 7.4 (Figures 4A and 4B).
Figure 4. Potentiometric measurements and dose-response curves for SARS-CoV-2 detection.
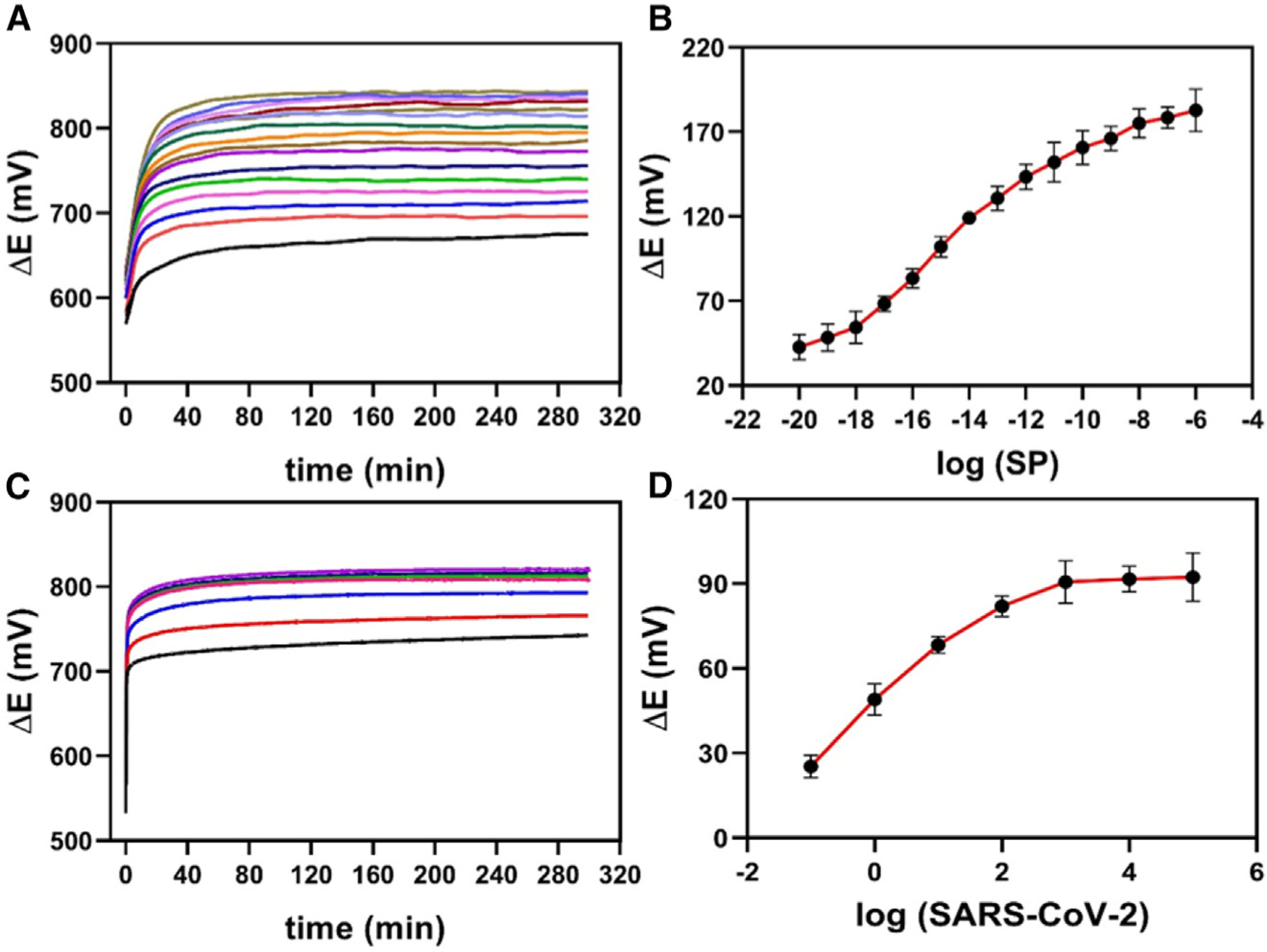
(A) Potentiometric responses of the BC-based biosensor for concentrations ranging from 10.0 zg mL−1 to 1.0 mg μL−1 SARS-CoV-2 SP in 0.1 mol L−1 PBS (pH = 7.4).
(B) Dose-response curve obtained from ΔE (V) values, subtracted from the blank values (ΔE (V) = Esample – Eblank), as a function of the logarithm of the SP concentration.
(C) Potentiometric responses for SARS-CoV-2 detection in a concentration range from 1 × 10−1 copies μL−1 to 1 × 105 copies μL−1.
(D) Dose-response curve obtained from ΔE (V) values, subtracted from the blank values (ΔE (V) = Esample — Eblank), as a function of the logarithm of the SARS-CoV-2 concentration. All potentiometric measurements were carried out after the incubation (7 min) of 10 mL of SP or SARS-CoV-2 samples on the surface of the BC-based biosensor. The measurements were recorded in triplicate (n = 3) in 0.1 mol L−1 PBS medium for 300 s, and the error bars correspond to the standard deviation.
Next, titered samples with B.1 SARS-CoV-2 concentrations ranging from 1 × 10−1 copies μL−1 to 1 × 105 copies μL−1 were analyzed (Figure 4C), and a dose-response curve was obtained by measuring ΔE as a function of the logarithm of the B.1 SARS-CoV-2 concentration (Figure 4D). The ΔE response increased from 10−1 to 103 copies μL−1 and after that reached a plateau, probably due to the limitation of recognition sites, leading to response saturation.
The LOD and limit of quantification (LOQ) of our electrochemical device were calculated based on the four-parameter logistic (4PL) method, which is commonly employed for bioassays that use binding interactions.46–48 Applying Equation 2 and 3,49 we obtained an LOD of 4.26 × 10−18 g mL−1 and an LOQ of 1.42 × 10−17 g mL−1 for SARS-CoV-2 SP (Figure 4B), and an LOD of 0.05 copies μL−1 and an LOQ of 0.17 viral RNA copies μL−1 (Figure 4D). These analytical parameters indicate that the sensitivity of our biosensing approach is similar to that of the RT-qPCR technique.36,37
| (Equation 2) |
where is a value of blank limit, is the mean of signal intensities for n blank (negative control) replicates, is the standard deviation of blank replicates, and t(1 – α; n − 1) is the 1 — α percentile of the t-distribution given n − 1 degrees of freedom, α = β = 0.05 significance levels.
| (Equation 3) |
where is the LOD in the signal domain, is the pooled standard deviation of n test replicates, and t[1 − β; m(n − 1)] is the 1 − β percentile of the t-distribution given m(n − 1) degrees of freedom. Again, we set α = β = 0.05, but these significance levels can be chosen properly for each study.
A side-by-side comparison of sensing methods for SARS-CoV-2 is given in Table 1. Our device provided the lowest LOD for SARS-CoV-2 SP solution (LOD = 4.26 × 10−18 g mL−1) and gave results in a short time. The testing time was set as 10 min, which included 7 min for incubation of the sample and 3 min for potentiometric analysis (ΔE sampled at 3 min). The main objective of our work was to develop a low-cost and reliable diagnostic method for COVID-19. Thus, we optimized the method to achieve the best analytical and clinical parameters possible (i.e., sensitivity, selectivity, and accuracy). The analytical parameters of our method were calculated to allow evaluation of the biosensor performance and establish the optimal conditions for its application. However, the primary purpose of our biosensor is as a rapid diagnostic (i.e., to determine whether clinical samples are positive or negative for COVID-19 and to enable frequent testing).
Table 1.
Comparison of analytical methods reported in the literature for SARS-CoV-2 detection
| Sensor | Technique | Target | LOD | Time (min) | Reference |
|---|---|---|---|---|---|
| BC/G-PEG/ACE2/BSA/Nafion | potentiometry | SP | 4.26 × 10−18 g mL−1 and 0.05 copies μL−1 |
10 | This study |
| GA/ACE2/BSA/Nafion | IES | SP | 2.80 × 10−15 g mL−1 | 4 | 43 |
| Paper/GO/SARS-CoV-2 antibody | SWV | IgG and IgM | 0.96 and 0.14 × 10−9 g mL−1 | 30 | 59 |
| Cotton-tipped electrode/SARS-CoV-2 antibody | SWV | N protein | 8.01 × 10−13 g mL−1 | 21 | 60 |
| GPE/GA/AuNP-cys/ACE2/BSA | SWV | SP | 1.96 × 10−13 g mL−1 | 6.5 | 4 |
| Graphene-ssDNA-AuNP/Au electrode | chronopotentiometry | N gene | 6.92 copies μL−1 | 5 | 61 |
| SARS-CoV-2 RapidFlex | DPV and OCP-EIS | viral antigen nucleo-capsid protein | 0.50 × 10−15 g mL−1 | 10 | 62 |
| AuNPs/CPP/AFB1 | colorimetric | SARS-CoV-2 RNA | 200 copies μL−1 | 5 | 63 |
| Cotton swab/AuNPs/ACE2 | colorimetric | SP | 0.15 × 10−12 g mL−1 | 5 | 3 |
| DETECTR | CRISPR technology | E gene and N gene | 10 copies μL−1 | 40 | 64 |
| AuNP/ASOs | colorimetric | N gene | 0.18 × 10−9 g mL−1 | 30 | 65 |
SWV, square wave voltammetry; DPV, differential pulse voltammetry; GPE, graphite pencil electrode; GA, glutaraldehyde; AuNPs, gold nanoparticles; cys, cysteamine; CPP, cationic perylene probe; AFB1, aptasensor for rapid detection of aflatoxin B1; ASOs, antisense oligonucleotides specific for N gene.
The cost to produce our biosensor was estimated at US$3.50 taking into account the chemicals and materials used to produce it (conductive inks, BC, G-PEG, ACE2, BSA, Nafion) and considering a lab-based production of 100 test units per batch, which requires mg/mL scale of the commercially available chemicals (Table S1). Because the electrodes used in our device can be rapidly mass-produced by industrial screen printers, our biosensor technology is highly scalable and the cost per biosensor is likely to go down when manufacturing is scaled up. Furthermore, large quantities of BC can be produced by genetically engineered bacterial strains grown in low-cost media, further reducing the final cost of the device.19
The biodegradability of BC16,17,20,50 and the eco-friendly features of our biosensor facilitate its use as a rapid test (i.e., a disposable device). However, the silver content of the RE remains a limitation for direct disposal of the device in landfills, which will need to be collected to recover and recycle the silver content through hydrometallurgical or similar technologies.51,52 We envision that the silver tracks can be easily separated from the test strips to be properly treated to allow the recovery of silver that can be used for other applications, ultimately enabling a circular economy.
Cross-reactivity, reproducibility, and potential stability assays
To investigate the specificity of our biosensing electrochemical device for SARS-CoV-2, we applied it to other viruses and viral antigens under the same optimized experimental conditions. In addition to SARS-CoV-2, we tested other viruses (influenza A (H1N1), strain A/California/2009; influenza A (H3N2), A/Nicaragua; influenza B [B/Colorado]; mouse hepatitis virus [MHV]; and herpes simplex virus-2 [HSV-2]) and three antigenic preparations (corresponding to heat-inactivated Zika virus and yellow fever, and gamma-irradiated Ebola virus). All the experiments were carried out in 0.1 mol L−1 PBS at pH = 7.4 for 300 s of analysis. Ten microliters of each virus or viral antigen was incubated on the biosensor surface for 7 min before the potentiometric measurements were taken (Figure S1). No cross-reactivity was detected in any of the seven samples, which highlights the utility of our sensor for SARS-CoV-2 detection and COVID-19 diagnosis. We hypothesize that the sensitivity and selectivity of our biosensors were achieved due to the use of graphene oxide nanomaterial conjugated with G-PEG amine functionalized to immobilize the biological receptor (ACE2) and due to the remarkable affinity of this species toward the SARS-CoV-2 SP.
Reproducibility assays were carried out to ensure that different test batches of biosensors performed similarly. For this study, we recorded potentiometric measurements of 1 × 101 copies μL−1 of SARS-CoV-2 prepared in a virus transportation medium (VTM) over 7 min of incubation time. The relative standard deviation (RSD) obtained with 10 biosensors representing different fabrication batches was 3.78%, indicating that our fabrication method and functionalization protocol were highly reproducible (Figure S2). The observed reproducibility indicates that the fabrication of our device is scalable and can be developed to provide on-demand testing at the POC.
The stability of the biosensor was evaluated potentiometrically using 0.1 mol L−1 PBS (pH = 7.4) and VTM for 60 min (Figure S3). The results indicate that the biosensor achieved high stability after 2 min, with a low drift response (<4%) over the evaluated period for PBS medium. When VTM was used, a drift response on the electrical potential was noted for a long analysis period (>500 s) (Figure S3), which may be related to the fouling of the electrode surface by proteins, antibiotics, or other biomolecules present in VTM. Therefore, we selected PBS as the optimal medium to carry out all potentiometric tests and sample analyses.
Detection of SARS-CoV-2 in clinical samples
Using the optimized experimental conditions, we applied our biodegradable electrochemical biosensor to the analysis of 15 OP/NP clinical samples, five of which had the original SARS-CoV-2 strain and 10 of which had the SARS-CoV-2 Delta variant. Viral loads in the samples ranged widely, with cycle threshold (Ct) values varying from 14.0 to 27.3 (Figure 5A; Table S2). Our biosensor detected SARS-CoV-2 samples and Delta variants in all clinical samples analyzed. Our ACE2-based biosensor provided a higher analytical response (i.e., increased potential difference) for the SARS-CoV-2 Delta variant samples compared with the original strain samples with similar Ct values, which may be associated with the higher affinity of their mutated SP toward the ACE2 receptor.53,54
Figure 5. Electrochemical detection of SARS-CoV-2 variants in human NP/OP biofluid samples.
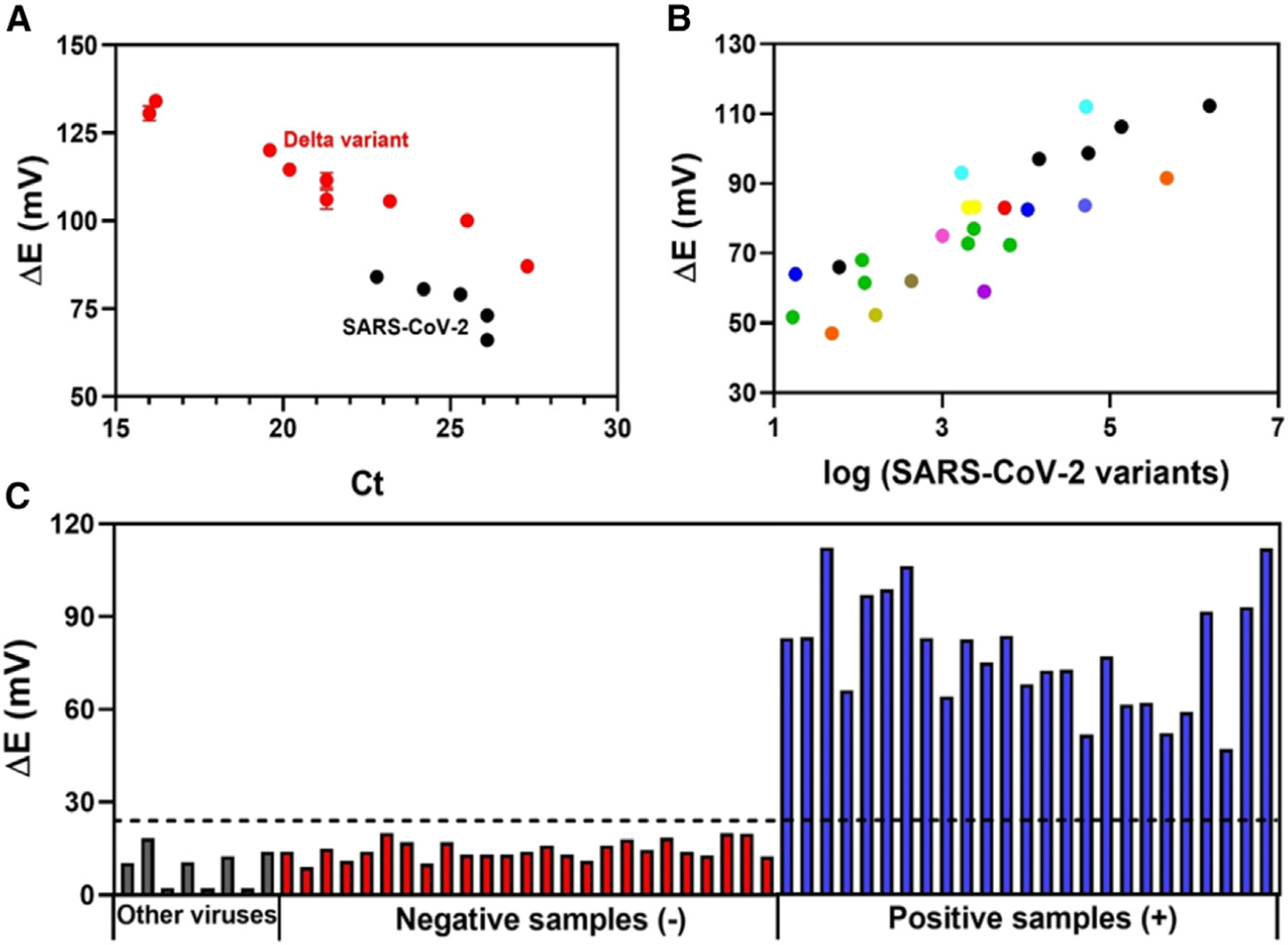
(A) Electrochemical response obtained for five clinical samples containing original SARS-CoV-2 strain (black circles) and 10 clinical samples containing SARS-CoV-2 Delta variant (B.1.617.2, red circles) as a function of Ct values. All measurements were recorded in triplicate (n = 3), and the error bars correspond to the standard deviation.
(B) Potential difference, ΔE, obtained using the modified electrode for another 12 lineages of SARS-CoV-2 as a function of the RNA concentration (copies μL−1) provided by the RT-PCR method,  B.1,
B.1,  B.1.291,
B.1.291,  B.1.369,
B.1.369,  B.1.340,
B.1.340,  B.1.243,
B.1.243,  B.1.311,
B.1.311,  B.1.1.304,
B.1.1.304,  B.1.1.317,
B.1.1.317,  B.1.2,
B.1.2,  B.1.1.7,
B.1.1.7,  B.1.240, and
B.1.240, and  B.1.350.
B.1.350.
(C) Comparison of the electrochemical response obtained by the cross-reactivity studies (gray bars), 25 SARS-CoV-2-negative clinical samples (red bars), and 25 positive SARS-CoV-2 clinical samples containing different lineages (blue bars). The dotted line indicates the cutoff value of ΔE (V) (ΔE (V) = Esample − Eblank) response established to indicate whether the sample was positive for SARS-CoV-2 variants as determined by our biosensor.
Emerging SARS-CoV-2 variants have been classified as variants of interest (VOIs) and variants of concern (VOCs), although there are also other classification schemes. Over the course of the pandemic, clinically relevant variants include the B.1.1.7 strain, first reported in the United Kingdom (Alpha); B.1.352, first reported in South Africa (Beta); P.1, first reported in Brazil (Gamma); B.1.617.2, first reported in India (Delta); and B.1.1.529, detected in multiple countries (Omicron).43,55 VOIs include B.1.427 and B.1.429, detected in the United States (California) (Epson); B.1.525, detected in the United States (New York)/Nigeria (Eta); B.1.526, detected in the United States (New York) (Iota); P.2, detected in Brazil (Zeta); P.3, detected in the Philippines (Theta); B.1.617.1, detected in India (Kappa); C.37, detected in Peru (Lambda); and B.1.621, detected in Colombia (Mu).56 As global immunity to currently circulating variants increases, a novel subvariant could originate from Delta or Alpha, for example. In addition, new variants of SARS-CoV-2 are likely to continue to emerge or re-emerge in the months and years ahead, highlighting the importance of inexpensive sensors capable of detecting and collecting data on multiple SARS-CoV-2 variants.57
In order to evaluate the efficacy and robustness of the biosensor for COVID-19 diagnosis, we tested another set of 50 NP/OP clinical samples, 25 of which were positive NP/OP samples containing 12 SARS-CoV-2 variants of different lineages and 25 of which were negative NP/OP clinical samples (Table S3) obtained, after heat-inactivation, from patients from the Hospital of the University of Pennsylvania (HUP). All the lineages were confirmed by RT-PCR. We set the cutoff value of our biosensor as ΔE > 25 mV as positive for SARS-CoV-2, and ΔE < 25 mV as negative (Figure 5C). The cutoff value was based on the analytical signal obtained for the lowest quantity of virus analyzed (Figure 4D). It is worth noting that we expressed the analytical response as the potential difference between the sample and the blank (Equation 1) to minimize eventual fluctuation in potential response due to temperature differences at the testing sites. This simple strategy can allow rapid discrimination of a positive or negative diagnostic outcome based on the response difference compared with the cutoff of the method (ΔE > 25 mV).
In addition to the NP/OP clinical samples of the Delta variant noted above (Figure 5A), our BC-based potentiometric biosensor accurately detected the virus in the 25 positive clinical samples containing 12 different SARS-CoV-2 lineages, which suggests that our method would not require additional adaptation for the detection of new SARS-CoV-2 variants, as long as ACE2 remains the entry point into human cells for the mutated virus. There was also a close correlation between the analytical response (ΔE) and the concentration of virus present in the clinical samples (Figure 5B), highlighting the potential of our biosensor for both rapid detection of COVID-19 and monitoring the infection status (viral loads) of patients.
Collectively, in this study, the robustness and accuracy of a BC-based biosensor were evaluated by analyzing relevant NP/OP clinical samples (40 positive NP/OP samples from 13 SARS-CoV-2 lineages and 25 negative NP/OP samples; Tables S2 and S3). The detection accuracy using these samples suggests that our device would not require additional adaptation to detect emerging SARS-CoV-2 variants as long as the newly mutated virus interacted with ACE2 to enable its entry into human cells. Based on its outstanding analytical parameters (high selectivity, reproducibility, specificity, and accuracy), low cost, and simplicity, our device is a potential candidate for frequent testing at the point of need. Thus, we envision that this technology may help to prevent outbreaks in countries where the SARS-CoV-2 vaccination rates are low but frequent testing is feasible and sanitary practices are adequate. In addition, the use of BC as a substrate overcomes several environmental issues associated with polymeric PCBs. Thus, we propose that biosensors composed of eco-friendly substrates, such as BC, may be better suited for widespread testing as they dramatically reduce the electronic waste (e-waste) and environmental impact compared with the use of nondegradable and nonrecyclable polymeric options such as PCBs.
In summary, we described a low-cost, eco-friendly, portable, and simple BC-based electrochemical biosensor for detecting SARS-CoV-2 rapidly, accurately, and with ultrasensitivity (LODs of 4.26 × 10−18 g mL−1 SP and 0.05 copies μL−1). The virus is detected in 10 min and amounts of only 10 μL of NP/OP clinical samples are needed. The detection of the SARS-CoV-2 virus with the BC-based biosensor was highly reproducible (RSD = 3.78%) and selective for SARS-CoV-2. All SARS-CoV-2 variants tested were detected without sophisticated instrumentation or additional reagents. Our BC-based device can be fabricated without large quantities of chemicals or toxic materials. In addition to the biocompatibility of BC, all materials used to fabricate this device are accessible and commercially available, yielding a test that costs around US$3.50 and that can be prepared in 3 h (functionalization steps). Our biosensor has the additional advantages of not requiring major advanced instruments, modern laboratory facilities, additional reagents, or sample pretreatment, which makes it ideal as a rapid test to be used at testing sites.
EXPERIMENTAL PROCEDURES
Resource availability
Lead contact
C.F.N. is the lead contact; e-mail address cfuente@upenn.edu.
Materials availability
Sensors generated in this study may be made available upon request from the research community.
Data and code availability
All other data supporting the findings of this study are available within the article and are described in the supplemental information or are available from the lead contact upon reasonable request.
Materials
All reagents used in the experiments were of analytical grade. Deionized water (resistivity ≥18 MΩ cm at 25°C) was obtained from a Milli-Q Advantage-0.10 purification system (Millipore). Human ACE2 Fc Chimera was obtained from GenScript. SP was kindly donated by Dr. Scott Hensley from the University of Pennsylvania. Graphene oxide conjugated with G-PEG amine functionalized, PEI, K3[Fe(CN)6], K4[Fe(CN)6], BSA, Nafion, EDAC, and NHS with a degree of purity ≥98% and phosphate buffer saline solution, pH = 7.4, were purchased from Sigma-Aldrich. Carbon and Ag/AgCl conductive inks of medical grade and dielectric ink were acquired from Creative Materials.
Viral strains and antigenic preparations
The following viral strains were used in the present study: SARS-CoV-2 OP/NP samples (B.1, B.1.617.2, B.1.291, B.1.369, B.1.340, B.1.243, B.1.311, B.1.1.304, B.1.1.317, B.1.2, B.1.1.7, B.1.240, B.1.350) quantified by the RT-PCR method; these strains were kindly provided by R.G.C.’s group from the University of Pennsylvania with sequence analysis by Andrew Marques and Dr. Frederic Bushman. Influenza A virus (H1N1), strain A/California; influenza A virus (H3N2), A/Nicaragua; influenza B, B/Colorado; and MHV were kindly donated by Dr. Susan Weiss’ Lab. Dr. Sita Awasthi and Dr. Harvey Friedman kindly donated HSV-2. Dr. Scott Hensley kindly donated the SARS-CoV-2 SP. Antigenic preparations of Zika virus and yellow fever virus, both already heat inactivated at 65°C for 30 min, and Zaire Ebolavirus (gamma irradiated) were provided by BEI Resources. The following reagents were obtained through BEI Resources, NIAID, NIH: Zaire Ebolavirus, Mayinga, infected cell lysate, gamma irradiated, NR-49809; yellow fever virus, 17D, heat inactivated, NR-51472; and Zika virus, PRVABC59, heat inactivated, NR-50369.
Fabrication of BC substrate
BC substrates were produced by G. hansenii (ATCC 53582) following the procedure described by Costa et al.,58 schematically illustrated in Figure 1A. First, the bacteria were inoculated in 1.0 L of HS medium, which had previously been autoclaved at 121°C for 15 min. Then, the mixture was transferred to a plastic container of approximately 40 × 20 cm and left at room temperature, 25°C ± 3°C, for 27 days in static conditions. Subsequently, the BC film formed was collected and cleaned using 5.0 mmol L−1 NaOH solution at 80°C for 4 h. Finally, the pretreated BC was washed with deionized water to remove alkalinity and kept at 80°C in an incubator until completely dry. This procedure provided a biodegradable substrate with a thickness of 90.0 ± 1.0 μm.
The electrochemical devices were manufactured by the screen-printing method with three-electrode configuration cells (dimensions 2.5 × 2.0 cm) on the biodegradable BC substrate. Carbon conductive ink was used to fabricate the WE and counter electrode (CE), and Ag/AgCl conductive ink was used to fabricate the RE. To cure the conductive tracks, the printed BC substrates were placed in a thermal oven at 70°C for 30 min. After the curing step, the devices were cut into small pieces (2.5 × 2.0 cm). To delimit the electrode area, a nonconductive ink was used, and the devices were submitted to an additional curing step under the same conditions as described above.
Modification of BC electrodes
To prepare the electrochemical biosensor, 5.0 μL of 2.0 mg mL−1 G-PEG solution was dropped on the carbon WE and allowed to dry for 60 min at 37°C. Next, 5.0 μL of a mixture of 0.33 mg mL−1 ACE2 receptor containing 25 mmol L−1 EDAC and 50 mmol L−1 of NHS solution were drop cast on the surface of the WE and incubated at 37°C for 60 min. The unmodified zones of the WE were blocked with 5.0 μL of 1.0% (m/v) BSA solution, and the WE was stored for 30 min at 37°C to dry, to avoid nonspecific interactions of other biomolecules present in the sample with the biosensor’s surface. Finally, 5.0 μL of 1.0% (m/v) Nafion was deposited onto the WE, and the WE was incubated at 37°C for 60 min. The biosensor was then washed with PBS 0.1 mol L−1 (pH = 7.4) before use.
Morphological, structural, and electrochemical characterization
The morphological characterization of the BC substrate was carried out using SEM (model TM3000) at an operating voltage of 15 kV. For SEM images, the BC samples were coated with an iridium layer applying 11.3 mA for 60 s. The structural characterizations of the BC substrate and BC-based carbon screen-printed electrodes with and without modification with G-PEG were recorded by Raman spectroscopy (model Horiba T64000) using a 532-nm laser with 30 mW of power.
For electrochemical characterization of the electrodes in each step of modification, the CV technique was used in a potential window ranging from 0.7 to −0.3 V and with a scan rate of 50 mV s−1. EIS experiments were carried out at frequencies ranging from 1 × 105 to 0.1 Hz using an amplitude of 10 mV, and under OCP. The electrochemical studies were recorded using different electrodes in 0.1 mol L−1 KCl solution containing 5.0 mmol L−1 of the redox probe [Fe(CN)6]3−/4− solution. Potentiometric measurements were carried out in a time interval of 300 s using 0.1 mol L−1 PBS (pH = 7.4). A MULTI AUTOLAB M101 potentiostat with six channels, controlled by the NOVA 2.1 software, was used for all the electrochemical measurements. Experiments were carried out at room temperature, 25°C ± 3°C.
SARS-CoV-2 biosensing
For SARS-CoV-2 biosensing, 10.0 μL of 0.1 mol L−1 PBS (pH = 7.4) or VTM containing either SP or SARS-CoV-2 samples was applied to the biosensor surface and the device was incubated at room temperature for 7 min. Following incubation, the electrochemical cell was gently washed with 0.1 mol L−1 PBS (pH = 7.4) to remove the unbound virus and sample. Then, 200 μL of the 0.1 mol L−1 PBS (pH = 7.4) was used for potentiometric measurements and the potential value (E) was obtained. The calibration curves were obtained in the concentration range from 10.0 zg mL−1 to 1.0 μg mL−1 SARS-CoV-2 SP in 0.1 mol L−1 PBS (pH = 7.4). All experiments were performed at room temperature and without purging the system to remove the dissolved oxygen in order to provide a simple protocol for POC testing.
Reproducibility, stability, and cross-reactivity studies
To carry out the reproducibility study, the potential response was obtained by exposing 10 electrodes (from different batches) to 1 × 101 copies μL−1 of SARS-CoV-2 prepared in VTM for 7 min. The stability of the electrode response was potentiometrically evaluated in both 0.1 mol L−1 PBS and VTM for 1 h. Cross-reactivity studies were performed with the following viral strains, all at 105 PFU mL−1: influenza A virus (H1N1), strain A/California/2009; influenza A virus (H3N2), A/Nicaragua; influenza B [B/Colorado]; MHV; and HSV-2. Cross-reactivity studies were also performed with heat-inactivated antigenic preparations of Zika virus (viral genome copy number 1.1 × 107 copies μL−1), yellow fever virus (viral genome copy number 1.8 × 104 copies μL−1), and Ebola virus (viral genome copy number 1.1 × 107 copies μL−1), obtained from BEI Resources. All the experiments were carried out by combining the viral sample with 0.1 mol L−1 PBS for 300 s of analysis, and 10 μL of each virus (or antigenic preparation) was incubated on the biosensor surface for 7 min before the potentiometric measurements were made.
Clinical sample analysis
NP/OP swab patient samples were heat inactivated prior to analysis. Of the 65 NP/OP samples analyzed in this study, 40 were positive and 25 were negative for SARS-CoV-2 when tested by the RT-PCR method. The 25 negative clinical samples were acquired from the HUP (institutional review board [IRB] protocol 844145) and were described in our previous paper.31 The 40 positive SARS-CoV-2 samples containing 13 variants (B.1.350, B.1.340, B.1, B.1.291, B.1.369, B.1.240, B.1.243, B.1.311, B.1.1.304, B.1.1.317, B.1.2, B.1.1.7 [Alpha variant], and B.1.617.2 [Delta variant]) were kindly provided by R.G.C.’s group from the University of Pennsylvania under IRB protocol 823392. We set a cutoff value of potential response (ΔE) higher than 25 mV to express a positive diagnostic result, in accordance with the analytical response obtained for the lowest detected concentration of SARS-CoV-2 (10−1 copies μL−1) in the dose-response curve (Figure 4D); i.e., samples that exhibited ΔE > 25 mV were considered positive for SARS-CoV-2 (Table S3; Figure 5C). The concentration range obtained by RT-PCR for the Delta variant and the other 12 SARS-CoV-2 variants in the clinical samples ranged from 14 to 27.3 Ct and from 1.67 × 101 to 1.53 × 106 RNA copies μL−1, respectively.
Supplementary Material
Highlights.
Electrochemical biosensor fabricated on biodegradable bacterial cellulose substrate
Label-free and ultrasensitive biosensor for COVID-19 diagnosis
Low-cost (US$3.50/test), rapid (10 min), and accurate detection of SARS-CoV-2 variants
ACKNOWLEDGMENTS
C.F.N. holds a Presidential Professorship atthe University of Pennsylvania, is a recipient of the Langer Prize by the AIChE Foundation, and acknowledges funding from the Procter & Gamble Company, United Therapeutics, a BBRF Young Investigator Grant, and the Defense Threat Reduction Agency (DTRA; HDTRA11810041 and HDTRA1-21-1-0014). Research reported in this publication was supported by the Nemirovsky Prize, the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM138201, Penn Health-Tech Accelerator Award, the IADR Innovation in Oral Care Award, and by funds provided by the Dean’s Innovation Fund from the Perelman School of Medicine at the University of Pennsylvania (all to C.F.N.). We thank Dr. Jon Epstein for his support, Dr. Karen Pepper for editing the manuscript, and de la Fuente Lab members for insightful discussions. All figures were prepared in BioRender.com. R.G.C. acknowledges grant R33-HL137063. W.R.d.A. acknowledges funding from Brazilian funding agencies CAPES (88887.479793/2020-00), FAPESP (2018/08782-1 and 2022/03250-7), FAEPEX/PRP/UNICAMP (3374/19), and CNPq (438828/2018-6) for supporting the research. We thank Dr. Sita Awasthi and Dr. Harvey Friedman for kindly donating HSV-2. We thank Dr. Susan Weiss’ Lab for kindly donating the following strains: influenza A virus (H1N1), strain A/California; influenza A (H3N2), A/Nicaragua; influenza B, B/Colorado; and MHV. We thank Dr. Benjamin Abella, Dr. Antonio Davila Jr, and Paul Callahan for their help collecting NP/OP clinical samples (25 negative samples used in this work) at the HUP (IRB 844145), all of which were described in our previous paper. The authors would like to also thank Dr. Scott Hensley for kindly donating SARS-CoV-2 SP, Dr. Kyle Rodino for help with residual specimens, Jevon Graham-Wooten for sample management, and Andrew Marques and Dr. Frederic Bushman for providing SARS-CoV-2 variant information on samples used in this study. The following reagents were obtained through BEI Resources, NIAID, NIH: Zaire Ebolavirus, Mayinga, infected cell lysate, gamma irradiated, NR-49809; yellow fever virus, 17D, heat inactivated, NR-51472; and Zika Virus, PRVABC59, heat inactivated, NR-50369.
INCLUSION AND DIVERSITY
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper self-identifies as a gender minority in their field of research. One or more of the authors of this paper received support from a program designed to increase minority representation in their field of research. We support inclusive, diverse, and equitable conduct of research.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.xcrp.2023.101476.
DECLARATION OF INTERESTS
A non-provisional patent application has been filed on the de la Fuente Lab’s related work (ID number 23-10301). C.F.N. provides consulting services to Invaio Sciences and is a member of the Scientific Advisory Boards of Nowture S.L. and Phare Bio. The de la Fuente Lab has received research funding or in-kind donations from United Therapeutics, Strata Manufacturing PJSC, and Procter & Gamble, none of which were used in support of this work. C.F.N. is on the Advisory Board of Cell Reports Physical Science.
REFERENCES
- 1.Coronavirus disease (COVID-19) World Health Organization. https://www.who.int/emergencies/diseases/novel-coronavirus-2019. [Google Scholar]
- 2.Kevadiya BD, Machhi J, Herskovitz J, Oleynikov MD, Blomberg WR, Bajwa N, Soni D, Das S, Hasan M, Patel M, et al. (2021). Diagnostics for SARS-CoV-2 infections. Nat. Mater 20, 593–605. 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferreira AL, de Lima LF, Torres MDT, de Araujo WR, and de la Fuente-Nunez C (2021). Low-cost optodiagnostic for minute-time scale detection of SARS-CoV-2. ACS Nano 15, 17453–17462. 10.1021/acsnano.1c03236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Lima LF, Ferreira AL, Torres MDT, de Araujo WR, and de la Fuente-Nunez C (2021). Minute-scale detection of SARS-CoV-2 using a low-cost biosensor composed of pencil graphite electrodes. Proc. Natl. Acad. Sci. USA 118, e2106724118. 10.1073/pnas.2106724118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moreira VM, Mascarenhas P, Machado V, Botelho J, Mendes JJ, Taveira N, and Almeida MG (2021). Diagnosis of SARS-cov-2 infection by RT-PCR using specimens other than naso- and oropharyngeal swabs: a systematic review and meta-analysis. Diagnostics 11, 363. 10.3390/diagnostics11020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seo G, Lee G, Kim MJ, Baek S-H, Choi M, Ku KB, Lee C-S, Jun S, Park D, Kim HG, et al. (2020). Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 14, 5135–5142. 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 7.Zhang N, Wang L, Deng X, Liang R, Su M, He C, Hu L, Su Y, Ren J, Yu F, et al. (2020). Recent advances in the detection of respiratory virus infection in humans. J. Med. Virol 92, 408–417. 10.1002/jmv.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Z, Huang C, Huang Z, Lin F, He Q, Tao D, Jaffrezic-Renault N, and Guo Z (2021). Advancements in electrochemical biosensing for respiratory virus detection: a review. Trends Anal. Chem 139, 116253. 10.1016/j.trac.2021.116253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Letko M, Marzi A, and Munster V (2020). Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol 5, 562–569. 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, et al. ; COVID-19 Genomics UK COG-UK Consortium (2021). SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol 19, 409–424. 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baldo TA, de Lima LF, Mendes LF, de Araujo WR, Paixão TRLC, and Coltro WKT (2021). Wearable and biodegradable sensors for clinical and environmental applications. ACS Appl. Electron. Mater 3, 68–100. 10.1021/acsaelm.0c00735. [DOI] [Google Scholar]
- 12.Guo J, Li J, Rao Q, and Xu Z (2008). Phenolic molding compound filled with nonmetals of waste PCBs. Environ. Sci. Technol 42, 624–628. 10.1021/es0712930. [DOI] [PubMed] [Google Scholar]
- 13.Millican JM, and Agarwal S (2021). Plastic pollution: a material problem? Macromolecules 54, 4455–4469. 10.1021/acs.macromol.0c02814. [DOI] [Google Scholar]
- 14.Swingler S, Gupta A, Gibson H, Kowalczuk M, Heaselgrave W, and Radecka I (2021). Recent advances and applications of bacterial cellulose in biomedicine. Polymers 13, 412–429. 10.3390/polym13030412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomes NO, Carrilho E, Machado SAS, and Sgobbi LF (2020). Bacterial cellulose-based electrochemical sensing platform: a smart material for miniaturized biosensors. Electrochim. Acta 349, 136341. 10.1016/j.electacta.2020.136341. [DOI] [Google Scholar]
- 16.Schröpfer SB, Bottene MK, Bianchin L, Robinson LC, Lima V.d., Jahno VD, Barud H.d.S., and Ribeiro SJL (2015). Biodegradation evaluation of bacterial cellulose, vegetable cellulose and poly (3-hydroxybutyrate) in soil. Polimeros 25, 154–160. 10.1590/0104-1428.1712. [DOI] [Google Scholar]
- 17.Popa L, Ghica MV, Tudoroiu E-E, Ionescu D-G, and Dinu-Pîrvu CE (2022). Bacterial cellulose—a remarkable polymer as a source for biomaterials tailoring. Materials 15, 1054. 10.3390/ma15031054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torres FG, Troncoso OP, Gonzales KN, Sari RM, and Gea S (2020). Bacterial cellulose-based biosensors. Med. Devices Sens 3, e10102. 10.1002/mds3.10102. [DOI] [Google Scholar]
- 19.Florea M, Hagemann H, Santosa G, Abbott J, Micklem CN, Spencer-Milnes X, de Arroyo Garcia L, Paschou D, Lazenbatt C, Kong D, et al. (2016). Engineering control of bacterial cellulose production using a genetic toolkit and a new cellulose-producing strain. Proc. Natl. Acad. Sci. USA 113, E3431–E3440. 10.1073/pnas.1522985113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shrivastav P, Pramanik S, Vaidya G, Abdelgawad MA, Ghoneim MM, Singh A, Abualsoud BM, Amaral LS, and Abourehab MAS (2022). Bacterial cellulose as a potential biopolymer in biomedical applications: a state-of-the-art review. J. Mater. Chem. B 10, 3199–3241. 10.1039/D1TB02709C. [DOI] [PubMed] [Google Scholar]
- 21.Sharma P, Mittal M, Yadav A, and Aggarwal NK (2023). Bacterial cellulose: nano-biomaterial for biodegradable face masks – a greener approach towards environment. Environ. Nanotechnol. Monit. Manag 19, 100759. 10.1016/j.enmm.2022.100759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kucińska-Lipka J, Gubanska I, and Janik H (2015). Bacterial cellulose in the field of wound healing and regenerative medicine of skin: recent trends and future prospectives. Polym. Bull 72, 2399–2419. 10.1007/s00289-015-1407-3. [DOI] [Google Scholar]
- 23.Belaustegui Y, Pantò F, Urbina L, Corcuera MA, Eceiza A, Palella A, Triolo C, and Santangelo S (2020). Bacterial-cellulose-derived carbonaceous electrode materials for water desalination via capacitive method: the crucial role of defect sites. Desalination 492, 114596. 10.1016/j.desal.2020.114596. [DOI] [Google Scholar]
- 24.Wu J-B, Lin M-L, Cong X, Liu H-N, and Tan P-H (2018). Raman spectroscopy of graphene-based materials and its applications in related devices. Chem. Soc. Rev 47, 1822–1873. 10.1039/C6CS00915H. [DOI] [PubMed] [Google Scholar]
- 25.Politano GG, Versace C, Vena C, Castriota M, Ciuchi F, Fasanella A, Desiderio G, and Cazzanelli E (2016). Physical investigation of electrophoretically deposited graphene oxide and reduced graphene oxide thin films. J. Appl. Phys 120, 195307. 10.1063/1.4968000. [DOI] [Google Scholar]
- 26.Malard LM, Pimenta MA, Dresselhaus G, and Dresselhaus MS (2009). Raman spectroscopy in graphene. Phys. Rep 473, 51–87. 10.1016/j.physrep.2009.02.003. [DOI] [Google Scholar]
- 27.Patel AN, Collignon MG, O’Connell MA, Hung WOY, McKelvey K, Macpherson JV, and Unwin PR (2012). A new view of electrochemistry at highly oriented pyrolytic graphite. J. Am. Chem. Soc 134, 20117–20130. 10.1021/ja308615h. [DOI] [PubMed] [Google Scholar]
- 28.Lounasvuori MM, Rosillo-Lopez M, Salzmann CG, Caruana DJ, and Holt KB (2015). The influence of acidic edge groups on the electrochemical performance of graphene nanoflakes. J. Electroanal. Chem 753, 28–34. 10.1016/j.jelechem.2015.05.010. [DOI] [Google Scholar]
- 29.Bishop GW, Ahiadu BK, Smith JL, and Patterson JD (2017). Use of redox probes for characterization of layer-by-layer gold nanoparticle-modified screen-printed carbon electrodes. J. Electrochem. Soc 164, B23–B28. 10.1149/2.0431702jes. [DOI] [Google Scholar]
- 30.Nasrollahzadeh M, Sajjadi M, Soufi GJ, Iravani S, and Varma RS (2020). Nanomaterials and nanotechnology-associated innovations against viral infections with a focus on coronaviruses. Nanomaterials 10, 1072. 10.3390/nano10061072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehner BAE, Benz D, Moshkalev SA, Meyer AS, Cotta MA, and Janissen R (2021). Biocompatible graphene oxide nanosheets densely functionalized with biologically active molecules for biosensing applications. ACS Appl. Nano Mater 4, 8334–8342. 10.1021/acsanm.1c01522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson CB, Farzan M, Chen B, and Choe H (2022). Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol 23, 3–20. 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, and Wang X (2020). Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220. 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 34.Ding J., and Qin W. (2020). Trends in analytical chemistry recent advances in potentiometric biosensors. Trends Anal. Chem 124, 115803. 10.1016/j.trac.2019.115803. [DOI] [Google Scholar]
- 35.Düzgün A, Maroto A, Mairal T, O’Sullivan C, and Rius FX (2010). Solid-contact potentiometric aptasensor based on aptamer functionalized carbon nanotubes for the direct determination of proteins. Analyst 135, 1037–1041. 10.1039/b926958d. [DOI] [PubMed] [Google Scholar]
- 36.Torres MDT, de Lima LF, Ferreira AL, de Araujo WR, Callahan P, Dávila A Jr., Abella BS, and de la Fuente-Nunez C (2022). Detection of SARS-CoV-2 with RAPID: a prospective cohort study. iScience 25, 104055. 10.1016/j.isci.2022.104055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zelada-Guillén GA, Bhosale SV, Riu J, and Rius FX (2010). Real-time potentiometric detection of bacteria in complex samples. Anal. Chem 82, 9254–9260. 10.1021/ac101739b. [DOI] [PubMed] [Google Scholar]
- 38.Zelada-Guillén GA, Riu J, Düzgün A, and Rius FX (2009). Immediate detection of living bacteria at ultralow concentrations using a carbon nanotube based potentiometric aptasensor. Angew Chem. Int. Ed. Engl 48, 7334–7337. 10.1002/anie.200902090. [DOI] [PubMed] [Google Scholar]
- 39.Abdelbasir SM, El-Sheikh SM, Morgan VL, Schmidt H, Casso-Hartmann LM, Vanegas DC, Velez-Torres I, and McLamore ES (2018). Graphene-anchored cuprous oxide nanoparticles from waste electric cables for electrochemical sensing. ACS Sustain. Chem. Eng 6, 12176–12186. 10.1021/acssuschemeng.8b02510. [DOI] [Google Scholar]
- 40.MacLeod MJ, and Johnson JA (2015). PEGylated N-heterocyclic carbene anchors designed to stabilize gold nanoparticles in biologically relevant media. J. Am. Chem. Soc 137, 7974–7977. 10.1021/jacs.5b02452. [DOI] [PubMed] [Google Scholar]
- 41.Ferreira AL, de Lima LF, Moraes AS, Rubira RJ, Constantino CJ, Leite FL, Delgado-silva AO, and Ferreira M (2021). Development of a novel biosensor for creatine kinase (CK-MB) using surface plasmon resonance (SPR). Appl. Surf. Sci 554, 149565. 10.1016/j.apsusc.2021.149565. [DOI] [Google Scholar]
- 42.Balint R, Cassidy NJ, and Cartmell SH (2014). Conductive polymers: towards a smart biomaterial for tissue engineering. Acta Biomater 10, 2341–2353. 10.1016/j.actbio.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 43.Torres MDT, de Araujo WR, de Lima LF, Ferreira AL, and de la Fuente-Nunez C (2021). Low-cost biosensor for rapid detection of SARS-CoV-2 at the point of care. Matter 4, 2403–2416. 10.1016/j.matt.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hlavatá L, Vyskočil V, Beníková K, Borbé lyová M, and Labuda J (2014). DNA-based biosensors with external Nafion and chitosan membranes for the evaluation of the antioxidant activity of beer, coffee, and tea. Open Chem 12, 604–611. 10.2478/s11532-014-0516-4. [DOI] [Google Scholar]
- 45.e Silva RF, Longo Cesar Paixão TR, Der Torossian Torres M, and de Araujo WR (2020). Simple and inexpensive electrochemical paper-based analytical device for sensitive detection of Pseudomonas aeruginosa. Sensor. Actuator. B Chem 308, 127669. 10.1016/j.snb.2020127669. [DOI] [Google Scholar]
- 46.Findlay JWA, and Dillard RF (2007). Appropriate calibration curve fitting in ligand binding assays. AAPS J 9, E260–E267. 10.1208/aapsj0902029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gottschalk PG, and Dunn JR (2005). The five-parameter logistic: a characterization and comparison with the four-parameter logistic. Anal. Biochem 343, 54–65. 10.1016/j.ab.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 48.Fong Y, Wakefield J, De Rosa S, and Frahm N (2012). A robust bayesian random effects model for nonlinear calibration problems. Biometrics 68, 1103–1112. 10.1111/j.1541-0420.2012.01762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holstein CA, Griffin M, Hong J, and Sampson PD (2015). Statistical method for determining and comparing limits of detection of bioassays. Anal. Chem 87, 9795–9801. 10.1021/acs.analchem.5b02082. [DOI] [PubMed] [Google Scholar]
- 50.Torgbo S, and Sukyai P (2020). Biodegradation and thermal stability of bacterial cellulose as biomaterial: the relevance in biomedical applications. Polym. Degrad. Stabil 179, 109232. 10.1016/j.polymdegradstab.2020.109232. [DOI] [Google Scholar]
- 51.Birloaga I, and Vegliò F (2018). Overview on hydrometallurgical procedures for silver recovery from various wastes. J. Environ. Chem. Eng 6, 2932–2938. 10.1016/j.jece.2018.04.040. [DOI] [Google Scholar]
- 52.Mishra G, Jha R, Rao MD, Meshram A, and Singh KK (2021). Recovery of silver from waste printed circuit boards (WPCBs) through hydrometallurgical route: a review. Environ. Challenges 4, 100073. 10.1016/j.envc.2021.100073. [DOI] [Google Scholar]
- 53.Goher SS, Ali F, and Amin M (2022). The Delta variant mutations in the receptor binding domain of SARS-CoV-2 show enhanced electrostatic interactions with the ACE2. Med. Drug Discov 13, 100114. 10.1016/j.medidd.2021.100114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu H, Wei P, Kappler JW, Marrack P, and Zhang G (2022). SARS-CoV-2 variants of concern and variants of interest receptor binding domain mutations and virus infectivity. Front. Immunol 13, 825256. 10.3389/fimmu.2022.825256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Konings F, Perkins MD, Kuhn JH, Pallen MJ, Alm EJ, Archer BN, Barakat A, Bedford T, Bhiman JN, Caly L, et al. (2021). SARS-CoV-2 Variants of Interest and Concern naming scheme conducive for global discourse. Nat. Microbiol 6, 821–823. 10.1038/s41564-021-00932-w. [DOI] [PubMed] [Google Scholar]
- 56.Otto SP, Day T, Arino J, Colijn C, Dushoff J, Li M, Mechai S, Van Domselaar G, Wu J, Earn DJD, and Ogden NH (2021). The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr. Biol 31, R918–R929. 10.1016/j.cub.2021.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Callaway E (2022). Are COVID surges becoming more predictable? New Omicron variants offer a hint. Nature 605, 204–206. 10.1038/d41586-022-01240-x. [DOI] [PubMed] [Google Scholar]
- 58.Costa AFS, Almeida FCG, Vinhas GM, and Sarubbo LA (2017). Production of bacterial cellulose by Gluconacetobacter hansenii using corn steep liquor as nutrient sources. Front. Microbiol 8, 2027–2112. 10.3389/fmicb.2017.02027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yakoh A, Pimpitak U, Rengpipat S, Hirankarn N, Chailapakul O, and Chaiyo S (2021). Paper-based electrochemical biosensor for diagnosing COVID-19: detection of SARS-CoV-2 antibodies and antigen. Biosens. Bioelectron 176, 112912. 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eissa S, and Zourob M (2021). Development of a low-cost cotton-tipped electrochemical immunosensor for the detection of SARS-CoV-2. Anal. Chem 93, 1826–1833. 10.1021/acs.analchem.0c04719. [DOI] [PubMed] [Google Scholar]
- 61.Alafeef M, Dighe K, Moitra P, and Pan D (2020). Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano 14, 17028–17045. 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Torrente-Rodríguez RM, Lukas H, Tu J, Min J, Yang Y, Xu C, Rossiter HB, and Gao W (2020). SARS-CoV-2 RapidPlex: a graphene-based multiplexed telemedicine platform for rapid and low-cost COVID-19 diagnosis and monitoring. Matter 3, 1981–1998. 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lerdsri J, Chananchana W, Upan J, Sridara T, and Jakmunee J (2020). Label-free colorimetric aptasensor for rapid detection of aflatoxin B1 by utilizing cationic perylene probe and localized surface plasmon resonance of gold nanoparticles. Sensor. Actuator. B Chem 320, 128356. 10.1016/j.snb.2020.128356. [DOI] [Google Scholar]
- 64.Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, Miao X, Streithorst JA, Granados A, Sotomayor-Gonzalez A, et al. (2020). CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol 38, 870–874. 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moitra P, Dighe K, Frieman MB, Alafeef M, Pan D, Frieman MB, Pan D, Pan D, and Pan D (2020). Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano 14, 7617–7627. 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All other data supporting the findings of this study are available within the article and are described in the supplemental information or are available from the lead contact upon reasonable request.


